Summary
Conventional type 1 dendritic cells (cDC1s) are critical for innate sensing of cancer, yet they are scarce in the tumor microenvironment (TME). Here, we present a protocol to identify and isolate cDC1 subsets from murine implantable tumors for subsequent transcriptomic profiling using a flow sorting-based strategy. We describe steps for cell culture of mouse tumors, tumoral growth, dissociation and isolation of tumoral cells, extracellular staining, and cell sorting. We then detail procedures for RNA isolation, mRNA library preparation, and sequencing.
For complete details on the use and execution of this protocol, please refer to Papadas et al.1
Subject areas: Cell Biology, Cell culture, Cell isolation, Flow Cytometry, Cancer, Sequencing, RNA-seq, Immunology, Model Organisms, Molecular Biology
Graphical abstract

Highlights
-
•
Protocol to identify and isolate rare intratumoral dendritic cells (DCs)
-
•
Tissue dissociation and preparation of single-cell suspensions from diverse tumor types
-
•
Flow-based enumeration of tumor-resident and migration-prone DC subsets
-
•
Low-input library construction and RNA-seq
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Conventional type 1 dendritic cells (cDC1s) are critical for innate sensing of cancer, yet they are scarce in the tumor microenvironment (TME). Here, we present a protocol to identify and isolate cDC1 subsets from murine implantable tumors for subsequent transcriptomic profiling using a flow sorting-based strategy. We describe steps for cell culture of mouse tumors, tumoral growth, dissociation and isolation of tumoral cells, extracellular staining, and cell sorting. We then detail procedures for RNA isolation, mRNA library preparation, and sequencing.
Before you begin
Timing: 1–5 h
In this protocol, we expand on our previously published procedures for tumor-resident dendritic cell (DC) identification, isolation by flow-sorting, RNA extraction, and library preparation for sequencing.1 This protocol delineates our cumulative experience in isolating, manipulating and profiling rare intratumoral DC subsets and refines our prior approaches by characterizing DC according to their maturation status/migratory propensity (tumor-resident non-migratory CCR7neg and tumor-resident, migratory-prone CCR7pos mregDC1). While we optimized this protocol using Lewis Lung Carcinoma (LLC) implantable, immunocompetent tumors, this protocol has been also tested in MC38 murine colon adenocarcinoma, CT26 murine colorectal carcinoma, 4T1 murine mammary cell carcinoma and B16 murine melanoma.
-
1.
Before you begin, make sure that you have institutional permissions and remain compliant with guidelines outlined by your local Institutional Animal Care and Use Committee (IACUC).
-
2.
Validate cell lines at least every 6 months and perform regular mycoplasma testing.
-
3.
Make sure that your injection techniques are reliable and produce consistent tumor growth.
-
4.
Maintain RNase-free environments when preparing RNA for sequencing.
Note: During cell culture, make sure you follow sterile techniques. Prepare culture media, harvest and passage cells under BSL-2 level conditions.
Note: For consistent and reproducible results in subsequent in vivo tumor growth kinetics, make sure your cell culture media are freshly prepared and not more than a month old when stored at 4°C.
Institutional permissions
All animal procedures in this protocol were approved by and performed according to the Guide for Care and Use of Laboratory Animals (NIH Publication 86–23) under IACUC-approved protocol #S19109 in the University of California, San Diego (UCSD).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| B220 PE (RA3-6B2) | BioLegend | Cat#103208 |
| CCR7 APC (4B12) | BioLegend | Cat#120108 |
| CD11b BV605 (M1/70) | BioLegend | Cat#101257 |
| CD11c Pacific Blue (N418) | BioLegend | Cat#117322 |
| CD3 FITC (17A2) | BioLegend | Cat#100204 |
| CD45 PE-Cy7 (30-F11) | BioLegend | Cat#103114 |
| MHC II Alexa 700 (M5/114.15.2) | BioLegend | Cat#107622 |
| TruStain Fc PLUS (anti-mouse CD16/32) (S17011E) | BioLegend | Cat#156604 |
| Chemicals, peptides, and recombinant proteins | ||
| 0.25% Trypsin-EDTA (1X) | Gibco | Cat#25200-56 |
| 2-Mercaptoethanol | Gibco | Cat#21985-023 |
| ACK lysing buffer | Quality Biological | Cat#118-156-721 |
