Abstract
Due to the intricate relationship between the small non-coding ribonucleic acid (miRNA) sequences, the classification of miRNA species, namely Human, Gorilla, Rat, and Mouse is challenging. Previous methods are not robust and accurate. In this study, we present AtheroPoint’s GeneAI 3.0, a powerful, novel, and generalized method for extracting features from the fixed patterns of purines and pyrimidines in each miRNA sequence in ensemble paradigms in machine learning (EML) and convolutional neural network (CNN)-based deep learning (EDL) frameworks. GeneAI 3.0 utilized five conventional (Entropy, Dissimilarity, Energy, Homogeneity, and Contrast), and three contemporary (Shannon entropy, Hurst exponent, Fractal dimension) features, to generate a composite feature set from given miRNA sequences which were then passed into our ML and DL classification framework. A set of 11 new classifiers was designed consisting of 5 EML and 6 EDL for binary/multiclass classification. It was benchmarked against 9 solo ML (SML), 6 solo DL (SDL), 12 hybrid DL (HDL) models, resulting in a total of 11 + 27 = 38 models were designed. Four hypotheses were formulated and validated using explainable AI (XAI) as well as reliability/statistical tests. The order of the mean performance using accuracy (ACC)/area-under-the-curve (AUC) of the 24 DL classifiers was: EDL > HDL > SDL. The mean performance of EDL models with CNN layers was superior to that without CNN layers by 0.73%/0.92%. Mean performance of EML models was superior to SML models with improvements of ACC/AUC by 6.24%/6.46%. EDL models performed significantly better than EML models, with a mean increase in ACC/AUC of 7.09%/6.96%. The GeneAI 3.0 tool produced expected XAI feature plots, and the statistical tests showed significant p-values. Ensemble models with composite features are highly effective and generalized models for effectively classifying miRNA sequences.
Keywords: miRNA classification, Ensemble deep learning, Ensemble machine learning, Composite feature extraction, Validation, Statistical tests
Subject terms: Computer science, Software, Statistics, Genetics, Genomics
Introduction
MicroRNAs (miRNAs) are short RNA molecules that play a crucial role in regulating gene expression1,2. Typically consisting of 20–25 nucleotides, they are formed through the transcription of longer RNA molecules by cellular enzymes. By binding to target messenger RNA (mRNA), miRNAs can inhibit mRNA’s translation, thereby controlling the expression of specific genes. This mechanism influences various biological processes such as proliferation3, apoptosis4, development5,6, and differentiation7. Disruptions in miRNA expression have been associated with diseases like cancer8–10 and cardiovascular disease11–13. Accurately classifying miRNA sequences based on their origin14–16 is crucial due to the diverse roles that miRNA sequences play in disease development across different species17–19. This classification enables the identification of conserved miRNA sequences and their target genes, contributing to a better understanding of miRNA function and the detection of potential threats20–22.
Machine learning’s application has been constantly observed in multiple bioinformatics studies23–33, including several tools have gained attention in the field of miRNA identification. These tools include Mipred25, Triplet34, HeteroMirPred35, micropred36, PlantMiRNAPred37, and mirnaDetect38. They have the ability to extract pre-miRNAs from protein-coding regions that exhibit stem-loop structures similar to genuine pre-miRNAs but have not been identified as such. In addition, numerous computational methods have been developed to enhance miRNA identification. These methods include MatureByes39, MiRMat40, MiRRim241, MiRdup42, MaturePred43, MiRPara44, mirExplorer45, Matpred46, and MiRduplexSVM47. MiRNA identification can be performed using de novo methods, which involve computational tools, or by utilizing next-generation sequencing data48,49. These methods focus on identifying pre-miRNA sequences that exhibit hairpin-like structures in the input data. They are categorized based on expression-based features or computed sequences.
The intricate nonlinear nature of miRNA sequences poses challenges for these methods, primary due to the high-dimensional feature spaces associated with the sequences50. To address these challenges, primitive methods like ensemble ML (EML) methods that employ voting mechanisms51–53 have been introduced. This was follwed by deep learning (DL) models, such as Convolutional Neural Networks (CNN) and Long Short-Term Memory (LSTM)54,55. DL models have the capability to capture the nonlinear complexity of miRNA sequences, making them well-suited for characterization and classification tasks54–57. Despite the promising results achieved by solo DL (SDL) models in miRNA classification, they often require large labeled datasets and are susceptible to overfitting58, which hinders their generalization capabilities59. To further improve classification performance, hybrid DL (HDL) and ensemble DL (EDL) models have been proposed60,61. These models leverage the strengths of multiple DL architectures62–64.
Extracting additional features from miRNA sequences is a valuable strategy for overcoming the aforementioned limitations. Although features like k-mer frequency and dinucleotide composition effectively capture sequence-specific details65–67, they have inherent limitations in extracting comprehensive information. To address these challenges, conventional features such as Energy, Contrast, and Entropy can be employed to capture structural characteristics68–71. Additionally, contemporary features like Shannon Entropy and Hurst Exponent can be derived to obtain additional insights. By combining both sequence-specific and structural features into a composite feature set, the effectiveness of DL models can be further enhanced, resulting in a more robust approach. Further, incorporation of CNN layers in this paradigm enhances classification by capturing local patterns and spatial dependencies. Hence usage of CNN-based EDL models with extracted composite features is paramount in building a robust and state-of-the-art framework for miRNA classification.
In the spirit of improving species classification by employing EDL and EML classifiers, along with novel composite feature extraction we built an extensive set of ensemble-based AI classifiers, focusing on four main hypotheses. First, we investigate the benefits of using EML models with voting compared to SML models for miRNA species classification in binary classification (BC) and multiclass classification (MCC) scenarios. Second, we validate the superiority of EDL models over HDL and SDL models. Additionally, we explore the advantages of incorporating CNN layers in miRNA species classification, comparing them to models without CNN layers. Lastly, we examine the advantage of transitioning from EDL models to EML models in ensemble-based species classification. By introducing composite features and enhancing ensemble learning, our approach brings a fresh perspective to design and improves the reliability of genomic sequence testing. Consequently, it enhances the accuracy of miRNA sequence classification, surpassing previous research that relied solely on statistical techniques.
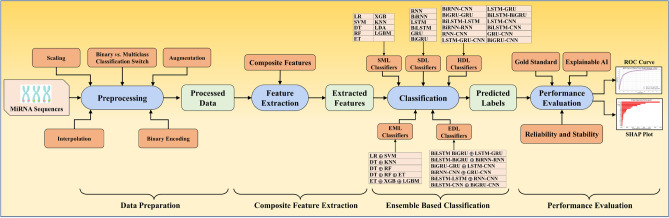
Figure 1 presents an overall block diagram of GeneAI 3.0 (AtheroPoint LLC, Roseville, CA, USA). With the input of miRNA species data containing gene sequences, the system performs an intensive data preparation (elliptical preprocessing block), which includes binary encoding of the gene sequence, scaling, augmentation using Adaptive Synthetic Sampling Approach for Imbalanced Learning (ADASYN)72, and interpolation. It then performs an elaborated feature extraction (elliptical feature extraction block), where it derives composite features from the binary miRNA sequence. GenAI 3.0 then incorporates 38 extensive AI models (classification block): nine SML, five EML, six SDL, twelve HDL and six EDL models, and classifies the species along with performance metrics (performance block), consisting of statistical tests and explainable AI (XAI) graphs.
Figure 1.
Global architecture of GenAI 3.0 (AtheroPoint LLC, CA, USA). SML: Solo machine learning; EML: Ensemble machine learning; SDL: Solo deep learning; HDL: Hybrid deep learning; EDL: Ensemble deep learning.
Our research findings validated the advantages of utilizing EDL models in gene classification by conducting experiments that establish the model order as EDL > HDL > SDL. We have also validated the benefits of EML models over SML models in the miRNA classification. Furthermore, we have assessed the performance improvements achieved using EDL models compared to EML models, as well as the advantages gained from incorporating CNN layers in DL models. Alongside our primary contributions, we have investigated the impact of training data size on the model's performance and validated the reliability and stability of our approach through statistical tests. Finally, we have employed XAI plots to interpret our classification findings and offer insights into species classification.
The paper starts elaboration on the methodology which discusses the extracted features, employed classifiers and optimization parameters along with experimental protocols in “Methodology” section. The results are presented in “Results” section, while “Performance evaluation” section provides a performance evaluation with Receiver Operating Characteristic (ROC) curves, and influence of training data size. “Reliability analysis using statistical tests” section demonstrates reliability using statistical tests, while “Explainable artificial intelligence” section uses XAI plots used to enhance the interpretability. “Discussion” section presents a discussion of the principal findings, a benchmarking with previous studies, and an overview of the study's strengths, weaknesses, and extensions. Finally, “Conclusion” section concludes the paper.
Methodology
In order to explore the connection between miRNA sequences and their corresponding species, we employed statistical ML and DL models for classification in our methodology. The initial stage involved collecting the primary dataset that would serve as the foundation for classification, ensuring its suitability for utilization in the classifiers. Next, we conducted quality control procedures, including categorical encoding of the miRNA sequences, data scaling, oversampling of the minority class, interpolation of missing sequences, and label encoding of the class labels. We also computed both conventional and contemporary features from the dataset. Subsequently, we meticulously designed the architecture of all the AI models used, along with the hyperparameter tuning approaches, loss functions, and training details employed to train these models. Lastly, we defined the performance metrics and experimental protocols utilized in our study.
Data and data preparation
This study utilized the miRNA Database available at http://www.mirbase.org/ for experimental design, data collection, and discussion purposes. The database encompasses miRNA sequences from various species, including Humans, Gorillas, Mouse, and Rat. Th dataset used in this study consisted of 2654 Human, 369 Gorilla, 1978 Mouse, and 764 Rat miRNA sequences. A ribonucleic acid (RNA) molecule is composed of a backbone comprising sugar ribose and phosphate groups. In contrast to deoxyribonucleic acid (DNA), the sugar ribose lacks deoxyribose and is connected to one of four bases: adenine (A), uracil (U), cytosine (C), or guanine (G). To convert miRNA sequences containing these four bases into binary sequences, we applied a set of rules that mapped each base to a corresponding binary digit73,74:
| 1 |
This was done using truncation of sequence75–77. This resulted in four datasets of binary sequences from the four species: Humans, Gorillas, Mouse, and Rat. Table 1 lists the specifications of each dataset.
Table 1.
Specifications of the miRNA dataset.
| Dataset name | Dataset size |
|---|---|
| Human | 2654 |
| Gorilla | 369 |
| Mouse | 1978 |
| Rat | 764 |
| Combined classes | 5765 |
The class labels for the four species (Human, Gorilla, Mouse, and Rat) were encoded between 0 and 3 to be used as target classes in the classifiers. This label encoding was employed to convert the category labels for each species into numerical values, allowing for the application of DL techniques to analyze the relationships between miRNA sequences and the different species.
To facilitate this analysis, six binary class datasets and four multiclass datasets were prepared. These datasets were carefully curated and preprocessed to cover various scenarios among the four species. In the binary class datasets, two species were compared using a binary classification approach. The objective was to accurately differentiate between the two species using the provided dataset features. We created multiple datasets with the aim of achieving generalization78–81 in species classification. The purpose behind this initiative was to train our model on a variety of datasets, ensuring its effectiveness in real-life scenarios. This approach allows any gene sequence to be pre-processed, features extracted, and utilized by our model. The binary datasets consisted of the following pairwise species comparisons: Human vs. Gorilla, Human vs. Rat, Human vs. Mouse, Mouse vs. Gorilla, Mouse vs. Rat, and Gorilla vs. Rat. For the multiclass datasets, the methodology used was "one vs. all." Each species was considered as one class, while the other three species were treated as the second class. The four multiclass datasets created were: Human vs. All, Rat vs. All, Gorilla vs. All, and Mouse vs. All. By utilizing these processed datasets, researchers could leverage DL techniques to gain insights into the relationships between miRNA sequences and different species.
Data availability/availability of data and materials
Due to its propriety nature, supporting data cannot be made available openly but are available from the corresponding author on reasonable request.
Quality Control
There is an unbalanced distribution of data points among the various classes in the dataset we acquired for our study. Data size in particular plays a vital role both in generalization vs. memorization protocols. When data size is low, we have seen studies where two types of data augmentation have been adopted74,79,82–87. If it is an image data, the data augmentation consisted of increasing the data size by flipping and rotating the images82–87. On the other hand, if the data is a point or tabular data, then the augmentation can be accomplished using SMOTE74 or ADASYN protocols88. To address this issue, we utilized the ADASYN technique, as depicted in Fig. 1. ADASYN is a method that generates synthetic samples for the minority class, thereby achieving a more balanced distribution of data points among the different classes. This approach is beneficial because imbalanced data can hinder the performance of supervised ML algorithms, which often prioritize the majority class and may exhibit poor performance on the minority classes. By employing ADASYN and balancing the representation of the classes in the dataset, we can enhance the performance of various classifiers. Some examples of classifiers that can benefit from this balanced data include Gradient Descent Boosting89, Support Vector Machine (SVM)77, and Logistic Regression (LR)88.
We also employed linear interpolation to handle missing values within the "Human" class. Linear interpolation is a method that estimates the missing values by assuming a linear relationship between the available data points. By applying linear interpolation to the four instances with missing values, we successfully completed the dataset and ensured the integrity of the data for further analysis.
Additionally, to improve the performance of our algorithms on the imbalanced dataset, we implemented data scaling techniques73,74 to standardize the features and ensure their similarity in scale. This enabled faster convergence of the algorithms and enhanced the accuracy of predictions. We specifically employed the Min–Max Scaler method, which rescales the data to a fixed range between 0 and 1. This is achieved by subtracting the minimum value and dividing by the range73,74. By utilizing this method, we standardized the features and reduced their values, which expedited the training process for both ML and DL models.
Feature representation and composite features extraction
The miRNA sequence St consists of four nucleotide bases: A, C, U, and G, which can be arranged in different combinations. The presence of these nucleotides in the miRNA sequence signifies their interdependencies, and through the analysis of their patterns, distinct characteristics can be identified to distinguish between various species. In order to differentiate species based on feature representations of miRNA sequences, we developed an innovative approach to uncover these nucleotide co-occurrences. To demonstrate the possible arrangements of these nucleotides in miRNA gene sequences, we utilized co-occurrence matrices generated through vector combinations, as depicted in the provided Table 2.
Table 2.
Possible sets of occurrences of nucleobases A, C, U, and G in an RNA sequence formed by the combination of vectors, where I, J, K, L, M, N, O, and P are the co-occurrence matrices.
| X | Y | XT*Y |
|---|---|---|
| X1 = (A, C, U, G) | (A, C, U, G) | I4x4 = (X1 T)4x1 x (Y)1x4 |
| X2 = (AA, CC, UU, GG) | (A, C, U, G) | J4x4 = (X2 T)4x1 x (Y)1x4 |
| X3 = (AC, AU, AG, CU, CG, UG) | (A, C, U, G) | K6x4 = (X3 T)6x1 x (Y)1x4 |
| X4 = (CA, UA, GA, UC, GC, GU) | (A, C, U, G) | L6x4 = (X4 T)6x1 x (Y)1x4 |
| X5 = (ACU, ACG, AUG, CUG) | (A, C, U, G) | M4x4 = (X5 T)4x1 x (Y)1x4 |
| X6 = (CAU, CAG, UAG, UCG) | (A, C, U, G) | N4x4 = (X6 T)4x1 x (Y)1x4 |
| X7 = (AUC, AGC, AGU, CGU) | (A, C, U, G) | O4x4 = (X7 T)4x1 x (Y)1x4 |
| X8 = (UCA, GCA, GUA, GUC) | (A, C, U, G) | P4x4 = (X8 T)4x1 x (Y)1x4 |
In order to gain insights into the inherent patterns of miRNA, it is essential to investigate the co-occurrences of nucleobases and analyze both their stationary and non-stationary patterns. To extract valuable information from these patterns, we employed the widely utilized grey-level co-occurrence matrix90, a technique commonly employed in texture analysis and pattern recognition91. We have adopted the same feature extraction namely entropy, contrast, energy, homogeneity, dissimilarity as previously published by our group92–96 Such algorithms are being used for tissue characterization in medical imaging97,98. For each miRNA sequence, we computed multiple co-occurrence matrices, namely I, J, K, L, M, N, O, and P. These matrices captured diverse patterns formed by the nucleobases A, C, U, and G. In Tables (ST1–ST8), we present these co-occurrence matrices, which offer an overview of the different nucleobase arrangements and their corresponding frequencies.
