Abstract
Invasive and superficial infections caused by the Candida species result in significant global morbidity and mortality. As the pathogenicity of these organisms is intimately intertwined with host immune response, therapies to target both the fungus and host inflammation may be warranted. Structural similarities exist between established inhibitors of the NLRP3 inflammasome and those of fungal acetohydroxyacid synthase (AHAS). Therefore, we leveraged this information to conduct an in silico molecular docking screen to find novel polypharmacologic inhibitors of these targets that resulted in the identification of 12 candidate molecules. Of these, compound 10 significantly attenuated activation of the NLPR3 inflammasome by LPS + ATP, while also demonstrating growth inhibitory activity against C. albicans that was alleviated in the presence of exogenous branched chain amino acids, consistent with targeting of fungal AHAS. SAR studies delineated an essential molecular scaffold required for dual activity. Ultimately, 10 and its analog 10a resulted in IC50 (IL-1β release) and MIC50 (fungal growth) values with low μM potency against several Candida species. Collectively, this work demonstrates promising potential of dual-target approaches for improved management of fungal infections.
Keywords: inflammasome, antifungal, AHAS, Candida, NLRP3, dual-target
Graphical Abstract:
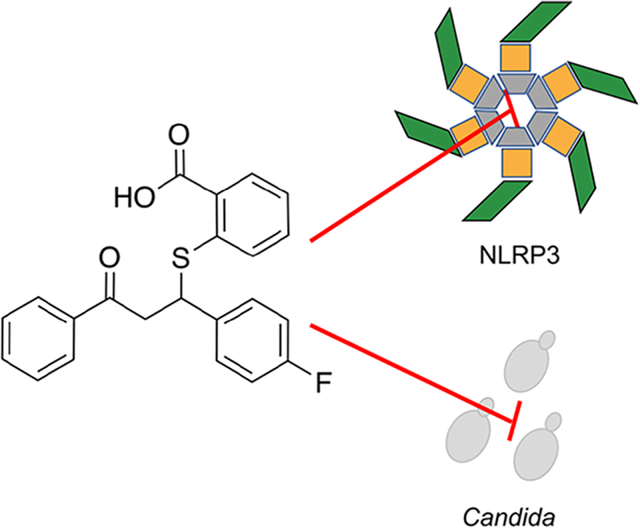
Collectively, Candida albicans and non-albicans Candida (NAC) species result in millions of debilitating invasive and superficial fungal infections each year.1 Systemic candidemia presents major obstacles in the nosocomial setting. Breach of innate biological barriers via invasive medical devices and translocation from the weakened gastrointestinal tract allow for dissemination of Candida throughout the patient, often resulting in multiorgan disease.2 Progression to fungal sepsis, or uncontrolled body-wide damaging inflammatory responses, severely impacts patient prognosis with mortality rates exceeding 40% even with appropriate treatment.3 Although nonlethal, vulvovaginal candidiasis, characterized by itching, burning, and soreness of the vaginal mucosa and skin, is the most prevalent candidal infection.4 In fact, it has been estimated that approximately 75% of women will experience at least one episode of VVC in their lifetime.5 While the disease is limited to the mucosal surface, the overwhelming incidence of such infections in immunocompetent hosts, disease chronicity, and negative impact on quality-of-life command better strategies to control symptomatic disease.6 Therefore, as the antifungal armamentarium remains limited to only three major drug classes (azoles, echinocandins, and polyenes) while the burden of Candida infections steadily increases, new therapeutic strategies are needed to both target the host response to curtail disease symptoms and also reduce fungal burden.7
Over the past decade, a growing body of literature has elucidated a crucial role for the NLRP3 inflammasome in mediating innate inflammatory responses to pathogenic fungi, including the Candida species.8 Inflammasomes are a collection of mammalian cytosolic receptors that, upon sensing of endogenous or exogenous DAMPs, undergo conformational changes to ultimately activate Caspase-1 which cleaves and releases mature interlukein-1 beta (IL-1β) to drive innate immune signaling and neutrophil recruitment. While previously believed to be unique to hematopoietic cells, recent work has shown inflammasomes, including the NLRP3 variety, are functionally expressed in other cell types such as epithelium.9–11 Complete genetic loss of NLRP3 predisposes mice to worsened disseminated candidiasis, while these same mice exhibit reduced immunopathology in a model of VVC.12,13 Moreover, pharmacological targeting of the NLRP3 inflammasome in a variety of infectious and non-infectious disease models has dramatically improved outcome by reducing damaging inflammation.14–17 Therefore, if finely restrained, the NLRP3 inflammasome may be an attractive target to attenuate immune responses to prevent uncontrolled inflammatory disease during fungal infection. However, its inhibition may also reduce fungal recognition and initial containment, so simultaneous control of fungal growth would be appealing.
Prior work in our laboratory demonstrated that the second-generation sulfonylureas used to manage type 2 diabetes mellitus (e.g., glyburide) exhibited the capacity to inhibit both sterile and C. albicans-mediated NLRP3 activation yet were completely ineffective at preventing fungal growth or filamentation.18 Recent work by others have shown that related, but distinct herbicidal sulfonylureas (e.g., chlorimuron ethyl) were effective at reducing growth of the Candida species via targeting of the nonmammalian enzyme acetohydroxyacid synthase (AHAS) that is required for branched chain amino acid biosynthesis.19 However, these drugs are completely ineffective at inhibiting NLRP3 (unpublished data). Given the structural similarities of these compounds, the objective of this study was to determine if chemical entities exist that could potentially inhibit both targets simultaneously. Using a combination of virtual chemical library screening and experimental hit validation approaches, a chemical series was identified (compound 10 and its analogs) and shown to inhibit IL-1β release from macrophage-like cells and statically inhibit the growth of multiple Candida species with reasonable potency. Collectively, these data establish a new class of inflammasome and potential AHAS inhibitor, demonstrate the feasibility of structure-guided dual-targeted drug discovery against these targets, and define scaffolds for further optimization as novel compounds with a desirable polypharmacological profile.
RESULTS
Experimental structures for both human NLRP3 and Saccharomyces cerevisiae AHAS were used to create models for in silico docking studies and known inhibitors employed to inform parameters used for docking algorithms (Figure 1A) and downstream workflow detailed below (Figure 1B).
Figure 1.
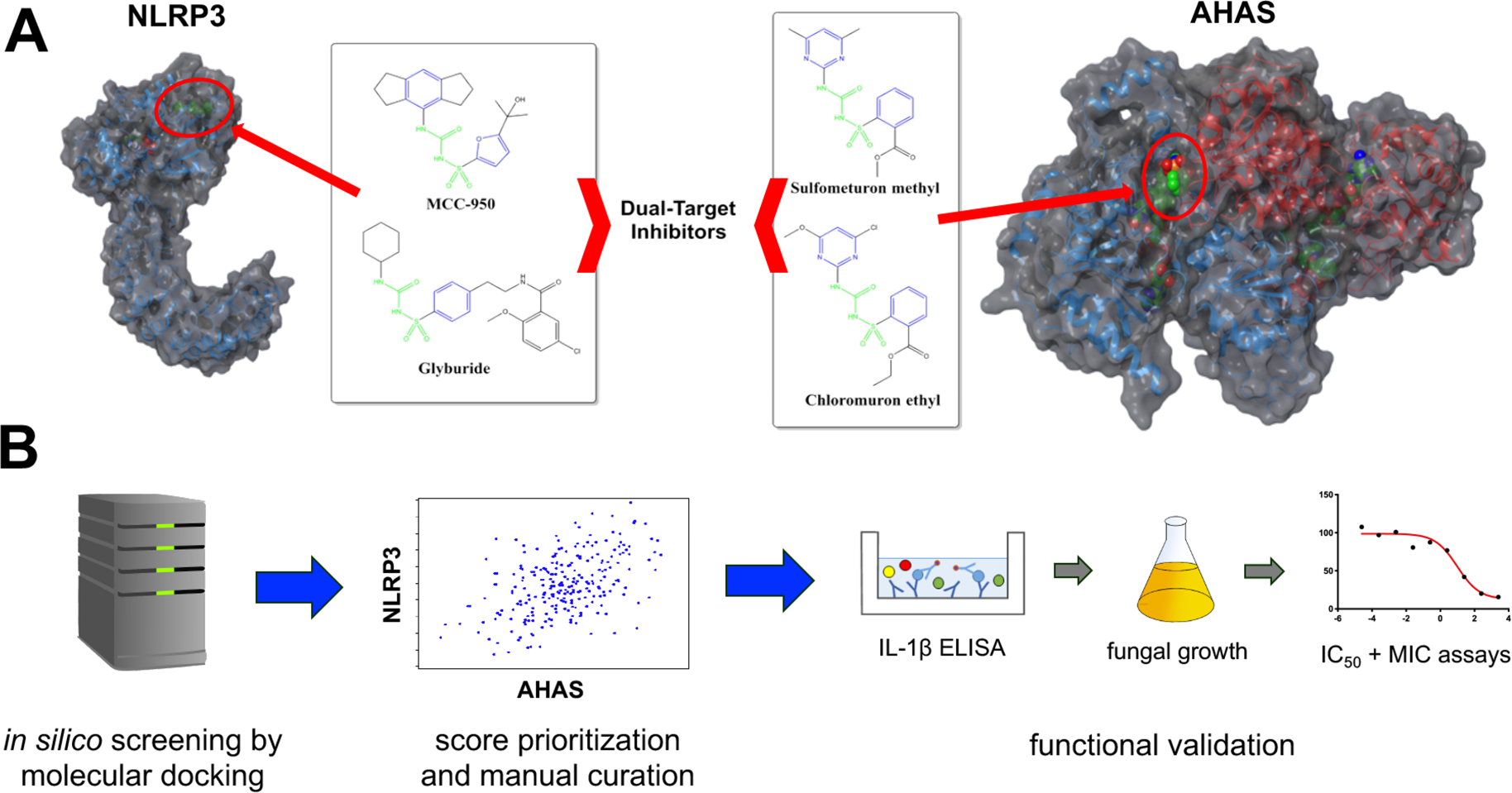
Approach used to identify dual-target inhibitors. (A) Established inhibitors of the NLRP3 inflammasome include sulfonylureas MCC950 and hypoglycemic agents such as glyburide. Inhibitors of yeast acetohydroxyacid synthase (AHAS) include a series of herbicidal sulfonylurea agents including chlorimuron ethyl and sulfometuron. Identical chemical features in both sets of compounds are highlighted in green and similar chemical features are highlighted in blue. The predicted NLRP3 and known AHAS binding sites are indicated by red circles. (B) The Maybridge screening collection of over 53000 compounds was used to perform in silico molecular docking for both NLRP3 and AHAS. Compounds scoring in the top 1% for each target were cross-referenced and common hits were ordered for experimental validation. Generally, compounds were first tested for their capacity to inhibit IL-1β release in THP1 cells stimulated with the inflammasome inducers LPS and ATP. Compounds showing good efficacy were tested for their capacity to inhibit C. albicans growth. Compounds possessing both anti-inflammatory and antifungal properties at a dose of 50 μM were further characterized by establishing inhibitory concentration 50 (IC50) and modified minimal inhibitory concentration (MIC) values.
The conserved aryl sulfonylurea moiety present in both the NLRP3 inhibitor MCC950 and the herbicide inhibitors of AHAS, with a predicted pKa of 3.4 (±2), suggests the compounds bind to their respective targets in an anionic state. Analysis of the experimental AHAS costructure active site and the predicted inhibitor binding pocket of NLRP3 confirmed the presence of a cationic pocket in both targets, bordered by Arg380 and Lys251 in AHAS and Arg268, Lys322, and Arg335 in NLRP3 (Figure 2).
Figure 2.
