Abstract
Cobalt (Co) and Nickel (Ni) are used nowadays in various industrial applications like lithium-ion batteries, raising concerns about their environmental release and public health threats. Both metals are potentially carcinogenic and may cause neurological and cardiovascular dysfunctions, though underlying toxicity mechanisms have to be further elucidated. This study employs untargeted transcriptomics to analyze downstream cellular effects of individual and combined Co and Ni toxicity in human liver carcinoma cells (HepG2). The results reveal a synergistic effect of Co and Ni, leading to significantly higher number of differentially expressed genes (DEGs) compared to individual exposure. There was a clear enrichment of Nrf2 regulated genes linked to pathways such as glycolysis, iron and glutathione metabolism, and sphingolipid metabolism, confirmed by targeted analysis. Co and Ni exposure alone and combined caused nuclear Nrf2 translocation, while only combined exposure significantly affects iron and glutathione metabolism, evidenced by upregulation of HMOX-1 and iron storage protein FTL. Both metals impact sphingolipid metabolism, increasing dihydroceramide levels and decreasing ceramides, sphingosine and lactosylceramides, along with diacylglycerol accumulation. By combining transcriptomics and analytical methods, this study provides valuable insights into molecular mechanisms of Co and Ni toxicity, paving the way for further understanding of metal stress.
Keywords: Cobalt, Nickel, Metal interactions, Transcriptomic analysis, Nrf2 signaling, Sphingolipid metabolism
Graphical abstract
Highlights
-
•
Combined exposure to Co and Ni led to higher number of differentially expressed genes.
-
•
Nrf2 plays a crucial role in cellular response to Co and Ni toxicity.
-
•
Co and Ni affect the cellular sphingolipid metabolism.
-
•
Both metals enriched Nrf2 regulated genes related to glycolysis, iron and glutathione metabolism.
Abbreviations
- CE-ICP-MS
capillary electrophoresis inductively coupled plasma mass spectrometry
- Cer
ceramide
- DAG
diacylglycerol
- DEG
differentially expressed genes
- dhCer
dihydroceramide
- dhSM
dihydrosphingomyelin
- dhSph
dihydrosphingosine
- FTH
ferritin heavy chain
- FTL
ferritin light chain
- GO
gene ontology
- GSH
glutathione
- GSSG
glutathione disulfide
- HIF-1
hypoxia-inducible factor-1
- HMOX-1
heme oxygenase 1
- KEAP1
Kelch like-ECH-associated protein 1
- LacCer
lactosylceramide
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LDHA
lactate dehydrogenase A
- NEM
N-ethylmaleimide
- Nrf2
nuclear factor erythroid 2-related factor 2
- PCA
principal component analysis
- RONS
reactive oxygen and nitrogen species
- SL
sphingolipid
- SM
sphingomyelin
- Sph
sphingosine
- SQSTM1
sequestosome 1
- TCA
tricarboxylic acid cycle
1. Introduction
Metal toxicity is a significant concern in environmental and occupational health, with cobalt (Co) and nickel (Ni) being significant contributors. Both metals find wide-ranging applications in the manufacturing of alloys, lithium-ion batteries for electric vehicles, and electronics, as well as essential components in catalysts or pigments [1,2]. Despite their industrial importance, Co and Ni seem to have a significant impact on the environmental and health, particularly in occupational settings where exposure levels can be elevated. As their usage in industrial processes continues to grow, environmental entry due to pollution rises, resulting in heightened human exposure through food sources or drinking water.
Co serves as an essential trace element primarily as a cofactor of vitamin B12 and the formation of amino acids and proteins, particularly in nerve cells. In contrast, the biological role of Ni in humans remains elusive [3]. However, concerns regarding potential adverse health effects from Co and Ni exposure are rising due to their carcinogenic potential upon inhalation, as classified by the International Agency for Research on Cancer (IARC; Co: group 2A; Ni: group 1) [4,5]. Oral exposure to these metals may lead to various risks, including neurological, cardiovascular, and thyroid dysfunction, but only limited data exist and the underlying mechanisms are still not conclusively clarified [6]. Hypotheses discussed in literature and targeted studies gave first mechanistic insights and point out the relevance of oxidative stress for both metals. Studies indicate that Co generates reactive oxygen and nitrogen species (RONS) on the cellular level via Fenton-like reactions, leading to oxidative stress, cell death and DNA damage. Similarly, Ni toxicity is associated with DNA crosslinks, inhibition of DNA repair, and mitochondrial dysfunction [7,8].
Most studies are targeted and focus on the toxicity of the individual metal. However, the combined exposure is the more realistic exposure scenario. A first study exposing liver carcinoma cells (HepG2) to the combination of Co and Ni pointed out alterations in the bioavailability, leading to heightened cellular Co levels and decreased Ni amount compared to individual treatment. Additionally, simultaneous treatment led to enhanced effects regarding RONS-induction, GSSG formation or cell death [9]. Although the relevance of combined exposure to elucidate toxicity is indicated, all former studies are focusing on specific endpoints. A transcriptomic analysis could provide a more global view of gene expression across the entire genome, allowing comparisons between alterations in individual and combined treatments. This untargeted approach will allow identifying the most relevant pathways of Co and/or Ni toxicity. The relevance of cellular responses identified by the transcriptome will be further supported by targeted analysis quantifying specific metabolites to gain a deeper understanding of pathways affected in HepG2 cells. By examining individual cellular pathways and identifying shared and distinct responses to individual and combined Co and Ni exposure, we aim to reveal on the complex interplay between gene regulation and cellular metabolism in metal toxicity.
2. Material and methods
2.1. Cell culture maintenance and treatment scenario
Human hepatocarcinoma cells (HepG2) were cultured in Eagle's Minimum Essential Medium (MEM; Sigma Aldrich) to which 10 % fetal bovine serum (FBS; Sigma Aldrich), 2 % (v/v) penicillin/streptomycin (Sigma Aldrich) and 1 % (v/v) non-essential amino acids (NEA; Sigma Aldrich) were added, as previously described [9]. Cells were incubated for 2 or 24 h with CoCl2 (Thermo Fischer Scientific; 99.9 %) and NiCl2 (Sigma Aldrich; 99.9 %) individually or in combination. For combined treatment of Co and Ni, two different combinations were selected, which are applied in an industrial relevant mixture of 6:1 (Ni:Co). The first combination, consisting of 12.5 μM Co + 75 μM Ni, demonstrated no cytotoxic effects when applied individually or in combination. The second, higher combination of 25 μM Co + 150 μM Ni, exhibited initial signs of cytotoxicity, though not statistically significant in individual and combined treatment. To compare the effects of combined and single treatments, cells were exposed to each concentration both together and individually. Cytotoxicity data for these treatments were published in our previous study [9].
2.2. Transcriptomics
For transcriptomic analysis, cells were pelletized using 0.25 % trypsin-EDTA for detaching, followed by washing with PBS and centrifugation at 2370×g and 4 °C for 4 min. After removing the supernatant, total RNA of the cell pellet was isolated using RNeasy Plus Mini Kit which includes DNAse digestion (Qiagen). Poly-A RNA libraries were prepared using the D-Plex mRNA-seq Kit for Illumina (Hologic Diagenode) according to the manufacturer's instructions with 2.5 μg total RNA as input for mRNA selection and 13 PCR cycles for amplification. Libraries were pooled and sequenced on the NovaSeq 6000 Illumina platform. De-multiplexing, quality trimming as well as removal of adapter-sequences and artificial poly-A-tails were performed as described by manufacturer using the cutadapt tool [10]. Resulting reads showed high quality, quantified by a mean per base quality phred score of 35.8, approx. 97 % of all reads having a quality phred score of 30 or higher (Q30) and a mean per base N content of 0.0005 % across all samples. Reads were mapped onto the human genome (hg38) using the bowtie2 plugin for Geneious Prime 2023.2 with a mean mapping ratio of approx. 85 % of reads across all samples mapping to annotated genes and gene expression levels were calculated using the “calculate expression levels” function of Geneious Prime [11]. Differences between the treatment groups were visualized by PCA on the base of transcripts per million (TPM) expression level data using the prcomp function and ggbiplot visualization function in R. Differentially expressed genes (DEGs) were calculated using the DESeq2 Plugin for Geneious Prime applying a threshold of p-value <0.01 and log2 fold change >1/<-1 for DEG characterization [12]. Comparisons were performed between treated and untreated cells. GO Term analysis of DEGs was performed using the geneontology.org website [13,14] which uses the PANTHER tool [15]. Sequencing data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-14292.
