Abstract
RNAs are often modified to invoke new activities. While many modifications are limited in frequency, restricted to non-coding RNAs, or present only in select organisms, 5-methylcytidine (m5C) is abundant across diverse RNAs and fitness-relevant across Domains of life, but the synthesis and impacts of m5C have yet to be fully investigated. Here, we map m5C in the model hyperthermophile, Thermococcus kodakarensis. We demonstrate that m5C is ~25x more abundant in T. kodakarensis than human cells, and the m5C epitranscriptome includes ~10% of unique transcripts. T. kodakarensis rRNAs harbor tenfold more m5C compared to Eukarya or Bacteria. We identify at least five RNA m5C methyltransferases (R5CMTs), and strains deleted for individual R5CMTs lack site-specific m5C modifications that limit hyperthermophilic growth. We show that m5C is likely generated through partial redundancy in target sites among R5CMTs. The complexity of the m5C epitranscriptome in T. kodakarensis argues that m5C supports life in the extremes.
Subject terms: RNA modification, Archaeal genes, RNA
The epitranscriptome is fitness-relevant across Domains. Here, the authors map m5C in the model hyperthermophile, Thermococcus kodakarensis. The abundance and complexity of the m5C epitranscriptome in T. kodakarensis argues that m5C supports life in the extremes.
Introduction
Studies of bacterial and eukaryotic epitranscriptomes have shown that chemical modifications to RNA are often abundant and dynamically modulated in response to physiological and environmental stimuli1–5, parasitic-6, pathogen-7,8 or viral-infection9,10 or are introduced within specific cells and lineages11–13. Modifications to even individual residues in long RNAs are known to elicit dramatic impacts on structure14, stability15–18, protein-interactions19–21, cellular localization22–24, translation efficiency16,25–27, half-life28, nuclear transport29, mitochondrial30 and plastid transport31. Specialized nucleotides within rRNAs and tRNAs often provide essential stability and importantly, these modified RNAs must be passed to daughter cells, providing a transgenerational role for epitranscriptomic modifications32–35. The conserved modification profiles defined for rRNA and tRNA demonstrate that the distribution of modifications are not random, but rather that modifications are strategically positioned to impact function at specific nucleotides36. Evidence suggests this regulatory paradigm extends to mRNAs and across Domains of life, with the epitranscriptome being essential for all life37,38.
mRNAs have been increasingly identified as modification targets. Site-specific modification of mRNA is linked to core biological functions and responses to environmental changes37,39. However, it remains contested whether modifications to mRNA coding sequences impart biological function and drive fitness or are simply a result of promiscuous activity of enzymes with intended specificity for non-coding RNA. Critical questions remain regarding how the epitranscriptome is generated, how modifications impact mRNA function, and how mRNAs are selected for modification. Despite increasingly sophisticated methods to site-specifically identify and quantify a variety of RNA modifications, the low abundance of mRNA transcripts and their low modification frequency hinders investigations into the regulation imposed by the epitranscriptome40.
Many archaea thrive in conditions inhospitable to most extant life. Maintaining RNA structure at high temperature, or at the extremes of salinity, pressure, or pH are facilitated, in part, by chemical modifications. Specific RNA modifications are critical for life at high temperatures, and loss of epitranscriptomic modifications is often lethal41–44. In Thermus thermophilus, a hyperthermophilic bacteria, tRNA 1-methyladenosie (m1A)43, 7-methylguanosine (m7G)45, and 2-Thioribothymidine42 are required for growth at high temperatures. Likewise, in Thermococcus kodakarensis, a hyperthermophilic archaeon, 2-dimethylguanosine (m22G)44 and 2’O-phosphouridine (p2U) in tRNAs are required for growth at high temperatures. Not only are individual modified residues in tRNAs essential for hyperthermophilic growth, 4-acetylcytidine (ac4C) and 2’O-methyl-ac4C in Pyrococcus furiosus and ac4C in T. kodakarensis are markedly increased with rising growth temperature2,46. 5-methyl-2-thiouridine (m5s2U), m22G, archaeosine (G+) and m1A have been shown to stabilize T. kodakarensis tRNA structure and enhance hyperthermophilic growth41,44,47.
Many RNA modifications have been identified and quantified in the model hyperthermophilic species T. kodakarensis, representing an order of magnitude higher absolute abundance and several orders of magnitude higher frequency than found in model eukaryotes2,48,49. The abundance of RNA modifications in T. kodakarensis, its tractable genetic system, and its range of growth temperatures permitting robust growth provide an ideal platform to resolve fundamental open questions regarding the phenotypic consequences of epitranscriptomic changes, including RNA targeting and RNA modifying enzymes.
m5C is one of the most abundant and conserved modifications that decorates the transcriptome across Domains50,51. m5C in most species is predicted to be generated through the coordinated, post-transcriptional activities of several RNA m5C methyltransferases (R5CMTs)38. Here we established the genesis, phenotypic impact, redundancy, and targeting mechanisms of the m5C epitranscriptome within T. kodakarensis. T. kodakarensis retains a ~25 fold higher abundance of m5C over human cell lines. With ultra-deep bisulfite sequencing, we map m5C sites across the T. kodakarensis transcriptome with single-nucleotide resolution under different biological conditions. We identify at least 232 candidate m5C sites in a diverse set of coding and non-coding RNAs, establish that ~10% of all unique and sufficiently expressed transcripts contain m5C, and show that mRNA represents the largest fraction of unique, m5C-modified RNAs. Moreover, we identify at least five R5CMTs responsible for installing individual m5C sites. Through bisulfite-sequencing of RNA recovered from 16 unique strains, each deleted for a non-essential, putative RNA methyltransferase (RMTases), we pair specific sites of m5C with unique R5CMTs, providing clues for targeting preferences of each R5CMT. R5CMT activity is further validated orthogonally through in vitro methylation assays and mass spectrometry. Dynamic changes in the m5C epitranscriptome were observed across distinct biological conditions. Strains deleted for individual (or pairs of) R5CMTs exhibit impaired growth at increasing temperatures, demonstrating that m5C modifications support life at high temperatures. Our results suggest overlap in R5CMT function, with many m5C sites being targeted for modification by at least two R5CMTs in vivo, indicating a partially redundant network of enzymes that maintain the m5C epitranscriptome. The combined single-nucleotide, quantitative mapping of m5C, phenotypic analyses of strains with abrogated m5C-profiles, in vitro biochemistry with multiple recombinant R5CMTs, and homology of T. kodakarensis R5CMTs with RMTases in each Domain provides a wealth of predictive power for understanding the impacts of the m5C epitranscriptome in extant life.
Results
m5C is abundant in T. kodakarensis coding and non-coding RNA
The nucleoside pools derived from total and rRNA-depleted RNAs isolated from T. kodakarensis strain TS559 were quantified and compared to that of 10 human cell lines (referred to here as the universal human reference (UHR)) by LC-MS/MS (Supplementary Fig. 1 and Supplementary Data 1). Total RNA preparations were dominated (~94%) by non-coding RNA (ncRNA) sequenced reads, while fractions enriched for mRNA through selective degradation of rRNA and size selection to remove RNAs <200 nt (see “Methods”) generated nearly ncRNA-free (~99.9%) mRNA sequencing results (Supplementary Fig. 1a). Within total RNA from the UHR, m5C constitutes ~0.1% (~1:1000) of cytidines (Supplementary Fig. 1b). The bulk of total m5C in the UHR is likely present in stable, abundant rRNAs and tRNAs, and the order of magnitude reduction in m5C levels upon mRNA selection confirms such. In stark contrast, total RNA preparations from T. kodakarensis revealed an unprecedented ~2.5% of total cytidines (~1:40) were replaced by m5C (Supplementary Fig. 1b). While degradation of T. kodakarensis rRNAs and size exclusion of tRNAs similarly reduced the levels of m5C nearly ten-fold, the resulting levels of m5C in mRNAs were still >6.5-times greater than m5C levels in the UHR mRNA pools. Analysis of small RNA fractions (<200 nt) indicated that small RNAs are also rich in m5C (Supplementary Fig. 1c and Supplementary Data 2).
The high levels of modified cytidines in total, size fractionated, and mRNA preparations from T. kodakarensis were confirmed to be nearly exclusively m5C and not 5-hydroxymethylcytidine, 3-methylcytidine, or 4-methylcytidine, by comparing retention times and mass transitions of corresponding modified nucleosides. While m5C levels are much higher in T. kodakarensis compared to the UHR, RNA modifications are not universally more prevalent in T. kodakarensis. Analysis of total and rRNA-depleted samples revealed that m6A is more abundant in the UHR than m5C and that m6A is exceptionally low in T. kodakarensis total RNA and undetectable in mRNA (Supplementary Fig. 1b). The selective employment of m5C over m6A in RNA is likely important for the biological functions of the T. kodakarensis epitranscriptome.
Bisulfite sequencing reveals abundant, high-confidence, and reproducible m5C modifications in the T. kodakarensis transcriptome
Sodium bisulfite treatment of RNA results in cytidine deamination, converting cytidines to uridines. Cytidines modified at the C5 position, however, are resistant to bisulfite-driven deamination and retain their cytidine identity. Cytidines retained after bisulfite treatment are therefore presumed to be m5C. However, we can not rule out the possibility that some retained cytidines are instead occupied by other bisulfite-resistant modifications or are otherwise resistant to deamination. Bisulfite treatment followed by cDNA synthesis, sequencing, and analysis (Supplementary Fig. 2a) permits quantitative assessment of m5C modification frequencies at single-nucleotide resolution. The cytidine conversion rates of six bisulfite-sequencing libraries, three from exponential (e1-3) and three from stationary (s1-3) cultures of strain TS559, ranged from ~ 99.8%–99.9% (Supplementary Fig. 2b), confirming near complete cytidine deamination which allows accurate identification of m5C sites.
RNA sequencing libraries were prepared for total RNA and rRNA-depleted fractions from T. kodakarensis TS559 cultures grown to early exponential or stationary growth phase. Sequenced reads were subjected to rigorous quality control pre- and post-alignment to the reference genome, and objective parameters were applied to call individual m5C sites (Supplementary Fig. 2). Initial examination of candidate methylation sites revealed obvious false-positives likely due to incomplete bisulfite conversion of some reads (i.e., sequencing reads wherein cytidine constitutes >3% of nucleotides), and these reads were removed from further analysis. To call a site as modified, we defined parameters for minimum overall sequencing coverage (≥47x), minimum m5C coverage (Supplementary Fig. 2c), minimum m5C modification frequency (Supplementary Fig. 2d), and reproducibility in two or three biological replicates (Supplementary Fig. 3a). m5C modification frequencies below 10% approached background levels in all libraries (Supplementary Fig. 2d), and therefore a 10% minimum modification frequency was applied as a requirement. Owing to the depth of high-quality sequencing, such as that of sites with ≥1000x coverage, the acceptable minimum m5C frequency was lowered to 5% in these cases. Total sequencing depth at high-confidence m5C sites typically ranged from ~100–1000x (Supplementary Fig. 2e), indicating that total coverage requirements were met for most m5C sites. Despite repeated attempts, tRNAs were not captured well in our sequencing libraries, and therefore a comprehensive analysis of tRNA m5C was not performed. Low abundance RNAs did not provide significant statistical representation to accurately define sites with m5C modification. For these reasons, the m5C profile reported here likely underrepresents the totality of the m5C epitranscriptome in T. kodakarensis. However, the extraordinarily high sequencing depth achieved as a result of the small genome (~2 Mbp) and high transcription levels typical for T. kodakarensis improves our ability to detect low stoichiometric modifications with greater accuracy and comparative power.
The position and frequency of m5C at each site proved to be highly reproducible between replicates, suggesting minimal false positives within the final data. The genomic coordinates and complete annotation of each m5C site is recorded in Supplementary Data 3. Linear regression of m5C sites between biological replicates revealed Pearson’s correlation coefficients average at ~0.90, indicating high reproducibility of m5C frequencies across the epitranscriptome (Supplementary Fig. 3a). Although we detected a minor proportion of m5C sites that appear as outliers among replicates (when the m5C site met our high-confidence thresholds in only two of three replicates), the vast majority of our reads met our high-confidence thresholds within each sample; these outliers form the small number of data points seen next to the x- or y-axis. Sites that were high-confidence in all 3 replicates produced Pearson’s correlation coefficients >0.90 (Supplementary Fig. 3b). To account for the discrepancy in reproducibility of some sites, we present a parallel analysis of datasets that include (1) m5C sites of high-confidence in at least two replicates (Fig. 1) and (2) m5C sites of high-confidence in all three replicates (Supplementary Fig. 4).
