Abstract
Microorganisms play an important role in the pathogenesis of inflammatory bowel disease (IBD). The oral cavity, the second-largest microbial niche, is connected to the gastro-intestinal tract. Ectopic gut colonization by oral microbes is a signature of IBD. Current studies suggest that patients with IBD often report more oral manifestations and these oral issues are closely linked with disease activity. Murine studies have indicated that several oral microbes exacerbate intestinal inflammation. Moreover, intestinal inflammation can promote oral microbial dysbiosis and the migration of oral microbes to the gastro-intestinal tract. The reciprocal consequences of oral microbial dysbiosis and IBD, specifically through metabolic alterations, have not yet been elucidated. In this review, we summarize the relationship between oral bacteria and IBD from multiple perspectives, including clinical manifestations, microbial dysbiosis, and metabolic alterations, and find that oral pathogens increase anti-inflammatory metabolites and decrease inflammation-related metabolites.
Keywords: inflammatory bowel disease, microbiota, oral bacteria, periodontitis, ectopic colonization
Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder affecting the digestive tract, encompassing Crohn’s disease (CD), ulcerative colitis (UC), and indeterminate colitis. Growing evidence indicates that bacterial dysbiosis plays a prominent role in IBD development. The gastro-intestinal tract, extending from the oral cavity to the anus, harbors a variety of bacteria that colonize distinct ecological niches along its length [1]. Oral and intestinal microbes interact structurally and functionally [2]. Under normal physiological conditions, interactions between bacteria in different ecological niches are limited [3]. However, a microbial imbalance exists in pathological states compared with healthy controls (HCs) [4]. Ectopic colonization is more common in pathological states and worsens pathology [3, 5, 6]. A recent study investigated the dynamic alterations in oral bacteria in patients with IBD (n = 22) compared with HCs (n = 8) and found that there was a notable rise in the levels of Saccharibacteria (TM7) and Absconditabacteria (SR1) in the saliva of patients with IBD [7]. TM7 and SR1 are positively correlated with altered inflammatory cytokines in IBD, indicating that oral microbial dysbiosis is associated with inflammatory immune responses in IBD [7, 8]. These findings imply that the mouth can act as a reservoir for pathogens. The ectopic translation from the mouth to the intestine may contribute to triggering inflammation in individuals with IBD [9].
Recently, several reviews have been published on oral bacteria and gastro-intestinal disease [2, 8, 10–16]. These reviews summarize the oral bacteria contributing to IBD and the related mechanisms; however, these mechanisms mainly focused on the host immune response (Table 1). For example, Klebsiella spp., Fusobacterium nucleatum, and Campylobacter concisus can drive pro-inflammatory cytokine release in the gut [14]. However, systematic summaries exploring the role of oral bacteria in IBD through metabolic alterations are limited.
Table 1.
The mechanisms of oral bacteria affecting gut inflammation summarized in previous reviews
| Oral bacteria | Pathway | References |
|---|---|---|
| Veillonella spp. | (i) Veillonella spp. correlate with increased levels of circulating lymphocytes (ii) V. parvula positively correlates with the inflammatory marker calprotectin (iii) Adhesive bacteria (involving Veillonella) increase Th17 cell activation and luminal secretory IgA (iv) Veillonella spp. positively coexist with Clostridioides in C. difficile colonization patients |
Read E, 2021 [14] Lu Y, 2023 [15] |
| Klebsiella spp. | (i) Klebsiella spp. interact with macrophages to promote the release of IL-1β via cysteine-11-mediated inflammatory vesicles, activating intestinal inflammasome (ii) Klebsiella spp. can activate Th1 cells via IFN-γ and Th17 cells (iii) K. pneumoniae invades the colonic mucus layer and intestinal epithelial cell lines, and translocates across the intestinal epithelium through Rho GTPase- and PI3K/Akt-dependent cell invasion (iv) The capsular polysaccharide of K. pneumoniae resists the opsonization and phagocytosis of macrophages, DCs, neutrophils, and epithelial cells (v) K. pneumoniae downregulates Lactobacillus reuteri and Bifidobacterium pseudolongum, and alters cecal metabolome (e.g. SCFAs) |
Read E, 2021 [14] Lu Y, 2023 [15] Kitamoto S, 2022 [2] Qi Y, 2022 [10] Imai J, 2021 [3] |
| Streptococcus mutans | (i) S. mutans exacerbates colitis though IFN-γ signaling (ii) S. mutans positively correlates with the inflammatory marker calprotectin |
Read E, 2021 [14] |
| Streptococcus salivarius | (i) S. salivarius downregulates the NF-κB pathway by LPS and inhibits PPARγ activation (ii) S. salivarius alters I-FABP and Angptl4 gene products regulating intracellular lipid accumulation (iii) S. salivarius reacts upon gastro-intestinal-derived CD4+ T cells and activates monocytes to secrete higher levels of IL-6, IL-12, and TNF (the Th1- and Th17-skewed cytokines) (iv) S. salivarius positively correlates with the inflammatory marker calprotectin |
Read E, 2021 [14] Lu Y, 2023 [15] |
| Streptococcus agalactiae | S. agalactiae reacts upon gastro-intestinal-derived CD4+ T cells and activates monocytes to secrete higher levels of IL-6, IL-12, and TNF (the Th1- and Th17-skewed cytokines) | Lu Y, 2023 [15] |
| Haemophilus parainfluenzae | H. parainfluenzae correlates with the inflammatory marker calprotectin | Read E, 2021 [14] |
| Rothia mucilaginosa | R. mucilaginosa correlates with the inflammatory marker calprotectin | Read E, 2021 [14] |
| Campylobacter concisus |
