Abstract
Background:
The mechanisms driving alopecia areata (AA) are still unclear, hindering development of targeted therapeutics. Specific Th2 targeting with dupilumab in AA provides a unique opportunity to dissect its pathogenesis and explore the role of Th2 pathway.
Methods:
We evaluated changes in scalp biomarkers in AA patients (with and without concomitant atopy) randomized to weekly dupilumab or placebo for 24 weeks, followed by open-label dupilumab for 24 weeks. Changes in biomarker levels were measured at weeks 12, 24, and 48 and were also correlated with clinical hair regrowth.
Results:
At week 24, preceding clinical hair regrowth outcomes, only dupilumab-treated patients presented significant suppression of cellular infiltrates, and multiple Th2-related, markers (CCL13/MCP-4, CCL18/PARC, CCL26/eotaxin-3, CCL24/Eotaxin-2), coupled with significant upregulation in the hair keratins. Th1-related suppression was evident later (week 48) when all patients received open-label dupilumab. Results were more pronounced in atopic AA patients, that showed 48% and 97% improvements in the lesional AA scalp profile at weeks 24 and 48, respectively, while 2% worsening was seen in the placebo arm at week 24. Moreover, placebo-treated patients presented 54% worsening in hair keratins when compared with baseline at week 24. At week 24, increases in hair keratins showed significant correlations only with decreases in Th2-related markers.
Conclusions:
Scalp biomarkers provide evidence of dupilumab efficacy in AA, detected even prior to clinical response, with exclusive correlations between early suppression of Th2 markers and increased hair keratins. These findings strengthen previous reports suggesting a possible role for Th2 cytokines as AA drivers.
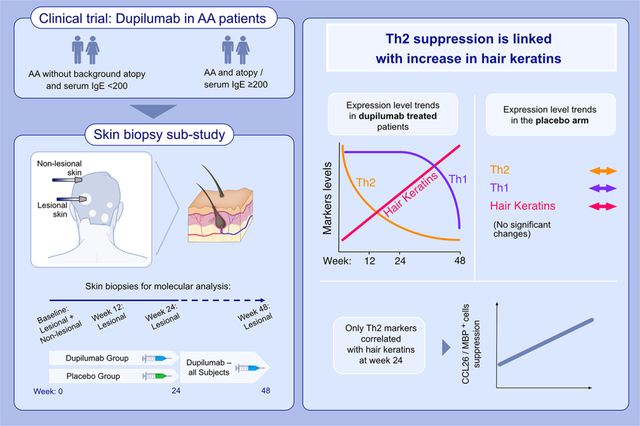
This biopsy sub-study provides a unique opportunity to explore the pathogenicity of Th2-related activation in alopecia areata. Only dupilumab-treated patients presented Th2 pathway suppression, coupled with significant upregulation in hair keratins. Th1-related suppression was evident later (week 48). At week 24, increases in hair keratins correlated only with decreases in Th2-related markers, underscoring the role of Th2 in AA.
Keywords: atopic dermatitis, biologics, clinical immunology, dermatology, IgE
Graphical Abstract
1 |. INTRODUCTION
Alopecia areata (AA) is the most common inflammatory skin disease resulting in reversible hair loss, affecting up to 2% of the population worldwide.1 The pathogenesis of AA is not fully understood,2 hindering the development of specific therapeutics. Thus, despite the great burden of AA on patients’ well-being3 and the increased prevalence of associated anxiety, depression, and even suicidal ideation,4–7 treatment options are limited and only baricitinib has been recently FDA-approved for moderate to severe AA.8
Alopecia areata is induced by a chain of events initiated by an unknown trigger, leading to compromised immune privilege of the hair follicle. The resultant immune attack and inflammatory milieu had been studied in AA animal models,9 but the players of this complex process in human AA are still elusive.
Although AA is traditionally considered as a Th1-related activation compromising immunological tolerability in the hair bulb,10–12 there is strong rationale to also test the role of Th2 antagonism in AA: AA patients have elevated Th2 markers in blood and skin13–16; GWAS studies suggest a role for Th2-related genes in AA pathogenesis17–19; large population studies show strong association between atopic diseases and AA20,21; some AA patients have high levels of serum IgE levels even without background atopy22–24; and anecdotal reports on hair regrowth in patients with concomitant AA and atopic dermatitis (AD) treated with dupilumab.25–27 We thus recently reported the results of a clinical trial in which AA patients were treated with dupilumab, an IL-4R inhibitor blocking Th2 signaling.28
Sixty adult AA patients with and without history of AD were randomized 2:1 to dupilumab or placebo arms for 24 weeks, following another 24 weeks of open-label dupilumab extension period. At week 24, worsening of AA was reported in the placebo arm while AA severity was stabilized in the dupilumab arm. After week 24, gradual, consistent improvement in AA was found across both study arms treated with weekly dupilumab. A sub-analysis on patients with background atopy or elevated serum IgE levels (≥200 IU/ml) showed significantly improved hair regrowth in this sub-population when compared with patients with no atopy and low IgE levels at baseline.28
Herein, to further explore the role of Th2 in AA pathogenesis and the effects of Th2 inhibition on tissue immune- and hair-related biomarkers, we report a biopsy sub-study of this clinical trial. This sub-study includes scalp skin biopsies collected at baseline, at weeks 12 and 24 (during the randomized, placebo-controlled phase), and at week 48 (where all patients were treated with dupilumab), to establish the molecular circuits underlying hair regrowth in AA patients treated with IL-4R antagonism.
2 |. METHODS
2.1 |. Patient characteristics
Sixty participants were randomized in a 2:1 ratio to receive dupilumab or placebo in a randomized, double-blind, multicenter study, which was approved by the institutional review board (IRB) at both study sites (NCT03359356). All patients signed IRB-approved informed consents. Scalp hair loss severity was assessed using SALT, ranging from 0 (no hair loss) to 100 (complete hair loss). Full trial protocol was previously published.28 Scalp skin punch biopsies (4.5 mm) from non-lesional and lesional skin were collected at baseline, and lesional scalp biopsies were collected at weeks 12, 24, and 48.
2.2 |. RNA-sequencing and quantitative real-time polymerase chain reaction
RNA was extracted from skin biopsy samples in Qiazol solution using tissue sonicator and processed downstream with QIAGEN miRNeasy Mini kit, as per company protocol (QIAGEN catalog#217004). Libraries were generated using TruSeq Stranded mRNA Library Prep kit (Illumina). mRNA was first extracted from 240 ng of total RNA using oligo-dT magnetic beads and fragmented at high temperature. A double-stranded cDNA library was then prepared by reverse transcription and second strand synthesis reagents. Indexing adaptors were ligated to the cDNA libraries and amplified on thermocycler. Next-generation sequencing was performed on the amplified libraries using Novaseq 6000 (Illumina Inc.) on 100 cycle single-ended read configuration. Our sequencing depth ranged between 35 and 45 M reads and the reference genome used was GRCh38/hg38.
For RT-PCR, reverse transcription to complementary DNA (cDNA) was carried out using the High-Capacity cDNA reverse transcription (Thermofisher). Pre-amplification was performed on all samples. TaqMan Low Density Array (TLDA) cards (Thermofisher) were used for quantitative RT-PCR (qRT-PCR). Primers are listed in Table S1. 100 ng total RNA was used for reverse transcription and resulting cDNA was used for PreAMP pool and TLDA. Eukaryotic 18 S recombinant RNA was used as an endogenous control. Expression values were normalized to Rplp0.
2.3 |. Immunohistochemistry
Immunohistochemistry was performed on frozen skin sections as previously described29,30 using antibodies as listed in Table S2. Counts were performed on full-depth biopsy sections including hair follicles (where applicable), and the average count per section across these X20 images was included for analysis. Cell counts were quantified using ImageJ V1.42 software (National Institutes of Health).
2.4 |. Statistical analysis
The statistical language R (www.r-project.org) was used to perform all analyses. Next-generation sequencing was performed on Illumina NovaSeq 6000 with single-ended 100 red cycles. RNA-seq sample quality was assessed using FastQC (Cambridge, United Kingdom) and MultiQC.31 Samples were aligned to the human reference genome using the STAR RNA-seq aligner,32 and sequencing reads were assigned to genomic features by featureCounts.33 Data were subsequently log2 transformed with voom transform34 and modeled using a mixed-effects model with time, treatment, and tissue as a fixed effect and random effect (using the R limma package) for each patient. The Benjamini-Hochberg procedure was used to adjust p values for multiple hypotheses by controlling the false discovery rate (FDR). Genes with fold change (|FCH|) > 1.5 and FDR < 0.05 in any post-versus pretreatment comparison were considered differentially expressed. qPCR values were normalized to Rplp0 by negatively transforming cycle threshold (Ct) values to dCt. Immune gene–specific subsets were quantified using gene-set variation analysis, a method of unsupervised sample-wise enrichment that results in an activity score for a subset of genes or pathway for each sample. Modeling was performed using the same approach described for single genes. The association of biomarkers and gene-sets with clinical responses was evaluated using Pearson correlation coefficients. Unsupervised clustering analysis using Pearson correlation and a McQuitty agglomeration scheme was performed.