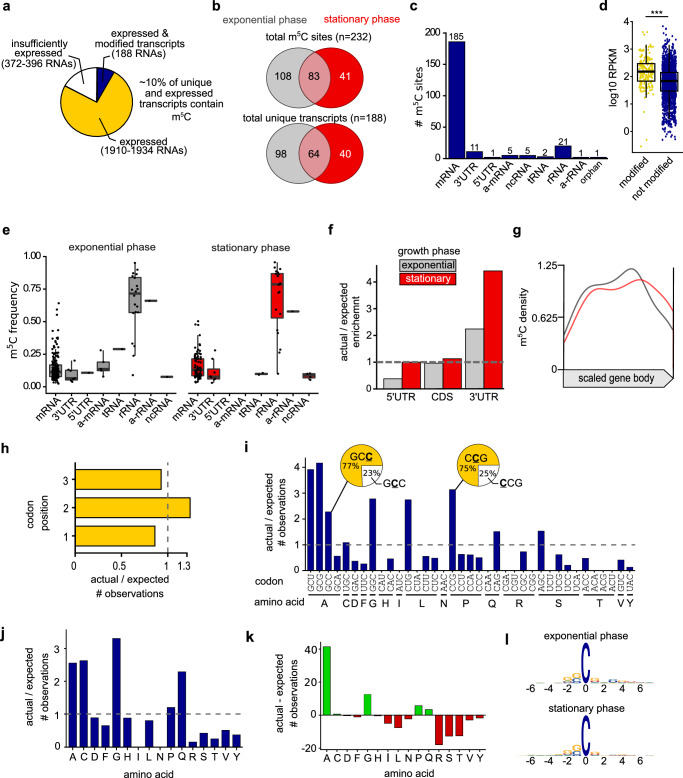
Fig. 1. T. kodakarensis has an extensive and dynamic m5C epitranscriptome in a diverse set of RNAs.
a Empirical analysis of the T. kodakarensis m5C epitranscriptome. Out of 1910–1934 expressed RNAs between two biological conditions, 188 contained at least one m5C site. b The number of distinct m5C sites and modified transcripts varied between exponential and stationary growth phase cells. c Number of m5C sites mapping to diverse RNAs, including some antisense (a-) RNAs. d Gene expression (log 10 mean RPKM) of modified (yellow) and unmodified (blue) mRNAs. The minima, maxima, 25th, 50th, and 75th percentiles are represented in the box-and-whiskers plot. e Modification frequencies across RNA types. The minima, maxima, 25th, 50th, and 75th percentiles are represented in the box-and-whiskers plot. f Comparison of the observed and expected number of m5C sites in different regions of an mRNA in exponential and stationary growth phase cells. The ratio (actual/expected) of m5C sites equals to 1 if the m5C sites are distributed randomly within the region. g Positional bias of m5C sites when mapped within coding sequences. h Positional bias of m5C sites at each codon position. i The number of m5C sites mapped to each codon compared to the number expected by random chance. Obvious deviations from expectation (actual/expected = 1) were observed. There were also apparent biases in codons that include more than one cytidine (see pie chart insets). j The actual/expected and (k) difference between actual and expected (actual–expected = 0 where no bias is detected) number of m5C sites mapping to particular amino acid codons. l Logo sequence analysis of nucleotides adjacent to m5C sites. Additional information is present in Supplementary Data 3.
m5C is highly abundant in a diverse set of RNAs with positional bias
Quantitative analysis of the m5C epitranscriptome revealed a dense modification profile where shifts in modification sites and frequencies were observed between two biological conditions: exponentially growing and stationary phase cells (Fig. 1 and Supplementary Fig. 4). T. kodakarensis encodes just 2306 open reading frames, and ~1900 unique transcripts (~82%) were expressed at or above our detection threshold (>47x average coverage across each nucleotide in the gene). Of those genes with sufficient expression to meet our statistical thresholds, ~10% (n = 188 unique transcripts) contain at least one m5C site (Fig. 1a); we identified a total of 232 unique positions that satisfied our minimum criteria for high-confidence and reproducibility, mapping to 188 unique transcripts (Fig. 1b). An identical analysis for sites reproducible in three of three replicates (Supplementary Fig. 4a), revealed 77 of ~1250 RNAs (~6%) retains at least one site of m5C modification. The total number of m5C sites, m5C frequencies, and modified transcripts changed between exponentially growing and stationary phase cells (Fig. 1b and Supplementary Fig. 4b), suggesting a dynamic m5C profile that shifts in response to environmental cues. Most m5C sites were detected within coding sequences (185 of 232, ~80% for minimally two biologically reproducible sites (Fig. 1c); 77 of 99, ~78% for three biologically reproducible sites (Supplementary Fig. 4c). The 16S and 23S ribosomal RNA fragments are densely modified, with 21 or 19 m5C sites detected in mature rRNA, in two or three biological replicates, respectively. No m5C sites were detected in either copy of the 5S rRNA. T. kodakarensis mRNAs often lack untranslated regions (UTRs) or encode very short UTRs52. We observed only sparse modification of UTRs or transcripts that mapped antisense (a-) to mRNAs, rRNAs, and other non-coding RNAs. Although tRNAs were not captured well during sequencing, one or two m5C sites were detected in tRNAs with 3/3 or 2/3 replicates, respectively.
Gene expression analysis from bisulfite-treated RNA is not feasible since bisulfite-driven RNA degradation introduces bias in what RNAs are sequenced. The identification of bisulfite-refractory sites may therefore be confounded by gene expression levels since highly expressed transcripts survive the harsh bisulfite treatment quantitatively better than lowly expressed transcripts. That is, transcripts existing in multiple copy numbers (highly expressed) will have a higher chance than lowly expressed transcripts to be recovered after the destruction of RNA due to sodium bisulfite treatment. We asked whether RNAs determined to be modified with m5C in this study may be better identified due to their expression levels. We retrieved RNA-seq data from NCBI Sequence Read Archive generated from mock-treated cultures grown under laboratory conditions similar to the present study2. The average RPKM levels of RNAs in which we detected a cytidine modification are about twofold higher compared to RNAs in which we did not detect a modified cytidine (Fig. 1d and Supplementary Fig. 4d). This is not to say m5C is enriched in highly expressed transcripts, rather it is likely cytidines resistant to bisulfite deamination (m5C or artifactual m5C) are better identified due to a higher abundance of these RNAs (and therefore better able to survive harsh bisulfite treatment), whereas m5C in lowly expressed RNAs may not be detectable at all.
Modification frequencies in mRNAs ranged from ~5–65% while averaging at ~15–20%, whereas modification frequencies in rRNA averaged at ~75% (Fig. 1e and Supplementary Fig. 4e). mRNA modifications in some model species are typically enriched in UTRs or near UTR-ORF boundaries53,54. We observed more m5C sites in 3’UTRs than expected if otherwise distributed randomly throughout an mRNA (Fig. 1f), but as no 3’UTR sites were reproducibly identified in all three replicates (Supplementary Fig. 4f), it is invalid to definitively suggest a 3’UTR bias. The distribution of modifications that mapped to coding sequences did not have an obvious positional bias over the open reading frame (Fig. 1g and Supplementary Fig. 4g).
As m5C sites accumulate within the coding sequence, it is possible that mRNA modifications may impact translation dynamics by statistically over impacting specific codons or codon positions. We did not observe a significant bias at any codon position relative to what would be expected if m5C sites were distributed at random (actual/expected = 1, Fig. 1h and Supplementary Fig. 4h). The absence of m5C enrichment at the third codon position likely rules out sole impacts on wobble base-pairing and therefore non-cognate amino acid incorporation. However, m5C is enriched in select codon sequences (Fig. 1i and Supplementary Fig. 4i). Complete analysis of codon and amino acid modification bias is recorded in Supplementary Data 4. We mapped m5C within GCG codons (alanine, n = 18) to an extent that is fourfold higher than expected if by random chance. Modifications to codons GCU (alanine, n = 13), GGC (glycine, n = 18), CCG (proline, n = 24), and CUG (leucine, n = 16) were similarly enriched. When codons include multiple cytidines, there is a strong bias for which cytidine is selected for modification. Out of 35 modified GCC codons, 77% of the modifications were to the third cytidine (Fig. 1i, pie chart inset). When considering sites reproducible in all three replicates, 100% of modifications to this codon occur at the third cytidine (Supplementary Fig. 4i, pie chart inset). Modifications mapping to leucine and proline codons indicate a strong bias for one of the four possible codons (CUG and CCG, respectively) whereas modifications are highly underrepresented within the other 3 codon possibilities. These data suggest select codons are targeted for modification within mRNAs and may impact the translatability of those codons.
A similar representation is observed at the amino acid level. m5C occurrences in particular amino acid sequences indicate 41 more m5C sites in alanine encoded codons than expected (actual–expected) (Fig. 1j, k). Codons encoding cysteine and glutamine are overrepresented with high-confidence m5C sites by two to sixfold (Fig. 1j). We expected to see 0.38 m5C sites in cysteine codons, but we detected 1. Even though this indicates a significant fold change (actual/expected) in the number of observations, less than 1 additional m5C site was detected compared to expected (actual–expected) (Fig. 1j, k). Therefore, overrepresentation of modified cysteine codons may not necessarily be biologically relevant. Near identical conclusions can be drawn in the analysis of m5C sites reproducible in all 3 replicates (Supplementary Fig. 4h–k). Taken together, these data indicate a strong positional bias in m5C sites in particular codon and amino acid contexts, possibly indicating m5C impacts the translatability of these codons. However, there does not appear to be a congruent nucleotide sequence context that describes all m5C sites (Fig. 1l). This observation is consistent with previous studies that failed to identify a single sequence motif associated with m5C modifications21,25,55. It is plausible that the T. kodakarensis m5C epitranscriptome is installed by a collection of RNA modifying enzymes, each targeting a unique sequence or RNA structure. In this case, a single nucleotide context would not be anticipated, rather, individual enzymes would be expected to target unique sequence motifs and RNA structures for the installation of m5C.
T. kodakarensis encodes a suite of RNA modifying enzymes
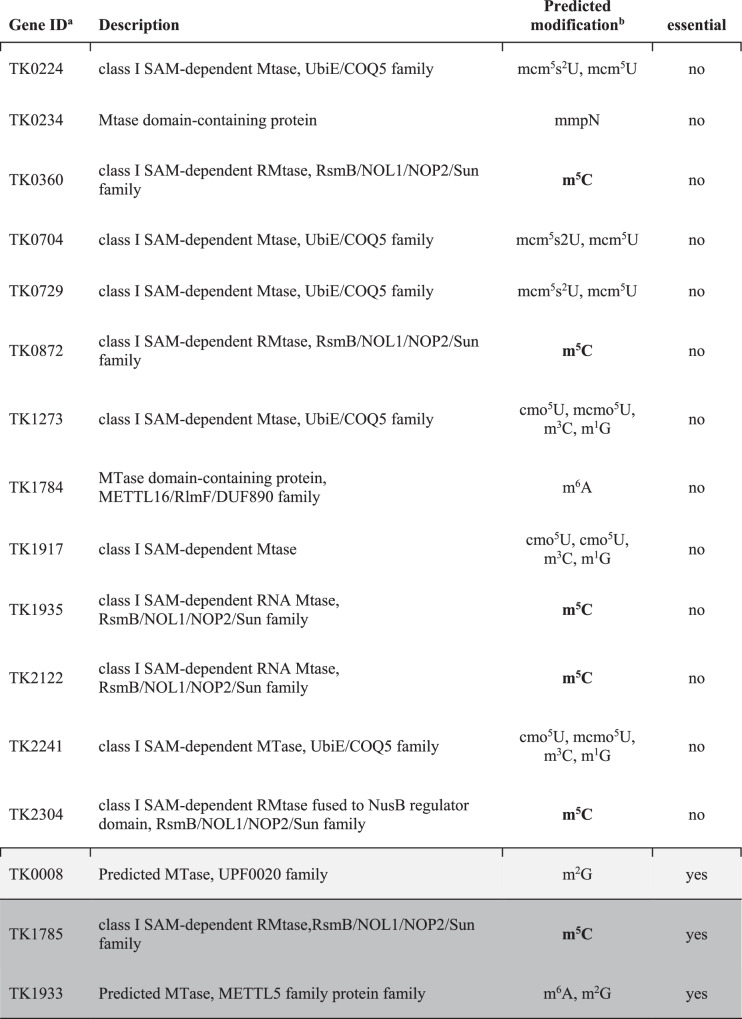
Publically available annotations of the T. kodakarensis genome revealed 16 ORFs that may encode R5CMTs (Table 1). Six of the sixteen putative RNA methyltransferase (RMTase) enzymes were predicted to install m5C based on domain comparisons with known RMTases in the Modomics Database but none have previously been determined to be bona fide RNA methyltransferases. Each of the six enzymes predicted to install m5C were also identified by pBLAST to have domain homology to the human NSUN family of proteins that are known to install the m5C epitranscriptome in mammals. Upon the deletion of any individual RMTases, any differences in the m5C epitranscriptome compared to the parental strain (TS559) could be rationalized to require the activities of the deleted enzyme. Using established techniques56, we thus attempted to individually, markerlessly delete sequences encoding the entirety of the ORF for each putative RMTase from the TS559 genome.
Table 1.
Putative RNA methyltransferases in T. kodakarensis
aTwo predicted methyltransferases (rows highlighted in gray and dark gray) are essential and cannot be deleted from the cell, while 14 are non-essential. It is unclear if TK0008 is essential.
bModification predictions are based on sequence and structural similarities to known RMTases from Modomics. The text is bolded where m5C is the predicted modification.
We were successful in generating 13 strains, each lacking a single putative RMTase. The sequences surrounding the targeted locus were first confirmed to lack the RMTase encoding sequences by several diagnostic PCRs (Fig. 2a). Then, we sequenced the entire genome of each deletion strain (typically at >15x coverage; Fig. 2b and Supplementary Fig. 5) to ensure the lack of second-site mutations. Sequencing of bisulfite-treated RNA (Fig. 2c and Supplementary Fig. 5) at ~100x to ~1600x coverage revealed, in all strains, a lack of transcripts from the loci targeted for deletion. By repeating our markerless genomic modification procedures, we were further able to construct double deletion strains wherein two putative RMTases were simultaneously deleted. Despite exhaustive efforts, two of the targeted putative RMTases (TK1785 and TK1933) could not be deleted. Attempts to delete gene TK0008 yielded mixed results; T. kodakarensis regularly maintains many copies of its genome57, and while we were able to generate strains that appeared to lack TK0008 via diagnostic PCR and whole genome sequencing, some genomes containing TK0008 must have been retained within the population that led to repetitive failures to yield strains that completely lacked TK0008 sequences after two culture passages. Thus, TK0008 was excluded from further analysis.