C. concisus can be classified into adherent toxigenic C. concisus (AToCC) and adherent invasive C. concisus (AICC) (i) AToCC possess a Zot, upregulate PAR2 expression, and break tight junctions and cytoskeletal remodeling; Zot can stimulate intestinal epithelial cells and macrophages to release pro-inflammatory cytokines and enhance the responses of macrophages to other enteric bacteria (ii) AICC can stimulate neutrophil cells by upregulating the neutrophil adherence molecule CD11b and oxidative burst response (iii) C. concisus flagellum-mediated attachment to and invasion of the colonic epithelial cell line Caco-2 (iv) C. concisus increases intestinal permeability through the downregulation of ZO-1, occludin, and claudin-5, together with apoptotic leaks (v) C. concisus impairs sodium (Na+) absorption in HT-29/B6 cells through the dysfunction of the epithelial Na+ channels, dependent on IL-32-regulated extracellular signal, regulated protein kinase (ERK)1/2, and claudin-8-dependent barrier dysfunction (vi) C. concisus increases levels of pattern-recognition receptors (e.g. TLR4, but not TLR2 or TLR5) (vii) C. concisus reduces autophagy-related genes, such as ATG9B (viii) Virulence factor membrane-bound hemolytic PLA2 exhibits cytolytic effects (ix) C. concisus enhances inflammatory response (IL-2, IL-5, IL-18, CCL2, IL-8, COX-2, IL-8 and TNF-α, CREB1, NF-κB, STAT, and interferon regulatory factor, IFI16 inflammasome, TLR3) |
Read E, 2021 [14] Qi Y, 2022 [10] Kitamoto S, 2022 [2] |
| Fusobacterium nucleatum | (i) F. nucleatum reduces the killing capacity of macrophages and NK cells (ii) F. nucleatum aggravates the progression of DSS-induced colitis by promoting M1 macrophage polarization through the activation of the AKT2 pathway (iii) F. nucleatum and its outer membrane vesicles activate the TLR/MyD88/NF-κB pathway, promoting the secretion of a series of pro-inflammatory cytokines, including IL-8, TNF, keratinocyte-derived chemokine (KC), IL-6, IFN-γ, and MCP-1 (iv) F. nucleatum activates the STAT3 signaling pathway, promoting Th1 and Th17 responses (v) F. nucleatum activates the CARD3/IL-17F/NF-κB cascade in epithelial cells (vi) Fap2 protein produced by F. nucleatum interacts with TIGIT, mediates NK-cell and T-cell inhibition, while T cell regulates inflammatory factors IL-10, IL-1β, and IL-6 (vii) F. nucleatum coexists with Clostridium through adhesin RadD, encouraging the bacterial biofilm formation of the intestinal mucus layer (viii) F. nucleatum invades human intestinal epithelial cell lines, disrupts the integrity of the epithelial barrier, reducing tight junction proteins such as ZO-1 and occluding, and stimulates the function change of MUC2 (ix) H2S produced by F. nucleatum inhibits the effective use of anti-inflammatory butyrate in colon cells |
Read E, 2021 [14] Lu Y, 2023 [15] Qi Y, 2022 [10] Kitamoto S, 2022 [2] |
| Porphyromonas gingivalis | (i) P. gingivalis disrupts the intestinal barrier by downregulating tjp-1 and Zo-1 (ii) P. gingivalis secretes gingipains (iii) P. gingivalis LPS activates Th2 response: significantly higher levels of IL-5, IL-10, and IL-13 but a lower level of IFN-γ, and activates Th17 cells (iv) P. gingivalis stimulates the overgrowth of commensal microbes in the intestine (generally upregulates Bacteroidetes and Deferribacteres, and downregulates Firmicutes) (v) P. gingivalis positively correlates with Pyricularia pennisetigena and Alternaria alternata (vi) P. gingivalis upregulates phenylalanine, tyrosine, and tryptophan in the intestinal microbiota, and alanine, glutamine, histidine, tyrosine, and phenylalanine in serum |
Imai J, 2021 [16] Lu Y, 2023 [15] Qi Y, 2022 [10] Kitamoto S, 2022 [2] |
| Fusobacterium varium | F. varium invade the intestinal epithelium and evoke the production of pro-inflammatory cytokines, such as IL-8 and TNF-α | Kitamoto S, 2022 [2] |
| Atopobium parvulum | A. parvulum increases expression of CXCL1 and IL-17 in the gut and induces pro-inflammatory molecules (e.g. COX-2, IL-8, and CEBPB) in epithelial cells and to promote T-cell activation by liberating H2S | Kitamoto S, 2022 [2] |
| Staphylococcus aureus | (i) S. aureus causes epithelial damage in the small, but not the large, intestine by producing SEB (ii) S. aureus dampens adheren junction protein expression (iii) S. aureus interacts with antigen‐presenting cells (e.g. macrophages and dendritic cells) |
Kitamoto S, 2022 [2] |
IL = interleukin, IFN = interferon, PI3K = phosphatidylinositol 3-kinase, DCs = dendritic cells, SCFA = short-chain fatty acid, NF-κB = nuclear factor-κB, LPS = lipopolysaccharides, PPARγ = peroxisome proliferator-activated receptor γ, Zot = zonula occludens toxin, Zo-1 = zonula occludens-1, TLR = Toll-like receptor, NK = natural killer, MCP-1 = monocyte chemoattractant protein-1, STAT = signal transducer and activator of transcription, CARD = caspase activation and recruitment domain, H2S = hydrogen sulfide, CXCL1 = chemokine (C–X–C motif) ligand 1, COX = cyclooxygenase, CEBPB = CCAAT enhancer-binding protein beta, SEB = staphylococcal enterotoxin B.
Thus, a more comprehensive and profound understanding of the oral–intestinal microbial axis will provide better insights into the progression of IBD. Here, we review the relationship between oral bacteria and IBD from multiple perspectives, including clinical manifestation, microbial dysbiosis, and metabolic alteration.
Clinical manifestations
Periodontal diseases, including gingivitis and periodontitis, have been linked to gastro-intestinal disorders. Periodontitis, a type of periodontal disease, is characterized by progressive destruction of the supporting structure of the teeth, such as the gum and bone surrounding the teeth [17, 18]. Its association with the accumulation of dental plaque and bacterial dysbiosis is well established [19]. Inflammation induced by bacteria present in dental plaque contributes to periodontitis [20]. Porphyromonas gingivalis is the major causative agent of periodontitis [4, 21]. A recent study showed that incipient periodontitis contributes to an increased short CD activity index [3]. A notable correlation was observed between periodontitis or tooth loss and increased IBD-related disability over the past 12 months [22]. Animal experiments demonstrate that saliva from patients with periodontitis exacerbates dextran sodium sulfate (DSS)-induced colitis [23] and ligature-induced periodontitis aggravates gut inflammation in a murine model of colitis [24]. IBD is accompanied by an increased incidence of periodontal disease and periodontal disease can affect the disease activity of IBD.
