Abstract
Purpose
To determine endothelial cell density (ECD) from real-world donor cornea endothelial cell (EC) images using a self-supervised deep learning segmentation model.
Methods
Two eye banks (Eversight, VisionGift) provided 15,138 single, unique EC images from 8169 donors along with their demographics, tissue characteristics, and ECD. This dataset was utilized for self-supervised training and deep learning inference. The Cornea Image Analysis Reading Center (CIARC) provided a second dataset of 174 donor EC images based on image and tissue quality. These images were used to train a supervised deep learning cell border segmentation model. Evaluation between manual and automated determination of ECD was restricted to the 1939 test EC images with at least 100 cells counted by both methods.
Results
The ECD measurements from both methods were in excellent agreement with rc of 0.77 (95% confidence interval [CI], 0.75–0.79; P < 0.001) and bias of 123 cells/mm2 (95% CI, 114–131; P < 0.001); 81% of the automated ECD values were within 10% of the manual ECD values. When the analysis was further restricted to the cropped image, the rc was 0.88 (95% CI, 0.87–0.89; P < 0.001), bias was 46 cells/mm2 (95% CI, 39–53; P < 0.001), and 93% of the automated ECD values were within 10% of the manual ECD values.
Conclusions
Deep learning analysis provides accurate ECDs of donor images, potentially reducing analysis time and training requirements.
Translational Relevance
The approach of this study, a robust methodology for automatically evaluating donor cornea EC images, could expand the quantitative determination of endothelial health beyond ECD.
Keywords: eye banking, deep learning, endothelial cells
Introduction
The determination of the central endothelial cell density (ECD) of the donor corneal endothelium has been a certification requirement for donor tissue suitability of the Eye Bank Association of America (EBAA) since 2001.1 However, the training and operating procedures for specular microscopic image capture, quality, and analysis for ECD determination have been left up to the individual eye bank medical directors. The impact of this variability in specular microscopic procedures on ECD measurement accuracy has been well demonstrated with comparisons to an image analysis reading center for standardized image analysis procedures.2,3
Nevertheless, eye-bank–determined ECD accuracy when compared to an image analysis reading center has significantly improved with training and a similar image analysis method.4 Tissue warming implementation prior to imaging has improved image quality,5,6 and wider field eye bank specular microscopy (Konan CellChek D; Konan Medical, Irvine, CA)7 has enabled the the analysis of more endothelial cells (ECs) from multiple areas within the image field. These improvements, however, may come at the expense of longer image analysis time and greater technician training. The EBAA has also recently established a standardized specular microscopic image capture technique to maximize image quality.8 Whether the central ECD is determined by the eye bank or an image analysis reading center, the number of ECs analyzed remains limited to usually no more than 100 to 150 cells using a manual method of counting, principally the center method.7 The accuracy of this approach can be influenced by cell selection, poor focusing, deficient image quality, improper application of the center method, Desçemet folds, and confounding image opacities (e.g., blood, debris, guttae).7 Furthermore, Huang et al.9 reported that the Konan CellChek fully automated analysis overestimated ECDs in central EC images with polymegethism and glaucomatous eyes.
Currently, a fully automated, commercially available donor cornea endothelial image analysis system is not on the market. Given the challenges of technician training, turnover, and work demands; image quality issues; and image analysis errors, an efficient, accurate, and automated analysis of donor EC images would be ideal. With the plentiful quantity of donor specular microscopic EC images routinely obtained as part of the standard operating procedure for eye banks, this is a promising opportunity for machine learning modeling.
Our group recently reported the application of deep learning segmentation to post-keratoplasty images and identified machine learning classifiers predicting rejections 1 to 24 months prior to a clinically apparent rejection.10 With lessons learned from our in vivo clinical machine learning image analysis work, we have directed efforts toward the more challenging machine learning image analysis of donor ex vivo endothelial images with higher ECDs with smaller cells, lower magnification, and optical interference from the plastic viewing storage chamber, confounded by other imaging issues outlined above. Our objective with this study was to develop a deep learning model that can accurately, rapidly, and objectively determine the ECD of the donor cornea endothelium to facilitate a more efficient determination of this key element in tissue suitability.
Methods
Unlabeled Data Cohort
Eversight (Ann Arbor, MI) and VisionGift (Portland, OR) eye banks provided central donor corneal EC specular microscopic images obtained at screenings between 2010 and 2023. A subset of only those donors that went on to keratoplasty with a valid manual ECD measurement or who were deemed keratoplasty ineligible based on slit-lamp findings were included. Utilizing the Konan CellChek D specular microscope, Eversight provided one analyzed image each with a single selected cell area from 13,942 donor corneas, and VisionGift provided one analyzed image with up to three selected cell areas from 1196 donor corneas.
Both eye banks participated during this period in the National Eye Institute–supported Specular Microscopy Ancillary Study (SMAS)3,11 and Cornea Preservation Time Study (CPTS),12 as well as the Diabetes Endothelial Keratoplasty Study (DEKS)13 involving defined donor specular microscopy procedures. Technician training at both eye banks has been previously described.4 Donor data obtained included donor age and sex, cause of death, diabetes status, lens type, death to preservation time, death to imaging time, tissue suitability evaluations, and the keratoplasty type the donor was used for. Both eye banks analyzed a minimum of 40 ECs per image and ideally more than 100 ECs, and they provided their final ECDs to our biostatistician (RCO) for inclusion in the statistical analyses.
