Abstract
During the progression of proliferative vitreoretinopathy (PVR) following ocular trauma, previously quiescent retinal pigment epithelial (RPE) cells transition into a state of rapid proliferation, migration, and secretion. The elusive molecular mechanisms behind these changes have hindered the development of effective pharmacological treatments, presenting a pressing clinical challenge. In this study, by monitoring the dynamic changes in chromatin accessibility and various histone modifications, we chart the comprehensive epigenetic landscape of RPE cells in male mice subjected to traumatic PVR. Coupled with transcriptomic analysis, we reveal a robust correlation between enhancer activation and the upregulation of the PVR-associated gene programs. Furthermore, by constructing transcription factor regulatory networks, we identify the aberrant activation of enhancer-driven RANK-NFATc1 pathway as PVR advanced. Importantly, we demonstrate that intraocular interventions, including nanomedicines inhibiting enhancer activity, gene therapies targeting NFATc1 and antibody therapeutics against RANK pathway, effectively mitigate PVR progression. Together, our findings elucidate the epigenetic basis underlying the activation of PVR-associated genes during RPE cell fate transitions and offer promising therapeutic avenues targeting epigenetic modulation and the RANK-NFATc1 axis for PVR management.
Subject terms: Retinal diseases, Epigenetics, Gene expression, Chromatin remodelling
Here the authors elucidate the epigenetic basis underlying the activation of PVR-associated genes during RPE cell fate transitions and offers potential therapeutic avenues targeting epigenetic modulation and the RANK-NFATc1 axis for PVR management.
Introduction
Open globe injury (OGI) is a critical ophthalmic emergency, often leading to blindness1. Traumatic proliferative vitreoretinopathy (PVR) is the major complication of OGI, observed in approximately 40–60% of post-OGI patients2. PVR is characterized by the formation of extensive fibrous proliferative membranes, whose contraction leads to retinal detachment or intraocular hemorrhage, severely compromising visual prognosis3. Despite advancements in vitreoretinal surgical techniques, clinical outcomes are still not optimal, and PVR persists as a primary cause of blindness4,5. The elusive mechanisms underlying PVR have hindered the development of effective pharmacological treatments, making it a pressing clinical challenge6.
The retinal pigment epithelial (RPE) cells play a central role in the pathogenesis of PVR7,8. Under physiological conditions, RPE cells are terminally differentiated and highly polarized, primarily serving to maintain visual function by phagocytosing shed photoreceptor outer segments and forming the outer retinal barrier. Following ocular injury, these cells undergo profound changes: they lose their polarity, migrate into the vitreous cavity or subretinal space, and proliferate5. As they undergo epithelial-mesenchymal transition (EMT), they contribute to the synthesis and secretion of extracellular matrix, leading to tissue remodeling and the formation of epiretinal and/or subretinal membranes, hallmarks of PVR9. While various factors, such as the disruption of cell-cell junctions, inflammatory responses, hypoxia, cytokines, and growth factors, have been identified to influence RPE cell fate transitions, the precise molecular mechanisms driving quiescent RPE cells towards a proliferative, migratory, and secretory phenotype remain unclear5,10.
Epigenetic mechanisms, such as chromatin accessibility and histone modifications, play pivotal roles in guiding gene expression and determining cell fate11. Transcription factors (TFs) often orchestrate transcriptional regulatory networks (TRNs) that are essential for defining and maintaining chromatin states12,13. Aberrant shifts in chromatin states are frequently linked with disease onset and progression14,15. In this study, we isolate primary RPE cells from both control mice and those post-PVR. Through profiling chromatin accessibility and various transcription-associated histone modifications, we delineate the chromatin dynamics during PVR progression. Our TRN analysis highlight a key role of the RANK-NFATc1 pathway in the advancement of PVR. Leveraging diverse intervention approaches, we propose PVR therapeutic strategies targeting epigenetic modifications and the RANK-NFATc1 pathway.
Results
Characterization of dynamic epigenetic landscape in RPE cells during PVR progression
To elucidate the epigenetic mechanisms underlying the pathogenesis of PVR after injury, we first established a traumatic PVR mouse model. By making a puncture posterior to the equator level, we induced a full-layer retinal injury, closely mimicking the penetrating wounds seen in patients with OGI. Subsequent Dispase injection aggravated the intraocular tissue damage, promoting the formation of PVR (Supplementary Fig. 1A). Compared to normal mice, those with PVR exhibited vitreous hemorrhage, extensive retinal detachment and formation of fibrous membrane (Supplementary Fig. 1B, C). Alpha-smooth muscle actin (αSMA), an EMT marker of RPE cell, was considered high-expressed in PVR. Notably, αSMA immunostaining was predominantly co-localized with the PVR lesion area (Supplementary Fig. 1D). Western blotting analysis revealed a pronounced upregulation of αSMA, in the neuroretina-RPE-choroid complex (eyecup) tissues of PVR mice (Supplementary Fig. 1E). The morphological characteristics and changes in marker genes of our PVR model are consistent with those described in prior studies using the same mouse model16–19. These data confirm the successful establishment of a traumatic PVR mouse model. Given the pivotal role of RPE cells in PVR membrane formation, we subsequently purified primary RPE cells from these mice. As depicted in Supplementary Fig. 1F, over 90% of the cells were RPE65-positive, a hallmark of RPE cell, and displayed melanin granules, indicating that we successfully isolated a high-purity population of primary RPE cells.
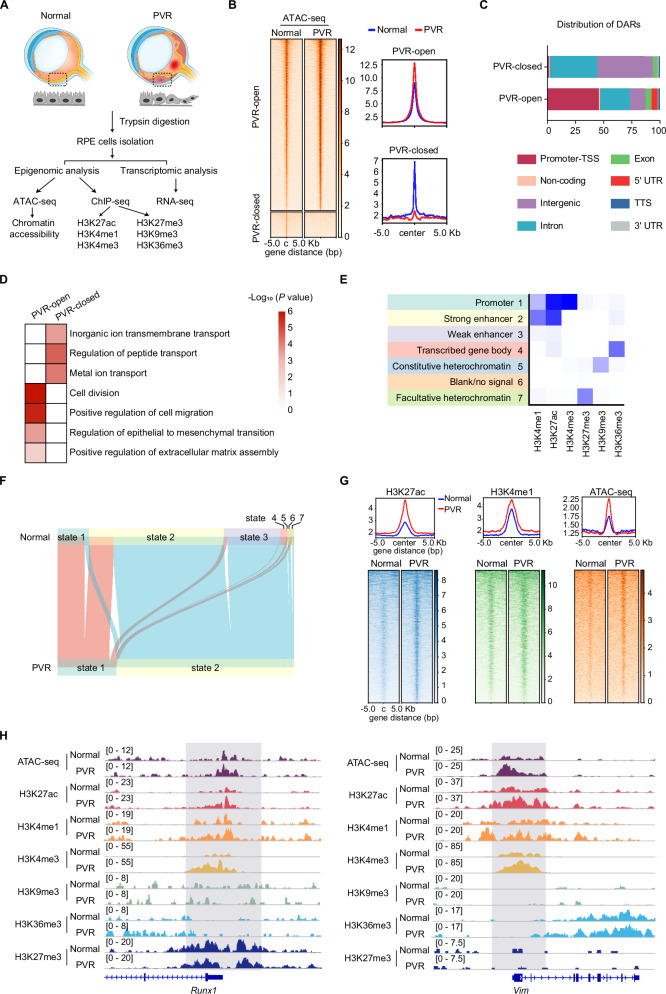
To map the genome-wide chromatin dynamics accompanying the RPE cell fate transition during PVR, we employed ATAC-seq (assay for transposase-accessible chromatin by sequencing), a robust technique to probe chromatin accessibility (Fig. 1A). The ATAC-seq data exhibited high reproducibility among three biological replicates (Supplementary Fig. 2). Using HOMER analysis, we identified 9,819 peaks showing increased accessibility (PVR-open), and 1,454 peaks with reduced accessibility (PVR-closed) in the PVR group. These differentially accessible regions (DARs) were predominantly localized in cis-regulatory regions, including promoters, intron, and intergenic regions (Fig. 1C). Gene Ontology (GO) analysis illuminated that the PVR-open genomic regions predominantly aligned with pathways governing cell migration, division, and epithelial to mesenchymal transition (Fig. 1D). This is consistent with existing literature that posits the EMT of RPE cells during PVR and emphasizes the role of RPE cells migration and proliferation in PVR progression6,10. Conversely, the PVR-closed genomic regions were enriched with genes pivotal for transport processes20, indicating that RPE cells may undergo a loss of their inherent cellular functions during PVR progression21. MDM2 (mouse double minute 2), an E3 ligase for P53, has previously been shown to facilitate the EMT of RPE cells22. PCNA (proliferating cell nuclear antigen) acts as a proliferation indicator, has been assessed in human traumatic PVR (TPVR) membranes3. In the PVR group, the promoter region of the Mdm2 and Pcna gene displayed enhanced chromatin accessibility (Supplementary Fig. 3A, B). These data highlight genome-wide shifts in cis-regulatory domains of RPE cells during PVR progression. To gain a deeper understanding of the epigenomic alterations during PVR progression, we charted several key histone modifications associated with transcription. These include markers indicative of transcriptional activation, namely H3K27ac, H3K4me1, H3K4me3 and H3K36me3; markers suggestive of transcriptional repression, such as H3K27me3 and H3K9me323. Given the limited availability of RPE cells, we adopted a low-input ChIP-seq (chromatin immunoprecipitation followed by sequencing) approach to assess histone modifications. All obtained data exhibited high reproducibility among three biological replicates (Supplementary Fig. 4A–F). Using ChromHMM, we integrated the ChIP-seq data to define chromatin states. As shown in Fig. 1E, ChromHMM algorithm identified four classes of chromatin states: heterochromatin (state 5 and 7), enhancers (state 2 and 3), transcribed genes (state 1 and 4), and blank/no signal state (state 6). Notably, we observed a pronounced shift from state 3 (weak enhancer) to state 2 (strong enhancer) within the PVR group, suggesting enhancer activation as PVR advanced (Fig. 1F).
Fig. 1. Profiling of dynamic epigenetic changes in RPE post-PVR.
