Abstract
In chicken, primordial germ cells (PGC) are crucial for the preservation and manipulation of genetic resources in poultry production. The HiS and FAcs culture systems are two important methods for the in vitro cultivation of chicken PGCs. The purpose of this study was to compare and analyze the two cultivation systems for PGCs (His and FAcs culture systems) to assess their efficacy and applicability in supporting PGC growth, maintaining PGC characteristics, and lineage transmission ability. The study found that both HiS and FAcs culture systems could maintain the basic biological characteristics of chicken PGCs, including the simultaneous expression of pluripotency and reproductive marker genes, as well as the presence of abundant glycogen granules. Subsequently, we identified 2,145 differentially expressed genes (DEG) through RNA sequencing. GO and KEGG analysis revealed a large number of DEGs enriched in the cell adhesion and calcium ion binding pathways, and the analysis found that these genes maintained a higher level in HiS-PGCs. Further personalized analysis found that the regulatory genes for maintaining PGC pluripotency were highly expressed in HiS-PGCs, while germ cell-related genes showed similar expression in both systems. Additionally, through RNA sequencing data and cell proliferation ability, it was found that PGCs in the FAcs system had a higher proliferation rate and a faster cell cycle. Finally, it was discovered that the expression of cell migration-related genes was maintained at a higher level in HiS-PGCs, but the migration efficiency of HiS-PGCs did not show a significant difference compared to FAcs-PGCs. These results suggest that both HiS and FAcs culture systems can maintain the proliferation and basic characteristics of chicken PGCs, but differences exist in cell proliferation, pluripotency regulation, and cell adhesion. These findings provide new information for optimizing PGC cultivation systems and are important for the preservation and genetic improvement of chicken PGCs.
Key words: chicken, primordial germ cell, culture system, gene expression, characteristic
INTRODUCTION
Primordial germ cells (PGCs) are important tools in the conservation of germplasm resources and gene editing in animal reproduction (Trefil et al., 2017, Lázár et al., 2021, Chen et al., 2023, Hamai et al., 2023). Particularly in chickens, PGCs are the most promising tools because the techniques used for mammalian gamete and embryo manipulation are not applicable to chickens due to their egg-laying nature. PGCs serve as the precursors of gametes and play a crucial role in the transmission of genetic information across generations (Park et al., 2003). Therefore, they are fundamental resources for preserving and manipulating genetic traits. PGCs provide a means to safeguard genetic diversity and protect rare or endangered avian species. By storing and maintaining PGCs from different individuals, researchers and breeders can safeguard genetic variability and prevent the loss of valuable traits. This is especially important for the conservation of rare and endangered species because genetic diversity is crucial for their long-term survival (Sun et al., 2022). Moreover, PGCs have vast potential in genetic editing for poultry production. Advanced biotechnologies such as CRISPR/Cas9 can be used to manipulate PGCs and introduce or eliminate specific genetic traits (Ballantyne et al., 2021; Lee et al., 2022; Pu et al., 2023). This has the potential to revolutionize poultry breeding by precisely modifying desired traits such as disease resistance, improved productivity, and enhanced welfare characteristics. Overall, PGCs are pivotal for preserving and manipulating genetic resources in poultry production. Understanding and optimizing the culture systems for PGCs, as well as their characteristics and behavior, are essential for advancing genetic technologies and ensuring sustainable poultry breeding programs.
Due to their characteristic of migrating with the bloodstream, chicken PGCs are easy to isolate and transplant. However, due to the limited number of PGCs obtained from in vivo isolation, it is difficult to meet the needs of actual production. Therefore, various chicken PGC culture systems have been developed (Karagenç and Petitte, 2000; van de Lavoir et al., 2006; Choi et al., 2010; Miyahara et al., 2014; Whyte et al., 2015; Chen et al., 2018). In recent years, the HiS and FAcs culture systems have become two important methods for the in vitro culture of chicken PGCs and have been increasingly used (van de Lavoir et al., 2006; Whyte et al., 2015). The FAcs culture system utilizes specific growth factors such as FGF2, Activin A, and insulin to support the proliferation and maintenance of PGCs in vitro. The system aims to create a controlled and defined environment to promote the survival and proliferation of PGCs while minimizing external influences that may alter their characteristics. On the other hand, the HiS culture system involves the preparation and addition of Buffalo Rat Liver (BRL)-conditioned medium and feeder layers to provide necessary support for the growth and development of PGCs. This system mimics the natural microenvironment by integrating the interactions between PGCs and somatic cells, thereby influencing the behavior and function of PGCs. It is worth noting that the preparation and addition of conditioned medium and feeder layers can lead to slight variations between batches and increase the workload. However, both these culture systems contribute to enhancing our understanding of PGC biology and pave the way for exploring applications such as germplasm preservation, genetic modification, and the production of transgenic animals. Studying and comparing these two culture systems is significant as it allows for the optimization of PGC culture conditions to enhance proliferation and genetic manipulation. By elucidating the differences between HiS and FAcs, researchers can refine and tailor the culture systems to better meet the specific needs of PGC research, ultimately promoting their utilization in chicken breeding programs and biotechnological applications.
Previous research has focused on developing culture systems for PGCs in order to better understand and utilize them (van de Lavoir et al., 2006). These culture systems aim to mimic the in vivo environment of PGCs and support their in vitro growth. However, comparative analyses of these culture systems are necessary to determine their efficacy and suitability for different research applications. Research has shown that adding cholesterol to the culture medium significantly enhances the proliferation of chicken PGCs and maintains their expression of germ-cell-related markers, thus retaining their capability to colonize the embryonic gonad (Chen et al., 2016). Additionally, the addition of Blebbistatin to the culture medium increases the survival and proliferation rates of chicken PGCs (Ezaki et al., 2020). When comparing defined and enriched (the defined medium with Knock-Out Serum Replacement) media for the growth of PGCs originating from the Hubbard JA57 broiler, the use of an enriched medium results in improved growth properties of chicken PGCs compared to a defined medium and also affects the gene expression of PGCs (Dehdilani et al., 2023). These studies emphasize the importance of comparing and analyzing PGC culture systems to determine the conditions most suitable for specific research objectives.
The aim of this study is to compare the effectiveness of FAcs and HiS culture systems in supporting the growth, maintaining the characteristics, and lineage transmission capability of PGCs. The research findings indicate that both the HiS-PGCs and FAcs-PGCs can maintain the essential biological traits of chicken PGCs. This includes the simultaneous expression of pluripotency and reproductive marker genes, the presence of abundant glycogen granules, and the ability to migrate and colonize the gonads. RNA sequencing analysis revealed an activation in cell adhesion, calcium ion binding pathways, and pluripotency regulation genes in HiS-PGCs, while the cell proliferation ability was lower. By comparing the efficacy of different culture systems in supporting PGC growth, maintaining PGC characteristics, and their lineage transmission capability, researchers can optimize their experimental methods and enhance our understanding of germ cell biology. These findings provide new information for optimizing PGC cultivation systems and are important for the preservation and genetic improvement of chicken PGCs.
MATERIALS AND METHODS
Isolation and Culture of PGCs
All experimental procedures were approved by the Experimental Animal Ethics Committee of Yangzhou University (approval code: 202103273). PGCs were cultured using previously established FAcs and HiS systems (van de Lavoir et al., 2006; Whyte et al., 2015). Rugao Yellow Chicken embryos were incubated until reaching Hamburger and Hamilton (HH) stages 27-28. Isolated gonads were processed, and the resulting cells were resuspended in FAcs or HiS culture medium. In the HiS group, mitotically inactivated BRL cells were added to the culture plate as feeder cells. Cultivation was conducted at 37°C with 5% CO2 and maximum humidity. In this study, the PGCs cultured in HiS and FAcs systems for approximately 90 to 150 d were used for experiments. HiS-PGCs and FAcs-PGCs were cultured for the same number of days in vitro when comparing the same parameters.
The FAcs medium was formulated by diluting calcium-free DMEM (Gibco, Carlsbad, CA) with sterile water at a 1:3 ratio to serve as the basal medium. Various components were supplemented, including 0.15 mM CaCl2 (C7902, Sigma, St. Louis, MO), 1× B-27 supplement (17504044, Gibco, Carlsbad, CA), 2mM GlutaMax (35050061, Gibco, Carlsbad, CA), 1× non-essential amino acids (11140050, Gibco, Carlsbad, CA), 0.1mM β-mercaptoethanol (31350010, Gibco, Carlsbad, CA), 1mM sodium pyruvate (11360070, Sigma, St. Louis, MO), 0.2% chicken serum (16110082, Gibco, Carlsbad, CA), 1× nucleosides (E8-008-D, Sigma, St. Louis, MO), 0.20% ovalbumin (A5503, Sigma, St. Louis, MO), 0.1 mg/mL sodium heparin (HY-17567A, MCE, Monmouth Junction, NJ), 25 ng/mL human Activin A (HY-P70311, MCE, Monmouth Junction, NJ), and 4 ng/mL human FGF2 (HY-P70600, MCE, Monmouth Junction, NJ).