| Antibiotic-antimycotic solution | Corning | Cat#30-004-CI |
| Dulbecco’s modified Eagle’s medium (DMEM) DMEM medium | Corning | Cat#10-013-CV |
| Ghost Dye Red 780 Live/Dead | Tonbo Biosciences | Cat#13-0865-T100 |
| GlutaMAX | Gibco | Cat#35050-061 |
| Heat activated fetal bovine serum one shot | Gibco | Cat#A38400-1 |
| HEPES buffer 1 M | Gibco | Cat#15630-080 |
| PBS (1X) | Corning | Cat#21-040-CV |
| Precision Count Beads | BioLegend | Cat#424902 |
| TRIzol reagent | Invitrogen | Cat#15596026 |
| Trypan blue stain (0.4%) | Gibco | Cat#15250-061 |
| UltraComp eBeads | Invitrogen | Cat#01-2222-41 |
| UltraPure 0.5 M EDTA, pH 8.0 | Invitrogen | Cat#15575020 |
| Critical commercial assays | ||
| Mouse tumor dissociation kit | Miltenyi Biotec | Cat#130-096-730 |
| Qubit 1X dsDNA High Sensitivity (HS) Assay Kit | Invitrogen | Q33230 |
| RNeasy Micro Kit | QIAGEN | Cat#74004 |
| Ultra-Low Input RNAseq cDNA prep (SMART-seq) kit | Takara Bio | Cat#634768 |
| Experimental models: Cell lines | ||
| MC38 Colon adenocarcinoma | Kerafast | Cat#ENH204-FP |
| CT26 Colon carcinoma | ATCC | Cat#CRL-2638 |
| Lewis lung carcinoma (LLC) | ATCC | Cat#CRL-1642 |
| B16F10 Melanoma | ATCC | Cat#CRL-6322 |
| 4T1 breast carcinoma | ATCC | Cat#CRL-2359 |
| Experimental models: Organisms/strains | ||
| 6- to 8-week-old male C57BL/6J mice | The Jackson Laboratory | Cat#000664 |
| Software and algorithms | ||
| BioRender | BioRender | https://www.biorender.com/ |
| GraphPad Prism version: 9.3.1 | Dotmatics | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo 10.7.1 | Tree Star | https://www.flowjo.com/ |
| Other | ||
| 100 mm tissue culture dish | Thermo Fisher Scientific | Cat#1301182 |
| 15 mL polystyrene conical tube | Falcon | Cat#352196 |
| 4°C Refrigerator | N/A | N/A |
| 4200 TapeStation System | Agilent | N/A |
| 50 mL polystyrene conical tube | Falcon | Cat#352070 |
| 5 mL polystyrene round-bottom tube | Falcon | Cat#352235 |
| BD 28G insulin syringe | BD | Cat#329461 |
| BD FACS Aria II, or alternative | BD Biosciences | N/A |
| Dual-chamber cell counting slides | Bio-Rad | Cat#145-0011 |
| Forceps | N/A | N/A |
| GentleMACS C tube | Miltenyi Biotec | Cat#130-093-237 |
| GentleMACS Octo dissociator with heaters | Miltenyi Biotec | Cat#130-096-427 |
| Illumina NextSeq 2000 platform | Illumina | N/A |
| Olympus PCR tubes 0.2 mL 8-tube strips | Genesee Scientific | Cat#27-125U |
| Pipettes P2, P20, P100, P200, and P1000 Kit | Gilson | Cat#F167360 |
| Qubit 4 Fluorometer | Invitrogen | N/A |
| Razor electric | Braun | N/A |
| Scissors | N/A | N/A |
| TipOne RPT filter tips | USA Scientific | Cat#1181 |
| Tissue-culture centrifuge (e.g., Sorvall X Pro Series) | Thermo Fisher Scientific | N/A |
Materials and equipment
-
•
Miltenyi mouse tumor dissociation kit (Miltenyi Biotec catalog #130-096-730) is recommended for tumor digestion. Lyophilized enzymes need to be reconstituted as shown in the following table and scaled up as required.
Note: Alternatively, if researchers do not have access to the Miltenyi kit, tumors fragmented in small pieces can be digested using a mixture of collagenase type IA (1 mg/mL) (Sigma, Cat C9891) and hyaluronidase type V (0.1 mg/mL) (Sigma, Cat. H6254) for 4–6 at 25°C as it is described in Waight et al.2
Tumor dissociation reagent reconstitution
| Reagent (1 vial) | Enzyme D | Enzyme R | Enzyme A |
|---|---|---|---|
| DMEM (mL) | 3 | 2.7 | – |
| Buffer A (mL) | – | – | 1 |
Prepare aliquots of appropriate volume to avoid repeated freeze-thaw cycles.
Tumor dissociation working solution (enzyme mix/each tumor)
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM (mL) | 1 X | 2.35 |
| Enzyme D (μL) | 4% | 100 |
| Enzyme R (μL) | 2% | 50 |
| Enzyme A (μL) | 0.5% | 12.5 |
Freshly prepare enzyme mix with each tumor in the gentleMACS Tube.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS (mL) | 1 X | 487.5 |
| FBS (mL) | 2% | 10 |
| 0.5 M EDTA, pH 8.0 | 2.5 mM | 2.5 |
Step-by-step method details
Thawing cryopreserved tumor cells- day 0
Timing: 30–45 min
This step describes the preparation of cell culture medium, thawing of cryopreserved tumor cells and cell plating in tissue-culture dishes.
-
1.
Generate and pre-warm medium (complete DMEM medium: 10–013 CV Corning DMEM with 10% fetal bovine serum, 50 mM 2-mercaptoethanol, 100 U/mL Penicillin, 100 mg/mL Streptomycin, 292 μg/mL L-Glutamine).
