Abstract
ODC (ornithine decarboxylase) is the rate-limiting enzyme in polyamine biosynthesis. Polyamines are essential for cellular growth and differentiation but enhanced ODC activity is associated with cell transformation. Post-translationally, ODC is negatively regulated through members of the antizyme family. Antizymes inhibit ODC activity, promote ODC degradation through the 26 S proteasome and regulate polyamine transport. Besides the ubiquitously expressed antizymes 1 and 2, there is the tissue-specific antizyme 3 and an yet uncharacterized antizyme 4. Antizyme 1 has been shown to be negatively regulated through the AZI (antizyme inhibitor) that binds antizyme 1 with higher affinity compared with ODC. In the present study, we show by yeast two- and three-hybrid protein–protein interaction studies that AZI interacts with all members of the antizyme family and is capable of disrupting the interaction between each antizyme and ODC. In a yeast-based ODC complementation assay, we show that human ODC is able to complement fully the function of the yeast homologue of ODC. Co-expression of antizymes resulted in ODC inhibition and cessation of yeast growth. The antizyme-induced growth inhibition could be reversed by addition of putrescine or by the co-expression of AZI. The protein interactions could be confirmed by immunoprecipitation of the human ODC–antizyme 2–AZI complexes. In summary, we conclude that human AZI is capable of acting as a general inhibitor for all members of the antizyme family and that the previously not yet characterized antizyme 4 is capable of binding ODC and inhibiting its enzymic activity similar to the other members of the antizyme family.
Keywords: antizyme inhibitor (AZI), ornithine decarboxylase (ODC), polyamine, protein interaction
Abbreviations: AD, activation domain; ADH, alcohol dehydrogenase; AZI, antizyme inhibitor; CPRG, chlorophenolred-β-D-galactopyranoside; GPD, glyceraldehyde-3-phosphate dehydrogenase; HA, haemagglutinin; HEK-293 cells, human embryonic kidney 293 cells; ODC, ornithine decarboxylase; SC, synthetic complete
INTRODUCTION
The polyamines putrescine, spermidine and spermine are abundant multivalent organic cations. They are largely bound to negatively charged molecules such as RNA and DNA [1]. Polyamines are essential for normal cell growth and differentiation but they are also known to play an important role in the regulation of cell proliferation and in the development of cancer. ODC (ornithine decarboxylase) is a key enzyme of polyamine biosynthesis. Elevated ODC activity is associated with neoplastic transformation caused by oncogenic Ras [2], v-Src [3], activated RhoA [4] or overexpression of the eukaryotic initiation factor 4E [5]. In addition, overexpression of ODC alone leads to neoplastic transformation of NIH/3T3 cells [6–8]. Inhibition of ODC activity can reverse the transformed phenotype. Therefore ODC activity as well as polyamine levels are tightly regulated at the level of transcription [9,10], translation [11] and post-translation [12].
At the post-translational level, ODC is regulated through a family of at least four inhibitory proteins called antizymes [13]. Antizymes bind ODC with high affinity and form ODC–antizyme heterodimers, thereby preventing the formation of the enzymically active ODC homodimer. Increased polyamine levels induce functional expression of antizymes through a unique ribosomal frameshift mechanism [14]. Antizyme 1 promotes the degradation of ODC through the 26 S proteasome in an ubiquitin-independent manner [15]. In addition, antizyme 1 also inhibits polyamine uptake and stimulates polyamine excretion [16,17]. Antizyme 2 has the same tissue distribution as antizyme 1 but is less abundantly expressed [18]. Although it is structurally similar to antizyme 1, it does not promote ODC degradation in vitro but inhibits, similar to antizyme 1, the cellular uptake of polyamines [19]. Antizyme 3 is testes-specific and its expression is restricted to certain stages of spermatogenesis [20]. A putative fourth member of the antizyme family, antizyme 4, was originally isolated from a human brain cDNA library and is functionally not characterized yet [13].
Antizyme 1 has been shown to act as a negative regulator of cell growth and possibly as a tumour suppressor. It has been demonstrated that overexpression of antizyme 1 leads to the inhibition of cell proliferation and to cell-cycle arrest [21,22]. Furthermore, antizyme 1 overexpression suppresses tumour growth in different mouse cancer models [23,24]. The overall decrease in polyamine levels through antizyme 1 has been regarded as the main reason for the inhibition of cell growth. However, the exact mechanism by which antizyme 1 influences the cell cycle has remained unclear.
The function of antizyme 1 is known to be modulated by an inhibitory protein, called AZI antizyme inhibitor. Binding of AZI to antizyme 1 prevents the binding of ODC to antizyme 1, thereby activating ODC [25]. However, the interactions between AZI and other members of the mammalian antizyme family have not yet been studied. AZI transcripts are quickly up-regulated on growth stimulation through serum or phorbol ester with kinetics that are much earlier than ODC transcripts [26], suggesting a possible role for AZI in cell-cycle progression. Furthermore, a study for differentially expressed genes in gastric cancer showed that AZI is up-regulated in gastric cancer when compared with normal tissue [27].
Saccharomyces cerevisiae yeast strains mutated in the endogenous ODC gene (SPE1) cannot synthesize putrescine and consequently fail to produce spermidine or spermine. As a consequence, deletion of the SPE1 gene leads to intracellular polyamine depletion and renders yeast growth of mutant cells dependent on exogenous polyamines [28]. In the present study, we show that human ODC fully complements the SPE1 deletion in yeast cells. Therefore we have been able to use a yeast system to monitor the influence of human antizymes or AZI on the activity of ODC.
Here we show that human AZI is capable of binding to all members of the antizyme family and thereby release active ODC from each antizyme complex. Taken together, these results suggest that AZI is a general inhibitor of antizyme function and might contribute to the high ODC activity and increased polyamine levels seen in most cases of neoplastic transformation.
MATERIALS AND METHODS
Materials
Oligonucleotides for cloning and sequencing were purchased from Metabion (Martinsried, Germany).
Culture of yeast cells
Yeast media and culture conditions were as described previously [29]. Yeast cells were grown at 30 °C in either YPD (yeast extract–phosphate–dextrose)-rich medium or polyamine-free SC (synthetic complete) minimal medium [0.67% yeast nitrogen base (Difco), 2% glucose or galactose and amino acids, except those required for selection]. Transformation of yeast cells was performed by the lithium acetate method [30]. After transformation, yeast cells were plated on to polyamine-free SC medium to deplete the cells of endogenous polyamines. The SC medium was sterilized by filtering and agar was replaced by agarose to avoid polyamine contamination.
