Abstract
Stroke causes death of brain tissue leading to long-term deficits. Behavioral evidence from neurorehabilitative therapies suggest learning-induced neuroplasticity can lead to beneficial outcomes. However, molecular and cellular mechanisms that link learning and stroke recovery are unknown. We show that in a mouse model of stroke, which exhibits enhanced recovery of function due to genetic perturbations of learning and memory genes, animals display activity-dependent transcriptional programs that are normally active during formation or storage of new memories. The expression of neuronal activity-dependent genes are predictive of recovery and occupy a molecular latent space unique to motor recovery. With motor recovery, networks of activity-dependent genes are co-expressed with their transcription factor targets forming gene regulatory networks that support activity-dependent transcription, that are normally diminished after stroke. Neuronal activity-dependent changes at the circuit level are influenced by interactions with microglia. At the molecular level, we show that enrichment of activity-dependent programs in neurons lead to transcriptional changes in microglia where they differentially interact to support intercellular signaling pathways for axon guidance, growth and synaptogenesis. Together, these studies identify activity-dependent transcriptional programs as a fundamental mechanism for neural repair post-stroke.
Subject terms: Stroke, Cellular neuroscience
A transcriptomic study at different time points with modulation of learning and memory genes after stroke shows robust expression of activity dependent transcriptional programs during motor recovery that are normally expressed during memory formation.
Introduction
Stroke leads to loss of brain tissue, causing persistent long-term motor, sensory, and cognitive impairments. Therapeutics for stroke recovery are limited to neurorehabilitative training. Patients, even after rehabilitation, live with severe impairments. Neurorehabilitation exploits behavioral principles of learning and adaption to induce functional gains after stroke1,2. Activities such as task-specific practice3,4, increased dosage5 and duration, and more recently virtual reality-aided feedback and training6,7 have been linked with increased neural plasticity and beneficial outcomes as a result. While much of our conclusions on the relationship between learning and stroke recovery have been drawn from behavioral evidence3,5,7 and functional MRI measures8,9, the exact cellular and molecular mechanisms in play are unknown; specifically—it is unclear if cellular and molecular substrates of learning and memory are recruited for recovery of function after stroke. An understanding of these pathways will allow for targeting the same cellular and molecular mechanisms in learning and memory to enhance recovery of function after stroke.
C–C chemokine receptor-5 (CCR5) and cAMP response element-binding protein (CREB) are critical for the formation of new memories in the normal adult brain10–14. While CREB expression is important for allocating neurons with information pertaining to a memory trace; CCR5 closes this temporal window for linking memories over time11. Allocated neurons that store a memory trace have been classically termed as the engram and have been causally linked with the expression and extinction of a memory15. Recent efforts using transgenic lines that label neurons based on the expression of immediate early genes when paired with behavioral learning paradigms in the adult healthy brain have been able to capture neurons that participate in the engram15–20. Transcriptional profiling of participating neurons has uncovered gene signatures that are unique to the engram. Many of the genes are activity-dependent genes that include immediate early genes (IEGs) that are expressed at the onset of an incoming stimulus, succeeded by a wave of late response genes (LRGs) that are normally targets of IEG transcription factors21. The expression of LRGs modulates plasticity at the synapse22–24. Existing data on the expression of these activity-dependent genes in the context of memory formation or novel experience allows us to ask if the same gene signatures are expressed under conditions of recovery after stroke.
We have recently shown that downregulation of CCR5 in cortical neurons adjacent to the infarct following a stroke to the motor cortex leads to robust improvements in motor recovery25. Recovery is associated with increased and differential sprouting of axons, preservation of dendritic spines in the secondary motor cortex, and increased signaling through the transcription factor CREB25. Overexpression of CREB alone after a stroke also improves recovery of motor function26. CREB-overexpressing neurons are allocated to a motor network such that chemogenetic inactivation of CREB-overexpressing neurons overturns recovery to a state of motor deficit26.
Here, we transcriptionally profiled neurons at different stages after stroke and under conditions of CCR5 knockdown or CREB overexpression, which enhances motor recovery. We show that under conditions of enhanced motor recovery, cortical neurons express activity-dependent gene sets that are unique to animals that recover and that these gene sets can predict functional outcomes in acute stroke. Low dimensional representations of the data suggest that activity-dependent genes occupy a molecular latent space unique to cortical neurons from animals that have CCR5 knockdown or CREB overexpression after stroke. Moreover, activity-dependent genes form robust gene co-expression networks and transcription factor-target co-expression networks when compared to untreated groups. In response to neuronal knockdown of CCR5 or overexpression of CREB after stroke, we also show that microglia express genes for ligands and receptors that support axonal guidance, dendritic signaling, and synaptogenesis.
Results
Cortical neurons after stroke express activity-dependent gene sets
We collected gene expression data with RNAseq from cohorts of animals with CCR5 knockdown (CCR5kd) or CREB overexpression (CREBoe) in healthy and stroke-induced mice. These two molecular perturbations robustly enhance motor recovery in mouse stroke models25,26 and are associated with improved cognitive function in human stroke25,27. Cortical neurons adjacent to the infarct (Supplementary Fig. S2c), from peri-infarct primary and secondary motor cortices were isolated with fluorescence-activated cell sorting (FACS) (Fig. 1b). Neurons were isolated from multiple cohorts with neuron-specific knockdown of CCR5 or overexpression of CREB in acute (7 days post-stroke) and chronic stroke (30 days post-stroke) along with controls that received the same viral backbone that lacked sequences to target CCR5 or CREB (timelines and groups in Fig. 1a, infarct sizes and location of virus expression relative to infarct in Supplementary Figs. S1 and S2).
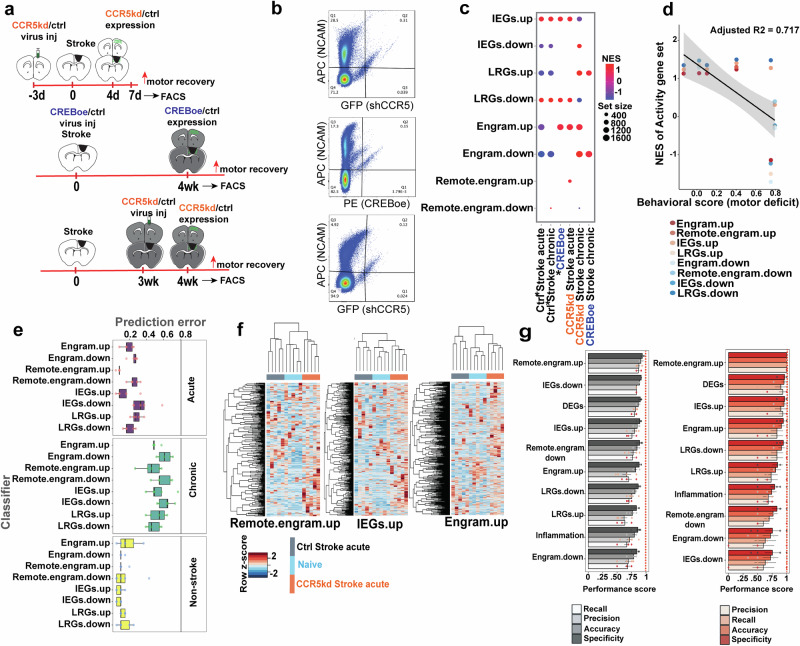
Fig. 1. Activity-dependent genes are expressed after stroke.
a Experimental timeline. b FACS plots from groups that correspond to treatments and timelines in (a), showing gating and selection of NCAM+ve events that are also positive for fluorescence expression carried by the viral construct (events that fall in the Q2 quadrant). c Gene set enrichment, FDR < 0.1, for various activity-dependent gene sets (y-axis) across conditions compared to Naïve (groups with *) or treatment groups compared with controls that received the same viral backbone. Set size denotes the number of genes in each gene set denoted by the size of the circle: positive enrichment in warmer colors and negative enrichment in cooler colors. d Scatter plot showing behavioral scores for each of the conditions compared to Naïve and their corresponding enrichment scores compared to Naïve for the various activity gene sets in (c). Increasing behavioral scores corresponds to increases in motor deficits. The line drawn represents the line of best fit and gray shaded region is the confidence interval around the slope of the regression line. e Classification of samples from untreated and treated groups before and after stroke using random forest classifiers trained on the various activity-dependent gene sets. Data are median ± SD of prediction error from five iterations. f Heatmap of row normalized gene expression (z-scores) of hierarchically clustered genes from gene sets from top-performing classifiers. Columns are individual samples from 3 groups (Naïve, Ctrl stroke acute, and CCR5kd stroke acute) and gene expression values in rows. g Model performance metrics for each classifier on training (gray) and test data (red). Data are mean values from each iteration with standard error.
The timing of neuronal isolations was aligned with when enhanced behavioral recovery was previously reported25,26. Briefly, with CCR5kd in acute stroke, robust enhancements in motor function were observed in week 1. Reducing CCR5 function in chronic stroke led to modest enhancements in function in week 4. In animals with CREBoe, enhancements in function were reported in week 4 with delivery of CREBoe in the first week of stroke. Hence, neurons were FAC-sorted at week 1 for CCR5kd in acute stroke and week 4 for cohorts of animals with the delivery of CCR5kd in chronic stroke or CREBoe in acute stroke (Fig. 1a, S1, S2 for timing of injections, location, and size of infarct, expression of virus relative to the infarct and cortical areas dissected). FAC-sorted neurons were subject to RNA sequencing. Sequencing data were aligned to the mouse reference transcriptome, filtered, and normalized to attain log2 counts per million (log2CPM), and differentially expressed genes were identified (Supplementary Fig. S3, Supplementary Data S1 and SI).
To determine if activity-dependent gene sets are expressed after stroke, we compiled datasets from 19 published studies16–20,22,28–40 (Supplementary Data S2 and SI) that measured gene expression changes with RNAseq captured from neurons tagged during learning a novel task or during long-term memory storage using activity-dependent transgenic mouse lines; or when exposed to a novel stimulus. These studies characterized genes differentially expressed in neurons of the engram during learning, remote engram during long-term memory storage, and neurons that express IEGs and LRGs when exposed to a novel stimulus. Genes from all studies were categorized into 8 classes: IEGs.up (i.e., IEGs differentially upregulated) and IEGs.down, LRGs.up, LRGs.down, engram.up, engram.down, remote.engram.up; remote.engram.down.
To determine enrichment of activity-dependent gene sets, we compared (FDR < 0.1) enrichment for several hundred genes per identified gene set between samples from animals with CCR5/CREB perturbations and their controls in normal (Supplementary Fig S3j), acute and chronic stroke (Fig. 1c). We found different classes of activity genes expressed in acute and chronic stroke when compared to Naïve (Fig. 1c, Supplementary Fig S3j). Many of these classes are negatively enriched (downregulated) after stroke, with the exception of gene sets for upregulated IEGs (IEGs.up) and downregulated LRGs (LRGs.down). The expression of IEGs is in alignment with previous studies, resulting from the ischemic event41. However, sustained downregulation of LRGs in acute to chronic stroke as seen with positive enrichment for LRGs.down and negative enrichment for LRGs.up in both acute and chronic phases, suggest downregulation or repression in the encoding of transcription factors whose targets are normally LRGs, the expression of which influences synaptic transmission and plasticity22–24. Additionally, we found that gene sets for the engram (engram.up) are negatively enriched in acute stroke and not enriched in chronic stroke. Both acute and chronic stroke groups are also negatively enriched for genes downregulated in the engram (engram.down), further supporting the overall downregulated expression of engram gene sets after stroke.