2.3. Isolation of cell nuclei
To isolate cell nuclei, HepG2 cells were washed with ice-cold PBS and scratched from cell culture plates using lysis buffer I (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF, 0.1 % NP-40 alternative) as described before [16]. Samples were shaken for 3 min, 4 °C at 1300 rpm, vortexed and shaken again for further 4 min. After centrifugation for 1 min at 4 °C and 6800×g, the supernatant containing the cytosolic fraction was removed and the remaining pellet was resuspended in lysis buffer II (40 mM HEPES, 400 mM KCl, 10 % glycerol, 1 mM DTT, 0.1 mM PMSF) and 294 mM NaCl followed by a sonification step (12 s, cycle: 0.5, amplitude: 80 %; Hielscher UP100H) and centrifugation for 30 min at 4 °C at 20000×g. The supernatant containing the nuclear fraction was used for protein determination, quantified via standard BCA assay (Sigma-Aldrich), and Western Blot analysis.
2.4. Western Blot analysis
For Western Blot analysis, cell pellets have been processed as previously described [17]. As primary antibodies, anti-β-actin (1:2500, ab115777, Abcam), anti-FTH (1:500, D1D4, Cell Signaling), anti-FTL (1:500, ab69090, Abcam), anti-HMOX-1 (1:500, sc-136960, Santa Cruz Biotechnology), anti-Lamin A (1:10000, L1293, Sigma Aldrich) and anti-NRF2 (1:1000, D1Z9C, Cell Signaling) were used. The secondary antibodies HRP-conjugated goat anti-mouse or goat anti-rabbit (1:10000, Bio-Rad Laboratories) were incubated for 1 h at RT. Quantified protein bands were normalized to β-actin (FTH, FTL, HMOX-1) or Lamin A (NRF2).
2.5. Immunofluorescence staining
HepG2 cells were seeded on glass cover slips, coated with Alcian Blue 8GX® (Sigma Aldrich). For Nrf2 staining, cells were fixed and permeabilized with PBS containing 4 % formaldehyde (Carl Roth) and 0.2 % Triton-X™ (Sigma Aldrich), washed with PBS and incubated 30 min at 37 °C with 5 % normal goat serum (Life Technologies) in PBS. The primary antibody anti-NRF2 was incubated for 2 h at room temperature (1:500 in 0.5 % BSA/PBS) followed by incubation of the second antibody Alexa Fluor® 488 goat anti-rabbit (Invitrogen) for 1 h (1:400 in 0.5 % BSA/PBS). Finally, cell nuclei were stained using 1 % Hoechst 33258 in MeOH (Sigma Aldrich). Images were taken with the Leica DM6 B fluorescence microscope and processed in LAS X imaging software.
2.6. Iron redox speciation
To investigate the iron redox speciation, HepG2 cells were pelletized and resuspended in RIPA buffer. Cells were lysed using an ultrasonic probe (20 s, cycle: 1, amplitude: 100 %; Hielscher UP100H), centrifuged at 10000×g for 10 min and the supernatant was transferred in a new tube. The samples were shipped on dry ice to the Helmholtz Center Munich in Germany for analysis. The speciation and quantification of Fe2+, Fe3+, and total iron were conducted using capillary electrophoresis inductively coupled plasma mass spectrometry (CE-ICP-MS), as previously detailed [18,19]. In brief, the analysis involved the use of a "PrinCe 706” CE system with an uncoated capillary (85 cm × 50 μm ID) and a custom-built CE-ICP-MS interface for the selective quantification of iron redox species at ICP-DRC (dynamic reaction cell)-MS. The DRC technology, using NH3 as the reaction gas, was employed for spectral interference-free detection of the 56Fe isotope. Fe2+/Fe3+ separation and quantification were conducted in a 20 mM HCl-electrolyte at +25 kV separation voltage, with 56Fe isotope detection at ICP-DRC-MS. To ensure quality control, total iron was additionally determined by ICP-sf-MS, and these values were compared to the sum of iron species quantified by CE-ICP-DRC-MS.
2.7. Quantification of metabolites related to glycolysis, TCA cycle and GSH metabolism by HPLC-MS/MS
Sample preparation was conducted and modified based on [20]. Cells were washed with PBS and scratched off the plate using 500 μL of 50 % methanol (MeOH) (−20 °C), containing 0.5 mM N-ethylmaleimide (NEM) and 10 mM formic acid (FA). Collected samples were sonicated for 20 s (cycle: 1, amplitude: 100 %; Hielscher UP100H) and an aliquot was taken for protein measurement. After adding 225 μL chloroform (−20 °C), samples were shaken for 10 min and then for another 2 min after adding 75 μL bidistilled water (4 °C), followed by 20 min incubation at −20 °C. They were centrifuged (4 °C, 9500×g, 15 min) and 350 μL of the upper layer was dried using a SpeedVac. The remaining pellet was dissolved in 50 μL 50 % ACN, centrifuged (16060×g, 10 min) and diluted before LC-MS/MS analysis. Isotope labelled standards (sodium l-Lactate-3,3,3-d3 (CDN Isotopes) and L-glutamic-2,3,3,4,4-d5 acid (Sigma Aldrich)) were added as internal standards. Metabolite quantification was performed on an Agilent 1290 Infinity II LC System coupled to a Sciex QTrap 6500 + triple-quadrupole mass spectrometer with an electrospray ion source operating in both positive and negative mode. Chromatographic separation was accomplished using a Hilicon iHilic®-Fusion column (150 × 2.1 mm, 3.5 μm) complemented by the appropriate pre-column (20 × 2.1 mm, 5 μm). Parameters for the LC-MS/MS measurement and mass transitions are listed in the supplementary (Tables S1 and S2).
2.8. Quantification of sphingolipids and diacylglycerols by HPLC-MS/MS
HepG2 cells were pelletized, washed with ice-cold PBS, and resuspended in MeOH. Sphingolipid (SL) and diacylglycerol (DAG) extraction was performed as previously described [21]. Briefly, 1.5 mL methanol/chloroform (2:1, v:v) containing eight internal standards (all from Avanti Polar Lipids, Alabaster, USA), seven for SL and one for DAG quantification (details see Supplementary Tables S3 and S4), was added to the samples (∼1x106 cells) that were then shaken overnight at 48 °C. Samples were worked up in two separate preparations, with (for SL) or without (for DAG) saponification of the lipid extract. The further procedure was analogous for both lipid classes. HPLC-MS/MS analyses were carried out under already published instrumental conditions [21,22]. The lipid extracts were chromatographically separated using a 1290 Infinity II HPLC (Agilent Technologies, Waldbronn, Germany) equipped with a Poroshell 120 EC-C8 column (3.0 × 150 mm, 2.7 μm; Agilent Technologies). Multiple reaction monitoring (MRM) was performed using a 6495C triple-quadrupole mass spectrometer (Agilent Technologies) in positive electrospray ionization mode (ESI+). The analyzed SL and DAG species, their mass transitions as well as their retention times are listed in Supplementary Tables S3 and S4. Analyte peak areas were normalized to those of their corresponding internal standards followed by external calibration. Peak integration and quantification were conducted with MassHunter Quantitative Analysis Software (version 10.1, Agilent Technologies). SL and DAG quantities were normalized to cell count and expressed as “pmol/1x106 cells”.
2.9. Statistics
Statistical analysis was performed with GraphPad Prism software 6.01 (GraphPad, La Jolla, CA, USA). Data are expressed as mean +SD, with significance stated as *p < 0.05, **p < 0.01 and ***p < 0.001 compared to untreated control.