Microbial dysbiosis
The gut bacteria of healthy individuals are dominated by four bacterial phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria, and ∼90% of them belong to Firmicutes and Bacteroidetes [25]. Patients with IBD have shown a significant reduction in microbial biodiversity (especially Firmicutes) and decreased stability. Increased levels of Ruminococcus gnavus, Escherichia coli, and Clostridium bolteae, and decreased levels of Faecalibacterium prausnitzii and C. hathewayi have been observed in IBD [26]. Furthermore, patients with IBD have shown a reduction in gut bacteria that produce short-chain fatty acids (SCFAs) [27]. The oral cavity, with >700 species, is the second-largest microbial library after the gastro-intestinal tract [28]. The diversity of species increases even more under diseases such as periodontitis [29]. IBD affects oral bacterial composition, with an abundance of bacteria such as TM7 and Veillonella in the oral cavity of patients with IBD compared with HCs [30]. Additionally, the oral bacterial community composition differs in patients with both IBD and periodontitis (n = 6) compared with those with periodontitis alone (n = 6). The periodontitis-only group also exhibited a greater prevalence of Actinobacteria, Bacteroidetes, and Fusobacteria, whereas the CD combined with periodontitis group showed higher levels of Firmicutes and Proteobacteria [31]. Chen et al. [32] found that, compared with the periodontitis-only group, Eikenella, Capnocytophaga, Prevotella_2, and Prevotella were more abundant in the CD combined with periodontitis (n = 14) group than in the periodontitis-only group (n = 14). Furthermore, Capnocytophaga and Streptococcus oralis were identified in CD combined with periodontitis-associated predominant microbial species, whereas Streptococcus and Bacillales were identified as periodontitis-associated predominant bacteria [31]. Elmaghrawy et al. [9] developed a prognostic model for IBD and achieved an area under the receiver-operating characteristic curve of 0.762 (IBD, n = 156; HCs, n = 102) based on IBD-related oral microbial dysbiosis. Thus, periodontal diseases that can cause oral bacterial dysbiosis are more prevalent in IBD, and IBD itself significantly influences the composition of oral bacteria.
Under normal conditions, interactions between oral and gut bacteria are limited. However, oral bacteria can ectopically colonize the gastro-intestinal tract and cause detrimental digestive diseases under certain circumstances [33]. Hematogenous and enteral dissemination have been proposed as potential routes of ectopic colonization from the mouth to the gut [2]. Oral bacteria can survive with dendritic cells and macrophages, aiding their movement from the oral cavity to the intestinal compartment. Oral bacteria easily and frequently disseminate into the blood through oral wounds, gingival crevices, and other untreated carious lesions [13, 34]. For instance, the presence of P. gingivalis has been identified in the bloodstream of patients with periodontal diseases [35]. An adult human is estimated to consume approximately 600 times daily and generate ∼1.5 L of spit containing 1.5 × 1012 bacteria [2, 34]. Balanced gastric acidity serves as the first barrier against ectopic colonization from the oral cavity to the gut [2, 36], but patients with IBD experience a decrease in gastric acidity, potentially enabling the transfer of oral bacteria, such as S. salivarius [2, 36]. Given that IBD is more susceptible to medications that affect pH, including proton-pump inhibitors (PPIs), it is plausible that the use of PPIs may also encourage the abnormal colonization of oral bacteria. Additionally, some oral pathological bacteria, such as P. gingivalis, can tolerate acidic environments and translocate to the gut [37]. Colonization resistance, provided by gut-resident bacteria, acts as a secondary barrier against ectopic colonization; however, this is attenuated by microbial dysbiosis and gut inflammation in IBD [6, 38]. The pathological changes in IBD establish the foundation for the migration of oral bacteria to the intestines. Consistently, the gut bacteria exhibited greater resemblance to the oral bacteria in patients with IBD (n = 60) compared with those with HCs (n = 45), indicating a higher prevalence of ectopic gut colonization by oral bacteria in IBD [3]. In vivo experiments with germ-free mice receiving saliva samples from patients with CD revealed that most fecal bacterial species were minor components of salivary bacteria [34]. These findings verify that oral bacteria can translocate to the gut. Furthermore, salivary bacteria can disrupt the balance of intestinal bacteria. Rats treated with periodontitis saliva samples exhibited increased levels of Roseburia and Lactobacillus, accompanied by lower relative abundances of Oscillibacter, Colidextribacter, and Bacteroides [39]. Saliva samples collected from both periodontitis patients and healthy individuals, when administered to mice groups, resulted in variations in the gut microbiota composition between the two groups. These differences were particularly notable in DSS-induced colitis, with significant changes in the genera Blautia, Aerococcus, Ruminococcus, and Helicobacter [23]. Oral-derived bacteria such as P. gingivalis, F. nucleatum, and Klebsiella spp. (e.g. K. pneumoniae) have been reported to exacerbate gut inflammation in vivo [2, 10]. Additionally, oral administration of P. gingivalis leads to alterations in the intestinal bacterial composition [40]. For instance, there was a notable increase in the proportions of unclassified Coriobacteriaceae, Gemellaceae, and Clostridiaceae, whereas the proportions of unclassified S24-7, Prevotellaceae, Mogibacteriaceae, Dorea, Butyricicoccus, and Bilophila showed a significant decrease after P. gingivalis administration [40]. In addition to bacteria, the presence of P. gingivalis in mice had an impact on the structure and diversity of the gut mycobiome [41]. Pyricularia pennisetigena and Alternaria alternata are positively correlated with P. gingivalis [41].