For growth measurements on plates, transformants were grown overnight in selective polyamine-free SC medium containing glucose. The overnight culture was then adjusted to an absorbance A600 3.0/ml and serial dilutions starting with A600 3.0/ml were spotted in 2 μl aliquots on to polyamine-free selective galactose plates or selective galactose plates containing 7.5 μM putrescine. The plates were then incubated for 3 days at 30 °C. Growth curves were measured in liquid culture by determining A600.
For protein expression analysis, transformants were grown in selective SC medium containing galactose to induce expression of the fusion proteins. The cells were resuspended in 5 ml at A600 0.2 and grown to mid- to late-exponential phase (A600=1–2). These cultures were diluted to A600 0.15. Final cultures were harvested at early-exponential phase (A600=0.5–0.7).
Strains and plasmids
The wild-type strain W303-1a (MATa ade2, ura3, his3, trp1, leu2, can1) was obtained from A.T.C.C. Strain EGY48 (MATα his3, trp1, ura3, leu2::2 LexAop-LEU2) was purchased from OriGene Technologies (Rockville, MD, U.S.A.). The spe1 deletion strain Y15034 (MATα his3, leu2, lys2, ura3, spe1Δ::kanMX4) and the corresponding wild-type strain Y10000 (matα, his3, leu2, lys2, ura3) were purchased from Euroscarf (Frankfurt, Germany). The centromeric yeast expression plasmids p413ADH (where ADH stands for alcohol dehydrogenase), p413GALL, p415ADH, p415GALL, p415GPD (where GPD stands for glyceraldehyde-3-phosphate dehydrogenase) or the 2μ plasmids p426ADH, p426GAL1 and p426GPD [31,32] were obtained from M. Funk (University Marburg, Marburg, Germany). The two-hybrid plasmids pEG202 and pJG4-5 and the LacZ (gene encoding β-galactosidase) reporter plasmid pRB1840 were part of the DupLex-A Yeast Two-Hybrid System purchased from OriGene Technologies (Rockville, MD, U.S.A.). The mammalian expression vector pcDNA3.1(+) was from Invitrogen.
The cDNA encoding ODC was amplified by PCR from a human liver cDNA library (OriGene Technologies) using the oligodeoxynucleotide primers listed in Table 1 (the EcoRI and XhoI sites are underlined). The PCR products were cloned into the EcoRI and XhoI sites of the yeast expression vectors p413GALL and p413ADH, the mammalian expression vector pcDNA3.1(+) and the yeast two-hybrid vectors pJG4-5 and pEG202 to yield p413GALL-ODC, p413ADH-ODC, pcDNA3.1-ODC, pJG-ODC and pEG202-ODC respectively.
Table 1. Nucleotide sequence of primers and vectors used for cloning of ODC, antizymes and AZI.
| Protein | Primer | Cloned into vector |
|---|---|---|
| ODC | Sense: 5′-GGAGAATTCATGAACAACTTTGGTAATGAAGAGTTTGACTGC-3′ | pEG202 p413GALL p413ADH pcDNA3.1(+) |
| Antisense: 5′-TAGCTCGAGCTACACATTAATACTAGCCGAAGCACAGGCTGC-3′ | ||
| AZ1 | Sense: 5′-GAGGAATTCATGGTGAAATCCTCCCTGCAGCG-3′ | pJG4-5, pEG202, p426ADH, p426GAL1, p426GPD |
| Antisense: 5′-GCACTCGAGCTACTCCTCCTCCTCTCCCGAAGACTCTCTC-3′ | ||
| Overlap extension: (A) 5′-GGTGCTCC-GATGCCCCTCACC-3′ | ||
| (B) 5′-GGTGAGGGGCATC-GGAGCACC-3′ | ||
| AZ2 | Sense: 5′-GAGGAATTCATGATAAACACCCAGGACAGTAGTATTTTGCC-3′ | pJG4-5 pEG202 p426GAL1 p426GPD |
| Antisense: 5′-GCACTCGAGTTAGTCCTCATCGGACAAGTTCTGGTC-3′ | ||
| Overlap extension: (A) 5′-GTGGTGCTCC-GATGCCCCTCAC-3′ | ||
| (B) 5′-GTGAGGGGCATC-GGAGCACCAC-3′ | ||
| HA tag, sense: 5′-GGAGAATTCATGTACCCTTATGATGTGCCAGATTATGCCTCT | pcDNA3.1(+) | |
| ATAAACACCCAGGACAGTAGTATTTTGCC-3′ | ||
| AZ3 | Sense: 5′-GAGGAATTCATGCTGCCTCGTTGTTATAAAAGCATC-3′ | pJG4-5 pEG202 p426GAL1 p426GPD |
| Antisense: 5′-GCACTCGAGTCAAGGAGGCTCACTGGGCAGG-3′ | ||
| Overlap extension: (A) 5′-TCCAGTGCTC-TGAGTCCCTAG-3′ | ||
| (B) 5′-CTAGGGACTCA-GAGCACTGGA-3′ | ||
| AZ4 | Sense: sense primer antizyme 1 | pJG4-5 pEG202 p426GPD |
| Antisense: 5′-GCACTCGAGCTAGCCCTGCAGCGAGTAGG-3′ | ||
| Overlap extension: (A) 5′-CGGTGGTGCTC-TGATGTCCCT-3′ | ||
| (B) 5′-AGGGACATCA-GAGCACCACCG-3′ | ||
| AZI | Sense (EcoRI): 5′-GGAGAATTCATGAAAGGATTTATTGATGATGCAAACTACTCCG-3′ | pJG4-5 p415GALL p415GPD |
| Sense (SpeI): 5′-GGAACTAGTATGAAAGGATTTATTGATGATGCAAACTACTCCG-3′ | ||
| Antisense: 5′-TAGCTCGAGTTAAGCTTCAGCGGAAAAGCTGTCTTCTTGCG-3′ | ||
| Myc tag, sense: 5′-GATGGATCCATGGAACAGAAGTTGATTTCCGAAGAAGACCTC | pcDNA3.1(+) | |
| ATGAAAGGATTTATTGATGATGCAAACTACTCCG-3′ |
The cDNAs encoding antizymes 1 and 2 were amplified from a human liver cDNA library (OriGene Technologies). For the amplification of human antizyme 3, a clone from Incyte (Palo Alto, CA, U.S.A.) was used as template. Human antizyme 4 was amplified using the IMAGE clone 1540076 as template. To express full-length functional antizymes, the antizyme-specific stop codon of the ribosomal frame-shifting site was deleted by site-directed mutagenesis using the overlap extension procedure [33]. Briefly, complementary primers [primer (A) and (B) in Table 1] were combined with sense and antisense primers to generate two DNA fragments with complementary sequences at the ends by PCR. These two PCR products were mixed and the single-stranded complementary DNA sequences anneal and act as primers for each other. The resulting fusion product was amplified further by PCR using sense and antisense primers. Deletions were introduced by incorporating the appropriate nucleotide changes into the overlapping primers. The nucleotide deletion within the sequence of each complementary primer is indicated by a hyphen in Table 1. The resulting antizyme fragments were cloned into the EcoRI and XhoI sites of the yeast expression vectors p426ADH, p426GAL1 and p426GPD and the two-hybrid vectors pEG202 and pJG4-5 to yield the corresponding constructs.
