Abstract
Purpose
Uveal melanoma (UM) is the most common primary intraocular malignancy with a high probability of metastatic disease. Although excellent treatment options for primary UM are available, therapy for metastatic disease remain limited. Drug discovery studies using mouse models have thus far failed to provide therapeutic solutions, highlighting the need for novel models. Here, we optimize zebrafish xenografts as a potential model for drug discovery by showcasing the behavior of multiple cell lines and novel findings on mutation-dependent compound synergism/antagonism using Z-Tada; an algorithm to objectively characterize output measurements.
Methods
Prognostic relevant primary (N = 4) and metastatic UM (N = 1) cell lines or healthy melanocytes (N = 2) were inoculated at three distinct inoculation sites. Standardized quantifications independent of inoculation site were obtained using Z-Tada; an algorithm to measure tumor burden and the number, size, and distance of disseminated tumor cells. Sequentially, we utilized this model to validate combinatorial synergism or antagonism seen in vitro.
Results
Detailed analysis of 691 zebrafish xenografts demonstrated perivitelline space inoculation provided robust data with high probability of cell dissemination. Cell lines with more invasive behavior (SF3B1mut and BAP1mut) behaved most aggressive in this model. Combinatorial drug treatment illustrated synergism or antagonism is mutation-dependent, which were confirmed in vivo. Combinatorial treatment differed per xenograft-model, as it either inhibited overall tumor burden or cell dissemination.
Conclusions
Perivitelline space inoculation provides robust zebrafish xenografts with the ability for high-throughput drug screening and robust data acquisition using Z-Tada. This model demonstrates that drug discovery for uveal melanoma must take mutational subclasses into account, especially in combinatorial treatment discoveries.
Keywords: uveal melanoma (UM), zebrafish, xenografts, drug screening, mutation dependent
Uveal melanoma (UM) is the most common primary intraocular malignancy with a poor prognosis, as 50% of patients develop metastasis within 5 years.1 UM etiology is characterized by driver mutations involved in the MEK-ERK pathway, where activating mutations in guanine nucleotide-binding protein Q (GNAQ), guanine nucleotide-binding protein 11 (GNA11), cysteinyl leukotriene receptor 2 (CYSLTR2), or phospholipase C beta 4 (PLCB4) induce uncontrolled cell proliferation.1 However, UM prognosis prediction relies on secondary mutations in eukaryotic translation initiation factor 1A X-linked (EIF1AX), splicing factor 3b subunit 1 (SF3B1) or BRCA1-associated protein-1 (BAP1). In addition, chromosomal aberrations predict patient outcome too, as loss of chromosome 3 and gain of 8q are associated with poor prognosis.2 Secondary driver mutations are typically mutually exclusive and determine whether patients are at low risk (EIF1AXmut), intermediate risk (SF3B1mut), or high risk (BAP1mut) of metastatic disease.3 Clinically, the most relevant UM to study are those with BAP1 mutations. Despite the availability of several BAP1mut cell lines, most mouse and zebrafish models have been generated with EIF1AXmut cells or UM cell lines with unknown driver or secondary mutations.4
Animal models play an important role in our understanding of cancer biology and drug discovery. Traditionally, mouse models dominate the realm of tumor-xenograft investigations,5,6 but since 2010, there has been a steep increase in the number of published zebrafish xenograft models.7 Zebrafish have several advantages over mice, as they yield a large number of offspring and are relatively cheap to maintain and time efficient. Additionally, these methods are easy to manipulate; only a small number of tumor cells are needed to generate zebrafish xenografts, and they have the potential for high-throughput drug screening. Zebrafish cancer avatars have been studied in multiple cancers, such as glioblastoma, breast cancer, hepatocellular carcinoma, prostate cancer, pediatric cancer, and UM.8–13 Although a promising Bap1-deficient mouse model was recently developed,14 mouse models have thus far failed to improve therapies for UM.4 Therefore, we are particularly interested in BAP1mut-based zebrafish xenograft models. To enhance drug discovery in UM research, we propose that zebrafish xenografts could serve as an initial model before progressing to more costly mammalian studies. Microinjection of UM tumor cells in zebrafish to generate xenografts has been accomplished by multiple groups. Unfortunately, inoculation of UM cells in zebrafish larvae is not consistent between laboratories, as UM cell lines have been inoculated at the yolk sac,13,15–19 perivitelline space,20,21 intraocular space,22 or duct of Cuvier.23,24 Moreover, analyses of zebrafish xenograft larvae are not always consistent. For example, tumor volume can be measured in different ways by multiplying the mean area by the total number of objects19 or evaluating tumor size at 3 days post-injection versus immediately after transplantation.20 In contrast, others use a script in image analysis software to acquire quantifications.25
To improve UM zebrafish xenograft models, we investigated zebrafish xenografts in detail using UM cell lines with representative mutations and chromosomal aberrations found in patients with UM, next to dermal melanocytes and a primary melanocyte cell line. We evaluated seven cell lines at two common inoculation sites and one novel inoculation site (retro-orbital) to identify the most efficient and informative site for obtaining robust data. Additionally, we developed publicly available analysis scripts to standardize quantification methods for both tumor volume and dissemination measurements, enabling the assessment of UM cell phenotypic behavior in vivo and the evaluation of compound efficacy.
Methods
Cell Culture Conditions
Five uveal melanoma cell lines, namely, 92.1 (established at the Leiden University Medical Center, Leiden, The Netherlands26), Mel202 (established at Schepens Eye Research Institute, Boston, MA, USA,27), MP38, MP46, and MM28 (established at Curie Institute, Paris, France28), immortalized dermal melanocytes CRL4059 and neonatal dermal melanocytes GM21808 (both obtained from American Type Culture Collection, Manassas, VA, USA), were used for this study. The chromosomal aberrations and mutation status and research resource identifiers of all the cell lines can be found in Supplementary Table S1. UM cell lines have been authenticated previously by single polymorphism analysis and AmpFLSTR Identifiler Plus PCR Amplification Kit (Thermo Fisher, Bleiswijk, The Netherlands) followed by sequencing. Additionally, we performed single nucleotide polymorphism analysis on CLR4059 and GM21808 with the Infinium Global Screening Array-24 version 3.0 BeadChip according to the manufacturer's protocols and guidelines (Illumina, San Diego, CA, USA) and analysis methods described previously29 (Supplementary Fig. S1). All uveal melanocytes were propagated in RPMI supplemented with 20% heat-inactivated fetal calf serum and 1% penicillin‒streptomycin; MP38 and MM28 media were supplemented with sodium pyruvate. Both dermal melanocyte strains were propagated in Medium 254 supplemented with human melanocyte growth supplements (Thermo Fisher Scientific, The Netherlands). All cell lines were incubated at 37°C in a humidified 5% CO2-enriched atmosphere and regularly checked for mycoplasma.
