Abstract
Objectives
Mutations in Tissue Inhibitor of Metalloproteinases 3 (TIMP3) cause Sorsby's Fundus Dystrophy (SFD), a dominantly inherited, rare form of macular degeneration that results in vision loss. TIMP3 is synthesized primarily by retinal pigment epithelial (RPE) cells, which constitute the outer blood-retinal barrier. One major function of RPE is the synthesis and transport of vital nutrients, such as glucose, to the retina. Recently, metabolic dysfunction in RPE cells has emerged as an important contributing factor in retinal degenerations. We set out to determine if RPE metabolic dysfunction was contributing to SFD pathogenesis.
Methods
Quantitative proteomics was conducted on RPE of mice expressing the S179C variant of TIMP3, known to be causative of SFD in humans. Proteins found to be differentially expressed (P < 0.05) were analyzed using statistical overrepresentation analysis to determine enriched pathways, processes, and protein classes using g:profiler and PANTHER Gene Ontology. We examined the effects of mutant TIMP3 on RPE metabolism using human ARPE-19 cells expressing mutant S179C TIMP3 and patient-derived induced pluripotent stem cell-derived RPE (iRPE) carrying the S204C TIMP3 mutation. RPE metabolism was directly probed using isotopic tracing coupled with GC/MS analysis. Steady state [U–13C6] glucose isotopic tracing was preliminarily conducted on S179C ARPE-19 followed by [U–13C6] glucose and [U–13C5] glutamine isotopic tracing in SFD iRPE cells.
Results
Quantitative proteomics and enrichment analysis conducted on RPE of mice expressing mutant S179C TIMP3 identified differentially expressed proteins that were enriched for metabolism-related pathways and processes. Notably these results highlighted dysregulated glycolysis and glucose metabolism. Stable isotope tracing experiments with [U–13C6] glucose demonstrated enhanced glucose utilization and glycolytic activity in S179C TIMP3 APRE-19 cells. Similarly, [U–13C6] glucose tracing in SFD iRPE revealed increased glucose contribution to glycolysis and the TCA cycle. Additionally, [U–13C5] glutamine tracing found evidence of altered malic enzyme activity.
Conclusions
This study provides important information on the dysregulation of RPE glucose metabolism in SFD and implicates a potential commonality with other retinal degenerative diseases, emphasizing RPE cellular metabolism as a therapeutic target.
Keywords: Retinal pigment epithelium metabolism, Central carbon metabolism, Isotopic tracer, RPE, TIMP3
Highlights
-
•
SFD mice display alterations in proteins associated with metabolism.
-
•
SFD RPE cells have increased glycolytic activity and glucose contribution to the TCA cycle.
-
•
Glutamine contribution to energy metabolism is unaltered in SFD RPE cells however there is reduced malic enzyme activity.
-
•
SFD RPE cells display metabolic dysfunction potentially implicating metabolism as a viable therapeutic target.
1. Introduction
Sorsby's Fundus Dystrophy (SFD) is a rare autosomal dominant form of macular degeneration which results in progressive vision loss starting in the 3rd to 4th decade of life. Mutations in the Tissue Inhibitor of Metalloproteinases 3 (TIMP3) gene cause the disease with 100% penetrance. TIMP3 is an extracellular inhibitor of matrix metalloproteinases (MMPs) which are involved in the degradation of extracellular matrix (ECM) proteins such as collagen and elastin. TIMP3 is synthesized by the retinal pigment epithelium (RPE) and endothelial cells of the choriocapillaris in the retina. The pathological features of SFD bear a striking similarity to the more common age-related macular degeneration (AMD) with defects in Bruch's membrane (BrM), subretinal deposits (“drusen”) and choroidal neovascularization (CNV) [[1], [2], [3], [4]]. Drusen in both AMD and SFD contain TIMP3, leading some to speculate that they share an underlying etiology [4,5] however, the mechanisms underlying the pathology of AMD and SFD remain unknown. It has been previously reported that TIMP3 mutations result in decreased MMP inhibitor function which could be contributing to the pathological mechanism of SFD [6].
Recently, metabolic dysfunction has emerged as an important contributor to AMD [[7], [8], [9]] as well as other forms of retinal degeneration including retinitis pigmentosa [10], diabetic retinopathy [11,12], retinopathy of prematurity [13,14], and macular telangiectasia (MacTel) [[15], [16], [17]]. Mutations in genes associated with metabolism (glycolysis [18,19], fatty acid beta oxidation [20], tri-carboxylic acid cycle (TCA) [19], and nucleotide synthesis [21,22]) have been shown to cause retinal degeneration. In addition, some system-wide metabolic diseases, are known to negatively impact the retina such as diabetic retinopathy (DR), a vision threatening complication of diabetes mellitus [21].
Metabolic perturbations such as hypoxia-induced metabolic stress, specifically in the RPE, cause photoreceptor degeneration in mice [23]. RPE cells constitute the outer blood retinal barrier and are essential for photoreceptor health and function. One important RPE function is the transport and production of vital nutrients, such as glucose, to the retina. Glucose is needed to support the heavy reliance of photoreceptors on aerobic glycolysis, where glucose is metabolized to lactate in the cytosol to generate ATP [24,25]. Under physiological conditions, RPE spares glucose for photoreceptors by utilizing alternative fuels including proline [[26], [27], [28]], fatty acids [29], glutamine [30] and lactate [31]. The disruption of metabolic coupling between RPE and photoreceptors result in the degeneration of photoreceptors [10]. Additionally, altered mitochondrial metabolism in the RPE of human AMD donor eyes underscores the importance of RPE metabolism in sustaining the retinal metabolic ecosystem [32].
We examined metabolic changes in the RPE expressing mutant TIMP3 that might contribute to the pathogenesis of SFD. Quantitative proteomics analysis on RPE of SFD mice carrying the TIMP3 S179C variant identified dysregulated glycolysis pathways. Using stable isotope labeling, we directly analyzed the effects of mutant TIMP3 on RPE metabolism using two in vitro models of SFD: ARPE-19 S179C TIMP3 and induced pluripotent stem cell (iPSC) derived RPE from patients with the S204C TIMP3 mutation (iRPE). Our data supports the hypothesis that TIMP3 mutations leads to increased glycolytic activity and glucose contribution to the TCA cycle in RPE cells.
2. Materials and methods
2.1. Animals
Heterozygous Timp3+/S179C, a generous gift from Dr. Bernhard Weber [33] were crossed to generate homozygous Timp3S179C6/S179C mice and their age-matched littermate controls. All experimental procedures were approved by the Animal Care and Use Committee (IACUC) guidelines of the Cleveland Clinic and conformed to the National Institutes of Health Guide for the Care and Use of Animals in Research and the ARVO statement for the use of animals in ophthalmic and vision research.
2.2. Proteomics preparation
After euthanasia, tissue was collected from 8 animals of each genotype (WT or TIMP3 S179C, 4 males and 4 females). Retina and RPE lysates were prepared from the enucleated eyes of 7 month old mice as described previously [34]. Protease buffer (200uL containing 100 mM TEAB, 2% SDS, 1 mM DDT) was added to each RPE/eye cup and incubated on ice for 1 h. RPE was dissociated from choroid/sclera with vigorous tapping of the tube. Choroid tissue was removed and RPE cells were lysed by drawing cells through a 26-27-gauge needle 15–20 times. Tissue was sonicated for 10–15 s at medium/frequency, centrifuged (14,000 rpm for 10 min at 4 °C) and the supernatant was used for analysis.
The protein concentration of the soluble protein fraction was estimated by the Bicinchoninic Acid assay (BCA) assay. Each fraction was reduced with DTT (10 mM at room temperature) alkylated with iodoacetamide (40 mM for 1h at room temperature) then quenched with DTT (40 mM) [35]. Two volumes of ice-cold acetone was used to precipitate proteins. Protein pellets were resuspended in triethylammonium bicarbonate buffer (TEAB) buffer (100 mM containing 0.5 mM CaCl2) and were digested overnight at 37 °C with trypsin (initially with 2% trypsin (w/w), followed in 2 h with another 2% (w/w), and the next day with another 1% (w/w) for 2h). AccQ-Tag amino acid analysis was used to quantify soluble peptides [36,37]. Gender-specific pooled WT samples were prepared for proteomic analyses by combining equal amounts of proteolyzed protein from 3 male and 4 female WT controls.
2.3. iTRAQ-labeling and peptide fractionation
iTRAQ-Labeling of tryptic peptides with an 8-plex iTRAQ kit was performed as previously described [37,38] Tryptic digests of the individual KI samples (21 ug each) and the pooled reference samples (21 ug each) were each labeled with a unique iTRAQ tag and combined in a total of 2 batches as follows. Batch 1 contained 6 iTRAQ tags among the pooled male and female reference samples, 2 individual male mutant samples and 2 female mutant samples. Batch 2 was a duplicate of Batch 1 but with different KI samples. Each batch was individually fractionated by reverse-phase high performance liquid chromatography (RP-HPLC) at pH 10 using methods detailed elsewhere [38] and chromatography fractions encompassing the entire elution were selectively combined and dried. Ten fractions per batch were analyzed by LC MS/MS.
2.4. Protein identification
RP-HPLC fractions were analyzed by LC- MS/MS on an Orbitrap Fusion Lumos Tribrid mass spectrometer [37,38]. Protein identification utilized the Mascot 2.6.2 search engine and the UniProt mouse database version 2019_07 (17,026 sequences). Database search parameters were restricted to three missed tryptic cleavage sites, a precursor ion mass tolerance of 10 ppm, a fragment ion mass tolerance of 20 mmu and a false discovery rate of ≤1%. Protein identification required detection of a minimum of two unique peptides per protein. Fixed protein modifications included N-terminal and ε-Lys iTRAQ modifications and S-carbamidomethyl-Cys. Variable protein modifications included Met oxidation, Asn and Gln deamidation and iTRAQ Tyr. A minimum Mascot ion score of 25 was used for accepting peptide MS/MS spectra.
2.5. Protein quantitation
The iTRAQ tag intensities on male and female KI mouse peptides versus their gender-specific pooled WT control were quantified by the weighted average method [39] using the Mascot 2.6.2 Summed Intensities Program. Protein quantitation required a minimum of two unique peptides per protein, utilized reporter ion tolerance of 10 ppm, and a Mascot peptide ion score ≥25. Protein ratios were determined in log space and transformed back to linear ratios for reporting.