The primary objective of constructing co-occurrence matrices from the miRNA sequence St is to analyze the occurrence frequency of specific combinations and offsets of the nucleobases A, C, U, and G. The co-occurrence matrix has a size of × 4 for a given offset, where represents the number of distinct nucleobase combinations found in sequence St. Each element in the co-occurrence matrices presented in Tables (ST1–ST8), denoted as the (, )th position, indicates the frequency of the th and th nucleobases occurring in the sequence St, which has a length of . This relationship can be mathematically expressed using the following equation:
| 2 |
The computation of matrix is contingent upon the spatial relationship defined by the offset (, ). These co-occurrence matrices are utilized to analyze the frequency of various combinations of the nucleobases A, C, U, and G in the sequence St. In order to extract distinctive and discriminative features, the matrices are subjected to normalization, resulting in the transformed matrices .
| 3 |
Subsequently, the normalized co-occurrence matrix is utilized to compute several properties, which include Entropy, Contrast, Energy, Homogeneity, and Dissimilarity95,99–101. The mathematical equations for these properties can be found in Table 3. These properties serve as quantitative measures to characterize different aspects of the co-occurrence patterns captured in the matrix . Afterwards, the features outlined in Table 3 are computed for each co-occurrence matrix Tables (ST1–ST8), and the corresponding feature vectors are presented in Table 4. Consequently, these feature vectors are utilized to construct the final feature set representation, denoted as fSet, for an RNA sequence of a miRNA sequence St:
Table 3.
Features extracted from a co-occurrence matrix of miRNA sequence St. .
| Feature | Mathematical formula |
|---|---|
| Energy | |
| Entropy | |
| Homogeneity | |
| Contrast | |
| Dissimilarity |
Table 4.
Extracted Feature vectors from the cooccurrence matrices.
| Feature vector | Co-occurrence matrix |
|---|---|
| fI = (f1, f2, f3, f4, f5) | I (Table ST1) |
| fJ = (f6, f7, f8, f9, f10) | J (Table ST2) |
| fK = (f11, f12, f13, f14, f15) | K (Table ST3) |
| fL = (f16, f17, f18, f19, f20) | L (Table ST4) |
| fM = (f21, f22, f23, f24, f25) | M (Table ST5) |
| fN = (f26, f27, f28, f29, f30) | N (Table ST6) |
| fO = (f31, f32, f33, f34, f35) | O (Table ST7) |
| fP = (f36, f37, f38, f39, f40) | P (Table ST8) |
Shannon entropy
Shannon Entropy (SE) is a valuable metric for quantifying the information content or uncertainty within a given sequence. It assesses the entropy of information in a Bernoulli process where two possibilities (0/1) occur with a probability of 102–105. The SE signifies the degree of uncertainty present in a binary string and can be computed using the following formula:
| 4 |
where represents the probability of a binary sequence having two distinct values. When = 0, indicating that the event is impossible, there is no ambiguity, and the SE is 0. Likewise, when = 1, indicating a certain outcome, the SE is also 0. In the case where = 1/2106, the level of uncertainty is at its highest, resulting in an SE value of 1.
Hurst exponent
Hurst Exponent (HE) is a measure that characterizes the autocorrelation properties of a time series107 and finds applications in applied mathematics. It takes values between 0 and 1, where values in the range of [0, 0.5] indicate negative autocorrelation in the time series108–110. Positive autocorrelation, on the other hand, is indicated by values in the range of [0.5, 1]. A HE value of 0.5 suggests that the variable is uncorrelated with its previous values, indicating a random series. HE score increases with the strength of the correlation between successive values. The following equation is used to calculate the HE of a binary sequence D of length , where represents the ith element of the binary sequence D.
| 5 |
where
| 6 |
| 7 |
| 8 |
| 9 |
Fractal dimension
The Fractal Dimension (FD) of miRNA sequences is a widely used feature for analyzing their structural complexity. The first step in calculating the FD involves transforming each miRNA sequence into indicator matrices111,112. The four nucleotides {A, U, C, G}c are represented by the symbol , and represents a miRNA sequence of length composed of four symbols chosen from . The indicator function for each miRNA sequence is defined by the following equation:
| 10 |
Here the indicator matrix will be:
| 11 |
To convert the miRNA sequence into a binary representation, a 2D dot-plot image is generated using the matrix, which consists of values 0 and 1. This binary image visually represents the distribution of zeros and ones in the sequence, where white dots represent 0 and black dots represent 1. The FD can be computed from an indicator matrix by averaging the sigma values of 1 randomly selected from an × indicator matrix112–114. The following equation is used to calculate the FD based on the sigma value:
| 12 |
Machine learning and deep learning classifiers
In this comprehensive data analysis, we developed a total of fourteen ML models, including nine SML models and five EML models. Additionally, we constructed 24 DL models, consisting of six SDL models, twelve HDL models, and six EDL models.
Machine learning classifiers
For simplicity and availability, we selected the following ML models: LR115,116, Linear SVM117–119, Decision Tree (DT)120, RF121–123, Extra Trees (ET)124,125, Extreme Gradient Boost (XGBoost)88,126, K-Nearest Neighbors (KNN)127,128, Linear Discriminant Analysis (LDA)129,130, Light Gradient Boosting Machine (LGBM)131,132, and Naive Bayes (NB)133. We specifically chose six nonlinear models (DT, RF, ET, XGBoost, KNN, LGBM) as they are suitable for nonlinear classification tasks, which is crucial for effectively classifying binary-encoded miRNA species. Each model possesses unique strengths and weaknesses, and by evaluating multiple models, we can compare their performances and select the most effective one. Furthermore, we created five EML models: (i) LR and SVM, (ii) DT and KNN, (iii) DT and RF, (iv) RF, DT, and ET, and (v) ET, XGBoost, and LGBM. These models were constructed using a voting-based ensemble classifier approach.
Solo deep learning classifiers
Among the DL models, we developed six SDL models: GRU (Gated Recurrent Unit), Bidirectional GRU (BiGRU), RNN (Recurrent neural network), Bidirectional RNN (BiRNN), LSTM, and Bidirectional LSTM (BiLSTM). These models were specifically designed to capture the temporal dependencies and intricate patterns present in the miRNA sequences, further enhancing the classification performance. We conducted rigorous evaluation and testing to assess the performance and effectiveness of each SDL model, for the selection of the most suitable architecture for miRNA species classification.
Hybrid deep learning classifiers
While these SDL models have shown limited success in miRNA classification, combining them into HDL models has proven to be beneficial in overcoming data scarcity and improving performance82,84,134,135. HDL models can effectively address domain-specific challenges and enhance accuracy in tasks such as miRNA classification by leveraging multiple architectural components. Considering these advantages, we constructed twelve HDL models: (i) LSTM-GRU, (ii) BiLSTM-BiGRU, (iii) LSTM-CNN, (iv) BiLSTM-CNN, (v) GRU-CNN, (vi) BiGRU-CNN, (vii) BiRNN-CNN, (viii) BiGRU-GRU, (ix) BiLSTM-LSTM, (x) BiRNN-RNN, (xi) RNN-CNN, and (xii) LSTM-GRU-CNN.
Ensemble deep learning classifiers
Furthermore, we created six EDL models: (i) BiLSTM-BiGRU and LSTM-GRU, (ii) BiLSTM-BiGRU and BiRNN-RNN, (iii) BiGRU-GRre U and LSTM-CNN, (iv) BiRNN-CNN and GRU-CNN, (v) BiLSTM-LSTM and RNN-CNN, and (vi) BiLSTM-CNN and BiGRU-CNN by concatenating their output vectors. By combining these multiple vectors, we can leverage the strengths and advantages of each individual model. The EDL models are depicted in Figures F1, F2, F3, F4, F5, and F6 in the supplementary material. All constituent models are utilized without their output layers and are truncated until the dropout layers. These model components are then concatenated using a concatenate layer and further employed as input to a dense layer network. Finally, the network is connected to a softmax layer for predicting the species.
Hypertuning parameters and optimization
During the study, the models were trained using a batch size of 64. The loss function chosen for training was categorical cross-entropy, which is commonly used for multi-class classification tasks. This loss function quantifies the dissimilarity between the predicted and actual probability distributions136,137.
The objective is to minimize the discrepancy between these distributions, leading to a reliable system that generates predicted probabilities that closely align with the true distribution. Categorical cross-entropy ensures that the differences between all probabilities are minimized. The mathematical equation for categorical cross-entropy is provided below:
| 13 |
where N represents the total number of miRNA sequences, TC denotes the number of species categories, and indicates that the h observation belongs to the cth category. Table ST9 in the supplementary material provides details on the number of epochs, initial learning rates, and optimizers utilized for each EDL model. The implementation of the study was carried out using Python 3.8 and the TensorFlow framework. The system execution occurred on a machine that featured a 12 GB NVIDIA P100 16 Graphics Processing Unit (GPU), an Intel Xeon Processors processor, and 12 GB of RAM.
Performance metrics
The proposed models were assessed for both binary and multiclass classification tasks, with the multiclass approach utilizing the "one vs. all" strategy138,139 for each species. To evaluate the models, several parameters were considered: true positive (TP), true negative (TN), false positive (FP), and false negative (FN). A sample belonging to a specific species is considered a TP if it is correctly classified as such. Likewise, a sample not belonging to the species is labeled as a TN if it is correctly classified as not belonging. However, if a sample not belonging to the species is incorrectly classified as belonging, sample not belonging to the species is incorrectly classified as belonging, it is a FP, and if a sample belonging to the species is incorrectly classified as not belonging, it is a FN. These parameters allow the derivation of various performance evaluation (PE) metrics, including: (i) Accuracy (η): Indicates the proportion of correct overall predictions out of the total predictions made. (ii) Recall (R): Represents the ratio of correctly predicted positive class instances to all positive members in the dataset. (iii) Precision (P): Measures the ratio of correctly predicted positive class instances to the total number of classified positive predictions. (iv) F1-score (F): The F1-score is the harmonic mean of precision and recall, serving as a valuable metric for evaluating model performance, especially on imbalanced datasets. (v) Area-under-the-curve (): It quantifies the two-dimensional area beneath the plotted ROC curve and is commonly used to assess model performance in both binary and multiclass classification problems.
In this study, we introduce formulations to measure the overall robustness of the model. To achieve this, six quantities are measured in this section, including , which represents the accuracy of model m summarized over all D datasets, , which indicates the accuracy of dataset d achieved by summarizing M models, , which represents the overall system accuracy achieved by averaging accuracy over M models and D datasets, , which summarizes the AUC of model m over all D datasets, , which indicates the robustness of dataset d achieved by summarizing the AUC over M models, and , which represents the overall system robustness achieved by averaging the AUC over M models and D datasets. These formulations are measured in each section for a combination of ML, SDL, HDL, and EDL models, as well as a combination of six binary and four multiclass datasets and their combinations. All these formulas were computed using the default K10 partition protocol.
| 14 |
| 15 |
| 16 |
| 17 |
| 18 |
| 19 |
| 20 |
| 21 |
| 22 |
| 23 |
Experimental protocols
To verify our hypothesis, we trained nine SML, six EML, six SDL, twelve HDL, and six EDL models, totalling 38 AI models, using a composite feature set. The feature set consisted of conventional features, including Entropy, Dissimilarity, Energy, Homogeneity, and Contrast, as well as contemporary features, such as Shannon entropy, Hurst exponent, and Fractal dimension. To test the resilience of the features on the AI models, we created various subsets of data with ten different datasets (six binary class and four multiclass).
Experiment 1: EDL Models vs. HDL Models vs. SDL Models
The main objective of this study is to examine and compare the effectiveness of SDL, HDL, and EDL models in classifying species using miRNA sequences. To achieve this, we trained and evaluated the performance of 24 AI models: six SDL, twelve HDL, and six EDL. The models were trained and tested using six binary and four multiclass balanced composite feature datasets. To evaluate the performance of these 24 AI models, their predictions were averaged across all ten datasets (6 binary class and 4 multiclass), and a comprehensive comparison was performed. To ensure the reliability of the results, the experiment utilized the K10 Cross-Validation protocols.
Experiment 2: EDL Models with CNN layers vs. without CNN layers
This study focuses on examining and comparing the impact of employing CNN layers into EDL models for species classification using miRNA sequences. The training and evaluation were conducted on twelve AI models, comprising four CNN-Based HDL models and two Non-CNN-Based HDL models. The models were trained and tested using six binary and four multiclass balanced composite feature datasets. To evaluate the performance of these 6 AI models, their predictions were averaged across all ten datasets (6 binary class and 4 multiclass), and a comprehensive comparison was performed. To ensure the reliability of the results, the experiment utilized the K10 Cross-Validation protocols.
Experiment 3: EML Models vs. SML Models
The primary aim of this study is to assess and contrast the efficacy of EML models versus SML models in the classification of species using miRNA sequences. The training and evaluation process involved 14 AI models, including nine SML models and five EML models. The models were trained and tested using six binary and four multiclass balanced composite feature datasets. To evaluate the performance of these 14 AI models, their predictions were averaged across all ten datasets (6 binary class and 4 multiclass), and a comprehensive comparison was performed. To ensure the reliability of the results, the experiment utilized the K10 Cross-Validation protocols.
Experiment 4: EDL Models vs. EML Models
The final objective of this study is to evaluate and compare the advantages offered by EDL models over EML models in stratifying species using miRNA sequences. A total of eleven AI models were trained and evaluated, including five EML models and six EDL models. The models were trained and tested using six binary and four multiclass balanced composite feature datasets. To evaluate the performance of these AI models, their predictions were averaged across all ten datasets (6 binary class and 4 multiclass), and a comprehensive comparison was performed. To ensure the reliability of the results, the experiment utilized the K10 Cross-Validation protocols.
Results
The protocols were employed to conduct tests on miRNA data from ten datasets, comprising of six binary class datasets and four multiclass datasets. The binary datasets included Human vs. Gorilla, Human vs. Rat, Human vs. Mouse, Mouse vs. Gorilla, Mouse vs. Rat, and Gorilla vs. Rat datasets. Additionally, there were four multiclass datasets, namely Human vs. All, Rat vs. All, Gorilla vs. All, and Mouse vs. All. To analyze the data, a total of fourteen ML models and eighteen DL models were utilized. The ML models consisted of nine SML models and five EML models. The DL models consisted of six SDL models, twelve HDL models and six EDL models The training process involved using the TensorFlow and Sklearn frameworks, and a Tesla P100 GPU on the K10 partition protocol was utilized for executing the training process. Experimental results were obtained based on these procedures.