The N-terminal HA (haemagglutinin) tag was introduced to antizyme 2 by PCR. The sequence for the HA epitope was added in front of the antizyme 2 sense primer (Table 1). HA-tagged antizyme 2 was cloned into the EcoRI and XhoI sites of pcDNA3.1(+).
The cDNA encoding AZI was amplified from a human liver cDNA library (OriGene Technologies) using the primers listed in Table 1 (the EcoRI, SpeI and XhoI sites are underlined) and cloned into the EcoRI and XhoI sites of pJG4-5 to yield pJG-AZI and the SpeI and XhoI sites of the yeast expression vectors p415GALL and p415GPD to yield plasmids p415GALL-AZI and p415GPD-AZI respectively. The N-terminal Myc tag of AZI was introduced by PCR using the Myc tag sense primer (the BamHI site is underlined) combined with the antisense primer (Table 1). The PCR fragment was cloned into pcDNA3.1(+), using BamHI and XhoI. All constructs were checked by sequencing.
Yeast two- and three-hybrid assays
To determine the β-galactosidase activity on solid medium, yeast transformants were transferred to SC selection plates, supplemented with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal, 100 μg/ml) buffered with 25 mM Na2HPO4/25 mM NaH2PO4.
To determine β-galactosidase activity in liquid culture, the chromogenic substrate CPRG (Chlorophenol Red-β-D-galactopyranoside; Indianapolis, IN, U.S.A.) was used. Yeast cells were diluted in SC selection medium (A600=0.05–0.1/ml) in a 96-well flat-bottomed microplate (100 μl/well). Cells were incubated for 6 h at 30 °C under constant shaking. Subsequently, 50 μl of the CPRG assay mixture (0.15 mg/ml digitonin, 0.3 mg/ml CPRG and 0.3 M sodium phosphate, pH 6.7) was added to each well and cells were further incubated at 30 °C under constant shaking. After 24 h, absorption was determined in a plate reader at 574 nm.
Cell culture and transient transfection
HEK-293 cells were grown in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% (v/v) fetal calf serum and penicillin/streptomycin (100 units in 100 μg/ml) in an atmosphere of 5% CO2. Cells were transfected using the calcium phosphate precipitation method [34] and incubated for an additional 48 h after transfection. Equal amounts of DNA were used for transfection. Lactacystin (Sigma) was added to the medium (final concentration, 8 μM) 16 h before the cells were harvested.
Cell lysis and immunoprecipitation
Yeast cells were lysed using Y-PER protein extraction reagent (Pierce, Rockford, IL, U.S.A.). Cells were pelleted (3000 g, 5 min, 4 °C) and the pellet was resuspended with the appropriate amount of Y-PER. The mixture was agitated at room temperature (21–23 °C) for 20 min and the cell debris was centrifuged at 14000 g for 10 min at 4 °C. The supernatant was reserved for protein concentration determination (bicinchoninic acid protein assay; Pierce) and Western-blot analyses.
HEK-293 cells were washed with cold PBS (0.2 g NaH2PO4, 1.15 g Na2HPO4, 8 g/l NaCl, 0.2 g/l KCl, pH 7.2) and lysed in cold lysis buffer (50 mM Hepes, 150 mM NaCl, 1% Triton X-100, pH 7.5) containing one tablet of the protease inhibitor Complete (Roche) per 25 ml of lysis buffer. The cell debris was centrifuged (14000 g, 10 min, 4 °C) and the supernatant was reserved for protein concentration determination (bicinchoninic acid protein assay; Pierce). The lysates were then subjected to immunoprecipitation and Western-blot analyses.
For the immunoprecipitation, an anti-HA affinity matrix was used with monoclonal anti-HA antibody (clone 3F10) covalently coupled with the agarose beads (Roche). All steps were performed at 4 °C. Aliquots (50 μl) of resuspended anti-HA affinity beads were washed twice with 1 ml of cold lysis buffer. Subsequently, 500 μl of cold lysate was added to the beads with the amount of total protein adjusted to 50 μg with lysis buffer. Samples were incubated on a rocker for 45 min. The beads were pelleted by centrifugation and the supernatants were carefully removed. The beads were washed three times with 1 ml of lysis buffer, solubilized in LDS sample buffer (Novex/Invitrogen) and the supernatant was analysed by gel electrophoresis and Western blotting.
Immunoblot assay of ODC, antizyme and AZI
Proteins were separated on 4–12% NuPAGE Bis-Tris gels using Mes/SDS running buffer (Novex/Invitrogen). After electrophoresis, proteins were transferred on to a PVDF membrane (Immobilon-P; Millipore). The membrane was blocked with TBST (50 mM Tris/HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) containing 8% dry milk for 1 h at room temperature. Proteins were detected with either peroxidase-conjugated anti-HA (clone 3F10, 1:500; Roche) or anti-HA 12CA5 (1:500; Roche), peroxidase-conjugated anti-Myc (clone 9E10, 1:500; Roche), anti-ODC (1:1000; Progen) or anti-LexA (1:250; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). As a secondary antibody, peroxidase-conjugated anti-rabbit or anti-mouse was used (1:10000; Pierce). Proteins were visualized using ECL®Plus enhanced chemiluminescence (Amersham Biosciences).
RESULTS
Protein–protein interactions between ODC, antizymes and AZI
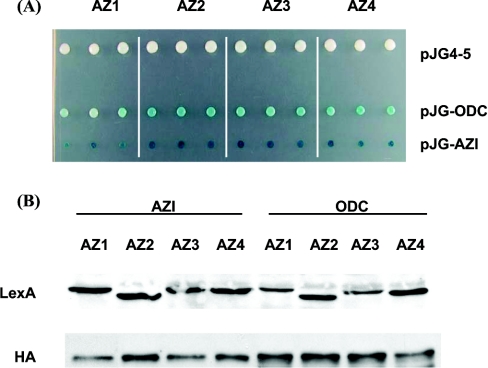
We have used the yeast two-hybrid system to analyse protein–protein interactions within the ODC–antizyme–AZI complex. Using this approach, we first examined whether these proteins showed self-activation when coupled with the C-terminus of the DNA-binding domain of LexA or the B42 AD (activation domain). None of these proteins were found to be self-active when fused to the B42 AD (results not shown). Moreover, when coupled with the LexA DNA-binding domain neither ODC nor antizymes 1–4 (Figure 1A) showed self-activation. However, we found strong self-activation when AZI was fused to the DNA-binding domain (results not shown). Therefore, we used AZI or ODC coupled with the AD and the antizyme constructs fused to the DNA-binding domain for the two-hybrid interaction analysis.