Drug Screening of Uveal Melanoma Cells
Dabrafenib, ricolinostat, withaferin A and quisinostat (Selleckchem, Berlin, Germany) were stored in DMSO at a 10 mM stock concentration at −80°C. E7107 (gifted by H3 Biomedicine Inc., Cambridge, MA, USA) was diluted in DMSO at a 10 mM stock concentration and stored at −20°C. To determine the IC50 values, a total of 6000 cells (92.1, Mel202, and MP46) were seeded per well in a 96-well plate. The following day, the medium was removed, the cells were washed once with PBS, and, subsequently, 100 µL of medium supplemented with the drug was added at various concentrations (ranging from 50 µM to 1 µM for dabrafenib, ricolinostat, and withaferin A or 500 nM to 1 nM for quisinostat and E7107). Identification of synergistic effects was performed after identifying the IC50 of each compound. A 6 × 6 matrix was used to treat cells at different concentrations based on the IC50 values. After 3 days, the medium was removed, the cells were washed once with PBS, and fixed in ice-cold methanol. After fixation, the cells were stained with 0.1% crystal violet for 30 minutes, washed with water, and air-dried. Finally, crystal violet was solubilized in methanol, and the absorption was measured at 545 nm. Samples were treated in triplicate and analyzed in GraphPad Prism version 9 using nonlinear regression. The data are presented as the means ± SD. Combinatorial treatments were analyzed for synergistic, noninteractive and antagonistic effects using SynergyFinder.30 A synergy score less than −10 was considered antagonistic, a score between −10 and 10 was considered additive, and a score greater than 10 was considered synergistic.
Zebrafish Husbandry and Xenograft Injections
Wild-type AB zebrafish were maintained under standard conditions with a 14-hour light/10-hour dark cycle. In this study, only larval zebrafish (no older than 120 hours post-fertilization) were used. The animal experiments were approved by the Animal Experimentation Committee at Erasmus MC, Rotterdam. Zebrafish embryos were raised in E3 medium supplemented with 0.003% phenylthiourea (PTU) in a Petri dish at 28°C. At 24 hours post-fertilization, the medium was refreshed following the dechlorination of the larvae. At 48 hours post-fertilization, the zebrafish larvae were anesthetized with 0.016% tricaine and used for injections. A total of 2.5 × 106 cells were harvested and stained with 2.5 µM CellTracker CM-Dil dye for 5 minutes at 37°C and subsequently for an additional 15 minutes at 4°C. After staining, CM-Dil dye was removed by centrifugation. The cells were then washed with PBS and resuspended in 2% PVP-40/PBS. A total of approximately 200 to 300 cells were injected into the yolk, perivitelline space, or retro-orbitally. For the retro-orbital injection, every larva was injected into the left eye. Correctly injected larvae were selected 1 hour post-injection under a fluorescence stereomicroscope, placed in E3 medium supplemented with PTU, and raised at 34°C. At 3 days post-injection, xenograft larvae were anesthetized and embedded in 1% low-melting agarose for live-cell imaging.
Confocal Microscopy of Zebrafish Xenografts
Zebrafish xenograft larvae were imaged using a Leica SP5 (Leica Microsystems, Mannheim, Germany) under standard conditions (561 nm, 35% laser power with additional bright field image). Tile scans of three images were generated to obtain full-body length images for analysis. The number of disseminated cells, the distance of dissemination, and the total tumor volume were calculated in FIJI with an in-house script (details can be found in the Results section and Supplementary Methods). The obtained values were then processed in GraphPad Prism version 9. Comparisons of the number of detected spots, distance of dissemination, and tumor volume between the different cell lines were statistically tested using ANOVA with Dunnett's multiple comparison method. The data are presented as the means ± standard deviation (SD).
Drug Screening in Zebrafish Xenografts
For zebrafish toxicity assays, the same range of concentrations used in vitro was used to treat the zebrafish larvae from 2 days post fertilization until 5 days post-fertilization. A total of 8 to 12 larvae per well were sorted in a 12-well plate with E3 medium supplemented with 0.003% phenylthiourea containing DMSO, ricolinostat, withaferin A, quisinostat, or E7107 at different concentrations. The medium was refreshed daily. At 1, 2, and 3 days post-treatment, larvae were inspected and registered; for each drug, this experiment was performed twice using 2 different wild-type AB parental zebrafish. The synergistic/antagonistic concentrations used per cell line were determined based on in vitro data where less than 20% of viable cells remained after single-compound inhibition. Drug treatment started at 3 hours post-injection, and the media containing the compounds were refreshed daily. At 3 days post-injection (dpi), the zebrafish xenografts were imaged and analyzed in the same fashion as described earlier. Zebrafish xenografts were statistically tested by comparing DMSO to drug A, drug B, or the combination of both drugs using Welch's t test. The data are presented as the means ± SD.
Results
Different Phenotypes of Cell Behavior Based on Inoculation Site
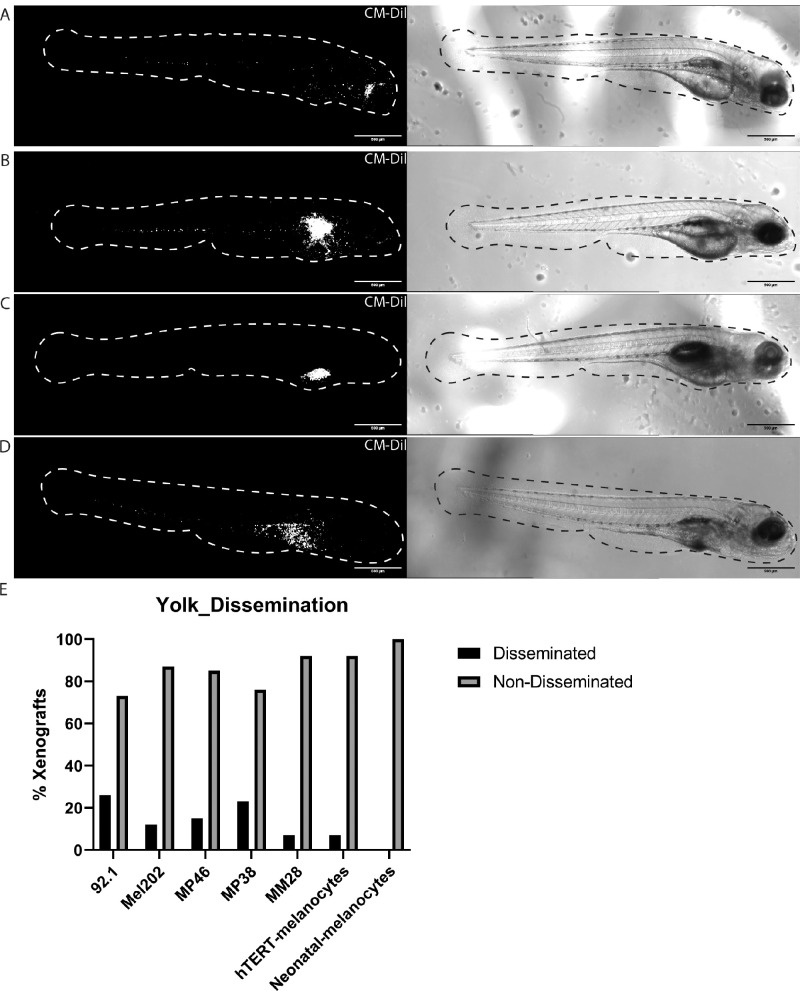
Gaining insights into the optimal inoculation site could differ depending on the research question or behavior of cells in vivo. Therefore, we used one metastatic UM cell line, five primary UM cell lines, and two dermal melanocyte lines to determine the optimal inoculation site. The images shown in Figure 1 depict zebrafish larvae with 92.1 cells inoculated at different sites and serve as a representative example of zebrafish larvae inoculated with other cell lines. Zebrafish larvae were sorted at 1 hour post-injection (1 hpi) for correctly injected cells; for example, larvae that had cells in the brain, intraocular space, or bloodstream were removed. At 3 days post-injection (3 dpi), we assessed cell behavior and found vastly different phenotypes. All cell lines grew and disseminated after retro-orbital (RO) and perivitelline space (PS) injections (see Figs. 1A, 1B). The most commonly used inoculation site, the yolk sac, demonstrated an inconsistent phenotype. The injected cells predominantly stayed within the yolk (see Figs. 1C, 1E), whereas a small number of larvae showed disseminated cells throughout the body (see Figs. 1D, 1E), which phenotypically resembled what was observed in PS injections.