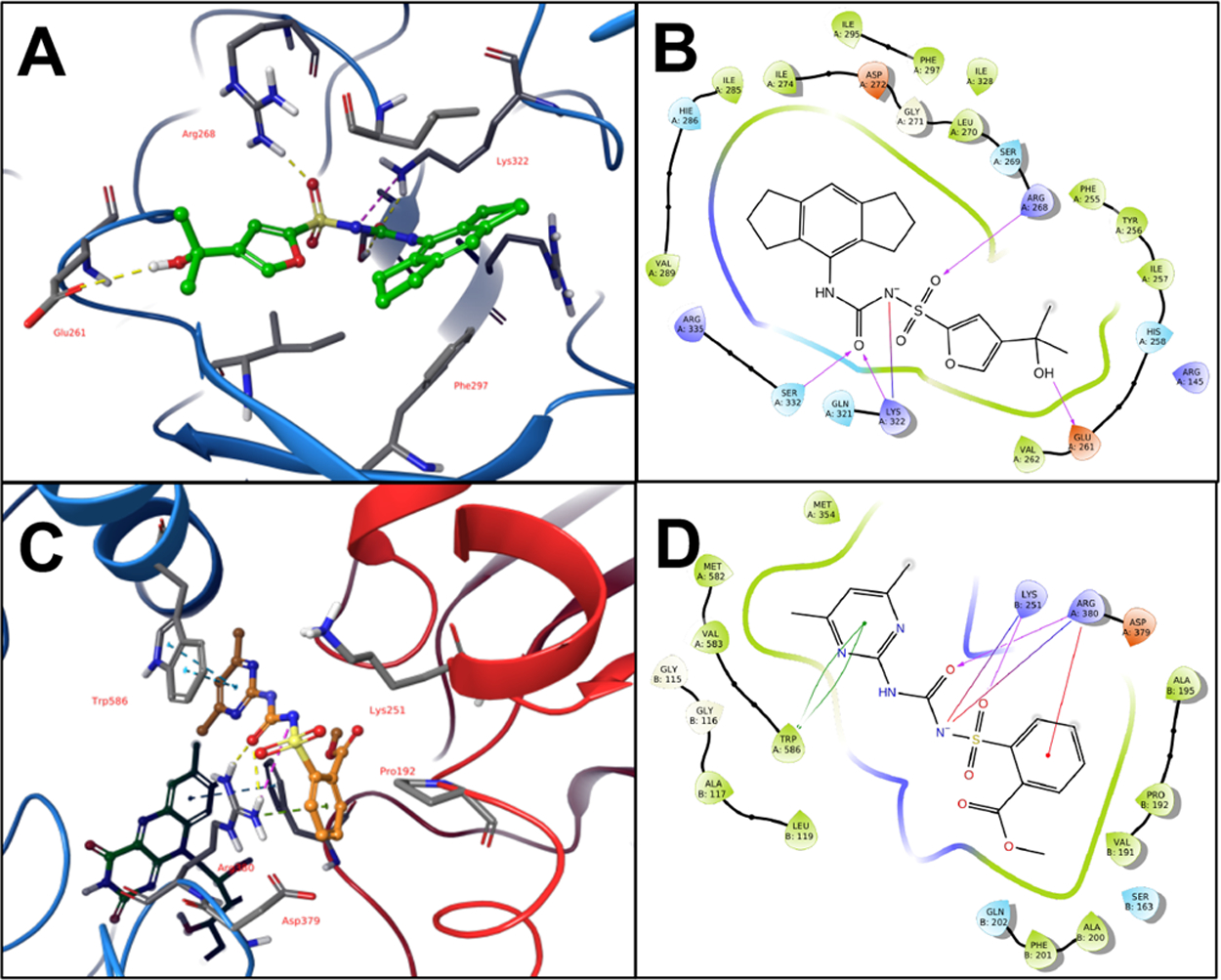
AHAS and NLRP3 Binding Sites. (A) MCC950 and key binding residues in proposed NLRP3 binding site. (B) 2D ligand interaction diagram of MCC950 in NLRP3 binding site. (C) Sulfometuron methyl and key binding residues in Sc-AHAS active site. FAD cofactor visible in background. (D) 2D ligand interaction diagram of sulfometuron methyl in AHAS active site.
Further, key aromatic residues in the binding pockets, Trp586 in AHAS and Phe297 in NLRP3, engage the aromatic rings of the respective inhibitors in pi-stacking interactions. Though the larger hexahydro-indacene ring of MCC950 prevents this compound from binding to the significantly smaller active site of AHAS, the conserved binding interactions and overlapping pharmacophore features of the two inhibitor classes suggested the potential of a structure-guided approach to the identification of dual-targeted compounds. Accordingly, we prepared both structures for molecular docking and screened the commercial Maybridge library of approximately 53000 compounds against each. Manual curation of the top 1% of scored compounds for each target was performed to identify diverse, top-scoring compounds common to both targets and to eliminate compounds with potential undesirable properties (reactive/toxic moieties, penetrability issues, assay interference). This resulted in a final list of 12 compounds predicted to inhibit both NLRP3 and AHAS (Table S1). The general experimental approach that followed was to first determine whether these compounds impacted ligand-based inflammasome activation at moderate concentration (50 μM) to rule out those that displayed no activity. If compounds passed this preliminary hurdle, they were used at the same concentration in growth assays to determine gross antifungal activity. Compounds that passed both parameters were utilized in standard dose–response IC50 and MIC50 assays to more finely resolve anti-inflammatory and antifungal activity, respectively (Figure 1B).
As we have established previously, IL-1β release in THP1 cells challenged with LPS and ATP is strongly NLRP3-dependent as both genetic deletion and inhibition with the known inflammasome inhibitor MCC950 significantly attenuates this response (Figure S1).18 Of the 12 compounds identified in silico, 4 compounds (1, 4, 7, 10) led to significantly decreased IL-1β release, while 5 compounds (2, 3, 8, 9, 12) led to exaggerated responses at 50 μM as compared to the vehicle-treated THP1 cells (Figure 3A). Importantly, treatment with LPS in the absence of ATP failed to release IL-1β demonstrating that release of this cytokine in this assay is dependent on danger sensing, further strengthening an inflammasome-based mechanism. Despite moderate inhibition with several compounds, compound 10 potently inhibited IL-1β release by approximately 90% (Figure 3A). In further support of an NLRP3-dependent inflammasome mechanism of inhibition, compound 10 impaired downstream Caspase-1 activation required for canonical IL-1β processing similar to treatment with the established inhibitor MCC950 (Figure 3B).20 Inflammasome inhibition can also be assessed by monitoring the more upstream process of ASC (an adaptor protein required by all inflammasomes) oligomerization via fluorescence microscopy of large ASC complexes termed “specks”.21 Compound 10 inhibited ASC-Speck formation similar to MCC950 (Figure 3C). Inclusion of compound 10 in a microplate growth assay demonstrated that C. albicans growth was completely inhibited by visible inspection (Figure 3D), confirming its predicted antifungal properties. In order to determine whether antifungal activity was potentially due to AHAS inhibition, growth media was supplemented with the branched chain amino acids isoleucine and valine and growth monitored kinetically. Impaired fungal growth exerted by compound 10 was alleviated in the presence of these amino acids, suggesting that this molecule interferes with branched chain amino acid biosynthesis, consistent with AHAS inhibition (Figure 3E). The ILV2 gene encodes for the putative C. albicans AHAS enzyme.22 We utilized a previously described target abundance-based fitness approach to further probe whether genetic alteration of the ILV2 locus impacts growth inhibition.23 Indeed, strains with altered ILV2 copy number (Δilv2/ILV2) or nonamino acid responsive variable strength promoters (Δ/Δilv2+PrACT1-ILV2 and Δ/Δilv2+Pr-TEF1-ILV2) driving ILV2 expression exhibited approximately 2- to 5-fold increased sensitivity to compound 10 as compared to the WT strain. Reduced ILV2 expression levels were confirmed by qRT-PCR and exhibited an inverse relationship with respect to growth inhibition (Figure 3F). Collectively, these data show that compound 10 has both anti-inflammatory and antifungal properties which can be reasonably inferred by its capacity to engage the binding pocket of both targets.
Figure 3.
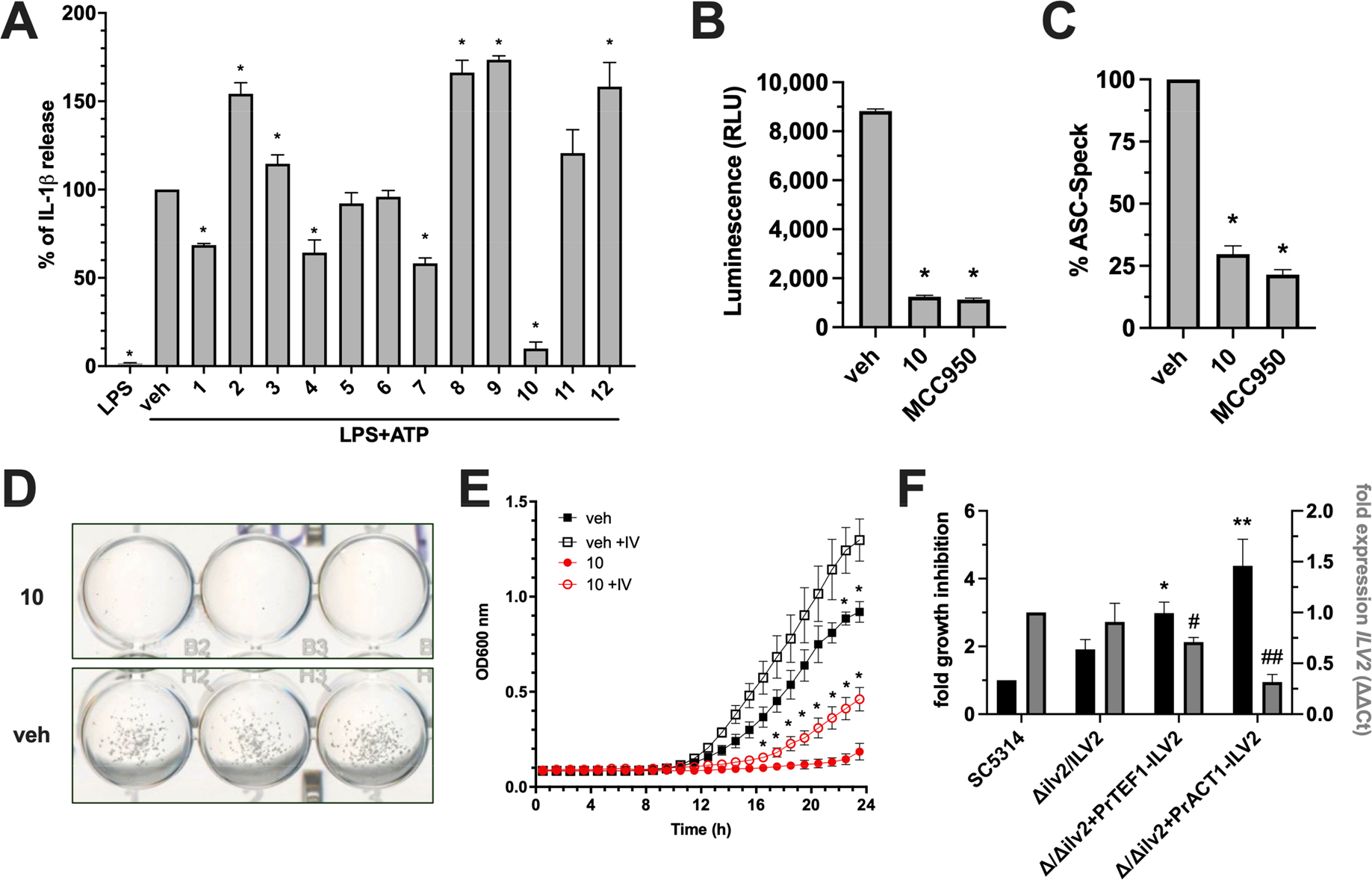
Compound 10 identified by molecular docking possesses both anti-inflammasome and antifungal activity. (A) THP1 cells were treated with 50 μM of each lead compound or vehicle alone (0.5% DMSO) for 1 h, followed by 20 ng of LPS for 3.5 h, and then 5 mM ATP for 30 min where indicated. IL-1β was measured by ELISA. (B) THP1 cells were used as described in panel A and treated with vehicle, 50 μM compound 10, or 0.1 μM MCC950. Processed Caspase-1 was measured by bioluminescence assay and values expressed as relative light units (RLU). (C) ASC-Speck GFP reporter cells were used and treated as described in panel B. The number of GFP+ Specks were enumerated in 10 random fields and calculated as percentage of the vehicle control. Cell culture experiments were conducted in technical quadruplicate (or duplicate for imaging) and results reported as the mean plus SD from independent experiments (n = 3). * indicates p < 0.05 using one-way ANOVA and Dunnet’s post-test. (D) A growth assay was performed by growing 2.5 × 103 C. albicans cells in YNB without amino acids for 24 h supplemented with 50 μM compound 10 or vehicle only. Images were captured on a digital scanner and are representative of 3 independent experiments. (E) Growth curves were conducted and OD600 nm monitored kinetically at 30 °C for 24 h in YNB media without (closed shapes) and with (open shapes) 10 mM each isoleucine and valine in the presence (red) or absence (black) of 50 μM compound 10. Experiments were conducted in biological triplicate and reported as mean plus SD * indicates p < 0.05 using multiple t test and Holm-Sidak post-test. (F) WT (SC5314) or genetically altered strains with respect to C. albicans AHAS expression (Δilv2/ILV2, Δ/Δilv2+PrTEF1-ILV2, Δ/Δilv2+PrACT1-ILV2) were grown in vehicle or 50 μM compound 10. Percent growth inhibition was calculated for each strain and normalized to WT values (black bars, left y-axis). Expression levels of ILV2 were assessed for each strain at the same time point using qRT-PCR (gray bars, right y-axis). Values were calculated using the ΔΔCT method by comparing to the housekeeping gene ACT1 and strain SC5314. Experiments were conducted in biological triplicate and reported as mean plus SD *, # p < 0.05, **, ## p < 0.01 using one-way ANOVA and Dunnett’s post-test.
