Abstract
During the past two decades evidence has been developed that indicates a handful of viruses with known oncogenic capacity, have potential roles in breast cancer. These viruses are mouse mammary tumour virus (MMTV - the cause of breast cancer in mice), high-risk human papilloma viruses (HPV-the cause of cervical cancer), Epstein Barr virus (EBV-the cause of lymphomas and naso-pharyngeal cancer) and bovine leukemia virus (BLV - the cause of cancers in cattle). These viruses may act alone or in combination. Each of these viruses are significantly more prevalent in breast cancers than in normal and benign breast tissue controls. The odds ratios for the prevalence of these viruses in breast cancer compared to normal and benign breast controls, are based on case control studies - MMTV 13·40, HPV 5.56, EBV 4·43 and BLV 2·57. The odds ratios for MMTV are much greater compared to the other three viruses. The evidence for a causal role for mouse mammary tumour virus and high risk for cancer human papilloma viruses in human breast cancer is increasingly comprehensive. The evidence for Epstein Barr virus and bovine leukemia virus is more limited. Overall the evidence is substantial in support of a viral cause of breast cancer.
Introduction
Viruses have long been suspected as having a causal role in breast cancer but the evidence has not been sufficient to establish causation. However, during the past two decades new evidence has been developed that indicates a handful of viruses with known oncogenic capacity, are the probable underlying causes of initiating breast cancer. These viruses are mouse mammary tumour virus (MMTV - the cause of breast cancer in mice), high-risk human papilloma viruses (HPV-the cause of cervical cancer), Epstein Barr virus (EBV-the cause of lymphomas and naso-pharyngeal cancer) and bovine leukemia virus (BLV - the cause of cancers in cattle). These viruses may act alone or in combination. Each of these viruses are significantly more prevalent in breast cancers than in normal and benign breast tissue controls.
95% of breast cancers are sporadic and historically have no known cause. Risks as compared to causal factors for breast cancer include familial susceptibility, early menarche, late menopause and late age first pregnancy. Radiation is an established risk factor. Genetics plays a small but important role in breast cancer. Inherited mutations in BRCA1 and 2 genes lead to an increased risk of 3 to 5% of breast cancer.
The evidence indicating a causal role for mouse mammary tumour virus is comprehensive but less so for the other viruses. In addition the odds ratios for MMTV are much greater compared to the other three viruses. There has been intense research interest in MMTV for over 90 years. For this reason there is more detailed information about MMTV in this review compared to the more recent studies of other viruses. However the role of high risk for cancer HPVs in breast cancer is also important and will be outlined in detail.
The history of research into MMTV and breast cancer has been published by Generoso Bevilacqua of Pisa, Italy [1]. The role of MMTV in breast cancer has been the dominant research interest. MMTV also appears to have a role in biliary cholangitis and autoimmune liver disease and has been identified in ovarian, prostate, endometrial and skin cancers [2–4].
The odds ratios (the ratio of the odds of the event in an exposed group versus a non-exposed group) for the prevalence of these viruses in breast cancer compared to normal and benign breast controls, are based on case control studies - MMTV 13·40, HPV 5.56, EBV 4·43 and BLV 2.57 [5–8].
Search strategy and selection criteria
PubMed Central is the main source of publications in this review. References listed in published articles have also been searched. We have used an extended version of the classic A.Bradford Hill causal criteria to assess the evidence concerning the role of these oncogenic viruses [9]. The extended Hill causal criteria include – identification of the causal pathogen, strength of association between the pathogen and the cancer, consistency, temporality (timing), experiment, analogy, means of transmission and oncogenic mechanisms.
Meta-analyses of case control studies of oncogenic viruses and breast cancer
MMTV, HPVs, EBV and BLV are significantly and consistently more prevalent in breast cancers than in normal and benign breast controls. These data are shown as odds ratios in Table 1 based on meta-analyses of case - control studies. Since the publication of these meta-analyses additional case control studies have been published for each of the four oncogenic viruses. These publications have been added to the tables.
Table 1.
Meta-analyses of virus positive breast cancers compared to normal / benign breast controls
| Author | Number of studies | Breast cancer Virus +/cases |
Normal / benign breast controls Virus +/cases |
Odds ratios (Confidence intervals) |
|---|---|---|---|---|
|
Wang 2021 [5] MMTV |
28 | 1287/4015 32% | 24/999 2.4% | 13·40 (11·44–15·36) |
|
Awan 2023 [6] HPV |
45 | 1145/4355 26.3% | 163/2361 6.9% | 5.56 (3.67–8.41) |
|
Agolli 2023 [7] EBV |
24 | 555/1989 28% | 83/1034 8% | 4·43 (3.47–5.66) |
|
Khatami 2020 [8] BLV |
9 | 334/826 40% | 215/898 24% | 2·57 (1·45–4·56) |
Mouse mammary tumour virus (MMTV)
Mouse mammary tumour virus (MMTV) is a retrovirus. In 1936, John Bittner discovered a pathogenic agent, which could be transmitted by milk from mouse mothers with breast cancer to their pups who as adults, later developed breast cancer [10]. Later, Samuel Graff and colleagues identified viral particles in mouse milk that could cause mammary cancer when intraperitoneally injected into laboratory strains of mice [11]. These retroviral RNA particles became known as mouse mammary tumour virus and are strongly linked with breast cancer in mice [12].
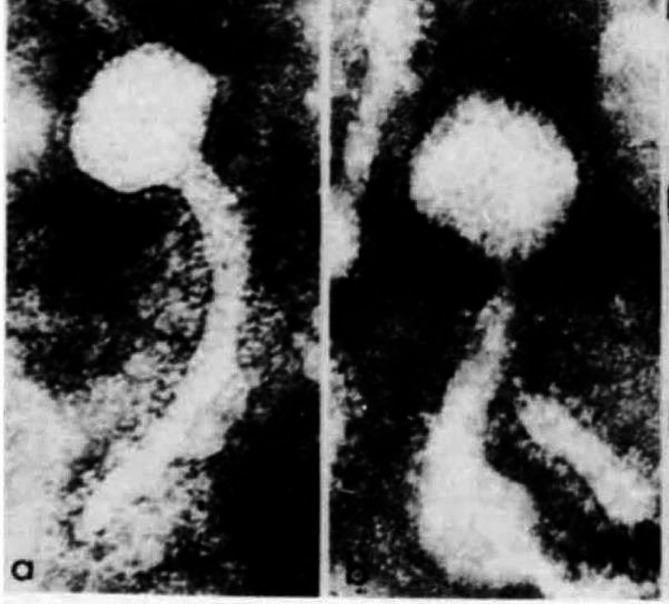
In 1971, Moore et al. using electron microscopy showed human milk containing viral particles with morphology identical to mouse mammary tumour virus in mouse milk [13] (Fig. 1). The images show an unusual shape for a virus including a long tail. This shape is probably because MMTVs begin as type A particles, later they become type B particles via a process of “budding” on the virus surface which takes place due to sophisticated molecular events [14].
Fig. 1.
MMTV particles identified in (a) human milk, (b) mouse milk (×18,000) by electron microscopy. Note the long curved tail [12]. (Republished with permission from Nature)
The nucleotide sequences and structure of MMTV-like viral sequences identified in human breast cancer tissues are virtually identical to the MMTV sequences identified in mouse mammary tumours [15]. The same 63 cancer-related genes have been identified in both human and mouse breast cancers [16]. The MMTV envelope and capsid protein expression is similar in both mouse and human breast cancers [17]. MMTV-associated tumour histology is similar in both mouse and human breast cancers [18]. In MMTV associated mouse breast cancers, the oncogene Wnt-1 is highly expressed. In humans, the influence of MMTV on human breast cells leads to abnormally high Wnt-1 expression [19].
In 1972 using molecular hybridisation Axel et al. demonstrated that MMTV was present in 62% of 29 human invasive breast cancers compared with 0% in benign and normal breast tissue controls [20]. In 1987, Moore et al. identified the complete nucleotide sequence of the mouse mammary tumour virus [21]. In 1981 Day et al. identified increased levels of the antibody to MMTV in 18.6% of US women with breast cancer compared to 2.8% of healthy women [22].
In 1995 using PCR, the Pogo group from New York identified the MMTV envelope gene in 38.5% of 314 human breast cancer specimens compared to 7% of 29 benign human breast specimens and in one of 27 normal human breast specimens [23]. These findings were confirmed by other research groups [24, 25]. The MMTV envelope and other cancer related gene sequences are identical in both human breast cancers and mouse mammary tumours [16, 26]. Accordingly, it is likely that MMTV is the same virus in both human and mouse breast cancers.
Strength and consistency of association between MMTV and human breast cancer
Case control studies
There are 26 case control studies in which MMTV has been identified. MMTV was not identified in 12 additional studies. The studies in which MMTV was identified, show a consistent pattern of outcomes, namely, positive identification of MMTV in 10 to 70% of breast cancers and zero to less than 5% in benign and normal breast tissue controls. In Western women, the prevalence of MMTV in breast cancers is approximately 30 to 40%. In China and Korea, the prevalence of MMTV is 10 to 20%. Positive MMTV case control studies are shown in Table 2. Negative MMTV breast cancer studies are shown in Table 3.
Table 2.
MMTV breast cancer case control studies. S = significant. Ns = not significant
| Study | Country | Method | Breast cancer | Breast control | Significance |
|---|---|---|---|---|---|
| Axel 1972 [20] | US | Molecular hybridisation | 19/29 66% | 0/15 0% | 0.004 s |
| Mesa-Tajada 1978 [30] | US | IHC | 15/131 39% | 0/18 0% | 0.001s |
| Wang 1995 [23] | US | PCR | 121/314 38.5% | 2/107 2% | 0.001s |
| Etkind 2000 [31] | US | PCR | 27/73 37% | 0/35 0% | 0.001s |
| Melana 2001 [32] | US | PCR | 32/106 30% | 1/106 1% | 0.001s |
| Melana 2002 [33] | Argentina | PCR | 23/74 31% | 1/10 10% | 0.003s |
| Ford 2003 [24] | Australia | PCR | 19/45 42% | 2/111 2% | 0.001s |
| Ford 2004 [34] | Australia | PCR | 45/144 31% | 0/111 0% | 0.001s |
| Zammarchi 2006 [25] | Italy | PCR | 15/45 33% | 0/8 0% | 0.008 s |
| Hachana 2008 [35] | Tunisia | PCR | 17/122 14% | 0/122 0% | 0.001s |
| Lawson 2010 [36] | Australia | In situ PCR | 33/74 45% | 0/29 0% | 0.001s |
| Mazzanti 2011 [29] | Italy | PCR | 47/69 68% | 0/20 0% | 0.001s |
| Glenn 2012 [37] | Australia | PCR | 39/50 78% | 13/40 33% | 0.045 s |
| Slaoui 2014 [38] | Morocco | PCR | 24/42 57% | 6/18 33% | 0.312 ns |
| Cedro-Tanda 2014 [39] | Mexico | PCR | 57/458 12% | 72/458 16% | 0.308 ns |
| Naushad 2014 [40] | Pakistan | PCR | 83/250 29% | 0/15 0% | 0.001s |
| Reza 2015 [41] | Iran | PCR | 12/100 12% | 0/100 0% | 0.001s |
| Shariatpanahi 2017 [42] | Iran | PCR | 19/59 32% | 3/59 5% | 0.002 s |
| Al Dossary 2018 [43] | Saudi Arabia | PCR | 6/101 6% | 0/51 0% | 0.082 ns |
| Seo 2019 [44] | Korea | PCR | 12/128 9% | 0/128 0% | 0.013 s |
| Al Hamad 2020 [45] | Jordan | PCR | 11/100 11% | 0/20 0% | 0.023 s |
| Loutfy 2021 [46] | Egypt | PCR | 38/50 76% | 0/10 0% | 0.001s |
| Wang 2021 [5] | China | PCR | 21/119 18% | 2/50 4% | 0.05 s |
| Khalid 2021 [47] | Pakistan | PCR | 69/105 66% | 2/15 13% | 0.023 s |
| Gupta 2021 [48] | Croatia | PCR | 5/70 7% | 0/16 0% | 0.056 ns |
| Gupta 2022 [49] | Qatar | PCR | 11/74 15% | 0/14 0% | 0.001s |
Table 3.
MMTV breast cancer – negative results
| Study | Country | Method | Negative MMTV breast cancer |
|---|---|---|---|
| Zangen 2002 [50] | Italy | PCR | 0/18 |
| Witt 2003 [51] | Austria | PCR | 0/50 |
| Mant 2004 [52] | United Kingdom | PCR | 0/44 |
| Bindra 2007 [53] | Sweden | PCR | 0/18 |
| Frank 2008 [54] | Germany | Hybridisation | 0/23 |
| Fukuoka 2008 [55] | Japan | PCR/hybridisation | 0/46 |
| Park 2011 [28] | Australia | PCR | 0/42 |
| Motamedifar 2012 [56] | Iran | PCR | 0/50 |
| Tabriz 2013 [57] | Iran | PCR | 0/40 |
| Morales-Sanchez 2013 [58] | Mexico | PCR | 0/65 |
| Perzova 2017 [59] | USA | PCR | 0/9 |
| Fekete 2023 [60] | Romania | PCR | 0/75 |
The implication is that there is a strong association between MMTV and human breast cancer.