Figure 1.
Different inoculation sites in zebrafish UM xenografts. (A) Zebrafish xenograft injected behind the eye (retro-orbital) with 92.1 cells at 3 dpi. (B) Zebrafish xenograft injected in the perivitelline space with 92.1 cells at 3 dpi. (C) Zebrafish xenograft injected in the yolk sac with 92.1 cells that did not show cell dissemination at 3 dpi. (D) Zebrafish xenograft injected in yolk sac with 92.1 cells showing cell dissemination at 3 dpi. (E) Amount of dissemination present in xenografts injected in the yolk with 92.1 (n = 42), Mel202 (n = 31), MP46 (n = 40), MP38 (n = 34), MM28 (n = 28), hTERT-melanocytes (CRL-4059, n = 26), and neonatal melanocytes (GM21807, n = 35).
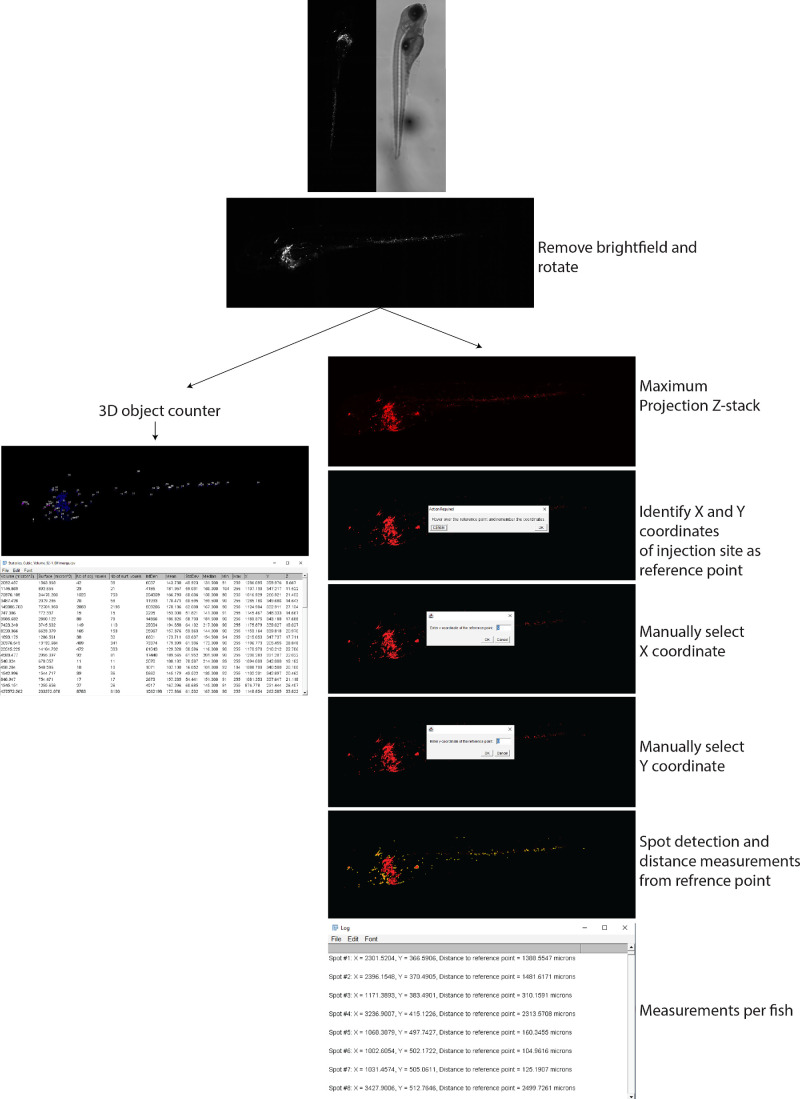
Z-Tada: Zebrafish Xenograft Tumor Volume and Dissemination Analysis
To quantify the tumor volume, number of disseminated cells, and distance of disseminated cells from the inoculation site, we developed Z-Tada, a system in which two scripts are used to analyze the tumor volume, dissemination distance, number of disseminated cells, and size of disseminated cell clusters in a uniform manner independent of the inoculation site (Fig. 2). By making confocal stack images, a simple yet robust method to measure tumor volume is by using the 3D counter plugin in FIJI. The first script is therefore quite simple because it splits the channels, removes the brightfield image, rotates the figure, and runs the 3D counter plugin to obtain the tumor volume in cubic microns (Supplementary Methods, Supplementary Fig. S2). This plugin identified spots and measured them in 3D; this allows 3D measurements of disseminated cells per identified object (in voxels) or total tumor volume by subsequently adding all objects together. The second script quantifies the number of disseminated cells and their migration distance using a single script. This script splits the channels, removes the brightfield image, rotates the figure, and makes a maximum Z-stack projection. The Z-stack projection is then subjected to thresholding and multiplied by a mask to reduce the background and obtain a binary figure. The user is then asked to hover over the image to identify the X and Y coordinates of the injection site, which can subsequently be entered manually in the pop-up screens. Next, disseminated cells were identified with the Find Maxima function and analyzed with the Analyze Particles function (see Supplementary Fig. S2, settings have a maximum particle size limitation to prevent measurement of the inoculation site, which is typically a large spot). For each spot, the X and Y coordinates are obtained and used to calculate a straight-line distance between the detected spot and reference point using the Pythagorean theorem (see Supplementary Methods, Supplementary Fig. S1). For example, a 92.1-xenograft inoculated retro-orbitally (Fig. 3A) or in the perivitelline space (Fig. 3B) illustrates the accuracy of this script independent of the inoculation site using an X-Y plot.
Figure 2.
Visual representation of quantifying zebrafish xenografts. Visual representation of quantifying zebrafish xenografts illustrating data acquirement in a step-by-step fashion.