Figure 1. Two-hybrid interaction assay for ODC, antizymes and AZI.
(A) Two-hybrid interactions between ODC, antizyme and AZI. The yeast two-hybrid strain EGY48 was transformed with plasmid pRB1840 (LacZ reporter), plasmid pEG202-AZ1, pEG202-AZ2, pEG202-AZ3 or pEG202-AZ4 (antizymes 1–4 respectively fused to the DNA-binding domain of LexA) and plasmid pJG4-5 (empty vector) or pJG-ODC or pJG-AZI (ODC or AZI respectively fused to the B42 AD). Transformants were grown on selective glucose medium and then analysed for reporter gene activation (LacZ) on selective galactose plates containing X-gal. Results are representative for multiple experiments with each interaction analysed in triplicate. (B) Expression of fusion proteins in two-hybrid interaction assay. The yeast two-hybrid strain EGY48 was transformed with antizyme–LexA fusion constructs (pEG202-AZ1, -AZ2, -AZ3, -AZ4 respectively) and co-transformed with either pJG-AZI or pJG-ODC AD vectors plus the LacZ reporter pRB1840. Protein expression of the AD-fusion proteins was induced by growing the transformants in galactose-containing medium. Equal amounts of protein were loaded in each lane, and expression levels of the LexA and AD-fusion proteins were monitored by immunoblot as described in the Materials and methods section using either anti-LexA or anti-HA antibodies. The immunoblot shown is representative for multiple experiments.
As illustrated in Figure 1(A), we were able to detect interactions between each of the antizyme constructs with ODC or AZI suggesting that all antizymes, including the putative antizyme 4, are capable of interacting with both ODC and AZI. Our analysis indicates, as illustrated by the intensity of the yeast colony staining (Figure 1A), that the interaction between AZI and the antizymes was stronger than the interaction between ODC and the antizymes although the expression levels of ODC and AZI were found to be equal (Figure 1B). These results are consistent with the current model that AZI binds to antizyme 1 with higher affinity than ODC, thereby preventing the formation of the antizyme–ODC complex.
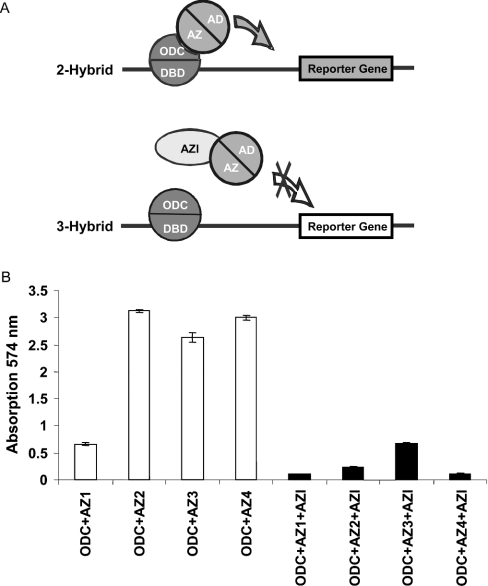
To analyse whether AZI is capable of competing with the binding of each antizyme to ODC, we then designed a yeast three-hybrid system.
As schematically described in Figure 2(A), we co-expressed AZI in the presence of ODC and antizymes 1–4. Since an additional selection marker was needed, we chose the yeast expression strain W303-1a. We found that co-expression of AZI blocked the interaction of ODC with each of the four antizymes (Figure 2B), suggesting that AZI is capable of disrupting the interactions between each member of the antizyme family and ODC. Interestingly, the interaction between antizyme 1 and ODC seemed to be weaker than the interaction between ODC and the other antizymes. However, it cannot be excluded that this is due to the expression level of the antizyme fusion proteins.
Figure 2. Three-hybrid interaction for ODC, antizymes and AZI.
(A) Schematic representation of the three-hybrid assay system for ODC, antizyme and AZI. The two-hybrid interaction between ODC and antizyme leads to the activation of a reporter gene. Co-expression of AZI disrupts this interaction and thereby prevents the activation of the reporter gene. AD, activation domain; DBD, DNA-binding domain. (B) Inhibition of ODC–antizyme interaction by AZI. W303-1a cells were transformed with plasmid pRB1840 (LacZ reporter), plasmid pEG202-ODC (ODC fused to the DNA-binding domain of LexA), the plasmids pJG-AZ1, pJG-AZ2, pJG-AZ3 or pJG-AZ4 (antizymes 1–4 respectively fused to the B42 AD), and either plasmid p415GPD (white bars) or p415GPD-AZI carrying AZI behind the GPD1 promoter (black bars). Transformants were grown on selective plates, and then analysed for reporter gene activation (LacZ) using the CPRG assay as described in the Materials and methods section. Data represent means±S.D. values for four independent experiments.
Functional expression of human ODC, antizyme and AZI in yeast
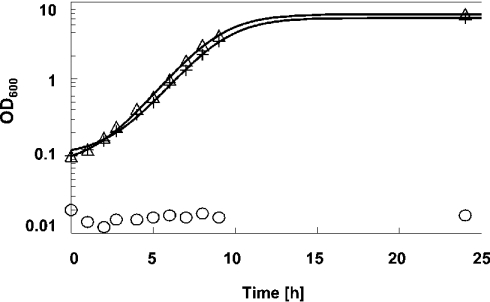
S. cerevisiae yeast cells deleted for the endogenous ODC gene (SPE1) are not capable of synthesizing polyamines. Since polyamines are essential for cell growth, these mutant cells do not grow in polyamine-free medium and arrest under polyamine-depleted conditions as large and mis-shaped cells [28]. We found that expression of human ODC can fully complement this phenotype and is sufficient to restore both the growth (Figure 3) and morphology defects (results not shown) of spe1 deleted cells. Moreover, the mutant phenotype associated with spe1 deletion was relieved by the addition of putrescine to the medium (results not shown), strongly suggesting that the growth defect is mainly caused by polyamine depletion.
Figure 3. Complementation of the growth defect of spe1 deleted yeast cells by human ODC.
Wild-type yeast cells (strain Y10000) were transformed with the control plasmid p413ADH (+), isogenic spe1 deleted cells (strain Y15034) were transformed with either the control plasmid p413ADH (O) or plasmid p413ADH-ODC carrying human ODC behind the yeast ADH1 promoter (Δ). Yeast cell growth in polyamine-free, selective glucose medium was followed by determination of A600. The data shown are representative of several experiments.