In order to establish a preliminary structure–activity relationship (SAR) of this hit scaffold, a series of 9 additional phenylpropyl-thio-benzoic acid analogs were ordered and assessed for anti-inflammatory and antifungal activity as per our pipeline (Figure 4A). Among these, treatment of THP1 cells prior to LPS + ATP challenge with compounds 10a, 10b, 10c, 10e, 10f, and 10g led to significantly decreased IL-1β release (Figure 4B). Regarding antifungal activity, all compounds led to significantly reduced growth of C. albicans, except for 10h and 10i (Figure 4C). However, compounds 10, 10a, 10c, and 10d exhibited growth inhibition that exceeded 80% of the vehicle control.
Figure 4.
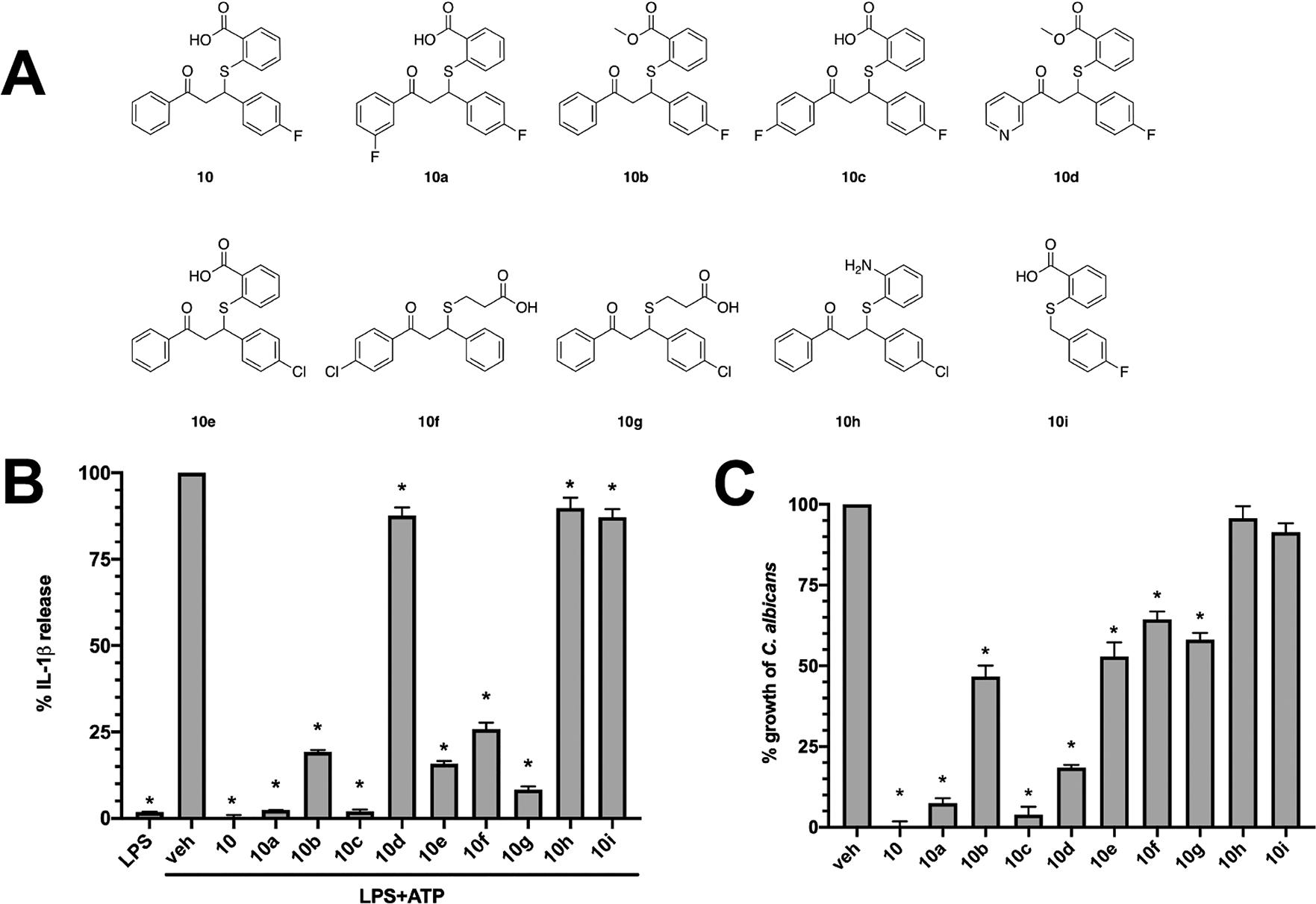
Structure activity relationship of compound 10 analogs with respect to IL-1β inhibition. (A) A series of compound 10 analogs were ordered containing substitutions in key groups deemed important for anti-inflammatory or antifungal activity. (B) THP1 cells were treated with 50 μM of each analog or vehicle alone (0.5% DMSO) for 1 h, followed by 20 ng of LPS for 3.5 h, and then 5 mM ATP for 30 min where indicated. Experiments were conducted in technical quadruplicate and results reported as the mean ± SD from independent experiments (n = 3). * indicates p < 0.05 using one-way ANOVA and Dunnett’s post-test. (C) C. albicans was grown in YNB medium at 30 °C for 24 h and in the presence of vehicle (0.5% DMSO) or each analog (50 μM). After incubation, wells were resuspended by pipetting, OD600 nm measured, and data expressed as percentage of vehicle treated control. Experiments were repeated in biological triplicate and are reported as mean + SD * indicates p < 0.05 using one-way ANOVA and Dunnet’s post-test.
Figure 5 summarizes our initial SAR observations for this scaffold, with essential features highlighted in bold. With respect to NLRP3-targeted anti-inflammatory activity, the acidic carboxyl group and the aromatic A-ring appear to be essential for activity.
Figure 5.
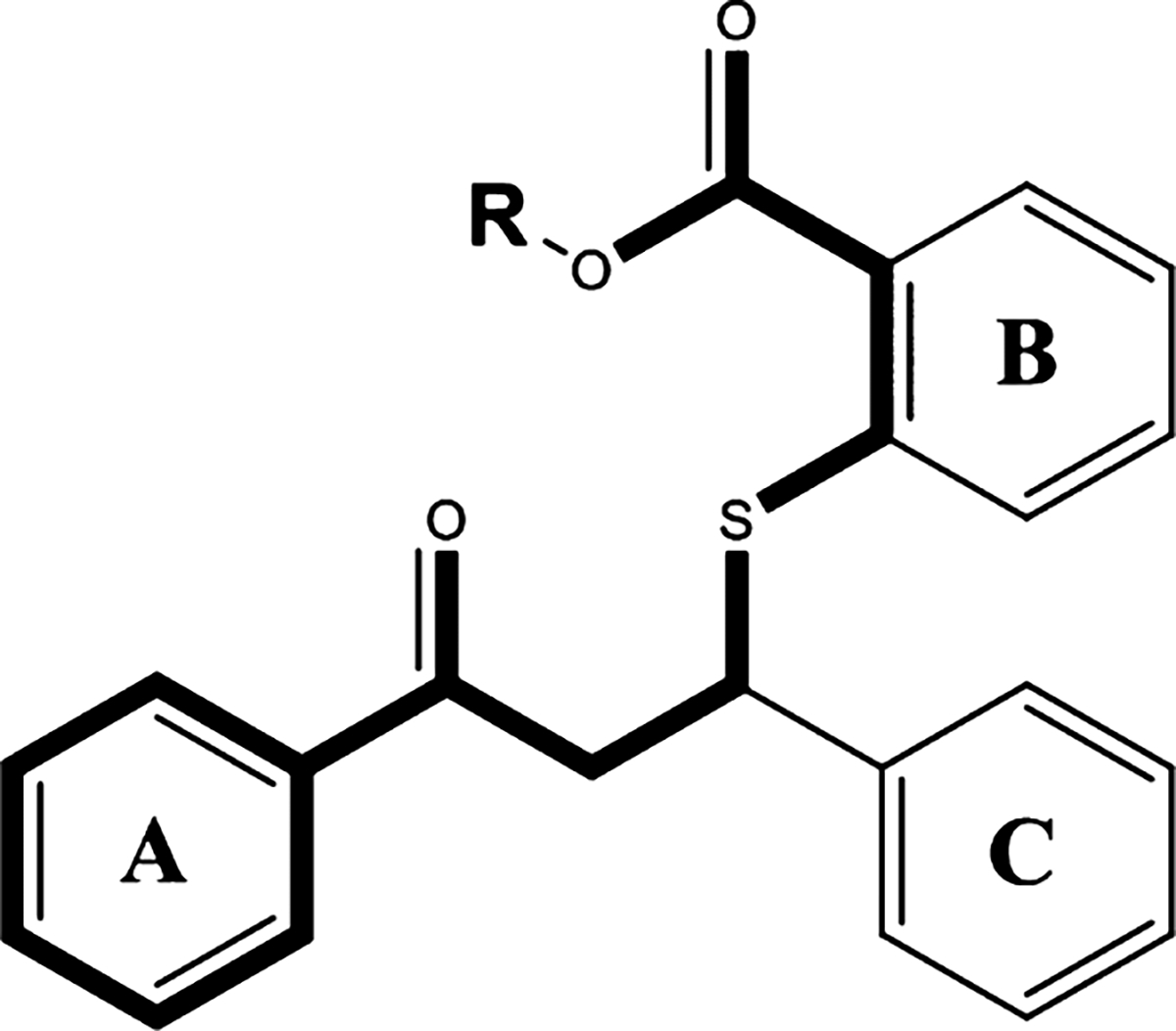
SAR analysis of the thiobenzoate scaffold. The highlighted portion of the thiobenzoate scaffold is required for both anti-NLRP3 and antifungal activity.
