Abstract
The steps governing healing with or without fibrosis within the same microenvironment are unclear. After acute kidney injury (AKI), injured proximal tubular epithelial cells activate SOX9 for self-restoration. Using a multimodal approach for a head-to-head comparison of injury-induced SOX9-lineages, we identified a dynamic SOX9 switch in repairing epithelia. Lineages that regenerated epithelia silenced SOX9 and healed without fibrosis (SOX9on-off). By contrast, lineages with unrestored apicobasal polarity maintained SOX9 activity in sustained efforts to regenerate, which were identified as a SOX9on-on Cadherin6pos cell state. These reprogrammed cells generated single-cell WNT activity to provoke a fibroproliferative response in adjacent fibroblasts, driving AKI to chronic kidney disease. Transplanted human kidneys displayed similar SOX9/CDH6/WNT2B responses. Thus, we have uncovered a sensor of epithelial repair status, the activity of which determines regeneration with or without fibrosis.
Depending upon the nature and severity of the initial injury, the damaged tissue microenvironment of adult mammalian organs can demonstrate progressive scarring, whereas other foci exhibit scarless tissue recovery (1–3). This spatial heterogeneity in interstitial fibrosis seen across diverse organs is also frequently observed during progression of acute kidney injury (AKI) to chronic kidney disease (CKD) (4, 5). AKI, defined by an abrupt decline in kidney function, is often caused by acute proximal tubular epithelial cell injury (6, 7) with selective damage per se able to drive interstitial fibrosis (8). Failed tubule recovery has been broadly linked with post-AKI fibrosis (9). However, the precise mechanism that determines why one focus progressively scars, whereas other initially injured foci that might only be a few cell distances away heal without fibrosis, have remained elusive.
Our previous work, substantiated by other studies, identified Sox9 (a SRY-related high-mobility-group box family of transcription factor) activation as a fundamental epithelial injury–induced repair response (Sox9+) (10, 11), and proposed that Sox9 activity observed 4 weeks after AKI might represent regions of unresolved injury and repair processes. However, the precise characterization of such processes, including their relationship to the initial Sox9+ cell, remained unexamined. To investigate this, we first tested our hypothesis that the descendants of Sox9+ cells would silence Sox9 upon regeneration, whereas persistence of Sox9 activity was predicted for the Sox9 lineage with features of unrestored epithelia. If true, then this would provide an opportunity to conduct head-to-head, spatiotemporal examination of the regenerated lineage compared with the progeny that could not fully restore the epithelia right from the onset of a single-inciting insult within with same microenvironment.
Identification of a dynamic SOX9/CDH6 switch within the repairing epithelia
First, injury-induced SOX9+ cells were fate-mapped, using previously validated Sox9IRES-CreERT2/+: R26RtdT/+ animals in a model of long-term survival–compatible bilateral ischemia reperfusion injury (IRI) that induced AKI to progress to CKD (5, 10). In human AKI, return of serum creatinine (a biochemical marker of kidney function) to normal baseline level reflects recovery from acute tubular necrosis through regeneration of tubular epithelial cells. In our model, serum creatinine returned to near normal levels by day 10. Therefore, the day 10 damaged tissue microenvironment, which correlated with clinically meaningful kidney function recovery, was investigated first (Fig. 1A and fig. S1, A and B). A strong basolateral expression of sodium-potassium ATPase ion pumps (ATP1A1basolateral/high) and a thick, apical lotus tetragonoglobus lectin–enriched brush border (LTLthick) characterized uninjured proximal tubular epithelia (fig. S1C). Consistent with our previous report (10), injured epithelial cells with disrupted basolateral polarity activated SOX9, with tdT+ cells reporting such cells at 48 hours after IRI (Fig. 1A). On day 10 after injury, the initially labeled tdT+ cells had expanded (fig. S1D), and nearly the entire Sox9 lineage, which regenerated the epithelia marked by restoration of apicobasolateral polarity, had silenced SOX9 (tdT+Sox9neg; Fig. 1A and fig. S1,E and F). Conversely, the Sox9 lineage that exhibited unrestored epithelia, marked by cytoplasmic flattening and disrupted apicobasolateral polarity, maintained the SOX9+ state, thereby demonstrating sustained Sox9 activity (IRI day 10: SOX9+tdT+; Fig. 1A; fig. S1, F to H; and tables S1 and S2). The unrestored lineages were mainly observed within the proximal tubules of the outer medullary region, which undergo relatively extensive cell loss due to exaggerated susceptibility to injury (7) (fig. S1H). A subset of such lineages displayed morphological features typically associated with tubulogenesis, such as prominent cytoplasmic projections with elongated nuclei suggestive of filopodia formation, likely reflecting an attempt to repopulate the extensively denuded tubule, as well as luminal clusters with long cellular axis oriented in all directions (12) (Fig. 1A and fig. S1, G and H). Similar to IRI-induced AKI, we observed distinct Sox9 lineages with dynamic Sox9 activity tightly linked to their restorative status in rhabdomyolysis-induced AKI (rhabdo-AKI), another distinct and clinically relevant model of toxic AKI (fig. S2, A and B, and table S2). Therefore, hereafter, the terms Sox9on, Sox9on-off, and Sox9on-on will be used to describe the following cells: Sox9on for the initially injured proximal tubular epithelial cells (PTECs) that activated SOX9 at 48 hours after injury, Sox9on-off for the Sox9on lineage that regenerated the epithelia, and Sox9on-on for the counterpart that weren’t able to restore the epithelia by day 10 after AKI.
Fig. 1. scRNA-seq reveals a dynamic SOX9/CDH6 switch within the repairing nephron epithelium.
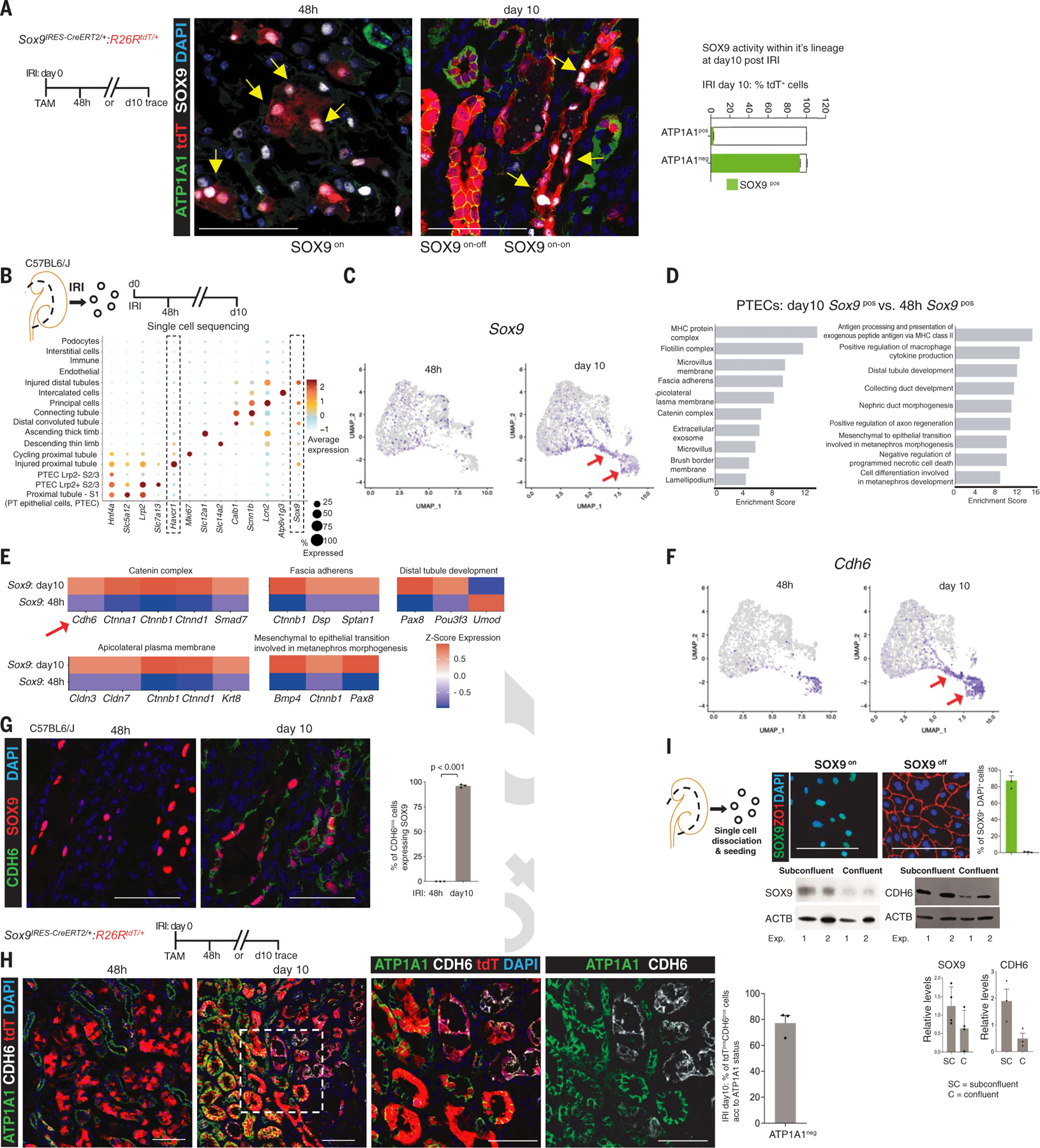
(A) Identification of a dynamic Sox9 switch. Shown are the schema of lineage-tracing of injury-induced Sox9pos cells and co-immunoanalysis for SOX9 and the basolateral polarity marker ATP1A1, showing that cells with disrupted basolateral polarity activated SOX9. Note the co-localization of tdT with SOX9pos cells at 48 hours after IRI (IRI 48 hours: Sox9on cells, yellow arrows). By day 10 after IRI, the lineage of Sox9on cells that restored basolateral polarity had silenced SOX9 (Sox9on-off cell state, regenerated epithelia), whereas the lineage with unrestored ATP1A1 maintained SOX9 activity (Sox9on-on cells, yellow arrows). (B) Schema of isolating single cells to compare IRI 48 hours Sox9pos (n = 2) versus day 10 Sox9pos cells, with dot plot showing average gene expression values and percentage of cells expressing cell-type- specific markers of differentiation, injury (Havcr1 and Lcn2), and repair (Sox9) response. (C) Time-resolved UMAP projection of Sox9pos cells within the proximal tubule cluster. Arrows show distinct clusters at day 10 after IRI containing Sox9pos cells compared with 48 hours after IRI. (D) Topmost enriched GO cellular components and biological processes in IRI day 10 Sox9pos cells versus 48 hour Sox9pos cells. (E) Heatmap of genes driving the GO terms. Shown is the enrichment of epithelial cell-cell adhesion machinery, including Cdh6, within day 10 Sox9pos cells. (F) Time-resolved UMAP projection of Cdh6pos cells (arrows) within the PT cluster. (G and H) Co-immunostaining for CDH6 and SOX9 (G) and ATP1A1 (H) showing CDH6 expression restricted to day 10 SOX9pos cells (G) and to the Sox9 lineage with unrestored ATP1A1-based basolateral polarity versus the lineage that restored polarity (H), indicating that CDH6 demarcates Sox9on-on cells (SOX9posCDH6pos cell state). (I) Co-immunostaining for SOX9 and the epithelial tight junction marker ZO-1 and immunoblot for SOX9 and CDH6 showing that cells that lacked cell-cell contact (subconfluent state) activated SOX9 and CDH6, which waned upon restoration of tight junctions (confluent monolayer). Box inset highlights the region depicted at high magnification. All n = 3 animals per time point unless otherwise stated. Data are shown as mean ± SEM. Scale bars, 100 μm. For total cells counted, see table S1.