Under conditions of enhanced motor recovery, with CCR5kd in acute stroke (Fig. 1c), we found positive enrichment of gene sets for upregulated IEGs (IEGs.up) showing a further increase in expression of IEGs when compared to its control in acute stroke, but also further downregulation of LRGs as seen with positive enrichment for LRGs.down. Unlike its control, CCR5kd in acute stroke is positively enriched for gene sets upregulated in the engram (engram.up) and remote engram (remote.engram.up). The opposite pattern of expression was seen in chronic stroke, where both CCR5kd and CREBoe when compared to its control, show increased expression of LRGs and reduced expression of IEGs. Positive enrichment for LRGs.up and negative enrichment for LRGs.down (with CCR5kd) show increased LRG expression and positive enrichment for IEGs.down and negative enrichment for IEGs.up show increased expression of IEGs in chronic stroke with treatment.
This trend in the expression of IEGs and LRGs in treated groups with enhanced motor recovery show that IEG expression is restricted to the earlier phases of stroke, associated with an increase in LRG expression in the chronic phases. Similar to CCR5 knockdown in acute stroke, the condition of CCR5kd in chronic stroke is enriched for gene sets upregulated in the engram (engram.up), but also downregulated in the engram (engram.down) suggesting an overall increase in genes differentially expressed in the engram.
Unexpectedly, with CREBoe in stroke we did not find enrichment for the gene set upregulated in the engram (engram.up) but found enrichment for the gene set downregulated in the engram, which is also in contrast with data from CREBoe in the normal brain that shows increased expression of engram genes (Fig. 1c and S3j). The timing of CREB activation plays a critical role in the allocation of active neurons to an engram42. The downregulation of engram genes and the upregulation of LRGs with CREBoe post-stroke at 4 weeks suggest that an earlier onset of expression of engram genes and IEGs may have taken place prior to when motor recovery was observed.
Next, to determine if activity-dependent gene expression is correlated with improved motor function, we used data from previously published studies25,26 that tested CCR5 and CREB signaling in functional motor recovery after stroke. Test scores that represent quantitation of motor deficit compared to naïve, were normalized from the different cohorts of mice across studies. Behavioral scores attained for each of the different conditions were plotted against the normalized enrichment score for the different activity-dependent gene sets for that condition when compared to naïve (Fig. 1d). With a linear regression model to determine the relationship between observed behavioral scores and enrichment of the different activity-dependent genes sets as the predictor variable, we find that 72% of the variance in the dataset was explained by the model (adjusted R2 = 0.717, p = 0.0001). The relationship also exhibited a Pearson’s correlation of −0.66, showing that an increase in motor deficit was associated with lower or negative enrichment of activity-dependent gene expression.
In summary, we show that IEGs are differentially enriched after stroke with downregulation of all other activity-dependent gene sets. However, during conditions of enhanced motor recovery, we show increased enrichment of many activity-dependent gene sets after stroke, further enhanced than its control, including increased enrichment of gene sets for the engram with CCR5kd post-stroke.
Given the enrichment of activity-dependent gene sets after stroke with differential enrichment profiles in conditions of recovery, particularly for sets represented in the engram, we asked if these expression profiles are unique enough to predict recovery. To determine if activity-dependent gene expression can be used to classify samples with enhanced motor recovery from CCR5kd or CREBoe, we trained random forest classifiers on the various activity-dependent gene sets expressed in normal and post-stroke conditions and tested if the classifiers could predict sample type (Fig. 1e–g, Supplementary Fig S3k, Supplementary Data 3). In non-stroke conditions, we found that all classifiers predicted which samples came from CCR5kd or CREBoe or naïve conditions with a prediction error of between 15 ± 12.6% (engram.up; median ± SD) to 10 ± 4.18% (IEGs.up; median ± SD), and aligns with previously determined roles of CCR5 and CREB in memory formation and learning11–15,43. In acute stroke, the highest performing classifiers are those that were trained on remote.engram.up with a prediction error of 8.3 ± 1.8% (median ± SD) and accuracy of 95.5 ± 5.1% (median ± SD); IEGs.up with a prediction error of 8.3 ± 11.4% and accuracy of 90.7 ± 9.6% and engram.up with a prediction error of 16.5 ± 6.6% and accuracy of 86.6 ± 10.2%, further proving that the enrichment of activity-dependent genes represented during memory formation and consolidation are uniquely expressed during motor recovery. Prediction rates of the above classifiers are comparable to classifiers trained on differentially expressed genes from groups with acute stroke (prediction error—12.5 ± 8.5%; Supplementary Fig. S3k) and are better than classifiers trained on inflammatory genes involved in interferon signaling pathways44 (prediction error—27.5 ± 1.6%, Fig. S3k) that are dominantly expressed in acute stroke. Model performances measured by sensitivity (recall) and specificity of the top-performing activity-dependent classifiers are in the range of 0.9–1 (Fig. 1g, Supplementary Data 3) and are superior to most previously reported models45 to predict stroke outcome based on clinical stroke scales and structural imaging data. Furthermore, to determine if individual samples clustered based on the expression of activity-dependent gene sets from the top-performing classifiers as a means to assess sample-to-sample variability in its classification, we performed hierarchical clustering, of samples. We found that all samples that belonged to one condition clustered in the same group based on the expression of gene sets from the top-performing classifiers (Fig. 1f).
On the contrary, all classifiers performed poorly in chronic stroke with prediction error rates between 45.8 ± 12.2% (remote.engram.up) to 64.1 ± 8.3% (IEGs.down) (Fig. 1e) and are worse compared to classifiers trained on differentially expressed genes in chronic stroke groups (13.6 ± 7.8%) (Fig. S3k); suggesting that classifiers trained on activity-dependent genes are poor predictors of recovered motor control in chronic stroke.
Collectively, these data show that activity-dependent gene sets are expressed after a stroke and can a priori identify the stroke condition and treatment category in acute stroke. Cortical neurons with CCR5kd are enriched with genes expressed in the engram, remote engram and immediate early genes and, the expression patterns of these gene sets can predict sample types in acute stroke. CREB-overexpressing neurons in chronic stroke are enriched with late response genes; however none of the classifiers trained on activity-dependent gene sets are able to predict sample-type in chronic stroke.
Activity-dependent genes post-stroke occupy a unique molecular latent space
Latent space representations that have been described as ‘an internal representation of externally observed events’, have been widely applied to high-dimensional neural activity datasets to extract low-dimensional features that best describe the data46,47. With respect to gene expression data, given the expression of thousands of genes across multiple conditions, applying compression methods to extract a latent space allow us to determine the most informative genes. We applied unsupervised generative modeling with variational autoencoders (VAE)48, to extract latent space information to identify genes that carry the highest weights in representing low-dimensional gene expression information after stroke (Fig. 2 and S2). VAEs can model complex relationships and non-linearities and retain biological patterns between conditions and have been shown to offer better biological insights48,49.
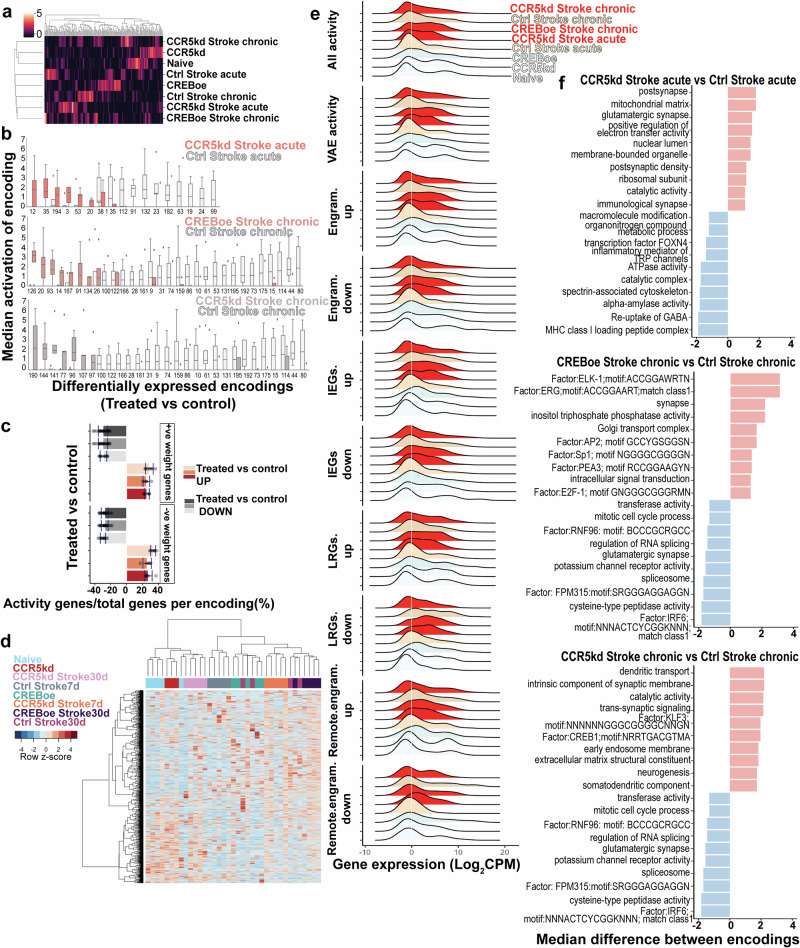
Fig. 2. Latent space representation of activity-dependent genes with VAEs.
a Heatmap of activation scores for each encoding hierarchically clustered. Rows correspond to groups and columns represent activation values for 200 encodings. b Median activation scores of encodings that are differentially expressed across treatment-control groups. c Percent fraction of highly weighted activity genes/total no. of activity genes, both positively and negatively weighted, across all differentially expressed encodings. Darker to lighter colors for either gray or red represent—CCR5kd acute, CREBoe chronic, and CCR5kd chronic vs post-stroke controls, respectively. Gray is for encodings downregulated and red for encodings upregulated. d Heatmap expression of highly weighted activity genes from encodings in hierarchically clustered samples from different conditions. e Ridge plots showing density distribution of expression of all activity-dependent genes expressed in different conditions, activity-dependent genes identified by the VAE, and expression of different classes of activity-dependent genes; showing higher and bimodal expression in CCR5kd acute and CREBoe chronic stroke groups. f Gene ontology terms for highly weighted genes from differentially expressed encodings; salmon bars are from encodings upregulated and blue are from encodings downregulated in treated vs control groups.
The VAE was programmed to encode gene expression data from 10,000 most variable genes across all samples in the dataset into 200 different feature encodings that occupied a latent space (Supplementary Fig. S4a, Methods). Activation of each encoding was determined by the weighted sum of genes in the encoding and differentially activated encodings were identified across treated vs control groups (Fig. 2a, b, Supplementary Fig. S4c). Based on the activation values, encodings distinct to each of the different groups were identified and samples per group could be hierarchically clustered based on activation scores (Fig. 2a). We identified 6 encodings differentially activated with CCR5kd when compared to its control in acute stroke; 5 encodings with CREBoe and 7 encodings activated with CCR5kd when compared to its control in chronic stroke (Fig. 2b, Supplementary Data S4). We also found 11 encodings differentially activated in acute and 22 encodings in chronic control stroke groups when compared to treated groups showing that the VAE was able to capture multiple features (encodings) unique to each of the different post-stroke conditions.