2.6. Statistical analyses of the proteomic data and enrichment analysis
Quantile normalization was used to normalize the mass spectrometric iTRAQ proteomics data. Batch effects were examined. Significantly elevated and decreased proteins were sought using the limma package in R, and the results were adjusted for multiple-testing using the Benjamini-Hochberg procedure [33,40]. Criteria for significantly elevated and decreased proteins included average protein ratios (TIMP3 mutant/gender specific WT) above or below the mean by at least one standard deviation with adjusted p-values ≤0.05. All Proteins found to be significantly altered (adjusted p-value <0.05) were used for enrichment analysis. KEGG enrichment analysis was conducted using G-profiler g:GOSt tool (https://biit.cs.ut.ee/gprofiler/gost, Version e111_eg58_p18_f463989d), implementing the g:SCS threshold with a significance of 0.05. PANTHER statistical overrepresentation test (PANTHER, Gene List Analysis v 18.0, https://www.pantherdb.org/) was used with the PANTHER pathway, GO Biological Process Complete and GO Protein Class Complete annotation set with the Fisher's exact test with False Discovery Rate (FDR) correction. Pathways (all significant parent terms, FDR<0.01), processes (top ten enriched parent term, FDR <0.01), classes (top eight enriched parent terms, FDR <0.01) were graphed using Flourish Studio (https://flourish.studio). Panther uses hierarchical relationships between terms which are broken down into sub classes. To determine the most enriched terms, only parent terms were considered so subclasses within the same pathway would not be over-represented. Data was visualized so that the annotations were on the the Y axis, fold enrichment was on the x-axis, FDR is represented by color and number of genes is represented by the size of the bubble.
2.7. SFD ARPE-19 cells
The generation of the SFD ARPE-19 cells has been published by our laboratory previously [41]. Briefly, a 550bp human S179C or WT TIMP3 cDNA insert was ligated into a pCEPT4 expression vector (Invitrogen). S179C TIMP3 or WT TIMP3 cDNA in pCPE4 vector was transfected into ARPE-19 cells (ATCC CRL-2302) using Effectene Transfection Reagent (Qiagen) using the manufacturer's protocol. Stable clones were isolated with Hygromycin selection. TIMP3 overexpression was confirmed with western blot followed by reverse zymography.
2.8. ARPE -19 cell culture
Cells were cultured using protocol adapted from Hazim et al., 2019 [42]. ARPE-19 were expanded using DMEM: F12, 10% FBS (Gibco #A526801, 1% Penicillin/Streptomycin until confluent within a T75 flask. After becoming confluent, cells were allowed to further differentiate in RPE media (MEM alpha with GlutaMAX (Thermo, #32561037), 1% FBS, 1% Penicillin/Streptomycin, 1% N1 supplement (Sigma, #N6530), taurine (0.25 mg/mL, Sigma #Y0625), hydrocortisone (20 ng/mL, Sigma #50396), triiodo-thyronin (0.013 ng/mL, Sigma, #T6397), and nicotinamide (10 mM Sigma #N0636) which yielded a homogenous and polarized monolayer for at least 3 weeks. Cells were dissociated with Trypsin/EDTA (0.025% EDTA) for 5 min at 37 °C. The reaction was quenched with DMEM: F12, 10% FBS, 1% Penicillin/Streptomycin before being spun down and resuspended in RPE media. Cells were plated in flat bottom 6-well tissue culture plates at a seeding density of 1.66 × 105 cells/cm2 in RPE media. Cells were maintained at 37 °C and 5% CO2. Media was changed 2 times per week for 4 weeks until isotopic tracing experiments.
2.9. ARPE19 glucose isotopic tracing
To ensure that the cells had adequate nutrients before labeling started, fresh (unlabeled) cell culture media was given to the cells 24 h prior. At the start of the experiment (time 0) unlabeled media was aspirated and wells were quickly washed with 3 mL warm normal saline (0.8% NaCl). Then, 2 mL of media containing [13C6] glucose was added. The media had a base of DMEM without glucose, without pyruvate, and without l-glutamine (Lerner Research Institute Central Cell Services Media Laboratory, 99AC500CUST) with 10 mM [13C6] labeled glucose (Cambridge Isotope Laboratories, 99%, Cat. # CLM-1396-PK.), 2.5 mM unlabeled glutamine, 10% dialyzed FBS, 1% Penicillin/Streptomycin antibiotics. Cells were incubated with labeled media at 37 °C for 24 h.
2.10. ARPE-19 metabolite extraction from cells and media
After 24 h, media was collected, flash frozen, and stored at −80 °C until analysis. Cells were rinsed with 1 mL warm normal saline (0.9% NaCl, Sigma S5886, Millipore ZEQ7000T0C). Cells were immediately put on ice and 1 mL of ice cold 80% Methanol (Fisher Scientific A454-4) containing 0.05 mg/mL 13C Ribitol internal standard was added to each well. Wells were scraped and all contents were transferred to a fresh 1.5 mL tube and immediately put on wet ice. Samples were briefly sonicated to ensure lysis (∼5 s) and then centrifuged (15,000×g for 5 min at 4 °C). 500 μL of supernatant was transferred to a fresh 1.5 mL tube. Samples were dried in a centrivap concentrator (Labconco 7310021) over-night at 4 °C and immediately derivatized the next morning.
Media samples stored in −80 °C were thawed on wet ice and vortexed. To 20 μL of media in a fresh tube, 70 μL of −20 °C Methanol and 10 μL of 0.05 mg/mL 13C Ribitol standard was added, vortexed, and placed on ice. Samples were centrifuged at 15,000×g for 10 min at 4 °C. 50 μL of supernatant was moved to a fresh tube and dried overnight in Centrivap at 4 °C.
2.11. ARPE-19 derivatization protocol
Samples were placed in Centrivap at 40 °C for 5 min to ensure removal of all moisture then immediately derivatized. 25 μL of Methoxyamine hydrochloride (Sigma 593-56-6)/pyridine (Sigma 110-86-1) solution (40 mg/mL) was added to each tube. The samples were mixed at 45 °C at 1,000 rpm for 30 min on the ThermoMixer (Eppendorf 5382000023). 75 μL of MSTFA +1% TMCS (Thermo Scientific 48915) was added to each tube. Samples were incubated at 45 °C at 1,000 rpm for 30-minutes on the ThermoMixer. Samples were centrifuged for 2 min at room temperature at 15,000×g. 68 μL of supernatant was put in GC/MS vials (Thomas Scientific 1152P35, Macherey–Nagel 702716).
2.12. ARPE-19 mass spectrometry/gas chromatography method
One microliter of each sample was injected into the 7890B GC connected to 5977 MSD Agilent GCMS system. Injections were made in splitless mode. The fused silica capillary column used was a DB-5MS 30 m × 0.25 mm × 0.25 μm with a 10 m of built in guard column (Agilent 122-5532G). The front inlet was set to 250 °C with septum purge flow of 3 mL/min of helium. Samples were analyzed in a constant flow mode with helium set to 1.1 mL/min. The GC method was 1 min at 60 °C, followed by 10 °C/min increments until 325 °C and finally held at 325 °C for 10 min. Pyruvate and lactate were measured by select ion monitoring (SIM). Ions 174–177 were monitored for pyruvate from 6.6 to 7.7 min while ions 219–222 were monitored for lactate from 7.7 to 9 min. Glucose in media was measured in full scan mode using electron ionization with a scan widow from 50 to 800 m/z. A solvent delay of 6.6 min was applied.
2.13. APRE-19 data analysis
Agilent Masshunter was used to quantify peak height for M0-Mn. Relative metabolite levels calculated by summing the isotopolouge heights. To account for differences in cell numbers between WT and mutant conditions, a correction factor was calculated using the ratio of WT to mutant (1) cells numbers, (2) total protein amounts (measured by BCA) and (3) total nucleic acid content. We averaged the ratios of 11 independent cell counts (0.79), total protein concentrations (Pierce BCA assays) from lysates of 6 experiments (0.89) and 1 measurement of total nucleic acids (0.85). We averaged all the ratios to get a final correction factor of 0.85. All mutant peak heights were multiplied by the correction factor and then fold change was calculated by dividing the peak height sums by the average of the control group. Isocor [43] V 2.0 was used to correct for natural isotopic abundances and calculate fractional enrichment.
2.14. Reprogramming of control and patient fibroblasts into iPSCs
iPSCs were reprogrammed from control and patient fibroblasts (confirmed for TIMP3 S204C mutation) as described previously [44,45] Briefly, fibroblasts were electroporated with non-integrating episomal vectors containing reprogramming factors (pCXLE-hOCT4-shP53, pCXLE-hSK and pCXLE-hUL) [[44], [45], [46]]. Electroporation was carried out using the nucleofection kit for primary fibroblasts (Lonza) and Nucleofector 2b Device (Lonza, Program T-016). Following this, the electroporated cells were cultivated in fibroblast medium (DMEM with high glucose, 10% fetal bovine serum, 2 mM glutamine, and 100 U/mL penicillin-streptomycin) for 6 days. After 6 days, reprogrammed fibroblasts were trypsinized and plated onto irradiated mouse embryonic fibroblast (MEF) feeder cells and maintained in DMEM/F12 (Gibco, ThermoFisher Scientific) with 20% Knockout serum replacement (Gibco, ThermoFisher Scientific), 1% MEM-NEAA (Gibco, ThermoFisher Scientific), 1% glutamax (Gibco, ThermoFisher Scientific), and 100 ng/mL FGF2; Peprotech). iPSC colonies appeared at ∼ day 17–30 from transfection and were manually collected for further expansion.
2.15. Differentiation of control and patient iPSCs into iRPE
iPSCs were routinely maintained on Matrigel (Corning) in mTeSRTM (Stemcell Technologies) and differentiated into iRPE using previously described protocol [44,45,47,48]. Briefly, confluent iPSC colonies were initially dissociated with ReLeSR™ and cultured as free-floating embryoid bodies (EBs) in T-25 flasks. Plating of EBs on laminin-coated 6 well plates was performed on day 6 and maintained in neural induction medium (NIM: DMEM/F12 containing 1% MEM-NEAA (Gibco, ThermoFisher Scientific), 1% Glutamax (Gibco, ThermoFisher Scientific), 1% N2 Supplement (Gibco, ThermoFisher Scientific) and 2 mg/mL heparin). On day 14, the EBs were switched to retinal differentiation medium (RDM). RDM was prepared by using 70% DMEM (Gibco, ThermoFisher Scientific, #11965-092), 30% F-12 (Cytiva #SH30026.01), 2% B-27 supplement without Vitamin A (Gibco, ThermoFisher Scientific) and 1% (v/v) GibcoTM Antibiotic-Antimycotic (ThermoFisher Scientific). RPE started to appear at around day 60–90 timepoint in the differentiated iPSC cultures. Patches of RPE were dissected out under a microscope followed by dissociation with 0.05% Trypsin–EDTA (Gibco, ThermoFisher Scientific) and further plated onto laminin-coated (Gibco, ThermoFisher Scientific) 24-well plates with RDM containing 10% FBS for 2 feedings and changed to RPE containing 2% FBS until they formed a confluent monolayer and then switched to RDM only. This was considered as passage 0 (P0) RPE monolayers. Control and patient iRPE cultures were passaged as needed. Cells were only used at passage 3 and below.
