Abstract
Glucuronoyl esterases (GEs) are carbohydrate active enzymes in carbohydrate esterase family 15 which are involved in the hydrolysis of lignin-carbohydrate complexes. They are encoded by a wide range of aerobic and anaerobic fungi and bacteria inhabiting diverse environments. The rumen microbiome is a complex microbial community with a wide array of enzymes that specialize in deconstructing plant cell wall carbohydrates. Enzymes from the rumen tend to show low similarity to homologues found in other environments, making the rumen microbiome a promising source for the discovery of novel enzymes. Using a combination of phylogenetic and structural analysis, we investigated the structure-function relationship of GEs from the rumen bacteria Fibrobacter succinogenes and Ruminococcus flavefaciens, and from the rumen fungus, Piromyces rhizinflata. All adopt a canonical α/β hydrolase fold and possess a structurally conserved Ser-His-Glu/Asp catalytic triad. Structural variations in the enzymes are localized to loops surrounding the active site. Analysis of the active site structures in these enzymes emphasized the importance of structural plasticity in GEs with non-canonical active site conformations. We hypothesize that interkingdom HGT events may have contributed to the diversity of GEs in the rumen, and this is demonstrated by the phylogenetic and structural similarity observed between rumen bacterial and fungal GEs. This study advances our understanding of the structure-function relationship in glucuronoyl esterases and illuminates the evolutionary dynamics that contribute to enzyme diversity in the rumen microbiome.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10930-024-10221-0.
Keywords: Glucuronoyl esterase, CAZyme, Carbohydrate esterase family 15, Neocallimastigomycota, Rumen
Introduction
Ruminants first evolved an estimated 50 million years ago (Mya) and are the most widely adapted livestock on earth, with ~ 200 species inhabiting environments ranging from the high arctic to the tropics [1]. The rumen is a large anaerobic chamber in the foregut of ruminants and is the principal site of feed digestion. The efficiency with which ruminants can utilize fibrous crop residues and by-products to produce milk and meat is unique amongst livestock, enabling the inclusion of plant materials in their diets that are unsuitable for human consumption. Like all mammalian herbivores, ruminants do not produce cellulolytic or hemi-cellulolytic enzymes to degrade ingested plant material; relying instead on a symbiotic association with rumen bacteria, fungi, methanogens, and protozoa to produce a diverse range of enzymes that break down the lignocellulose matrix in plant cell walls [2–4]. The enzymes produced by rumen microbes convert plant polysaccharides into sugars which are subsequently fermented to the volatile fatty acids (VFAs): acetate, propionate, and butyrate [2, 3]. The resulting VFAs and microbial protein are utilized by the animal for maintenance energy, growth, and lactation [5].
Plant biomass makes up the largest component of the feed consumed by ruminant livestock [6]. The lignocellulose matrix of plant cell walls is composed of a complex network of cellulose, hemicellulose, pectin, and the phenolic polymer lignin. The diversity of chemical groups and linkages in plant cell walls, and the extensive network of covalent linkages with lignin, make lignocellulose highly recalcitrant [7]. Substantial effort has been made to understand the role that carbohydrate active enzymes (CAZymes) play in breaking down the plant cell wall, however, the mechanisms employed by microbes to break down lignin-carbohydrate complexes (LCCs) are less well defined [8]. Microbes in aerobic environments express lignin-modifying enzymes including laccases, manganese peroxidases, and lignin peroxidases, which together with carbohydrate esterases, digest LCCs [9, 10]. The hydrolysis of LCCs in the gut is primarily attributed to the action of carbohydrate esterases [8, 11], and while it is generally thought that lignin digestion does not occur in anaerobic environments, recent evidence is challenging this dogmatic view [12].
Glucuronoyl esterases (GEs) within the carbohydrate esterase family 15 (CE15) esterase family, in a range of aerobic and anaerobic fungi and bacteria, have been identified as playing an important role in breaking down LCCs [8, 13, 14] by cleaving the ester bond between 4-O-methyl-d-glucuronyl in xylan and alcohol groups in lignin [15, 16]. There is mounting evidence that the removal of glucuronic acid residues significantly improves the saccharification of lignocellulosic feed stocks [17–19], suggesting that the activity of these enzymes acts in synergy with glycosyl hydrolases. As of Aug 10 2023, of the ~ 747 unique carbohydrate esterase family 15 esterases identified in the CAZy database, only 22 have been biochemically characterized and have generally been found to exhibit narrow substrate specificity, with little to no activity against the model substrates typically hydrolyzed by esterases [15, 20]. The structures of 9 GEs have been determined from the carbohydrate esterase family 15 esterase family and all share a conserved α/β-hydrolase fold. Outside of the core scaffold, GEs display structural variability in the loops surrounding the catalytic site, and this hypothesized to contribute to their in-vivo specificity against LCCs [8]. Bacterial GEs typically possess variable insertions in an N-terminal loop (RegN) and at 3 regions surrounding the active site referred to as Reg1-Reg3. These insertions make bacterial GE active sites deeper, and more contoured than the active sites observed in fungal counterparts [15, 16, 21]. GEs share an invariant Ser-His-Glu/Asp catalytic triad and employ a conserved catalytic mechanism that involves the formation of an acyl-enzyme covalent intermediate, which is subsequently cleaved through acid-base hydrolysis. Both the acylation and deacylation steps involve the formation of a tetrahedral transition state and are stabilized by the catalytic histidine and a conserved arginine residue that is found adjacent to the catalytic serine [22, 23]. The position of the catalytic nucleophile and general base is highly conserved; however, the general acid has been observed at two distinct locations in the structure [24]. To reflect this variability, GEs are classified as possessing a canonical (CE15-B) or non-canonical (CE15-A) catalytic triad. This classification is based on the structural arrangement of the catalytic acid and whether they are located on loops following β7 or β8, respectively. It is unclear how these differences in the arrangement of the catalytic triad impact enzyme function, but it may be related to differences in the structure of the natural substrate targeted by the enzymes [24]. Catalytic residues of classical α/β-hydrolase serine esterases have also been classified as having a canonical (A-type) and non-canonical (B-type) arrangement based on the position of the general acid and whether they are located on a loop following β7 or β6, respectively [23–25]. Unfortunately, the discrepancy between GE and classical α/β-hydrolase nomenclature can cause confusion. We note that we are considering the spatial arrangement of catalytic residues using the GE specific classification scheme put forth by Ernst and colleagues [24].
The microbial diversity of the rumen represents a vast genetic pool that is ideal for the discovery of novel enzymes targeting plant cell wall carbohydrates. Many of the enzymes encoded by rumen microbes have low homology to related proteins found in other environments, which makes the rumen microbiome a valuable resource for the discovery of novel biocatalysts [26]. To this end, we have conducted an analysis of GE diversity within rumen microbes and examined the structure-function relationship of three GEs from Fibrobacter succinogenes, Ruminococcus flavefaciens, and Piromyces rhizinflata. This work aimed to contribute additional knowledge about the diversity of GEs within the carbohydrate esterase family 15 esterase family and to expand our understanding of the structure-function relationship within these enzymes.
Methods
Phylogenetic analysis of the carbohydrate esterase family 15 family of esterases: A phylogenetic tree of the carbohydrate esterase family 15 family of esterases was built using SACCHARIS on August 10 2023 [27]. GEs from rumen bacteria and fungi were identified within the Hungate 1000 dataset [28] and the genomes of Neocallimastigomycota that are available in Mycocosm [29]. The carbohydrate esterase family 15 HMM model was obtained from dbCAN2 [30], and HMMER3 [31] was used to search the rumen microbe genome dataset. Carbohydrate esterase family 15 sequences in the CAZy database (http://www.cazy.org/; [32, 33]) were downloaded (August 10 2023), combined with the rumen carbohydrate esterase family 15 esterase dataset, and dereplicated using CD-HIT [34, 35] using a sequence identity cut-off of 100% to remove duplicate sequences. The resulting dereplicated sequences were used as input to SACCHARIS for phylogenetic analyses [27] (https://github.com/saccharis/SACCHARIS_2). Briefly, CD-HIT parsed sequences were pruned to the carbohydrate esterase family 15 domain using dbCAN2 [30] and then aligned using MUSCLE [36]. ModelTest-NG [37] was used for best-fit model selection using the sequence alignment, and FastTree [38] was used to generate the phylogenetic tree.