Relationship between oral microbes and IBD from a metabolic perspective
Metabolic nature of IBD
Metabolic alterations have also been described in patients with IBD [42]. Metabolites play important roles in the pathogenesis of IBD [43–45]. Metabolomics refers to the systematic profiling of small-molecule metabolites in biological samples, encompassing sugars, amino acids, organic acids, nucleotides, and lipids [46–48]. Proton nuclear magnetic resonance spectroscopy and mass spectrometry including liquid chromatography and gas chromatography are the most widely used tools in metabolomics [49]. Metabolomics primarily involves two approaches: targeted and untargeted. Targeted metabolomic studies are hypothesis-driven, focusing on the precise and accurate measurement of predefined sets of metabolites. In contrast, untargeted metabolomic studies involve the simultaneous measurement of a wide spectrum of metabolites within the sample. The samples utilized in metabolomics in IBD mainly include serum, plasma, feces, and urine [42, 50–52]. Different samples provide different types of biochemical information. Serum and plasma provide an insightful snapshot of systemic metabolism, whereas feces reflect digestive metabolism, including microbiota-related metabolites, host–microbial co-metabolites, and microbial transformation of dietary components [50]. Urine is a better indicator of host, microbial, and exogenous metabolites such as drugs [50]. Several major pathways affected by bacteria in IBD include lipid, amino acid, and bile acid metabolism [53]. Lipids include fatty acyl glycerophospholipids and sphingolipids. Fatty acids are classified as SCFAs, medium-chain fatty acids (MCFAs), and long-chain fatty acids (LCFAs). SCFAs are found at lower levels in the feces of patients with IBD than in that of patients with HCs and are related to disease activity [27, 54, 55]. MCFAs, such as pentanoate, hexanoate, heptanoate, octanoate, and nonanoate, were significantly decreased in patients with IBD (IBD, n = 151; HCs, n = 40) [56]. Polyunsaturated fatty acids (PUFAs), a subtype of LCFAs, are classified as n-3 and n-6 PUFAs. Consumption of a diet characterized by an elevated ratio of n-6 to n-3 PUFAs is linked to increased susceptibility to CD [57]. Moreover, a higher intake of n-6 PUFAs increased the risk of UC [58, 59]. Conversely, the intake of n-3 PUFAs has been linked to a reduced risk of UC [58, 59]. Monounsaturated fatty acids (MUFAs), a subtype of LCFAs, exhibit decreased levels in both the blood and intestinal mucosa. However, the therapeutic effectiveness of MUFAs for IBD remains controversial [60–62]. Lysophosphatidylcholine (LysoPC) and lysophosphatidylcholine (LysoPS) levels are elevated in the stool and blood of patients with IBD [63–65]. LysoPC and LysoPS have been identified to impair the epithelial and immune barrier [62]. Sphingolipids are the most differentially abundant metabolites in the stool of IBD patients and perturbed sphingolipid metabolism contributes to the inflammation process [63, 66]. Disordered amino acid and bile acid metabolic pathways have also been observed in IBD. Tryptophan levels in both serum and feces were also reduced in IBD [67, 68]. A pivotal study conducted by Vantrappen et al. [69] involving 13 non-operated patients with CD, 10 patients with UC, and 10 HCs revealed that patients with CD, but not UC, displayed a reduced size of the bile acid pool in comparison with those with HCs. Furthermore, a reduction in the size of the bile acid reservoir showed an inverse correlation with the Colitis Disease Activity Index and the percentage of unconjugated bile acids increased in IBD [69]. Perturbation of metabolites in IBD is believed to be associated with gut microbial dysbiosis [70, 71]. Additionally, the existence of Akkermansia municiphila, Oscillibacter spp., and Bilophila wadsworthia was related to the levels of dicarboxylic acids, sebacate, dodecanedioate, taurine, and N,N,N-trimethyl-alanyl-proline betaine [72]. Caprylic acid was positively correlated with Alistipes shahii, A. putredinis, and A. finegoldii, whereas it was significantly and negatively correlated with the abundance of R. gnavus [66]. Recent studies have demonstrated the interaction between oral microbes and IBD through the metabolic pathway.
Inflammatory metabolites provide the basis for ectopic colonization
Microbiota-mediated colonization resistance has been implicated as a mechanism for separating oral and colonic niches in gnotobiotic murine experiments [6]. However, whether this mechanism is essential in humans remains unknown [73]. Rashidi et al. [73] recruited healthy volunteers (n = 43) exposed to a short course of a single antibiotic, patients with acute leukemia (n = 39), and stem cell transplant recipients (n = 29). The latter two groups had only one oral species colonizing the gut, even considering the influence of antibiotics and the extent of damage to the gut microbiota. This suggests that colonization resistance was dispensable in oral and gut microbiota segregation [73]. The colon possesses certain physicochemical traits (such as reduced oxygen levels and fecal toxins) and there are various antimicrobial defenses from the mouth to the colon (including gastric acid, bile salts, mucosal immunoglobulins, and antimicrobial peptides) [73]. By eliminating microbiota-mediated colonization resistance, these barriers may be effective enough to prevent ectopic colonization [73]. However, within the context of persistent inflammation such as IBD, the chemical barrier against ectopic colonization is destroyed, leading to dysbiotic microbiota and creating opportunities for ectopic colonization. For instance, Veillonella parvula, an oral microbe that derives its energy from organic acids, was abundant in the gut of patients with IBD [5]. Subsequent investigations revealed that nitrate, as a specific metabolite of inflammation, could provide fundamental conditions for oral microbes, such as V. parvula, to translocate from the oral cavity to the gut because nitrate respiration allows oral microbes to use amino acids and peptides as carbon sources [5].