There appear to be two reasons for the negative outcomes in 12 case control studies. One, MMTV may not be present in some human and mouse populations (see the epidemiology section below), and second the use of inadequate PCR techniques. This problem has been highlighted by Pogo et al. who demonstrated that the techniques used by Park et al. gave negative outcomes as compared to positive outcomes using alternative techniques [27, 28]. A further difficulty is the low MMTV viral load. Using quantitative PCR Mazzanti et al. (2011) demonstrated there is a dramatic decrease in MMTV viral load as the cancer progresses from ductal carcinoma in situ to invasive carcinoma [29]. They demonstrated there can be a complete loss of the virus in invasive ductal carcinoma.
Contamination issues
It has been argued that some MMTV positive human breast cancer results may be due to contamination with mouse DNA. Both Mazzanti et al. and Nartey et al. excluded mouse DNA contamination by performing murine mitochondrial DNA and intracisternal A-particle long terminal repeats by PCR [61, 62]. In addition MMTV has been independently identified in breast cancers in over 22 countries. It is most unlikely that contamination occurred in each of these laboratories.
MMTV and human breast cancer histology
Some human breast cancer specimens, in which MMTV-like env DNA sequences have been identified, were shown to have histological characteristics (morphology) similar to MMTV-associated mouse mammary tumours [18]. These observations are compatible with an association between the presence of MMTV-like env DNA sequences and some human breast cancers.
Epidemiology
The prevalence of breast cancer varies greatly between countries [63]. The US, Australia, the Netherlands, France and Germany have high rates - over 95 cases per 100,000 age-adjusted females. Japan, China, and India have low rates—between 30 and 45 cases per 100,000 age-adjusted females. There are marked differences in the prevalence of breast cancer between Western and Eastern Europe. In the United Kingdom, the Netherlands, France and Italy, the prevalence of breast cancer is over 95 as compared to Russia and the Baltic countries, with a prevalence of less than 50 per 100,000 age-adjusted females. There are also differences in parts of northern as compared to southern China [5].
While there are several reasons including food consumption patterns, which may account for the differences in the prevalence of breast cancer, an additional reason is the distribution of MMTV-infected mice [64, 65]. Mus domesticus, which is the common house mouse in Western countries, carries more infectious MMTV than does Mus musculus [64]. Mus domesticus mice are mainly located in Western Europe and Mus musculus in Eastern Europe. Mus domesticus is more prevalent in northern as compared to southern China [5]. The prevalence of MMTV positive human breast cancer is significantly higher in Western as compared to eastern Europe and in northern China (23%) as compared to southern China. (6%). In a meta – analysis Wang et al. also showed there was a significant correlation between the prevalence of MMTV positive breast cancer in different countries and the global distribution of Mus domesticus [5]. Recent developments provide support for this hypothesis [65]. Mus domesticus mouse population outbreaks in Australia and New Zealand are associated with a modest increase in breast cancer incidence rates with a lag of approximately 3 years [65].
Serology studies
The prevalence of MMTV antibodies in the serum of women with breast cancer is consistently five-fold higher than in the sera of women with benign breast conditions or in normal women (Table 4). An exception is the study by Goerdert et al. (2006) who did not identify any MMTV antibodies in the sera of 92 women with breast cancer (however, MMTV gp52 was not identified in the positive controls) [66]. Zhang et al. more recently used ELISA gp 52 to demonstrate MMTV antibodies in 10% of sera from women with breast cancer compared to 2% of controls (p = 0.017) [67].
Table 4.
Mouse mammary tumour virus breast cancer serology
| Study | Country | Breast cancer | Controls | Significance |
|---|---|---|---|---|
| Muller 1972 [68] | Germany | 75/228 33% | 11/95 12% | 0.002 s |
| Ogawa 1978 [69] | Japan | 26/43 60% | 4/37 11% | 0.001s |
| Mehta 1978 [70] | India | 26/34 76% | 0/10 0% | 0.001s |
| Witkin 1979 [71] | US | 11/65 17% | 2/60 3% | 0.001 s |
| Imai 1979 [72] | Japan | 49/89 55% | 18/68 27% | 0.020 s |
| Witkin 1980 [73] | US | 14/54 26% | 5/63 8% | 0.026s |
| Day 1981 [22] | US | 27/145 19% | 1/36 3% | 0.026 s |
| Nagayoshi 1981 [74] | Japan | 34/96 36% | 3/59 5% | 0.026 s |
| Tomana 1981 [75] | US | 56/137 41% | 2/56 4% | 0.001 s |
| Zotter 1981 [76] | Germany | 84/367 23% | 11/184 6% | 0.001 s |
| Holder 1983 [77] | US | 41/52 79% | 2/18 11% | 0.004 s |
| Litvinov 1984 [78] | Russia | 51/92 55% | 3/94 3% | 0.001 s |
| Chattopadhyah 1984 [79] | India | 14/14 100% | 0/13 0% | 0.004 s |
| Kovarik 1989 [80] | Czech | 2/60 3% | 0/60 0% | 0.226 ns |
| Goerdert 2006 [66] | US | 0/92 | ||
| Zhang 2020 [67] | Canada | 10/98 10% | 2/98 2% | 0.017 s |
Genetics
The Bevilacqua group in Pisa, Italy, have shown that MMTV-like sequences are not significantly associated with hereditary breast carcinoma [81]. 30% of 56 sporadic breast cancers contain MMTV sequences compared to 4% of 47 BRCA hereditary breast cancers (p < 0.001) [81].
Temporality (timing) of the Association
MMTV has been identified in normal and benign human breast tissues up to 10 years prior to the development of MMTV positive human breast cancers in the same patient [62]. This is an important causal criteria. MMTV has also been identified in the sputum of children [61]. The implication is that breast cancer may take many decades to develop following infections with MMTV. On the other hand Stewart and Chen (2022) have hypothesised that MMTV positive breast cancer can develop within 3 years following mouse epidemics [65]. There is no obvious explanation for these differences.
MMTV transmission
Human saliva is a likely means of transmission of MMTV between humans [61]. MMTV gene sequences are present in saliva in 27% of normal children, 11% of normal adults and 57% of women with breast cancer [61]. MMTV gene sequences have been identified in human parotid glands - the source of saliva [61]. Humans have well developed lymphatic structures in the mouth and nose which are possible entry points for MMTV. That MMTV can infect adult mice via nasal lymphoid tissue has been demonstrated experimentally [82]. Accordingly, MMTV can enter mammals, including humans, from a range of sites.
Because MMTV is transmitted via milk from mouse mothers to their pups, the presence of MMTV in human milk has been investigated. MMTV has been identified in 5% of breast milk samples from healthy lactating Australian women [83]. In addition it has been shown that the prevalence of MMTV in human milk is significantly higher among women who are at greater than normal risk of breast cancer [84]. However MMTV transmission via human milk is unlikely to be an influential means of transmission in humans. There are two reasons. One, is the destructive effect of human milk. This was shown experimentally by Sarkar et al. (1973) [85]. Two, because epidemiological studies have not shown associations between breast feeding and breast cancer [86].
MMTV has been identified in mammary tumours in dogs and cats [87, 88]. It is possible, but not proven, that transmission could occur between cats, dogs and humans, as has been shown between mice and humans.
In many countries it is permissible for 1% in weight of cereals to contain mouse or rat faecal material. US regulatory food standards allow up to two pellets of rodent excreta per pint of wheat (US pint = 551 cm3) [89]. Because MMTV is endemic in many rodent populations, transmission via rodent faecal material by consumption of uncooked cereals and other foods is possible.
Experimental evidence
MMTVs can infect human breast cells in culture and can randomly integrate into the human genome [90]. MMTV proteins have been shown to be capable of malignantly transforming normal human breast epithelial cells [91].
Analogy
The life cycle of MMTV is similar in mice and humans. MMTV infects T and B lymphocytes in the Peyer’s lymphocyte patches. MMTV is activated by super antigens which circulate in lymphocytes and enters breast epithelial cells, where they integrate into the mouse and human genome [92].
Oncogenic mechanisms
The underlying causal mechanisms by which MMTV may cause human breast cancer are far from clear. MMTV proteins are capable of malignant transformation of normal human breast epithelial cells [91]. An additional mechanism involves the APOBEC enzyme family. APOBEC3B is an enzyme which inhibits retrovirus replication. In humans, inactivating mutations and deletions in APOBEC3B appear to play a role in breast cancer development [93, 94]. Human papilloma viruses have been shown to alter the expression of APOBEC3B, which may reduce its protective effects against MMTV [95].
Conclusion
The evidence meets the extended Hill causal criteria. A causal role for MMTV-like viruses in human breast cancer is probable.
Human papilloma virus
High risk for cancer human papilloma viruses (HPV) in breast cancer were first identified in 1992 by Anna Di Lonardo and colleagues working in Rome [96]. Their findings were later confirmed in case control studies all of which demonstrated that the prevalence of high- risk HPVs was consistently higher in breast cancer than in normal and benign breast controls [7].
High-risk HPVs have been identified in breast cancer in 20 countries [6]. High risk HPV types 16 and 18 are predominant in Western women. HPV types 16, 18, 33, 52 and 58 are common in Chinese, Korean, Japanese and Qatari women. In studies in which high-risk HPVs were identified, there were no correlations with breast cancer grade, survival, or steroid receptor expression [97]. The reason for the variations in outcomes is partly due to the difficulty of identifying the extremely low HPV viral loads in breast cancer as compared to cervical cancer [98].
Strength and consistency of association between HPVs and breast cancer
There have been 46 case control studies in which HPVs have been identified [6]. The prevalence of high risk for cancer HPVs was 1335 (31%) of breast cancers as compared to 163 (9%) of 1838 normal and benign breast tissue controls (p = 0.001). A meta-analysis of these studies indicated an odds ratio of 5.56 (p = 0.01) for HPV positive breast cancer cases compared to HPV positive controls [6] [Awan 2023]. The outcomes of these case control studies are consistent. These case control studies are shown in Table 5.
Table 5.