For the HiS medium, KO-DMEM supplemented 5% FBS, 2mM GlutaMax was conditioned with BRL cells. The HiS medium was prepared by the KO-DMEM medium was supplemented with 40% BRL-conditioned medium, 7.5% fetal calf serum (FCS), 2.5% chicken serum, 2 mM GlutaMax, 1 mM sodium pyruvate, 1× nucleosides, 1× non-essential amino acids and 0.1 mM β-mercaptoethanol, 6 ng/mL SCF and 4 ng/mL human recombinant FGF2.
PAS Staining
We conducted PAS staining using the PAS Staining kit (G1360, Solarbio, Beijing, China) according to the manufacturer's instructions. Chicken PGCs were fixed on slides using a fixing solution until adherent, followed by 2 washes with water. Subsequently, oxidant was added dropwise, and the slides were oxidized at room temperature for 15 to 20 min. After washing the slides 4 times with distilled water, they were allowed to dry. Shiff Reagent was then applied, and the slides were incubated in a humid chamber for 8 to 15 min. Following removal from the chamber, the slides were briefly rinsed twice with sodium sulfite solution for 10 s each, followed by a 2-min rinse with running water. Mayer's hematoxylin staining solution was applied, and the slides were stained for 10 to 30 s. After washing with water, the slides were sealed with cedar oleoresin and examined microscopically.
Immunofluorescence and Confocal Microscopy
Following three washes with PBS, PGCs were fixed on slides with 4% paraformaldehyde (Solarbio, Beijing, China) for 30 min at room temperature until adherence occurred, followed by 2 PBS washes. Subsequently, PGCs were permeabilized with PBS containing 0.5% Triton X-100 (Solarbio, Beijing, China) for 15 min at room temperature and then blocked in PBS containing 1.0% BSA for 1 h at room temperature. Overnight, PGCs were incubated at 4°C with primary antibodies, including anti-SSEA-1 (1:100; MC-480, DSHB, Iowa), anti-EMA-1 (1:100; MC-480, DSHB, Iowa), anti-C-KIT (1:100; 8380-01, SouthernBiotech, Birmingham, AL), anti-CVH (1:100; orb5967, Biorbyt, Cambridge, United Kingdom), anti-SOX2 (1:100; sc-398254, Santa Cruz, TX) diluted in the blocking solution. After 3 PBS washes, PGCs were incubated for 1 h at room temperature with either donkey anti-rabbit IgG (HuaBio, China), or goat anti-mouse IgG (HuaBio, China). Subsequently, PGCs were stained with DAPI (Solarbio, Beijing, China) for 10 min and washed three times with PBS. Finally, 10 μL of anti-fluorescence quenching agent (Solarbio, Beijing, China) was added, the slide was sealed with cedar oleoresin, and observation was performed using a confocal microscope (Zeiss LSM 710 Meta). Image processing was conducted using Olympus Fluoview 4.1a viewer (Olympus).
RNA Isolation and Library Preparation
RNA extraction was carried out using the TRIzol reagent (Vazyme, China) following the manufacturer's protocol, followed by assessment of purity and quantification using the NanoDrop 2000 spectrophotometer. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer. Libraries were prepared using the VAHTS Universal V6 RNA-seq Library Prep Kit according to the manufacturer's instructions. Transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd.
RNA Sequencing and Differentially Expressed Genes Analysis
The libraries underwent sequencing on an Illumina NovaSeq 6000 platform, generating 150 bp paired-end reads. Raw reads in fastq format were processed using fastp (Chen et al., 2018) to eliminate low-quality reads, resulting in clean reads for subsequent analyses. The clean reads were then aligned to the reference genome (GRCg6a/galGal6) using HISAT2 (Kim et al., 2015). FPKM (Roberts et al., 2011) values for each gene were computed, and read counts for each gene were obtained using HTSeq-count (Anders et al., 2015). To assess sample duplication, principal component analysis (PCA) analysis was conducted using R (v 3.2.0).
Differential expression analysis was carried out using DESeq2 (Love et al., 2014), employing a threshold of q-value <0.05 and |log2FC|>1 to identify significantly DEGs. Hierarchical cluster analysis of DEGs was performed using R (v 3.2.0) to illustrate expression patterns across various groups and samples. Gene ontology (GO) (Consortium, 2019) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2007) pathway enrichment analyses of DEGs were conducted using R (v 3.2.0) based on the hypergeometric distribution to identify significantly enriched terms.
For Gene Set Enrichment Analysis (GSEA), GSEA software (Mootha et al., 2003; Subramanian et al., 2005) was utilized. This approach employed a predefined gene set to rank genes according to their degree of differential expression between the two sample types. Subsequently, it was determined whether the predefined gene set was enriched at either the top or bottom of the ranking list.
RNA Isolation and Reverse Transcription
One million PGCs were collected and mixed with 1 mL of Trizol reagent. After a 5-min incubation at room temperature, chloroform was added, followed by vigorous shaking for 15 s. The mixture was then centrifuged at 10,000 r/min for 15 min at 4°C. After adding an equal volume of isopropanol and incubating for 10 min, the sample was centrifuged again at 10,000 r/min for 10 min at 4°C. The supernatant was discarded, and the RNA pellet was washed with 75% ethanol before being dried. Finally, 30 µL of DNase/RNase-free distilled water was added to dissolve the RNA, and its concentration was measured using a NanoDrop 2000 spectrophotometer.
For reverse transcription, DNase I-treated total RNA was mixed with 4 µL of 4×gDNA wiper Mix and RNase-free ddH2O to a total volume of 16 µL. The mixture was incubated at 42°C for 2 min, followed by the addition of 4 µL of 5 × HiScript III qRT SuperMix. Reverse transcription was carried out at 37°C for 15 min, followed by 85°C for 5 s to synthesize the cDNA.
PCR and Agarose Gel Electrophoresis
PCR was conducted using PrimeSTAR Max DNA Polymerase (Takara, R045A, Takara Bio, Kusatsu, Shiga, Japan) with the following thermal cycling conditions: initial denaturation at 98°C for 5 min, followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 15 s, extension at 72°C for 30 s, and a final extension step at 72°C for 5 min. The specific primer sequences are listed in Table 1. The PCR products were resolved by electrophoresis on a 1.5% agarose gel, stained with RedSafe Nucleic Acid Staining Solution (INtRON biotechnology), and visualized under UV illumination.
Table 1.
The primers used in the present study.