EDL models vs. HDL models vs. SDL models
In this experiment, we conducted a comparison of six SDL classifiers, twelve HDL models and six EDL models. The performance evaluation involved calculating the average mean accuracy (ACC) and area-under-the-curve (AUC) for all the models across ten datasets, consisting of six binary class datasets and four multiclass datasets. The binary datasets comprised Human vs. Gorilla, Human vs. Rat, Human vs. Mouse, Mouse vs. Gorilla, Mouse vs. Rat, and Gorilla vs. Rat, while the multiclass datasets included Human vs. All, Rat vs. All, Gorilla vs. All, and Mouse vs. All. The results of the experiment are presented in Tables ST10, ST11, ST12, and ST13 given in the supplementary material.
Table ST10 shows that the SDL4 classifier (BiLSTM) achieved the best performance among all SDL models, with an ACC/AUC of 90.06%/0.9112. In Tables ST11 and ST12, the HDL2 classifier (BiLSTM-BiGRU) performed the best among all HDL models, with an ACC/AUC of 92.53%/0.9306. Furthermore, in Table ST13, the EDL6 classifier (BiLSTM-CNN ⊕ BiGRU-CNN) achieved the highest performance among all HDL/EDL models, with an ACC/AUC of 93.38%/0.9407. Table 5 presents the mean comparison, indicating that EDL/HDL classifiers outperformed SDL classifiers on all datasets. The mean accuracy and AUC differences between HDL and SDL across all datasets were 2.17% and 2.4%, respectively. The mean accuracy and AUC differences between EDL and HDL across all datasets were 2.01% and 1.52%, respectively. Additionally, the mean accuracy and AUC differences between EDL and SDL across all datasets were 4.18% and 3.92%, respectively.
Table 5.
Comparison of SDL vs. HDL vs. EDL models.
| Comparison for six binary classifiers and four multiclass classifiers of SDL, HDL and EDL Models | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | SDL | HDL | EDL | Absolute difference (%) | ||||||||
| (%) | [0–1] | (%) | [0–1] | (%) | [0–1] | |||||||
| Binary Class (BC) Classification | ||||||||||||
| Human vs. Gorilla | 89.31 | 0.8998 | 92.19 | 0.9336 | 95.29 | 0.9575 | 2.88 | 3.38 | 3.1 | 2.39 | 5.98 | 5.77 |
| Human vs. Mouse | 81.21 | 0.8203 | 82.99 | 0.8384 | 85.77 | 0.8636 | 1.78 | 1.81 | 2.78 | 2.52 | 4.56 | 4.33 |
| Human vs. Rat | 83.19 | 0.8397 | 85.57 | 0.8766 | 87.92 | 0.8851 | 2.38 | 3.69 | 2.35 | 0.85 | 4.73 | 4.54 |
| Mouse vs. Gorilla | 94.11 | 0.9518 | 96.68 | 0.9704 | 97.28 | 0.9737 | 2.57 | 1.86 | 0.6 | 0.33 | 3.17 | 2.19 |
| Mouse vs. Rat | 91.23 | 0.9212 | 94.3 | 0.9575 | 95.66 | 0.9735 | 3.07 | 3.63 | 1.36 | 1.6 | 4.43 | 5.23 |
| Rat vs. Gorilla | 92.31 | 0.9295 | 95.38 | 0.9611 | 96.03 | 0.9696 | 3.07 | 3.16 | 0.65 | 0.85 | 3.72 | 4.01 |
| Mean of 6 BC | 88.56 | 0.8937 | 91.19 | 0.923 | 92.99 | 0.9372 | 2.63 | 2.93 | 1.8 | 1.42 | 4.43 | 4.35 |
| Multiclass (MC) Classification | ||||||||||||
| Human vs. All | 85.1 | 0.8545 | 85.88 | 0.8738 | 88.65 | 0.8934 | 0.78 | 1.93 | 2.77 | 1.96 | 3.55 | 3.89 |
| Gorilla vs. All | 93.53 | 0.9421 | 95.95 | 0.9702 | 97.45 | 0.9765 | 2.42 | 2.81 | 1.5 | 0.63 | 3.92 | 3.44 |
| Rat vs. All | 89.43 | 0.9104 | 90.54 | 0.9174 | 93.06 | 0.939 | 1.11 | 0.7 | 2.52 | 2.16 | 3.63 | 2.86 |
| Mouse vs. All | 87.68 | 0.8947 | 89.3 | 0.9046 | 91.74 | 0.924 | 1.62 | 0.99 | 2.44 | 1.94 | 4.06 | 2.93 |
| Mean of 4 MCC | 88.94 | 0.9004 | 90.42 | 0.9165 | 92.73 | 0.9332 | 1.48 | 1.61 | 2.31 | 1.67 | 3.79 | 3.28 |
| Binary class + Multiclass Classification | ||||||||||||
| Mean of 10 Classifiers | 88.71 | 0.8964 | 90.88 | 0.9204 | 92.89 | 0.9356 | 2.17 | 2.4 | 2.01 | 1.52 | 4.18 | 3.92 |
(%) represents accuracy and (0-1) represents AUC. : Mean accuracy of SDL models; : Mean AUC of SDL models; : Mean accuracy of HDL models; : Mean AUC of HDL models; : Mean accuracy of EDL models; : Mean AUC of EDL models;: Mean absolute accuracy difference (HDL vs. SDL); ; : Mean absolute AUC difference (HDL vs. SDL); ; : Mean absolute accuracy difference (EDL vs. HDL); : Mean absolute AUC difference (EDL vs. HDL); ; : Mean absolute accuracy difference (EDL vs. SDL); ; : Mean absolute AUC difference (EDL vs. SDL); .
Significant values are in [bold].
These results validate our hypothesis that HDL classifiers perform better due to the complex nature of miRNA. HDL models can capture intricate nonlinear relationships between input features and output labels by recursively splitting the data into smaller subsets, enabling accurate predictions. Furthermore, combining multiple models in EDL/HDL classifiers allows them to leverage the strengths of different models, leading to improved performance. The ability to customize and adjust these models based on specific problem domains further enhances their effectiveness.
EDL models with CNN layers vs. EDL models without CNN layers
In this experiment, we conducted a comparison to assess the impact of adding CNN layers in the architecture of EDL models. Specifically, we evaluated the performance of four CNN-based EDL classifiers (EDL3, EDL4, EDL5, and EDL6) and two non-CNN-based EDL classifiers (EDL1 and EDL2) on ten datasets, comprising of six binary class datasets and four multiclass datasets. The evaluation metrics of average mean accuracy and AUC were calculated and reported in Table 6. The results of our experiment demonstrated that incorporating CNN layers in the EDL models significantly enhanced their classification performance. By utilizing feature extraction techniques, the models exhibited improved accuracy and AUC scores. The mean absolute difference in accuracy and AUC across all datasets, resulting from the feature extraction process using contemporary features, was found to be 0.73% and 0.92%, respectively. These findings validated our hypothesis that incorporating CNN layers in DL models can enhance their effectiveness in classifying miRNA sequences. This improvement stems from the ability of CNN layers to capture both temporal and spatial dependencies within the data, enabling the models to learn hierarchical representations. The combination of temporal and spatial information allows for more comprehensive and accurate classification of miRNA sequences.
Table 6.
Comparison of EDL models with CNN vs. without CNN layers.
| Comparison for six binary classifiers and four multiclass classifiers of DL Models with and without CNN Layers | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dataset | woCNN | wCNN | Absolute difference (%) | wCNN > woCNN | ||||
| (%) | [0–1] | (%) | [0–1] | Acccuracy (%) | AUC (%) | |||
| Binary Class (BC) Classification | ||||||||
| Human vs. Gorilla | 95.21 | 0.9528 | 95.33 | 0.9598 | 0.12 | 0.7 | 0.12% Increase | 0.7% Increase |
| Human vs. Mouse | 83.89 | 0.8435 | 86.7 | 0.8737 | 2.81 | 3.02 | 2.81% Increase | 3.02% Increase |
| Human vs. Rat | 86.89 | 0.8727 | 88.44 | 0.8914 | 1.55 | 1.87 | 1.55% Increase | 1.87% Increase |
| Mouse vs. Gorilla | 96.44 | 0.9712 | 97.7 | 0.9749 | 1.26 | 0.37 | 1.26% Increase | 0.37% Increase |
| Mouse vs. Rat | 95.58 | 0.9674 | 95.7 | 0.9765 | 0.12 | 0.91 | 0.12% Increase | 0.91% Increase |
| Rat vs. Gorilla | 95.85 | 0.9693 | 96.12 | 0.9697 | 0.27 | 0.04 | 0.27% Increase | 0.04% Increase |
| Mean of 6 BC | 92.31 | 0.9295 | 93.33 | 0.941 | 1.02 | 1.15 | 1.02% Increase | 1.15% Increase |
| Multiclass (MC) Classification | ||||||||
| Human vs. All | 88.44 | 0.8904 | 88.75 | 0.895 | 0.31 | 0.46 | 0.31% Increase | 0.46% Increase |
| Gorilla vs. All | 97.18 | 0.975 | 97.58 | 0.9772 | 0.4 | 0.22 | 0.4% Increase | 0.22% Increase |
| Rat vs. All | 92.9 | 0.932 | 93.14 | 0.9426 | 0.24 | 1.06 | 0.24% Increase | 1.06% Increase |
| Mouse vs. All | 91.61 | 0.9207 | 91.81 | 0.9256 | 0.2 | 0.49 | 0.2% Increase | 0.49% Increase |
| Mean of 4 MCC | 92.53 | 0.9295 | 92.82 | 0.9351 | 0.29 | 0.56 | 0.29% Increase | 0.56% Increase |
| Binary class + Multiclass Classification | ||||||||
| Mean of 10 Classifiers | 92.4 | 0.9295 | 93.13 | 0.9387 | 0.73 | 0.92 | 0.73% Increase | 0.92% Increase |
(%) represents accuracy and (0-1) represents AUC. : Mean accuracy of EDL models without CNN layers; : Mean AUC of EDL models without CNN layers; : Mean accuracy of EDL models with CNN layers; : Mean AUC of EDL models with CNN layers; : Mean absolute accuracy difference (with vs. without CNN layers); ; : Mean absolute AUC difference (with vs. without CNN layers); .
Significant values are in [bold].
EML models vs. SML models
In this experiment, we conducted a comparison of nine SML classifiers and five EML models. The performance evaluation involved calculating the average mean accuracy and AUC for all the models across ten datasets, consisting of six binary class datasets and four multiclass datasets. The binary datasets comprised Human vs. Gorilla, Human vs. Rat, Human vs. Mouse, Mouse vs. Gorilla, Mouse vs. Rat, and Gorilla vs. Rat, while the multiclass datasets included Human vs. All, Rat vs. All, Gorilla vs. All, and Mouse vs. All. The results obtained from the experiment are presented in Tables ST14 and ST15 in the supplementary material. Table ST14 displays the performance results of the SML models, where the ET classifier achieved the highest performance with an ACC/AUC of 90.33%/0.9049. It was followed by RF with an ACC/AUC of 89.31%/0.8922 and LGBM with an ACC/AUC of 88.06%/0.8896. In Table ST15, the EML4 classifier (DT ⊕ RF ⊕ ET) demonstrated the best performance among all the EML models, achieving an ACC/AUC of 91.14%/0.9171.
Table 7 presents the mean comparison, indicating that the EML classifiers outperformed the SML classifiers on all datasets. The average accuracy and AUC differences between EML and SML across all datasets were 6.24% and 6.46%, respectively. These findings validate our hypothesis that EML models perform better due to the complex nature of miRNA, as they can capture intricate nonlinear relationships by recursively partitioning the data into smaller subsets, enabling accurate predictions. The use of a voting classifier in EML models allows them to combine the strengths of different models, leading to improved performance.
Table 7.
Comparison of SML vs. EML models.
| Comparison for six binary classifiers and four multiclass classifiers of SML and EML Models | ||||||
|---|---|---|---|---|---|---|
| Dataset | SML | EML | Absolute difference (%) | |||
| (%) | [0–1] | (%) | [0–1] | |||
| Binary Class (BC) Classification | ||||||
| Human vs. Gorilla | 79.82 | 0.8044 | 88.32 | 0.8893 | 8.5 | 8.49 |
| Human vs. Mouse | 67.82 | 0.6975 | 74.69 | 0.7431 | 6.87 | 4.56 |
| Human vs. Rat | 76.13 | 0.7792 | 83.69 | 0.846 | 7.56 | 6.68 |
| Mouse vs. Gorilla | 84.23 | 0.8603 | 90.84 | 0.9179 | 6.61 | 5.76 |
| Mouse vs. Rat | 81.6 | 0.826 | 86.97 | 0.8792 | 5.37 | 5.32 |
| Rat vs. Gorilla | 85.18 | 0.8552 | 92.03 | 0.9273 | 6.85 | 7.21 |
| Mean of 6 BC | 79.13 | 0.8037 | 86.09 | 0.8672 | 6.96 | 6.35 |
| Multiclass (MC) Classification | ||||||
| Human vs. All | 75.73 | 0.7514 | 79.12 | 0.8094 | 3.39 | 5.8 |
| Gorilla vs. All | 84.91 | 0.8464 | 91.39 | 0.9096 | 6.48 | 6.32 |
| Rat vs. All | 81.24 | 0.8113 | 86.94 | 0.8829 | 5.7 | 7.16 |
| Mouse vs. All | 78.95 | 0.7825 | 84.04 | 0.8546 | 5.09 | 7.21 |
| Mean of 4 MCC | 80.21 | 0.7979 | 85.37 | 0.8641 | 5.16 | 6.62 |
| Binary class + Multiclass Classification | ||||||
| Mean of 10 Classifiers | 79.56 | 0.8014 | 85.8 | 0.866 | 6.24 | 6.46 |
(%) represents accuracy and (0-1) represents AUC. : Mean accuracy of SML models; : Mean AUC of SML models; : Mean accuracy of EML models; : Mean AUC of EML models; : Mean absolute accuracy difference (EML vs. SML); ; : Mean absolute AUC difference (EML vs. SML); .
Significant values are in [bold].
EDL models vs. EML models
In this experiment, we conducted a comparison of five EML classifiers and six EDL models. The performance evaluation involved calculating the average mean accuracy and AUC for all the models across ten datasets, consisting of six binary class datasets and four multiclass datasets. The binary datasets comprised Human vs. Gorilla, Human vs. Rat, Human vs. Mouse, Mouse vs. Gorilla, Mouse vs. Rat, and Gorilla vs. Rat, while the multiclass datasets included Human vs. All, Rat vs. All, Gorilla vs. All, and Mouse vs. All. Table 8 presents the mean comparison, indicating that the EDL classifiers outperformed the EML classifiers on all datasets. The average accuracy and AUC differences between EDL and EML across all datasets were 7.09% and 6.96%, respectively.
Table 8.