Ectopic colonization of oral microbes promotes metabolic disorders
Oral bacteria have been demonstrated to affect gut and serum metabolites (Table 2). Salivary bacteria of periodontitis, when administered to rats, significantly affected metabolites related to lipids, indoles, and their derivatives [39]. When saliva samples from periodontitis patients were administered to DSS-treated mice via gastric gavage, it was found that salivary bacteria exacerbated DSS-induced colitis [23]. Periodontitis salivary bacteria alter the levels of anti-inflammatory metabolites, including SCFAs and tryptophan-related metabolites, and increase metabolites related to inflammation, such as arachidonic acid, in DSS-induced colitis [23]. SCFAs derived from bacteria play crucial roles in preserving host immune homeostasis and reinforcing epithelial integrity [62]. This is achieved through their interactions with receptors such as GPR41, GPR43, GPR109a, and OLFR78 (in mice) or OR51E2 (in humans), as well as via host epigenetic modifications, in both a G protein-coupled receptor (GPCR)-independent and -dependent manner [62]. Tryptophan, an essential aromatic amino acid, is primarily metabolized through three major pathways. Gut bacteria facilitate the conversion of tryptophan into different compounds, which include ligands of the aryl hydrocarbon receptor [77]. Additionally, the kynurenine pathway functions in immune and epithelial cells through indoleamine 2,3-dioxygenase (IDO) 1 [77]. The third pathway involves the production of serotonin (5-hydroxytryptamine) in enterochromaffin cells via tryptophan hydroxylase 1 [77]. The kynurenine pathway comprises ≥90% of tryptophan catabolism [78]. Inflammatory cytokines such as IFN-γ, TNF-α, and IL1β are abundant in IBD and induce the upregulation of IDO 1 [78]. Therefore, we hypothesized that oral bacteria might affect tryptophan levels through microbial dysbiosis or by exacerbating inflammation. For arachidonic acid metabolism, the upregulation of prostaglandin I2, prostaglandin F2α, and dihydroxyeicosatrienoic acid was observed following the administration of saliva in periodontitis [23]. Aerococcus and Ruminococcus exhibited a strong positive correlation with arachidonic acid metabolism but a negative correlation with the biosynthesis of unsaturated fatty acids [23]. In contrast, Blautia and Helicobacter displayed a contrasting association [23]. Periodontitis salivary bacteria exacerbate DSS-induced colitis and enhance colitis-induced anxiety-like behaviors via metabolic pathways [23, 74]. The histidine metabolism plays a critical role in the underlying pathway in this process because periodontitis salivary bacteria alter histidine metabolism in both gut and brain metabolomics [74]. Additionally, the supplementation of histidine-related metabolites had a similar anxiety-worsening impact to periodontitis salivary bacteria [74]. The serum metabolome of mice was modified by administering P. gingivalis orally, resulting in increased levels of alanine, glutamine, histidine, tyrosine, and phenylalanine [40]. In addition, P. gingivalis can alter the serum metabolome by influencing gut mycobiomes such as Amphiamblys, P. pennisetigena, and Valsa malicola. For instance, P. gingivalis administration enriched the presence of P. pennisetigena, which showed a positive connection to metabolites related to lipid metabolism-related metabolites, such as LysoPC. Additionally, it displayed a negative relationship with indole-3-acetamide, 5-hydroxy-tryptophan, and indoleacetaldehyde [41]. Certain oral pathogens, such as Klebsiella, were positively correlated with primary and conjugated bile acids, including cholic acid, taurocholic acid, and glycochenodeoxycholic acid, or taurochenodeoxycholic acid (IBD, n = 32; HC, n = 23) [12, 76]. Certain oral bacteria including Atopobium, Fusobacterium, Veillonella, Prevotella, Streptococcus, and Aggregatibacter which are positively correlated with the severity of intestinal disease, metabolize sulfur-containing amino acids into hydrogen sulfide (H2S), an inflammatory mediator [2]. Atopobium parvulum is recognized as the principal pathobiont and plays a pivotal role as a central hub within the H2S network (IBD, n = 131; HC, n = 63) [75]. Atopobium parvulum-induced colitis in interleukin-10-deficient (IL10–/–) mice was mitigated by the H2S scavenger bismuth [75]. In contrast, germ-free IL10–/– mice monocolonized with A. parvulum did not experience significant colitis, indicating that other microbes or their byproducts are necessary for the development of A. parvulum-driven colitis [75]. It is possible that A. parvulum promotes the growth of colitogenic pathogens by inducing H2S, which can stimulate pro-inflammatory molecules such as cyclooxygenase-2, IL-8, and CCAAT enhancer-binding protein beta in epithelial cells and facilitate T-cell activation [79]. Other indigenous oral bacteria, such as Streptococcus and Neisseria, can generate acetaldehyde through the catabolism of ethanol and glucose [80]. Considering the pro-inflammatory potential of acetaldehyde, which can disrupt epithelial barrier function [2, 81], it is conceivable that the ectopic colonization of the gut by these oral bacteria may trigger gut inflammation. These findings revealed that oral pathogens elevate anti-inflammatory metabolites and decrease inflammation-related metabolites.
Table 2.
The effect of oral bacteria in IBD via the metabolic pathway
| Author, year | Subjects | Oral microbiota | Sample | Metabolites | Detection methods | Action |
|---|---|---|---|---|---|---|
| Rojas-Tapias DF, 2022 [5] | Mouse, vitro | V. parvula | – | Nitrate | – | Nitrate facilitates ectopic colonization of oral V. parvula in the intestine |
| Qian J, 2022 [23] | Mouse | Periodontitis salivary microbiota | Gut |
|
LC–MS (Thermo Ultimate 3000 system) |
|
| Kato T, 2018 [40] | Mouse | P. gingivalis | Serum | Alanine, Glutamine, Histidine, Tyrosine, and Phenylalanine | NMR spectrometer (Bruker Avance II 700; Bruker Biospin, Rheinstetten, Germany) | P. gingivalis administration elevated alanine, glutamine, histidine, tyrosine, and phenylalanine in the serum |
| Chen S, 2022 [41] | Mouse | P. gingivalis | Serum |
|
Untargeted metabolomics profiling (Thermo UHPLC-Q Exactive Mass Spectrometer) |
|
| Qian J, 2023 [74] | Mouse | Periodontitis salivary microbiota | Caecum content | Histidine metabolism | LC–MS, UHPLC, and AB SCIEX TripleTOF 6600 |
|
| Mottawea W, 2016 [75] | Human, mouse | A. parvulum | Mucosal-luminal interface samples | H2S | 16S rDNA sequencing, qPCR, HPLC-ESI-MS/MS | A. parvulum playing a pivotal role as the central hub within the H2S network |
| Yang ZH, 2021 [76] | Human | Klebsiella | Feces | Cholic acid, taurocholic acid, and glycochenodeoxycholic acid or taurochenodeoxycholic acid | Targeted metabolomics profiling (Waters XEVO TQ-S mass spectrometer) | Klebsiella was positively correlated with the primary and conjugated bile acids including cholic acid, taurocholic acid, and glycochenodeoxycholic acid or taurochenodeoxycholic acid |
SCFAs = short-chain fatty acids, PGI2 = prostaglandin I2, PGF2α = prostaglandin F2α, LC–MS = liquid chromatography–mass spectrometry.