| Gene | Sequence | Annealing temp. | Product size (bp) | Reference |
|---|---|---|---|---|
| CVH F | CAGACCGCATGCTTGATATG | 60°C | 135 | (Chen et al., 2018) |
| CVH R | CAGCCAGCCTCTGAACTTCT | |||
| DAZL F | TCACTGACAGGACTGGTGTTTC | 60°C | 127 | (Chen et al., 2018) |
| DAZL R | ATTGCTGGTCCCAGTTTCAG | |||
| POUV F | GTTGTCCGGGTCTGGTTCT | 60°C | 189 | (Chen et al., 2018) |
| POUV R | GTGGAAAGGTGGCATGTAGAC | |||
| NANOG F | GGTTTCAGAACCAACGGATG | 60°C | 121 | (Chen et al., 2018) |
| NANOG R | GTGGGGGGTCATATCCAGGTA | |||
| SOX2 F | GTGAACCAGAGGATGGACAGTTACG | 60°C | 185 | (Whyte et al., 2015) |
| SOX2 R | TGCGAGCTGGTCATGGAGTTG | |||
| PRDM1 F | CCCACGAGTGTCAGGTTTGT | 60°C | 133 | (Chen et al., 2018) |
| PRDM1 R | AGGTGCACAAACTGGGTGAA | |||
| CD63 F | CAGGAGGACTTCCACTGCTG | 60°C | 232 | — |
| CD63 R | AGAAGGCAATACCCAACGCA | |||
| DLL1 F | CGACGACCTCACCACAGAAA | 60°C | 296 | — |
| DLL1 R | CACACCCAGGCAAGCAAATC | |||
| EDN1 F | CTCACGTCACCGCAACAAAG | 60°C | 104 | — |
| EDN1 R | TGTTACCAACAGCCTCCAGC | |||
| LAMA5 F | CAGCTCCTGCATTCCCTCAA | 60°C | 238 | — |
| LAMA5 R | CAGGTTCTGAATCCGGCTGT | |||
| MYLK F | GTTCCCCGCAGCTCTGTC | 60°C | 283 | — |
| MYLK R | AAATCTTGCTGTGGCTCCCA | |||
| OVOL3 F | CTTTGATCTGAAGCGCCACG | 60°C | 220 | — |
| OVOL3 R | ATGCAGGTACAGGTCCTCCT | |||
| RET F | TAGATCTTGCGGCTTCCACC | 60°C | 164 | — |
| RET R | AGTAAATGCATGTGAAATTCTACCA | |||
| S1PR1 F | GCTTACACTGCCAACCTCCT | 60°C | 123 | — |
| S1PR1 R | GGCCAACAAGCTGAACACAG | |||
| SEMA4G F | TGAACACCACGCACCTCTAC | 60°C | 160 | — |
| SEMA4G R | CCATCCACAATGAGGCCAGT | |||
| SPP1 F | AGCTCATTGAGGATGACGCC | 60°C | 261 | — |
| SPP1 R | CATCCTCAGCGCTCTCTAGC | |||
| EMP1 F | TCTTCGTCTCCACCATTGCC | 60°C | 158 | — |
| EMP1 R | GATCATGAAGGCTTGCGCTG | |||
| GAPDH F | GAGGGTAGTGAAGGCTGCTG | 60°C | 113 | (Chen et al., 2018) |
| GAPDH R | CATCAAAGGTGGAGGAATGG |
Quantitative Real-Time PCR
qRT-PCR was conducted using the ChamQ Universal SYBR qPCR Master Mix kit (Vazyme, Q711) following the manufacturer's protocol. The reaction mixture consisted of 10 µL of 2× ChamQ Universal SYBR qPCR Master Mix, 2 µL of cDNA, 0.4 µl of forward primer, 0.4 µL of reverse primer, and DNase/RNase-free distilled water to adjust the final volume to 20 µL. Amplification was performed with the following cycling conditions: initial denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 72°C for 15 s. A final extension step was carried out at 72°C for 5 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the reference gene. Primers used for amplification are listed in Table 1. mRNA quantification data were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Cell Proliferation Assay
PGCs were seeded into 6-well plates at a density of 1 × 106 cells per well and passaged every 3 d in FAcs medium or every 5 d in HiS medium. The cells were cultured for up to 15 d, and the cell density was measured using a cell counting plate at each passage. After 3 repetitions, the cell proliferation was calculated and a proliferation curve was plotted using GraphPad Prism.
Cell Cycle Assay
Cell cycle analysis was conducted utilizing a cell cycle detection kit (40301ES50, Yeasen) in accordance with the manufacturer's guidelines. Initially, 1 × 106 PGCs were harvested and rinsed with cold PBS, followed by gentle mixing with 1 mL of cold 70% ethanol for overnight fixation at 4°C. On the subsequent day, after centrifugation, ethanol was aspirated, and cells were washed with 1 mL of cold PBS. Then, propidium iodide staining solution was prepared by combining 10 μL of propidium iodide storage solution (40301-B) and 10 μL of RNase A (40301-A) solution with 0.5 mL of staining buffer (40301-C). Each cell sample was incubated with 0.5 mL of the prepared staining solution, thoroughly suspended, and incubated in darkness at 37°C for 30 min. The stained cells were analyzed using a flow cytometer (FACSAria SORP, BD), and subsequent data analysis was performed utilizing FlowJo and Prism analysis software.
EdU Detection
Cell proliferation capacity was assessed using the EdU Cell Proliferation Assay Kit (RiboBio, Guangzhou, Guangduong, China). HiS-PGCs and FAcs-PGCs were incubated with 50 μM EdU culture medium at 37°C for 2 h. The cells were then washed twice with PBS and fixed with 4% paraformaldehyde for 15 min. After fixation, cells were incubated with 2 mg/mL glycine for 5 min and washed twice with PBS. They were then permeabilized with 0.5% TritonX-100 at room temperature for 10 min. Subsequently, cells were incubated with 100 μl of 1×Apollo staining reaction solution at room temperature in the dark for 10 min, followed by centrifugation and removal of the staining solution. The PGCs were stained with DAPI (Solarbio, Beijing, China) for 10 min, washed 3 times with PBS, and sealed with cedar oleoresin before observation under a fluorescence microscope.
Detection of PGC Migration
Three days before injection, incubate recipient chicken embryos. Prior to injection, adjust cell density to 3,000 cells/μl. At 2.5 d of development (HH 14-16), disinfect embryos and create a small hole (approximately 1 cm in diameter) at the blunt end using fine forceps. Under a stereomicroscope, locate the blood vessel and inject 1 to 2 μL of cloned GFP-PGCs suspension into the dorsal aorta blood vessel. Apply 20 μL of Penicillin/Streptomycin into the opening, gently seal with medical breathable tape, and return eggs to the incubator with the blunt end facing up. At 7.5 d of development (HH 32), isolate embryos using fine forceps under a stereomicroscope and observe GFP-PGC migration to the gonads using a stereoscopic fluorescence microscope.
Statistical Analysis
The experiments were repeated at least three times to ensure robustness and reliability of the findings. Statistical analysis was conducted using Student's t-test to assess the significance of differences. Prior to analysis, all percentage data underwent arcsine transformation to meet the assumptions of the statistical tests. The results are presented as the mean ±SEM to provide a measure of the variability within the data. Significance was defined at p < 0.05. All statistical calculations and analyses were performed using SPSS software version 19 (SPSS, Inc., Chicago, IL), a widely accepted statistical tool in research.
RESULTS
Both the HiS and FAcs Culture Systems are Capable of Maintaining the Basic Biological Characteristics of Chicken PGCs
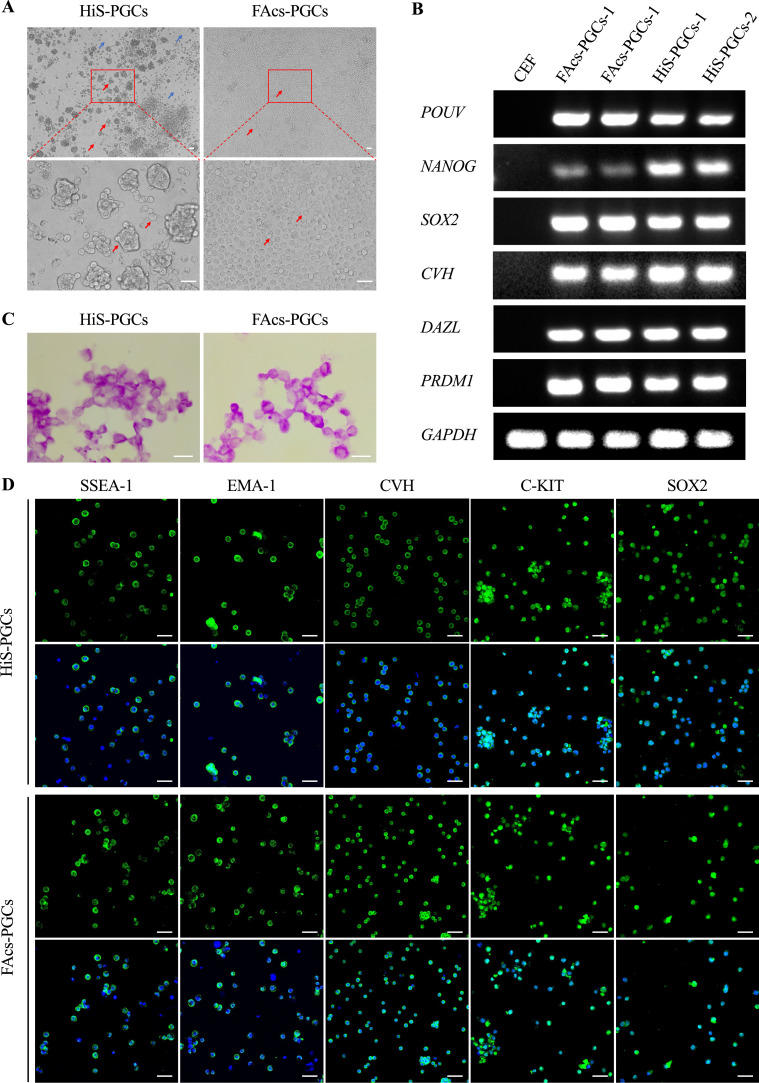
To compare the differences between the HiS and FAcs culture systems, we first evaluated their efficacy in maintaining the basic characteristics of chicken PGCs. Microscopic observation revealed that HiS-PGCs display a clustered growth pattern, while FAcs-PGCs tend to grow in a more dispersed manner (Figure 1A). Subsequently, through RT-PCR and agarose gel electrophoresis experiments, we found that both HiS-PGCs and FAcs-PGCs expressed germ cell-specific genes (CVH and DAZL), pluripotency genes (POUV and NANOG), PGC critical regulatory gene (PRDM1), and telomerase reverse transcriptase gene (TERT) in PGCs, while these genes were not expressed in chicken embryo fibroblasts (CEF) (Figure 1B). Furthermore, PAS staining results showed that both HiS-PGCs and FAcs-PGCs exhibited positive staining (Figure 1C). Immunofluorescent detection of HiS-PGCs and FAcs-PGCs demonstrated sustained expression of SSEA-1 and germ cell markers CVH, as well as pluripotent cell markers SOX2, EMA-1, and C-KIT (Figure 1D).