3 |. RESULTS
3.1 |. Patient characteristics and clinical response
Of 60 patients included in the clinical trial (40 in the dupilumab arm and 20 in the matching placebo arm),28 56 were included in the biopsy sub-analysis at baseline: 38 and 18 in the dupilumab and placebo arms, respectively. Of these, 27 and 10 participants (in the corresponding arms) also donated baseline non-lesional scalp sample (not collected from patients with alopecia totalis or universalis). Participants’ characteristics are presented in Table 1.
TABLE 1.
Characteristics of patients with baseline lesional scalp biopsies
| Placebo (N = 18) | Dupilumab (N = 38) | p-value | |
|---|---|---|---|
| Mean age, years (SD) | 46.3 (12.5) | 41.7 (14.0) | .24 |
| Female sex, N (%) | 11 (61.1) | 28 (73.7) | .34 |
| Race, N (%) | |||
| White | 13 (72.2) | 29 (76.3) | .66 |
| African American | 2 (10.5) | 3 (7.9) | |
| Asian | 2 (10.5) | 6 (15.8) | |
| Other | 1 (5.3) | 0 (0) | |
| Mean duration since last hair regrowth, years (SD) | 3.4 (3.1) | 3.8 (2.9) | .7 |
| Mean SALT (SD) | 76.3 (24.4) | 68.9 (27.1) | .34 |
| Patients with BL SALT > 75, N (%) | 10 (55.6) | 18 (47.4) | .57 |
| Patients with AD history, N (%) | 4 (22.2) | 16 (42.1) | .15 |
| Patients with family history of atopy, N (%) | 8 (44.4) | 17 (44.7) | .98 |
| Mean IgE, IU/ml (SD) | 179.3 (309.8) | 508.4 (1209.7) | .27 |
| Patients with IgE > 200 IU/ml, N (%) | 6 (33.3) | 12 (31.6) | .9 |
| Any atopic background (personal/family/ high IgE) | 10 (55.6) | 21 (55.3) | .98 |
| Additional biopsies | |||
| Baseline–non-lesional | 10 | 27 | |
| Week 12–lesional | 36 | 15 | |
| Week 24–lesional | 34 | 14 | |
| Week 48–lesional | 9 | ||
In this clinical trial, while the placebo arm presented AA worsening (a least-squares mean SALT change of −6.5) at week 24, the dupilumab arm presented disease stabilization (SALT change of 2.2) (p < .05).28 At week 48, 32.5%, 22.5%, and 15% of patients achieved SALT30/SALT50/SALT75 improvement, respectively. Baseline serum total IgE levels predicted treatment response with 83% accuracy; of patients with baseline IgE ≥ 200 IU/ml, 53.8%, 46.2%, and 38.5%, achieved SALT30/SALT50/SALT75 improvement, respectively. As this biopsy sub-study includes most of the participants of the original clinical trial (95% and 90% of the dupilumab and placebo arms, respectively), it presents similar clinical results as the full study population.28
3.2 |. Th2-related cells and eosinophil counts are suppressed with dupilumab treatment
Immunostaining was performed to demonstrate changes in inflammatory infiltrates of T-cell (CD3+, CD8+ cells), dendritic cell (DCs) (CD11c+ cells), and Th2-related cell counts (FcER1+ and OX40+ cells, and eosinophils, marked by major basic protein/MBP+) during placebo and dupilumab treatment (Figure 1).
FIGURE 1.
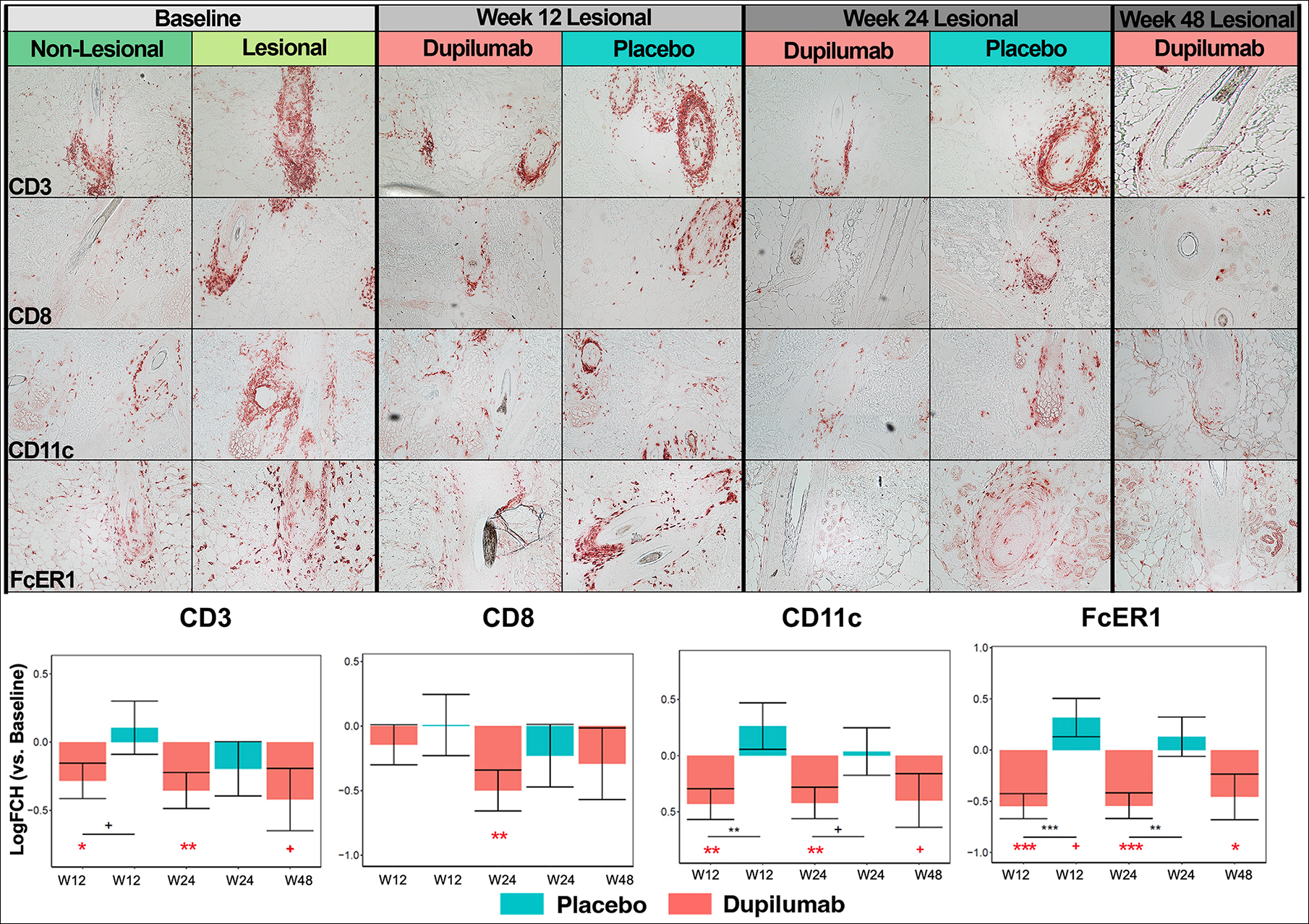
Immunohistochemistry (IHC) of scalp cellular infiltrates in alopecia areata (AA) at baseline and during placebo and dupilumab treatment. Representative images of CD3+ T-cells, CD8+ T-cells, CD11c+ dendritic cells, FcER1+ cells, OX40+ cells, and major basic protein (MBP)+ eosinophils are presented along with cell count plots at the bottom of the figure. W12, week 12; W24, week 24; W48, week 48. +/*/**/*** denote significance (p < .1/.05/.01/.001, respectively) of difference in comparison with baseline (when in red) and in dupilumab versus placebo at a specific timepoint (when in black).
Overall, cellular infiltrates were most commonly identified in proximity to hair follicles, extending into the deep dermis and intra-follicular regions in samples with extensive infiltrates. While CD3+ and CD8+ T-cells showed decreasing counts with dupilumab treatment, no significant changes in counts were recorded in the placebo arm. CD11c+ DC and FcER1+ cell counts decreased as early as week 12 of dupilumab treatment, with significant decreases noted in comparison with baseline counts as well as with counts in the placebo arm at weeks 12 and 24. Similar results were found for OX40+ cells and eosinophils, where infiltrates were largely found in intra-follicular skin (Figure 1).