3. Results
3.1. Transcriptomic analysis derives the impact of Nrf2
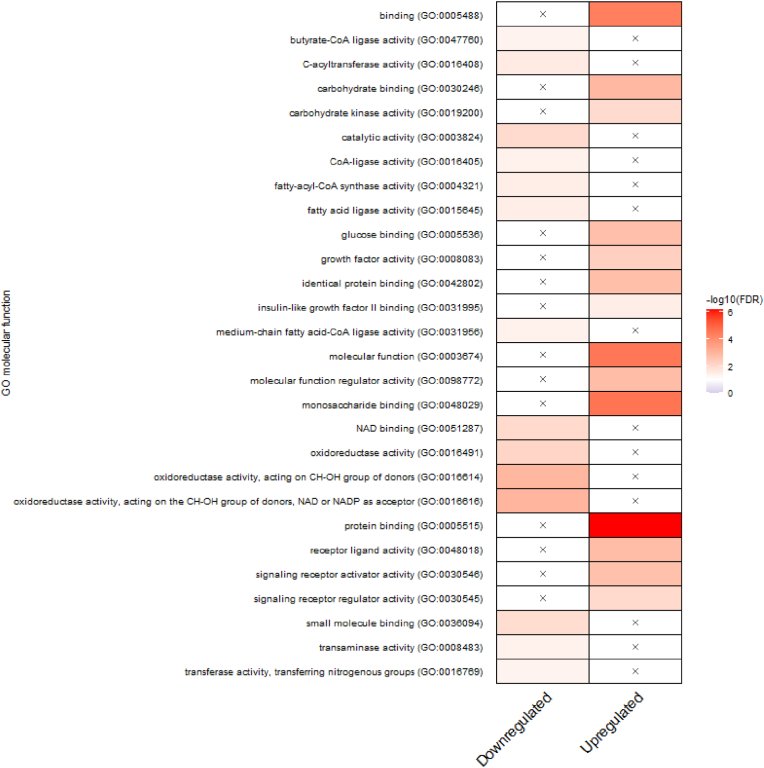
A transcriptomic analysis was conducted to gain deeper insights into the toxicity mechanisms of Co and Ni in HepG2 cells. Both metals were exposed in subtoxic concentrations, as well as the combination of these doses, which were examined in our previous study [9]. Principal component analysis (PCA) revealed different gene expression patterns between the treatments compared to the untreated control, with PC1 explaining 38.8 % variance, indicating differences from the control, and PC2 explaining 12.7 % variance, illustrating the separation of the different exposure scenarios (Fig. 1). Fig. 1B further indicates that Co and Ni in case of combined exposure result in a significantly higher number of differentially expressed genes compared to individual exposure. Subsequently, the DEGs in response to the combination (25 μM Co + 150 μM Ni) compared to the untreated control were used for Gene Ontology (GO) term enrichment (Fig. 2). Combining both metals seems to influence a variety of molecular functions in HepG2 cells, including the upregulation of carbohydrate metabolism, protein binding or receptor activity. Conversely, downregulated pathways primarily involve lipid metabolism and oxidoreductase activity. Fig. 3 highlights the connections between the most affected DEGs and their associated metabolic pathways in response to the combined treatment of Co and Ni. Both metals seem to have an impact on crucial cellular processes, particularly glycolysis, iron homeostasis, and glutathione metabolism. The identified target DEGs are connected to conserved cellular responses driven by the transcription factor Nrf2.
Fig. 1.
A) Principal component analysis (PCA plot) of PC1 and PC2, comparing the different Co(II) and Ni(II) treatments and the untreated control in HepG2 cells. Individual samples of each treatment group are highlighted by different letters. WT = untreated control. B) Venn-diagrams showing the overlap of DEGs comparing 25 μM Co(II), 150 μM Ni(II) and the combination of both treatments in HepG2 cells.
Fig. 2.
Summary of over-represented GO terms related to molecular function, comparing combined treatment (25 μM Co + 150 μM Ni) with untreated control in HepG2 cells. The x means no significant representation. Colors display intensity of enrichment (-log10 (FDR-corrected p-values)) with p value cutoff of 0.01. Visualization by an R-script from Ref. [23]. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Affected pathways (transcriptome) by Co(II) and Ni(II) in HepG2 cells upon 24 h combined exposure (25 μM Co + 150 μM Ni). Genes in green font represent upregulation, magenta font shows downregulation, and grey font indicates no alterations compared to untreated control. More detailed data is shown in Supplementary Table S5. Abbreviations: GLUT glucose transporter; HK hexokinase; G6PC glucose-6-phosphatase catalytic subunit 1; GPI glucose-6-phosphatase isomerase; FBP fructose-bisphosphatase; ALDOA aldolase A; ENO enolase; PKM pyruvate kinase M; LDHA lactate dehydrogenase A; PGM phosphoglucomutase; GBE 1,4-alpha-glucan branching enzyme; G6PD glucose-6-phosphate dehydrogenase; PGD phosphogluconate dehydrogenase; TALDO transaldolase; TKT transketolase; FTH ferritin heavy chain; FTL ferritin light chain; FECH ferrochelatase; HMOX1 heme oxygenase 1; BLVRB biliverdin reductase B; PDK pyruvate dehydrogenase kinase; PDH pyruvate dehydrogenase; PCK phosphoenolpyruvate carboxykinase; CS citrate synthase; AH aconitate hydratase; IDH isocitrate dehydrogenase; SCS succinyl-CoA synthetase; SDH succinate dehydrogenase; FH fumarate hydratase; MDH malate dehydrogenase; ACL ATP citrate lyase; ACC acetyl-CoA carboxylase; FASN fatty acid synthase; HMGCS HMG-CoA-synthase; HMGCR HMG-CoA-reductase; GLUD glutamate dehydrogenase; xCT solute carrier family 7 member 11; GCL glutamate-cysteine ligase; GSS glutathione synthetase. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Nuclear translocation of Nrf2 induced by Co and Ni
Nuclear factor erythroid 2-related factor 2 (Nrf2) is an important transcription factor for regulating cellular defense mechanisms against environmental stressors, for detoxification of xenobiotics and for maintaining redox homeostasis [24]. The nuclear translocation and therefore activation of Nrf2 was assessed by quantifying the protein concentration in nuclear lysates after 2 h of treatment of HepG2 cells with Co and Ni individually or in combination (Fig. 4A). Given the rapid activation and subsequent degradation of Nrf2, a shorter incubation time was selected compared to the other experiments investigating the consequences of Nrf2 activation [25]. Both metals, except of 75 μM Ni, led to elevated amounts of nuclear Nrf2 compared to the untreated control, with 150 μM Ni showing the strongest effect. Concurrent incubation of Co and Ni also resulted in increased nuclear Nrf2 translocation (Fig. 4A). These effects could also be demonstrated by immunofluorescence staining against nuclear Nrf2 (Fig. 4B).
Fig. 4.
Nuclear Nrf2 translocation in HepG2 cells after 2 h treatment with Co(II) and Ni(II). A) Protein concentration was quantified via Western Blot and normalized to Lamin A. B) Nrf2 translocation visualized by immunofluorescence staining in 63x magnification. The pictures are processed by Leica Thunder Imager. Blue: Hoechst staining, Green: Nrf2 staining. Data is presented as mean +SD of n = 3 independent experiments. Statistical significance was tested by an unpaired t-test depicted as *p ≤ 0.05, **p ≤ 0.01: compared to untreated control. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Impact on iron metabolism
Based on the transcriptome data, target genes of Nrf2 connected to the iron metabolism are studied further. Transcriptomic analysis revealed an upregulation in the gene expression of HMOX-1, FTL, FTH and BLVRB, induced by the high Co and Ni combination (Fig. 3). The higher combination of Co and Ni (25 μM Co + 150 μM Ni) resulted in elevated protein expression of HMOX-1 compared to control (Fig. 5A). HMOX-1 plays a crucial role in metabolizing the prooxidant heme into iron and biliverdin [26]. Similarly, there was a significant effect on the expression of the iron storage protein ferritin which was more pronounced for FTL (Fig. 5B), with a trend also observed for FTH (Fig. S1). A shift in the Fe(II)/Fe(III) ratio towards the divalent species is an indicator for a redox imbalance caused by higher RONS formation within the cell [27]. After 24 h treatment with Co and/or Ni, there were no significant alterations in this ratio, however, a trend towards Fe(II) was observed (Fig. 5C).