Figure 1.
Characterization of PGCs cultured in HiS and FAcs medium. (A) Morphology observation of PGCs after 90 d of in vitro culture in HiS and FAcs media. Scale bar: 50 μm. Original magnification, ×4. The red arrows indicate PGCs, and the blue arrows indicate feeder layer cells. (B) RT-PCR and agarose gel electrophoresis analyses demonstrating the expression of germ cell and pluripotency marker genes in HiS-PGCs and FAcs-PGCs. CEF was used as a negative control. (C) PAS staining showing positive results for HiS-PGCs and FAcs-PGCs. Scale bar: 25 μm. (D) Immunofluorescence staining analyses showing the expression of SSEA-1, EMA-1, CVH, C-KIT, and SOX2 proteins in both HiS-PGCs and FAcs-PGCs. The green fluorescence in the upper panels represents the proteins, while the lower panels show the merged images of the proteins with DAPI. Scale bar: 50 μm.
RNA Sequencing Analysis Reveals DEGs and Functional Annotation of PGCs in HiS and FAcs Culture Systems
To comprehensively compare the HiS and FAcs culture systems in cultivating PGCs, we conducted RNA sequencing on HiS-PGCs and FAcs-PGCs. Six cDNA libraries were generated, each comprising three biological replicates for both conditions. We obtained raw reads ranging from 48.62-53.40 million, resulting in approximately 49.03 million clean reads per sample (ranging from 47.35–51.97 million, Table S1), with an average quality control pass rate of 97.16%. Subsequently, 90.44–94.17% of the clean reads successfully mapped to the chicken reference genome, of which 86.15–89.35% were uniquely mapped (Table S1). Additionally, PCA illustrated tight clustering of biological replicates within each group, while distinctly separating HiS-PGCs and FAcs-PGCs (Figure S1A). The heatmap of sample correlation confirmed the grouping of HiS-PGCs and FAcs-PGCs within their respective categories (Figure S1B). Expression values were calculated and visualized as box plots of logarithmically transformed RPKM values for each sample (Figure S1C), with RPKM density distribution depicted in Figure S1D.
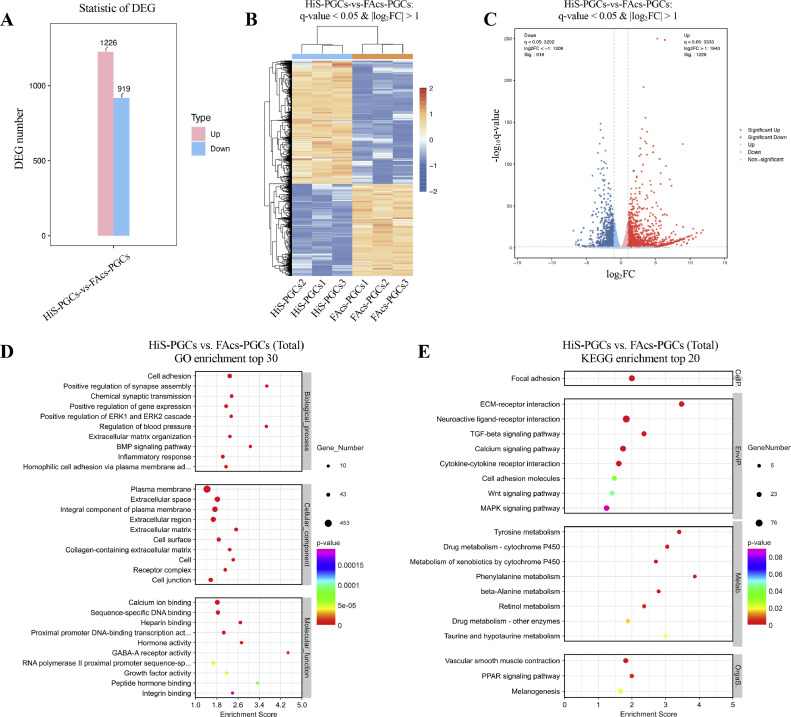
To elucidate the similarities and differences between the HiS and FAcs culture systems in PGC cultivation, we analyzed DEGs. A total of 2145 DEGs were identified using q-value <0.05 and |log2FC| >1 (Figures 2A–2C). Among them, 919 DEGs were down-regulated, and 1226 DEGs were up-regulated (Figures 2A–2C). Functional classification of DEGs was conducted using GO and KEGG classification systems. GO analysis revealed significant enrichment in categories such as cell adhesion, calcium ion binding, and BMP signaling pathway (Figure 2D, Table S2). Similarly, KEGG analysis showed significant enrichment in pathways such as focal adhesion, TGF-β signaling pathway, calcium signaling pathway, and cell adhesion molecules (Figure 2E, Table S3). These findings suggest discrepancies between the HiS and FAcs culture systems in terms of preserving PGC adhesion and calcium ion binding processes.
Figure 2.
DEGs screening, GO, and KEGG analysis of PGCs cultured in HiS and FAcS medium. Statistics (A and B) and volcanic map (C) of DEGs between HiS-PGCs and FAcs-PGCs. GO (D) and KEGG (E) enrichment top pathways of DEGs between HiS-PGCs and FAcs-PGCs. HiS-PGCs and FAcs-PGCs cultured in vitro for 105 d were used for RNA sequencing analysis.
Validation and Comparative Analysis of Biological Characteristics related Genes in RNA Sequencing Data from PGCs Cultured in HiS and FAcs System
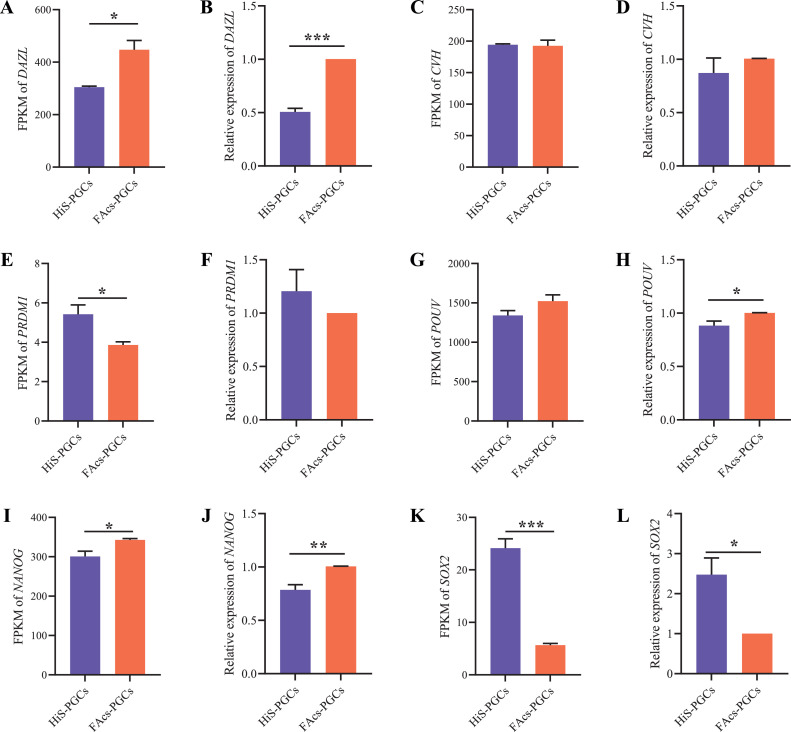
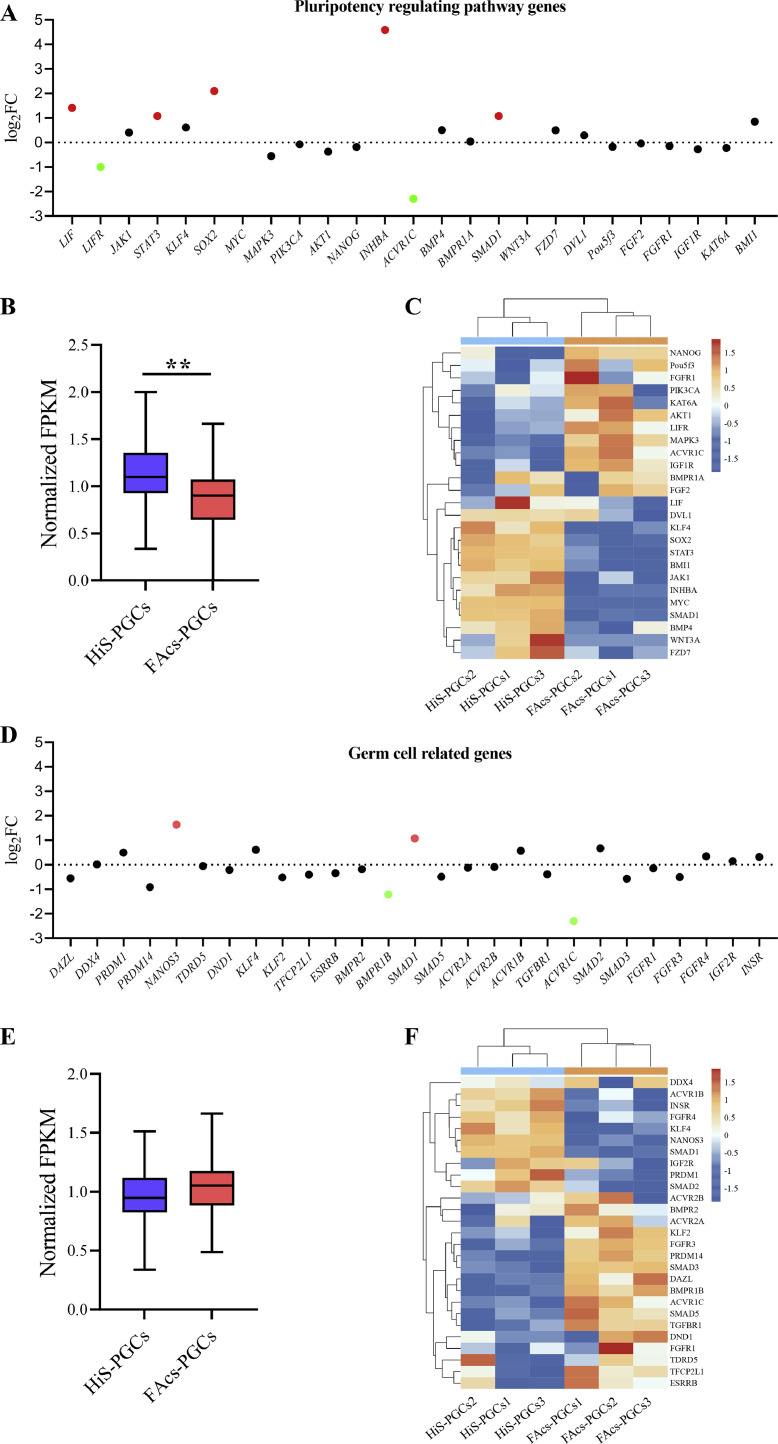