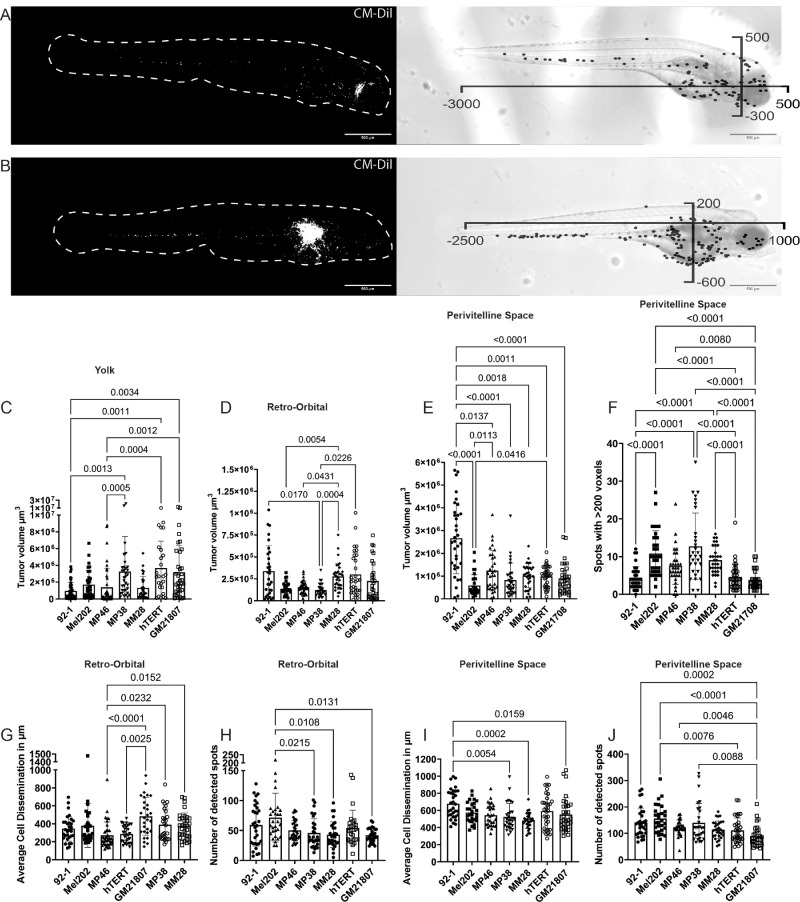
Figure 3.
Quantification of UM zebrafish xenografts. (A) Max projected Z-stack of retro-orbital injected zebrafish xenograft at 3 dpi with cells in white followed by an X-Y plot overlaying the corresponding bright field image. (B) Max projected Z-stack of perivitelline space-injected zebrafish xenograft at 3 dpi with cells in white followed by an X-Y plot overlaying the corresponding bright field image. (C, D, E) Overview of tumor volume analysis per cell line in the yolk sac (92.1, n = 42, Mel202, n = 31, MP46, n = 40, MP38, n = 34, MM28, n = 28, hTERT-melanocytes, n = 26, and GM21807, n = 35), retro-orbital region (92.1, n = 30, Mel202, n = 30, MP46, n = 38, MP38, n = 28, MM28, n = 33, hTERT-melanocytes, n = 32, and GM21807, n = 31), or perivitelline space injections (92.1, n = 35, Mel202, n = 31, MP46, n = 33, MP38, n = 31, MM28, n = 33, hTERT-melanocytes, n = 38, and GM21807, n = 33). (F) Number of detected objects larger than 200 voxels in perivitelline space-inoculated zebrafish xenografts. (G) Cell dissemination distance per cell line in retro-orbital inoculated zebrafish larvae at 3 dpi. (H) Number of disseminated cells detected per cell line in retro-orbital inoculated zebrafish larvae at 3 dpi. (I) Cell dissemination distance per cell line in perivitelline space-inoculated zebrafish larvae at 3 dpi. (J) Number of detected disseminated cells per cell line in perivitelline space-inoculated zebrafish larvae at 3 dpi.
Tumor volume analysis revealed vastly different behaviors based on the inoculation site (Figs. 3C–E). In general, yolk sac inoculation yielded the greatest tumor volume at 3 dpi (see Supplementary Figs. S2A–G), with MP38, hTERT-melanocytes, and neonatal melanocytes demonstrating the greatest tumor volume (see Fig. 3C). Retro-orbital inoculation yielded the lowest tumor volume of all inoculation sites (see Supplementary Figs. S2A–G). MP38 yielded the lowest tumor volume, which was comparable to that of MP46 and Mel202, whereas 92.1, hTERT-melanocytes, and neonatal melanocytes harbored the greatest tumor volumes at 3 dpi (see Fig. 3D). Perivitelline space inoculation resulted in intermediate tumor volumes between inoculation sites but was highly consistent between cell lines, whereas yolk sac inoculation was more dependent on the cell line (see Supplementary Figs. S2A–G). Cell line 92.1 thrived in perivitelline inoculations with the highest tumor volume, whereas Mel202 harbored the lowest tumor volume in perivitelline space injections. All other cell lines yielded similar tumor volumes (see Fig. 3E). Additionally, the size of the detected objects was inspected to investigate whether there were size differences per cell line in perivitelline space-inoculated larvae. An arbitrary threshold of >200 voxels was used to count the number of disseminated cell clusters that could be considered to be more than 1 cell. In the 92.1 cell line, hTERT-melanocyte and neonatal melanocyte-xenografts yielded a low number of disseminated cells >200 voxels, whereas Mel202, MP46, MP38, and MM28 had a greater abundance of large disseminated cell objects (Fig. 3F).
Retro-orbital and perivitelline space inoculation consistently promoted cell dissemination in all the cell lines. The average dissemination distance per larva was similar between cell lines after retro-orbital injection, where MP38, MM28, and neonatal melanocytes tended to disseminate the furthest (Fig. 3G). The cell dissemination distance after perivitelline space inoculation was highly consistent between the cell lines, with only 92.1 indicating a slightly greater distance (Fig. 3I). The number of spots was significantly greater in the perivitelline space injection group than in the retro-orbital inoculation group (Supplementary Fig. S2H). Mel202-xenografts had the greatest number of spots in both models, whereas 92.1 had a slight increase in the perivitelline space model compared to the other cell lines used (Figs. 3H, 3J). Due to the consistent tumor volume, high number of disseminated cells, differences in disseminated cell sizes, and largest dissemination distance in perivitelline space inoculations, we argue that this site is the most useful site for use in UM zebrafish–larvae xenograft systems.