To determine the biological significance of genes expressed in differentially activated encodings, we ran a Gene Ontology analysis on highly weighted genes (Fig. 2f, Supplementary Data S4) from the top 5 differentially activated encodings. Highly weighted genes were defined as having weights 2 standard deviations above or below the mean of gene weights within the encoding48. Differentially expressed encodings in CCR5kd in acute stroke vs control treatment in acute stroke identified upregulated pathways that map to cellular components of the postsynapse (GO:0098794, adjusted p value = 0.033) and mitochondrial matrix (GO:0005759, adjusted p value = 0.024), and downregulated pathways that include MHC class1 signaling (GO:0042824, adjusted p value = 0.023), GABA reuptake (REAC:R-MMU-888593, adjusted p value = 0.049) and alpha amylase activity (GO:0004556, adjusted p value = 0.008). In chronic stroke, with CREBoe vs control treatment, many IEG transcription factors and binding targets such as ELK-1 (TF:M01981, adjusted p value = 0.014), ERG1 (Zif268) (TF:M01752_1, adjusted p value = 0.012) and cellular synaptic components (GO:0045202, adjusted p value = 0.001) are overrepresented whereas pathways in IRF6 signaling (TF:M02874_1, adjusted p value = 0.036), potassium channel activity (GO:0015459, adjusted p value = 0.005) and spliceosome (KEGG:03040, adjusted p value = 0.024) are downregulated. In the case of CCR5kd in chronic stroke vs control treatment, upregulated pathways include dendritic transport (GO:0098935, adjusted p value = 0.001), trans-synaptic signaling (GO:0099537, adjusted p value = 0.027) and CREB signaling (TF:M10200, adjusted p value = 0.024); whereas downregulated pathways map to similar terms such as IRF6 signaling and spliceosome. In summary, we find that encodings in the latent space differentially activated in treated (recovery-enhanced) conditions and control conditions map to meaningful biological pathways that support neuronal development, immune signaling and transcription factors that regulate neuronal activity.
Additionally, we found that encoding 20 was differentially encoded and common to both CCR5kd in acute stroke and CREBoe in chronic stroke (Supplementary Fig. S4c, d). Genes in this encoding mapped to several components including synapse (GO:0045202, adjusted p value = 0.001), axon (GO:0030424, adjusted p value = 0.0003), and growth cone (GO:0030426, adjusted p value = 0.043) (Supplementary Fig. S4c, d Supplementary Data S4) suggesting a convergence between the two conditions that employ the same genes that may be involved in synaptic signaling, as a learnt biological feature.
Given that many of the encodings map to biological pathways that support neuronal plasticity, we asked if activity-dependent genes are encoded in the latent space. We assessed the weights of activity-dependent genes within differentially expressed encodings, i.e., encodings that have higher activation scores in treated than control and vice-versa (Fig. 2b, Supplementary Data S4). Of a total of 3042 activity-dependent genes expressed in the dataset, 2030 activity-dependent genes are highly weighted genes expressed across all differentially activated encodings across all groups, representing 26–31 ± 4% of highly weighted genes within each differentially activated encoding per group (Fig. 2c). The expression of these genes is sufficient to cluster sample-type, with the exception of control chronic stroke (Fig. 2d) and have collectively higher and bimodal expression with CCR5 knockdown in acute stroke and CREBoe in chronic stroke (Fig. 2e, Supplementary Fig. S4b).
The assignment of high weights to activity-dependent genes that occupy multiple differentially activated encodings in the latent space, the mappability of differentially expressed encodings to biological pathways for neuronal plasticity, the higher expression of these genes in groups with CCR5kd or CREBoe, and the ability to cluster samples based on the expression of these highly weighted activity genes, show that the expression of activity-dependent genes is a defining molecular feature of motor recovery.
Motor recovery is associated with the expression of robust co-expression networks of activity genes and their transcription factors
With the overrepresentation of activity-dependent genes in conditions of motor recovery, we next investigated if they form co-expression networks, as the co-expression of genes is indicative of regulation by the same transcriptional programs50. We identified networks of co-expressed activity-dependent genes in groups with CCR5kd and its control in acute stroke and CREBoe and its control in chronic stroke, using a previously described method51 for identification of tight co-expression clusters. We identified two clusters of 1000–1500 genes in each treatment—control pair where one cluster is highly expressed with treatment and downregulated in the control group and vice versa (Fig. 3c). Within each cluster, we further screened for genes that were activity-dependent and differentially expressed compared to non-stroke (naïve) conditions to identify a co-expression network of activity-dependent genes unique to post-stroke conditions with high connectivity (i.e., Pearson’s correlation with absolute values >0.8) (Supplementary Data S5).
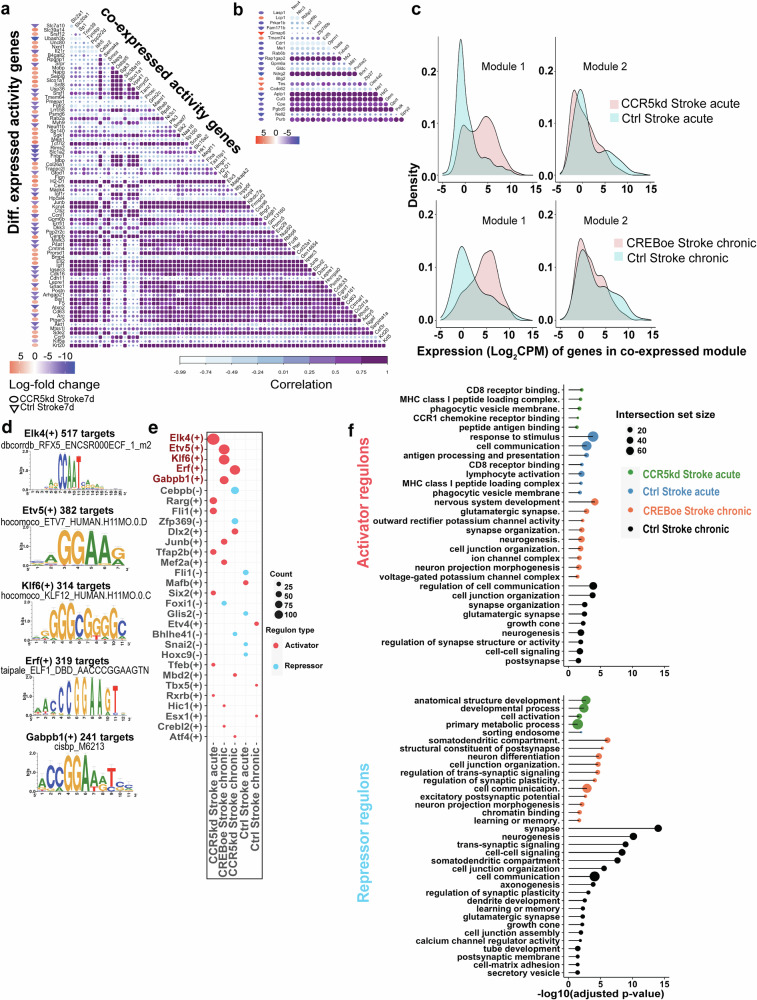
Fig. 3. Co-expression networks of activity-dependent genes after stroke.
Co-expression networks of differentially expressed activity-dependent genes post-stroke and their co-expression partners with a CCR5kd in acute stroke and b control acute stroke group. All gene names and correlation values are available in Supplementary Data S5. Nodes are genes and Pearson’s correlation values are the edges in the network. c Density distributions of genes expressed in the co-expression modules identified in treated and control groups, where module 1 is highly expressed in the treated groups. d Sequence logos for TF binding sites from the top five TFs that bind activity-dependent genes in neurons from treated conditions. e Expression of neuronal regulons (activators and repressors) with the highest numbers of activity-dependent genes across conditions post-post stroke. f Gene ontology terms from genes expressed in activator and repressor regulons expressed in microglia across conditions.
In acute stroke, we found lower numbers of activity-dependent genes that are co-expressed (44 genes) compared to CCR5kd in acute stroke, which showed robust expression of a co-expression network with 144 genes (Fig. 3a, b). The smaller co-expression network of activity-dependent genes in control vs treated samples in acute stroke is in alignment with our data on the negative enrichment of activity-dependent genes and high predictability of motor recovery in acute stroke based on the expression of activity-dependent gene signatures with treatment. In contrast, in chronic stroke, CREBoe and its control had similar numbers of co-expressed activity-dependent genes (155 and 152 respectively) (Supplementary Fig. S5a, b)
Given that we had similar-sized co-expression networks with CREBoe and control groups in chronic stroke, we next asked if these networks represent functional pathways that are distinct to treatment. Biological pathways represented by co-expressed genes in the network for each condition were extracted with Gene Ontology (Supplementary Fig. S5c, Supplementary Data S5).
No biological domains were represented by co-expressed activity-dependent genes in acute stroke. However, with CCR5kd in acute stroke, which is a condition of enhanced recovery, several pathways such as neuronal development (GO:0007399, adjusted p value = 0.016), transport (GO:0006810, adjusted p value = 0.001), and kinase binding (GO:0019900, adjusted p value = 0.009) are represented. In chronic stroke, while similar sized activity-dependent gene co-expression networks are expressed, there are only two broad biological domains—protein binding (cellular component, GO:0005515, adjusted p value = 0.005)) and intracellular anatomical structure (molecular function, GO:0005622, adjusted p value = 0.048) that are represented in the control group.
However, with CREBoe in chronic stroke, which also enhances recovery, many biological domains inclusive of molecular function, cellular components, and biological processes such as neuronal development (GO:0048666, adjusted p value = 0.005), differentiation (GO:0030154, adjusted p value = 0.024), somatodendritic compartment (GO:0036477, adjusted p value = 0.001) and cytoskeletal signaling (GO:0008092, adjusted p value = 0.032) are enriched. This suggests that while there is co-expression of a small number (acute stroke) or similar numbers (chronic stroke) of activity-dependent genes, the product of this expression does not map to any known and annotated biological processes except in conditions of enhanced stroke recovery, in which CCR5kd or CREBoe activate classes of neuronal development, differentiation, and specific intracellular signaling cascades.
As co-expression network analysis identified the expression of a robust network in acute stroke and a network that supported neuronal development and plasticity in chronic stroke under conditions of motor recovery, we reasoned that an activity-dependent molecular program might be regulated by distinct transcriptional programs during recovery. To identify transcription factors that control activity-dependent molecular programs after stroke, we inferred gene regulatory networks using a gradient-boost learning algorithm to identify transcription factors and their co-expressed targets52,53. Co-expressed genes with binding motifs for a transcription factor comprised a regulon. Regulons are either repressive—where expression of target genes is inhibited, or activators—where expression of target genes is enhanced. Identified regulons were scored52 and the top 20 regulons were screened for enrichment in target genes that are activity-dependent genes (Supplementary Data S6). Across all top 20 regulons, CCR5kd in acute stroke has 300 target genes that are activity-dependent, of which 297 come from activator regulons and 3 from repressor regulons; in contrast with the control group with 122 activity-dependent target genes of which 81 are from repressor regulons (Fig. 3f, Supplementary Data S6). The identification of targets of transcription factors that are activity-dependent shows that these transcription factors regulate activity programs after stroke54.