To ensure the reliability of RNA sequencing analysis results, we validated the data obtained from RNA sequencing analysis for reproductive marker genes (DAZL, CVH, and PRDM1) and pluripotency-maintaining genes (POUV, NANOG, and SOX2) through qRT-PCR. The results showed consistency between the RNA sequencing analysis and qRT-PCR validation, demonstrating high accuracy of RNA sequencing analysis (Figure 3). Compared to HiS-PGCs, the expression levels of DAZL, POUV, and NANOG were significantly higher in FAcs-PGCs, while the expression levels of PRDM1 and SOX2 were significantly lower, and there were no significant differences in the other genes (Figure 3). Subsequently, we conducted a systematic comparison of 25 genes related to pluripotency regulation and 27 genes related to germ cells between HiS-PGCs and FAcs-PGCs using RNA sequencing analysis results. Our findings, with a q-value<0.05 and logFC cutoff, showed that 5 pluripotency-regulating pathway genes and 2 germ cell-related genes were upregulated by more than 1-fold in HiS-PGCs compared to FAcs-PGCs (Figure 4A and D). Conversely, only 2 pluripotency-regulating pathway genes and 2 germ cell-related genes were downregulated by less than -1-fold in HiS-PGCs compared to FAcs-PGCs (Figures 4A and 4D). By normalizing the FPKM values of these genes, we observed that the overall expression levels of pluripotency-regulating pathway genes in HiS-PGCs were significantly higher than in FAcs-PGCs, while the overall expression levels of germ cell-related genes showed no significant difference (Figures 4B–4C and 4E–4F). These results suggest that the expression levels of pluripotency-regulating pathway genes in HiS-PGCs are generally higher, while the expression levels of germ cell-related genes do not show significant differences.
Figure 3.
Validation of biological characteristics related genes in RNA sequencing data from PGCs cultured in HiS and FAcs system. FPKM comparison analysis of DAZL (A), CVH (C), PRDM1 (E), POUV (G), NANOG (I), and SOX2 (K) gene between HiS-PGCs and FAcs-PGCs. qRT-PCR analysis of DAZL (A), CVH (C), PRDM1 (E), POUV (G), NANOG (I), and SOX2 (K) gene between HiS-PGCs and FAcs-PGCs after 110 d of in vitro culture. * indicates p < 0.05. ** indicates p < 0.01. *** indicates p < 0.001.
Figure 4.
Comparative analysis of biological characteristics related genes in RNA sequencing data from PGCs cultured in HiS and FAcs system. DEGs of pluripotency regulating pathway (A) and germ cell related genes (D) between HiS-PGCs and FAcs-PGCs. Red: significantly upregulated. Green: significantly downregulated. Black: unregulated. Comparative Analysis of normalized FPKM of pluripotency regulating pathway (B) and germ cell related genes (E) between HiS-PGCs and FAcs-PGCs. Heatmap of pluripotency regulating pathway (C) and germ cell related genes (F) between HiS-PGCs and FAcs-PGCs.
It has been reported that high expression of DNA repair genes and lower expression of apoptosis genes are also important characteristics of PGCs (Rengaraj et al., 2022). Therefore, we also compared the differences in the BER pathway genes, NER pathway genes, MMR pathway genes, NHEJ pathway genes, HR pathway genes, and apoptosis pathway genes. The results showed that only 5/38 BER pathway genes, 1/38 NER pathway genes, 0/19 MMR pathway genes, 1/10 NHEJ pathway genes, 2/36 HR pathway genes, and 1/22 apoptosis pathway genes were shown to be 1-fold upregulated or downregulated in HiS-PGCs compared to FAcs-PGCs (Figure S2), indicating that the DNA repair genes and apoptosis genes pathways did not show significant differences between HiS-PGCs and FAcs-PGCs.
