Abstract
In vivo, muscle and neuronal cells are post-mitotic, and their function is predominantly regulated by proteostasis, a multilayer molecular process that maintains a delicate balance of protein homeostasis. The ubiquitin-proteasome system (UPS) is a key regulator of proteostasis. A dysfunctional UPS is a hallmark of muscle ageing and is often impacted in neuromuscular disorders (NMDs). Malfunction of the UPS often results in aberrant protein accumulation which can lead to protein aggregation and/or mis-localization affecting its function. Deubiquitinating enzymes (DUBs) are key players in the UPS, controlling protein turnover and maintaining the free ubiquitin pool. Several mutations in DUB encoding genes are linked to human NMDs, such as ATXN3, OTUD7A, UCHL1 and USP14, whilst other NMDs are associated with dysregulation of DUB expression. USP5, USP9X and USP14 are implicated in synaptic transmission and remodeling at the neuromuscular junction. Mice lacking USP19 show increased maintenance of lean muscle mass. In this review, we highlight the involvement of DUBs in muscle physiology and NMDs, particularly in processes affecting muscle regeneration, degeneration and inflammation following muscle injury. DUBs have recently garnered much respect as promising drug targets, and their roles in muscle maturation, regeneration and degeneration may provide the framework for novel therapeutics to treat muscular disorders including NMDs, sarcopenia and cachexia.
Keywords: deubiquitinase, muscle atrophy, muscle regeneration, myopathies, neuromuscular disorders, ubiquitin
Ubiquitin is a small protein of 76 amino acids that is highly conserved in eukaryotes [1]. The post-translational modification of protein substrates by ubiquitin is key to most cellular processes impacted by proteostasis. Ubiquitination primarily occurs via covalent isopeptide bond formation to lysine residues via a cascade of enzymatic activities involving the E1 ubiquitin-activating, E2 ubiquitin-conjugating, and E3 ubiquitin–protein ligase enzymes. Notably, ubiquitination can also be facilitated through ester bond formation on serine and threonine, and thioester linkages to cysteines [1]. Next to targeting substrate proteins, carbohydrates, lipopolysaccharides and ribosyl-moieties can also be ubiquitinated, of which the latter is predominantly involved in host-pathogen interactions [2,3]. Through its involvement in the ubiquitin proteasome system (UPS), ubiquitin itself can also be ubiquitinated, forming polyubiquitin chains that tag misfolded, defective or redundant proteins for degradation by the proteasomal system. In contrast with E3 ligases, deubiquitinating enzymes (DUBs) remove ubiquitin moieties and thereby counteract ubiquitination signaling. The widespread nature of ubiquitin-dependent biological processes is reflected by the ∼700 known E3 ligases, and the ∼100 DUB species. DUBs are divided in different families based on distinct mechanisms of action, including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), JAB1/MPN/Mov34 metalloenzyme, Machado–Joseph deubiquitinases (MJDs), motif interacting with Ub-containing novel DUB family, and zinc finger containing ubiquitin peptidase 1 (ZUP1) [4,5].
Regulation of proteostasis by the UPS is essential for tissue maintenance, in particular for those of post-mitotic cells, such as neurons and muscle fibers. Over 10% of E3 ligases are implicated in common and rare neurological disorders [6]. In skeletal muscles, two E3 ligases, TRIM63 and FBXO32, also known as MuRF1 and Atrogin-1, regulate muscle atrophy [7], a key pathological feature in muscle degeneration and muscle wasting [8–10]. Here, we surveyed most recent literature in addition to public databases for information on the role of DUBs in a diverse range of biochemical pathways which have been implicated in muscle physiology, in neuromuscular disorders (NMDs) and specifically in muscle wasting. This is particularly timely, as DUBs are emerging as attractive drug targets [11], offering potential inroads into novel therapeutics for NMDs, including myopathies and muscular dystrophies as well as cachexia and age-associated wasting as seen in sarcopenia.
Ubiquitin processing is linked to muscle biology and physiology
Skeletal muscle, the largest tissue in vertebrates by mass, provides stability and facilitates mobility and balance of the skeleton. All are vital for normal daily functioning. Muscle contraction is voluntarily controlled by motor neurons connecting the central nerve system to nerve terminals and neuromuscular junctions (NMJs) at the muscle plasma membrane. A wide range of movements are accommodated through the activity of over 600 different skeletal muscles which vary in size, shape, architecture, and molecular composition [12,13]. Myofibers are characterized by the sarcomeric protein architecture that facilitates contraction and stability. Myofibers are long, multinucleated cells that are formed during embryo development, but become post-mitotic after birth. Muscle degeneration is characterized by loss of muscle mass (atrophy) and loss of contraction, together negatively affecting mobility and stability in both acute and chronic dystrophy. Repair of damaged muscles is through regeneration, by which muscle stem cells proliferate and fuse into new myofibers. The UPS plays a pivotal role in controlling protein homeostasis in myofibers. Out of the E1, E2 and E3 ligases, it is chiefly the E3 ligases that are implicated in muscle atrophy (reviewed in [14–16]). In contrast with E3 ubiquitin ligases, the role of DUBs in skeletal muscle biology, physiology and muscle disorders is less well-understood [17]. Age-associated loss of muscle mass is accompanied by an up-regulation of many DUBs [18], suggesting multiple roles for DUBs in muscle physiology and pathology. A genetic screen in muscle cells indicates that dysregulation of several DUBs including USP7, USP10, USP13, USP16 and USP18 expression abrogates muscle cell differentiation [19]. The expression profile of four muscle related USPs (USP10, USP14, USP19 and USP45) also differs between fast- and slow-twitch myofibers [20]. For most of these DUBs, their molecular role in the context of muscle biology remains obscure.
NMDs involving DUB pathogenic variants or aberrant expression associated with muscle integrity and function
DUB mutations associated with muscle pathology are often linked to neurological disorders and inflammation [21,22]. For instance, a biallelic loss of OTUD7A causes severe muscular hypotonia associated with intellectual disability and seizures in humans [23] (Table 1). In a separate study, Otud7a knockout mice displayed muscle hypotonia, impaired dendritic spine density, and reduced glutamatergic synaptic transmission [39]. Although these studies highlight the importance of OTUD7A with regard to neuromuscular transmission, which likely underlies muscle weakness, the exact molecular mechanisms behind this remain to be determined. Interestingly, Garret et al. [40] found that mutated OTUD7A impairs proteasomal activity by regulating proteasomal assembly in a yet unknown manner. Nevertheless, this could indicate that muscle weakness in pathological conditions caused by a OTUD7A mutation could, in part, be a result of imbalanced proteostasis.
Table 1. Most common DUB pathogenic variants associated with human disease linked muscle pathology.
| Family | DUB | Human/mouse pathology | Muscle disease trait | References |
|---|---|---|---|---|
| UCH |
UCHL1 Loss of function variants Deletions 2-223/25-223/insertion 52L > LL E7A I93M R178Q A216D |
Processing of ubiquitin precursors and ubiquitinated protein substrates. Its absence impairs the synaptic transmission at the NMJ, inducing profound structural defects at the presynaptic nerve terminals and muscle denervation. Spastic paraplegia 79–autosomal recessive (SP79-AR) | Neurodegeneration Spastic paraplegia 79A Muscle denervation Difficulty with balance, weakness and stiffness in the legs, muscle spasms |
[24,25,26] |
| USP | USP9X FAF-X |
Antagonizes ubiquitin-mediated proteolysis, preventing protein degradation and controlling synapse development. Defects in the synaptic transmission at the NMJ | Defects in the synaptic transmission at the neuromuscular junction | [27] [28] |
| USP |
USP14 4 bp deletion 233_236delTTCC; p.Leu78Glnfs*11 |
Crucial for synaptic development and function at the NMJ. Its catalytically inactive form causes developmental deficits in the NMJ structure and synaptic transmission. Its loss causes presynaptic defects | Severe tremors, hind limb paralysis, and postnatal lethality Muscle wasting, cachexia |
[29] [30] [31] |
| USP | USP18 USP18 truncation at 652C > T (N218) |
Pseudo-TORCH syndrome (PTS) microcephaly, enlarged ventricles, cerebral calcification due to severe interferonopathy Mutations linked to multiple sclerosis susceptibility |
Muscle spasms, stiffness and weakness Bradycardia Muscle differentiation |
[32] [19] |
| USP | USP19 | Usp19 KO mice have lean muscle phenotype and less myofiber atrophy | Muscular atrophy Muscle wasting, cachexia |
[33] [34] [31] [35] [36] |
| OTU |
OTUD7A/Cezanne 15q13.3 deletion including the OTUD7A locus, frameshift OTUD7A variant c.1125del, p.(Glu375Aspfs*11) |
Intellectual disability, and seizures | Severe muscular hypotonia | [23] |
| MJD | ATXN3 polyglutamine expansion in ataxin-3 (ATX3; MJD1) | Machado–Joseph disease (MJD), autosomal-dominant neurodegenerative disorder Affects Ca2+ signaling |
Muscle spasticity | [37,38] |
Mutations recorded in DUBs and their associations with pathologies in humans are described. DUB knockout mice and other organisms also show phenotypes including muscle specific traits.
Abbreviations: ATXN, ataxin; DUB, deubiquitinase; OTU, ovarian tumor domain containing protease; MJD, Machado–Joseph-disease; UCH, ubiquitin C-terminal hydrolase; USP, ubiquitin specific protease; NMJ, neuro-muscular junction; FAF, fat facets (fly).