3.3 |. Global transcriptomic improvement toward the non-lesional signature is only detected in dupilumab-treated AA
RNA-sequencing was performed to assess the global transcriptomic changes in lesional skin versus baseline non-lesional skin across study time-points, using criteria of fold change (FCH) > |1.5| and FDR < 0.05. A gradual improvement was observed in the dupilumab arm from week 12 onward, as indicated by the transition toward the baseline non-lesional signature. However, the placebo arm did not show molecular reversal, as seen in Figure 2A,B, where a minor profile worsening was detected in placebo patients at both weeks 12 and 24 (−3% and −6%, respectively), versus improvements of 17% and 30% in the dupilumab arm at the corresponding weeks, respectively. During the open label extension at week 48 (including those patients treated with placebo in the blinded period, until week 24), an improvement of 63% was detected. Furthermore, while similar dysregulation was seen at baseline in scalp profiles of both dupilumab- and placebo-treated patients, the up- and downregulated lesional profiles of the dupilumab arm has been largely reversed toward that of the non-lesional profile (marked by dotted line), in contrast to the placebo arm (Figure 2C).
FIGURE 2.
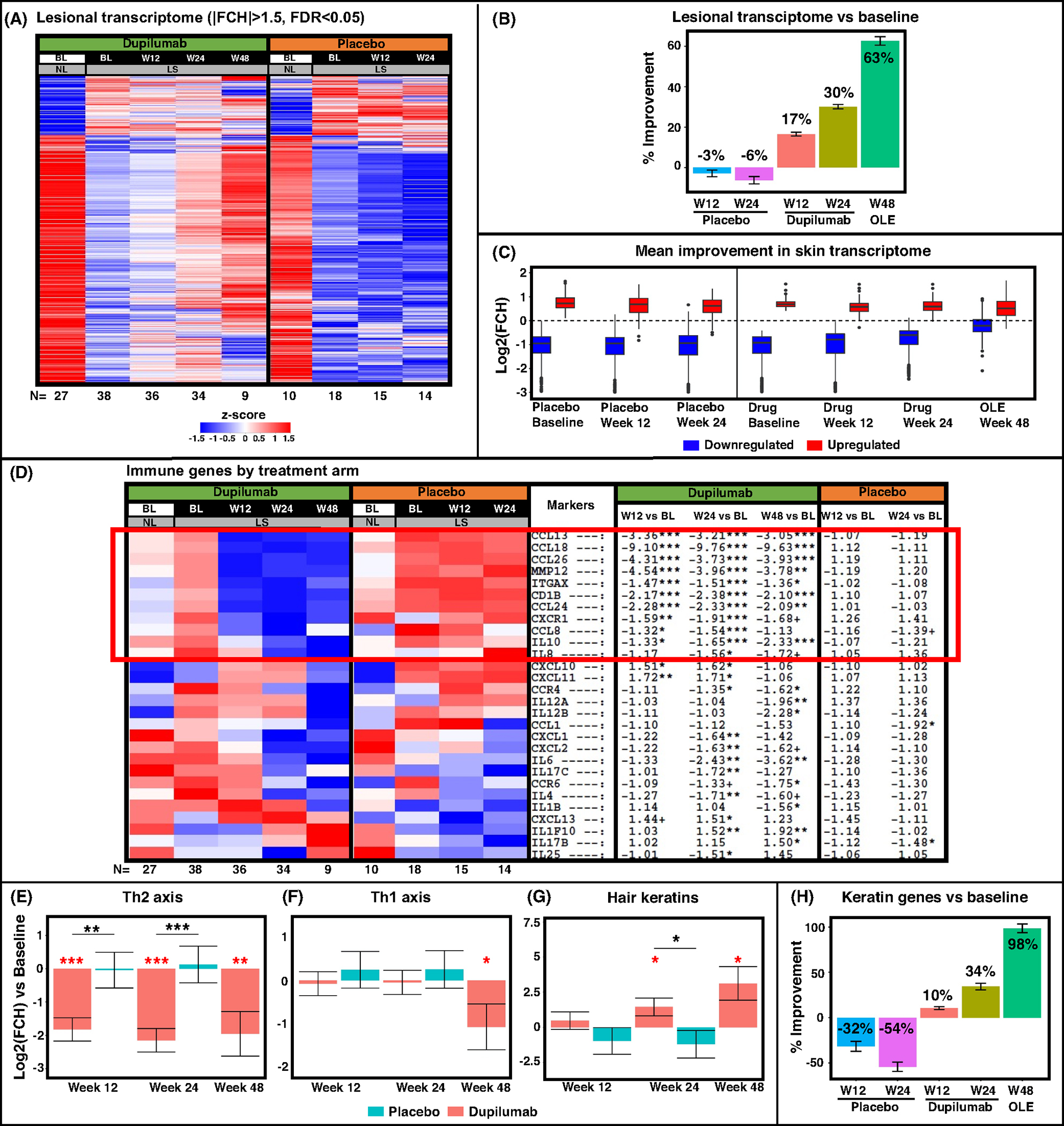
RNA-sequencing results of scalp samples in alopecia areata (AA) at baseline and during placebo and dupilumab treatment. Lesional transcriptome of all differentially expressed genes (DEGs) presented as a heatmap with respective number of participants at the bottom (A), and as a plot indicating percent of improvement in DEGs during treatment (B). Blue and Red boxes denote down- and upregulated DEGs, respectively, across treatment arms during the trial, with a dashed line representing non-lesional scalp at baseline (C). Immune and keratin pathway-specific results are presented as a heatmap of preselected immune genes, including a red box highlighting genes related with dupilumab’s mechanism of action (D), gene-set variation analysis (E, F, G), and a plot of keratin gene-set percent improvement (H). BL, baseline; LS, lesional; NL, non-lesional; OLE, open label extension; W12, week 12; W24, week 24; W48, week 48. +/*/**/*** denote significance (p < .1/.05/.01/.001, respectively) of difference in comparison with baseline (when in red) and in dupilumab versus placebo at a specific timepoint (when in black).
We next evaluated changes in lesional scalp with dupilumab and placebo using a previously published immune gene-set35–37 as shown in a heatmap comparing FCHs versus baseline (Figure 2D). General inflammation (MMP12) and cellular activation (CD1B, ITGAX/CD11c, CXCR1/IL-8RA) markers, multiple Th2-related chemokines (CCL13/MCP-4, CCL18/PARC, CCL26/eotaxin-3, CCL24/Eotaxin-2), and the Th2/regulatory marker IL-10 were all significantly downregulated starting from week 12 in the dupilumab arm (highlighted with a red box, FCH < −1.5, p < .05), with no changes recorded with placebo in these genes. Other markers, representing both innate immunity (e.g., IL-6, IL-8/CXCL8, IL-1B) and cellular activation (e.g., CD69, LAT) showed later response to dupiluamb, starting at week 24 or 48 (Figure 2D and Table S3). Th1-related responses varied as well; few Th1-related markers (e.g., CXCL10/11) showed minimal initial increase with dupilumab treatment, diminished at week 48, while multiple other Th1 markers, including CXCL9, IFNγ, IFNγR1, and CCL2/3/4, were not modulated by dupilumab or placebo at any time point (Table S3). Th17-related markers showed late (e.g., IL-17RA/RB) or no response (e.g., IL-17A, LCN2, PI3/Elafin) to dupilumab treatment. Similarly, most Janus kinase (JAK)/STAT markers (JAK1/2/3, TYK2, STAT3/4/6) were not significantly modulated by the drug (Table S3).
Using Gene-Set Variation Analysis (GSVA) of previously reported gene-sets,35–38 we assessed changes in Th1 and Th2 pathways as well as in hair keratin gene-sets across study arms (Figure 2E–G). While the Th2 axis was significantly downregulated at all timepoints in dupilumab-treated participants (when compared with both placebo and baseline, p < .001), significant suppression of Th1 axis was only detected at week 48 (p < .05). Hair keratins showed upregulation only in the dupilumab arm already at week 24, with further increase at week 48 (p < .05; Figure 2G). More specifically, a 98% improvement in hair keratins was detected on week 48 when compared with baseline, versus a worsening of 54% in the placebo arm at week 24 (Figure 2H).
3.4 |. RT-PCR further delineates Th2 suppression with hair keratins upregulations
To validate RNA-seq data and evaluate mRNA expressions of key inflammatory and hair keratin markers, some of which are not well detected by RNA-seq due to relatively low levels, we performed RT-PCR on a large panel of inflammatory and hair keratin genes (Figure 3 and S1).
