Abstract
Introduction
Chronic infection with hepatitis C virus (HCV) is a leading cause of liver disease and infectious disease deaths. While recent and emerging treatment options for HCV patients have enabled higher rates of sustained virologic response (SVR), the demographic, clinical, geographic, and payer characteristics of the estimated 3.4 million chronic HCV patients in the USA are poorly understood. The goal of this study was to create a dataset describing the current HCV patient landscape in the USA.
Methods
Data from two large national laboratory companies representing the majority of US patients screened for HCV antibody and/or tested for HCV RNA from 2013 through 2016 were organized into the present study dataset. Age, gender, payer channel, 3-digit ZIP code and ordering physician specialty, and 3-digit ZIP code information were available for all patients. Among RNA-positive patients, additional clinical characteristics included HCV genotype, fibrosis stage, renal function, and HIV status. Initiating treatment and attaining cure were imputed using data-driven algorithms based on successive RNA viral load measurements.
Results
The number of RNA-positive HCV patients increased from 200,066 patients in 2013 to 469,550 in 2016. The availability of clinical data measurements and rates of treatment initiation increased over the study period, indicating improved care engagement for HCV patients. Treatment and cure rates varied by age, disease severity, geographic location, and payer channel. Sensitivity and specificity of the cure prediction algorithms were consistently above 0.90, validating the robustness of the data imputation approach.
Conclusion
This is the largest, most comprehensive dataset available to describe the current US HCV patient landscape. Our results highlight that the epidemiology of HCV is evolving with an increasing number of patients who are younger and have milder disease than described in previous years. Results of this study should help guide efforts toward the elimination of HCV in this country. Future work will focus on factors associated with varying treatment and cure patterns and describing recent changes in the HCV patient landscape.
Funding
AbbVie.
Plain Language Summary
Plain language summary available for this article.
Electronic supplementary material
The online version of this article (10.1007/s12325-018-0721-1) contains supplementary material, which is available to authorized users.
Keywords: Hepatitis C, Direct-acting antivirals, Infectious disease, Real-world evidence, Sustained virologic response, Treatment algorithm
Plain Language Summary
Hepatitis C virus (HCV) infection may cause serious health problems and death. Unfortunately, the health care community does not have complete identification of patients with HCV. This study describes the creation of a dataset that combines information for HCV patients and shows relevant information about HCV patients’ age, geographic location, disease severity, and treatment and cure status from 2013 through 2016. This dataset helps the health care community understand the HCV patient landscape and make informed decisions about how to best treat this population.
Introduction
At least 3.4 million people in the USA exhibit past or current infection with hepatitis C virus (HCV) [1]. Chronic HCV infection is a leading cause of liver fibrosis that may progress to liver cirrhosis, increasing the risk of hepatocellular carcinoma or hepatic decompensation [2, 3]. Deaths from chronic HCV in the USA currently outnumber deaths from 60 other nationally notifiable infectious diseases combined, including human immunodeficiency virus (HIV), tuberculosis, and hepatitis B [4]. Modeling projections estimate over 300,000 deaths, over 150,000 cases of hepatocellular carcinoma, and over 200,000 cases of decompensated cirrhosis among US HCV patients by 2050 with current disease management practices [5]. National strategies targeting HCV disease transmission and treatment access have been recommended to address this urgent public health threat [6, 7]. The World Health Organization established a screening goal of 90% by 2030, but most states in the USA are not on course to meet this goal [8, 9].
There is no vaccine to prevent chronic HCV infection [10], but antiviral therapies offer the potential of achieving sustained virologic response (SVR) [11]. The first direct-acting antivirals (DAAs) were introduced in 2011 [11, 12]. The earliest DAAs enabled SVR in up to 75% of patients, and SVR rates have improved to more than 95% in more recently introduced DAAs [11, 12]. DAAs introduced in 2017 address several unmet needs, including shorter treatment duration, efficacy in patients who have failed previous DAA therapies, indication in patients with chronic kidney disease (CKD) or compensated cirrhosis, and pan-genomic activity [13–16].
The advent of DAAs transformed the HCV landscape by shifting patients away from less effective interferon-based therapies [17]. Clinical practice guidelines updated in 2017 recommend that all HCV patients receive DAAs except for patients with short life expectancies that will not be improved through HCV treatment [18]. However, evidence suggests that treatment is prioritized for patients who have more severe HCV disease, are older, and have comorbidities, changing the profile of the untreated HCV patient population [17].
There are several limitations in understanding the epidemiological and economic burdens of chronic HCV. Acute and chronic HCV cases are not uniformly reported to the Centers for Disease Control (CDC) by all states, and it is therefore difficult to accurately portray the US HCV population [19]. Many chronic HCV patients were infected in prior decades [20], and not all acute cases will transition to chronic infection [21], so acute case trends may not be proportionate with chronic HCV disease burden. National HCV prevalence is often estimated from the National Health and Nutrition Examination Survey (NHANES), which is based on subjects surveyed in only 15 counties [22]. Medical claims studies often do not involve nationwide data stratified by state. Claims studies may provide HCV cost burden and some clinical information, but claims studies are often limited to a population from a single payer [17, 23]. Lastly, the HCV treatment landscape has changed dramatically since the highly effective therapies were released in 2011 [11, 12].
Two web-based sources of information on the prevalence of HCV are HepVu and Polaris Observatory [24, 25]. HepVu [25] is an online dashboard providing year 2010 estimates of HCV antibody prevalence and HCV mortality within the USA at the state level based on data from NHANES, the National Vital Statistics System, and US Census data [26]. Polaris Observatory uses a variety of sources, including expert opinion, to display the projected future global burden of HCV infections, treatment, and mortality [27]. While these sources provide useful information on HCV epidemiology, a current comprehensive source of information that combines patient information and clinical characteristics stratified by year, payer, and state would provide important insights into trends in access to care, treatment, and cure rates across payer and patient groups. This paper describes a robust methodology to develop the largest available dataset of HCV patients to date, which includes the majority of US HCV patients and describes longitudinal data from 2013 to 2016.