Conversion of the carboxylate to a methyl ester, as with 10b, resulted in a partial loss of activity and substitution with a basic amino group (10h) resulted in complete loss of activity. Removal of the A-ring (10i) resulted in complete loss of activity, while addition of fluorine at meta or para positions (10a, 10c) did not significantly affect activity. Activity data suggests that a larger chlorine substitution may be result in loss of activity (10f vs 10g), possibly due to steric hindrance. Interestingly, conversion of the A-ring phenyl ring to a pyridine ring (10d) resulted in almost complete loss of activity. Regarding anti-inflammatory activity, the B-ring does not appear to be essential, as analogs 10f and 10g retained partial activity. The SAR contribution of the C-ring is less clear as we were unable to acquire analogs lacking this ring, however we note the partial loss of activity with the substitution of the bulkier chlorine for fluorine at the para position. These SAR observations are consistent with our earlier observations of key, overlapping features in the NLRP3 and AHAS active sites as well as the molecular docking results (Figure 6). As shown in Figure 6AB, the benzoic acid moiety of 10 is predicted to engage Arg268 and Lys322 in the cationic pocket of the binding site, while the A-ring engages Phe297 in a pi-stacking interaction.
Figure 6.
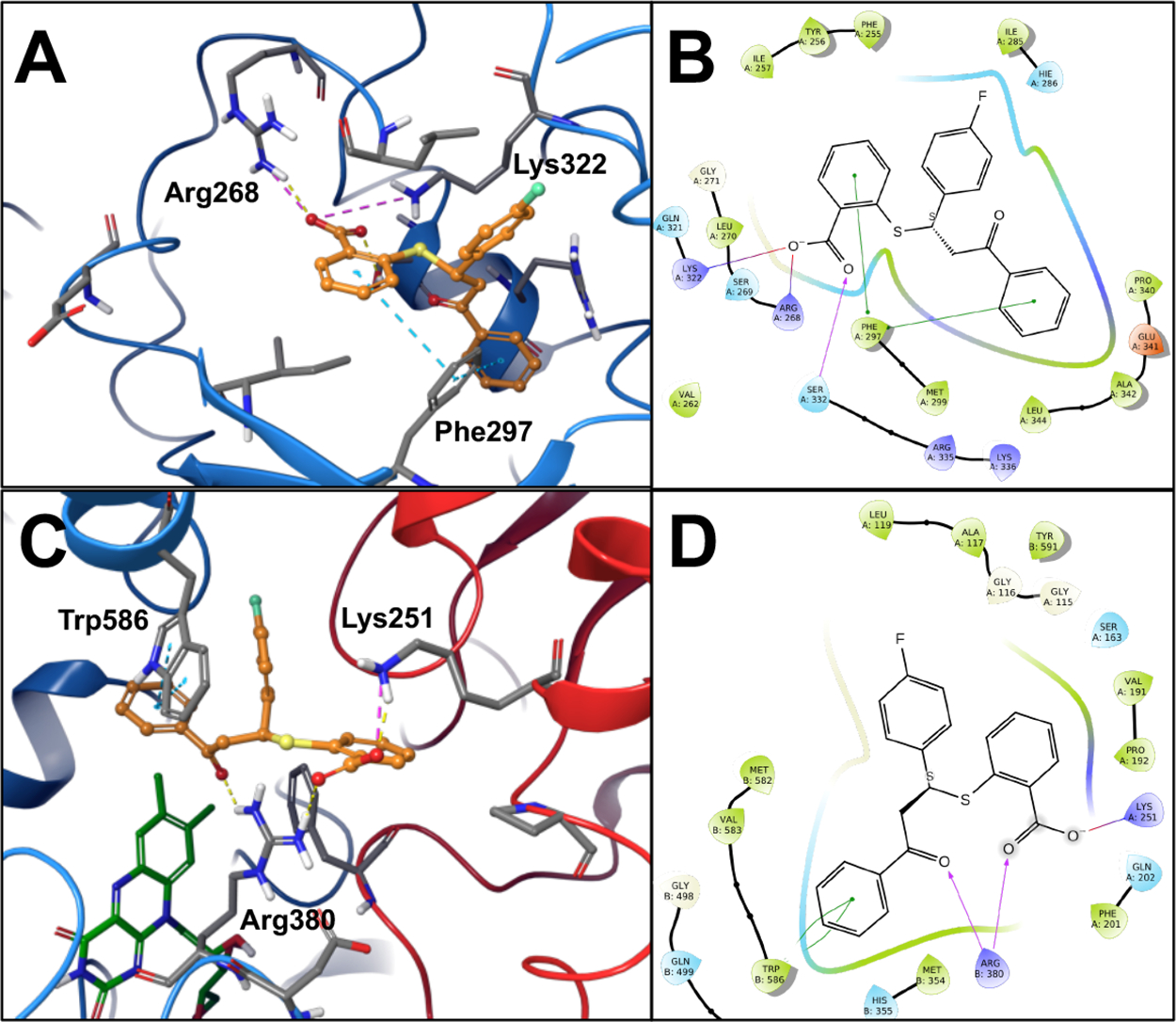
Binding Site Interactions of compound 10. (A) Compound 10 docking pose in AHAS active site. (B) 2D ligand interaction diagram of compound 10 docked into AHAS active site. (C) compound 10 docking pose in NLRP3 binding site. (D) 2D ligand interaction diagram of compound 10 in NLRP3 binding site.
Similar SAR observations can be made based on experimental antifungal activity (Figure 4C) of the same analog series. The anionic carboxyl group and the aromatic A-ring appear to be essential for activity, with partial loss of activity noted with conversion of the acid to a methyl ester (10b), complete loss of activity with substitution of the carboxyl group to an amine (10h), and complete loss of activity with removal of the A-ring altogether (10i). Similar to the anti-inflammatory SAR observations, we noted that substitutions of bulkier chlorines on the A- and C-rings (10e, 10f, 10g) resulted in notable loss of activity, likely due to steric hindrance in the smaller AHAS active site. Interestingly, 10d, with the methyl ester and pyridine A-ring, still retained moderate antifungal activity, whereas the anti-inflammatory activity of this compound was almost completely abrogated. Loss of the B-ring, as with 10f and 10g, also affected antifungal activity to a larger extent than anti-inflammatory activity. This suggests that the B-ring may play a more important role for binding to the AHAS target than the NLRP3 target. These SAR observations can be rationalized by the predicted binding poses of compound 10 from our molecular docking studies (Figure 6CD). The carboxylate group is predicted to interact with Arg380 and Lys251 in the cationic pocket, while the aromatic A-ring engages Trp586 in a pi-stacking interaction. Overall, the anti-inflammatory and antifungal SAR observations support our hypothesis that key, conserved binding site features in the two targets can be leveraged for rational design of dual-targeted agents.
Of these compounds, 10, 10a, 10c, and 10g were among the most potent inhibitors and were selected, along with compound 13 (i.e., chlorimuron ethyl), for more precise quantitation of activity. Importantly, none of these compounds resulted in reduced mammalian cell viability as compared to vehicle-treated controls, demonstrating that anti-inflammatory effects observed were not due to broad toxicity (Table 1).
Table 1.
IC50 for IL-1β Release, Mammalian Cell Viability, and C. albicans MIC50 Values for Compound 10, analogs 10a, 10c, 10g, and the Herbicidal Sulfonylurea Chlorimuron Ethyl (Compound 13)
| Compound | IC50(μM) ± SD | %viability ± SD | MIC50 (μM) ± SD |
|---|---|---|---|
| 10 | 2.3 ± 0.8 | 101.5 ± 1.4 | 6.4 ± 2.6 |
| 10a | 5.9 ± 0.5 | 106.5 ± 4.5 | 5.3 ± 1.1 |
| 10c | 2.9 ± 0.2 | 102.6 ± 6.0 | 27.3 ± 4.3 |
| 10g | 12.4 ± 1.7 | 99.0 ± 6.2 | >50 |
| 13 | >50 | 103.2 ± 7.3 | 0.5 ± 0.2 |
Regarding anti-inflammatory activity, dose–response studies revealed that IC50 values were approximately 2.3, 5.9, 2.9, and 12.4 μM for compounds 10, 10a, 10c, and 10g respectively. Unsurprisingly, compound 13 showed no attenuation of inflammasome activity at doses up to 50 μM (Table 1). MIC50 assays revealed antifungal activity against C. albicans at doses of 6.4, 5.3, and 26.7 μM for compounds 10, 10a, and 10c. Compound 13 potently inhibited C. albicans growth at a dose of 0.5 μM as described previously.19 However, compound 10g failed to exhibit notable antifungal activity at doses up to 50 μM and was not selected for further study (Table 1). Resuspension of wells containing no visible growth were plated on YPD agar. Resultant fungal growth indicated that all compounds tested had static antifungal activity. Interference with branched chain amino acid biosynthesis by compounds 10, 10a and 10c at MIC50 doses was inferred by restoration of fungal growth in culture media supplemented with isoleucine and valine and results were similar to growth patterns obtained using the established AHAS inhibitor compound 13 (Figure 7).
Figure 7.
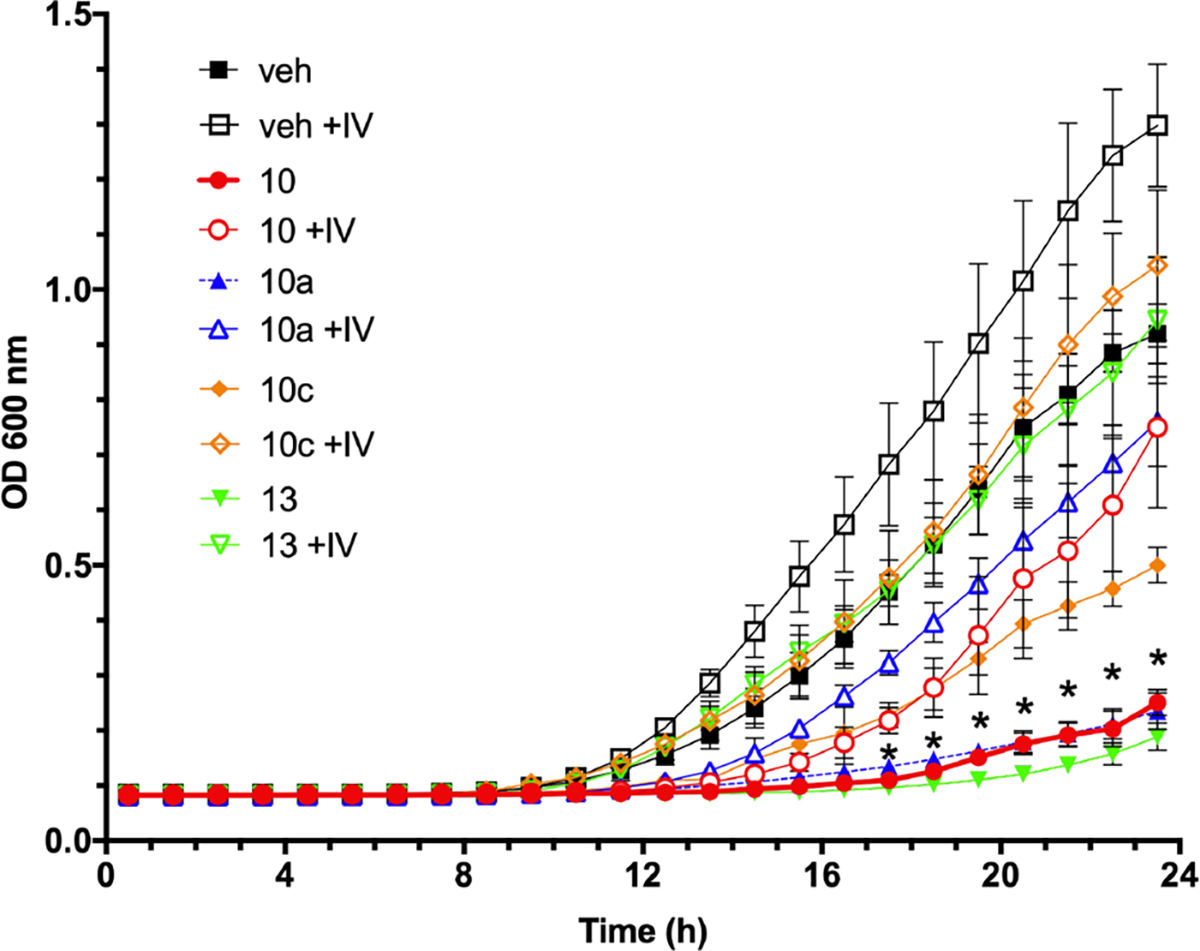
Identified dual-target inhibitors demonstrate antifungal activity that is rescued by branched chain amino acid supplementation. C. albicans cells were diluted to 2.53 cells/mL in YNB without amino acids with (open shapes) or without (closed shapes) 10 mM each isoleucine and valine. Cultures were treated with 0.5% vehicle alone (diamonds), 6.4 μM compound 10 (squares), 5.3 μM compound 10a (circles), or 0.5 μM compound 13 (triangles). Microtiter plates were incubated with orbital shaking at 200 rpm and optical density values at 600 nm collected over 24 h. * indicates p < 0.05 for all compounds when comparing amino acid supplemented and unsupplemented readings at each time point using multiple t tests and Holm-Sidak post-test.