To study the Sox9on and Sox9on-on cells, we performed single-cell RNA sequencing (scRNA-seq) 48 hours and 10 days after IRI (Fig. 1B and figs. S3 and S4). After confirming cell-type-specific distinct clustering of single-cell datasets (Fig. 1B and figs. S3 and S4), we focused on the proximal tubule (fig. S5A). Single-cell analysis showed heterogeneous Sox9+ cells at 48 hours, whereas the day 10 population demonstrated relative homogeneity (Fig. 1C and fig. S5B). Gene ontology (GO) term analysis of the differentially expressed genes showed enrichment of terms related to the formation of proximal tubular epithelia and function in Sox9+ cells at 10 days versus 48 hours (Fig. 1D). For example, in the category “cellular component,” the terms “lamellipodium,” “fascia adherens,” and “catenin complex” were enriched, in addition to “microvillus,” the latter a characteristic of PTECs. The cadherin-catenin-actin complex and lamellipodium are essential for the formation and maturation of epithelia (13, 14) (Fig. 1D and fig. S6A). In addition to Ctnn1a and Ctnnb1, Cdh6, which encodes cadherin 6 (k-cadherin), also contributed to the enriched term “catenin complex” (Fig. 1, D to F). Cdh6 is essential for the formation of a fully polarized nephron epithelium during nephrogenesis (15). Spatiotemporal mapping confirmed that CDH6 expression was restricted to IRI day 10 SOX9+ cells and to the Sox9 lineage with Sox9on-on activity, which exhibited disrupted or absent apicobasolateral polarity (Fig. 1, G and H, and table S1). Consistent with the observed LTLlow/absent apical brush border of Sox9on-on cells, our single-cell analysis also predicted low Lrp2 (encoding the apical brush border component megalin) status of Cdh6+ cells (fig. S6A). The presence of CDH6+LRP2low cells was confirmed at day 10 after injury (fig. S6B). Additionally, the genes associated with apicobasolateral polarity (Fat1, Myo9a), both linked with proximal tubule formation and function (16), were enriched in Sox9on-on versus Sox9on cells (fig. S6C). Complementing the GO category cellular component, the biological processes terms linked with nephrogenesis (Bmp4, Ctnnb1, and Pax8) were enriched within Sox9on-on versus Sox9on cells (Fig. 1E). Haploinsufficiency of SOX9, BMP4, CTNNB1, and PAX8, respectively, led to murine and human kidney hypoplasia and/or malformations. We validated Bmp4 enrichment within Sox9on-on cells (fig. S6, D and E).
Next, we fate-mapped Sox9on-on cells by injecting tamoxifen at day 10 after IRI (fig. S7). Consistent with the results of a co-immunoanalysis study (Fig. 1G), ~90% of the initially labeled cells were CDH6+. Two weeks later, at least a subset of Sox9on-on cells regenerated the epithelia, consistent with the “ongoing attempt to regenerate” prediction of the scRNAseq analysis (fig. S7, A to D, and table S2). Although the proportion of regenerated tubules in the damaged outer medullary region was smaller compared with the outer and inner cortices, the regenerated lineage silenced SOX9/CDH6 activity. Therefore, this finding supports a continuous, dynamic SOX9/CDH6 axis despite the interstitium becoming progressively fibrotic and inflamed. A similar dynamic activity was also observed in rhabdo-AKI (fig. S8). Further, on similar lines, in vitro replicating subconfluent cells lacking cell-cell contact activated SOX9 and CDH6, which returned to baseline upon quiescent, confluent, monolayer formation, the latter characterized by acquisition of ZO-1, a tight junction protein (Sox9on-off cell state; Fig. 1I).
We also leveraged published databases that used single-nuclear sequencing to cross-validate the Sox9/Cdh6 switch (fig. S9A) (17, 18). Consistent with our scRNAseq analysis, single-nuclear trajectory analysis of Sox9+ nuclei demonstrated distinct Sox9 lineages. One Sox9 lineage restored the transcriptome back to normal PTECs and switched off Sox9. By contrast, another lineage, which was marked by sustained Sox9 activity, acquired a distinct transcriptome characterized by Cdh6+ nuclei (fig. S9, B to E). Thus, snRNA-seq analysis further substantiated our findings. We have therefore identified a dynamic Sox9/Cdh6 switch tightly linked with the restorative status of the proximal tubule epithelia, with Sox9on-on cell state (SOX9posCDH6pos) most probably highlighting ongoing attempts to regenerate the epithelia.
SOX9posCDH6pos cells form a central hub of myofibroblast generation and maintenance
In IRI- and rhabdo-induced AKI to CKD models, head-to-head comparison between the two lineages showed that most of the αSMA+ myofibroblasts were conspicuously located adjacent to Sox9on-on cells (Fig. 2A; fig. S10, A to D; and tables S1 and S2). A similar robust, intimate association was observed with the CDH6pos lineage (Fig. 2B). The outer medullary region exhibited most of the αSMA response paralleling Sox9on-on activity (fig. S10, A to D). Substantial juxtaposition at single-cell spatial distance was confirmed by Sox9IRES-CreERT2/+:R26RtdT/+: Acta2-GFP [Acta2 encoding αSMA, green fluorescent protein (GFP)] animals (Fig. 2B, fig. S10C, and tables S1 and S2). To ascertain whether a distinct Acta2negative profibrotic celltype, for example, ACTA2neg COL1A1+ cells, might encase the CDH6neg cells, we searched our scRNAseq datasets. Consistent with known responses in fibrotic kidneys, Acta2-, Col1a1-, and Col3a1-expressing cells were observed within the Pdgfrb+ interstitial cluster, the latter signature predominantly contributed by the day 10 Pdgfrb+cells (fig. S11A). Unlike the lung, no distinct Acta2negCol1a1+ or Col3a1+ demarcated cell clusters were observed (fig. S11A). These findings were cross-validated by searching published snRNA-seq datasets of post-AKI kidneys (fig. S11B). Co-immunoanalysis confirmed that interstitial cells co-expressed ACTA2 and COL1A1, with a marked paucity of COL1A1+ cells around the regenerated lineage (fig. S11C). Up to 88% of Pdgfrb+ cells that activated αSMA were the resident Pdgfrb+ fibroblasts located within single-cell distance of Sox9on-on cells (fig. S11, D and E, and table S2). The above findings highlight that Sox9on-off lineages heal without fibrosis and implicate the CDH6pos lineage (SOX9posCDH6pos) as a possible cell state that generates myofibroblasts through a short-range secretory ligand.
Fig. 2. CDH6 status is tightly linked to repair with or without αSMApos myofibroblast response through single-cell Wnt activity.
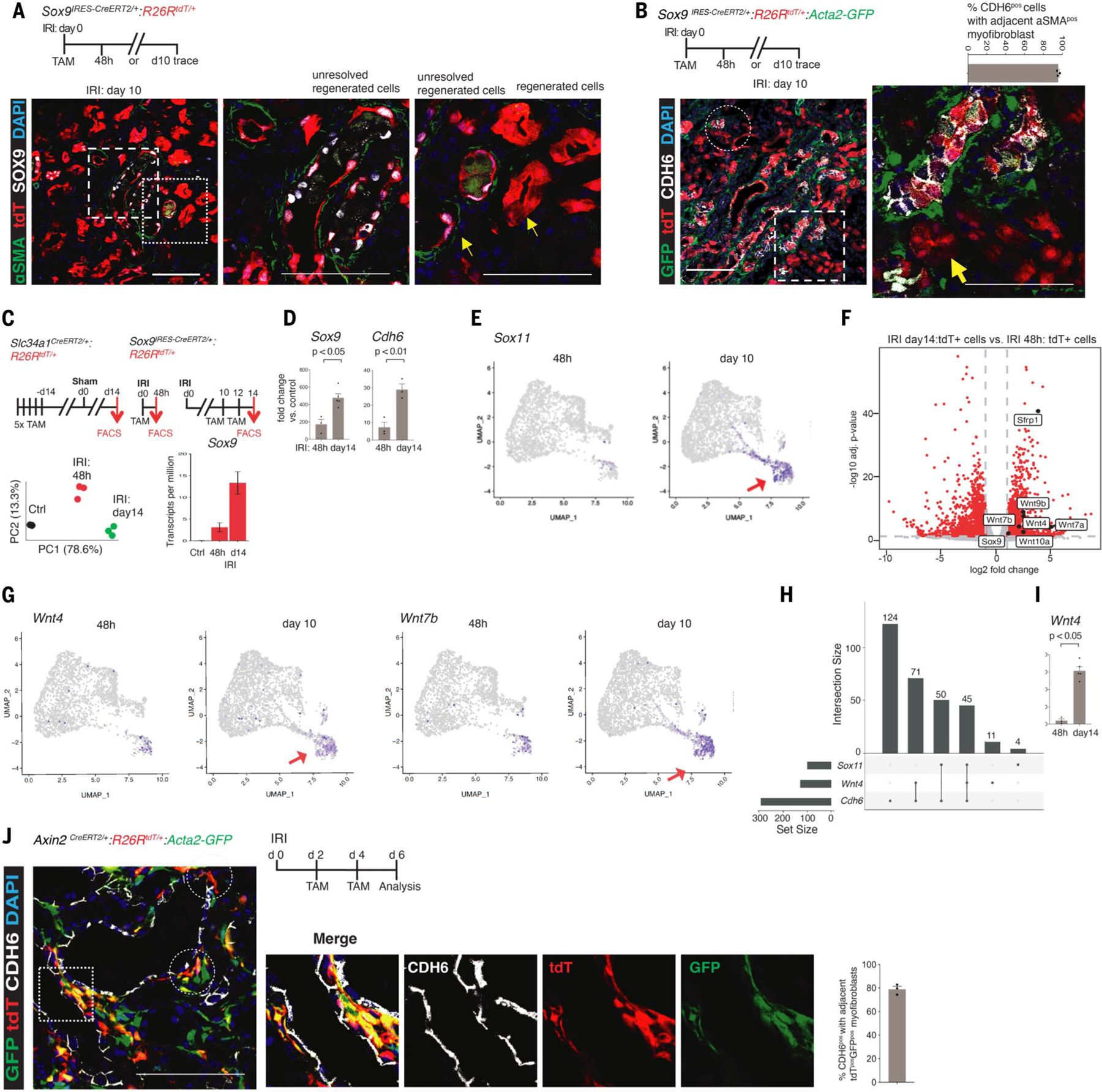
(A) Schema of lineage-tracing of IRI-induced Sox9pos cells and co-immunostaining showing αSMApos myofibroblasts adjacent to SOX9postdTpos cells (Sox9on-on cells, arrows) but no detectable αSMA activity around the Sox9on-off cells (arrow). Left and right magnified panels correspond to the foci highlighted by corresponding left and right box insets. (B) Co-immunoanalysis showing αSMApos myofibroblasts encasing the CDH6pos Sox9 lineages at single-cell spatial distance; by contrast, the paucity of such response around the CDH6negSox9-lineage can be observed (arrow). Circle and box highlight such foci, with the magnified panel corresponding to the foci highlighted by the box inset. (C) Schemas for purifying Slc34a1pos normal PTECs (control tdTpos cells) and Sox9pos cells 48 hours and day 14 after IRI and principal components analysis (PCA) plot showing distinct transcriptomic profiles of purified tdTpos cells. Sox9 transcripts were enriched in purified tdTpos cells. (D) qPCR of purified tdTpos cells confirming Sox9 and Cdh6 enrichment. (E) UMAP projections for Sox11 at 48 hours and day 10 after IRI. Arrows highlight the distinct day 10 cluster composed of these cells. Single Sox9pos and Cdh6pos cells also contributed to this cluster (Fig. 1, B and E). (F) Volcano plot demonstrating enrichment of Wnt ligands within day 14 versus 48 hour tdTpos cells. (G) UMAP projections for Wnt4 and Wnt7b at 48 hours and day 10 after IRI. Arrows highlight the distinct day 10 cluster, which contained such single Wnt4pos and Wnt7bpos cells. Note that the same distinct cluster was composed of day 10 Sox9pos, Cdh6pos, and Sox11pos cells [see also Fig. 1, B and E, and this figure (E)]. (H) Upset plot analysis of scRNAseq datasets showing subset of single Cdh6pos cells enriched with Wnt4, including Cdh6posSox11pos subsets. (I) qPCR of purified tdTpos cells confirming Wnt4 enrichment in day 14 post-IRI tdTpos cells versus their 48-hour counterparts. (J) Schema for labeling early Axin2pos cells after IRI and immunoanalysis for CDH6 showing co-localization of tdTpos and GFPpos cells with tdTposGFPpos cells located adjacent to CDH6pos subsets. Circles and box highlight single-cell, biologically active Wnt-enriched niches robustly linked with subsets of CDH6pos cells. Magnified panels correspond to the foci highlighted by box inset. All n = 3 animals per time point unless otherwise stated. Data are shown as mean ± SEM. Scale bars, 100 μm. For total cells counted, see table S1.