2.16. Generation of gene corrected iRPE control line
The day before transfection, hiPSCs were dissociated into single cells using TrypLE and seeded onto Matrigel coated 96-well plate at a density of ∼3–4 x 104 cells per well in mTeSR plus medium supplemented with 10 μM ROCK inhibitor (Y-27632; STEMCELL Technologies). Alt-R CRISPR-Cas9 crRNA(5′-TGATGATGCATTTATCCGGG-3′), tracrRNA ATTOTM 550 (Integrated DNA technologies) were resuspended and crRNA: tracrRNA duplexes were prepared according to the manufacturer's instructions. A transfection complex was prepared with 6 nmol of Alt-R™ SpCas9 nuclease V3 (Integrated DNA technologies), 12 nmol crRNA: tracrRNA and 50 nmol of ssODN donor construct (5′GCATCCGGCAGAAGGGCGGCTACTGCAGCTGGTACCGAGGATGGGCCCCTCCGGATAAAAGCATCATCAATGCCACAGACCCCTGAGCGCCAGACCCTGCCCCACCTCAC3') using Lipofectamine Stem Transfection Reagent (Cat. No. STEM00001; Thermo Fisher Scientific) according to manufacturer's instructions and added dropwise to the well. The media was replaced after 24 h and on day 3 post-transfection, cells were dissociated using TrypLE and seeded at low density (∼300–500 cells) in each well of 6-well plate). The clones were manually picked and expanded. For screening, gDNA PCR was performed using primers TIMP3-F (5′CCTGCTACTACCTGCCTTGC) and TIMP3-R (5′GGGTGGGAATTACAATCCTCAAATG) followed by restriction digestion using enzyme Nsil (New England Biolabs, Cat no. R0127L). Sanger sequencing was performed to verify the mutation correction followed by karyotyping to confirm that no chromosomal abnormalities were present.
2.17. iRPE transportation
Cells were transported by car in a portable incubator (VEVOR XHC-25) from The University of Rochester (Rochester, NY) directly to Cleveland Clinic Cole Eye Institute (Cleveland, OH). Cells were maintained at 37 °C +/− 1 °C during the 5 h of transit. Media was supplemented with 25 mM of HEPES buffer (Gibco #15630080) to maintain media buffering capacity without CO2 supplementation. Immediately upon arrival, media was replaced with fresh RDM media. Cells were maintained at 37 °C, 5% CO2, 95% humidity for at least one month before experiments were conducted.
2.18. iRPE isotopic tracing
Forty-eight hours before experiments, fresh RDM media was added to ensure that the cells had adequate nutrients before labeling was initiated. Isotopic tracing was conducted in DMEM media without glucose, without pyruvate and without glutamine, (Cleveland Clinic Custom) supplemented with 10 mM labeled or unlabeled glucose and 2.5 mM labeled or unlabeled glutamine. Glucose tracing experiments contained 10 mM uniformly labeled [13C6] Glucose and 2.5 mM unlabeled glutamine. Glutamine tracing experiments contained 10 mM unlabeled glucose and 2.5 mM uniformly labeled [13C5] glutamine. Cells were incubated (37 °C, 5% CO2, 95% humidity) with 1 mL of glucose or glutamine labeled media for 24 h.
2.19. iRPE metabolite extraction
After 24 h, the media was collected, and flash frozen for extracellular metabolomics. Cells were quickly washed once with 1 mL of chilled normal saline (0.9% NaCl). Intracellular metabolism was quenched, and metabolites extracted by adding 200 μL of ice cold 80% methanol containing 13C ribitol and Norvaline (Waters 186009301) as internal standards. Plates were immediately put on ice and cells were detached using plastic cell scraper. To ensure full cell lysis, cells were sonicated at 4 °C for 30–40 s. Proteins were precipitated out through centrifugation at 15,000×g for 5 min at 4 °C. 25 μL of supernatant was transferred to a fresh 1.5 mL tube before being dried at room temperature using a SpeedVac concentrator (Savant SC110-120). Samples subsequently were stored at −80 °C.
Extracellular media samples were thawed on ice and metabolites were extracted from 5 μL of media with 95 μL extraction buffer (ice cold 80% methanol containing 5 μL of 0.05 mg/mL ribitol and 5 μL of 5.86 pg/μL Norvaline as internal standards) Samples were centrifuged at 15,000×g for 10 min at 4 °C. 50 μL of supernatant was moved to a fresh tube and dried at room temperature for 20 min with Savant SpeedVac concentrator.
2.20. iRPE derivatization
Derivatization protocol was adapted from previously published protocols [49]. Briefly, to ensure samples were completely dry, they were put into the SpeedVac concentrator for 5 min 10 μL Methoxyamine/pyridine (40mg/1 mL) was added to dry samples. Using an Eppendorf ThermoMixer® C, samples were mixed at 300 rpms for 1.5 h at 37 °C. 30 μL MTBSTFA (N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide, MilliPore Sigma, 394882) was added and mixed on the thermomixer at 300rpms for 30 min at 70 °C. Samples were centrifuged for 10 min at room temperature before 30 μL of supernatant was moved to Crimp Vials with glass inserts and capped.
2.21. GC/MS analysis
Methods were adapted from previous publications [49]. An auto sampler (Agilent 7693A ASL) was used to inject 1 μL in splitless mode using a 5 μL syringe (Agilent G4153-80213) into an Agilent 8890 GC followed by an Agilent 5977C MSD. Inlet was lined with a splitless fritted liner (Agilent 5190-5112-025) at a temperature of 250 °C and pressure of 14.067 psi. To avoid carry-over, the syringe was washed twice before injection and three times after injection. Each wash consisted of 2 μL hexane followed by 2 μL acetone. The total inlet flow was 54 mL/min and septum purge flow was 3 mL/min in standard mode. Purge flow to split vent was 50 mL/min at 0.5 min. The column used was an ultra-inert DB-5ms (UI) with 30m length x 0.25 mm inner diameter x 0.25 μm film thickness (Agilent 122-5532UI) with a constant flow of 1 mL/min. Initial oven temperature was 95 °C with a 2-minute hold time and was ramped to 270 °C at a rate of 10 °C/min. Oven was ramped a second time at 5 °C/min until 300 °C was reached and held for 6 min. MSD transfer line was constantly kept at 290 °C. MSD was run full scan mode (50–600m/z) with a solvent delay of 5.95 min. MSD Source and quad were set to 250 °C and 150 °C, respectively.
2.22. GC/MS quality control
Each run contained 2 solvent blanks to assess carry-over, 2 derivatization blanks to access background, and 2 biological quality control samples made from pooled porcine RPE that were extracted the same way as experimental samples. After derivatization, biological QC samples were pooled once again to ensure uniformity before being separated into two separate vials for GC/MS analysis. One QC samples was injected 3 times at the beginning of the sequence and the other QC sample was injected 3 times at the end to confirm reproducibility and evaluate machine drift over the run. Each metabolite was evaluated separately for reducibility by quantitating the area under the curve (AUC) normalized to total sum. Only metabolites that had a relative standard deviation under 7% across all six QC injections were included in results.
2.23. iRPE extracellular glucose concentration assay (Hexokinase/G6PDH method)
Glucose concentration was measured using a modified protocol by Hass et., al [50]. Media was collected after 24 h and stored at −80 °C for later use. The final concentration of the assay buffer consisted of 50 mM Tris (Thermofisher, AM9855G), 1 mM MgCl2 (Fisher, M33-500), 500 μM NADP+ (Sigma, N0505), 500 μM ATP (Sigma, A2383), 0.2 U/mL Hexokinase (Sigma, H4502), 0.08 U/m, and glucose-6-phosphate dehydrogenase (Sigma, G6378) with a final pH of 8.1. Standards ranging from 2 to 10 mM glucose were prepared and the spectrophotometer (SpectraMax Mini) was pre-warmed to 37 °C. 5 μL of standards/samples were added to a clear bottom 96 well plates and 195 μL of assay buffer was added to the plate. Absorbance was read at multiple times over 60 min at 340 nm. Sample concentration was determined using a standard curve with a linear fit.
2.24. iRPE data analysis and statistics
The area under the curve was quantified with Agilent MassHunter Quantitative Analysis. Compounds, retention times, and quantitative m/z can be found in Supplemental methods 1. Total relative metabolite abundance was calculated by summing the area together of all isotopologues (M0-Mn) normalized with total sum of measured metabolites (total spectral area of the total ion chromatogram). Fold change relative to Crispr corrected controls was calculated by taking the normalized metabolite level divided by the average of the control samples. Multiple two-tailed, unpaired Mann Whitney test were performed using GraphPad Prism version 10.0.0 for Windows, GraphPad Software, Boston, Massachusetts USA, www.graphpad.com. A non-parametric test was chosen because metabolites were not always normally distributed, and variances were not always equal between the control and experimental groups.
3. Results
3.1. Quantitative proteomics on RPE of mice expressing TIMP3S179C reveal abnormalities in metabolic processing
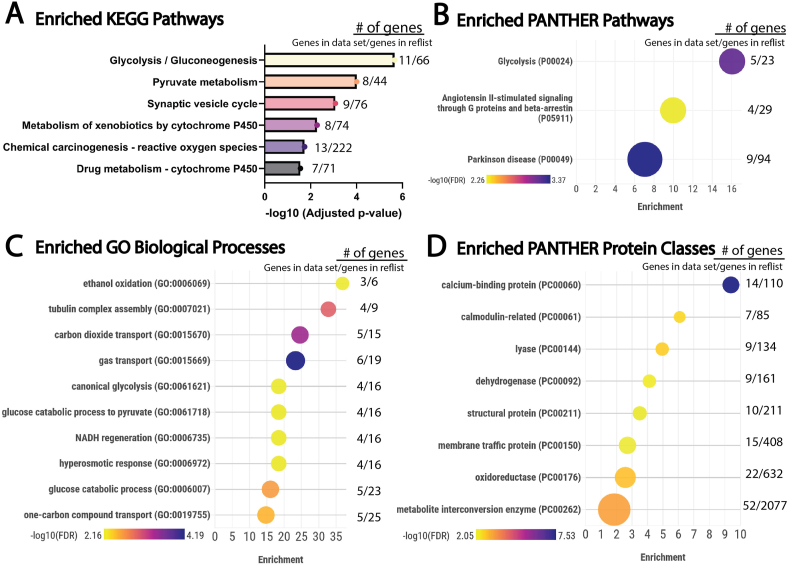
To understand the molecular and cellular processes involved in SFD, quantitative proteomics was performed on RPE isolated from the posterior eye cups of mice that globally expressed the S179C variant of TIMP3 and compared with wild-type littermates. Adult mice (7 months old) were used knowing that in humans with SFD, the onset of disease is usually seen in early-mid adulthood. We identified 298 proteins that were differentially expressed in the RPE of mutant mice, (adjusted p < 0.05, Supplemental Material 1). Differentially expressed proteins were analyzed using g:Profiler g:GOSt to find overrepresented KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Figure 1A). This tool performs functional analysis and then uses Fisher's one-tailed test to determine if pathways are overrepresented compared to a reference database. Glycolysis/gluconeogenesis was the most statistically significant enriched pathway (adjusted p-value = 2.17E-06) followed by pyruvate metabolism (adjusted p-value = 9.78E-05). Other enriched pathways identified included xenobiotic metabolism, reactive oxygen species and drug metabolism indicating that these pathways are dysregulated within the SFD mouse RPE.