Fig. 5.
Impact on Fe metabolism in HepG2 cells after 24 h treatment with Co(II) and Ni(II). Protein expression of A) HMOX-1 and B) FTL were quantified via Western Blot and normalized to β-actin. C) Ratio of Fe2+ and Fe3+ normalized to protein content and untreated control. Data is presented as mean + SD of n ≥ 3 independent experiments. Statistical significance was tested by an unpaired t-test depicted as *p ≤ 0.05, **p ≤ 0.01: compared to untreated control.
3.4. Glycolysis, TCA cycle, and GSH metabolism
Nrf2 is also directing the transcription of genes encoding for enzymes involved in glycolysis, TCA cycle, and GSH metabolism, thereby influencing the synthesis of numerous metabolites [28]. LC-MS/MS analysis revealed that Co and Ni did not affect the glucose levels in HepG2 after 24 h treatment (Fig. 6A). However, the pyruvate content increased significantly with the higher combination (Fig. 6C), and lactate levels were enhanced across all applied Co and/or Ni concentrations (Fig. 6B). TCA cycle metabolites α-ketoglutarate and succinate showed elevated amounts only with the high combination (Fig. 6D/E), while malate content increased significantly with 150 μM Ni or the combination of 12.5 μM Co + 75 μM Ni (Fig. 6F). All applied concentrations except for 75 μM Ni led to decreased glutamate levels, while glutamine was enhanced by the high combination of Co and Ni (Fig. 6G/H). Furthermore, the cellular cysteine amount was lower compared to control after treatment with Co or the combination of Co and Ni, whereas Ni individually did not show this effect (Fig. 6I).
Fig. 6.
Levels of metabolites related to glycolysis, TCA cycle and GSH metabolism in HepG2 cells after 24 h treatment with Co(II) and Ni(II). Amount of A) glucose, B) lactate, C) pyruvate, D) α-ketoglutarate, E) succinate, F) malate, G) glutamate, H) glutamine and I) cysteine were quantified by LC-MS/MS and normalized to protein content and untreated control. Data is presented as mean +SD of n ≥ 3 independent experiments. Statistical significance was tested by an unpaired t-test depicted as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001: compared to untreated control.
3.5. Sphingolipids and diacylglycerols
Transcriptomic analysis also revealed an impact on sphingolipid metabolism and diacylglycerols (DAGs) induced by Co and Ni exposure. Specifically, the combination of both metals (25 μM Co + 150 μM Ni) led to an increased expression of CERS1, whose encoded protein is responsible for converting dihydrosphingosine (dhSph) to dihydroceramides (dhCer) and sphingosine (Sph) to ceramides (Cer) (Fig. 7) [29]. CERS1, which belongs to a group of six isoforms, specifically forms dhCer or Cer species with a C18:0 fatty acid side chain [30]. In line with this, LC-MS/MS analysis revealed increased C18:0 dhCer levels (Fig. S2A) and a trend for elevated C18:0 Cer (Fig. S2B) compared to the control after treatment with Co, Ni or a combination of both. Interestingly, all other dhCer species measured were also highly elevated (Fig. S2A), leading to significantly increased dhCer/Cer ratios after treatment with Ni and/or Co (Fig. 8A). DhCer and Cer can be converted by sphingomyelin synthases (SGMS, Fig. 7) to dihydrosphingomyelins (dhSM) and sphingomyelins (SM), respectively. Consequently, we also detected increased dhSM/SM ratios (Fig. 8B, Fig. S2C/D) after Co and/or Ni treatment. Furthermore, Co and Ni individually and combined resulted in a decrease in Sph (Fig. 8C) as well as in lactosylceramides (LacCer) (Fig. 8D). Additionally, Co and Ni caused elevated levels of various DAG species (Fig. 8E). In particular, those representatives that have two saturated fatty acids in the molecule (e.g. 16:0_16:0; 16:0_18:0 and 18:0_18:0) accumulated. The effect was reduced in the case of monounsaturated DAGs (e.g. 16:0_18:1 and 18:0_18:1) and eliminated or even reversed in the case of polyunsaturated DAGs (e.g. 18:1_18:1).
Fig. 7.
Effected pathways (transcriptome) related to sphingolipid metabolism by Co(II) and Ni(II) in HepG2 cells after 24 h combined exposure (25 μM Co + 150 μM Ni). Genes in green font represent upregulation, magenta font shows downregulation, and grey font indicates no alterations compared to untreated control. More detailed data is shown in Supplementary Table S6. Abbreviations: SPT serine palmitoyltransferase; KDSR 3-ketodihydrosphingosine reductase; CERS ceramide synthase; ASAH N-acylsphingosine amidohydrolase; DEGS sphingolipid delta 4-desaturase; SMPD sphingomyelin phosphodiesterase; SGMS sphingomyelin synthase; GALC galactosylceramidase; UGT8 UDP glycosyltransferase 8; UGCG UDP-glucose ceramide glucosyltransferase; GBA glucosylceramidase; B4GALT6 beta-1,4-galactosyltransferase 6; CERK ceramide kinase; S1PPase sphingosine-1-phosphatase; SPHK sphingosine kinase; SGPL sphingosine-1-phosphate lyase. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 8.
Quantification of A) dihydroceramides/ceramides ratio, B) dihydrosphingomyelins/sphingomyelins ratio, C) long chain bases, D) glycosylceramides, and E) diacylglycerols after individual or combined treatment with Co(II) and Ni(II) for 24 h in HepG2 cells. Sphingolipid and diacylglycerol levels were quantified using LC-MS/MS. Data is presented as mean +SD of n ≥ 3 independent experiments. Statistical significance was tested using Two-way ANOVA depicted as *p ≤ 0.05, ***p ≤ 0.001: compared to untreated control. Abbreviations: dhCer dihydroceramides; Cer ceramides; dhSM dihydrosphingomyelins; SM sphingomyelins; dhSph dihydrosphingosine; Sph sphingosine; S1P sphingosine-1-phosphate; HexCer hexosylceramides; LacCer lactosylceramides.
4. Discussion and conclusions
This study focuses on elucidating the toxicity mechanisms of Co and Ni and exploring possible effects of their combined presence, providing insights into their environmental and public health threats. Since Co and Ni are distributed in tissues, in particular the liver and kidneys upon oral intake and hence are responsible for hepatotoxicity, human liver carcinoma cells (HepG2) are applied in this study. The untargeted transcriptomic approach allows identifying the most relevant cellular responses in case of individual or combined exposure to Co and Ni. The relevance of the pathways is validated and verified by further targeted analysis.
Both Co and Ni led to a variety of differentially expressed genes when compared to the untreated HepG2 cells, with a notable overlap in gene expression between the two metals individually. Sun et al. investigated the effects of single Co and Ni treatment in Japanese flounder, observing a greater number of DEGs induced by Co, contrary to the findings in HepG2 cells where Ni triggered a larger response [31]. Transcriptomic analysis following Co and Ni exposure in human cell lines are limited to lung cells and monocytes and therefore barely comparable [32,33]. In this study we demonstrated that combined treatment resulted in considerably higher number of DEGs, suggesting possible interactions between Co and Ni, leading to distinct gene regulation pathways compared to individual exposure. Interestingly, the identified target DEGs merge in cellular responses driven by the transcription factor Nrf2. Nrf2 plays a pivotal role in regulating cellular metabolism pathways, such as iron or glutathione homeostasis, glycolysis, TCA cycle, and lipid metabolism, thus underscoring its significance in cellular protection and pathogenesis [34].