Identification of high risk for cancer human papilloma virus in breast cancers and controls (case control studies)
| Study | Country | HPV Breast cancer | HPV Non cancer breast controls | Main HPV types | Significance |
|---|---|---|---|---|---|
| Yu 2000 [99] | Japan / China | 18/52 35% | 0/15 0% | 18,33 | 0.001s |
| Ren 2003 [100] | China | 45/80 56% | 2/30 7% | 16,18 | 0.002s |
| Damin 2004 [101] | Brazil | 25/101 25% | 0/41 0% | 16,18 | 0.001s |
| Tsai 2007 [102] | Taiwan | 8/62 13% | 2/32 6% | 0.004s | |
| Choi 2007 [103] | Korea | 8/123 7% | 0/31 0% | 16, 18,58 | 0.001s |
| Gumus 2006 [104] | Turkey | 37/50 74% | 16/50 32% | 18,33 | 0.162 ns |
| Fan 2008 [105] | China | 23/52 44% | 1/16 6% | 16 | 0.001s |
| He 2009 [106] | China | 24/40 60% | 1/20 5% | 16 | 0.001s |
| De Leon 2009 [107] | Mexico | 15/41 37% | 0/43 0% | 16,18 | 0.001s |
| Mendizabal 2009 [108] | Mexico | 3/67 4% | 0/40 0% | 16,18,33 | 0.157ns |
| Heng 2009 [109] | Australia | 8/26 31% | 3/28 11% | 16,18 | 0.611ns |
| Mou 2011 [110] | China | 4/62 6% | 0/46 0% | 16,18 | 0.025s |
| Sigaroodi 2012 [111] | Iran | 15/58 26% | 1/41 2% | 16,18 | 0.002s |
| Frega 2012 [112] | Italy | 9/31 29% | 0/12 0% | 16,18 | 0.005s |
| Divani 2012 [113] | Greece | 6/35 17% | 0/35 0% | 16,18 | 0.025s |
| Glenn 2012 [37] | Australia | 25/50 50% | 8/40 20% | 16,18 | 0.006s |
| Liang 2013 [114] | China | 48/224 21% | 6/37 16% | 16,18,33,58 | 0.001s |
| Ahangar2014 [115] | Iran | 22/65 34% | 0/65 0% | 16 | 0.001s |
| Ali 2014 [116] | Iraq | 60/129 47% | 3/41 7% | 16,18,33 | 0.001s |
| Hong 2014 [117] | China | 23/45 51% | 1/20 5% | 16,18 | 0.001s |
| Manzouri 2014 [118] | Iran | 10/55 18% | 7/51 14% | 16 | 0.083 ns |
| Fu 2015 [119] | China | 25/169 15% | 1/83 1% | 58 | 0.001s |
| Li 2015 [120] | China | 3/187 2% | 0/92 0% | 6,16,18 | 0.157 ns |
| Gannon 2015 [121] | Australia | 13/78 17% | 1/10 10% | 0.002s | |
| Lawson 2015 [97] | Australia | 27/40 66% | 6/21 29% | 16,18,58 | 0.001s |
| Doosti 2016 [122] | Iran | 20/87 23% | 0/84 0% | 16,18 | 0.001s |
| Wang 2016 [123] | China | 52/146 36% | 3/83 3.6% | 16,18,58 | 0.001s |
| Zhang 2016 [124] | China | 34/325 11% | 4/100 4% | 16,18 | 0.001s |
| Delgardo 2017 [125] | Spain | 130/251 52% | 49/186 26% | 16 | 0.001s |
| Ladera 2017 [126] | Venezuela | 14/22 64% | 1/22 4.5% | 16,18,52,56 | 0.001s |
| Naushad 2017 [127] | Pakistan | 45/250 18% | 0/15 0% | 0.001s | |
| Islam 2017 [128] | India | 203/313 65% | 2/21 10% | 16,18,33 | 0.001s |
| Salman 2017 [129] | United Kingdom | 35/74 47% | 11/36 18% | 16,18,35,45,59 | 0.001s |
| Malekpour 2018 [130] | Iran | 8/98 8% | 0/40 0% | 16,18 | 0.008s |
| ElAmrani 2018 [131] | Morocco | 19/76 25% | 1/12 8% | 51,52,58 | 0.001s |
| Cavalcante 2018 [132] | Brazil | 51/103 50% | 15/95 16% | 6,11,18,31 | 0.001s |
| Khodabandehlou 2019 [133] | Iran | 35/72 48.6% | 5/3116.1% | 18 | 0.003s |
| Al Hamad 2020 [45] | Jordan | 21/100 21% | 0/20 0% | 16, 18 | 0.007s |
| Mofrad 2021 [134] | Iran | 7/59 12% | 0/11 0% | 18 | 0.004s |
| El-Seik 2021 [135] | Egypt | 16/72 22.2% | 0/15 0% | 16,18 | 0.003s |
| Golrokh 2021 [136] | Iran | 7/59 12% | 0/15 0% | 6,18 | 0.014s |
| Guo 2021 [137] | US | 25/56 45% | 3/9 33% | 11, 18 | 0.005s |
| Nagi 2021 [138] | Lebanon | 66/102 67% | 5/14 36% | 16,35,45,52,58 | 0.001s |
| Alinezhadi 2022 [139] | Iran | 7/8 88% | 2/9 22% | 16 | 0.025s |
| Tavakolian 2023 [140] | Iran | 9/40 23% | 3/ 50 6% | 18 | 0.014s |
| Khasawneh 2024 [141] | Jordan | 27/110 25% | 0/30 0% | 16,18 | 0.001s |
The prevalence of high risk HPV is consistently higher in all studies of breast cancers as compared to controls
s = significant at 0.05 level ns = not significant at 0.05 level
In a prospective cohort study involving 61,872 subjects in Taiwan, patients with HPV were 1.4 times more likely to develop breast cancer than patients of the same age without HPV [142].
Women who develop HPV associated cervical cancer have a higher risk of developing HPV associated breast cancer [143]. These women are on average 10 years younger at the age of developing breast cancer than older women who develop breast cancer. Younger women are more sexually active and at greater risk of sexually transmitted HPV infections.
Oncogenic mechanisms
High-risk HPVs encode proteins, several of which have an oncogenic capacity. The expression of HPV E6 and E7 proteins leads to malignant changes in normal epithelial cells. HPV E6 proteins degrade p53 (a cancer suppressing gene). HPV E7 enhances viral replication and malignant transformation from normal to cancer cells by upregulating Cox-2 which increases inflammation. HPV associated koilocytes have been identified in breast cancers [144]. Koilocytes are large squamous cells with acentric nuclei surrounded by a halo. Koilocytes are the basis of cervical Pap smears. HPV E5 and E6 act early in transformation and lead to the formation of koilocytes. Although several of these HPV oncogenic mechanisms in cervical cancer appear to be involved in breast cancer, additional mechanisms are involved. These may involve the antiviral enzyme APOBEC3B. HPV infections upregulate and lead to mutations in APOBEC3B which increase the risk of breast cancer [95]. Experimental evidence shows that exposure to HPV E6 and E7 proteins can transform and immortalise normal human breast epithelial cells [96]. High risk HPVs of the same type have been identified in benign breast tissues 1 to 11 years before the development of HPV positive breast cancers in the same women [145].
Transmission
Sexual intercourse is regarded as the primary route of human papillomavirus (HPV) transmission. However, HPVs are stable viruses which can stay on tissue and other surfaces for many days. There is evidence which indicates HPVs can also be transmitted by non- sexual means from mother to child by fomites, from health care workers to patients and in an unknown manner to adolescent girls with no sexual experience [146]. There is also evidence that HPVs may be transmitted via saliva and blood and from the cervix to the breast by circulating extra cellular vesicles – also known as exosomes [147].
Conclusions
High risk for cancer HPVs have been consistently identified and are significantly more prevalent in breast cancers than normal and benign breast cancers. In conclusion it is likely that HPVs have a causal role in breast cancer.
Epstein Barr virus (EBV)
In 1957 surgeon Dennis Burkitt, working in Uganda, identified acute malignant lymphomas in children [148]. Anthony Epstein in collaboration with Burkitt, Bert Achong and Yvonne Barr, identified viral particles in the lymphoma specimens [149]. They later identified the particles as human herpes virus 4 (Epstein Barr virus – EBV). This pioneering work was of universal value because it was the first demonstration that viruses could cause cancer in humans.
In 1995, working in London, Louise Labreque and colleagues made the first identification of EBV in breast cancer [150]. Various EBV genes have since been identified in breast cancer in a wide range of countries.
Strength and consistency of association between EBVs and breast cancer
EBV positive lymphocytes commonly infiltrate breast cancer cells. Studies which do not identify breast cancer cells separately from infiltrating lymphocytes are not valid. Therefore a careful assessment of each study is required before inclusion in meta-analyses. In a meta-analysis of 24 case control studies by Agolli et al. 2023 the odds ratio of EBV positive breast cancer compared to normal and benign breast controls was 4.43 (p = 0.01) [7]. The evidence is consistent that EBVs are significantly more prevalent in breast cancers than controls. These case control studies are shown in Table 6.
Table 6.
Case control studies Epstein Barr virus and breast cancer
| Study | Country | Identification method | EBV positive breast cancer | EBV positive breast controls | Significance |
|---|---|---|---|---|---|
| Labreque 1995 [150] | United Kingdom | PCR, ISH | 19/91 21% | 0/21 0% | 0.001 |
| Luqmani 1995 [151] | United Kingdom | PCR, IHC | 15/28 54% | 0/12 0% | 0.001 |
| Bonnet 1999 [152] | France | PCR, IHC | 51/100 50% | 0/30 0% | 0.001 |
| Fina 2001 [153] |
Algeria Europe |
PCR, ISH, microdissection | 162/509 32% | 0/10 0% | 0.001 |
| Grinstein 2002 [154] | United States | PCR, IHC | 14/33 42% | 3/26 12% | 0.039 |
| Preciado 2005 [155] | Argentina | PCR, IHC | 24/69 35% | 0/17 0% | 0.001 |
| Fawzy 2008 [156] | Egypt | PCR, IHC | 10/40 25% | 0/20 0% | 0.001 |
| Joshi 2009 [157] | India | IHC | 28/51 55% | 0/30 0% | 0.001 |
| Lorenzetti 2010 [158] | Argentina | PCR, ISH, IHC | 22/71 31% | 0/48 0% | 0.001 |
| Kadivar 2011 [159] | Iran | PCR, IHC | 0/100 0% | 0/42 0% | |
| Mazouni 2011 [160] | France |
PCR microdissection |
65/196 33% | 1/15 7% | 0.001 |
| Hachana 2011 [161] | Tunisia | PCR, IHC | 33/90 | 0/123 | 0.001s |
| Glenn 2012 [37] | Australia | PCR, in situ PCR, IHC | 34/50 68% | 14/40 35% | 0.011s |
| Zekri 2012 [162] | Iraq | PCR, IHC, ISH | 32/90 35% | 0/20 0% | 0.001s |
| Khabaz 2013 [163] | Jordan | PCR, IHC | 24/92 26% | 3/49 6% | 0.001s |
| Yahia 2014 [164] | Sudan | PCR, ISH | 49/92 53% | 12/50 24% | 0.001s |
| Mohammadizadeh 2014 [165] | Iran | PCR, IHC | 6/74 8% | 0/80 0% | 0.001s |
| Richardson 2015 [166] | New Zealand | PCR | 24/70 34% | 9/70 13% | 0.253 ns |
| Ahmed 2016 [167] | Egypt | IHC | 11/10,710% | 0/107 0% | 0.001s |
| El Naby 2017 [168] | Egypt | PCR, IHC | 10/42 24% | 6/42 14% | 0.689 ns |
| Fessahaye 2017 [169] | Eritrea | PCR, ISH, IHC | 40/144 28% | 4/33 12% | 0.003s |
| Pai 2018 [170] | India | ISH | 25/83 30% | 0/7 0% | 0.001s |
| Al Hamad 2020 [45] | Jordan | ISH | 24/100 24% | 0/20 0% | 0.007s |
| Alinezhad 2021 [171] | Iran | PCR | 9/80 11.2% | 0/80 0% | 0.009s |
| Nagi 2021 [138] | Lebanon | PCR | 41/102 40% | 0/14 0% | 0.001s |
| Zhang W. 2022 [172] | China | PCR | 54/140 41% | 0/25 0% | 0.001s |
| Khasawneh 2024 [141] | Jordan | PCR | 18/110 16% | 1/30 3% | 0.001s |
PCR = polymerase chain reaction, IHC = immunohistochemistry, ISH = in situ hybridisation, ns = not significant at 0.05 level
Epstein–Barr virus gene sequences have been identified in benign breast tissues 1 to 11 years prior to the development of EBV-positive breast cancer [145]. This is an important causal criteria.
In economically developed countries EBV associated infectious mononucleosis occurs most commonly among teenagers. This is in contrast to developing countries where EBV infections mainly occur in early childhood. EBV infections in teenagers and young adults is associated with Hodgkins lymphomas. There is a strong correlation between EBV associated Hodgkins lymphoma and breast cancer [173]. Epstein–Barr virus is mostly transmitted from person to person via saliva. EBV has been found in 61% of blood samples from healthy donors, which may explain its transmission through the body [174].
Oncogenic mechanisms
EBV infections predispose human breast epithelial cells to malignant transformation [175]. EBV EBNA-1 has been associated with BRCA- 1 gene defects which in turn is associated with breast cancer [176]. The precise oncogenic mechanisms for EBV are not known.
Conclusions
The evidence for a role of EBV in breast cancer while consistent, needs to be further developed.
Bovine leukemia virus
Bovine leukemia virus is an oncogenic retrovirus capable of integrating into a host’s DNA causing a lifetime infection. Janice Miller and colleagues of the US were the first to identify virus like particles in cattle lymphosarcoma in 1969 [177]. These particles became known as bovine leukemia virus. Only a small proportion of infected animals develop cancer – most of which are lymphomas.
BLV infects cattle in the Americas, some parts of Europe and Asia plus the Middle East. Breast cancer is more prevalent in red meat eating and cow’s milk consuming populations as compared to those with a high prevalence of lactose intolerance such as China [178]. In cattle BLV is mainly located in lymphocytes and mammary epithelial cells which can exfoliate into milk.
In 2003 Gertrude Buehring and her colleagues at the University of California at Berkeley were able to identify BLV antibodies in human blood serum [179]. Later, Buehring and her colleagues, identified BLV in 44% of US breast cancers [180]. BLV has since been identified in breast cancers in women from Australia, Argentina, Columbia, Brazil, Iran and Pakistan but not in Europe, China or Japan. The nucleotide sequences of the BLV env gene are 97.8 to 99.7% the same in both human breast tissues and cattle blood [181]. This indicates that BLV is probably a zoonotic infection.
Using whole genome sequencing Gillet and Willems did not identify BLV DNA in any of 51 human breast cancer sequences based on the US National Center for Biotechnology and Information [182]. This is contrary to the outcomes based on serology and PCR. A possible explanation suggested by Vinner et al. 2015 is the extremely low BLV load in human breast cancer [183]. Amato et al. using PCR did not identify BLV in US breast cancers [184]. The reason is not known.
Based on 9 case control studies, the prevalence of BLV was 334 (40%) of 826 breast cancers.
compared to 215 (24.0%) of 898 normal and benign breast controls [8]. With two exceptions the prevalence of BLV is significantly higher in breast cancers as compared to controls. In a meta-analysis of case control studies BLV was associated with an odds ratio of 2.6 increased risk of breast cancer [8]. These case control studies are shown in Table 7.
Table 7.