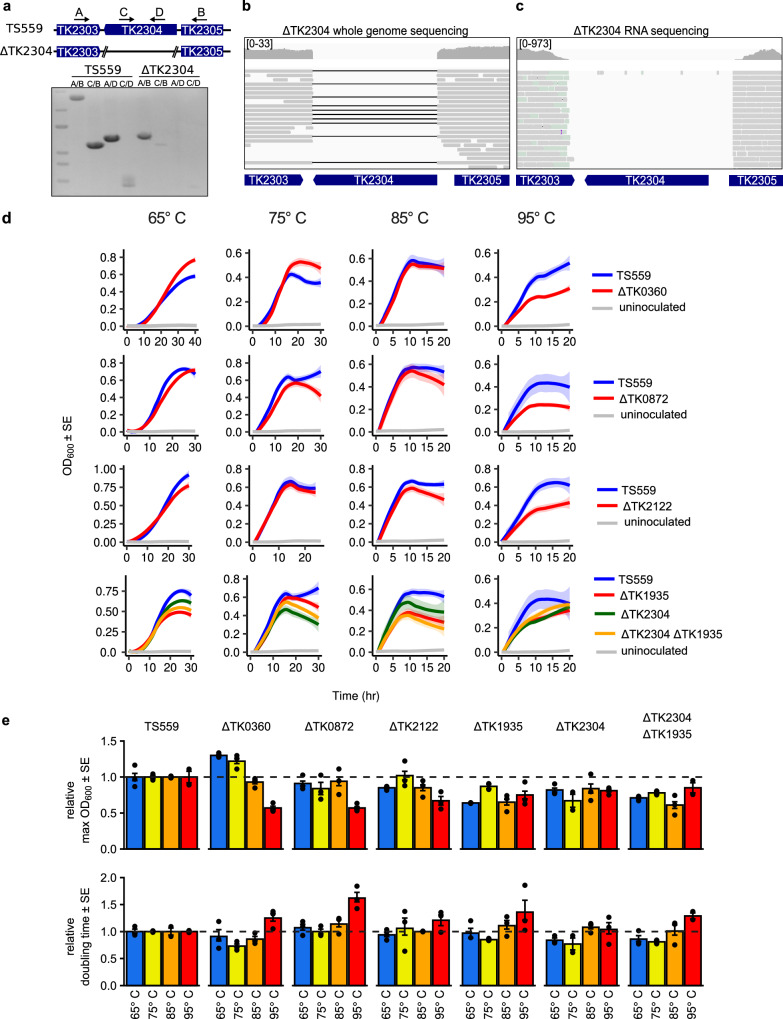
Fig. 2. The m5C epitranscriptome supports hyperthermophilic growth.
a The deletion of gene TK2304 was initially confirmed by PCR using 4 sets of primers. External primers (A/B) amplify across the deleted locus and result in a full length amplicon in parent strain TS559 and an amplicon reduced by the size of gene TK2304 in the ΔTK2304 strain. Internal primers (C/D) or a combination of internal and external primers (A/D, C/B) result in amplification in strain TS559 but not ΔTK2304. An uncropped gel image is provided in Source Data. Final confirmation was performed using (b) Minion whole genome sequencing (DNA-seq) and (c) Illumina RNA bisulfite sequencing (RNA-seq). Visual inspection of DNA and RNA sequences aligned to the T. kodakarensis reference genome using Integrative Genomics Viewer show the absence of reads from the deleted loci. In the DNA-seq windows, black lines connect contiguous reads and indicate a gapped alignment. The coverage range for each window is listed in brackets. d Head-to-head growth competitions were performed at 65°, 75°, 85°, and 95 °C for each strain deleted for an R5CMT (ΔTK0360, ΔTK0872, ΔTK2122, ΔTK1935, ΔTK2304), the double deletion (ΔTK1935 ΔTK2304), and parent strain TS559. e The maximum optical density (Max OD600) and the rate of growth (doubling time) for each culture is illustrated relative to parent strain TS559. ± 1 standard error (SE) is represented by error bars for n = 3 biological replicates.
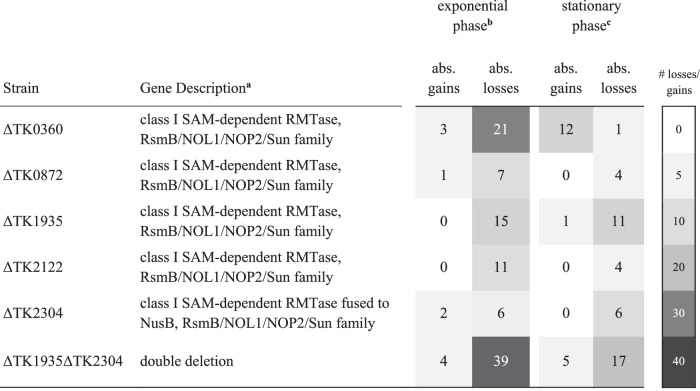
We collected and bisulfite-sequenced RNA from each single or double deletion strain grown to early exponential or late stationary phase in duplicate. High-confidence and reproducible m5C sites were identified in each deletion strain (see linear regression analysis in Supplementary Fig. 6) following the pipeline described in Supplementary Fig. 2 and compared with m5C sites established in the parental strain, TS559. Differential m5C frequencies demanded minimally a greater than twofold change and a p value cutoff of 0.01 to be deemed statistically relevant. The number of sites identified in parental and deletion strains where m5C frequencies were completely lost (Table 2) or differed sufficiently to meet our stringent criteria (Supplementary Table 2) are reported. We identified multiple sites where an absolute loss in m5C modification frequency (≤2%) was observed in one of five deletion strains; ΔTK0360, ΔTK0872, ΔTK1935, ΔTK2122, and ΔTK2304 (Table 1), indicating these genes encode enzymes that likely directly target RNA for m5C installation. Bisulfite-sequencing of RNAs from the remaining ten single deletion strains did not reveal any absolute losses within the m5C epitranscriptome (Supplementary Table 2), suggesting that these enzymes either do not install m5C or were not active under our experimental conditions; note that RNA transcripts for each putative RMTase gene targeted for deletion were present in the TS559 parent stain, indicating these genes are expressed under our experimental conditions. Our results are thus consistent with the predicted modification potentials listed in Table 1 and pBLAST homology searches for R5CMTs. To our surprise, certain strains deleted for putative m5C-specific and alternative RMTase enzymes showed gains in m5C sites or increased modification frequencies. Sites that show new modification status in deletion strains compared to the parental strain met our stringent statistical rigors for reproducibility. The m5C gains imply that the loss of selected m5C sites due to deletion of distinct RMTases from T. kodakarensis results in compensatory modifications throughout the epitranscriptome. This level of complexity and intertanglement of enzyme activities reinforces that the epitranscriptome is dynamic and responsive to changes in gene expression and environmental cues.
Table 2.
Absolute losses and gains in m5C sites in strains deleted for individual R5CMTs
aThe NCBI description and protein family (Uniprot and InterPro predictions) are provided.
b,cBS-seq of individual or double deletion strains identified absolute losses and gains in m5C sites between exponential and stationary growth phase cells.
A redundant network of R5CMTs likely generates the m5C epitranscriptome
We detected 232 high-confidence and reproducible m5C sites (Fig. 1a) and correlated the absolute loss of ~46 of these sites with the loss of one of five R5CMTs (Table 2). Out of 21 sites mapping to mature rRNA, we only identified enzymes likely to install 9 m5C sites (Fig. 3). It is possible that essential enzymes from Table 1, or enzymes missed by our annotations of the genome, are responsible for the installation of the remaining m5C sites detected here. Alternatively, multiple R5CMTs may target identical sites for installation of m5C. To explore the latter possibility, we deleted both TK1935 and TK2304 within the same cell. Bisulfite sequencing of the double deletion strain (ΔTK1935 ΔTK2304) during exponential or stationary growth phases revealed a synergistic absolute 39 losses in m5C sites compared to the 17 and 6 losses identified in the ΔTK1935 and ΔTK2304 single deletion strains, respectively (Table 2 and Fig. 4i). This suggests that T. kodakarensis may generate epitranscriptomes with overlapping and complex activities for distinct RNA modifications.
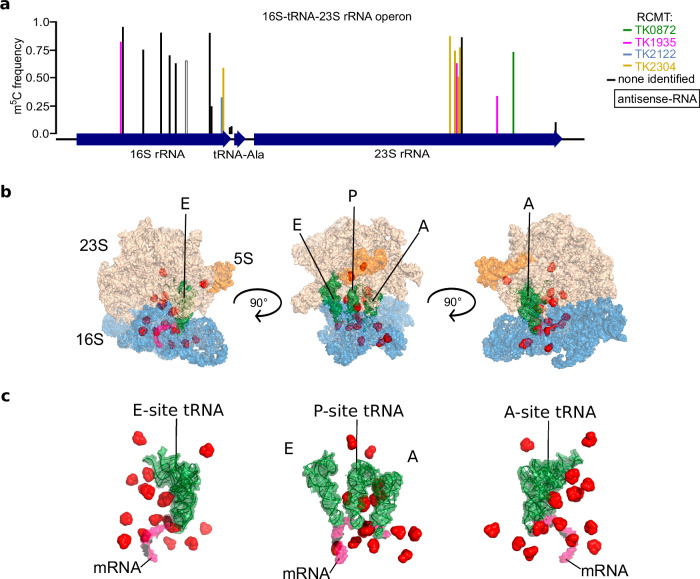
Fig. 3. The T. kodakarensis ribosome is densely modified at key functional regions involved in translation.
a Each bar represents an m5C site mapped to the 16S-tRNAAla-23S operon; bar height represents modification frequency and its color corresponds to the enzyme responsible for its installation. One m5C site was mapped antisense to the 16S rRNA (black outline). Additional information is provided in Supplementary Data 5. b, c m5C sites (red spheres) are not randomly distributed. 16S subunit (blue), 23S subunit (nude), 5S subunit (orange), A, P, and E site tRNAs (green), and the mRNA (pink) are shown at 3 angles rotated at 90°.
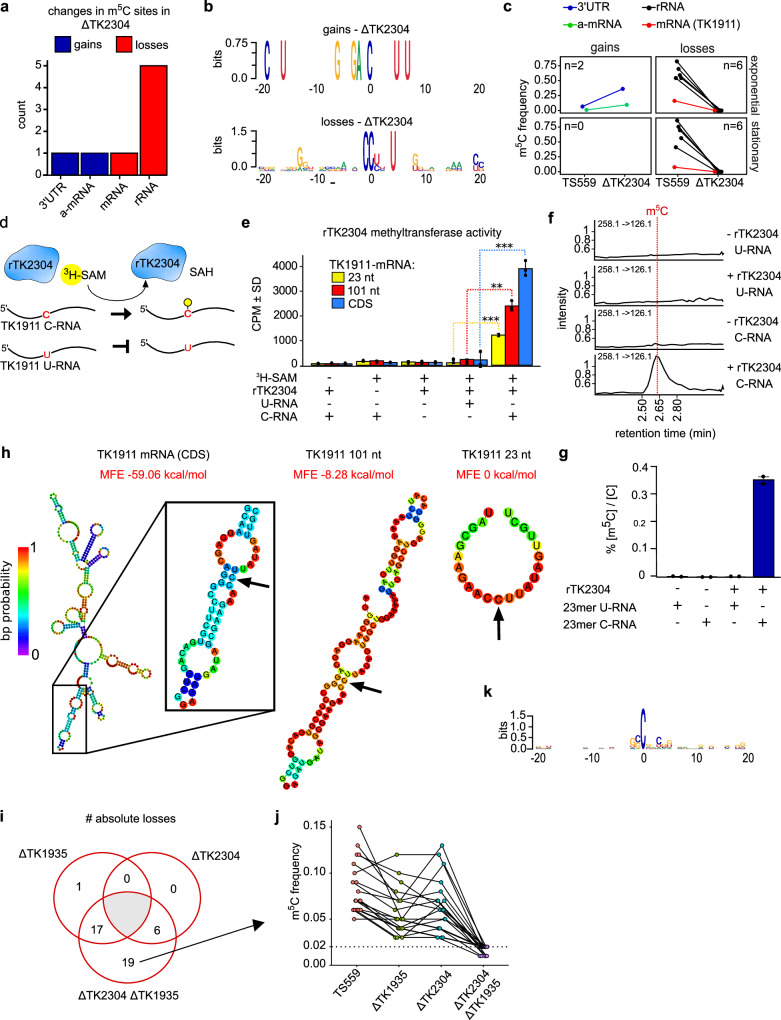
Fig. 4. TK2304 encodes a bona fide R5CMT with apparent RNA sequence and structural specificity.
a Bisulfite sequencing of strains deleted for the gene TK2304 reveals losses and gains in m5C sites. b Logo sequence analysis of adjacent nucleotides surrounding sites where m5C was lost or gained. c The change in modification frequency between parent strain TS559 and ΔTK2304 is faceted by growth phase and direction of regulation. The number of sites (n) is listed within each window and the color corresponds to RNA type. d The methyltransferase assay schematic is shown. Recombinant enzyme (rTK2304), radiolabeled methyl co-factor (3H-SAM), and RNA substrate are incubated together. The enzyme will transfer the labeled methyl group to the cytidine target in the C-RNA but not the U-RNA where the cytidine is replaced with a uridine. e The results of the methyltransferase assays are displayed in counts per minute (CPM) which measures the β-decay of the radiolabeled methyl group covalently bound to the RNA substrate. A two-side, two-sample t-test was used to compare enzymatic activity on the 23 nt, 101, and full length substrates, producing p values of 0.0001, 0.0024, and 0.0001, respectively. n = 3 biological replicates. **p < 0.01, ***p < 0.001. f, g Mass spectrometry analysis of RNAs methylated by rTK2304. The 101 nt oligonucleotide was digested to single nucleosides and the m5C levels were quantified relative to cytidine across controls. n = 2 biological replicates. h The secondary structures of the 23 nt, 101 nt, and full coding sequence (CDS) RNAs encoding the gene TK1911 were predicted using the Vienna RNAFold webserver83. The arrow points to the cytidine targeted for methylation. The color of each base corresponds to the base pair (bp) probability, and the minimum free energy (MFE) for each structure is listed. i The number of unique and absolute losses in m5C sites between single and double deletion strains ΔTK2304 and ΔTK1935. j The change in modification frequencies between strains of the 19 m5C sites completely lost (m5C frequency ≤2%) in ΔTK2304 ΔTK1935. k Logo sequence analysis of the 19 sites uniquely and completely lost in ΔTK2304 ΔTK1935.
The m5C epitranscriptome enhances cellular thermophily
T. kodakarensis grows over a wide temperature range (~50 to ~98 °C) with a growth optimum of 85 °C resulting in a ~40-min doubling time under ideal conditions. When constructing the single and double deletion R5CMT strains, we noted an obvious growth impairment of some deletion strains. Growth of the parental and deletion strains were then monitored at 65°, 75°, 85°, and 95 °C to present strains with different environments (Fig. 2d). At 65° and 75 °C, the growth of strains deleted for one of the five non-essential R5CMTs is largely comparable to the parent strain TS559 (Fig. 2d), but deletion strains fared more poorly at increasing culture temperatures. Notable at 85 °C and more obviously at 95 °C, each R5CMT deletion strain displayed significant growth defects compared to the parental strain, and in many cases exhibit slower growth rates and reduced end-point culture densities (Fig. 2e). The strain deleted for both TK1935 and TK2304 (ΔTK2304 ΔTK1935) experienced a compounding growth defect compared to either individual deletion strain, suggesting the m5C epitranscriptome holistically supports cell viability and survivability at high temperatures.