To identify the secretory ligand(s), we next performed RNA-seq–based profiling of IRI day 14 tdT+ versus 48 hour tdT+ cells, with Slc34a1+tdT+ cells serving as normal, uninjured control PTECs (Fig. 2C and fig. S12, A and B). Sox9 transcripts were enriched within both tdT+ populations versus normal control PTECs (~3-fold and ~13-fold), respectively (Fig. 2C and fig. S12C). Quantitative polymerase chain reaction (qPCR) validated Sox9 enrichment and confirmed Cdh6 activation specifically within IRI day 14 tdT+ cells versus 48 hour tdT+ cells (Fig. 2D), demonstrating the fidelity of the Sox9 reporter animals for Sox9 activity. Genes such as Ctnnb1, Ctnnd1, Pou3f3, and Smad7, which contributed to the topmost enriched GO terms in single day 10 Sox9+cells (Fig. 1E), were also enriched within IRI day 14 tdT+ versus 48 hour tdT+ cells (fig. S12, D and E). The GO analysis enriched term “kidney development” showed Sox11 enrichment within IRI day 14 tdT+ versus 48 hour tdT+ cells (fig. S12E). The renal role of Sox11 is relatively unknown, except for its role in human and murine nephrogenesis (19). scRNA-seq analysis revealed that Sox11+ cells constituted a subset of the Cdh6pos population (Fig. 2E; see also Fig. 1, C and F). Thus, the above validatory studies confirmed that our bulk Sox9 cell-type–specific RNA-seq study could be used reliably to identify possible secretory ligands that might engage the adjacent fibroblast.
The Wnt signaling pathway, consisting of Wnt ligands, was among the top enriched GO terms within IRI day 14 tdT+ cells versus 48 hour tdT+ cells, although it was conspicuously devoid of the canonical Wnt-β catenin pathway activity reporter Axin2 (Fig. 2F and fig. S12E). Mammalian Wnt proteins are a family of lipid-modified glycoproteins that signal within a typical range of just one or two cells in a juxtracrine or autocrine manner (20, 21). Integration with scRNA-seq datasets identified single cells with de novo activation of Wnt4, and Wnt7b specifically within Cdh6pos cells, including a Cdh6+Sox11+ subset that also expressed Wnt4 (Fig. 2, G and H). qPCR and RNA-scope studies confirmed Wnt4+ induction within CDH6pos cells, thus validating RNA-seq findings (Fig. 2I and fig. S12F). Using a previously validated and published WNT4 antibody (22, 23), co-immunostainings substantiated the RNA scope studies. A strong WNT4 expression was strictly restricted to Sox9on-on cells, with such cells displaying a single-cell spatial and tight association with GFPpos myofibroblasts, indicating that Sox9on-on cells maintain biologically active Wnt niches during the progression of post-AKI fibrosis (fig. S13, A and B, and table S2). Thus, the SOX9posCDH6pos cell state might be the secretory cells that generate and maintain Wnt-enriched niches after AKI.
Next, we used Axin2CreERT2/+:R26RmT/mG animals, in which Axin2 activity reports cells with ongoing canonical Wnt signaling (20), to ascertain the identity of Wnt-responsive cells (WRCs). In a healthy adult kidney, a subset of interstitial αSMAneg PDGFRB+ cells residing within the inner medulla region expressed membrane GFP (mGFP), demarcating resident WRCs (fig. S14, A to C). Contrary to the previous report (24); no resident WRCs were detected within the nephron epithelia of the cortices and outer medulla (fig. S14, A to C). After AKI, the animals treated with only corn oil (the vehicle for tamoxifen) did not demonstrate mGFP+ cells, confirming tamoxifen dependence and ruling out injury-induced spontaneous Cre activation (fig. S14, D and E). The kidneys of the tamoxifen-treated animals showed that subsets of αSMA+ myofibroblasts mounted Axin2 activity, as shown by mGFP. By contrast, the resident Axin2+ cells remained αSMAneg (fig. S14, F to I). Thus, myofibroblasts represent de novo WRCs after injury. To determine the spatiotemporal relationship between the earliest WRCs, CDH6pos cells and myofibroblasts, we scrutinized Axin2CreERT2/+:R26RtdT/+:Acta2-GFP mice. Examination uncovered subsets of CDH6pos cells forming a biologically active Wnt niche at the single-cell level (Fig. 2J and table S1).
Genetic lineage studies showed that the descendants of Axin2+cells formed the bulk of the scar tissue by4 weeks after IRI (fig. S15A). Nearly half of the replicating myofibroblasts were Axin2+ (fig. S15B). Further, comparative blinded analysis of the β-catenin–deficient lineage of the earliest αSMA+ myofibroblasts (Ctnnb1-cKO) versus the lineage with intact β-catenin activity (Ctnnb1-wt, controls) showed that Ctnnb1-cKO displayed significantly less expansion (P < 0.01) and lacked the intense αSMA response at day 28 after IRI (fig. S15C). Moreover, the characteristic strong αSMA response adjacent to Sox9on-on cells was conspicuously deficient in most of the Ctnnb1-cKO cells (8.3 ± 2.4% versus 79.2 ± 2.6%, Ctnnb1-cKO versus Ctnnb1-wt, P < 0.01; fig. S15C and table S2). Thus, canonical β-catenin signaling is essential for the maintenance of αSMA+ activity within the myofibroblasts after AKI.
To determine whether SOX9on-onCDH6pos cells secrete Wnt and drive fibrosis, we performed a head-to-head comparison between tdT-demarcated Sox9+cells lacking Wls versus and the intact Wls secretory apparatus (Fig. 3A). Wls encodes Wntless, a highly conserved, transmembrane protein essential for Wnt secretion and function (25). Successful recombination of the floxed Wls allele was confirmed by two different approaches (fig. S16, A to D). Blinded analysis revealed a substantial reduction in αSMA response around the tdT+SOX9: Wls-cKO versus controls (tdT+SOX9: Wls-Het; Fig. 3A and table S1). Both groups had a similar proportion of SOX9on-on cells (Fig. 3B). Further scrutiny of Sox9: Wls-cKO animals showed that a proportion of SOX9+ cells underwent successful recombination, as shown by tdT expression, thus generating a microenvironment mosaic for Wls activity: SOX9+tdTpos (Wls-cKO) and SOX9+tdTneg (Wls-WT) cells, respectively. Leveraging the observed mosaicism, a direct-blinded comparison for αSMA activity around SOX9+tdTpos and SOX9+tdTneg tubules revealed a significant (P < 0.01) reduction in myofibroblasts around the SOX9+tdTpos tubules compared with their SOX9+tdTneg counterparts (Fig. 3C, fig. S16E, and table S1). Macrophages could be a collateral source of Wnts. We did not detect Wnt7b, Wnt9b, Wnt11, or Wnt4 induction in fluorescence-activated cell sorting (FACS)–purified LyzM Cre-derived tdT+F4/80+cells compared with uninjured resident tdT+F4/80+cells despite an ~5-fold increase in F4/80+cells (fig. S17, A to B). Consistent with this finding, blinded analysis for αSMA+ myofibroblasts or overall fibrotic responses showed no effect of Wls removal within the LyzM Cre-derived immune cells (fig. S17, C to F). Although kidney stromal cells express Wnts, removal of Wnt4 within these cells had no effect on overall fibrosis (26), thus ruling out stromal Wnt4 as the main driver of post-AKI fibrosis. The above findings reveal the SOX9posCDH6pos cell state as the central source of continuous, biologically active Wnt signal driving fibrosis after AKI.
Fig. 3. Sox9on-on cell state is the main driver of myofibroblast formation and maintenance.