In chronic stroke, neurons with CREBoe expressed 322 genes that are activity-dependent, of which 300 are from activator regulons and, with CCR5kd, 236 target genes are activity-dependent and 111 genes are from activator regulons. However, in the control group in chronic stroke, 101 genes are activity-dependent with 73 genes from repressor regulons (Fig. 3e, Supplementary Data 6). These data show increased proportions of activity-dependent genes under conditions of motor recovery, that are targets of transcription factors that positively modulate their expression, whereas in control stroke conditions, there are lower numbers of activity gene targets and expression of transcription factors that repress their expression. Moreover, these data provide further insight to previous data on the enrichment of activity-dependent genes and expression of large co-expression networks during recovery, showing that these are the result of programs that are transcriptionally regulated by transcription factors that support the expression of activity-dependent genes during recovery, that are normally repressed after stroke.
Next, we screened gene targets that are activity-dependent genes and identified regulons with the highest proportion of activity-dependent genes as their targets. With CCR5kd in acute stroke, we found that the transcription factor ETS domain-containing protein Elk-4 (Elk-4) has the highest number of activity-dependent gene targets—102 genes; whereas with CREBoe, ETS variant transcription factor 5 (Etv5) and Kruppel-like factor 6 (Klf6) were enriched with activity-dependent gene targets (76 and 73 activity-dependent gene targets); and with CCR5kd in chronic stroke, ETS2 Repressor Factor (Erf) has 68 targets (Fig. 3d,e, Supplementary Data 6). However, in control stroke groups, many of the regulons have fewer activity-dependent target genes (<18) with most regulons being repressors with the exception of V-maf musculoaponeurotic fibrosarcoma oncogene homolog B (Mafb2). In chronic stroke, expression of regulons that are activators have far fewer activity-dependent target genes (<15 genes) when compared to treated groups.
The expression of robust activity-dependent programs through co-expression networks and transcription factor-target expression networks during conditions of motor recovery not only shows that activity-dependent genes are expressed but they are uniquely regulated by programs that support the expression of large networks of activity-dependent genes that biologically map to neuronal development and dendritic signaling genes. Contrarily, animals with poor recovery have diminished activity-dependent programs as seen both at the level of co-expression and the expression of transcription factor modules (repressor regulons) that either negatively regulate activity-dependent gene expression or have diminished modules of fewer activity-dependent genes.
Neuronal knockdown of CCR5 or overexpression of CREB results in transcriptional changes in microglia and neuro-microglial ligand–receptor interactions unique to motor recovery
Microglial interactions with neurons shape network connectivity in an activity-dependent manner and can remodel synapses depending on context55–60. Microglia sense cellular excitability changes in neurons and can act as negative feedback regulators, where microglial contact can dampen neuronal activity to regulate neuronal excitability56. Neuronal excitability is dampened after stroke and we show the expression of robust activity-dependent gene expression programs that map to changes in neuronal excitability26,61,62 as being expressed in motor recovery. Moreover, CCR5 and CREB signal between neurons and microglia in normal and disease states63,64 in controlling memory function65,66. Hence, we reasoned that neuronal activity changes at the gene expression level could be influenced by microglia that may express a different transcriptional profile in states of recovery.
To address this, we measured gene expression changes in microglia through RNAseq where we FAC-sorted microglia from the same cell suspensions from which neurons were isolated in naïve, acute, chronic, and treated groups post-stroke (groups as described in Fig. 1a, Supplementary Fig. S6). To determine what gene sets are enriched across treated vs control conditions post-stroke, we analyzed enrichments for various pathways from the molecular signature database (FDR < 0.1, Supplementary Fig. S6c). Many molecular pathways pertaining to translational initiation, ribosomal proteins and cellular response to stimuli are negatively enriched in microglia where neurons carried knockdown to CCR5 in acute stroke compared to control stroke. Similarly, with neuronal CREBoe in chronic stroke, we found many negatively enriched pathways in microglia with the exception of neurotransmitter release and hippocampal neuronal developmental pathways (Supplementary Fig. S6c).
The enrichment for several downregulated pathways under conditions of motor recovery suggests a repressive transcriptional state in microglia with neuronal CCR5kd or CREBoe after stroke. To determine these transcriptional programs regulated by transcription factor-target co-expression networks, we inferred gene regulatory networks in microglia and identified the top 20 regulons across conditions (Supplementary Fig. S6d, Supplementary Data S7). In acute stroke, we found many regulons that are repressors than activators when compared to Naïve, suggesting an overall dampening of microglia and has been shown in other disease states where homeostatic signaling is lowered in microglia under pathological conditions67,68. However, with CCR5kd in acute stroke, we find more repressive regulons compared with its control where there are 12 repressors compared with 8, suggesting further downregulation of programs in microglia (Supplementary Fig. S6d, Supplementary Data S7). The size of the regulons also varied, where we found that the largest regulon in the CCR5kd group is a repressor with 110 target genes (peroxisome proliferator-activated receptor gamma, PPARG) compared with 11 targets from the largest repressor (Homeobox D4, Hoxd4) in the control group in acute stroke that express activators with smaller numbers of targets. In chronic stroke, the trend in repression continued when compared to Naïve and with comparable numbers of activators and repressors with CREBoe or CCR5kd in chronic stroke. While both acute and chronic stroke groups show dampened transcriptional programs, further dampening is seen with the CCR5kd in the acute phase and not with the treated groups in the chronic phase (Supplementary Fig. S6d). Further repression with CCR5kd in acute stroke could also be a result of disrupted chemokine signaling in microglia active during a reactive state69.
To determine biological pathways that are represented by these regulons, we mapped targets from repressors and activators separately using Gene Ontology (Fig. 3f, Supplementary Data S7)). We found that the target genes from the repressor regulons with CCR5kd in acute stroke map to cell activation (GO:0001775, adjusted p value = 0.02) and metabolic processes (GO:0044238, adjusted p value = 0.036) whereas the activator regulons map to binding (GO:0005488, adjusted p value = 0.002). This is the opposite trend in acute stroke without treatment, where activator regulons map to cell communication (GO:0007154, adjusted p value = 0.001), lymphocyte activation (GO:0046649, adjusted p value = 0.009) and response to stimuli (GO:0051716, adjusted p value = 0.004); whereas the repressors map to a single term pertaining to endosome trafficking (GO:0097443, adjusted p value = 0.01), suggesting that microglia with CCR5kd have lower cellular activity compared to its control. With CREBoe and its control in chronic stroke, both activator and repressor regulons map to several processes involved in synaptic plasticity and neuronal development (Fig. 3f, Supplementary Data S7). These data further illustrate the dampening of biological pathways in microglia in acute stroke with CCR5kd. It also suggests that this dampening is specific to CCR5 knockdown in the acute phase, whereas repressive programs expressed in the chronic phase are agnostic to treatment.
Given the overall repression in microglial signaling post-stroke, we next explored intercellular interactions with neurons through ligand–receptor signaling (Fig. 4, Supplementary Fig. S7, Supplementary Data S8). To determine cell–cell interactions, we used a previously developed method to identify differentially over-expressed ligands and their receptors from a curated database between neurons and microglia, and intercellular communication probabilities were computed70. 857 interactions were detected between neurons and microglia in the naïve condition (Fig. 4a). In acute stroke, 684 interactions were detected in the control group whereas these interactions are reduced to a third with CCR5kd with 262 interactions. These data are in alignment with an expression of repressor regulons after stroke with further repression seen with CCR5kd in acute stroke. In chronic stroke, similar trends in reduced numbers of interactions were detected, with further modest reduction with CREBoe (410 and 304, respectively). This was in contrast with CCR5kd in chronic stroke where 666 interactions were detected, being higher than its control in chronic stroke.
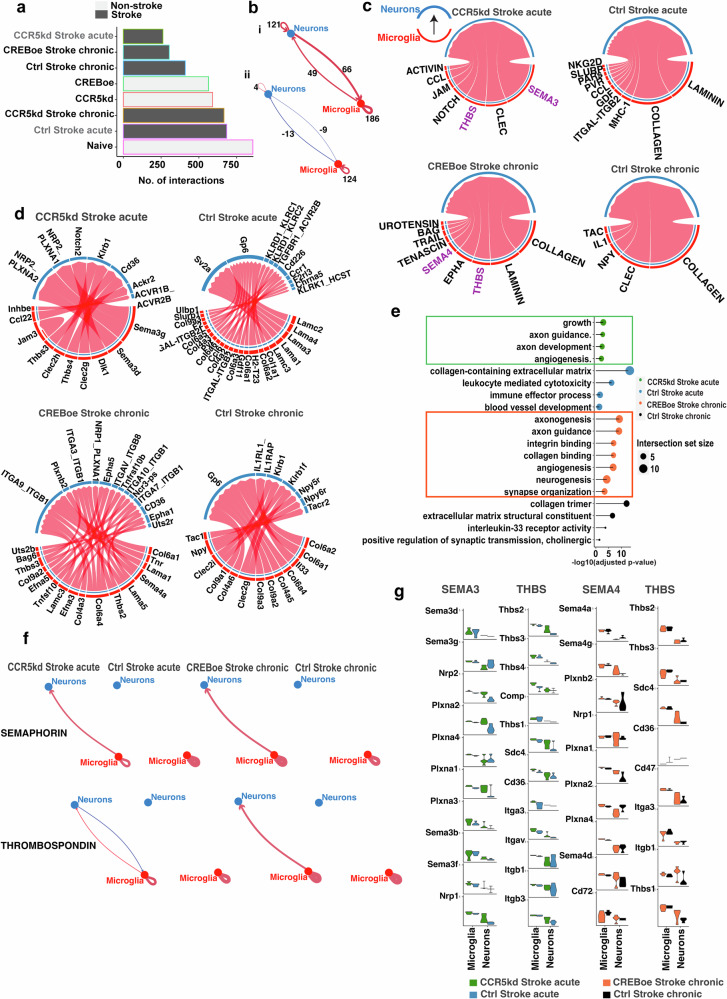
Fig. 4. Differential neuro-microglial interactions post-stroke.
a Number of ligand–receptor interactions between neurons and microglia in different conditions. b Differential interactions in (i) control stroke acute vs CCR5kd acute (ii) control stroke chronic vs CREBoe chronic stroke. Red arrows denote increased no. of interactions, and blue denotes reduced no. of interactions; line thickness is proportional to no. of interactions. c, d Ligand–receptor interactions (d) from microglia to neurons and signaling pathways (c) represented by ligand–receptor interactions in treated vs control conditions. e Gene ontology terms mapped to expressed ligands and their receptors expressed across conditions. f Weighted-edge graphs showing directionality of semaphorin and thrombospondin signaling. Blue arrows from microglia to neurons and red from neurons to microglia; line thickness denotes signaling strength. g Expression of ligand–receptor genes from thrombospondin and semaphoring signaling pathways across cell types and post-stroke conditions.
Differential interactions across treatment-control pairs were computed taking into account differential ligand–receptor interactions between and within the two cell-types and the directionality of this interaction i.e., from neurons to microglia or microglia to neurons were inferred based on weighted directed graphs (Fig. 4b). We found 115 interactions that are increased between neurons and microglia and 186 autocrine microglial interactions in control vs CCR5kd in acute stroke. In chronic stroke, we found 22 interactions that are downregulated between neurons and microglia and 124 autocrine interactions that upregulated in microglia in the control group compared to CREBoe after stroke (Fig. 4b).