Under physiological conditions, Nrf2 is degraded in the cytoplasm by Kelch like-ECH-associated protein 1 (KEAP1) and Cullin 3 [34]. Upon oxidative stress or exposure to electrophiles, this regulatory system is disrupted, leading to the translocation of Nrf2 into the nucleus. This translocation of Nrf2 could be verified in this study upon Co and Ni exposure (alone and combined). There are only few studies reporting the Nrf2 activation in various cell lines following exposure to Co [35,36] or Ni [37,38]. However, they are primarily focusing on individual treatment, but lacking combined treatment or associated impacts of Nrf2 activation on cellular metabolism. The main mechanism triggering Nrf2 activation and subsequent translocation is oxidative stress related, by modifying the cysteine residues of KEAP1 and disrupting the KEAP1-NRF2 complex [28]. In a recent investigation involving HepG2 cells, it was observed that Co treatment led to an increase in RONS production, along with enhanced levels of oxidized glutathione (GSSG). Conversely, Ni exposure did not induce any significant alterations in these parameters [9]. Despite this, Ni led to a Nrf2 translocation, suggesting an oxidative stress independent activation of Nrf2 by Ni. It has been discussed that sequestosome 1 (SQSTM1), also known as p62, can bind to KEAP1 leading to the stabilization of Nrf2 [39]. In the present study, transcriptomic analysis revealed an upregulation of sqstm1, especially in response to the individual treatment of Ni or the combined exposure of Co and Ni (Table S5). Additionally, the exposure of Ni to human lung epithelial cells resulted in a significant upregulation in SQSTM1 protein expression [40]. Given the differences in their stress induction, it is plausible that Co and Ni engage distinct mechanisms in activating Nrf2 signaling pathways.
The Nrf2-regulated pathways governing iron and GSH metabolism were affected by Co and Ni. Key proteins involved in these pathways, including HMOX-1, GCLC, and GCLM were identified as downstream targets in a human epithelial cell line using knockdown cells of Nrf2 and Keap1. Furthermore, it was reported that Nrf2 expression reached its highest amount 6 h following Co treatment, while its targets reached their maximum expression levels after 24 h exposure [24,35]. From targeted studies investigating the toxicity of both metals, especially Co is associated to hypoxia-inducible factor-1 (HIF-1) and evidence suggests an association between the HIF-1 and Nrf2 signaling pathways, with Nrf2 downstream targets potentially acting as regulators of HIF-1 [41,42]. Knockdown of Nrf2 revealed reduced expression of HIF-1α in both human glioma cells and human colon cancer cells, even under hypoxic conditions or following Co treatment as a hypoxia-mimicking agent [43,44]. Even if Co is used as a hypoxia-mimicking agent, its toxicity mechanism seems to differ from that of physical hypoxia concerning Nrf2 signaling, particularly at transcriptional and post-transcriptional levels. Notably, Co treatment led to increased FTH mRNA expression in human erythroleukemia cells, whereas incubation under hypoxia showed no alterations in iron metabolism [45]. In our study, the combined exposure to Co and Ni resulted in elevated levels of FTL and FTH gene- and protein expression, leading to increased iron storage capacity, consequently preventing the potential for production of Fe3+ during Fenton reaction and radical formation. Despite Ni inducing Nrf2 activation, HMOX-1 protein expression was only increased in combined treatment with Co rather than when Ni was exposed individually. Lewis et al. reported similar findings in human monocytic cells treated with Ni, hypothesizing that this could result from either a sub-threshold level of activation or a form of Nrf2 incapable to promote HMOX-1 transcription [38]. Additionally, they also noted the absence of RONS induction which is consistent with the previous investigation in HepG2 cells [9]. In a human bronchial epithelial cell line, Ni caused an upregulation in HMOX-1 mRNA levels, along with elevated RONS levels. However, it is important to consider that the cells were exposed to a notably higher Ni concentration, which complicates direct comparison [46]. The two amino acids cysteine and glutamate, which are components of GSH, were decreased by Co and Ni, measured via LC-MS/MS. Transcriptomic analysis revealed an increased expression of GCLC and GCLM, induced by both Co and Ni, catalyzing the initial step of GSH synthesis. Following treatment with Co alone or in combination with Ni, HepG2 cells showed elevated GSSG levels. Notably, the GSH amount remained unchanged with both metals, indicating more GSH synthesis and direct oxidation [9]. Nrf2 also regulates transport and metabolism of glutamine, essential for GSH synthesis as it is converted to glutamate via the catalysis of glutaminase [24]. Upon combined treatment with Co and Ni, HepG2 cells exhibited elevated glutamine levels, indicating heightened transport into the cell likely driven by increased demand for glutamate. Across various cancer cell lines, it was observed that Nrf2 activation heightened the dependency on exogenous glutamine, primarily directed towards GSH synthesis [47,48]. Notably, Nrf2 plays a role in the expression of the glutamine transporter SLC1A5, as evidenced in a study using Keap1-mutant cells [49]. Next to this, glutamate is also indirectly involved in GSH synthesis by serving as an exchange molecule for cystine transporter SLC7A11 (also known as xCT), which is positively regulated by Nrf2 [50]. Enhanced expression of SLC7A11 was observed after combined treatment with Co and Ni in HepG2 cells. Enhanced glutamate secretion limited its availability for the TCA cycle or other pathways [47]. The effects of Co and Ni on TCA cycle metabolites α-ketoglutarate, succinate, or malate, showed only a slight increase, indicating that this pathway may not be the main route of toxicity for Co and Ni in cellular metabolism. Nrf2 enhances glycolysis by increasing the glucose import and inducing the expression of key enzymes along the glycolytic pathway [34]. Particularly under hypoxic conditions, cells prioritize converting most of the glucose to lactate rather than utilizing it in oxidative phosphorylation [51]. Treatment with Co and Ni resulted in increased lactate levels compared to untreated HepG2 cells, indicating heightened conversion of pyruvate to lactate, thus ensuring cellular energy supply. Lactate production is catalyzed by lactate dehydrogenase A (LDHA), which is notably enhanced in tumor growth and increased cell proliferation. Knockdown of Nrf2 in breast cancer cells led to a decrease in LDHA protein expression, correlating with reduced expression of HIF-1α [52].
Cellular metabolism of bioactive sphingolipids is highly regulated by a variety of enzymes, contributing to essential cellular functions such as cell growth, cell cycle regulation, cell death, inflammation, response to stress stimuli, and autophagy [53]. A previous metabolomic study in liver cells pointed out an impact especially of Co on the sphingolipid metabolism and synthesis [54]. The accumulation of dhCer, an intermediate in the de novo synthesis of Cer, is discussed to be stimulated by hypoxia or oxidative stress [55]. In mouse embryonic fibroblasts, knockdown of dihydroceramide desaturase 1 (DEGS1) resulted in elevated levels of dhCer and dhSM, while Cer, SM, Sph, and LacCer were decreased [56]. Similar results were observed in this study in HepG2 cells following treatment with Co and Ni in HepG2 cells, with more intense effects after combined incubation. Exposing different breast cancer cell lines to hypoxia resulted in DEGS1 inhibition with concurrent increase of dhCer [57], suggesting that Co and Ni may inhibit DEGS1 through their hypoxia-mimicking properties. The accumulation of dhCers is associated with altered bioenergetics, notably evidenced by decreased ATP/AMP ratios [56]. This alteration is believed to activate AMPK, which in turn stimulates autophagy through the activation of ULK1/2 [58]. Hernández-Tiedra et al. revealed the link between an increased dhCer/Cer ratio and autophagy-mediated apoptosis in human glioma cells [59]. An association between AMPK and Nrf2 signaling pathways is, among others, given by the earlier mentioned p62, which interacts with KEAP1 to activate Nrf2. Elevated levels of p62, in turn, facilitate AMPK-mediated activation of ULK [60]. Both ULK1/2 and p62 are overexpressed in HepG2 cells treated with Ni individually or in combination with Co (Table S5). In contrast to their less abundant saturated analogs (dhSM), SM are major components of biological membranes. Both sphingolipid subclasses have different effects on membrane fluidity; SM favors it, while dhSM increases membrane rigidity [61]. An increase in the cellular dhSM/SM ratio, as observed in this study after Co and/or Ni treatment, could facilitate the formation of assemblies in SM-rich lipid domains, which could play an important role in membrane-related biological processes [62].