Protein production and purification: Genes encoding GEs from Ruminococcus flavefaciens (the C-terminal carbohydrate esterase family 15 esterase domain of CesA herein referred to as RfCE15), Fibrobacter succinogenes (FsCE15), and the rumen fungus Piromyces rhizinflata (PrCE15) were synthesized and codon optimized for protein production in Escherichia coli (Thermo Fisher). The Gateway cloning system was used to generate both C- and N-terminal His6 tagged destination vectors from the Champion™ pET300/NT-DEST and pET301/CT-DEST Gateway™ Vector Kit (Thermo Fisher). Protein production constructs were sequence verified and transformed into BL21(DE3) E. coli (New England Biolabs) for protein production. Cells were grown in Lysogeny Broth (LB) supplemented with 100 µL mL-1 ampicillin to an optical density (600 nm) of 0.6–0.8 at which point protein production was induced with the addition of Isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Protein production was carried out at 25 °C for 18 h in an incubated shaker. Cells were harvested by centrifugation at 4,000 × g for 20 min at 4 °C. The resulting cell pellet was resuspended in 20 mL of lysis buffer (0.5 M NaCl and 20 mM HEPES pH 7.5) and a protease inhibitor tablet (Roche) was dissolved in the cell suspension. Cells were ruptured by sonication and cell debris was removed by centrifugation at 30,000 × g for 45 min at 4 °C. Protein was purified to homogeneity via a combination of Ni-IMAC (nickel immobilized metal-affinity chromatography) and size exclusion chromatography with a Superdex 200 column. Protein concentration was determined by measuring absorbance at 280 nm using the molar absorption coefficient calculated by ProtParam [39]. Purified protein was stored in 20 mM HEPES (pH 7.5) and 200 mM NaCl. Protein was used immediately, or flash frozen in liquid nitrogen and stored at -80 °C.
Protein crystallization: Initial crystallization trials were carried out with Crystal Screen 1 + 2 (Hampton Research) and PACT premier™/JCSG-plus™ (Molecular Dimensions) screens in Intelli-Plate 96 − 2 sitting drop crystallization plates (Hampton Research). Protein was mixed in a 1:1 ratio of protein: reservoir with a final drop volume of 2 µl. 100 µL of crystallization solution was added to reservoirs. Crystallization hits were optimized in a 24 well sitting drop Cryschem crystallization plate (Hampton Research). Diffraction quality crystals were obtained for the C-terminally His-tagged construct of FsCE15 and RfCE15 and the N-terminally His-tagged construct of PrCE15. Optimized crystallization conditions were: 25 mg/ml FsCE15, 10 mM ZnCl2, 18% PEG 6000, and 100 mM sodium acetate-acetic acid buffer (pH 5.0); 30 mg/ml RfCE15, 11-13% 2-propanol, 18-20% PEG 4000; 30 mg/ml PrCE15, 100 mM HEPES (pH 7.0), 20% PEG 6000. Crystals were mounted in nylon loops in the presence of 18% glycerol as a cryoprotectant and flash frozen in liquid nitrogen.
Data collection and structure determination: Diffraction data was collected at -173 °C on beamline 08ID-1 at the Canadian Light Source (Saskatoon, SK) at a wavelength of 0.97949 Å. Data was integrated with XDS [40] and scaled with Scala and the CCP4 suite of programs [41]. The structure was solved by molecular replacement with Phaser-MR in Phenix [42]. The structure of the GE from Trichoderma reesei, Cip2 (PDB: 3PIC, [43]) was trimmed to a poly-ala backbone and used as a search model to solve the structures of PrCE15 and FsCE15. Using a similar approach, the structure of RfCE15 was solved using the GE from Sporotrichum thermophile (PDB: 4G4G, [44]) as a search model. The resulting models served as a starting point for iterative cycles of TLS (Translation–Libration–Screw-rotation), positional, real space and B-factor refinement in Phenix [42], followed by manual model building with Coot [45]. Refinement was monitored by flagging 5% of all reflections as “free” [46]. Stereochemistry of the model was monitored throughout model building and refinement using the Phenix Validation tools. Statistics for data collection and refinement are shown in Table 1.
Table 1.
X-ray data collection and refinement statistics for the structures of the glucuronoyl esterases from F. succinogenes (FsCE15), P. rhizinflata (PrCE15), and R. flavefaciens (RfCE15). Statistics for the highest-resolution shell are shown in parentheses
| Data Collection | FsCE15 | PrCE15 | RfCE15 |
|---|---|---|---|
| Beamline | CLSI 08ID-1 | CLSI 08ID-1 | CLSI 08ID-1 |
| Wavelength (Å) | 1.0334 | 1.0334 | 1.0334 |
| Space group | P 41 21 2 | P 61 | P 21 21 21 |
| Cell Dimensions | |||
| a, b, c (Å) | 57.28 57.28 222.16 | 139.68 139.68 125.83 | 51.95 60.87 124.99 |
| α, β, γ | 90.00, 90.00, 90.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 45.31–1.93 (1.99–1.93) | 46.74–2.54 (2.63–2.54) | 43.6–1.25 (1.30–1.25) |
| Rmerge | 0.167 (1.02) | 0.0813 (0.933) | 0.0882 (1.05) |
| Rpim | 0.0494 (0.291) | 0.0265 (0.300) | 0.0358 (0.427) |
| CC1/2 | 0.998 (0.8940) | 0.999 (0.858) | 0.999 (0.724) |
| Wilson B-factor | 26.4 | 59.7 | 12.8 |
| < I/σI> | 10.99 (2.86) | 19.03 (2.76) | 11.27 (1.85) |
| Completeness (%) | 99.97 (100.0) | 99.96 (100.0) | 99.96 (100.0) |
| Redundancy | 12.5 (13.2) | 10.4 (10.6) | 7.0 (6.9) |
| Total reflections | 362,382 (36,927) | 478,327 (48,424) | 773,121 (74,487) |
| Unique reflections | 28,949 (2791) | 45,952 (4555) | 110,196 (10,867) |
| Refinement | |||
| Resolution (Å) | 1.93 | 2.54 | 1.25 |
| Rwork/Rfree | 0.228/0.285 | 0.229/0.269 | 0.168/0.190 |
| Protein residues | 380 | 773 | 409 |
| Non-hydrogen atoms | |||
| Protein | 2858 | 5835 | 3222 |
| Ligands | 0 | 0 | 12 (GOL) |
| Water | 108 | 83 | 335 |
| RMSD | |||
| Bond lengths (Å) | 0.007 | 0.01 | 0.005 |
| Bond angles (°) | 0.84 | 1.08 | 0.81 |
| Ramachandran (%) | |||
| Preferred | 95.99 | 92.85 | 96.56 |
| Allowed | 4.01 | 7.15 | 3.44 |
| Disallowed | 0 | 0 | 0 |
| B-factors (Å2) | |||
| Protein | 33.77 | 62.99 | 14.89 |
| Ligands | 0 | 0 | 20.91 (GOL) |
| Water | 32.1 | 54.59 | 23.04 |
| PDB ID | 8TRU | 8TRX | 8TSE |
Enzyme Assays: d-glucuronic acid methyl ester (MeGlcA), d-galacturonic acid methyl ester (MeGalA), Benzyl d-glucuronate (BnzGlcA), and Allyl D-glucuronate (AllylGlcA) (all from Carbosynth) were dissolved in 99% DMSO and filtered with a 0.45 μm syringe filter. A 3-component buffer system (25 mM acetic acid, 25 mM MES, 50 mM Tris-HCl, pH variable) was used for all assays as previously described [16]. Uronic acid formation was measured continuously in a BioTec Synergy™ HT multi-detection microplate reader using the d-Glucuronic Acid/d-Galacturonic Acid (K-Uronic) Assay Kit (Megazyme). Due to substrate instability, a control that did not contain enzyme served as an auto-hydrolysis blank. The blank reaction was measured continuously in parallel with the enzyme containing reactions, and the rate of auto-hydrolysis was subtracted from enzyme catalyzed hydrolysis. Assays to examine the effect of pH on enzyme activity were conducted using BnzGlcA at a concentration of 2 mM and varying the pH of the 3-component buffer. Kinetic assays were performed with BnzGlcA (0.5–10 mM) at pH 7.0 and kinetic parameters were calculated using SigmaPlot. To examine substrate specificity, the specific activity of the enzymes against 2 mM MeGlcA, MeGalA, and AllylGlcA was measured at pH 7.0 and the activity against these substrates was compared relative to BnzGlcA and expressed as a percentage of 100%.