Methods
Data Source and Available Characteristics
This study is based solely on laboratory, administrative claims, and payer data and does not contain any new studies with human or animal subjects performed by any of the authors. This study dataset represents the largest available HCV dataset in the USA. The study dataset was derived by combining clinical laboratory tests from two large national laboratory companies. Each laboratory dataset included results for patients screened for HCV antibody and/or tested for HCV RNA from 2013 through 2016; not all patients had both antibody and RNA tests. Overall, the merged study dataset contained antibody screening results for 17,149,480 patients and HCV RNA test results for 1,592,984 patients, for whom age, gender, payer channel (Medicaid, Medicare, commercial, out of pocket), ordering physician, and 3-digit ZIP code information were available (Fig. 1). Of those tested for HCV RNA, 914,285 patients were identified as testing HCV-positive, for whom more detailed characteristics are available: HCV genotype (including sub-genotype 1A versus 1B); measures of fibrosis (i.e., FibroSure/METAVIR scores): liver enzymes ALT/AST, platelets; renal status: serum creatinine, estimated glomerular filtration rate (eGFR, results for African-Americans and non-African-Americans), urine albumin; Child–Pugh score: serum albumin, total bilirubin, prothrombin time, international normalized ratio (INR), ascites and encephalopathy diagnosis codes; HIV diagnosis or HIV RNA-positive test; and resistance-associated substitutions (RAS). The HCV RNA positivity rate of 60% in this dataset is consistent with other reports of HCV RNA positivity ranging from 43% to 72% among HCV antibody-positive patients undergoing RNA testing [28–30].
Fig. 1.
Description of laboratory data contents. ALT alanine aminotransferase, AST aspartate aminotransferase, eGFR estimated glomerular filtration rate, HIV human immunodeficiency virus, INR international normalized ratio, PT prothrombin time, RAS resistance-associated substitutions. Not all patients had both antibody and RNA tests
Each of the clinical characteristics included in the dataset represents an important element in the management of HCV patients. There are six major HCV genotypes (1–6); mixed genotypes are also possible [18]. While sub-genotype testing was included in the laboratory data, only sub-genotypes 1A and 1B were extracted for this analysis.
Fibrosis is indicated by five stages of severity (F0–F4) and describes the accumulation of nonfunctional liver scar tissue that may eventually progress to cirrhosis, designated as stage F4 [31]. For this dataset, fibrosis stage was derived using a hierarchical approach dependent on a single F-stage availability, listed in order of priority: (1) FibroSure/METAVIR stage, (2) modified FIB-4 score [32], or (3) APRI score [33] (Table 1). The modified FIB-4 scoring utilized the population median scores to separate the fibrosis stages into the following categories: F0 < 0.97, F1 = 0.97–1.44, F2 = 1.45–3.25, F3 = 3.26–5.20, and F4 > 5.20. A sensitivity analysis of modified FIB-4 scoring to identify cirrhotic patients compared to a definition based on APRI scoring (F4 defined as APRI score > 2) resulted in a negligible difference of 0.6% (10.2% versus 9.6% of patients identified as F4 stage, respectively).
Table 1.
| Clinical characteristic | Equation | References |
|---|---|---|
| FIB-4 | FIB-4 = (age, years × AST, U/L)/(platelet count, 109/L × √ALT, U/L) | Sterling et al. [32] |
| APRI | APRI = ([AST, IU/L/AST ULN, IU/L]/platelet count, 109/L) × 100 | Lin et al. [33] |
| eGFR | eGFR = 141 × min(serum creatinine, mg/dL)/κ,1)α × max(serum creatinine, mg/dL/κ, 1)− 1.209 × 0.993Age × 1.018 (if female) × 1.159 (if African-American) | Levey et al. [50] |
The Child–Pugh (or Child–Turcotte–Pugh) score is used to grade cirrhosis severity as A (5–6 points), B (7–9 points), or C (10–15 points); A is considered compensated cirrhosis, and B and C are considered decompensated cirrhosis [18]. Child–Pugh scores were calculated using total bilirubin, serum albumin, INR, ascites diagnosis and severity rating, and encephalopathy diagnosis and severity rating. Because it was not possible to assess the severity of ascites (mild/moderate versus severe) or encephalopathy (grade 1–2 versus grade 3–4) in the laboratory datasets, the worse severity grade was selected. This assumption maximized the potential proportion of patients with Child–Pugh B or C, but a sensitivity analysis assessing the impact of this assumption showed that the increase results in only 1% more patients attributed to Child–Pugh B–C.
Serum creatinine is used to calculate the eGFR, a measure of kidney function [34]. eGFR values are used to categorize stages of renal disease; patients with eGFR < 15 ml/min/1.73 m2 are likely requiring renal dialysis, and patients with eGFR 15–60 ml/min/1.73 m2 can be considered to have some stage of CKD [18]. Both laboratories utilized the CKD-EPI equation to derive the eGFR, reporting both African-American and non-African-American eGFR results. Since the laboratory datasets do not include race, the present analysis used non-African-American eGFR scores because previous publications suggest that approximately 75% of the US HCV population is non-African-American [35]. Sensitivity analysis of using African-American versus non-African-American eGFR values resulted in 3.47%, 0.20%, and 0.09% lower absolute percentage points for eGFR categories of 30–59, 15–29, and < 15 ml/min/1.73 m2, respectively.
Discussion with laboratory vendors indicated that the study dataset largely did not include dialysis patients. On the basis of internal calculations, HCV RNA-positive patients with an eGFR < 15 ml/min/1.73 m2 represent approximately 6% of the total HCV RNA-positive population.
Urine albumin is an additional marker of kidney damage [34]. Only 3.5–4.9% of patients with a positive HCV RNA test had a urine albumin record from 2013 through 2016 in the study dataset, and therefore urine albumin was not explored further for trend analysis in this study.
HIV co-infection as a dichotomous variable was determined on the basis of diagnosis code or positive HIV RNA test.