Figure 1.
Enrichment analysis of proteins found to be significantly altered in S179C TIMP3 mice RPE when compared to WT controls. Proteomics was conducted on TIMP3 S179C RPE and WT controls (n = 8). All Proteins found to be significantly altered (Adjusted p-value < 0.05) were used for enrichment analysis. A, Enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathways. All enriched KEGG pathways (FDR<0.01) that were found using G-profiler g:GOSt (https://biit.cs.ut.ee/gprofiler/gost, Version e111_eg58_p18_f463989d). FDR is represented on the x-axis as -log10 of the adjusted p-value. B, All Significantly enriched PANTHER pathways, C, Enriched GO biological processes (Top 10 enriched, FDR<0.01), and D, Enriched GO Protein Classes (Top 8 enriched, FDR<0.01) utilized the PANTHER statistical overrepresentation test (PANTHER, Gene List Analysis v 18.0, https://www.pantherdb.org/) utilizing the Fisher's exact test with FDR correction. Data were visualized using Flourish Studio (https://flourish.studio) where annotations are on the y-axis, fold enrichment is on the x-axis, FDR is represented by color and number of differentially expressed proteins is represented by the size of the bubble. To the right is listed the number of proteins we found to be differentially expressed/the total number of proteins in the reference list. Complete gene list, functional analysis, and enrichment results can be found in supplemental materials 1 and 2. (For interpretation of the references to color/colour in this figure legend, the reader is referred to the Web version of this article.)
Overrepresented pathways were independently tested using Protein Analysis Through Evolutionary Relationships (PANTHER) overrepresentation test. Like g:profiler, PANTHER compares a list of differentially expressed proteins to a reference gene list, and determines whether a particular class (i.e. GO biological process, pathway, protein class) of genes is over- or under-represented [51]. This outputs not only the statistical significance (FDR) but also calculates fold enrichment (number of genes from input list/the expected number of gene if there is no enrichment). We ran analyses using the PANTHER pathways (Figure 1B), GO Biological Process Complete (Figure 1C), GO Protein Class Complete (Figure 1D), annotation sets. In agreement with KEGG analysis, glycolysis was the most enriched pathway with an enrichment fold of 16 (FDR = 6.54E-04). Multiple pathways related to glucose metabolism (in addition to others) were enriched within the biological processes data set. This includes ethanol oxidation (FDR = 6.58E-03), carbon dioxide transport (FDR = 3.52E-04), canonical glycolysis (FDR = 6.74E-03), glucose catabolic process to pyruvate (FDR = 6.80E-03), NADH regeneration (FDR = 6.85E-03), glucose catabolic process (FDR = 2.29E-03), and one-carbon compound transport (FDR = 3.19E-03). When PANTHER protein class was evaluated, metabolite interconversion enzymes were enriched (FDR = 5.52E-04). Together, this suggests a metabolic dysregulation that includes glucose metabolic pathways in RPE that express SFD-related mutant TIMP3.
3.2. TIMP3S179C ARPE-19 cells have increased glycolytic activity
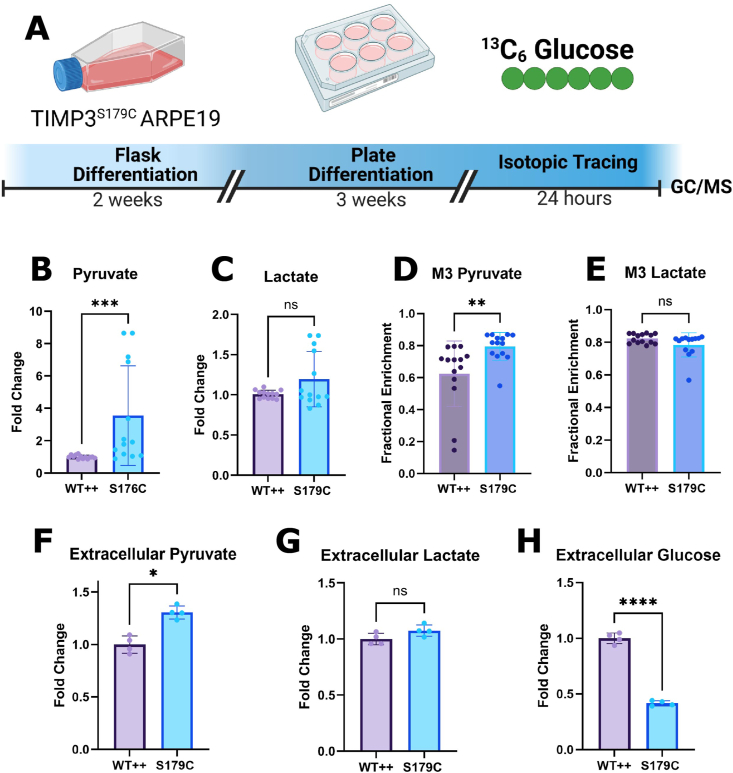
Uniformly labeled [U–13C6] glucose isotopic tracing was performed on S179C TIMP3 ARPE-19 cells [52], an immortalized RPE human cell line that was designed in our laboratory previously to expressed mutant TIMP3 known to be causative of SFD in humans [6].Comparisons were made with control cells engineered to express equivalent amounts of wild-type (WT++) and mutant (S179C) TIMP3 [6,41] (Figure 2A). We have previously reported that RPE cells expressing S179C TIMP3 exhibited altered cell adhesion, migration, and invasion; however metabolism had yet to be explored [6]. ARPE-19 cells were cultured using a method previously shown to increase epithelial cell morphology, cytoskeletal organization and RPE-related functions [42]. All non-labeled glucose in the cell culture media was replaced by 10 mM [U–13C6] glucose. Cells were incubated with the labeled media for 24 h to ensure that isotopic steady state was reached before metabolite extraction. To study glycolysis specifically, relative levels of intracellular pyruvate (Figure 2B) and lactate (Figure 2C) were evaluated and compared to controls. An increase in intracellular pyruvate in TIMP3 S179C mutant RPE suggests increased glycolytic activity. Extracellular lactate levels were increased however this was not statistically significant.
Figure 2.
TIMP3S179C ARPE-19 cells have increased glycolytic activity. A, ARPE19 cells overexpressing S179C TIMP3 (S179C) or wild type TIMP3 (WT++) were fully differentiated before steady state [U–13C6] Glucose isotopic tracing using GC/MS was conducted for 24 h. B, C, Total intracellular pyruvate but not lactate levels were increased in S179C cells (n = 13) D, M3 Pyruvate enrichment was increased in S179C cells while E, M3 Lactate remained unchanged. (n = 13) F, G, The conditioned media was evaluated for extracellular pyruvate and lactate. Pyruvate was increased indicating increased production (n = 4). H, Extracellular glucose levels were evaluated and found to be reduced by ∼50% in the S179C ARPE-19 cells indicating increased glucose utilization. (n = 4). Statistics were performed using a two-tailed, unpaired, Mann–Whitney test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Experimental schematic was generated using BioRender. Graphs were made with GraphPad Prism.
Fully labeled M6 glucose (all 6 carbons labeled) is broken down via glycolysis to generate fully labeled M3 (3 carbons labeled) pyruvate. Under steady state conditions, an average of 64% of pyruvate was M3 labeled in WT APRE19 while 79% was M3 labeled in TIMP3 S179C cells (Figure 2D). Increased M3 pyruvate enrichment supports increased glucose utilization and contribution to glycolysis in TIMP3 S179C cells. Pyruvate can also undergo aerobic glycolysis to produce M3 lactate (Figure 2E) which was comparable in the mutant and WT++ cells. After 18 h, the spent media was collected for extracellular analysis. Consistent with results obtained from the intracellular analysis, extracellular pyruvate (Figure 1F) was elevated further supporting an increase in glucose contribution to glycolysis. Likewise, lactate levels were not altered (Figure 1G).Finally, glucose levels were measured in conditioned medium to evaluate glucose utilization. Glucose levels were significantly decreased in TIMP3 S179C cells, confirming increased glucose utilization (Figure 2H). Collectively, these data suggest metabolic dysregulation in RPE cells expressing the TIMP3 S179C mutation. Specifically, mutant TIMP3 results in increased glucose utilization and glycolytic activity in RPE cells.
3.3. SFD iRPE display dysregulated central carbon metabolism
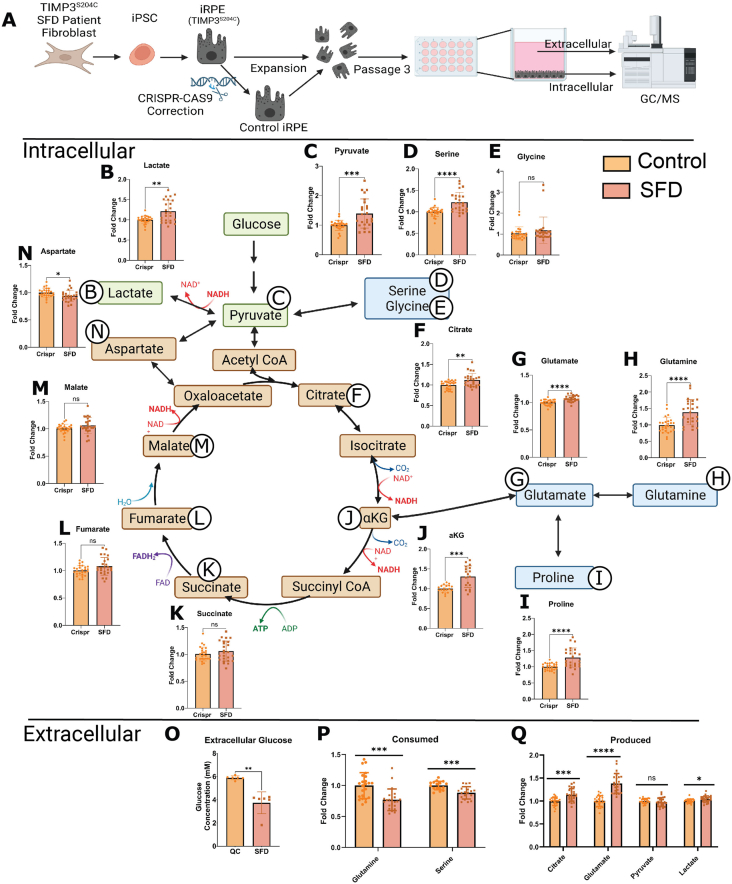
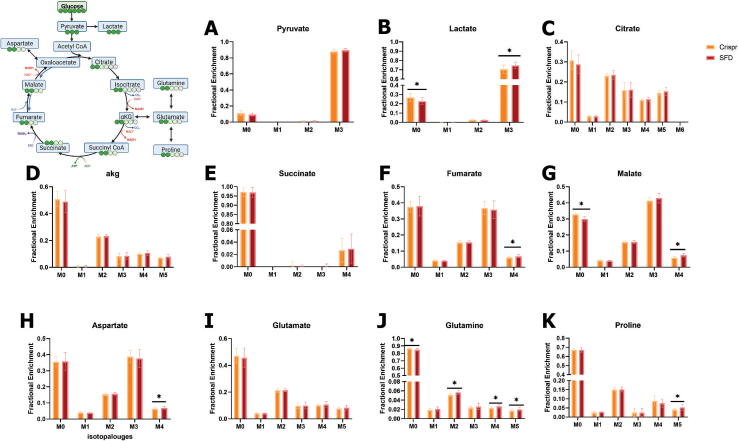
To confirm that these findings were not limited to the immortalized ARPE-19 cell line, we performed similar studies on an human induced pluripotent stem cell (iPSC) model of SFD [44] (iRPE) to validate our results and further characterize RPE central carbon metabolism. Previous studies utilizing these cells has confirmed SFD-like pathology including lipoproteinaceous deposits (drusen-like) and ECM alterations including increased collagen deposition2.iPSC-RPE cells have previously been shown to recapitulate hallmarks of healthy RPE [45,53]. Cells were originally collected through skin biopsies from a male patient with the TIMP3 S204C variant of SFD. Limited access to SFD patients due to the rarity of this disease was the determining factor for the mutation and sex of the patient. To our knowledge, there is no evidence of sex discrepancies associated with SFD. Patient fibroblasts were dedifferentiated to iPSC cells and then re-differentiated into iRPE cells (Figure 3A). There is significant evidence that reprogramming of somatic cells to a pluripotent state induces genomic instability and a wide range of heterogeneity and therefore absence of uniformity necessary for studying mechanisms [[54], [55], [56], [57]]. We therefore rationalized the use of cells from multiple (3) embryoid bodies from a single iPSC line and compared this to control cells where the mutation was corrected using CRISPR-Cas9. Correction of this mutation was confirmed by sequencing and karyotyping was conducted to confirm no genetic abnormalities in the corrected line.