-
2.
Retrieve vial of cryopreserved Lewis Lung Carcinoma (LLC, ATCC CRL-1642) from long term storage in liquid N2.
-
3.
Rapidly thaw frozen cells in 37°C water bath (1–2 min).
-
4.
Add cell suspension from vial dropwise to a conical tube containing 10 mL of warmed media.
-
5.
Spin down cells for 5 min at 300 × g; carefully aspirate supernatant.
-
6.
Add 8–10 mL of pre-warmed media and plate into 10 cm2 sterile tissue culture-treated dish.
Note: Initial plating density of LLC cells is 0.5–1 million cells per 10 cm plate, however, be mindful that other tumor cell lines cells may be sensitive to alternative plating densities and may display different rates of expansion; thus follow the specific originating investigator or vendor’s instructions regarding recommended culture conditions.
Culturing and passaging tumor cell lines- days 0-2
Timing: 1–3 days
This step describes the culturing and passaging of tumor cell lines under sterile conditions prior to in vivo inoculation.
-
7.
Monitor cells until 60%–80% confluency has been reached, replacing cell medium if necessary.
Note: Initial plating density (high vs low) will impact the duration of this step. As mentioned above, the number of cells seeded will determine the time needed to reach 60%–80% confluency in the culture dish.
-
8.
Aspirate spent media.
-
9.
Gently wash cells with appropriate volume of endotoxin-free PBS.
-
10.
Aspirate PBS.
-
11.
Add enough 0.25% trypsin solution to cover the tissue culture dish and incubate dish for 1–2 min at 37°C until cells have detached.
Note: Alternatively, a solution of accutase 400–600 units/mL could be used. In fact, it would be highly recommended to try accutase, aiming to reduce cell stress and preserve cell viability. However, cell lines with high adherent capacity could be quite resistant to accutase. For example, 4T1 cells are expected to show resistance to accutase and detachment process could be too long (thus affecting viability) or inefficient. Accutase may be used for B16 melanoma cells as long as incubation occurs for 10 min at 25°C instead of 37°C.
-
12.
Neutralize trypsin with an equal volume of FBS-containing cell culture medium.
-
13.
Collect cells and spin down for 5 min at 300 × g using Sorvall X Pro Series (Thermo Fisher) centrifuge; carefully pour off supernatant.
-
14.
Divide cell suspension to desired concentration, add media, and re-plate cells.
CRITICAL: Passage your cells at least twice before harvesting to ensure appropriate growth kinetics.
Note: Calculate the number of cells you will need for your in vivo experiments at this stage and plan to have an excess of 2–4 million to account for losses during harvesting and centrifugation.
Harvesting tumor cells for injection-days 3–4
Timing: 30–60 min
This step describes the harvesting of tumor cells from tissue culture treated plates in preparation for in vivo inoculation.
-
15.
On day of in vivo inoculation, tissue culture plates should be approximately 70%–90% confluent.
-
16.
Repeat steps 8–13.
-
17.
Resuspend cells in PBS.
-
18.
Count number of cells in suspension.
Note: We recommend counting the cells 2x then averaging the 2 counts to ensure consistency and avoid errors.
-
19.
Spin down cells for 5 min at 300 × g.
-
20.
Resuspend cells in PBS to reach a final concentration of 5×105 cells/100 μL PBS.
Note: The use of PBS in combination with Matrigel Matrix basement membrane at a 1:1 can improve subcutaneous tumor engraftment. However, Matrigel was not necessary in our hands when working with the aforementioned tumor models.
CRITICAL: Matrigel when used should always be kept at 4°C. Transport only on ice.
CRITICAL: As you resuspend the cells in PBS, make sure PBS is pre-chilled at 4°C to maintain cell viability during transportation and the subsequent potentially lengthy inoculation process, depending on the number of recipient animals.
Subcutaneous injection of tumor cells- days 3–4
Timing: 2–4 h
This step describes the subcutaneous inoculation of tumor cells in syngeneic mice recipients under general anesthesia utilizing isoflurane.
-
21.
Induce and maintain general anesthesia in recipient mice through inhalation using isoflurane.
Note: For anesthesia induction, dial the oxygen flow rate to 2 L/min and set the isoflurane flow at 4%. For anesthesia maintenance, turn the isoflurane rate down to 2%.
CRITICAL: Most animals will become unconscious within 1–2 min following exposure to the isoflurane inhalant. Therefore, animals should be constantly monitored to guard against overdose.
-
22.
Prepare tumor cell injections using 28G needle insulin syringe containing 100 μL of tumor cell suspension. Make sure no bubbles are present in the injectable solution.
Note: It is useful to use 1.5 mL Eppendorf tubes for your single cell suspensions, as it allows easier access for the syringe, as well as better handling when it comes to flicking the tube prior to aspiration of the solution by your syringe.
CRITICAL: LLC cell suspensions are rather viscous, thus cell suspensions may reform a pellet while in solution (particularly when >3×106 cells). In this situation, make sure you pipet up down and flick gently the tube multiple times to ensure appropriate re-suspension.