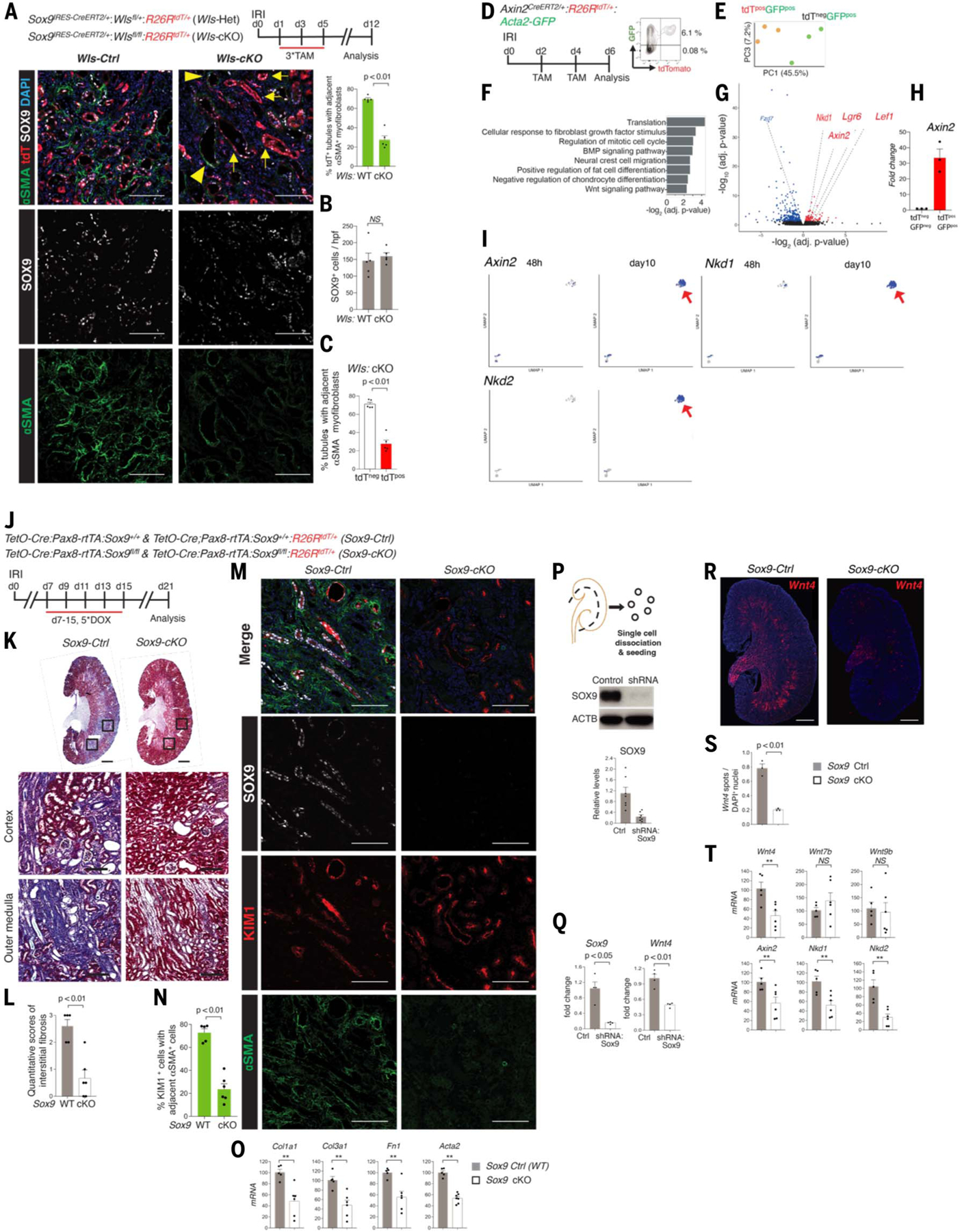
(A to C) Schema of Wls removal from Sox9on-on cells, outer medullary region representative image, and blinded co-immunoanalysis for SOX9 and αSMA (n = 5 animals/group). No difference can be seen in Sox9on-on cells between the two groups (B). The mosaic tissue–damaged microenvironment displayed significantly (P < 0.01) reduced αSMApos myofibroblasts adjacent to tdTpos demarcated Wls-KO tubules (arrows) versus tdTneg tubules with intact Wls [(C), arrowheads]. (D) Schema of isolating early Axin2pos cells after IRI with FACS plot showing that nearly all tdTpos cells co-expressed GFP. (E) PCA plot of purified tdTposGFPpos and tdTnegGFPpos cells. (F) GO analysis showing Wnt pathway among the top 10 terms, confirming that Axin2pos cells demarcated early WRCs. (G and H) Volcano plot showing the molecular signature of early WRCs, with qPCR confirming Axin2 enrichment within tdTposGFPpos cells, thus validating reporter animals. (I) UMAP projection of Axin2, Nkd1, and Nkd2 cells. Such cells were within the same PdgfrbposCola1posCol3a1pos cluster (see also fig. S9). (J) Experimental outline of removal of Sox9on-on activity during the AKI to CKD transition. (K and L) Trichrome staining (K) and quantitative scores of interstitial fibrosis (L) (blinded analysis, n = 5 Sox9-Ctrl and 6 Sox9-cKO animals). (M and N) Co-immunostaining for SOX9, KIM1, and αSMA (M) and blinded co-immunoanalysis showing head-head comparison between KIM1+ regions for αSMA activity (N) in Sox9-cKO versus Sox9-Ctrl animals. (O) qPCR analysis of genes associated with fibrosis in the kidneys from Sox9-cKO versus Sox9-Ctrl animals. (P and Q) Western blot confirming SOX9 knock-down in subconfluent primary TECs (P), with qPCR showing Sox9 and Wnt4 down-regulation (Q). (R and S) RNAscope image (R) and analysis (S) showing significant (P < 0.01) Wnt4 reduction. (T) qPCR analysis of Wnts under scrutiny and Axin2, Nkd1, and Nkd2 in the kidneys from Sox9-cKO versus Sox9-Ctrl. All images are representative images. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01 unpaired two-sided Student’s t test; ***P < 0.01, paired Student’s t test. Scale bars in whole scanned images [(K) and (R)], 1000 μm; all others, 100 μm. For total cells counted, see table S1.
Next, we sought to determine whether Sox9on-on activity per se is the critical component of such profibrotic niches in vivo. Our previous work highlighted the essential role of Sox9 activation in PTEC regeneration. Proximal tubule–specific removal of Sox9 before injury led to impaired tubular repair and renal function recovery compared with animals with intact Sox9 activity (10). Based on the findings of our scRNA-seq and Sox9+ cell-type-specific bulk RNA-seq studies, we hypothesized that SOX9posCDH6pos cells with Sox9on-on likely recruited essential downstream proregenerative programs by day 10 after AKI. Therefore, it might be feasible to remove Sox9on-on activity during the progression of AKI to CKD. The precise molecular signature of early WRCs after AKI remains unknown. Therefore, to determine whether removal of Sox9on-on activity leads to the abrogation of fibrosis and the obliteration of Wnt niches, we established a molecular signature of WRCs.
To this end, we used Axin2CreERT2/+:R26RtdT/+: Acta2-GFP animals (Fig. 3D). Comparison of the transcriptome between the earliest WRCs (tdTpos GFP GFPpos) versus other myofibroblasts (tdTneg GFPpos) cells confirmed the Wnt signaling pathway activation as one of the top-most enriched GO terms within the tdTpos cells; Axin2, Lef1, Nkd1, and Lgr6 were enriched, whereas Fzd7, encoding Frizzled 7, a Wnt receptor, was down-regulated (Fig. 3, E to G, and table S3), suggesting that Fzd7 might be the Wnt-sensing receptor activating Pdgfrb+ cells. Other top enriched GO terms included “response to fibroblast growth factor” and “regulation of mitotic cell cycle,” suggesting that the early Axin2+ myofibroblast subset may have greater proliferative properties compared with other myofibroblast populations, consistent with lineage-tracing and Ki67-based co-immunoanalysis studies (Fig. 3F and fig. S15, A and B). qPCR validated Axin2 enrichment within tdTposGFPpos versus tdTnegGFPpos cells (Fig. 3H). Our scRNA-seq study analysis not only further endorsed induction of Axin2 activity within the same cluster containing Acta2+ Col1a1+Col3a1+Pdgfrb+ cells at day 10 after IRI, but consistent with the above findings, also showed induction of Nkd1+ cells specifically restricted to the above day 10 postIRI cluster (Fig. 3I). Nkd2, reported as a marker of terminal myofibroblasts in a previous study (27) and in our molecular profiling study of AKI to CKD (5), was also enriched in early Acta2+Col1a1+ Pdgfrb+cluster (Fig. 3I). Thus, these findings not only validated the Axin2 reporter but also established a molecular signature of early WRCs.
To remove Sox9on-on activity during the transition from AKI to CKD, doxycycline injections to Sox9 knock-out (Sox9-cKO) and control wild-type (Sox9-WT, Ctrl) animals were administered 1 week after AKI. The tdT+ cells highlighted cells with successful recombination. First, doxycycline dependence of the system was confirmed (fig. S18A). Further, in the absence of doxycycline, Sox9-cKO animals displayed an injury-induced, early SOX9 activation response, with nearly half of SOX9+ cells expressing Ki67 akin to animals with intact Sox9 activity, thus ruling out spontaneous injury-induced Cre activation and preservation of early Sox9 repair responses (fig. S18B). Doxycycline led to obliteration of Sox9on-on activity in the Sox9-cKO (tdT+Sox9KO) animals; by contrast, Sox9-WT animals displayed intact Sox9on-on activity (tdT+Sox9+) (fig. S18C).
To ensure rigorous comparison between similar damaged tissue microenvironments, a blinded, head-to-head comparison between Havcr1+ (also called kidney injury molecule-1, Kim1) regions of Sox9-cKO and Sox9-WT animals was performed. The kidneys lacking Sox9on-on activity displayed a significantly reduced fibrotic signature, including αSMA+ myofibroblasts (Kim1+Sox9WT versus Kim1+Sox9KO, 69.0 ± 3.6% versus 28.2 ± 5.8%, respectively, P < 0.01; Fig. 3, J to O, and table S1). Before the removal of Sox9on-on activity, the proportions of SOX9 and αSMA cells were similar in Sox9-WT and Sox9-cKO animals (fig. S18D). Using previously validated shRNA to knock down Sox9 (28), we found a marked reduction in Wnt4 in primary nephron tubular epithelial cells in Sox9 knock down cells versus controls (Fig. 3, P and Q). Next, we confirmed Sox9-Wnt4 link in vivo (Fig. 3, R to T). The outer medulla region, the predominant site of Sox9on-on activity and the main site of de novo injury-induced Wnt4 response, displayed maximal blunting of the Wnt4 response upon removal of Sox9on-on activity (Fig. 3R). These findings provided further evidence for Sox9on-on activity in generating biologically active Wnt4 niches. The resident Wnt4 expression within the papilla and inner medulla region (a region with no Sox9on-on cells) remained intact and thus served as a robust, internal positive control (Fig. 3, R and S). Further, in addition to the panel of profibrotic genes, the herein identified early molecular signature of injury-induced WRCs (Axin2, Nkd1, and Nkd2) showed reductions in Sox9-cKO versus Sox9-WT animals (Fig. 3T). Genetic fate mapping of the Sox9on-on-deficient cells revealed that a significantly (P < 0.01) larger proportion of Sox9on-on-deficient cells had restored polarity compared with their counterparts with intact activity (fig. S19). This finding suggests that the reduced fibrosis secondary to the removal of Sox9on-on activity might serve as a relatively favorable milieu for the epithelium to restore itself. Thus, Sox9on-on activity transforms the SOX9posCDH6pos cell state into Wnt-secreting cells, a central hub for myofibroblast formation and maintenance.
The SOX9on-onCDH6poscell state highlights sustained efforts to regenerate the epithelium by attaining a progenitor-like cell state
Having identified the biological relevance of Sox9on-on activity, we next aimed to better define and understand the epigenetic features of the Sox9on-on cell state. To this end, we generated single-nuclei assay for transposase-accessible chromatin sequencing (snATAC-seq) profiles of day 10 Sox9 descendants after injury. To ensure sufficient availability of single Sox9on-on nuclei for meaningful analysis, we dissected the inner cortices and outer medulla region, the site of predominant Sox9on-on cells, and the single-nuclei isolated from freshly enriched lineage-traced tdTpos cells were subjected to snATAC-seq (Fig. 4A). Integration of snATAC-seq with the scRNA-seq profile demonstrated that the nuclei subclustered in a nephron cell-type-specific manner based on differentially open or closed chromatin accessibility state (Fig. 4B and fig. S20, A and B). The differential chromatin accessibility of Sox9 led to the clustering of lineage-traced tdTpos nuclei into “Sox9on-on” and “Sox9on-off” nuclei (Fig. 4C).