Growth defects were also observed in strains deleted for the other 8 putative RMTases. The growth rates for most strains were generally within expectations at 65°, 75°, and 85 °C, but longer doubling times were observed at 95 °C in many strains (Supplementary Fig. 7). Many deletion strains also displayed a lower end-point culture density compared to the parent strain, and this defect becomes increasingly more apparent at elevated temperatures (Supplementary Fig. 7). Unsurprisingly, strains deleted for multiple RMTases experienced a compounding effect compared to their single deletion counterparts.
The T. kodakarensis ribosome is hyper modified at key regions involved in mRNA decoding
Ribosomal RNA across bacterial and eukaryotic model species typically harbor 2–3 m5C sites. In E. coli, each of the three m5C sites is installed by a unique Rsm MTase58, whereas human cells rely on the NSUN family of MTases to install both ribosomal m5C sites59. Surprisingly, we detected an order of magnitude increase (n = 23) in m5C sites mapping to the single T. kodakarensis 16S-tRNAAla-23S rRNA operon (Fig. 3a and Supplementary Fig. 7). An additional m5C site was mapped antisense to the 16S rRNA, two m5C sites map to the region between the 16S and tRNAAla, and 21 m5C sites are incorporated into the final 16S (n = 12) or 23S (n = 9) sequences. The publically available CryoEM structure of the T. kodakarensis ribosome (PDB: 6TH6) resolves 18 candidate m5C sites, and 12 (66.7%) show densities consistent with methylation at the C5 position (Supplementary Data 5). There are no noticeable differences in m5C sites or frequencies between exponential or stationary growth phases, and no m5C residues are detectable in either copy of the 5S fragment (Supplementary Fig. 7a, b). Genomic coordinates and modification frequencies in the 16S-tRNAAla-23S rRNA operon across all strains are recorded in Supplementary Data 5. A previous study demonstrated that T. kodakarensis ribosomes are hypermodified with ac4C, implying that additional modifications support rRNA function and perhaps ribosome performance at high temperatures2. Our data provides evidence that the T. kodakarensis ribosome is also hypermodified with m5C, implying that RNA modifications may support ribosome function and performance at high temperatures.
We were able to correlate 9 m5C sites in the 16S and 23S rRNA fragments with specific R5CMT activities, based on the absolute losses in m5C sites due to deletion of specific enzymes. Four of the five identified R5CMTs appear to target rRNAs (Fig. 3a), to the exclusion of TK0360. The m5C sites mapping to rRNA are generally distributed throughout the 16S and 23S rRNA fragments with a linear clustering of sites in the 23S fragment. However, when viewed in the atomic structure of the T. kodakarensis 70S ribosome, m5C sites obviously crowd the A, P, and E sites and are positioned along the path where the mRNA is fed into the decoding center (Fig. 3b, c). It is therefore clear that m5C sites in mature ribosomes are not randomly distributed but enriched in highly conserved and functional regions.
R5CMTs display sequence and structural specificity for substrate RNAs
The mechanisms that target MTases to distinct positions in the transcriptome are unclear. To elucidate the targeting strategies of R5CMTs identified here, we analyzed the sites where differential m5C modification between deletion and parental strain were detected. A representative analysis is shown for the deletion of TK2304 (Fig. 4) while analysis for deletions of TK0360, TK0872, TK1935, and TK2122 are recorded in Supplementary Figs. 8 and 9. Deletion of TK2304 resulted in 6 absolute losses and 2 gains in m5C sites (Fig. 4a). Logos sequence analysis indicates some conservation of sequence contexts for gains and losses, but the totality of sequence context alone is insufficiently complex to completely explain targeting of these sites. Three definitive nucleotides (CCnnnU) constitute a partial RNA sequence motif targeted by TK2304, while the two gains in m5C sites follow an eight nucleotide motif (Fig. 4b).Sites where m5C was gained or lost were mapped mainly to rRNAs (Fig. 4c, black lines), but one m5C site was detected in a single mRNA (Fig. 4c, red line). Gains and losses found within distinct sequence contexts were identified for all other R5CMTs (Supplementary Fig. 9a–d.II), but in nearly all cases, no sequence context alone would suffice as the sole targeting mechanism for a single R5CMT. The only exception, interestingly, identifies more complex sequence motifs targeted by TK1935, but, surprisingly, the motif varied depending on whether the m5C occurs in rRNA or mRNA (Supplementary Fig. 9b.II). Our analyses suggest that R5CMTs partially rely, to varying extents, on primary sequence for site-specific methylation. However, three to six nucleotides are not unique enough to encode the specificity required to target an enzyme without producing thousands of off-target methylation events throughout the transcriptome, even as small as that of T. kodakarensis.
Modification frequencies in mRNAs are typically low (~15–20%, Fig. 1e) and questions remain regarding whether any biological relevance of substoichiometric mRNA modifications is merited given the low modification frequencies. mRNAs may be intentional targets of MTases or be occasionally mistargeted by MTases with primary roles in rRNA or tRNA. To add credence to specific mRNA targeting, we identified a high-confidence, but low modification frequency m5C within the TK1911 mRNA that was completely lost due to deletion of TK2304. Gene TK1911 encodes a hypothetical protein with an unknown function. During exponential growth, the TK1911 mRNA is modified at a frequency of ~15–18%, while in stationary phase, modification frequency of the same site drops to just ~8–13% (Fig. 4c), thus representing a site that minimally but convincingly falls within our criteria for m5C modification calls. To directly correlate the in vivo loss of m5C sites with enzymatic activity, we designed an in vitro assay where a recombinant TK2304 enzyme (rTK2304) was challenged to site-specifically methylate the mRNA encoding TK1911 (Fig. 4d). When rTK2304 was provided with a radio-labeled, methyl-donor S-adenosyl-L-methionine (3H-SAM) and an RNA substrate identical in sequence to TK1911 (C-RNA), an obvious transfer of the radiolabeled methyl group from SAM to the RNA was detected (Fig. 4e). Reactions lacking SAM, rTK2304, or an RNA substrate displayed background levels of radioactivity recovered on RNA substrates. As an idealized negative control, we also generated an mRNA with the sequence of TK1911 that contained a single nucleotide substitution, replacing the C targeted for m5C modification with a U (U-RNA) (Fig. 4d); note that all other cytidines within the U-RNA were retained. Replacing the native TK1911 RNA (C-RNA) with an otherwise identical RNA with a single U substitution (U-RNA) resulted in only background levels of methyl transfer, equivalent to that of no SAM, no enzyme, or no RNA controls (Fig. 4e).
To elucidate the necessary and sufficient sequences targeting the TK1911 mRNA for modification by rTK2304, we generated RNA substrates of decreasing lengths (444 nt full length mRNA, 101 and 23 nt truncated RNAs; Fig. 4e) with both a C or U at the site of modification identified in vivo. While rTK2304 is unable to methylate any of the U-containing RNAs above background levels, rTK2304 methylates all C-RNAs with increasing activity on longer RNAs. As the sequence immediately surrounding the target cytidine is identical between substrates, other sequence and structural elements within the RNA must elicit differential activity of rTK2304 under these in vitro conditions. Structural comparisons between the three RNA substrates (Fig. 4h) adumbrate that the secondary and tertiary structures between these RNAs may play a role in the substrate recognition or catalytic activity of rTK2304 in vitro. Our results demonstrate that rTK2304 activity in vitro is improved by supplying more RNA sequences and structures to guide site-specific methylation of the TK1911 mRNA and are in contrast to expectations of spurious recognition and methylation of the TK1911 mRNA by TK2304.
To confirm modification of the TK1911 mRNA at the site predicted based on the loss of in vivo modification due to deletion of TK2304, the 101 nt substrate containing a C or U at the central position was mixed with unlabeled SAM and rTK2304 in vitro, purified, then digested to single nucleosides for LC-MS/MS analysis. We observed 0.37% of m5C/C ratio (or 5% oligo modification frequency) on the C-containing 101-nt RNA (Fig. 4f, g and Supplementary Data 6). The U-containing 101 nt RNA did not reveal any mass or m5C/C ratio changes following incubation with SAM and rTK2304. Intact oligonucleotide mass spectrometry analysis of RNase T1 digested 101 nt substrate also indicated the presence of a methyl group at the predicted cytidine target site (Supplementary Fig. 8a, b and Supplementary Data 7).
To better understand substrate recognition, we analyzed the activity levels of recombinant R5CMTs on several RNAs. Like rTK2304, the other R5CMTs were challenged to methylate different 23 nt RNAs corresponding to sequences identified as likely in vivo targets based on the complete loss of m5C frequencies in deletion strains. Statistically significant, and often highly disparate activity levels were observed for rTK2122 and rTK1935 on C- versus U-containing substrates despite all substrates likely lacking base-paired structures (Supplementary Fig. 9a–d.IV). rTK1935 displayed robust activity on RNAs encoding both identified sequence motifs (Supplementary Fig. 10b.II and IV). When each 23 nt was aligned (Supplementary Fig. 9a–d.IV insets), greater sequence consensus was observed with gapped alignments, which was not captured in the logos sequence analysis. These data indicate nucleotide sequence does play a role in specificity, and analysis of non-contiguous sequence contexts may be more appropriate for identifying motifs most ideal for enzymatic targeting.
In vitro experimentation employing recombinant forms of five R5CMTs with C- and U-containing RNA substrates demonstrated specific MTase activity for three enzymes. While methyltransferase activities for rTK2304, rTK1935, and rTK2122 were specific and validated by LC-MS/MS (Fig. 4, Supplementary Fig. 8 and Supplementary Data 7), rTK0872 and rTK0360 displayed low and non-specific in vitro methyltransferase activities on a host of substrates (Supplementary Fig. 9). It is likely that TK0872 and TK0360 activities are regulated in vivo by additional factors to ensure faithful modifications within the epitranscriptome.
Analysis of m5C sites differentially modified in the double deletion strain, ΔTK2304 ΔTK1935, indicate that all but one site in ΔTK1935 and all sites in ΔTK2304 were likewise lost in ΔTK2304 ΔTK1935 (Fig. 4i). We identified 74 sites with reduced m5C frequencies and 12 sites with increased m5C frequencies (greater than twofold change). Thirty-nine of these sites exhibited an absolute loss in modification frequency, and 19 of these sites were uniquely lost in ΔTK2304 ΔTK1935; these 19 sites displayed robust methylation frequencies in TS559 and either single deletion strain, but a complete loss in m5C signal above background is observed when both enzymes are deleted (Fig. 4j). All overlapping sites map to coding sequences and little-to-no logos sequence could be discerned (Fig. 4k) nor does a sequence alignment produce a recognition motif. This would indicate the targeting strategies for these overlapping sites rely on other factors such as RNA structure. Taken together, we have identified a suite of R5CMTs, likely with partial redundancy, that install the m5C epitranscriptome.
R5CMTs likely target RNAs with structures not easily predicted in silico
Targeting strategies that rely on structural features in the substrate RNA were first reported in yeast, where Trm4 was shown to methylate tRNA-like structures in mRNA60. Clarification of the RNA structural recognition mechanism of R5CMTs is difficult. We performed structural analysis of the 3 bona fide RNA m5C methyltransferases reported here (Supplementary Figs. 10–12). We surveyed the available CryoEM structure of the mature T. kodakarensis ribosome for secondary and tertiary structures at m5C sites as well as secondary structure predictions of all sites using the Vienna RNAfold package. We anticipated to see structural similarities between bona fide methylation sites, however, we did not see obvious consistencies in RNA structures targeted by each enzyme.
The predicted structures analyzed here are that of mature RNAs and do not represent intermediate/co-transcriptional structures that may be optimal targets for these enzymes. RNA structures may also be difficult to predict at high temperatures. It is also difficult to predict tertiary RNA structures, which is likely important for binding to RNA. Emerging evidence suggests the T. kodakarensis epitranscriptome is densely modified with ac4C, so structural predictions are surely confounded by other modifications or in vivo factors not easily accommodated in silico. We are not confident that our structural predictions are truly reflective of in vivo structures at m5C sites at the time of methylation. Further studies are necessary to clarify structural elements present in RNAs that contribute to the specificity of methyltransferases.
R5CMTs identified here likely methylate tRNAs
tRNAs are the most chemically complex RNAs in a cell and are major targets for m5C modification in Eukarya and Archaea50. tRNAs were not captured well in our sequencing libraries despite repeated attempts, and therefore a comprehensive analysis of tRNA m5C profiles was not performed. However, transcripts with TKt41 (tRNATrp) and TKt30 (tRNAIle) sequences persisted through sequencing, likely in their immature form when their transcript lengths were longer (Supplementary Fig. 13a) and before their modification profiles were fully realized. The genomic regions that encode TKt41 and TKt30 are 140 nt and 169 nt long, respectively. Transcript positions corresponding to C113 in tRNATrp and C51 in tRNAIle are modified across growth conditions, but their modification frequencies across 16 strains and biological replicates are skewed. Modifications to these RNAs are completely lost in ΔTK0360 (Supplementary Fig. 13b). Therefore, TK0360 likely encodes a tRNA methyltransferase.
Mass spectrometry analysis of small and large RNA fractions indicates a robust signal for m5C in RNAs <200 nt (Supplementary Fig. 1c, small fraction), which is largely composed of tRNAs. To identify potential tRNA methyltransferases, bulk assays of total small and large RNA preparation with radiolabeled SAM transfer driven by in vitro activities of purified enzymes were carried out with five recombinant R5CMTs. In support of likely tRNA targeting activity, rTK2122 shows considerable activity on small (tRNA) and large (rRNA) RNA fractions derived from the strain ΔTK2122 but not the parent strain where the modification sites are already occupied (Supplementary Fig. 14a). These data indicate that the protein product of gene TK2122 likely targets tRNAs and rRNAs in addition to the 2 bona fide mRNA targets described above. rTK2304 shows significant activity on the large RNA fraction, but not small fractions, derived from ΔTK2304 but not from the parent strain (Supplementary Fig. 14b). These data would suggest TK2304 encodes a methyltransferase that targets rRNA, in addition to the single bona fide mRNA target described. Interestingly, there is no detectable rTK1935 activity on either small or large RNA pools (Supplementary Fig. 14c), despite high activity recovered on small synthetic oligos. We speculate that the pool of RNA isolated for these experiments is dominated by mature rRNA and tRNA, and the protein encoded by TK1935 may act co-transcriptionally, relying more heavily on nucleotide sequence rather than structure. This may explain the more complex sequence motif detected at m5C sites lost in ΔTK1935.