Labeled Data Cohort
A second subset of 174 images (81 Eversight, 93 VisionGift) selected from the same dataset reported by Huang et al.4 was utilized for supervised training of the deep learning algorithm. The selection criterion for this subset was analyzability as determined by an expert reader analyst (BMB). These images underwent automatic cell border segmentation as previously described.14 Following postprocessing, the predictions were manually edited using an in-house centroid-selection algorithm.15 The centroid-selection algorithm started with identifying cellular connected components and their centroid locations. These were visualized by plotting the centroids as red circles and overlaying the binarized segmentation prediction in green on the original EC image displayed in the software viewing window. From here, the image analyst clicked on visible centroids to exclude them from the final segmentation if the corresponding cell borders were incorrectly annotated. The final ground-truth labels were binary segmentations with single-pixel-width white borders (pixel value 1) and black cells and background (pixel value 0). To enhance the deep learning training performance, the borders were dilated with a 3 × 3 square kernel for one iteration.
Deep Learning Networks
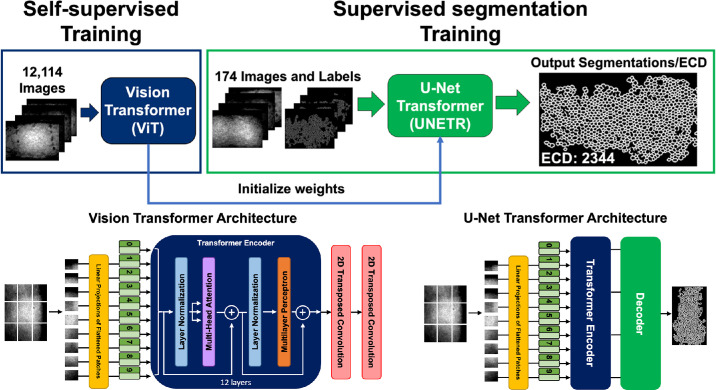
In this study, training was conducted in two stages and employed two network architectures (Fig. 1). A self-supervised learning technique was implemented in the first stage using the Vision Transformer (ViT) architecture.16 The second stage involved using supervised learning to train a U-Net Transformer (UNETR) model.17 Self-supervised learning enables the model to learn important imaging features by masking areas of the training images and evaluating the attempt of the model to recreate these masked areas via comparison with the ground-truth input images. Supervised learning requires labeled datasets to train deep learning algorithms to perform a desired task—in this study, cell-border segmentation. The benefit of combining these deep learning techniques is that the model can achieve high segmentation accuracy performance despite a limited labeled dataset by utilizing thousands of unlabeled images to learn pre-existing features.
Figure 1.
Training workflow for deep learning–based endothelial cell segmentation. Self-supervised training was performed using 12,114 images through a ViT to learn feature representations without labeled data. Learned weights were then used to initialize a UNETR for supervised cell border segmentation training with 174 images and labels. Final output after postprocessing was a single-pixel-width cell border segmentation from which ECD was automatically calculated. The illustrations of the ViT and UNETR are based on work by Dosovitskiy et al.16 and Hatamizadeh et al.,17 respectively.
The ViT and UNETR architectures first deconstructed an image into a sequence of patches imitating natural language processing (NLP) transformer models that interpret sentences as a sequence of words. Each patch of the input image was 16 × 16 pixels. The ViT is composed of a transformer encoder with two convolutional transpose layers to output a reconstructed image of the same size as the input image.16 Because the ViT architecture is the backbone of the UNETR, the weights learned during the ViT training initialized the UNETR encoder weights. The UNETR decoder weights remained randomly initialized, refined thereafter during supervised learning.
Data augmentations, network parameters, and training parameters (i.e., loss functions, validation metrics, and learning rates) described by Tang et al.18 were utilized and are described briefly. Prior to ViT self-supervised training, input images were scaled to [0, 1] and randomly cropped into 10 samples of 128 × 128 pixels before an additional copy of the cropped image was generated. From here, random patches no larger than 64 × 64 pixels were either dropped out or mutated from both image copies independently. The ViT was trained with two loss functions: contrastive loss19 and mean absolute error. The contrastive loss maximized agreement between the reconstructed image patches of the original input and the reconstructed image patches of the copied input. The mean absolute error loss was used to minimize the absolute pixel value difference between the reconstructed original input image patch and the ground-truth input image patch. The data augmentations of the supervised learning task with UNETR shared intensity scaling and random cropping of input images into 10 samples of 128 × 128 pixels. Thereafter, images were randomly flipped, rotated, or underwent a 10% shift in intensity values. The UNETR model was trained using a combined Dice and cross-entropy loss.
Deep Learning Experiments
The 15,138 unlabeled donor EC images from 8169 donors were rescaled from 1296 × 972 to 1004 × 1338 pixels to maintain a 0.8-µm resolution shared with the 174 EC images of the labeled dataset. For self-supervised learning, a type of unsupervised learning where the model predicts parts of its input data from other features of the same data, 9085 randomly selected EC images (8327 from Eversight, 758 from VisionGift) were used to train our ViT model, and the remaining 3029 EC images (2807 from Eversight, 222 from VisionGift) were used for validation. The ViT model was trained for 300 epochs with batch size of 48 and 1e-4 learning rate.
For supervised learning, the best ViT model weights were loaded onto a UNETR model before fine-tuning on 174 labeled donor EC images (139 for training and 35 for validation) for 215 epochs, equivalently 30,000 iterations, with a batch size of 1. Finally, 3024 unlabeled donor EC images (2808 from Eversight, 216 from VisionGift) were used as the held-out testing dataset for the final UNETR model to conduct inference. The final model binarized each segmentation prediction by determining which class (border or cell/background) received the higher probability. The deep learning training workflow is illustrated in Figure 1.