Fig. 4. Epigenetic reprogramming of single Sox9on-on nuclei to a nephron progenitor–like state contrasts with the Sox9on-off counterpart, which reverts to normal PTEC.
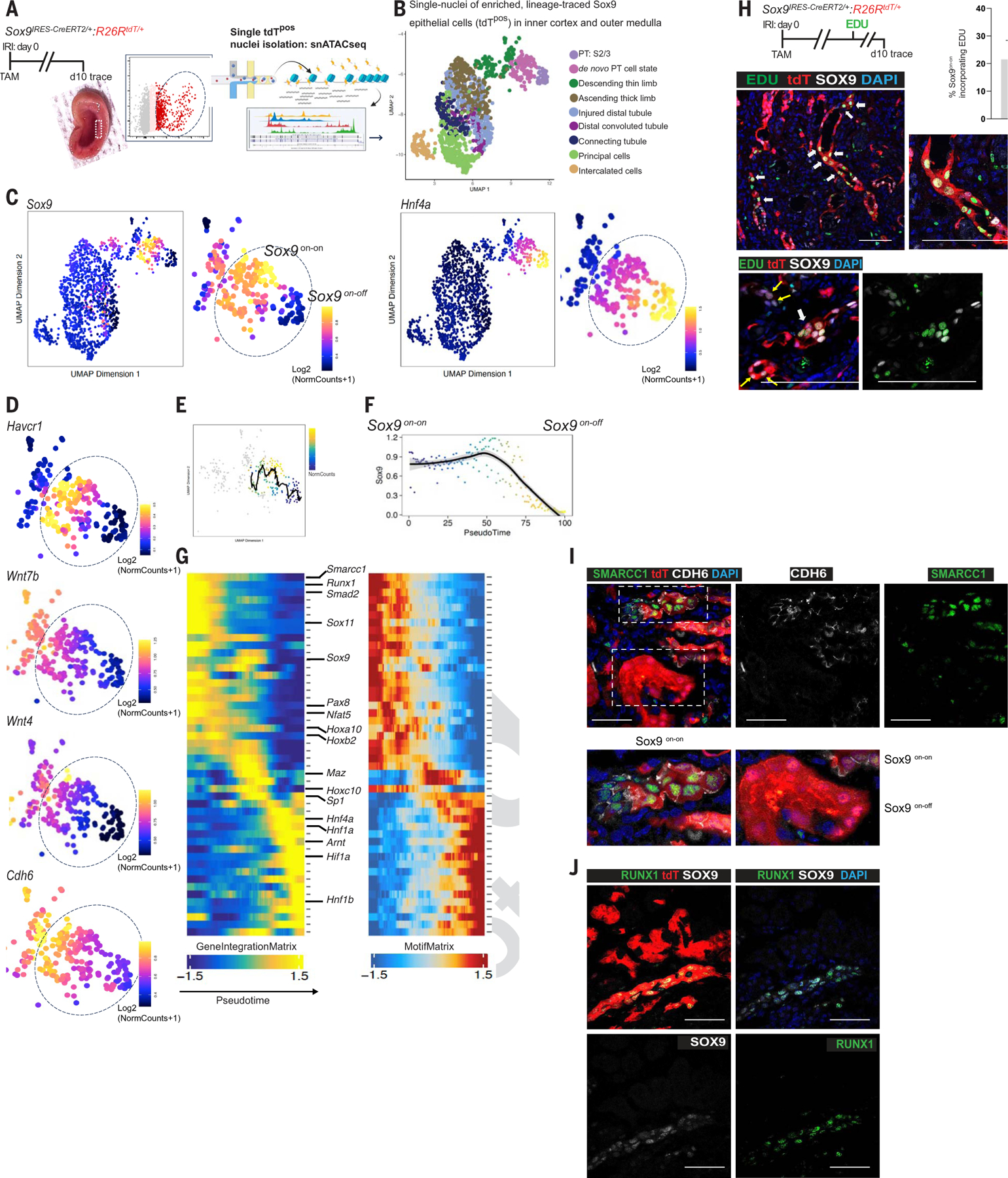
(A) Schema of the workflow to obtain single nuclei of the FACS-enriched tdTpos Sox9-descendants at day 10 after injury. Rectangle (inset) highlights the dissected region showing the inner cortices and outer medulla, the site of relatively extensive PTEC loss. (B) Integration of scRNA-seq and snATAC-seq showing the cellular identity of single-tdTpos nuclei (n = 2 animals). (C) Identification of Sox9on-on and Sox9on-off single nuclei: Chromatin accessibility revealing clustering of tdTpos descendants based on relatively open and closed chromatin accessibility state of Sox9. Sox9on-off nuclei exhibited open chromatin accessibility for Hnf4a, a known marker of healthy, mature PTECs, suggesting that the Sox9-lineage that regenerated normal PTECs closed chromatin accessibility for Sox9. (D) UMAP representation of chromatin accessibility analysis showing the relatively open chromatin state of Havcr1, Wnt7b, Wnt4, and Cdh6 versus their accessibility state in Sox9on-off nuclei. (E and F) Sox9 trajectory analysis with scRNAseq-imputed gene expression (E) and pseudotime (F) scale showing the Sox9on-on → Sox9on-off transition. (G) Heatmaps of cross-platform linked genes involved in the transcriptional cascade during Sox9on-on → Sox9on-off transition. Note that the highlighted genes in Sox9on-on nuclei are linked with tissue development and/or nephrogenesis, and the Sox9on-off nuclei displayed open chromatin accessibility for Hnf4β, another known marker of healthy, mature PTECs in addition to Hnf4a. (H) Schema of EDU regime and co-immunostaining for SOX9 and EDU showing SOX9postdTposEDUpos cells. Two representative images are shown, with the lower panel demonstrating a cluster of such cells. n = 3 animals. (I and J) Co-immunostaining for SMARCC1 and CDH6 showing SMARCC1 expression confined to day 10 CDH6pos Sox9 lineage cells (I), and RUNX1 and SOX9 co-immunostaining showing RUNX1 restricted to Sox9on-on versus Sox9on-off lineage (J) as predicted by (G), thus validating Sox9 lineage-specific snATAC-seq datasets. n = 2 animals. Scale bars, 100 μm.
The Sox9on-off nuclei exhibited a relatively open chromatin accessibility state of Hnf4a (Fig. 4C) and its downstream gene, Lrp2 (fig. S20B), the markers of mature functional PTECs, thus providing strong evidence that the Sox9 lineage regenerates functional PTECs. By contrast, the Sox9on-on nuclei demonstrated a relatively open chromatin accessibility state of Cdh6, Wnt4, and Wnt7b, in addition to Sox11 (Fig. 4, C and D, and fig. S20B), further showing the Sox9on-onCdh6pos cell state to be Wnt enriched (Fig. 4D). Consistent with scRNAseq analysis (Fig. 1B), snATACseq analysis also provided evidence for UMOD+ thick ascending limb of Loop of Henle and AQP2+ collecting duct epithelial cells to activate SOX9 after injury (fig. S20, C and D). To uncover the transcriptional regulators that characterize the distinct outcomes of Sox9-descendants, the on-on and on-off nuclei were subjected to trajectory analysis (Fig. 4, E and F). Transcriptional regulators with an essential role in nephrogenesis, such as Sox11, Nfat5, Maz, and Pax8, displayed dynamic transcriptional changes along the pseudotime in the Sox9on-on cell state (29–31) (Fig. 4G). The dynamic reparative process was associated with cell proliferation, with ~22% of Sox9on-on descendants in the S-phase of the cell cycle (Fig. 4H). Moreover, consistent with the prediction of snATAC-seq pseudotemporal analysis, the expression of both SMARCC1 and RUNX1 was restricted to the Sox9on-on lineage compared with its on-off counterpart (Fig. 4, I and J). Smarcc1, an ATP-dependent chromatin-remodeling complex, maintains proliferation, pluripotency, and self-renewal of embryonic stem cells (32), whereas Runx1 drives muscle regeneration and hematopoietic stem and progenitor cell specification (33). Thus, these findings not only validated snATAC-seq pseudotemporal analysis, but also identified them as potential candidates upstream of Sox9on-on activity.
snATAC-seq analysis of the inner cortices and outer medulla region (Fig. 4A) revealed a sufficient population of Sox9on-on nuclei. Therefore, Sox9 CUT&RUN genomic occupancy assay in the day 10 lineage versus its parent Sox9on cells (48 hours after injury) provided an opportunity to examine the direct effects of sustained Sox9 activity within its lineage in vivo. We conducted a time-resolved, lineage-specific SOX9 CUT&RUN genomic occupancy assay (fig. S21A). An H3K4me3 genomic occupancy assay was used as a control. Because of the technical challenges involved in isolating high-quality, >90% viable fragile tdTpos-enriched cells under low-flow FACS conditions and the limited labeling possible by a single tamoxifen injection, a pellet of ~30,000 to 100,000 cells was subjected to a genomic occupancy assay/antibody/time point. The cell pellet was expected to contain Sox9on-off cells, therefore, to reduce these cells, the lineage cells were enriched from the IC/OM region.
Despite these limitations, (i) validation of appropriate insert size for a transcription factor (fig. S21B), (ii) distinct time- and antibody-specific clustering of biological samples using Pearson correlation approaches and UMAP (uniform manifold approximation and projection) approaches (fig. S21, C and D), (iii) signal of SOX9 peak relative to the transcription start site (fig. S21E), and (iv) significant optimal Sox9 motif enrichment with Sox9 antibody versus input (P < 1E-300 at day 10, P < 0.02 at 48 hours; fig. S21F) or versus H3K4me3 antibody (P < 1E-300 at day 10) suggested that the data could be reliably mined for meaningful biological conclusions with respect to SOX9-specific target binding. Consistent with the previous studies of genome-wide chromatin binding by SOX9, 10 to 15% of SOX9-bound regions occurred within transcriptional start or promoter sites (34, 35). Indeed, the enriched GO terms across both time points showed that the significantly bound region and the corresponding significantly called peaks and genes compared with their respective inputs [false discovery rate (FDR)–adjusted P value < 0.01; fig. S21G), were linked with the biological processes involved in epithelial regeneration in Sox9 48 hour and day 10 cells (fig. S21, H and I), consistent with the genetic-lineage tracing studies and scRNA-seq analysis of such cells (Fig. 1D). Calmodulin binding was one of the top GO enriched “molecular function” terms (fig. S21, H and I). SOX factors are known to contain a calmodulin-binding domain, and this calcium ion–enabled interaction imports SOX9 to the nucleus, with subsequent target gene activation (36, 37). Most Sox9 target genes were specific either to 48 hours (91.2%) or day 10 (85.11%) after injury, indicative of distinct time-dependent genomic occupancy (fig. S21J). The top enriched GO terms at both time points showed links with epithelial development processes consistent with its role in epithelial restoration. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed the top terms linked with the RhoGTPase, calcium, and epidermal growth factor receptor (EGFR) pathways, whereas by day 10, metabolic pathways were uncovered as the top pathway. Indeed, Sox9 is a critical mediator of metabolic processes in chondrogenic cells (38) and regulates components of the EGFR pathway in malignant cells (39). The maintenance of the pluripotent stem cell state, along with chromatin organization, appeared in day 10 SOX9+ cells as shown by both KEGG pathway and GO analysis, thus directly implicating sustained Sox9 activity in endogenous reprogramming to a progenitor-like state in at least a subset of Sox9on-on cells.