A Experimental strategy of epigenomic and transcriptomic analysis of RPE cells isolated from normal and PVR mice. The ATAC-seq, ChIP-seq, and RNA-seq experiments were each conducted in triplicate. B Heatmap showing the chromatin accessibility in RPE cells from normal and PVR mice. C Bar charts showing the genome-wide distribution of differentially accessible regions (DARs) in RPE cells. D Gene ontology (GO) analysis (performed using Metascape) of DARs in RPE cells. Statistical analysis was conducted with Metascape (https://metascape.org/gp/index.html), using the two-sided cumulative hypergeometric distribution. E Seven chromatin states inference based on the ChromHMM algorithm. F Alluvial plot showing the dynamics of chromatin states. G Average ATAC-seq and ChIP-seq signals of H3K4me1 around pronounced H3K27ac signal regions (upper), and heatmaps visualization of histone marks and ATAC-seq signals (lower). H Representative ChIP-seq tracks of H3K27ac, H3K4me1, H3K4me3, H3K9me3, H3K36me3, H3K27me3 and ATAC-seq signals on representative genes. Source data are provided as a Source Data file.
Histone modifications and chromatin accessibility are intricately intertwined, collaboratively regulating the expression of pivotal genes involved in cell fate determination24. To discern if chromatin state alterations align with chromatin accessibility, we probed the signals of H3K4me1 and chromatin accessibility within pronounced H3K27ac signal regions (enhancer markers). A marked increase in chromatin enrichment of H3K4me1 and enhanced chromatin accessibility was evident in regions associated with increased H3K27ac (Fig. 1G), suggesting that enhanced enhancer activity is congruent with increased chromatin accessibility in RPE cells during PVR progression. RUNX1 (Runt-related transcription factor 1) has previously been shown to regulate cell proliferation and migration in PVR25. Vimentin, a marker of fibrosis previously identified in the PVR membrane, plays a crucial role in the EMT of RPE cells26,27. The Runx1 and Vim gene in PVR RPE cells exhibited heightened chromatin accessibility and elevated levels of H3K27ac, H3K4me1, and H3K4me3 compared to normal RPE cells, while other histone modifications remained relatively unchanged (Fig. 1H). Collectively, these findings uncover the epigenetic underpinnings that drive the development of PVR following injury, with a marked activation of enhancers and enhanced chromatin accessibility in RPE cells during PVR progression.
Active chromatin states correlate with the upregulation of PVR-associated gene expression programs
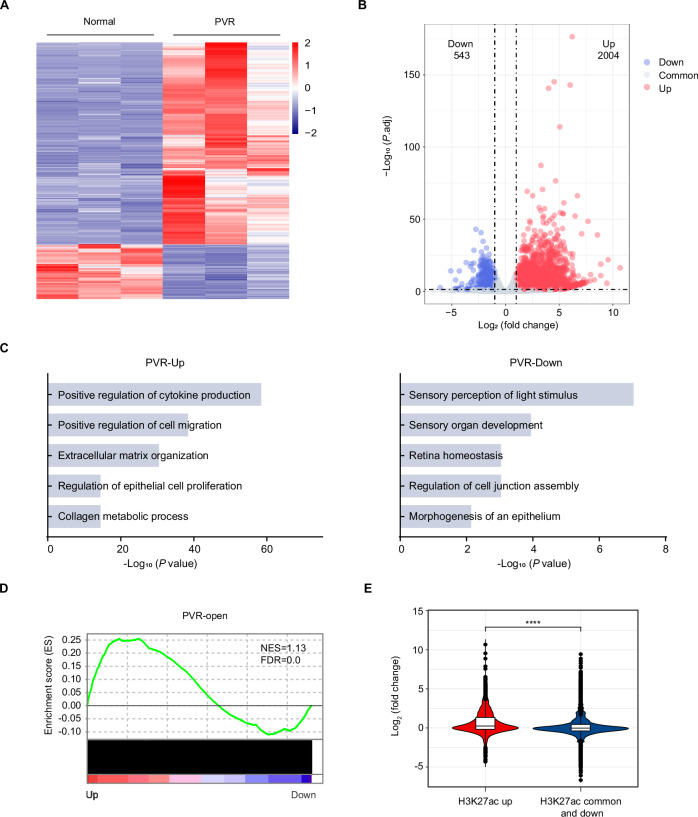
Increased chromatin accessibility combined with enhancer activation acts as a crucial regulatory mechanism for activating gene expression28,29. Given the pivotal role of the activation of key genes in cell fate transitions, we sought to explore the connection between these epigenetic changes in PVR and the corresponding PVR-related gene expression programs. To this end, we performed RNA sequencing (RNA-seq) on RPE cells purified from both normal and PVR mice. The DESEQ2 algorithms analysis revealed marked differences in the transcriptomic profiles between normal RPE cells and PVR RPE cells (Fig. 2A). A total of 2004 genes were found to be up-regulated, and 543 genes were down-regulated over 2-fold in PVR compared to normal control (Fig. 2B). GO analysis revealed that the downregulated genes predominantly encompassed retina function-related genes, such as those involved in the sensory perception of light stimulus, sensory organ development and retina homeostasis (Fig. 2C), suggesting a compromised functional state of RPE cells during PVR progression. Conversely, upregulated genes were enriched in pathways including extracellular matrix organization, collagen metabolic process, and cytokine production regulation (Fig. 2C), indicating enhanced fibrotic and inflammatory activities in PVR. Gene set enrichment analysis (GSEA) showed that genes from regions with PVR-open chromatin regions were notably prevalent among the upregulated genes in PVR RPE cells (Fig. 2D). An integrated analysis of RNA-seq and ChIP-seq data showed that regions with a pronounced H3K27ac signal were associated with gene activation, in contrast to regions with diminished or stable H3K27ac signals (Fig. 2E). Together, the above findings underscore a robust correlation between active chromatin states and the enhanced expression of PVR-associated genes.
Fig. 2. Active chromatin states are associated with the elevated expression of PVR-related genes.
A Heatmap of differentially gene expression in RPE cells. B Volcano plots showing differentially expressed genes (normal versus PVR). P values were calculated by two-sided Wald test in DESeq2 package. C Gene ontology (GO) analysis of differentially genes. Statistical analysis was conducted with Metascape (https://metascape.org/gp/index.html), using the two-sided cumulative hypergeometric distribution. D Gene set enrichment analysis (GSEA) displaying PVR-open chromatin regions enriched for genes upregulated in PVR RPE cells. FDR: false discovery rate; NES: normalized enrichment score. E Violin plots show that genes marked with higher levels of H3K27ac are significantly associated with gene activation. n = 3 biologically independent experiments. The black horizontal lines within the boxes indicate the median values, the edges of the boxes represent the first and third quartiles of the dataset. The whiskers extend to 1.5 times the interquartile range. P < 2.22 × 10−16 was determined by two-sided Wilcoxon test. ****P < 0.0001. Source data are provided as a Source Data file.
BET inhibition alleviates PVR progression
The H3K27 acetyl mark, present in active enhancer and open chromatin regions, serves as a docking site for the recruitment of epigenomic reader proteins30. Bromodomain and extra terminal domain (BET) proteins recognize acetylated histones and orchestrates the assembly of the transcriptional machinery at active enhancers31–33. Small-molecule inhibitors of BET protein BRD4, such as JQ1, bind to the acetyl-lysine binding pocket of BRD4 bromodomains, displacing BRD4 from chromatin34. This displacement leads to the removal of the Mediator complex and RNA Polymerase II from enhancers, subsequently diminishing associated gene expression35. Given the significant correlation between enhancer-activated genes and PVR progression, we sought to explore the potential therapeutic effect of JQ1-mediated enhancer inhibition in treating PVR.
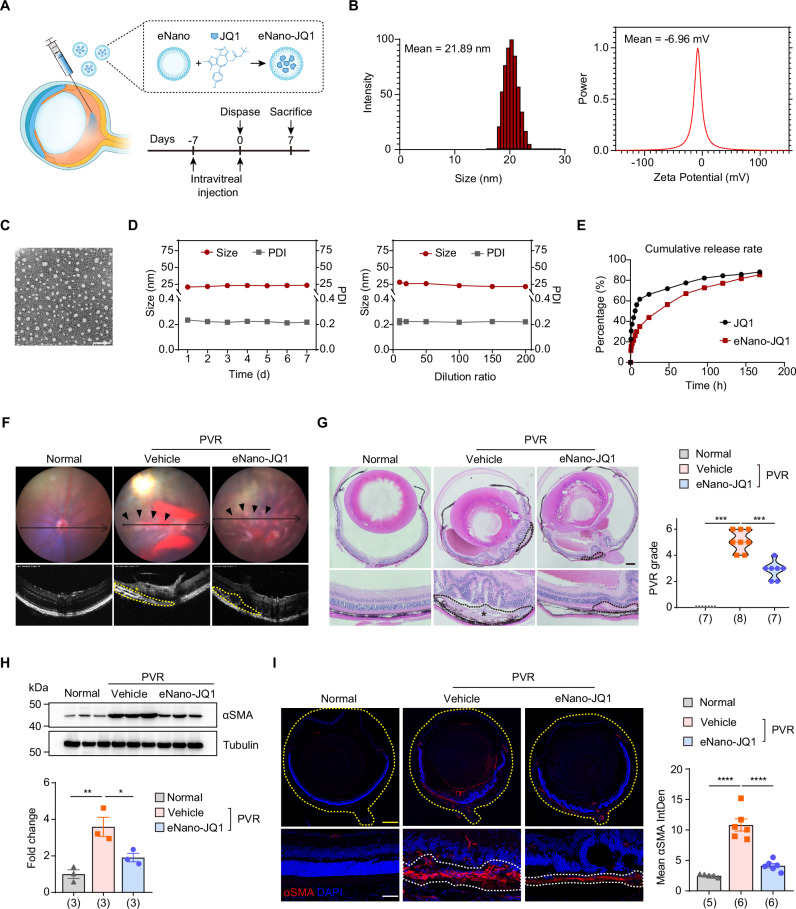
Upon administering JQ1 via intraperitoneal injection, we observed a moderate decrease in PVR membrane formation, but there was no marked improvement in retinal detachment or vitreous hemorrhage (Supplementary Fig. 5). However, the required daily dosage of 50 mg/kg resulted in significant side effects, as previously reported36, constraining its practical use. Furthermore, given JQ1’s limited water solubility, it requires dissolution in DMSO34,37. Yet, even at minimal concentrations, DMSO can induce retinal toxicity38. To circumvent the systemic side effects and to provide an alternative to DMSO as a solvent, we formulated a nanoemulsion tailored for the intraocular delivery of JQ1 (eNano-JQ1) (Fig. 3A). The eNano-JQ1 was formulated using a low-energy emulsification technique, employing tricaprylin as the oil phase and PEG-40 hydrogenated castor oil as the emulsifier. Both components are non-toxic and have been previously used in ocular applications39,40. Nanoemulsion formulations were typically characterized based on particle size distribution, polydispersity index (PDI) and zeta potential measurement. Zeta potential is a key physical property of particles in suspension, reflecting the surface charge that significantly affects their stability and interactions41,42. Figure 3B illustrates the size distribution and zeta potential measurements of the eNano-JQ1 after a ten-fold dilution, with average sizes of 21.89 ± 0.36 nm, and average zeta potentials of −6.96 ± 1.36 mV. The microstructural morphology, depicted in Fig. 3C, displayed uniform spherical shapes and dispersion. The average size and PDI remained consistent across 1–7 days, as well as following a 10–200-fold dilution, indicating reliable storage stability (Fig. 3D). Subsequent in vitro release studies, conducted seven days post-synthesis, revealed that eNano-JQ1 provided a more consistent and sustained release than free JQ1, reaching a cumulative release rate over 80% (Fig. 3E).