Post-Processing
Predicted segmentations contained thick cell borders and stray “branches,” a consequence of fine-tuning with labels that contained 3-pixel-wide cell borders and varying image quality and physiological artifacts in donor EC images. Thus, a postprocessing step was implemented (see Supplementary Fig. S1). The prediction segmentations were first skeletonized to single-pixel width, after which stray borders were opened and subsequently closed with a 3 × 3 square kernel to remove smaller disconnected segments and fill small cell border gaps, respectively. Finally, all external, stray “branches” or borders were removed by exploiting an eight-way connected component function from Bolelli et al.20 to find endothelial cells fully enclosed by a segmented border.
Clinical Morphometrics
ECD was determined for the whole image and a cropped image. To calculate the ECD for the whole image, the predictions underwent a filling operation to convert each fully enclosed cell from black pixels to white. This area of white borders and enclosed cells was deemed as the cellular area. The total number of pixels in the cellular area was multiplied by the pixel area (e.g., 0.8 µm × 0.8 µm) to obtain the physical cell area. The number of enclosed cells, determined by a connected components function, was divided by the physical cell area to obtain the ECD for the segmentation. ECD was calculated for the cropped image by using a circular region to encompass the manually annotated cells after postprocessing for the whole image analysis was complete.
Statistical Analyses
Statistical analyses were performed using R 4.3.2 (R Foundation for Statistical Computing, Vienna, Austria) with the SimplyAgree, boot, boot.pval, and tidyverse packages.21–25 Agreement between manual and automated determinations of ECD was evaluated with Bland–Altman analysis and Lin's concordance correlation coefficient (rc).26,27 Associations among donor age, diabetes, death to preservation time, and ECD (average of manual and automated measurements) and differences in ECD between automated and manual methods were explored via linear regression models. Analysis was restricted to the test EC images with at least 100 cells counted by both methods to ensure reliable results.2 Cluster bootstrap resampling to account for clustering of cornea EC images from the same donor was used to assess the statistical significance of differences between the automated and manual methods. P values were obtained via confidence interval inversion, and a two-sided P < 0.05 was considered statistically significant.
Computational Resources
All image processing and deep learning algorithm development were conducted using Python 3.9 libraries such as MONAI, PyTorch (version 1.13.1), numpy, opencv, scikit-learn, and PyQt. ViT and UNETR were trained using four 48-GB NVIDIA A600 GPUs (Nvidia Corporation, Santa Clara, CA). The overall architecture required approximately 87.1 million learning parameters and approximately 520 GFLOPs per image. Self-training took approximately 3 hours, and fine-tuning using one NVIDIA GPU took approximately 2 hours for training. The inference pipeline of segmentation, postprocessing, and ECD calculation required less than 2 seconds.
Results
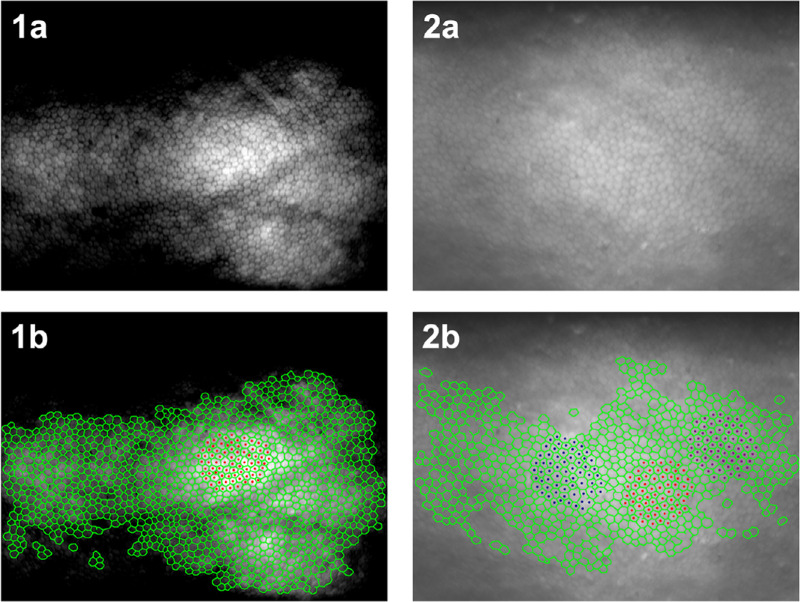
Of the 3024 EC test images, there were 1939 (66%) with at least 100 ECs counted by both methods (see Supplementary Fig. S2). Within this cohort, the proposed automated approach identified between 100 and 1263 ECs per image, whereas the manual approach identified between 100 and 371 ECs per image. This difference in cell identification is illustrated in Figure 2, where example images from Eversight and VisionGift have undergone both automated and manual analyses. Furthermore, it is evident that the automated analysis is not limited to the brightest region of the image, nor a contiguous region of cells. In fact, the automated approach identified many cells in the darker regions of the Eversight image.
Figure 2.
Example of donor cornea endothelial cell images and comparative display of corresponding manual and automatic annotations overlay. Top row: (1a) and (2a) are specular microscopic images from donor cornea endothelium provided by Eversight and VisionGift, respectively. Bottom row: (1b) and (2b) depict automated cell border annotations (green) and manual cell centroid annotations (red, blue, and purple).