-
23.
Use a shaver to remove hair in the desired area of injections. The most common subcutaneous injection sites are the right or left flank.
-
24.
Introduce needle under the skin of the desired area without penetrating peritoneal space and inject 100 μL of cell suspension slowly (Figure 1).
Note: After injection, remove the needle slowly to avoid leaking of the cells and pinch injection site with forceps for 30 s to avoid potential cell leakage.
Note: Forceps can be used to lift the skin for needle access.
Note: The formation of a palpable “papule” beneath the skin indicates successful subcutaneous inoculation (Figure 1).
-
25.
Maintain mice in standard housing conditions during tumor development; growth kinetics of the grafts depend on the nature and number of the cells injected. Typically, engrafted tumors are visible, palpable and can be monitored starting around Day 3 post-implantation. At this point, engrafted tumors are roughly 25 mm3 in volume. For this protocol, 21 days is generally enough for average tumor growth (approximately 650 mm3).
Note: Monitor the tumor-bearing mice at least every other day to ensure clinical stability and appropriate tumor growth kinetics, as well as to observe mouse health including any drop in body weight. If a mouse fails to grow a subcutaneous tumor or you feel that the tumor may be invading the peritoneal layer and/or it can be palpated intra-abdominally, it is prudent to exclude that animal from the cohort.
Figure 1.

Tumor inoculation through subcutaneous injection in the mouse flank
Needle must be introduced under the skin at a 45-degree angle avoiding peritoneal membrane perforation.
Harvesting tumors- days 24–25
Timing: 2–4 h
This step describes the process of mouse euthanasia and tumor harvesting prior to enzymatic and mechanical digestion using the gentleMACS dissociator.
-
26.
Euthanize mice using a CO2 chamber in line with institutional guidelines.
Note: Other approved modes of euthanasia including intraperitoneal sodium pentobarbital may be used in accordance with specific institutional policies and guidelines.
-
27.
Harvest tumors using forceps and surgical scissors and place in a 3 × 3 cm Petri dish filled with cold sterile PBS resting on ice.
Note: Avoid harvesting hair, fat, or necrotic tumor tissue during tumor harvest. Forceps can be used to gently separate non-tumor tissue.
-
28.
Place tumors in 2.35 mL of DMEM media containing 100 μL enzyme D, 50 μL enzyme R and 12.5 μL enzyme A per Miltenyi Biotec Kit #130-096-730 instructions (enzymes should be prepared by reconstitution of the lyophilized powder with 3 mL of DMEM according to manufacturer instructions). Enzyme solutions can be stored at −20°C for up to 6 months.
Note: Either DMEM or RPMI is compatible with the experimental models utilized here.
Note: For tumor types generating less viscous suspensions than LLC, enzyme R may be used at 20% of the aforementioned amount to preserve critical cell surface epitopes.
Note: It is useful to make aliquots (in 1.5 mL Eppendorf tubes) of enzyme stocks A, D and R in advance. Multiple freeze-thaw cycles may compromise enzyme activity and should be avoided.
Note: From this point onward, keep all containers on ice to preserve cell viability.
-
29.
Transfer tumors to the designated gentleMACS C tubes and cut tumors in small pieces using fine surgical scissors while being in DMEM/enzyme replete buffer to facilitate digestion.
CRITICAL: Make sure tumor pieces are small enough (2–4 mm) for complete digestion.
-
30.
Place tumors in the Miltenyi tumor dissociator and digest them at 37°C using the “37C_m_TDK-2” protocol for 42 min.
CRITICAL: When placed the tube C into the gentleMACS Octo Dissociator, it has to be ensured that the sample material is located in the area of the rotor/stator.
Note: The machine has 8 available spaces for tumor digestion, if more tumors need to be processed, they can be kept at 4°C or even 25°C in PBS. We highly recommended to euthanize mice in groups of up to 8 mice. Tumor cell suspensions should be kept at 4°C.
-
31.
Post-digestion, detach the C tube from the gentleMACS dissociator.
-
32.
Filter the digested tumor cell suspension through a cell strainer (70 μm) into a 50 mL conical tube.
CRITICAL: If the tumor cell suspension is still viscous and hard to pipet after passing through the filter, you may repeat the straining once more. LLC tumor suspensions are especially viscous, thus proper cell straining is key to successful completion of the experiment. If the suspension remains stubbornly viscous, a syringe plug can be used to mash up cell suspension and facilitate the straining.
-
33.
Centrifuge at 300 × g for 7 min at 4°C.
-
34.
After aspirating the supernatant, apply RBC lysis (ACK Lysing) buffer (1–2 mL, depending on tumor size) for 2 min at 25°C. Use an additional 20–30 mL PBS to top up the suspension.
Note: In the case of hypervascular tumors, you may still visualize a red cell pellet after one round of RBC lysis. You may repeat the RBC lysis step. Be mindful that additional RBC lysis may have a negative impact on cell viability, thus a maximum of 2x RBC lysis rounds is recommended.
-
35.
Centrifuge again at 300 × g for 7 min and resuspend in convenient volume prior to cell count.