Homozygous missense and splice-site mutations in UCHL1 have been reported in cases with spastic paraplegia and progressive ataxia (Table 1). Interestingly, genome screening illuminated different frame-shift or in-frame shifts that are predicted to cause a loss-of-function. A p.Glu7Ala and p.Ile983Met missense mutation caused reduced binding affinity and consequently a lowered catalytical activity of <10% and ∼50% as compared with wild-type UCHL1 [24,25]. These loss-of-function mutations often lead to nerve fiber loss, axonal swelling, degeneration of the spinal cord, and impaired neuromuscular denervation and synaptic transmission. Interestingly, one case showed a heterozygous variant with a missense mutation p.Ala216Asp, leading to an insoluble protein variant, and a p.Arg178Gln mutation that showed a four-fold increased hydrolytic activity [24,25]. The maintenance of the NMJ structure and function is also associated with UCHL1, as Uchl1 knockout mice develop selective motor neuropathy [41,42]. UCHL1 also regulates lipid and perilipin 2 levels [43] and mitochondrial oxidative activity in skeletal muscles [44]. However, a mode of action of UCHL1 remains unexplored in these KO studies. Guillain–Barré syndrome (GBS), a condition affecting the myelin sheath of nerves, leading to muscle weakness and sometimes paralysis, has been linked with altered UCHL1 levels. Higher levels of UCHL1 were detected in the cerebrospinal fluid of GBS patients, when compared with controls, with a possible association between UCHL1 levels and disease severity [45]. Furthermore, UCHL1 has been found to be significantly higher in the plasma of Parkinson's disease (PD) patients at moderate stages of disease, when compared with early-stage counterparts and healthy controls [46], reflecting a possible correlation with PD pathology [47]. Altogether, it appears that several NMDs are characterized by a change in UCHL1 expression levels, including spinal muscular atrophy (SMA) [48]. However, a positive correlation was found between increased UCHL1 levels and total protein levels in GBS patients [45], indicating that any interpretation of expression levels should be interpreted with caution. Interestingly, UCHL1 has been found to bind the proteasome, and in case of point mutations, such as I93M, C90S and E7A, that have reduced hydrolytic activity, UCHL1 binds the 20S core subunit of the proteasome and negatively affects its activity [49]. The effect of various mutations and deletions in human UCHL1 gene on, for instance, the mitochondrial respiration and proteasomal activity, suggests that both the catalytic and scaffolding functions of UCHL1 are of importance in muscle biology. The study of UCHL1 pathogenic variants have contributed to the understanding of its function in multiple cellular processes, which is key to revealing its mode of action in disease mechanisms.
Spinocerebellar ataxia type 3, also known as Machado–Joseph disease (MJD), is a neurodegenerative disorder caused by a polyglutamine-coding CAG trinucleotide repeat expansion in the MJD domain protease subfamily member ATXN3 (Table 1) [37,50]. Mechanistically, it is not yet fully understood how the ATXN3 variants contribute to ataxia. However, next to nuclear aggregation of mutated ATXN3, it was found that mutated ATXN3 directly binds InsP3R1, a calcium channel, causing it to open more easily, leading to aberrant calcium signaling, which contributes to pathogenicity [38,51]. ATXN3 also interacts with VCP (/p97), an ATPase complex, that thereby facilitates the deubiquitination of BECN1 by ATXN3 [52]. Interestingly, reduced proteasomal degradation due to mutations in p97 have been found in the muscle wasting phenotype of amyotrophic lateral sclerosis (ALS) and cachexia, suggesting an important role for ATXN3 and the p97 complex in muscle physiology [53].
SMA is caused by a mutation in the survival motor neuron 1 gene (SMN1), which results in low levels of SMN protein, motor neuron degeneration, and ultimately, to lethality in infants. USP9X stabilizes SMN protein by a direct removal of ubiquitin moieties, which might prove to be an interesting target. However, whereas USP9X deubiquitinates wild-type SMN protein, its mutated form is unaffected by USP9X, causing its rapid degradation, and thereby contributes to impaired neuronal growth and development [27]. Elucidating why USP9X does not target mutated SMN protein could prove a valuable avenue in treating SMA, possibly for adult-onset disease. Aberrant morphology of NMJs has been found in Drosophila with USP5 mutations and is thought to be linked to altered ubiquitin homeostasis through ubiquilin, a ubiquitin-like protein [54] (Table 1). Nevertheless, a role for USP5 does not seem to exclusively affect NMJs [55].
ALS, also known as motor neurone disease (MND) or Lou Gehrig's disease, is a rare, progressive, and fatal type of degeneration of the spinal cord and motor neurons that control voluntary muscles [56]. Cases of ALS are primarily sporadic, but hereditary forms also exist. Components of the UPS play a pivotal role in ALS [57]. A key hallmark of ALS is the pathogenic deposition of TAR DNA-binding protein 43 (TDP-43) in the spinal cord and brain of both sporadic and familial cases [58]. Ubiquitinated TDP-43 is enriched in ALS brain inclusions [59], which is likely due to the co-ordinated action of UBE2E3 conjugation and either Parkin, VHL/CUL2, Znf179, or Praja 1 ligation [57,60]. Counteracting this, several DUBs have been shown to regulate TDP-43 protein turnover and stress granule clearance, including CYLD [61], USP5/13 [62], USP7 [63], USP8/UBPY [60], USP10 [64], USP14 [65], and USP47 [66]. Moreover, a rare variant of CYLD has been found in a cohort of Chinese ALS patients [67]. Finally, UCHL1 has been proposed as an ALS candidate biomarker in CSF and serum [68,69]. Although these events may affect skeletal muscle indirectly, targeting these DUBs might lead to alleviation of the muscle pathology.
In contrast with NMDs, the role of DUBs in myopathies is less explored. A transcriptomic study of degenerated muscles of oculopharyngeal muscular dystrophy (OPMD) patients and OPMD animal models showed significant dysregulation in DUB expression. OPMD is a late-onset myopathy, caused by an expansion mutation in PABPN1 that leads to nuclear aggregation of PABPN1. The reduced functional levels of PABPN1 and dysregulated DUB expression implies a role for the UPS [70]. Reduced PABPN1 levels lead to muscle atrophy by direct regulation of FBXO32 level [71]. PABPN1 has also been shown to directly regulate the transcript levels via alternative polyadenylation site usage of PSMD14/POH1, a metalloprotease that is part of the 19S proteasome that is directly involved in the degradation of polyubiquitinated proteins [70,72]. Another DUB that is associated with the proteasome, USP14, has been implicated in muscle atrophy due to increased expression, but its functional role remains unclear [73]. USP14 has been shown to be essential for the maintenance of synaptic ubiquitin levels and the development of NMJs [29]. A 4-bp biallelic deletion induced loss of USP14 leads to distal arthrogryposis (multiple congenital contractures) in human individuals that is characterized by abnormal development and function of skeletal muscles [30]. Mouse models with no or reduced USP14 expression levels show neurological and muscular abnormalities similar to humans, supporting the notion that USP14 plays a crucial role in development of the neuromuscular system [74,75]. Together, several ubiquitin E3 ligases, DUBs and the biogenesis of the 26S proteasome, have been demonstrated to be involved in enhanced degradation of muscular proteins [31].
USP19 has been shown to be overexpressed in mammalian skeletal muscle in a plethora of degenerative disorders associated with muscle atrophy, including fasting and caloric restriction, diabetes, cancer, and smoking [76]. USP19 knockout mice are viable, and in response to fasting exert less muscle wasting as compared with WT littermates. Lower levels of brown and white adipose tissue have also been found [35,36]. In skeletal muscles of patients with lung and gastrointestinal cancers, a potential link is observed between the expression of USP19, TRIM63 and FBXO32 mRNA, further strengthening the link between USP19 and human muscle wasting [77]. Mechanistically, a functional role linking USP19 activity and expression to diet-induced insulin responsiveness, glucocorticoid signaling, adipogenesis, but also fat and muscle distribution has been demonstrated in mice [36], although the exact molecular details remain to be determined.
Links to insulin responsiveness and skeletal muscle mass have also been demonstrated for USP21 [78]. Removal of USP21 resulted in an increased mitochondrial activity in skeletal muscle, leading to promotion of an oxidative myofiber phenotype, and inhibition of obesity and type 2 diabetes [78]. This phenomenon could be reversed by overexpression of this DUB. Similarly, in skeletal muscles, USP21 is up-regulated in individuals with obesity, and correlates with fasting glucose levels in animals on a high-fat diet [78]. In skeletal muscle, USP9X has been proposed to play a key role in the anti-diabetic effect of calorie restriction through its increase and stabilization of AMPKα2 receptors [79]. Altering the expression levels of USP21 and USP9X have therefore been suggested to have therapeutic potential in the treatment of obesity and impaired insulin sensitivity, which are both associated with muscle physiology.
As introduced earlier, links have also been made between NMDs and the levels of DUBs detected in patient biofluids and tissues. For instance, multiple sclerosis (MS), an autoimmune disorder leading to muscle spasms, stiffness and weakness, has been linked to changes in USP18, TNFAIP3 (A20) and USP16 expression levels. Low levels of USP18 mRNA were detected in the peripheral blood mononuclear cells of patients with relapsing-remitting MS (RRMS), which could possibly lead to enhanced TNF and type-I interferon (IFN) signaling as these DUBs are major negative regulators of immune signaling [80]. A reduced expression level of TNFAIP3 is observed in monocytes of RRMS patient [81], whereas an increased expression of TNFAIP3 has been found in the lesions of post-mortem brain tissue of MS patients [82]. Also, increased expression of USP16 was recorded in CD3+ T cells of MS patients [83]. Furthermore, two USP18 polymorphisms have been associated with MS susceptibility [84] (Table 1). USP18 inactivating mutations in human individuals lead to severe type 1 interferonopathies through a lack of USP18-mediated negative regulation of the IFN signaling and thereby causing severe chronic inflammation, which predominantly affects the CNS, but also other organ systems including the heart (bradycardia) [32].