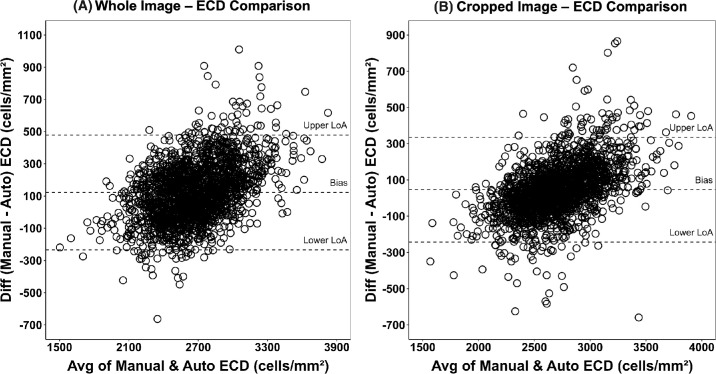
The ECD measurements from the two methods were in excellent agreement with rc of 0.77 (95% confidence interval [CI], 0.75–0.79; P < 0.001) and bias (mean difference) of 123 cells/mm2 (95% CI, 114–131; P < 0.001). Eighty-one percent of the automated ECDs were within 10% of the manual ECDs. When the analysis was further restricted to the cropped image, the rc was 0.88 (95% CI, 0.87–0.89; P < 0.001), bias was 46 cells/mm2 (95% CI, 39–53; P < 0.001), and 93% of the automated ECDs were within 10% of the manual ECDs (Table). The Bland–Altman plots with limits of agreement for the whole and cropped images are displayed in Figures 3A and 3B, respectively.
Table.
Comparison of Whole and Cropped Image ECD for Test Images by Manual and Automated Methods (N = 1939 EC Images)
| ECD (cells/mm2) | Manual Test, Mean ± SD [Min, Max] | Automated Test, Mean ± SD [Min, Max] | Bias (95% CI)a | rc (95% CI) |
|---|---|---|---|---|
| Whole image | 2730 ± 349) | 2607 ± 272) | 123 (114–131)* | 0.77 (0.75–0.79)* |
| [1393, 4132] | [1612, 3607] | |||
| Cropped image | 2730 ± 349) | 2683 ± 277) | 46 (39–53)* | 0.88 (0.87–0.89)* |
| [1393, 4132] | [1655, 3765] |
Cluster bootstrap percentile confidence intervals are based on 100,000 bootstrap samples.
P < 0.001.
Figure 3.
(A) Bland–Altman plot of whole image manual versus automated ECD (N = 1939). Bland–Altman plot of whole image ECDs (cells/mm2) determined by study eye banks versus the ECDs determined by the deep learning model. The difference in ECDs is plotted against their average value. The Bland–Altman plot shows a small, estimated bias (mean difference) of 123 (95% CI, 114–131), with a lower limit of agreement (LoA) of −234 (95% CI, −248 to −220) and an upper limit of agreement of 479 (95% CI, 462–497). Auto, automated. (B) Bland–Altman plot of cropped images of manual versus automated ECDs (N = 1939). Bland–Altman plot of cropped image ECDs (cells/mm2) was determined by study eye banks versus the ECDs determined by the deep learning model. The difference in ECDs is plotted against their average value. The Bland–Altman plot shows a small, estimated bias (mean difference) of 46 (95% CI, 39–53), with a lower limit of agreement of −243 (95% CI, −258 to −228) and an upper limit of agreement of 335 (95% CI, 319–352).
In the univariable and multivariable linear models predicting differences in ECD between manual and automated methods, only donor age was statistically significant (P < 0.001) but not clinically significant. The estimated coefficient for donor age in the multivariable model was −25 cells/mm2 (95% CI, −33 to −17) per decade. For the cropped image, the estimated coefficient for donor age in the multivariable model was −20 cells/mm2 (95% CI, −27 to −14) per decade (see Supplementary Table S1). Differences in ECDs between the manual and automated methods monotonically increased with increasing ECD (P < 0.001); the estimated increase from the linear model was 134 cells/mm2 (95% CI, 122–147) per 500 cells/mm2 for whole images and 120 cells/mm2 (95% CI, 110–131) per 500 cells/mm2 for cropped images (see Supplementary Table S2). This “proportional bias” results in limits of agreement that tend to be too wide for donor corneas with lower ECDs and too narrow for corneas with higher ECD values.28
Discussion
We have shown that the donor cornea ECD determined by our automated, deep learning algorithm compares favorably to the real-world eye-bank–determined ECDs from two major U.S. eye banks. The two methods only differed by a mean of 123 cells/mm2, and 81% of the automated ECDs were within 10% of the eye-bank–determined ECD; the agreement was even tighter when the analysis was conducted within the comparable cropped area of cells that the eye bank had analyzed. Only a mean difference of 46 cells/mm2 and 93% of the automated ECDs were within 10% of the eye-bank–determined ECD. Although these differences were statistically significant, from a medical director standpoint these differences would not impact tissue suitability. Not surprisingly, the difference between the automated-determined and eye-bank–determined ECDs increased with increasing ECD, whether with the whole or cropped image. Similar findings have been reported comparing an automated approach for cell segmentation of in vivo EC images to a flex-center manual analysis.29 Other authors, however, have reported a decrease in the difference between automated and manual ECD methods as more cells in post-keratoplasty EC images were identified.30
Notably, for the analysis of donor factors that could affect the accuracy of automated analysis (specifically, a more polymegethetic endothelial cell population with increasing donor age7,31 and diabetes32), only donor age was found to have a statistically but not clinically significant difference (difference of −25 cells/mm2; 95% CI, −33 to −17) per decade in the multivariable model). This difference may be due to technician cell selection with greater polymegethism and pleomorphism with increasing age.33,34 Conversely, the automated approach is applicable across the entire spectrum of donor conditions. With comparable ECDs between the two ECD analysis methods, the advantages of the automated analysis are its rapidity for ECD determination within seconds and technician training being primarily limited to image acquisition.