Note: Alternatively, you may use “37C_m_TDK-1” dissociation program on the gentleMACS instrument for “softer” (less desmoplastic) tumors.
Preparation for FACS analysis and FACS cell sorting day 25
Timing: 4–6 h
This step describes tumor cell surface staining using fluorescently-labeled antibodies and viability dye prior to DC subset flow-sorting.
(Before proceeding with staining, review step 46 regarding compensation controls).
-
36.
After cell count, stain 10–15 million cells with dead cell exclusion buffer (Ghost Dye 780) in 15 mL Falcon tube. Resuspend cell pellet in 1000 μL PBS containing 1 μL Ghost 780 Dye.
Note: Remove Ghost Dye vial from freezer and allow to equilibrate at 25°C prior to use.
-
37.
Cover cell suspensions with aluminum foil to protect from light and incubate at 4°C for 30 min.
Note: 30-min incubation may occur either in the cold room (in dark) or on ice. However, if on ice, make sure that the tubes are protected from light.
-
38.
Wash cell suspension with 5 mL of FACS buffer and centrifuge for 5 min at 300 × g.
-
39.
Aspirate supernatants and resuspend cell pellets in FACS Sorting Buffer.
Note: FACS Sorting Buffer: PBS 1x supplemented with 2% FBS or 0.1% BSA and 5 mM EDTA. It may be stored at 4°C for up to 1 week.
-
40.
Perform Fc block staining using anti-CD16/CD32 mouse antibody in a 1:50 concentration. 50 μL is an optimal blocking volume for up to 10 million cells.
-
41.
Incubate cell suspensions for 5 min at 4°C before proceeding to cell surface staining.
-
42.
Stain cells with a “cocktail” antibody mix containing fluorescent antibodies for CD45, MHC II, CD11b, B220, CD3, CCR7, CD11c, using 1 μL of antibody per 106 cells. 100 μL is an optimal staining volume for up to 10 million cells.
Note: Samples with more than 10 million cells should be scaled accordingly to obtain an equal staining among samples.
CRITICAL: For reliable results, do not pipette less than 1 μL of antibody. If small volumes are required make a dilution of the antibody first.
-
43.
Incubate cell suspensions for 45 min, protected from light, at 4°C.
-
44.
Centrifuge the stained cells at 300 × g for 5 min, followed by a wash (2–10 mL) with FACS buffer.
-
45.
Aspirate the supernatant and finally resuspend in FACS buffer to achieve single cell suspensions before proceeding to the flow sorting instrument. We usually resuspend in 106–1.5×106 cells/100 μL. Final volumes of resuspension will be determined by the total number of live cells.
Note: To improve cell viability, you can add additional FBS up to 5%.
Note: Absolute quantification of DC density in the tumor (# DC/μg of tumor) can be achieved using Precision Count Beads. Vigorously vortex the precision count beads container for 30–40 s to ensure complete mixing and break up any aggregates that may have occurred during storage. While your samples are ready to be analyzed or sorting, add 100 μL of precision count beads per sample. Accurate pipetting is crucial at this stage. Using reverse pipetting to ensure the exact amount of beads is recommended. Detect fluorescent beads and cell population in the flow cytometer for cell counting. After data acquisition, use the following formula for DC quantification:
From the absolute DC count per μL tumor cell suspension (# DC/μL), extrapolate the absolute DC count per μg tumor weight by using the following formula:
-
46.
Prepare compensation controls. Use an unstained sample as well as single-fluorochrome labeled samples. For Ghost 780 dye compensation, heat at least 10,000 cells at 65°C for 5 min and incubate them with Ghost 780 dye as mentioned in step 36.
Note: The use of ultra compensation mouse beads (see key resources table) will avoid wasting your samples. In this case, per each fluorochrome, prepare a few drops of beads with 1 μL of antibody and use them as single-fluorochrome labeled samples. Beads are not suitable for L/D Ghost 780 compensation; use cells instead.
Note: Use compensation set up option in FACS Aria software to calculate automatic compensation.
-
47.
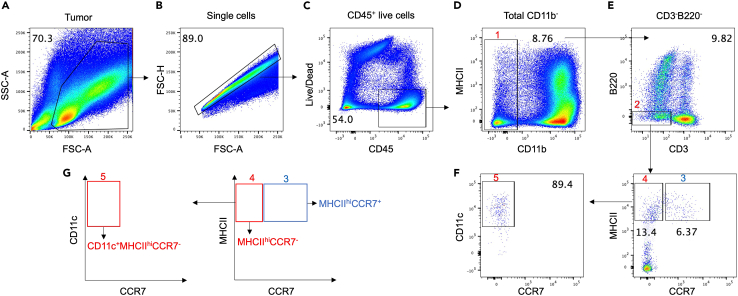
Follow gating strategy as delineated in Figure 2 to sort CCR7pos and CCR7neg DC subsets.