Thermal stability of enzymes: A Protein Thermal Shift™ qPCR assay (Thermo Fisher) was performed in a qPCR plate MicroAmp™ Fast Optical 96-Well Reaction Plate, using a Quantstudio 6 Flex qPCR system (Applied Biosystems). Stability assays were carried out in 20 µL reactions containing: 2 µL 200 X diluted SYPRO™ Orange Protein Gel Stain (Thermo Fisher), 1 µL 20 X three component buffer pH 7, 15 µL of 14 µM enzyme (Cf = 10.5 µM) (replaced with d2H2O for blanks), and 2 µL of additive(s). Additives in the assay were d2H2O (Control), 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 100 mM and 500 mM NaCl. Four technical replicates were included for each unique condition examined in the assay and the experiment was repeated twice. The Tm was calculated using the program TSA CRAFT [47].
Results
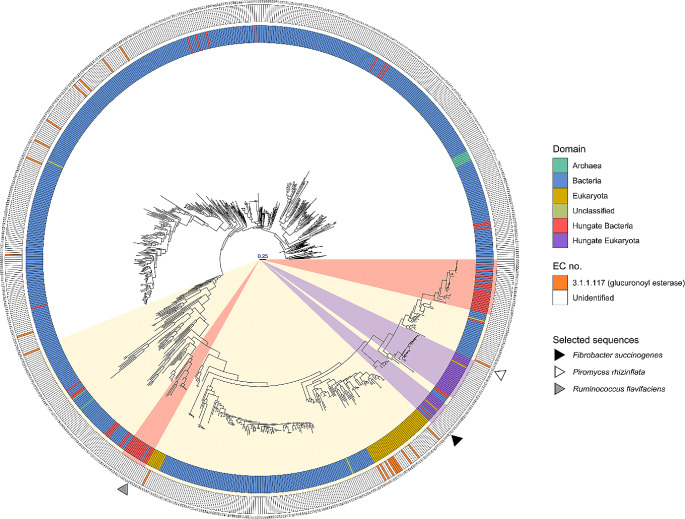
Phylogenetic analysis of carbohydrate esterase family 15 esterases in the rumen: A search of the Hungate 1000 collection identified 65 carbohydrate esterase family 15 esterases encoded in 53 of the 418 bacterial genomes. The majority (45/53) of the rumen bacteria which encoded a GE tended to possess a single GE. Multiple copies of carbohydrate esterase family 15 esterases were primarily identified in bacteria within the phylum Bacteroidota (Bacteroides sp, Porphyromonadaceae, and Prevotella). An unknown Lachnospiraceae was the only bacteria outside of the Bacteroidota with multiple carbohydrate esterase family 15 esterase containing ORFs. A search of the 10 genome sequences of anaerobic gut fungi (AGF) within the phylum Neocallimastigomycota identified an additional 36 carbohydrate esterase family 15 esterases. All AGF genomes were found to encode multiple genes for GEs, ranging in number from 2 to 6 orthologues, with an average of 4 per genome (Supplementary Data 1). In agreement with previous phylogenetic analysis of the carbohydrate esterase family 15 family, we observed that the proteins formed 2 super clades (Fig. 1). Clade 1 consisted mainly of bacterial carbohydrate esterase family 15 esterases, with a small cluster of archaeal sequences, while clade 2 (highlighted in light yellow) consisted of bacterial, archaeal, and fungal enzymes. Despite the identification of carbohydrate esterase family 15 esterases in archaea, they were notably scarce (8 out of 826), all of which originated from a family of extreme halophiles within the order Halobacteriales. The rumen carbohydrate esterase family 15 esterases were found predominantly in 4 main clusters within clade 2 (Fig. 1; rumen eukaryotic clusters highlighted in purple, rumen bacterial clusters highlighted in red).
Fig. 1.
Phylogenetic analysis of rumen carbohydrate esterase family 15 esterases. Glucuronoyl esterases in the carbohydrate esterase family 15 esterase family from the CAZy database (accessed August 10, 2023) and the rumen collection were clustered by their sequence identity, used as query sequence inputs for SACCHARIS [27], and embedded into a mid-point rooted phylogenetic tree. Coloured rings surrounding the tree highlight sequence characteristics by domain (inner) and EC number and CAZy database annotated function (outer). Carbohydrate esterase family 15 esterases from anaerobic gut fungi and rumen bacteria are highlighted in purple (inner ring) and red, respectively. Proteins that have been biochemically characterized are shown in orange (outer ring). Proteins characterized in this study are highlighted with a triangle on the outside of the circle. Sequences used to generate the tree are numbered and accession codes provided in Supplementary Data 1
Structure of rumen GEs: Although we attempted to express genes and crystallize proteins of GEs from a range of rumen microbes, only FsCE15, PrCE15, and RfCE15 produced diffraction quality crystals. Statistics for the data collection and refinement are shown in Table 1. Crystals of FsCE15 belonged to space group P41212 with a single protein molecule in the asymmetric unit. The structure of FsCE15 was refined to Rwork/Rfree of 22.8%/28.5% (respectively) using data to a maximum resolution of 1.93 Å. Crystals of PrCE15 belonged to space group P61 with a single protein molecule in the asymmetric unit. The structure of PrCE15 was refined to Rwork/Rfree of 22.9%/26.9% using data with a maximum resolution of 2.54 Å. Crystals of RfCE15 belonged to space group P212121 with 2 protein molecules in the asymmetric unit. The structure of RfCE15 was refined to Rwork/Rfree of 16.8%/19.0% using data with a maximum resolution of 1.25 Å. All of the residues in the three models were found in allowable regions of the Ramachandran plot. Structures have been deposited to the protein databank with accession codes: 8TRU (FsCE15), 8TRX (PrCE15), 8TSE (RfCE15).
