Abstract
ZiPD (zinc phosphodiesterase; synonyms are ElaC, ecoZ, RNaseZ and 3′ tRNase) and the iron-dependent redox enzyme FlRd (flavorubredoxin) from Escherichia coli represent prototypical cases of proteins sharing the metallo-β-lactamase fold that require strict metal selectivity for catalytic activity, yet their metal selectivity has only been partially understood. In contrast with hydrolytic metallo-β-lactamase proteins, iron-dependent FlRd-like enzymes have an atypical glutamate ligand, which replaces one otherwise conserved histidine ligand. X-ray absorption spectroscopy revealed that the FlRd metallo-β-lactamase domain is capable of incorporating two zinc ions into the binuclear metal-binding site. Zinc dissociation constants, determined by isothermal titration calorimetry are similar for zinc binding to E. coli ZiPD (Kd1=2.2±0.2 μM and Kd2=23.0±0.6 μM) and to the E. coli FlRd metallo-β-lactamase domain (Kd1=0.7±0.1 μM and Kd2=26.0±0.1 μM). In good correspondence, apo-ZiPD requires incubation with 10 μM zinc for full reconstitution of the phosphodiesterase activity. Accordingly, metal selectivity of ZiPD and FlRd only partially relies on first shell metal ligands. Back mutation of the atypical glutamate in FlRd to a histidine unexpectedly resulted in an increased first zinc dissociation constant (Kd1=30±4 μM and Kd2=23±2 μM). In combination with a recent mutational study on ZiPD [Vogel, Schilling and Meyer-Klaucke (2004) Biochemistry 43, 10379–10386], we conclude that the atypical glutamate does not guide metal selectivity of the FlRd metallo-β-lactamase domain but suppresses possible hydrolytic cross-activity.
Keywords: isothermal titration calorimetry, metal binding, metallo-β-lactamase domain, phosphodiesterase, X-ray absorption spectroscopy, zinc affinity
Abbreviations: bpNPP, bis(p-nitrophenyl)phosphate; DTT, dithiothreitol; FlRd, flavorubredoxin; FMN, flavin mononucleotide; ITC, isothermal titration calorimetry; Ni-NTA, Ni2+-nitrilotriacetate; PIXE, proton induced X-ray emission; ROO, rubredoxin–oxygen oxidoreductase; TEV, tobacco etch virus; ZiPD, zinc phosphodiesterase
INTRODUCTION
Metal ions, as part of metalloproteins, are essential for numerous biocatalytic processes. In many cases, metalloenzymes require specific metal ions to achieve catalytic functionality, e.g. zinc for hydrolytic activities or iron for redox proteins. Highly selective binding of metal ions is therefore a presumption in the biology of metalloproteins, yet the structural and functional basis of this process often remains ambiguous. As a recent example, virtually identical metal co-ordination spheres were found for manganese- or iron-dependent homoprotocatechuate 2,3-dioxygenases, and the metal ion specificity of these enzymes could not be elucidated [1].
Another example is the metallo-β-lactamase superfamily, which comprises intra- and extracellular metalloproteins present in all the three domains of life [2]. The characteristic metallo-β-lactamase fold consists of external α-helices and two internal layers of β-sheets [3]. A second characteristic feature is the active site with a binuclear metal-binding site. Its metal-binding ligands are part of loops and turns that connect the β-strands of the internal β-sheets [4]. Proteins of the metallo-β-lactamase superfamily catalyse a wide variety of hydrolytic reactions but also include enzymes with redox activity [2,5]. While the hydrolytic enzymes mostly use zinc for catalysis, iron is required for the redox enzymes. However, metal selectivity of metallo-β-lactamase proteins has not been entirely understood. For example, glyoxalase II, a thiolesterase with a metallo-β-lactamase fold [6], is catalytically active with various ratios of in vivo bound iron, manganese and zinc [7]. The proteins for which this family is named are extracellular enzymes providing antibiotic resistance by hydrolysing the amide bond of β-lactam antibiotics. For this subgroup, zinc dependence of the catalytic activity was the object of intense research. The metallo-β-lactamase AE036 from Aeromonas hydrophila is active with one zinc and inhibited by the presence of a second zinc ion in the active site [8]. The metallo-β-lactamases from Bacteroides fragilis and Bacillus cereus (strain 569/H/9) have comparable catalytic activities for both the mono- and bi-zinc forms [9,10].