Note: We define non-migratory cDC1s as CD45+, MHCII high, CD11b-, CD3-, B220-, CD11c+ CCR7neg, whereas migratory cDC1s are CD45+, MHCII high, CD11b-, CD3-, B220-, CCR7pos. CD11c surface expression in migratory/mregDC is dim-variable.3
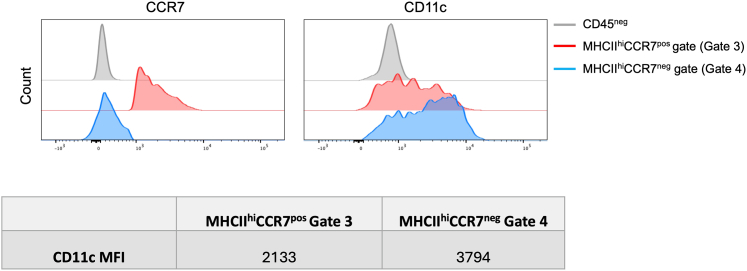
Note: We only use a final CD11c gating step for the CCR7neg cDC1 population (Figures 2 and 3, Gate 5). This is because CD11c is downregulated in CCR7pos DC (Figure 3).3
Note: Antibodies that mark excluded lineages from the desired cell populations can be conjugated with same fluorochromes. This will help to reduce the number of fluorochromes in the flow panel and facilitate compensation. For example, the use of CD3 and B220 antibodies with PE fluorochrome would not influence cDC subset gating and will streamline the experiment.
-
48.
Sort DCs on FACS Aria or equivalent (FACS Jazz/FACS Melody/Aurora CS) under low pressure (20–25 psi) using 100 microns nozzle size.
Note: Pooling several samples together is highly recommended to facilitate the downstream process if less than 1000 cDC cells are obtained per each tumor.
CRITICAL: Gently vortex each sample prior to acquisition/sorting to ensure adequate suspension of the cell and bead populations.
Note: This approach maximizes recovery and viability, while it minimizes nozzle clogs.
Note: If you are running samples across multiple experimental arms, it may be wise to book 2 or 3 sorters simultaneously in order to minimize total sorting time and maximize yield.
Note: In case the sorting time is limited, prior enrichment of hematopoietic (CD45+) cells may be considered in order to reduce cell sorting duration.
-
49.
Collect sorted populations in a conical tube containing 700 μL TRIzol.
Note: 1.5 mL tubes can be also used to collect the sorted population.
Pause point: Sorted DC in TRIzol could be safely frozen at −80°C for several weeks and RNA extraction may take place at a later time point.
Figure 2.
FACS gating strategy to delineate CCR7pos and CCR7neg intratumoral cDC1 subsets
(A–G) Flow cytometry gating strategy from tumors harvested at 3 weeks after LLC tumor cell inoculation. Cells were first gated based on forward scatter (FSC) and side scatter (SSC) (A), then gated for single live cells by size (FSC-A and FSC-H) (B) and Ghost Dye Red 780- viable cells (C), sequential gating of CD11b- (Gate 1) (D), CD3-B220- (Gate 2) (E), CCR7posMHCIIhi (Gate 3) and CCR7negMHCIIhi (Gate 4). CCR7negMHCIIhi DC were then further subjected to a final gating step according to CD11c expression (Gate 5) (F). (G) Diagram showing gate 3, 4, and 5 based in CCR7, MHCII and CD11c expression.
Figure 3.
Modulation of CD11c surface expression in migratory versus non-migratory cDC1
CCR7 and CD11c staining intensities for CCR7pos and CCR7neg fractions (corresponding to Gates 3 and 4, respectively, in Figure 2) are shown as histogram plots (top). CCR7neg subset is represented in blue and CCR7pos subset is in red. CCR7 staining of CD45neg non-hematopoietic cells is shown for comparison. CD11c staining intensity is also provided as mean fluorescence intensity (MFI) for each Gate (bottom).
RNA extraction- day 25+
Timing: 2 h
This step describes the extraction of RNA from Trizol-resuspended sorted DC using the QIAGEN RNeasy Micro Kit.
-
50.
Add 200 μL of chloroform to each sample and mix gently.
-
51.
Centrifuge at 12000 × g for 15 min at 4°C.
-
52.
Recover the supernatant into a new Eppendorf.
-
53.
Add 350 μL of 100% ethanol to each sample, mix.
-
54.
Transfer all the solution into the column (QIAGEN RNeasy Micro Kit): 700 μL at a time.
Note: Steps: 6–19 closely follow the instructions of the QIAGEN RNAeasy Micro Kit (cat: 74004) on purifying RNA from mammalian cells (modified version of the RNAeasy Micro Kit Handbook).
-
55.
Centrifuge at 12000 × g for 30 s at 25°C, discard the flow-through.
-
56.
Add 350 μL of RW1 Buffer.
-
57.
Centrifuge at 12000 × g for 30 s at 25°C, discard the flow-through.
-
58.
For each sample, mix 10 μL of DNase (RNase-free DNase Set-QIAGEN) with 70 μL RDD Buffer, add 80 μL of this mixture to each sample (add to the center of the column).
-
59.
Wait for 15 min at 25°C.
-
60.