Drug Synergism Differs Among UM Subtypes
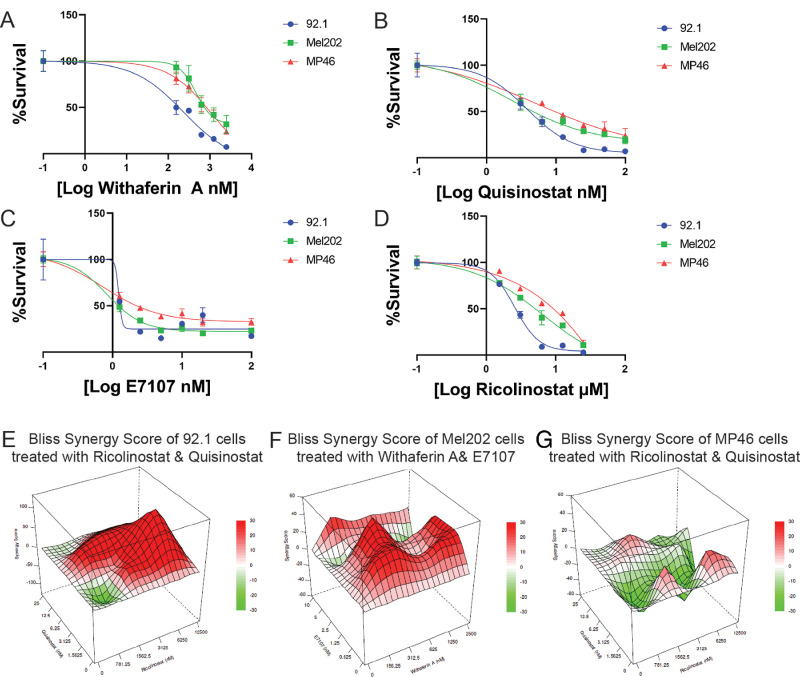
To evaluate the robustness and abilities of zebrafish xenografts as a drug discovery model, we screened compounds previously tested in mouse or zebrafish xenografts. A total of 4 compounds were evaluated: quisinostat, ricolinostat, withaferin A (tested in mice31), and E7107 (tested ex vivo32). Quisinostat and ricolinostat are both histone acetyl deaminase inhibitors (HDACis), which were previously shown to be effective in MM28- and MP46-xenografts33 (quisinostat) and OMM2.5-xenografts21 (ricolinostat). Withaferin A is an MAPK/PI3K-AKT inhibitor which was able to effectively inhibit Mel290-, 92.1-, and OMM2.3-xenografts showcasing its universal application independent of molecular subclasses.31 E7107 is a spliceosome inhibitor,34 which has recently been studied using ex vivo UM tissue. Both SF3B1wt and SF3B1mut demonstrated decreased cell viability, however SF3B1mut cells are more sensitive to E7107.32 Unlike in previous studies, in this study, compound screening was performed on the EIF1AXmut, SF3B1mut, and BAP1neg UM cell lines. All the compounds inhibited UM cell lines in vitro (Figs. 4A–D). As UM typically shares activation of the MEK-ERK pathway due to driver mutations in GNAQ, GNA11, PLCB4, or CYSLTR21 but differs in prognosis based on secondary mutations,3 we hypothesize that combinatorial inhibition could improve the inhibitory response dependent on UM subtype. To evaluate this hypothesis, we combined each compound with each other to identify synergistic or antagonistic effects. Surprisingly, we detected synergistic effects of the histone deacetylase inhibitors quisinostat and ricolinostat on EIF1AXmut cells, whereas this combination had antagonistic effects on BAP1mut cells (see the Table, Figs. 4E, 4G). On the other hand, we found strong synergistic effects of the combination of the MAPK/PI3K-AKT inhibitor withaferin A and the spliceosome inhibitor E7107 in Mel202 cells (see the Table, Fig. 4F). Despite all the compounds being able to inhibit UM cells regardless of secondary mutation status (see Figs. 4A–C), synergism during combinatorial inhibition highly differed among UM subtypes (see the Table).
Figure 4.
Dose‒response curve and synergy plots for UM cells. (A) Dose‒response curve of withaferin A. (B) Dose response curve of quisinostat. (C) Dose‒response curve of E7107. (D) Dose‒response curve of ricolinostat. Synergy scores are plotted in a 3D graph, with synergy scores on the Z-axis, the concentration of drug A on the Y-axis and the concentration of drug B on the X-axis. (E) Bliss synergy score plot of ricolinostat and quisinostat in 92.1 cells. (F) Bliss synergy score plot of Mel202 cells treated with withaferin A and E7107. (G) Bliss synergy score plot of ricolinostat and quisinostat in MP46 cells.
Table.
Synergy Scores Based on SynergyFinder
| Drug A | Drug B | ZIP | Loewe | H SA | Bliss |
|---|---|---|---|---|---|
| 92.1 (EIF1AXmut) | |||||
| E7107 | Quisinostat | −5.03 | −0.41 | 2.89 | −5.43 |
| E7107 | Ricolinostat | −7.42 | −1.29 | 2.68 | −7.45 |
| Ricolinostat | Quisinostat | 21.96 | −3.23 | 7.09 | 24.01 |
| Withaferin A | E7107 | 2.03 | 3.01 | 7.91 | 1.97 |
| Withaferin A | Quisinostat | 1.47 | −2.33 | 5.25 | 2.21 |
| Withaferin A | Ricolinostat | 6.38 | 3.28 | 12.4 | 6.92 |
| Mel202 (SF3B1mut) | |||||
| E7107 | Quisinostat | −4.71 | 9.02 | 11 | −4.35 |
| E7107 | Ricolinostat | 1.34 | −4.15 | −5.21 | −3.46 |
| Ricolinostat | Quisinostat | −1.6 | −2.43 | 0.4 | −2.5 |
| Withaferin A | E7107 | 13.39 | 11.95 | 15.05 | 16.26 |
| Withaferin A | Quisinostat | −8.33 | 4.05 | 3.12 | −8.97 |
| Withaferin A | Ricolinostat | 4.76 | 17.04 | 18.36 | 4.43 |
| MP46 (BAP1mut) | |||||
| E7107 | Quisinostat | −1.33 | 5.39 | 7.57 | −2.38 |
| E7107 | Ricolinostat | 6.69 | −8.35 | −9 | 3.5 |
| Ricolinostat | Quisinostat | −10.73 | 4.38 | 3.02 | −11.65 |
| Withaferin A | E7107 | −1.52 | −3.85 | 2.67 | −2.4 |
| Withaferin A | Quisinostat | −10 | 2.74 | 0.02 | −8.23 |
| Withaferin A | Ricolinostat | 9.58 | −0.01 | −3.85 | 2.98 |
A total of four tools are used in SynergFinder: ZIP, Loewe, has, and Bliss. For each combination, the score is given and divided among 92.1, Mel202, and MP46. Synergism or antagonism is highlighted in bold.