FIGURE 3.
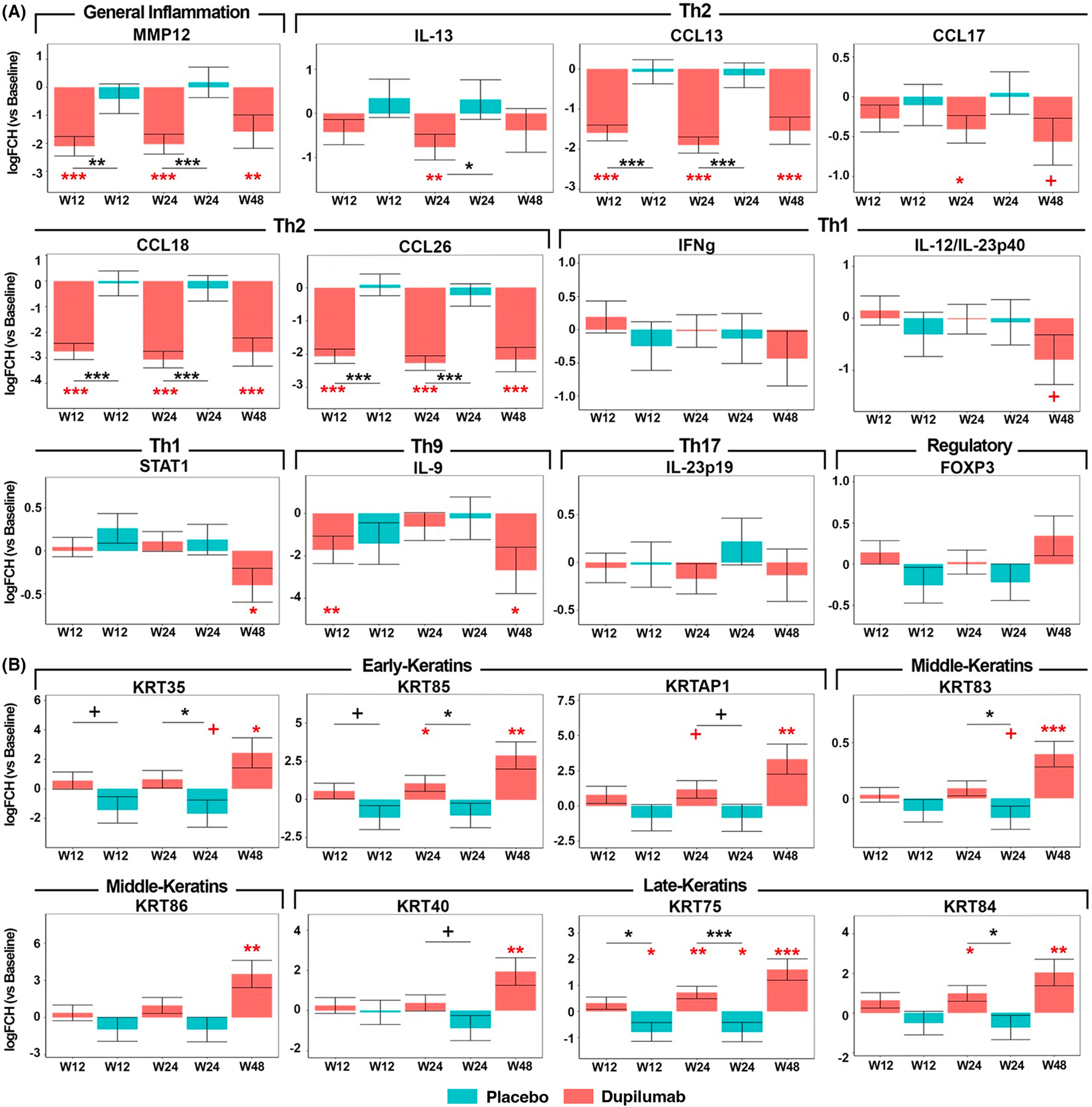
qRT-PCR analysis of selected immune (A) and hair keratin (B) genes in alopecia areata (AA) across treatment arms. Values show log2 FCH expression and are presented as means ± SEMs. W12, week 12; W24, week 24; and W48, week 48. +/*/**/*** denote significance (p < .1/.05/.01/.001, respectively) of difference in comparison with baseline (when in red) and in dupilumab versus placebo at a specific timepoint (when in black).
Overall, PCR data were in line with RNA-seq results, showing early (already at week 12 for most markers) and significant suppression of Th2 cytokines only in dupilumab-treated scalp (CCL13/MCP-4, CCL17/TARC, CCL18/PARC, CCL26/eotaxin-3, IL-13; p < .05). However, Th1 markers, such as IFNγ and IL-12/IL-23p40, showed mild, non-significant and late (at week 48) decreases, except for STAT1, that was significantly downregulated at that timepoint. The Treg marker FOXP3 was minimally modulated across treatment arms, with mild increase in dupilumab-treated participants versus mild decrease in the placebo arm.
Hair keratins showed gradual and significant increases in the dupilumab arm while decreases were seen in the placebo arm (i.e., KRT85, KRT75, and KRT84; p < .05).
3.5 |. Correlation analysis suggests early Th2 response versus late multi-faceted immune modulation in association with SALT change
To associate changes across immune and hair keratin markers with changes in disease severity (measured by change in SALT) and baseline serum IgE levels (a marker predictive of dupilumab response),28 we conducted a Pearson correlation analysis.
At week 24, despite the minimal change in hair regrowth,28 we already detected significant correlations between decreases in Th2 biomarkers (CCL26/eotaxin-3 and eosinophil/MBP+ cell counts) and increases in early, middle and late hair keratins (e.g., KRT85/86/75; r < −.35; p < .05; Figure S2).
Figure 4 presents a heatmap and table of selected robust correlations at week 48 (r > |.8|), showing significant, strong correlations between SALT change and decrease in markers related to general inflammation (MMP12), cellular activation (IL-2RA, IL-15), Th2 (IL-10, CCL18/PARC, CCL13/MCP-4), Th1 (IFNγ, CXCL10), and Tregs (FOXP3) (r > .8, p < .1 for all, highlighted in a green box). Hair keratins were negatively correlated with SALT changes, including KRTAP1, KRT86, depicting early and middle hair keratins, respectively (r ≤ −.8, p ≤ .1). Baseline serum IgE levels were strongly correlated with increase in hair keratins (r ≥ .92, p < .05) and decrease in Th1 (CXCL10, IL-12/IL-23p40), cellular activation (IL-2RA), and Th2 (eosinophils/MBP+ cell counts, CCL18/PARC, CCL13/MCP-4) markers (r ≤ −.84, p < .1).
FIGURE 4.