Taken together, human mutations in DUBs, knockout animal models or their altered expression profiles affecting skeletal muscle pathology and myopathies are often linked to complex phenotypes, predominantly associated with NMDs, in addition to inflammation, cachexia and cancer.
DUBs in muscle injury and regeneration
Inflammation
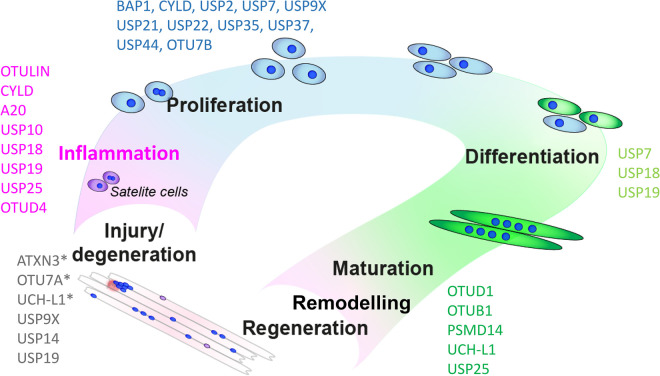
Skeletal muscles have a strong capacity to regenerate injured tissue via a cascade of interconnected events [85,86] (Figure 1). In damaged and diseased muscle tissues, an inflammatory response is activated to mediate regeneration and tissue repair. This inflammatory response in muscles involves the secretion of cytokines, such as IFNs, IL-1β and TNF-α, resulting in the attraction of immune cells, macrophages, neutrophils, T-cells and B-cells [85]. This immune response leads to the removal of damaged cells, after which new migrating cell populations will repair the damaged tissue regions [86]. However, in case of an excessive or a chronic inflammatory response as seen in inflammatory myopathies, regeneration fails and results in muscle wasting instead. In other muscular dystrophies, such as Duchenne muscular dystrophy (DMD), a prolonged inflammatory response has also been shown to exacerbate muscle damage, impair regeneration and associate with disease severity [87,88]. Inflammatory myopathies (myositis), such as polymyositis, inclusion body myositis and dermatomyositis, often involve excessive cytokine [89] and type 1 or 2 IFN expression that lead to muscle weakness (reviewed in [90]). Cytokine levels are relevant in the diagnostics and differentiate between various types of myositis [91,92]. IFN production and the respective response pathways are tightly regulated by ubiquitination and deubiquitination events, and as a consequence, regulatory functions for several DUBs have been described. For example, TNFAIP3, CYLD, USP3, USP4, USP15, USP25, and OTUD5 deubiquitinate key components of the IFN production pathway, including RIG-I, TRAF2, TRAF3, TRAF6, RIP1, and TRIF (reviewed in [93,94]). As for IFN response pathways, USP18 attenuates the type 1 IFN response via a negative feedback loop, and it is the main enzyme involved in the catalytical removal of the interferon stimulated gene 15 (ISG15) a di-ubiquitin analogue, from substrate proteins [95].
Figure 1. DUBs associated with muscle physiology and pathology.
Deubiquitinases (DUBs) have been linked to various stages of muscle development, homeostasis and neuromuscular disorders affected by mutations (indicated with an asterix *, see also Table 1) and altered expression profiles. DUBs associate with diverse stages of muscle physiology, such as muscle injury/damage, muscle inflammation (e.g. myositis), proliferation, myogenic differentiation, muscle regeneration, remodeling and maturation. DUBs highlighted in black (bold) have been studied in skeletal muscles directly. It is likely that DUBs highlighted in gray might have an indirect effect on skeletal muscle physiology and pathology. See text for details.
IRAK signaling is part of the TLR/MYD88 cascade which is controlled by the DUBs OTULIN, OTUD4, CYLD, TNFAIP3, USP19 and USP25, all of which edit linear and K63-linked ubiquitin chains [96,97]. The MYD88 cascade has been found to mediate muscle wasting in cachexia as well in a model for muscle denervation, highlighting its importance in muscle physiology [98,99]. Furthermore, a long non-coding RNA, referred to as INKILN (inflammatory MKL1 interacting lncRNA), can induce vascular smooth muscle inflammation via scaffolding of MKL1 (myocardin-like protein) and USP10 [100]. Similarly, the long-noncoding RNA Atrolnc-1 that binds to A20, can induce muscle wasting in CKD mice [101].
IL-1β, another cytokine essential for muscle regeneration [102], is initially expressed by activated monocytes and macrophages in response to priming signals from receptors, such as Toll-like receptors (TLRs) or nucleotide-binding oligomerization domain-like receptors (NLRs). It is then processed and activated through the inflammasome system, a critical process in muscle inflammation that is tightly regulated by several DUBs including USP7/47, BRCC3, TNFAIP3, STAMBP, UCHL1 [103] and other members of the UCH family (reviewed in [104]). Interestingly, NLRP3 removal attenuates inflammation induced skeletal muscle atrophy [105]. Down-regulation of NLRP3 and IL-1β levels by adiponectin treatment protect dystrophic mice muscles from excessive inflammation, which ultimately might prove therapeutically relevant for DMD patients [106]. IL-1β signaling was also found to be promoted by IRAK1 induced phosphorylation of USP20 in vascular smooth muscle cells and vascular inflammation [107,108].
USP19 attenuates the TNF-α– and IL-1β–triggered inflammatory response via deubiquitination of TGF-β-activated kinase 1. Mice lacking USP19 were shown to have increased levels of cytokines and elevated inflammatory responses [109]. Taken together, DUBs are essential in regulating cytokine expression and signaling, and thereby ultimately in regulating the balance between a necessary immune response for muscle tissue regeneration or an excessive inflammatory response that causes more damage to the muscle. Despite this essential role and thus also therapeutic potential, the role of DUBs on inflammation of skeletal muscles remains poorly studied (Figure 1).
Proliferation and myogenic fusion
Cytokines that regulate the progression of removal of damaged tissue can activate quiescent satellite cells (SCs). The activated SCs will provide new cells for tissue regeneration (Figure 1). Muscle regulatory transcription factors (MRFs) regulate the myogenic program. MYF5 and MYOD stimulate muscle cell proliferation and subsequent expression of MYF6 and myogenin initiate the terminal differentiation into multinucleated myofibers. The dynamic expression of these MRFs is crucial and a function for DUBs is becoming more evident. A DUB siRNA screen using myogenic fusion in tissue culture as a readout has revealed several DUBs that affect cell proliferation, differentiation and maturation [19]. In this study, USP7, USP10, USP13, USP18, USP45, UCHL1 and SENP2 were all found to reduce myogenesis and may therefore all be essential for the switch from proliferation to differentiation [19]. More generally, many DUBs, such as BAP1, CYLD, USP2, USP7, USP9X, USP21, USP22, USP35, USP37, USP44 and Cezanne/OTUD7B, have been implicated in cellular proliferation across different stages of the cell cycle [110]. As these DUBs are ubiquitously expressed, they may also affect myocyte and cardiomyocyte proliferation, by controlling the turnover of MyoD [111] and TATA-binding protein, another transcription regulator that is stabilized via direct deubiquitination by USP10 [112]. A second regulator of muscle specific transcripts involved in muscle cell differentiation, Myocyte-specific enhancer factor 2A, was found to be targeted by SENP2. SENP2 promotes myostatin expression and was found to inhibit muscle cell differentiation [113]. Down-regulation of USP7 or pharmacological inhibition impairs muscle differentiation by affecting myogenin stability, thereby enhancing SC myogenic progression [114].
USP19 is perhaps one of the most studied DUBs in pathological conditions characterized by muscle atrophy, but also with regards to muscle cell differentiation and maturation [34,115]. USP19 may interfere with hormone homeostasis in skeletal muscle [116], possibly through a 17β-estradiol (E2) and (o)estrogen receptor (ER)-dependent feedback loop [117]. Only the ER-localized isoform of USP19 (USP19-ER) modulated myoblast fusion as well as the expression of myogenin and myofibrillar proteins in a USP19 catalytic activity-dependent manner [33,118]. This may involve ER related functions of USP19 including stabilization of endoplasmic-reticulum-associated protein degradation substrates [119–121] and the HRD1 ubiquitin E3 ligase [120]. Also, USP19 activates NRF1 (encoded by Nfe2l1), thereby altering glucose deprivation sensing, cholesterol abundance, proteasomal inhibition and oxidative stress [122]. Underscoring its importance in muscle physiology, the Almac Group have recently described the first-in-class inhibitor of USP19 for the treatment of cancer-induced muscle atrophy [121].
USP18 was recently identified to be a key regulator of muscle cell proliferation, differentiation and maturation [19]. USP18 enzymatic function typically regulates the fate of proteins modified by ISG15 in the context of innate immune activation by removing ISG15 from protein substrates. However, in muscle cells, USP18 may have an additional role, independent from ISG15, in regulating the timing of differentiation and subsequently maturation. The way USP18 achieves this is perhaps through altering the expression of myogenic transcription (co) factors. In this model, a predominantly nuclear isoform of USP18 seems to regulate gene transcription in a non-canonical fashion. Similar functions of USP18 have been previously described in the context of cancer and involve the association with STAT2, IRF9, and the formation of an active transcription complex that binds to specific IFN-responsive elements [123,124]. Inhibition of either USP18 catalytic activity or scaffolding functions [125] may result in different outcomes as the catalytic activity predominantly affects the removal of ISG15 (deISG15ylation) of protein substrates, most of which are ISGs, whereas scaffolding negatively regulates IFN signaling, thereby possibly affecting cell viability [126–128]. Although future research is required to fully appreciate how USP18 regulates muscle differentiation and maturation, USP18, and its nuclear isoform in particular, might represent an attractive therapeutic target [83].