Identification of bovine leukemia virus in human breast cancer (case control studies)
| Study | Location | BLV positive breast cancer | BLV positive normal benign breast | Significance |
|---|---|---|---|---|
| Giovanna 2013 [185] | Columbia | 19/53 36% | 24/53 45% | 0.682 ns |
| Buehring 2015 [186] | US | 67/114 59% | 30/104 29% | 0.001s |
| Zhang R 2016 [187] | China | 0/91 0% | 0/100 0% | |
| Buehring 2017 [188] | Australia | 40/50 80% | 19/46 41% | 0.001s |
| Baltzell 2017 [189] | US | 35/61 57% | 20/103 20% | 0.059 ns |
| Khalilian 2019 [190] | Iran | 57/172 30% | 5/28 8% | 0.001s |
| Schwingel 2019 [191] | Brazil | 22/72 30.5% | 10/72 13.9% | 0.017s |
| Delamelina 2020 [192] | Brazil | 47/49 96% | 23/39 59% | 0.001s |
| Canova 2021 [181] | Brazil | 51/59 86% | ||
| Khan 2022 [193] | Pakistan | 728 /2710 27% | 10/80 13% | 0.005s |
| Yamanaka 2022 [194] | Japan | 0/23 0% | ||
| Amato 2023 [184] | US | 0/56 0% |
ns = not significant at 0.05 level
Of special interest is a study of BLV in human breast cancers in south eastern Brazil where the people traditionally ingest raw (unpasteurized) milk and cheese. BLV was present in 90% of the dairy cattle and 96% of human breast cancer cases [192].
In a study of Australian women with breast cancer, BLV was identified in benign breast tissue 3–10 years before BLV positive malignancy was diagnosed [188]. This observation is in accord with the causal criteria of a prior infection before the development of the same pathogen related cancer. In this same study BLV was identified in high proportions in both breast and benign controls (80% and 41% respectively). As only a small proportion of BLV infected animals develop cancer it is possible that only a small proportion of BLV infections in humans also progress to breast cancer.
BLV has not been identified based on PCR in breast cancers in several studies including Japan [194] and the US [184]. Canova et al. (2021) suggest that these conflicting results might be related to differing PCR methods [181]. It is possible that the viral DNA sequences targeted by the PCR may not have been present in the genomes that were analysed. Partial genome deletions following integration into the host cells are common and can be an important mechanism to avoid the host immune response. Such deletions have been observed in studies of BLV-related primate T lymphotropic virus type 1 (PTLV-1), including deletions in the gag region, followed by deletions of the pol and env genes. In contrast, the tax and LTR regions were the less frequently deleted genes [195].
Transmission
BLV has been identified in up to 49% of fresh milk and raw beef available for human consumption [196, 197]. Further, BLV RNA has been identified in the air and on surfaces at dairy workplaces, which may be a source of occupational infection [198].
Oncogenic mechanisms
Due to the economic importance of BLV in the cattle industry there have been detailed investigations into its oncogenic mechanisms. BLV encodes the regulatory protein Tax. It is the key protein involved in viral replication. In animals BLV is a three stage process. (i) BLV infection of cells, (ii) immortalisation of cells by the influence of Tax proteins and (iii) malignant transformation following p53 and other mutations [199]. Only a small proportion of infected animals develop cancer – most of which are lymphomas. Although it is likely that the oncogenic mechanisms of BLV are similar in humans, there is no evidence available.
Conclusion
It is likely that BLV has a causal role in some human breast cancers but additional evidence is required before any conclusions can be made.
Inter-relationship between MMTV, HPV, EBV and BLV in human breast cancer
MMTV, HPV and EBV have been identified in the same Australian breast cancers [37].
In addition these multiple viruses have been identified in benign breast specimens 10 years before the development of the same multiple virus associated breast cancers in the same women [145]. Co-infection of high-risk HPV and EBV, has been observed in Lebanese and other breast cancers [138]. As outlined above HPV appears to influence the oncogenicity of MMTV via its influence on APOBEC enzymes [95]. The oncogenic influences between HPV and EBV are not known.
Discussion and conclusions
Mouse mammary tumour virus
The evidence that MMTV has a causal role in human breast cancer is increasingly comprehensive. The oncogenic influences of MMTV in human breast cancer appears to be almost identical to MMTV in mice. In Western women, the prevalence of MMTV in breast cancers is approximately 30 to 40%. In China, Korea and Vietnam, the prevalence of MMTV is 10 to 20%. Overall the odds ratio between MMTV in human breast cancer and normal and benign breast tissues is very high at 13·40. There appears to be an association between MMTV positive breast cancer and the location of MMTV infected Mus domesticus mice. The life cycle of MMTV in humans is similar that of mice although the means of transmission probably differs. In humans the most likely means of transmission is via sputum whereas in mice transmission is via mouse milk from infected mother to pup.
While it is likely that MMTV has a causal role in human breast cancer the development of additional evidence is of advantage. In addition to the above evidence there is a need to replicate the Graff et al. studies on mice conducted in 1949 in which MMTV particles isolated from mouse milk were injected into the perineum of healthy mice. Approximately half of these mice developed breast cancers [11]. It should be possible to isolate MMTV particles from human breast cancers and inject them into experimental mice.
Conclusion
The evidence meets the extended Hill causal criteria. A causal role for MMTV-like viruses in human breast cancer is probable.
Human papilloma viruses
High risk for cancer HPVs have been consistently identified and are significantly more prevalent in breast cancers than normal and benign breast cancers. There have been 46 case control studies in 20 countries in which high risk for cancer HPVs have been identified [6]. The prevalence of high risk for cancer HPVs was 1335 (31%) of breast cancers as compared to 163 (9%) of 1838 normal and benign breast tissue controls (p = 0.001). Overall the odds ratio between high risk HPVs in breast cancer and normal and benign breast tissues is 5.56. While sexual intercourse is accepted as the main means of HPV transmission, there is evidence that HPVs may be transmitted via saliva and blood and from the cervix to the breast by circulating extra cellular vesicles – also known as exosomes.
The causal mechanisms for HPV related breast cancer appear to differ from cervical cancer. While HPV proteins E6 and E7 probably have a causal role as demonstrated by the presence of HPV related koilocytes (characteristic cells used as the basis of Pap smears) in breast cancer, HPVs in breast cancer appear to have two additional causal mechanisms by (i) influencing APOBEC mechanisms (APOBEC enzymes offer antiviral protection) and (ii) combining with EBV. In conclusion it is likely that HPVs have a causal role in breast cancer.
Conclusion
In conclusion it is likely that HPVs have a causal role in breast cancer.
Epstein Barr virus
EBV has been consistently identified and is significantly more prevalent in breast cancers than normal and benign breast cancers. The prevalence of EBV was 844 (31%) of 2754 breast cancers as compared to 53 (5%) of 1061 normal and benign breast tissue controls (p = 0.001). Overall the odds ratio between EBV in breast cancer and normal and benign breast tissues is 4.43. The underlying mechanisms for a role of EBV in breast cancer is not clear.
Conclusion
The evidence for a role of EBV in breast cancer while consistent, needs to be further developed.
Bovine leukemia virus
Based on 9 case control studies, the prevalence of BLV was 334 (40%) of 826 breast cancers compared to 215 (24.0%) of 898 normal and benign breast controls. Overall the odds ratio between BLV in breast cancer and normal and benign breast tissues is 2.26. The associations between the presence of BLV in fresh meat and milk and increased prevalence of BLV positive breast cancer is suggestive of a causal role for BLV.
Conclusion
It is likely that BLV has a causal role in some human breast cancers however additional evidence is required before any conclusions can be made.
Susceptibility to virus associated breast cancer
Only a small proportion of women exposed to the established risk factors for breast cancer – early age menarche, late age menopause, late age first pregnancy, excess weight, genetics, develop breast cancer. There is no available evidence relevant to viruses and breast cancer. The most plausible reason why some women exposed to these risk factors, including virus infections, develop breast cancer is genetic susceptibility [200, 201]. This susceptibility may be familial or sporadic.
Prevention of viral induced breast cancer
Effective vaccines against HPV infections are widely available for both girls and boys [202]. Recently a successful vaccine against BLV in cattle has been developed [203]. This is a crucial development. In future the use of culling (killing) BLV infected cattle should no longer be necessary.
Vaccines against MMTV and EBV for use in humans are not available and require urgent development. Using traditional methods vaccines have been successful in the prevention of MMTV associated breast cancer in mice [204].
Author contributions
JL and WG wrote the manuscript.
Funding
There was no financial support for this review.
Data availability
All the relevant data are within the paper.
Declarations
Ethics approval and consent to participate
Ethics are not applicable to this review paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bevilacqua G. The viral origin of human breast Cancer: from the mouse mammary tumor virus (MMTV) to the human Betaretrovirus (HBRV). Viruses. 2022;14(8):1704. 10.3390/v14081704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Syed H, Penner T, Mason AL. Linking human betaretrovirus with autoimmunity and liver disease in patients with primary biliary cholangitis. Viruses. 2022;14(9):1941. 10.3390/v14091941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johal H, Faedo M, Faltas J, Lau A, Mousina R, Cozzi P, Defazio A, Rawlinson WD. DNA of mouse mammary tumor virus-like virus is present in human tumors influenced by hormones. J Med Virol. 2010;82(6):1044–50. 10.1002/jmv.21754 [DOI] [PubMed] [Google Scholar]