Fig. 3. Inhibition of the BET bromodomain mitigates the progression of PVR.
A Schematic view of experimental strategy in the PVR mouse model. B The size and zeta potential of eNano-JQ1. C Transmission electron microscopy image of eNano-JQ1. Scale bar: 100 nm. This was repeated at least three times independently with similar results. D The stability was evaluated by measuring changes in size and polydispersity index (PDI) at different time (left) and dilution ratio (right). n = 3 independent experiments. Data were represented as means ± SEMs. E Release profile of JQ1 from eNano-JQ1. F Representative fundus imaging and OCT image of mice. The pathological changes were indicated with black arrows. The retinal folds or tractional area was circled with yellow dotted lines, and PVR membrane was marked with an asterisk (*). G Representative H&E staining of eye sections from mice with indicated treatment (left). The black dotted lines indicated pathological changes, and PVR membrane was marked with an asterisk (*). Scale bar: 250 μm. Quantification of PVR severity (right). n = 7, 8, 7 samples, respectively. P = 0.0002, 0.0009 were determined by two-tailed Mann–Whitney test. ***P < 0.001. H Western blot analysis (up) and quantification (down) of αSMA in eyecup tissues from normal and PVR mice treated with vehicle or eNano-JQ1. n = 3 samples. Data are represented as means ± SEMs. P values = 0.005, 0.0356 were determined by one-way ANOVA multiple comparisons test. *P < 0.05, **P < 0.01. I Representative immunofluorescent staining of αSMA in mouse eye sections (left). The yellow dotted lines indicated the whole eye. The white dotted lines indicated PVR membrane. Scale bars: yellow 100 μm, white 20 μm. Quantification of mean fluorescence integrated density of αSMA (right). n = 5, 6, 6 samples, respectively. Data are represented as means ± SEMs. P values < 0.0001 were determined by one-way ANOVA multiple comparisons test. ****P < 0.0001. Source data are provided as a Source Data file.
Subsequently, we evaluated the safety of ophthalmic administration of eNano-JQ1. Optical coherence tomography (OCT) imaging revealed that both the retinal structure and thickness remained normal after intraocular administration of vehicle and eNano-JQ1 (Supplementary Fig. 6A, B). Electroretinogram (ERG) analysis, used to assess visual function, indicated no significant decrease in a-wave (attributed to photoreceptor rods/cones) and b-wave (reflecting function of inner retina, mainly Müller and bipolar cells) (Supplementary Fig. 6C). Histological evaluations confirmed the retina’s integrity (Supplementary Fig. 6D) and the TUNEL assay further underscored the absence of notable cell death (Supplementary Fig. 6E). Collectively, a seven-day slow-release intraocular administration of vehicle and eNano-JQ1 demonstrated a high safety profile.
We next evaluate the therapeutic potential of eNano-JQ1 in PVR mouse model. As shown in Fig. 3F, OCT imaging confirmed improved retinal thickness in the eNano-JQ1 group, with an absence of proliferative membranes in elevated retinal region. Further histological analysis revealed reduced vitreous blood accumulation, clearer retinal structure, and fewer fibrotic-like lesions in the eNano-JQ1 group compared to vehicle control. Utilizing established grading criteria, we found that eyes receiving eNano-JQ1 treatment displayed a milder PVR phenotype compared to those treated with vehicle (Fig. 3G). The reduced αSMA expression in mouse eyecup tissues (Fig. 3H) and its decreased localization in frozen mouse eye sections (Fig. 3I) highlight the pronounced anti-fibrotic efficacy of eNano-JQ1. The PVR membranes were notably thinner and less widespread, with a more preserved retinal structure. Overall, our results underscore the potent inhibitory effect of eNano-JQ1 on PVR progression in mice.
Identification of the transcriptional regulatory network in PVR
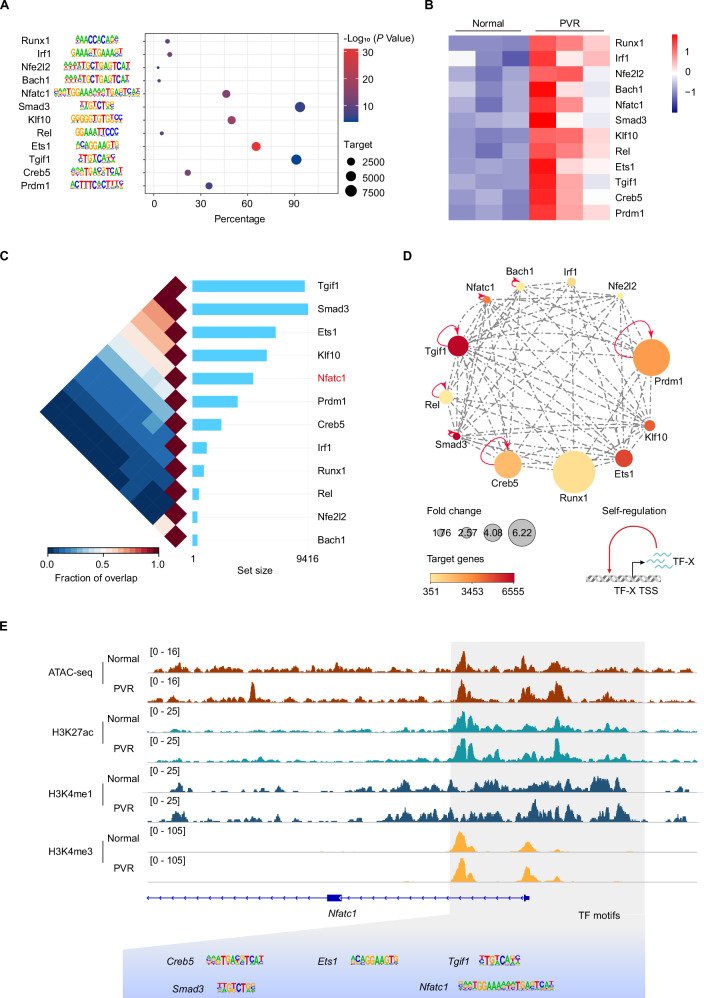
Chromatin states are largely dictated by transcription factor-mediated chromatin remodeling13. To identify key TFs involved in reshaping the epigenetic landscapes in PVR, we assessed the enrichment of TF motifs within PVR-open regions. By integrating transcriptomic data, we pinpointed TFs that were upregulated during PVR progression. We identified 12 TFs whose binding motifs were enriched and their expression was elevated in PVR RPE cells (Fig. 4A, B). TFs often interact with each other to form regulatory networks and thus enhance transcriptional activity43. We next mapped TRNs based on their interconnectivity through binding motif and expression level (Fig. 4C). Notably, Nfatc1, Tgif1, Rel, Creb5, Prdm1, Bach1 and Smad3 are TFs were discovered as self-regulated TFs (Fig. 4D), which may work synergistically as amplifiers to reinforce PVR-associated gene expression program. As shown in Fig. 4E, the accessible region of Nfatc1 contains motifs of these TRN TFs. The TGF-β signaling pathway is known to be instrumental in PVR progression, and Smad3 inhibition can attenuate the disease’s course44,45. Previous studies have highlighted RUNX1’s role in regulating cell proliferation and migration, with its inhibition mitigating PVR25. Therefore, the discovery of these previously reported PVR-related TFs validates the feasibility of our TRN analysis in identifying potential TFs involved in PVR. Among the TFs pinpointed within the TRN TFs, NFATc1 is notably recognized for its central role in modulating immune responses and directing osteoclast differentiation46,47. Given its undefined role in PVR, we aim to investigate the specific function of NFATc1 in RPE cells during PVR progression.
Fig. 4. Unraveling the transcriptional regulatory networks underlying PVR.
A Bubble chart showing the enrichment of transcription factor (TF) binding motifs in PVR-open region. P values were calculated by HOMER based on binomial distribution. B Heatmap depicting the FPKM values of TFs. C The triangular heatmap displays the co-localization of TFs. Fraction of overlap refers to the proportion of shared elements between two TF, where 0 indicates no overlap and 1 means full overlap. The set size shows the number of genomic regions in each TF. D The TF regulatory network for PVR. Node color represents number of TF target genes and node size represents changes in TF expression. E IGV track profiles of ATAC-seq and ChIP-seq for H3K27ac, H3K4me1 and H3K4me3 on a representative TF gene. Source data are provided as a Source Data file.