Next, we identified differentially regulated signaling pathways based on differential ligand–receptor interactions in treatment-control pairs. Pathways were clustered using manifold learning to produce clusters of signaling pairs expressed on the two cell-types and the euclidean distances served as a measure to identify pathways that are furthest apart in treatment vs control groups (Supplementary Fig. S7a,b) as a means of identifying pathways that are most active in each condition. In acute stroke, we found many ligand–receptor pairs for laminin and collagen signaling from microglia to neurons; whereas with CCR5kd in acute stroke, we found ligand-receptors pairs that correspond to semaphorin, CLEC and thrombospondin signaling (Fig. 4c, d, Supplementary Data S8). In chronic stroke, similar to acute, we found many signaling pairs for collagen signaling that were also expressed with CREBoe in chronic stroke. Interestingly, similar to both CCR5kd in acute stroke, CREBoe in chronic stroke also differentially expressed signaling pairs for semaphorin and thrombospondin signaling (Fig. 4f, g).
With respect to signaling from neurons to microglia, in acute stroke with CCR5kd, we found signaling via various neuropeptides including NPY, CCK, PACAP and thrombospondin whereas with CREBoe in chronic stroke, pathways that correspond to WNT, interferon, FGF and GDNF signaling are expressed (Supplementary Fig. S7c, d).
A Gene Ontology analysis (Fig. 4e, Supplementary Data S8), of ligand receptor pairs that signal from microglia to neurons show overrepresentation for pathways in axon guidance (GO:0007411, CREBoe adjusted p value = 9.9E−10; CCR5kd, adjusted p value = 0.002) axon development (GO:0061564, CREBoe adjusted p value = 2.05E−9; CCR5kd adjusted p value = 0.003), synapse organization (GO:0050808, CREBoe adjusted p value = 0.0003) and angiogenesis (GO:0001525, CREBoe adjusted p value = 1.79E−7; CCR5kd adjusted p value = 0.006) in groups with CCR5kd or CREBoe compared to control acute or chronic groups that were enriched in terms for collagen (GO:0062023, Ctrl acute adjusted p value = 9.65E−14; chronic acute adjusted p value = 0.0001) and extracellular matrix components (GO:0005201, Ctrl acute adjusted p value = 3.95E−14; chronic acute adjusted p value = 3.15E−7), immune signaling (GO:0002252, Ctrl acute adjusted p value = 0.02) and synaptic transmission (GO:0032224, chronic acute adjusted p value = 0.04).
Taken together, our data show that microglia differentially interact with neurons under conditions of motor recovery. Transcriptional repressor programs are active with CCR5kd reducing cell activation states with reduced numbers of ligand–receptor interactions between neurons and microglia. However, with CREBoe, transcriptional programs that activate and repress synaptic plasticity and development are expressed suggesting bidirectional control of signaling events. Common to both CCR5kd and CREB are the expression of semaphorins and thrombospondin signaling pathways from microglia to neurons that support axon guidance and growth. Distinct to the two treatments are the expression of signaling pathways from neurons to microglia where signaling via neuropeptides with CCR5kd and growth factor signaling with CREBoe are predominant.
Discussion
To determine transcriptional programs expressed under conditions of motor recovery, we measured gene expression with RNAseq, in cortical neurons and microglia in animals with varying levels of enhancement in motor function from knockdown of CCR525 or overexpression of CREB26. The modulation of the expression of these genes and the timing of when these genes were perturbed maps to graded changes in behavioral performance. For e.g.,—with CCR5kd in the acute phase of stroke, robust enhancements in motor function have been reported whereas knockdown of CCR5 in the chronic phase leads to modest improvements. With CREBoe in acute stroke, recovery of function is observed in the chronic phase of stroke. To identify cellular and molecular processes that represent a recovery state, we sequenced neurons with different gene perturbations at times when motor recovery was first observed. We show that neurons are enriched with activity-dependent genes that are expressed when exposed to a novel stimulus in the normal animal, or neurons that participate in formation of new memories.
With supervised classification and unsupervised generative models, we show that these activity-dependent programs, in particular, programs that are differentially expressed during formation or long-term storage of memories are highly enriched and predictive of recovery. While with supervised classification, predictions are limited to recovery in acute stroke, from generative modeling, we learn that activity-dependent gene expression is a fundamental feature, occupying low-dimensional space in conditions of motor recovery irrespective of the phase of stroke.
Many of these activity-dependent genes form robust co-expression networks that map to several biological processes that support neuronal development and dendritic signaling. Both processes play significant roles in neural repair in stroke54 and other models of injury71 as well as in learning and memory72. At the gene regulatory level, transcription factor co-expression modules, regulons, that support transcription of activity-dependent genes are expressed under conditions of motor recovery. Many of these transcription factors, such as Elk-4, Etv-5, Erf, Gabpb1 belong to the E-twenty six, Ets family of transcription factors that are evolutionary conserved with roles in development73,74 and synaptic plasticity75–77. For eg, Elk-4 belongs to the ternary complex factor subfamily of the Ets family that binds serum response element in c-Fos; an immediate early gene and marker of neuronal activity.
Neurons from animals with motor deficits, without treatment, in acute and chronic stroke, also co-express activity-dependent genes, but with key differences in their expression. In acute stroke, a smaller co-expression program is expressed that expands in chronic stroke. Moreover, transcriptional programs that regulate activity-dependent gene expression are repressed in acute stroke, with the exception of Mafb; whereas in chronic stroke smaller programs are expressed. Mafb signaling has been associated with reduction in post-stroke inflammation78 and is in alignment with cellular events where the acute phase of stroke transitions into a phase marked by endogenous plasticity79,80. The expression of activity-dependent programs in chronic stroke is indicative of spontaneous rewiring from endogenous plasticity80 that underlies incomplete or compensated forms of motor recovery. It can be inferred that these programs are not robust enough to induce recovery of function as seen with CCR5/CREB perturbations and also from a lack of biological pathways that map to the expression of these activity-dependent genes.
Moreover, as activity-dependent gene expression programs are tied to excitability levels in neurons, the ability of the classifiers to predict with higher accuracies in acute stroke could be reflective of neuronal activity changes that are most distinct in the acute phase of stroke. The acute phase of stroke is marked by post-stroke depression, with lost synchronization of neuronal firing across cortical neurons81 as well as reduced amplitude of firing26. However, motor recovery is associated with increased neuronal firing and synchronization82. In chronic stroke, neuronal network activity resumes to baseline levels83, presumably due to spontaneous rewiring, and hence the lack of sensitivity of the classifiers to distinguish between treated as well as control groups. It can be inferred that activity-dependent molecular programs track neural activity changes at the functional level with depressed activity associated with diminished activity dependent transcriptional programs and increased neural activity being associated with the expression of robust activity-dependent co-expression networks.
Given increasing evidence on the roles of microglia in tuning neuronal connectivity in the healthy and diseased brain55–60, we investigated differential changes in microglial signaling with neuronal perturbations of CCR5/CREB and under conditions of motor recovery. Given that activity-dependent programs are enriched within neurons with CCR5kd/CREBoe, we hypothesized that this could lead to changes in neuronal activity patterns, that maybe shaped by its interactions with microglia, as has been reported in other systems55–60. We show with CCR5kd in acute stroke, microglia appear to be in a dampened cellular state as seen with gene set enrichment and transcription factor—target co-expression analyses. This dampening extends to its intercellular communication with neurons where the number of ligand–receptor interactions with neurons were reduced to a third when compared with its control in acute stroke. Unlike, CCR5kd, microglial interactions with neurons that overexpress CREB show modest reductions in the number of interactions and express transcriptional programs that activate and repress multiple signaling process in synaptic plasticity. Microglia were isolated based on TMEM119 expression, originally reported as being expressed with high cell-type specificity84. However, more recent work suggests a downregulation in the expression of many microglial homeostatic genes in diseased states85–87. It can be inferred, that the signaling process captured here unique to recovery could be confined to certain subsets of TMEM119-expressing microglia and, future work is required to determine if different subsets express differential recovery responses in relation to neuronal expression of activity-dependent genes.
In addition to its roles in learning and memory, knockdown or overexpression of CCR5 and CREB induces recovery of motor function with differences in the timing of which this recovery is manifested-early with CCR5kd and delayed with CREBoe; yet CCR5kd in acute stroke and CREBoe in chronic stroke have similar molecular signaling features. Given that the samples were isolated at different cellular phases of stroke, there exists close proximity in their transcriptional profiles, as seen with hierarchical clustering of sample encodings with VAEs, its bimodal expression pattern of activity-dependent genes and the expression of robust transcriptional co-expression programs for activity-dependent genes, although they differ in the transcription factors that modulate these programs. Common and distinct themes also emerge with neuro-microglial interactions where thrombospondin and semaphorin signaling are common to both groups that support axon guidance, axonogenesis and development whereas signaling from neurons to microglia differ based on whether neurons carry CCR5kd or CREBoe. Commonalities with CREBoe/CCR5kd, that surpass the cellular phases from which neurons or microglia were isolated but have similarities at the behavioral level with induction of motor recovery, show that enrichment of activity-dependent transcriptional programs in memory is a fundamental molecular program of neural repair for post-stroke motor recovery.
Methods
In vivo animal studies
All procedures were performed in accordance with the University of California, Los Angeles and the Jackson Laboratory’s Institutional Animal Care and Use Committee (IACUC) guidelines. Authorization numbers are JAX AUS: 23-059; UCLA ARC-2000-159. Adult male C57BL/6 J, 3–5 months of age were procured from Jackson Laboratories (strain: 000664). Animals were housed in the animal facility with ad-libitum food and water, controlled temperature, humidity and 12 h of day–light cycles. Animals were allowed to acclimatize for 3–7 days prior to experiments. We have complied with all relevant ethical regulations for animal use.
Stroke model
A focal ischemic stroke was induced using a well-established method25,26. Briefly, animals were anesthetized with isoflurane and body temperature was maintained between 36.5–37 °C with a rectal probe. Using aseptic surgical techniques, an incision to the skin over the skull was made. Light from a cold light source (KL1500 LCD; Carl Zeiss MicroImaging) attached to a 40× objective, or from a pigtailed laser diode (Thorlabs) was positioned and illuminated at 1.5 mm lateral to bregma, anterior–posterior coordinates at 0. Five minutes prior to illumination, animals were injected with a light-sensitive dye—Rose Bengal (Acros Organics), i.p., at a concentration of 10 mg/ml and volume of 1% body weight. Light intensities and duration of illumination were optimized to attain an infarct with area that extends across the anterior–posterior axes upto approx. 1.6 mm2 in the region that corresponds to the motor cortex and is contained within the cortex without infracting underlying sub-cortical structures (Fig. S1).
Cranial injections
Previously validated AAVs that carry an shRNA to target neuronal CCR525 (AAV2/5-hU6-shCCR5-EF1-GFP; control virus: AAV2/5-hU6-dsRED-EF1-GFP) or neuronal overexpression of CREB26 (LV-Cam2a-tdTomato_ns-mCREB; control virus: LV-Cam2a-tdTomato_ns) with lentiviruses were used. The timelines for stroke induction and AAV/lentiviral injections are shown in Fig. 1a and S2. AAVs were injected at 1.5 mm A/P, 1 mm M/L, 0.75 D/V, 3 days prior to a stroke to enable the delivery of knockdown within the first week and lentiviruses were delivered at 1.5 mm A/P, 1.0 mm A/P, 1 mm M/L, 0.75 D/V on the day of stroke. For knockdown of CCR5 in chronic stroke, AAV2/5-hU6-shCCR5-EF1-GFP was injected 3 weeks post-stroke.