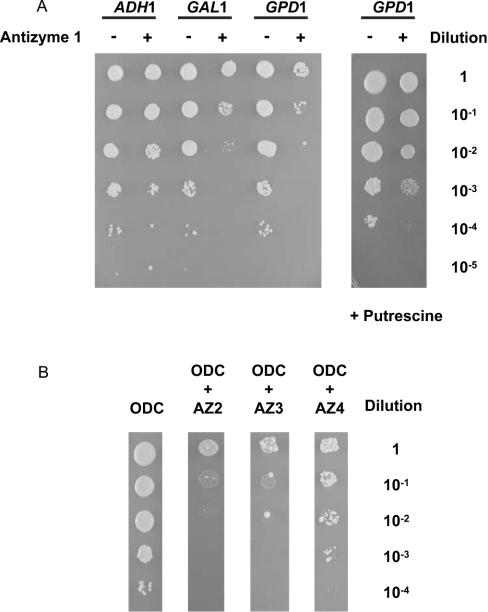
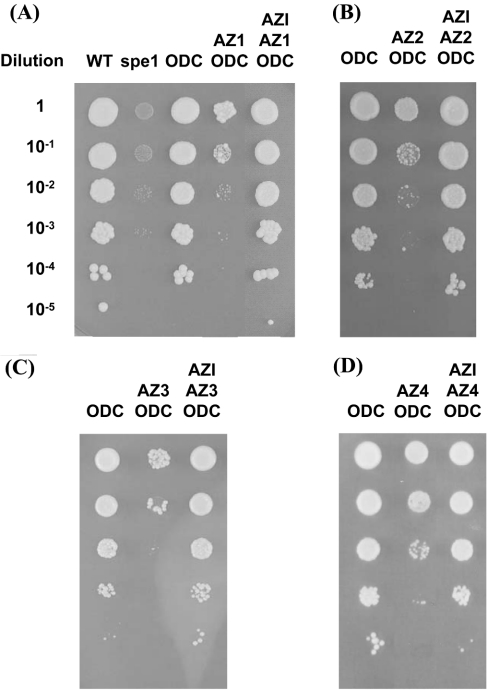
For the functional characterization of the ODC–antizyme–AZI complex, we made use of the capability of human ODC to complement the growth defect of spe1 deleted cells and designed an assay system where ODC is co-expressed with antizyme or antizyme and AZI. Overexpression of antizyme 1 prevented the ODC-mediated complementation of the growth defect (Figure 4A). The extent of growth inhibition was consistent with the strength of the yeast promoter that was used to drive the expression of antizyme 1 (Figure 4A). The growth defect of cells expressing both human ODC and antizyme 1 was relieved by the addition of putrescine (Figure 4A), suggesting that the detrimental effect of antizyme 1 overexpression on spe1 deleted cells expressing human ODC is mainly caused by polyamine depletion. Moreover, we found that the other members of the antizyme family, antizymes 2–4, were also able to inhibit yeast cell growth (Figure 4B). The experiments were repeated several times to confirm inhibition of ODC by antizymes. Taken together, these results suggest that all members of the antizyme family, including the not yet characterized antizyme 4, can be functionally expressed in yeast and block ODC activity.
Figure 4. Functional expression of antizymes in yeast.
(A) Functional expression of antizyme 1 in yeast. Yeast cells deleted for spe1 (strain Y15034) were transformed with plasmid p413GALL-ODC (carrying human ODC behind the yeast GALL promoter) and either the control plasmids p426ADH (–), p426GAL1 (–) and p426GPD (–) or plasmids p426ADH-AZ1 (+), p426GAL1-AZ1 (+) or p426GPD-AZ1 (+) (carrying human antizyme 1 behind the yeast ADH1, GAL1 and GPD1 promoters respectively). Liquid cultures were grown overnight in selective polyamine-free glucose medium. The overnight cultures were adjusted to A600 3.0/ml and serial dilutions starting with 3.0/ml were spotted (2 μl aliquots) as indicated on polyamine-free selective galactose plates (left panel) or selective galactose plates containing 7.5 μM of putrescine (right panel). The plates were then incubated for 3 days at 30 °C. The colony assays shown are representative for multiple experiments. (B) Functional expression of antizymes 2–4 in yeast. Yeast cells deleted for spe1 (strain Y15034) were transformed with plasmid p413GALL-ODC alone (lane 1, carrying human ODC behind the yeast GALL promoter) or co-transformed with plasmid p426GAL1-AZ2 (lane 2, carrying antizyme 2 behind the yeast GAL1 promoter), p426GAL1-AZ3 (lane 3, carrying antizyme 3 behind the yeast GAL1 promoter) or p426GPD-AZ4 (lane 4, carrying antizyme 4 behind the yeast GPD1 promoter). Liquid cultures were grown overnight in selective polyamine-free glucose medium. The overnight culture was adjusted to A600 3.0/ml and serial dilutions were spotted (2 μl of aliquots) as indicated on polyamine-free selective galactose plates. The plates were then incubated for 3 days at 30 °C. The results shown are representative for multiple experiments.
We next tested if the antizyme-mediated growth defect of spe1 mutants can be relieved by the co-expression of human AZI. At the amino acid level, AZI is highly homologous with ODC [25]. Therefore we first addressed if AZI by itself has any ODC-like activity in the yeast system. Spe1 cells transfected with AZI constructs alone were not able to grow on polyamine-deficient medium even if AZI was expressed under the strong GPD promoter (results not shown). Thus AZI did not show any ODC-like activity in spe1 cells. However, we found that AZI could promote cell growth through inactivation of antizymes.
As shown in Figure 5, we found in the complementation assay that AZI was capable of relieving the inhibition of ODC caused by co-expression of either antizymes 1, 2, 3 or 4 (Figure 5). In these experiments, expressions of both AZI and ODC were under control of the galactose-inducible GALL promoter. Interestingly, spe1 mutants, which were co-expressing ODC, antizyme and AZI showed identical growth behaviour as spe1 cells complemented with ODC alone or the isogenic wild-type cells (Figure 5A). Spe1 cells, which co-expressed ODC and antizyme alone showed severe growth inhibition confirming the results seen previously (Figure 4B).
Figure 5. Inhibition of antizyme activity by AZI in yeast.