Remodeling and functional recovery
Ubiquitin-proteasome mediated proteolysis has been associated with muscle regeneration and remodeling [129]. In particular, the expression levels of DUBs appear to be induced after passive leg cycling in persons with spinal cord injury [130], but also more generally after acute tendon/muscle injury [131] as well as other proteases [132]. Muscle atrophy is observed in muscle wasting, cachexia and muscular diseases such as OPMD. Mechanistically, UPS-associated DUBs may play a role, such as POH1/PSMD14, a DUB metalloprotease associated with the 26S proteasome complex and involved in regulating muscle size/proteostasis via the PI3K-PKB/Akt-mTORC1-FoxO pathway [133]. Generally, a key feature is the development and restoration of synaptic connections to the muscle, which is also regulated by ubiquitin-dependent mechanisms [28]. On the other end of the spectrum, possibly different from skeletal muscles, USP25, UCHL1 and OTUB1 were suggested to attenuate pathological hypertrophy in cardiomyocytes by stabilizing SERCA2a, EGFR and DEPTOR, respectively [134–136]. In addition, OTUD1 and USP15 appear to modulate cardiac myocyte remodeling in the context of heart failure, the former by targeting STAT3 [137,138]. Taken together, DUBs have specific roles in regenerating skeletal muscle mass, in particular in NMJ integrity, maintenance and development.
Translational opportunities
The ubiquitin system has long been considered an attractive target area for the treatment of muscle atrophies and wasting diseases, with varying degrees of success. Targeting of disease-causing proteins by using small molecules called proteolysis-targeting chimera (PROTAC) protein degraders that molecularly link E3 ligases with those target proteins, has for example been promising with various PROTACs currently in clinical trials [139]. Recent insights into molecular mechanisms now also reveal many DUBs as potential therapeutic targets, with selective small molecule inhibitors in development [11,140]. In fact, USP1 and USP30 inhibitors are currently being tested in the clinics for the treatment of solid tumors and chronic kidney disease, respectively [141–143]. UCHL1 could also prove a promising therapeutic target, since impaired UCHL1 expression and function have been found in various NMDs (Table 1). However, further investigation is required to pinpoint whether UCHL1 catalytical activity or its scaffolding role or both should be targeted. We recently found transient inhibition of USP18 expression to enhance muscle differentiation, and thereby possibly providing a therapeutically interesting target with regards to muscle recovery, after injury. However, the interplay between the type 1 IFN immune response and the newly-described type 1 IFN-independent role of USP18 [19] in muscle cell differentiation should be clarified first, especially in an in vivo context. Notably, USP19 inhibition could potentially ameliorate maintenance of muscle mass in muscle atrophy, obesity-linked loss of lean muscle mass [144] and sarcopenia, all of which reflect major comorbidity traits in ageing. Alternatively, as an example, stabilization of critical elements of myogenic transcription, such as MyoD, by DUB activation or recruitment-enhanced deubiquitination may also represent a viable strategy to maintain muscle mass. RESTORACs or deubiquitinase-targeting chimera molecules, in contrast with PROTACs, prevent aberrant protein degradation by selectively recruiting DUBs that remove ubiquitin from the respective target proteins and might thereby be considered for muscle wasting conditions [145,146].
In summary, the pivotal role of the ubiquitin system in various aspects of muscle pathologies, and more recent insights into the role of DUBs in these processes offer potential pharmacological inroads for the design and development of novel therapeutics.
Perspectives
Highlight importance of the field: human mutations in DUBs, knockouts in animal models or their altered expression profiles all contribute to skeletal muscle physiology and myopathies that are often associated with aberrant neuronal development, NMDs and inflammation.
Summary of the current thinking: the ubiquitin system has long been considered an attractive target area for the treatment of muscle atrophies and wasting diseases. In addition to opportunities for protein degraders, recent insights into molecular mechanisms increasingly reveal many DUBs as potential therapeutic targets, with selective small molecule inhibitors in development.
Future directions: regarding skeletal muscle and muscle-associated diseases, several DUBs including USP14, USP18, USP19 and UCHL1 represent attractive therapeutic modalities, although not only through inhibition, but also activation (restoring function) mechanisms. Preserving muscle mass and function is relevant in the context of myopathies and NMDs, inflammation, cachexia, cancer and ageing (sarcopenia).
Acknowledgements
We thank members of the Kessler and Raz labs for helpful discussions. Also, we apologize to the many researchers whose work we could not cite here due to space restrictions.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- DMD
Duchenne muscular dystrophy
- DUB
deubiquitinase
- ER
(o)estrogen receptor
- GBS
Guillain-Barré syndrome
- IFN
Interferon
- INKILN
inflammatory MKL1 interacting lncRNA
- ISG
interferon-stimulating gene
- MJD
Machado–Joseph deubiquitinases
- MRF
muscle regulatory transcription factors
- MS
multiple sclerosis
- NMD
neuromuscular dystrophy
- OPMD
oculopharyngeal muscular dystrophy
- OTU
ovarian tumor
- PD
Parkinson's disease
- PROTAC
proteolysis-targeting chimera
- RRMS
relapsing-remitting multiple sclerosis
- SC
satellite cell
- SMA
spinal muscular atrophy
- TDP-43
TAR DNA-binding protein 43
- TLR
Toll-like receptor
- UCH
ubiquitin C-terminal hydrolases
- UPS
ubiquitin proteasome system
- USP
ubiquitin-specific protease
- ZUP1
zinc finger containing ubiquitin peptidase 1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Work in the BMK lab was funded by FORMA Therapeutics, Bayer AG, Pfizer Inc., by an EPSRC grant EP/N034295/1, and by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Science (CIFMS), China [grant number: 2018-I2M-2-002].
Open Access
Open access for this article was enabled by the participation of University of Oxford in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with JISC.
References
- 1.Hershko, A. and Ciechanover, A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 2.Dikic, I. and Schulman, B.A. (2023) An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 24, 273–287 10.1038/s41580-022-00543-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Squair, D.R. and Virdee, S. (2022) A new dawn beyond lysine ubiquitination. Nat. Chem. Biol. 18, 802–811 10.1038/s41589-022-01088-2 [DOI] [PubMed] [Google Scholar]
- 4.Lange, S.M., Armstrong, L.A. and Kulathu, Y. (2022) Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol. Cell 82, 15–29 10.1016/j.molcel.2021.10.027 [DOI] [PubMed] [Google Scholar]
- 5.Clague, M.J., Urbé, S. and Komander, D. (2019) Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 10.1038/s41580-019-0099-1 [DOI] [PubMed] [Google Scholar]
- 6.George, A.J., Hoffiz, Y.C., Charles, A.J., Zhu, Y. and Mabb, A.M. (2018) A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front. Genet. 9, 29 10.3389/fgene.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes, D.C., Goodman, C.A., Baehr, L.M., Gregorevic, P. and Bodine, S.C. (2023) A critical discussion on the relationship between E3 ubiquitin ligases, protein degradation, and skeletal muscle wasting: it's not that simple. Am. J. Physiol. Cell Physiol. 325, C1567–C1582 10.1152/ajpcell.00457.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilodeau, P.A., Coyne, E.S. and Wing, S.S. (2016) The ubiquitin proteasome system in atrophying skeletal muscle: roles and regulation. Am. J. Physiol. Cell Physiol. 311, C392–C403 10.1152/ajpcell.00125.2016 [DOI] [PubMed] [Google Scholar]
- 9.Attaix, D., Combaret, L., Béchet, D. and Taillandier, D. (2008) Role of the ubiquitin-proteasome pathway in muscle atrophy in cachexia. Curr. Opin. Support. Palliat. Care 2, 262–266 10.1097/SPC.0b013e3283196ac2 [DOI] [PubMed] [Google Scholar]
- 10.Sandri, M. (2013) Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 45, 2121–2129 10.1016/j.biocel.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrigan, J.A., Jacq, X., Martin, N.M. and Jackson, S.P. (2018) Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 10.1038/nrd.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbassi-Daloii, T., el Abdellaoui, S., Voortman, L.M., Veeger, T.T., Cats, D., Mei, H.et al. (2023) A transcriptome atlas of leg muscles from healthy human volunteers reveals molecular and cellular signatures associated with muscle location. Elife 12, e80500 10.7554/eLife.80500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dave, H.D, Shook, M. and Varacallo, M. (2023) Anatomy, Skeletal Muscle, pp. 1–9, StatPearls, Treasure Island (FL) [PubMed] [Google Scholar]