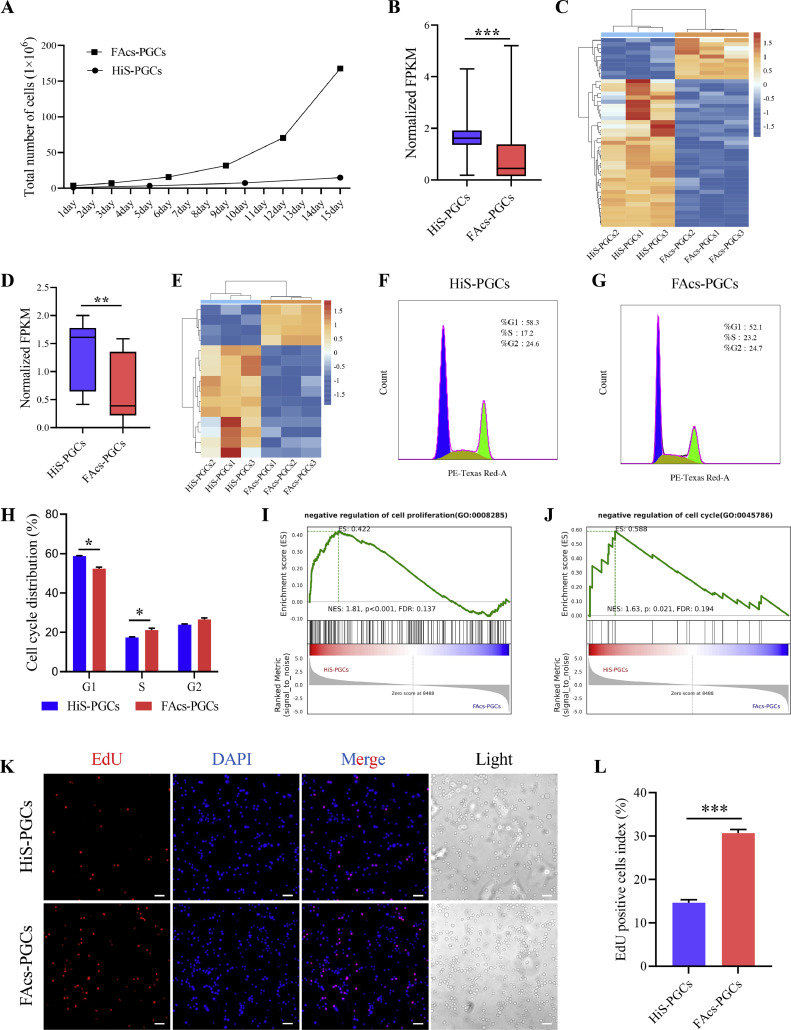
The FAcs Culture System Maintains a Higher Proliferation Capacity of PGCs
In order to compare the proliferation ability of PGCs in 2 culture systems, initially, 1 × 106 PGCs were seeded in each well and cultured continuously for 15 d to observe the proliferation of cells. During the culture process, it was observed that PGCs in the FAcs culture system required approximately 3 d for passaging, whereas PGCs in the HiS culture system required about 5 d for passaging. It was indeed revealed that PGCs exhibited a faster proliferation ability in the FAcs culture system (Figure 5A). By analyzing the GO entries related to the quantity of cells in the DEGs, we discovered a significant enrichment of genes involved in pathways associated with the negative regulation of cell proliferation and the cell cycle. Moreover, after normalizing the FPKM values of DEGs, we observed significantly higher overall expression levels of genes associated with negative regulation of cell proliferation and the cell cycle in HiS-PGCs compared to FAcs-PGCs (Figures 5B–5E). This suggests that the proliferation ability and cell cycle of PGCs in the FAcs culture system were faster. Furthermore, it was observed through PI staining and flow cytometry that the proportion of cells in the S phase was significantly higher, while the proportion of cells in the G1 phase was significantly lower in FAcs-PGCs compared to HiS-PGCs, suggesting a faster cell cycle in FAcs-PGCs (Figures 5F–5H). GSEA further confirmed these results, showing enrichment in pathways related to negative regulation of cell proliferation and the cell cycle in HiS-PGCs (Figure 5I and J). To further confirm this conclusion, we analyzed the proliferation ability of cells through EdU experiments. The results showed that the proportion of EdU-positive cells in FAcs-PGCs was significantly higher than that in HiS-PGCs (Figures 5K and 5L), indicating that FAcs-PGCs have a faster proliferation ability.
Figure 5.
The HiS culture system maintains a higher proliferation capacity of PGCs. (A) Total number of PGCs after 15 d of culture in HiS and FAcs medium. Comparative Analysis of normalized FPKM of genes that negatively regulate cell proliferation (B) and cell cycle (D) between HiS-PGCs and FAcs-PGCs. Heatmap of genes that negatively regulate cell proliferation (C) and cell cycle (E) between HiS-PGCs and FAcs-PGCs. (F-H) Cell cycle analysis of HiS-PGCs compared to FAcs-PGCs after 120 d of in vitro culture. GSEA of genes that negatively regulate cell proliferation (I) and cell cycle (J) at HiS-PGCs versus FAcs-PGCs. (K and L) Evaluation of cell proliferation capacity using the EdU assay in HiS-PGCs and FAcs-PGCs after 140 d of in vitro culture. Scale bar: 50μm. * indicates p < 0.05. *** indicates p < 0.001.
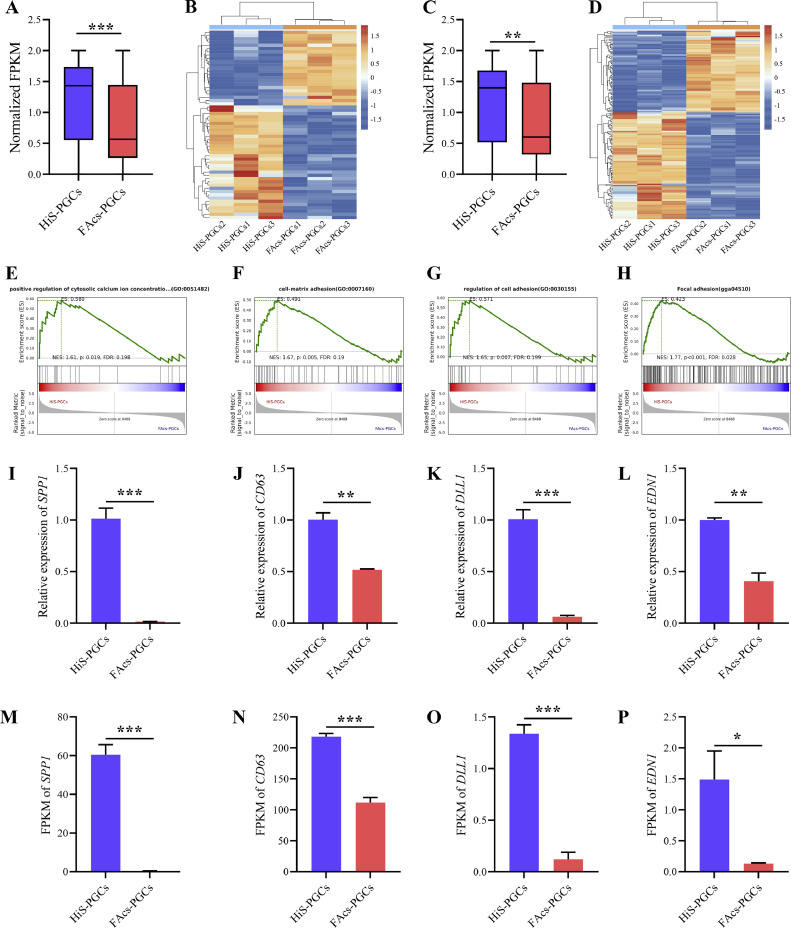
The HiS-PGCs Maintain Higher Levels of Cell Adhesion and Calcium Ion Binding
Previous study has indicated that the phenomenon of cell clustering is closely associated with pathways such as calcium-dependent cell adhesion (van de Lavoir et al., 2006; Whyte et al., 2015). The enrichment of DEGs in the pathways of cell adhesion and calcium ion binding in HiS-PGCs and FAcs-PGCs may be related to the clustering phenomenon of PGCs in the HiS system (Figures 2D–2E). In order to explain the reasons for this phenomenon, GO entries related to calcium ion binding and transport, and cell adhesion were selected (Figures 6A–6D). The FPKM values of these genes were normalized and statistically analyzed, revealing that the overall expression levels of genes involved in calcium ion binding and transport, and cell adhesion in HiS-PGCs were significantly higher compared to FAcs-PGCs (Figure 6A-D). This is consistent with the high calcium ion levels in the HiS system and the increased tendency of cells to cluster (van de Lavoir et al., 2006; Whyte et al., 2015). Additionally, GSEA analysis also showed enrichment of pathways such as positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway, cell-matrix adhesion, regulation of cell adhesion, and focal adhesion in HiS-PGCs (Figures 6E–6H). Subsequently, key genes related to calcium ion binding and transport, and cell adhesion were selected, and their expression was measured using qRT-PCR. The qRT-PCR results were consistent with the RNA sequencing data, confirming the elevated expression of these genes in HiS-PGCs and demonstrating the reliability and accuracy of our RNA sequencing analysis (Figures 6I–6P, Figure S3).
Figure 6.
The HiS-PGCs maintain higher levels of cell adhesion and calcium ion binding. Comparative Analysis of normalized FPKM of calcium ion binding and transport genes (A) and cell adhesion genes (C) between HiS-PGCs and FAcs-PGCs. Heatmap of calcium ion binding and transport genes (B) and cell adhesion genes (D) between HiS-PGCs and FAcs-PGCs. GSEA of calcium ion binding and transport genes (E) and cell adhesion genes (F-H) at HiS-PGCs versus FAcs-PGCs. qRT-PCR analysis of SPP1 (I), CD63 (J), DLL1 (K), and EDN1 (L) gene between HiS-PGCs and FAcs-PGCs. FPKM comparison analysis of SPP1 (M), CD63 (N), DLL1 (O), and EDN1 (P) gene between HiS-PGCs and FAcs-PGCs. * indicates p < 0.05. ** indicates p < 0.01. *** indicates p < 0.001.