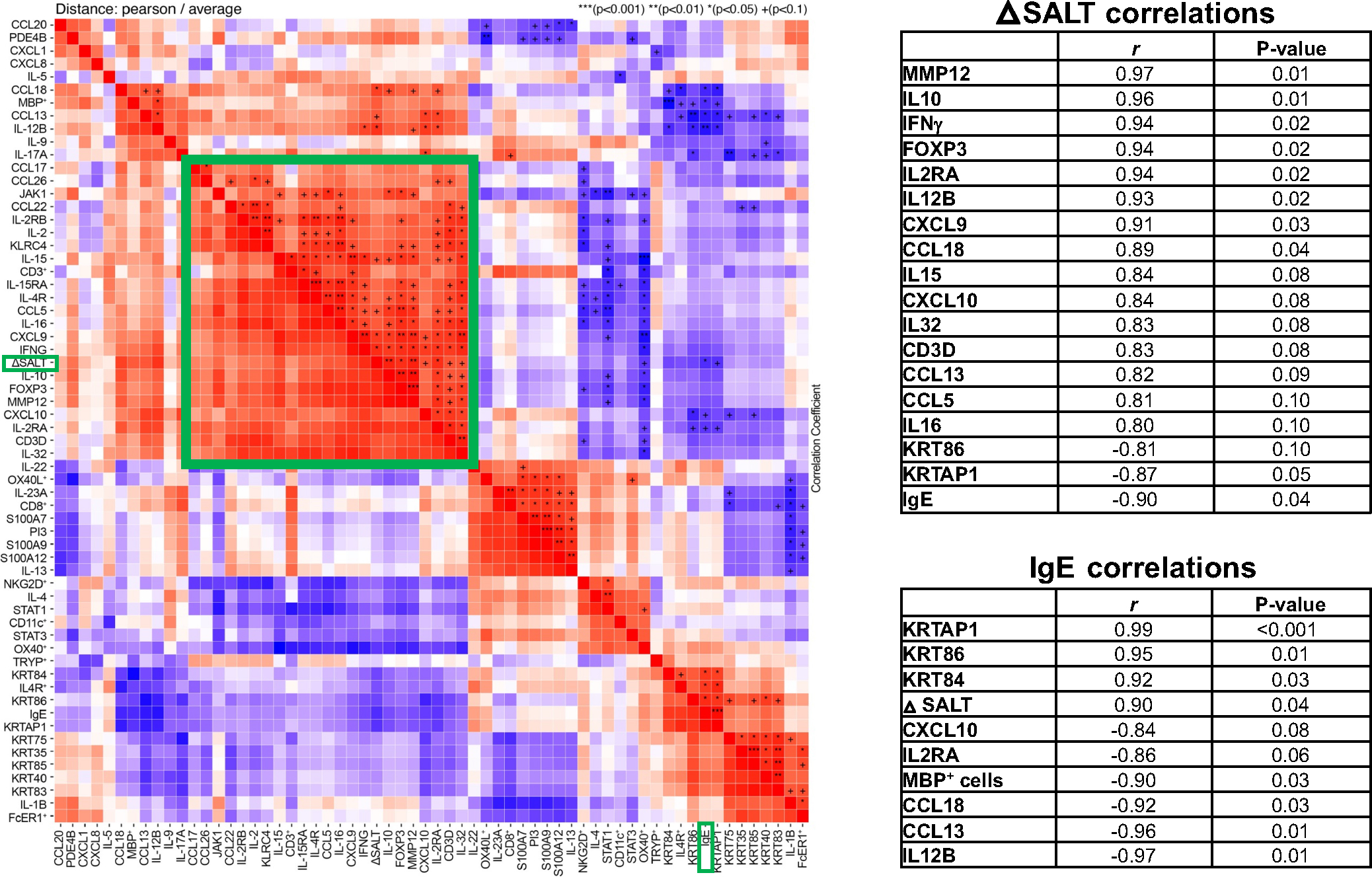
Correlation analysis between qRT-PCR markers, immunohistochemistry (IHC) cell counts, baseline serum IgE levels, and severity of alopecia tool (SALT) change at week 48. Presented as a heatmap (left) with a cluster of tissue markers correlating with SALT highlighted in a green box, and as tables (right) of top correlations with SALT change and baseline serum IgE levels (r ≥ |.8|, p ≤ .1). +/*/**/*** denote significance (p < .1/.05/.01/.001, respectively).
3.6 |. Atopic AA patients present greater tissue improvements with dupilumab
To establish the role of background atopy on dupilumab response in AA, we also stratified patients based on existence of atopy, defined as personal or family atopy and/or baseline serum IgE levels ≥200 IU/ml.
As could be appreciated by the blue to red transition, atopic patients presented a 97% transcriptomic improvement toward non-lesional baseline levels (Figure 5A,B). Moreover, modulation of downregulated genes exceeding baseline non-lesional levels (dashed line, Figure 5C) only in dupilumab-treated participants. In contrast, placebo-treated atopic participants presented an overall worsening, resembling the placebo response found in the entire cohort (Figure 5B).
FIGURE 5.
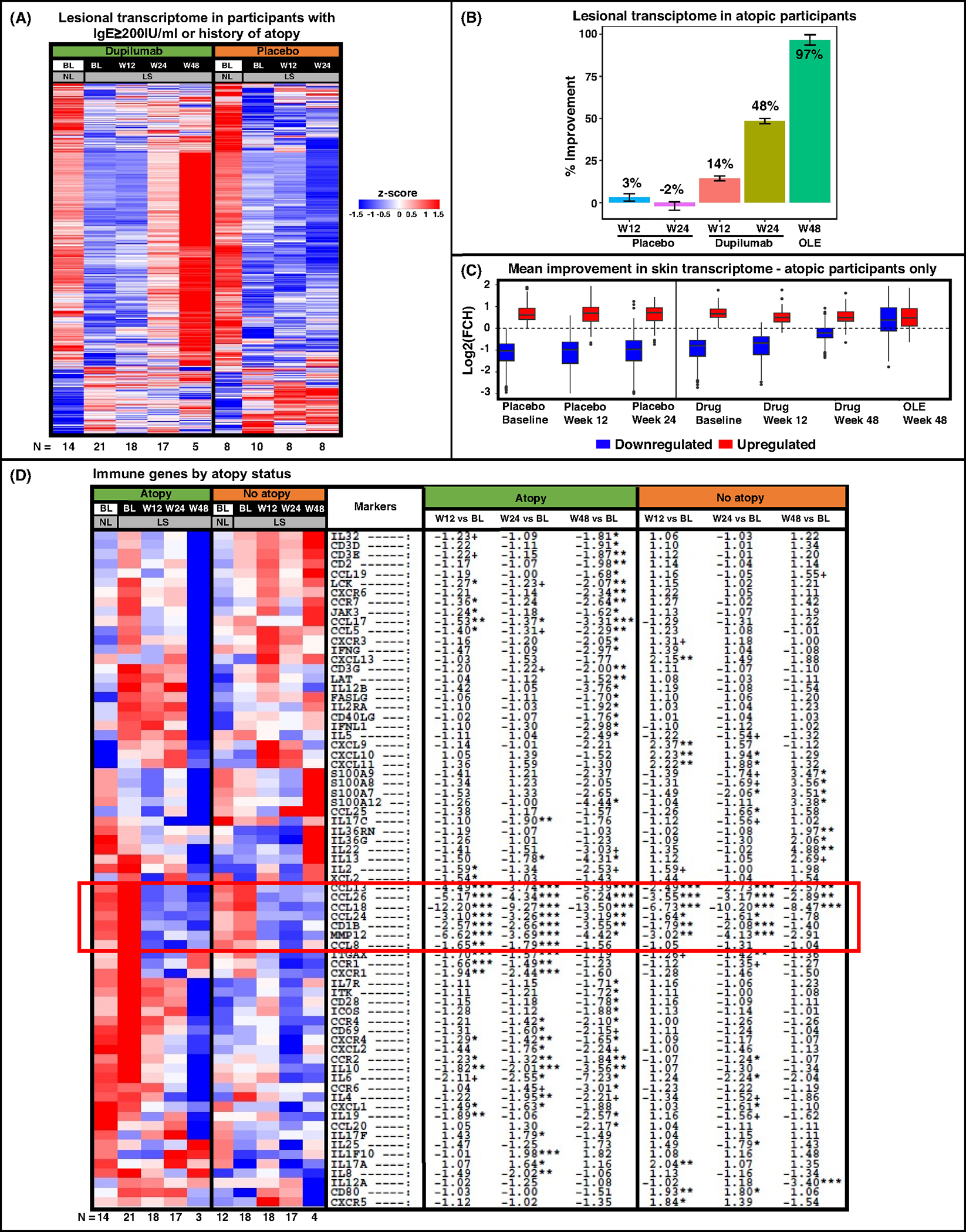
RNA-sequencing results of scalp samples in alopecia areata (AA) by atopic background (determined as personal/family history of atopy or elevated serum IgE levels ≥200 IU/ml). Lesional transcriptome of all differentially expressed genes (DEGs) in atopic participants only presented as a heatmap with respective number of participants at the bottom (A), and as a plot indicating percent of improvement in DEGs during treatment (B). Blue and Red boxes denote down- and upregulated DEGs, respectively, across treatment arms during the trial in atopic participants, with a dashed line representing non-lesional scalp at baseline (C). Immune gene-set presented as a heatmap stratified by atopic background, including a red box highlighting genes related with dupilumab’s mechanism of action (D). BL, baseline; LS, lesional; NL, non-lesional; OLE, open label extension; W12, week 12; W24, week 24; W48, week 48. +/*/**/*** denote significance (p < .1/.05/.01/.001, respectively).
We further evaluated changes in our previously published immune gene-set35–37 in the group of dupilumab-treated patients, stratified by background atopy (Figure 5D). While both atopic and non-atopic participants displayed downregulations of Th2-related markers (CCL13/MCP-4, CCL18/PARC, CCL26/eotaxin-3, CCL24/eotaxin-2) starting from week 12 (highlighted in a red box), other gene modulations differed between the sub-groups. For example, atopic participants presented broader suppression of Th2-related markers (including CCL17/TARC, IL-13, IL-4, and IL-5), along with greater downregulations of innate immunity (IL-6, IL-8/CXCL8) and cellular maturation/activation (LCK, LAT, CD40LG, CD69, ICOS) markers. Moreover, while atopic participants showed late Th1 response (at week 48), including IFNγ, IL-12/IL-23p40, and IFNL1/IL-29, non-atopic participants showed early upregulations of some Th1-related markers (e.g., CXCL9/10/11) with no significant Th1-related downregulations throughout the treatment period. Using GSVA, Th2 suppression was demonstrated across both study arms at weeks 12 and 24 (p < .05), but not at week 48, where only atopic participants showed Th2 attenuation. We found decreased Th1 modulation (approaching significance for the atopic group) only at week 48 (p < .1; Figure 6A).