- 14.Hughes, D.C., Baehr, L.M., Waddell, D.S., Sharples, A.P. and Bodine, S.C. (2022) Ubiquitin ligases in longevity and aging skeletal muscle. Int. J. Mol. Sci. 23, 7602 10.3390/ijms23147602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peris-Moreno, D., Cussonneau, L., Combaret, L., Polge, C. and Taillandier, D. (2021) Ubiquitin ligases at the heart of skeletal muscle atrophy control. Molecules 26, 407 10.3390/molecules26020407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitajima, Y., Yoshioka, K. and Suzuki, N. (2020) The ubiquitin–proteasome system in regulation of the skeletal muscle homeostasis and atrophy: from basic science to disorders. J. Physiol. Sci. 70, 40 10.1186/s12576-020-00768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wing, S.S. (2013) Deubiquitinases in skeletal muscle atrophy. Int. J. Biochem. Cell Biol. 45, 2130–2135 10.1016/j.biocel.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altun, M., Besche, H.C., Overkleeft, H.S., Piccirillo, R., Edelmann, M.J., Kessler, B.M.et al. (2010) Muscle wasting in aged, sarcopenic rats is associated with enhanced activity of the ubiquitin proteasome pathway. J. Biol. Chem. 285, 39597–39608 10.1074/jbc.M110.129718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olie, C.S., Pinto-Fernández, A., Damianou, A., Vendrell, I., Mei, H., den Hamer, B.et al. (2023) USP18 is an essential regulator of muscle cell differentiation and maturation. Cell Death Dis. 14, 231 10.1038/s41419-023-05725-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, F., Li, Y., Chen, L., Cheng, J., Wu, P., Chu, W.et al. (2018) Characterization of the ubiquitin specific protease (USP) family members in the fast and slow muscle fibers from Chinese perch (Siniperca chuatsi). Gene 677, 1–9 10.1016/j.gene.2018.07.013 [DOI] [PubMed] [Google Scholar]
- 21.Liu, N., Lin, M.-M. and Wang, Y. (2023) The emerging roles of E3 ligases and DUBs in neurodegenerative diseases. Mol. Neurobiol. 60, 247–263 10.1007/s12035-022-03063-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolly, L.A., Kumar, R., Penzes, P., Piper, M. and Gecz, J. (2022) The DUB club: deubiquitinating enzymes and neurodevelopmental disorders. Biol. Psychiatry 92, 614–625 10.1016/j.biopsych.2022.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki, H., Inaba, M., Yamada, M., Uehara, T., Takenouchi, T., Mizuno, S.et al. (2021) Biallelic loss of OTUD7A causes severe muscular hypotonia, intellectual disability, and seizures. Am. J. Med. Genet. A 185, 1182–1186 10.1002/ajmg.a.62054 [DOI] [PubMed] [Google Scholar]
- 24.Rydning, S.L., Backe, P.H., Sousa, M.M.L., Iqbal, Z., Øye, A.-M., Sheng, Y.et al. (2017) Novel UCHL1 mutations reveal new insights into ubiquitin processing. Hum. Mol. Genet. 26, 1031–1040 10.1093/hmg/ddx072 [DOI] [PubMed] [Google Scholar]
- 25.Bilguvar, K., Tyagi, N.K., Ozkara, C., Tuysuz, B., Bakircioglu, M., Choi, M.et al. (2013) Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc. Natl Acad. Sci. U.S.A. 110, 3489–3494 10.1073/pnas.1222732110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, J., Tucci, A., Cipriani, V., Demidov, G., Rocca, C., Senderek, J.et al. (2022) Heterozygous UCHL1 loss-of-function variants cause a neurodegenerative disorder with spasticity, ataxia, neuropathy, and optic atrophy. Genet. Med. 24, 2079–2090 10.1016/j.gim.2022.07.006 [DOI] [PubMed] [Google Scholar]
- 27.Han, K.-J., Foster, D.G., Zhang, N.-Y., Kanisha, K., Dzieciatkowska, M., Sclafani, R.A.et al. (2012) Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron. J. Biol. Chemi. 287, 43741–43752 10.1074/jbc.M112.372318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiAntonio, A., Haghighi, A.P., Portman, S.L., Lee, J.D., Amaranto, A.M. and Goodman, C.S. (2001) Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412, 449–452 10.1038/35086595 [DOI] [PubMed] [Google Scholar]
- 29.Chen, P.-C., Qin, L.-N., Li, X.-M., Walters, B.J., Wilson, J.A., Mei, L.et al. (2009) The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J. Neurosci. 29, 10909 10.1523/JNEUROSCI.2635-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turgut, G.T., Altunoglu, U., Sivrikoz, T.S., Toksoy, G., Kalaycı, T., Avcı, Ş.et al. (2022) Functional loss of ubiquitin-specific protease 14 may lead to a novel distal arthrogryposis phenotype. Clin. Genet. 101, 421–428 10.1111/cge.14117 [DOI] [PubMed] [Google Scholar]
- 31.Bowen, T.S., Schuler, G. and Adams, V. (2015) Skeletal muscle wasting in cachexia and sarcopenia: molecular pathophysiology and impact of exercise training. J. Cachexia Sarcopenia Muscle 6, 197–207 10.1002/jcsm.12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meuwissen, M.E.C., Schot, R., Buta, S., Oudesluijs, G., Tinschert, S., Speer, S.D.et al. (2016) Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 213, 1163–1174 10.1084/jem.20151529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundaram, P., Pang, Z., Miao, M., Yu, L. and Wing, S.S. (2009) USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells. Am. J. Physiol. Endocrinol. Metab. 297, E1283–E1290 10.1152/ajpendo.00409.2009 [DOI] [PubMed] [Google Scholar]
- 34.Combaret, L., Adegoke, O.A.J., Bedard, N., Baracos, V., Attaix, D. and Wing, S.S. (2004) USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am. J. Physiol. Endocrinol. Metab. 288, E693–E700 10.1152/ajpendo.00281.2004 [DOI] [PubMed] [Google Scholar]
- 35.Coyne, E.S., Bedard, N., Wykes, L., Stretch, C., Jammoul, S., Li, S.et al. (2018) Knockout of USP19 deubiquitinating enzyme prevents muscle wasting by modulating insulin and glucocorticoid signaling. Endocrinology 159, 2966–2977 10.1210/en.2018-00290 [DOI] [PubMed] [Google Scholar]
- 36.Coyne, E.S., Bédard, N., Gong, Y.J., Faraj, M., Tchernof, A. and Wing, S.S. (2019) The deubiquitinating enzyme USP19 modulates adipogenesis and potentiates high-fat-diet-induced obesity and glucose intolerance in mice. Diabetologia 62, 136–146 10.1007/s00125-018-4754-4 [DOI] [PubMed] [Google Scholar]
- 37.Berciano, J., Infante, J., García, A., De Pablos, C., Amer, G., Miguel Polo, J.et al. (2006) Stiff man–like syndrome and generalized myokymia in spinocerebellar ataxia type 3. Mov. Disord. 21, 1031–1035 10.1002/mds.20865 [DOI] [PubMed] [Google Scholar]
- 38.Chen, X., Tang, T.-S., Tu, H., Nelson, O., Pook, M., Hammer, R.et al. (2008) Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 3. J. Neurosci. 28, 12713 10.1523/JNEUROSCI.3909-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin, J., Chen, W., Chao, E.S., Soriano, S., Wang, L., Wang, W.et al. (2018) Otud7a knockout mice recapitulate many neurological features of 15q13.3 microdeletion syndrome. Am. J. Hum. Genet. 102, 296–308 10.1016/j.ajhg.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garret, P., Ebstein, F., Delplancq, G., Dozieres-Puyravel, B., Boughalem, A., Auvin, S.et al. (2020) Report of the first patient with a homozygous OTUD7A variant responsible for epileptic encephalopathy and related proteasome dysfunction. Clin. Genet. 97, 567–575 10.1111/cge.13709 [DOI] [PubMed] [Google Scholar]
- 41.Genç, B., Jara, J.H., Schultz, M.C., Manuel, M., Stanford, M.J., Gautam, M.et al. (2016) Absence of UCHL 1 function leads to selective motor neuropathy. Ann. Clin. Transl. Neurol. 3, 331–345 10.1002/acn3.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen, F., Sugiura, Y., Myers, K.G., Liu, Y. and Lin, W. (2010) Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc. Natl Acad. Sci. U.S.A. 107, 1636–1641 10.1073/pnas.0911516107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antony, R., Aby, K., Gao, H., Eichholz, M., Srinivasan, R. and Li, Y. (2022) UCHL1 regulates lipid and perilipin 2 level in skeletal muscle. Front. Physiol. 13, 855193 10.3389/fphys.2022.855193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao, H., Antony, R., Srinivasan, R., Wu, P., Wang, X. and Li, Y. (2020) UCHL1 regulates oxidative activity in skeletal muscle. PLoS One 15, e0241716 10.1371/journal.pone.0241716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagamine, S., Fujiwara, Y., Shimizu, T., Kawata, A., Wada, K., Isozaki, E.et al. (2015) Association of ubiquitin carboxy-terminal hydrolase-L1 in cerebrospinal fluid with clinical severity in a cohort of patients with Guillain–Barré syndrome. Neurol. Sci. 36, 921–926 10.1007/s10072-015-2137-x [DOI] [PubMed] [Google Scholar]
- 46.Lee, Y.-T.C. and Hsu, S.-T.D. (2017) Familial mutations and post-translational modifications of UCH-L1 in Parkinson's disease and neurodegenerative disorders. Curr. Protein Pept. Sci. 18, 733–745 10.2174/1389203717666160217143721 [DOI] [PubMed] [Google Scholar]