RAS refer to mutations in the HCV viral genome that may reduce response to antiviral therapies [18]. Results on NS5A, NS5B, and NS3 polymorphisms (i.e., wild-type and substitutions) were available in the study dataset. Approximately 6% of patients in 2016 had a RAS test reading.
Imputation Algorithms for Treatment Receipt and Attaining Sustained Virologic Response
Continuity of medical or pharmacy benefit enrollment was not available in the data, nor was there direct information on treatment timing, type, or duration. To address this data limitation, data-driven imputation algorithms were used to identify patients who initiated treatment and patients who achieved virologic cure (Fig. 2). The algorithms were built and validated against another cohort of 49,421 treated HCV patients, identified in Symphony Health Solutions (SHS) medical and pharmacy claims dataset from 2013 through 2016. The SHS database is nationally representative and directly captures claims from commercial and government (e.g., Medicare, Medicaid) claims-processing intermediaries independently of a patient’s participation in a health plan or payer type. The SHS cohort consisted of a subset of the same patients as the study dataset and had linked HCV RNA lab measurements; however, as a result of HIPAA compliance restrictions, the patient identifier commonality between the study and SHS datasets was unknown. The SHS cohort data therefore reflected the same laboratory data structure of the study RNA dataset and offered the benefit of detailing the temporal profile of the decline in RNA viral load from beginning to after end of treatment.
Fig. 2.
Conceptual description of treatment and cure imputation algorithms
The first steps of the treatment algorithm involved (1) exploring the relationship between RNA viral load decline and time since initiation in the SHS database and (2) defining a minimum meaningful RNA decline attributable to HCV treatment that could then be applied to the study dataset to flag patients who can be assumed to have initiated therapy. It was estimated that a viral load decline of at least 1.2 × log10 units (equal to the threshold of identification for most HCV RNA tests) indicated that treatment was initiated in the immediate period prior to the decline. This decision is supported by review of the HCV RNA kinetics computational modeling literature, suggesting that HCV RNA decline is observable in the immediate days after starting treatment (shown as 0.6 × log10 decline per day) [36]. A detailed description of the treatment imputation algorithm is presented in the Supplementary Materials in Appendix A.
For the cure algorithm, achieving SVR was defined in the SHS dataset for patients who continued to have negative HCV RNA for 4–30 weeks after the end of treatment. Using data for SHS patients with known SVR status only, four ensembles (one for each year from 2013 through 2016) of machine learning models predicting SVR as an outcome were developed and optimized via iterative resampling in training and hold-out testing datasets. For each ensemble, various individual types of machine learning models (i.e., random forest, decision tree, neural network, elastic net, ridge regression, lasso regression, and logistic regression) were trained on the same patient sample, and then the predictions of each individual model were combined using the ensembling technique model stacking, implemented via gradient boosting. The ensembling approach helped increase predictive performance in terms of balanced sensitivity, specificity, and area under the receiver operating characteristic curve. The use of machine learning methods has improved predictive modelling in other studies in HCV, as well [37–39].
Variables describing the relationship between RNA decline and timing or RNA measurement since the first positive RNA were found to be the most predictive of SVR. Such variables included the RNA value at the last observable week, median and mean RNA values over the follow-up, covariance and correlation metrics of timing of RNA measurement and RNA viral load, and the linear slope between RNA viral load and time since the first positive RNA. Additional descriptors of high predictive power included being on Medicaid or Medicare insurance, age, fibrosis stage F4, genotype 1B, and others. The sensitivity and specificity of the developed algorithm were consistently above 0.90 for each of the 4 years. A detailed description of the SVR imputation algorithm and associated performance metrics is presented in Appendix B in the Supplementary Material.
Patient Classification Scheme in Study Dataset
The process of categorization into the treatment-naïve or treatment-experienced categories was based on whether patients exhibited a detectable RNA decline for the first time or had another RNA decline in previous years. On the basis of the initial flags for whether patients exhibited a sizeable RNA decline (i.e., were treated) or achieved SVR, a patient classification scheme categorized patients into one of five mutually exclusive categories for each year of observation from 2013 through 2016: treatment-naïve patients not initiating therapy, treatment-naïve patients initiating therapy, treatment-experienced patients who were not retreated, treatment-experienced patients who were retreated, and cured patients treated in the prior year. Patients who were predicted to have achieved SVR on the basis of the cure algorithm were classified as cured and attributed to the year following the year of treatment (e.g., if patient has sizeable RNA decline in 2014, the patient is classified as treated in 2014 and cured in 2015). By default, this approach indicated that no patients in 2013 could be classified as cured given the lack of data for 2012.
As a result of the longitudinal nature of the study data from 2013 through 2016, patients may not have consistent measurements in each of the 4 years and may have gap years in which no HCV RNA measurement was available. Potential gap years were addressed in the following manner: for those who did not have any RNA measurement in a particular year, patients were assumed to still be HCV-infected (i.e., still in the health care system) as long as they had another clinical test measurement during the year (e.g., genotype) and had tested positive for HCV RNA in previous years.
Another objective of the study dataset was to estimate the epidemiological characteristics of the HCV patient population on the basis of all commercial, Medicare, and Medicaid payers’ geographic footprint in the USA in every 3-digit ZIP code. To achieve this goal, a separate detailed dataset from Decision Resources Group (DRG) with lives covered by various payers in 2016 was used to derive each payer’s market share. The datasets generated and/or analyzed during the current study are not publicly available as they were obtained from a proprietary database through a license agreement. The market share for each payer per 3-digit ZIP was then used as a random sampling statistic through which HCV-positive patients from the study dataset were attributed to each particular payer. Results from this research effort may be presented in a future publication.
Software
Data cleaning and manipulation were conducted with SAS 9.4 (Cary, NC, USA). The machine learning algorithm was implemented in R software (R Foundation for Statistical Computing, Vienna, Austria). Mapping of HCV prevalence rates was conducted in ESRI ArcGIS Desktop 10.5 (Environmental Systems Research Institute, Redlands, CA, USA).