Add 350 μL of RW1 Buffer to the column.
-
61.
Centrifuge at 12000 × g for 30 s, discard the flow-through.
-
62.
Add 500 μL of RPE Buffer (RPE Buffer is supplied as a concentrate. Ensure that 220 mL ethanol is added to RPE Buffer before use).
-
63.
Centrifuge at 12000 × g for 30 s at 25°C, discard the flow-through.
-
64.
Add 500 μL of RPE Buffer.
-
65.
Centrifuge at 12000 × g for 30 s at 25°C, change to Collection Tube (2 mL, without caps).
-
66.
Centrifuge at 12000 × g for 1 min at 25°C, change to Collection Tube (1.5 mL, with caps).
-
67.
Add 12–25 μL of RNase-free water, centrifuge at 12000 × g for 30 s 25°C (don’t through away the filtrate).
-
68.
Add 12–25 μL of RNase-free water, centrifuge at 12000 × g for 1 min at 25°C.
Alternative: When a small number of cells has been sorted (<5,000) you may skip step 19 to avoid low final RNA concentration and very dilute sample.
-
69.
Keep the RNA in freezer at −20°C.
RNA validation, QC, library preparation and sequencing- day 25+
Timing: 2–3 days
This step describes the determination of RNA yield and purity and the preparation of cDNA libraries using Ultra Low Input RNA-seq reagents.
-
70.
Quantify RNA through the Agilent TapeStation system (TapeStation 4200, High Sensitivity RNA reagents) (Figure 4).
Note: The RNA yield post-extraction is relatively low given the rarity of intratumoral DC subsets. Thus using TapeStation analysis provides efficient and reliable separation of total RNA samples of a concentration as low as 100 pg/uL. Typical RNA yields range between 500–900 pg/sample.
-
71.
Prepare libraries using Ultra Low Input RNA-seq cDNA prep (SMART-seq) kit (Takara) as per manufacturer’s instructions.
-
72.
Assess libraries’ quality and quantity using the TapeStation system (TapeStation 4200, High Sensitivity D1000 reagents) and the Qubit 4 (dsDNA Quantification High Sensitivity reagents) (Figure 4).
-
73.
Pool and sequence libraries on the Illumina NextSeq 2000 platform using the P1 flow cell (sequencing depth PE100) and 2% PhiX resulting in more than 60 M reads per sample.
Figure 4.
Typical electropherograms obtained by Agilent TapeStation system
(A) RNA quality control. 18s and 28 rRNA bands were detected to determine the quality of samples. The intensity of these peaks were correlated to the RNA concentration in samples.
(B) cDNA library quality control.
Expected outcomes
The migratory-prone MHCIIhi CCR7pos CD11bneg intratumoral DC population (mregDC1) (see gating in Figure 2) is smaller than the non-migratory CCR7neg cDC1 population at baseline in LLC tumors (Figure 5; Table 1).4 In a typical experiment comprising 5 individual LLC tumors, an average of 5680 CCR7neg cDC1 and 1630 CCR7pos cDC1 were flow-sorted per 650 mm3 tumor harvested on Day 21 post-tumor cell inoculation.
Figure 5.
Relative frequencies of MHCIIhiCCR7neg and MHCIIhiCCR7pos gate populations in LLC tumors
CCR7 and MHCII staining was used to delineate both populations within live CD45+ CD11b- CD3- B220- gate (see also Figure 2).
Table 1.
Quantification of tumor-resident CCR7neg cDC1 and CCR7pos mregDC1 (Lewis Lung Carcinoma tumors)
| CCR7neg cDC1 | CCR7pos mregDC1 | |
|---|---|---|
| DC count per 105 tumor-resident CD45+ cells | 413 | 151 |
DC subset cell numbers/105 intratumoral hematopoietic CD45+ cells were estimated using precision count beads as delineated in step 45. Alternatively, absolute DC subset counts can be expressed as per unit tumor tissue weight, which permits the direct comparison of DC subset density across different tumor models (Step 45).
CCR7, a 7-fold transmembrane G protein-coupled receptor (GPCR), mediates the migration of cDC1s to the draining lymphoid node to prime T cells.5 cDCs undergo maturation after receptor-mediated endocytosis, phagocytosis, and macropinocytosis of antigens.6,7,8,9,10,11 Maturation results in upregulation of MHCII as well as increased expression of co-stimulatory molecules, such as CD80, and migration through CCR7 expression.3,4,12 The tumor DC maturation/activation module is balanced by a conserved regulatory program (mregDC program) that limits anti-tumor immunity.3
Limitations
DCs comprise a relatively small fraction of the intratumoral immune milieu (<5%). Depending on the tumor type and the degree of tumor inflammation, this percentage could vary. Although this study was performed and validated in the LLC tumor model, the RNA yield and quality may differ in other tumor types, which may pose challenges in obtaining comparable data across tumor models. Therefore, prior to planning RNA-seq steps, it is highly recommended to conduct pilot experiments to confirm the identification and quantification of DC subtypes by conventional flow cytometry in the particular tumor model desired.