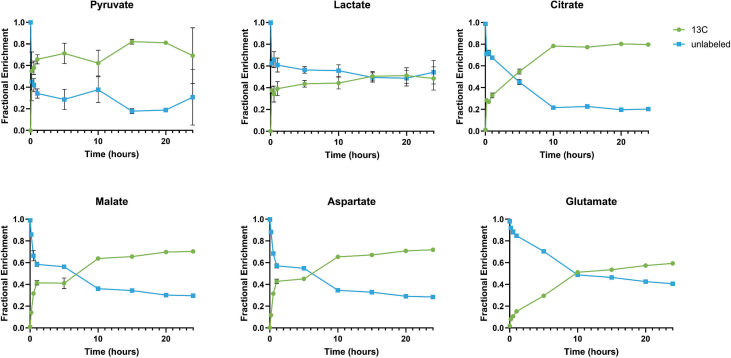
Figure 3.
SFD iRPE cells display dysregulated central carbon metabolism A, Fibroblasts from SFD patients with the S204C mutation were de-differentiated into iPSCs and then differentiated into iPSC-RPE cells (iRPE). Cells were expanded until passage 3 and then cultured for a minimum 30 days. Relative intracellular and extracellular (from conditioned media) metabolites were quantified with GC/MS. B–N, Relative abundance of total metabolite levels were compared between SFD iRPE (SFD) and Crispr corrected controls (Control) (n = 25 for all metabolites except for αKG where n = 18). O, Extracellular glucose concentration in conditioned media was quantified after 24 h using an enzymatic assay (n = 6). P, relative extracellular metabolites levels featuring metabolites that were decreased after 24 h compared to time 0 (i.e. consumed). P. metabolites that were increased after 24 h compared with time 0 (i.e. produced) (n = 25). Statistics performed were two-tailed unpaired Mann–Whitney. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. TCA cycle schematic was generated using BioRender. Graphs were made with GraphPad Prism.
As with ARPE-19, we found evidence of dysregulated central carbon metabolism in SFD iRPE cells. GC/MS was used to compare relative levels of intracellular and extracellular metabolites. To account for any potential differences in cell count, we calculated the total metabolite levels (sum of all isotopologues) and normalized them using total sum normalization by using the total ion chromatogram (TIC). Consistently, there were increased levels of intracellular lactate (Figure 3B) and pyruvate (Figure 3C) supporting increased glycolytic activity. Serine and glycine (Figure 3D,E) are metabolites generated when the glycolytic intermediate 3-phosphoglycerate (3-PG) is diverted by a 3-step enzymatic reaction that generates serine which can then be converted directly to glycine to feed carbon into one-carbon pathway. Serine levels (Figure 3D) but not glycine levels (Figure 3E) were found to be significantly increased indicative of increased glycolysis with higher production of serine with no change in serine utilization downstream into the one-carbon pathway. Citrate and aKG were both significantly increased.
iRPE had increased intracellular alpha-ketoglutarate (αKG) (Figure 3J), proline (Figure 3I) and glutamine (Figure 3H). αKG is primarily generated within the TCA cycle from citrate but can also be generated from glutamate through the process of anaplerosis. Similarly, glutamine can be generated from αKG in a process referred to as cataplerosis in which TCA intermediates exit the TCA cycle and are redirected for biosynthesis. Intermediates that follow αKG in the TCA cycle were unchanged in SFD iRPE, including succinate (Figure 3K), fumarate (Figure 3L), and malate (Figure 3M). Aspartate was slightly decreased(Figure 3N).
Extracellular metabolomics was conducted on the media to evaluate nutrient/metabolite uptake and release in SFD iRPE cells. After 24 h, glucose concentration was examined using an enzymatic assay (Figure 3O). SFD iRPE cells had less glucose in the media indicating increased glucose uptake. In addition, we compared the metabolites utilized by the cells (present in the original media composition at time 0 and decreased after 24 h). There was increased utilization of glutamine and serine by SFD iRPE cells (Figure 3P). We also evaluated metabolites released by RPE cells (metabolite levels increased after 24 h compared with time 0) and observed an increased production/release of citrate, glutamate and lactate by SFD iRPE cells (Figure 3Q).
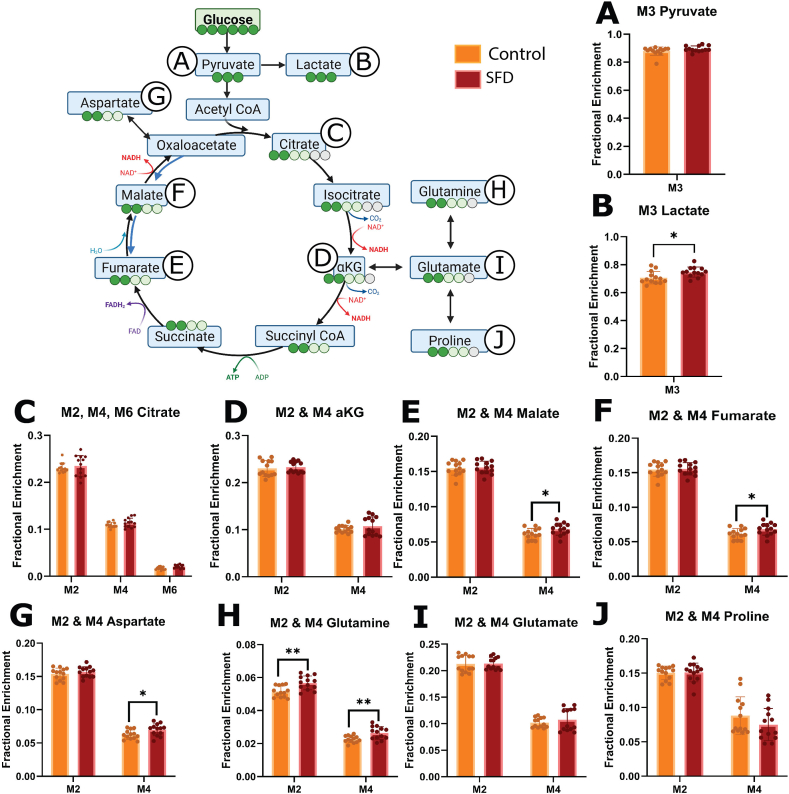
3.4. SFD iRPE have increased glucose contribution to the TCA cycle and increased cataplerosis to glutamine
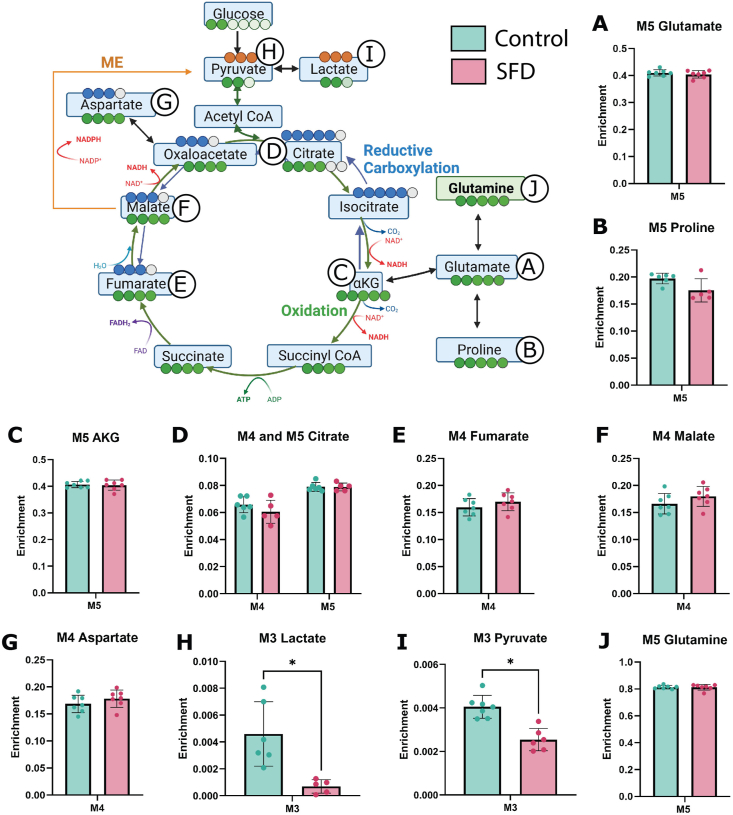
Since the metabolite pool size alone is insufficient to deduce pathway activity or nutrient contribution, we used stable isotope tracing with [13C6] glucose to further characterize central carbon metabolism. When [13C6] glucose undergoes glycolysis, M3 lactate and M3 pyruvate are produced. Although intracellular M3 pyruvate enrichment was unchanged, Lactate enrichment was increased in SFD iRPE cells compared with the Crispr corrected control (Figure 4 A, B). This is consistent with increased glucose contribution to glycolysis in SFD iRPE cells relative to control. Uniformly labeled [13C6] glucose produces M2, M4 and M6 citrate during the first, second, and third round of the TCA cycle respectively while M2 and M4 αKG, fumarate, malate, and aspartate are generated through the first and second round of the TCA cycle respectively. Increased M6 citrate indicates a slight increase in glucose contribution to the TCA cycle via Acetyl CoA (Figure 4C). This is supported by a concurrent increase in M4 fumarate, malate and aspartate enrichment (Figure 4G–I), suggesting increased TCA cycle activity. Pyruvate can alternatively enter the TCA cycle via pyruvate carboxylase (PC) which generates M3 aspartate, malate and fumarate (Supporting Information 4). No significant changes of these isotopologues were found, indicating that there is no change in pyruvate anaplerosis.