- 4.Goubran M, Wang W, Indik S, Faschinger A, Wasilenko ST, Bintner J, Carpenter EJ, Zhang G, Nuin P, Macintyre G, Wong GK, Mason AL. Isolation of a human betaretrovirus from patients with primary biliary cholangitis. Viruses. 2022;14(5):886. 10.3390/v14050886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang FL, Zhang XL, Yang M, Lin J, Yue YF, Li YD, et al. Prevalence and characteristics of mouse mammary tumor virus-like virus associated breast cancer in China. Infect Agent Cancer. 2021;16:47. 10.1186/s13027-021-00383-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awan UA, Khattak AA, Ahmed N, Guo X, Akhtar S, Kamran S, Yongjing Z, Liu J, Khan S. An updated systemic review and meta-analysis on human papillomavirus in breast carcinogenesis. Front Oncol. 2023;13:1219161. 10.3389/fonc.2023.1219161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agolli A, Ishak A, Viswanathan M, Co EL, Shivakumar J, Agolli O. Epstein-Barr viral infection and the risk for breast Cancer: a systematic review. Int J Hematol Oncol Stem Cell Res. 2023;17(2):114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khatami A, Pormohammad A, Farzi R, Saadati H, Mehrabi M, Kiani SJ, et al. Bovine Leukemia virus (BLV) and risk of breast cancer: a systematic review and meta-analysis of case-control studies. Infect Agent Cancer. 2020;15:48. 10.1186/s13027-020-00314-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill AB. The environment and disease: association or causation? Proc R Soc Med. 1965;58(5):295–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bittner JJ. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162. 10.1126/science.84.2172.162.a [DOI] [PubMed] [Google Scholar]
- 11.Graff S, Moore DH, Stanley WM, Randall HT, Haagense CD. Isolation of mouse mammary carcinoma virus. Cancer. 1949;2:755–62. [DOI] [PubMed] [Google Scholar]
- 12.Duesberg PH, Blair PB. Isolation of the nucleic acid of mouse mammary tumor virus (MMTV). Proc Natl Acad Sci USA. 1966;55:1490–7. 10.1073/pnas.55.6.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore DH, Charney J, Kramarsky B, Lasfargues EY, Sarkar NH, Brennan MJ, et al. Search for a human breast Cancer Virus. Nature. 1971;229:611–5. 10.1038/229611a0 [DOI] [PubMed] [Google Scholar]
- 14.Bevilacqua G. Unusual findings of viral production (MuMTV) in murine mammary tumors (BALB/cfC3H). Exp Mol Pathol. 1983;39(3):271–81. 10.1016/0014-4800(83)90057-6 [DOI] [PubMed] [Google Scholar]
- 15.Melana SM, Nepomnaschy I, Sakalian M, Abbott A, Hasa J, Holland JF, et al. Characterization of viral particles isolated from primary cultures of human breast Cancer cells. Cancer Res. 2007;67:8960–5. 10.1158/0008-5472.CAN-06-3892 [DOI] [PubMed] [Google Scholar]
- 16.Callahan R, Mudunuri U, Bargo S, Raafat A, McCurdy D, Boulanger C, et al. Genes affected by mouse mammary tumor virus (MMTV) proviral insertions in mouse mammary tumors are deregulated or mutated in primary human mammary tumors. Oncotarget. 2012;3:1320–34. 10.18632/oncotarget.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melana SM, Nepomnaschy I, Hasa J, Djougarian A, Holland JF, Pogo BG, et al. Detection of human mammary tumor virus proteins in human breast cancer cells. J Virol Methods. 2010;163:157–61. 10.1016/j.jviromet.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 18.Glenn WK, Lawson JS, Whitaker NJ. Mouse mammary tumour-like virus gene sequences and specific breast cancer morphology. J Clin Pathol. 2007;60(9):1071. 10.1136/jcp.2006.044487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson JS, Glenn WK, Salmons B, Ye Y, Heng RB, Moody P, et al. Mouse mammary tumor virus–like sequences in human breast Cancer. Cancer Res. 2010;70:3576–85. 10.1158/0008-5472.CAN-09-4160 [DOI] [PubMed] [Google Scholar]
- 20.Axel R, Schlom J, Spiegelman S. Presence in human breast Cancer of RNA homologous to mouse mammary tumour virus RNA. Nature. 1972;235:32–6. 10.1038/235032a0 [DOI] [PubMed] [Google Scholar]
- 21.Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–90. 10.1128/jvi.61.2.480-490.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day NK, Witkin SS, Sarkar NH, Kinne D, Jussawalla DJ, Levin A, et al. Antibodies reactive with murine mammary tumor virus in sera of patients with breast cancer: Geographic and family studies. Proc Natl Acad Sci USA. 1981;78:2483–7. 10.1073/pnas.78.4.2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, et al. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–9. [PubMed] [Google Scholar]
- 24.Ford CE, Tran D, Deng Y, Ta VT, Rawlinson WD, Lawson JS. Mouse mammary tumor virus-like gene sequences in breast tumors of Australian and Vietnamese women. Clin Cancer Res. 2003;9:1118–20. [PubMed] [Google Scholar]
- 25.Zammarchi F, Pistello M, Piersigilli A, Murr R, Di Cristofano C, Naccarato AG, et al. MMTV-like sequences in human breast cancer: a fluorescent PCR/laser microdissection approach. J Pathol. 2006;209:436–44. 10.1002/path.1997 [DOI] [PubMed] [Google Scholar]
- 26.Ahmad W, Khader TA, Panicker NG, Akhlaq S, Baby J, Gull B, Mustafa F. MMTV-like env sequences from human breast Cancer patients cannot yet be considered as a separate species. Hamdan Med J. 2022;15:155–63. 10.4103/hmj.hmj_35_22 [DOI] [Google Scholar]
- 27.Pogo BG, Melana SM, Moran H, Holland JF. Presence of MMTV-like env gene sequences in human breast cancer. Breast Cancer Res Treat. 2011;125:295–7. 10.1007/s10549-010-1179-2 [DOI] [PubMed] [Google Scholar]
- 28.Park DJ, Southey MC, Giles GG, Hopper JL. No evidence of MMTV-like env sequences in specimens from the Australian breast Cancer Family Study. Breast Cancer Res Treat. 2011;125:229–35. 10.1007/s10549-010-0946-4 [DOI] [PubMed] [Google Scholar]
- 29.Mazzanti CM, Al Hamad M, Fanelli G, Scatena C, Zammarchi F, Zavaglia K, Lessi F, Pistello M, Naccarato AG, Bevilacqua G. A mouse mammary tumor virus env-like exogenous sequence is strictly related to progression of human sporadic breast carcinoma. Am J Pathol. 2011;179(4):2083–90. 10.1016/j.ajpath.2011.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mesa-Tejada R, Keydar I, Ramanarayanan M, Ohno T, Fenoglio C, Spiegelman S. Detection in human breast carcinomas of an antigen immunologically related to a group-specific antigen of mouse mammary tumor virus. Proc Natl Acad Sci USA. 1978;75:1529–33. 10.1073/pnas.75.3.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6:1273–8. [PubMed] [Google Scholar]
- 32.Melana S, Holland JF, Pogo BG. Search for mouse mammary tumor virus like env sequences in cancer and normal breast from the same individuals. Clin Cancer Res. 2001;7:283–4. [PubMed] [Google Scholar]
- 33.Melana SM, Picconi MA, Rossi C, Mural J, Alonio LV, Teyssié A, Holland JF, Pogo BG. Detection of murine mammary tumor virus (MMTV) env gene like sequences in breast cancer from Argentine patients. Med (B Aires). 2002;62:323–7. (Spanish). [PubMed] [Google Scholar]
- 34.Ford CE, Faedo M, Crouch R, Lawson JS, Rawlinson WD. Progression from normal breast pathology to breast cancer is associated with increasing prevalence of mouse mammary tumor virus-like sequences in men and women. Cancer Res. 2004;64:4755–9. 10.1158/0008-5472.CAN-03-3804 [DOI] [PubMed] [Google Scholar]
- 35.Hachana M, Trimeche M, Ziadi S, Amara K, Gaddas N, Mokni M, Korbi S. Prevalence and characteristics of the MMTV-like associated breast carcinomas in Tunisia. Cancer Lett. 2008;271:222–30. 10.1016/j.canlet.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 36.Lawson JS, Glenn WK, Salmons B, Ye Y, Heng B, Moody P, Johal H, Rawlinson WD, Delprado W, Lutze-Mann L, Whitaker NJ. Mouse mammary tumor virus-like sequences in human breast cancer. Cancer Res. 2010;70:3576–85. 10.1158/0008-5472.CAN-09-4160 [DOI] [PubMed] [Google Scholar]
- 37.Glenn WK, Heng B, Delprado W, Iacopetta B, Whitaker NJ, Lawson JS. Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS ONE. 2012;7:e48788. 10.1371/journal.pone.0048788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slaoui M, Mzibri ME, Razine R, Qmichou Z, Attaleb M, Amrani M. Detection of MMTV-Like sequences in Moroccan breast cancer cases. Infect Agent Cancer. 2014;9:37. 10.1186/1750-9378-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cedro-Tanda A, Córdova-Solis A, Juárez-Cedillo T, Pina-Jiménez E, Hernández-Caballero ME, Moctezuma-Meza C, et al. Prevalence of HMTV in breast carcinomas and unaffected tissue from Mexican women. BMC Cancer. 2014;14:942. 10.1186/1471-2407-14-942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naushad W, Bin Rahat T, Gomez MK, Ashiq MT, Younas M, Sadia H. Detection and identification of mouse mammary tumor virus like DNA sequences in blood and breast tissues of breast cancer patients. Tumour Biol. 2014;35:8077–86. 10.1007/s13277-014-1972-3 [DOI] [PubMed] [Google Scholar]
- 41.Reza MA, Reza MH, Mahdiyeh L, Mehdi F, Hamid ZN. Evaluation frequency of Merkel Cell Polyoma, Epstein-Barr and Mouse Mammary Tumor viruses in patients with breast Cancer in Kerman, Southeast of Iran. Asian Pac J Cancer Prev. 2015;16:7351–7. 10.7314/APJCP.2015.16.16.7351 [DOI] [PubMed] [Google Scholar]
- 42.Shariatpanahi S, Farahani N, Salehi AR, Salehi R. High prevalence of mouse mammary tumor virus-like gene sequences in breast cancer samples of Iranian women. Nucleosides Nucleotides Nucleic Acids. 2017;36:621–30. 10.1080/15257770.2017.1360498 [DOI] [PubMed] [Google Scholar]
- 43.Al Dossary R, Alkharsah KR, Kussaibi H. Prevalence of mouse mammary tumor virus (MMTV)-like sequences in human breast cancer tissues and adjacent normal breast tissues in Saudi Arabia. BMC Cancer. 2018;18:170. 10.1186/s12885-018-4074-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo I, Cho JH, Lee MH, Park WJ, Kwon SY, Lee JH. Clinical and prognostic value of human mammary tumor virus in Korean patients with breast carcinoma. Annals Clin Lab Sci. 2019;49:171–4. [PubMed] [Google Scholar]
- 45.Al Hamad M, Matalka I, Al Zoubi MS, Armogida I, Khasawneh R, Al-Husaini M, Sughayer M, Jaradat S, Al-Nasser AD, Mazzanti CM. Human mammary tumor virus, human papilloma virus, and Epstein-Barr Virus infection are associated with sporadic breast cancer metastasis. Breast Cancer (Auckl). 2020;14:1178223420976388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loutfy SA, Abdallah ZF, Shaalan M, Moneer M, Karam A, Moneer MM, Sayed IM, Abd El-Hafeez AA, Ghosh P, Zekri AN. Prevalence of MMTV-Like env sequences and its Association with BRCA1/2 genes mutations among Egyptian breast Cancer patients. Cancer Manag Res. 2021;13:2835–48. 10.2147/CMAR.S294584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khalid HF, Ali A, Fawad N, Rafique S, Ullah I, Rehman G, Shams MU, Idrees M. MMTV-LIKE virus and c-myc over-expression are associated with invasive breast cancer. Infect Genet Evol. 2021;91:104827. 10.1016/j.meegid.2021.104827 [DOI] [PubMed] [Google Scholar]
- 48.Gupta I, Ulamec M, Peric-Balja M, Ramic S, Al Moustafa AE, Vranic S, Al-Farsi HF. Presence of high-risk HPVs, EBV, and MMTV in human triple-negative breast cancer. Hum Vaccin Immunother. 2021;17(11):4457–66. 10.1080/21645515.2021.1975452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta I, Al-Sarraf R, Farghaly H, Vranic S, Sultan AA, Al-Thawadi H, Al Moustafa AE, Al-Farsi HF. Incidence of HPVs, EBV, and MMTV-Like Virus in breast Cancer in Qatar. Intervirology. 2022;65(4):188–94. 10.1159/000525277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zangen R, Harden S, Cohen D, Parrella P, Sidransky D. Mouse mammary tumor-like env gene as a molecular marker for breast cancer? Int J Cancer. 2002;102(3):304–7. 10.1002/ijc.10702 [DOI] [PubMed] [Google Scholar]
- 51.Witt A, Hartmann B, Marton E, Zeillinger R, Schreiber M, Kubista E. The mouse mammary tumor virus-like env gene sequence is not detectable in breast cancer tissue of Austrian patients. Oncol Rep. 2003;10:1025–9. [PubMed] [Google Scholar]
- 52.Mant C, Cason J. A human murine mammary tumour virus-like agent is an unconvincing aetiological agent for human breast cancer. Rev Med Virol. 2004;14:169–77. 10.1002/rmv.427 [DOI] [PubMed] [Google Scholar]