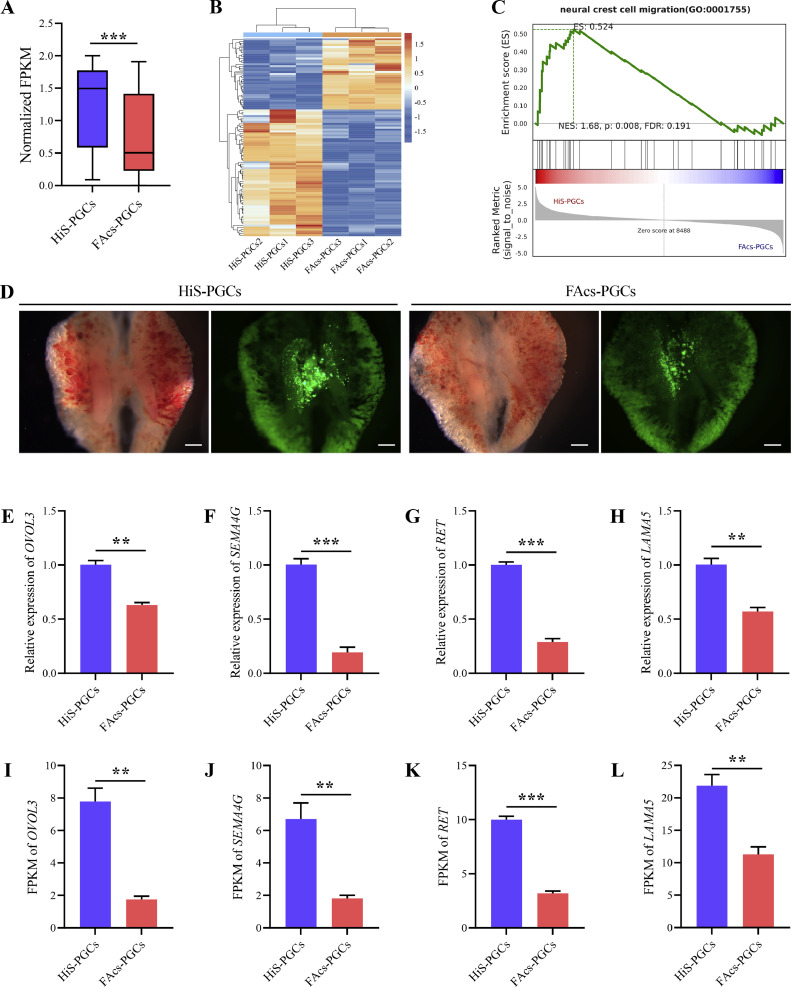
HiS-PGCs Maintain a Higher Level of Expression of Cell Migration-Related Genes
The ability of PGCs to migrate and colonize the gonads is one of their most important features. Therefore, we further compared the expression of cell migration-related genes in HiS-PGCs and FAcs-PGCs. By filtering the GO entries related to cell migration and analyzing the normalized FPKM values of DEGs under these entries, we found that the overall expression level of cell migration-related genes in HiS-PGCs was significantly higher compared to FAcs-PGCs (Figures 7A and 7B). Additionally, in the GSEA analysis, the neural crest cell migration pathway was enriched in HiS-PGCs (Figure 7C), suggesting a higher migration capability of HiS-PGCs. Subsequently, we administered GFP-labeled HiS-PGCs and FAcs-PGCs into the gonads of recipient chick embryos at 2.5-days-old and allowed them to incubate until 7.5 d. Our observations revealed that both HiS-PGCs and FAcs-PGCs migrated to the recipient gonads effectively, with no significant variance in migration efficiency between the two cell types (Figure 7D, Table 2). Furthermore, qRT-PCR analysis confirmed that the expression levels of key genes related to cell migration were consistent with the RNA sequencing data (Figures 7E–7L).
Figure 7.
HiS-PGCs maintain a higher level of expression of cell migration-related genes. (A) Comparative Analysis of normalized FPKM of cell migration genes between HiS-PGCs and FAcs-PGCs. (B) Heatmap of cell migration genes between HiS-PGCs and FAcs-PGCs. (C) GSEA of cell migration genes at HiS-PGCs versus FAcs-PGCs. (D) Representative images showing the migration of HiS-PGCs and FAcs-PGCs to recipient chicken embryonic gonads. Scale bar: 250μm. qRT-PCR analysis of OVOL3 (E), SEMA4G (F), RET (G), and LAMA5 (H) gene between HiS-PGCs and FAcs-PGCs. FPKM comparison analysis of OVOL3 (I), SEMA4G (J), RET (K), and LAMA5 (L) gene between HiS-PGCs and FAcs-PGCs. ** indicates p < 0.01. *** indicates p < 0.001.
Table 2.
The migration efficiency of HiS-PGCs and FAcs-PGCs.
| Cell line | No. of embryos injections | No. of live embryos (%) | No. of colonization (%) |
|---|---|---|---|
| HiS-PGCs | 40 | 27 (67.5%) | 25 (92.6%) |
| FAcs-PGCs | 41 | 25 (61.0%) | 23 (92.0%) |
DISCUSSION
This article aims to compare the HiS and FAcs culture systems in terms of their ability to support the growth, maintenance of characteristics, and lineage transfer of PGCs. It was found that HiS-PGCs tend to exhibit more clustered growth, whereas FAcs-PGCs show a more dispersed growth pattern. The study results suggest that while both HiS and FAcs culture systems can support the proliferation and basic characteristics of chicken PGCs, there are differences in terms of proliferation, pluripotency regulation, and cell adhesion processes. Although the migration efficiency between the two systems is similar, HiS-PGCs maintain cell migration-related genes at a higher level. Further research into the potential molecular mechanisms underlying these differences is crucial in order to optimize the in vitro culture of PGCs and enhance their potential application in genetic resource preservation and manipulation in animal production.
PGCs, like pluripotent stem cells, express high levels of genes related to pluripotency regulation, and as ancestral cells of gametes, the expression of early reproductive cell marker genes is also an important feature (van de Lavoir et al., 2006; Whyte et al., 2015). In this study, pluripotency and reproduction-related genes in HiS-PGCs and FAcs-PGCs were detected at both mRNA and protein levels, and it was found that both HiS-PGCs and FAcs-PGCs can efficiently maintain the expression of pluripotency and reproduction-related genes. Furthermore, both culture systems can maintain the continuous presence of glycogen granules in PGCs, which is also one of the basic characteristics of PGCs. These results are consistent with previous research (van de Lavoir et al., 2006, Whyte et al., 2015) and indicate that the HiS-PGCs and FAcs-PGCs used in this study have the basic characteristics of PGCs, ensuring that the samples used for subsequent RNA sequencing represent the influence of the HiS and FAcs systems on PGC culture. Through RNA sequencing analysis, 2,145 DEGs were identified, and the consistent results of qRT-PCR and RNA sequencing analysis confirmed the accuracy of the sequencing results. The comparative analysis of genes regulating pluripotency and reproduction-related genes at the RNA sequencing data level showed an overall higher in the expression of genes regulating pluripotency in HiS-PGCs, while there was no significant difference in the expression of reproduction-related genes in the two systems. This result also indicates that the reason PGCs in the HiS system are prone to dedifferentiation into EGCs is due to the upregulation of genes regulating pluripotency (van de Lavoir et al., 2006). Maintaining a high level of DNA repair genes and lower expression of apoptosis genes is also an important feature of PGCs as germ cells, and the overall expression levels of these genes in HiS-PGCs and FAcs-PGCs did not show significant differences, indicating the consistency of the two culture systems in protecting genetic stability. This is essential for PGCs as the tool cells for germplasm preservation and genetic improvement.
The FAcs system decreases the concentration of calcium ions, alleviating the clustering growth pattern of PGCs in the HiS system[11,12]. In this study, it was observed that HiS-PGCs show clustered growth, while FAcs-PGCs show dispersed growth, consistent with previous research (van de Lavoir et al., 2006; Whyte et al., 2015; Altgilbers et al., 2021). The GO and KEGG analyses highlighted that DEGs were significantly enriched in pathways associated with cell adhesion and calcium ion binding in HiS-PGCs. In addition, comparative analysis indicated that these genes are maintained at higher levels in HiS-PGCs, and GSEA analysis also showed enrichment in pathways such as focal adhesion, cell-matrix adhesion, regulation of cell adhesion, and positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway. These findings further elucidate the differences in the growth characteristics of PGCs due to variations in the concentration of calcium ions added in the HiS and FAcs culture systems.
During the PGC culture process, it was observed that the proliferation ability of PGCs in the FAcs system is faster than that in the HiS system. Analysis of RNA sequencing data found that a large number of DEGs are enriched in pathways related to negative regulation of cell proliferation and cell cycle, and the expression levels of these genes are significantly higher in HiS-PGCs. These results were validated by cell cycle detection, EdU assay and GSEA, indicating that in the FAcs system, the proliferation ability and cell cycle of PGCs are at a higher level. The faster proliferation rate of PGCs in the FAcs system may mean that they are more easily expandable, providing more cell numbers for research and application. This is crucial for research and production applications in genetic resource preservation and genetic manipulation (Trefil et al., 2017; Lázár et al., 2021; Chen et al., 2023; Hamai et al., 2023), as it saves time and economic costs by obtaining a large number of cells more quickly. In addition, the improved proliferation rate may also mean that it is easier to observe the growth and differentiation process of cells in research, leading to a better understanding of their biological characteristics. Therefore, fast-proliferating chicken PGCs can provide more possibilities for chicken breeding and contribute to improving the reproductive efficiency and quality of livestock.