- 47.Ng, A.S.L., Tan, Y.J., Lu, Z., Ng, E.Y., Ng, S.Y.E., Chia, N.S.Y.et al. (2020) Plasma ubiquitin C-terminal hydrolase L1 levels reflect disease stage and motor severity in Parkinson's disease. Aging 12, 1488–1495 10.18632/aging.102695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powis, R.A., Mutsaers, C.A., Wishart, T.M., Hunter, G., Wirth, B. and Gillingwater, T.H. (2014) Increased levels of UCHL1 are a compensatory response to disrupted ubiquitin homeostasis in spinal muscular atrophy and do not represent a viable therapeutic target. Neuropathol. Appl. Neurobiol. 40, 873–887 10.1111/nan.12168 [DOI] [PubMed] [Google Scholar]
- 49.Reichelt, J., Sachs, W., Frömbling, S., Fehlert, J., Studencka-Turski, M., Betz, A.et al. (2023) Non-functional ubiquitin C-terminal hydrolase L1 drives podocyte injury through impairing proteasomes in autoimmune glomerulonephritis. Nat. Commun. 14, 2114 10.1038/s41467-023-37836-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLoughlin, H.S., Moore, L.R. and Paulson, H.L. (2020) Pathogenesis of SCA3 and implications for other polyglutamine diseases. Neurobiol. Dis. 134, 104635 10.1016/j.nbd.2019.104635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wrobel, L., Hill, S.M., Ashkenazi, A. and Rubinsztein, D.C. (2021) VCP/p97 modulates PtdIns3P production and autophagy initiation. Autophagy 17, 1052–1053 10.1080/15548627.2021.1898742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao, M.V., Williams, D.R., Cocklin, S. and Loll, P.J. (2017) Interaction between the AAA+ ATPase p97 and its cofactor ataxin3 in health and disease: nucleotide-induced conformational changes regulate cofactor binding. J. Biol. Chem. 292, 18392–18407 10.1074/jbc.M117.806281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Re Cecconi, A.D., Barone, M., Gaspari, S., Tortarolo, M., Bendotti, C., Porcu, L.et al. (2022) The p97-Nploc4 ATPase complex plays a role in muscle atrophy during cancer and amyotrophic lateral sclerosis. J. Cachexia Sarcopenia Muscle 13, 2225–2241 10.1002/jcsm.13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, C.-H., Huang, Y.-C., Chen, P.-Y., Cheng, Y.-J., Kao, H.-H., Pi, H.et al. (2017) USP5/Leon deubiquitinase confines postsynaptic growth by maintaining ubiquitin homeostasis through Ubiquilin. Elife 6, e26886 10.7554/eLife.26886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, C.-H., Chen, G.-C. and Chien, C.-T. (2014) The deubiquitinase Leon/USP5 regulates ubiquitin homeostasis during Drosophila development. Biochem. Biophys. Res. Commun. 452, 369–375 10.1016/j.bbrc.2014.08.069 [DOI] [PubMed] [Google Scholar]
- 56.Ilieva, H., Vullaganti, M. and Kwan, J. (2023) Advances in molecular pathology, diagnosis, and treatment of amyotrophic lateral sclerosis. BMJ 383, e075037 10.1136/bmj-2023-075037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran, N.-N. and Lee, B.-H. (2022) Functional implication of ubiquitinating and deubiquitinating mechanisms in TDP-43 proteinopathies. Front. Cell Dev. Biol. 10, 931968 10.3389/fcell.2022.931968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Boer, E.M.J., Orie, V.K., Williams, T., Baker, M.R., De Oliveira, H.M., Polvikoski, T.et al. (2020) TDP-43 proteinopathies: a new wave of neurodegenerative diseases. J. Neurol. Neurosurg. Psychiatry 92, 86–95 10.1136/jnnp-2020-322983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arai, T., Hasegawa, M., Akiyama, H., Ikeda, K., Nonaka, T., Mori, H.et al. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 10.1016/j.bbrc.2006.10.093 [DOI] [PubMed] [Google Scholar]
- 60.Hans, F., Fiesel, F.C., Strong, J.C., Jäckel, S., Rasse, T.M., Geisler, S.et al. (2014) UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y regulate TDP-43 protein ubiquitination. J. Biol. Chem. 289, 19164–19179 10.1074/jbc.M114.561704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobson-Stone, C., Hallupp, M., Shahheydari, H., Ragagnin, A.M.G., Chatterton, Z., Carew-Jones, F.et al. (2020) CYLD is a causative gene for frontotemporal dementia - amyotrophic lateral sclerosis. Brain 143, 783–799 10.1093/brain/awaa039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie, X., Matsumoto, S., Endo, A., Fukushima, T., Kawahara, H., Saeki, Y.et al. (2018) Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J. Cell Sci. 131, jcs210856 10.1242/jcs.210856 [DOI] [PubMed] [Google Scholar]
- 63.Zhang, T., Periz, G., Lu, Y.-N. and Wang, J. (2020) USP7 regulates ALS-associated proteotoxicity and quality control through the NEDD4L-SMAD pathway. Proc. Natl Acad. Sci. U.S.A. 117, 28114–28125 10.1073/pnas.2014349117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi, M., Kitaura, H., Kakita, A., Kakihana, T., Katsuragi, Y., Onodera, O.et al. (2022) USP10 inhibits aberrant cytoplasmic aggregation of TDP-43 by promoting stress granule clearance. Mol. Cell. Biol. 42, e0039321 10.1128/mcb.00393-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee, B.-H., Lee, M.J., Park, S., Oh, D.-C., Elsasser, S., Chen, P.-C.et al. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregory, J.M., McDade, K., Livesey, M.R., Croy, I., Marion de Proce, S., Aitman, T.et al. (2020) Spatial transcriptomics identifies spatially dysregulated expression of GRM3 and USP47 in amyotrophic lateral sclerosis. Neuropathol. Appl. Neurobiol. 46, 441–457 10.1111/nan.12597 [DOI] [PubMed] [Google Scholar]
- 67.Gu, X., Chen, Y., Wei, Q., Hou, Y., Cao, B., Zhang, L.et al. (2021) Rare CYLD variants in Chinese patients with amyotrophic lateral sclerosis. Front. Genet. 12, 740052 10.3389/fgene.2021.740052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu, S., Wuolikainen, A., Wu, J., Öhman, A., Wingsle, G., Moritz, T.et al. (2019) Targeted multiple reaction monitoring analysis of CSF identifies UCHL1 and GPNMB as candidate biomarkers for ALS. J. Mol. Neurosci. 69, 643–657 10.1007/s12031-019-01411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li, R., Wang, J., Xie, W., Liu, J. and Wang, C. (2020) UCHL1 from serum and CSF is a candidate biomarker for amyotrophic lateral sclerosis. Ann. Clin. Transl. Neurol. 7, 1420–1428 10.1002/acn3.51141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anvar, S.Y., ‘t Hoen, P.A.C., Venema, A., van der Sluijs, B., van Engelen, B., Snoeck, M.et al. (2011) Deregulation of the ubiquitin-proteasome system is the predominant molecular pathology in OPMD animal models and patients. Skelet. Muscle 1, 15 10.1186/2044-5040-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riaz, M., Raz, Y., van Putten, M., Paniagua-Soriano, G., Krom, Y.D., Florea, B.I.et al. (2016) PABPN1-dependent mRNA processing induces muscle wasting. PLoS Genet. 12, e1006031 10.1371/journal.pgen.1006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anvar, S.Y., Tucker, A., Vinciotti, V., Venema, A., van Ommen, G.-J.B., van der Maarel, S.M.et al. (2011) Interspecies translation of disease networks increases robustness and predictive accuracy. PLoS Comput. Biol. 7, e1002258 10.1371/journal.pcbi.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lecker, S.H., Jagoe, R.T., Gilbert, A., Gomes, M., Baracos, V., Bailey, J.et al. (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 18, 39–51 10.1096/fj.03-0610com [DOI] [PubMed] [Google Scholar]
- 74.Wilson, S.M., Bhattacharyya, B., Rachel, R.A., Coppola, V., Tessarollo, L., Householder, D.B.et al. (2002) Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 32, 420–425 10.1038/ng1006 [DOI] [PubMed] [Google Scholar]
- 75.Marshall, A.G., Watson, J.A., Hallengren, J.J., Walters, B.J., Dobrunz, L.E., Francillon, L.et al. (2013) Genetic background alters the severity and onset of neuromuscular disease caused by the loss of ubiquitin-specific protease 14 (Usp14). PLoS One 8, e84042 10.1371/journal.pone.0084042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu, Q., Xu, W.-G., Luo, Y., Han, F.-F., Yao, X.-H., Yang, T.-Y.et al. (2011) Cigarette smoke-induced skeletal muscle atrophy is associated with up-regulation of USP-19 via p38 and ERK MAPKs. J. Cell. Biochem. 112, 2307–2316 10.1002/jcb.23151 [DOI] [PubMed] [Google Scholar]
- 77.Bédard, N., Jammoul, S., Moore, T., Wykes, L., Hallauer, P.L., Hastings, K.E.M.et al. (2015) Inactivation of the ubiquitin-specific protease 19 deubiquitinating enzyme protects against muscle wasting. FASEB J. 29, 3889–3898 10.1096/fj.15-270579 [DOI] [PubMed] [Google Scholar]