Various pathways facilitate the intracellular formation of DAGs, including their production during triacylglycerol and phospholipid biosynthesis, or as a byproduct in the SM synthesis using Cer and phosphatidylcholine catalyzed by sphingomyelin synthase [63,64]. Exposure of Co and Ni individually and combined to HepG2 cells resulted in an increase of intracellular DAG levels. Lakatos et al. demonstrated in mesenchymal stromal cells that DAG content was elevated under hypoxic conditions and in the presence of Co as a hypoxia-mimicking agent. Inhibiting phosphatidylcholine-specific phospholipase C and sphingomyelin synthase reduced DAG levels under hypoxia, suggesting that increased DAGs result from enhanced enzyme activity [65]. Accumulation of DAGs is associated with the activation of protein kinase C (PKC) and their translocation to membranes [64]. This activation leads to the phosphorylation of various enzymes, including Nrf2, which is subsequently stabilized for translocation into the nucleus [66].
In conclusion, this study delves into the toxicity mechanisms of Cobalt (Co) and Nickel (Ni), shedding light on their individual and combined effects on cellular metabolism. Transcriptomic analysis provided a comprehensive understanding of gene regulation pathways impacted by both metals in HepG2 cells, revealing both shared and distinct responses. Notably, Co and Ni showed a synergistic effect, resulting in a significantly higher number of differentially expressed genes compared to individual exposure. Especially the activation of the Nrf2 signaling pathway seems to play an important role in cellular response to Co and Ni exposure. Additionally, alterations in sphingolipid metabolism and diacylglycerol accumulation underscore the complex cellular responses induced by these metals. Combining transcriptomic analysis and analytical methods provide an important and unconventional approach to gain insights into the intricate interplay between gene regulation and cellular metabolism underlying metal toxicity.
CRediT authorship contribution statement
Alicia Thiel: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Conceptualization. Franziska Drews: Writing – review & editing, Visualization, Formal analysis. Marcello Pirritano: Writing – review & editing, Visualization, Formal analysis. Fabian Schumacher: Writing – review & editing, Investigation, Formal analysis. Vivien Michaelis: Writing – review & editing, Methodology, Conceptualization. Maria Schwarz: Writing – review & editing, Methodology. Sören Franzenburg: Investigation. Tanja Schwerdtle: Writing – review & editing. Bernhard Michalke: Methodology, Investigation. Anna P. Kipp: Writing – review & editing. Burkhard Kleuser: Writing – review & editing. Martin Simon: Writing – review & editing. Julia Bornhorst: Writing – review & editing, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the DFG Research Unit TraceAge (FOR 2558, BO4103/4–2), the initiative “Metal based compounds (MeBaCo)” supported by the University of Wuppertal and the Faculty of Mathematics and Natural Sciences and by the DFG Research Infrastructure NGS_CC (project 407495230) as part of the Next Generation Sequencing Competence Network (project 423957469). NGS analyses were carried out at the Competence Center for Genomic Analysis (Kiel). The authors thank Daniel Herrmann for excellent technical assistance with LC-MS/MS analyses of sphingolipids and DAGs.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103290.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Genchi G., Carocci A., Lauria G., Sinicropi M.S., Catalano A. Nickel: human health and environmental toxicology. Int. J. Environ. Res. Publ. Health. 2020;17 doi: 10.3390/ijerph17030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leyssens L., Vinck B., van der Straeten C., Wuyts F., Maes L. Cobalt toxicity in humans-A review of the potential sources and systemic health effects. Toxicology. 2017;387:43–56. doi: 10.1016/j.tox.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Sule K., Umbsaar J., Prenner E.J. Mechanisms of Co, Ni, and Mn toxicity: from exposure and homeostasis to their interactions with and impact on lipids and biomembranes. Biochim. Biophys. Acta Biomembr. 2020;1862 doi: 10.1016/j.bbamem.2020.183250. [DOI] [PubMed] [Google Scholar]
- 4.Karagas M.R., Wang A., Dorman D.C., Hall A.L., Pi J., Sergi C.M., Symanski E., Ward E.M., Arrandale V.H., Azuma K., Brambila E., Calaf G.M., Fritz J.M., Fukushima S., Gaitens J.M., Grimsrud T.K., Guo L., Lynge E., Marinho-Reis A.P., McDiarmid M.A., Middleton D.R.S., Ong T.P., Polya D.A., Quintanilla-Vega B., Roberts G.K., Santonen T., Sauni R., Silva M.J., Wild P., Zhang C.W., Zhang Q., Grosse Y., Benbrahim-Tallaa L., de Conti A., DeBono N.L., El Ghissassi F., Madia F., Reisfeld B., Stayner L.T., Suonio E., Viegas S., Wedekind R., Ahmadi S., Mattock H., Gwinn W.M., Schubauer-Berigan M.K. Carcinogenicity of cobalt, antimony compounds, and weapons-grade tungsten alloy. Lancet Oncol. 2022;23:577–578. doi: 10.1016/S1470-2045(22)00219-4. [DOI] [PubMed] [Google Scholar]
- 5.IARC, Arsenic . 2012. Metals, Fibres, and Dusts, No. tenth0C, IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [Place of publication not identified] [PMC free article] [PubMed] [Google Scholar]
- 6.Tvermoes B.E., Paustenbach D.J., Kerger B.D., Finley B.L., Unice K.M. Review of cobalt toxicokinetics following oral dosing: implications for health risk assessments and metal-on-metal hip implant patients. Crit. Rev. Toxicol. 2015;45:367–387. doi: 10.3109/10408444.2014.985818. [DOI] [PubMed] [Google Scholar]
- 7.Simonsen L.O., Harbak H., Bennekou P. Cobalt metabolism and toxicology--a brief update. Sci. Total Environ. 2012;432:210–215. doi: 10.1016/j.scitotenv.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Beyersmann D., Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008;82:493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- 9.Thiel A., Michaelis V., Restle M., Figge S., Simon M., Schwerdtle T., Bornhorst J. Single is not combined: the role of Co and Ni bioavailability on toxicity mechanisms in liver and brain cells. Chemosphere. 2024;357 doi: 10.1016/j.chemosphere.2024.142091. [DOI] [PubMed] [Google Scholar]