Lastly, similar MIC50 assays were utilized to determine antifungal activity of the most potent compounds (10, 10a, and 13) across the spectrum of clinically relevant NAC species. MIC50 values for C. auris (2-fold higher), C. glabrata (2-fold lower), and C. parapsilosis were fairly similar to those obtained for C. albicans, with the exception of 10a having reduced activity against C. parapsilosis (Table 2). C. dubliniensis, and C. tropicalis are the most closely related NAC species to C. albicans. While C. dubliniensis was similarly susceptible to these compounds, growth of C. tropicalis appeared to be much less impacted (Table 2).
Table 2.
MIC50 Values (n = 3) for Compounds 10, 10a, and 13 (Chlorimuron Ethyl) against Candida Species
| MIC50(μM) ± SD | |||
|---|---|---|---|
|
| |||
| 10 | 10a | 13 | |
| C. albicans | 6.4 ± 2.6 | 5.3 ± 1.1 | 0.5 ± 0.2 |
| C. auris | 11.7 ± 2.3 | 13.1 ± 4.2 | 0.8 ± 0.3 |
| C. dubliniensis | 5.4 ± 2.8 | 8.2 ± 3.1 | 1.3 ± 0.7 |
| C. glabrata a | 45.7 ± 4.2 | 23.8 ± 8.3 | 5.6 ± 2.4 |
| C. krusei | 47.1 ± 0.6 | 24.0 ± 8.9 | 22.0 ± 4.3 |
| C. parapsilosis | 6.4 ± 1.6 | 25.0 ± 4.3 | 1.1 ± 0.4 |
| C. tropicalis | 49.9 ± 0.5 | 44.0 ± 5.5 | 21.1 ± 3.0 |
Values for all species were obtained at 24 h postgrowth, except for C. glabrata which required an incubation time of 48 h for adequate visible growth.
This apparent tolerance was also reflected in C. krusei. Importantly, reduced susceptibility for these species was also noted in the presence of compound 13, suggesting potentially divergent AHAS orthologs. However, alignment of consensus amino acids sequences from each species revealed near total conservation at residues predicted to interact with the herbicidal sulfonylurea metosulam (Figure S2), indicating that this target in each species is conserved. Thus, susceptibility differences may be driven by other mechanisms such as engagement of alternate biosynthetic pathways, differential expression of the target, or disparate compound entry or efflux.
DISCUSSION
Infections caused by the Candida species remain an unmet clinical need. Among other factors, increased use of life-saving or -extending invasive medical devices, new immunomodulatory chemotherapeutics, acquired immunodeficiencies, comorbidities, and an aging population, contribute to an increasing rise in candidiasis prevalence.24 Currently, there are three major classes of drugs to treat such infections, including the azoles, echinocandins, and polyenes which target ergosterol biosynthesis, beta-glucan synthesis, and the fungal membrane, respectively.25 While these drugs show relatively good efficacy against many Candida species, widespread or extended use of the azoles and echinocandins has driven the emergence of isolates that exhibit high level genetically encoded resistance.26–28 Moreover, concerns regarding patient toxicity, especially for the polyenes limit their use as a drug of last resort.25 Even the azoles, regarded as highly safe and efficacious front-line therapeutics to treat a variety of fungal infections, have been associated with increased risk of stillbirth and spontaneous abortion.29 Inherent resistance exhibited by some of the Candida species, especially to the azole class, further complicates effective treatment and disease management.25 Even more worrisome is the advent of multi- or pan-antifungal resistant C. auris, an emerging fungus responsible for nosocomial outbreaks with high mortality rates.30–32 Given recent rising rates of antifungal resistance of the Candida species and an increasing pool of susceptible patients, new therapeutics are urgently needed to combat these pathogens.33
Using in silico and experimental approaches, we have identified new molecules (compound 10 and its analogs) which exhibit relatively good potency against many of the clinically relevant Candida species by presumably targeting the AHAS enzyme. AHAS condenses two pyruvate molecules to form acetolactate to synthesize the branched chain amino acids valine or leucine or pyruvate with 2-ketobutyrate to form acetohydroxybutyrate to synthesize isoleucine.34 AHAS inhibition results in arrested growth in environments where branched chain amino acids cannot be imported or efficiently synthesized from precursor molecules.35 Therefore, it is reasonable to speculate whether molecules like compound 10 would be effective in reducing growth in vivo. Evidence suggests two-fold that AHAS likely serves as a rational drug target. In C. albicans, the ILV2 gene encodes for AHAS and an Δ/Δilv2 strain is avirulent in a murine model of disseminated candidiasis.22 This suggests that insufficient freely available isoleucine, leucine, or valine exist in vivo to complement AHAS deficiency. Second, a series of studies by Guddat and colleagues have shown that the sulfonylurea herbicides such as chlorimuron ethyl (e.g., compound 13) and others are potent inhibitors of purified C. albicans AHAS protein and inhibit the in vitro growth of several fungi, including a number of Candida species and Cryptococcus neoformans.19,36,37 While our MIC50 data obtained with compound 13 closely match those published previously, other values were rather disparate which may suggest significant strain-to-strain variation with respect to disruption of branched chain amino acid biosynthesis.36 Moreover, prophylactic administration of chlorimuron ethyl to mice provided protection against disseminated C. albicans infection and significantly reduced fungal burden in the kidney, spleen, liver, and lungs.36 As established AHAS inhibitors show low toxicity and the AHAS enzyme is absent in humans, this family of compounds becomes even more attractive to treat microbial diseases. Given the generally broad conservation of AHAS across bacterial, fungal, and archaeal organisms, compound 10 may have utility against a diverse array of pathogens.34 While compound-mediated growth inhibition is alleviated in the presence of branched chain amino acids, more detailed biochemical work using purified AHAS enzyme is required to fully validate on-target activity of this compound series.
While the Candida species are armed with traditional virulence determinants akin to bacterial pathogens, their pathogenicity is more nuanced and intimately intertwined with the host inflammatory response. This association has been elegantly conceptualized as what is known as the “Damage Response Framework” (DRF), a spectrum of disease states characterized by both pathogen and/or host-mediated damage.38 Classical opportunists fall into class 1 where disease is only mediated in the absence of an immune response. Classes 2–4 include pathogens that damage in hosts with weak, normal, or strong immune responses, respectively. Class 5 and 6 pathogens induce damage that is aided by a strong host immune response or mediated entirely by host immunopathology. Given the plasticity and robustness of Candida, it fits into all DRF classes depending on disease state and patient status.39 Inhibition of strong immune responses, especially in these latter DRF classes including immune reconstitution syndrome (IRS)-related infection, some cases of sepsis, and VVC, may offer protection against collateral damage driven by host immune cells responding to the fungus.2,4,40 A single drug, to both eliminate the pathogen while controlling innate danger-mediated inflammation, could prove immensely useful in specific disease states.
The NLRP3 inflammasome plays a crucial role in detecting and responding to danger signals, including those elicited by infectious fungi. During murine models of invasive candidiasis, clearly, NLRP3 plays a protective function, as mice with genetic deletion of this or other inflammasome components are hypersusceptible.13,41 However, pharmacologic attenuation of NLRP3 in vivo during systemic candidiasis has not yet been reported. Targeting of NLRP3 using the sulfonylurea glyburide or potent inhibitor MCC950 offers protection in models of melioidosis, bronchopulmonary dysplasia, allergic asthma, cystitis, endotoxemia, autoimmune encephalomyelitis, subarachnoid hemorrhage, and bacterial sepsis.15,42–48 Given body-wide dysregulated immune responses and multiorgan nature of sepsis, inhibition of NLRP3 may offer protection at renal, respiratory, cardiovascular, gastrointestinal, and central nervous systems via reduced influx of inflammatory cells and reversion to homeostatic physiological responses.49 Therefore, it is reasonable to hypothesize that inhibition of the NLRP3 inflammasome, or its downstream signals, may offer protection against fungal disease. However, finely tuned control of these pathways must not tip the balance to immunosuppresive hyporesponsiveness. Such concerns may be greatly mitigated by using drugs similar to compound 10 which can control fungal burden and inflammation simultaneously.
Although NLRP3 attenuation during fungal sepsis may be challenging, more superficial diseases like mucosal infections may be more amenable to such treatment. One such infection with overwhelming prevalence estimated to be in the hundreds of millions of cases per year is VVC, a disease of the lower female reproductive tract that is mediated by robust recruitment of neutrophils and production of inflammatory cytokines and chemokines.4 However, recruited neutrophils appear to be anergic in the vaginal environment with respect to C. albicans clearance, yet contribute to high levels of inflammation.50,51 Use of NLRP3−/− mice demonstrated that neutrophil recruitment during murine VVC is largely inflammasome-dependent.12,52 In further support of this, systemic treatment with glyburide or MCC950 to pharmacologically target NLRP3 significantly inhibits neutrophil recruitment in this model.12 Importantly, neutrophil depletion using anti-Ly6G antibodies did not alter fungal burden as compared to untreated mice, further confirming their dispensable role in controlling vaginal candidiasis.53,54 Targeted and genome-wide approaches have determined that polymorphisms in human NLRP3 are associated with hyperactive IL-1β responses and increased incidence of recurrent VVC.55 Thus, a dual-target inhibitor such as compound 10 may be a very attractive molecule for treatment of this fungal disease in particular, considering potential ease of administration in a topical gel or cream to exert effects specifically at the vaginal mucosa. Comprehensive preclinical testing to assess efficacy of compound 10 and its analogs against both systemic and local candidiasis is warranted and currently underway in our laboratories.
In summary, using a series of computational and biological approaches, we have identified and characterized a novel molecular structure that exhibits both anti-inflammatory and antifungal activity through inhibition of host NLRP3 and likely fungal AHAS, respectively. Although an excellent proof-of-principle, further synthetic optimization of these compounds is needed to improve the potency of each relative activity while ensuring optimal physicochemical properties and metabolic stability. Included in these next steps will be the characterization of the relative activities of the purified enantiomers of this hit series. Future studies planned will also involve screening of additional small-molecule libraries focused using the SAR observations noted herein. Weak acids, such as aryl sulfonamides and aryl sulfonylureas, will be prioritized to identify additional active scaffolds with improved PK potential. While not pan-candidal, the compounds reported here target an array of clinically relevant Candida species and may offer new scaffolds for future drug discovery and ultimately better clinical management of devastating fungal infections.
METHODS
Docking Site and Library Preparation.
The cocrystal structure of S. cerevisiae acetohydroxyacid synthase with bound sulfometuron methyl (PDB ID 1T9C) was used to prepare the AHAS docking grid used for all molecular docking runs.37 We note the high similarity between the S. cerevisiae and the C. albicans AHAS proteins (72% identity overall with 100% identity within 10 Å of the bound inhibitor). The AHAS enzyme is a functional homodimer, with the active site falling at the interface of the two chains near a bound FMN cofactor. Schrödinger’s Glide program was used to create a 10 Å cubic receptor grid based upon centroid of the bound inhibitor.56–59 All crystallographic waters were removed, no waters were retained in the docking grid. No constraints, rotatable side chains, or excluded volumes were added to the grid.