Machine learning techniques to automate methodologies for determining endothelial characteristics and morphometrics with in vivo corneal endothelial image analysis have been more commonly described.9,30,35–43 These efforts have included U-Net–based convolutional neural networks to automatically identify cell edges within in vivo EC images42 and a sliding window technique in similar network architectures for cell border segmentation.37,39 ECD calculation improvement was achieved with DenseUNets for cell border segmentation of in vivo EC images and watershed-based postprocessing to clean corresponding deep learning predictions.41 Most recently, efforts to reduce computational resources of deep learning algorithms while maintaining high performance have included parallel networks for cell border segmentation and region of interest extraction followed by a postprocessing pipeline.43 Our efforts build on these works by using self-supervised learning to train transformer network cell border segmentation of donor endothelial images. These images have distinct tissue morphologies, imaging artifacts, and pathologies in comparison to clinical in vivo EC images.
The number of ECs that must be analyzed to be reflective of the central ECD has been debated.33,44–50 Estimates have ranged from at least 30 cells per image33 to as many cells as possible,45 all influenced by the practical limitations of technician time and analysis method accuracy.7 Accurate automated ECD analysis with or without manual intervention is commonly performed clinically with images of good to excellent image quality for a homogeneous cell population with normal cell structure and in the absence of endothelial pathology.9,51–55 However, unlike clinical endothelial image analysis, a fully automated analysis of donor endothelium for ECD has not proven successful,56 whereas semi-automated analytical approaches currently employed in eye banking have raised questions of automation error, human bias, and technique.2,3,57 The EBAA Procedures Manual does not specify the ideal number of endothelial cells to be analyzed; it simply states that, to obtain the most accurate analysis, a large field and/or multiple fields should be captured, counted, and/or averaged.8 In our study, we chose a minimum of 100 ECs minimum to be analyzed by the eye bank, as is commonly practiced in eye banking. Our proposed automated approach for image analysis and donor central ECD determination would more than satisfy EBAA procedural standards, with efficiency and without bias.8
Donor specular microscopy and central ECD analysis have been crucial in determining a minimum ECD of at least 2000 cells/mm2 as part of donor tissue suitability over the past 30 years. The contribution of the determination of donor central ECD toward donor tissue suitability has paid off, as primary graft failure is uncommon in the United States, with a rate of 0.2% annually between 2019 and 2022 according to the EBAA Online Adverse Reaction Reporting System (Jennifer DeMatteo, personal communication, 2024). Notably, long-term graft success (e.g., at 5 years for Fuchs dystrophy at a single site) following primary penetrating keratoplasty, Desçemet stripping automated endothelial keratoplasty (DSAEK), and Desçemet membrane endothelial keratoplasty (DMEK) has been excellent: 95%,58 93%,59 and 93%,59 respectively. However, for the use of deep learning–determined donor central ECD as part of donor tissue suitability to be accepted by the eye banking and surgeon community, there will have to be data relating the deep learning–determined ECD to short-term primary graft failures and long-term graft outcomes (graft success, endothelial cell loss).
Other fields of ophthalmology with large quantities of diagnostic images have already taken advantage of machine learning modeling and their AI applications toward management decisions.60–68 Initial diagnostic AI models developed using in vivo clinical images have been trained to predict corneal disease such as microbial keratitis, keratoconus, dry eye syndrome, and Fuchs dystrophy.36,69 One of the most expeditious ways for graft outcomes to be related to deep learning–determined donor ECDs is for single-site and multicenter study groups with access to the donor images of their recipients to compare automatically determined ECDs to eye-bank–determined ECDs and the graft outcomes of corresponding recipients.
Our study had several limitations. First, our two eye banks differed in the number of areas analyzed for each donor's central endothelium, so their representation of the central endothelium may have differed; however, the specular microscope utilized, technician training, and image analysis techniques did not differ. In this study, the cell border segmentations of the test set donor cornea endothelial images were not manually edited, as were the 174 donor cornea endothelial images used for supervised deep learning training. Although our study proposes an automatic approach for ECD determination, a brief manual check of the image segmentation would not be unprecedented. Furthermore, we are continuing our efforts to optimize and prove the consistency, robustness, and generalizability of our proposed deep learning segmentation algorithm by investigating other transformer network architectures and self-supervised learning approaches such as generative adversarial learning (e.g., boundary attention learning). Also, the influence of polymegethism and pleomorphism on the comparison between the eye-bank–determined ECD and the deep learning–determined ECD was not directly measured. As McCarey et al.49 pointed out, due to the greater variation in cell area and cell shape, cell selection has a greater effect on ECD determination; thus, cell selection and bias by the technician to analyze an image area with possibly smaller cells resulting in a higher ECD could have an influence on the central ECD determined. This influence may have been present, as we noted higher agreement between the eye-bank–determined ECD and deep learning–determined ECD when the deep learning analysis was cropped to approximately the same area of cells that the eye bank analyzed. Examining the coefficient of variation and percentage of hexagonal cells could perhaps shed further light on this finding but was beyond the scope of our study.