- 53.Bindra A, Muradrasoli S, Kisekka R, Nordgren H, Wärnberg F, Blomberg J. Search for DNA of exogenous mouse mammary tumor virus-related virus in human breast cancer samples. J Gen Virol. 2007;88:1806–9. 10.1099/vir.0.82767-0 [DOI] [PubMed] [Google Scholar]
- 54.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, Leib-Mösch C, Seifarth W. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J Virol. 2008;82:1808–18. 10.1128/JVI.02115-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuoka H, Moriuchi M, Yano H, Nagayasu T, Moriuchi H. No association of mouse mammary tumor virus-related retrovirus with Japanese cases of breast cancer. J Med Virol. 2008;80:1447–51. 10.1002/jmv.21247 [DOI] [PubMed] [Google Scholar]
- 56.Motamedifar M, Saki M, Ghaderi A. Lack of association of mouse mammary tumor virus-like sequences in Iranian breast cancer patients. Med Princ Pract. 2012;21:244–8. 10.1159/000334572 [DOI] [PubMed] [Google Scholar]
- 57.Tabriz HM, Zendehdel K, Shahsiah R, Fereidooni F, Mehdipour B, Hosseini ZM. Lack of detection of the mouse mammarytumor-like virus (MMTV) env gene in Iranian women breast cancer using real time PCR. Asian Pac J Cancer Prev. 2013;14:2945–8. 10.7314/APJCP.2013.14.5.2945 [DOI] [PubMed] [Google Scholar]
- 58.Morales-Sánchez A, Molina-Muñoz T, Martínez-López JLE, Hernández-Sancén P, Mantilla A, Leal YA, Torres J, Fuentes-Pananá EM. No association between Epstein-Barr Virus and Mouse Mammary Tumor Virus with breast cancer in Mexican women. Sci Rep. 2013;3:2970. 10.1038/srep02970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perzova R, Abbott L, Benz P, Landas S, Khan S, Glaser J, Cunningham CK, Poiesz B. Is MMTV associated with humanbreast cancer? Maybe, but probably not. Virol J. 2017;14:196. 10.1186/s12985-017-0862-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fekete Z, Tertan BO, Raduly L, Eniu DT, Buiga R, Galatar M, Berindan-Neagoe I. Prevalence of MMTV-like sequences in breast cancer samples in Romanian patients-there is a geographic difference compared to the western world. Infect Agent Cancer. 2023;18(1):39. 10.1186/s13027-023-00486-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazzanti CM, Lessi F, Armogida I, Zavaglia K, Franceschi S, Al Hamad M, Roncella M, Ghilli M, Boldrini A, Aretini P, Fanelli G, Marchetti I, Scatena C, Hochman J, Naccarato AG, Bevilacqua G. Human saliva as route of inter-human infection for mouse mammary tumor virus. Oncotarget. 2015;6(21):18355–63. 10.18632/oncotarget.4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nartey T, Mazzanti CM, Melana S, Glenn WK, Bevilacqua G, Holland JF, Whitaker NJ, Lawson JS, Pogo BGT. Mouse mammary tumor-like virus (MMTV) is present in human breast tissue before development of virally associated breast cancer. Infect Agents Cancer. 2017;12:1. 10.1186/s13027-016-0113-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149:778–89. 10.1002/ijc.33588 [DOI] [PubMed] [Google Scholar]
- 64.Stewart TH, Sage RD, Stewart AF, Cameron DW. Breast cancer incidence highest in the range of one species of house mouse, Mus domesticus. Br J Cancer. 2000;82:446–51. 10.1054/bjoc.1999.0941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stewart AFR, Chen H-H. Revisiting the MMTV Zoonotic Hypothesis to Account for Geographic Variation in Breast Cancer Incidence. Viruses 2022;14:559. [DOI] [PMC free article] [PubMed]
- 66.Goedert JJ, Rabkin CS, Ross SR. Prevalence of serologic reactivity against four strains of mouse mammary tumour virus among US women with breast cancer. Br J Cancer. 2006;94:548–51. 10.1038/sj.bjc.6602977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang G, Bashiri K, Kneteman M, Cave K, Hong Y, Mackey JR, Alter HJ, Mason AL. Seroprevalence of human Betaretrovirus surface protein antibodies in patients with breast Cancer and liver disease. J Oncol. 2020:7406146. [DOI] [PMC free article] [PubMed]
- 68.Müller M, Grossman H. An antigen in human breast cancer sera related to the murine mammary tumour virus. Nat New Biol. 1972;237:116–7. 10.1038/newbio237116a0 [DOI] [PubMed] [Google Scholar]
- 69.Ogawa H, Tanaka H. Occurrence of antibody against intracytoplasmic A-particles of mouse mammary tumor virus in sera from breast cancer patients. Gan. 1978;69:539–44. [PubMed] [Google Scholar]
- 70.Mehta SP, Sirsat SM, Jussawalla DJ. Humoral antibodies to mouse mammary tumor virus in sera from breast cancer patients. Indian J Exp Biol. 1978;16:1126–30. [PubMed] [Google Scholar]
- 71.Witkin SS, Egeli RA, Sarkar NH, Good RA, Day NK. Virolysis of mouse mammary tumor virus by sera from breast cancer patients. Proc. Natl. Acad. Sci. USA 1979;76:2984–2987. [DOI] [PMC free article] [PubMed]
- 72.Imai M, Yamada C, Saga S, Nagayoshi S, Hoshino M. Immunological cross reaction between sera from patients with breastcancer and mouse mammary tumor virus. Gan. 1979;70:63–74. [PubMed] [Google Scholar]
- 73.Witkin SS, Sarkar NH, Good RA, Day NK. An enzyme-linked immunoassay for the detection of antibodies to the mouse mammary tumor virus: application to human breast cancer. J Immunol Methods. 1980;32(1):85–91. 10.1016/0022-1759(80)90119-2 [DOI] [PubMed] [Google Scholar]
- 74.Nagayoshi S, Imai M, Tsutsui Y, Saga S, Takahashi M, Hoshino M. Use of the immune adherence hemagglutination test for titration of breast cancer patients’ sea cross-reacting with purified mouse mammary tumor virus. Gan. 1981;72:98–103. [PubMed] [Google Scholar]
- 75.Tomana M, Kajdos AH, Niedermeier W, Durkin WJ, Mestecky J. Antibodies to mouse mammary tumor virus-related antigen in sera of patients with breast carcinoma. Cancer. 1981;47:2696–703. [DOI] [PubMed] [Google Scholar]
- 76.Zotter S, Grossmann H, Johannsen BA, Pilz C. Is there any diagnostic relevance of human antibodies which react with mouse mammary tumor virus (MuMTV)? Arch. Geschwulstforsch. 1981;51:338–43. [PubMed] [Google Scholar]
- 77.Holder WD Jr., Wells SA. Jr. Antibody reacting with the murine mammary tumor virus in the serum of patients with breast carcinoma: a possible serological detection method for breast carcinoma. Cancer Res. 1983;43:239–44. [PubMed] [Google Scholar]
- 78.Litvinov SV, Remennik IV, Kriukova IN. Humoral antibodies to the antigens related to the structural protein antigens of the mammary cancer virus (MTV) in breast cancer patients and in healthy donors. Bull Exp Biol Med. 1984;97:600–3. 10.1007/BF00808256 [DOI] [PubMed] [Google Scholar]
- 79.Chattopadhyay J, Chattopadhyay U, Chowdhury JR. Immunological relatedness of a murine mammary tumor-associatedantigen and human breast cancer. Gan. 1984;75:342–8. [PubMed] [Google Scholar]
- 80.Kovarík A, Hlubinová K, Prachar J, Simkovic D, Knotek J. No significant correlation between specific antibodies to mouse mammary tumour virus and human cancer. Br J Cancer. 1989;60(4):572–5. 10.1038/bjc.1989.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naccarato AG, Lessi F, Zavaglia K, Scatena C, Al Hamad MA, Aretini P, Menicagli M, Roncella M, Ghilli M, Caligo MA, et al. Mouse mammary tumor virus (MMTV)-like exogenous sequences are associated with sporadic but not hereditary human breast carcinoma. Aging. 2019;11:7236–41. 10.18632/aging.102252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Velin D, Fotopoulos G, Luthi F, Kraehenbuhl J-P. The nasal-associated lymphoid tissue of adult mice acts as an entry site for the mouse mammary tumor Retrovirus. J Exp Med. 1997;185:1871–6. 10.1084/jem.185.10.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johal H, Ford C, Glenn W, Heads J, Lawson J, Rawlinson W. Mouse mammary tumor like virus sequences in breast milk from healthy lactating women. Breast Cancer Res Treat. 2011;129:149–55. 10.1007/s10549-011-1421-6 [DOI] [PubMed] [Google Scholar]
- 84.Nartey T, Moran H, Marin T, Arcaro KF, Anderton DL, Etkind P, Holland JF, Melana SM, Pogo BG-T. Human mammary tumor virus (HMTV) sequences in human milk. Infect Agents Cancer. 2014;9:20–20. 10.1186/1750-9378-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarkar NH, Charney J, Dion AS, Moore DH. Effect of human milk on the mouse mammary tumor virus. Cancer Res. 1973;33(3):626–9. [PubMed] [Google Scholar]
- 86.Zhou Y, Chen J, Li Q, Huang W, Lan H, Jiang H. Association between breastfeeding and breast cancer risk: evidence from a meta-analysis. Breastfeed Med. 2015;10(3):175–82. 10.1089/bfm.2014.0141 [DOI] [PubMed] [Google Scholar]
- 87.Szabo S, Haislip AM, Garry RF. Of mice, cats, and men: is human breast cancer a zoonosis? Microsc Res Tech. 2005;68(3–4):197–208. 10.1002/jemt.20232 [DOI] [PubMed] [Google Scholar]
- 88.Hsu W-L, Lin H-Y, Chiou S-S, Chang C-C, Wang S-P, Lin K-H, Chulakasian S, Wong M-L, Chang S-C. Mouse mammary Tumor Virus-Like nucleotide sequences in Canine and Feline Mammary tumors. J Clin Microbiol. 2010;48,:4354–62. 10.1128/JCM.01157-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.US Food & Drug Administration. The Food defect action levels. Handbook; 2005.
- 90.Konstantoulas CJ, Indik S. C3H strain of mouse mammary tumour virus, like GR strain, infects human mammary epithelial cells, albeit less efficiently than murine mammary epithelial cells. J Gen Virol. 2015;96:650–62. 10.1099/jgv.0.000006 [DOI] [PubMed] [Google Scholar]
- 91.Katz E, Lareef MH, Rassa JC, Grande SM, King LB, Russo J, Ross SR, Monroe JG. MMTV env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J Exp Med. 2005;201:431–9. 10.1084/jem.20041471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Jiang J-D, Xu D, Li Y, Qu C, Holland JF, Pogo BG. T. A mouse mammary tumor virus-like long terminal repeat superantigen in human breast Cancer. Cancer Res. 2004;64:4105–11. 10.1158/0008-5472.CAN-03-3880 [DOI] [PubMed] [Google Scholar]
- 93.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–37096. 10.1038/nature11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pereira de Sousa N, Vitiello GAF, Amarante MK. Involvement of APOBEC3A/B deletion in mouse mammary tumor virus (MMTV)-like positive human breast Cancer. Diagnostics (Basel). 2023;13(6):1196. 10.3390/diagnostics13061196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ohba K, Ichiyama K, Yajima M, Gemma N, Nikaido M, Wu, et al. In vivo and in Vitro studies Suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLoS ONE. 2014;9:e97787. 10.1371/journal.pone.0097787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Lonardo A, Venuti A, Marcante ML. Human papillomavirus in breast cancer. Breast Cancer Res Treat. 1992;21(2):95–100. 10.1007/BF01836955 [DOI] [PubMed] [Google Scholar]
- 97.Lawson JS, Glenn WK, Salyakina D, Delprado W, Clay R, Antonsson A, Heng B, Miyauchi S, Tran DD, Ngan CC, Lutze-Mann L, Whitaker NJ. Human papilloma viruses and breast Cancer. Front Oncol. 2015;5:277. 10.3389/fonc.2015.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khan NA, Castillo A, Koriyama C, Kijima Y, Umekita Y, Ohi Y. Human papillomavirus detected in female breast carcinomas in Japan. Br J Cancer. 2008;99(3):408–14. 10.1038/sj.bjc.6604502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99..Yu Y, Morimoto T, Sasa M, Okazaki K, Harada Y, Fujiwara T, et al. Human papillomavirus type 33 DNA in breast cancer in Chinese. Breast Cancer. 2000;7:33–6. 10.1007/BF02967185 [DOI] [PubMed] [Google Scholar]
- 100.Ren Z, Huang J, Shi Z et al. Detection of human papillomavirus types 16 and 18 infection in breast cancer tissues by Primed in situ labeling. Zhongguo Zhongliu Linchuang 30:243–246 (2003). (Data from Ren 2019).