Comparison of EML vs. EDL models.
| Comparison for six binary classifier.s and four multiclass classifiers of EML and EDL Models | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dataset | EML | EDL | Absolute difference (%) | EDL > EML | ||||
| (%) | [0–1] | (%) | [0–1] | Acccuracy (%) | AUC (%) | |||
| Binary Class (BC) Classification | ||||||||
| Human vs. Gorilla | 88.32 | 0.8893 | 95.29 | 0.9575 | 6.97 | 6.82 | 6.97% Increase | 6.82% Increase |
| Human vs. Mouse | 74.69 | 0.7431 | 85.77 | 0.8636 | 11.08 | 12.05 | 11.08% Increase | 12.05% Increase |
| Human vs. Rat | 83.69 | 0.846 | 87.92 | 0.8851 | 4.23 | 3.91 | 4.23% Increase | 3.91% Increase |
| Mouse vs. Gorilla | 90.84 | 0.9179 | 97.28 | 0.9737 | 6.44 | 5.58 | 6.44% Increase | 5.58% Increase |
| Mouse vs. Rat | 86.97 | 0.8792 | 95.66 | 0.9735 | 8.69 | 9.43 | 8.69% Increase | 9.43% Increase |
| Rat vs. Gorilla | 92.03 | 0.9273 | 96.03 | 0.9696 | 4 | 4.23 | 4% Increase | 4.23% Increase |
| Mean of 6 BC | 86.09 | 0.8672 | 92.99 | 0.9372 | 6.9 | 7 | 6.9% Increase | 7% Increase |
| Multiclass (MC) Classification | ||||||||
| Human vs. All | 79.12 | 0.8094 | 88.65 | 0.8934 | 9.53 | 8.4 | 9.53% Increase | 8.4% Increase |
| Gorilla vs. All | 91.39 | 0.9096 | 97.45 | 0.9765 | 6.06 | 6.69 | 6.06% Increase | 6.69% Increase |
| Rat vs. All | 86.94 | 0.8829 | 93.06 | 0.939 | 6.12 | 5.61 | 6.12% Increase | 5.61% Increase |
| Mouse vs. All | 84.04 | 0.8546 | 91.74 | 0.924 | 7.7 | 6.94 | 7.7% Increase | 6.94% Increase |
| Mean of 4 MCC | 85.37 | 0.8641 | 92.73 | 0.9332 | 7.36 | 6.91 | 7.36% Increase | 6.91% Increase |
| Binary class + Multiclass Classification | ||||||||
| Mean of 10 Classifiers | 85.8 | 0.866 | 92.89 | 0.9356 | 7.09 | 6.96 | 7.09% Increase | 6.96% Increase |
(%) represents accuracy and (0-1) represents AUC. : Mean accuracy of EML models; : Mean AUC of EML models; : Mean accuracy of EDL models; : Mean AUC of EDL models; : Mean absolute accuracy difference (EML vs. EDL); ; : Mean absolute AUC difference (EML vs. EDL); .
Significant values are in [bold].
These findings validate our hypothesis that EDL models outperform EML models due to their ability to capture complex patterns and relationships in the data through multiple layers of non-linear transformations. This can be attributed to their complex architecture, which allows them to automatically learn hierarchical representations of miRNA data, capturing both local and global patterns.
Performance evaluation
The evaluation process encompassed a comprehensive analysis of the models' performance, employing various visualization techniques such as ROC curves and bar charts to visualize the performance of the models. To ensure the system's stability, its robustness and model stability are evaluated through observing effect of training data size on classifiers. This allowed us to provides insight into the reliability and stability of the models and identify areas for improvement.
Receiver operating curves, mean accuracy curves, and mean AUC for classifier models
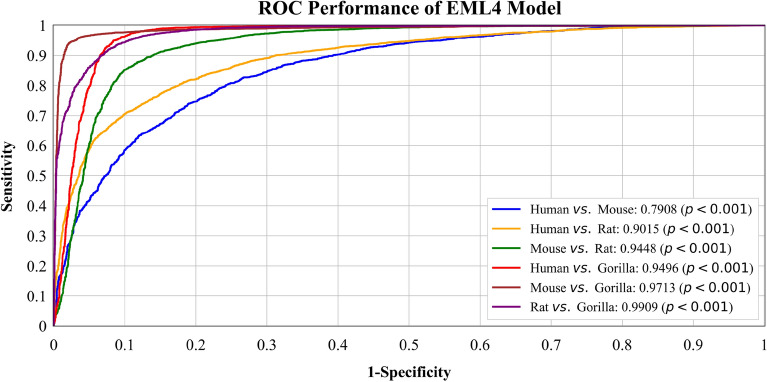
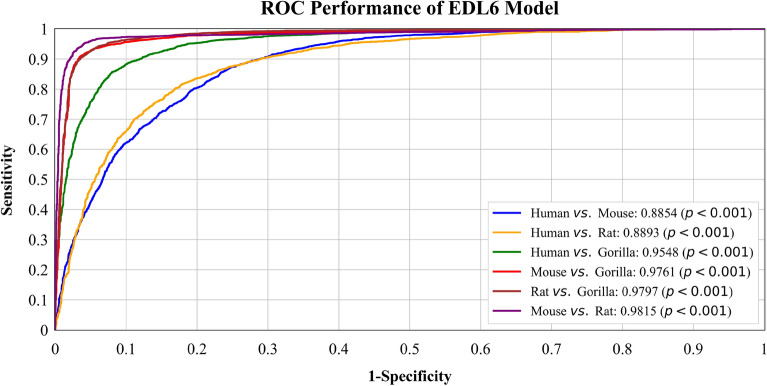
We plotted ROC curves of two best models, with all their classifiers, on all six binary datasets: Human vs. Gorilla, Human vs. Rat, Human vs. Mouse, Mouse vs. Gorilla, Mouse vs. Rat, and Gorilla vs. Rat. The performance of the models across their complete operating range was thoroughly evaluated, as shown in Fig. 1. In Fig. 2, the ROC curve for the EML4 Model is presented. The AUC score for Rat vs. Gorilla is the highest at 0.9909, followed by Mouse vs. Gorilla with an AUC score of 0.9713. This is followed by Human vs. Gorilla with an AUC of 0.9496 and Mouse vs. Rat with an AUC of 0.9448. The AUC score for Human vs. Rat is 0.9015, and Human vs. Mouse has the lowest AUC score of 0.7908. Figure 3 displays the ROC curve for the best-performing EDL (EDL6) Model. Among the comparisons, Mouse vs. Rat has the highest AUC score of 0.9815, followed by Rat vs. Gorilla with an AUC score of 0.9797. The AUC score for Mouse vs. Gorilla is 0.9761, and Human vs. Gorilla has an AUC of 0.9548. The AUC score for Human vs. Rat is 0.8893, and Human vs. Mouse has the lowest AUC score of 0.8854.
Figure 2.
ROC curves for EML (EML4) model using K10 protocol.
Figure 3.
ROC curves for EDL (EDL6) model using K10 protocol.
Furthermore, to establish the statistical significance of our results, p-values were computed for all species in each dataset. Our findings indicate that the p-values were less than 0.01, signifying a high confidence level in the observed differences between the species.
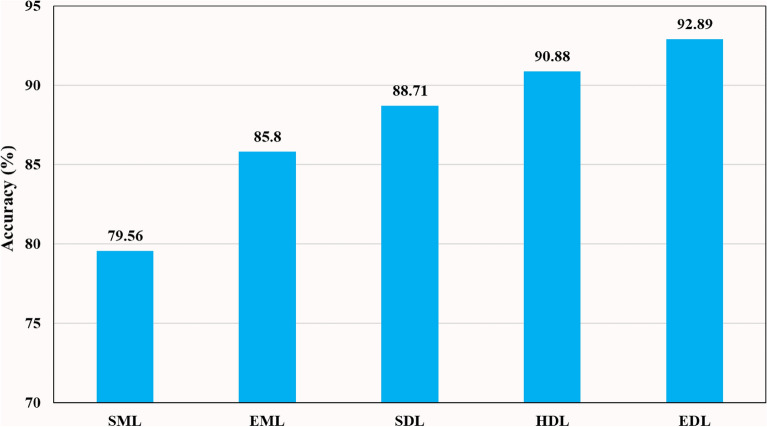
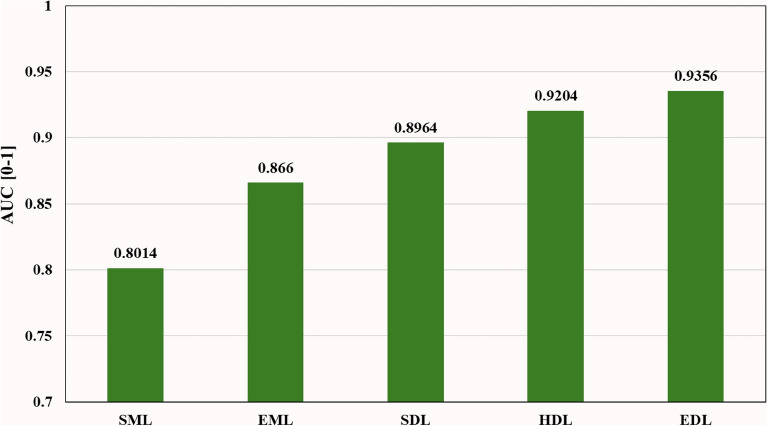
Bar charts are effective visual tools for presenting table data. Figure 4 illustrates the accuracy of nine SML, five EML, six SDL, twelve HDL, and six EDL models averaged across multiple binary and multiclass datasets. The mean accuracy increased progressively from 79.56% (SML) to 85.8% (EML), 88.71% (SDL), 90.88% (HDL), and 92.83% (EDL) models. Additionally, Fig. 5 depicts the AUC of the same models, showing a similar progressive increase in mean accuracy from 0.8014 (SML) to 0.866 (EML), 0.8964 (SDL), 0.9204 (HDL), and 0.933 (EDL) models when averaged across multiple binary and multiclass datasets.
Figure 4.
Comparison of Accuracy of SML vs. EML vs. SDL vs. HDL vs. EDL models using K10 protocol.
Figure 5.
Comparison of AUC of SML vs. EML vs. SDL vs. HDL vs. EDL models using K10 protocol.
Effect of training data size on classifier performance: varying partitional protocols
In this experimental study, we investigated the influence of varying training data sizes on the performance of DL models. Performance metrics were evaluated using different Cross-Validation protocols, namely K10 (default), K5, K4, and K2. Our analysis, presented in Table 9, revealed a gradual decline in performance metrics across these protocols. The evaluation included 24 DL classifiers, consisting of 6 SDL, 12 HDL, and 6 EDL models, applied to ten datasets encompassing both binary class and multiclass datasets. The average mean accuracy and AUC were computed, indicating a decrease in mean accuracy from 90.82% (K10) to 85.96% (K2), corresponding to a 4.86% reduction. Similarly, the AUC decreased from 0.9175 (K10) to 0.8634 (K2), indicating a 5.41% decline. Despite the reduced amount of training data in the K2 (50:50) validation protocol, our DL models demonstrated reliable performance metrics. This finding emphasizes the effectiveness of our approach, particularly the benefits gained from using ensemble models along with feature extraction. Hence, our models exhibit strong performance even in scenarios with limited training data, demonstrating their ability to maintain consistent performance under such conditions.
Table 9.
Mean performance of 24 DL models on different Cross-Validation protocols.
| Performance Metrics | K2 | Cross-Validation Results | Absolute Difference (%) | ||||
|---|---|---|---|---|---|---|---|
| K4 | K5 | K10 | D1 | D2 | D3 | ||
| Accuracy (%) | 85.96 | 87.2 | 88.54 | 90.82 | 2.28 | 3.62 | 4.86 |
| AUC [0–1] | 0.8634 | 0.8863 | 0.9019 | 0.9175 | 1.56 | 3.12 | 5.41 |
| Recall (%) | 86.06 | 87.73 | 88.98 | 91.39 | 2.41 | 3.66 | 5.33 |
| Precision (%) | 85.18 | 86.59 | 87.67 | 90.01 | 2.34 | 3.42 | 4.83 |
| F1 Score (%) | 85.62 | 87.16 | 88.32 | 90.69 | 2.37 | 3.53 | 5.07 |
D1: Absolute difference in performance metrics between K10 and K5 Cross-Validation protocols. D1 = K10 -K5.
D2: Absolute difference in performance metrics between K10 and K4 Cross-Validation protocols. D2 = K10 -K4.
D3: Absolute difference in performance metrics between K10 and K2 Cross-Validation protocols. D3 = K10 -K2.
Significant values are in [bold].
Reliability analysis using statistical tests
The stability of the system was thoroughly assessed and validated using three statistical tests conducted on the EDL models across all ten testing sets. There are several published studies which uses statistical tests for establishing the reliability and stability of the AI system80,81,140,141. These tests are conducted on the employed models, and the specific tests we carried out are all showcased in the manuscript, namely Adjusted R2, Z (Two-Tailed), and ANOVA tests. The purpose of these tests was to determine the significance of the predicted data and monitor the p-value in the ANOVA test, ensuring it was less than 0.01 (p < 0.01). Detailed results of these tests, conducted following the methodology outlined in96,142–145, are presented in Table ST16 in the supplementary material. The outcomes revealed that all six EDL models (EDL1, EDL2, EDL3, EDL4, EDL5, and EDL6) exhibited statistical significance with p < 0.01 in the ANOVA test, indicating strong outcomes and highlighting the models' reliability, stability, and clinical importance. The adjusted R-squared test evaluated the accuracy of the models by measuring the extent of feature variance, while the Z-score in the two-tailed tests indicated the deviation of the score from the mean population in terms of standard deviation. Therefore, these statistically validated findings reinforce the significance of our results and provide strong support for the reliability of the EDL models in this study.
Explainable artificial intelligence
To gain further insights into the decision-making process of the ML algorithms, we employed XAI techniques, specifically utilizing the SHapley Additive exPlanations (SHAP) method146–150. By leveraging SHAP, we were able to delve into the impact of different features on the classification outcomes, enhancing our understanding of species-specific information and the distinctive effects of individual features on each species. This invaluable information contributes to a deeper comprehension and differentiation among the various species.
Using the SHAP explainer151, we developed an interpretable AI classifier as discussed in Fig. 1 that provided insights into the significance of different features for each species. The SHAP-generated graphs presented in Figs. 6, 7, 8 and 9 revealed that the "Fractal" feature played a crucial role in classifying all species except for the Mouse. In the case of the Mouse species, the most important feature was "f3," followed by "Hurst" and "f9." For the other three species, "Fractal" was the primary feature, accompanied by "Shannon" for Humans and "f4" for Gorilla and Rat. The importance of the remaining features, derived from the co-occurrence matrix as detailed in Table 4, gradually decreased. These findings emphasize the significance of feature selection when constructing accurate and dependable classifiers, particularly in biology and ecology152.
Figure 6.
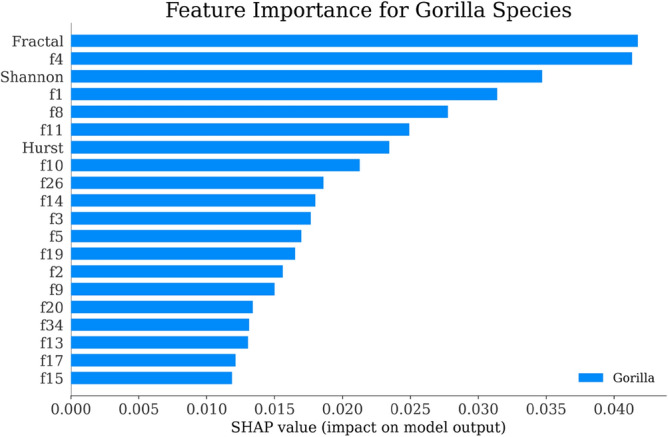
Feature Importance of Gorilla Species by SHAP explainer for EDL (EDL6) model using K10 protocol.
Figure 7.