In summary, an automated image analysis approach involving deep learning techniques provided accurate ECDs of donor images comparable to real-world eye-bank–determined ECDs for donors suitable for keratoplasty and achieved a significantly reduced analysis time. This analytical approach would also eliminate cell selection bias and limit training requirements to the best conditions and techniques for achieving excellent donor endothelial image quality. It also offers the potential for future studies that would go beyond ECD as a tissue suitability marker with the discovery of machine learning morphologic features, which, as we have shown with postoperative image analysis and graft rejection,10 may be even more predictive of graft success.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (NIH R21EY02949801 to BAMB and DLW; PHS 5 T32 EB 7509-15 to NMJ; U10 EY12728 to JHL; U10 EY012358 to JHL; U10 EY020798 to JHL); Eye Bank Association of America Richard Lindstrom Research Grant (BAMB), Eversight Eye & Vision Research Grant (BAMB); and Cleveland Eye Bank Foundation (JHL). The content of this report is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, Eye Bank Association of America, Eversight, or Cleveland Eye Bank Foundation. All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. This work made use of the High-Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. This research was conducted in space renovated using funds from a National Institutes of Health construction grant (C06 RR12463) awarded to Case Western Reserve University.
Disclosure: B.A.M. Benetz, None; V.S. Shivade, None; N.M. Joseph, None; N.J. Romig, None; J.C. McCormick, None; J. Chen, None; M.S. Titus, Eversight (E); O.B. Sawant, Eversight (E); J.M. Clover, VisionGift (E); N. Yoganathan, VisionGift (E); H.J. Menegay, None; R.C. O'Brien, None; D.L. Wilson, None; J.H. Lass, Cleveland Eye Bank Foundation (S), Eversight (S)
References
- 1. Eye Bank Association of America. Medical standards. Available at: https://restoresight.org/wp-content/uploads/2020/07/Med-Standards-June-20-2020_7_23.pdf. Accessed July 29, 2024.
- 2. Benetz BA, Stoeger CG, Patel SV.. Comparison of donor cornea endothelial cell density determined by eye banks and by a central reading center in the cornea preservation time study. Cornea. 2019; 38(4): 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lass JH, Gal RL, Ruedy KJ.. An evaluation of image quality and accuracy of eye bank measurement of donor cornea endothelial cell density in the Specular Microscopy Ancillary Study. Ophthalmology. 2005; 112(3): 431–440. [DOI] [PubMed] [Google Scholar]
- 4. Huang H, Benetz BA, Clover JM, et al.. Comparison of donor corneal endothelial cell density determined by eye banks and by a central image analysis reading center using the same image analysis method. Cornea. 2022; 41(5): 664–668. [DOI] [PubMed] [Google Scholar]
- 5. Clover J, Ansin A, Tran KD.. A protocol for implementation and use of a tissue incubator for rapid corneal warming at the eye bank. Int J Eye Banking. 2018; 6(1): 1–7. [Google Scholar]
- 6. Tran KD, Clover J, Ansin A, Stoeger CG, Terry MA.. Rapid warming of donor corneas is safe and improves specular image quality. Cornea. 2017; 36(5): 581–587. [DOI] [PubMed] [Google Scholar]
- 7. Sayegh RR, Benetz BA, Lass JH.. Specular microscopy. In: Mannis MJ, Holland EJ, eds. Cornea: Fundamentals, Diagnosis, Management. New York: Elsevier. 2016: 160–179. [Google Scholar]
- 8. Eye Bank Association of America. Procedures manual. Available at: https://restoresight.org/wp-content/uploads/2017/12/EBAA-Procedures-Manual-2017.pdf. Accessed July 29, 2024.
- 9. Huang J, Maram J, Tepelus TC, et al.. Comparison of manual & automated analysis methods for corneal endothelial cell density measurements by specular microscopy. J Optom. 2018; 11(3): 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joseph N, Benetz BA, Chirra P, et al.. Machine learning analysis of postkeratoplasty endothelial cell images for the prediction of future graft rejection. Transl Vis Sci Technol. 2023; 12(2): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lass JH, Benetz BA, Gal RL.. Donor age and factors related to endothelial cell loss 10 years after penetrating keratoplasty: specular microscopy ancillary study. Ophthalmology. 2013; 120(12): 2428–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lass JH, Benetz BA, Verdier DD, et al.. Corneal endothelial cell loss 3 years after successful Descemet stripping automated endothelial keratoplasty in the cornea preservation time study: a randomized clinical trial. JAMA Ophthalmol. 2017; 135(12): 1394–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lass JH. Impact of donor diabetes on DMEK success and endothelial cell loss (DEKS). Available at: https://classic.clinicaltrials.gov/ct2/show/NCT05134480. Accessed July 29, 2024.