The NLRP3 docking grid was built using the published cryo-EM structure of the inactive human NLRP3 inflammasome bound to NEK7 (PDB ID 6NPY).60 As there is no experimental costructure of NLRP3 with a small-molecule inhibitor, it was necessary to identify and validate the inhibitor binding site prior to docking. Building up on the work of Mekni et al. and previous publications characterizing the mechanism of action of the NLRP3 inhibitor MCC950, we investigated the proposed MCC950 binding site in the globular NACHT domain, proximal to the Walker B motif.61–63 The Schrödinger Induced Fit Docking (IFD) Protocol was used to generate an initial pose of MCC950 bound to NLRP3 using a docking grid encompassing residues 255, 257, 261–270, 273, 275–289, 297, 299, 300, 305, 316, 322, 331, 333, 338, 340, and 342.64,65 The IFD pose was subsequently refined using a 100 ns MD simulation (Schrödinger, DESMOND) with an average costructure calculated from the resulting trajectory used for the generation of the final docking grid.66,67 The final Glide docking grid was generated, as above, using a 10 Å cubic region centroid to the bound MCC950 inhibitor. No active site waters, docking constraints, rotatable side chains, or excluded volumes were added to the docking grid.
The Maybridge Screening Collection of approximately 53000 small molecule compounds was downloaded and prepared for molecular docking using Schrödinger’s LigPrep application.68 The raw 2D structures were desalted and 3D structures were generated using the OPLS3 force field with ionization states generated for target pH 6.0 to 8.0 using Epik.69,70 Stereoisomers and tautomers were fully enumerated. A custom filter was applied to remove compounds with known chemically reactive or toxic functional groups and molecular weight outside of a 250 to 650 Da range. The resulting library was saved in Maestro file format for subsequent docking and scoring.
Molecular Docking and Compound Selection.
The prepared Maybridge library was docked into each of the target grids using Schrödinger’s Glide docking program and a virtual screening workflow that involved three iterative docking stages for each target.56–59 In the first stage, flexible docking of the library against each target was performed using Glide with HTVS (high throughput) settings. In the second stage, the top 10% of scored compounds (selected by ligand efficiency normalized Glide docking scores) were redocked into each target grid using Glide with SP (standard precision) settings. In the final stage, the top 10% of the Glide SP docked compounds (selected again by ligand efficiency normalized Glide scores) were docked using Glide with XP (extra precision) settings. Default Glide settings were used for each stage of docking, against each target. Scaling of van der Waals radii for nonpolar atoms was employed to soften the potential for nonpolar parts of the docked compounds using a scaling factor of 0.8 and partial charge cutoff of 0.15. Epik was used to add state penalties to docking scores; nitrogen inversions and ring conformations were sampled. The molecular docking protocol was validated for each target by successful redocking of the known inhibitors from the experimental (AHAS) or IFD/MD (NLRP3) structures to a non-hydrogen RMSD of less than 1.5 Å as we have described in a previous publication.71 The top scoring compounds from the final docking stage for each target were compared and high-scoring compounds present on both score-ranked lists were identified, visually inspected (binding pose, desirable physicochemical features), and selected for ordering and testing in experimental assays. Twelve compounds were ultimately chosen for experimental testing (Table S1).
Growth of Microorganisms.
All strains used or created in this study can be found in Table S2. When possible, genome-sequenced reference isolates were used. Strains included C. albicans SC5314, C. auris [429]0382, C. dubliniensis CD36, C. glabrata CBS138, C. krusei 81-B-5, C. parapsilosis CDC317, and C. tropicalis MYA3404.72 Strains were maintained as glycerol stocks stored at −80 °C. A small amount of stock was streaked onto YPD agar and incubated at 30 °C for 48 h to yield isolated colonies. A single colony was transferred to 5 mL of liquid YPD medium and incubated at 30 °C with shaking (200 rpm) for 18 h prior to validation assays. Candida was then washed 3X with cell culture grade water by centrifugation at 8000 rpm and resuspended in an equivalent volume of appropriate culture medium prior to downstream assays.
Vector Construction.
All primers used for vector and strain construction can be found in Table S3. In order to create constitutive expression strains, the ILV2 open reading frame was PCR amplified from C. albicans SC5314 genomic DNA using SuperFi high fidelity polymerase (Thermo Fisher) and primers ILV2-ORF-F-SalI and ILV2-ORF-R-MluI. Amplicons were purified by GeneJet column (Thermo Fisher) and digested with restriction nucleases SalI and MluI. Plasmids pKE1 and pKE4, harboring the CaACT1 or CaTEF1 promoters, respectively and ADH1 terminator, were similarly digested. Amplicons were ligated into these cut vectors to yield plasmids pKE1-ILV2 and pKE4-ILV2. After transformation and propagation in E. coli DH5α, reisolated plasmids were verified by gel electrophoresis following restriction digestion with SalI and MluI. All sequences were verified by Sanger methodology (Genewiz) using primers ACT1PRSEQF, TEF1PRSEQF, ILV2DETF, and ADH13SEQR.
Strain construction.
Plasmids pGEMHIS1 and pRSARG4ΔSpe were PCR amplified using primers ILV2DISF and ILV2DISR to generate disruption cassettes containing HIS1 or ARG4 loci, respectively, and ILV2 flanking regions. Using the lithium acetate method, C. albicans strain BWP17 was first transformed with the ARG4-containing disruption cassette and plated on selective media to generate strain JM01.73,74 Integration of ARG4 at the first ILV2 locus was confirmed by PCR of genomic DNA using primer pairs ILV2AMPF and ARG4INTF and ILV2AMPR and AR-G4INTR. Strain JM01 was then transformed with either NheI-linearized pLUX, pKE1-ILV2, or pKE4-ILV2 all containing the URA3 selectable marker and plated onto media lacking uridine to generate strains JM02, JM03, and JM04, respectively. Integration of these plasmids at the IRO1-URA3 locus was confirmed by PCR of genomic DNA using primers LUXINTDETF and LUXINTDETR. Strain JM02 was sequentially transformed with NruI-linearized pGEMHIS1 to restore histidine prototrophy to yield strain JM05 (Δilv2/ILV2). Integration of pGEMHIS1 was confirmed by PCR using primers HIS1DETF and HIST1INTR5. Strains JM03 and JM04 were transformed with a disruption cassette containing HIS1 to generate strains JM06 (Δ/Δilv2+PrACT1-ILV2) and JM07 (Δ/Δilv2+PrTEF1-ILV2), respectively. Integration of HIS1 at the second ILV2 locus was confirmed by PCR using primer pairs ILV2AMPF and HIS1INTF and ILV2AMPR and HIS1INTR. Loss of both native ILV2 alleles was confirmed by absence of an amplicon using PCR using primer pairs ILV2AMPF and ILV2DETR and ILV2AMPR and ILV2DETF.
Growth of THP1 Cells.
WT (THP1-null), NLRP3−/− (THP1-KO-NLPR3), and ASC-Speck reporter (THP1-ASC-GFP) THP1 monocyte-like cells (Invivogen) were cultured according to the manufacturer’s protocol in RPMI 1640 medium containing 25 mM HEPES supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin-streptomycin, and 100 ug/mL normocin as described previously.18 Cells underwent routine testing for lack of mycoplasma contamination. THP1 cells were enumerated on the Countess II FL (Life Technologies) and frozen as aliquots of ~5 × 106 cells in liquid nitrogen. Upon cryorecovery, cells were incubated for 3 d at 37 °C and 5% CO2 in complete culture medium (RPMI 1640, 10% heat-inactivated FBS, and 100 U/mL penicillin-streptomycin). THP1 cells were counted, assessed for viability by exclusionary Trypan Blue staining, and diluted to 5.5 × 105 cells/mL in complete culture medium. Aliquots of 180 μL were seeded at a final density of 1 × 105 cells/well of a 96-well tissue culture-treated polystyrene plate. ASC-Speck GFP reporter cells were similarly cultivated and plated to a density of 5 × 105 cells/well on Falcon vessel tissue culture-treated glass slides (Corning). PMA was added at 100 nM final concentration, and cells were incubated for an additional 24 h to differentiate into a macrophage phenotype.
Preparation of Compound Stocks.
Lead compounds, analogs, and established inhibitors were ordered from Maybridge (Fisher Scientific) (1–12, 10a–g), ChemDiv (10h), Vitascreen LLC (10i), Fisher Scientific (13), or Invivogen (14) with purities ≥95% as provided in the certificates of analysis. Using sterilized instruments, compounds were prepared as 200X (50 μM final) or 400X (100 μM final) working stocks in 100% DMSO. Stocks were further diluted in either phenol red-free RPMI containing 25 mM HEPES, pH 7.0 for cell culture work or YNB without amino acids and ammonium sulfate containing 2% glucose, 165 mM MOPS, pH 7.0 for respective initial validation experiments. In all assays, final DMSO concentrations remained at 0.5%.
LC/MS of Lead Compounds.
LCMS was acquired for lead compounds in negative ionization mode using a Waters Xevo G2-S QTof (Figure S3). The high resolution mass spectrometer (Waters, Milford, MA) was equipped with an mESI source and coupled with a Waters Acquity I-Class UPLC with a PDA detector. A BEH C18 column (2.1 mm × 50 mm, 1.7 μm, Waters, Milford, USA) was used. The mobile phase for LC was A: 95% H2O/5% acetonitrile, and B: 100% acetonitrile. Data were collected and processed using Masslynx 4.1 software.
Inflammasome Activation Assay.
Following differentiation, spent culture medium was replaced with 180 μL of compound working stocks or serial dilutions prepared in phenol red-free RPMI and incubated for 1 h at 37 °C. Controls containing vehicle only (0.5% DMSO) were also included. Cells were then challenged with 20 ng of Escherichia coli 0111:B4 LPS for 3.5 h, followed by addition of 5 mM ATP for 30 min prior to elicit inflammasome activation. Mock-activation controls without ATP addition were also included. Plates were gently centrifuged (200g for 2 min) to settle contents and 100 μL of culture supernatant transferred to polystyrene microtiter plate containing an equal volume of 2X ELISA diluent (Invitrogen). Optical density values from mock-activated controls were subtracted from those of LPS+ATP challenge. Experiments were conducted in technical quadruplicate and repeated independently in triplicate. Data are reported as mean values plus SD.
IC50 Calculation for IL-1β Release.
The inflammasome activation assay was conducted as described above, except that serial dilutions of compound stocks in phenol red-free RPMI containing 0.5% DMSO were used. All values were compared to the vehicle-treated control and calculated as a percentage of maximum IL-1β release. Concentrations were log transformed, plotted in GraphPad Prism, and IC50 values were obtained using a four-parameter variable slope and best-fit algorithm. Experiments were conducted in technical quadruplicate and independently repeated in triplicate. Data are reported as mean plus SD.
Caspase-1 activation assay.
THP1 cells were treated exactly as described in the inflammasome activation assay above. The Caspase-Glo assay (Promega) was conducted according to manufacturer’s protocol. Following the challenge, 50 μL of culture supernatant was transferred to white microtiter plates, 50 μL of Caspase-Glo 1 reagent was immediately added, and the mixture was incubated for 1 h. Luminescence was captured using a BioTek Synergy H1 microplate reader. Experiments were conducted in technical quadruplicate and repeated independently in triplicate. Data are reported as mean values plus SD.
ASC-Speck Reporter Assay.
THP1-ASC-GFP cells were prepared as described and similarly treated according to the inflammasome activation assay above. Immediately following a 30 min ATP challenge, the media was carefully removed and cells were fixed with 2% buffered formalin and gently rinsed 3X in PBS. Following the removal of the chamber, Vectashield (Vector Laboratories, Inc.) was added to prevent photo-bleaching. ASC-Speck formation was assessed by fluorescence microscopy (Nikon Elcipse Ni) using FITC and DIC filter sets. Specks were enumerated in 10 random fields per condition. Data are reported as percent of Speck formation relative to the vehicle-treated control. Experiments were independently repeated in triplicate. Data are reported as mean values plus SD.