We were not able to detect rTK0360 or rTK0872 activity on RNA fractions (Supplementary Fig. 14d, e) or synthetic RNAs, likely indicating these enzymes require additional factors we have not identified (such as RNA secondary/tertiary structures) that are not easily achieved in vitro. It is unclear whether rTK2304, rTK1935, rTK0360, or rTK0872 do not show activity on the small RNA fraction due to our in vitro reactions conditions, whether other in vivo factors may be necessary for activity, or if these enzymes truly do not target tRNAs. TK1785, an essential gene which we could not delete, is predicted to install m5C and could be the dominant tRNA methyltransferase.
Discussion
New and sensitive techniques that combine bioinformatics, biochemistry, next-generation sequencing, genetic manipulations, mass spectrometry, and evolutionary comparisons continue to add to our understanding of temporal changes to the epitranscriptome and how these changes impart phenotype. How m5C residues impact the fate and functions of mRNAs at the individual transcript level has been historically challenging to address due to low abundance RNAs and substoichiometric modification frequencies in conventional model organisms. Given the generally low abundance and substoichiometry of many internal mRNA modifications, it stands to reason how a few transcripts containing a particular RNA modification would exert biological impact among a majority of unmodified transcripts. It remains contested whether mRNAs are specifically targeted for modification and whether m5C in coding regions are functionally relevant. Here, we achieved exceedingly deep coverage of the m5C epitranscriptome and have detected substoichiometric m5C sites with high-confidence and reproducibility in low abundance RNAs, as well as discovered the enzymes responsible for modification at many of these sites. As such, we have identified several ideal candidate m5C sites for future mechanistic studies.
Using a combination of RNA-bisulfite sequencing and mass spectrometry, we showed that T. kodakarensis has a densely modified m5C epitranscriptome that includes diverse RNAs. In bacterial model species, m5C is largely exclusive to rRNA50,61, and unlike eukaryotic models, we have shown that mRNAs dominate the pool of unique, m5C-containing RNAs in T. kodakarensis. Although mass spectrometry analysis indicated that tRNAs are rich in m5C, our sequencing approach excluded tRNAs and other small RNAs, so many additional m5C sites are likely present in the epitranscriptome than what is presented here. In T. kodakarensis, differential m5C sites were detected across exponential or stationary growing cells where media nutrients are plentiful or depleted, respectively. These data would indicate that m5C may be dynamically responding to metabolic cues. Likely due to approximately half of T. kodakarensis genes having no annotated function, we were unable to observe enrichment of any particular gene ontology. However, gene ontology enrichment analysis of m5C-containing mRNAs in human cell lines tend to correlate m5C with metabolic pathways21,62. Numerous studies have observed shifting m5C-profiles dependent of stress response63,64, viral infection65, cell types and tissues22,53,66–68, and even organellar localization53,69, repeatedly indicating that the m5C epitranscriptome is highly responsive to external and internal factors.
The regional position of m5C within an mRNA likely has differential effects on its function. In mammalian species, m5C in mRNA tends to be enriched at 5’ or 3’ UTRs54. In Arabidopsis, the evidence is conflicting with several studies placing m5C in 3’ UTRs, 5’ UTRs or reporting a slight enrichment within coding sequences22,63,70. In zebrafish and rice, an enrichment has been observed within coding sequences71,72. In T. kodakarensis, we observed a slight enrichment in 3’ UTRs. However, this enrichment was only apparent when our strict high-confidence thresholds were met in at least 2/3 replicates and not when datasets were limited to only include sites detected in all 3 replicates. Otherwise, m5C appears to be distributed evenly throughout mRNAs. We also did not observe a strong bias in codon position, likely eliminating the possibility of m5C in mRNA having substantial impacts on wobble base pairing. We did observe an m5C enrichment in select codon sequences, leading us to speculate whether m5C residues regulate the translatability of select codons. In mice, NSUN2-mediated methylation of mRNA encoding interleukin-17a had no observable impact on RNA half-life, but led to increased translation output, indicating a direct role of m5C to regulate the translatability of interleukin-17a73. The methyl group in m5C strengthens the hydrophobic interaction (stacking effect) in stem structures36; the stacking effect may be important for the ribosome decoding process at high temperatures. Further research is needed to discern whether the positional distribution of m5C within an mRNA leads to unique regulatory effects at specific codons or within specific regions.
Previous work has indicated that some m5C sites in eukaryal models increase the stability of RNAs74. The exact nature of these stabilizing effects are ill-defined, but studies have demonstrated that “reader” proteins recognize some m5C sites, leading to specific regulatory outcomes. It has been shown that these intermolecular partnerships can protect mRNAs from nucleolytic cleavage75, prolong half-life15,76, relocate mRNAs within a cell29, and/or increase protein yield25. Apart from acting as a substrate for RNA-binding proteins, m5C has inherent properties distinct from a naked cytidine. m5C does not impact Watson-Crick base pairing, but it does increase hydrophobicity and the melting temperature of CG hydrogen bonds36, and therefore may affect base stacking and overall RNA structure. High temperatures are a constant insult to T. kodakarensis, and it is possible if not likely that the dense m5C epitranscriptome provides much needed structural stability to RNAs.
Two reports have demonstrated that deletion of genes encoding R5CMTs leads to hypersensitivity to heat stress in rice and worms69,71. T. kodakarensis is a heat-loving archaeon, but the rules of thermophily are poorly understood. The selective employment of m5C over m6A (Supplementary Fig. 1b), and its unprecedented abundance leads us to speculate that m5C may increase the thermal stability of RNAs. T. kodakarensis encodes at least 5 R5CMTs, and we showed that deletion of genes encoding one of 5 R5CMTs results in a temperature-dependent growth defect with increasing severity at 95 °C compared to lower temperatures. When multiple R5CMTs were deleted from the same cell, a compounding growth defect was apparent. These data indicate that the m5C epitranscriptome supports life at extreme temperatures.
The strategies that grant RNA modifying enzymes specificity for distinct positions in the transcriptome have yet to be entirely defined. Previous work by us and others have shown some RNA modifications to have defined sequence contexts surrounding the site of modification. Nat10 in T. kodakarensis was shown to target GCC motifs for acetylation2, and most R5CMTs identified here show small ~3–6 nucleotide motifs. Previous studies have also shown that many m5C sites where the R5CMT has been identified are present in a sequence context of ~4 nucleotides22,63,64,77. These short motifs are perplexing, as these sequences do not encode enough complexity to target an enzyme to a select cytidine in the transcriptome without producing thousands of off target modifications. Yet, we see highly precise targeting of RNA modifications in vivo. It is possible that non-contiguous sequences may serve as a better benchmark for identifying sequence motifs because RNA is flexible to the extent as to allow nucleotides to bulge out in a way that would permit non-contiguous nucleotides to contact the enzyme. However, this does not explain why we observed differential enzymatic activity levels on the same RNA substrate of varying lengths (Fig. 4e). Further studies are necessary to clarify the structural elements present in RNAs that contribute to the specificity of methyltransferases.
Here we have shown that the extensive m5C epitranscriptome is established and maintained by a suite of enzymes. We first showed that the individual loss of these 5 enzymes leads to the in vivo loss of site-specific m5C via RNA BS-seq. Then, we demonstrated the in vitro activity of 3 R5CMTs to install m5C using radiolabelling assays and mass spectrometry. Of the >230 m5C sites detected in the parent strain, and since we were only able to correlate the installation of a quarter of these sites with an R5CMT, there is likely at least one more R5CMT encoded in T. kodakarensis. Two MTases we screened, TK1785 and TK1933, are essential and could not be deleted. Annotations would strongly suggest TK1785 encodes a R5CMT, and its essentiality would likewise indicate the essentiality of the m5C epitranscriptome. It is also possible that cytidines may be redundantly targeted for modification and deletion of all R5CMT would be necessary to remove all m5C sites. It has been speculated but sparse evidence would indicate whether methyltransferases share partial redundancy in target sites. To test this, we generated a strain deleted for both TK1935 and TK2304, two genes that encode bona fide R5CMTs (Fig. 4). The in vivo data shows vast changes in the m5C-profile and many additional losses in m5C sites not detected in either individual deletion strain, indicating a partially redundant network of R5CMTs with overlapping methylation targets.
We observed hundreds of m5C sites in T. kodakarensis in diverse RNAs and detected changes in m5C sites in strains deleted for putative RMTases as well as differential m5C-profiles between growth phases. The dynamic nature of the m5C profile reinforces that the epitranscriptome is highly responsive to physiological changes. Deletion of R5CMTs result in temperature-dependent growth defects with increasing severity at higher temperatures, consistent with the hypothesis that RNA m5C supports hyperthermophilic growth. Taken together these data suggest that a suite of R5CMTs with partial redundancy maintain the m5C epitranscriptome and support life at extreme temperatures.
Methods
Cell growth
All T. kodakarensis strains were grown anaerobically at 85 °C in artificial sea-water with yeast, tryptone (ASW-YT) and supplemented with agmatine78. Cultures grown for total ribonucleoside analysis by LC-MS/MS were grown to mid exponential growth phase (OD600 = ~0.3) in duplicate before harvested by centrifugation. Culture prepared for bisulfite sequencing were harvested during early exponential growth (OD600 0.1–0.2) and then left to continue growing before harvesting the remaining culture after 2 h after reaching its maximum optical density (0.6–0.8, stationary growth phase); 300 ml of culture were harvested by centrifugation during early exponential growth phase and 200 ml of culture were harvested during stationary phase. Cell pellets were stored at −20 °C.
Strain construction
Procedures for generating deletion plasmids are previously reported in Hileman et al.79. For each putative RMTase, the gene and ~700 bp up and downstream were PCR amplified from genomic DNA and gel purified (Qiagen, cat# 28706×4). Primers were modified to include 13 bp terminal extensions with homology to pTS700 at SwaI. pTS700 was linearized with SwaI (NEB, cat# R0604S) before the insert was cloned into pTS700 by ligation independent cloning. The coding sequence of the target gene was then removed from the plasmid using QuickChange site directed mutagenesis Agilent cat# 200516). Regions where the target gene overlapped with other genomic elements (i.e., other genes or known promoter sequences) were retained in the plasmid. Deletion-plasmid sequences were confirmed using sanger sequencing.
Procedures for generating deletion strains are published in Gehring et al.56. For each RMTase gene targeted for deletion, the corresponding deletion-plasmid was transformed into T. kodakarensis strain TS559 and cells were plated on agmatine-free, rich medium. After 2–5 days of growth at 85 °C, transformants were picked into rich liquid media lacking agmatine and grown overnight. For each isolate, genomic DNA was extracted from 1 ml of fully grown culture by phenol/chloroform/isoamyl alcohol (25/24/1; v/v/v) followed by alcohol precipitation in an equal volume of ethanol. Plasmid integration into the genome was confirmed by PCR using two primer pairs, with one primer from each pair having homology to the genome and the other primer to the plasmid. This ensures that the amplicon originates from a genomically integrated sequence.
Isolates where the plasmid had integrated at the predicted loci were plated on minimal medium with the counter selectable marker, 6-methyl purine, and grown anaerobically for 2–5 days at 85 °C. Colonies were then picked into rich liquid media and grown overnight. Genomic DNA was purified from each strain and screened for deletion of the target loci. Candidate deletion strains were identified via PCR using four sets of primers. First, PCR using primer pairs that flanked the deletion loci (A/B primers) resulted in a reduced amplicon size equivalent to the size of the deleted region compared to the parent strain. Primer sets both with homology to a region internal to the deletion loci (C/D primers) resulted in PCR amplification in the parent strain but not the deletion strain. Two combinations of external and internal primers (A/D and C/B primer pairs), which should not produce an amplicon in the deletion strain were used to ensure the deletion loci was absent in the deleted strain. Candidate deletion strains where PCR indicated a successful deletion were confirmed via whole genome sequencing.
To ensure the region targeted for deletion was correctly excised and to ensure no off-target genome modifications were present, each candidate strain was screened by whole genome sequencing on a MinIon platform. Each strain was grown anaerobically overnight in 5 ml of rich medium with agmatine at 85 °C. Cultures were pelleted and genomic DNA purified using the Monarch Genomic DNA Purification kit (NEB, cat# T3010S). Library preparation for each strain was completed using the Rapid Barcoding kit 96 (ONT, cat# SQK-RBK110.96). Sequencing was done on the MinIon Mk1C with the R9.4.1 flow cell. Fast5 files were converted to Fastq files using Guppy default settings. Fastq files were aligned to the custom TS559 reference genome using MiniMap2 v2.26-r117 and alignment files were compared to the reference genome using Medaka Variant Calling Pipeline via Neural Networks. Visual inspection of the deletion loci on Integrated Genomics Viewer confirmed deletion of the loci. As a third confirmatory measure, the bisulfite sequencing of RNA (described below) was checked for RNA that aligned to the deleted loci. For all libraries, we observed virtually zero coverage at the deleted loci, fully indicating the gene had been cleanly deleted.
RNA preparation
The universal human reference (UHR) RNA (Agilent, cat# 740000) was generated by pooling 10 cell lines to reduce cell type bias. T. kodakarensis cells were resuspended in 1 mL TRIzol and 200 μL chloroform with a 5–10 min incubation at room temperature proceeding each reagent. Cellular debris were removed via centrifugation at 21,000 × g for 15 min. The aqueous phase was alcohol precipitated in 2.7x the volume of 100% ethanol and incubated at −80 °C for 30 min. RNA pellets were collected via centrifugation at 21,000 × g for 30 min at 4 °C. RNA was resuspended in nuclease-free water and treated with DNaseI (NEB, cat# M0303) for 30 min in 1X DNase buffer at 37 °C. RNA from each sample was purified using the Monarch RNA Clean Up kit (NEB, cat# T2040S or T2030S) or the Zymo RNA Clean and Concentrator kit (Zymo, cat# R1017). A fraction of total RNA was depleted for tRNAs using the Zymo RNA Clean and Concentrator Kit following the manufacturer’s protocol for selective recovery of RNA > 200 nt.
rRNA depletion
rRNA was depleted from a fraction of the RNA samples using reagents provided in the NEBNext rRNA Depletion Kit (NEB, cat# E6310) and custom DNA oligos80. The manufacturer’s protocol was followed with the following changes: The NEBNext rRNA Depletion Solution provided in the kit was substituted for a mixture of 85 oligonucleotides at 1 µM concentration for each oligo whose sequences were complementary to T. kodakarensis rRNA. All volumes for the probe hybridization, RNase H treatment and DNase I treatment sections of the protocol were scaled up twofold and 24 µl of 62.5 ng/µl T. kodakarensis RNA was used as the starting material.
