Abstract
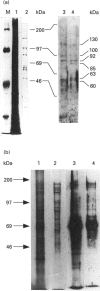
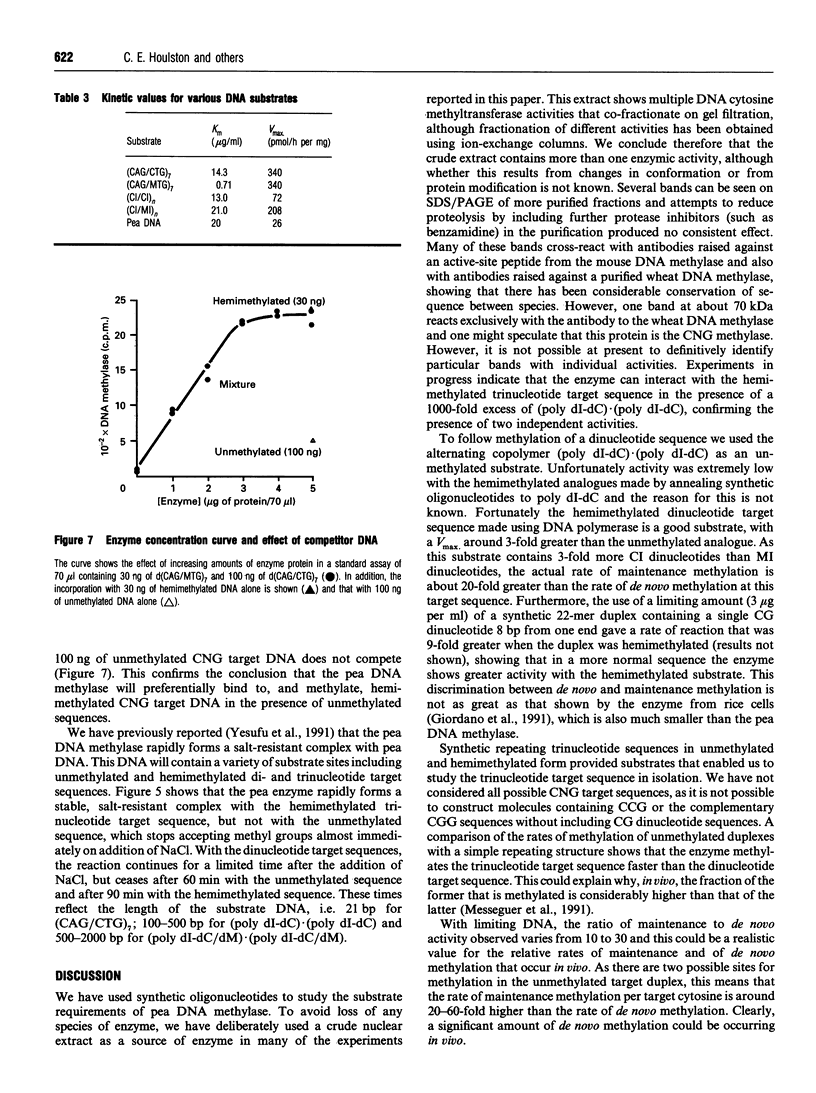
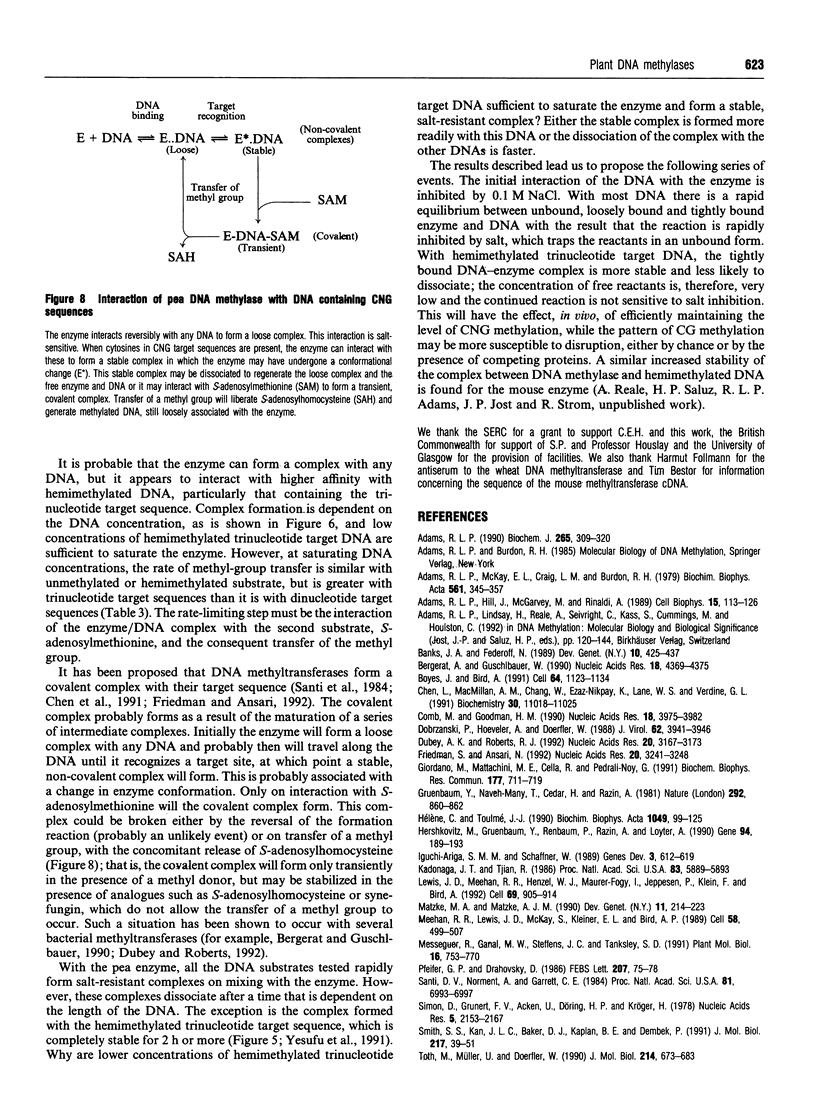
DNA methylase, present in low-salt extracts of nuclei prepared from Pisum sativum shoot tips, methylates model DNA substrates containing CNG trinucleotides or CI dinucleotides only. The binding to the hemimethylated trinucleotide substrates is very much stronger and more persistent than the binding to the unmethylated substrates or to the hemimethylated dinucleotide substrate. When the DNA concentration is limiting, the rate of methyl-group transfer with the hemimethylated CNG substrate is much greater than that with the unmethylated CNG. However, the Vmax. is similar for the two CNG substrates. On fractionation using Q-Sepharose, two peaks of activity are seen with different relative activities using the di- and trinucleotide substrates. The relative activity with these substrates changes during purification, during plant growth and on heating at 35 degrees C as well, indicating that more than one enzyme or more than one form of the enzyme may be present.