Results
Overview of Patient Dataset
A summary of characteristics from all patients in the study dataset is shown (Table 2). The total number of patients in the study dataset increased from 200,066 in 2013 to 469,550 in 2016. The proportion of all patients with genotype, fibrosis stage, eGFR, and Child–Pugh score information availability rose from 2013 to 2016, indicating improved collection of clinical information.
Table 2.
Data availability of all patients in the study dataset
| Variable description, N (%) | 2013 | 2014 | 2015 | 2016 |
|---|---|---|---|---|
| Patients | 200,066 (100) | 339,836 (100) | 426,510 (100) | 469,550 (100) |
| Untreated—TX naïve | 186,823 (93.38) | 285,696 (84.07) | 297,466 (69.74) | 308,126 (65.62) |
| Initiated therapy—TX naïve | 13,109 (6.55) | 44,669 (13.14) | 93,775 (21.99) | 89,020 (18.96) |
| Not retreated—TX experienced | 0 (0.00) | 2188 (0.64) | 6114 (1.43) | 5296 (1.13) |
| Retreated—TX experienced | 134 (0.07) | 463 (0.14) | 1179 (0.28) | 1075 (0.23) |
| Cured | 0 (0.00) | 6820 (2.01) | 27,976 (6.56) | 66,033 (14.06) |
| Age | 181,777 (90.86) | 313,267 (92.18) | 413,403 (96.93) | 445,682 (94.92) |
| Gender | 181,607 (90.77) | 312,999 (92.10) | 413,133 (96.86) | 445,267 (94.83) |
| Genotype | 91,752 (45.86) | 178,129 (52.42) | 247,331 (57.99) | 258,228 (54.99) |
| Fibrosis stage | 82,549 (41.26) | 166,119 (48.88) | 252,199 (59.13) | 288,775 (61.50) |
| eGFRa | 114,875 (57.42) | 201,619 (59.33) | 270,581 (63.44) | 293,441 (62.49) |
| Child–Pugh class (% of F4 patients) | 6450 (56.83) | 14,649 (56.57) | 24,132 (55.54) | 27,008 (54.92) |
Percentages represent proportion of total patients with non-missing data for a respective year unless otherwise stated
aDialysis patients not included
Using the detection of RNA decline over time as a proxy for flagging patients as having initiated therapy, the treatment algorithm identified 6.62% of patients being treated in 2013, 13.55% in 2014, 25.83% in 2015, and 22.33% in 2016 (among HCV-positive patients in that year who were eligible for therapy). Treatment rates were higher among those on Medicare, followed by patients with commercial insurance, and substantially lower among those on Medicaid.
A summary of the predictive performance of the cure algorithm and predicted SVR proportions in patients flagged as treated in this dataset is shown (Table 3). Predictions in datasets closely followed observed cure rates in the nationally representative SHS. As expected, with the introduction of more effective DAA treatments each year, SVR rates increased over the study period from at least 70% in 2013 to approximately 95% in 2016. Cure rates generally were similar across patients with different insurance, although treated Medicaid patients consistently had slightly lower SVR.
Table 3.
Predictive performance of the cure algorithm and summary of SVR predictions
| Parameter | Year 2013 | Year 2014 | Year 2015 | Year 2016 |
|---|---|---|---|---|
| Accuracy | 0.982 | 0.987 | 0.994 | 0.997 |
| Sensitivity | 0.995 | 0.997 | 0.999 | 0.999 |
| Specificity | 0.953 | 0.916 | 0.914 | 0.956 |
| SVR in each dataset | ||||
| SHS—as observed | 70% | 87% | 94% | 94% |
| SHS—predicted | 71% | 88% | 94% | 94% |
| Laboratory dataset 1—predicted | 78% | 85% | 94% | 95% |
| Laboratory dataset 2—predicted | 71% | 83% | 94% | 94% |
Trend Analysis for Care Engagement with Respect to Age
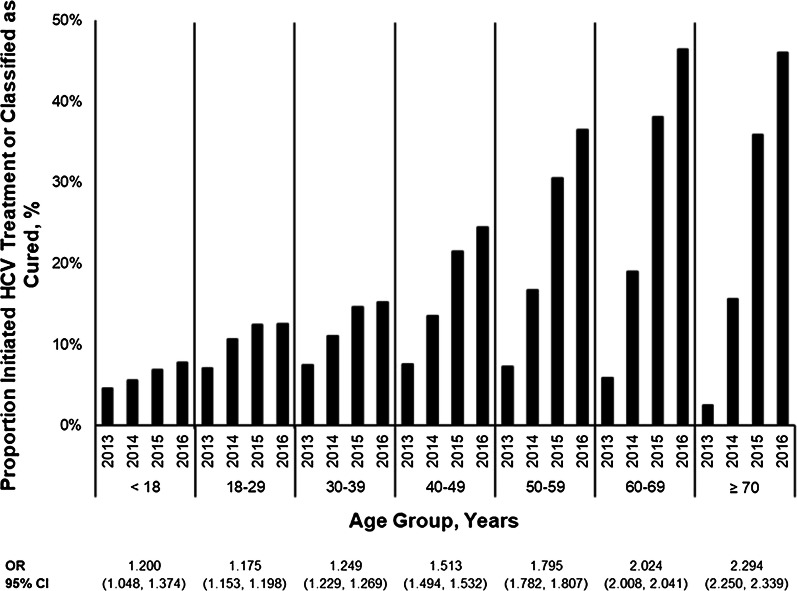
Previous analyses have demonstrated that older patients may be prioritized for HCV treatment [17, 40]. To confirm any evidence of care engagement and prioritization in this study, the cumulative proportions of patients who initiated treatment, were retreated, or were classified as cured in a given year were combined and compared across age strata. The average annual increase in the odds of treatment or cure was calculated for patients attributed to each year from 2013 through 2016 using logistic regression. Results demonstrate that the odds of being treated/cured increased over time for all patients, regardless of age (Fig. 3). However, the odds were highest for patients ages 70 years and older (OR = 2.294; 95% CI = 2.250, 2.339) and lowest for ages 18 through 29 years (OR = 1.175; 95% CI = 1.153, 1.198).