LC-MS/MS
Cellular total RNA, rRNA-depleted samples and RNA substrates for the in vitro methyltransferase assays (see below) were digested to nucleosides at 37°C overnight using Nucleoside Digestion Mix (NEB, cat# M0649S). Tandem liquid chromatography-mass spectrometry (LC-MS/MS) analysis was performed by injecting digested RNAs on an Agilent 1290 Infinity II UHPLC equipped with a G7117A diode array detector and a 6495C triple quadrupole mass detector operating in the positive electrospray ionization mode (+ESI). UHPLC was carried out on a Waters XSelect HSS T3 XP column (2.1 × 100 mm, 2.5 µm) with a gradient mobile phase consisting of methanol and 10 mM aqueous ammonium acetate (pH 4.5). MS data acquisition was performed in the dynamic multiple reaction monitoring (DMRM) mode. Each nucleoside was identified in the extracted chromatogram associated with its specific MS/MS transition (m5C precursor ion m/z: 258.1; product ion m/z: 126.1). To resolve m4C and m5C positional isomers, 0.1% formic acid was used as aqueous mobile phase. The abundance of each nucleoside derived from the digested samples were quantified based on chromatogram peak integration standard curves. Relative abundance of each modified nucleoside was determined by further dividing the peak integrations of modified and unmodified nucleosides (for example: m5C/C). Nucleoside quantification was performed with Agilent MassHunter WorkStation Quantitative Analysis for QQQ version 11.1.
For site-specific analysis of the 101 nt rTK2304 substrate, the fragment was instead digested with RNase T1 (Thermo, cat# EN0542) at 37 °C for 1 h and subject to an Eclipse Fusion Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with a Vanquish UHPLC (Thermo Fisher Scientific). Digested oligonucleotides were separated in a mobile phase gradient consists of buffer A (1% hexafluoroisopropanol (HFIP), 0.1% N,N-diisopropylethylamine (DIEA), 1 μM EDTA) and buffer B (90% Methanol, 10% water, 0.075% HFIP, 0.0375% DIEA, 1 μM EDTA) on a C18 column (Waters ACQUITY Premier Oligonucleotide, 1.7 µm, 2.1 × 100 mm). The obtained chromatogram peaks were deconvoluted by a ProMass HR software package 3.0 revision 12 (Novatia LLC, USA). Methylated oligonucleotide search was performed with NucleicAcidSearchEngine 2.7.081. MS1 setting: Resolution: 60 K; Scan range: 450–2000 m/z; RF lens: 50%. ddMS2 setting: Resolution: 30 K; 7 s dynamic exclusion; Charge state: 2–20; Isolation window: 3 Da; Stepped HCD: 22, 24, 26%. MS1 setting for 23-nt oligonucleotide analysis: Resolution: 120 K; Scan range: 500–2000 m/z; RF lens: 50%.
RNA library preparation and bisulfite sequencing
Frozen cell pellets were resuspended in a total volume of 7.5 ml TRI reagent RT (MRC Inc. Cincinnati, OH), vortexed and incubated at room temperature for 5–10 min prior to cooling on ice. A total of 375 µl of 4-bromoanisole (MRC Inc.) was added and tubes were mixed by inversion prior to centrifugation at 12,000 × g for 15 min at 4 °C. The aqueous layer (~4.5 ml) was removed, 6.75 ml of 100% isopropanol was added, tubes were mixed by inversion and the centrifugation step was repeated for 30 min. Liquid was removed from the tubes, each pellet was washed with 75% ethanol and the centrifugation was repeated for 10 min. The 75% ethanol was removed and pellets were air dried for 5 min or less. Pellets were resuspended in 450 µl nuclease-free water (Thermo Fisher), 50 µl DNase I buffer, 5 µl DNase I (NEB, cat# M0303) and incubated at 37 °C for 30 min. An equal volume of acid-phenol:chloroform, pH4.5 with IAA 125:24:1 (Thermo Fisher) was added, tubes were mixed by inversion, centrifuged and aqueous layer removed and precipitated similarly as above. After the 75% ethanol wash step, each pellet was dissolved in nuclease-free water and stored at −80 °C.
rRNA depletion was performed as described above. Bisulfite treatment of RNA recovered from rRNA depletion or 400–750 ng of total RNA was carried out using an EZ RNA Methylation Kit (Zymo Research Irvine, CA) with a 65 °C for 120 min incubation in place of the 54 °C for 45 min incubation in the protocol. Eluted RNA was either immediately used for construction of sequencing libraries or stored at −80 °C. Libraries for sequencing were prepared using the NEBNext® Ultra™ or Ultra™ II Directional RNA Library Prep Kit for Illumina® (NEB, cat# E7420 or E7760) using the “protocol for use with purified mRNA or rRNA-depleted RNA”. The recommended fragmentation conditions for partially degraded RNA were used in the fragmentation step. Bisulfite-treated libraries were sequenced on one of three Illumina platforms, Replicate 2 of strain TS559 and replicate 1 of all single deletion strains were sequenced on the Next-Seq, with the exception that both replicate 1 and 2 for single deletion strains ΔTK0360 and ΔTK1784 were all sequenced on the Nova-seq. Replicate 3 of strain TS559, replicate 2 for single deletion strains and all double deletion strains were sequenced on the Nova-seq.
Raw data processing
Fastq reads were subjected to adapter trimming and quality control using Trimmomatic v0.39 using the following command: PE <sampleID>_R1.fastq.gz strain_R2.fastq.gz ILLUMINACLIP:adapters.fa:5:15:10 SLIDINGWINDOW:1:20 MINLEN:20. Reads where any base was sequenced with a PHRED score <20 were removed from further analysis. Reads less than 15 nt were also removed. Using the custom python3 script, BSfilter.py, fastq reads with >3% cytosine retention (# cytosines/read length) were removed. Fastq files for each pair end were independently mapped to our custom reference genome (Thermococcus kodakarensis strain TS559) using BSseeker2 v2.1.8. Fastq reads for pair R1 were reverse complemented prior to mapping using the following command: python2/BSseeker2/Antisense.py -i strain_R1_C.fastq -o <sampleID>_R1_CA.fastq. Mapping was completed using the following command: python2 bs_seeker2-align.py -i <sampleID>_R1_CA.fastq -g TS559_reference_genome.fasta -temp_dir = /tmp -m 2 -XS = 0.03,3 -bt2-mm -bt-p 10 -aligner=bowtie2 -p /bowtie2/. For mapping, bowtie2 v2.2.14 was used to support BSseeker2. Two mismatches per alignment were allowed (excluding C-to-T mismatches). Bam files for each pair end were merged using Samtools v1.17. Using Samtools, alignments were removed from bam files where MAPQ score was <20 and when detected as a PCR duplicate according to the Samtools manual using the following series of commands: samtools view -h -b -q 20 <sampleID>_unsorted.bam | samtools sort -n -o <sampleID>_mapq_nsorted.bam; samtools fixmate -rm strain_mapq_nsorted.bam; <sampleID>_fixmate.bam; samtools sort <sampleID>_fixmate.bam><sampleID>_fixmate_csorted.bam; samtools markdup -r <sampleID>_fixmate_csorted.bam <sampleID>_rmdup.bam. The Samtools rmdup identifies PCR duplicates by identifying pairs of reads that align the 5’ end of mate 1 and the 3’ end of mate 2 to the same exact positions in the genome.
To reduce file size and retain as much data as possible prior to generating CGmaps, each bam file was pseudoreplicated (split) into 10 bam files using the custom python3 script, pseudoreplicate_paired_samfile.py and the following command: python3 pseudoreplicate_paired_alignment_file.py -r 10 -b -s <strainID>-i <sampleID>_rmdup.bam. For each strain, the seed (-s) corresponds to the 4 digit TK gene ID (i.e., TS559, 2304, etc). CGmaptools v0.1.2 was used to obtain CGmaps where C/T coverage and C coverage are calculated at each genomically encoded cytidine where total coverage > 0. The following command was used to generate CGmaps for each pseudoreplicate: CGmaptools convert bam2cgmap -b <sampleID>_rmdup_split10.bam -g TS559_genome.fasta -rmOverlap -o <sampleID>_rmdup_split10. Pseudoreplicate CGmaps were then merged into a single CGmap using the following command: cgmaptools mergelist tosingle -i <sampleID>_rmdup_split1.CGmap.gz, <sampleID>_rmdup_split2.CGmap.gz… <sampleID>_rmdup_split10.CGmap.gz -o <sampleID>.CGmap.gz.
R software
CGmaps were then analyzed using R v4.2.2 software. Jupyter notebooks are provided (see data availability statement) for analysis and figure generation (see Data Availability). The tidyverse v2.0.0 collection of packages include dplyr v1.1.0, readr v2.1.4, forcats v1.0.0, stringr v1.5.0, ggplot2 v3.4.1, tibble v3.1.8, lubridate v1.9.2, tidyr v1.3.0, and purrr v1.0.1.
Power calculations for coverage requirements
Statistical power was calculated using the R software and the pwr v1.3.0 library (one-sample t-test with effect size (d) = 0.5). Effects sizes were estimated with a significance level of 0.01. For comparison of m5C frequencies where a 2x fold change is observed between parent and deletion strains, a total coverage of 47x is needed to achieve an 80% power. We therefore required all candidate m5C sites to have a total coverage of greater than or equal to 47 reads.
Cytosine deamination ratio
The data analysis pipeline for detection of high-confidence and reproducible m5C sites was written using R software and the Tidyverse collection of packages. CGmaps provide coverage information at all genomically encoded cytidines and were the initial input into the pipeline. We first calculated total cytidine conversion across all sequenced cytosines. We queried the total T or C coverage at all cytidine positions in the reference genome and divided T coverage by C + T coverage (T/(C + T)) at all genomically encoded cytidines, which gave us conversion rates ≥ ~99.8% for all libraries (Fig. 2b). These metrics indicate that cytidines were adequately deaminated.
High-confidence m5C detection
Three metrics were implemented to call high-confidence m5C sites; total coverage, m5C coverage and m5C frequency. Based on a power calculation, a total coverage of 47X at individual sites of modification were necessary for adequate m5C frequency comparisons between strains. In addition to total coverage, m5C coverage (cytidine coverage) was parameterized based on m5C conversion rates at each genomically encoded cytosine and is therefore different for each library. Cytidine coverage at each reference cytosine was to calculate the 99% percentile for cytosine coverage (visually represented as a histogram in Fig. 2c), and as most sites have very few remaining cytidine due to bisulfite conversion to uridine, we required that for a true modification site, m5C coverage must be ≥ the 99% percentile. Across 6 parent strain libraries, the minimum m5C coverage was calculated to be between 5X and 18X, depending on the library. A minimum m5C frequency was also applied to capture high-confidence and biologically relevant modifications. Modification frequency was calculated by dividing cytidine coverage by cytidine + thymidine coverage (C/(C + T)). From modification sites that met both total and m5C coverage minima, m5C frequencies were visualized via histogram (Fig. 2d). Visual inspection indicates that modification frequencies level-off at ~10%, indicating many modifications with a frequency below 10% may be false-positives. We determined that a high-confidence m5C site must reach a 10% modification frequency. For sites with exceedingly high total coverage ≥1000X, we lowered the modification frequency minima to 5%. These three criteria define high-confidence m5C sites and were applied to all libraries.
Reproducible m5C detection
The reproducibility of m5C sites was also considered. Our main analysis includes sites that met our high-confidence thresholds in at least 2 replicates. Parallel analysis was performed for sites that were determined to be high-confidence in all 3 replicates.
Linear regression of m5C frequencies
We performed linear regression on modification frequencies between replicates where sites were high-confidence in at least 2 or 3 replicates (Supplementary Fig. 2a, b). Linear regression was performed using R software; the function geom_smooth(method = “lm”) was used to visualize modification frequencies across replicates. Pearson’s correlation coefficients were calculated in R using cor(x,y method = c(“person”)) where x and y are modification frequencies between replicates.
Unique transcript expression threshold
To determine the proportion of unique transcripts with and without modification as represented in Fig. 1a, we calculated the number of unique transcripts that met an average of 47x coverage across all nucleotides in each unique transcript (including genes and non-coding RNAs). As such, unique transcripts with an average coverage ≥47x at each nucleotide were considered to be expressed at or above our expression thresholds. The expression threshold was applied in ≥2 or 3 replicates for m5C sites reproducible in ≥2 replicates or 3 replicates (Supplementary Fig. 4a). Most unique transcripts had >47x average coverage—and most m5C sites exceeded 47x coverage (Supplementary Fig. 2e)—so they are deemed sufficiently expressed in these datasets to allow a 47x coverage requirement for identifying high-confidence m5C sites. We did not perform differential gene expression analysis as bisulfite treatment degrades RNA and results in sequencing datasets biased for longer RNAs, and therefore can not be quantitatively reliable.
m5C site annotation
The complete annotation of each m5C site was performed using the two custom Python scripts; annotation_pipeline.py and helpers.py. In addition, the reference genome for T. kodakarensis strain TS559 and its corresponding gtf annotation file as well as the wildtype KOD1 reference genome were queried. All sites where a m5C modification was detected were recorded in a text file, each separated by a new line. This line separated file was inputted into annotation_pipeline.py where the complete annotation for each site was generated. All additional analysis and figure generation were performed using R software.
Amino acid and codon bias
The transcriptome-wide occurrence of each codon sequence and amino acid identity were queried using a custom Python script, amino_acid_probability.py. Probability of each codon or amino acid were calculated from known or suspected open reading frames for strain TS559. These calculations are recorded in Supplementary Data 4.
m5C mRNA distribution
Modifications sites that mapped to mRNAs were assigned to a region: CDS, 5’ UTR52 or 3’ UTR (unpublished, or within 20 nt of the gene end when not indicated). To determine the enrichment of m5C sites in each region, the number of m5C sites in each region was divided by the average length of each region; 5’UTRs average 17 nt, 3’UTRs average 33 nt, and coding sequences average 1000 nt. When a m5C site maps within a coding sequence, the percent position within the transcript was determined by calculating distance from the transcription start site and divided by the length of the gene. The percent position in the transcript was graphed via histogram in R using geom_density().