In Vivo Evaluation of Synergistic Treatment Therapies
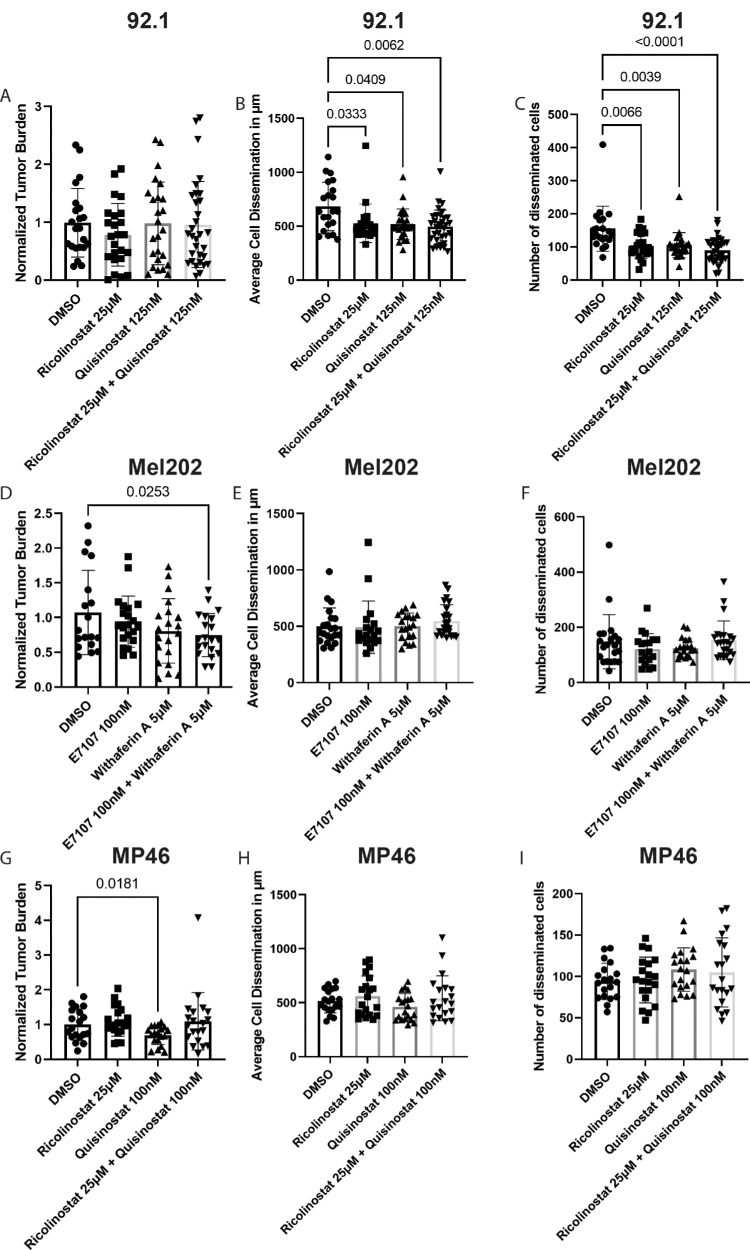
To evaluate the synergistic effect of compounds, we first investigated the toxicity tolerance of single compounds in zebrafish larvae (Supplementary Fig. S3). None of the compounds exhibited toxicity in wild-type zebrafish larvae, even at the highest concentration tested in vitro. Therefore, we were able to utilize compounds at concentrations at which the in vitro survival of UM cells was <20% without toxic effects in zebrafish xenografts. Interestingly, compared with DMSO, ricolinostat, quisinostat, or combined treatment of 92.1-xenografts did not decrease the overall tumor burden (Fig. 5A). However, all the compounds inhibited the migration and number of disseminated cells, and dual treatment had the strongest inhibitory effect (Figs. 5B, 5C). Treatment of Mel202-xenografts with E7107, withaferin A, or their combination significantly inhibited the overall tumor burden in dual-treated xenografts (Fig. 5D), supporting our synergistic in vitro findings. However, these compounds were unable to inhibit cell dissemination (Figs. 5E, 5F). Finally, the overall tumor burden of MP46-xenografts was lower in quisinostat-treated xenografts than in ricolinostat-treated or DMSO-treated xenografts. Notably, combined treatment prevented the inhibitory effect of quisinostat, as these xenografts harbored similar tumor burdens as those of the DMSO controls (Fig. 5G), supporting our antagonistic in vitro findings. Additionally, neither quisinostat, ricolinostat, nor the combination treatment reduced cell dissemination in MP46-xenografts (Figs. 5H, 5I).
Figure 5.
In vivo drug screening of zebrafish xenografts. (A) Normalized tumor burden of 92.1-xenografts after 3 days of treatment with DMSO (n = 21), ricolinostat (n = 22), quisinostat (n = 25), or their combination (n = 33). (B) Average cell dissemination distance of treated 92.1-xenografts. (C) Number of disseminated cells in treated 92.1-xenografts. (D) Normalized tumor burden of Mel202-xenografts after 3 days of treatment with DMSO (n = 21), E7107 (n = 19), withaferin A (n = 20), or the combination of both (n = 23). (E) Average cell dissemination distance of treated Mel202-xenografts. (F) Number of disseminated cells in treated Mel202-xenografts. (G) Normalized tumor burden of MP46-xenografts after 3 days of treatment with DMSO (n = 19), ricolinostat (n = 20), quisinostat (n = 20), or their combination (n = 20). (H) Average cell dissemination distance of treated MP46-xenografts. (I) Number of disseminated MP46-xenograft-bearing cells.
Discussion
UM remains a devastating disease with a high propensity for metastasis and limited therapeutic options. To discover novel therapeutics, we optimized our xenograft zebrafish model, developed a robust algorithm to consistently determine model output measures, and carried out a detailed analysis of zebrafish xenografts using multiple cell lines, which, to our knowledge, has not been previously investigated in this model.4 Previous UM-zebrafish xenografts typically utilized cell lines without known secondary mutations (OMM1,13 OMM2.3,13,16,20,21 Mel270,13 and Mel28520) or patient-derived spheroids (spUM-LB00824 and spXmm6623). The main differences between previously described models and the model described in this paper are the method of quantification, cell line models, and evaluated compounds. Numerous UM-zebrafish xenografts have been generated by the Snaar Laboratory, which uses an automated quantification platform based on body axis that provided cell dissemination distance and overall tumor burden.35 The method presented here provides an ImageJ script based quantification that works in a similar fashion, but allows the user to determine the inoculation site to be a broader applicable method while also providing the number of disseminated cells next to cell dissemination distance and overall tumor burden. Recent models from the Snaar Laboratory have utilized patient-derived spheroids xenografts and have shown the applicability of this model for patient-specific drug screening.23 However, there is no genetic characterization of these new cellular models available for public use, which are available for established cell lines. Here, we describe the novel cellular behavior of established cell lines with known secondary mutations in all three clinically relevant molecular subclasses (EIF1AX, SF3B1, and BAP1) in both primary and metastasis-derived cell lines with single and combinatorial drug screening. Notably, all established cell lines were derived from patients who eventually developed metastatic disease, including the 92.1 cell line, which harbors an EIF1AX mutation.
Due to the variety of inoculation sites in zebrafish xenografts, we first evaluated which inoculation site would provide robust data that would allow investigation of the cellular behavior and effectiveness of therapeutic compounds. By generating 691 xenografts with primary and metastatic UM cell lines via retro-orbital, perivitelline space, or yolk-sac injections, this study revealed that the perivitelline space is the most robust site for UM zebrafish xenografts. The quantification of zebrafish xenografts has varied across laboratories, for which we have developed Z-Tada. This provides a method for standardized analysis of the tumor volume and cell dissemination of zebrafish xenograft larvae, which accurately detects tumor cells and provides migration distances. Although yolk sac inoculation typically yielded the highest tumor burden (see Supplementary Fig. S2), this site lacked robust cell dissemination (see Fig. 1). Retro-orbital inoculation provided robust cell dissemination, yet the number of disseminated cells and overall tumor burden were significantly lower than perivitelline space inoculation (see Supplementary Fig. S2). Interestingly, cell lines behave differently depending on the molecular subclass. The cell line 92.1 (primary EIF1AXmut) typically yielded a high tumor burden, but the size of the disseminated cell clusters was relatively small, whereas MP38 (primary BAP1mut) and MM28 (metastatic BAP1mut) developed more disseminated cell clusters with a large size (see Fig. 3). Although this model provides robust data on tumor volume and cell dissemination, healthy and hTERT-immortalized melanocytes were also able to proliferate and disseminate, suggesting that this model is highly prone to cell dissemination regardless of cell type. Nonetheless, molecular subclasses with a greater probability of developing metastatic disease (SF3B1mut and BAP1mut) behave more aggressively in this model. Although we utilized a total of 5 UM cell lines in a large number of xenografts, this study investigated only 1 metastatic BAP1mut cell line; which can limit translatability and reproducibility. Increasing the number of metastatic BAP1mut cell lines could provide a more robust and translatable view of this platform. However, reliable BAP1mut cell lines are scarce and notoriously hard to establish, resulting in a low number of available cellular models. Development of more BAP1mut cell lines is necessary in UM research in order to increase reproducibility of cellular models.