Fig. 3.
Proportion and odds ratio of evidence of care engagement by age group, 2013–2016. CI confidence interval, OR odds ratio
Analysis of Treated Patients by State
To investigate treatment trends further, the prevalence of nearly 90,000 treated HCV patients in 2016 was determined on a state-by-state basis. States with the highest prevalence of treated HCV patients were mainly found on the West Coast, Appalachia, the Northeast, and the Southeast, while much of the Upper Midwest and Great Plains had the lowest prevalence of treated patients (Fig. 4).
Fig. 4.
Treated HCV RNA-positive patients per 100,000 residents, 2016
Analysis of Untreated Patient Population
The identification of treated patients also conversely enabled a review of the untreated HCV patient landscape. Patient classification and derived clinical and demographic characteristics for all patients who had a positive HCV RNA test and were untreated or not retreated in the study dataset are shown in Table 4. Notably, the total number of untreated or not retreated patients nearly doubled from 2013 (N = 186,823) to 2016 (N = 313,422). The proportion of patients with genotype 3 increased from 2013 (10.14%) to 2016 (12.64%). Patients with a fibrosis stage of F0 composed a larger proportion of the total untreated/not retreated patient group in 2016 (30.92%) as compared to 2013 (19.04%). The median untreated patient age decreased from 55 years in 2013 to 53 years in 2016, as patients under age 40 composed a larger proportion of the total patient population in 2016 (26.24%) as compared to 2013 (16.58%).
Table 4.
Untreated patients classification and characteristics
| Variable description | Statistic or category | 2013 (N = 186,823) | 2014 (N = 287,884) | 2015 (N = 303,580) | 2016 (N = 313,422) |
|---|---|---|---|---|---|
| Patient classification, N (%) | Untreated—TX naïve | 186,823 (100.00) | 285,696 (99.24) | 297,466 (97.99) | 308,126 (98.31) |
| Not retreated—TX experienced | 0 (0.00) | 2188 (0.76) | 6114 (2.01) | 5296 (1.69) | |
| Patient age, years | Median (Q1 to Q3) | 55.00 (46.00 to 60.00) | 55.00 (47.00 to 61.00) | 55.00 (45.00 to 61.00) | 53.00 (39.00 to 60.00) |
| Patient age, N (%) | Less than 18 years | 579 (0.34) | 705 (0.27) | 770 (0.26) | 822 (0.28) |
| 18–29 years | 12,203 (7.20) | 18,833 (7.15) | 24,147 (8.26) | 32,726 (11.11) | |
| 30–39 years | 15,312 (9.04) | 24,662 (9.36) | 32,482 (11.11) | 43,710 (14.84) | |
| 40–49 years | 26,318 (15.53) | 36,831 (13.98) | 39,325 (13.45) | 41,210 (14.00) | |
| 50–59 years | 67,697 (39.95) | 101,009 (38.34) | 104,270 (35.67) | 92,410 (31.38) | |
| 60–69 years | 38,895 (22.95) | 67,728 (25.71) | 76,216 (26.07) | 69,977 (23.77) | |
| 70+ years | 8453 (4.99) | 13,699 (5.20) | 15,138 (5.18) | 13,597 (4.62) | |
| Gender, N (%) | Female | 65,859 (35.25) | 103,654 (36.01) | 115,224 (37.96) | 113,522 (36.22) |
| Male | 103,433 (55.36) | 159,572 (55.43) | 176,916 (58.28) | 180,741 (57.67) | |
| Unknown | 17,531 (9.38) | 24,658 (8.57) | 11,440 (3.77) | 19,159 (6.11) | |
| HCV genotype, N (%) | Genotype 1 | 545 (0.63) | 966 (0.64) | 876 (0.51) | 628 (0.40) |
| Genotype 1A | 52,302 (60.78) | 92,313 (61.20) | 104,508 (60.36) | 93,729 (60.35) | |
| Genotype 1B | 13,991 (16.26) | 24,762 (16.42) | 26,399 (15.25) | 21,407 (13.78) | |
| Genotype 2 | 8888 (10.33) | 14,514 (9.62) | 18,335 (10.59) | 17,204 (11.08) | |
| Genotype 3 | 8727 (10.14) | 15,367 (10.19) | 19,735 (11.40) | 19,628 (12.64) | |
| Genotype 4 | 1097 (1.27) | 1806 (1.20) | 2025 (1.17) | 1614 (1.04) | |
| Genotype 5/6 | 487 (0.57) | 929 (0.62) | 992 (0.57) | 767 (0.49) | |
| Genotype mix | 11 (0.01) | 189 (0.13) | 271 (0.16) | 338 (0.22) | |
| Fibrosis stage, N (%) | F0 | 14,477 (19.04) | 28,170 (20.81) | 42,362 (25.06) | 54,024 (30.92) |
| F1 | 14,315 (18.83) | 24,683 (18.24) | 29,173 (17.26) | 29,818 (17.07) | |
| F2 | 27,485 (36.15) | 45,854 (33.88) | 51,813 (30.65) | 49,465 (28.31) | |
| F3 | 9243 (12.16) | 16,444 (12.15) | 19,164 (11.34) | 17,169 (9.83) | |
| F4 | 10,504 (13.82) | 20,191 (14.92) | 26,554 (15.71) | 24,236 (13.87) | |
| Child–Pugh class, N (%) | A | 4588 (10.72) | 8452 (11.92) | 10,966 (12.79) | 9185 (11.05) |
| B–C | 1327 (3.10) | 2126 (3.00) | 2503 (2.92) | 2340 (2.82) | |
| eGFR non-AA levels, N (%) | eGFR < 15a | 906 (0.85) | 1510 (0.92) | 1995 (1.10) | 1957 (1.08) |
| eGFR15–29 | 970 (0.91) | 1509 (0.92) | 1803 (1.00) | 1713 (0.95) | |
| eGFR 30–44 | 2154 (2.03) | 3659 (2.23) | 4054 (2.24) | 3698 (2.05) | |
| eGFR 45–59 | 5590 (5.26) | 9545 (5.81) | 10,586 (5.85) | 8999 (4.98) | |
| eGFR 60–89 | 37,828 (35.60) | 60,861 (37.04) | 65,846 (36.40) | 58,863 (32.57) | |
| eGFR ≥ 90 | 58,819 (55.35) | 87,207 (53.08) | 96,599 (53.40) | 105,483 (58.37) | |
| HIV, N (%) | Yes | 6943 (3.72) | 8594 (2.99) | 8206 (2.70) | 6494 (2.07) |
Percentages represent proportion of total patients with non-missing data for a respective year unless otherwise stated
aDialysis patients not included
Discussion
To the best of our knowledge, this is the largest study to date to describe the changes in the HCV epidemiology and patient characteristics in the USA. Strengths of this study over other available data sources include the most current available data, use of HCV RNA-confirmed cases, geographic and population expansion beyond the NHANES dataset, and the ability to stratify these epidemiological and clinical data by patient characteristics, payer, disease severity, comorbidities, and year [25–27]. The use of HCV RNA-confirmed test results, as opposed to HCV antibody-positive test results, is of particular importance because of the high false positive rate associated with HCV antibody tests [41]. Immediate future work will focus on identifying trends as stratified within various payer populations.