Logos sequence analysis
A 41 nt region surrounding each m5C site was queried using the annotation pipeline described above. The RNA sequences were analyzed in R using geom_logo() and theme_logo() from the third party package, ggseqlogo v0.1. Sequence alignments as illustrated in Supplementary Fig. 9 were performed using the Clustal Omega Webserver hosted by EMBL-EBI.
Gene ontology enrichment analysis
Gene ontology (GO) analysis of m5C-containing mRNAs was performed using the PANTHER bioinformatics database82. GO terms with p value of less than 0.05 were considered as statistically significant. No enrichment was detected.
Gene expression analysis
RNA-seq data used to measure gene expression were queries from NCBI Sequence Read Archive (SRR9966705, SRR9966714, SRR9966715, SRR9966721). RNA was purified from Thermococcus kodakarensis grown under laboratory conditions similar to the present study, depleted of rRNA, and sequenced on the Illumina NextSeq 500 in a paired-end format2. Using Fastp (version 0.23.4), Fastq reads were trimmed for adapters, deduplicated, and filtered for reads with PHRED score >20. Quality control was confirmed using Fastqc (version 0.11.9). Paired reads were aligned to the reference genome using BWA (version 0.7.17-r1188) using the aln and sampe commands. Read pair coverage at each genomic element was calculated using FeatureCounts (version 2.0.0), and RPKMs were averaged over 4 replicates.
Phenotypic growth analysis
Individual cultures were passages at least twice before inoculated into 10 mL rich medium with agmatine and sulfur in triplicate. Cultures were grown at 65°, 75°, 85°, or 95 °C in a water or oil bath. The optical density at 600 nm (OD600) was measured for each culture every 60 (85°/95°) or 120 (65°/75°) minutes using UV-Vis spectroscopy. The OD600 and the standard error was plotted in R using geom_smooth(method = ‘loess’, SE = TRUE). The maximum OD600 of each of the three replicates was divided by that of the average maximum OD600 of the control TS559 strain to graph the relative proportion to the control. The doubling time of each replicate was calculated using the following equation: (Δtime * ln(2)) / ln(final OD600 / initial OD600). Generally, the initial OD600 was taken where the OD600 first doubled (~0.05) and the final OD600 was taken where the OD600 was that of half the maximum OD600 (~0.3–0.4). This ensures calculation of the quickest doubling time of the culture. The doubling time of each culture was averaged across 3 replicates, divided by that of the control TS559 strain, and graphed as a relative proportion to the control.
Recombinant protein expression and purification
The gene sequences for each RNA MTase were PCR amplified from the T. kodakarensis genome using primers with 17 bp terminal extensions homologous to the expression vector. The coding sequences along with an E. coli ribosome entry site were cloned into a pQE80 expression vectors at the EcoRI restriction site using Infusion cloning (Takara Bio., Cat # 638910). For genes where the start codon is GTG in T. kodakarensis, we exchanged the start codon for ATG for expression in E. coli. Gene sequences were C-terminally tagged with 6 histidines (6xHis) to aid in affinity purification. Transcriptional expression is controlled by the lac promoter, and the plasmid confers ampicillin resistance. Expression vectors were transformed into the Nico21 E. coli cell line (NEB, cat# C2529H). We additionally transformed the pRARE plasmid (confers chloramphenicol resistance) into the Nico21 cell line to overcome the codon bias between T. kodakarensis and E. coli. Cells were grown on ampicillin (100 μg/mL)- and chloramphenicol (25 μg/mL)-containing solid LB medium. A single colony was picked into liquid broth containing the necessary antibiotics for plasmid retention and grown overnight. Overnight cultures were used to inoculate larger cultures in a 1:100 ratio and grown at 37 °C with shaking. At an OD600 of 0.3–0.5, cultures were spiked with isopropyl β-d-1-thiogalactopyranoside (IPTG, 200–400 μM final concentration) to induce protein expression and D-sorbitol (2–3% w/v final concentration) to improve protein solubility. Cultures were shaken at 37 °C for an additional 3 h before they were harvested via centrifugation. Cell pellets were stored at −20 °C.
Cell pellets were thawed and resuspended in buffer A (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 10% glycerol) and sonicated on ice for 30 min to lyse the cells. Cellular debris were removed by centrifugation at 21,000 × g for 15 min. The supernatant was heated to 65 °C for 15 min to remove most of the E. coli proteins before centrifugation at 21,000 × g for 15 min. The 6xHis-tagged protein in the heat-treated supernatant was purified on an AKTA system using a HiTrap Chelating HP column (Cytiva Life Sciences, cat# 17040901). The column was charged with Ni2+ before passing through the head-treated cell lysate. After the protein bound to the column, the column was washed with buffer A until the UV spectra returned to base-line, indicating the removal of unbound E. coli proteins. The column was then washed in 20–40 mM imidazole to further remove non-specific proteins. The protein was eluted from the nickel at increasing concentrations of buffer B (25 mM Tris-HCl pH 8.0, 100 mM NaCl, 10% glycerol, 200 mM imidazole). Protein elution typically peaked at 165 mM imidazole. Elution fractions were analyzed by SDS-PAGE (BioRad, cat# 5678095) under denaturing conditions followed by Western blotting against the 6xHis tag (1°: Mouse anti-6x-His Tag Monoclonal antibody, Invitrogen, cat # MA1-21315; 2°: Goat anti-Mouse Phosphatase, KPL, cat # 05-18-18). Uncropped gel images are provided in Source Data. Fractions where the target protein was cleanly eluted were pooled and dialyzed into storage buffer (25 mM Tris-HCl pH 8.0, 100 mM NaCl, 50% glycerol v/v), and diluted to a 10 μM concentration in storage buffer.
Generation of RNA substrates for in vitro methyltransferase assays
RNA substrates used in methyltransferase activity assays (Supplementary Table 2) were selected where the RNA was modified in vivo, but the m5C was lost in a strain deleted for an R5CMT. All 23 nt RNA substrates were ordered from IDT and resuspended in nuclease-free water (Thermo Fisher Scientific, cat# AM9932). Full length TK1911 mRNA (TK1911 coding sequence without UTRs) was amplified from the T. kodakarensis genome by PCR with primers 5’-atttaggtgacactatagATGACTGACAAGAAGAAGC and 5’-TTATCCCTCACTGCCCCTAAG. The equivalent U-RNA (where the target cytidine is substituted for a uracil) was purchased from Twist Biosciences and PCR amplified using the same primers. The forward primer is tailed with the Sp6 Polymerase promoter sequence. The PCR reaction was run on a 1% TBE agarose gel and gel purified (Quiagen, cat# 28706×4). A total of 500 ng of DNA amplicon was used in an in vitro transcription reaction according to the manufacturer’s protocol for the HiScribe Sp6 RNA synthesis kit (NEB, cat# E2070S). Reactions were quenched with 2x volume of TRizol reagent (Thermo Fisher Scientific, cat# 15596026) and a 1:5 ratio of chloroform (Thermo Fisher Scientific, Inc). Reactions were mixed by pulse vortex and separated by centrifugation at 21,000 × g for 1 min. The aqueous layer was taken into 1 μL GlycoBlue (Thermo Fisher Scientific, cat# AM9515) and 3x the volume of 100% ethanol. RNA samples were incubated at −80 °C for 30 min then pelleted by centrifugation at 21,000 × g for 30 min. Reactions were aspirated completely, resuspended in nuclease-free water (Thermo Fisher Scientific, cat# AM9932) and quantified using the Qubit RNA broad range kit (Thermo Fisher Scientific, cat# Q10210). RNA samples were concentrated by alcohol precipitation or diluted to 10 μM in nuclease-free water.
Methyltransferase activity assay
To confirm the suspected methyltransferase activity of R5CMTs, we performed enzymatic reactions with the recombinantly expressed and purified enzyme and RNA substrates generated in vitro or purchased from IDT. Complete reactions were done in a 20 μL volume with 1 µM enzyme in storage buffer, 1 μM RNA, ~1 μM [3H-methyl]-SAM (PerkinElmer, cat# NET155V001MC) and 1x MTase buffer (25 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM DTT). Negative control reactions where 1 reaction component was excluded in place of water or the U-RNA was substituted in place of the C-substrate were performed in parallel. Reactions were incubated at 95 °C for 30 s to denature the RNA and then 65 °C for 15 min to facilitate enzymatic activity. All reactions were completed in biological triplicate.
Reaction temperatures were lowered to 4 °C before 15 μL of each reaction was spotted onto cut 1”x1” squares of Hybond-N+ paper (Cytiva, cat # RPN2020B), where the RNA strongly binds to the membrane. The squares were left to dry for 10 min before submerged in 200 mL 5% w/v Trichloroacetic acid (TCA, Thermo Fisher Scientific, cat# 421455000) for 5–10 min with rocking. The TCA was exchanged for fresh TCA 5 times to wash away reaction components while 3H-methylated RNA remained bound to the membrane. After 5 TCA washes, the squares were air dried for 20 min, then placed in a scintillation vial containing 5 mL of liquid EcoScint scintillation fluid (Fisher Scientific, cat#50-899-90184). The β-decay of each reaction was measured in counts per minute (CPM) using the Liquid Scintillation counter TRI-CARB 2900TR (Pickard). CPMs between reactions with C- and U-RNA were compared and p values were calculated using a two-sample unpaired t-test. Comparisons resulting in a p value < 0.05 earned one star (*) while p values < 0.01 earned two stars (**).
RNA structural analysis
RNA secondary structures and minimum free energies were computed using Vienna RNAfold 2.4.18 webserver or the Vienna RNAfold package v2.4.14. RNA fold predictions for in vitro RNAs were done at 65 °C, while in vivo RNA folds were predicted at 85 °C. For local RNA folds, a 41 nt region surrounding the target cytidine was queried from the full length fold. Nucleotides that are base paired are represented by a | symbol while single stranded nucleotides are represented by periods in Supplementary Data 3. The predicted secondary structures at m5C sites indicate that roughly 50% of m5C sites are base paired or single stranded, suggesting that the dataset is likely not contaminated with false positives due to denaturation resistance.
Methyltransferase assays on small and large RNA fractions
RNA was purified from T. kodakarensis cells growth to mid exponential growth phase. Cells were resuspended in TRIzol and 1/5 volume chloroform with a 5–10 min incubation at room temperature proceeding each reagent. Cellular debris were removed via centrifugation at 21,000 × g for 15 min. The RNA contained within the aqueous phase was purified using the RNA Clean and Concentrator kit (Zymo, cat# R1017), following the manufacturers protocol for tRNA depleted by selective recovery of RNA > 200 nt. Briefly, the aqueous phase was mixed with an equal volume of RNA-binding buffer and an equal volume of 100% ethanol before passed through a column, where the large RNA fraction binds. The flow through was mixed with 2 volumes of 100% ethanol and passed over a column where the small RNAs bind. The manufacturer’s protocol was followed for DNase I treatment and purification of each size fraction. Methyltransferase assays were performed as described above with 1 µg or RNA from either the small or large fraction.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank members of the NSF-funded archaeal epitranscriptomics consortium for helpful comments that improved the manuscript and figures. This work was supported by funding (to T.J.S.) from the USA National Science Foundation, award #2022065, and the USA National Institutes of Health, R35-GM143963. K.A.F. received financial support from a T32 training grant from the National Institutes of Health, GM132057 and a departmental GAANN fellowship. This study was also privately funded from New England Biolabs, Inc. Authors R.T.F., Y.T., N.D., E.J.W., I.R.C., and G.B.R. are employees of New England Biolabs, Inc. This affiliation does not affect the authors’ impartiality, objectivity of data generation or its interpretation, adherence to journal standards and policies, or availability of data.
Author contributions
K.A.F., V.T., H.P.F., B.W.B., and J.S. generated, genotyped, and phenotyped T. kodakarensis strains, inclusive of biomass collections at different growth phases and temperatures for RNA analyses. K.A.F. and L.E. cloned, expressed, and purified enzymes, generated RNAs substrates, and characterized bulk in vitro activities of RNA modification enzymes. R.T.F. was responsible for RNA purifications from T. kodakarensis strains, depletion of rRNA and size selection, bisulfite conversion reactions, library preparations, and Illumina sequencing. The bisulfite sequencing data pipeline and statistical analyses were conceived, implemented, and resolved primarily by K.A.F. with inputs from all authors. RNA modification levels in bulk and select samples, inclusive of HPLC and quantitative mass spectrometry, were determined by R.T.F., Y.T., N.D., E.J.W., G.B.R., and I.R.C. T.J.S., I.R.C., and G.B.R. supervised research. K.A.F. and T.J.S. drafted initial figures and text that were improved and approved by all authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The Bisulfite-sequencing fastq files generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA937301. Whole genome sequencing fastq files generated in this study have been deposited in the NCBI SRA under BioProject PRJNA1125032. RNA-seq Data presented in Fig. 1d and Supplementary Fig. 1d were queried from NCBI Sequence Read Archive under accession codes SRR9966705, SRR9966714, SRR9966715, and SRR9966721. The CryoEM structure of the T. kodakarensis ribosome was queried from the Protein Data Bank under accession code 6TH6. The original agarose gel electrophoresis image represented in Fig. 2a is provided in Source Data. Source data are provided with this paper.
Code availability
Custom R and Python scripts and associated files are provided on Github at https://github.com/kscott94/The_m5C-epitranscriptome_of_Thermococcus_kodakarensis, or on Zenodo at 10.5281/zenodo.11556635.
Competing interests
All authors have approved the final version of the manuscript. K.A.F., V.T., J.S., H.P.F., B.W.B., L.E., and T.J.S. do not have competing financial interests or conflicts of interest to report. R.T.F., Y.T., N.D., E.J.W., I.R.C., and G.B.R. are employed and funded by New England Biolabs, Inc., a manufacturer and vendor of molecular biology reagents, including nucleic acid modifying and synthesis enzymes. The authors state that this affiliation does not affect their impartiality, objectivity of data generation or interpretation, adherence to journal standards and policies, or availability of data.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51410-w.