Given the very low amount of RNA utilized for the experiment, researchers performing these experiments may be limited to using an ultra-low input RNA-seq kit for library preparation, which increases the cost of the study significantly, particularly when multiple samples are analyzed.
Troubleshooting
Problem 1
Cell suspension appears extremely viscous after tumor dissociation, making it difficult to be manually pipetted up and down.
Potential solution
Perform DNase treatment: centrifuge cell suspension at 300 × g for 5 min, remove supernatant completely, and resuspend pellet in 5 mL DMEM containing 200 U/mL DNase, then incubate for 5 min at 25°C.
Problem 2
Clogs during sorting.
Potential solution
Remove the sample from the sorter and re-filter through a 70 micron filter.
Problem 3
Fluorescent antibody staining irreproducible in scaled-up cell preps.
Potential solution
For reproducible staining using fluorescently-labeled antibodies, antibody concentration in the staining mix is critically important. Perform pilot experiments to determine the optimal concentration of each antibody in the staining mix. Scale up as appropriate in the composite staining experiment.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fotis Asimakopoulos (fotis@health.ucsd.edu).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be fulfilled by the technical contact, Athanasios Papadas (apapadas@health.ucsd.edu).
Materials availability
Mouse lines and all other reagents reported in this paper are available from the lead contact upon execution of a Materials Transfer Agreement (MTA).
Data and code availability
This study did not generate or analyze datasets or code.
Acknowledgments
This work was supported through the NIH/National Cancer Institute (R01CA252937) and the Robert J. Shillman Foundation. This work was supported by the NGS Core Facility of the Salk Institute with funding from NIH-NCI CCSG: P30 CA01495, NIH-NlA San Diego Nathan Shock Center P30 AG068635, the Chapman Foundation, and the Helmsley Charitable Trust.
We thank Cheryl Kim, Denise Hinz, and Chris Dillingham (La Jolla Institute of Immunology Flow Cytometry Facility) for help with flow-based assays.
Author contributions
A.P., D.J.L., Y.D., D.H., A.G., and E.M. designed and performed the experiments and wrote the first draft of the manuscript. A.C., Y.H., B.Z., and F.A. edited the manuscript. F.A. was overall responsible for design and conduct of the study and for securing funding.
Declaration of interests
A.P., A.C., and F.A. are named inventors on patents focused on the manipulation of dendritic cells in the tumor microenvironment.
Contributor Information
Athanasios Papadas, Email: apapadas@health.ucsd.edu.
Fotis Asimakopoulos, Email: fotis@health.ucsd.edu.
References
- 1.Papadas A., Deb G., Cicala A., Officer A., Hope C., Pagenkopf A., Flietner E., Morrow Z.T., Emmerich P., Wiesner J., et al. Stromal remodeling regulates dendritic cell abundance and activity in the tumor microenvironment. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waight J.D., Netherby C., Hensen M.L., Miller A., Hu Q., Liu S., Bogner P.N., Farren M.R., Lee K.P., Liu K., Abrams S.I. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J. Clin. Invest. 2013;123:4464–4478. doi: 10.1172/JCI68189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maier B., Leader A.M., Chen S.T., Tung N., Chang C., LeBerichel J., Chudnovskiy A., Maskey S., Walker L., Finnigan J.P., et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580:257–262. doi: 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meiser P., Knolle M.A., Hirschberger A., de Almeida G.P., Bayerl F., Lacher S., Pedde A.M., Flommersfeld S., Hönninger J., Stark L., et al. A distinct stimulatory cDC1 subpopulation amplifies CD8(+) T cell responses in tumors for protective anti-cancer immunity. Cancer Cell. 2023;41:1498–1515.e10. doi: 10.1016/j.ccell.2023.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Forster R., Schubel A., Breitfeld D., Kremmer E., Renner-Muller I., Wolf E., Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 6.Cabeza-Cabrerizo M., Cardoso A., Minutti C.M., Pereira da Costa M., Reis e Sousa C. Dendritic Cells Revisited. Annu. Rev. Immunol. 2021;39:131–166. doi: 10.1146/annurev-immunol-061020-053707. [DOI] [PubMed] [Google Scholar]
- 7.Conejo-Garcia J.R., Rutkowski M.R., Cubillos-Ruiz J.R. State-of-the-art of regulatory dendritic cells in cancer. Pharmacol. Ther. 2016;164:97–104. doi: 10.1016/j.pharmthera.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gajewski T.F., Cron K.R. cDC1 dysregulation in cancer: An opportunity for intervention. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabado R.L., Balan S., Bhardwaj N. Dendritic cell-based immunotherapy. Cell Res. 2017;27:74–95. doi: 10.1038/cr.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veglia F., Gabrilovich D.I. Dendritic cells in cancer: the role revisited. Curr. Opin. Immunol. 2017;45:43–51. doi: 10.1016/j.coi.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy K.M. Transcriptional control of dendritic cell development. Adv. Immunol. 2013;120:239–267. doi: 10.1016/B978-0-12-417028-5.00009-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze datasets or code.