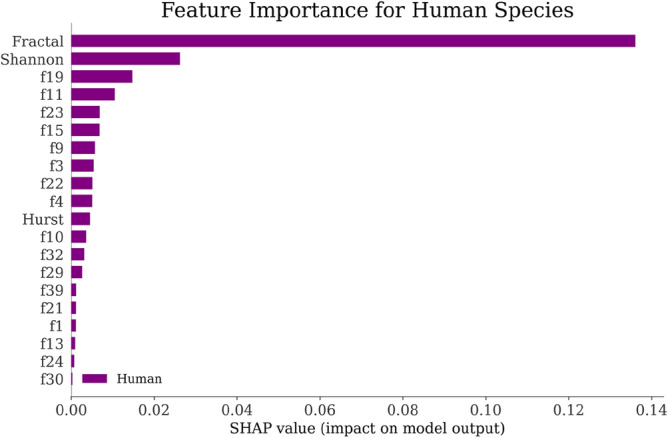
Feature Importance of Human Species by SHAP explainer for EDL (EDL6) model using K10 protocol.
Figure 8.
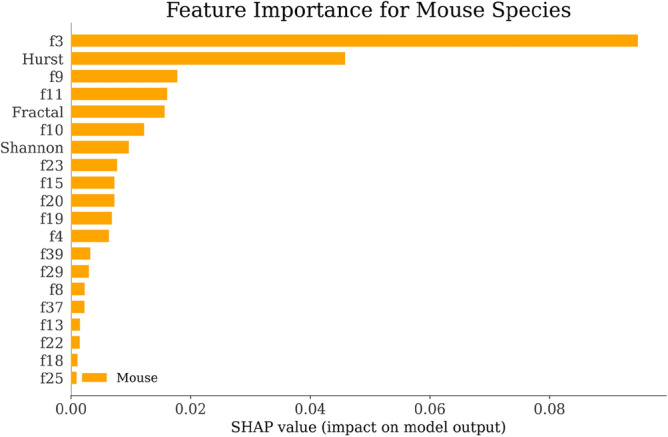
Feature Importance of Mouse Species by SHAP explainer for EDL (EDL6) model using K10 protocol.
Figure 9.
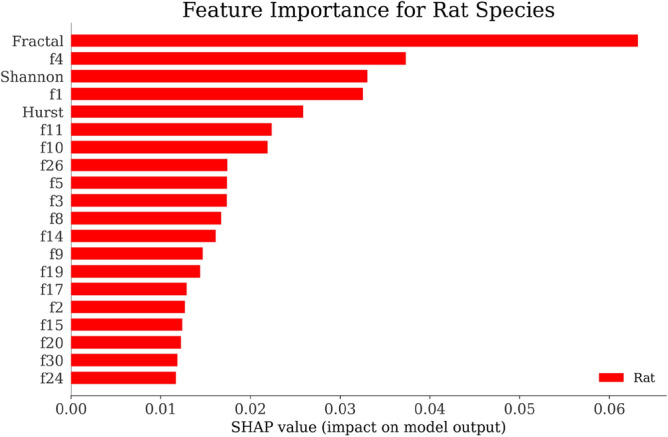
Feature Importance of Rat Species by SHAP explainer for EDL (EDL6) model using K10 protocol.
Discussion
Principal findings
After conducting an extensive study, we obtained valuable insights and drew conclusions pertaining to our research problem: (i) We devised four hypotheses and developed a total of 38 AI classifiers, which consisted of nine SML classifiers, five EML classifiers, six SDL classifiers, twelve HDL classifiers, and six EDL classifiers, in order to test them. (ii) For our experimental analysis, we utilized ten pre-processed datasets, comprising six binary classification datasets and four multiclass classification datasets. (iii) To enhance the processing and conversion of miRNA sequences into co-occurrence features, we implemented a novel quality control phase for our system. This involved performing scaling and binary encoding of the sequences. (iv) Our findings indicate that EML classifiers outperformed SML classifiers, yielding a mean accuracy increase of 6.24% and a 6.46% increase in AUC. Furthermore, HDL classifiers exhibited a significant advantage over SDL classifiers, with an increase in accuracy and AUC of 2.17% and 2.4%, respectively. (v) Also, EDL classifiers further improved upon HDL classifiers, with a mean accuracy of 2.01% and an AUC of 1.52%. (vi) Additionally, EDL classifiers significantly improved upon EML classifiers, with a mean accuracy increase of 7.09% and an AUC of 6.96%. (vii) We also observed that utilizing CNN-based HDL models with a feature extraction methodology greatly improved performance compared to non-CNN-based HDL models, yielding a mean accuracy increase of 0.73% and a 0.92% increase in AUC. (viii) We ensured the reliability and stability of our system by subjecting the classifiers to statistical tests. (ix) In order to verify the system's stability with smaller gene data sizes, we conducted a power analysis on the six binary class and four multiclass datasets, thereby validating the precision of the GeneAI 3.0 system. (x) We evaluated the impact of training data size by implementing Cross-Validation protocols in an increasing order. (xi) Finally, we utilized the SHAP explainer to interpret the classification results of the best (EDL6) model. This allowed us to gain insights into the significance of each species' features in their respective classifications.
Benchmarking: a comparative analysis
Numerous methods have been suggested for miRNA classifiers and species-independent lncRNA predictors, such as Precursor miRNAs classification, Non-coding RNA classification, and cross-species miRNA identification. These methods have undergone extensive validation and proven effective in identifying and categorizing miRNA and lncRNA. In contrast, this study introduces a unique approach to classify miRNA based on stationary patterns derived from gene sequences. The primary aim is to determine the species of origin by analyzing specific parameters associated with each species family. While this approach is innovative, its effectiveness and practicality need to be assessed through a comparative analysis with existing methods. Comparing different approaches is crucial for advancing the field of miRNA classification and enhancing our comprehension of miRNA biology. Therefore, it is essential to evaluate the proposed approach's accuracy, efficiency, and generalizability in comparison to established methods.
Table 10 focused on six studies that focused on developing classifiers for miRNA and lncRNA. Yousef et al.153 employed a RF classifier and created a specific feature set called k-mer, which consisted of k-mer Distance, k-mer location distance, and k-mer first-last distance. These features were added to the basic k-mer features to classify Precursor miRNA. The evaluation of their method was conducted using a database obtained from USEARCH. Cao et al.154 explored the utilization of an RF model with incremental feature selection and the Pearson correlation coefficient. Their objective was to predict lncRNA from both lncRNA and mRNA transcripts in a dataset consisting of six species. The dataset used in their study was sourced from Ensemble data repository.
Table 10.
Benchmarking table showing studies that were implemented for miRNA and lncRNA classification.
| Author & Year | Objective | Method | Model | Feature Extraction | Dataset | Class Type | Performance | CVP* | Clinical Validation |
|---|---|---|---|---|---|---|---|---|---|
| Yousef et al.153 | Precursor miRNAs classification | Addition of K-mer Distance, K-mer Location Distance, and K-mer First-Last Distance to the core K-mer Features for Classification | RF | K-mer Distance Features | USEARCH (16 Species) | BC & MC |
ACC: 93% ACC: 86% (Laurasiatheria) |
K100 Monte Carlo | × |
| Jiang et al.157 | Precursor miRNA classification | Backpropagation Neural network model was used to identify microRNA precursors using 98-dimensional novel features | ANN | Conventional Features | Carleton (SMIRP) | BC | ACC: 93.42% | K5 | × |
| Cao et al.154 | Predicting lncRNAs | Predicting lncRNA from lncRNA and mRNA transcripts, a RF classifier with incremental feature selection and the Pearson correlation coefficient was used | RF | Incremental feature selection | Ensembl v97, GreeNC (6 Species)CD-hit | Class Specific | ACC: 91.09% | K10 | Adjusted p-value; Z-value |
| Zhao et al.156 | Predicting lncRNAs | Improvement paradigm for miRNA target prediction using DT-based meta-strategy and multi-threshold sequential-voting | DT-based voting sytem | Multi-threshold sequential-voting for meta-prediction | MiRTarBase | BC | ACC: 91.09% | × | × |
| Gu et al.155 | Predicting lncRNAs | Ensemble Learning based approach using a multi-classifer based system to miRNA related to disease by discovering new potential associations | Multi-classifiers voting | Similarity and structural feature data | HMDD V2.0 | BC | AUC: 0.9229 | K5 | × |
| Amin et al.158 | Non-coding RNA classification | Development of a Feature database of Peptide, ORF, and Whole sequence and selection using separate classifiers with Hierarchical majority voting |
LR RF |
Extensive Feature selection based on database, species and ncRNA type | RNACentral (16 Species) | BC |
ACC: 91.928%(All Features) F1-score: 94.885% (All Features) |
Nested K10 | Chi-squared (in model) |
| Singh et al. Proposed | miRNA classification | Using Shannon Entropy, Hurst Exponent and Fractal Dimension along with contemporary features like Entropy, and Diversity to predict Species from MicroRNA gene sequence |
ML SDL HDL EDL |
Stationary Patterns of Nucleotides | miRNA Database (4 Species) | BC & MC | Best ACC (EDL): 97.41% Best EDL AUC: 0.97 (EDL1 in supplementary material) | K2, K4, K5, and K10 (Default) | R2, Z-two tailed, ANOVA |
*CVP Cross-Validation Protocol; ACC Accuracy (%); BC Binary Class; MC Multiclass Classification; XAI Explainable AI; ML Machine Learning; SDL Solo Deep Learning; HDL Hybrid Deep Learning; EDL Ensemble Deep Learning; LR Logistic Regression; RF Random Forest; CNN Convolutional Neural Networks; ANN Artificial Neural Networks; K#: Cross-Validation protocol having the ratio of training: testing data sets; K2: 50%:50%; K4: 75%:25%; K5:80%:20%; K10: 90%:10%.
Gu et al.155 introduced an Ensemble Learning approach for miRNA-related disease classification using a multi-classifier system based on associated probabilities. Their method aimed to discover new potential associations between miRNA and diseases. The results were validated using various versions of the HMDD database, making it a reliable approach that does not rely on known associations between miRNA and diseases. Zhao et al.156, introduces an improved paradigm for miRNA target prediction was presented. They utilized a DT-based meta-strategy and a multi-threshold sequential voting method for meta-prediction. This approach aimed to enhance the accuracy of existing miRNA target prediction schemes.
Jiang et al.157 implemented a neural network-based scheme for end-to-end classification of pre-miRNA. They utilized a database consisting of 98 features, including n-gram frequency, structural sequence, structural diversity, and energy. The approach incorporated primary and secondary structure information to identify pre-miRNA in seven different species. Amin et al.158 employed a comprehensive feature extraction approach for non-coding RNA classification. They constructed an extensive feature database and trained it using LR and RF models. The database consisted of peptide features, open reading frame (ORF) features, and whole sequence features, with classifiers individually applied to each feature class. A hierarchical majority voting mechanism was utilized to combine the features.
In our proposed work (R7), we introduce a novel approach for miRNA classification based on species of origin. Multiple LSTM, GRU, CNN, and RNN-based SML, EML, SDL, HDL, and EDL models are employed. A feature extraction module is used to extract both conventional features like entropy and energy, as well as contemporary features such as Shannon entropy, Hurst exponent, and fractal dimension. This integration of different features helps build a more robust model. Our study focuses on achieving generalization, employing XAI as part of scientific validation, and conducting thorough testing to ensure the reliability and stability of the GeneAI 3.0 system.
Special note on ensemble-based feature extraction in miRNA classification
Ensemble-based feature extraction techniques have emerged as a powerful approach in miRNA classification tasks. By combining multiple feature extraction methods, using concatenation and splitting, these ensembles can effectively capture diverse aspects of miRNA sequences, leading to improved classification performance. The ensemble architecture allows for the fusion of features extracted from different methods, such as structural and compositional information, enabling the neural network to leverage complementary information and capture complex patterns in miRNA data. This approach not only enhances the classification accuracy but also helps mitigate overfitting by providing a regularization effect. Additionally, by incorporating different ensemble architectures, including completely different paradigms, the ensemble-based feature extraction further enriches the classification process, allowing for a more comprehensive and robust miRNA classification.
The effectiveness of ensemble-based techniques in miRNA classification is not limited to DL but also observed in traditional ML approaches. Techniques like RF and stacked ML models employ ensembles of multiple ML models to enhance classification performance. The ensemble architectures, such as weighted averaging, hard voting, and soft voting, play a crucial role in combining the predictions or features extracted from different models, leveraging their complementary strengths, and achieving better classification outcomes in miRNA analysis. By harnessing the collective intelligence of multiple models, ensemble-based feature extraction offers a powerful framework to improve the accuracy, sensitivity, and specificity of miRNA classification models. These ensemble-based approaches pave the way for more reliable and robust miRNA classification, enabling researchers to gain deeper insights into the complex world of gene expression and regulation.
Special note on generalization
For generalizations, the models have to undergo training and testing on multiple datasets. Our group has done several methods for generalization78–81. In81, we developed an ensemble-based transfer learning paradigm, successfully classifying skin lesion images from two different and diverse datasets. We trained on one set and classified lesions from the other set. Study80 focused on our work in depression detection, where we developed a generalized model for text classification with the primary goal of detecting depression. Study79 attempted to achieve generalization in Covid-19 patients' lung segmentation across five different combinations of data by employing unseen data tests and statistical analyses. Finally, Study78, focusing on COVID-19 lung computed tomography segmentation, achieved generalization by testing on two unseen datasets, pairing 72 Italian and 80 Croatian patients.
We achieved generalization in these systems by simplifying the model, enabling it to work across multiple domains effectively in various situations through the mixing of domains. In Study80, we trained a model to be robust enough for depression detection as well as sentiment analysis by facilitating inter-dataset (cross-domain) training and leveraging knowledge from a multi-domain dataset. Our model demonstrated the capability to detect depression even when trained on a sentiment dataset, while also analysing sentiment when trained on a depression dataset. Likewise in this study, we conducted both multi-class and binary class classification, comprising a total of 10 datasets, where the model demonstrated satisfactory performance. This generalization ensures the effectiveness of our models for use in real-life scenarios, as any gene sequence can be pre-processed, features extracted and utilized by our model.
Strengths, weakness, and extensions
Our study presents a novel approach to gene dataset analysis using 38 AI classifiers, which consisted of nine SML classifiers, five EML classifiers, six SDL classifiers, twelve HDL classifiers, and six EDL classifiers. Through rigorous evaluation, we found that these models demonstrated exceptional performance in both binary and multiclass classification tasks. Furthermore, our study involved building an extensive composite feature set, generating new features such as Shannon entropy, Hurst exponent, and Fractal dimension, which were incorporated with existing co-occurrence features to enhance the AI system's performance. Additionally, our study addressed the challenge of interpretability by incorporating XAI techniques, allowing us to gain insights into the inner workings of the models. This enables us to leverage feature-specific knowledge and concentrate on further research for each species independently. It provides a critical overview of the important features that individually impact the likelihood of a miRNA sequence belonging to a specific species. This knowledge, derived from the feature plots, is crucial for the practical implementation of the machine learning model in our study. It will significantly influence how we hypertune our model and, on a biological level, understand which features (both conventional and contemporary) matter more for each species. Notably, our proposed methodology demonstrated robustness through its consistent performance in multiple statistical tests, including the Adjusted R2 Test, paired T-test, ANOVA, and null-hypothesis significance testing (p-value). Across all six binary and four multiclass datasets, our methodology consistently provided interpretable, reliable and accurate results, highlighting its potential to improve classification accuracy in gene species classification.
One limitation of our gene classification approach is the potential for model generalization and overfitting due to the limited size of the available training data, especially in binary class classification tasks. Although ensemble-based models have been employed to mitigate this issue, there is room for improvement by utilizing Generative Adversarial training-based mechanisms to synthesize additional data. Another weakness is the absence of attention mechanisms, which could hinder the model's ability to mitigate overfitting and enhance its overall robustness. To address these limitations, incorporating attention-based techniques can offer a more focused and streamlined classification of species, ultimately improving the accuracy and reliability of the gene classification scheme.