FACS
Tissue anterior to the stroke site, containing peri-infarct cortex and primary-secondary motor cortices, spanning 2.5 mm anterior to bregma (Supplementary Fig. S2c) were harvested at 1 week or 1 month post-stroke. Tissue was kept in ice-cold hibernation, sliced and enzymatically digested with Neurocult dissociation (STEMCELL Technologies) medium at 37 °C for 8 min. The reaction was stopped with the addition of Neurocult inhibition medium, centrifuged for 10 min at 300 g and triturated to form a single cell suspension. Undigested tissue was filtered out with a 70 μm mesh.
Neuronal enrichment
Cells were resuspended in 80 μl of hibernation and treated with a neuronal isolation antibody cocktail that contains antibodies against non-neuronal cells and additional anti-GLAST and CD11b microbeads for 15 min on ice. Cells were passed through magnetic columns causing non-neuronal cells to be trapped in the columns and the flow-through to be enriched in neurons. The flow-through was treated with surface markers for neurons- APC-conjugated NCAM and PE-conjugated CD200 for 30 min on ice; washed and cells resuspended in 300 μl of ice-cold hibernate.
Microglial enrichment
The columns were flushed to release non-neuronal cells, treated with mouse Fc block for 5 min on ice followed by rabbit TMEM119 antibody for 30 min on ice. Cells were washed once and then resuspended in 80 μl of hibernate and treated with APC-conjugated CD11b and goat-anti rabbit Alexa-flour 594 for 25 min on ice. Cells were washed and then resuspended in 300 μl of hibernate.
Sorting
Cells were sorted using the ARIA cell sorter (BD, UCLA FACS core). Forward and side scatters along with DAPI staining allowed for elimination of doublets, debris and selection of single viable events. Further gates were drawn to select events that were triple positive for GFP, APC and PE to select GFP-expressing CCR5kd/control neurons or for double positives for APC and td-tomato to select CREBoe/control neurons. For sorting of microglia, events that were double-positive for red fluorescence (emission spectra of Alexa-flour 594, TMEM199+ve) and APC (CD11b+ve) were selected. As AAVs transduce more cells than lentiviruses and to ensure that number of events across samples are comparable i.e within a difference of a log fold of 10, events were capped at 1,500 positive events per sample. Selected cells were sorted into 50–100 μl of RNA lysis buffer.
RNA isolation and sequencing
RNA was extracted using a Zymo RNA micro extraction kit in accordance with manufacturer’s guidelines. cDNA was generated and amplified using the SMART seq v4 ultra low input and SSv4 PLUS kits from Clonetech and libraries were prepared with Nextera XT DNA Library Preparation by the UCLA Technology Center for Genomics and Bioinformatics core, followed by sequencing with Novaseq 6000 (Illumina) to attain 50 bp paired-end reads at a depth of 40–50 M reads.
Immunohistochemistry, infarct size quantification, and fluorescence intensity measurements
Animals were perfused with 4% paraformaldehyde, cryoprotected in 30% sucrose overnight and sectioned with a cryostat to obtain 40um thick sections. Sections were then blocked with blocking solution which was a cocktail of MOM (VectorLabs), BSA (0.05%), goat serum (0.05%), triton X(0.00001%), tween (0.005%) and PBS (0.1 M), for 1 h on a shaker at room temperature. Sections were then treated with primary antibodies diluted in blocking solution overnight at 4 °C on a shaker. Primary antibodies used were mouse NeuN (1:1000, Abcam) and chicken GFAP (1:500,Origene). Sections were then washed 3 times with PBS and treated with secondary antibodies-goat anti-mouse (1:500) and goat anti-chicken (1:500) and incubated at room temperature for 2 h with shaking. Sections were then washed three times with PBS, mounted on superfrost plus slides (Fisher), dehydrated with alcohols (50%, 75%, 95%, 100%) for 30 s each, followed by Xylene for 1 min twice, and mounted with DPX. Sections were then imaged with a fluorescence widefield microscope (THUNDER, Leica microsystems) at 5×, 10×, or 20× magnification.
To quantify infarct sizes, sections that represent different anterior–posterior coordinates, using the Paxinos atlas as a guide, were sampled and sections that contained an infract, primarily marked by an absence of NeuN staining were selected and the region of the infarct was marked with Fiji to compute area of the infarct. To compute distance from the corpus callosum, the deepest point was marked and line measurements to the corpus callosum were taken.
To quantify fluorescence intensity from expression of shCCR5 AAV, images were processed in Fiji. Briefly ROI boxes were drawn around an area of positive signal as well as no signal to measure intensity from the AAV and background fluoresce using the integrated density function in Fiji across sections from different anterior–posterior coordinates. Data were then normalized to its background fluorescence for each section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Quantification, statistics, and reproducibility
Sequence read alignment, filtering, and normalization
Paired-end reads were aligned to the mouse reference transcriptome, EnsDb.Mmusculus.v79, using Kallisto88 resulting in 7.6 ± 4.3 million uniquely mapped reads per sample. Quality control parameters such as GC bias and per sequence quality were assessed with fastQC. Count data expressed as transcripts per million (TPM) for each transcript was estimated by Kallisto and the abundance files generated were read into R using the txi import function and the EnsDb package to produce a count matrix of genes and TPMs. Using EdgeR89, counts per million (CPM) for each gene was generated and genes with less than 1 CPM in 2 samples per condition were filtered out. The total number of genes in the datasets detected following filtering were 16,662 genes for neurons and 27,830 for microglia. Normalization factors using the trimmed mean of M-values method were calculated on log2 transformed CPM data as a scaling factor for library sizes. Any batch effects from experimental day and FACS counts from samples with low events were corrected for using the removeBatchEffect function (in-built into the limma package, Table S1). The expression data are thus expressed as TMM normalized log2CPM. Samples that did not pass the quality metrics, had lower than 100 K reads per sample, had median gene expression of >|2| standard deviations in per sample expression distributions, and were outliers detected with principal component analysis were removed.
Each sample came from one animal and the total number of samples per group that were selected for downstream processing include:
Neurons
Naïve (n = 5), CCR5kd (n = 3), CREBoe (n = 5), Ctrl acute stroke (n = 5), CCR5kd acute stroke (n = 5), Ctrl chronic stroke (n = 4), CREBoe chronic stroke (n = 5), CCR5kd chronic stroke (n = 5).
Microglia
Naïve (n = 4), CCR5kd (n = 3), CREBoe (n = 2), Ctrl acute stroke (n = 5), CCR5kd acute stroke (n = 4), Ctrl chronic stroke (n = 5), CREBoe chronic stroke (n = 5), CCR5kd chronic stroke (n = 4).
For all comparisons, groups were either compared to Naïve, or, to groups that received the same viral backbone as in groups with CCR5kd or CREBoe, but lack the sequences to target these genes, also termed as associative controls.
Differential gene expression analysis
Differentially expressed genes (DEGs) were identified using linear modeling with an open-source package, limma90. Briefly, a design matrix of groups and batch information was inputted. Contrast matrices, with groups for comparisons, were either all groups compared to naïve, or pairwise comparisons between treated-control pairs. The mean–variance relationship was calculated, and a linear model was fitted to each gene. Statistical testing with the empirical Bayes method allowed for precise estimates of gene-wise variability of log-fold changes in gene expression. Differentially expressed genes from all groups compared to naïve were used for further analysis, such as to identify overlapping DEGs and activity-dependent genes and for the construction of co-expression networks. Data can be found in Supplementary Data S1.
Gene set enrichment of activity-dependent genes
Compilation of activity-dependent gene expression set : Data from 19 studies that reported differentially expressed genes during learning or exposure to a novel stimulus were selected (Supplementary Data S2). These included:
(a) Studies on learning, memory formation, and consolidation that collected RNAseq data in tagged neurons that formed the engram using activity-dependent transgenic lines. Gene expression in the ‘remote engram’ refers to genes expressed following consolidation of memory, expressed a month after learning.
(b) Studies on experience-dependent plasticity that used activity-dependent lines or synthetic activity-dependent promoters or in cultured neurons with neuronal expression of differentially regulated genes prior to and after exposure to a new stimulus to identify ERGs and LRGs.
Gene expression data from the above studies were then categorized into up and downregulated sets of engram genes, remote engram genes, ERGs and LRGs. Notably, these gene sets were not mutually exclusive. For, e.g., engram gene sets also expressed ERGs and LRGs, as well as genes unique to the engram. As these datasets were collected from studies in different brain regions, compiled datasets were then filtered for its expression in the current data set in this study. Following filtering, 3042 activity-dependent genes were detected in the current dataset.
Gene set enrichment analysis
GSEA91 was performed with gene expression data, normalized log2CPM, where every gene was averaged across samples per condition and the difference in gene expression per gene across treated and control pairs was calculated. Genes were then ranked in descending order of the difference in gene expression in treated vs control groups. Given an a priori set of activity-dependent genes, we computed if the set was randomly distributed or occupied the top or bottom of the ranked set of genes. Enrichment scores, negative or positive, were computed using a running sum statistic. Statistical significance of the enrichment scores was computed by bootstrapping with shuffled permutations of conditions 10,000 times, creating a null distribution of enrichment scores, and the p-value of the observed enrichment scores was calculated. Normalized enrichment scores (NES) were then calculated by normalizing the enrichment score for each activity-dependent gene set (e.g., engram.up and engram.dowm) to account for the size of the gene set. Multiple hypothesis testing for false discovery rates of less than 0.1 were calculated by comparing the tails of the observed and null distributions for the NES.
Linear modeling with behavioral data
Behavioral data containing deficit scores measured with grid walk were taken from published studies. Specifically data presented in Fig. 2d (ref. 25) and Fig. 2b (ref. 26). Min–max scaling was applied to all experimental groups from each dataset, for comparability on the same scale. For each of the experimental conditions time points that had both behavioral scores and normalized enrichment scores (NES) were selected and plotted. To determine the relationship between gene expression and motor deficit, a linear regression analysis was performed where enrichment scores and the different activity groups were entered as the independent variables to predict behavioral scores. R2 values adjusted for the number of variables in the model were computed, as well as the p-values for the regression coefficients. Additionally, to determine the degree of correlation, a person’s correlation was computed from the dataset with behavioral and NES scores.
Random Forest classification
Classifiers were trained with the R package, Ranger on each of the different activity-dependent gene sets. Data were split into train and test sets (80/20) with condition (i.e., Ctrl acute stroke and CCR5kd acute stroke) as the dependent variable (Supplementary Data S3). A random forest model, trained with bootstrap aggregation from 1000 to 3000 decision trees, was made from the training dataset, and the out-of-bag (OOB) error was calculated as the average of the prediction error from the trees during training. Hyperparameters such as ‘mtry’ i.e., the number of variables at each split were selected to get the least possible OOB error. The random forest model obtained was then used to predict conditions from the test dataset. The mean of the prediction error from the test data and the OOB error during training was calculated as the prediction error for that classifier. Training and testing were iterated 5 times and the data points are prediction errors from each iteration expressed as mean ± SD. To determine model performance for multiclass classification, precision, recall (sensitivity), specificity, and accuracy were calculated from confusion matrices in both training and testing data based on true positive, true negative, false positive, and false negative rates from the prediction of each of the different labels and were averaged across labels for each classifier. The data were then plotted as mean performance across all iterations with standard error.