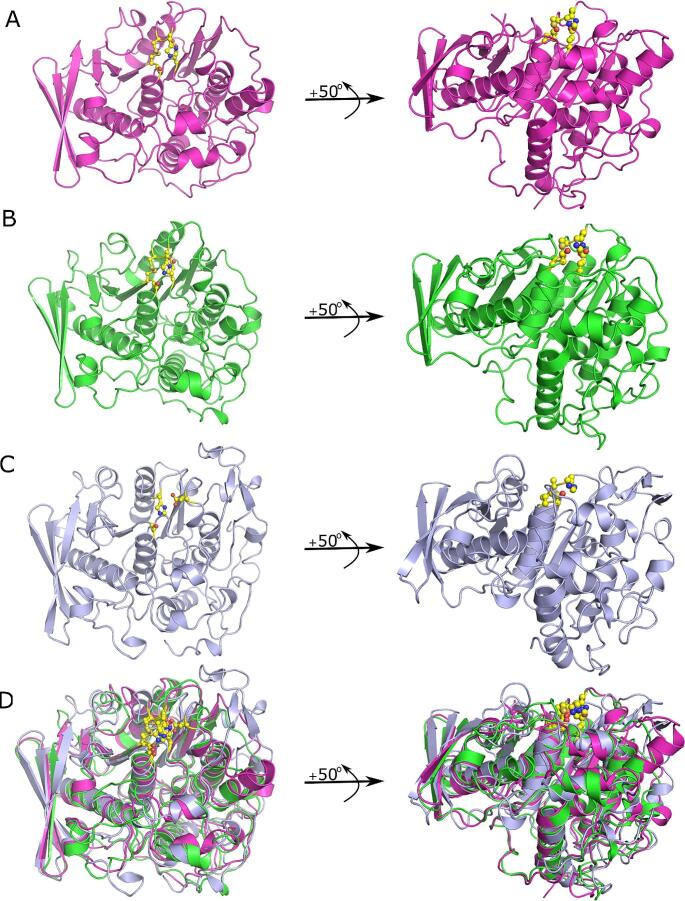
Rumen GEs adopt a canonical α/β hydrolase fold: FsCE15, PrCE15, and RfCE15 adopted the canonical α/β-hydrolase fold that is seen in all of the GE structures solved to date (Fig. 2). The fold of all three enzymes consists of a central twisted 10 stranded mixed β-sheet flanked by α-helices on either side. Structure-based least square superpositions of PrCE15, FsCE15, and RfCE15 had Cα root mean square deviations (RMSD) of < 1.3 Å with the structural variation being limited to surface exposed loops (Fig. 2D). All of the enzymes lacked insertions at the RegN and Reg1-Reg3 variable regions that are typically observed in bacterial GEs and displayed a flat active site cleft common with fungal GEs.
Fig. 2.
Comparison of the fold of rumen glucuronoyl esterases within the carbohydrate esterase family 15 esterase family. (A) Fibrobacter succinogenes (FsCE15), (B) Piromyces rhizinflata (PrCE15), and (C) Ruminococcus flavefaciens (RfCE15) adopt the canonical α/β-hydrolase fold that is seen in all of the GE structures solved to date. (D) Structure-based least squares structural superposition of FsCE15, PrCE15 and RfCE15. Catalytic residues, and the conserved disulfide bridge in the active sites of FsCE15 and PrCE15 are shown as yellow ball and sticks. Figures were generated with PyMol
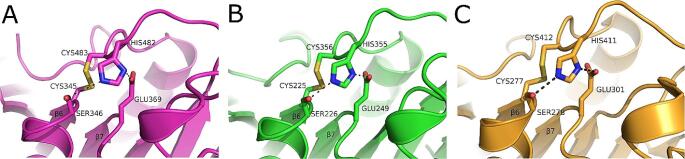
Comparative Analysis of the FsCE15 and PrCE15 Active Sites: To identify the putative catalytic residues in FsCE15 and PrCE15, a structure based superposition with the T. reesei GE Cip2 was generated. FsCE15 and PrCE15 had an RMSD of 0.87 Å and 1.1 Å with Cip2, respectively. The location and identity of the experimentally validated catalytic residues of Cip2 were conserved in both FsCE15 and PrCE15. The putative catalytic nucleophile, general base, and general acid of FsCE15 are Ser346, His482, and Glu369, respectively (Fig. 3A). A similar examination of these resides in PrCE15 putatively identifies the catalytic residues as Ser226, His355, and Glu249 (Fig. 3B). The catalytic residues of FsCE15 and PrCE15 are positioned in the canonical conformation characteristic of CE15-B subfamily [24] with the general acid located on a loop at the end of β7 (Fig. 3). FsCE15 and PrCE15 have a conserved disulfide bond between residues Cys345 - Cys483 and Cys225 - Cys356, respectively, which anchors the general base within optimal hydrogen bonding distance (2.7 Å) to the catalytic serine. This disulfide is conserved amongst CE15-B subfamily GEs that adopt a canonical catalytic triad and can be seen in the glucuronoyl esterase Cip2 from (Fig. 3C), Lentithecium fluviatile LfCE15C [48], Cerrena unicolor CuGE [24], and S. thermophile StCE15 [44]. The glycine and lysine residues of the nucleophilic elbow GXSRXGK motif [48] are conserved in both FsCE15 and PrCE15. The arginine following the catalytic serine in the motif contributes to the stabilization of the transition state, and while the arginine is conserved in FsCE15 it is replaced with a tyrosine in PrCE15. Interestingly, it was more common for this residue to be tyrosine in AGF GEs (26 out of 35 sequences) with arginine found at this position in the nine remaining rumen enzymes.
Fig. 3.
Canonical configuration of catalytic residues in the active site of glucuronoyl esterases within the CE15-B subfamily. (A) Glucuronoyl esterase from Fibrobacter succinogenes, FsCE15, (B) Glucuronoyl esterase from Piromyces rhizinflata, PrCE15, (C) Glucuronoyl esterase from Trichoderma reesei, Cip2. All figures were generated with PyMOL
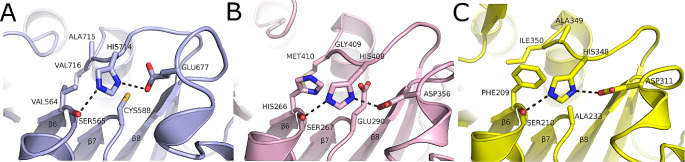
RfCE15 catalytic residues adopt a non-canonical conformation: Similar to FsCE15 and PrCE15, RfCE15 adopts the same α/β-hydrolase fold that is highly conserved amongst GEs in the carbohydrate esterase family 15 CAZy family (Fig. 2D). RfCE15 is the C-terminal GE domain of a multi-modular enzyme from R. flavefaciens called CesA. CesA was first described by Aurilia an colleagues as an acetylxylan esterase with a domain of unknown function (DUF) [47]. The DUF was later characterized as a GE belonging to the carbohydrate esterase family 15 CAZy family [48]. The catalytic residues of RfCE15 are observed in a non-canonical configuration, with the putative general acid Glu677 located on a loop at the end of β8; making RfCE15 a member of the CE15-A GE subfamily. The nucleophilic serine (Ser565) is positioned similarly to the conformation observed in all other carbohydrate esterase family 15 esterases and located at the base of the active site in the characteristic GXSRXGK sequence motif. The glycine, lysine, and arginine residues within this motif were all conserved in RfCE15. The general base (His714) is located on a loop directly above the catalytic serine, but is shifted by 2.2 Å relative to the position of this loop in FsCE15 and PrCE15. The shift in this position accommodates the non-canonical conformation of the general acid residue (Fig. 4A). Despite the shift in Cα of His714, the imidazole ring occupies a similar location as that found in the other GEs, with His714 Nε2 making a 2.8 Å hydrogen bond with Oγ of Ser565, and a 2.9 Å bidentate hydrogen bond with the carboxyl functional group of Glu677 (Fig. 4A).
Fig. 4.