Figure 4.
Stable isotope tracing using [U–13C6] glucose in SFD iRPE cells compared with controls. SFD iRPE cells have increased glucose contribution to the TCA cycle. Steady state isotopic tracing using [U–13C6] glucose was conducted for 24 h, fractional enrichment is displayed on the Y axis. Isotopologues generated through glycolysis and TCA cycle are displayed on the X axis. A, B, When [U–13C6] glucose is broken down via glycolysis, M3 pyruvate (3-carbon labeled Pyruvate) and M3 lactate are generated. C–I, M2 and M4 TCA cycle metabolites are generated during the first and second round of the TCA cycle respectively when pyruvate enters through Acetyl CoA. M2 and M4 glutamate, glutamine and proline are generated when cataplerosis occurs, and TCA intermediates are depleted. Statistics performed were two-tailed unpaired Mann–Whitney test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. TCA pathway schematic was generated using BioRender. Graphs were made with GraphPad Prism (n = 13).
One of the main cataplerotic pathways from the TCA cycle involves the conversion of αKG to glutamate then glutamine. Increased M2 and M4 glutamine enrichment in SFD iRPE implies that glucose is being used to generate glutamine by leaving the TCA cycle. This is further supported by an increase in M5 glutamine and M5 proline (Supporting Information 4) which are generated from fully labeled citrate. In total, this demonstrates that SFD iRPE cells display increased TCA cycle activity and cataplerosis. This is likely contributing to the intracellular glutamine accumulation. It remains unclear why SFD iRPE cells would redirect nutrients allocated for energy production towards glutamine biosynthesis and how this additional glutamine will be utilized.
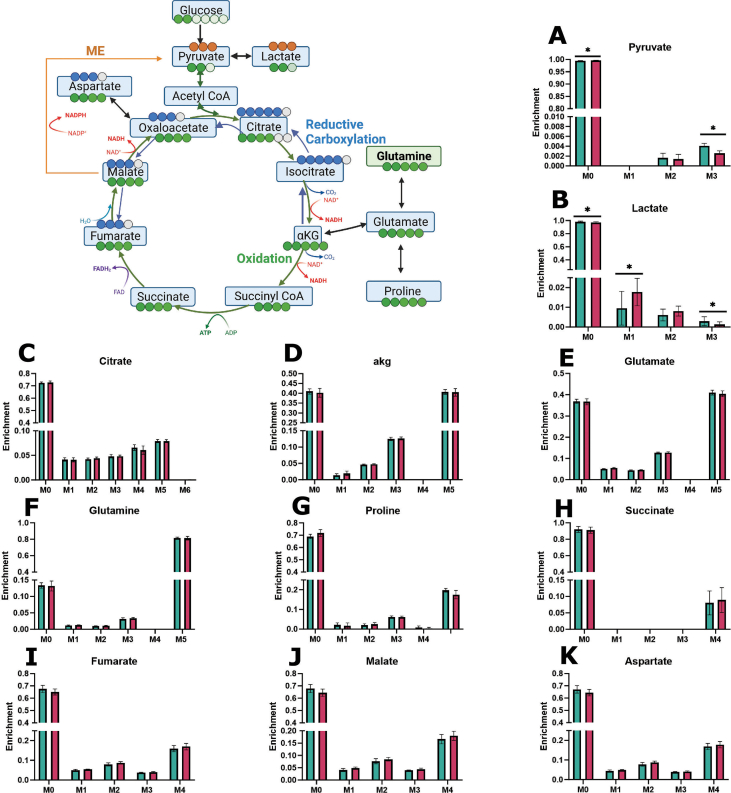
3.5. SFD iRPE has no change in glutamine contribution to the TCA cycle and impaired malic enzyme activity
We demonstrated increased glutamine cataplerosis, or the removal of TCA intermediates, in SFD iRPE cells, however, anaplerosis, or the replenishment of TCA cycle intermediates, is a process that happens in parallel. Anaplerosis can be an important fate for intracellular glutamine. It involves its integration into the TCA cycle through αKG. Furthermore, increased anaplerosis has previously been identified in another model of retinal degeneration [58]. Therefore, glutamine metabolism was probed directly using uniformly labeled [U–13C5] glutamine isotopic tracing (Figure 5, Supporting Information 5). SFD iRPE and Crispr corrected controls cells were incubated for 24 h with media that contained labeled glutamine. Relative metabolite levels between the mutant and control cells were determined via GC/MS and mass isotopomer distributions (MIDs) were calculated after correction for natural isotopic abundance. M5 glutamine can be converted to M5 glutamate (Figure 5A). M5 glutamate enrichment was similar in control and SFD iRPE indicating that glutamine entry and conversion to glutamate was unchanged in SFD iRPE. M5 glutamate can also be converted to M5 proline. Like glutamate, there was no change in glutamine contribution to proline (Figure 5B). When M5 glutamine is utilized by the TCA cycle, it generates M5 αKG along with M4 intermediates such as fumarate, malate, and aspartate (Figure 5C–G). We found no change in TCA cycle M4 intermediate metabolite enrichment, indicating that glutamine anaplerosis remained unchanged in SFD iRPE cells. We observed a significant decrease in M3 lactate and pyruvate isotopic enrichment in SFD iRPE cells (Figure H–I) despite equivalent labeled glutamine uptake (Figure 5J). M3 pyruvate and lactate is generated from uniformly labeled glutamine via mitochondrial malic enzyme, indicating potential inhibition of malic enzyme (ME) activity in iRPE cells This is consistent with a previous study using SFD iRPE cells suggesting decreased ME activity [2]. ME plays a pivotal role in converting malate to pyruvate while producing NADPH, a crucial factor for energy metabolism and maintaining cellular redox balance. Malic enzyme can also catalyze the reverse reaction of pyruvate to malate making it relevant for anaplerosis. It important to note that the amount of pyruvate and lactate generated from malic enzyme, even under control conditions, is relatively low at 0.5%. The biological relevance of this change will need to be confirmed. We have previously shown that SFD iRPE cells have high oxidative stress at baseline and increased susceptibility to oxidative stress damage [2]. This together with our current data suggests that there is a potential loss of redox balance in SFD iRPE cells. Further research is required to ascertain the significance of ME activity and its role in the generation of NADPH within the RPE during healthy and diseased conditions.
Figure 5.
Stable isotope tracing using [U–13C5] glutamine in SFD iRPE cells compared with controls. Steady state isotopic tracing with [U–13C5] Glutamine was conducted for 24 h, and fractional enrichment was calculated and is displayed on the Y axis. M5 glutamine is metabolized to M5 glutamate (A) which converted to either M5 proline (B) or M5 αkg (C). M4 fumarate or M5 citrate (D) or M4 fumarate (E) are produced from reductive carboxylation or oxidation of αKG respectively. Forward flow of the TCA cycle results in M4 Malate (F), M4 Aspartate (G), and M4 Citrate (D). Carbons can exit the TCA cycle to generate M2 pyruvate (H) and M2 lactate (I) via Acetyl CoA or M3 pyruvate and M3 lactate via malic enzyme (ME). (J) M5 glutamine represents glutamine taken up from the media. Individual data points represent biological replicates(n = 7). A two-tailed unpaired Mann–Whitney test was used to evaluate statistical significance. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Experimental schematic was generated using BioRender. Graphs were made with GraphPad Prism.
4. Discussion
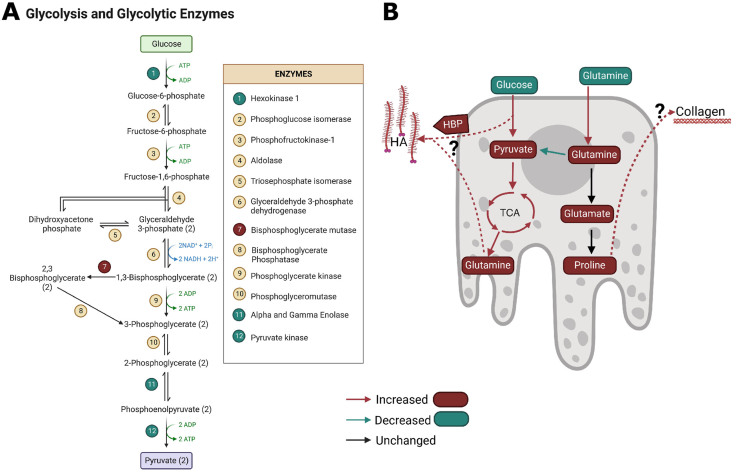
Our data showing increased glycolytic activity and glucose consumption in RPE cells expressing mutant TIMP3 supports the hypothesis that TIMP3 mutations lead to altered glucose metabolism. The abnormalities of glycolysis in TIMP3 mutant RPE was corroborated by proteomics data from RPE of SFD mutant mice. PANTHER functional analysis identified five dysregulated enzymes involved in glycolysis (Figure 6A). Furthermore, enrichment analysis determined that glycolytic enzymes were enriched among all the differentially expressed protein. Together, this strongly suggests that TIMP3 mutations result in dysfunction of glucose metabolism. Increased glycolysis in the RPE has been shown to perturb retinal metabolism in retinitis pigmentosa [10]. While SFD and retinitis pigmentosa are caused by mutations in unique genes, our data suggests the possibility that altered glucose metabolism could be a common feature of these two retinal degenerative diseases and possibly others. RPE central carbon metabolism could be an attractive therapeutic target that could be used for both conditions.
Figure 6.
Dysregulated glycolytic enzymes and a summary of metabolic dysregulation in SFD RPE. A, Schematic showing glycolytic intermediates and the altered glycolytic enzymes identified via proteomics. Enzymes in green circles were found to be decreased while the enzymes in red circles were found to be increased. B) Summary of metabolic dysregulation in SFD RPE. Red arrows and bubbles indicate an increase in abundance/activity while green represents a decrease in abundance/activity. Glucose utilization increases and intracellular pyruvate accumulate while glucose contribution to the TCA cycle and glutamine cataplerosis increases. Glutamine utilization increases and extracellular glutamate accumulates as intracellular glutamate decreases and intracellular glutamine accumulates. Glutamines conversion to pyruvate via malic enzyme is decreased while glutamine anaplerosis, or contribution to the TCA cycle, is unchanged. It remains to be determined which pathway is metabolizing the additional glutamine taken up from the media and generated from glucose. We hypothesize that excess glutamine and glycolytic intermediates could be used to generate hyaluronic acid, through the hexosamine biosynthetic pathway or could be converted to proline and utilized for collagen synthesis. (For interpretation of the references to color/colour in this figure legend, the reader is referred to the Web version of this article.)