FIGURE 6.
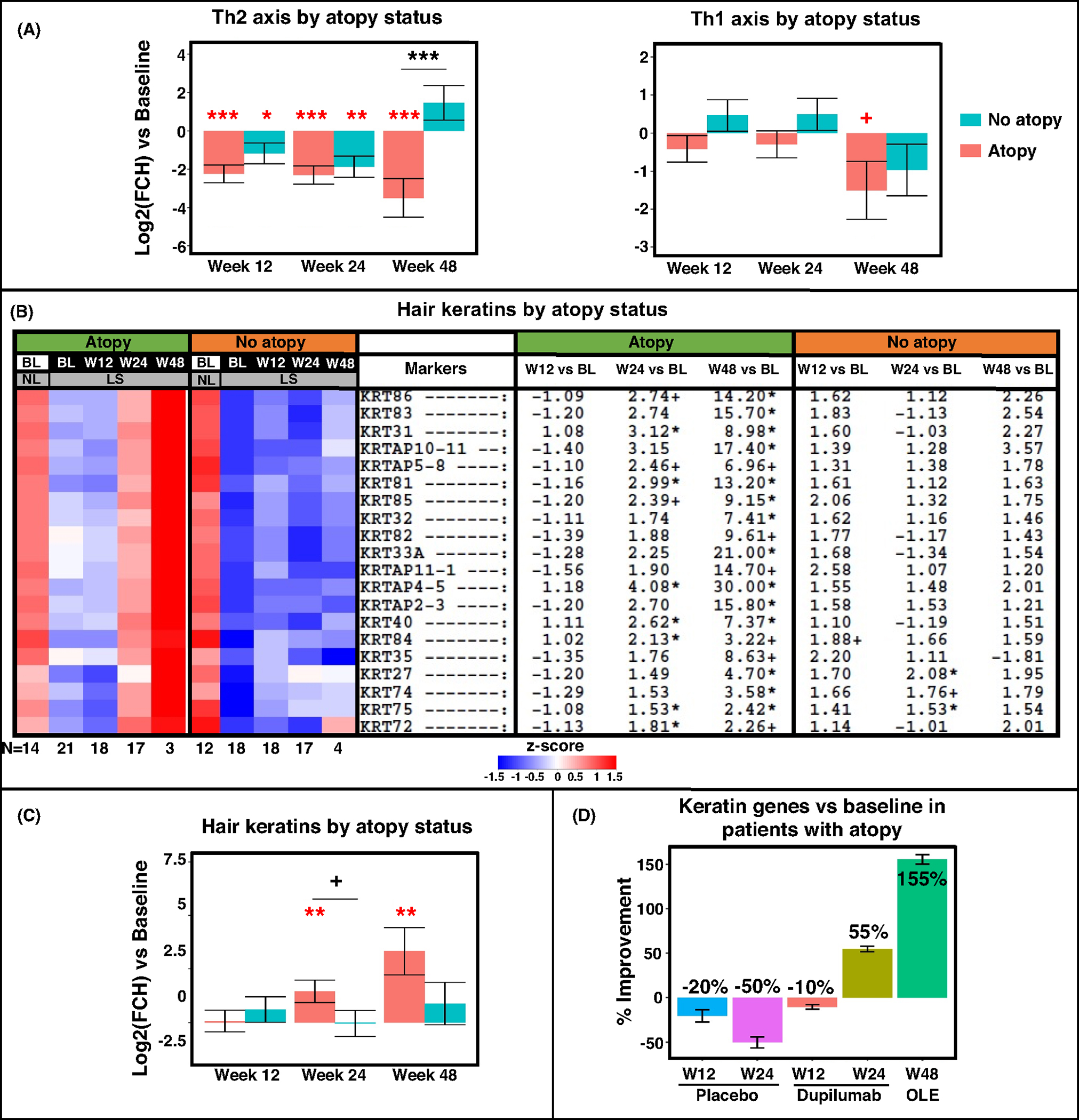
RNA-sequencing results by gene-set pathways from scalp samples by atopic background (determined as personal/family history of atopy or elevated serum IgE levels ≥200 IU/ml). Presented as a gene-set variation analysis (GSVA) of Th2 and Th1 pathways (A), a heatmap and GSVA of hair keratin genes (B, C), and a plot of keratin gene-set percent improvement in atopic participant only (D). BL, baseline; LS, lesional; NL, non-lesional; OLE, open label extension; W12, week 12; W24, week 24; W48, week 48. +/*/**/*** denote significance (p < .1/.05/.01/.001, respectively) of difference in comparison with baseline (when in red) and in dupilumab versus placebo at a specific timepoint (when in black).
A heatmap and GSVA depicting hair keratins by atopic background (Figure 6B,C) show significant upregulation of hair keratins with dupilumab treatment occurring primarily in atopic individuals (e.g., KRT85/86/84), already starting week 24. These trends were not seen in atopic patients treated with placebo. While improvements of 55% and 155% were recorded in dupilumab-treated atopic participants at weeks 24 and 48, respectively, worsening of 50% was recorded in the placebo arm at week 24 (Figure 6D).
4 |. DISCUSSION
While clinical studies with broader-acting agents, such as corticosteroids and even JAK inhibitors, can only inform on the effects of non-specific immune suppression on AA-affected hair follicles, our clinical trial with dupilumab, specifically inhibiting the Th2-pathway, provides a unique opportunity to further dissect the pathogenesis of AA and the role of Th2 skewing in patients with moderate-to-severe AA.
We found that Th2 targeting with dupilumab was able to largely reverse the AA phenotype, while minimal changes and even worsening was noted in the placebo arm at both 24 and 48 weeks. While we observed a global change of 63% in the AA scalp profile, we detected a nearly complete reversal toward non-lesional state (98% improvement) in a previously reported hair keratin gene-subset38 at week 48.
At week 24, the last timepoint in which the control arm was still treated with placebo, only dupilumab-treated patients presented suppression of cellular infiltrates including T-cells, atopic DCs presenting markers related to IgE/Th2 (FcER1+ and OX40+), and eosinophils. Suppression of Th2-related markers (CCL13/MCP-4, CCL18/PARC, CCL26/eotaxin-3, CCL24/Eotaxin-2), but also general inflammation (MMP12), T-cell and DCs (CD1B, ITGAX/CD11c, CXCR1/IL-8RA), and other products were also exclusively detected in the dupilumab arm, starting at week 12. Further, while clinical responses to dupilumab were primarily evident at week 48, significant upregulation in the hair keratin gene-set39 was already evident by week 24, only in dupilumab-treated patients. These results were more pronounced in atopic AA patients (which showed better clinical response to dupilumab),28 who presented with deeper, broader Th2 attenuation when compared with non-atopic patients, along with greater upregulations of hair keratins. Also at week 24, only Th2-related markers were significantly correlated with increases in hair keratins, including CCL26/eotaxin-3 (associated with dupilumab response in skin of AD patients treated with dupilumab)40 and eosinophil scalp infiltrates. On the other hand, at week 24, placebo-treated patients showed a 54% worsening of hair keratins when compared with baseline. The suppression of Th1 immune pathway was only evident at week 48 (with no changes in this pathway seen at week 24), primarily in atopic individuals. The inhibition beyond the Th2 pathway may likely be explained by the fact that although dupilumab is targeting only the IL-4Rα, this receptor can be found on multiple cell types and thus allows wider inhibition of immune cells, including DCs, T-cells, eosinophils, and keratinocytes.41 Taken together, the increase in hair keratins at 24 weeks in dupilumab-treated patients in the setting of a selective Th2 inhibition, along with the exclusive correlations between Th2-related markers and increase in hair keratins and hair regrowth, suggest that Th2 cytokines are likely pathogenic in AA and may contribute to the suppression of hair keratins in AA.16,39
At week 48, robust correlations were found between SALT improvement and changes in multiple scalp markers, including both Th2- (IL-10, CCL18/PARC, CCL13/MCP-4) and Th1-related (IFNγ and CXCL9) markers. These observations also support the role for Th1/IFNγ in perpetuating the AA phenotype,11,12 with a possible scenario where early Th2 inhibition allows restoration of the follicular immune privilege and thus prevents further follicular damage, which is contributed by Th1 activation.