Non-canonical configuration of catalytic residues in the active site of glucuronoyl esterases within the CE15-A subfamily (A) C-terminal glucuronoyl esterase domain from Ruminococcus flavefaciens CesA, RfCE15 (B) Glucuronoyl esterase from Optitus terrae, OtCE15A and (C) Glucuronoyl esterase from Caldicellulosiruptor kristjansonii, CkCE15A. All figures were generated with PyMOL
Structural differences in RfCE15 were also observed in flexible loops located on the enzyme’s surface. Most notably, a 16 amino acid insertion from residues Gly603-Tyr619 forms an extended Ω-loop that wraps around the backside of the general base loop. Despite the presence of this large insertion, the surface profile of RfCE15’s active site is a flat similar to that observed in previously characterized fungal GEs and in FsCE15 and PrCE15 (Fig. 2C). As observed in other CE15-A subfamily GEs, the conserved disulfide bond found in CE15-B GEs, was not observed in RfCE15. Instead, these residues are Val564 and Val715 and form a hydrophobic pocket that is composed of an extended network of Van der Waals interactions between Ile114, Ile121, Met228, Phe314, ILE316, Ile366, and Met369. The contacts between the Ω-loop, the residues in the hydrophobic pocket, and the general base loop may function to stabilize the position of the general base in RfCE15. Examining the equivalent region in other structurally characterized GEs in the CE15-A subfamily from Optitus terrae (PDB: 6gs0, OtCE15A) (Fig. 4B) and Caldicellulosiruptor kristjansonii (PDB: 7nn3, CkCE15A) (Fig. 4C) revealed that the residues in this pocket are not conserved. Despite the lack of sequence conservation, this region displays extensive non-covalent contacts with the general base loop, which may function to stabilize the general base loop, fulfilling a role similar to that of the disulfide bond in CE15-B GEs. Notably, OtCE15A and CkCE15A have Reg2 insertions typical of bacterial GEs, but that is not observed in RfCE15, that also appears to contribute to stabilizing the general base loop in place of the conserved disulfide.
Biochemical Characterization of FsCE15, PrCE15, and RfCE15: The biochemical activity of FsCE15, PrCE15, and RfCE15 was examined using the model substrates: MeGlcA, MeGalA, BnzGlcA, and AllylGlcA. All of the enzymes cleaved BnzGlcA with specific activities ranging from 2.66 U/min to 14.9 U/min (Table 2). The specific activity of PrCE15 and RfCE15 against BnzGlcA was approximately 5X higher than that observed with FsCE15. None of the enzymes were active against MeGlcA or MeGalA. All of the enzymes cleaved AllylGlcA, however the activity was 85–90% lower than the activity observed with BnzGlcA. Based on the substrate preference of all three enzymes, all subsequent biochemical assays were conducted using BnzGlcA. The optimal pH for each enzyme was determined (Supplementary Fig. 1). FsCE15 has a broad pH profile with maximal activity at pH 8.0. It retains ≥ 90% relative activity against BnzGlcA over a pH range from 6.5 to 8.5. RfCE15 and PrCE15 had optimal activity at slightly lower pH values, 6.5-7.0 and 7.0-7.5, respectively. We determined apparent steady state kinetic parameters for PrCE15 and RfCE15. Both enzymes displayed similar affinities for BnzGlcA; with PrCE15 and RfCE15 having apparent Km values of 5.0 ± 0.4 mM and 6.0 ± 0.4 mM, respectively (Table 2). The apparent kcat of PrCE15 was determined to be 22.3 ± 0.7 s-1, compared to an apparent kcat of 91.2 ± 0.8 s-1 for RfCE15. Unfortunately, due to high background absorbance levels above 10 mM substrate, the reaction between FsCE15 and BnzGlcA could not be adequately saturated and we were unable to determine steady-state kinetic parameters for this enzyme. It should be noted that substrate concentrations used to determine steady-state kinetic parameters should ideally reach 10x the concentration of the enzymes Km. Unfortunately, due to the low affinity that all three enzymes displayed for the synthetic substrate BnzGlcA, and the high background absorbance, it was not possible to reach saturating substrate concentrations in any of our assays. Given this limitation, the reported kinetic constants should be considered apparent values and interpreted cautiously. Consistent with previously characterized GEs, none of the enzymes had detectable activity against para-nitrophenyl linked substrates.
Table 2.
Catalytic activity of FsCE15, PrCE15 and RfCE15 against Benzyl D-glucuronate
| Enzyme | pH optimum | Specific activity (µmol min-1mg-1) | Apparent Km (mM) | Apparent kcat (s-1) | Apparent kcat/ Km (s-1M-1) |
|---|---|---|---|---|---|
| PrCE15 | 7.5 | 12.5 ± 0.8 | 5.0 ± 0.4 | 22.3 ± 0.8 | 4485 ± 376 |
| RfCE15 | 6.5 | 14.9 ± 0.3 | 6.0 ± 0.4 | 29.7 ± 1.0 | 4950 ± 392 |
| FsCE15 | 8.0 | 2.66 ± 0.08 | ND*, Reaction could not be saturated | ||
*Not determined
Thermal stability of glucuronoyl esterases: The thermal stability of FsCE15, PrCE15, and RfCE15 was examined using a Thermofluor assay to investigate the role of the conserved disulfide bond in stabilizing the structure of GEs (Table 3). Melting curves can be found in supplementary Fig. 2. PrCE15 displayed a multiphasic unfolding curve however, upon reduction of the disulfide bond, PrCE15 unfolding was monophasic with the higher Tm unfolding event no longer detectable. Similarly, the addition of a reducing agent lowered the Tm of FsCE15 by 11.5 °C. These results demonstrates the important role that the disulfide bond in the active site of CE15-B GEs plays in stabilizing tertiary structure. In contrast, the addition of reducing agent to RfCE15 did not alter its thermal stability, an observation consistent with the lack of disulfide bonds. The influence of ionic strength on protein stability was also examined, but the thermal stability of the GEs was not altered by the addition of up to 500 mM NaCl.
Table 3.
Mean unfolding temperature (Tm) of FsCE15, PrCE15, and RfCE15 in the presence and absence of reducing agent (TCEP) and at increasing ionic strength. The standard error of the mean Tm calculated in duplicate experiments is shown
| Control buffer* | 10 mM TCEP | 100mM NaCl | 500 mM NaCl | |
|---|---|---|---|---|
| PrCE15 | 44.5 ± 0.6/59.9 ± 0.3 | 49.5 ± 0.47 | 41.2 ± 0.3/59.8 ± 0.4 | 42.5 ± 0.3/59.4 ± 0.2 |
| RfCE15 | 44.4 ± 0.04 | 46.3 ± 0.18 | 44.5 ± 0.19 | 44.2 ± 0.04 |
| FsCE15 | 48.7 ± 0.2 | 37.2 ± 0.37 | 47.9 ± 0.23 | 47.5 ± 0.1 |
* 3-component buffer system: 25 mM acetic acid, 25 mM MES, 50 mM Tris-HCl pH 7.0
Discussion
Glucuronoyl Esterases in the Rumen Share Characteristics of Both Prokaryotic and Eukaryotic Enzymes: GEs can be broadly defined in terms of “bacterial“ and “fungal“ origin however, our characterization of these enzymes indicates that this clear differentiation for members of the rumen microbiome is questionable. Sequence based phylogenetic analysis of the carbohydrate esterase family 15 CAZy family has previously found that bacterial GEs display more sequence diversity than fungal GEs [8, 15]; a conclusion supported by our analysis (Fig. 1). When the structural features of GEs are considered alongside sequence, although the GEs from F. succinogenes and R. flavefaciens are bacterial in origin, they both exhibit structural features that are typically observed in fungal GEs. This observation brings up the question as to whether the observed similarity between bacterial and fungal GEs in the rumen arose as a result of an inter-kingdom horizontal gene transfer (HGT) event(s). AGF diverged from a common ancestor ~ 66 Mya, coinciding with the emergence of terrestrial grasses [49]. At this time, there was a shift in the diets of terrestrial mammals towards herbivory and the emergence of ruminants ~ 50 Mya [1]. The intense selective pressures, and unique physicochemical properties that are found in the rumen likely influenced the evolution of AGF.