- 78.Kim, A., Koo, J.H., Jin, X., Kim, W., Park, S.-Y., Park, S.et al. (2021) Ablation of USP21 in skeletal muscle promotes oxidative fibre phenotype, inhibiting obesity and type 2 diabetes. J. Cachexia Sarcopenia Muscle 12, 1669–1689 10.1002/jcsm.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, P., Zhang, R.-Y., Song, J., Guan, Y.-F., Xu, T.-Y., Du, H.et al. (2012) Loss of AMP-activated protein kinase-α2 impairs the insulin-sensitizing effect of calorie restriction in skeletal muscle. Diabetes 61, 1051–1061 10.2337/db11-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malhotra, S., Bustamante, M.F., Pérez-Miralles, F., Rio, J., Ruiz de Villa, M.C., Vegas, E.et al. (2011) Search for specific biomarkers of IFNβ bioactivity in patients with multiple sclerosis. PLoS One 6, e23634 10.1371/journal.pone.0023634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Navone, N.D., Perga, S., Martire, S., Berchialla, P., Malucchi, S. and Bertolotto, A. (2014) Monocytes and CD4+ T cells contribution to the under-expression of NR4A2 and TNFAIP3 genes in patients with multiple sclerosis. J. Neuroimmunol. 272, 99–102 10.1016/j.jneuroim.2014.04.017 [DOI] [PubMed] [Google Scholar]
- 82.Perga, S., Montarolo, F., Martire, S., Bonaldo, B., Bono, G., Bertolo, J.et al. (2021) Overexpression of the ubiquitin-editing enzyme A20 in the brain lesions of multiple sclerosis patients: moving from systemic to central nervous system inflammation. Brain Pathol. 31, 283–296 10.1111/bpa.12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, Y., Liu, R., Cao, Q., Fan, K., Huang, L., Yu, J.et al. (2019) USP16-mediated deubiquitination of calcineurin A controls peripheral T cell maintenance. J. Clin. Invest. 129, 2856–2871 10.1172/JCI123801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malhotra, S., Morcillo-Suárez, C., Nurtdinov, R., Rio, J., Sarro, E., Moreno, M.et al. (2013) Roles of the ubiquitin peptidase USP18 in multiple sclerosis and the response to interferon-β treatment. Eur. J. Neurol. 20, 1390–1397 10.1111/ene.12193 [DOI] [PubMed] [Google Scholar]
- 85.Ziemkiewicz, N., Hilliard, G., Pullen, N.A. and Garg, K. (2021) The role of innate and adaptive immune cells in skeletal muscle regeneration. Int. J. Mol. Sci. 22, 3265 10.3390/ijms22063265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Forcina, L., Cosentino, M. and Musarò, A. (2020) Mechanisms regulating muscle regeneration: insights into the interrelated and time-dependent phases of tissue healing. Cells 9, 1297 10.3390/cells9051297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Howard, Z.M., Lowe, J., Blatnik, A.J., Roberts, D., Burghes, A.H.M., Bansal, S.S.et al. (2021) Early inflammation in muscular dystrophy differs between limb and respiratory muscles and increases with dystrophic severity. Am. J. Pathol. 191, 730–747 10.1016/j.ajpath.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tulangekar, A. and Sztal, T.E. (2021) Inflammation in duchenne muscular dystrophy-exploring the role of neutrophils in muscle damage and regeneration. Biomedicines 9, 1366 10.3390/biomedicines9101366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cerezo, L.A., Vencovský, J. and Šenolt, L. (2020) Cytokines and inflammatory mediators as promising markers of polymyositis/dermatomyositis. Curr. Opin. Rheumatol. 32, 534–541 10.1097/BOR.0000000000000744 [DOI] [PubMed] [Google Scholar]
- 90.Lundberg, I.E. Fujimoto, M., Vencovsky, J., Aggarwal, R., Holmqvist, M., Christopher-Stine, L.et al. (2021) Idiopathic inflammatory myopathies. Nat. Rev. Dis. Primers 7, 86 10.1038/s41572-021-00321-x [DOI] [PubMed] [Google Scholar]
- 91.Wang, Y., Zheng, Y., Zhao, Y., Liu, Y., Zhang, W., Yu, M.et al. (2022) Comparison of cytokine/chemokine profiles between dermatomyositis and anti-synthetase syndrome. Front. Neurol. 13, 1042580 10.3389/fneur.2022.1042580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Badrising, U.A., Tsonaka, R., Hiller, M., Niks, E.H., Evangelista, T., Lochmüller, H.et al. (2017) Cytokine profiling of serum allows monitoring of disease progression in inclusion body myositis. J. Neuromuscul. Dis. 4, 327–335 10.3233/JND-170234 [DOI] [PubMed] [Google Scholar]
- 93.Vere, G., Alam, M.R., Farrar, S., Kealy, R., Kessler, B.M., O'Brien, D.P.et al. (2022) Targeting the ubiquitylation and ISGylation machinery for the treatment of COVID-19. Biomolecules 12, 300 10.3390/biom12020300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Porritt, R.A. and Hertzog, P.J. (2015) Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 36, 150–160 10.1016/j.it.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 95.Hou, C., Durrleman, C., Periou, B., Barnerias, C., Bodemer, C., Desguerre, I.et al. (2021) From diagnosis to prognosis: revisiting the meaning of muscle ISG15 overexpression in juvenile inflammatory myopathies. Arthritis Rheumatol. 73, 1044–1052 10.1002/art.41625 [DOI] [PubMed] [Google Scholar]
- 96.Lork, M., Verhelst, K. and Beyaert, R. (2017) A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 24, 1172–1183 10.1038/cdd.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Newton, K. and Gitlin, A.D. (2022) Deubiquitinases in cell death and inflammation. Biochem. J. 479, 1103–1119 10.1042/BCJ20210735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bohnert, K.R., Goli, P., Roy, A., Sharma, A.K., Xiong, G., Gallot, Y.S.et al. (2019) The Toll-like receptor/MyD88/XBP1 signaling axis mediates skeletal muscle wasting during cancer cachexia. Mol. Cell. Biol. 39, e00184-19 10.1128/MCB.00184-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parveen, A., Bohnert, K.R., Tomaz da Silva, M., Wen, Y., Bhat, R., Roy, A.et al. (2021) MyD88-mediated signaling intercedes in neurogenic muscle atrophy through multiple mechanisms. FASEB J. 35, e21821 10.1096/fj.202100777RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang, W., Zhao, J., Deng, L., Ishimwe, N., Pauli, J., Wu, W.et al. (2023) INKILN is a novel long noncoding RNA promoting vascular smooth muscle inflammation via scaffolding MKL1 and USP10. Circulation 148, 47–67 10.1161/CIRCULATIONAHA.123.063760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun, L., Si, M., Liu, X., Choi, J.M., Wang, Y., Thomas, S.S.et al. (2018) Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachexia Sarcopenia Muscle 9, 962–974 10.1002/jcsm.12321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang, W. and Hu, P. (2018) Skeletal muscle regeneration is modulated by inflammation. J. Orthop. Transl. 13, 25–32 10.1016/j.jot.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang, Z., Damianou, A., Vendrell, I.et al. (2024) Proximity proteomics reveals UCH-L1 as an essential regulator of NLRP3-mediated IL-1β production in human macrophages and microglia. Cell Rep. 10.1016/j.celrep.2024.114152 [DOI] [PubMed] [Google Scholar]
- 104.Lopez-Castejon, G. (2020) Control of the inflammasome by the ubiquitin system. FEBS J. 287, 11–26 10.1111/febs.15118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang, N., Kny, M., Riediger, F., Busch, K., Schmidt, S., Luft, F.C.et al. (2017) Deletion of Nlrp3 protects from inflammation-induced skeletal muscle atrophy. Intensive Care Med. Exp. 5, 3 10.1186/s40635-016-0115-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boursereau, R., Abou-Samra, M., Lecompte, S., Noel, L. and Brichard, S.M. (2018) Downregulation of the NLRP3 inflammasome by adiponectin rescues Duchenne muscular dystrophy. BMC Biol. 16, 33 10.1186/s12915-018-0501-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liang, Z., Damianou, A., Di Daniel, E. and Kessler, B.M. (2021) Inflammasome activation controlled by the interplay between post-translational modifications: emerging drug target opportunities. Cell Commun. Signal. 19, 23 10.1186/s12964-020-00688-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang, L., Wu, J.-H., Jean-Charles, P.-Y., Murali, P., Zhang, W., Jazic, A.et al. (2023) Phosphorylation of USP20 on Ser334 by IRAK1 promotes IL-1β-evoked signaling in vascular smooth muscle cells and vascular inflammation. J. Biol. Chem. 299, 104911 10.1016/j.jbc.2023.104911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lei, C.-Q., Wu, X., Zhong, X., Jiang, L., Zhong, B. and Shu, H.-B. (2019) USP19 inhibits TNF-α- and IL-1β-triggered NF-κB activation by deubiquitinating TAK1. J Immunol. 203, 259–268 10.4049/jimmunol.1900083 [DOI] [PubMed] [Google Scholar]
- 110.Bonacci, T. and Emanuele, M.J. (2020) Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin. Cancer Biol. 67, 145–158 10.1016/j.semcancer.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ciechanover, A., Breitschopf, K., Hatoum, O.A. and Bengal, E. (1999) Degradation of MyoD by the ubiquitin pathway: regulation by specific DNA-binding and identification of a novel site for ubiquitination. Mol. Biol. Rep. 26, 59–64 10.1023/A:1006964122190 [DOI] [PubMed] [Google Scholar]
- 112.Li, L., Martinez, S.S., Hu, W., Liu, Z. and Tjian, R. (2015) A specific E3 ligase/deubiquitinase pair modulates TBP protein levels during muscle differentiation. Elife 4, e08536 10.7554/eLife.08536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qi, Y., Zuo, Y., Yeh, E.T.H. and Cheng, J. (2014) An essential role of small ubiquitin-like modifier (SUMO)-specific protease 2 in myostatin expression and myogenesis. J. Biol. Chem. 289, 3288–3293 10.1074/jbc.M113.518282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de la Vega, E., González, N., Cabezas, F., Montecino, F., Blanco, N. and Olguín, H. (2020) USP7-dependent control of myogenin stability is required for terminal differentiation in skeletal muscle progenitors. FEBS J. 287, 4659–4677 10.1111/febs.15269 [DOI] [PubMed] [Google Scholar]