- 10.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 11.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aleksander S.A., Balhoff J., Carbon S., Cherry J.M., Drabkin H.J., Ebert D., Feuermann M., Gaudet P., Harris N.L., Hill D.P., Lee R., Mi H., Moxon S., Mungall C.J., Muruganugan A., Mushayahama T., Sternberg P.W., Thomas P.D., van Auken K., Ramsey J., Siegele D.A., Chisholm R.L., Fey P., Aspromonte M.C., Nugnes M.V., Quaglia F., Tosatto S., Giglio M., Nadendla S., Antonazzo G., Attrill H., Dos Santos G., Marygold S., Strelets V., Tabone C.J., Thurmond J., Zhou P., Ahmed S.H., Asanitthong P., Luna Buitrago D., Erdol M.N., Gage M.C., Ali Kadhum M., Li K.Y.C., Long M., Michalak A., Pesala A., Pritazahra A., Saverimuttu S.C.C., Su R., Thurlow K.E., Lovering R.C., Logie C., Oliferenko S., Blake J., Christie K., Corbani L., Dolan M.E., Ni L., Sitnikov D., Smith C., Cuzick A., Seager J., Cooper L., Elser J., Jaiswal P., Gupta P., Naithani S., Lera-Ramirez M., Rutherford K., Wood V., de Pons J.L., Dwinell M.R., Hayman G.T., Kaldunski M.L., Kwitek A.E., Laulederkind S.J.F., Tutaj M.A., Vedi M., Wang S.-J., D'Eustachio P., Aimo L., Axelsen K., Bridge A., Hyka-Nouspikel N., Morgat A., Engel S.R., Karra K., Miyasato S.R., Nash R.S., Skrzypek M.S., Weng S., Wong E.D., Bakker E., Berardini T.Z., Reiser L., Auchincloss A., Argoud-Puy G., Blatter M.-C., Boutet E., Breuza L., Casals-Casas C., Coudert E., Estreicher A., Livia Famiglietti M., Gos A., Gruaz-Gumowski N., Hulo C., Jungo F., Le Mercier P., Lieberherr D., Masson P., Pedruzzi I., Pourcel L., Poux S., Rivoire C., Sundaram S., Bateman A., Bowler-Barnett E., Bye-A-Jee H., Denny P., Ignatchenko A., Ishtiaq R., Lock A., Lussi Y., Magrane M., Martin M.J., Orchard S., Raposo P., Speretta E., Tyagi N., Warner K., Zaru R., Diehl A.D., Chan J., Diamantakis S., Raciti D., Zarowiecki M., Fisher M., James-Zorn C., Ponferrada V., Zorn A., Ramachandran S., Ruzicka L., Westerfield M. The gene ontology knowledgebase in 2023. Genetics. 2023;224 doi: 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas P.D., Ebert D., Muruganujan A., Mushayahama T., Albou L.-P., Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31:8–22. doi: 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfram T., Schwarz M., Reuß M., Lossow K., Ost M., Klaus S., Schwerdtle T., Kipp A.P. N-acetylcysteine as modulator of the essential trace elements copper and zinc. Antioxidants. 2020;9 doi: 10.3390/antiox9111117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaelis V., Aengenheister L., Tuchtenhagen M., Rinklebe J., Ebert F., Schwerdtle T., Buerki-Thurnherr T., Bornhorst J. Differences and interactions in placental manganese and iron transfer across an in vitro model of human villous trophoblasts. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23063296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalke B., Willkommen D., Venkataramani V. Iron redox speciation analysis using capillary electrophoresis coupled to inductively coupled plasma mass spectrometry (CE-ICP-MS) Front. Chem. 2019;7:136. doi: 10.3389/fchem.2019.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalke B., Willkommen D., Venkataramani V. Setup of capillary electrophoresis-inductively coupled plasma mass spectrometry (CE-ICP-MS) for quantification of iron redox species (Fe(II), Fe(III)) JoVE. 2020 doi: 10.3791/61055-v. [DOI] [PubMed] [Google Scholar]
- 20.Dong X.-M., Wu P., Cheng L.-H., Shou L., Dong H., Chen X.-Y., Gao H.-J., Chen J.-X., Xiang F., Zhang Q., Zhang D.-H., Zhou J.-L., Xie T. Targeted profiling of polar metabolites in cancer metabolic reprogramming by hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2022;1686 doi: 10.1016/j.chroma.2022.463654. [DOI] [PubMed] [Google Scholar]
- 21.Zeitler S., Schumacher F., Monti J., Anni D., Guhathakurta D., Kleuser B., Friedland K., Fejtová A., Kornhuber J., Rhein C. Acid sphingomyelinase impacts canonical transient receptor potential channels 6 (TRPC6) activity in primary neuronal systems. Cells. 2020;9 doi: 10.3390/cells9112502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naser E., Kadow S., Schumacher F., Mohamed Z.H., Kappe C., Hessler G., Pollmeier B., Kleuser B., Arenz C., Becker K.A., Gulbins E., Carpinteiro A. Characterization of the small molecule ARC39, a direct and specific inhibitor of acid sphingomyelinase in vitro. J. Lipid Res. 2020;61:896–910. doi: 10.1194/jlr.RA120000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnot T., Gillard M., Nagel D. A simple protocol for informative visualization of enriched gene ontology terms. BIO-PROTOCOL. 2019;9 doi: 10.21769/BioProtoc.3429. [DOI] [Google Scholar]
- 24.Lin T.-Y., Cantley L.C., DeNicola G.M. In: A Master Regulator of Oxidative Stress - the Transcription Factor Nrf2. Morales-Gonzalez J.A., Morales-Gonzalez A., Madrigal-Santillan E.O., editors. InTech; 2016. NRF2 rewires cellular metabolism to support the antioxidant response. [Google Scholar]
- 25.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Oliveira J., Denadai M.B., Costa D.L. Crosstalk between heme oxygenase-1 and iron metabolism in macrophages: implications for the modulation of inflammation and immunity. Antioxidants. 2022;11 doi: 10.3390/antiox11050861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 28.He F., Ru X., Wen T. NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21134777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan-Chen S., Guitton J., Bourron O., Le Stunff H., Hajduch E. Sphingolipid metabolism and signaling in skeletal muscle: from physiology to physiopathology. Front. Endocrinol. 2020;11:491. doi: 10.3389/fendo.2020.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy M., Futerman A.H. Mammalian ceramide synthases. IUBMB Life. 2010;62:347–356. doi: 10.1002/iub.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z., Gong C., Ren J., Zhang X., Wang G., Liu Y., Ren Y., Zhao Y., Yu Q., Wang Y., Hou J. Toxicity of nickel and cobalt in Japanese flounder. Environ. Pollut. 2020;263 doi: 10.1016/j.envpol.2020.114516. [DOI] [PubMed] [Google Scholar]
- 32.Gölz L., Buerfent B.C., Hofmann A., Rühl H., Fricker N., Stamminger W., Oldenburg J., Deschner J., Hoerauf A., Nöthen M.M., Schumacher J., Hübner M.P., Jäger A. Genome-wide transcriptome induced by nickel in human monocytes. Acta Biomater. 2016;43:369–382. doi: 10.1016/j.actbio.2016.07.047. [DOI] [PubMed] [Google Scholar]
- 33.Malard V., Berenguer F., Prat O., Ruat S., Steinmetz G., Quemeneur E. Global gene expression profiling in human lung cells exposed to cobalt. BMC Genom. 2007;8:147. doi: 10.1186/1471-2164-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Lin X., Meng Di, Zeng L., Zhuang R., Huang S., Lv W., Hu J. Nrf2 mediates metabolic reprogramming in non-small cell lung cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.578315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B., Cheng H., Wang L., Fu J., Zhang G., Guan D., Qi R., Gao X., Zhao R. Protective roles of NRF2 signaling pathway in cobalt chloride-induced hypoxic cytotoxicity in human HaCaT keratinocytes. Toxicol. Appl. Pharmacol. 2018;355:189–197. doi: 10.1016/j.taap.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 36.Yu J., Yang H., Fang B., Zhang Z., Wang Y., Dai Y. mfat-1 transgene protects cultured adult neural stem cells against cobalt chloride-mediated hypoxic injury by activating Nrf2/ARE pathways. J. Neurosci. Res. 2018;96:87–102. doi: 10.1002/jnr.24096. [DOI] [PubMed] [Google Scholar]
- 37.Lewis J.B., Messer R.L.W., Pitts L., Hsu S.D., Hansen J.M., Wataha J.C. Ni(II) ions dysregulate cytokine secretion from human monocytes. J. Biomed. Mater. Res. B Appl. Biomater. 2009;88:358–365. doi: 10.1002/jbm.b.31063. [DOI] [PubMed] [Google Scholar]