Suppression of NFATc1 in RPE cells mitigates PVR progression
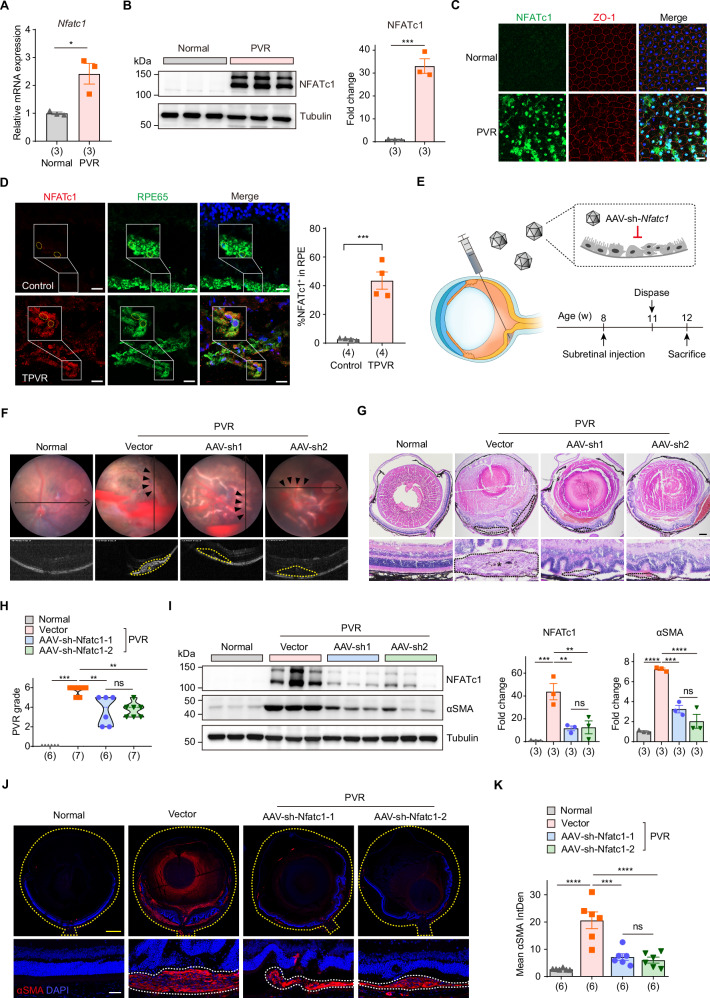
We first verified the expression of NFATc1 after PVR. Both mRNA and protein levels of NFATc1 substantially increased in PVR (Fig. 5A, B). Subsequent immunofluorescence assays on RPE cells, co-stained with NFATc1 and the tight junction protein ZO-1, revealed that while ZO-1 typically demarcates the regular hexagonal structure of healthy RPE cells, its integrity is compromised post-PVR. Notably, the fluorescence intensity of NFATc1 in PVR mice is markedly elevated, with pronounced nuclear translocation (Fig. 5C and Supplementary Fig. 7A). Importantly, we also observed an elevated expression of NFATc1 in the RPE cells of TPVR patients compared to donors (Fig. 5D). The clinical characteristics of these patients were detailed in Supplementary Fig. 7B. In response to external signals, NFATc1 can translocate from the cytoplasm to the nucleus, and subsequently activate downstream target genes46. We thus assessed the expression of NFATc1’s target genes, including Mmp9 and Ctsk. As showed in Supplementary Fig. 7C, the mRNA levels of these genes were all increased post-PVR. Analysis of publicly available RNA-seq data (GSE179603)26 revealed that NFATC1, MMP9, and CTSK in human PVR membrane specimens were all substantially upregulated at the RNA level (Supplementary Fig. 7D).
Fig. 5. Suppressing NFATc1 in RPE slows PVR progression.
A RT-qPCR analysis of Nfatc1 expression in primary RPE cells. n = 3 biologically independent experiments. Data are represented as means ± SEMs. P = 0.0188 was determined by two-tailed unpaired T test. *P < 0.05. B Western blot analysis (left) and quantification (right) of NFATc1 in eyecup tissues from normal and PVR mice. n = 3 samples. Data are represented as means ± SEMs. P = 0.0005 was determined by two-tailed unpaired T test. ***P < 0.001. C Immunofluorescence staining of NFATc1 in RPE flat from normal and PVR mice. Scale bar: 20 μm. This was repeated at least three times independently with similar results. D Immunofluorescence staining of NFATc1 and RPE65 in human PVR membrane and donor eye sections. Scale bars: 20 μm. n = 4 samples. Data are represented as means ± SEMs. P = 0.0005 was determined by two-tailed unpaired T test. ***P < 0.001. E Schematic view of experimental strategy in the PVR mouse model. F Representative fundus imaging and OCT image of normal and PVR mice treated with vector, rAAV2/2-sh-Nfatc1-1 (AAV-sh1) or rAAV2/2-sh-Nfatc1-2 (AAV-sh2). The pathological changes were indicated with black arrows. The retinal folds or tractional area was circled with yellow dotted lines, and PVR membrane was marked with an asterisk (*). G Representative H&E staining of eye sections from mice with indicated treatment. The black dotted lines indicated pathological changes, and PVR membrane was marked with an asterisk (*). Scale bar: 250 μm. H Quantification of PVR severity. n = 6, 7, 6, 7 samples, respectively. P = 0.0006, 0.0087, 0.0017 were determined by two-tailed Mann–Whitney test. **P < 0.01, ***P < 0.001. I Western blot analysis (left) and quantification (right) of NFATc1, αSMA in eyecup tissues from normal and PVR mice treated with vectors, AAV-sh1 or AAV-sh2. n = 3 samples. Data are represented as means ± SEMs. For NFATc1, P = 0.0009, 0.0055, 0.0066 were determined by one-way ANOVA multiple comparisons test. For αSMA, P < 0.0001, P = 0.0005, P < 0.0001 were determined by one-way ANOVA multiple comparisons test. **P < 0.01, ***P < 0.001, ****P < 0.0001. J Representative immunofluorescent staining of αSMA in mouse eye sections. The yellow dotted lines indicated the whole eye. The white dotted lines indicated PVR membrane. Scale bars: yellow 100 μm, white 20 μm. K Quantification of mean fluorescence integrated density of αSMA. n = 6 samples. Data are represented as means ± SEMs. P < 0.0001, P = 0.0001, P < 0.0001 were determined by one-way ANOVA multiple comparisons test. ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
The elevated expression and transcriptional activity of NFATc1 in PVR promoted us to postulate whether its suppression could slow PVR progression. Subretinal injection of recombinant adeno-associated virus type 2/2 (rAAV2/2) has been well characterized and widely used for delivering therapeutic genes specifically to RPE cells48,49. Thus, we engineered two rAAV2/2 vectors bearing shRNA sequences against Nfatc1 (rAAV2/2-sh-Nfatc1) and assessed their impacts on the progression of PVR (Fig. 5E). PVR mice treated with rAAV2/2-sh-Nfatc1 exhibited a hazy fundus, small patches of hemorrhage, and fewer retinal folds, yet without noticeable retinal detachment or extensive PVR membranes being detected through OCT scans (Fig. 5F). Histological examination of eye sections showed that silencing Nfatc1 inhibited the proliferation of the subretinal membrane, and mitigated tractional damage to the retina, leading to decreased PVR severity (Fig. 5G, H). Immunoblotting analysis showed a marked reduction of NFATc1 total protein levels, validating NFATc1 knocking down efficiency. Concurrently, a decline in αSMA protein expression indicates diminished fibrosis within ocular tissues (Fig. 5I). Immunostaining of αSMA protein in mouse eye sections revealed that post-administration of rAAV2/2-sh-Nfatc1, the thickness and extent of the αSMA-positive region beneath the retina in PVR mice were reduced, correlating with fewer retinal folds and a confined area of retinal detachment (Fig. 5J, K). Collectively, these findings suggest that inhibiting NFATc1 specifically in RPE cells mitigates PVR progression.
Blocking the RANK-NFATc1 axis alleviates PVR progression
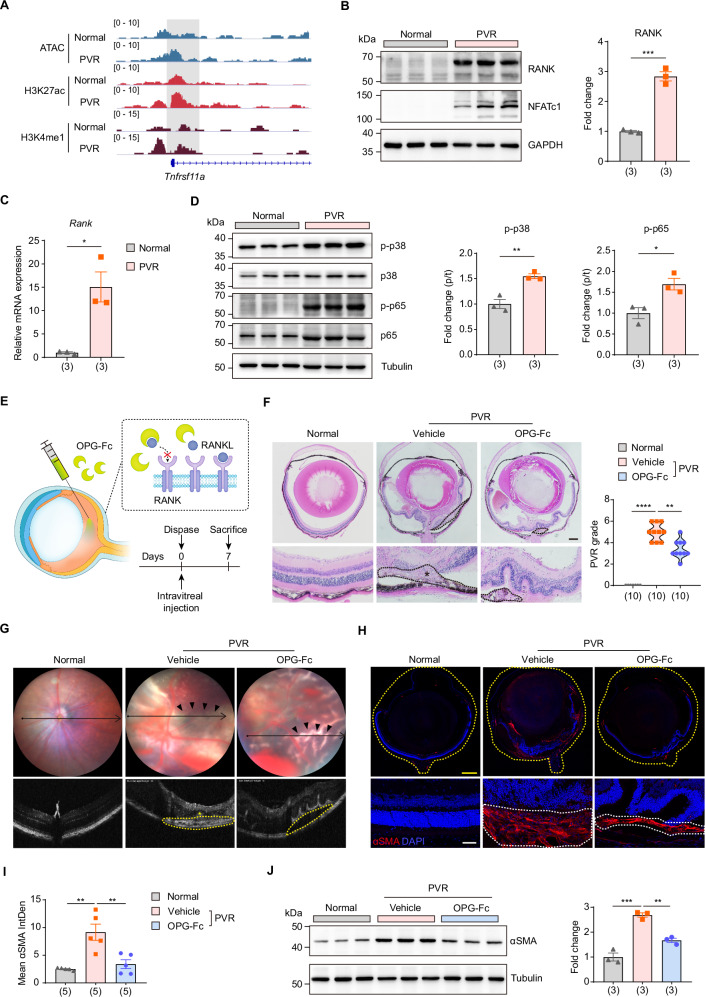
RANK (Receptor Activator of Nuclear Factor κB) is a cell surface receptor that, upon binding with its ligand RANKL, initiates a cascade of intracellular signaling events, ultimately leading to the activation and subsequent nuclear translocation of NFATc146,50. There was an increase in chromatin accessibility in the upstream region of the Tnfrsf11a (Rank) promoter, accompanied by elevated H3K27ac and H3K4me1 marks in the PVR group compared to the normal controls (Fig. 6A). Furthermore, the mRNA and protein levels of RANK were both increased post-PVR (Fig. 6B, C). RANK is believed to potentiate NFATc1 activation through the modulation of intermediary pathways51. In line with this, phosphorylation of both p38 MAPK and p65 NF-κB were elevated in PVR mouse (Fig. 6D). These observations collectively suggest that the RANK pathway is activated during the progression of PVR.