- 115.Wing, S.S. (2016) Deubiquitinating enzymes in skeletal muscle atrophy—an essential role for USP19. Int. J. Biochem. Cell Biol. 79, 462–468 10.1016/j.biocel.2016.07.028 [DOI] [PubMed] [Google Scholar]
- 116.Ogawa, M., Kitakaze, T., Harada, N. and Yamaji, R. (2015) Female-specific regulation of skeletal muscle mass by USP19 in young mice. J. Endocrinol. 225, 135–145 10.1530/JOE-15-0128 [DOI] [PubMed] [Google Scholar]
- 117.Ogawa, M., Yamaji, R., Higashimura, Y., Harada, N., Ashida, H., Nakano, Y.et al. (2011) 17β-Estradiol represses myogenic differentiation by increasing ubiquitin-specific peptidase 19 through estrogen receptor α. J. Biol. Chem. 286, 41455–41465 10.1074/jbc.M111.276824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wiles, B., Miao, M., Coyne, E., Larose, L., Cybulsky, A.V. and Wing, S.S. (2015) USP19 deubiquitinating enzyme inhibits muscle cell differentiation by suppressing unfolded-protein response signaling. Mol. Biol. Cell 26, 913–923 10.1091/mbc.E14-06-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hassink, G.C., Zhao, B., Sompallae, R., Altun, M., Gastaldello, S., Zinin N, V.et al. (2009) The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 10, 755–761 10.1038/embor.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harada, K., Kato, M. and Nakamura, N. (2016) USP19-mediated deubiquitination facilitates the stabilization of HRD1 ubiquitin ligase. Int. J. Mol. Sci. 17, 1829 10.3390/ijms17111829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Page, N., Karthikaisamy, V., O'Hara, D., Cranston, A.N., O'Dowd, C.R., Jacq, X.et al. (2023) Abstract LB022: a novel first-in-class USP19 inhibitor for the treatment of cancer-induced muscle atrophy. Cancer Res. 83, LB022 10.1158/1538-7445.AM2023-LB022 [DOI] [Google Scholar]
- 122.Hu, S., Xiang, Y., Qiu, L., Wang, M. and Zhang, Y. (2022) Activation of the membrane-bound Nrf1 transcription factor by USP19, a ubiquitin-specific protease C-terminally anchored in the endoplasmic reticulum. Biochim. Biophys. Acta Mol. Cell Res. 1869, 119299 10.1016/j.bbamcr.2022.119299 [DOI] [PubMed] [Google Scholar]
- 123.Arimoto, K., Löchte, S., Stoner, S.A., Burkart, C., Zhang, Y., Miyauchi, S.et al. (2017) STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol. 24, 279–289 10.1038/nsmb.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Arimoto, K., Miyauchi, S., Troutman, T.D., Zhang, Y., Liu, M., Stoner, S.A.et al. (2023) Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis. Nat. Commun. 14, 251 10.1038/s41467-022-35348-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Campos Alonso, M. and Knobeloch, K.-P. (2024) In the moonlight: non-catalytic functions of ubiquitin and ubiquitin-like proteases. Front. Mol. Biosci. 11, 1349509 10.3389/fmolb.2024.1349509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jové, V., Wheeler, H., Lee, C.W., Healy, D.R., Levine, K., Ralph, E.C.et al. (2024) Type I interferon regulation by USP18 is a key vulnerability in cancer. iScience 27, 109593 10.1016/j.isci.2024.109593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Clancy, A., Rusilowicz-Jones E, V., Wallace, I., Swatek, K.N., Urbé, S. and Clague, M.J. (2023) ISGylation-independent protection of cell growth by USP18 following interferon stimulation. Biochem. J. 480, 1571–1581 10.1042/BCJ20230301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pinto-Fernandez, A., Salio, M., Partridge, T., Chen, J., Vere, G., Greenwood, H.et al. (2021) Deletion of the deISGylating enzyme USP18 enhances tumour cell antigenicity and radiosensitivity. Br. J. Cancer 124, 817–830 10.1038/s41416-020-01167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Taillandier, D., Combaret, L., Pouch, M.-N., Samuels, S.E., Béchet, D. and Attaix, D. (2004) The role of ubiquitin-proteasome-dependent proteolysis in the remodelling of skeletal muscle. Proc. Nutr. Soc. 63, 357–361 10.1079/PAR2004358 [DOI] [PubMed] [Google Scholar]
- 130.Willoughby, D.S., Priest, J.W. and Jennings, R.A. (2000) Myosin heavy chain isoform and ubiquitin protease mRNA expression after passive leg cycling in persons with spinal cord injury. Arch. Phys. Med. Rehabil. 81, 157–163 10.1016/S0003-9993(00)90134-5 [DOI] [PubMed] [Google Scholar]
- 131.Kaokhum, N., Pinto-Fernández, A., Wilkinson, M., Kessler, B.M. and Ismail, H.M. (2022) The mechano-ubiquitinome of articular cartilage: differential ubiquitination and activation of a group of ER-associated DUBs and ER stress regulators. Mol. Cell. Proteomics 21, 100419 10.1016/j.mcpro.2022.100419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Duguez, S., Le Bihan, M.-C., Gouttefangeas, D., Féasson, L. and Freyssenet, D. (2003) Myogenic and nonmyogenic cells differentially express proteinases, Hsc/Hsp70, and BAG-1 during skeletal muscle regeneration. Am. J. Physiol. Endocrinol. Metab. 285, E206–E215 10.1152/ajpendo.00331.2002 [DOI] [PubMed] [Google Scholar]
- 133.Kaiser, M.S., Milan, G., Ham, D.J., Lin, S., Oliveri, F., Chojnowska, K.et al. (2022) Dual roles of mTORC1-dependent activation of the ubiquitin-proteasome system in muscle proteostasis. Commun. Biol. 5, 1141 10.1038/s42003-022-04097-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ye, B., Zhou, H., Chen, Y., Luo, W., Lin, W., Zhao, Y.et al. (2023) USP25 ameliorates pathological cardiac hypertrophy by stabilizing SERCA2a in cardiomyocytes. Circ. Res. 132, 465–480 10.1161/CIRCRESAHA.122.321849 [DOI] [PubMed] [Google Scholar]
- 135.Wang, W., Li, J., Zou, J., Ayala, J., Sun, S.-C. and Su, H. (2021) Abstract 12936: a critical role of deubiquitinase OTUB1 in adaptive cardiac hypertrophy. Circulation 144, A12936 10.1161/circ.144.suppl_1.12936 [DOI] [Google Scholar]
- 136.Bi, H.-L., Zhang, X.-L., Zhang, Y.-L., Xie, X., Xia, Y.-L., Du, J.et al. (2023) The deubiquitinase UCHL1 regulates cardiac hypertrophy by stabilizing epidermal growth factor receptor. Sci. Adv. 6, eaax4826 10.1126/sciadv.aax4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang, M., Han, X., Yu, T., Wang, M., Luo, W., Zou, C.et al. (2023) OTUD1 promotes pathological cardiac remodeling and heart failure by targeting STAT3 in cardiomyocytes. Theranostics 13, 2263–2280 10.7150/thno.83340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Isumi, Y., Hirata, T., Saitoh, H., Miyakawa, T., Murakami, K., Kudoh, G.et al. (2011) Transgenic overexpression of USP15 in the heart induces cardiac remodeling in mice. Biochem. Biophys. Res. Commun. 405, 216–221 10.1016/j.bbrc.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 139.Békés, M., Langley, D.R. and Crews, C.M. (2022) PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 10.1038/s41573-021-00371-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Antao, A.M., Tyagi, A., Kim, K.-S. and Ramakrishna, S. (2020) Advances in deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers (Basel) 12, 1579 10.3390/cancers12061579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cadzow, L., Tobin, E., Sullivan, P., Shenker, S., Nayak, S., Ali, J.et al. (2022) Abstract ND01: KSQ-4279: a first-in-class USP1 inhibitor for the treatment of cancers with homologous recombination deficiencies. Cancer Res. 82, ND01 10.1158/1538-7445.AM2022-ND01 [DOI] [Google Scholar]
- 142.Li, Y., Wu, J., Liu, J., Qin, L., Cai, X., Qiao, J.et al. (2023) Abstract 502: ISM3091, a novel selective USP1 inhibitor as a targeted anticancer therapy. Cancer Res. 83, 502 10.1158/1538-7445.AM2023-502 [DOI] [Google Scholar]
- 143.Simoneau, A., Wu, H.-J., Bandi, M., Lazarides, K., Sun, S., Liu, S.et al. (2023) Abstract 4968: characterization of the clinical development candidate TNG348 as a potent and selective inhibitor of USP1 for the treatment of BRCA1/2mut cancers. Cancer Res. 83, 4968 10.1158/1538-7445.AM2023-4968 [DOI] [Google Scholar]
- 144.Murdock, D.J., Wu, N., Grimsby, J.S., Calle, R.A., Donahue, S., Glass, D.J.et al. (2022) The prevalence of low muscle mass associated with obesity in the USA. Skelet. Muscle 12, 26 10.1186/s13395-022-00309-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Teh, W.P., Zhu, H., Marto, J.A. and Buhrlage, S.J. (2022) DUB to the rescue. Mol. Cell 82, 1411–1413 10.1016/j.molcel.2022.03.039 [DOI] [PubMed] [Google Scholar]
- 146.Peng, Y., Liu, J., Inuzuka, H. and Wei, W. (2023) Targeted protein posttranslational modifications by chemically induced proximity for cancer therapy. J. Biol. Chem. 299, 104572 10.1016/j.jbc.2023.104572 [DOI] [PMC free article] [PubMed] [Google Scholar]