Two well-known SOX9 target genes (40, 41) showed distinct time-specific binding. At 48 hours but not at day 10, Sox9 bound Col1a1; by contrast, Col2a1 was bound at day 10 but not at 48 hours (fig. S21K). Fgf/Fgfr/Sox9 forms a feed-forward loop to maintain cell identity and growth (42). Conversely, our data showed that Fgfr1 was targeted at both time points, and a higher engagement was observed by day 10. In day 10–specific target gene analysis, the Wnt signaling pathway, consisting of genes such as Wnt2, Wnt5a, Wnt5b, Tcf7, and Tcf7l2, and others, featured among the top pathways. Tcf7 and Tcf7l2 are known to cooperate with β-catenin in committed nephron progenitor cells (43). To confirm whether Wnt2 represents a direct transcriptional target of SOX9, we examined the effect of removal of Sox9on-on activity on Wnt2 mRNA expression. Wnt2 mRNA was markedly reduced in Sox9on-on cKO versus intact Sox9on-on activity (fig. S21L). These findings validate the CUT&RUN SOX9 genomic occupancy assay and uncover a direct Sox9/Wnt2 link, which might contribute to maintaining fibroproliferative response after AKI. Thus,we have provided further evidence for the direct role of Sox9on-on activity in the generation and maintenance of Wnt-enriched niches.
Integration of HOMER analysis of consensus sequence motifs associated with sites targeted by SOX9 CUT&RUN assay and transcription regulators uncovered by pseudotime trajectory analysis in our snATAC-seq datasets revealed top cofactors operating specifically in the Sox9on-on cell state (fig. S21M). These include binding motifs for Yy1, Nfat5, Sp1, Arnt::Hif1a, and Tcf4. Yy1, a structural regulator of enhancer-promotor loops and gene expression, is essential for embryonic stem cell viability (44), and Nfat5 is essential for nephrogenesis and protects against stress (29). Tcf4, which is essential for skin epithelia repair and homeostasis (45), was noted to be co-enriched with Wnt4 in nephron progenitors, and Hif1a-Sox9 axis is involved in chondrogenesis (46). Thus, our findings indicate that Sox9on-on activity might cooperate with these factors to regulate downstream gene expression.
CDH6posWNT2Bpos cells marks fibrotic foci in human kidneys
To determine the clinical relevance of our findings, we studied transplanted human kidneys. Immediately after kidney transplantation, ischemia reperfusion injury–induced AKI frequently leads to delayed graft function, which is an independent predictor of subsequent renal allograft loss and dysfunction (47). Renal allograft biopsies from such patients (n = 3, day 7 after kidney transplantation) and pre-implantation biopsies obtained from the same allograft (controls) demonstrated a marked increase in SOX9+ cells within the AQP1+ PTECs (Fig. 5A). This was also confirmed at the transcriptional level in protocol biopsies obtained after reperfusion (48) (Fig. 5B).
Fig. 5. Human renal allografts display dynamic SOX9/CDH6/WNT2B activity, with CDH6pos cells demarcating SOX9on-on activity and fibrotic foci.
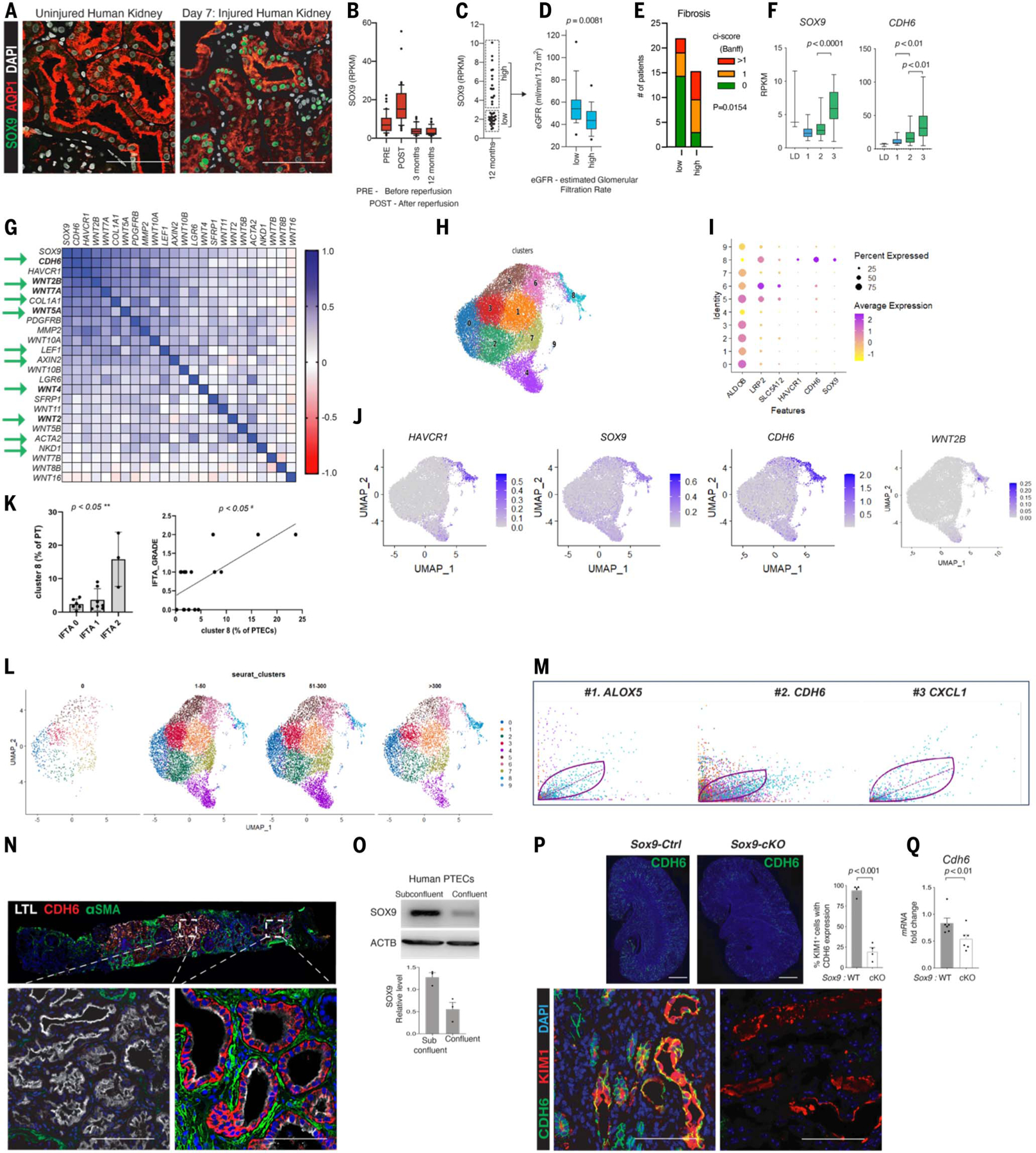
(A) Co-immunostaining for SOX9 and LTL in biopsies obtained before implantation (uninjured) and IRI-induced AKI after transplantation (same allograft) showing early SOX9 activation. (B) Box plot showing SOX9 levels at different time points within kidney transplant protocol biopsies (n = 163). (C and D) Dot plots (C) and box plots (D) showing categorization of patients and kidney function according to SOX9 levels at 1 year (comparison by Mann-Whitney U test; n = 35). (E) Histograms showing the number of patients with different degrees of kidney fibrosis, as estimated by ci-score according to Banff classification (comparison by chi-square test; n = 39). (F) Box plot showing SOX9 and CDH6 levels categorized according to a previously reported model discriminating the transcriptome of kidney transplant biopsies in successful repair (1), transition to chronic injury (2), or CKD (3). LD, biopsies obtained from living donors at the time of transplantation (49). (G) Heatmap showing expression correlation of genes of interest at 3 and 12 months after transplantation (Spearman r correlation coefficient; n = 72). Arrows highlight the identified molecular signatures of SOX9/CDH6/WNT, including AXIN2 and NKD1 in human kidneys. Note that CDH6 is the topmost correlated gene. (H) UMAP showing distinct PTEC clustering (29,180 genes × 24,070 cells). (I) Dot plot showing average gene expression values and percentage of cells expressing markers of differentiation and injury, SOX9 and CDH6, by each identified cluster of PTECs. Cluster 8 consisted of SOX9-, CDH6-, and WNT2B-expressing cells (see also figs. S22, C to E, and S23). (J) Feature plot displaying the normalized transcript expression for the respective genes. Magic mRNAassay of renal cells is depicted. (K) Analysis comparing the percentage of cluster 8 and the interstitial fibrosis and tubular atrophy (IFTA) grade for each patient. Percentage of cluster 8 is calculated with respect to the PTEC number in each patient. P < 0.05 based on the Kruskal-Wallis test and Pearson correlation analysis. (L) UMAP showing time-resolved PTECs clustering after transplantation (shown as days after transplantation). Note the emergence of cluster 8 with time after transplantation. (M) Cluster-type-specific genes analysis revealed CDH6 as being among the top two driver genes that underlie the dynamic activity in cluster 8. (N) Co-immunostaining showing that CDH6pos cells displayed a tight intimate association with ACTA2pos myofibroblasts within human kidney allograft, with CDH6neg foci showing no ACTA2pos myofibroblasts. (O) Immunoblot showing that subconfluent human primary PTECs activated SOX9, which waned upon confluency. (P and Q) Co-immunoanalysis (P) and qPCR (Q) showing reduction in CDH6pos cells and Cdh6 mRNA, respectively, upon removal of Sox9on-on activity (blinded analysis, unpaired two-sided Student’s t test; data are shown as mean ± SEM). Scale bars in whole scanned image (P), 1000 μm; all others, 100 μm.
Further, a stratification of kidney allograft protocol biopsies performed 1 year after transplantation showing SOX9 levels demonstrated that patients with persistent SOX9 expression displayed increased interstitial fibrosis and reduced renal function compared with the cohort that displayed return of Sox9 activity to baseline levels (Fig. 5, C to E). SOX9 and CDH6 levels correlated with the transition from AKI to CKD in transplant biopsies according to a previously reported model (49) (Fig. 5F). In the same kidney transplant cohort, protocol biopsies obtained 3 and 12 months after transplantation revealed CDH6 as the topmost gene correlated with SOX9 activity (Fig. 5G). Such strong correlation was also noted with fibrosis-associated genes such as COL1A1 and ACTA2. Although strong correlation was noted with WNT4, the correlation was more pronounced with other members of the WNT family in humans, particularly with WNT2B. Patients with a low immunological risk profile were uniformly managed in a single center using a protocol comprising tacrolimus-, mycophenolate mofetil–, and prednisone-based immuno-suppression. Rejection episodes were rare.