Through metabolomics, glutamine was found to be to be the most altered metabolite intracellularly and extracellularly. Intracellular levels of glutamine were increased while extracellular glutamine was decreased in SFD iRPE cells. Furthermore, uniformly labeled glucose isotopic tracing found that SFD iRPE cells have an increased propensity to synthesize glutamine from glucose. However, when glutamine metabolism was probed specifically with uniformly labeled glutamine, there was no change in contribution to intracellular glutamate or anaplerosis. It remains unclear why SFD iRPE cells have increased glutamine uptake despite our results suggesting that SFD RPE do not display changes in glutamine anaplerosis or proline synthesis. This strongly suggests that glutamine is being utilized in a way that has yet to be identified in SFD iRPE cells.
It is possible that RPE glutamine and glucose utilization is related to extracellular deposition of proteins, lipids, and glycosaminoglycan in both physiological and pathological contexts. Thickened Bruch's membrane is a common pathological feature in patients with SFD [2,3,59]. Collagen is the primary component of BrM [60] and is reported to accumulate in AMD [44,[61], [62], [63]]. Furthermore, the SFD iRPE cells used in this study have been previously shown to accumulate Collagen IV [44]. Collagen is synthesized from proline, hydroxyproline and glycine. Proline is derived from glutamine [64] while glycine can be synthesized from glycolytic intermediates. Therefore, metabolic perturbations to glucose and glutamine have the potential to alter ECM deposition by modulating collagen synthesis.
Collagen is not the only ECM component controlled by glucose and glutamine metabolism. HA is an extracellular glycosaminoglycan that is synthesized via the hexosamine biosynthetic pathway (HBP) which branches off from glycolysis and requires the conversion to glutamine to glutamate. Glutamine: fructose-6-phosphate amido transferase (GFAT) utilize glutamine to catalyze the first rate-limiting step of the HBP pathway.We have previously reported that HA deposition increases in SFD and AMD RPE cells from patients [59]. It's possible that RPE cells utilize glutamine and glucose to support increase HBP flux (Figure 6B). Alternatively, increased HA production could be a byproduct of glycolytic spillover from glycolytic intermediate accumulation. Regardless, either scenario would require glutamine utilization and could explain why SFD iRPE cells are generating more glucose-derived glutamine.
While our results clearly demonstrate that TIMP3 mutations associated with SFD result in changes to RPE metabolism, the molecular mechanism by which this occurs are unclear. Mutant TIMP3 regulates the extracellular matrix via matrix degrading enzymes which has the potential for changing cellular metabolism. Patients with SFD and AMD demonstrate significant alterations in Bruch's membrane structure. SFD TIMP3 mutations result in highly dysregulated ECM [2] which has the potential to alter metabolism in various ways. ECM biomechanics has previously been demonstrated to regulate different aspects of metabolism [65]. Furthermore, ECM components, including HA, can act as ligands to cell surface receptors such as CD44, that trigger signaling cascades that regulate metabolism [66]. ECM constituents can also potentially be used as a nutrient source [67,68]. On the other hand, metabolism is known to influence ECM deposition. For example, proline availability can regulate collagen deposition [69]. Another possibility is that TIMP3 can directly interact with metabolic enzymes. A high throughput study identified 18 TIMP3 binding partners, one of which was ADP dependent glucose kinase which is responsible for converting glucose to glucose-6-phosphate [70]. Exploring the relationship between ECM and metabolism could provide valuable insights into the molecular mechanisms underlying retinal degeneration.
Funding and additional information
This research was supported by NIH 5 T32 EY 7157-19, R01 EY 027083, P30 EY 025585, 1F31EY033223-01, RPB Center Grant, Cleveland Eye Bank Foundation, Timken Foundation and Cleveland Clinic Foundation support.
CRediT authorship contribution statement
Allison Grenell: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Charandeep Singh: Methodology, Writing – review & editing. Monisha Raju: Investigation. Alyson Wolk: Investigation. Sonal Dalvi: Resources. Geeng-Fu Jang: Investigation. John S. Crabb: Formal analysis, Investigation. Courtney E. Hershberger: Formal analysis. Kannan V. Manian: Investigation. Karen Hernandez: Investigation. John W. Crabb: Data curation, Formal analysis. Ruchira Singh: Resources. Jianhai Du: Formal analysis, Methodology. Bela Anand-Apte: Conceptualization, Data curation, Formal analysis, Funding acquisition, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work the author(s) used OpenAI's ChatGPT3 to improve clarity and check grammar. After using this tool/service, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Dr. Henri Brunengraber and the Mouse Metabolic Phenotyping Center for guidance on isotopic tracing experiments, Noah Chernosky for his assistance with transporting iRPE cells, and the laboratory of Dr. Jonathan Sears for sharing supplies and analytical standards for GC/MS analysis.
Abbreviations:
- AMD
Age-related Macular degeneration
- BrM
Bruch's membrane
- ECM
Extracellular matrix
- HA
Hyaluronan
- RPE
retinal pigment epithelium
- SFD
Sorsby's fundus dystrophy
- TIMP3
Tissue inhibitor of metalloproteinase 3
- TCA cycle
tri-carboxylic acid cycle
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101995.
Contributor Information
Allison Grenell, Email: abg77@case.edu.
Bela Anand-Apte, Email: anandab@ccf.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
Data availability
Data will be made available on request.
References
- 1.Capon M.R., Marshall J., Krafft J.I., Alexander R.A., Hiscott P.S., Bird A.C. Sorsby's fundus dystrophy. A light and electron microscopic study. Ophthalmology. 1989;96(12):1769–1777. doi: 10.1016/s0161-6420(89)32664-9. [DOI] [PubMed] [Google Scholar]
- 2.Engel A.L., Wang Y., Khuu T.H., Worrall E., Manson M.A., Lim R.R., et al. Extracellular matrix dysfunction in Sorsby patient-derived retinal pigment epithelium. Exp Eye Res. 2022;215 doi: 10.1016/j.exer.2021.108899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gourier H.C.Y., Chong N.V. Can novel treatment of age-related macular degeneration Be developed by better understanding of Sorsby's fundus dystrophy. J Clin Med. 2015;4(5):874–883. doi: 10.3390/jcm4050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollyfield J.G., Salomon R.G., Crabb J.W. Proteomic approaches to understanding age-related macular degeneration. Adv Exp Med Biol. 2003;533:83–89. doi: 10.1007/978-1-4615-0067-4_11. [DOI] [PubMed] [Google Scholar]
- 5.Fariss R., Apte S., Luthert P., Bird A., Milam A. Accumulation of tissue inhibitor of metalloproteinases-3 in human eyes with Sorsby's fundus dystrophy or retinitis pigmentosa. Br J Ophthalmol. 1998;82(11):1329–1334. doi: 10.1136/bjo.82.11.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi J.H., Ebrahem Q., Yeow K., Edwards D.R., Fox P.L., Anand-Apte B. Expression of Sorsby's fundus dystrophy mutations in human retinal pigment epithelial cells reduces matrix metalloproteinase inhibition and may promote angiogenesis. J Biol Chem. 2002;277(16):13394–13400. doi: 10.1074/jbc.M110870200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M., Jiang N., Chu Y., Postnikova O., Varghese R., Horvath A., et al. Dysregulated metabolic pathways in age-related macular degeneration. Sci Rep. 2020;10(1):2464. doi: 10.1038/s41598-020-59244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou X.-W., Wang Y., Pan C.-W. Metabolomics in age-related macular degeneration: a systematic review. Invest Ophthalmol Vis Sci. 2020;61(14) doi: 10.1167/iovs.61.14.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher C.R., Ferrington D.A. Perspective on AMD pathobiology: a bioenergetic crisis in the RPE. Invest Ophthalmol Vis Sci. 2018;59(4):AMD41–A47. doi: 10.1167/iovs.18-24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W., Kini A., Wang Y., Liu T., Chen Y., Vukmanic E., et al. Metabolic deregulation of the blood-outer retinal barrier in retinitis pigmentosa. Cell Rep. 2019;28(5):1323–1334.e4. doi: 10.1016/j.celrep.2019.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen T.T., Wong T.Y. Retinal vascular manifestations of metabolic disorders. Trends Endocrinol Metabol: TEM (Trends Endocrinol Metab) 2006;17(7):262–268. doi: 10.1016/j.tem.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Abcouwer S.F., Gardner T.W. Diabetic retinopathy: loss of neuroretinal adaptation to the diabetic metabolic environment. Ann N Y Acad Sci. 2014;1311(1):174–190. doi: 10.1111/nyas.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh C. Metabolism and vascular retinopathies: current perspectives and future directions. Diagnostics. 2022;12(4):903. doi: 10.3390/diagnostics12040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh C., Hoppe G., Tran V., McCollum L., Bolok Y., Song W., et al. Serine and 1-carbon metabolism are required for HIF-mediated protection against retinopathy of prematurity. JCI Insight. 2019;4(14) doi: 10.1172/jci.insight.129398. 129398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scerri T.S., Quaglieri A., Cai C., Zernant J., Matsunami N., Baird L., et al. Genome-wide analyses identify common variants associated with macular telangiectasia type 2. Nat Genet. 2017;49(4):559–567. doi: 10.1038/ng.3799. [DOI] [PubMed] [Google Scholar]
- 16.Gantner M.L., Eade K., Wallace M., Handzlik M.K., Fallon R., Trombley J., et al. Serine and lipid metabolism in macular disease and peripheral neuropathy. N Engl J Med. 2019;381(15):1422–1433. doi: 10.1056/NEJMoa1815111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonelli R., Ansell B.R.E., Lotta L., Scerri T., Clemons T.E., Leung I., et al. Genetic disruption of serine biosynthesis is a key driver of macular telangiectasia type 2 aetiology and progression. Genome Med. 2021;13(1):39. doi: 10.1186/s13073-021-00848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Wang Y., Zhang B., Zhao L., Lyubasyuk V., Wang K., et al. A missense mutation in HK1 leads to autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014;55(11):7159–7164. doi: 10.1167/iovs.14-15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Sun Z., Zhao P., Huang L., Xu M., Yang Y., et al. Whole-exome sequencing revealed HKDC1 as a candidate gene associated with autosomal-recessive retinitis pigmentosa. Hum Mol Genet. 2018;27(23):4157–4168. doi: 10.1093/hmg/ddy281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C.T., Shao Z., Fu Z. Dysfunctional peroxisomal lipid metabolisms and their ocular manifestations. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.982564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Coorey N.J., Zhang M., Zeng S., Madigan M.C., Zhang X., et al. Metabolism dysregulation in retinal diseases and related therapies. Antioxidants. 2022;11(5) doi: 10.3390/antiox11050942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan W.W., Wubben T.J., Besirli C.G. Photoreceptor metabolic reprogramming: current understanding and therapeutic implications. Commun Biol. 2021;4:245. doi: 10.1038/s42003-021-01765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurihara, T., Westenskow, P.D., Gantner, M.L., Usui, Y., Schultz, A., Bravo, S., et al., n.d. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. eLife 5, Doi: 10.7554/eLife.14319. [DOI] [PMC free article] [PubMed]