In the future, we can further enhance our gene classification scheme by addressing limitations and implementing potential improvements. One major limitation is the lack of diversity in the dataset, which can hinder the model's ability to generalize. To overcome this, we can incorporate a wider range of gene species and sequences into the dataset. This can be achieved by leveraging big data sources159 or exploring other public data repositories160,161. By expanding the dataset, we can train more complex models that exhibit improved accuracy and generalization performance. In dealing with gene sequence data, graph neural networks and attention-enabled mechanisms show promise162–164. These approaches can better capture the intricate relationships between gene sequences and the species of origin. By leveraging these techniques, we can enhance the accuracy and interpretability of our gene classification scheme. To address the scarcity of data available for training models, we can consider employing Generative Adversarial training-based schemes. These schemes can generate synthetic data, thereby augmenting the training set and helping to overcome the data dearth165–167. We also plan to enhance our model by employing a cross-domain-based framework. This involves training on one gene sequence dataset and testing on another from a different database. More gene data can be selected, evaluated to prove the deep learning methods. Another avenue to explore is the utilization of Autoencoders in gene classification. Autoencoders have the ability to reduce dimensionality and extract essential features from the data. By incorporating an Autoencoder-based paradigm, we can improve the efficiency and accuracy of gene classification tasks168–170. Additionally, applying pruning strategies for AI models141 and studying the comorbidity effect in genomics can contribute to enhancing the classification system. Pruning techniques optimize the model's architecture and computational efficiency, while investigating comorbidity sheds light on the interconnected nature of genetic factors and disease manifestation171. By implementing these potential improvements, we can develop more accurate and robust models with broader applicability in the fields of genetics and bioinformatics.
Conclusion
This study presents a novel paradigm for feature extraction in miRNA classification using EDL and EML models. Specifically, we utilized 38 types of AI models (nine SML, six EML, six SDL and twelve HDL and six EDL) architectures, to extract features from co-occurrence-based binary-coded sequences. The extracted composite features combined contemporary and conventional features, resulting in a total of 43 generated features. We conducted a thorough data analysis using 10 classification algorithms, including binary and multiclass classifiers, and four experimental protocols to evaluate the effectiveness of our proposed scheme. Our results showed that our proposed scheme outperformed existing methods regarding accuracy, sensitivity, and specificity. Furthermore, we conducted Cross-Validation to ensure the robustness of our model, and our results demonstrated that our model was highly reliable even with limited training data. Finally, we conducted statistical tests to demonstrate the reliability and stability of our Artificial Intelligence system.
Supplementary Information
Abbreviations
- AI
Artificial intelligence
- ACC
Accuracy
- ADASYN
Adaptive synthetic sampling approach for imbalanced leaning
- ANOVA
Analysis of variance
- AUC
Area-under-the-curve
- BC
Binary classification
- BiGRU
Bidirectional GRU
- BiLSTM
Bidirectional LSTM
- BiRNN
Bidirectional RNN
- CNN
Convolutional neural network
- DL
Deep learning
- DT
Decision trees
- ET
Extra trees
- EDL
Ensemble deep learning
- EML
Ensemble machine learning
- FD
Fractal dimension
- FN
False negative
- FP
False positive
- GPU
Graphics processing unit
- GRU
Gated recurrent unit
- HDL
Hybrid deep learning
- HE
Hurst exponent
- HINN
Hierarchical input neural networks
- KNN
K-nearest neighbors
- LDA
Linear discriminant analysis
- LGBM
Light gradient boosting model
- lncRNA
Long non-coding RNAs
- LR
Logistic regression
- LSTM
Long short-term memory
- MCC
Multiclass classification
- miRNA
MicroRNA
- ML
Machine learning
- mRNA
Messenger RNA
- NB
Naïve Bayes
- ORF
Open reading frame
- RF
Random forest
- ROC
Receiver operating curves
- RNA
Ribonucleic acid
- RNN
Recurrent neural network
- SDL
Solo deep learning
- SHAP
Shapley additive explanations
- SE
Shannon entropy
- SML
Solo machine learning
- SVM
Support vector machine
- TP
True positive
- TN
True negative
- XAI
Explainable AI
- Xgboost
Extreme gradient boost
Symbols
Accuracy
- R
Recall
- P
Precision
- F
F1-score
Arithmetic mean
- A
Adenine
- U
Uracil
- C
Cytosine
- G
Guanine
Co-occurrence matrix
Normalized co-occurrence matrix
- St
MiRNA sequence
Length of miRNA sequence
Probability of Bernoulli process
Binary miRNA sequence
- fSet
Final feature set representation
Difference between maximum and minimum instances of the binary miRNA sequence
Standard deviation of the Binary miRNA sequence
Nucleotides representation: {A, U, C, G}
Categorical cross-entropy Loss
Accuracy of model ‘m’ summarized over all D datasets over K10 protocol
Accuracy achieved over dataset ‘d’ over all M Models over K10 protocol
Overall system accuracy over M models and D datasets
AUC of model m summarized over all D datasets
AUC achieved over dataset d over all M Models
Overall system AUC over M models and D datasets
- M
Total number of Models used in study
- D
Total number of Datasets used in study
- ⊕
Concatenation of two models
Author contributions
Conceptualization, J.S., N.N.K., and J.S.S.; methodology, J.S., R.K.R., J.R.L., L.E.M., and J.S.S.; investigation, N.N.K., J.R.L., I.M.S., L.S., and J.S.S.; resources, M.M.F., L.S.; writing-original draft preparation, J.S., R.K.R., and M.K.K.; writing-review and editing, J.S., A.M.J., E.R.I., and J.S.S.; visualization, J.S., N.S., E.R.I., L.S., and J.S.S.; supervision, J.R.L., I.M.S., A.M.J., M.M.F., M.F., L.S., and J.S.S.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Data availability
The datasets generated during and analyzed during the current study are not publicly available due to their propriety nature but are available from the corresponding author on reasonable request.
Code availability
The code used during the current study are not publicly available due to due to their propriety nature but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-56786-9.
References
- 1.Anglicheau, D., Muthukumar, T. & Suthanthiran, M. MicroRNAs: Small RNAs with big effects. Transplantation90(2), 105 (2010). 10.1097/TP.0b013e3181e913c2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson, P., Kiriakidou, M., Sharma, A., Maniataki, E. & Mourelatos, Z. The microRNA world: Small is mighty. Trends Biochem. Sci.28(10), 534–540 (2003). 10.1016/j.tibs.2003.08.005 [DOI] [PubMed] [Google Scholar]
- 3.Pogue, A. et al. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci. Lett.476(1), 18–22 (2010). 10.1016/j.neulet.2010.03.054 [DOI] [PubMed] [Google Scholar]
- 4.Cheng, A. M., Byrom, M. W., Shelton, J. & Ford, L. P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res.33(4), 1290–1297 (2005). 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Torre, A., Georgi, S. & Reh, T. A. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci.110(26), E2362–E2370 (2013). 10.1073/pnas.1301837110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren, Z. & Ambros, V. R. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl. Acad. Sci.112(18), E2366–E2375 (2015). 10.1073/pnas.1422858112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto, T. et al. Cell cycle-targeting microRNAs promote differentiation by enforcing cell-cycle exit. Proc. Natl. Acad. Sci.114(40), 10660–10665 (2017). 10.1073/pnas.1702914114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, H. S. et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget6(10), 8089 (2015). 10.18632/oncotarget.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo, Q. et al. Tumor-suppressive microRNA-195-5p regulates cell growth and inhibits cell cycle by targeting cyclin dependent kinase 8 in colon cancer. Am. J. Transl. Res.8(5), 2088 (2016). [PMC free article] [PubMed] [Google Scholar]
- 10.Karatas, O. F. et al. miR-33a is a tumor suppressor microRNA that is decreased in prostate cancer. Oncotarget8(36), 60243 (2017). 10.18632/oncotarget.19521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barwari, T., Joshi, A. & Mayr, M. MicroRNAs in cardiovascular disease. J. Am. College Cardiol.68(23), 2577–2584 (2016). 10.1016/j.jacc.2016.09.945 [DOI] [PubMed] [Google Scholar]
- 12.Small, E. M., Frost, R. J. & Olson, E. N. MicroRNAs add a new dimension to cardiovascular disease. Circulation121(8), 1022–1032 (2010). 10.1161/CIRCULATIONAHA.109.889048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, Y. & Zhang, C. MicroRNA-21 in cardiovascular disease. J. Cardiovasc. Transl. Res.3, 251–255 (2010). 10.1007/s12265-010-9169-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloosterman, W. P. & Plasterk, R. H. The diverse functions of microRNAs in animal development and disease. Dev. Cell11(4), 441–450 (2006). 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 15.Bhayani, M. K., Calin, G. A. & Lai, S. Y. Functional relevance of miRNA* sequences in human disease. Mutation Res./Fundam. Mol. Mech. Mutagenesis731(1–2), 14–19 (2012). 10.1016/j.mrfmmm.2011.10.014 [DOI] [PubMed] [Google Scholar]
- 16.Chen, X. et al. WBSMDA: Within and between score for MiRNA-disease association prediction. Sci. Rep.6(1), 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, X., Wu, Q.-F. & Yan, G.-Y. RKNNMDA: Ranking-based KNN for MiRNA-disease association prediction. RNA Biol.14(7), 952–962 (2017). 10.1080/15476286.2017.1312226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You, Z.-H. et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput. Biol.13(3), e1005455 (2017). 10.1371/journal.pcbi.1005455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backes, C., Meese, E. & Keller, A. Specific miRNA disease biomarkers in blood, serum and plasma: Challenges and prospects. Mol. Diagn. Ther.20, 509–518 (2016). 10.1007/s40291-016-0221-4 [DOI] [PubMed] [Google Scholar]
- 20.Jadideslam, G. et al. The MicroRNA-326: Autoimmune diseases, diagnostic biomarker, and therapeutic target. J. Cell. Physiol.233(12), 9209–9222 (2018). 10.1002/jcp.26949 [DOI] [PubMed] [Google Scholar]
- 21.Shah, M. Y. & Calin, G. A. MicroRNAs as therapeutic targets in human cancers. Wiley Interdisci. Rev. RNA5(4), 537–548 (2014). 10.1002/wrna.1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, C.-S. et al. Catalog of Erycina pusilla miRNA and categorization of reproductive phase-related miRNAs and their target gene families. Plant Mol. Biol.82, 193–204 (2013). 10.1007/s11103-013-0055-y [DOI] [PubMed] [Google Scholar]
- 23.Kleftogiannis, D. et al. Where we stand, where we are moving: Surveying computational techniques for identifying miRNA genes and uncovering their regulatory role. J. Biomed. Inform.46(3), 563–573 (2013). 10.1016/j.jbi.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 24.Eszlinger, M. et al. Molecular profiling of thyroid nodule fine-needle aspiration cytology. Nat. Rev. Endocrinol.13(7), 415–424 (2017). 10.1038/nrendo.2017.24 [DOI] [PubMed] [Google Scholar]
- 25.Jiang, P. et al. MiPred: Classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res.35(2), W339–W344 (2007). 10.1093/nar/gkm368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, Y. et al. A support vector machine and a random forest classifier indicates a 15-miRNA set related to osteosarcoma recurrence. OncoTargets Ther.15, 253–269 (2018). 10.2147/OTT.S148394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghobadi, M. Z., Emamzadeh, R. & Afsaneh, E. Exploration of mRNAs and miRNA classifiers for various ATLL cancer subtypes using machine learning. BMC Cancer22(1), 1–8 (2022). 10.1186/s12885-022-09540-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jha, A. & Shankar, R. Employing machine learning for reliable miRNA target identification in plants. BMC Genomics12, 1–18 (2011). 10.1186/1471-2164-12-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stegmayer, G. et al. Predicting novel microRNA: A comprehensive comparison of machine learning approaches. Briefings Bioinform.20(5), 1607–1620 (2019). 10.1093/bib/bby037 [DOI] [PubMed] [Google Scholar]
- 30.Rahman, M. H. et al. Bioinformatics and machine learning methodologies to identify the effects of central nervous system disorders on glioblastoma progression. Brief. Bioinform.22(5), bbaa365 (2021). 10.1093/bib/bbaa365 [DOI] [PubMed] [Google Scholar]
- 31.Wang, C. A modified machine learning method used in protein prediction in bioinformatics. Int. J. Bioautom.19, 1 (2015). [Google Scholar]
- 32.Le, N. Q. K., Li, W. & Cao, Y. Sequence-based prediction model of protein crystallization propensity using machine learning and two-level feature selection. Brief. Bioinform.24(5), bbad319 (2023). 10.1093/bib/bbad319 [DOI] [PubMed] [Google Scholar]
- 33.Ou, Y.-Y. Identifying the molecular functions of electron transport proteins using radial basis function networks and biochemical properties. J. Mol. Graph. Model.73, 166–178 (2017). 10.1016/j.jmgm.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 34.Xue, C. et al. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinform.6, 1–7 (2005). 10.1186/1471-2105-6-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lertampaiporn, S., Thammarongtham, C., Nukoolkit, C., Kaewkamnerdpong, B. & Ruengjitchatchawalya, M. Heterogeneous ensemble approach with discriminative features and modified-SMOTEbagging for pre-miRNA classification. Nucleic Acids Res.41(1), e21–e21 (2013). 10.1093/nar/gks878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batuwita, R. & Palade, V. microPred: Effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics25(8), 989–995 (2009). 10.1093/bioinformatics/btp107 [DOI] [PubMed] [Google Scholar]
- 37.Xuan, P. et al. PlantMiRNAPred: Efficient classification of real and pseudo plant pre-miRNAs. Bioinformatics27(10), 1368–1376 (2011). 10.1093/bioinformatics/btr153 [DOI] [PubMed] [Google Scholar]
- 38.Wei, L. et al. Improved and promising identification of human microRNAs by incorporating a high-quality negative set. IEEE/ACM Trans. Comput. Biol. Bioinform.11(1), 192–201 (2013). 10.1109/TCBB.2013.146 [DOI] [PubMed] [Google Scholar]
- 39.Blum, A. & Mitchell, T. Combining labeled and unlabeled data with co-training. in Proceedings of the Eleventh Annual Conference on Computational Learning Theory, 1998, pp. 92–100.
- 40.He, C. et al. MiRmat: Mature microRNA sequence prediction. PLoS One7(12), e51673 (2012). 10.1371/journal.pone.0051673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terai, G., Okida, H., Asai, K. & Mituyama, T. Prediction of conserved precursors of miRNAs and their mature forms by integrating position-specific structural features (2012). [DOI] [PMC free article] [PubMed]
- 42.Leclercq, M., Diallo, A. B. & Blanchette, M. Computational prediction of the localization of microRNAs within their pre-miRNA. Nucleic Acids Res.41(15), 7200–7211 (2013). 10.1093/nar/gkt466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xuan, P., Guo, M., Huang, Y., Li, W. & Huang, Y. MaturePred: Efficient identification of microRNAs within novel plant pre-miRNAs. PloS One6(11), e27422 (2011). 10.1371/journal.pone.0027422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, Y., Wei, B., Liu, H., Li, T. & Rayner, S. MiRPara: A SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinform.12(1), 1–14 (2011). 10.1186/1471-2105-12-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan, D.-G., Liao, J.-Y., Qu, Z.-H., Zhang, Y. & Qu, L.-H. mirExplorer: Detecting microRNAs from genome and next generation sequencing data using the AdaBoost method with transition probability matrix and combined features. RNA Biol.8(5), 922–934 (2011). 10.4161/rna.8.5.16026 [DOI] [PubMed] [Google Scholar]
- 46.Li, J. et al. MatPred: Computational identification of mature micrornas within novel pre-MicroRNAs. BioMed Res. Int.2015, 23 (2015). 10.1155/2015/546763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karathanasis, N., Tsamardinos, I. & Poirazi, P. MiRduplexSVM: A high-performing miRNA-duplex prediction and evaluation methodology. PloS One10(5), e0126151 (2015). 10.1371/journal.pone.0126151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peace, R. & Green, J. R. Computational sequence-and NGS-based microRNA prediction. In Signal Processing and Machine Learning for Biomedical Big Data: CRC Press, 2018, pp. 381–410.