Full text
PDF

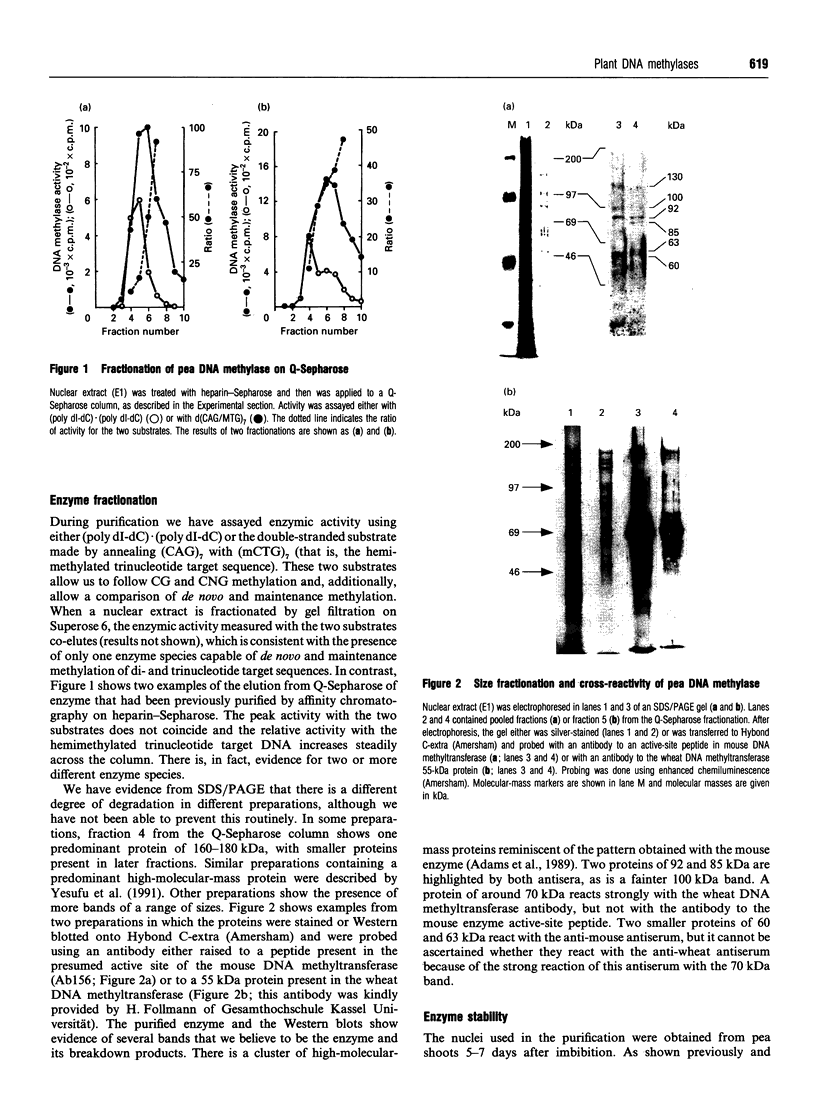
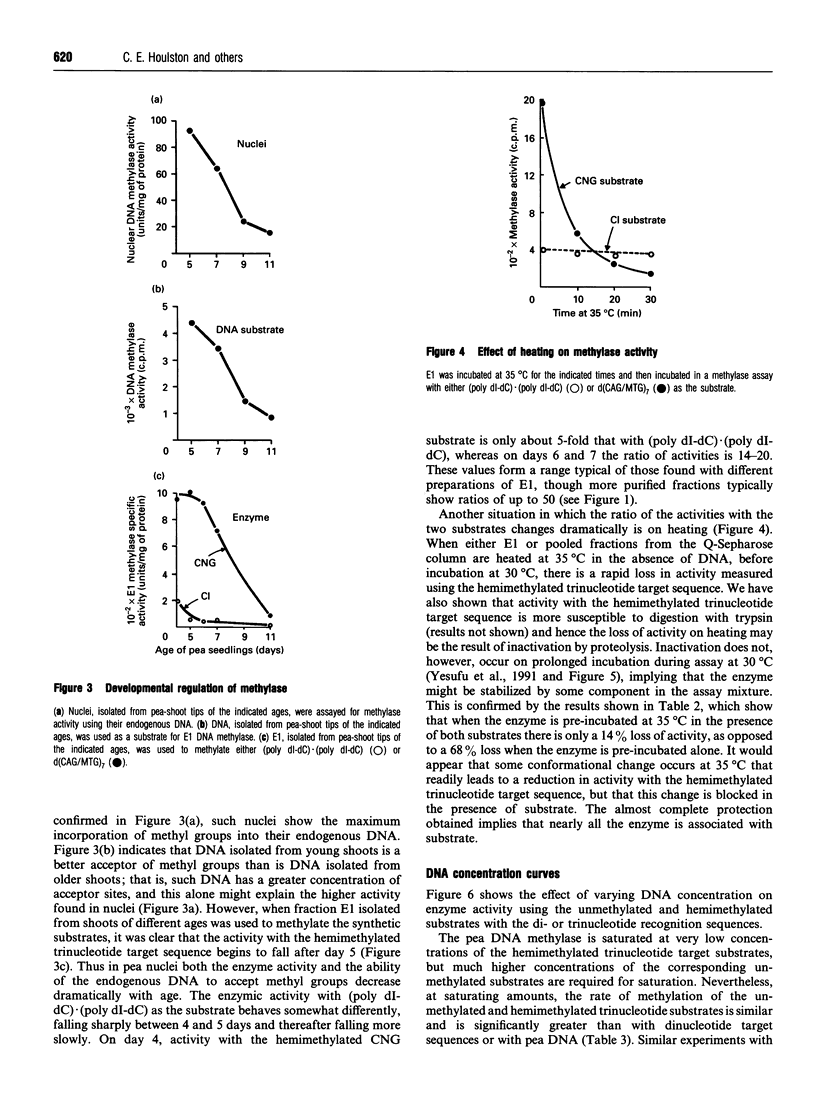
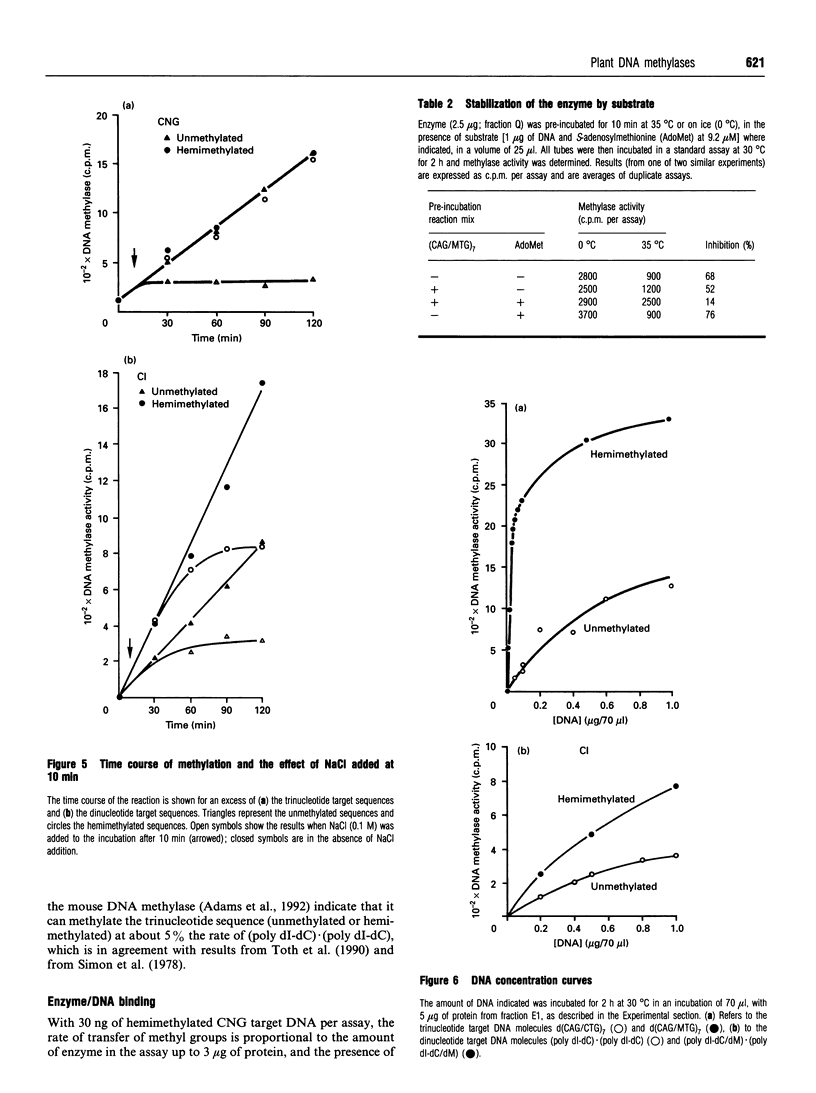

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L. DNA methylation. The effect of minor bases on DNA-protein interactions. Biochem J. 1990 Jan 15;265(2):309–320. doi: 10.1042/bj2650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. L., Hill J., McGarvey J. M., Rinaldi A. Mouse DNA methylase. Intracellular location and degradation. Cell Biophys. 1989 Aug-Oct;15(1-2):113–126. doi: 10.1007/BF02991584. [DOI] [PubMed] [Google Scholar]
- Adams R. L., McKay E. L., Craig L. M., Burdon R. H. Mouse DNA methylase: methylation of native DNA. Biochim Biophys Acta. 1979 Feb 27;561(2):345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- Bergerat A., Guschlbauer W. The double role of methyl donor and allosteric effector of S-adenosyl-methionine for Dam methylase of E. coli. Nucleic Acids Res. 1990 Aug 11;18(15):4369–4375. doi: 10.1093/nar/18.15.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Comb M., Goodman H. M. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990 Jul 11;18(13):3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzanski P., Hoeveler A., Doerfler W. Inactivation by sequence-specific methylations of adenovirus promoters in a cell-free transcription system. J Virol. 1988 Nov;62(11):3941–3946. doi: 10.1128/jvi.62.11.3941-3946.