To evaluate whether this model system can reliably be used for novel drug screening for UM, we investigated previously used compounds that were shown to effectively inhibit tumor growth or cell dissemination in UM cells. A potential therapeutic option for high-risk UM is HDACis, which are able to differentiate UM into a more melanocyte-like state.36 In mice, the HDACis inhibitor quisinostat (targeting HDAC3, 5, 8, and 9) was shown to selectively inhibit BAP1mut xenografts (MP46 and MM28).33 However, in zebrafish xenografts, quisinostat was also found to effectively inhibit the dissemination of metastatic cell lines (OMM2.3) or in combination with flavopiridol against spheroids (spXmm66) that do not harbor BAP1 mutations.13,23 In this study, we confirmed inhibition of overall tumor burden in quisinostat-treated BAP1mut zebrafish xenografts (MP46). However, quisinostat was also able to inhibit the cell dissemination of 92.1 cells (EIF1AXmut) but failed to reduce the overall tumor burden (see Fig. 5); suggesting quisinostat effectivity is dependent on molecular subclass. Another HDAC inhibitor, ricolinostat (targeting HDAC6), was previously shown to inhibit tumor burden in zebrafish xenografts using the metastatic UM cell line OMM2.3.21 Here, we showed that ricolinostat was also able to inhibit other types of UM cells in vitro (see Fig. 4) but was effective only in 92.1-based xenografts. Ricolinostat was not effective against BAP1mut-cells (MP46) in vivo. Interestingly, combining HDACis was shown to be either synergistic (92.1) or antagonistic (MP46) based on the molecular subclass (Fig. 4, see the Table). Our zebrafish xenografts demonstrated that the synergistic effects of quisinostat and ricolinostat on 92.1 cells strongly inhibited cell dissemination but did not affect overall tumor burden. However, its antagonistic effect seen in vitro was also reproduced in MP46-xenografts, as the inhibitory effect of quisinostat was lost in the combination-treated xenografts (see Fig. 5). Targeting HDACs is a promising therapy for many cancers and has been combined with several compounds that target other pathways37; however, combining multiple HDACs is not a strategy that has been studied in detail before. Our study suggests combining multiple HDACis can be mutation-dependent; and should be investigated preclinically in detail to validate efficacy. In addition to HDACis, we evaluated withaferin A, a compound tested only in mice. Withaferin A is able to inhibit the MET and MEK1/2 pathways, making this an interesting compound for UM because it acts on the primary driver pathways.31 In vitro, withaferin A inhibited the growth of all the tested cell lines. However, withaferin A in combination with the spliceosome inhibitor E7107 had synergistic effects on Mel202 cells, yet failed to illustrate synergistic effects on other cell lines tested (Fig. 4, see the Table). The synergistic effect was reproduced in Mel202-xenografts, suggesting that this combination could provide novel therapeutic options for patients with SF3B1mut UM (see Fig. 5).
Zebrafish xenografts hold potential as a drug screening platform for UM, where synergism and antagonism can be studied in detail. This study only treated xenografts for a total of 3 days, which could be too short to see a more defined inhibitory effect. However, this methodology has the advantage that utilized unique aspects of zebrafish larvae. Zebrafish larvae develop rapidly, have a heart beat at 24 hours post-fertilization, and all major organs are functional at 5 days post-fertilization.38 In combination with their small size (approximately 4 millimeter at 5 days post-fertilization), this method allows for high-throughput screening as single zebrafish larvae can grow in a 96-wells plate. Other important elements in this platform that could affect drug efficiency are the number of fish treated per well and the efficiency of compound uptake by zebrafish larvae. However, an important advantage of this model is functional liver metabolism in zebrafish larvae,39 which can metabolize compounds and therefore alter their effects, which is lacking in in vitro systems. Additionally, zebrafish larvae contain a functional innate immune system at this point of development; yet lack adaptive immune cells.40 Therefore, this system is more complex than traditional xenograft models that lack an immune system completely; but not suitable for adaptive therapeutics. Nonetheless, this platform illustrates that molecular subclasses of UM are important parameters for discovering novel therapeutic compounds as a first-tier screening model. More defined mechanisms should therefore be studied in more complex models, such as adult xenograft models or endogenous animal models.
In summary, this study illustrates that optimal inoculation of UM cells in the perivitelline space allows for robust tumor burden and cell dissemination analysis in a short time span of 3 days. Using Z-Tada, this model is able to identify differences in cellular behavior depending on genetic background in a standardized fashion. Furthermore, this methodology allows standardized read-out parameters and can identify synergistic or antagonistic effects. Ultimately, the large number of offspring and rapid development of major organs in zebrafish larvae allows for high-throughput drug discovery with standardized read-out using Z-Tada. Additionally, injection of fresh patient material could be used in this platform to generate a high-throughput personalized drug-discovery platform. Using this model, we provide evidence that clinically prognostic subclasses of UM are key to developing effective therapies. Synergistic compound screenings for UM should take this into account, as the same combination can be synergistic in low-risk UM but antagonistic in high-risk UM.
Supplementary Material
Acknowledgments
The authors thank Thomas Huizer, BSc, and Nikki Könemann, BSc, for their contributions to the data acquisition and technical support.
Supported by “Rotterdamse Stichting Blindenbelangen” and “Stichting voor Ooglijders.”
Author Contributions: Q.B. and E.B. contributed to the conceptualization, experimental design, and interpretation of the data. Q.B. performed the data acquisition, script development, and initial analysis. Q.B. drafted the manuscript, followed by reviewing and editing by Q.B., E.B., and E.K. Funding was acquired by E.B. and E.K. Supervision was carried out by E.B. and E.K.
Ethics Approval and Consent to Participate: In this study, only larval zebrafish (no older than 120 hours post-fertilization) were used. The animal experiments were approved by the Animal Experimentation Committee at Erasmus MC, Rotterdam, The Netherlands.