- 24.Kumagai A.K. Glucose transport in brain and retina: implications in the management and complications of diabetes. Diabetes Metabol Res Rev. 1999;15(4):261–273. doi: 10.1002/(SICI)1520-7560(199907/08)15:4<261::AID-DMRR43>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Tang J., Zhu X.W., Lust W.D., Kern T.S. Retina accumulates more glucose than does the embryologically similar cerebral cortex in diabetic rats. Diabetologia. 2000;43(11):1417–1423. doi: 10.1007/s001250051548. [DOI] [PubMed] [Google Scholar]
- 26.Chao J.R., Knight K., Engel A.L., Jankowski C., Wang Y., Manson M.A., et al. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J Biol Chem. 2017;292(31):12895–12905. doi: 10.1074/jbc.M117.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yam M., Engel A.L., Wang Y., Zhu S., Hauer A., Zhang R., et al. Proline mediates metabolic communication between retinal pigment epithelial cells and the retina. J Biol Chem. 2019;294(26):10278–10289. doi: 10.1074/jbc.RA119.007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swarup A., Samuels I.S., Bell B.A., Han J.Y.S., Du J., Massenzio E., et al. Modulating GLUT1 expression in retinal pigment epithelium decreases glucose levels in the retina: impact on photoreceptors and Müller glial cells. Am J Physiol Cell Physiol. 2019;316(1):C121–C133. doi: 10.1152/ajpcell.00410.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adijanto J., Du J., Moffat C., Seifert E.L., Hurley J.B., Philp N.J. The retinal pigment epithelium utilizes fatty acids for ketogenesis. J Biol Chem. 2014;289(30):20570–20582. doi: 10.1074/jbc.M114.565457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du J., Yanagida A., Knight K., Engel A.L., Vo A.H., Jankowski C., et al. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc Natl Acad Sci USA. 2016;113(51):14710–14715. doi: 10.1073/pnas.1604572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanow M.A., Giarmarco M.M., Jankowski C.S., Tsantilas K., Engel A.L., Du J., et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. eLife. 2017;6 doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrington D.A., Ebeling M.C., Kapphahn R.J., Terluk M.R., Fisher C.R., Polanco J.R., et al. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol. 2017;13:255–265. doi: 10.1016/j.redox.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber B.H.F., Lin B., White K., Kohler K., Soboleva G., Herterich S., et al. A mouse model for Sorsby fundus dystrophy. Invest Ophthalmol Vis Sci. 2002;43(8):2732–2740. [PubMed] [Google Scholar]
- 34.Wei H., Xun Z., Granado H., Wu A., Handa J.T. An easy, rapid method to isolate RPE cell protein from the mouse eye. Exp Eye Res. 2016;145:450–455. doi: 10.1016/j.exer.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng K.-P., Gugiu B., Renganathan K., Davies M.W., Gu X., Crabb J.S., et al. Retinal pigment epithelium lipofuscin proteomics. Mol Cell Proteomics : MCP. 2008;7(7):1397–1405. doi: 10.1074/mcp.M700525-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crabb J.W., West K.A., Dodson W.S., Hulmes J.D. Amino acid analysis. Current Protocols in Protein Science. 1997;7(1):11.9.1–11.9.42. doi: 10.1002/0471140864.ps1109s07. [DOI] [PubMed] [Google Scholar]
- 37.Saikia P., Crabb J.S., Dibbin L.L., Juszczak M.J., Willard B., Jang G.-F., et al. Quantitative proteomic comparison of myofibroblasts derived from bone marrow and cornea. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-73686-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang G.-F., Crabb J.S., Hu B., Willard B., Kalirai H., Singh A.D., et al. Proteomics of primary uveal melanoma: insights into metastasis and protein biomarkers. Cancers. 2021;13(14):3520. doi: 10.3390/cancers13143520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hultin-Rosenberg L., Forshed J., Branca R.M.M., Lehtiö J., Johansson H.J. Defining, comparing, and improving iTRAQ quantification in mass spectrometry proteomics data. Mol Cell Proteomics. 2013;12(7):2021–2031. doi: 10.1074/mcp.M112.021592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R-Development-Team . R; Vienna, Austria: 2019. R: a language and environment for statistical computing. [Google Scholar]
- 41.Qi J.H., Bell B., Singh R., Batoki J., Wolk A., Cutler A., et al. Sorsby fundus dystrophy mutation in tissue inhibitor of metalloproteinase 3 (TIMP3) promotes choroidal neovascularization via a fibroblast growth factor-dependent mechanism. Sci Rep. 2019;9 doi: 10.1038/s41598-019-53433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazim R.A., Volland S., Yen A., Burgess B.L., Williams D.S. Rapid differentiation of the human RPE cell line, ARPE-19, induced by nicotinamide. Exp Eye Res. 2019;179:18–24. doi: 10.1016/j.exer.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millard P., Delépine B., Guionnet M., Heuillet M., Bellvert F., Létisse F. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics. 2019;35(21):4484–4487. doi: 10.1093/bioinformatics/btz209. (Oxford, England) [DOI] [PubMed] [Google Scholar]
- 44.Galloway C.A., Dalvi S., Hung S.S.C., MacDonald L.A., Latchney L.R., Wong R.C.B., et al. Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc Natl Acad Sci USA. 2017;114(39):E8214–E8223. doi: 10.1073/pnas.1710430114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manian K.V., Galloway C.A., Dalvi S., Emanuel A.A., Mereness J.A., Spencer W., et al. 3D iPSC modelling of retinal pigment epithelium-choriocapillaris complex identifies factors involved in the pathology of macular degeneration. Cell Stem Cell. 2021;28(5):846–862.e8. doi: 10.1016/j.stem.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crombie D.E., Curl C.L., Raaijmakers A.J., Sivakumaran P., Kulkarni T., Wong R.C., et al. Friedreich's ataxia induced pluripotent stem cell-derived cardiomyocytes display electrophysiological abnormalities and calcium handling deficiency. Aging (Albany NY) 2017;9(5):1440–1449. doi: 10.18632/aging.101247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang C., Han J., Dalvi S., Manian K., Winschel L., Volland S., et al. A human model of Batten disease shows role of CLN3 in phagocytosis at the photoreceptor–RPE interface. Commun Biol. 2021;4(1):1–18. doi: 10.1038/s42003-021-01682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalvi S., Galloway C.A., Winschel L., Hashim A., Soto C., Tang C., et al. Environmental stress impairs photoreceptor outer segment (POS) phagocytosis and degradation and induces autofluorescent material accumulation in hiPSC-RPE cells. Cell Death Discovery. 2019;5(1):1–16. doi: 10.1038/s41420-019-0171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu R., Wang Y., Du J. Tracing nitrogen metabolism in mouse tissues with gas chromatography-mass spectrometry. Bio-Protocol. 2021;11(4) doi: 10.21769/BioProtoc.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hass D., Hurley J. 2023. Glucose concentration assay (Hexokinase/G6PDH method) [Google Scholar]
- 51.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with PANTHER Classification System. Nat Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunn K.C., Aotaki-Keen A.E., Putkey F.R., Hjelmeland L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62(2):155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 53.Singh R., Phillips M.J., Kuai D., Meyer J., Martin J.M., Smith M.A., et al. Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest Ophthalmol Vis Sci. 2013;54(10):6767–6778. doi: 10.1167/iovs.13-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hussein S.M., Batada N.N., Vuoristo S., Ching R.W., Autio R., Närvä E., et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471(7336):58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 55.Laurent L.C., Ulitsky I., Slavin I., Tran H., Schork A., Morey R., et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8(1):106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C., Klco J.M., Helton N.M., George D.R., Mudd J.L., Miller C.A., et al. Genetic heterogeneity of induced pluripotent stem cells: results from 24 clones derived from a single C57bl/6 mouse. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grenell A., Wang Y., Yam M., Swarup A., Dilan T.L., Hauer A., et al. Loss of MPC1 reprograms retinal metabolism to impair visual function. Proc Natl Acad Sci USA. 2019;116(9):3530–3535. doi: 10.1073/pnas.1812941116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolk A., Hatipoglu D., Cutler A., Ali M., Bell L., Hua Qi J., et al. Role of FGF and hyaluronan in choroidal neovascularization in Sorsby fundus dystrophy. Cells. 2020;9(3):608. doi: 10.3390/cells9030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newsome D.A., Huh W., Green W.R. Bruch's membrane age-related changes vary by region. Curr Eye Res. 1987;6(10):1211–1221. doi: 10.3109/02713688709025231. [DOI] [PubMed] [Google Scholar]
- 61.Marshall G.E., Konstas A.G., Reid G.G., Edwards J.G., Lee W.R. Type IV collagen and laminin in Bruch's membrane and basal linear deposit in the human macula. Br J Ophthalmol. 1992;76(10):607–614. doi: 10.1136/bjo.76.10.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green W.R., Enger C. Age-related macular degeneration histopathologic studies. The 1992 Lorenz E. Zimmerman Lecture. Ophthalmology. 1993;100(10):1519–1535. doi: 10.1016/s0161-6420(93)31466-1. [DOI] [PubMed] [Google Scholar]
- 63.Yuan X., Gu X., Crabb J.S., Yue X., Shadrach K., Hollyfield J.G., et al. Quantitative proteomics: comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol Cell Proteomics: MCP. 2010;9(6):1031–1046. doi: 10.1074/mcp.M900523-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phang J.M., Liu W., Hancock C.N., Fischer J.W. Proline metabolism and cancer: emerging links to glutamine and collagen. Curr Opin Clin Nutr Metab Care. 2015;18(1):71–77. doi: 10.1097/MCO.0000000000000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deville S.S., Cordes N. The extracellular, cellular, and nuclear stiffness, a trinity in the cancer resistome—a review. Front Oncol. 2019;9 doi: 10.3389/fonc.2019.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weng X., Maxwell-Warburton S., Hasib A., Ma L., Kang L. The membrane receptor CD44: novel insights into metabolism. Trends Endocrinol Metabol. 2022;33(5):318–332. doi: 10.1016/j.tem.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Muranen T., Iwanicki M.P., Curry N.L., Hwang J., DuBois C.D., Coloff J.L., et al. Starved epithelial cells uptake extracellular matrix for survival. Nat Commun. 2017;8(1) doi: 10.1038/ncomms13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rainero E. Extracellular matrix internalization links nutrient signalling to invasive migration. Int J Exp Pathol. 2018;99(1):4–9. doi: 10.1111/iep.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karna E., Szoka L., Huynh T.Y.L., Palka J.A. Proline-dependent regulation of collagen metabolism. Cell Mol Life Sci. 2020;77(10):1911–1918. doi: 10.1007/s00018-019-03363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.