Perspectives
Microbial studies on IBD have focused on the bacteria in the gut. The translocation of oral bacteria to the gut in IBD highlights the role of the oral–intestinal microbial axis. We hypothesized that ectopic colonization of oral pathological bacteria could promote IBD, which in turn boosts oral bacterial dysbiosis (Figure 1). Accumulating evidence supports an underlying association between oral microbes and IBD. Additionally, the oral cavity is an easily accessible site for microbial community assessment. The convenience and non-invasiveness of saliva collection compared with blood and feces make it an ideal candidate for the diagnosis or monitoring of IBD. However, prediction models based on a combination of oral and gut bacteria have yet to be developed. However, the association between oral bacteria and IBD remains unclear. Most studies on the oral–intestinal microbial axis are based predominantly on second-generation sequencing of 16S rRNA, yet this is insufficient for species identification in most bacteria. With advancements in sequencing technology, more precise and comprehensive sequencing technologies such as metagenomic techniques can be used to explore the association between oral bacteria and IBD. Additionally, the effects of oral bacteria on IBD have mainly been studied from the perspective of the immune response. Research on metabolic shifts resulting from ectopic colonization of oral bacteria is in its nascent stages and deserves further investigation.
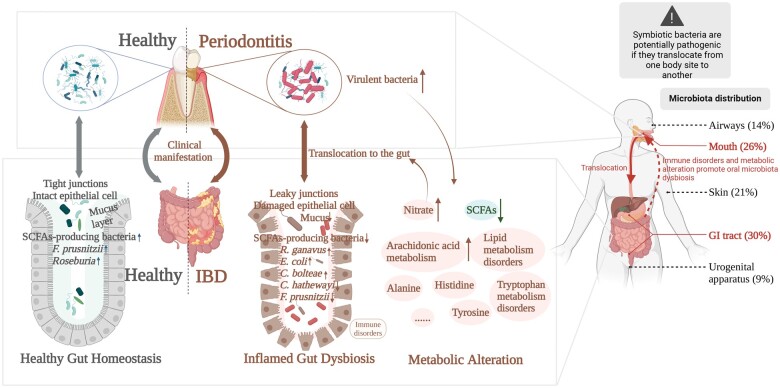
Figure 1.
Mutual interaction between oral microbiota dysbiosis and IBD. Patients with IBD have more pronounced oral clinical manifestations and periodontitis is associated with active disease. Virulent oral bacteria worsen IBD and contribute to oral microbiota dysbiosis, leading to metabolic alteration and immune disorders. Translocation of oral bacteria results in metabolic alteration such as increased arachidonic acid metabolism and decreased SCFAs in IBD. Specific metabolites in IBD such as nitrate facilitate ectopic colonization of oral bacteria in the gut. (Created using BioRender.com.) IBD = inflammatory bowel disease, SCFA = short-chain fatty acid.
Authors’ Contributions
B.X. and J.H. conceived the original idea for this study. B.X. performed a literature search and formulated the study protocol. M.Z., J.H., and M.Z. revised the manuscript. All authors reviewed and approved the final draft of the manuscript.
Acknowledgements
We thank for the National Key Clinical Discipline for its support.
Contributor Information
Bingjie Xiang, Department of Gastroenterology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Disease, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Key Laboratory of Human Microbiome and Chronic Diseases (Sun Yat-sen University), Ministry of Education, Guangzhou, Guangdong, P. R. China; Biomedical Innovation Center, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Jun Hu, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Disease, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Key Laboratory of Human Microbiome and Chronic Diseases (Sun Yat-sen University), Ministry of Education, Guangzhou, Guangdong, P. R. China; Biomedical Innovation Center, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Guangdong Institute of Gastroenterology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Min Zhang, Department of Gastroenterology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Disease, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Key Laboratory of Human Microbiome and Chronic Diseases (Sun Yat-sen University), Ministry of Education, Guangzhou, Guangdong, P. R. China; Biomedical Innovation Center, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Min Zhi, Department of Gastroenterology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Disease, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Key Laboratory of Human Microbiome and Chronic Diseases (Sun Yat-sen University), Ministry of Education, Guangzhou, Guangdong, P. R. China; Biomedical Innovation Center, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China; Guangdong Institute of Gastroenterology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Funding
This work was supported by the Sun Yat-sen University Clinical Research 5010 Program 2014008 (M.Z.), the National Natural Science Foundation of China [82270544], the Bureau of Science and Technology of Guangzhou Municipality [SL2022B03J00237], the “Jie Bang Gua Shuai” project of the Sixth Affiliated Hospital of Sun Yat-sen University [2022JBGS06], the program of Guangdong Provincial Clinical Research Center for Digestive Diseases [2020B1111170004], and the China Crohn's & Colitis Foundation [grant number CCCF.QF-2022A53-2].
Conflicts of Interest
The authors declare that there is no conflict of interests in this study.