- 101.Damin AP, Karam R, Zettler CG, Caleffi M, Alexandre CO. Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Res Treat. 2004;84:131–7. 10.1023/B:BREA.0000018411.89667.0d [DOI] [PubMed] [Google Scholar]
- 102.Tsai JH, Hsu CS, Tsai CH, Su JM, Liu YT, Cheng MH, et al. Relationship between viral factors, axillary lymph node status and survival in breast cancer. J Cancer Res Clin Oncol. 2007;133:13–21. 10.1007/s00432-006-0141-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Choi YL, Cho EY, Kim JH, Nam SJ, Oh YL, Song SY, et al. Detection of human papillomavirus DNA by DNA chip in breast carcinomas of Korean women. Tumour Biol. 2007;28:327–32. 10.1159/000124238 [DOI] [PubMed] [Google Scholar]
- 104.Gumus M, Yumuk PF, Salepci T, Aliustaoglu M, Dane F, Ekenel M, et al. HPV DNA frequency and subset analysis in human breast cancer patients’ normal and tumoral tissue samples. J Exp Clin Cancer Res. 2006;25:515–21. [PubMed] [Google Scholar]
- 105.Fan CL, Zhou JH, Hu CY. Expression of human papillomavirus in mammary carcinoma and its possible mechanism in carcinogenesis. Virol Sin. 2008;23:226–31. 10.1007/s12250-008-2914-2 [DOI] [Google Scholar]
- 106.He Q, Zhang SQ, Chu YL, Jia XL, Wang XL. The correlations between HPV16 infection and expressions of c-erbB-2 and bcl-2 in breast carcinoma. Mol Biol Rep. 2009;36:807–12. 10.1007/s11033-008-9249-9 [DOI] [PubMed] [Google Scholar]
- 107.de León DC, Montiel DP, Nemcova J, Mykyskova I, Turcios E, Villavicencio V, et al. Human papillomavirus (HPV) in breast tumors: prevalence in a group of Mexican patients. BMC Cancer. 2009;9:26. 10.1186/1471-2407-9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mendizabal-Ruiz AP, Morales JA, Ramírez-Jirano LJ, Padilla-Rosas M, Morán-Moguel MC, Montoya-Fuentes H. Low frequency of human papillomavirus DNA in breast cancer tissue. Breast Cancer Res Treat. 2009;114:189–94. 10.1007/s10549-008-9989-1 [DOI] [PubMed] [Google Scholar]
- 109.Heng B, Glenn WK, Ye Y, Tran B, Delprado W, Lutze-Mann L, et al. Human papilloma virus is associated with breast cancer. Br J Cancer. 2009;101:1345–50. 10.1038/sj.bjc.6605282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mou X, Chen L, Liu F, Shen Y, Wang H, Li Y, et al. Low prevalence of human papillomavirus (HPV) in Chinese patients with breast cancer. J Int Med Res. 2011;39:1636–44. 10.1177/147323001103900506 [DOI] [PubMed] [Google Scholar]
- 111.Sigaroodi A, Nadji SA, Naghshvar F, Nategh R, Emami H, Velayati AA. Human papillomavirus is associated with breast cancer in the north part of Iran. Sci World J. 2012:837191. [DOI] [PMC free article] [PubMed]
- 112.Frega A, Lorenzon L, Bononi M, De Cesare A, Ciardi A, Lombardi D, et al. Evaluation of E6 and E7 mRNA expression in HPV DNA positive breast cancer. Eur J Gynaecol Oncol. 2012;33:164–7. [PubMed] [Google Scholar]
- 113.Divani SN, Giovani AM. Detection of human papillomavirus DNA in fine needle aspirates of women with breast cancer. Arch Oncol. 2012;20:12–4. 10.2298/AOO1202012D [DOI] [Google Scholar]
- 114.Liang W, Wang J, Wang C, Lv Y, Gao H, Zhang K, et al. Detection of high-risk human papillomaviruses in fresh breast cancer samples using the hybrid capture 2 assay. J Med Virol. 2013;85:2087–92. 10.1002/jmv.23703 [DOI] [PubMed] [Google Scholar]
- 115.Ahangar-Oskouee M, Shahmahmoodi S, Jalilvand S, Mahmoodi M, Ziaee AA, Esmaeili HA, et al. No detection of ‘high-risk’ human papillomaviruses in a group of Iranian women with breast cancer. Asian Pac J Cancer Prev. 2014;15:4061–5. 10.7314/APJCP.2014.15.9.4061 [DOI] [PubMed] [Google Scholar]
- 116.Ali SH, Al-Alwan NA, Al-Alwany SH. Detection and genotyping of human papillomavirus in breast cancer tissues from Iraqi patients. East Mediterr Health J. 2014;20:372–7. 10.26719/2014.20.6.372 [DOI] [PubMed] [Google Scholar]
- 117.Hong L, Tang S, Does. HPV 16/18 infection affect p53 expression in invasive ductal carcinoma? An experimental study. Pak J Med Sci. 2014;30:789–92. 10.12669/pjms.304.4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Manzouri L, Salehi R, Shariatpanahi S, Rezaie P. Prevalence of human papilloma virus among women with breast cancer since 2005–2009 in Isfahan. Adv Biomed Res. 2014;3:75. 10.4103/2277-9175.125873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fu L, Wang D, Shah W, Wang Y, Zhang G, He J. Association of human papillomavirus type 58 with breast cancer in Shaanxi Province of China. J Med Virol. 2015;87:1034–40. 10.1002/jmv.24142 [DOI] [PubMed] [Google Scholar]
- 120.Li J, Ding J, Zhai K. Detection of human papillomavirus DNA in patients with breast tumor in China. PLoS ONE. 2015;10:e0136050. 10.1371/journal.pone.0136050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gannon OM, Antonsson A, Milevskiy M, Brown MA, Saunders NA, Bennett IC. No association between HPV positive breast cancer and expression of human papilloma viral transcripts. Sci Rep. 2015;5:18081. 10.1038/srep18081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Doosti M, Bakhshesh M, Zahir ST, Shayestehpour M, Karimi-Zarchi M. Lack of evidence for a relationship between high risk human papillomaviruses and breast Cancer in Iranian patients. Asian Pac J Cancer Prev. 2016;17:4357–61. [PubMed] [Google Scholar]
- 123.Wang D, Fu L, Shah W, Zhang J, Yan Y, Ge X, He J, Wang Y, Li X. Presence of high risk HPV DNA but indolent transcription of E6/E7 oncogenes in invasive ductal carcinoma of breast. Pathol Res Pract. 2016;212:1151–6. 10.1016/j.prp.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 124.Zhang N, Ma ZP, Wang J, Bai HL, Li YX, Sun Q et al. Human papillomavirus infection correlates with inflammatory Stat3 signaling activity and IL-17 expression in patients with breast cancer. Am J Transl Res. 2016;8:3214-3226.29. [PMC free article] [PubMed]
- 125.Delgado-García S, Martínez-Escoriza JC, Alba A, Martín-Bayón TA, Ballester-Galiana H, Peiró G, et al. Presence of human papillomavirus DNA in breast cancer: a Spanish case-control study. BMC Cancer. 2017;17:320. 10.1186/s12885-017-3308-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ladera M, Fernandes A, López M, Pesci-Feltri A, Ávila M, Correnti M. Presence of human papillomavirus and Epstein-Barr virus in breast cancer biopsies as potential risk factors. Gaceta Mexicana De Oncologia. 2017;16:107–12. [Google Scholar]
- 127.Naushad W, Surriya O, Sadia H. Prevalence of EBV, HPV and MMTV in Pakistani breast cancer patients: a possible etiological role of viruses in breast cancer. Infect Genet Evol. 2017;54:230–7. 10.1016/j.meegid.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 128.Islam S, Dasgupta H, Roychowdhury A, Bhattacharya R, Mukherjee N, Roy A, et al. Study of association and molecular analysis of human papillomavirus in breast cancer of Indian patients: clinical and prognostic implication. PLoS ONE. 2017;12:e0172760. 10.1371/journal.pone.0172760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salman NA, Davies G, Majidy F, Shakir F, Akinrinade H, Perumal D, et al. Association of High Risk Human Papillomavirus and breast cancer: a UK based Study. Sci Rep. 2017;7:43591. 10.1038/srep43591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Malekpour AR, Deldar Z, Mollaei HR, Arabzadeh SA, Iranpour M. Evaluation of HPV DNA positivity in colorectal cancer patients in Kerman, Southeast Iran. Asian Pac J Cancer Prev. 2018;19:193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.ElAmrani A, Gheit T, Benhessou M, McKay-Chopin S, Attaleb M, Sahraoui S, et al. Prevalence of mucosal and cutaneous human papillomavirus in Moroccan breast cancer. Papillomavirus Res. 2018;5:150–5. 10.1016/j.pvr.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cavalcante JR, Pinheiro LGP, Almeida PRC, Ferreira MVP, Cruz GA, Campelo TA, et al. Association of breast cancer with human papillomavirus (HPV) infection in Northeast Brazil: molecular evidence. Clin (Sao Paulo). 2018;73:e465. 10.6061/clinics/2018/e465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khodabandehlou N, Mostafaei S, Etemadi A, Ghasemi A, Payandeh M, Hadifar S, et al. Human papilloma virus and breast cancer: the role of inflammation and viral expressed proteins. BMC Cancer. 2019;19:61. 10.1186/s12885-019-5286-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mofrad MG, Sadigh ZA, Ainechi S, Faghihloo E. Detection of human papillomavirus genotypes, herpes simplex, varicella zoster and cytomegalovirus in breast cancer patients. Virol J. 2021;18:25. 10.1186/s12985-021-01498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.El-Sheik N, Mousa NO, Tawfeik AM, Saleh AM, Elshikh I, Deyab M, et al. Assessment of Human Papillomavirus infection and risk factors in Egyptian women with breast Cancer. Breast Cancer (Auckl). 2021;15:1178223421996279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Golrokh Mofrad M, Sadigh ZA, Ainechi S, Faghihloo E. Detection of human papillomavirus genotypes, herpes simplex, varicella zoster and cytomegalovirus in breast cancer patients. Virol J. 2021;18(1):25. 10.1186/s12985-021-01498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guo H, Idrovo JP, Cao J, Roychoudhury S, Navale P, Auguste LJ, Bhuiya T, Sheikh-Fayyaz S. Human papillomavirus (HPV) detection by Chromogenic in situ hybridization (CISH) and p16 immunohistochemistry (IHC) in breast intraductal papilloma and breast carcinoma. Clin Breast Cancer. 2021;21(6):e638–46. 10.1016/j.clbc.2021.04.006 [DOI] [PubMed] [Google Scholar]
- 138.Nagi K, Gupta I, Jurdi N, Jabeen A, Yasmeen A, Batist G, Vranic S, Al-Moustafa AE. High-risk human papillomaviruses and Epstein-Barr virus in breast cancer in Lebanese women and their association with tumor grade: a molecular and tissue microarray study. Cancer Cell Int. 2021;21(1):308. 10.1186/s12935-021-02009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alinezhadi M, Makvandi M, Kaydani GA, Jazayeri SN, Charostad J, Talaeizadeh AT, Angali KA. Detection of high-risk human papillomavirus DNA in invasive ductal carcinoma specimens. Asian Pac J Cancer Prev. 2022;23(9):3201–7. 10.31557/APJCP.2022.23.9.3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tavakolian S, Faghihloo E. The prevalence of varicella zoster virus, herpes simplex virus type 2, and human papillomavirus in breast cancerous tissues and their adjacent ones in Iran. J Res Med Sci. 2023;28:65. 10.4103/jrms.jrms_475_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Khasawneh AI, Himsawi N, Sammour A, Al Shboul S, Alorjani M, Al-Momani H, Shahin U, Al-Momani H, Alotaibi MR, Saleh T. Association of Human Papilloma Virus, Cytomegalovirus, and Epstein-Barr virus with breast Cancer in Jordanian women. Med (Kaunas). 2024;60(5):699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu CH, Liao CY, Yeh MH, Wei JC. Risk role of breast Cancer in Association with Human Papilloma Virus among Female Population in Taiwan: a Nationwide Population-based Cohort Study. Healthc (Basel). 2022;10(11):2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lawson JS, Glenn WK, Salyakina D, Clay R, Delprado W, Cheerala B, Tran DD, Ngan CC, Miyauchi S, Karim M, Antonsson A, Whitaker NJ. Human papilloma virus identification in breast Cancer patients with previous cervical neoplasia. Front Oncol. 2016;5:298. 10.3389/fonc.2015.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lawson JS, Glenn WK, Heng B, Ye Y, Tran B, Lutze-Mann L, Whitaker NJ. Koilocytes indicate a role for human papilloma virus in breast cancer. Br J Cancer. 2009;101(8):1351–6. 10.1038/sj.bjc.6605328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lawson JS, Glenn WK. Multiple oncogenic viruses are present in human breast tissues before development of virus associated breast cancer. Infect Agents Cancer. 2017;12:55. 10.1186/s13027-017-0165-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Budukh A, Maheshwari A, Palayekar V, Bagal S, Purwar P, Deodhar K, et al. Prevalence and nonsexual transmission of human papilloma virus (HPV) in the adolescence girls from rural area of Maharashtra state, India. Indian J Cancer. 2018;55(4):336–9. 10.4103/ijc.IJC_188_18 [DOI] [PubMed] [Google Scholar]
- 147.De Carolis S, Storci G, Ceccarelli C, Savini C, Gallucci L, Sansone P, et al. HPV DNA associates with breast cancer malignancy and it is transferred to breast cancer stromal cells by extracellular vesicles. Front Oncol. 2019;9:860. 10.3389/fonc.2019.00860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Burkitt D, O’Conor GT. Malignant lymphoma in African children. I. A clinical syndrome. Cancer. 1961;14:258–69. [DOI] [PubMed] [Google Scholar]
- 149..Epstein A. Burkitt lymphoma and the discovery of Epstein-Barr virus. Br J Haematol. 2012;156(6):777–9. 10.1111/j.1365-2141.2011.09008.x [DOI] [PubMed] [Google Scholar]
- 150.Labrecque LG, Barnes DM, Fentiman IS, Griffin BE. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 1995;55(1):39–45. [PubMed] [Google Scholar]
- 151.Luqmani Y, Shousha S. Presence of Epstein-Barr-virus in breast-carcinoma. Int J Oncol. 1995;6:899–903. [DOI] [PubMed] [Google Scholar]
- 152.Bonnet M, Guinebretiere JM, Kremmer E, Grunewald V, Benhamou E, Contesso G, et al. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–81. 10.1093/jnci/91.16.1376 [DOI] [PubMed] [Google Scholar]