- 14. Joseph N, Kolluru C, Benetz BAM, Menegay HJ, Lass JH, Wilson DL.. Quantitative and qualitative evaluation of deep learning automatic segmentations of corneal endothelial cell images of reduced image quality obtained following cornea transplant. J Med Imaging (Bellingham). 2020; 7(1): 014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joseph N, Marshall I, Fitzpatrick E.. Deep learning segmentation of endothelial cell images using an active learning paradigm with guided label corrections. J Med Imaging. 2024; 11(1): 014006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dosovitskiy A, Beyer L, Kolesnikov A.. An image is worth 16x16 words: transformers for image recognition at scale. arXiv. 2021, 10.48550/arXiv.2010.11929. [DOI] [Google Scholar]
- 17. Hatamizadeh A, Tang Y, Nath V, et al.. UNETR: transformers for 3D medical image segmentation. arXiv. 2021, 10.48550/arXiv.2103.10504. [DOI] [Google Scholar]
- 18. Tang Y, Yang D, Li W, et al.. Self-supervised pre-training of Swin transformers for 3D medical image analysis. arXiv. 2022, 10.48550/arXiv.2111.14791. [DOI] [Google Scholar]
- 19. Chen T, Kornblith S, Norouzi M, Hinton G.. A simple framework for contrastive learning of visual representations. In: Daume H, Singh A, eds. ICML’20: Proceedings of the 37th International Conference on Machine Learning. New York: Association for Computing Machinery; 2020: 1597–1607. [Google Scholar]
- 20. Bolelli F, Cancilla M, Grana C.. Two more strategies to speed up connected components labeling algorithms. In: Battiato S, Gallo G, Schettini R, Stanco F, eds. Image Analysis and Processing—ICIAP 2017. Cham: Springer; 2017. [Google Scholar]
- 21. Caldwell AR. SimplyAgree: an R package and jamovi module for simplifying agreement and reliability analyses. J Open Source Software. 2022; 7(71): 4148. [Google Scholar]
- 22. Canty A. boot: bootstrap functions. Available at: https://cran.r-project.org/web/packages/boot/index.html. Accessed July 29, 2024.
- 23. Davison AC, Hinkley DV.. Bootstrap Methods and Their Applications. Cambridge, UK: Cambridge University Press; 1997. [Google Scholar]
- 24. Thulin M. Package ‘boot.pval’. Available at: https://cran.r-project.org/web/packages/boot.pval/boot.pval.pdf. Accessed July 29, 2024.
- 25. Wickham H, Averick M, Bryan J, et al.. Welcome to the tidyverse. J Open Source Software. 2019; 4(43): 1686. [Google Scholar]
- 26. Bland JM, Altman DG.. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986; 1(8476): 307–310. [PubMed] [Google Scholar]
- 27. Lawrence I, Lin K.. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989; 1: 255–268. [PubMed] [Google Scholar]
- 28. Bland JM, Altman DG.. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999; 8(2): 135–160. [DOI] [PubMed] [Google Scholar]
- 29. Karmakar R, Nooshabadi S, Eghrari A.. An automatic approach for cell detection and segmentation of corneal endothelium in specular microscope. Graefes Arch Clin Exp Ophthalmol. 2022; 260(4): 1215–1224. [DOI] [PubMed] [Google Scholar]
- 30. Vigueras-Guillén JP, van Rooij J, Engel A, Lemij HG, van Vliet LJ, Vermeer KA.. Deep learning for assessing the corneal endothelium from specular microscopy images up to 1 year after ultrathin-DSAEK surgery. Transl Vis Sci Technol. 2020; 9(2): 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Islam QU, Saeed MK, Mehboob MA.. Age related changes in corneal morphological characteristics of healthy Pakistani eyes. Saudi J Ophthalmol. 2017; 31(2): 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Del Buey MA, Casas P, Caramello C. An update on corneal biomechanics and architecture in diabetes. J Ophthalmol. 2019; 2019: 7645352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laing RA, Sanstrom MM, Berrospi AR, Leibowitz HM.. Changes in the corneal endothelium as a function of age. Exp Eye Res. 1976; 22(6): 587–594. [DOI] [PubMed] [Google Scholar]
- 34. Ohara K, Tsuru T, Inoda S.. Morphometric parameters of the corneal endothelial cells. Nippon Ganka Gakkai Zasshi. 1987; 91(11): 1073–1078. [PubMed] [Google Scholar]
- 35. Daniel MC, Atzrodt L, Bucher F, et al.. Automated segmentation of the corneal endothelium in a large set of ‘real-world’ specular microscopy images using the U-Net architecture. Sci Rep. 2019; 9(1): 4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kang L, Ballouz D, Woodward MA.. Artificial intelligence and corneal diseases. Curr Opin Ophthalmol. 2022; 33(5): 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kucharski A, Fabijańska A.. CNN-watershed: a watershed transform with predicted markers for corneal endothelium image segmentation. Biomed Signal Process Control. 2021; 68: 102805. [Google Scholar]
- 38. Selig B, Vermeer KA, Rieger B, Hillenaar T, Luengo Hendriks CL. Fully automatic evaluation of the corneal endothelium from in vivo confocal microscopy. BMC Med Imaging. 2015; 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vigueras-Guillén JP, Sari B, Goes SF, et al.. Fully convolutional architecture vs sliding-window CNN for corneal endothelium cell segmentation. BMC Biomed Eng. 2019; 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vigueras-Guillen JP, Rooij J, Lemij HG, Vermeer KA, Vliet LJ.. Convolutional neural network-based regression for biomarker estimation in corneal endothelium microscopy images. Annu Int Conf IEEE Eng Med Biol Soc. 2019; 2019: 876–881. [DOI] [PubMed] [Google Scholar]
- 41. Vigueras-Guillen JP, van Rooij J, van Dooren BTH, et al.. DenseUNets with feedback non-local attention for the segmentation of specular microscopy images of the corneal endothelium with guttae. Sci Rep. 2022; 12(1): 14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fabijańska A. Segmentation of corneal endothelium images using a U-Net-based convolutional neural network. Artif Intell Med. 2018; 88: 1–13. [DOI] [PubMed] [Google Scholar]