Neocallimastigomycota are known to possess some of the highest densities of carbohydrate-active enzymes CAZymes of any characterized microbes, with many of themse CAZymes showing low homology to characterized enzymes. AGF also tend to have larger genomes than aerobic fungi due to high levels of gene duplication. This is particularly true of CAZymes, with 25–40% of these genes in AGF having undergone duplication [50–52]. This is clearly evident in GEs, with AGF possessing an average of four GE containing genes in there genomes. It has been hypothesized that microbes encode multiple GE orthologues as a result of each one having distinct biological roles [15]. The rumen microbiome is inhabited by multiple species of AGF and the multiplicity in GE genes observed here may have evolved to enable AGF to effectively complement the enzyme activities of the rumen bacteria in the presence of diverse substrates. This is known to occur within members of Neocallimastigomycota which have evolved mechanisms to target components of the plant cell wall distinct from those of bacteria, thereby facilitating their co-existence in the highly competitive rumen environment [53, 54]. It is possible that HGT between rumen bacteria and AGF also contributed to the expansion of the number of GEs found in the Neocallimastigomycota genome. There is evidence of extensive HGT between CAZymes from rumen bacteria to AGF with over 50% of the carbohydrate esterase family 15 genes in AGF showing evidence that they originated in rumen bacteria [52]. Interestingly, HGT events between Fibrobacterota and AGF were one of the most commonly observed events by Murphey at al. [52]. This observation is supported by our phylogenetic analysis showing that FsCE15 clustered closely with Neocallimastigomycota carbohydrate esterase family 15 esterases (Fig. 1). FsCE15 also possesses structural features that are typically observed in fungal GEs including: a shallow active site lacking any of the insertions in the Reg1-Reg3 regions, catalytic residues that adopt a canonical CE15-A active site conformation, and a conserved disulfide bond in the active site. Although we cannot say which organism this gene may have originated in, the similarity between FsCE15 and PrCE15 may have resulted due to an HGT event that occurred between F. succinogenes and P. rhizinflata at some point in the evolutionary past. FsCE15 is therefore unique amongst bacterial GEs characterized to date and may represent an intermediate between fungal and bacterial GEs.
GEs Play a Role in the Metabolism of Lignocellulose by Rumen Microbes But Their Physiological Substrate Is Unclear: The biochemical properties and kinetic parameters of FsCE15, PrCE15, and RfCE15 are consistent with previously characterized GEs. All three enzymes showed specificity for BnzGlcA but, displayed low affinity and poor catalytic efficiency against synthetic substrates. It is likely that FsCE15, PrCE15, and RfCE15 would show higher levels of catalytic activity against physiological substrates that they target in vivo [13, 15]. Unfortunately, the role of glucuronic acid – lignin cross-linkages in the structural integrity of the cell wall of the crops and grasses typically consumed by ruminants is unclear. Recently, it has been suggested that GEs not only target the LCC-hemicellulose complex but may also play a role in breaking down pectin-lignin LCCs [55]. It is known that genes encoding GEs are expressed during the hydrolysis of lignocellulosic substrates in the rumen [56]. Furthermore, transcriptomic analysis of lignocellulose digestion by P. rhizinflata found that genes encoding GEs are expressed during growth on corn stover, barley straw, and alfalfa; suggesting that these enzymes play a role in the utilization of lignocellulose by the rumen microbiome [53, 56].
Although it has been shown that supplementing enzyme cocktails with GEs can result in improved saccharification [15], no improvements in glucose or xylose release from corn stover or barley straw was observed by the GEs characterized in this work (data not shown). Due to the challenges associated with isolating natural LCC substrates, almost all GEs characterized to date have relied on the use of model substrates. Further research is needed to identify potential LCC substrates in ruminant feeds which would enable GEs from rumen microbes to be characterized using physiologically relevant substrates.
The Structural Properties of Rumen GEs Are Consistent with Previously Characterized Members of the Carbohydrate Esterase 15 CAZy Family: Consistent with other GEs, FsCE15, PrCE15, and RfCE15 adopt a canonical α/β hydrolase fold (Fig. 2), and possess a structurally conserved Ser-His-Glu/Asp catalytic triad (Figs. 3 and 4). Structural variation between FsCE15, PrCE15, and RfCE15 was limited to surface loops surrounding the active site, and these differences were correlated with the presence or absence of a disulfide bond in the active site. Although the position of the catalytic nucleophile and general base is highly conserved in GEs, the general acid has been observed at 2 distinct locations which has led to GEs being classified as possessing a canonical (CE15-B) or non-canonical (CE15-A) catalytic triad [24]. The canonical configuration of the catalytic residues has a Glu located on a loop after β7, whereas the non-canonical configuration has a Glu/Asp located on a flexible loop after β8. There are also examples of GEs that have an acidic residue at both sites, providing functional redundancy in these enzymes [22]. All structurally characterized GEs within the CE15-B subfamily are bacterial in origin and lack the disulfide bond that anchors the general base into position to enable optimal hydrogen bonding with the nucleophilic serine (Fig. 3). In contrast, all of the previously structurally characterized GEs within the CE15-A subfamily posses this covalent bond and originate from fungi. The presence of this disulphide bond also provides stability to GEs, demonstrated by the structural destabilization of the protein structure and lowering of its melting temperature upon reduction of this covalent bond (Table 3). The physiological temperature of the rumen is 39 °C, so it does not seem likely that high thermal stability would be important for these enzymes in vivo. This feature may also contribute to the stability of these enzymes in other ways including protease stability however, this has not been confirmed in vitro.
In the absence of the disulfide bond within the non-canonical CE15-B GE subfamily, extensive networks of Van der Waals interactions and H-bonding likely play a similar stabilizing role. The non-covalent nature of the interactions with the general base loop in CE15-B subfamily enzymes would provide stability to the general base loop, while still providing conformational flexibility. It is likely that the lack of a covalent anchor in the active site provides these enzymes with the structural plasticity required to bring the non-canonical general acid within hydrogen bonding distance of the general base. This conformational flexibility has also been suggested to play a role in the substrate specificity of GEs [15, 16, 21].
CAZymes commonly adopt multi-domain structures composed of complementary enzyme activities that function synergistically to hydrolyze complex polysaccharides. We were unable to express a multidomain construct of CesA, however a search of the AlphaFold database identified a model of the full length R. flavefaciens CesA (AlphaFold model ID: Q9RLB8) [57]. Interestingly, AlphaFold predicts that the active sites of the N-terminal carbohydrate active enzyme 3 family, and C-terminal carbohydrate active enzyme 15 family, domains could adopt a low energy conformation in which the GE and acetylxylan esterase catalytic sites are oriented towards each other (supplementary Fig. 3). This tertiary arrangement could facilitate the simultaneous cleavage of an ester bond between 4-O-methyl-d-glucuronyl in xylan and lignin alcohol groups in LCCs by the GE domain, and deacetylation of the xylan backbone of D-glucuronylxylan by the acetylxylan esterase domain. We hypothesize that CesA may be an interesting model system to study enzyme cooperativity in multi-domain CAZymes [47, 48]. Due to the theoretical nature of the CesA AlphaFold model, experimental validation is required to draw any such conclusions.
Conclusion
We have experimentally determined the 3-dimensional structures and biochemical properties of three glucuronoyl esterases; two encoded by rumen prokaryotes and one encoded by a rumen fungus. Furthermore, we have examined the diversity of the carbohydrate esterase family 15 esterase family found in the rumen. Phylogenetic and structural analysis of rumen GEs lead to the hypothesis that interkingdom HGT events may have contributed to the diversity of GEs in the rumen, and that FsCE15 may represent an intermediate between fungal and bacterial GEs. This examination of the GE active site structures highlights the importance of structural plasticity in carbohydrate esterase family 15 esterases with a non-canonical active site conformation, and suggests that this flexibility may play a role in defining GE substrate specificity. These data expand our current understanding of the structure-function relationship in glucuronoyl esterases and illuminates the evolutionary dynamics that contribute to enzyme diversity in the rumen microbiome.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
All X-ray diffraction data were collected at the Canadian Light Source at the Canadian Light Source, a national research facility of the University of Saskatchewan, which is supported by the Canada Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council (NRC), the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan. Beamline access was enabled through proposal #: 009909 - Mining the rumen and rumen microbes to discover novel biocatalysts for use in agriculture, awarded to RJG. The beamline staff of 08ID-1 is graciously acknowledged for facilitating mail-in data collection. Structures of FsCE15, PrCE15, and RfCE15 have been deposited in the PDB under the accession codes 8TRU, 8TRX, and 8TSE, respectively.