We noted an increase in the total number of untreated or not retreated HCV RNA-positive patients from 2013 through 2016. Substantial efforts supported by the CDC are focused on screening patients born before 1966 [42, 43]. However, acute HCV cases in patients 20 through 39 years of age have risen dramatically since 2009 [19]. Up to 70% of these new HCV cases in younger patients are attributed to injection drug use [7, 44, 45], particularly associated with the rise in heroin use over the last decade [46]. Our study identifies a change in HCV demographics in 2016 compared to other years, with proportionally more untreated and treated patients younger than 40 years of age as compared to previous years. As the study dataset represents the majority of US HCV cases, we speculate that the shift in demographics among untreated/not retreated patients may be partially explained by (1) the rise in HCV caused by injection drug use, (2) the accumulation of younger patients in the health care system as older patients are prioritized for treatment, and (3) a growing focus on evaluating risk factors and screening for HCV.
While prior evidence suggests that a substantial proportion of chronic HCV patients do not successfully transition through the multi-step HCV care continuum [29, 47], we observed significant improvements in the proportion of patients being tested for genotype and liver fibrosis staging over the study years. This observation is consistent with increased awareness of HCV, improved screening efforts supported by the CDC, and nationwide efforts to improve the outcomes of the linkage to care process in recent years [29, 42, 43, 48]. Future work will also examine HCV antibody screening rates and outcomes across the testing-to-care continuum for patients. Also, differences in treatment rates with respect to insurance may be related to issues of affordability, coverage, and underlying disease severity; these factors will separately be explored and presented in forthcoming individual papers based on this study.
While it is challenging to compare national treatment rates reported here to other nationally representative sources, a comparison can be made with respect to HCV patients with commercial insurance. Published treatment rates from a retrospective analysis of administrative claims of 56,000 HCV adults with commercial insurance from 2013 to 2014 suggest that average treatment rates during the 2013 boceprevir/telaprevir era were 8%, and average treatment rates were 18% during the 2014 sofosbuvir ± simeprevir era [17]. Those estimates are slightly higher than the 7.16% in 2013 and 14.10% in 2014 for patients on commercial insurance flagged as treated in our study, which could be because the treatment algorithm employed for this study required the presence of at least two HCV RNA measurements to detect a decline in viral loads. It is possible that some patients could have undergone treatment but had only one available HCV RNA laboratory measurement observed in the data, and our approach would have failed to flag these patients as treated. As the laboratory data used for this study covered the majority of all HCV laboratory tests in the USA, and treated patients already represent a select group of patients who have gone through the HCV care cascade and are likely to return for more than one HCV RNA measurement, this misclassification bias is assumed to be small and to make up for the difference between this study and previously published estimates.
The developed cure algorithm, which captures the full potential of recent developments in advanced predictive analytics and their application to big data such as our study [49], adequately predicted SVR in these patients. The advantage of using machine learning over standard regression techniques is that it enables modeling of non-linear and non-monotone relationships between variables that are determined in a data-driven way [38]. This analytic approach addressed our limited structural knowledge of the laboratory data generation process in the two datasets and factors that might contribute to any missing measurements.
Limitations
One limitation of this study was the inability to fully capture patients’ past treatment history prior to 2013. A patient whose previous treatment failed might make decisions regarding second treatment initiation and therapy options differently than a treatment-naïve patient. Similarly, specific treatment regimens were not captured by the study dataset. Additionally, there was a possibility of prediction error in identifying cured patients, as viral load may be missing because of insufficient follow-up time. As a result of data truncation, no cured patients were attributed to 2013. There may be HCV-positive patients who are not captured in the study dataset as a result of diagnosis prior to 2013 with no follow-up tests. There is also a chance that a patient may appear in both datasets provided by the national laboratory companies represented. However, the majority of the data is limited to those with a positive HCV RNA viral load test. As stated above, dialysis patients were largely not captured in the underlying laboratory dataset, but compose a minority proportion of the total HCV RNA-positive patient population.
Conclusion
This strategic initiative addressed key gaps in the evidence regarding the evolving HCV epidemiology and treatment landscape. The evidence generated in this study supports the development of holistic model approaches that integrate current strategies for linkage to care and treatment programs for HCV patients. The results of this study can be shared with physicians, payers, and government programs to describe HCV burden from patient and payer standpoints and to inform stakeholders of unmet needs in HCV treatment.
Our results highlight that the epidemiology of HCV is evolving. There are an increasing number of young patients and patients with milder disease than described in previous years. Results of this study should help guide efforts toward the elimination of HCV in this country.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Funding
Sponsorship for this study, article processing charges and open access fee were funded by AbbVie. All authors had full access to all of the data in the study and take complete responsibility for the integrity of the data and accuracy of the data analysis.