Viability Assays.
The XTT reduction assay was used to determine the metabolic activity of THP1 cells after treatment with 50 μM of each compound prepared in a phenol-red free RPMI medium.75 An additional treatment group containing 1% SDS was also utilized as a positive control. After a 5 h incubation, medium was removed, cells were gently washed in PBS, and 200 μL of XTT working reagent (0.5 mg/mL XTT and 1 μM menadione) was added for 2 h. Following incubation, cells were gently centrifuged to settle contents, 100 μL of supernatant was transferred to a fresh microplate, and the OD490 nm was recorded. Toxicity of each compound or SDS was expressed as the percentage relative to vehicle only controls. Experiments were conducted in technical quadruplicate and performed in biological triplicate. Data are expressed as the mean plus SD.
Modified MIC50 Assays.
MIC50 assays were conducted as described by the established CLSI broth microdilution method with slight modification.76 Compound stocks (400X) were prepared in YNB as described above to 100 μM, followed by serial dilution in similar medium containing 0.5% DMSO vehicle, and 100 μL was transferred to a microtiter plate. Control wells containing media with vehicle only were also included. Candida species were diluted according to CSLI methodology in YNB media to eventually obtain a density of 5 × 103 cells/mL and an equivalent volume mixed with diluted compound. Plates were incubated at 37 °C for 24 to 48 h and imaged using a digital scanner. Quantitative determinations were made by mixing wells via pipetting and measuring OD600 nm. Experimental values were blank subtracted using OD600 nm readings of control wells. MIC50 values were calculated using GraphPad Prism by using a four-parameter variable slope and best-fit algorithm. MIC50 assays were conducted in technical duplicate and biological triplicate. Data is expressed as mean plus-minus SD. Contents of wells exhibiting no growth with the most dilute compound were spread on YPD agar and incubated overnight at 30 °C to determine static or cidal activity.
Growth Curves.
Candida species were grown as described above, washed, diluted to 5 × 103 cells/mL in YNB without amino acids and ammonium sulfate containing 2% glucose, and 100 μL was transferred to wells of a microtiter plate. In some cases, YNB containing 10 mM each isoleucine and valine was used. Compounds were prepared at 2X the determined MIC50 value (Table 1) in similar YNB media containing 1% DMSO final. An equivalent volume of compound (100 μL) was mixed with cells by pipetting. OD600 nm readings were captured at 60 min intervals using a BioTek Synergy spectrophotometer with an incubation temperature of 30 °C and orbital shaking at 200 rpm. Experiments were repeated in technical quadruplicate and biological triplicate. Data is expressed as the mean plusminus SD.
Growth Inhibition Using ILV2 Mutant Strains.
Growth curves were conducted as described above in the presence of vehicle or 50 μM compound 10 using strains with altered ILV2 (encodes for C. albicans AHAS) copy number (Δilv2/ILV2) or constitutive, nonamino acid stress responsive promoters (Δ/Δilv2+PrTEF1-ILV2, Δ/Δilv2+PrACT1-ILV2). Percent inhibition was calculated by the following formula: (1-(OD600 compound 10/OD600 vehicle))*100 at 36 h and normalized to the WT (SC5314) control to generate fold inhibition. Experiments were repeated in biological triplicate and data expressed as mean plus SD.
Quantitation of ILV2 Expression.
Strains SC5314, Δilv2/ILV2, Δ/Δilv2+PrTEF1-ILV2, and Δ/Δilv2+PrACT1-ILV2 were cultivated in YNB without amino acids and ammonium sulfate as described above for 36 h. RNA was extracted by the hot acid phenol method, quantitated by NanoDrop spectrophotometer, and purity assessed by A260/A280 ratio. RNA (200 ng) was treated with RNase-free DNAase (Thermo Fisher) and RNA reverse transcribed using the RevertAid kit (Thermo Fisher) according to manufacturer’s instructions. Primer pairs ILV2DETF and ILV2QPCR-R and CaACT1QPCR-F and CaACT1QPCR-R were used with 2X Maxima SYBR Green mix (Thermo Fisher) to amplify approximately 150 bp fragments from 20 ng of cDNA. Quantitative real-time PCR (qRT-PCR) reactions were monitored using the Applied Biosystems 7500 platform and software. Expression levels of ILV2 were compared to expression of the housekeeping gene ACT1 and strain SC5314 using the ΔΔCT method as described previously.72 Experiments were repeated in biological triplicate and data expressed as mean plus SD.
Statistics.
All experiments were performed in biological triplicate. Statistical tests were performed using GraphPad Prism v8.4.3 software. ELISA data were compared using one-way ANOVA followed by Dunnet’s post-test. Growth curve data was analyzed using multiple t test followed by Holm-Sidak post-test. Growth inhibition and qRT-PCR data was analyzed using a one-way ANOVA and Dunnet’s post-test.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Disease grants (R21AI127942 and R01AI134796) awarded to BMP. This work was also supported by the UTHSC College of Pharmacy Dean’s Enhancement Program awarded to KEH and BMP. J.M. received financial support from the China Scholarship Council award 201906150153.We also thank Dr. Dejian Ma and the UTHSC Analytical Facility for help with compound quality analyses and use of the Waters Xevo G2-S QTof MS instrument supported by National Institutes of Health grant (1S10OD010678–01) awarded to Dr. Wei Li.
ABBREVIATIONS
- AHAS
acetohydroxyacid synthase
- ANOVA
analysis of variance
- ASC
apoptosis associated speck-like protein containing a CARD
- ATP
adenosine triphosphate
- CLSI
Clinical and Laboratory Standards Institute
- DAMP
danger-associated molecular pattern
- DMSO
dimethyl sulfoxide
- DRF
Damage Response Framework
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)
- IC50
half maximal inhibitory concentration
- LPS
lipopolysaccharide
- MIC
minimal inhibitory concentration
- MOPS
[3-(N- morpholino) propanesulfonic acid]
- NAC
non-albicans Candida
- NLRP3
nucleotide-binding oligomerization domain-like receptor family pyrin domain containing 3
- OD
optical density
- PBS
phosphate buffered saline
- PMA
Phorbol 12-myristate 13-acetate
- RPMI
Roswell Park Memorial Institute
- SD
standard deviation
- VVC
vulvovaginal candidiasis
- WT
wild-type
- XTT
[2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt]
- YNB
yeast nitrogen base
- YPD
yeast peptone dextrose
Footnotes
The authors declare no competing financial interest.
Images were minimally processed and any adjustments applied evenly across the entire image. Microsoft Powerpoint v16.39 and Adobe Photoshop v21.1.1 were used for all image manipulation. All graphs were constructed and exported using GraphPad Prism v8.4.3.
Molecular Formula Strings were uploaded as a csv file.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.1c00270.
Maybridge compounds, strains used or created in this study, primers used in this study, IL-1β release in THP1 cells is largely NLRP3-dependent, alignment of AHAS orthologs from several Candida species, LC/MS confirmation of purity/identity of lead compounds 10 and 10a Files A,B,C (PDF)
PDB costructures used for in silico docking (PDB)
PDB costructures used for in silico docking (PDB)
PDB costructures used for in silico docking (PDB)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.1c00270
Contributor Information
David J Lowes, Department of Clinical Pharmacy and Translational Science, College of Pharmacy, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States.
Jian Miao, Graduate Program in Pharmaceutical Sciences, College of Graduate Health Sciences, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States.
Rand A Al-waqfi, Graduate Program in Pharmaceutical Sciences, College of Graduate Health Sciences, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States.
Kristiana A. Avad, Graduate Program in Pharmaceutical Sciences, College of Graduate Health Sciences and Doctor of Pharmacy Program, College of Pharmacy, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States
Kirk E Hevener, Department of Pharmaceutical Sciences, College of Pharmacy, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States.
Brian M Peters, Department of Clinical Pharmacy and Translational Science, College of Pharmacy and Department of Microbiology, Immunology, and Biochemistry, College of Medicine, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States.
REFERENCES
- (1).Bongomin F, Gago S, Oladele RO, and Denning DW (2017) Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi (Basel) 3, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Duggan S, Leonhardt I, Hunniger K, and Kurzai O (2015) Host response to Candida albicans bloodstream infection and sepsis. Virulence 6, 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Labelle AJ, Micek ST, Roubinian N, and Kollef MH (2008) Treatment-related risk factors for hospital mortality in Candida bloodstream infections. Crit. Care Med. 36, 2967–2972. [DOI] [PubMed] [Google Scholar]
- (4).Willems HME, Ahmed SS, Liu J, Xu Z, and Peters BM (2020) Vulvovaginal candidiasis: A current understanding and burning questions. J. Fungi (Basel) 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sobel JD (1997) Vaginitis. N. Engl. J. Med. 337, 1896–1903. [DOI] [PubMed] [Google Scholar]
- (6).Yano J, Sobel JD, Nyirjesy P, Sobel R, Williams VL, Yu Q, Noverr MC, and Fidel PL Jr (2019) Current patient perspectives of vulvovaginal candidiasis: incidence, symptoms, management and post-treatment outcomes. BMC Women’s Health 19, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wiederhold NP (2017) Antifungal resistance: current trends and future strategies to combat. Infect. Drug Resist. 10, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Camilli G, Griffiths JS, Ho J, Richardson JP, and Naglik JR (2020) Some like it hot: Candida activation of inflammasomes. PLoS Pathog. 16, e1008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bui FQ, Johnson L, Roberts J, Hung SC, Lee J, Atanasova KR, Huang PR, Yilmaz O, and Ojcius DM (2016) Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1beta and the danger signals ASC and HMGB1. Cell. Microbiol. 18, 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Thinwa J, Segovia JA, Bose S, and Dube PH (2014) Integrin-mediated first signal for inflammasome activation in intestinal epithelial cells. J. Immunol. 193, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wu R, Hogberg J, Adner M, Ramos-Ramirez P, Stenius U, and Zheng H (2020) Crystalline silica particles cause rapid NLRP3-dependent mitochondrial depolarization and DNA damage in airway epithelial cells. Part. Fibre Toxicol. 17, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bruno VM, Shetty AC, Yano J, Fidel PL Jr., Noverr MC, and Peters BM (2015) Transcriptomic analysis of vulvovaginal candidiasis identifies a role for the NLRP3 inflammasome. mBio 6, 00182–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, and Fitzgerald KA (2009) An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 5, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hull C, Dekeryte R, Buchanan H, Kamli-Salino S, Robertson A, Delibegovic M, and Platt B (2020) NLRP3 inflammasome inhibition with MCC950 improves insulin sensitivity and inflammation in a mouse model of frontotemporal dementia. Neuropharmacology 180, 108305. [DOI] [PubMed] [Google Scholar]
- (15).Koh GC, Maude RR, Schreiber MF, Limmathurotsakul D, Wiersinga WJ, Wuthiekanun V, Lee SJ, Mahavanakul W, Chaowagul W, Chierakul W, White NJ, van der Poll T, Day NP, Dougan G, and Peacock SJ (2011) Glyburide is anti-inflammatory and associated with reduced mortality in melioidosis. Clin. Infect. Dis. 52, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Primiano MJ, Lefker BA, Bowman MR, Bree AG, Hubeau C, Bonin PD, Mangan M, Dower K, Monks BG, Cushing L, Wang S, Guzova J, Jiao A, Lin LL, Latz E, Hepworth D, and Hall JP (2016) Efficacy and pharmacology of the NLRP3 inflammasome inhibitor CP-456,773 (CRID3) in murine models of dermal and pulmonary inflammation. J. Immunol. 197, 2421–2433. [DOI] [PubMed] [Google Scholar]