Variational autoencoders
This method was adapted from the use of VAEs trained on The Cancer Genome Atlas48. Ten thousand most variable genes, selected by median absolute deviation in the dataset, were identified, and data were augmented by a factor of 2 by adding Gaussian noise with a mean of 0 and standard deviation of 0.1. Data were then scaled using 0–1 normalization and split into train and test data (80/20). 10,000 input genes were encoded into 200 features/encodings and reconstructed to their original dataset (decoding). Kullback–Leibler (KL) divergence was added to minimize reconstruction loss. Batch normalization was applied, and hyperparameters, including a batch size of 500, with 2000 epochs and a learning rate of 0.0002 were applied. The activation values for each encoding represent learned distributions and were used to identify differentially expressed encodings between treated and control samples. Differentially expressed encodings were calculated as the median difference in their activation scores between treated and control, and encoding with a difference of 0.5 and above was selected (Supplementary Data S4). Within each of these differentially expressed encodings, high positively or negatively weighted genes with 2 times the standard deviation were selected. Activity-dependent genes expressed within highly weighted genes for each differentially expressed encoding were identified.
To determine biologically meaningful terms that map to highly weighted genes (Supplementary Data S4), Gene Ontology analysis with g.profiler (gprofiler2) was performed. g.profiler performs overrepresentation analysis to identify statistically significant enriched terms. This allowed for identification of GO terms that map to cellular (CC), molecular function (MF) and biological processes (BP); as well as pathways from the KEGG, REACTOME, TRANSFAC and CORUM databases. For reporting GO terms from the top five differentially expressed encodings, GO terms were selected based on the youngest child term and the size of the intersection. For terms with comparable intersection sizes, terms with the lowest p values were selected. If only child terms were present, then child terms with the highest intersection were selected. In cases where there were no GO:CC terms, then pathways from REACTOME/KEGG were considered.
Gene co-expression and regulatory networks
We used a previously reported method51 to identify tight co-expression clusters of activity-dependent genes with higher co-expression in treated vs control groups (Supplementary Data S5). Briefly, k-means clustering was applied to all activity-dependent genes expressed with different k-values to produce a pool of seed clusters that are then evaluated and selected by M–N scatter plots to select large non-overlapping clusters with low dispersion. Unlike most other co-expression methods that partition the entire dataset, this allowed us to identify tightly co-expressed genes that were up in the treated condition and cluster of genes that were up in the control condition. Each cluster contained 1000–1500 genes. Connectivity between genes was computed with Pearson’s correlation and the nodes in the network were further pruned to only include genes with high correlation absolute value >0.8 and genes that are differentially expressed from DGE analysis of all groups compared to Naïve. This led to the construction of a network of highly co-expressed activity-dependent genes that are unique to post-stroke and differentially expressed.
Gene regulatory networks of transcription factor-target co-expression modules were inferred using a gradient-boost learning algorithm52,53. Genes co-expressed with transcription factors were identified and were pruned for enrichment with binding motifs near the transcriptional start site. This produces regulons of transcription factors and their co-expressed targets that are either negatively correlated (repressor regulons) or positively correlated (activator regulons) measured by Pearson product–moment correlation coefficient of their expression values. The enrichment of a regulon for that condition was determined by the area under the curve for genes expressed in the regulon. Regulons were then ranked by a regulon specificity score92 measured by Jensen–Shannon divergence with scores between 0 and 1, identifying condition-specific regulons. The top 20 regulons in each condition were identified (Supplementary Data S6 and S7). In the neuronal dataset, from the top 20, regulons with activity-dependent genes as targets were screened, and the top 5 regulons with the highest number of activity-dependent genes as targets were plotted. For the microglia dataset, target genes were mapped to Gene Ontology terms with g.profiler.
Ligand–receptor interactions
Ligand–receptor interactions between neurons and microglia were inferred with CellChat70. Briefly, over-expressed ligand–receptors across neurons and microglia were identified, and communication probabilities between the two cell types were computed. Probability values were assigned to an interaction by integrating gene expression data and known interactions of ligands with their receptors and co-factors. Gene expression per condition with a trimmed mean of 25% was used for calculating the number of ligand–receptor interactions. Functional clustering and manifold learning for signaling pathways were calculated using functions within CellChat (Supplementary Data S7).
Supplementary information
Description of additional supplementary files
Acknowledgements
These studies were funded by Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (S.T.C) and JAX—institutional start-up funds (M.T.J.). We thank Dr. Xinmin Li and the UCLA Technology Center for Genomics and Bioinformatics core for library preparation and RNA sequencing and Felicia Codrea, Jessica Scholes, and Jeff Calimlim from the UCLA BSCRC FACS core for fluorescence activated cell sorting of all samples used in this study.
Author contributions
S.T.C. and M.T.J. conceived the project and designed experiments. M.T.J. conducted experiments and collected data. M.T.J. and S.T.C. analyzed and interpreted data and wrote the paper.
Peer review
Peer review information
Communications Biology thanks Rajkumar Verma, Richard Kopchok III, and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Mireya Plass and David Favero. A peer review file is available.
Data availability
RNAseq data with metadata are deposited in GEO (GEO accession: GSE270766). All source data for the figures are available in Supplementary Data S1–S8. All other data are available from the corresponding author (or other sources, as applicable) on reasonable request.
Code availability
Source packages have been listed in the resources table—Table S1, Supplementary Information—and code can be made available on reasonable request.
Competing interests
The authors declare no competing interests. Mary T. Joy is an Editorial Board Member for Communications Biology but was not involved in the editorial review of nor the decision to publish this article.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06723-3.
References
- 1.Roemmich, R. T. & Bastian, A. J. Closing the loop: from motor neuroscience to neurorehabilitation. Annu. Rev. Neurosci.41, 415–429 (2018). 10.1146/annurev-neuro-080317-062245 [DOI] [PubMed] [Google Scholar]
- 2.Dimyan, M. A. & Cohen, L. G. Neuroplasticity in the context of motor rehabilitation after stroke. Nat. Rev. Neurol.7, 76–85 (2011). 10.1038/nrneurol.2010.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwakkel, G., Veerbeek, J. M., van Wegen, E. E. & Wolf, S. L. Constraint-induced movement therapy after stroke. Lancet Neurol.14, 224–234 (2015). 10.1016/S1474-4422(14)70160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livingston-Thomas, J. et al. Exercise and environmental enrichment as enablers of task-specific neuroplasticity and stroke recovery. Neurotherapeutics13, 395–402 (2016). 10.1007/s13311-016-0423-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward, N. S., Brander, F. & Kelly, K. Intensive upper limb neurorehabilitation in chronic stroke: outcomes from the Queen Square programme. J. Neurol. Neurosurg. Psychiatry90, 498–506 (2019). 10.1136/jnnp-2018-319954 [DOI] [PubMed] [Google Scholar]
- 6.Laver, K. E. et al. Virtual reality for stroke rehabilitation. Cochrane Database Syst. Rev.11, CD008349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krakauer, J. W. et al. Comparing a novel neuroanimation experience to conventional therapy for high-dose intensive upper-limb training in subacute stroke: the SMARTS2 randomized trial. Neurorehabil. Neural Repair35, 393–405 (2021). 10.1177/15459683211000730 [DOI] [PubMed] [Google Scholar]
- 8.Grefkes, C. & Fink, G. R. Connectivity-based approaches in stroke and recovery of function. Lancet Neurol.13, 206–216 (2014). 10.1016/S1474-4422(13)70264-3 [DOI] [PubMed] [Google Scholar]
- 9.Liew, S. L. et al. The ENIGMA Stroke Recovery Working Group: big data neuroimaging to study brain-behavior relationships after stroke. Hum. Brain Mapp.43, 129–148 (2022). 10.1002/hbm.25015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, A. C. et al. Hotspots of dendritic spine turnover facilitate clustered spine addition and learning and memory. Nat. Commun.9, 422 (2018). 10.1038/s41467-017-02751-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen, Y. et al. CCR5 closes the temporal window for memory linking. Nature606, 146–152 (2022). 10.1038/s41586-022-04783-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou, M. et al. CCR5 is a suppressor for cortical plasticity and hippocampal learning and memory. Elife5, e20985 (2016). [DOI] [PMC free article] [PubMed]
- 13.Sano, Y. et al. CREB regulates memory allocation in the insular cortex. Curr. Biol.24, 2833–2837 (2014). 10.1016/j.cub.2014.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou, Y. et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat. Neurosci.12, 1438–1443 (2009). 10.1038/nn.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josselyn, S. A. & Tonegawa, S. Memory engrams: Recalling the past and imagining the future. Science367, eaaw4325 (2020). [DOI] [PMC free article] [PubMed]
- 16.Chen, M. B., Jiang, X., Quake, S. R. & Sudhof, T. C. Persistent transcriptional programmes are associated with remote memory. Nature587, 437–442 (2020). 10.1038/s41586-020-2905-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marco, A. et al. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci.23, 1606–1617 (2020). 10.1038/s41593-020-00717-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao-Ruiz, P. et al. Engram-specific transcriptome profiling of contextual memory consolidation. Nat. Commun.10, 2232 (2019). 10.1038/s41467-019-09960-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho, J. H., Huang, B. S. & Gray, J. M. RNA sequencing from neural ensembles activated during fear conditioning in the mouse temporal association cortex. Sci. Rep.6, 31753 (2016). 10.1038/srep31753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, Y. et al. Class IIa HDACs regulate learning and memory through dynamic experience-dependent repression of transcription. Nat. Commun.10, 3469 (2019). 10.1038/s41467-019-11409-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yap, E. L. & Greenberg, M. E. Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron100, 330–348 (2018). 10.1016/j.neuron.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardinly, A. R. et al. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature531, 371–375 (2016). 10.1038/nature17187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloodgood, B. L., Sharma, N., Browne, H. A., Trepman, A. Z. & Greenberg, M. E. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature503, 121–125 (2013). 10.1038/nature12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, M. et al. A specific requirement of Arc/Arg3.1 for visual experience-induced homeostatic synaptic plasticity in mouse primary visual cortex. J. Neurosci.30, 7168–7178 (2010). 10.1523/JNEUROSCI.1067-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joy, M. T. et al. CCR5 is a therapeutic target for recovery after stroke and traumatic brain injury. Cell176, 1143–1157.e1113 (2019). 10.1016/j.cell.2019.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caracciolo, L. et al. CREB controls cortical circuit plasticity and functional recovery after stroke. Nat. Commun.9, 2250 (2018). 10.1038/s41467-018-04445-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assayag, E. B. et al. Preventing post-stroke dementia. The MARCH Trial. Protocol and statistical analysis plan of a randomized clinical trial testing the safety and efficacy of Maraviroc in post-stroke cognitive impairment. Eur. Stroke J.7, 314–322 (2022). 10.1177/23969873221098857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hrvatin, S. et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci.21, 120–129 (2018). 10.1038/s41593-017-0029-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheadle, L. et al. Visual experience-dependent expression of Fn14 is required for retinogeniculate refinement. Neuron99, 525–539 e510 (2018). 10.1016/j.neuron.2018.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyssowski, K. M. et al. Different neuronal activity patterns induce different gene expression programs. Neuron98, 530–546 e511 (2018). 10.1016/j.neuron.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatzi, C. et al. Exercise-induced enhancement of synaptic function triggered by the inverse BAR protein, Mtss1L. Elife10.7554/eLife.45920 (2019). [DOI] [PMC free article] [PubMed]