Finally, through the comparison of the migration efficiency of PGCs in the two systems, it was found that although the migration efficiency between the two systems is similar, the cell migration-related genes in His-PGCs are maintained at a higher level. Factors in the cell culture system, including culture medium components, extracellular matrix, and growth factors, may affect cell behavior (Choi et al., 2010; Whyte et al., 2015; Chen et al., 2018; Collarini et al., 2019). When the expression of cell migration-related genes is maintained at a higher level in His-PGCs, it may be due to a specific extracellular signal or environment provided by the His culture system, which may be more conducive to the expression and maintenance of cell migration-related genes. Further consideration and verification are needed to understand the factors influencing the expression and maintenance of cell migration-related genes in PGCs in order to provide new ideas for the continuous optimization of PGC culture systems. PGCs that migrate to the gonads can further develop into germline stem cells and ultimately form gametes, producing offspring. It will be necessary to systematically test and evaluate the offspring production capabilities of FAcs-PGCs and HiS-PGCs in future studies. At the same time, as PGCs can be cultured for long-term passaging (van de Lavoir et al., 2006; Chen et al., 2018), whether the FAcs and HiS culture systems can permanently maintain their proliferation and fundamental characteristics in vitro remains an important question for further investigation.
In conclusion, the results of the study indicate that although the HiS and FAcs culture systems can support the proliferation and basic characteristics of chicken PGCs, there are differences in proliferation, pluripotency regulation, and cell adhesion processes. Further research on the potential molecular mechanisms controlling these differences is crucial to optimizing the in vitro culture of PGCs and enhancing their potential application in genetic resource preservation and manipulation in animal production.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2021YFD1200301, 2021YFD1200302), the National Natural Science Foundation of China (32102534), and the Natural Science Foundation of Jiangsu Province (BK20210813).
Data Availability Statement: The data that support the findings of this study are available from the corresponding author upon reasonable request. The raw sequence reads have been archived in the NCBI SRA database with the accession number PRJNA1086231 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA1086231).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2024.104058.
Contributor Information
Ying-Jie Niu, Email: niuyj@yzu.edu.cn.
Bichun Li, Email: yubcli@yzu.edu.cn.
Appendix. Supplementary materials
REFERENCES
- Altgilbers S., Klein S., Dierks C., Weigend S., Kues W.A. Cultivation and characterization of primordial germ cells from blue layer hybrids (Araucana crossbreeds) and generation of germline chimeric chickens. Sci Rep. 2021;11:12923. doi: 10.1038/s41598-021-91490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne M., Woodcock M., Doddamani D., Hu T., Taylor L., Hawken R.J., McGrew M.J. Direct allele introgression into pure chicken breeds using Sire Dam Surrogate (SDS) mating. Nat Commun. 2021;12:659. doi: 10.1038/s41467-020-20812-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.- C., Saito D., Suzuki T., Takemoto T. An inducible germ cell ablation chicken model for high-grade germline chimeras. Development. 2023;150 doi: 10.1242/dev.202079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-C., Chang W.-C., Lin S.-P., Minami M., Jean C., Hayashi H., Rival-Gervier S., Kanaki T., Wu S.C., Pain B. Three-dimensional culture of chicken primordial germ cells (cPGCs) in defined media containing the functional polymer FP003. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Chen M., Lu Z., Yang M., Xie L., Zhang W., Xu H., Lu K., Lu Y. Cholesterol induces proliferation of chicken primordial germ cells. Anim. Reprod. Sci. 2016;171:36–40. doi: 10.1016/j.anireprosci.2016.05.011. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.W., Kim S., Kim T.M., Kim Y.M., Seo H.W., Park T.S., Jeong J.W., Song G., Han J.Y. Basic fibroblast growth factor activates MEK/ERK cell signaling pathway and stimulates the proliferation of chicken primordial germ cells. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collarini E.J., Leighton P.A., Van de Lavoir M.- C. In: Pages 403–430 in Microinjection. Liu C., Du Y., editors. Springer; New York, NY, USA: 2019. Production of transgenic chickens using cultured primordial germ cells and gonocytes. [DOI] [PubMed] [Google Scholar]
- Consortium T.G.O. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehdilani N., Yousefi Taemeh S., Rival-Gervier S., Montillet G., Kress C., Jean C., Goshayeshi L., Dehghani H., Pain B. Enhanced cultivation of chicken primordial germ cells. Sci. Rep. 2023;13:12323. doi: 10.1038/s41598-023-39536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki R., Hirose F., Furusawa S., Horiuchi H. An improved protocol for stable and efficient culturing of chicken primordial germ cells using small-molecule inhibitors. Cytotechnology. 2020;72:397–405. doi: 10.1007/s10616-020-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamai N., Koide C., Tansho Y., Ooka Y., Hirano M., Fatira E., Tsudzuki M., Nakamura Y. Development of cryopreservation media for the slow-freezing of cultured primordial germ cells in chicken. J. Reprod. Dev. 2023;69:109–117. doi: 10.1262/jrd.2022-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagenç L., Petitte J.N. Soluble factors and the emergence of chick primordial germ cells in vitro. Poultry Science. 2000;79:80–85. doi: 10.1093/ps/79.1.80. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár B., Molnár M., Sztán N., Végi B., Drobnyák Á., Tóth R., Tokodyné Szabadi N., McGrew M.J., Gócza E., Patakiné Várkonyi E. Successful cryopreservation and regeneration of a partridge colored Hungarian native chicken breed using primordial germ cells. Poultry Science. 2021;100 doi: 10.1016/j.psj.2021.101207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Choi H.J., Park K.J., Woo S.J., Kim Y.M., Han J.Y. Development and characterization of a CRISPR/Cas9-mediated RAG1 knockout chicken model lacking mature B and T cells. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.892476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara D., Mori T., Makino R., Nakamura Y., Oishi I., Ono T., Nirasawa K., Tagami T., Kagami H. Culture conditions for maintain propagation, long-term survival and germline transmission of chicken primordial germ cell-like cells. J. Poult. Sci. 2014;51:87–95. [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.- F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M.J., Patterson N., Mesirov J.P., Golub T.R., Tamayo P., Spiegelman B., Lander E.S., Hirschhorn J.N., Altshuler D., Groop L.C. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Park T.S., Jeong D.K., Kim J.N., Song G.H., Hong Y.H., Lim J.M., Han J.Y. Improved germline transmission in chicken chimeras produced by transplantation of gonadal primordial germ cells into recipient embryos. Biol. Reprod. 2003;68:1657–1662. doi: 10.1095/biolreprod.102.006825. [DOI] [PubMed] [Google Scholar]
- Pu L., Xie L., Chen J., Sun H., Huang Z., Xu T., Tian K., Zhong J., Xu H., Liu X., Lu Y. Tracking the dynamics of female germ cell development during peri-hatch periods using a gene-edited chicken model. Poultry Science. 2023;102 doi: 10.1016/j.psj.2022.102377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengaraj D., Won S., Jung K.M., Woo S.J., Lee H., Kim Y.M., Kim H., Han J.Y. Chicken blastoderms and primordial germ cells possess a higher expression of DNA repair genes and lower expression of apoptosis genes to preserve their genome stability. Sci Rep. 2022;12:49. doi: 10.1038/s41598-021-04417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li Y., Zong Y., Mehaisen G.M.K., Chen J. Poultry genetic heritage cryopreservation and reconstruction: advancement and future challenges. J. Anim. Sci. Biotechnol. 2022;13:115. doi: 10.1186/s40104-022-00768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefil P., Aumann D., Koslová A., Mucksová J., Benešová B., Kalina J., Wurmser C., Fries R., Elleder D., Schusser B., Hejnar J. Male fertility restored by transplanting primordial germ cells into testes: a new way towards efficient transgenesis in chicken. Sci. Rep. 2017;7:14246. doi: 10.1038/s41598-017-14475-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lavoir M.-C., Diamond J.H., Leighton P.A., Mather-Love C., Heyer B.S., Bradshaw R., Kerchner A., Hooi L.T., Gessaro T.M., Swanberg S.E., Delany M.E., Etches R.J. Germline transmission of genetically modified primordial germ cells. Nature. 2006;441:766–769. doi: 10.1038/nature04831. [DOI] [PubMed] [Google Scholar]
- Whyte J., Glover J.D., Woodcock M., Brzeszczynska J., Taylor L., Sherman A., Kaiser P., McGrew M.J. FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Rep. 2015;5:1171–1182. doi: 10.1016/j.stemcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.