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A. K., Roberts R. J. Sequence-specific DNA binding by the MspI DNA methyltransferase. Nucleic Acids Res. 1992 Jun 25;20(12):3167–3173. doi: 10.1093/nar/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S., Ansari N. Binding of the EcoRII methyltransferase to 5-fluorocytosine-containing DNA. Isolation of a bound peptide. Nucleic Acids Res. 1992 Jun 25;20(12):3241–3248. doi: 10.1093/nar/20.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M., Mattachini M. E., Cella R., Pedrali-Noy G. Purification and properties of a novel DNA methyltransferase from cultured rice cells. Biochem Biophys Res Commun. 1991 Jun 14;177(2):711–719. doi: 10.1016/0006-291x(91)91846-5. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Hershkovitz M., Gruenbaum Y., Renbaum P., Razin A., Loyter A. Effect of CpG methylation on gene expression in transfected plant protoplasts. Gene. 1990 Oct 15;94(2):189–193. doi: 10.1016/0378-1119(90)90386-6. [DOI] [PubMed] [Google Scholar]
- Hélène C., Toulmé J. J. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim Biophys Acta. 1990 Jun 21;1049(2):99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989 May;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Messeguer R., Ganal M. W., Steffens J. C., Tanksley S. D. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol Biol. 1991 May;16(5):753–770. doi: 10.1007/BF00015069. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Drahovsky D. Preferential binding of DNA methyltransferase and increased de novo methylation of deoxyinosine containing DNA. FEBS Lett. 1986 Oct 20;207(1):75–78. doi: 10.1016/0014-5793(86)80015-1. [DOI] [PubMed] [Google Scholar]