- 153.Fina F, Romain S, Ouafik L, Palmari J, Ben Ayed F, Benharkat S, et al. Frequency and genome load of Epstein-Barr virus in 509 breast cancers from different geographical areas. Br J Cancer. 2001;84:783–90. 10.1054/bjoc.2000.1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grinstein S, Preciado MV, Gattuso P, Chabay PA, Warren WH, De Matteo E, et al. Demonstration of Epstein-Barr virus in carcinomas of various sites. Cancer Res. 2002;62:4876–8. [PubMed] [Google Scholar]
- 155.Preciado MV, Chabay PA, De Matteo EN, Gonzalez P, Grinstein S, Actis A, et al. Epstein-Barr virus in breast carcinoma in Argentina. Arch Pathol Lab Med. 2005;129:377–81. 10.5858/2005-129-377-EVIBCI [DOI] [PubMed] [Google Scholar]
- 156.Fawzy S, Sallam M, Awad NM. Detection of Epstein-Barr virus in breast carcinoma in Egyptian women. Clin Biochem. 2008;41:486–92. 10.1016/j.clinbiochem.2007.12.017 [DOI] [PubMed] [Google Scholar]
- 157.Joshi D, Quadri M, Gangane N, Joshi R, Gangane N. Association of Epstein Barr virus infection (EBV) with breast cancer in rural Indian women. PLoS ONE. 2009;4:e8180. 10.1371/journal.pone.0008180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lorenzetti MA, De Matteo E, Gass H, Martinez Vazquez P, Lara J, Gonzalez P, et al. Characterization of Epstein Barr virus latency pattern in Argentine breast carcinoma. PLoS ONE. 2010;5:e13603. 10.1371/journal.pone.0013603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kadivar M, Monabati A, Joulaee A, Hosseini N. Epstein-Barr virus and breast cancer: lack of evidence for an association in Iranian women. Pathol Oncol Res. 2011;17:489–92. 10.1007/s12253-010-9325-z [DOI] [PubMed] [Google Scholar]
- 160.Mazouni C, Fina F, Romain S, Ouafik L, Bonnier P, Brandone JM, Martin PM. Epstein-Barr virus as a marker of biological aggressiveness in breast cancer. Br J Cancer. 2011;104:332–7. 10.1038/sj.bjc.6606048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hachana M, Amara K, Ziadi S, Romdhane E, Gacem RB, Trimeche M. Investigation of Epstein-Barr virus in breast carcinomas in Tunisia. Pathol Res Pract. 2011;207:695–700. 10.1016/j.prp.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 162.Zekri AR, Bahnassy AA, Mohamed WS, El-Kassem FA, El-Khalidi SJ, Hafez MM, et al. Epstein-Barr virus and breast cancer: epidemiological and molecular study on Egyptian and Iraqi women. J Egypt Natl Canc Inst. 2012;24:123–31. 10.1016/j.jnci.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 163.Khabaz MN. Association of Epstein-Barr virus infection and breast carcinoma. Arch Med Sci. 2013;9:745–51. 10.5114/aoms.2013.37274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yahia ZA, Adam AA, Elgizouli M, Hussein A, Masri MA, Kamal M, et al. Epstein Barr virus: a prime candidate of breast cancer aetiology in Sudanese patients. Infect Agent Cancer. 2014;9:9. 10.1186/1750-9378-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mohammadizadeh F, Zarean M, Abbasi M. Association of Epstein-Barr virus with invasive breast carcinoma and its impact on well-known clinicopathologic parameters in Iranian women. Adv Biomed Res. 2014;3:141. 10.4103/2277-9175.135158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Richardson AK, Currie MJ, Robinson BA, Morrin H, Phung Y, Pearson JF, Anderson TP, Potter JD, Walker LC. Cytomegalovirus and Epstein-Barr virus in breast cancer. PLoS ONE. 2015;10:e0118989. 10.1371/journal.pone.0118989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ahmed RA, Yussif SM. Immunohistochemical detection of human cytomegalovirus, Epstein-Barr virus and human papillomavirus in invasive breast carcinoma in Egyptian women: a tissue microarray study. J Solid Tumors. 2016;6:6–16. 10.5430/jst.v6n2p8 [DOI] [Google Scholar]
- 168.El-Naby NEH, Hassan Mohamed H, Mohamed Goda A, El Sayed Mohamed A. Epstein-Barr virus infection and breast invasive ductal carcinoma in Egyptian women: a single center experience. J Egypt Natl Canc Inst. 2017;29:77–82. 10.1016/j.jnci.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 169.Fessahaye G, Elhassan AM, Elamin EM, Adam AAM, Ghebremedhin A, Ibrahim ME. Association of Epstein - Barr virus and breast cancer in Eritrea. Infect Agent Cancer. 2017;12:62. 10.1186/s13027-017-0173-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pai T, Gupta S, Gurav M, Nag S, Shet T, Patil A, et al. Evidence for the association of Epstein-Barr virus in breast cancer in Indian patients using in-situ hybridization technique. Breast J. 2018;24:16–22. 10.1111/tbj.12828 [DOI] [PubMed] [Google Scholar]
- 171.Alinezhad F, Ahangar Oskouee M, Bannazadeh Baghi H, Tamiri Oskouee S, Esmaeili HA. Evidence of Epstein-Barr Virus in female breast Cancer. Iran J Public Health. 2021;50(2):425–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang WT, Zhu GL, Xu WQ, Zhang W, Wang HZ, Wang YB, Li YX. Association of PD-1/PD-L1 expression and Epstein–Barr virus infection in patients with invasive breast cancer. Diagn Pathol. 2022;17(1):61. 10.1186/s13000-022-01234-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yasui Y, Potter JD, Stanford JL, Rossing MA, Winget MD, et al. Breast cancer risk and delayed primary Epstein-Barr virus infection. Cancer Epidemiol Biomark Prev. 2001;10:9–16. [PubMed] [Google Scholar]
- 174.Gupta I, Nasrallah GK, Sharma A, Jabeen A, Smatti MK, Al-Thawadi HA, et al. Co-prevalence of human papillomaviruses (HPV) and Epstein–Barr virus (EBV) in healthy blood donors from diverse nationalities in Qatar. Cancer Cell Int. 2020;20(1):107. 10.1186/s12935-020-01190-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hu H, Luo ML, Desmedt C, Nabavi S, Yadegarynia S, Hong A, et al. Epstein-Barr Virus infection of mammary epithelial cells promotes Malignant Transformation. EBioMedicine. 2016;9:148–60. 10.1016/j.ebiom.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ali SHM, Shakir H, Al-Alwany M, Al-Wadi GLA, Huda Q, AL MA, et al. Coexpressional protein products of BRCA-1 and EBV-EBNA-1 genes in tissues from human female patients with breast cancers: an immunohistochemicalscreening study. Int J Chem Tech Res. 2017;10:709–19. [Google Scholar]
- 177.Miller JM, Miller LD, Olson C, Gillette KG. Virus-like particles in phytohemagglutinin-stimulated lymphocyte cultures with reference to bovine lymphosarcoma. J Natl Cancer Inst. 1969;43(6):1297–305. [PubMed] [Google Scholar]
- 178.Zur Hausen H, Bund T, de Villiers EM. Infectious agents in bovine red meat and milk and their potential role in Cancer and other Chronic diseases. Curr Top Microbiol Immunol. 2017;407:83–116. [DOI] [PubMed] [Google Scholar]
- 179.Buehring GC, Philpott SM, Choi KY. Humans have antibodies reactive with bovine leukemia virus. AIDS Res Hum Retrovir. 2003;19(12):1105–13. 10.1089/088922203771881202 [DOI] [PubMed] [Google Scholar]
- 180.Buehring GC, Shen HM, Jensen HM, Choi KY, Sun D, Nuovo G. Bovine leukemia virus DNA in human breast tissue. Emerg Infect Dis. 2014;20(5):772–82. 10.3201/eid2005.131298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Canova R, Weber MN, Budaszewski RF, da Silva MS, Schwingel D, Canal CW, Kreutz LC. Bovine leukemia viral DNA found on human breast tissue is genetically related to the cattle virus. One Health. 2021;13:100252. 10.1016/j.onehlt.2021.100252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gillet NA, Willems L. Whole genome sequencing of 51 breast cancers reveals that tumors are devoid of bovine leukemia virus DNA. Retrovirology. 2016;13(1):75. 10.1186/s12977-016-0308-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vinner L, Mourier T, Friis-Nielsen J, Gniadecki R, Dybkaer K, Rosenberg J, Langhoff JL, Cruz DFS, Fonager J, Izarzugaza JMG, et al. Investigation of human cancers for Retrovirus by Low-Stringency Target Enrichment and High-Throughput sequencing. Sci Rep. 2015;5:13201. 10.1038/srep13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Amato S, Ramsey J, Ahern TP, Rovnak J, Barlow J, Weaver D, Eyasu L, Singh R, Cintolo-Gonzalez J. Exploring the presence of bovine leukemia virus among breast cancer tumors in a rural state. Breast Cancer Res Treat. 2023;202(2):325–34. 10.1007/s10549-023-07061-4 [DOI] [PubMed] [Google Scholar]
- 185.Giovanna M, Carlos UJ, María UA, Gutierrez MF. Bovine leukemia virus gene segment detected in human breast tissue Open. J Med Microbiol. 2013;3:84–90. [Google Scholar]
- 186.Buehring GC, Shen HM, Jensen HM, Jin DL, Hudes M, Block G. Exposure to bovine leukemia virus is Associated with breast Cancer: a case-control study. PLoS ONE. 2015;10:e0134304. 10.1371/journal.pone.0134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang R, Jiang J, Sun W, Zhang J, Huang K, Gu X, Yang Y, Xu X, Shi Y, Wang C. Lack of association between bovine leukemia virus and breast cancer in Chinese patients. Breast Cancer Res. 2016;18:101. 10.1186/s13058-016-0763-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Buehring GC, Shen H, Schwartz DA, Lawson JS. Bovine leukemia virus linked to breast cancer in Australian women and identified before breast cancer development. PLoS ONE. 2017;12:e0179367. 10.1371/journal.pone.0179367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Baltzell KA, Shen HM, Krishnamurthy S, Sison JD, Nuovo GJ, Buehring GC. Bovine leukemia virus linked to breast cancer but not coinfection with human papillomavirus: case-control study of women in Texas. Cancer. 2018;124:1342–9. 10.1002/cncr.31169 [DOI] [PubMed] [Google Scholar]
- 190.Khalilian M, Hosseini SM, Madadgar O. Bovine leukemia virus detected in the breast tissue and blood of Iranian women. Microb Pathog. 2019;135:103566. 10.1016/j.micpath.2019.103566 [DOI] [PubMed] [Google Scholar]
- 191.Schwingel D, Andreolla AP, Erpen LMS, Frandoloso R, Kreutz LC. Bovine leukemia virus DNA associated with breast cancer in women from South Brazil. Sci Rep. 2019;9:2949. 10.1038/s41598-019-39834-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Delarmelina E, Buzelin MA, Souza BS, Souto FM, Bicalho JM, Câmara RJF, et al. High positivity values for bovine leukemia virus in human breast cancer cases from Minas Gerais, Brazil. PLoS ONE. 2020;15:e0239745. 10.1371/journal.pone.0239745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Khan Z, Abubakar M, Arshed MJ, Aslam R, Sattar S, Shah NA, Javed S, Tariq A, Bostan N, Manzoor S. Molecular investigation of possible relationships concerning bovine leukemia virus and breast cancer. Sci Rep. 2022;12(1):4161. 10.1038/s41598-022-08181-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yamanaka MP, Saito S, Hara Y, Matsuura R, Takeshima SN, Hosomichi K, Matsumoto Y, Furuta RA, Takei M, Aida Y. No evidence of bovine leukemia virus proviral DNA and antibodies in human specimens from Japan. Retrovirology. 2022;19(1):7. 10.1186/s12977-022-00592-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kamihira K, Sugahara K, Tsuruda S, Minami A, Uemura N, Akamatsu H, Nagai K, Murata H, Hasegawa Y, Hirakata Y, Takasaki K, Tsukasaki Y, Yamada. Proviral status of HTLV-1 integrated into the host genomic DNA of adult T-cell leukemia cells. Clin Lab Haematol. 2005;27:235–41. 10.1111/j.1365-2257.2005.00698.x [DOI] [PubMed] [Google Scholar]
- 196.Olaya-Galán NN, Corredor-Figueroa AP, Guzmán-Garzón TC, Ríos-Hernandez KS, Salas-Cárdenas SP, Patarroyo MA, Gutierrez MF. Bovine leukaemia virus DNA in fresh milk and raw beef for human consumption. Epidemiol Infect. 2017;145(15):3125–30. 10.1017/S0950268817002229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Barzegar H, Mirshahabi H, Motamed N, Yavarmanesh M, Mahdavi Poor B, Moaddab SR, Asgharzadeh M. Identification of bovine leukemia virus in raw milk samples in North-West of Iran. Vet Res Forum. 2021 Spring;12(2):223–7. [DOI] [PMC free article] [PubMed]
- 198.Stobnicka-Kupiec A, Gołofit-Szymczak M, G´orny RL, Cyprowski M. Prevalence of bovine leukemia virus (BLV) and bovine adenovirus (BAdV) genomes among air and surface samples in dairy production. J Occup Environ Hyg. 2020;17(6):312–23. 10.1080/15459624.2020.1742914 [DOI] [PubMed] [Google Scholar]
- 199.Aida Y, Murakami H, Takahashi M, Takeshima SN. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front Microbiol. 2013;4:328. 10.3389/fmicb.2013.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Milne RL, Gaudet MM, Spurdle AB, Fasching PA, Couch FJ, Benítez J, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res. 2010;12(6):R110. 10.1186/bcr2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wendt C, Margolin S. Identifying breast cancer susceptibility genes - a review of the genetic background in familial breast cancer. Acta Oncol. 2019;58(2):135–46. 10.1080/0284186X.2018.1529428 [DOI] [PubMed] [Google Scholar]
- 202.Kamolratanakul S, Pitisuttithum P. Human papillomavirus vaccine efficacy and effectiveness against Cancer. Vaccines (Basel). 2021;9(12):1413. 10.3390/vaccines9121413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Archilla G, Gutiérrez G, Camussone C, Calvinho L, Abdala A, Alvarez I, Petersen M, Franco L, Destefano G, Monti G, Jacques JR, Joris T, Willems L, Trono K. A safe and effective vaccine against bovine leukemia virus. Front Immunol. 2022;13:980514. 10.3389/fimmu.2022.980514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Charney J, Holben JA, Cody CM, Moore DH. Further immunization studies with mammary tumor virus. Cancer Res. 1976;36(2 pt 2):777–80. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the relevant data are within the paper.