- 43. Karmakar R, Nooshabadi SV, Eghrari AO.. Mobile-CellNet: automatic segmentation of corneal endothelium using an efficient hybrid deep learning model. Cornea. 2023; 42(4): 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abib FC, Holzchuh R, Schaefer A, Schaefer T, Godois R.. The endothelial sample size analysis in corneal specular microscopy clinical examinations. Cornea. 2012; 31(5): 546–550. [DOI] [PubMed] [Google Scholar]
- 45. Binder PS, Akers P, Zavala EY.. Endothelial cell density determined by specular microscopy and scanning electron microscopy. Ophthalmology. 1979; 86(10): 1831–1847. [DOI] [PubMed] [Google Scholar]
- 46. Hirst LW, Auer C, Abbey H, Cohn J, Kues H.. Quantitative analysis of wide-field endothelial specular photomicrographs. Am J Ophthalmol. 1984; 97(4): 488–495. [DOI] [PubMed] [Google Scholar]
- 47. Hirst LW, Yamauchi K, Enger C, Vogelpohl W, Whittington V.. Quantitative analysis of wide-field specular microscopy. II. Precision of sampling from the central corneal endothelium. Invest Ophthalmol Vis Sci. 1989; 30(9): 1972–1979. [PubMed] [Google Scholar]
- 48. Inaba M, Matsuda M, Shiozaki Y, Kosaki H.. Regional specular microscopy of endothelial cell loss after intracapsular cataract extraction: a preliminary report. Acta Ophthalmol (Copenh). 1985; 63(2): 232–235. [DOI] [PubMed] [Google Scholar]
- 49. McCarey BE, Edelhauser HF, Lynn MJ.. Review of corneal endothelial specular microscopy for FDA clinical trials of refractive procedures, surgical devices and new intraocular drugs and solutions. Cornea. 2008; 27(1): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Doughty MJ, Müller A, Zaman ML.. Assessment of the reliability of human corneal endothelial cell-density estimates using a noncontact specular microscope. Cornea. 2000; 19(2): 148–158. [DOI] [PubMed] [Google Scholar]
- 51. Goldich Y, Marcovich AL, Barkana Y.. Comparison of corneal endothelial cell density estimated with 2 noncontact specular microscopes. Eur J Ophthalmol. 2010; 20(5): 825–830. [DOI] [PubMed] [Google Scholar]
- 52. Maruoka S, Nakakura S, Matsuo N.. Comparison of semi-automated center-dot and fully automated endothelial cell analyses from specular microscopy images. Int Ophthalmol. 2018; 38(6): 2495–2507. [DOI] [PubMed] [Google Scholar]
- 53. Ohno K, Nelson LR, McLaren JW, Hodge DO, Bourne WM.. Comparison of recording systems and analysis methods in specular microscopy. Cornea. 1999; 18(4): 416–423. [DOI] [PubMed] [Google Scholar]
- 54. van Schaick W, van Dooren BTH, Mulder PGH, Völker-Dieben HJM. Validity of endothelial cell analysis methods and recommendations for calibration in Topcon SP-2000P specular microscopy. Cornea. 2005; 24(5): 538–544. [DOI] [PubMed] [Google Scholar]
- 55. Cheung SW, Cho P.. Endothelial cells analysis with the TOPCON specular microscope SP-2000P and IMAGEnet system. Curr Eye Res. 2000; 21(4): 788–798. [DOI] [PubMed] [Google Scholar]
- 56. Hirneiss C, Schumann RG, Grüterich M, Welge-Luessen UC, Kampik A, Neubauer AS.. Endothelial cell density in donor corneas: a comparison of automatic software programs with manual counting. Cornea. 2007; 26(1): 80–83. [DOI] [PubMed] [Google Scholar]
- 57. Munir WM, Munir SZ. Characteristics of semiautomated endothelial cell-density measurements among corneal donor eyes. JAMA Ophthalmol. 2022; 140(9): 885–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Price FW Jr, Whitson WE, Collins KS, Marks RG.. Five-year corneal graft survival. A large, single-center patient cohort. Arch Ophthalmol. 1993; 111(6): 799–805. [DOI] [PubMed] [Google Scholar]
- 59. Price DA, Kelley M, Price FW, MO Price. Five-year graft survival of Descemet membrane endothelial keratoplasty (EK) versus Descemet stripping EK and the effect of donor sex matching. Ophthalmology. 2018; 125(10): 1508–1514. [DOI] [PubMed] [Google Scholar]
- 60. Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC.. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit Med. 2018; 1(1): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Abràmoff MD, Lou Y, Erginay A, et al.. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci. 2016; 57(13): 5200–5206. [DOI] [PubMed] [Google Scholar]
- 62. Brown JM, Campbell JP, Beers A, et al.. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018; 136(7): 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM.. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017; 135(11): 1170–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. De Fauw J, Ledsam JR, Romera-Paredes B, et al.. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018; 24(9): 1342–1350. [DOI] [PubMed] [Google Scholar]
- 65. Gargeya R, Leng T.. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017; 124(7): 962–969. [DOI] [PubMed] [Google Scholar]
- 66. Grassmann F, Mengelkamp J, Brandl C, et al.. A deep learning algorithm for prediction of Age-Related Eye Disease Study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology. 2018; 125(9): 1410–1420. [DOI] [PubMed] [Google Scholar]
- 67. Li Z, He Y, Keel S, Meng W, Chang RT, He M.. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018; 125(8): 1199–1206. [DOI] [PubMed] [Google Scholar]
- 68. Ting DSW, Cheung CYL, Lim G, et al.. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017; 318(22): 2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eleiwa T, Elsawy A, Özcan E, Abou Shousha M. Automated diagnosis and staging of Fuchs’ endothelial cell corneal dystrophy using deep learning. Eye Vis (Lond). 2020; 7: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.