Author Contributions
RJG conceived of and designed the study, provided resources, solved and refined structures, interpreted data, generated figures and wrote the paper. MK performed biochemical characterization, purified proteins, and conducted crystallization trials. KEL carried out phylogenetic analysis and generated Fig. 1. DRJ solved and refined structures and contributed to the carbohydrate esterase family 15 phylogenetic analysis. LW solved structures and provided advice on model refinement. TAM provided resources and helped co-ordinate the study. WA provided resources, helped to co-ordinate the study, and interpret results. All authors contributed to manuscript writing, and have read and approved the submitted manuscript.
Funding
Open access funding provided by Agriculture & Agri-Food Canada library. This work was supported by Agriculture and Agri-Food Canada (Grant ID: J-002872 to RJG) and the Beef Cattle Research Council (Grant ID: FDE.07.20 to RJG). The funders played no role in the design of the experiment or interpretation of the results.
Open access funding provided by Agriculture & Agri-Food Canada library.
Data Availability
Structures of FsCE15, PrCE15, and RfCE15 have been deposited in the PDB under the accession codes 8TRU, 8TRX, and 8TSE, respectively.
Declarations
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hackmann TJ, Spain JN (2010) Invited review: ruminant ecology and evolution: perspectives useful to ruminant livestock research and production. J Dairy Sci 93(4):1320–1334. 10.3168/jds.2009-2071 10.3168/jds.2009-2071 [DOI] [PubMed] [Google Scholar]
- 2.Gruninger RJ, Ribeiro GO, Cameron A, McAllister TA (2019) Invited review: application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal 13(9):1843–1854. 10.1017/S1751731119000752 10.1017/S1751731119000752 [DOI] [PubMed] [Google Scholar]
- 3.Mizrahi I, Wallace RJ, Morais S (2021) The rumen microbiome: balancing food security and environmental impacts. Nat Rev Microbiol 19(9):553–566. 10.1038/s41579-021-00543-6 10.1038/s41579-021-00543-6 [DOI] [PubMed] [Google Scholar]
- 4.Huws SA, Creevey CJ, Oyama LB, Mizrahi I, Denman SE, Popova M et al (2018) Addressing Global Ruminant Agricultural challenges through understanding the rumen Microbiome: past, Present, and Future. Front Microbiol 9:2161. 10.3389/fmicb.2018.02161 10.3389/fmicb.2018.02161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell JB, Rychlik JL (2001) Factors that alter rumen microbial ecology. Science 292(5519):1119–1122. 10.1126/science.1058830 10.1126/science.1058830 [DOI] [PubMed] [Google Scholar]
- 6.Low KE, Tingley JP, Klassen L, King ML, Xing X, Watt C, Hoover SER, Gorzelak M, Abbott DW (2023) Carbohydrate flow through agricultural ecosystems: implications for synthesis and microbial conversion of carbohydrates. Biotechnol Adv 69:108245. 10.1016/j.biotechadv.2023.108245 10.1016/j.biotechadv.2023.108245 [DOI] [PubMed] [Google Scholar]
- 7.Dos Santos AC, Ximenes E, Kim Y, Ladisch MR (2019) Lignin-enzyme interactions in the hydrolysis of lignocellulosic biomass. Trends Biotechnol 37(5):518–531. 10.1016/j.tibtech.2018.10.010 10.1016/j.tibtech.2018.10.010 [DOI] [PubMed] [Google Scholar]
- 8.Larsbrink J, Lo Leggio L (2023) Glucuronoyl esterases - enzymes to decouple lignin and carbohydrates and enable better utilization of renewable plant biomass. Essays Biochem 67(3):493–503. 10.1042/EBC20220155 10.1042/EBC20220155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B et al (2012) The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336(6089):1715–1719. 10.1126/science.1221748 10.1126/science.1221748 [DOI] [PubMed] [Google Scholar]
- 10.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6(1):41. 10.1186/1754-6834-6-41 10.1186/1754-6834-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puchart V, Biely P (2023) Microbial xylanolytic carbohydrate esterases. Essays Biochem 67(3):479–491. 10.1042/EBC20220129 10.1042/EBC20220129 [DOI] [PubMed] [Google Scholar]
- 12.Lankiewicz TS, Choudhary H, Gao Y, Amer B, Lillington SP, Leggieri PA et al (2023) Lignin deconstruction by anaerobic fungi. Nat Microbiol 8(4):596–610. 10.1038/s41564-023-01336-8 10.1038/s41564-023-01336-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosbech C, Holck J, Meyer A, Agger JW (2019) Enzyme kinetics of fungal glucuronoyl esterases on natural lignin-carbohydrate complexes. Appl Microbiol Biotechnol 103(10):4065–4075. 10.1007/s00253-019-09797-w 10.1007/s00253-019-09797-w [DOI] [PubMed] [Google Scholar]
- 14.Spanikova S, Biely P (2006) Glucuronoyl esterase–novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett 580(19):4597–4601. 10.1016/j.febslet.2006.07.033 10.1016/j.febslet.2006.07.033 [DOI] [PubMed] [Google Scholar]
- 15.Arnling Baath J, Mazurkewich S, Knudsen RM, Poulsen JN, Olsson L, Lo Leggio L, Larsbrink J (2018) Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion. Biotechnol Biofuels 11:213. 10.1186/s13068-018-1213-x 10.1186/s13068-018-1213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnling Baath J, Mazurkewich S, Poulsen JN, Olsson L, Lo Leggio L, Larsbrink J (2019) Structure-function analyses reveal that a glucuronoyl esterase from Teredinibacter turnerae interacts with carbohydrates and aromatic compounds. J Biol Chem 294(16):6635–6644. 10.1074/jbc.RA119.007831 10.1074/jbc.RA119.007831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyczakowski JJ, Wicher KB, Terrett OM, Faria-Blanc N, Yu X, Brown D, Krogh K, Dupree P, Busse-Wicher M (2017) Removal of glucuronic acid from xylan is a strategy to improve the conversion of plant biomass to sugars for bioenergy. Biotechnol Biofuels 10:224. 10.1186/s13068-017-0902-1 10.1186/s13068-017-0902-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.d’Errico C, Jorgensen JO, Krogh KB, Spodsberg N, Madsen R, Monrad RN (2015) Enzymatic degradation of lignin-carbohydrate complexes (LCCs): model studies using a fungal glucuronoyl esterase from Cerrena Unicolor. Biotechnol Bioeng 112(5):914–922. 10.1002/bit.25508 10.1002/bit.25508 [DOI] [PubMed] [Google Scholar]
- 19.Latha Gandla M, Derba-Maceluch M, Liu X, Gerber L, Master ER, Mellerowicz EJ, Jonsson LJ (2015) Expression of a fungal glucuronoyl esterase in Populus: effects on wood properties and saccharification efficiency. Phytochemistry 112:210–220. 10.1016/j.phytochem.2014.06.002 10.1016/j.phytochem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 20.Duranova M, Hirsch J, Kolenova K, Biely P (2009) Fungal glucuronoyl esterases and substrate uronic acid recognition. Biosci Biotechnol Biochem 73(11):2483–2487. 10.1271/bbb.90486 10.1271/bbb.90486 [DOI] [PubMed] [Google Scholar]
- 21.De Santi C, Gani OA, Helland R, Williamson A (2017) Structural insight into a CE15 esterase from the marine bacterial metagenome. Sci Rep 7(1):17278. 10.1038/s41598-017-17677-4 10.1038/s41598-017-17677-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazurkewich S, Poulsen JN, Lo Leggio L, Larsbrink J (2019) Structural and biochemical studies of the glucuronoyl esterase OtCE15A illuminate its interaction with lignocellulosic components. J Biol Chem 294(52):19978–19987. 10.1074/jbc.RA119.011435 10.1074/jbc.RA119.011435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zong Z, Mazurkewich S, Pereira CS, Fu H, Cai W, Shao X, Skaf MS, Larsbrink J, Lo Leggio L (2022) Mechanism and biomass association of glucuronoyl esterase: an alpha/beta hydrolase with potential in biomass conversion. Nat Commun 13(1):1449. 10.1038/s41467-022-28938-w 10.1038/s41467-022-28938-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst HA, Mosbech C, Langkilde AE, Westh P, Meyer AS, Agger JW, Larsen S (2020) The structural basis of fungal glucuronoyl esterase activity on natural substrates. Nat Commun 11(1):1026. 10.1038/s41467-020-14833-9 10.1038/s41467-020-14833-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitriou PS, Denesyuk A, Takahashi S, Yamashita S, Johnson MS, Nakayama T, Denessiouk K (2017) Alpha/beta-hydrolases: a unique structural motif coordinates catalytic acid residue in 40 protein fold families. Proteins 85(10):1845–1855. 10.1002/prot.25338 10.1002/prot.25338 [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro GO, Gruninger RJ, Badhan A, McAllister TA (2016) Mining the rumen for fibrolytic feed enzymes. Anim Front 6(2):20–26. 10.2527/af.2016-0019 10.2527/af.2016-0019 [DOI] [Google Scholar]