- (17).Ren P, Wu D, Appel R, Zhang L, Zhang C, Luo W, Robertson AAB, Cooper MA, Coselli JS, Milewicz DM, Shen YH, and LeMaire SA (2020) Targeting the NLRP3 inflammasome with inhibitor MCC950 prevents aortic aneurysms and dissections in mice. J. Am. Heart Assoc. 9, No. e014044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lowes DJ, Hevener KE, and Peters BM (2020) The second-generation anti-diabetic sulfonylureas inhibit Candida albicans and Candidalysin mediated activation of the NLRP3 inflammasome. Antimicrob. Agents Chemother. 64, e01777–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lee YT, Cui CJ, Chow EW, Pue N, Lonhienne T, Wang JG, Fraser JA, and Guddat LW (2013) Sulfonylureas have antifungal activity and are potent inhibitors of Candida albicans acetohydroxyacid synthase. J. Med. Chem. 56, 210–219. [DOI] [PubMed] [Google Scholar]
- (20).O’Brien M, Moehring D, Munoz-Planillo R, Nunez G, Callaway J, Ting J, Scurria M, Ugo T, Bernad L, Cali J, and Lazar D (2017) A bioluminescent caspase-1 activity assay rapidly monitors inflammasome activation in cells. J. Immunol. Methods 447, 1–13. [DOI] [PubMed] [Google Scholar]
- (21).Stutz A, Horvath GL, Monks BG, and Latz E (2013) ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040, 91–101. [DOI] [PubMed] [Google Scholar]
- (22).Kingsbury JM, and McCusker JH (2010) Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase ilv2 mutants is influenced by the carbon source and rapamycin. Microbiology (London, U. K.) 156, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Butts A, DeJarnette C, Peters TL, Parker JE, Kerns ME, Eberle KE, Kelly SL, and Palmer GE (2017) Target Abundance-Based Fitness Screening (TAFiS) Facilitates Rapid Identification of Target-Specific and Physiologically Active Chemical Probes. mSphere 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Brandt ME, and Park BJ (2013) Think fungus-prevention and control of fungal infections. Emerging Infect. Dis. 19, 1688–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Scorzoni L, de Paula ESAC, Marcos CM, Assato PA, de Melo WC, de Oliveira HC, Costa-Orlandi CB, Mendes-Giannini MJ, and Fusco-Almeida AM (2017) Antifungal therapy: new advances in the understanding and treatment of mycosis. Front. Microbiol. 8, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Balashov SV, Park S, and Perlin DS (2006) Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50, 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lopez-Ribot JL, McAtee RK, Lee LN, Kirkpatrick WR, White TC, Sanglard D, and Patterson TF (1998) Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42, 2932–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sanglard D, Ischer F, Koymans L, and Bille J (1998) Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Molgaard-Nielsen D, Svanstrom H, Melbye M, Hviid A, and Pasternak B (2016) Association Between Use of Oral Fluconazole During Pregnancy and Risk of Spontaneous Abortion and Stillbirth. JAMA 315, 58–67. [DOI] [PubMed] [Google Scholar]
- (30).Arendrup MC, and Patterson TF (2017) Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216, S445–S451. [DOI] [PubMed] [Google Scholar]
- (31).Clancy CJ, and Nguyen MH (2017) Emergence of Candida auris: an international call to arms. Clin. Infect. Dis. 64, 141–143. [DOI] [PubMed] [Google Scholar]
- (32).Ostrowsky B, Greenko J, Adams E, Quinn M, O’Brien B, Chaturvedi V, Berkow E, Vallabhaneni S, Forsberg K, Chaturvedi S, Lutterloh E, and Blog D (2020) Candida auris isolates resistant to three classes of antifungal medications - New York, 2019. MMWR Morb Mortal Wkly Rep 69, 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, and Jones RN (2012) Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J. Clin Microbiol 50, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Liu Y, Li Y, and Wang X (2016) Acetohydroxyacid synthases: evolution, structure, and function. Appl. Microbiol. Biotechnol. 100, 8633–8649. [DOI] [PubMed] [Google Scholar]
- (35).McCourt JA, and Duggleby RG (2006) Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids 31, 173–210. [DOI] [PubMed] [Google Scholar]
- (36).Garcia MD, Chua SMH, Low YS, Lee YT, Agnew-Francis K, Wang JG, Nouwens A, Lonhienne T, Williams CM, Fraser JA, and Guddat LW (2018) Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections. Proc. Natl. Acad. Sci. U. S. A. 115, E9649–E9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).McCourt JA, Pang SS, Guddat LW, and Duggleby RG (2005) Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase. Biochemistry 44, 2330–2338. [DOI] [PubMed] [Google Scholar]
- (38).Casadevall A, and Pirofski LA (2003) The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jabra-Rizk MA, Kong E, Tsui C, Nguyen M, Clancy CJ, Fidel PL Jr, and Noverr M (2016) Candida albicans pathogenesis: fitting within the “host-microbe damage response framework. Infect. Immun. 84, 2724–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Perfect JR (2012) The impact of the host on fungal infections. Am. J. Med. 125, S39–51. [DOI] [PubMed] [Google Scholar]
- (41).van de Veerdonk FL, Joosten LA, Shaw PJ, Smeekens SP, Malireddi RK, van der Meer JW, Kullberg BJ, Netea MG, and Kanneganti TD (2011) The inflammasome drives protective Th1 and Th17 cellular responses in disseminated candidiasis. Eur. J. Immunol. 41, 2260–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Abdallah DM, Nassar NN, and Abd-El-Salam RM (2011) Glibenclamide ameliorates ischemia-reperfusion injury via modulating oxidative stress and inflammatory mediators in the rat hippocampus. Brain Res. 1385, 257–262. [DOI] [PubMed] [Google Scholar]
- (43).Cui W, Zhang S, Cai Z, Hu X, Zhang R, Wang Y, Li N, Chen Z, and Zhang G (2015) The antidiabetic agent glibenclamide protects airway hyperresponsiveness and inflammation in mice. Inflammation 38, 835–845. [DOI] [PubMed] [Google Scholar]
- (44).Huang K, Gu Y, Hu Y, Ji Z, Wang S, Lin Z, Li X, Xie Z, and Pan S (2015) Glibenclamide Improves Survival and Neurologic Outcome After Cardiac Arrest in Rats. Crit. Care Med. 43, e341–9. [DOI] [PubMed] [Google Scholar]
- (45).Hughes FM Jr., Hill HM, Wood CM, Edmondson AT, Dumas A, Foo WC, Oelsen JM, Rac G, and Purves JT (2016) The NLRP3 Inflammasome Mediates Inflammation Produced by Bladder Outlet Obstruction. J. Urol. 195, 1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Liao J, Kapadia VS, Brown LS, Cheong N, Longoria C, Mija D, Ramgopal M, Mirpuri J, McCurnin DC, and Savani RC (2015) The NLRP3 inflammasome is critically involved in the development of bronchopulmonary dysplasia. Nat. Commun. 6, 8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Schattling B, Steinbach K, Thies E, Kruse M, Menigoz A, Ufer F, Flockerzi V, Bruck W, Pongs O, Vennekens R, Kneussel M, Freichel M, Merkler D, and Friese MA (2012) TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat. Med. 18, 1805–1811. [DOI] [PubMed] [Google Scholar]
- (48).Sheth KN, Simard JM, Elm J, Kronenberg G, Kunte H, and Kimberly WT (2016) Human Data Supporting Glyburide in Ischemic Stroke. Acta Neurochir Suppl 121, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Danielski LG, Giustina AD, Bonfante S, Barichello T, and Petronilho F (2020) The NLRP3 Inflammasome and Its Role in Sepsis Development. Inflammation 43, 24–31. [DOI] [PubMed] [Google Scholar]
- (50).Fidel PL Jr., Barousse M, Espinosa T, Ficarra M, Sturtevant J, Martin DH, Quayle AJ, and Dunlap K (2004) An intravaginal live Candida challenge in humans leads to new hypotheses for the immunopathogenesis of vulvovaginal candidiasis. Infect. Immun. 72, 2939–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yano J, Noverr MC, and Fidel PL Jr. (2017) Vaginal heparan sulfate linked to neutrophil dysfunction in the acute inflammatory response associated with experimental vulvovaginal candidiasis. mBio 8, 00211–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Borghi M, De Luca A, Puccetti M, Jaeger M, Mencacci A, Oikonomou V, Pariano M, Garlanda C, Moretti S, Bartoli A, Sobel J, van de Veerdonk FL, Dinarello CA, Netea MG, and Romani L (2015) Pathogenic NLRP3 inflammasome activity during Candida infection is negatively regulated by IL-22 via activation of NLRC4 and IL-1Ra. Cell Host Microbe 18, 198–209. [DOI] [PubMed] [Google Scholar]
- (53).Black CA, Eyers FM, Russell A, Dunkley ML, Clancy RL, and Beagley KW (1998) Acute neutropenia decreases inflammation associated with murine vaginal candidiasis but has no effect on the course of infection. Infect. Immun. 66, 1273–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Peters BM, Palmer GE, Nash AK, Lilly EA, Fidel PL Jr., and Noverr MC (2014) Fungal morphogenetic pathways are required for the hallmark inflammatory response during Candida albicans vaginitis. Infect. Immun. 82, 532–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Jaeger M, Carvalho A, Cunha C, Plantinga TS, van de Veerdonk F, Puccetti M, Galosi C, Joosten LA, Dupont B, Kullberg BJ, Sobel JD, Romani L, and Netea MG (2016) Association of a variable number tandem repeat in the NLRP3 gene in women with susceptibility to RVVC. Eur. J. Clin. Microbiol. Infect. Dis. 35, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Schrödinger Release 2020–3: Glide; Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- (57).Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, and Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749. [DOI] [PubMed] [Google Scholar]
- (58).Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, and Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- (59).Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, and Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759. [DOI] [PubMed] [Google Scholar]
- (60).Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, Hauenstein AV, Wu Z, Nunez G, Mao Y, and Wu H (2019) Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Coll RC, Hill JR, Day CJ, Zamoshnikova A, Boucher D, Massey NL, Chitty JL, Fraser JA, Jennings MP, Robertson AAB, and Schroder K (2019) MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol. 15, 556–559. [DOI] [PubMed] [Google Scholar]
- (62).Mekni N, De Rosa M, Cipollina C, Gulotta MR, De Simone G, Lombino J, Padova A, and Perricone U (2019) In silico insights towards the identification of NLRP3 druggable hot spots. Int. J. Mol. Sci. 20, 4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Tapia-Abellan A, Angosto-Bazarra D, Martinez-Banaclocha H, de Torre-Minguela C, Ceron-Carrasco JP, Perez-Sanchez H, Arostegui JI, and Pelegrin P (2019) MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 15, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Schrödinger Release 2020–3: Induced Fit Docking protocol; Glide; Prime, Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- (65).Sherman W, Beard HS, and Farid R (2006) Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 67, 83–84. [DOI] [PubMed] [Google Scholar]
- (66).Schrödinger Release 2020–3: Desmond Molecular Dynamics System, D. E. Shaw Research: New York, NY, 2020. [Google Scholar]
- (67).Schrödinger Release 2020–3: Maestro-Desmond Interoperability Tools, Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- (68).Schrödinger Release 2020–3: LigPrep, Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- (69).Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, and Friesner RA (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296. [DOI] [PubMed] [Google Scholar]
- (70).Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, and Uchimaya M (2007) Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput.-Aided Mol. Des. 21, 681–691. [DOI] [PubMed] [Google Scholar]
- (71).Hevener KE, Zhao W, Ball DM, Babaoglu K, Qi J, White SW, and Lee RE (2009) Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 49, 444–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Willems HME, Lowes DJ, Barker KS, Palmer GE, and Peters BM (2018) Comparative Analysis of the Capacity of the Candida Species To Elicit Vaginal Immunopathology. Infect. Immun. 86, e00527–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Gietz RD, and Woods RA (2005) Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 313, 107–120. [DOI] [PubMed] [Google Scholar]
- (74).Wilson RB, Davis D, and Mitchell AP (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Roehm NW, Rodgers GH, Hatfield SM, and Glasebrook AL (1991) An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J. Immunol. Methods 142, 257–265. [DOI] [PubMed] [Google Scholar]
- (76).Noake T, Kuriyama T, White PL, Potts AJ, Lewis MA, Williams DW, and Barnes RA (2007) Antifungal susceptibility of Candida species using the Clinical and Laboratory Standards Institute disk diffusion and broth microdilution methods. J. Chemother. 19, 283–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