Disclosure: Q.C.C. van den Bosch, None; E. Kiliç, None; E. Brosens, None
References
- 1. Jager MJ, Shields CL, Cebulla CM, et al.. Uveal melanoma. Nat Rev Dis Primers. 2020; 6: 24. [DOI] [PubMed] [Google Scholar]
- 2. van den Bosch T, van Beek JGM, Vaarwater J, et al.. Higher percentage of FISH-determined monosomy 3 and 8q amplification in uveal melanoma cells relate to poor patient prognosis. Invest Ophthalmol Vis Sci. 2012; 53: 2668–2674. [DOI] [PubMed] [Google Scholar]
- 3. Yavuzyigitoglu S, Koopmans AE, Verdijk RM, et al.. Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology. 2016; 123: 1118–1128. [DOI] [PubMed] [Google Scholar]
- 4. van den Bosch QCC, de Klein A, Verdijk RM, Kiliç E, Brosens E.. Uveal melanoma modeling in mice and zebrafish. Biochim Biophys Acta Rev Cancer. 2024; 1879: 189055. [DOI] [PubMed] [Google Scholar]
- 5. Nadeau JH, Auwerx J.. The virtuous cycle of human genetics and mouse models in drug discovery. Nat Rev Drug Discov. 2019; 18: 255–272. [DOI] [PubMed] [Google Scholar]
- 6. Ireson CR, Alavijeh MS, Palmer AM, Fowler ER, Jones HJ.. The role of mouse tumour models in the discovery and development of anticancer drugs. Br J Cancer. 2019; 121: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Groenewoud A, Forn-Cuní G, Engel FB, Snaar-Jagalska BE. XePhIR: the zebrafish xenograft phenotype interactive repository. Database (Oxford). 2022; 2022: baac028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pliakopanou A, Antonopoulos I, Darzenta N, et al.. Glioblastoma research on zebrafish xenograft models: a systematic review. Clin Transl Oncol. 2024; 26: 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wawruszak A, Okoń E, Dudziak K.. Advancements in zebrafish models for breast cancer research: unveiling biomarkers, targeted therapies, and personalized medicine. Med Sci Monit. 2023; 29: e940550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Targen S, Konu O.. Zebrafish xenotransplantation models for studying gene function and drug treatment in hepatocellular carcinoma. J Gastrointest Cancer. 2021; 52: 1248–1265. [DOI] [PubMed] [Google Scholar]
- 11. Amawi H, Aljabali AAA, Boddu SHS, et al.. The use of zebrafish model in prostate cancer therapeutic development and discovery. Cancer Chemother Pharmacol. 2021; 87: 311–325. [DOI] [PubMed] [Google Scholar]
- 12. Casey MJ, Stewart RA.. Pediatric cancer models in zebrafish. Trends Cancer. 2020; 6: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der Ent W, Burrello C, Teunisse AFAS, et al.. Modeling of human uveal melanoma in zebrafish xenograft embryos. Invest Ophthalmol Vis Sci. 2014; 55: 6612–6622. [DOI] [PubMed] [Google Scholar]
- 14. Roula F, Fagun J, Anne Nathalie L, Amy L, Catherine DVR. BAP1 deficient human and mouse uveal melanomas up-regulate a shared EMT pathway. bioRxiv 2023.05.24.542173, 10.1101/2023.05.24.542173. [DOI]
- 15. Fornabaio G, Barnhill RL, Lugassy C, et al.. Angiotropism and extravascular migratory metastasis in cutaneous and uveal melanoma progression in a zebrafish model. Sci Rep. 2018; 8: 10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Ent W, Burrello C, de Lange MJ, et al.. Embryonic zebrafish: different phenotypes after injection of human uveal melanoma cells. Ocul Oncol Pathol. 2015; 1: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding Y, Yu J, Chen X, et al.. Dose-dependent carbon-dot-induced ROS promote uveal melanoma cell tumorigenicity via activation of mTOR signaling and glutamine metabolism. Adv Sci (Weinh). 2021; 8: 2002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu L, Zhou D, Zhang G, et al.. Co-occurrence of BAP1 and SF3B1 mutations in uveal melanoma induces cellular senescence. Mol Oncol. 2022; 16: 607–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Yang J, Zhang Q, et al.. Copper ionophore elesclomol selectively targets GNAQ/11-mutant uveal melanoma. Oncogene. 2022; 41: 3539–3553. [DOI] [PubMed] [Google Scholar]
- 20. Slater K, Heeran AB, Garcia-Mulero S, et al.. High cysteinyl leukotriene receptor 1 expression correlates with poor survival of uveal melanoma patients and cognate antagonist drugs modulate the growth, cancer secretome, and metabolism of uveal melanoma cells. Cancers (Basel). 2020; 12: 2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sundaramurthi H, García-Mulero S, Tonelotto V, et al.. Uveal melanoma cell line proliferation is inhibited by ricolinostat, a histone deacetylase inhibitor. Cancers (Basel). 2022; 14: 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tobia C, Coltrini D, Ronca R, et al.. An orthotopic model of uveal melanoma in zebrafish embryo: a novel platform for drug evaluation. Biomedicines. 2021; 9: 1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Groenewoud A, Yin J, Gelmi MC, et al.. Patient-derived zebrafish xenografts of uveal melanoma reveal ferroptosis as a drug target. Cell Death Discov. 2023; 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin J, Zhao G, Kalirai H, et al.. Zebrafish patient-derived xenograft model as a preclinical platform for uveal melanoma drug discovery. Pharmaceuticals (Basel). 2023; 16: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Groenewoud A, Yin J, Snaar-Jagalska BE. Ortho- and ectopic zebrafish xeno-engraftment of ocular melanoma to recapitulate primary tumor and experimental metastasis development. J Vis Exp. 2021; 175: e62356. [DOI] [PubMed] [Google Scholar]
- 26. De Waard-Siebinga I, Blom DJ, Griffioen M, et al.. Establishment and characterization of an uveal-melanoma cell line. Int J Cancer. 1995; 62: 155–161. [DOI] [PubMed] [Google Scholar]
- 27. Verbik DJ, Murray TG, Tran JM, Ksander BR.. Melanomas that develop within the eye inhibit lymphocyte proliferation. Int J Cancer. 1997; 73: 470–478. [DOI] [PubMed] [Google Scholar]
- 28. Amirouchene-Angelozzi N, Nemati F, Gentien D, et al.. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol Oncol. 2014; 8: 1508–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuil LE, MacKenzie KC, Tang CS, et al.. Size matters: large copy number losses in Hirschsprung disease patients reveal genes involved in enteric nervous system development. PLoS Genet. 2021; 17: e1009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng S, Wang W, Aldahdooh J, et al.. SynergyFinder Plus: toward better interpretation and annotation of drug combination screening datasets. Genomics Proteomics Bioinformatics. 2022; 20: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samadi AK, Cohen SM, Mukerji R, et al.. Natural withanolide withaferin A induces apoptosis in uveal melanoma cells by suppression of Akt and c-MET activation. Tumour Biol. 2012; 33: 1179–1189. [DOI] [PubMed] [Google Scholar]
- 32. Nguyen JQN, Drabarek W, Leeflang AMCHJ, et al.. Spliceosome inhibition in SF3B1-mutated uveal melanoma. medRxiv 2022; 2022.2009.2020.22280164. Available at: https://www.medrxiv.org/content/10.1101/2022.09.20.22280164v1. [Google Scholar]
- 33. Kuznetsoff JN, Owens DA, Lopez A, et al.. Dual screen for efficacy and toxicity identifies HDAC inhibitor with distinctive activity spectrum for BAP1-mutant uveal melanoma. Mol Cancer Res. 2021; 19: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Folco EG, Coil KE, Reed R.. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011; 25: 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghotra VPS, He S, de Bont H, et al.. Automated whole animal bio-imaging assay for human cancer dissemination. PLoS One. 2012; 7: e31281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Landreville S, Agapova OA, Matatall KA, et al.. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res. 2012; 18: 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang T, Wang F, Elhassan RM, et al.. Targeting histone deacetylases for cancer therapy: trends and challenges. Acta Pharm Sin B. 2023; 13: 2425–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Veldman MB, Lin S.. Zebrafish as a developmental model organism for pediatric research. Pediatric Research. 2008; 64: 470–476. [DOI] [PubMed] [Google Scholar]
- 39. Goessling W, Sadler KC.. Zebrafish: an important tool for liver disease research. Gastroenterology. 2015; 149: 1361–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM.. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol. 2004; 28: 9–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.