References
- 1. Chichlowski M, German JB, Lebrilla CB. et al. The influence of milk oligosaccharides on microbiota of infants: opportunities for formulas. Annu Rev Food Sci Technol 2011;2:331–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kitamoto S, Kamada N.. Periodontal connection with intestinal inflammation: microbiological and immunological mechanisms. Periodontol 2000 2022;89:142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Imai J, Ichikawa H, Kitamoto S. et al. A potential pathogenic association between periodontal disease and Crohn's disease. JCI Insight 2021;6:e148543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wei MY, Shi S, Liang C. et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer 2019;18:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rojas-Tapias DF, Brown EM, Temple ER. et al. Inflammation-associated nitrate facilitates ectopic colonization of oral bacterium Veillonella parvula in the intestine. Nat Microbiol 2022;7:1673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li B, Ge Y, Cheng L. et al. Oral bacteria colonize and compete with gut microbiota in gnotobiotic mice. Int J Oral Sci 2019;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi Y, Zang SQ, Wei J. et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics 2021;113:664–76. [DOI] [PubMed] [Google Scholar]
- 8. Lam GA, Albarrak H, McColl CJ. et al. The oral-gut axis: periodontal diseases and gastrointestinal disorders. Inflamm Bowel Dis 2023;29:1153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elmaghrawy K, Fleming P, Fitzgerald K. et al. The oral microbiome in treatment-naïve paediatric IBD patients exhibits dysbiosis related to disease severity that resolves following therapy. J Crohns Colitis 2023;17:553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qi Y, Wu HM, Yang Z. et al. New insights into the role of oral microbiota dysbiosis in the pathogenesis of inflammatory bowel disease. Dig Dis Sci 2022;67:42–55. [DOI] [PubMed] [Google Scholar]
- 11. Elmaghrawy K, Hussey S, Moran GP.. The oral microbiome in pediatric IBD: a source of pathobionts or biomarkers? Front Pediatr 2020;8:620254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kitamoto S, Kamada N.. Untangling the oral-gut axis in the pathogenesis of intestinal inflammation. Int Immunol 2022;34:485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wade WG. The oral microbiome in health and disease. Pharmacol Res 2013;69:137–43. [DOI] [PubMed] [Google Scholar]
- 14. Read E, Curtis MA, Neves JF.. The role of oral bacteria in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2021;18:731–42. [DOI] [PubMed] [Google Scholar]
- 15. Lu Y, Li Z, Peng X.. Regulatory effects of oral microbe on intestinal microbiota and the illness. Front Cell Infect Microbiol 2023;13:1093967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imai J, Kitamoto S, Kamada N.. The pathogenic oral-gut-liver axis: new understandings and clinical implications. Expert Rev Clin Immunol 2021;17:727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng X, Tizzano M, Redding K. et al. Gingival solitary chemosensory cells are immune sentinels for periodontitis. Nat Commun 2019;10:4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen Z, Kuang S, Zhang Y. et al. Chitosan hydrogel incorporated with dental pulp stem cell-derived exosomes alleviates periodontitis in mice via a macrophage-dependent mechanism. Bioact Mater 2020;5:1113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamimi F, Altigani S, Sanz M.. Periodontitis and coronavirus disease 2019. Periodontol 2000 2022;89:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L, Jiang H, Chen R. et al. Human β-defensin 3 gene modification promotes the osteogenic differentiation of human periodontal ligament cells and bone repair in periodontitis. Int J Oral Sci 2020;12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leone P, Roche J, Vincent MS. et al. Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat Commun 2018;9:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madsen GR, Bertl K, Pandis N. et al. The impact of periodontitis on inflammatory bowel disease activity. Inflamm Bowel Dis 2023;29:396–404. [DOI] [PubMed] [Google Scholar]
- 23. Qian J, Lu J, Huang Y. et al. Periodontitis salivary microbiota worsens colitis. J Dent Res 2022;101:559–68. [DOI] [PubMed] [Google Scholar]
- 24. Kitamoto S, Nagao-Kitamoto H, Jiao Y. et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 2020;182:447–62.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee M, Chang EB.. Inflammatory Bowel Diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology 2021;160:524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haneishi Y, Furuya Y, Hasegawa M. et al. Inflammatory bowel diseases and gut microbiota. Int J Mol Sci 2023;24:3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu J, Cheng S, Yao J. et al. Correlation between altered gut microbiota and elevated inflammation markers in patients with Crohn’s disease. Front Immunol 2022;13:947313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Y, Wang M, Liu Q. et al. Recent advances in the medical applications of hemostatic materials. Theranostics 2023;13:161–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abusleme L, Hoare A, Hong BY. et al. Microbial signatures of health, gingivitis, and periodontitis. Periodontol 2000 2021;86:57–78. [DOI] [PubMed] [Google Scholar]
- 30. Baima G, Massano A, Squillace E. et al. Shared microbiological and immunological patterns in periodontitis and IBD: A scoping review. Oral Dis 2022;28:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun B, Liu B, Gao X. et al. Metagenomic analysis of saliva reveals disease-associated microbiotas in patients with periodontitis and crohn's disease-associated periodontitis. Front Cell Infect Microbiol 2021;11:719411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X, Sun B, Li L. et al. The oral microbiome analysis reveals the similarities and differences between periodontitis and Crohn's disease-associated periodontitis. FEMS Microbiol Lett 2022;369:fnac054. [DOI] [PubMed] [Google Scholar]
- 33. Kitamoto S, Nagao-Kitamoto H, Hein R. et al. The Bacterial Connection between the Oral Cavity and the Gut Diseases. J Dent Res 2020;99:1021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Atarashi K, Suda W, Luo C. et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science 2017;358:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horliana AC, Chambrone L, Foz AM. et al. Dissemination of periodontal pathogens in the bloodstream after periodontal procedures: a systematic review. PLoS One 2014;9:e98271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Press AG, Hauptmann IA, Hauptmann L. et al. Gastrointestinal pH profiles in patients with inflammatory bowel disease. Aliment Pharmacol Ther 1998;12:673–8. [DOI] [PubMed] [Google Scholar]
- 37. Walker MY, Pratap S, Southerland JH. et al. Role of oral and gut microbiome in nitric oxide-mediated colon motility. Nitric Oxide 2018;73:81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seedorf H, Griffin NW, Ridaura VK. et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 2014;159:253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang N, Zheng L, Qian J. et al. Salivary microbiota of periodontitis aggravates bone loss in ovariectomized rats. Front Cell Infect Microbiol 2022;12:983608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kato T, Yamazaki K, Nakajima M. et al. Oral administration of porphyromonas gingivalis alters the gut microbiome and serum metabolome. mSphere 2018;3:e00460-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen S, Niu C, Lv W.. Multi-omics insights reveal the remodeling of gut mycobiome with P. gingivalis. Front Cell Infect Microbiol 2022;12:937725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adolph TE, Meyer M, Schwärzler J. et al. The metabolic nature of inflammatory bowel diseases. Nat Rev Gastroenterol Hepatol 2022;19:753–67. [DOI] [PubMed] [Google Scholar]