- 49.Chen, L. et al. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform.20(5), 1836–1852 (2019). 10.1093/bib/bby054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page, J., Brenner, M. P. & Kerswell, R. R. Revealing the state space of turbulence using machine learning. Phys. Rev. Fluids6(3), 034402 (2021). 10.1103/PhysRevFluids.6.034402 [DOI] [Google Scholar]
- 51.Paul, T. K. & Iba, H. Prediction of cancer class with majority voting genetic programming classifier using gene expression data. IEEE/ACM Trans. Comput. Biol. Bioinform.6(2), 353–367 (2008). 10.1109/TCBB.2007.70245 [DOI] [PubMed] [Google Scholar]
- 52.Hassan, M. R. et al. A voting approach to identify a small number of highly predictive genes using multiple classifiers. BMC Bioinform.10, 1–12 (2009). 10.1186/1471-2105-10-S1-S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li, Y. & Luo, Y. Performance-weighted-voting model: An ensemble machine learning method for cancer type classification using whole-exome sequencing mutation. Quant. Biol.8, 347–358 (2020). 10.1007/s40484-020-0226-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, X., Xu, S., Zhang, Y. & Huang, X. Nucleotide-level convolutional neural networks for pre-miRNA classification. Sci. Rep.9(1), 628 (2019). 10.1038/s41598-018-36946-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, X. & Sun, Y. Fast and accurate microRNA search using CNN. BMC Bioinform.20(23), 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park, S., Min, S., Choi, H.-S. & Yoon, S. Deep recurrent neural network-based identification of precursor micrornas. Adv. Neural Inf. Process. Syst.30, 30 (2017). [Google Scholar]
- 57.Amin, N., McGrath, A. & Chen, Y.-P.P. Evaluation of deep learning in non-coding RNA classification. Nat. Mach. Intell.1(5), 246–256 (2019). 10.1038/s42256-019-0051-2 [DOI] [Google Scholar]
- 58.Kleftogiannis, D., Theofilatos, K., Likothanassis, S. & Mavroudi, S. YamiPred: A novel evolutionary method for predicting pre-miRNAs and selecting relevant features. IEEE/ACM Trans. Comput. Biol. Bioinform.12(5), 1183–1192 (2015). 10.1109/TCBB.2014.2388227 [DOI] [PubMed] [Google Scholar]
- 59.Suri, J. S. et al. A powerful paradigm for cardiovascular risk stratification using multiclass, multi-label, and ensemble-based machine learning paradigms: A narrative review. Diagnostics12(3), 722 (2022). 10.3390/diagnostics12030722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jamthikar, A. D. et al. Ensemble machine learning and its validation for prediction of coronary artery disease and acute coronary syndrome using focused carotid ultrasound. IEEE Trans. Instrum. Meas.71, 1–10 (2021). 10.1109/TIM.2021.3139693 [DOI] [Google Scholar]
- 61.Tandel, G. S. et al. Role of ensemble deep learning for brain tumor classification in multiple magnetic resonance imaging sequence data. Diagnostics13(3), 481 (2023). 10.3390/diagnostics13030481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, H. et al. CL-PMI: A precursor MicroRNA identification method based on convolutional and long short-term memory networks. Front. Genet.10, 967 (2019). 10.3389/fgene.2019.00967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tasdelen, A. & Sen, B. A hybrid CNN-LSTM model for pre-miRNA classification. Sci. Rep.11(1), 1–9 (2021). 10.1038/s41598-021-93656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakraborty, R. & Hasija, Y. Predicting MicroRNA sequence using CNN and LSTM stacked in Seq2Seq architecture. IEEE/ACM Trans. Comput. Biol. Bioinform.17(6), 2183–2188 (2019). 10.1109/TCBB.2019.2936186 [DOI] [PubMed] [Google Scholar]
- 65.Ru, X., Cao, P., Li, L. & Zou, Q. Selecting essential MicroRNAs using a novel voting method. Mol. Therapy-Nucleic Acids18, 16–23 (2019). 10.1016/j.omtn.2019.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas, J., Thomas, S. & Sael, L. DP-miRNA: An improved prediction of precursor microRNA using deep learning model. In 2017 IEEE International Conference on Big Data and Smart Computing (BigComp), 2017: IEEE, pp. 96–99.
- 67.Asim, M. N. et al. MirLocPredictor: A ConvNet-based multi-label MicroRNA subcellular localization predictor by incorporating k-Mer positional information. Genes11(12), 1475 (2020). 10.3390/genes11121475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu, X. et al. Improved pre-miRNAs identification through mutual information of pre-miRNA sequences and structures. Front. Genet.10, 119 (2019). 10.3389/fgene.2019.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan, L. et al. Radiotranscriptomics signature-based predictive nomograms for radiotherapy response in patients with nonsmall cell lung cancer: Combination and association of CT features and serum miRNAs levels. Cancer Med.9(14), 5065–5074 (2020). 10.1002/cam4.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, S., Tu, J., Wang, L. & Lu, Z. Entropy-based model for miRNA isoform analysis. PLoS One10(3), e0118856 (2015). 10.1371/journal.pone.0118856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thakur, V. et al. Characterization of statistical features for plant microRNA prediction. BMC Genomics12(1), 1–12 (2011). 10.1186/1471-2164-12-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He, H., Bai, Y., Garcia, E. A. & Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In 2008 IEEE international joint conference on neural networks (IEEE world congress on computational intelligence), 2008: IEEE, pp. 1322–1328.
- 73.Johri, A. M. et al. Deep learning artificial intelligence framework for multiclass coronary artery disease prediction using combination of conventional risk factors, carotid ultrasound, and intraplaque neovascularization. Comput. Biol. Med.150, 106018 (2022). 10.1016/j.compbiomed.2022.106018 [DOI] [PubMed] [Google Scholar]
- 74.Konstantonis, G. et al. Cardiovascular disease detection using machine learning and carotid/femoral arterial imaging frameworks in rheumatoid arthritis patients. Rheumatol. Int.42(2), 215–239 (2022). 10.1007/s00296-021-05062-4 [DOI] [PubMed] [Google Scholar]
- 75.Saba L. et al., Plaque tissue morphology-based stroke risk stratification using carotid ultrasound: A polling-based PCA learning paradigm. In Vascular and Intravascular Imaging Trends, Analysis, and Challenges, Volume 2: Plaque characterization: IOP Publishing Bristol, UK, 2019, pp. 9–1–9–45.
- 76.Araki, T. et al. PCA-based polling strategy in machine learning framework for coronary artery disease risk assessment in intravascular ultrasound: A link between carotid and coronary grayscale plaque morphology. Comput. Methods Prog. Biomed.128, 137–158 (2016). 10.1016/j.cmpb.2016.02.004 [DOI] [PubMed] [Google Scholar]
- 77.Maniruzzaman, M. et al. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput. Methods Prog. Biomed.176, 173–193 (2019). 10.1016/j.cmpb.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 78.Suri, J. S. et al. Multicenter study on COVID-19 lung computed tomography segmentation with varying glass ground opacities using unseen deep learning artificial intelligence paradigms: COVLIAS 1.0 validation. J. Med. Syst.46(10), 62 (2022). 10.1007/s10916-022-01850-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dubey, A. K. et al. Ensemble deep learning derived from transfer learning for classification of COVID-19 patients on hybrid deep-learning-based lung segmentation: A data augmentation and balancing framework. Diagnostics13(11), 1954 (2023). 10.3390/diagnostics13111954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh, J., Singh, N., Fouda, M. M., Saba, L. & Suri, J. S. Attention-enabled ensemble deep learning models and their validation for depression detection: A domain adoption paradigm. Diagnostics13(12), 2092 (2023). 10.3390/diagnostics13122092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanga, P. et al. DermAI 1.0: A robust, generalized, and novel attention-enabled ensemble-based transfer learning paradigm for multiclass classification of skin lesion images. Diagnostics13(19), 3159 (2023). 10.3390/diagnostics13193159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skandha, S. S. et al. A hybrid deep learning paradigm for carotid plaque tissue characterization and its validation in multicenter cohorts using a supercomputer framework. Comput. Biol. Med.141, 105131 (2022). 10.1016/j.compbiomed.2021.105131 [DOI] [PubMed] [Google Scholar]
- 83.Sanagala, S. S. et al. Ten fast transfer learning models for carotid ultrasound plaque tissue characterization in augmentation framework embedded with heatmaps for stroke risk stratification. Diagnostics11(11), 2109 (2021). 10.3390/diagnostics11112109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jain, P. K. et al. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput. Biol. Med.136, 104721 (2021). 10.1016/j.compbiomed.2021.104721 [DOI] [PubMed] [Google Scholar]
- 85.Agarwal, M. et al. Wilson disease tissue classification and characterization using seven artificial intelligence models embedded with 3D optimization paradigm on a weak training brain magnetic resonance imaging datasets: A supercomputer application. Med. Biol. Eng. Comput.59, 511–533 (2021). 10.1007/s11517-021-02322-0 [DOI] [PubMed] [Google Scholar]
- 86.Saba, L. et al. Ultrasound-based internal carotid artery plaque characterization using deep learning paradigm on a supercomputer: A cardiovascular disease/stroke risk assessment system. Int. J. Cardiovasc. Imaging37, 1511–1528 (2021). 10.1007/s10554-020-02124-9 [DOI] [PubMed] [Google Scholar]
- 87.Skandha, S. S. et al. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: Atheromatic™ 2.0. Comput. Biol. Med.125, 103958 (2020). 10.1016/j.compbiomed.2020.103958 [DOI] [PubMed] [Google Scholar]
- 88.Teji, J. S., Jain, S., Gupta, S. K. & Suri, J. S. NeoAI 1.0: Machine learning-based paradigm for prediction of neonatal and infant risk of death. Comput. Biol. Med.147, 105639 (2022). 10.1016/j.compbiomed.2022.105639 [DOI] [PubMed] [Google Scholar]
- 89.Saxena, S. et al. Fused deep learning paradigm for the prediction of o6-methylguanine-DNA methyltransferase genotype in glioblastoma patients: A neuro-oncological investigation. Comput. Biol. Med.10, 106492 (2023). 10.1016/j.compbiomed.2022.106492 [DOI] [PubMed] [Google Scholar]
- 90.Acharya, U. R. et al. GyneScan: An improved online paradigm for screening of ovarian cancer via tissue characterization. Technol. Cancer Res. Treatm.13(6), 529–539 (2014). 10.7785/tcrtexpress.2013.600273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Umer, S., Dhara, B. C. & Chanda, B. Texture code matrix-based multi-instance iris recognition. Pattern Anal. Appl.19, 283–295 (2016). 10.1007/s10044-015-0482-2 [DOI] [Google Scholar]
- 92.Acharya, U. R. et al. Ovarian tumor characterization using 3D ultrasound. Technol. Cancer Research Treatm.11(6), 543–552 (2012). 10.7785/tcrt.2012.500272 [DOI] [PubMed] [Google Scholar]
- 93.Acharya, U. R., Faust, O., Sree, S. V., Molinari, F. & Suri, J. S. ThyroScreen system: High resolution ultrasound thyroid image characterization into benign and malignant classes using novel combination of texture and discrete wavelet transform. Comput. Methods Prog. Biomed.107(2), 233–241 (2012). 10.1016/j.cmpb.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 94.Acharya, U. R. et al. Cost-effective and non-invasive automated benign & malignant thyroid lesion classification in 3D contrast-enhanced ultrasound using combination of wavelets and textures: A class of ThyroScan™ algorithms. Technol. Cancer Res. Treatm.10(4), 371–380 (2011). 10.7785/tcrt.2012.500214 [DOI] [PubMed] [Google Scholar]
- 95.Suri, J. S. et al., Symptomatic vs. asymptomatic plaque classification in carotid ultrasound (2011). [DOI] [PubMed]
- 96.Acharya, U. R. et al. Data mining framework for fatty liver disease classification in ultrasound: A hybrid feature extraction paradigm. Med. Phys.39(7), 4255–4264 (2012). 10.1118/1.4725759 [DOI] [PubMed] [Google Scholar]
- 97.Shrivastava, V. K., Londhe, N. D., Sonawane, R. S. & Suri, J. S. Reliable and accurate psoriasis disease classification in dermatology images using comprehensive feature space in machine learning paradigm. Expert Syst. Appl.42(15–16), 6184–6195 (2015). 10.1016/j.eswa.2015.03.014 [DOI] [Google Scholar]
- 98.Acharya, U. R. et al. Evolutionary algorithm-based classifier parameter tuning for automatic ovarian cancer tissue characterization and classification. Ultraschall in der Medizin-Eur. J. Ultrasound35(03), 237–245 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Biswas, M. et al. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput. Methods Prog. Biomed.155, 165–177 (2018). 10.1016/j.cmpb.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 100.Acharya, U. et al. Diagnosis of Hashimoto’s thyroiditis in ultrasound using tissue characterization and pixel classification. Proc. Inst. Mech. Eng. Part H: J. Eng. Med.227(7), 788–798 (2013). 10.1177/0954411913483637 [DOI] [PubMed] [Google Scholar]
- 101.Rodrigues, P. S., Giraldi, G. A., Provenzano, M., Faria, M. D., Chang, R. F. & Suri, J. S. A new methodology based on q-entropy for breast lesion classification in 3-D ultrasound images. In 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, 2006: IEEE, pp. 1048–1051. [DOI] [PubMed]
- 102.Burgin, M. Inductive complexity and shannon entropy. In Information and Complexity: World Scientific, 2017, pp. 16–32.