To obtain single-cell-level resolution of the above observed responses, we leveraged scRNA-seq datasets involving 16 kidney transplant biopsies (50) (Fig. 5H and fig. S22, A and B). scRNA-seq showed that SOX9/CDH6/WNT2B–expressing cells predominantly resided in the same cluster (cluster 8), which also contained HAVCR1+ cells (Fig. 5, I and J, and fig. S22,C to E). This cluster demonstrated significant correlation with fibrosis (P < 0.05) (Fig. 5K), and time-resolved PTECs showed de novo emergence of cluster 8 after transplantation (Fig. 5L). Unlike Wnt2 (fig. S21L), Wnt2b did not exhibit a significant reduction upon removal of Sox9on-on activity (fig. S22F). Cluster 8 displayed dynamic activity and greater latent time compared with other PTEC clusters (fig. S23, A to E). Further, in another unbiased analysis to identify the driver gene that confers cluster-specific dynamic behavior, CDH6 was found to be the gene with the second-highest likelihood of underlying dynamic activity in HAVCR1+ cluster 8 (Fig. 5M). Akin to our findings in mice, CDH6pos LTLlow PTECs (Fig. 5N) and persistent SOX9 activity (fig. S24) displayed intimate association with αSMA+ myofibroblasts. Thus, these findings validated the identified dynamic axis in human kidneys at the single-cell level and revealed WNT2B as the likely WNT that may drive human fibrosis.
Akin to mouse PTECs, subconfluent primary human PTECs mounted SOX9 activity, which subsided upon tight monolayer formation, further emphasizing the tight link of SOX9 activity with cell-cell contact status (Fig. 5O). Our transcriptomic profiling and spatiotemporal mapping studies involving both murine and human kidneys uncovered a robust link between dynamic Sox9on-on activity and Cdh6 response. Because cell-cell contact and/or adhesion disruption leads to Sox9 activation, with silencing of Sox9 upon cell-cell contact restoration, it would make biological sense that in the setting of prolonged cell-cell contact disruption, the persistent Sox9 transcriptional activity would induce a cadherin in its attempt to restore cell-cell adhesion. Therefore, we verified that Cdh6 not only demarcates Sox9on-on cells but might also be regulated by Sox9. We detected significant reduction of CDH6pos cells and Cdh6 mRNA (P < 0.01) in Sox9-cKO kidneys compared with the WT (Fig. 5, P and Q). Thus, SOX9posCDH6pos cells demarcate cells with SOX9on-on activity–driven WNT signaling niches and fibrosis after human AKI.
Discussion
In this study, by establishing a model system that facilitated head-to-head comparison between the two initially committed lineages to regenerate the injured proximal tubular nephron epithelia but with divergent outcomes (one that expeditiously regenerated tissue versus the other that was unable to do so), we identified the unifying mechanism underpinning scarless versus fibrotic tissue repair at the single-cell level within the same microenvironment. We also identified how precisely Wnt-enriched niches are formed and maintained after injury. Until now, despite the prominent link between Wnt and tissue fibrosis after injury, this question has remained unanswered (51, 52). For example, in the setting of lung injury, the Wnt-inducing factor was highlighted as the “unknown factor” (52). Our study highlights the potential for abrogating fibrotic responses through a cell-state-specific intervention, even when introduced 1 week after injury, during the progression of AKI to CKD. This was illustrated by precise cell-state-specific genetic perturbation of Sox9on-on activity or removal of Wntless within the Sox9on-on cells.
Our data suggest that the Soxon-on CDH6pos cell state signifies an ongoing regenerating phase during the transition from AKI to CKD. This was supported by (i) enrichment of biological and cellular processes linked with PTEC formation at the single-cell level; (ii) integrated analysis of scRNA-seq and Sox9 lineage-specific snATAC-seq datasets, unveiling a cascade of transcriptional regulators linked to nephrogenesis in the Sox9on-on nuclei; and (iii) distinct, time-specific SOX9 genomic occupancy within its lineage, indicating direct involvement in activating programs for nephron epithelia formation. Indeed, it would make biological sense for Sox9 in its sustained attempt to regenerate the unrestored epithelia to recruit Wnts, specifically Wnt4. Unexpectedly, Wnt2, which is essential for murine lung development but understudied in nephrogenesis, emerged as direct target gene of sustained SOX9 activity. Wnt2 was substantially decreased upon Sox9 activity removal, highlighting the direct role of Sox9on-on in provoking fibrosis during a sustained effort to regenerate the epithelia. WNT2B demonstrated stronger correlation with sustained SOX9 activity in transplanted human kidneys then did than WNT2. These findings hint at differential deployment of paralogous Wnt2 genes in the progression from AKI to CKD between humans and mice. Therefore, we have uncovered a link that explains how the tissue regeneration process culminates in fibrosis.
The SOX9posCDH6pos cell state contrasts with other known maladaptive cell types implicated in postinjury organ fibrosis, including senescent, partial epithelial to mesenchymal, or cell-cycle-arrested cells (2, 53, 54). However, it remains a possibility that with time these cell states might be subsequently attained due to the secondary effects of severely fibrotic adverse milieu. Sox9-expressing myofibroblasts have been linked with renal fibrosis (55). Our study, which used varied orthogonal approaches, did not reveal such cells in post-AKI fibrotic kidneys, although this does not completely rule out the possibility that this population exists.
Across phyla, damage-induced repair response represents a fundamental tissue survival mechanism. How does an injured tubular epithelial cell temporally sense its reparative state in vivo in damaged-tissue microenvironment that lacks resident stem or progenitor cell population? Is there a unifying, central “on-off” molecular switch? For example, in yeast, a single transcriptional control mechanism enables cells to respond to fluctuating nutrient concentrations (56). Herein, we identified SOX9 as a dynamic, fundamental, intrinsic transcriptional link between loss of epithelial integrity and regenerative response in vivo. Recently, Yamanaka and colleagues linked apicobasolateral polarity, maintained by the tight junction protein ZO-1, with receptivity to signaling proteins and multicellular patterning in an in vitro human gastrulation platform (57). Genetic removal of ZO-1 led to sustained BMP4 signaling pathway (BMP4/pSMAD1/5) activation and the ensuing distinct cell-state specification. We found that the Sox9on-on cell state is Bmp4/Smads enriched (Fig. 1E and figs. S6D and S20B) and isa subset attaininga nephron progenitor-like cell state with time. It is tempting to speculate that a similar enhanced receptivity to the as-yet elusive signaling proteins in such disrupted epithelial cells drives Sox9 activity and resultsg in a distinct cell state. However, how precisely such disruption might regulate Sox9 remains to be established, which is a limitation of the current study. Further, a pharmacological approach to perturb the identified pathway remains unidentified, but our findings lay the ground for drug discovery and precise cell-state-specific genetic perturbation strategies to retard fibrosis.
In the present study, we have shown that the duration of the regeneration response is a key determinant of healing with or without fibrosis, with the SOX9posCDH6pos cell state interconnecting the transition from AKI to CKD.
Methods summary
All animal procedures were approved by the Cedars-Sinai Medical Center Institutional Animal Care and Use Committee, and institutional review board–approved human kidney biopsies were used. All animals used in the study are described in table S4. To induce the IRI-induced transition from AKI to CKD, 9- to 12-week-old weight-matched (25 to 30 g) mice were subjected to long-term survival compatible bilateral renal IRI surgery. To induce the transition from rhabdo-AKI to CKD, adult anesthetized mice (9 to 12 weeks old) were administered an intramuscular injection of 50% hypertonic glycerol solution in each quadriceps muscle (total dose, 8 mg/kg). For induction of CreERT2 protein, mice were injected with tamoxifen dissolved in corn oil through an intraperitoneal injection. For induction of doxycycline-inducible Cre protein, mice were injected with doxycycline dissolved in 0.9% normal saline intraperitoneally. Enzymatic digestion, which was conducted on ice to isolate cells from the kidney for RNA or nuclei extraction studies, used B. Licheniformis Cold Active Protease, DNase1, and Liberase TL. Cell-type-specific bulk RNAseq libraries were constructed using the Universal plus mRNA-seq with NuQuant kit from NuGEN. Sample libraries were sequenced on the NovaSeq platform (Illumina) using 150–base pair paired-end sequencing. Single-cell RNA libraries were obtained using the Chromium platform. Sox9-lineage–specific single nuclei for snATAC-Seq studies were isolated according to the 10xGe-nomics protocol using the low-input version and with a 1:5 diluted lysis buffer in nuclease-free water. Isolated cell samples were immediately processed with a Chromium Next GEM Single Cell ATAC Kit v2 (10xGenomics). Cell pellets of ~100,000 FACS-enriched Sox9 lineage cells were subjected to Sox9 genomic occupancy assay, and the libraries generated from the immune-enriched DNA samples using the Illumina kit were analyzed using Partek Flow software v10. RNAscope based in situ hydridization assay was performed on 12-μm-cut, optimal cutting temperature (OCT)–embedded cryosections according to the manufacturer’s protocol. The qPCR primers and primary and secondary antibodies used in this study are detailed in tables S5 to S7. Most of the imaging was performed on the Zeiss 780 confocal system. Unpaired, two-sided Student’s t test was used to compare two independent groups.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Chiamthamachinda for technical assistance with quantification, immunostaining, and genotyping; and I. Zafar for technical assistance with immunostaining and genotyping; C. Santiskulvong at the Applied Genomics, Computation, and Translational Core and flow core facility of Cedars-Sinai Medical Center; the Technology Center for Genomics & Bioinformatics (TCGB) at David Geffen School of Medicine, University of California, Los Angeles (UCLA) for cell-type-specific RNA-sequencing; and the UAB-UCSD O’Brien Center, Birmingham, AL, for serum creatinine measurements.
Funding:
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (grant R01 DK118265 to S.K.); the American Heart Association (grant 18CDA34110416 to S.K.); the American Society of Nephrology (John Merrill Transplant Scholar Grant to S.K.); UCLA CTSI (S.K.); the One Legacy Foundation (S.K.); and the Department of Defense (grant CDMRP KC200178 to S.K and J.B.). Work in the P.E.C. laboratory is supported by the Swiss National Foundation (Sinergia grant CRSII5_202302) and by the Balli and Gianella foundations. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Competing interests: S.K., S.C.J., and S.A. are inventors on provisional patent application (US63/605, 777) submitted by Cedars-Sinai Medical Center that covers “Cadherin6 expression status in determination of renal fibrosis and related uses thereof.” The remaining authors declare no competing interests.
Data and materials availability:
The datasets generated during this study are available in the Gene Expression Omnibus (GEO) database under accession no. GSE249781. These include single-cell sequencing (scRNA-seq) datasets (GSE196929); Sox9 cell-type-specific bulk RNAseq datasets (GSE249778); Axin2CreERT2/+: R26RtdT/+:Acta2-GFP cell-type-specific bulk RNAseq datasets (GSE249777); Sox9-lineage specific, time-resolved genomic occupancy assay datasets (GSE249776); and Sox9-lineage specific snATACseq data (GSE249780). Accession codes of the published data in GEO used in this study are as follows: RNAseq data for human kidney transplant biopsies: GSE126805 and single-nuclei RNAseq: GSE151167, GSE139107, and GSE163863). The accession codes for the published human scRNAseq data have been deposited in the European repository Biostudies under accession code E-MTAB-12051.