- 43. Ala M. Tryptophan metabolites modulate inflammatory bowel disease and colorectal cancer by affecting immune system. Int Rev Immunol 2022;41:326–45. [DOI] [PubMed] [Google Scholar]
- 44. Sun M, Ma N, He T. et al. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr 2020;60:1760–8. [DOI] [PubMed] [Google Scholar]
- 45. Li M, Yang L, Mu C. et al. Gut microbial metabolome in inflammatory bowel disease: From association to therapeutic perspectives. Comput Struct Biotechnol J 2022;20:2402–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guasch-Ferré M, Hruby A, Toledo E. et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care 2016;39:833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nan W, Xiong F, Zheng H. et al. Myristoyl lysophosphatidylcholine is a biomarker and potential therapeutic target for community-acquired pneumonia. Redox Biol 2022;58:102556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin Y, Yuan Y, Ouyang Y. et al. Metabolome-wide association study of multiple plasma metals with serum metabolomic profile among middle-to-older-aged chinese adults. Environ Sci Technol 2022;56:16001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y, Bouza M, Wu C. et al. Sub-nanoliter metabolomics via mass spectrometry to characterize volume-limited samples. Nat Commun 2020;11:5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gallagher K, Catesson A, Griffin JL. et al. Metabolomic analysis in inflammatory bowel disease: a systematic review. J Crohns Colitis 2021;15:813–26. [DOI] [PubMed] [Google Scholar]
- 51. Yamamoto M, Shanmuganathan M, Hart L. et al. Urinary metabolites enable differential diagnosis and therapeutic monitoring of pediatric inflammatory bowel disease. Metabolites 2021;11:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin FP, Ezri J, Cominetti O. et al. Urinary metabolic phenotyping reveals differences in the metabolic status of healthy and inflammatory bowel disease (IBD) children in relation to growth and disease activity. Int J Mol Sci 2016;17:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Upadhyay KG, Desai DC, Ashavaid TF. et al. Microbiome and metabolome in inflammatory bowel disease. J Gastroenterol Hepatol 2023;38:34–43. [DOI] [PubMed] [Google Scholar]
- 54. Kumari R, Ahuja V, Paul J.. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J Gastroenterol 2013;19:3404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Machiels K, Joossens M, Sabino J. et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–83. [DOI] [PubMed] [Google Scholar]
- 56. De Preter V, Machiels K, Joossens M. et al. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut 2015;64:447–58. [DOI] [PubMed] [Google Scholar]
- 57. Guerreiro CS, Ferreira P, Tavares L. et al. Fatty acids, IL6, and TNFalpha polymorphisms: an example of nutrigenetics in Crohn's disease. Am J Gastroenterol 2009;104:2241–9. [DOI] [PubMed] [Google Scholar]
- 58. Tjonneland A, Overvad K, Bergmann MM. et al. ; IBD in EPIC Study Investigators. Linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: a nested case-control study within a European prospective cohort study. Gut 2009;58:1606–11. [DOI] [PubMed] [Google Scholar]
- 59. de Silva PS, Luben R, Shrestha SS. et al. Dietary arachidonic and oleic acid intake in ulcerative colitis etiology: a prospective cohort study using 7-day food diaries. Eur J Gastroenterol Hepatol 2014;26:11–8. [DOI] [PubMed] [Google Scholar]
- 60. Esteve-Comas M, Núñez MC, Fernández-Bañares F. et al. Abnormal plasma polyunsaturated fatty acid pattern in non-active inflammatory bowel disease. Gut 1993;34:1370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fernández-Bañares F, Esteve-Comas M, Mañé J. et al. Changes in mucosal fatty acid profile in inflammatorybowel disease and in experimental colitis: a common response to bowel inflammation. Clin Nutr 1997;16:177–83. [DOI] [PubMed] [Google Scholar]
- 62. Kayama H, Takeda K.. Emerging roles of host and microbial bioactive lipids in inflammatory bowel diseases. Eur J Immunol 2023;53:e2249866. [DOI] [PubMed] [Google Scholar]
- 63. Brown EM, Ke X, Hitchcock D. et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 2019;25:668–80.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Otake-Kasamoto Y, Kayama H, Kishikawa T. et al. Lysophosphatidylserines derived from microbiota in Crohn's disease elicit pathological Th1 response. J Exp Med 2022;219:e20211291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Iwatani S, Iijima H, Otake Y. et al. Novel mass spectrometry-based comprehensive lipidomic analysis of plasma from patients with inflammatory bowel disease. J Gastroenterol Hepatol 2020;35:1355–64. [DOI] [PubMed] [Google Scholar]
- 66. Franzosa EA, Sirota-Madi A, Avila-Pacheco J. et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ooi M, Nishiumi S, Yoshie T. et al. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm Res 2011;60:831–40. [DOI] [PubMed] [Google Scholar]
- 68. Scoville EA, Allaman MM, Brown CT. et al. Alterations in lipid, amino acid, and energy metabolism distinguish crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics 2018;14:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vantrappen G, Ghoos Y, Rutgeerts P. et al. Bile acid studies in uncomplicated Crohn’s disease. Gut 1977;18:730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lavelle A, Sokol H.. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223–37. [DOI] [PubMed] [Google Scholar]
- 71. Dowdell AS, Colgan SP.. Metabolic Host-Microbiota Interactions in Autophagy and the Pathogenesis of Inflammatory Bowel Disease (IBD). Pharmaceuticals (Basel) 2021;14:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vich Vila A, Hu S, Andreu-Sánchez S. et al. Faecal metabolome and its determinants in inflammatory bowel disease. Gut 2023;72:1472–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rashidi A, Koyama M, Dey N. et al. Colonization resistance is dispensable for segregation of oral and gut microbiota. BMC Med Genomics 2023;16:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qian J, Lu J, Cheng S. et al. Periodontitis salivary microbiota exacerbates colitis-induced anxiety-like behavior via gut microbiota. NPJ Biofilms Microbiomes 2023;9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mottawea W, Chiang CK, Mühlbauer M. et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun 2016;7:13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang ZH, Liu F, Zhu XR. et al. Altered profiles of fecal bile acids correlate with gut microbiota and inflammatory responses in patients with ulcerative colitis. World J Gastroenterol 2021;27:3609–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Agus A, Planchais J, Sokol H.. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018;23:716–24. [DOI] [PubMed] [Google Scholar]
- 78. Nikolaus S, Schulte B, Al-Massad N. et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 2017;153:1504–16.e2. [DOI] [PubMed] [Google Scholar]
- 79. Attene-Ramos MS, Nava GM, Muellner MG. et al. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen 2010;51:304–14. [DOI] [PubMed] [Google Scholar]
- 80. Tagaino R, Washio J, Abiko Y. et al. Metabolic property of acetaldehyde production from ethanol and glucose by oral Streptococcus and Neisseria. Sci Rep 2019;9:10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dunagan M, Chaudhry K, Samak G. et al. Acetaldehyde disrupts tight junctions in Caco-2 cell monolayers by a protein phosphatase 2A-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 2012;303:G1356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]