Medical Writing Assistance
Medical writing assistance in the preparation of this article was provided by Sarah Ronnebaum of Pharmerit International. This assistance was funded by AbbVie.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work, and have given their approval for this version to be published.
Authorship Contributions
Analytic support for this project was provided by Srimoyee Bose, Isabelle Robert, and Julia Wilkerson, who, at the time of the study, were affiliated with Pharmerit International.
Disclosures
Viktor Chirikov is an employee of Pharmerit International, which received funding by AbbVie to conduct the study. Shivaji R. Manthena is an employee of AbbVie and may own AbbVie stock. Johnathan P. Strezewski is an employee of AbbVie and may own AbbVie stock. Steven E. Marx is an employee of AbbVie and may own AbbVie stock. Sam Saab is a consultant and on the speaker bureau for AbbVie, Gilead, Dova, Salix, Bayer, Merck, and Intercept, and is on the speaker bureau for Bristol-Myers Squibb. AbbVie participated in the interpretation of data, review, and approval of the publication.
Compliance with Ethics Guidelines
This study is based on laboratory, administrative claims, and payer data and does not contain any new studies with human or animal subjects performed by any of the authors.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Footnotes
Enhanced digital features
To view enhanced digital features for this article go to 10.6084/m9.figshare.6280820.
References
- 1.Messina JP, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61(1):77–87. 10.1002/hep.27259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razavi H, et al. Chronic hepatitis C virus (HCV) disease burden and cost in the United States. Hepatology. 2013;57(6):2164–70. 10.1002/hep.26218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61(1 Suppl):S58–68. 10.1016/j.jhep.2014.07.012 [DOI] [PubMed] [Google Scholar]
- 4.Ly KN, et al. Rising mortality associated with hepatitis C virus in the United States, 2003–2013. Clin Infect Dis. 2016;62(10):1287–8. 10.1093/cid/ciw111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chhatwal J, et al. Hepatitis C disease burden in the United States in the era of oral direct-acting antivirals. Hepatology. 2016;64(5):1442–50. 10.1002/hep.28571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley GJ, Strom BL. A national strategy for the elimination of viral hepatitis emphasizes prevention, screening, and universal treatment of hepatitis C. Ann Intern Med. 2017;166(12):895–6. 10.7326/M17-0766 [DOI] [PubMed] [Google Scholar]
- 7.US Centers for Disease Control and Prevention. National viral hepatitis action plan. 2017. https://www.hhs.gov/sites/default/files/National%20Viral%20Hepatitis%20Action%20Plan%202017-2020.pdf. Accessed 24 Oct 2017.
- 8.World Health Organization. Combating hepatitis B and C to reach elimination by 2030. 2016; http://www.who.int/hepatitis/publications/hep-elimination-by-2030-brief/en/. Accessed 8 May 2018.
- 9.Mehta D, Mccombs J, Sanchez Y, Marx S, Saab S. Effectiveness of hepatitis C virus screening laws in United States: evidence from paid claims data from 2010 to 2016. In: The International Liver Congress. 2018. Paris, France.
- 10.Islam N, et al. Hepatitis C cross-genotype immunity and implications for vaccine development. Sci Rep. 2017;7(1):12326. 10.1038/s41598-017-10190-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burstow NJ, et al. Hepatitis C treatment: where are we now? Int J Gen Med. 2017;10:39–52. 10.2147/IJGM.S127689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asselah T, et al. Direct-acting antivirals for the treatment of hepatitis C virus infection: optimizing current IFN-free treatment and future perspectives. Liver Int. 2016;36(Suppl 1):47–57. 10.1111/liv.13027 [DOI] [PubMed] [Google Scholar]
- 13.Alkhouri N, Lawitz E, Poordad F. Novel treatments for chronic hepatitis C: closing the remaining gaps. Curr Opin Pharmacol. 2017;37:107–11. 10.1016/j.coph.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 14.Gane E et al. High efficacy of ABT-493 and ABT-530 treatment in patients with HCV genotype 1 or 3 infection and compensated cirrhosis. Gastroenterology. 2016;151(4):651–659.e1. [DOI] [PubMed]
- 15.Hubbard H, Lawitz E. Glecaprevir + pibrentasvir (ABT493 + ABT-530) for the treatment of Hepatitis C. Expert Rev Gastroenterol Hepatol. 2018;12(1):9–17. 10.1080/17474124.2018.1411802 [DOI] [PubMed] [Google Scholar]
- 16.Kwo PY, et al. Glecaprevir and pibrentasvir yield high response rates in patients with HCV genotype 1–6 without cirrhosis. J Hepatol. 2017;67(2):263–71. 10.1016/j.jhep.2017.03.039 [DOI] [PubMed] [Google Scholar]
- 17.Yao X, et al. Adoption of new agents and changes in treatment patterns for hepatitis C: 2010-2014. Am J Manag Care. 2016;22(6):e224–32. [PubMed] [Google Scholar]
- 18.American Association for the Study of Liver Disease and Infectious Disease Society of America. HCV guidance: recommendations for testing, managing, and treating hepatitis C. 2017. https://www.hcvguidelines.org/sites/default/files/full-guidance-pdf/HCVGuidance_September_21_2017_f.pdf. Accessed 24 Jan 2018.
- 19.Centers for Disease Control and Prevention. Viral hepatitis surveillance, United States, 2015. 2015; https://www.cdc.gov/hepatitis/statistics/2015surveillance/pdfs/2015HepSurveillanceRpt.pdf. Accessed 24 Oct 2017.
- 20.Joy JB, et al. The spread of hepatitis C virus genotype 1a in North America: a retrospective phylogenetic study. Lancet Infect Dis. 2016;16(6):698–702. 10.1016/S1473-3099(16)00124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong GL, et al. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144(10):705–14. 10.7326/0003-4819-144-10-200605160-00004 [DOI] [PubMed] [Google Scholar]
- 22.Edlin BR, et al. Toward a more accurate estimate of the prevalence of hepatitis C in the United States. Hepatology. 2015;62(5):1353–63. 10.1002/hep.27978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goolsby Hunter A, et al. Clinical characteristics, healthcare costs, and resource utilization in hepatitis C vary by genotype. Curr Med Res Opin. 2017;33(5):829–36. 10.1080/03007995.2017.1288613 [DOI] [PubMed] [Google Scholar]
- 24.CDA Foundation. Polaris Observatory. http://polarisobservatory.org/polaris_view/hepC.htm. Accessed 7 Dec 2017.