(A) Wild-type (WT) yeast cells (strain Y10000, lane 1) or cells deleted for spe1 (strain Y15034, lane 2) were transformed with the control plasmids p413GALL, p426GPD and p415GALL. Cells deleted for spe1 (strain Y15034) were transformed with plasmid p413GALL-ODC (carrying human ODC behind the GALL promoter) and the control plasmids p426GPD and p415GALL (lane 3) or transformed with p413GALL-ODC and plasmid p426GPD-AZ1 (carrying human antizyme 1 behind the GPD1 promoter) and the control plasmid p415GALL (lane 4) or transformed with plasmids p413GALL-ODC, p426GPD-AZ1 and p415GALL-AZI (carrying human AZI behind the GALL promoter, lane 5). (B) Cells deleted for spe1 (strain Y15034) were transformed with either p413GALL-ODC and the control plasmids p426GPD and p415GALL (lane 1) or p413GALL-ODC and plasmid p426GPD-AZ2 (carrying human antizyme 2 behind the yeast GPD1 promoter) and the control plasmid p415GALL (lane 2) or plasmids p413GALL-ODC, p426GPD-AZ2 and p415GALL-AZI (lane 3). (C) Cells deleted for spe1 (strain Y15034) were transformed with either p413GALL-ODC and the control plasmids p426GPD and p415GALL (lane 1) or p413GALL-ODC and plasmid p426GPD-AZ3 (carrying human antizyme 3 behind the yeast GPD1 promoter) and the control plasmid p415GALL (lane 2) or plasmids p413GALL-ODC, p426GPD-AZ3 and p415GALL-AZI (lane 3). (D) Cells deleted for spe1 (strain Y15034) were transformed with either p413GALL-ODC and the control plasmids p426GPD and p415GALL (lane 1) or p413GALL-ODC, plasmid p426GPD-AZ4 (carrying human antizyme 4 behind the GPD1 promoter) and the control plasmid p415GALL (lane 2) or plasmids p413GALL-ODC, p426GPD-AZ4 and p415GALL-AZI (lane 3). Liquid cultures were grown overnight in selective polyamine-free glucose medium. The overnight cultures were adjusted to A600 3.0/ml and 2 μl aliquots of the overnight culture were spotted at the indicated serial dilutions on to polyamine-free selective galactose plates. The plates were then incubated for 3 days at 30 °C. The results shown are representative for multiple experiments.
Association of ODC, antizyme 2 and AZI in HEK-293 cells
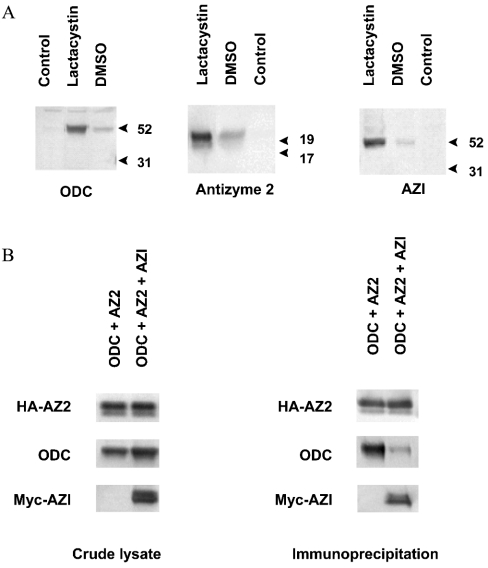
To confirm the interaction analyses performed in yeast in a mammalian system, ODC, antizyme 2 and AZI were transiently expressed in HEK-293 cells and the associations were examined by immunoprecipitation. We confirmed using the yeast complementation assay that the N-terminal introduction of an HA tag to antizyme or a Myc tag to AZI did not change the activity of the corresponding proteins (results not shown). Consistent with the finding that ODC is degraded through the 26 S proteasome [15], the expression of ODC could be stabilized by the addition of the proteasome inhibitor lactacystin to the cell culture (Figure 6A). Similarly, lactacystin was capable of stabilizing the expression of antizyme 2 (Figure 6A). These findings support the recent report that antizyme 1 is rapidly degraded through a mechanism that requires functional ubiquitin-dependent proteolytic activity [35]. Moreover, we also found a stabilizing effect of lactacystin on the expression of AZI (Figure 6A), suggesting that not only the degradation of ODC and antizyme family members but also that of AZI is regulated by a proteasomal mechanism.
Figure 6. Association of ODC antizyme 2 and AZI in HEK-293 cells.
(A) Stabilizing effect of lactacystin on ODC, antizyme 2 and AZI expression. HEK-293 cells were transiently transfected with either pcDNA3.1-ODC, pcDNA3.1-HA-AZ2, pcDNA3.1-Myc-AZI or pcDNA3.1 alone (control). Lactacystin (8 μM) or an equal volume of DMSO was added. Antizyme 2 was detected with an anti-HA antibody (clone 3F10). AZI was detected with an anti-Myc antibody (clone 9E10). ODC was detected using a polyclonal anti-ODC antibody. Proteins were visualized with ECL®Plus enhanced chemiluminescence. The immunoblots shown are representative for multiple experiments. (B) In vivo association of ODC, antizyme 2 (AZ2) and AZI. HEK-293 cells were transiently transfected with either pcDNA3.1-ODC and pcDNA3.1-HA-AZ2 or with pcDNA3.1-ODC, pcDNA3.1-HA-AZ2 and pcDNA3.1-Myc-AZI. Left panel: expression of ODC, HA-tagged antizyme 2 and Myc-tagged AZI in the lysate. Immunoblot analysis with anti-HA, ODC or anti-Myc antibody. Right panel: association of ODC and antizyme 2 in the absence or presence of AZI. HA-tagged antizyme 2 was immunoprecipitated with anti-HA affinity matrix. Antizyme 2 interacting proteins were detected with antibodies against ODC or Myc. The immunoblots shown are representative for multiple experiments.
As shown in the immunoprecipitation experiments (Figure 6B), we found that ODC strongly binds to antizyme 2. However, in the presence of AZI, the amount of ODC bound to antizyme 2 was much smaller when compared with the amount bound in the absence of AZI although the same amount of ODC and antizyme 2 were present in the crude lysate. This suggests that AZI prevents binding of ODC to antizyme 2, which is consistent with the results obtained in the yeast three-hybrid system and the yeast complementation assay.
DISCUSSION
By using a yeast two-hybrid interaction assay we demonstrate that ODC binds to all members of the antizyme family (Figures 1A and 2B) including the previously not yet characterized antizyme 4. In the three-hybrid interaction assay, we found that these interactions including the interaction of antizyme 4 with ODC are prevented by AZI (Figure 2B). Moreover, in our yeast-based complementation assay, we could demonstrate that antizyme 4, similar to the other members of the antizyme family, is capable of inhibiting the enzymic activity of ODC (Figure 4B) and that this inhibition is reversed by AZI (Figure 5). These results strongly suggest that antizyme 4 is a functional member of the antizyme family and hence may play an important role in ODC regulation. Further analysis should determine whether antizyme 4 accelerates, similar to antizyme 1, the degradation of ODC and the inhibition of polyamine uptake.