Fig. 6. Suppression of the RANK-NFATc1 axis delays PVR progression.
A IGV track profiles of ATAC-seq, ChIP-seq of H3K27ac and H3K4me1 on Tnfrsf11a (Rank) gene. B Western blot analysis (left) and quantification (right) of RANK in eyecup tissues from normal and PVR mice. n = 3 samples. Data are represented as means ± SEMs. P = 0.0003 was determined by two-tailed unpaired T test. ***P < 0.001. C RT-qPCR analysis of Rank expression in primary RPE cells from normal and PVR mice. n = 3 biologically independent experiments. Data are represented as means ± SEMs. P = 0.0118 was determined by two-tailed unpaired T test. *P < 0.05. D Western blot analysis (left) and quantification (right) of phosphorylated p38 and p65 in eyecup tissues from normal and PVR mice. n = 3 samples. Data are represented as means ± SEMs. P = 0.0059 for p-p38, P = 0.0221 for p-p65 were determined by two-tailed unpaired T test. *P < 0.05, **P < 0.01. E Schematic view of experimental strategy in the PVR mouse model. F Representative H&E staining of eye sections from mice with indicated treatment (left). The black dotted lines indicated pathological changes, and PVR membrane was marked with an asterisk (*). Scale bar: 250 μm. Quantification of PVR severity (right). n = 10 samples. P < 0.0001, P = 0.0016 were determined by two-tailed Mann–Whitney test. **P < 0.01, ****P < 0.0001. G Representative fundus imaging and OCT image of mice. The pathological changes were indicated with black arrows. The retinal folds or tractional area was circled with yellow dotted lines, and PVR membrane was marked with an asterisk (*). (H) Representative immunofluorescent staining of αSMA in mouse eye sections. The yellow dotted lines indicated the whole eye. The white dotted lines indicated PVR membrane. Scale bars: yellow 100 μm, white 20 μm. I Quantification of mean fluorescence integrated density of αSMA. n = 5 samples. Data are represented as means ± SEMs. P = 0.001, 0.003 were determined by one-way ANOVA multiple comparisons test. **P < 0.01. J Western blot analysis (left) and quantification (right) of αSMA in eyecup tissues from normal and PVR mice treated with vehicle or OPG-Fc. n = 3 samples. Data are represented as means ± SEMs. P = 0.0001, 0.002 were determined by one-way ANOVA multiple comparisons test. **P < 0.01, ***P < 0.001. Source data are provided as a Source Data file.
Osteoprotegerin (OPG) acts as a soluble decoy receptor that binds to RANKL, preventing its interaction with RANK and thereby inhibiting the RANK pathway52–54. Thus, we employed OPG-Fc to inhibit the RANK pathway and examined its influence on PVR progression (Fig. 6E). We first assessed the safety of intraocular administration of OPG-Fc. Fundus imaging and OCT image showed consistent retinal structure and thickness between the normal control and OPG-treated mice (Supplementary Fig. 8A, B). ERG analysis further demonstrated similar a-wave and b-wave amplitudes in both groups (Supplementary Fig. 8C). Histological evaluations confirmed the retina’s integrity (Supplementary Fig. 8D). Additionally, the TUNEL assay revealed no elevated cell death in either the normal control or OPG-treated groups (Supplementary Fig. 8E). These findings suggest that intraocular administration of OPG is well-tolerated and safe.
Subsequent OCT scans and histological evaluations demonstrated that OPG-Fc treatment led to diminished anterior retinal hemorrhages, fewer retinal fold formations, and reduced retinal detachment, resulting in a lower severity score for PVR (Fig. 6F, G). Furthermore, both immunoblotting and immunofluorescence assays highlighted a decline in αSMA levels, indicative of attenuated intraocular fibrosis post-OPG treatment (Fig. 6H–J). Collectively, these findings underscore the protective role of inhibiting RANK, an upstream regulator of NFATc1, against PVR progression.
Discussion
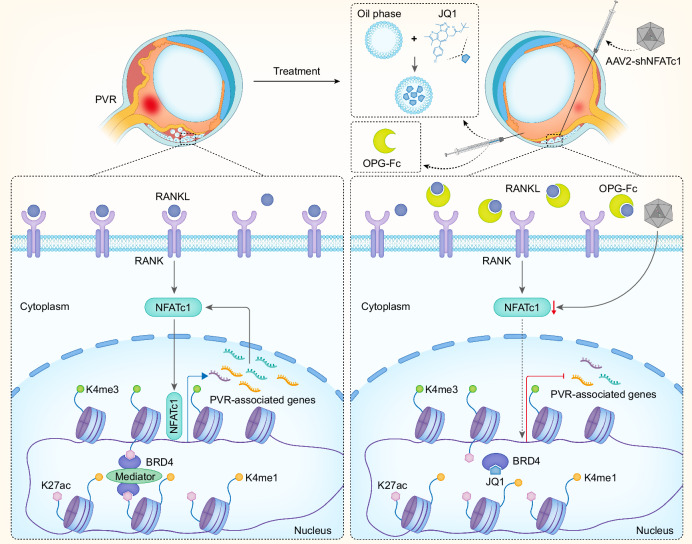
Previous studies have identified RPE cells as the primary cells implicated in the progression of PVR7. In a healthy state, RPE cells are differentiated and polarized, crucial for visual function and forming the outer retinal barrier. However, post-ocular injury, these cells lose polarity, migrate, proliferate, and undergo EMT, contributing to extracellular matrix synthesis and the formation of characteristic PVR membranes55. Epigenetic modifications, which include DNA methylation, histone modifications, and chromatin remodeling, are increasingly recognized as pivotal regulators of cell fate decisions11. Previous study indicates that inhibition of DNA methylation and MeCP2, an DNA methylation reader protein, suppresses the EMT of RPE cells and their transdifferentiation into myofibroblasts56. Boles et al. shows that co-treatment with TGF-β and TNF-α accelerates EMT in RPE cultures derived from adult human RPE stem cells and identified potential active enhancers linked to actively transcribed genes in these cells57. Recent studies suggest that histone deacetylases expression is increased in RPE cells exposed to TGF-β, and HDAC inhibitors can attenuate TGF-β2-induced EMT55,58,59. Yet, many of these insights stem from in vitro cell culture models, which do not fully capture the unique in vivo environment where RPE cells reside during PVR progression. As PVR advances, the surrounding microenvironment undergoes marked alterations, with factors like inflammation, hypoxia, matrix modifications, various growth factors and cytokines profoundly impacting RPE cell behavior5,10. Leveraging our optimized approaches for assessing chromatin accessibility and multiple histone modifications in low inputs of RPE cells, we conducted a comprehensive epigenomic analysis on primary RPE cells directly isolated from a PVR mouse model. This multi-omics strategy allowed us to map the evolving chromatin landscape that characterizes RPE cell fate transitions during PVR progression. Our data suggest that genomic regions gaining accessibility post-PVR predominantly associate with pathways governing cell migration, proliferation, and secretion. In contrast, regions losing accessibility are enriched in genes essential for transport processes, suggesting a potential loss of inherent RPE cellular functions as PVR advances. Our findings further reveal that elevated enhancer activity aligns with increased chromatin accessibility in RPE cells as PVR progresses, underscoring the pronounced activation of enhancers in these cells. Collectively, these findings highlight genome-wide shifts in the cis-regulatory domains of RPE cells during PVR, indicating that dynamic epigenetic remodeling is instrumental in guiding RPE cell fate transitions (Fig. 7).
Fig. 7. Working model.
A schematic illustration presents the epigenetic mechanisms activating RANK-NFATc1 signaling during the progression of PVR and outlines the strategies for targeted intervention.
Our transcriptomic analysis revealed that, following PVR, RPE cells exhibited activation of numerous pathways associated with disease progression. Notably, these cells showed enrichment in pathways governing extracellular matrix organization, cytokine production, and inflammatory responses, underscoring increased fibrotic and inflammatory activities in PVR. In conjunction with these transcriptomic data, we identified a pronounced correlation between active chromatin states and the upregulated expression of PVR-associated genes. By employing the BRD4 inhibitor, JQ1, we targeted and suppressed active transcriptional enhancers. This approach, unlike traditional strategies targeting singular genes or pathways, offers a broader genomic modulation of PVR-associated genes. To enhance the therapeutic efficacy, we encapsulated JQ1 in a nanoemulsion and administered intraocular injection. This localized drug delivery not only minimized side effects but also substantially improved therapeutic outcomes. While surgery is the conventional treatment for patients with ocular trauma5,60, the incorporation of intraocular drug intervention during surgical procedures offers a promising strategy to prevent PVR onset. This aligns with current clinical protocols and paves the way for potential clinical applications.
Through TRN analysis, we identified key TFs involved in the dynamic changes of chromatin states during PVR progression, including previously reported TFs such as Runx1 and Smad325,44,45. Drawing from our PVR mouse model and further validated with clinical patient samples, we confirmed the aberrant activation of the RANK-NFATc1 pathway post-PVR. Targeted knockdown of NFATc1 in RPE cells or the use of OPG-Fc, a specific antibody against RANKL, effectively slowed disease progression. Denosumab, a monoclonal antibody that mimics the function of OPG in human, binds to and suppress RANKL, inhibiting bone resorption61,62. Denosumab has been approved by the FDA for the treatment of osteoporosis, prevention of skeletal-related events in patients with bone metastases from solid tumors, and for patients with giant cell tumor of bone54. Given its mechanism of action and established safety profile, our research indicates a promising avenue for repurposing Denosumab in the clinical management of PVR.
Methods
Study approval
Our study conformed to the Declaration of Helsinki. The Ethical Committee of Tianjin Medical University General Hospital approved both the protocol pertaining to patient sample usage (IRB2023-KY-307), and the conduct of all mouse experiments (IRB2023-DWFL-369) in this study. The usage of control human samples followed the approval of Ethics Committee of First Affiliated Hospital of Harbin Medical University (2023JS35). Sex and/or gender are not relevant for any findings in this study and were therefore not considered in our study design, and participants was not determined based on self-report nor assigned. Informed consent was obtained from all participants. Participants did not receive compensation. The guidelines of the ARVO (The Association for Research in Vision and Ophthalmology) Statement for the Use of Animals in Ophthalmic and Vision Research were followed.