REFERENCES AND NOTES
- 1.Rockey DC, Bell PD, Hill JA, Fibrosis—A common pathway to organ injury and failure. N. Engl. J. Med 372, 1138–1149 (2015). doi: 10.1056/NEJMra1300575; pmid: [DOI] [PubMed] [Google Scholar]
- 2.Henderson NC, Rieder F, Wynn TA, Fibrosis: From mechanisms to medicines. Nature 587, 555–566 (2020). doi: 10.1038/s41586-020-2938-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konieczny P, Naik S, Healing without scarring. Science 372, 346–347 (2021). doi: 10.1126/science.abi5770 [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Cellular and molecular pathways of renal repair after acute kidney injury. Kidney Int 93, 27–40 (2018). doi: 10.1016/j.kint.2017.07.030; [DOI] [PubMed] [Google Scholar]
- 5.Liu J et al. , Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2, e94716 (2017). doi: 10.1172/jci.insight.94716; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abuelo JG, Normotensive ischemic acute renal failure. N. Engl. J. Med 357, 797–805 (2007). doi: 10.1056/NEJMra064398; [DOI] [PubMed] [Google Scholar]
- 7.Bonventre JV, Yang L, Cellular pathophysiology of ischemic acute kidney injury. J. Clin. Invest 121, 4210–4221 (2011). doi: 10.1172/JCI45161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grgic I et al. , Targeted proximal tubule injury triggers interstitial fibrosis and glomerulosclerosis. Kidney Int 82, 172–183 (2012). doi: 10.1038/ki.2012.20; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatachalam MA et al. , Acute kidney injury: A springboard for progression in chronic kidney disease. Am. J. Physiol. Renal Physiol 298, F1078–F1094 (2010). doi: 10.1152/ajprenal.00017.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S et al. , Sox9 activation highlights a cellular pathway of renal repair in the acutely injured mammalian kidney. Cell Rep 12, 1325–1338 (2015). doi: 10.1016/j.celrep.2015.07.034 [DOI] [PubMed] [Google Scholar]
- 11.Zhang K et al. , In vivo two-photon microscopy reveals the contribution of Sox9+ cell to kidney regeneration in a mouse model with extracellular vesicle treatment. J. Biol. Chem 295, 12203–12213 (2020). doi: 10.1074/jbc.RA120.012732; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan BL, Kolodziej PA, Organogenesis: Molecular mechanisms of tubulogenesis. Nat. Rev. Genet 3, 513–523 (2002). doi: 10.1038/nrg840; [DOI] [PubMed] [Google Scholar]
- 13.Nelson WJ, Nusse R, Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303, 1483–1487 (2004). doi: 10.1126/science.1094291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasioukhin V, Bauer C, Yin M, Fuchs E, Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100, 209–219 (2000). doi: 10.1016/S0092-8674(00)81559-7; [DOI] [PubMed] [Google Scholar]
- 15.Mah SP, Saueressig H, Goulding M, Kintner C, Dressler GR, Kidney development in cadherin-6 mutants: Delayed mesenchyme-to-epithelial conversion and loss of nephrons. Dev. Biol 223, 38–53 (2000). doi: 10.1006/dbio.2000.9738; [DOI] [PubMed] [Google Scholar]
- 16.Thelen S, Abouhamed M, Ciarimboli G, Edemir B, Bähler M, Rho GAP myosin IXa is a regulator of kidney tubule function. Am. J. Physiol. Renal Physiol 309, F501–F513 (2015). doi: 10.1152/ajprenal.00220.2014 [DOI] [PubMed] [Google Scholar]
- 17.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD, Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. U.S.A 117, 15874–15883 (2020). doi: 10.1073/pnas.2005477117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legouis D et al. , Altered proximal tubular cell glucose metabolism during acute kidney injury is associated with mortality. Nat. Metab 2, 732–743 (2020). doi: 10.1038/s42255-020-0238-1 [DOI] [PubMed] [Google Scholar]
- 19.Neirijnck Y et al. , Sox11 gene disruption causes congenital anomalies of the kidney and urinary tract (CAKUT). Kidney Int 93, 1142–1153 (2018). doi: 10.1016/j.kint.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clevers H, Loh KM, Nusse R, An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014). doi: 10.1126/science.1248012 [DOI] [PubMed] [Google Scholar]
- 21.Farin HF et al. , Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016). doi: 10.1038/nature16937; [DOI] [PubMed] [Google Scholar]
- 22.Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, Basler K, GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453 (2018). doi: 10.1038/s41586-018-0190-3 [DOI] [PubMed] [Google Scholar]
- 23.Kim S et al. , The polycystin complex mediates Wnt/Ca(2+) signalling. Nat. Cell Biol 18, 752–764 (2016). doi: 10.1038/ncb3363; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinkevich Y et al. , In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell Rep 7, 1270–1283 (2014). doi: 10.1016/j.celrep.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bänziger C et al. , Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509–522 (2006). doi: 10.1016/j.cell.2006.02.049; pmid: [DOI] [PubMed] [Google Scholar]
- 26.DiRocco DP, Kobayashi A, Taketo MM, McMahon AP, Humphreys BD, Wnt4/β-catenin signaling in medullary kidney myofibroblasts. J. Am. Soc. Nephrol 24, 1399–1412 (2013). doi: 10.1681/ASN.2012050512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuppe C et al. , Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021). doi: 10.1038/s41586-020-2941-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo W et al. , Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 (2012). doi: 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Rodríguez C et al. , Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A 101, 2392–2397 (2004). doi: 10.1073/pnas.0308703100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haller M, Au J, O’Neill M, Lamb DJ, 16p11.2 transcription factor MAZ is a dosage-sensitive regulator of genitourinary development. Proc. Natl. Acad. Sci. U.S.A 115, E1849–E1858 (2018). doi: 10.1073/pnas.1716092115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouchard M, Souabni A, Mandler M, Neubüser A, Busslinger M, Nephric lineage specification by Pax2 and Pax8. Genes Dev 16, 2958–2970 (2002). doi: 10.1101/gad.240102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho L et al. , An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A 106, 5181–5186 (2009). doi: 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P et al. , Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature 523, 468–471 (2015). doi: 10.1038/nature14569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohba S, He X, Hojo H, McMahon AP, Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep 12, 229–243 (2015). doi: 10.1016/j.celrep.2015.06.013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuglerud BM et al. , SOX9 reprograms endothelial cells by altering the chromatin landscape. Nucleic Acids Res 50, 8547–8565 (2022). doi: 10.1093/nar/gkac652; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harley VR, Lovell-Badge R, Goodfellow PN, Hextall PJ, The HMG box of SRY is a calmodulin binding domain. FEBS Lett 391, 24–28 (1996). doi: 10.1016/0014-5793(96)00694-1 [DOI] [PubMed] [Google Scholar]
- 37.Argentaro A et al. , A SOX9 defect of calmodulin-dependent nuclear import in campomelic dysplasia/autosomal sex reversal. J. Biol. Chem 278, 33839–33847 (2003). doi: 10.1074/jbc.M302078200 [DOI] [PubMed] [Google Scholar]
- 38.van Gastel N et al. , Lipid availability determines fate of skeletal progenitor cells via SOX9. Nature 579, 111–117 (2020). doi: 10.1038/s41586-020-2050-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F et al. , EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol. Cell 60, 307–318 (2015). doi: 10.1016/j.molcel.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell DM et al. , SOX9 directly regulates the type-II collagen gene. Nat. Genet 16, 174–178 (1997). doi: 10.1038/ng0697-174 [DOI] [PubMed] [Google Scholar]
- 41.Oh CD et al. , Identification of SOX9 interaction sites in the genome of chondrocytes. PLOS ONE 5, e10113 (2010). doi: 10.1371/journal.pone.0010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seymour PA et al. , A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development 139, 3363–3372 (2012). doi: 10.1242/dev.078733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Q et al. , A β-catenin-driven switch in TCF/LEF transcription factor binding to DNA target sites promotes commitment of mammalian nephron progenitor cells. eLife 10, e64444 (2021). doi: 10.7554/eLife.64444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weintraub AS et al. , YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588.e28 (2017). doi: 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen H et al. , Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat. Genet 41, 1068–1075 (2009). doi: 10.1038/ng.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robins JC et al. , Hypoxia induces chondrocyte-specific gene expression in mesenchymal cells in association with transcriptional activation of Sox9. Bone 37, 313–322 (2005). doi: 10.1016/j.bone.2005.04.040 [DOI] [PubMed] [Google Scholar]
- 47.Huang E et al. , Three-year outcomes of a randomized, double-blind, placebo-controlled study assessing safety and efficacy of C1 esterase inhibitor for prevention of delayed graft function in deceased donor kidney transplant recipients. Clin. J. Am. Soc. Nephrol 15, 109–116 (2020). doi: 10.2215/CJN.04840419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cippà PE et al. , A late B lymphocyte action in dysfunctional tissue repair following kidney injury and transplantation. Nat. Commun 10, 1157 (2019). doi: 10.1038/s41467-019-09092-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cippà PE et al. , Transcriptional trajectories of human kidney injury progression. JCI Insight 3, e123151 (2018). doi: 10.1172/jci.insight.123151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamarthée B et al. , Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection. Nat. Commun 14, 4359 (2023). doi: 10.1038/s41467-023-39859-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphreys BD, Mechanisms of renal fibrosis. Annu. Rev. Physiol 80, 309–326 (2018). doi: 10.1146/annurev-physiol-022516-034227 [DOI] [PubMed] [Google Scholar]
- 52.Burgy O, Königshoff M, The WNT signaling pathways in wound healing and fibrosis. Matrix Biol 68-69, 67–80 (2018). doi: 10.1016/j.matbio.2018.03.017 [DOI] [PubMed] [Google Scholar]
- 53.Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV, Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med 16, 535–543, 1p, 143 (2010). doi: 10.1038/nm.2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolstein JM et al. , INK4a knockout mice exhibit increased fibrosis under normal conditions and in response to unilateral ureteral obstruction. Am. J. Physiol. Renal Physiol 299, F1486–F1495 (2010). doi: 10.1152/ajprenal.00378.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raza S et al. , SOX9 is required for kidney fibrosis and activates NAV3 to drive renal myofibroblast function. Sci. Signal 14, eabb4282 (2021). doi: 10.1126/scisignal.abb4282 [DOI] [PubMed] [Google Scholar]
- 56.Ricci-Tam C et al. , Decoupling transcription factor expression and activity enables dimmer switch gene regulation. Science 372, 292–295 (2021). doi: 10.1126/science.aba7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasic I et al. , Loss of TJP1 disrupts gastrulation patterning and increases differentiation toward the germ cell lineage in human pluripotent stem cells. Dev. Cell 58, 1477–1488.e5 (2023). doi: 10.1016/j.devcel.2023.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available in the Gene Expression Omnibus (GEO) database under accession no. GSE249781. These include single-cell sequencing (scRNA-seq) datasets (GSE196929); Sox9 cell-type-specific bulk RNAseq datasets (GSE249778); Axin2CreERT2/+: R26RtdT/+:Acta2-GFP cell-type-specific bulk RNAseq datasets (GSE249777); Sox9-lineage specific, time-resolved genomic occupancy assay datasets (GSE249776); and Sox9-lineage specific snATACseq data (GSE249780). Accession codes of the published data in GEO used in this study are as follows: RNAseq data for human kidney transplant biopsies: GSE126805 and single-nuclei RNAseq: GSE151167, GSE139107, and GSE163863). The accession codes for the published human scRNAseq data have been deposited in the European repository Biostudies under accession code E-MTAB-12051.