The JAK–STAT family, including JAK1/2/3 and tyrosine kinase 2 (TYK2), had been increasingly studied in AA.42,43 Within this class, JAK1/JAK3 mediate signaling for γ-common cytokines, including IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21.39,44 While JAK inhibition presents promising clinical results in AA, safety concerns were raised due to broad effects on the immune system, especially in the setting of long-term treatment, as needed in AA.45 Recently, a JAK3 inhibitor (ritlecitinib) showed pronounced Th2 suppression, including CCL13/MCP-4, CCL26/eotaxin-3, and CCL18/PARC in an AA clinical trial.39 Furthermore, Th2 markers, including CCL17/TARC, CCL18/PARC, IL-5, and IL-13, showed the strongest correlations with SALT improvement in ritlecitinib-treated AA patients, in both scalp and serum. Interestingly, JAK–S TAT markers, including JAK1, JAK2, JAK3, and TYK2, did not show significant downregulations with dupilumab in our cohort. However, given the mechanism of action of JAK inhibition, blocking γ-common cytokines, it is possible that JAK1/3 inhibition will block IL-4R-related signaling in a way that may be comparable with dupilumab, explaining the similar results obtained across both therapeutics.
The role of Tregs in AA is unknown, with some data showing Treg deficiency,46,47 while others demonstrate mild increase in scalp Tregs in AA,48 perhaps indicating some Treg dysfunction instead of absence, similar to AD.49 Moreover, the lack of efficacy of low-dose IL-2 treatment (which increases Treg population) in AA may support the latter.50 Indeed, the response to dupilumab treatment was not associated with increased Tregs, with no significant changes observed in FOXP3 and other regulatory markers (except IL-10 that is also a Th2-related cytokine).
Limitations of this study include the following: lack of placebo arm after week 24, lack of tissue collections after week 48, and inability to fully dissect the roles of Th2 vs. Th1 immunity to AA pathogenesis.
In conclusion, the results of this study provide insights into the effects of Th2 modulation with dupilumab on the immune and hair keratin dysregulation in AA scalp, particularly in patients with an atopic background. In addition to broad attenuation of Th2 immunity, dupilumab also inhibits additional immune cells and pathways, including inhibition of Th1-related markers as a delayed event. Larger and longer clinical trials with dupilumab in both adults and children with AA are needed to help determine the role of specific Th2 inhibition in the therapeutic paradigm of AA. These studies will also help to fully understand the relative contributions of Th1 and Th2 immune pathways to hair keratin differentiation and synthesis in AA.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank the participats in the study.
FUNDING INFORMATION
YRY was supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health, through Rockefeller University, Grant #UL1TR001866. This investigator-initiated study was supported by a grant from Regeneron/Sanofi.
ROLE OF THE FUNDING SOURCE
This investigator-initiated study was supported by a grant from Regeneron/Sanofi. The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Funding information
National Center for Advancing Translational Sciences; Regeneron Pharmaceuticals
Abbreviations:
- AA
Alopecia areata
- AD
Atopic dermatitis
- DC
Dendritic cell
- IgE
Immunoglobulin E
- JAK
Janus kinase
- SALT
Severity of alopecia tool
- TYK2
tyrosine kinase 2
Footnotes
CONFLICT OF INTEREST
EGY has served as a consultant for AbbVie, Amgen, Allergan, Asana Bioscience, Celgene, Concert, Dermira, DS Biopharma, Escalier, Galderma, Glenmark, Kyowa Kirin, LEO Pharmaceuticals, Lilly, Mitsubishi Tanabe, Novartis, Pfizer, Regeneron, Sanofi, and Union Therapeutics; a member of advisory boards of Allergan, Asana Bioscience, Celgene, DBV, Dermavant, Dermira, Escalier, Galderma, Glenmark, Kyowa Kirin, LEO Pharma, Lilly, Novartis, Pfizer, Regeneron, and Sanofi; and a recipient of research grants from AbbVie, AnaptysBio, AntibioTx, Asana Bioscience, Boehringer-Ingelheim, Celgene, DBV, Dermavant, DS Biopharma, Galderma, Glenmark, Innovaderm, Janssen Biotech, Kiniska Pharma, LEO Pharmaceuticals, Lilly, Medimmune, Sienna Biopharmaceuticals, Novan, Novartis, Ralexar, Regeneron, Pfizer, UCB, and Union Therapeutics. MGL is an employee of Mount Sinai and receives research funds from: Abbvie, Amgen, Arcutis, Avotres, Boehringer Ingelheim, Cara therapeutics, Dermavant Sciences, Eli Lilly, Incyte, Janssen Research & Development, LLC, Ortho Dermatologics, Regeneron, and UCB, Inc., and is a consultant for Aditum Bio, Almirall, AltruBio Inc., AnaptysBio, Arcutis, Inc., Arena Pharmaceuticals, Aristea Therapeutics, Avotres Therapeutics, BiomX, Brickell Biotech, Boehringer-Ingelheim, Bristol-Myers Squibb, Cara Therapeutics, Castle Biosciences, Celltrion, Corevitas, Dermavant Sciences, Evommune, Inc., Facilitatation of International Dermatology Education, Forte Biosciences, Foundation for Research and Education in Dermatology, Hexima Ltd., LEO Pharma, Meiji Seika Pharma, Mindera, Pfizer, Seanergy, Trevi, and Verrica. JGK has received grants paid to The Rockefeller University from Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Dermira, Innovaderm, Janssen, Kadmon, Kineta, Kyowa, LEO Pharma, Lilly, Novartis, Paraxel, Pfizer, Provectus, Regeneron, and Vitae and personal fees from AbbVie, Baxter, Biogen Idec, Boehringer Ingelheim, Bristol Myers Squibb, Delenex, Dermira, Janssen, Kadmon, Kineta, Lilly, Merck, Novartis, Pfizer, Sanofi, Serono, and XenoPort. All other authors declare no competing interests.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Mirzoyev SA, Schrum AG, Davis MDP, Torgerson RR. Lifetime incidence risk of alopecia areata estimated at 2.1% by Rochester epidemiology project, 1990–2009. J Invest Dermatol. 2014;134(4):1141–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajabi F, Drake LA, Senna MM, Rezaei N. Alopecia areata: a review of disease pathogenesis. Br J Dermatol. 2018;179(5):1033–1048. [DOI] [PubMed] [Google Scholar]
- 3.Renert-Yuval Y, Correa da Rosa J, Garcet S, et al. Analysis of alopecia areata surveys suggests a threshold for improved patient-reported outcomes. Br J Dermatol. 2022;187:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velez-Muniz RDC, Peralta-Pedrero ML, Jurado-Santa Cruz F, Morales-Sanchez MA. Psychological profile and quality of life of patients with alopecia Areata. Skin Appendage Disord. 2019;5(5):293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toussi A, Barton VR, Le ST, Agbai ON, Kiuru M. Psychosocial and psychiatric comorbidities and health-related quality of life in alopecia areata: a systematic review. J Am Acad Dermatol. 2020;85:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Titeca G, Goudetsidis L, Francq B, et al. ‘The psychosocial burden of alopecia areata and androgenetica’: a cross-sectional multicentre study among dermatological out-patients in 13 European countries. J Eur Acad Dermatol Venereol. 2020;34(2):406–411. [DOI] [PubMed] [Google Scholar]
- 7.Rencz F, Gulacsi L, Pentek M, Wikonkal N, Baji P, Brodszky V. Alopecia areata and health-related quality of life: a systematic review and meta-analysis. Br J Dermatol. 2016;175(3):561–571. [DOI] [PubMed] [Google Scholar]
- 8.FDA.gov. FDA Approves First Systemic Treatment for Alopecia Areata. https://www.fda.gov/news-events/press-announcements/fda-approves-first-systemic-treatment-alopecia-areata. Accessed on June 29, 2022. [Google Scholar]
- 9.Bertolini M, McElwee K, Gilhar A, Bulfone-Paus S, Paus R. Hair follicle immune privilege and its collapse in alopecia areata. Exp Dermatol. 2020;29(8):703–725. [DOI] [PubMed] [Google Scholar]
- 10.Xing L, Dai Z, Jabbari A, et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med. 2014;20(9):1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freyschmidt-Paul P, McElwee KJ, Hoffmann R, et al. Interferon-gamma-deficient mice are resistant to the development of alopecia areata. Br J Dermatol. 2006;155(3):515–521. [DOI] [PubMed] [Google Scholar]