The results presented here strongly suggest that AZI may act as a general inactivator of antizyme function. Our results demonstrate that AZI is capable of binding to all members of the antizyme family (Figures 1A and 2B) and thereby interferes with their ODC interaction. It is reasonable to assume that the binding of AZI to antizymes induces the release of enzymically active ODC from each ODC–antizyme complex. In this respect, it is interesting to note that AZI can inactivate all antizymes since antizymes 1 and 2 share only 55% amino acid identity and antizyme 3 is even more divergent when compared with antizyme 1 (26% amino acid identity) and antizyme 2 (29% amino acid identity) respectively [13]. The two-hybrid results combined with the expression data (Figure 1) also suggest that the protein interactions between AZI and antizymes are stronger than the corresponding interaction of ODC with the antizymes, which implies that additional conserved amino acids of AZI might contribute to the interaction with members of the antizyme family.
AZI shares almost 50% amino acid identity with ODC but previous studies have shown that AZI has no enzymic activity under in vitro conditions. When compared with ODC, AZI has conserved all four amino acid residues required for the formation of the active site [25] and additional acidic residues, which have been shown to contribute to the active site [36]. Therefore it was conceivable that AZI might possess enzymic activity under in vivo conditions. However, here we confirm that AZI does not have ODC-like activity in the yeast complementation assay strongly suggesting that AZI acts as an inactive homologue of ODC. This supports the current model that the stimulatory effect of AZI on ODC activity is mediated through the competitive binding of the enzymically inactive AZI to antizyme thereby releasing enzymically active ODC from the inhibitory complex. This interpretation is corroborated by the results presented in the present paper suggesting that the interaction between AZI and the members of the antizyme family is stronger than their corresponding interaction with ODC. Together with the previously reported observation that AZI was found to be highly overexpressed in gastric tumour tissue and in the gastric cancer cell line SNU5 [27] and that increased ODC activity has been linked to human tumour progression [37–40], our results suggest that AZI may contribute to the elevated ODC activity observed in many types of cancer.
The growth defect of spe1 deleted yeast cells or spe1 deleted cells, in which ODC and antizyme 1 were co-expressed, was not completely complemented by putrescine (results not shown). This incomplete rescue might be due to the fact that putrescine has to be taken up into the cells to reverse polyamine depletion. It was reported that polyamine depleted spe1 cells need up to 6 h to regain nearly normal growth rates after polyamine addition [28]. Alternatively, there might be an additional impact, e.g. on the cell cycle, which cannot be reversed by the addition of polyamines alone. A similar effect has been observed in S. pombe where, in contrast with S. cerevisiae, an antizyme homologue has been discovered. Overexpression of S. pombe antizyme leads to growth arrest that cannot be completely reversed by addition of putrescine even at a very high concentration (1 mM) [41]. In this study, we found that in contrast with the effect of exogenous putrescine, co-expression of AZI completely reversed antizyme-induced growth inhibition. In addition, there was no difference when compared with the growth of the isogenic wild-type strain (Figure 5).
A recent study has shown that antizyme 1 is rapidly degraded through the proteasome in a mechanism that requires a functional ubiquitin system and that this degradation is inhibited by specific proteasome inhibitors [35]. In our mammalian cell expression system (Figure 6A), we found that the expression of antizyme 2 is greatly stabilized by the specific proteasome inhibitor lactacystin strongly suggesting that, similar to antizyme 1, antizyme 2 is also degraded through a proteasome-mediated mechanism. Although further experiments are needed to confirm this conclusion for other members of the antizyme family, it is quite probable that such a proteasome-dependent degradation mechanism may apply for the regulation of every member of the antizyme family.
The mammalian expression system also demonstrated that the expression of AZI is greatly stabilized through lactacystin indicating that not only antizymes but also AZI is degraded through a proteasomal mechanism (Figure 6A). In contrast with ODC, the degradation of AZI is not stimulated by antizyme 1 [25]. Therefore it will be interesting to test whether a functional ubiquitin system is required for the degradation of AZI.
Our yeast complementation assay allows us to analyse the functions of ODC, antizyme and AZI under easy, non-radioactive in vivo model conditions. It can be used in combination with the two- and three-hybrid assays to map regions that are important for the multiple interactions among ODC, antizyme and AZI in the regulatory complex. This in vivo assay does not require protein purification thereby facilitating the rapid examination of mutated or truncated variants of the components of the ODC–antizyme–AZI complex or the functional characterization of novel ODC-like proteins [42].
Acknowledgments
We thank C. Nesti and X. Luo for their support in bioinformatics and R. Soellner and S. Ohage for helpful discussions.