Animal models
Eight-week-old C57BL/6J male mice were purchased from Charles River (Beijing, China). Sex and gender are not relevant for any findings in this study and were therefore not incorporated in the study design. Mice were maintained under a 12-hour light-dark cycle at a temperature of 21–25 °C with humidity level between 30 to 70%, and were provided unrestricted access to food and water. The mice were then randomly designated into experimental groups. For the experimental PVR model, mice were anesthetized with Isoflurane inhalations and then received intravitreal injection of 0.02 U/μL Dispase® (10269638001, Roche). PVR model was performed on one eye of each animal, designating the untreated contralateral eye as the normal control. This approach ensured that each animal served as its own control, thereby reducing variability. After seven days, fundus photography and optical coherence tomography (OCT) were carried out using MicroIV (Phoenix Research Labs, USA). The eyes were later enucleated for subsequent analysis. The severity of PVR observed was rated in accordance to the grading criteria previously reported63.
Isolation of RPE cells
Primary RPE cells were procured following the established protocol64,65. A brief review of the steps includes the euthanasia of mice through CO2 asphyxiation, followed by immediate enucleation of eyes. Posterior eyecups were collected in HBSS-H– buffer (10 mM HEPES and HBSS– (without Ca/Mg) buffer) on ice within 45 min, and later incubated in hyaluronidase solution (H3506, Sigma) at 37 °C for 30 min. Each eyecup was then transferred to another tube with 1.5 mL of ice-cold HBSS-H+ buffer (10 mM HEPES and HBSS+ (with Ca/Mg) buffer) for 30 min on ice to stop the hyaluronidase activity. Subsequent steps included removal of the optic nerves and the detachment of the RPE-choroid complex from the neural retina, with the detached complex being incubated in trypsin (25200114, Thermo Fisher Scientific) at 37 °C for 30 min. The digestion process was terminated by adding a solution containing fetal bovine serum (FBS). Place the eyecup face down and shake it. The detached RPE sheets were collected in 20% FBS in HBSS-H+ buffer, followed by centrifugation at 340 g for 2 min at room temperature. The RPE pellet was re-suspended in 1 mL of 0.05% trypsin and 0.02% EDTA solution, incubated for one min at 37 °C, and then the process was terminated with 9 mL of solution containing FBS. Primary RPE cells purity was observed using a light microscope and immunofluorescence staining for the RPE cell marker RPE65 (Abcam, Cat. #ab231782, Lot #1001368-30, 1:100 dilution). The freshly prepared primary RPE cells was used immediately for subsequent experiments. For sequencing analyses such as ATAC-seq and RNA-seq, cells from three animals were pooled for each replicate. For the ChIP-seq experiments of histone modifications (H3K27ac, H3K4me1, H3K4me3, H3K27me3, H3K9me3 and H3K36me3), cells from nine animals were pooled for each replicate. Each sequencing experiment was conducted in triplicate.
ATAC-seq and data analysis
A total of 20,000 RPE cells were lysed in cold lysis buffer (10 mM NaCl, 10 mM Tris-HCl pH 7.4, 3 mM MgCl2, 0.1% (v/v) IGPAL CA-630) for 20 min on ice. The nuclei were collected by centrifuged at 500 g for 10 min and re-suspend with 50 μL reaction buffer (10 μL TruePrep Tagment Buffer L, 3 μL TruePrep Tagment Enzyme, and 35 μL ddH2O from Vazyme, TD501) at 37 °C for 30 min. After tagmentation, DNA was purified with QIAquick PCR Purification Kit (28106, QIAGEN). And the DNA fragmentation was subjected to library preparation according to the manufacturer’s instructions (TD501, Vazyme). The DNA libraries were sequenced on Illumina platforms. The reads were aligned to the M. musculus genome (mm9) by bowtie2 (v2.3.5.1). The differential ATAC-seq peaks between the normal control and PVR were identified using HOMER, and annotatePeaks.pl from HOMER was utilized to associate the peaks with genomic regions and nearby genes. Using deepTools (v3.4.3), the ATAC signals were visualized as a heatmap.
Low-input ChIP-seq and data analysis
Low-input ChIP-seq was conducted following previous protocols, with some modifications66. 100,000 cells were lysed in 19 μL lysis buffer (0.5% Tween-20, 0.1% SDS, 0.5% NP-40, and Protease Inhibitor Cocktail) for 10 min on ice. Then, 19 μL MNase buffer (2 mM CaCl2, 100 mM Tris-HCl pH 8.0) containing 0.02 U MNase (N3755, Sigma) was added and incubated at 37 °C for 5 min. The reaction was terminated by adding 5 μL stop buffer (55 mM EDTA, 110 mM Tris-HCl pH 8.0) and incubated with 45 μL 2× RIPA buffer (280 mM NaCl, 0.1% SDS, 1% Triton X-100, 5 mM EGTA, 0.2% sodium deoxycholate, and Protease Inhibitor Cocktail) for 10 min on ice. Then the sample was centrifuged at 19,000 g for 15 min, the supernatant was collected and added with 40 μL RIPA buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 140 mM NaCl, 0.1% SDS, 1% Triton X-100, and 0.1% sodium deoxycholate). Immunoprecipitation was performed using 1 μg antibody of H3K27ac (Abcam, Cat. #ab4729, Lot #GR3448944-1), H3K4me1 (Abcam, Cat. #8895, Lot #GR3426435-2), H3K4me3 (Millipore, Cat. #05-745 R, Lot #4070079), H3K36me3 (Abcam, Cat. #9050, Lot #GR3382010-2), H3K9me3 (Abcam, Cat. #8898, Lot #GR3444658-1) or H3K27me3 (Abcam, Cat. #192985, Lot #GR3264827-11) with Dynabeads (10004D, Thermo Fisher Scientific). Then beads were collected and washed with 150 μL RIPA buffer five times, followed by 150 μL LiCl buffer (250 mM LiCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.5% sodium deoxycholate, and 0.5% NP-40). The beads were re-suspended in buffer containing 29 μL of ddH2O and 20 μg of proteinase K, and then subjected to shaking at 55 °C for 90 min. The supernatant was collected and incubated at 72 °C for 40 min. The DNA was purified and followed by library preparation according to the manufacturer’s instructions (ND607, Vazyme). The DNA libraries were then sequenced on Illumina platforms. The reads were aligned to the M. musculus genome (mm9) by bowtie2 (v2.3.5.1) and SAMtools (v1.9) was used to remove duplicated reads. HOMER was used for peak calling, annotation, and motif enrichment analysis. The deepTools (v3.4.3) was used to produce the BigWig files.
RNA-seq and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were extracted from primary RPE cells using TRIzol (15596018; Invitrogen), and the mRNA was used for RNA-seq library construction. The libraries were then sequenced on Illumina platforms. HISAT2 (v2.1.0) was used to align reads to mouse reference genome, and reads were counted using featureCounts (v1.6.0). DESeq2 algorithms were used to calculate genes with differential expression. Genes with fold changes of ≤ −2 or ≥2, adjusted P < 0.05 were considered as significantly differentially expressed. For RT-qPCR, cDNA Synthesis Kit (05081955001, Roche) was used to reverse transcribe RNA into cDNA. SYBR-based RT-qPCR was performed to detect the mRNA expression of Nfatc1, Mmp9, Ctsk, and Rank. The primer sequences are listed in Supplementary Table 1.
Nanoemulsion (eNano-JQ1) preparation and characterization
Nanoemulsion loaded with JQ1 (eNano-JQ1) was synthesized using a simple sonication method. Briefly, 15% tricaprylin, 58% PEG-40 hydrogenated castor oil which serves as a surfactant, 26% ethanol which acts as a co-surfactant were accurately weighed and mixed. When the mixture was sonicated until it fully dissolved, 1% of the JQ1 raw material was added and completely stirred to obtain the eNano-JQ1 (10 mg/mL). Dynamic Light Scattering (DLS) was used to measure the hydrodynamic diameter and polydispersity index (PDI) of the eNano-JQ1. The Zeta potential was measured by ZetaPALS (Brookhaven Instruments, USA) at room temperature. Transmission electron microscopy (TEM) was used to assess the morphologies of the eNano-JQ1. Briefly, the nanoemulsion was deposited on carbon-copper coated grids for 2 min, and then stained with phosphotungstic acid solution, followed by visualization using a high-resolution TEM (Hitachi HT7700, Japan).
Drug treatment
The protocol for systemic administration of the bromodomain-containing protein 4 (BRD4) inhibitor entailed daily intraperitoneal injections of JQ1 (HY13030, MedChemExpress) at a dosage of 50 mg per kilogram of body weight for a continuous period of seven days, initiated immediately post-Dispase injection. For sustained intraocular administration, 1 mg/mL of eNano-JQ1 in 1 µL volume was administered intravitreally seven days prior and immediately post-Dispase injection. For the gene therapy procedure, rAAV2/2 vectors, rAAV2/2-shNfatc1-Mus-1 (GCCGCAGAACACTACAGTT, AACTGTAGTGTTCTGCGGC) and rAAV2/2-shNfatc1-Mus-2 (GCTCAGAAACTCTGATAT, AATATCAGAGTTTCTGAGC) were obtained from PackGene Biotech (Guangzhou, China) and were delivered via a subretinal injection containing 4.9 × 109 viral genome copies in 1 μL volume. Three weeks post-rAAV2/2 virus injection, mice received Dispase for model. For competitive blocking of RANK, Osteoprotegerin (OPG)-Fc (459-MO, R&D Systems) was injected intravitreally in 1 µL volume immediately post-Dispase injection. Three eyes in each group were used for live fundus photography and OCT analysis, then were enucleated for protein extraction. In eNano-JQ1 part, eight eyes were used for H&E stain, six eyes were used for immunofluorescence assay. In NFATc1 gene therapy part, seven eyes were used for H&E stain, six eyes were used for immunofluorescence assay. In OPG-Fc part, ten eyes were used for H&E stain, five eyes were used for immunofluorescence assay.