- Santi D. V., Norment A., Garrett C. E. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Kan J. L., Baker D. J., Kaplan B. E., Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991 Jan 5;217(1):39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- Toth M., Müller U., Doerfler W. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J Mol Biol. 1990 Aug 5;214(3):673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- Van Lier J. J., Smits M. T., Buck H. M. B-Z transition in methylated DNA. A quantum-chemical study. Eur J Biochem. 1983 Apr 15;132(1):55–62. doi: 10.1111/j.1432-1033.1983.tb07324.x. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F., Molloy P. L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988 Sep;2(9):1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Weber H., Ziechmann C., Graessmann A. In vitro DNA methylation inhibits gene expression in transgenic tobacco. EMBO J. 1990 Dec;9(13):4409–4415. doi: 10.1002/j.1460-2075.1990.tb07891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesufu H. M., Hanley A., Rinaldi A., Adams R. L. DNA methylase from Pisum sativum. Biochem J. 1991 Jan 15;273(Pt 2):469–475. doi: 10.1042/bj2730469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. L., Ehrlich K. C., Supakar P. C., Ehrlich M. A plant DNA-binding protein that recognizes 5-methylcytosine residues. Mol Cell Biol. 1989 Mar;9(3):1351–1356. doi: 10.1128/mcb.9.3.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]