- 38.Lewis J.B., Messer R.L., McCloud V.V., Lockwood P.E., Hsu S.D., Wataha J.C. Ni(II) activates the Nrf2 signaling pathway in human monocytic cells. Biomaterials. 2006;27:5348–5356. doi: 10.1016/j.biomaterials.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Saito T., Ichimura Y., Taguchi K., Suzuki T., Mizushima T., Takagi K., Hirose Y., Nagahashi M., Iso T., Fukutomi T., Ohishi M., Endo K., Uemura T., Nishito Y., Okuda S., Obata M., Kouno T., Imamura R., Tada Y., Obata R., Yasuda D., Takahashi K., Fujimura T., Pi J., Lee M.-S., Ueno T., Ohe T., Mashino T., Wakai T., Kojima H., Okabe T., Nagano T., Motohashi H., Waguri S., Soga T., Yamamoto M., Tanaka K., Komatsu M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016;7 doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang H., Zhu J., Li Y., Zhang L., Gu J., Xie Q., Jin H., Che X., Li J., Huang C., Chen L.-C., Lyu J., Gao J., Huang C. Upregulation of SQSTM1/p62 contributes to nickel-induced malignant transformation of human bronchial epithelial cells. Autophagy. 2016;12:1687–1703. doi: 10.1080/15548627.2016.1196313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth R.K., Warfel N.A. Strange bedfellows: nuclear factor, erythroid 2-like 2 (Nrf2) and hypoxia-inducible factor 1 (HIF-1) in tumor hypoxia. Antioxidants. 2017;6 doi: 10.3390/antiox6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxwell P., Salnikow K. HIF-1: an oxygen and metal responsive transcription factor. Cancer Biol. Ther. 2004;3:29–35. doi: 10.4161/cbt.3.1.547. [DOI] [PubMed] [Google Scholar]
- 43.Ji X., Wang H., Zhu J., Zhu L., Pan H., Li W., Zhou Y., Cong Z., Yan F., Chen S. Knockdown of Nrf2 suppresses glioblastoma angiogenesis by inhibiting hypoxia-induced activation of HIF-1α. Int. J. Cancer. 2014;135:574–584. doi: 10.1002/ijc.28699. [DOI] [PubMed] [Google Scholar]
- 44.Kim T.-H., Hur E., Kang S.-J., Kim J.-A., Thapa D., Lee Y.M., Ku S.K., Jung Y., Kwak M.-K. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1α. Cancer Res. 2011;71:2260–2275. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- 45.Huang B.-W., Miyazawa M., Tsuji Y. Distinct regulatory mechanisms of the human ferritin gene by hypoxia and hypoxia mimetic cobalt chloride at the transcriptional and post-transcriptional levels. Cell. Signal. 2014;26:2702–2709. doi: 10.1016/j.cellsig.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forti E., Salovaara S., Cetin Y., Bulgheroni A., Tessadri R., Jennings P., Pfaller W., Prieto P. In vitro evaluation of the toxicity induced by nickel soluble and particulate forms in human airway epithelial cells. Toxicol. Vitro. 2011;25:454–461. doi: 10.1016/j.tiv.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Sayin V.I., LeBoeuf S.E., Singh S.X., Davidson S.M., Biancur D., Guzelhan B.S., Alvarez S.W., Wu W.L., Karakousi T.R., Zavitsanou A.M., Ubriaco J., Muir A., Karagiannis D., Morris P.J., Thomas C.J., Possemato R., Vander Heiden M.G., Papagiannakopoulos T. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife. 2017;6 doi: 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sánchez-Rivera F.J., Subbaraj L., Martinez B., Bronson R.T., Prigge J.R., Schmidt E.E., Thomas C.J., Goparaju C., Davies A., Dolgalev I., Heguy A., Allaj V., Poirier J.T., Moreira A.L., Rudin C.M., Pass H.I., Vander Heiden M.G., Jacks T., Papagiannakopoulos T. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin C.-S., Mishra P., Watrous J.D., Carelli V., D'Aurelio M., Jain M., Chan D.C. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat. Commun. 2017;8 doi: 10.1038/ncomms15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeBerardinis R.J., Chandel N.S. We need to talk about the Warburg effect. Nat. Metab. 2020;2:127–129. doi: 10.1038/s42255-020-0172-2. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H.-S., Du G.-Y., Zhang Z.-G., Zhou Z., Sun H.-L., Yu X.-Y., Shi Y.-T., Xiong D.-N., Li H., Huang Y.-H. NRF2 facilitates breast cancer cell growth via HIF1ɑ-mediated metabolic reprogramming. Int. J. Biochem. Cell Biol. 2018;95:85–92. doi: 10.1016/j.biocel.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 53.Hannun Y.A., Obeid L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellouard M., Gasser M., Lenglet S., Gilardi F., Bararpour N., Augsburger M., Thomas A., Alvarez J.-C. Toxicity and metabolomic impact of cobalt, chromium, and nickel exposure on HepaRG hepatocytes. Chem. Res. Toxicol. 2022;35:807–816. doi: 10.1021/acs.chemrestox.1c00429. [DOI] [PubMed] [Google Scholar]
- 55.Lachkar F., Ferré P., Foufelle F., Papaioannou A. Dihydroceramides: their emerging physiological roles and functions in cancer and metabolic diseases. Am. J. Physiol. Endocrinol. Metab. 2021;320:E122–E130. doi: 10.1152/ajpendo.00330.2020. [DOI] [PubMed] [Google Scholar]
- 56.Siddique M.M., Li Y., Wang L., Ching J., Mal M., Ilkayeva O., Wu Y.J., Bay B.H., Summers S.A. Ablation of dihydroceramide desaturase 1, a therapeutic target for the treatment of metabolic diseases, simultaneously stimulates anabolic and catabolic signaling. Mol. Cell Biol. 2013;33:2353–2369. doi: 10.1128/MCB.00226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Devlin C.M., Lahm T., Hubbard W.C., van Demark M., Wang K.C., Wu X., Bielawska A., Obeid L.M., Ivan M., Petrache I. Dihydroceramide-based response to hypoxia. J. Biol. Chem. 2011;286:38069–38078. doi: 10.1074/jbc.M111.297994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alers S., Löffler A.S., Wesselborg S., Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernández-Tiedra S., Fabriàs G., Dávila D., Salanueva Í.J., Casas J., Montes L.R., Antón Z., García-Taboada E., Salazar-Roa M., Lorente M., Nylandsted J., Armstrong J., López-Valero I., McKee C.S., Serrano-Puebla A., García-López R., González-Martínez J., Abad J.L., Hanada K., Boya P., Goñi F., Guzmán M., Lovat P., Jäättelä M., Alonso A., Velasco G. Dihydroceramide accumulation mediates cytotoxic autophagy of cancer cells via autolysosome destabilization. Autophagy. 2016;12:2213–2229. doi: 10.1080/15548627.2016.1213927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petsouki E., Cabrera S.N.S., Heiss E.H. AMPK and NRF2: interactive players in the same team for cellular homeostasis? Free Radic. Biol. Med. 2022;190:75–93. doi: 10.1016/j.freeradbiomed.2022.07.014. [DOI] [PubMed] [Google Scholar]
- 61.Vieira C.R., Munoz-Olaya J.M., Sot J., Jiménez-Baranda S., Izquierdo-Useros N., Abad J.L., Apellániz B., Delgado R., Martinez-Picado J., Alonso A., Casas J., Nieva J.L., Fabriás G., Mañes S., Goñi F.M. Dihydrosphingomyelin impairs HIV-1 infection by rigidifying liquid-ordered membrane domains. Chem. Biol. 2010;17:766–775. doi: 10.1016/j.chembiol.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 62.Kinoshita M., Kyo T., Matsumori N. Assembly formation of minor dihydrosphingomyelin in sphingomyelin-rich ordered membrane domains. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-68688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tallima H., Azzazy H.M.E., El Ridi R. Cell surface sphingomyelin: key role in cancer initiation, progression, and immune evasion. Lipids Health Dis. 2021;20:150. doi: 10.1186/s12944-021-01581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolczynska K., Loza-Valdes A., Hawro I., Sumara G. Diacylglycerol-evoked activation of PKC and PKD isoforms in regulation of glucose and lipid metabolism: a review. Lipids Health Dis. 2020;19:113. doi: 10.1186/s12944-020-01286-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lakatos K., Kalomoiris S., Merkely B., Nolta J.A., Fierro F.A. Mesenchymal stem cells respond to hypoxia by increasing diacylglycerols. J. Cell. Biochem. 2016;117:300–307. doi: 10.1002/jcb.25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matzinger M., Fischhuber K., Heiss E.H. Activation of Nrf2 signaling by natural products-can it alleviate diabetes? Biotechnol. Adv. 2018;36:1738–1767. doi: 10.1016/j.biotechadv.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.