Western blot analysis
The eyes of mice were enucleated, followed by the removal of the cornea, lens, and optic nerves. The neuroretina-RPE-choroid-sclera complex (eyecup) tissues were lysed in RIPA buffer. Subsequently, it was centrifuged at 13,000 g for 20 min at 4 °C. The total protein concentration in the samples was then quantified using the Nanodrop (Thermo Fisher, USA). For comparative analysis, equal amounts of protein were isolated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes. These PVDF membranes were then left to incubate with primary antibodies overnight at 4 °C, which included α-SMA (Sigma, Cat. #A2547, Lot #235414), NFATc1 (Abcam, Cat. #ab25916, Lot #GR3424521-2), RANK (Abcam, Cat. #ab200369, Lot #GR224490-39), p38 (CST, Cat. #8690, Lot #9), p-p38 (CST, Cat. #4511, Lot #13), p65 (CST, Cat. #8242, Lot #16) and p-p65 (CST, Cat. #3033, Lot #19), 1:1000 diluted; Tubulin (Utibody, Cat. #UM4003, Lot #A0220) and GAPDH (Utibody, Cat. #UM4002, Lot #A0819), 1:2000 diluted. The western blots were finally quantified using Fiji software (ImageJ v1.54f).
Histology and immunofluorescence
Mouse eyes used for histological analysis underwent fixation in 4% PFA overnight at 4 °C, followed by paraffin embedding. Subsequently, an eight-micrometer section was utilized for H&E staining. For immunofluorescence analysis, mouse eyes were processed to fifteen-micrometer thick frozen sections. Briefly, the dried slices were permeabilized with 0.3% Triton X-100, blocked with 2% BSA in 0.3% Triton X-100, and then incubated with α-SMA (Sigma, Cat. #C6198, Lot #0000214427, 1:200 dilution). For the preparation of RPE flats, the eyes were fixed in 4% PFA for 2 h, followed by removal of the cornea and lens, and peeling of the neural retina from RPE-choroid complex. These RPE flats were permeabilized with 1% Triton X-100, blocked with 5% BSA in 0.5% Triton X-100 at 4 °C, and incubated with NFATc1 (Abcam, Cat. #ab25916. Lot #GR3424521-2, 1:50 dilution) and ZO-1 (ProteinTech, Cat. #21773-1-AP, Lot #00129646, 1:100 dilution), followed by secondary antibodies (Jacksonimmuno, 1:200 dilution). Human PVR membranes and donor eyecups were formalin-fixed for 30 min, dehydrated with 30% sucrose overnight at 4 °C. After snap frozen, sections of 10 μm thickness were obtained, and then immunofluorescence was performed using antibodies against NFATc1 (Santa Cruz Biotechnology, Cat. #sc-7294, Lot #H3022, 1:50 dilution) and RPE65 (Abcam, Cat. #ab231782, Lot #1001368-30, 1:100 dilution). The analysis for all samples above was performed under a confocal microscope (LSM-800, Carl Zeiss, Germany).
Intraocular toxicity evaluation
Post 7 days of intraocular administration, the thickness of retinal layers were evaluated through fundus and OCT images, utilizing the InSight software. Three eyes in each group of normal control, vehicle, and drug were used for live fundus photography and OCT analysis. After that, eyes were enucleated for paraffin embedding. Electroretinogram (ERG) measurement was conducted on the Celeris D430 rodent ERG testing system (Diagnosys LLC, MA, USA), recording both dark adapted a-wave and b-wave. Three eyes in each group were used for ERG under dark adaptation, and then were enucleated for TUNEL assay. For histological analysis, eyes were enucleated to prepare paraffin sections with H&E staining. For TUNEL assay on the eye frozen sections; it involved washing the sections in PBS for 10 min and incubating them in a solution of 2% BSA and 0.3% Triton X-100 for 5 min at room temperature. For a positive control, incubated the normal control section with DNase I (20 U/mL) for 10 min at room temperature. The assay was performed with TUNEL Apoptosis Assay Kit (T6014, UElandy, China) in accordance with the manufacturer’s instructions. Finally, the nuclei were stained with DAPI (blue). Sections from each mouse were imaged using a confocal microscope.
Statistics
The data reported from all studies represent the mean ± SEM across a minimum of three independent experiments. Statistical analyses were conducted using the T-test, One-way ANOVA, and Mann–Whitney analysis, with the level of significance set at P < 0.05. All statistical analyses were performed using the GraphPad Prism software. Moreover, we conducted a Power analysis to ensure the statistical robustness of our animal experiments67.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by grants from the National Key R&D Program (2021YFC2401404 to H.Y.), the National Natural Science Foundation of China (32241018 and 82070689 to Y.C., 82330031 to H.Y., 82401296 to M.L., and 82271896 to X.Z.), and Postdoctoral Fellowship Program of CPSF (GZC20231924 to M.L.).
Author contributions
M.L. and Y.L. performed animal and biochemistry studies and wrote the manuscript. X.Z. performed bioinformatic analysis. X.Y. contributed to animal studies. Y.H. prepared and characterized eNano-JQ1, Y.Z., Z.Y., X.G. and M.L. performed biochemistry studies, H.Z. and X.J. provided control human eye sections, X.W. provided expertise on animal studies and edited the manuscript, Z.Z prepared and characterized eNano-JQ1, and edited the manuscript, Y.C. and H.Y. conceived and supervised the project, analyzed data, and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Li Jia Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The ATAC-seq, ChIP-seq and RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE244812. The processed ATAC-seq, ChIP-seq and RNA-seq data are available at GEO database (GSE244812). The relevant raw data from each figure generated in this study are provided in the Supplementary Information/Source Data file. The cited data used in this study are available in the GEO database under accession code GSE179603. Source data are provided with this paper.
Competing interests
The authors declare the following competing interests: A patent application (P.R.C application number: 202410504090.1) was filed by Tianjin Medical University General Hospital. H.Y., Y.C., M.L., Y.L., Y.Z., and Z.Y. from Tianjin Medical University, Z.Z. and Y.H. from Xuzhou Medical University are inventors. The patent application covered the preparation methods for eNano-JQ1 and their intraocular application. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mengyu Liao, Xu Zhu, Yumei Lu.
Contributor Information
Ziming Zhao, Email: zmzhao@xzhmu.edu.cn.
Yupeng Chen, Email: ychen@tmu.edu.cn.
Hua Yan, Email: zyyyanhua@tmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51624-y.
References
- 1.Knyazer, B. et al. Prognostic factors in posterior open globe injuries (zone-III injuries). Clin. Exp. Ophthalmol.36, 836–841 (2008). 10.1111/j.1442-9071.2009.01922.x [DOI] [PubMed] [Google Scholar]
- 2.Colyer, M. H., Chun, D. W., Bower, K. S., Dick, J. S. & Weichel, E. D. Perforating globe injuries during operation Iraqi Freedom. Ophthalmology115, 2087–2093 (2008). 10.1016/j.ophtha.2008.05.013 [DOI] [PubMed] [Google Scholar]
- 3.Jin, Y. et al. TRAUMATIC PROLIFERATIVE VITREORETINOPATHY: Clinical and Histopathological Observations. Retina37, 1236–1245 (2017). 10.1097/IAE.0000000000001350 [DOI] [PubMed] [Google Scholar]
- 4.Cardillo, J. A. et al. Post-traumatic proliferative vitreoretinopathy. The epidemiologic profile, onset, risk factors, and visual outcome. Ophthalmology104, 1166–1173 (1997). 10.1016/S0161-6420(97)30167-5 [DOI] [PubMed] [Google Scholar]
- 5.Pennock, S., Haddock, L. J., Eliott, D., Mukai, S. & Kazlauskas, A. Is neutralizing vitreal growth factors a viable strategy to prevent proliferative vitreoretinopathy? Prog. Retin Eye Res.40, 16–34 (2014). 10.1016/j.preteyeres.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 6.Pastor, J. C. et al. Proliferative vitreoretinopathy: A new concept of disease pathogenesis and practical consequences. Prog. Retin Eye Res.51, 125–155 (2016). 10.1016/j.preteyeres.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 7.Li, X., Zhao, M. & He, S. RPE epithelial-mesenchymal transition plays a critical role in the pathogenesis of proliferative vitreoretinopathy. Ann. Transl. Med.8, 263 (2020). 10.21037/atm.2020.03.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastor, J. C., de la Rúa, E. R. & Martín, F. Proliferative vitreoretinopathy: risk factors and pathobiology. Prog. Retin Eye Res.21, 127–144 (2002). 10.1016/S1350-9462(01)00023-4 [DOI] [PubMed] [Google Scholar]
- 9.Tamiya, S. & Kaplan, H. J. Role of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Exp. Eye Res142, 26–31 (2016). 10.1016/j.exer.2015.02.008 [DOI] [PubMed] [Google Scholar]
- 10.Morescalchi, F. et al. Proliferative vitreoretinopathy after eye injuries: an overexpression of growth factors and cytokines leading to a retinal keloid. Mediators Inflamm.2013, 269787 (2013). 10.1155/2013/269787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perino, M. & Veenstra, G. J. Chromatin Control of Developmental Dynamics and Plasticity. Dev. Cell38, 610–620 (2016). 10.1016/j.devcel.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 12.Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature489, 91–100 (2012). 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss, T. C. & Hager, G. L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet.15, 69–81 (2014). 10.1038/nrg3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nacev, B. A. et al. The epigenomics of sarcoma. Nat. Rev. Cancer20, 608–623 (2020). 10.1038/s41568-020-0288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh, E. S. & Petronis, A. Origins of human disease: the chrono-epigenetic perspective. Nat. Rev. Genet.22, 533–546 (2021). 10.1038/s41576-021-00348-6 [DOI] [PubMed] [Google Scholar]
- 16.Cantó Soler, M. V., Gallo, J. E., Dodds, R. A. & Suburo, A. M. A mouse model of proliferative vitreoretinopathy induced by dispase. Exp. Eye Res.75, 491–504 (2002). 10.1006/exer.2002.2031 [DOI] [PubMed] [Google Scholar]
- 17.Szczesniak, A. M. et al. Cannabinoid 2 receptor is a novel anti-inflammatory target in experimental proliferative vitreoretinopathy. Neuropharmacology113, 627–638 (2017). 10.1016/j.neuropharm.2016.08.030 [DOI] [PubMed] [Google Scholar]
- 18.Yoo, K. et al. Substance P prevents development of proliferative vitreoretinopathy in mice by modulating TNF-α. Mol. Vis.23, 933–943 (2017). [PMC free article] [PubMed] [Google Scholar]