- 103.Zurek, W. H. Algorithmic randomness and physical entropy. Phys. Rev. A40(8), 4731 (1989). 10.1103/PhysRevA.40.4731 [DOI] [PubMed] [Google Scholar]
- 104.Roach, T. N., Nulton, J., Sibani, P., Rohwer, F. & Salamon, P. Entropy in the tangled nature model of evolution. Entropy19(5), 192 (2017). 10.3390/e19050192 [DOI] [Google Scholar]
- 105.Acharya, U. R. et al. Linear and nonlinear analysis of normal and CAD-affected heart rate signals. Comput. Methods Prog. Biomed.113(1), 55–68 (2014). 10.1016/j.cmpb.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 106.Rout, R. K., Hassan, S. S., Sindhwani, S., Pandey, H. M. & Umer, S. Intelligent classification and analysis of essential genes using quantitative methods. ACM Trans. Multimedia Comput. Commun. Appl. (TOMM)16(1), 1–21 (2020). [Google Scholar]
- 107.Acharya, U. R., Sree, S. V., Ang, P. C. A., Yanti, R. & Suri, J. S. Application of non-linear and wavelet based features for the automated identification of epileptic EEG signals. Int. J. Neural Syst.22(02), 1250002 (2012). 10.1142/S0129065712500025 [DOI] [PubMed] [Google Scholar]
- 108.Li, W. & Kaneko, K. Long-range correlation and partial 1/fα spectrum in a noncoding DNA sequence. Europhys. Lett.17(7), 655 (1992). 10.1209/0295-5075/17/7/014 [DOI] [Google Scholar]
- 109.Arneodo, A. et al. What can we learn with wavelets about DNA sequences?. Phys. A Stat. Mech. Appl.249(1–4), 439–448 (1998). 10.1016/S0378-4371(97)00504-9 [DOI] [Google Scholar]
- 110.Carbone, A., Castelli, G. & Stanley, H. E. Time-dependent Hurst exponent in financial time series. Phys. A Stat. Mech. Appl.344(1–2), 267–271 (2004). 10.1016/j.physa.2004.06.130 [DOI] [Google Scholar]
- 111.Rout, R. K., Pal Choudhury, P., Maity, S. P., Daya Sagar, B. & Hassan, S. S. Fractal and mathematical morphology in intricate comparison between tertiary protein structures. Comput. Methods Biomech. Biomed. Eng. Imaging Vis.6(2), 192–203 (2018). 10.1080/21681163.2016.1214850 [DOI] [Google Scholar]
- 112.Upadhayay, P. D., Agarwal, R. C., Rout, R. K. & Agrawal, A. P. Mathematical Characterization of Membrane Protein Sequences of Homo-Sapiens. in 2019 9th International Conference on Cloud Computing, Data Science & Engineering (Confluence), 2019: IEEE, pp. 382–386.
- 113.Cattani, C. Fractals and hidden symmetries in DNA. Math. Probl. Eng.20, 10 (2010). [Google Scholar]
- 114.Rout, R. K., Ghosh, S. & Choudhury, P. P. Classification of mer proteins in a quantitative manner. Int. J. Comput. Appl. Eng. Sci.10, 2 (2014). [Google Scholar]
- 115.Cuadrado-Godia, E. et al. Ranking of stroke and cardiovascular risk factors for an optimal risk calculator design: Logistic regression approach. Comput. Biol. Med.108, 182–195 (2019). 10.1016/j.compbiomed.2019.03.020 [DOI] [PubMed] [Google Scholar]
- 116.Jamthikar, A. et al. Cardiovascular/stroke risk prevention: A new machine learning framework integrating carotid ultrasound image-based phenotypes and its harmonics with conventional risk factors. Indian Heart J.72(4), 258–264 (2020). 10.1016/j.ihj.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shrivastava, V. K., Londhe, N. D., Sonawane, R. S. & Suri, J. S. Exploring the color feature power for psoriasis risk stratification and classification: A data mining paradigm. Comput. Biol. Med.65, 54–68 (2015). 10.1016/j.compbiomed.2015.07.021 [DOI] [PubMed] [Google Scholar]
- 118.Huang, S. et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics15(1), 41–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu, Y., Guo, J., Hu, G. & Zhu, H. Gene prediction in metagenomic fragments based on the SVM algorithm. BMC Bioinform.14, 1–12 (2013). 10.1186/1471-2105-14-S5-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tandel, G. S. et al. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med.122, 103804 (2020). 10.1016/j.compbiomed.2020.103804 [DOI] [PubMed] [Google Scholar]
- 121.Devetyarov, D. & Nouretdinov, I. Prediction with Confidence Based on a Random Forest Classifier. In AIAI 37–44 (Springer, 2010). [Google Scholar]
- 122.Kursa, M. B. Robustness of Random Forest-based gene selection methods. BMC Bioinform.15, 1–8 (2014). 10.1186/1471-2105-15-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goldstein, B. A., Polley, E. C. & Briggs, F. B. Random forests for genetic association studies. Stat. Appl. Genet. Mol. Biol.10, 1 (2011). 10.2202/1544-6115.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sharaff, A. & Gupta, H. Extra-tree classifier with metaheuristics approach for email classification. In Advances in Computer Communication and Computational Sciences: Proceedings of IC4S 2018, 2019: Springer, pp. 189–197.
- 125.Lanjewar, M. G., Parab, J. S., Shaikh, A. Y. & Sequeira, M. CNN with machine learning approaches using ExtraTreesClassifier and MRMR feature selection techniques to detect liver diseases on cloud. Cluster Comput.1, 16 (2022). [Google Scholar]
- 126.Jamthikar, A. D. et al. Multiclass machine learning vs. conventional calculators for stroke/CVD risk assessment using carotid plaque predictors with coronary angiography scores as gold standard: A 500 participants study. Int. J. Cardiovasc. Imaging37, 1171–1187 (2021). 10.1007/s10554-020-02099-7 [DOI] [PubMed] [Google Scholar]
- 127.Pan, F., Wang, B., Hu, X. & Perrizo, W. Comprehensive vertical sample-based KNN/LSVM classification for gene expression analysis. J. Biomed. Inform.37(4), 240–248 (2004). 10.1016/j.jbi.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 128.Li, L., Darden, T. A., Weingberg, C., Levine, A. & Pedersen, L. G. Gene assessment and sample classification for gene expression data using a genetic algorithm/k-nearest neighbor method. Comb. Chem. High Throughput Screen.4(8), 727–739 (2001). 10.2174/1386207013330733 [DOI] [PubMed] [Google Scholar]
- 129.Sharma, A. & Paliwal, K. K. Linear discriminant analysis for the small sample size problem: An overview. Int. J. Mach. Learn. Cybern.6, 443–454 (2015). 10.1007/s13042-013-0226-9 [DOI] [Google Scholar]
- 130.Park, C. H. & Park, H. A comparison of generalized linear discriminant analysis algorithms. Pattern Recogn.41(3), 1083–1097 (2008). 10.1016/j.patcog.2007.07.022 [DOI] [Google Scholar]
- 131.Ahamed, B. S. & Arya, S. LGBM classifier based technique for predicting type-2 diabetes. Eur. J. Mol. Clin. Med.8(3), 454–467 (2021). [Google Scholar]
- 132.Liu, T., Zhang, X., Chen, R., Deng, X. & Fu, B. Development, comparison, and validation of four intelligent, practical machine learning models for patients with prostate-specific antigen in the gray zone. Front. Oncol.13, 1157384 (2023). 10.3389/fonc.2023.1157384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Ferrari, L. & Aitken, S. Mining housekeeping genes with a Naive Bayes classifier. BMC Genomics7(1), 1–14 (2006). 10.1186/1471-2164-7-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jena, B. et al. Artificial intelligence-based hybrid deep learning models for image classification: The first narrative review. Comput. Biol. Med.137, 104803 (2021). 10.1016/j.compbiomed.2021.104803 [DOI] [PubMed] [Google Scholar]
- 135.Das, S. et al. An artificial intelligence framework and its bias for brain tumor segmentation: A narrative review. Comput. Biol. Med.10, 5273 (2022). [DOI] [PubMed] [Google Scholar]
- 136.Sharma, N. et al. Segmentation-based classification deep learning model embedded with explainable AI for COVID-19 detection in chest X-ray scans. Diagnostics12(9), 2132 (2022). 10.3390/diagnostics12092132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Divate, M. et al. Deep learning-based pan-cancer classification model reveals tissue-of-Origin specific gene expression signatures. Cancers14(5), 1185 (2022). 10.3390/cancers14051185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu, Y. & Zheng, Y. F. One-against-all multi-class SVM classification using reliability measures. In Proceedings. 2005 IEEE International Joint Conference on Neural Networks, 2005., 2005, vol. 2: IEEE, pp. 849–854.
- 139.Aly, M. Survey on multiclass classification methods. Neural Netw.19(1–9), 2 (2005). [Google Scholar]
- 140.Suri, J. S. et al. COVLIAS 2.0-cXAI: Cloud-based explainable deep learning system for COVID-19 lesion localization in computed tomography scans. Diagnostics12(6), 1482 (2022). 10.3390/diagnostics12061482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Agarwal, M. et al. Eight pruning deep learning models for low storage and high-speed COVID-19 computed tomography lung segmentation and heatmap-based lesion localization: A multicenter study using COVLIAS 2.0. Comput. Biol. Med.146, 105571 (2022). 10.1016/j.compbiomed.2022.105571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Saba, L. et al. Intra-and inter-operator reproducibility analysis of automated cloud-based carotid intima media thickness ultrasound measurement. J. Clin. Diagn. Res.12, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Biswas, M. et al. Deep learning strategy for accurate carotid intima-media thickness measurement: An ultrasound study on Japanese diabetic cohort. Comput. Biol. Med.98, 100–117 (2018). 10.1016/j.compbiomed.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 144.Huang, S.-F. et al. Analysis of tumor vascularity using three-dimensional power Doppler ultrasound images. IEEE Trans. Med. Imaging27(3), 320–330 (2008). 10.1109/TMI.2007.904665 [DOI] [PubMed] [Google Scholar]
- 145.Maniruzzaman, M. et al. Accurate diabetes risk stratification using machine learning: Role of missing value and outliers. J. Med. Syst.42, 1–17 (2018). 10.1007/s10916-018-0940-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kamal, M. S. et al. Alzheimer’s patient analysis using image and gene expression data and explainable-AI to present associated genes. IEEE Trans. Instrum. Meas.70, 1–7 (2021).33776080 10.1109/TIM.2021.3107056 [DOI] [Google Scholar]
- 147.Kamal, M. S., Dey, N., Chowdhury, L., Hasan, S. I. & Santosh, K. Explainable AI for glaucoma prediction analysis to understand risk factors in treatment planning. IEEE Trans. Instrum. Meas.71, 1–9 (2022). 10.1109/TIM.2022.3171613 [DOI] [Google Scholar]
- 148.Marcílio, W. E. & Eler, D. M. From explanations to feature selection: Assessing SHAP values as feature selection mechanism. in 2020 33rd SIBGRAPI conference on Graphics, Patterns and Images (SIBGRAPI), 2020: Ieee, pp. 340–347.
- 149.Lubo-Robles, D., Devegowda, D., Jayaram, V., Bedle, H., Marfurt, K. J. & Pranter, M. J. Machine learning model interpretability using SHAP values: Application to a seismic facies classification task. In SEG International Exposition and Annual Meeting, 2020: SEG, p. D021S008R006.
- 150.Meng, Y., Yang, N., Qian, Z. & Zhang, G. What makes an online review more helpful: An interpretation framework using XGBoost and SHAP values. J. Theor. Appl. Electron. Commerce Res.16(3), 466–490 (2020). 10.3390/jtaer16030029 [DOI] [Google Scholar]
- 151.Cau, R. et al. Machine learning approach in diagnosing Takotsubo cardiomyopathy: The role of the combined evaluation of atrial and ventricular strain, and parametric mapping. Int. J. Cardiol.373, 124–133 (2023). 10.1016/j.ijcard.2022.11.021 [DOI] [PubMed] [Google Scholar]
- 152.Singh, P. & Sharma, A. Interpretation and classification of arrhythmia using deep convolutional network. IEEE Trans. Instrum. Meas.71, 1–12 (2022). [Google Scholar]
- 153.Yousef, M. & Allmer, J. Classification of precursor MicroRNAs from different species based on K-mer distance features. Algorithms14(5), 132 (2021). 10.3390/a14050132 [DOI] [Google Scholar]
- 154.Cao, L. et al. PreLnc: An accurate tool for predicting lncRNAs based on multiple features. Genes11(9), 981 (2020). 10.3390/genes11090981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gu, C. & Li, X. Prediction of disease-related miRNAs by voting with multiple classifiers. BMC Bioinform.24(1), 1–17 (2023). 10.1186/s12859-023-05308-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao, B. & Xue, B. Improving prediction accuracy using decision-tree-based meta-strategy and multi-threshold sequential-voting exemplified by miRNA target prediction. Genomics109(3–4), 227–232 (2017). 10.1016/j.ygeno.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 157.Jiang, L., Zhang, J., Xuan, P. & Zou, Q. BP neural network could help improve pre-miRNA identification in various species. BioMed Res. Int.2016, 2 (2016). 10.1155/2016/9565689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Amin, N., McGrath, A. & Chen, Y.-P.P. FexRNA: Exploratory data analysis and feature selection of non-coding RNA. IEEE/ACM Trans. Comput. Biol. Bioinform.18(6), 2795–2801 (2021). 10.1109/TCBB.2021.3057128 [DOI] [PubMed] [Google Scholar]
- 159.El-Baz, A. & Suri, J. S. Big Data in Multimodal Medical Imaging (CRC Press, 2019). [Google Scholar]
- 160.Project MinE: Study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur. J. Human Genet.26(10), 1537–1546 (2018). [DOI] [PMC free article] [PubMed]
- 161.Moore, A. C., Winkjer, J. S. & Tseng, T.-T. Bioinformatics resources for microRNA discovery. Biomark. Insights10, 29513 (2015). 10.4137/BMI.S29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang, Z.-Y. et al. iLoc-miRNA: Extracellular/intracellular miRNA prediction using deep BiLSTM with attention mechanism. Brief. Bioinform.23(5), bbac395 (2022). 10.1093/bib/bbac395 [DOI] [PubMed] [Google Scholar]
- 163.Li, Z., Zhong, T., Huang, D., You, Z.-H. & Nie, R. Hierarchical graph attention network for miRNA-disease association prediction. Mol. Therapy30(4), 1775–1786 (2022). 10.1016/j.ymthe.2022.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yan, C. et al. PDMDA: Predicting deep-level miRNA–disease associations with graph neural networks and sequence features. Bioinformatics38(8), 2226–2234 (2022). 10.1093/bioinformatics/btac077 [DOI] [PubMed] [Google Scholar]
- 165.Wan, C. & Jones, D. T. Protein function prediction is improved by creating synthetic feature samples with generative adversarial networks. Nat. Mach. Intell.2(9), 540–550 (2020). 10.1038/s42256-020-0222-1 [DOI] [Google Scholar]
- 166.Lan, L. et al. Generative adversarial networks and its applications in biomedical informatics. Front. Public Health8, 164 (2020). 10.3389/fpubh.2020.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wei, K., Li, T., Huang, F., Chen, J. & He, Z. Cancer classification with data augmentation based on generative adversarial networks. Front. Comput. Sci.16, 1–11 (2022). 10.1007/s11704-020-0025-x [DOI] [Google Scholar]
- 168.Wei, R. & Mahmood, A. Recent advances in variational autoencoders with representation learning for biomedical informatics: A survey. Ieee Access9, 4939–4956 (2020). 10.1109/ACCESS.2020.3048309 [DOI] [Google Scholar]
- 169.Gokhale, M., Mohanty, S. K. & Ojha, A. A stacked autoencoder based gene selection and cancer classification framework. Biomed. Signal Process. Control78, 103999 (2022). 10.1016/j.bspc.2022.103999 [DOI] [Google Scholar]
- 170.Betechuoh, B. L., Marwala, T. & Tettey, T. Autoencoder networks for HIV classification. Curr. Sci.91, 11 (2006). [Google Scholar]
- 171.Suri, J. S. et al. COVID-19 pathways for brain and heart injury in comorbidity patients: A role of medical imaging and artificial intelligence-based COVID severity classification: A review. Comput. Biol. Med.124, 103960 (2020). 10.1016/j.compbiomed.2020.103960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to its propriety nature, supporting data cannot be made available openly but are available from the corresponding author on reasonable request.
The datasets generated during and analyzed during the current study are not publicly available due to their propriety nature but are available from the corresponding author on reasonable request.
The code used during the current study are not publicly available due to due to their propriety nature but are available from the corresponding author on reasonable request.