- 27.Jones DR, Thomas D, Alger N, Ghavidel A, Inglis GD, Abbott DW (2018) SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol Biofuels 11:27. 10.1186/s13068-018-1027-x 10.1186/s13068-018-1027-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seshadri R, Leahy SC, Attwood GT, Teh KH, Lambie SC, Cookson AL et al (2018) Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat Biotechnol 36(4):359–367. 10.1038/nbt.4110 10.1038/nbt.4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R et al (2014) MycoCosm Portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42(Database issue):D699–704. 10.1093/nar/gkt1183 10.1093/nar/gkt1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46(W1):W95–W101. 10.1093/nar/gky418 10.1093/nar/gky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–37 (Web Server issue). 10.1093/nar/gkr367 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N (2022) The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50(D1):D571–D577. 10.1093/nar/gkab1045 10.1093/nar/gkab1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B (2010) A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J 432(3):437–444. 10.1042/BJ20101185 10.1042/BJ20101185 [DOI] [PubMed] [Google Scholar]
- 34.Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. 10.1093/bioinformatics/bts565 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. 10.1093/bioinformatics/btl158 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 36.Edgar RC (2022) Muscle5: high-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun 13(1):6968. 10.1038/s41467-022-34630-w 10.1038/s41467-022-34630-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T (2020) ModelTest-NG: a New and Scalable Tool for the selection of DNA and protein evolutionary models. Mol Biol Evol 37(1):291–294. 10.1093/molbev/msz189 10.1093/molbev/msz189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):e9490. 10.1371/journal.pone.0009490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkens MR, Appel RD, Bairoch A (2005) Protein Identification and Analysis Tools on the ExPASy server. In: Walker JM (ed) The Proteomics protocols Handbook. Humana, pp 571–607
- 40.Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66 (Pt 2):125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed]
- 41.Krissinel E, Lebedev AA, Uski V, Ballard CB, Keegan RM, Kovalevskiy O et al (2022) CCP4 cloud for structure determination and project management in macromolecular crystallography. Acta Crystallogr D Struct Biol 78(Pt 9):1079–1089. 10.1107/S2059798322007987 10.1107/S2059798322007987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI et al (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75(Pt 10):861–877. 10.1107/S2059798319011471 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pokkuluri PR, Duke NE, Wood SJ, Cotta MA, Li XL, Biely P, Schiffer M (2011) Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea Jecorina. Proteins 79(8):2588–2592. 10.1002/prot.23088 10.1002/prot.23088 [DOI] [PubMed] [Google Scholar]
- 44.Charavgi MD, Dimarogona M, Topakas E, Christakopoulos P, Chrysina ED (2013) The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst. Acta Crystallogr D Biol Crystallogr 69(Pt 1):63–73. 10.1107/S0907444912042400 10.1107/S0907444912042400 [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4):486–501. 10.1107/S0907444910007493 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunger AT (1997) Free R value: cross-validation in crystallography. Methods Enzymol 277:366–396. 10.1016/s0076-6879(97)77021-6 10.1016/s0076-6879(97)77021-6 [DOI] [PubMed] [Google Scholar]
- 47.Lee PH, Huang XX, Teh BT, Ng LM (2019) TSA-CRAFT: a Free Software for Automatic and Robust Thermal Shift Assay Data Analysis. SLAS Discov 24(5):606–612. 10.1177/2472555218823547 10.1177/2472555218823547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazurkewich S, Scholzen KC, Brusch RH, Poulsen JCN, Theibich Y, Huttner S, Olsson L, Larsbrink J, Lo Leggio L (2023) Structural and functional investigation of a fungal member of carbohydrate esterase family 15 with potential specificity for rare xylans. Acta Crystallogr D Struct Biol 79(Pt 6):545–555. 10.1107/S205979832300325X 10.1107/S205979832300325X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Youssef NH, Couger MB, Hanafy RA, Elshahed MS, Stajich JE (2019) Molecular dating of the emergence of anaerobic Rumen Fungi and the impact of laterally acquired genes. mSystems 4(4). 10.1128/mSystems.00247-19 [DOI] [PMC free article] [PubMed]
- 50.Youssef NH, Couger MB, Struchtemeyer CG, Liggenstoffer AS, Prade RA, Najar FZ, Atiyeh HK, Wilkins MR, Elshahed MS (2013) The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl Environ Microbiol 79(15):4620–4634. 10.1128/AEM.00821-13 10.1128/AEM.00821-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haitjema CH, Gilmore SP, Henske JK, Solomon KV, de Groot R, Kuo A et al (2017) A parts list for fungal cellulosomes revealed by comparative genomics. Nat Microbiol 2:17087. 10.1038/nmicrobiol.2017.87 10.1038/nmicrobiol.2017.87 [DOI] [PubMed] [Google Scholar]
- 52.Murphy CL, Youssef NH, Hanafy RA, Couger MB, Stajich JE, Wang Y et al (2019) Horizontal gene transfer as an indispensable driver for evolution of Neocallimastigomycota into a distinct Gut-Dwelling Fungal Lineage. Appl Environ Microbiol 85(15). 10.1128/AEM.00988-19 [DOI] [PMC free article] [PubMed]
- 53.Gruninger RJ, Nguyen TTM, Reid ID, Yanke JL, Wang P, Abbott DW, Tsang A, McAllister T (2018) Application of Transcriptomics to compare the carbohydrate active enzymes that are expressed by Diverse Genera of Anaerobic Fungi to Degrade Plant Cell Wall Carbohydrates. Front Microbiol 9:1581. 10.3389/fmicb.2018.01581 10.3389/fmicb.2018.01581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng X, Wilken SE, Lankiewicz TS, Gilmore SP, Brown JL, Henske JK et al (2021) Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat Microbiol 6(4):499–511. 10.1038/s41564-020-00861-0 10.1038/s41564-020-00861-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seveso A, Mazurkewich S, Banerjee S, Poulsen JC, Lo Leggio L, Larsbrink J (2024) Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds. Appl Environ Microbiol:e0176823. 10.1128/aem.01768-23 10.1128/aem.01768-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen TTM, Badhan AK, Reid ID, Ribeiro G, Gruninger R, Tsang A, Guan LL, McAllister T (2023) Comparative analysis of functional diversity of rumen microbiome in bison and beef heifers. Appl Environ Microbiol 89(12):e0132023. 10.1128/aem.01320-23 10.1128/aem.01320-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589. 10.1038/s41586-021-03819-2 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structures of FsCE15, PrCE15, and RfCE15 have been deposited in the PDB under the accession codes 8TRU, 8TRX, and 8TSE, respectively.