- 12.Gilhar A, Kam Y, Assy B, Kalish RS. Alopecia areata induced in C3H/HeJ mice by interferon-gamma: evidence for loss of immune privilege. J Invest Dermatol. 2005;124(1):288–289. [DOI] [PubMed] [Google Scholar]
- 13.Song T, Pavel AB, Wen HC, et al. An integrated model of alopecia areata biomarkers highlights both TH1 and TH2 upregulation. J Allergy Clin Immunol. 2018;142(5):1631–1634 e1613. [DOI] [PubMed] [Google Scholar]
- 14.Bain KA, McDonald E, Moffat F, et al. Alopecia areata is characterized by dysregulation in systemic type 17 and type 2 cytokines, which may contribute to disease-associated psychological morbidity. Br J Dermatol. 2020;182(1):130–137. [DOI] [PubMed] [Google Scholar]
- 15.Czarnowicki T, He HY, Wen HC, et al. Alopecia areata is characterized by expansion of circulating Th2/Tc2/Th22, within the skin-homing and systemic T-cell populations. Allergy. 2018;73(3):713–723. [DOI] [PubMed] [Google Scholar]
- 16.Suarez-Farinas M, Ungar B, Noda S, et al. Alopecia areata profiling shows TH1, TH2, and IL-23 cytokine activation without parallel TH17/TH22 skewing. J Allergy Clin Immunol. 2015;136(5):1277–1287. [DOI] [PubMed] [Google Scholar]
- 17.Jagielska D, Redler S, Brockschmidt FF, et al. Follow-up study of the first genome-wide association scan in alopecia areata: IL13 and KIAA0350 as susceptibility loci supported with genome-wide significance. J Invest Dermatol. 2012;132(9):2192–2197. [DOI] [PubMed] [Google Scholar]
- 18.Betz RC, Pforr J, Flaquer A, et al. Loss-of-function mutations in the filaggrin gene and alopecia areata: strong risk factor for a severe course of disease in patients comorbid for atopic disease. J Invest Dermatol. 2007;127(11):2539–2543. [DOI] [PubMed] [Google Scholar]
- 19.Petukhova L, Christiano AM. Functional interpretation of genome-wide association study evidence in alopecia Areata. J Invest Dermatol. 2016;136(1):314–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kridin K, Renert-Yuval Y, Guttman-Yassky E, Cohen AD. Alopecia Areata is associated with atopic diathesis: results from a population-based study of 51,561 patients. J Allergy Clin Immunol Pract. 2020;8(4):1323–1328 e1321. [DOI] [PubMed] [Google Scholar]
- 21.Barahmani N, Schabath MB, Duvic M, National Alopecia Areata Registry. History of atopy or autoimmunity increases risk of alopecia areata. J Am Acad Dermatol. 2009;61(4):581–591. [DOI] [PubMed] [Google Scholar]
- 22.Kasumagic-Halilovic E, Prohic A. Serum levels of total immunoglobulin e in patients with alopecia areata: relationship with clinical type of the disease. Acta Dermatovenerol Croat. 2006;14(3):149–152. [PubMed] [Google Scholar]
- 23.Zhao Y, Zhang B, Caulloo S, Chen X, Li Y, Zhang X. Diffuse alopecia areata is associated with intense inflammatory infiltration and CD8+ T cells in hair loss regions and an increase in serum IgE level. Indian J Dermatol Venereol Leprol. 2012;78(6):709–714. [DOI] [PubMed] [Google Scholar]
- 24.Attia EA, El Shennawy D, Sefin A. Serum interleukin-4 and total immunoglobulin E in nonatopic alopecia Areata patients and HLA-DRB1 typing. Dermatol Res Pract. 2010;2010:503587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada K, Irisawa R, Ito T, Uchiyama M, Tsuboi R. The effectiveness of dupilumab in patients with alopecia areata who have atopic dermatitis: a case series of seven patients. Br J Dermatol. 2020;183(2):396–397. [DOI] [PubMed] [Google Scholar]
- 26.Penzi LR, Yasuda M, Manatis-Lornell A, Hagigeorges D, Senna MM. Hair regrowth in a patient with long-standing alopecia Totalis and atopic dermatitis treated with Dupilumab. JAMA Dermatol. 2018;154(11):1358–1360. [DOI] [PubMed] [Google Scholar]
- 27.Kageyama R, Ito T, Hanai S, et al. Immunological properties of atopic dermatitis-associated alopecia Areata. Int J Mol Sci. 2021;22(5):2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman-Yassky E, Renert-Yuval Y, Bares J, et al. Phase 2a randomized clinical trial of dupilumab (anti-IL-4Ralpha) for alopecia areata patients. Allergy. 2021;77:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tintle S, Shemer A, Suarez-Farinas M, et al. Reversal of atopic dermatitis with narrow-band UVB phototherapy and biomarkers for therapeutic response. J Allergy Clin Immunol. 2011;128(3):583–593. e581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suarez-Farinas M, Tintle SJ, Shemer A, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127(4):954–964. e951–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- 34.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhingra N, Shemer A, Correa da Rosa J, et al. Molecular profiling of contact dermatitis skin identifies allergen-dependent differences in immune response. J Allergy Clin Immunol. 2014;134(2):362–372. [DOI] [PubMed] [Google Scholar]
- 36.Guttman-Yassky E, Ungar B, Noda S, et al. Extensive alopecia areata is reversed by IL-12/IL-23p40 cytokine antagonism. J Allergy Clin Immunol. 2016;137(1):301–304. [DOI] [PubMed] [Google Scholar]
- 37.Ewald DA, Malajian D, Krueger JG, et al. Meta-analysis derived atopic dermatitis (MADAD) transcriptome defines a robust AD signature highlighting the involvement of atherosclerosis and lipid metabolism pathways. BMC Med Genomics. 2015;8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glickman JW, Dubin C, Dahabreh D, et al. An integrated scalp and blood biomarker approach suggests the systemic nature of alopecia areata. Allergy. 2021;76(10):3053–3065. [DOI] [PubMed] [Google Scholar]
- 39.Guttman-Yassky E, Pavel AB, Diaz A, et al. Ritlecitinib and brepocitinib demonstrate significant improvement in scalp alopecia areata biomarkers. J Allergy Clin Immunol. 2022;149(4):1318–1328. [DOI] [PubMed] [Google Scholar]
- 40.Renert-Yuval Y, Thyssen JP, Bissonnette R, et al. Biomarkers in atopic dermatitis-a review on behalf of the international eczema council. J Allergy Clin Immunol. 2021;147(4):1174–1190 e1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman-Yassky E, Bissonnette R, Ungar B, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in atopic dermatitis patients. J Allergy Clin Immunol. 2018;143:155–172. [DOI] [PubMed] [Google Scholar]
- 42.King B, Guttman-Yassky E, Peeva E, et al. A phase 2a randomized, placebo-controlled study to evaluate the efficacy and safety of the oral Janus kinase inhibitors ritlecitinib and brepocitinib in alopecia areata: 24-week results. J Am Acad Dermatol. 2021;85(2):379–387. [DOI] [PubMed] [Google Scholar]
- 43.Phan K, Sebaratnam DF. JAK inhibitors for alopecia areata: a systematic review and meta-analysis. J Eur Acad Dermatol Venereol. 2019;33(5):850–856. [DOI] [PubMed] [Google Scholar]
- 44.Renert-Yuval Y, Guttman-Yassky E. A novel therapeutic paradigm for patients with extensive alopecia areata. Expert Opin Biol Ther. 2016;16(8):1005–1014. [DOI] [PubMed] [Google Scholar]
- 45.Gilhar A, Keren A, Paus R. JAK inhibitors and alopecia areata. Lancet. 2019;393(10169):318–319. [DOI] [PubMed] [Google Scholar]
- 46.Conteduca G, Rossi A, Megiorni F, et al. Single nucleotide polymorphisms in the promoter regions of Foxp3 and ICOSLG genes are associated with alopecia areata. Clin Exp Med. 2014;14(1):91–97. [DOI] [PubMed] [Google Scholar]
- 47.Hamed FN, Astrand A, Bertolini M, et al. Alopecia areata patients show deficiency of FOXP3+CD39+ T regulatory cells and clonotypic restriction of Treg TCRbeta-chain, which highlights the immunopathological aspect of the disease. PLoS ONE. 2019;14(7):e0210308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Duca E, Ruano Ruiz J, Pavel AB, et al. Frontal fibrosing alopecia shows robust T helper 1 and Janus kinase 3 skewing. Br J Dermatol. 2020;183(6):1083–1093. [DOI] [PubMed] [Google Scholar]
- 49.He H, Del Duca E, Diaz A, et al. Mild atopic dermatitis lacks systemic inflammation and shows reduced nonlesional skin abnormalities. J Allergy Clin Immunol. 2020;147:1369–1380. [DOI] [PubMed] [Google Scholar]
- 50.Le Duff F, Bouaziz JD, Fontas E, et al. Low-dose IL-2 for treating moderate to severe alopecia Areata: a 52-week multicenter prospective placebo-controlled study assessing its impact on T regulatory cell and NK cell populations. J Invest Dermatol. 2021;141(4):933–936 e936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