References
- 1.Thomas, J. M., Batista, P. J. & Meier, J. L. Metabolic regulation of the epitranscriptome. ACS Chem. Biol.14, 316–324 (2019). 10.1021/acschembio.8b00951 [DOI] [PubMed] [Google Scholar]
- 2.Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature583, 638–643 (2020). 10.1038/s41586-020-2418-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birk, M. A. et al. Temperature-dependent RNA editing in octopus extensively recodes the neural proteome. Cell186, 2544–2555.e13 (2023). 10.1016/j.cell.2023.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, C. & Jia, G. Reversible RNA modification N1-methyladenosine (m1A) in mRNA and tRNA. Genom. Proteom. Bioinforma.16, 155–161 (2018). 10.1016/j.gpb.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohira, T. et al. Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance. Nature605, 372–379 (2022). 10.1038/s41586-022-04677-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikne, V. et al. A pseudouridylation switch in rRNA is implicated in ribosome function during the life cycle of Trypanosoma brucei. Sci. Rep.6, 25296 (2016). 10.1038/srep25296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He, Y. et al. Novel insights into the role of 5-Methylcytosine RNA methylation in human abdominal aortic aneurysm. Front. Biosci.26, 1147–1165 (2021). 10.52586/5016 [DOI] [PubMed] [Google Scholar]
- 8.Shen, Q. et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature554, 123–127 (2018). 10.1038/nature25434 [DOI] [PubMed] [Google Scholar]
- 9.Gokhale, N. S. et al. Altered m6A modification of specific cellular transcripts affects Flaviviridae infection. Mol. Cell77, 542–555.e8 (2020). 10.1016/j.molcel.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gokhale, N. S. et al. N6-Methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe20, 654–665 (2016). 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Șelaru, A., Costache, M. & Dinescu, S. Epitranscriptomic signatures in stem cell differentiation to the neuronal lineage. RNA Biol.18, 51–60 (2021). 10.1080/15476286.2021.1985348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heck, A. M. & Wilusz, C. J. Small changes, big implications: the impact of m6A RNA methylation on gene expression in pluripotency and development. Biochim. Biophys. Acta Gene Regul. Mech.1862, 194402 (2019). 10.1016/j.bbagrm.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, M. et al. The demethylase activity of FTO (fat mass and obesity associated protein) is required for preadipocyte differentiation. PLoS ONE10, e0133788 (2015). 10.1371/journal.pone.0133788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature518, 560–564 (2015). 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang, Y. et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell75, 1188–1202.e11 (2019). 10.1016/j.molcel.2019.06.033 [DOI] [PubMed] [Google Scholar]
- 16.Arango, D. et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell175, 1872–1886.e24 (2018). 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun.7, 12626 (2016). 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature505, 117–120 (2014). 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, Y., Hsu, P. J., Chen, Y.-S. & Yang, Y.-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res.28, 616–624 (2018). 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro, I. C. et al. Identification of putative reader proteins of 5-methylcytosine and its derivatives in Caenorhabditis elegans RNA. Wellcome Open Res. 7, 282 (2022). 10.12688/wellcomeopenres.17893.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, X. et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27, 606–625 (2017). 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, L. et al. m5C Methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol.29, 2465–2476.e5 (2019). 10.1016/j.cub.2019.06.042 [DOI] [PubMed] [Google Scholar]
- 23.Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell61, 507–519 (2016). 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 24.Vallecillo-Viejo, I. C. et al. Spatially regulated editing of genetic information within a neuron. Nucleic Acids Res.48, 3999–4012 (2020). 10.1093/nar/gkaa172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumann, U. et al. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol.18, 40 (2020). 10.1186/s12915-020-00769-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, A. et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res.27, 444–447 (2017). 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res.27, 315–328 (2017). 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boo, S. H. & Kim, Y. K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med.52, 400–408 (2020). 10.1038/s12276-020-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominissini, D. & Rechavi, G. 5-methylcytosine mediates nuclear export of mRNA. Cell Res.27, 717–719 (2017). 10.1038/cr.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohnsack, M. T. & Sloan, K. E. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci.75, 241–260 (2018). 10.1007/s00018-017-2598-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin, R. M. RNA editing implicated in chloroplast-to-nucleus communication. Proc. Natl Acad. Sci.116, 9701–9703 (2019). 10.1073/pnas.1905566116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brachova, P. et al. Inosine RNA modifications are enriched at the codon wobble position in mouse oocytes and eggs†. Biol. Reprod.101, 938–949 (2019). 10.1093/biolre/ioz130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, Q. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science351, 397–400 (2016). 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- 34.Kiani, J. et al. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet.9, e1003498 (2013). 10.1371/journal.pgen.1003498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson, V. R., Heaney, J. D., Tesar, P. J., Davidson, N. O. & Nadeau, J. H. Transgenerational epigenetic effects of the Apobec1 cytidine deaminase deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc. Natl Acad. Sci. USA109, E2766–E2773 (2012). 10.1073/pnas.1207169109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ontiveros, R. J., Stoute, J. & Liu, K. F. The chemical diversity of RNA modifications. Biochem. J.476, 1227–1245 (2019). 10.1042/BCJ20180445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Höfer, K. & Jäschke, A. Epitranscriptomics: RNA modifications in bacteria and archaea. Microbiol. Spectr.6, 10.1128/microbiolspec.RWR-0015-2017 (2018). [DOI] [PubMed]
- 38.Jonkhout, N. et al. The RNA modification landscape in human disease. RNA23, 1754–1769 (2017). 10.1261/rna.063503.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson, E., Cui, Y.-H. & He, Y.-Y. Roles of RNA modifications in diverse cellular functions. Front. Cell Dev. Biol.10, 828683 (2022). 10.3389/fcell.2022.828683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer, M., Kapoor, U. & Jantsch, M. F. Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol.7, 170077 (2017). 10.1098/rsob.170077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orita, I. et al. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance. Nucleic Acids Res.47, 1964–1976 (2019). 10.1093/nar/gky1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shigi, N. et al. Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem.281, 2104–2113 (2006). 10.1074/jbc.M510771200 [DOI] [PubMed] [Google Scholar]
- 43.Droogmans, L. et al. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res.31, 2148–2156 (2003). 10.1093/nar/gkg314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirata, A. et al. Distinct modified nucleosides in tRNATrp from the hyperthermophilic archaeon Thermococcus kodakarensis and requirement of tRNA m2G10/m22G10 methyltransferase (archaeal Trm11) for survival at high temperatures. J. Bacteriol. 201, 10.1128/jb.00448-19 (2019). [DOI] [PMC free article] [PubMed]
- 45.Tomikawa, C., Yokogawa, T., Kanai, T. & Hori, H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res.38, 942–957 (2010). 10.1093/nar/gkp1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowalak, J. A., Dalluge, J. J., McCloskey, J. A. & Stetter, K. O. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry33, 7869–7876 (1994). 10.1021/bi00191a014 [DOI] [PubMed] [Google Scholar]
- 47.Turner, B. et al. Archaeosine modification of archaeal tRNA: role in structural stabilization. J. Bacteriol.202, e00748–19 (2020). 10.1128/JB.00748-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennis, P. P., Tripp, V., Lui, L., Lowe, T. & Randau, L. C/D box sRNA-guided 2’-O-methylation patterns of archaeal rRNA molecules. BMC Genomics16, 632 (2015). 10.1186/s12864-015-1839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Legrand, C. et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res.27, 1589–1596 (2017). 10.1101/gr.210666.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenz, C., Lünse, C. E. & Mörl, M. tRNA modifications: impact on structure and thermal adaptation. Biomolecules7, 35 (2017). 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trixl, L. & Lusser, A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA10, e1510 (2019). 10.1002/wrna.1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jäger, D., Förstner, K. U., Sharma, C. M., Santangelo, T. J. & Reeve, J. N. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genomics15, 684 (2014). 10.1186/1471-2164-15-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amort, T. et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol.18, 1 (2017). 10.1186/s13059-016-1139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sibbritt, T., Patel, H. R. & Preiss, T. Mapping and significance of the mRNA methylome. Wiley Interdiscip. Rev. RNA4, 397–422 (2013). 10.1002/wrna.1166 [DOI] [PubMed] [Google Scholar]
- 55.Liu, M. et al. 5-methylcytosine modification by Plasmodium NSUN2 stabilizes mRNA and mediates the development of gametocytes. Proc. Natl Acad. Sci. USA119, e2110713119 (2022). [DOI] [PMC free article] [PubMed]
- 56.Gehring, A. M., Sanders, T. J. & Santangelo, T. J. Markerless gene editing in the hyperthermophilic archaeon Thermococcus kodakarensis. Bio Protoc.7, e2604 (2017). 10.21769/BioProtoc.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spaans, S. K., van der Oost, J. & Kengen, S. W. M. The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles19, 741–750 (2015). 10.1007/s00792-015-0750-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sergeeva, O. V., Bogdanov, A. A. & Sergiev, P. V. What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie117, 110–118 (2015). 10.1016/j.biochi.2014.11.019 [DOI] [PubMed] [Google Scholar]
- 59.Sharma, S. & Entian, K.-D. Chemical modifications of ribosomal RNA. Methods Mol. Biol.2533, 149–166 (2022). 10.1007/978-1-0716-2501-9_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Motorin, Y. & Grosjean, H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA5, 1105–1118 (1999). 10.1017/S1355838299982201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet.9, e1003602 (2013). 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song, Y. et al. Comprehensive analysis of key m5C modification-related genes in type 2 diabetes. Front. Genet.13, 1015879 (2022). 10.3389/fgene.2022.1015879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui, X. et al. 5-Methylcytosine RNA methylation in Arabidopsis Thaliana. Mol. Plant10, 1387–1399 (2017). 10.1016/j.molp.2017.09.013 [DOI] [PubMed] [Google Scholar]
- 64.Jian, H. et al. Alteration of mRNA 5-methylcytosine modification in neurons after OGD/R and potential roles in cell stress response and apoptosis. Front. Genet.12, 633681 (2021). 10.3389/fgene.2021.633681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wnuk, M., Slipek, P., Dziedzic, M. & Lewinska, A. The roles of host 5-methylcytosine RNA methyltransferases during viral infections. Int. J. Mol. Sci.21, 8176 (2020). 10.3390/ijms21218176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin, Y. et al. Overview of distinct 5-methylcytosine profiles of messenger RNA in normal and knock-down NSUN2 colorectal cancer cells. Front. Genet.14, 1121063 (2023). 10.3389/fgene.2023.1121063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He, Y., Zhang, Q., Zheng, Q., Yu, X. & Guo, W. Distinct 5-methylcytosine profiles of circular RNA in human hepatocellular carcinoma. Am. J. Transl. Res.12, 5719–5729 (2020). [PMC free article] [PubMed] [Google Scholar]
- 68.Bataglia, L., Simões, Z. L. P. & Nunes, F. M. F. Transcriptional expression of m6A and m5C RNA methyltransferase genes in the brain and fat body of honey bee adult workers. Front. Cell Dev. Biol.10, 921503 (2022). 10.3389/fcell.2022.921503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Navarro, I. C. et al. Translational adaptation to heat stress is mediated by RNA 5-methylcytosine in Caenorhabditis elegans. EMBO J.40, e105496 (2021). 10.15252/embj.2020105496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.David, R. et al. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell29, 445–460 (2017). 10.1105/tpc.16.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang, Y. et al. OsNSUN2-mediated 5-methylcytosine mRNA modification enhances rice adaptation to high temperature. Dev. Cell53, 272–286.e7 (2020). 10.1016/j.devcel.2020.03.009 [DOI] [PubMed] [Google Scholar]
- 72.Xue, S. et al. Depletion of TRDMT1 affects 5-methylcytosine modification of mRNA and inhibits HEK293 cell proliferation and migration. Biochem. Biophys. Res. Commun.520, 60–66 (2019). 10.1016/j.bbrc.2019.09.098 [DOI] [PubMed] [Google Scholar]
- 73.Wang, N., Tang, H., Wang, X., Wang, W. & Feng, J. Homocysteine upregulates interleukin-17A expression via NSun2-mediated RNA methylation in T lymphocytes. Biochem. Biophys. Res. Commun.493, 94–99 (2017). 10.1016/j.bbrc.2017.09.069 [DOI] [PubMed] [Google Scholar]
- 74.Bohnsack, K. E., Höbartner, C. & Bohnsack, M. T. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes10, 102 (2019). 10.3390/genes10020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature534, 335–340 (2016). 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen, X. et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol.21, 978–990 (2019). 10.1038/s41556-019-0361-y [DOI] [PubMed] [Google Scholar]
- 77.Huang, T., Chen, W., Liu, J., Gu, N. & Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol.26, 380–388 (2019). 10.1038/s41594-019-0218-x [DOI] [PubMed] [Google Scholar]
- 78.Scott, K. A., Williams, S. A. & Santangelo, T. J. Thermococcus kodakarensis provides a versatile hyperthermophilic archaeal platform for protein expression. Methods Enzymol.659, 243–273 (2021). 10.1016/bs.mie.2021.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hileman, T. H. & Santangelo, T. J. Genetics techniques for Thermococcus kodakarensis. Front. Microbiol.3, 195 (2012). 10.3389/fmicb.2012.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morlan, J. D., Qu, K. & Sinicropi, D. V. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS ONE7, e42882 (2012). 10.1371/journal.pone.0042882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wein, S. et al. A computational platform for high-throughput analysis of RNA sequences and modifications by mass spectrometry. Nat. Commun.11, 926 (2020). 10.1038/s41467-020-14665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mi, H. & Thomas, P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol.563, 123–140 (2009). 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gruber, A. R., Lorenz, R., Bernhart, S. H., Neuböck, R. & Hofacker, I. L. The Vienna RNA websuite. Nucleic Acids Res.36, W70–W74 (2008). 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The Bisulfite-sequencing fastq files generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA937301. Whole genome sequencing fastq files generated in this study have been deposited in the NCBI SRA under BioProject PRJNA1125032. RNA-seq Data presented in Fig. 1d and Supplementary Fig. 1d were queried from NCBI Sequence Read Archive under accession codes SRR9966705, SRR9966714, SRR9966715, and SRR9966721. The CryoEM structure of the T. kodakarensis ribosome was queried from the Protein Data Bank under accession code 6TH6. The original agarose gel electrophoresis image represented in Fig. 2a is provided in Source Data. Source data are provided with this paper.
Custom R and Python scripts and associated files are provided on Github at https://github.com/kscott94/The_m5C-epitranscriptome_of_Thermococcus_kodakarensis, or on Zenodo at 10.5281/zenodo.11556635.