References
- 1.Igarashi K., Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 2.Holtta E., Sistonen L., Alitalo K. The mechanisms of ornithine decarboxylase deregulation in c-Ha-ras oncogene-transformed NIH 3T3 cells. J. Biol. Chem. 1988;263:4500–4507. [PubMed] [Google Scholar]
- 3.Holtta E., Auvinen M., Andersson L. C. Polyamines are essential for cell transformation by pp60v-src: delineation of molecular events relevant for the transformed phenotype. J. Cell Biol. 1993;122:903–914. doi: 10.1083/jcb.122.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shantz L. M., Pegg A. E. Ornithine decarboxylase induction in transformation by H-Ras and RhoA. Cancer Res. 1998;58:2748–2753. [PubMed] [Google Scholar]
- 5.Shantz L. M., Coleman C. S., Pegg A. E. Expression of an ornithine decarboxylase dominant-negative mutant reverses eukaryotic initiation factor 4E-induced cell transformation. Cancer Res. 1996;56:5136–5140. [PubMed] [Google Scholar]
- 6.Auvinen M., Paasinen A., Andersson L. C., Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature (London) 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 7.Auvinen M., Laine A., Paasinen-Sohns A., Kangas A., Kangas L., Saksela O., Andersson L. C., Holtta E. Human ornithine decarboxylase-overproducing NIH3T3 cells induce rapidly growing, highly vascularized tumors in nude mice. Cancer Res. 1997;57:3016–3025. [PubMed] [Google Scholar]
- 8.Moshier J. A., Dosescu J., Skunca M., Luk G. D. Transformation of NIH/3T3 cells by ornithine decarboxylase overexpression. Cancer Res. 1993;53:2618–2622. [PubMed] [Google Scholar]
- 9.Li R. S., Law G. L., Seifert R. A., Romaniuk P. J., Morris D. R. Ornithine decarboxylase is a transcriptional target of tumor suppressor WT1. Exp. Cell Res. 1999;247:257–266. doi: 10.1006/excr.1998.4361. [DOI] [PubMed] [Google Scholar]
- 10.Bello-Fernandez C., Packham G., Cleveland J. L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shantz L. M., Pegg A. E. Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int. J. Biochem. Cell Biol. 1999;31:107–122. doi: 10.1016/s1357-2725(98)00135-6. [DOI] [PubMed] [Google Scholar]
- 12.Coffino P. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2001;2:188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov I. P., Gesteland R. F., Atkins J. F. Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28:3185–3196. doi: 10.1093/nar/28.17.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsufuji S., Matsufuji T., Miyazaki Y., Murakami Y., Atkins J. F., Gesteland R. F., Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell (Cambridge, Mass.) 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature (London) 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell J. L., Judd G. G., Bareyal-Leyser A., Ling S. Y. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. Biochem. J. 1994;299:19–22. doi: 10.1042/bj2990019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakata K., Kashiwagi K., Igarashi K. Properties of a polyamine transporter regulated by antizyme. Biochem. J. 2000;347:297–303. [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov I. P., Gesteland R. F., Atkins J. F. A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics. 1998;52:119–129. doi: 10.1006/geno.1998.5434. [DOI] [PubMed] [Google Scholar]
- 19.Zhu C., Lang D. W., Coffino P. Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J. Biol. Chem. 1999;274:26425–26430. doi: 10.1074/jbc.274.37.26425. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov I. P., Rohrwasser A., Terreros D. A., Gesteland R. F., Atkins J. F. Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4808–4813. doi: 10.1073/pnas.070055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwata S., Sato Y., Asada M., Takagi M., Tsujimoto A., Inaba T., Yamada T., Sakamoto S., Yata J., Shimogori T., et al. Anti-tumor activity of antizyme which targets the ornithine decarboxylase (ODC) required for cell growth and transformation. Oncogene. 1999;18:165–172. doi: 10.1038/sj.onc.1202275. [DOI] [PubMed] [Google Scholar]
- 22.Koike C., Chao D. T., Zetter B. R. Sensitivity to polyamine-induced growth arrest correlates with antizyme induction in prostate carcinoma cells. Cancer Res. 1999;59:6109–6112. [PubMed] [Google Scholar]
- 23.Feith D. J., Shantz L. M., Pegg A. E. Targeted antizyme expression in the skin of transgenic mice reduces tumour promoter induction of ornithine decarboxylase and decreases sensitivity to chemical carcinogenesis. Cancer Res. 2001;61:6073–6081. [PubMed] [Google Scholar]
- 24.Fong L. Y., Feith D. J., Pegg A. E. Antizyme overexpression in transgenic mice reduces cell proliferation, increases apoptosis, and reduces N-nitrosomethylbenzylamine-induced forestomach carcinogenesis. Cancer Res. 2003;63:3945–3954. [PubMed] [Google Scholar]
- 25.Murakami Y., Ichiba T., Matsufuji S., Hayashi S. Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J. Biol. Chem. 1996;271:3340–3342. doi: 10.1074/jbc.271.7.3340. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson J., Grahn B., Heby O. Antizyme inhibitor is rapidly induced in growth-stimulated mouse fibroblasts and releases ornithine decarboxylase from antizyme suppression. Biochem. J. 2000;346:699–704. [PMC free article] [PubMed] [Google Scholar]
- 27.Jung M. H., Kim S. C., Jeon G. A., Kim S. H., Kim Y., Choi K. S., Park S. I., Joe M. K., Kimm K. Identification of differentially expressed genes in normal and tumor human gastric tissue. Genomics. 2000;69:281–286. doi: 10.1006/geno.2000.6338. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz B., Hittelman A., Daneshvar L., Basu H. S., Marton L. J., Feuerstein B. G. A new model for disruption of the ornithine decarboxylase gene, SPE1, in Saccharomyces cerevisiae exhibits growth arrest and genetic instability at the MAT locus. Biochem. J. 1995;312:83–90. doi: 10.1042/bj3120083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M. D., Winston F. M., Hieter P., Sherman F., Fink G. R., Hicks J. Cold Spring Harbor Laboratory. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. Methods in Yeast Genetics: A Laboratory Course Manual. [Google Scholar]
- 30.Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 31.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 32.Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 34.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gandre S., Bercovich Z., Kahana C. Ornithine decarboxylase-antizyme is rapidly degraded through a mechanism that requires functional ubiquitin-dependent proteolytic activity. Eur. J. Biochem. 2002;269:1316–1322. doi: 10.1046/j.1432-1033.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- 36.Osterman A. L., Kinch L. N., Grishin N. V., Phillips M. A. Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase identified by alanine scanning mutagenesis. J. Biol. Chem. 1995;270:11797–11802. doi: 10.1074/jbc.270.20.11797. [DOI] [PubMed] [Google Scholar]
- 37.Bettuzzi S., Davalli P., Astancolle S., Carani C., Madeo B., Tampieri A., Corti A., Saverio B., Pierpaola D., Serenella A., et al. Tumor progression is accompanied by significant changes in the levels of expression of polyamine metabolism regulatory genes and clusterin (sulfated glycoprotein 2) in human prostate cancer specimens. Cancer Res. 2000;60:28–34. [PubMed] [Google Scholar]
- 38.Duranton B., Holl V., Schneider Y., Carnesecchi S., Gosse F., Raul F., Seiler N. Polyamine metabolism in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620) Amino Acids. 2003;24:63–72. doi: 10.1007/s00726-002-0333-5. [DOI] [PubMed] [Google Scholar]
- 39.Manni A., Astrow S. H., Gammon S., Thompson J., Mauger D., Washington S. Immunohistochemical detection of ornithine-decarboxylase in primary and metastatic human breast cancer specimens. Breast Cancer Res. Treat. 2001;67:147–156. doi: 10.1023/a:1010697218986. [DOI] [PubMed] [Google Scholar]
- 40.Manni A., Washington S., Griffith J. W., Verderame M. F., Mauger D., Demers L. M., Samant R. S., Welch D. R. Influence of polyamines on in vitro and in vivo features of aggressive and metastatic behavior by human breast cancer cells. Clin. Exp. Metastasis. 2002;19:95–105. doi: 10.1023/a:1014536909007. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov I. P., Matsufuji S., Murakami Y., Gesteland R. F., Atkins J. F. Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J. 2000;19:1907–1917. doi: 10.1093/emboj/19.8.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pitkanen L. T., Heiskala M., Andersson L. C. Expression of a novel human ornithine decarboxylase-like protein in the central nervous system and testes. Biochem. Biophys. Res. Commun. 2001;287:1051–1057. doi: 10.1006/bbrc.2001.5703. [DOI] [PubMed] [Google Scholar]