- 25.Emory University, Rollins School of Public Health, in partnership with Gilead Sciences, Inc. HepVu. https://hepvu.org/. Accessed 7 Dec 2017.
- 26.Rosenberg ES, et al. Estimation of state-level prevalence of hepatitis C virus infection, US States and District of Columbia, 2010. Clin Infect Dis. 2017;64(11):1573–81. 10.1093/cid/cix202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razavi H, et al. The present and future disease burden of hepatitis C virus (HCV) infection with today’s treatment paradigm. J Viral Hepat. 2014;21(Suppl 1):34–59. 10.1111/jvh.12248 [DOI] [PubMed] [Google Scholar]
- 28.Cornett JK, et al. Results of a hepatitis C virus screening program of the 1945–1965 birth cohort in a large emergency department in New Jersey. Open Forum Infect Dis. 2018;5(4):5. 10.1093/ofid/ofy065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez G, et al. Early identification and linkage to care for people with chronic HBV and HCV infection: the HepTLC Initiative. Public Health Rep. 2016;131(Suppl 2):5–11. 10.1177/00333549161310S202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castrejon M, et al. Implementation of a large system-wide hepatitis C virus screening and linkage to care program for baby boomers. Open Forum Infect Dis. 2017;4(3):09. 10.1093/ofid/ofx109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merck Manual. Fibrosis of the liver. https://www.merckmanuals.com/home/liver-and-gallbladder-disorders/fibrosis-and-cirrhosis-of-the-liver/fibrosis-of-the-liver. Accessed 10 Jan 2018.
- 32.Sterling RK, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–25. 10.1002/hep.21178 [DOI] [PubMed] [Google Scholar]
- 33.Lin ZH, et al. Performance of the aspartate aminotransferase-to-platelet ratio index for the staging of hepatitis C-related fibrosis: an updated meta-analysis. Hepatology. 2011;53(3):726–36. 10.1002/hep.24105 [DOI] [PubMed] [Google Scholar]
- 34.Malkina A. Merck Manual. Chronic kidney disease. https://www.merckmanuals.com/professional/genitourinary-disorders/chronic-kidney-disease/chronic-kidney-disease. Accessed 1 Feb 2018.
- 35.Gordon SC et al. Race, age, and geography impact hepatitis C genotype distribution in the United States. J Clin Gastroenterol. 2017. 10.1097/MCG.0000000000000872. [DOI] [PMC free article] [PubMed]
- 36.Perelson AS, Guedj J. Modelling hepatitis C therapy-predicting effects of treatment. Nat Rev Gastroenterol Hepatol. 2015;12(8):437–45. 10.1038/nrgastro.2015.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konerman MA, et al. Improvement of predictive models of risk of disease progression in chronic hepatitis C by incorporating longitudinal data. Hepatology. 2015;61(6):1832–41. 10.1002/hep.27750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singal AG, et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013;108(11):1723–30. 10.1038/ajg.2013.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chirikov VV et al. Tree-based claims algorithm for measuring pretreatment quality of care in Medicare disabled hepatitis C patients. Med Care. 2017;55(12):e104–e112. [DOI] [PubMed]
- 40.Lo Re V 3rd, et al. Disparities in absolute denial of modern hepatitis C therapy by type of insurance. Clin Gastroenterol Hepatol. 2016;14(7):1035–43. 10.1016/j.cgh.2016.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moorman AC, Drobenuic J, Kamili S. Prevalence of false-positive hepatitis C antibody results, National Health and Nutrition Examination Study (NHANES) 2007–2012. J Clin Virol. 2017;89:1–4. 10.1016/j.jcv.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flanigan CA, et al. Evaluation of the impact of mandating health care providers to offer hepatitis C virus screening to all persons born during 1945–1965—New York, 2014. MMWR Morb Mortal Wkly Rep. 2017;66(38):1023–6. 10.15585/mmwr.mm6638a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen C, Carnes CA, Downs A. Know more hepatitis: CDC’s national education campaign to increase hepatitis C testing among people born between 1945 and 1965. Public Health Rep. 2016;131(Suppl 2):29–34. 10.1177/00333549161310S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suryaprasad AG, et al. Emerging epidemic of hepatitis C virus infections among young nonurban persons who inject drugs in the United States, 2006–2012. Clin Infect Dis. 2014;59(10):1411–9. 10.1093/cid/ciu643 [DOI] [PubMed] [Google Scholar]
- 45.Zibbell JE, et al. Increases in hepatitis C virus infection related to injection drug use among persons aged ≤ 30 years—Kentucky, Tennessee, Virginia, and West Virginia, 2006–2012. MMWR Morb Mortal Wkly Rep. 2015;64(17):453–8. [PMC free article] [PubMed] [Google Scholar]
- 46.Center for Behavioral Health Statistics and Quality. 2015 national survey on drug use and health: detailed tables. Rockville: Substance Abuse and Mental Health Services Administration; 2016.
- 47.Blackburn NA, Patel RC, Zibbell JE. Improving screening methods for hepatitis C among people who inject drugs: findings from the HepTLC Initiative, 2012–2014. Public Health Rep. 2016;131(Suppl 2):91–7. 10.1177/00333549161310S214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel RC, Vellozzi C, Smith BD. Results of hepatitis C birth-cohort testing and linkage to care in selected U.S. sites, 2012–2014. Public Health Rep. 2016;131(Suppl 2):12–9. 10.1177/00333549161310S203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.National Academies of Sciences, Engineering, and Medicine. Big data and analytics for infectious disease research, operations, and policy: proceedings of a workshop. 2016, Washington DC: The National Academies Press. [PubMed]
- 50.Levey AS, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–12. 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.