- 19.Iribarne, M. et al. Blockade of endothelinergic receptors prevents development of proliferative vitreoretinopathy in mice. Am. J. Pathol.172, 1030–1042 (2008). 10.2353/ajpath.2008.070605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storm, T., Burgoyne, T. & Futter, C. E. Membrane trafficking in the retinal pigment epithelium at a glance. J. Cell Sci.133, jcs238279 (2020). 10.1242/jcs.238279 [DOI] [PubMed] [Google Scholar]
- 21.Zhou, M. et al. Role of Epithelial-Mesenchymal Transition in Retinal Pigment Epithelium Dysfunction. Front. Cell Dev. Biol.8, 501 (2020). 10.3389/fcell.2020.00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, B. et al. Blockade of MDM2 with inactive Cas9 prevents epithelial to mesenchymal transition in retinal pigment epithelial cells. Lab. Invest.99, 1874–1886 (2019). 10.1038/s41374-019-0307-9 [DOI] [PubMed] [Google Scholar]
- 23.Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu, R., Li, C., Liu, X. & Gao, S. Insights into epigenetic patterns in mammalian early embryos. Protein Cell12, 7–28 (2021). 10.1007/s13238-020-00757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delgado-Tirado, S. et al. Topical delivery of a small molecule RUNX1 transcription factor inhibitor for the treatment of proliferative vitreoretinopathy. Sci. Rep.10, 20554 (2020). 10.1038/s41598-020-77254-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laich, Y. et al. Single-Cell Protein and Transcriptional Characterization of Epiretinal Membranes From Patients With Proliferative Vitreoretinopathy. Invest. Ophthalmol. Vis. Sci.63, 17 (2022). 10.1167/iovs.63.5.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wada, I. et al. Mechanisms of Epithelial-Mesenchymal Transition and Prevention of Dispase-Induced PVR by Delivery of an Antioxidant αB Crystallin Peptide. Antioxid. (Basel)11, 2080 (2022). 10.3390/antiox11102080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, Y. et al. Active enhancer and chromatin accessibility landscapes chart the regulatory network of primary multiple myeloma. Blood131, 2138–2150 (2018). 10.1182/blood-2017-09-808063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Q. et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab.35, 943–960.e949 (2023). 10.1016/j.cmet.2023.04.015 [DOI] [PubMed] [Google Scholar]
- 30.Wu, T., Kamikawa, Y. F. & Donohoe, M. E. Brd4’s Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep.25, 1756–1771 (2018). 10.1016/j.celrep.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 31.Jang, M. K. et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell19, 523–534 (2005). 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- 32.Yang, Z. et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell19, 535–545 (2005). 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- 33.Fujisawa, T. & Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol.18, 246–262 (2017). 10.1038/nrm.2016.143 [DOI] [PubMed] [Google Scholar]
- 34.Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature468, 1067–1073 (2010). 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhagwat, A. S. et al. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep.15, 519–530 (2016). 10.1016/j.celrep.2016.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X. et al. BET bromodomain inhibitor JQ1 regulates spermatid development by changing chromatin conformation in mouse spermatogenesis. Genes Dis.9, 1062–1073 (2022). 10.1016/j.gendis.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azegami, N. et al. Native Mass Spectrometry of BRD4 Bromodomains Linked to a Long Disordered Region. Mass Spectrom11, A0110 (2022). 10.5702/massspectrometry.A0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galvao, J. et al. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J.28, 1317–1330 (2014). 10.1096/fj.13-235440 [DOI] [PubMed] [Google Scholar]
- 39.Nagai, N. & Otake, H. Novel drug delivery systems for the management of dry eye. Adv. Drug Deliv. Rev.191, 114582 (2022). 10.1016/j.addr.2022.114582 [DOI] [PubMed] [Google Scholar]
- 40.El-Gendy, M. A., Mansour, M., El-Assal, M. I. A., Ishak, R. A. H. & Mortada, N. D. Delineating penetration enhancer-enriched liquid crystalline nanostructures as novel platforms for improved ophthalmic delivery. Int. J. Pharm.582, 119313 (2020). 10.1016/j.ijpharm.2020.119313 [DOI] [PubMed] [Google Scholar]
- 41.Nawaz, A. et al. Synthesis and Characterization of Chitosan-Decorated Nanoemulsion Gel of 5-Fluorouracil for Topical Delivery. Gels8, 412 (2022). 10.3390/gels8070412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasdev, N., Handa, M., Kesharwani, P. & Shukla, R. Rosemary oil low energy nanoemulsion: optimization, µrheology, in silico, in vitro, and ex vivo characterization. J. Biomater. Sci. Polym. Ed.33, 1901–1923 (2022). 10.1080/09205063.2022.2088527 [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson, A. C., Nakauchi, H. & Göttgens, B. Mammalian Transcription Factor Networks: Recent Advances in Interrogating Biological Complexity. Cell Syst.5, 319–331 (2017). 10.1016/j.cels.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Z. Y. et al. Artesunate inhibits the development of PVR by suppressing the TGF-β/Smad signaling pathway. Exp. Eye Res.213, 108859 (2021). 10.1016/j.exer.2021.108859 [DOI] [PubMed] [Google Scholar]
- 45.Saika, S. et al. Smad3 is required for dedifferentiation of retinal pigment epithelium following retinal detachment in mice. Lab. Invest.84, 1245–1258 (2004). 10.1038/labinvest.3700156 [DOI] [PubMed] [Google Scholar]
- 46.Takayanagi, H. et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell3, 889–901 (2002). 10.1016/S1534-5807(02)00369-6 [DOI] [PubMed] [Google Scholar]
- 47.Asagiri, M. et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med.202, 1261–1269 (2005). 10.1084/jem.20051150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipinski, D. M., Thake, M. & MacLaren, R. E. Clinical applications of retinal gene therapy. Prog. Retin Eye Res.32, 22–47 (2013). 10.1016/j.preteyeres.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov.18, 358–378 (2019). 10.1038/s41573-019-0012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson, D. M. et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature390, 175–179 (1997). 10.1038/36593 [DOI] [PubMed] [Google Scholar]
- 51.Tsukasaki, M. & Takayanagi, H. Osteoimmunology: evolving concepts in bone-immune interactions in health and disease. Nat. Rev. Immunol.19, 626–642 (2019). 10.1038/s41577-019-0178-8 [DOI] [PubMed] [Google Scholar]
- 52.Yasuda, H. et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl Acad. Sci. USA95, 3597–3602 (1998). 10.1073/pnas.95.7.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacey, D. L. et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell93, 165–176 (1998). 10.1016/S0092-8674(00)81569-X [DOI] [PubMed] [Google Scholar]
- 54.Lacey, D. L. et al. Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nat. Rev. Drug Discov.11, 401–419 (2012). 10.1038/nrd3705 [DOI] [PubMed] [Google Scholar]
- 55.Zou, H. et al. Polarity and epithelial-mesenchymal transition of retinal pigment epithelial cells in proliferative vitreoretinopathy. PeerJ8, e10136 (2020). 10.7717/peerj.10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He, S. et al. Inhibition of DNA Methylation and Methyl-CpG-Binding Protein 2 Suppresses RPE Transdifferentiation: Relevance to Proliferative Vitreoretinopathy. Invest. Ophthalmol. Vis. Sci.56, 5579–5589 (2015). 10.1167/iovs.14-16258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boles, N. C. et al. Epigenomic and Transcriptomic Changes During Human RPE EMT in a Stem Cell Model of Epiretinal Membrane Pathogenesis and Prevention by Nicotinamide. Stem Cell Rep.14, 631–647 (2020). 10.1016/j.stemcr.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao, W. et al. Trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition in retinal pigment epithelium cells. J. Cell Mol. Med.18, 646–655 (2014). 10.1111/jcmm.12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatanaka, H. et al. Epigenetic regulation of the epithelial mesenchymal transition induced by synergistic action of TNF-α and TGF-β in retinal pigment epithelial cells. Biochem. Biophys. Res. Commun.544, 31–37 (2021). 10.1016/j.bbrc.2021.01.060 [DOI] [PubMed] [Google Scholar]
- 60.Khan, M. A., Brady, C. J. & Kaiser, R. S. Clinical management of proliferative vitreoretinopathy: an update. Retina35, 165–175 (2015). 10.1097/IAE.0000000000000447 [DOI] [PubMed] [Google Scholar]
- 61.Bekker, P. J. et al. A single-dose placebo-controlled study of AMG 162, a fully human monoclonal antibody to RANKL, in postmenopausal women. J. Bone Min. Res.19, 1059–1066 (2004). 10.1359/JBMR.040305 [DOI] [PubMed] [Google Scholar]
- 62.Kostenuik, P. J. et al. Denosumab, a fully human monoclonal antibody to RANKL, inhibits bone resorption and increases BMD in knock-in mice that express chimeric (murine/human) RANKL. J. Bone Min. Res.24, 182–195 (2009). 10.1359/jbmr.081112 [DOI] [PubMed] [Google Scholar]
- 63.Han, H. et al. Activated Blood Coagulation Factor X (FXa) Contributes to the Development of Traumatic PVR Through Promoting RPE Epithelial-Mesenchymal Transition. Invest. Ophthalmol. Vis. Sci.62, 29 (2021). 10.1167/iovs.62.9.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez-Godino, R., Garland, D. L. & Pierce, E. A. Isolation, culture and characterization of primary mouse RPE cells. Nat. Protoc.11, 1206–1218 (2016). 10.1038/nprot.2016.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shang, P. et al. Primary Cell Cultures from the Mouse Retinal Pigment Epithelium. J. Vis. Exp.133, 56997 (2018). [DOI] [PMC free article] [PubMed]
- 66.Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature537, 553–557 (2016). 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- 67.Charan, J. & Kantharia, N. D. How to calculate sample size in animal studies? J. Pharm. Pharmacother.4, 303–306 (2013). 10.4103/0976-500X.119726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ATAC-seq, ChIP-seq and RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE244812. The processed ATAC-seq, ChIP-seq and RNA-seq data are available at GEO database (GSE244812). The relevant raw data from each figure generated in this study are provided in the Supplementary Information/Source Data file. The cited data used in this study are available in the GEO database under accession code GSE179603. Source data are provided with this paper.