- 32.Lacar, B. et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun.7, 11022 (2016). 10.1038/ncomms11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeger, B. N. et al. A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun.9, 3084 (2018). 10.1038/s41467-018-05418-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap, E. L. et al. Bidirectional perisomatic inhibitory plasticity of a Fos neuronal network. Nature590, 115–121 (2021). 10.1038/s41586-020-3031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tepe, B. et al. Single-cell RNA-seq of mouse olfactory bulb reveals cellular heterogeneity and activity-dependent molecular census of adult-born neurons. Cell Rep.25, 2689–2703.e2683 (2018). 10.1016/j.celrep.2018.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garay, P. M. et al. RAI1 regulates activity-dependent nascent transcription and synaptic scaling. Cell Rep.32, 108002 (2020). 10.1016/j.celrep.2020.108002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel, I. et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell157, 1216–1229 (2014). 10.1016/j.cell.2014.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha, R. N. et al. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci.14, 848–856 (2011). 10.1038/nn.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madabhushi, R. et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell161, 1592–1605 (2015). 10.1016/j.cell.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sando, R. et al. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell151, 821–834 (2012). 10.1016/j.cell.2012.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akins, P. T., Liu, P. K. & Hsu, C. Y. Immediate early gene expression in response to cerebral ischemia. Friend or foe? Stroke27, 1682–1687 (1996). 10.1161/01.STR.27.9.1682 [DOI] [PubMed] [Google Scholar]
- 42.Park, A. et al. A time-dependent role for the transcription factor CREB in neuronal allocation to an engram underlying a fear memory revealed using a novel in vivo optogenetic tool to modulate CREB function. Neuropsychopharmacology45, 916–924 (2020). 10.1038/s41386-019-0588-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han, J. H. et al. Neuronal competition and selection during memory formation. Science316, 457–460 (2007). 10.1126/science.1139438 [DOI] [PubMed] [Google Scholar]
- 44.Androvic, P. et al. Decoding the transcriptional response to ischemic stroke in young and aged mouse brain. Cell Rep.31, 107777 (2020). 10.1016/j.celrep.2020.107777 [DOI] [PubMed] [Google Scholar]
- 45.Bonkhoff, A. K. & Grefkes, C. Precision medicine in stroke: towards personalized outcome predictions using artificial intelligence. Brain145, 457–475 (2022). 10.1093/brain/awab439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Churchland, M. M., Cunningham, J. P., Kaufman, M. T., Ryu, S. I. & Shenoy, K. V. Cortical preparatory activity: representation of movement or first cog in a dynamical machine? Neuron68, 387–400 (2010). 10.1016/j.neuron.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandarinath, C. et al. Latent factors and dynamics in motor cortex and their application to brain-machine interfaces. J. Neurosci.38, 9390–9401 (2018). 10.1523/JNEUROSCI.1669-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Way, G. P. & Greene, C. S. Extracting a biologically relevant latent space from cancer transcriptomes with variational autoencoders. Pac. Symp. Biocomput.23, 80–91 (2018). [PMC free article] [PubMed] [Google Scholar]
- 49.Seninge, L., Anastopoulos, I., Ding, H. & Stuart, J. VEGA is an interpretable generative model for inferring biological network activity in single-cell transcriptomics. Nat. Commun.12, 5684 (2021). 10.1038/s41467-021-26017-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart, J. M., Segal, E., Koller, D. & Kim, S. K. A gene-coexpression network for global discovery of conserved genetic modules. Science302, 249–255 (2003). 10.1126/science.1087447 [DOI] [PubMed] [Google Scholar]
- 51.Abu-Jamous, B. & Kelly, S. Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol.19, 172 (2018). 10.1186/s13059-018-1536-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE10.1371/journal.pone.0012776 (2010). [DOI] [PMC free article] [PubMed]
- 54.Li, S. et al. An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat. Neurosci.13, 1496–1504 (2010). 10.1038/nn.2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akiyoshi, R. et al. Microglia enhance synapse activity to promote local network synchronization. eNeuro10.1523/ENEURO.0088-18.2018 (2018). [DOI] [PMC free article] [PubMed]
- 56.Badimon, A. et al. Negative feedback control of neuronal activity by microglia. Nature586, 417–423 (2020). 10.1038/s41586-020-2777-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cserep, C. et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science367, 528–537 (2020). 10.1126/science.aax6752 [DOI] [PubMed] [Google Scholar]
- 58.Favuzzi, E. et al. GABA-receptive microglia selectively sculpt developing inhibitory circuits. Cell184, 5686 (2021). 10.1016/j.cell.2021.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parkhurst, C. N. et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell155, 1596–1609 (2013). 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schafer, D. P. et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron74, 691–705 (2012). 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clarkson, A. N., Huang, B. S., Macisaac, S. E., Mody, I. & Carmichael, S. T. Reducing excessive GABA-mediated tonic inhibition promotes functional recovery after stroke. Nature468, 305–309 (2010). 10.1038/nature09511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kraft, A. W., Bauer, A. Q., Culver, J. P. & Lee, J. M. Sensory deprivation after focal ischemia in mice accelerates brain remapping and improves functional recovery through Arc-dependent synaptic plasticity. Sci. Transl. Med.10.1126/scitranslmed.aag1328 (2018). [DOI] [PubMed]
- 63.Festa, B. P. et al. Microglial-to-neuronal CCR5 signaling regulates autophagy in neurodegeneration. Neuron10.1016/j.neuron.2023.04.006 (2023). 10.1016/j.neuron.2023.04.006 [DOI] [PubMed] [Google Scholar]
- 64.Gao, Y. et al. Microglia CREB-Phosphorylation Mediates Amyloid-beta-Induced Neuronal Toxicity. J. Alzheimers Dis.66, 333–345 (2018). 10.3233/JAD-180286 [DOI] [PubMed] [Google Scholar]
- 65.Sanguino-Gomez, J. et al. An emerging role for microglia in stress-effects on memory. Eur. J. Neurosci.55, 2491–2518 (2022). 10.1111/ejn.15188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartolotti, N. & Lazarov, O. CREB signals as PBMC-based biomarkers of cognitive dysfunction: a novel perspective of the brain-immune axis. Brain Behav. Immun.78, 9–20 (2019). 10.1016/j.bbi.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sobue, A. et al. Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol. Commun.9, 1 (2021). 10.1186/s40478-020-01099-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chitu, V. et al. Microglial homeostasis requires balanced CSF-1/CSF-2 receptor signaling. Cell Rep.30, 3004–3019.e3005 (2020). 10.1016/j.celrep.2020.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosito, M. et al. Microglia reactivity entails microtubule remodeling from acentrosomal to centrosomal arrays. Cell Rep.42, 112104 (2023). 10.1016/j.celrep.2023.112104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poplawski, G. H. D. et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nature581, 77–82 (2020). 10.1038/s41586-020-2200-5 [DOI] [PubMed] [Google Scholar]
- 72.d’Aquin, S. et al. Compartmentalized dendritic plasticity during associative learning. Science376, eabf7052 (2022). 10.1126/science.abf7052 [DOI] [PubMed] [Google Scholar]
- 73.Hollenhorst, P. C., McIntosh, L. P. & Graves, B. J. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem.80, 437–471 (2011). 10.1146/annurev.biochem.79.081507.103945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fontanet, P., Irala, D., Alsina, F. C., Paratcha, G. & Ledda, F. Pea3 transcription factor family members Etv4 and Etv5 mediate retrograde signaling and axonal growth of DRG sensory neurons in response to NGF. J. Neurosci.33, 15940–15951 (2013). 10.1523/JNEUROSCI.0928-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Besnard, A., Galan-Rodriguez, B., Vanhoutte, P. & Caboche, J. Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci.5, 35 (2011). 10.3389/fnins.2011.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Briguet, A. & Ruegg, M. A. The Ets transcription factor GABP is required for postsynaptic differentiation in vivo. J. Neurosci.20, 5989–5996 (2000). 10.1523/JNEUROSCI.20-16-05989.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaeffer, L., de Kerchove d’Exaerde, A. & Changeux, J. P. Targeting transcription to the neuromuscular synapse. Neuron31, 15–22 (2001). 10.1016/S0896-6273(01)00353-1 [DOI] [PubMed] [Google Scholar]
- 78.Shichita, T. et al. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat. Med.23, 723–732 (2017). 10.1038/nm.4312 [DOI] [PubMed] [Google Scholar]
- 79.Corbett, D. et al. Enhancing the alignment of the preclinical and clinical stroke recovery research pipeline: consensus-based core recommendations from the stroke recovery and rehabilitation roundtable translational working group. Neurorehabil. Neural Repair31, 699–707 (2017). 10.1177/1545968317724285 [DOI] [PubMed] [Google Scholar]
- 80.Joy, M. T. & Carmichael, S. T. Encouraging an excitable brain state: mechanisms of brain repair in stroke. Nat. Rev. Neurosci.22, 38–53 (2021). 10.1038/s41583-020-00396-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mischa V. & Bandet, I. R. W. Aberrant cortical activity, functional connectivity, and neural assembly architecture after photothrombotic stroke in mice. eLife12, RP90080 (2023). [DOI] [PMC free article] [PubMed]
- 82.Ganguly, K., Khanna, P., Morecraft, R. J. & Lin, D. J. Modulation of neural co-firing to enhance network transmission and improve motor function after stroke. Neuron110, 2363–2385 (2022). 10.1016/j.neuron.2022.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo, L., Kondapavulur, S., Lemke, S. M., Won, S. J. & Ganguly, K. Coordinated increase of reliable cortical and striatal ensemble activations during recovery after stroke. Cell Rep.36, 109370 (2021). 10.1016/j.celrep.2021.109370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bennett, M. L. et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA113, E1738–E1746 (2016). 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zrzavy, T. et al. Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain140, 1900–1913 (2017). 10.1093/brain/awx113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mercurio, D. et al. Protein expression of the microglial marker Tmem119 decreases in association with morphological changes and location in a mouse model of traumatic brain injury. Front. Cell Neurosci.16, 820127 (2022). 10.3389/fncel.2022.820127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kenkhuis, B. et al. Co-expression patterns of microglia markers Iba1, TMEM119 and P2RY12 in Alzheimer’s disease. Neurobiol. Dis.167, 105684 (2022). 10.1016/j.nbd.2022.105684 [DOI] [PubMed] [Google Scholar]
- 88.Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol.34, 525–527 (2016). 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 89.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47 (2015). 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suo, S. et al. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep.25, 1436–1445.e1433 (2018). 10.1016/j.celrep.2018.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
RNAseq data with metadata are deposited in GEO (GEO accession: GSE270766). All source data for the figures are available in Supplementary Data S1–S8. All other data are available from the corresponding author (or other sources, as applicable) on reasonable request.
Source packages have been listed in the resources table—Table S1, Supplementary Information—and code can be made available on reasonable request.