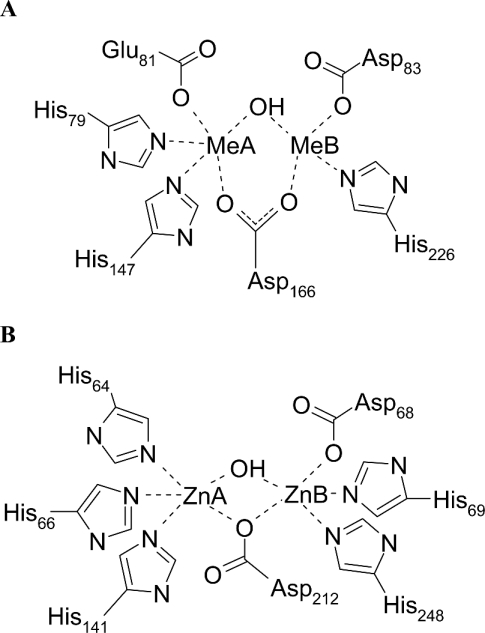
ElaC proteins form a new family of cytosolic enzymes sharing the metallo-β-lactamase fold. ElaC from Escherichia coli was recently characterized as an effective phosphodiesterase with a bi-zinc centre [11]. The protein was named ZiPD (zinc phosphodiesterase; synonyms are ecoZ, RNase Z and 3′ tRNase) since no explanation for the name ElaC had been published. ZiPD is exclusively activated by zinc but not by other transition metals, such as iron and manganese [11]. ZiPD homologues are present in all the three domains of life. A number of eukaryotes possess two ElaC genes, one of which (ElaC2) has approximately twice the size of ZiPD and is regarded as a fusion of two ZiPD monomers [12]. ElaC2 was identified as a prostate cancer susceptibility gene [12] and its Caenorhabditis elegans homologue is involved in germline proliferation [13]. ZiPD proteins from various sources act as tRNA 3′-processing endoribonucleases, and include ZiPD homologues from Arabidopsis thaliana and Methanococcus janaschii [14], human and yeast [15] as well as Drosophila melanogaster [16], Thermotoga maritime [17], Haloferax volcanii [18] and Bacillus subtilis [19]. ZiPD from E. coli relies on the presence of two zinc ions in its active site for phosphodiesterase activity [11]. Recently, the ZiPD zinc co-ordination was elucidated [20]. One zinc-binding site is composed of three histidine residues, whereas the other zinc-binding site consists of two histidine residues and one aspartate residue. Both zinc ions, with a metal–metal distance of approx. 3.3 Å (1 Å=0.1 nm), are bridged by an additional aspartate residue (Figure 1B).
Figure 1. The binuclear metal-binding sites of D. gigas ROO/E. coli FlRd and E. coli ZiPD.
(A) The bi-metal site of E. coli FlRd. The scheme is based on the D. gigas ROO structure, whose metal ligands are conserved in E. coli FlRd [5,22]. The present Zn K-edge EXAFS points out that the FlRd metallo-β-lactamase domain is also capable of binding zinc (Figure 2, Table 1). Me, metal (zinc or iron). (B) The model for ZiPD is taken from the literature [20]; however, the present data do not unequivocally distinguish whether Asp-212 bridges the two zinc ions in a monodentate or bidentate manner.
The metallo-β-lactamase fold is also found in a distinct group of cytosolic redox proteins [5]. These multidomain proteins are found in archaea and bacteria and comprise an N-terminal metallo-β-lactamase-like domain with a bi-iron site, a flavodoxin domain with an FMN (flavin mononucleotide) cofactor and in some cases, such as in E. coli, also a C-terminal rubredoxin domain [5,21,22]. The ROO (rubredoxin–oxygen oxidoreductase) from Desulfovibrio gigas is the terminal oxygen reductase of a cytosolic oxygen-scavenging pathway, which simultaneously allows for NAD+ regeneration and oxygen removal [23,24]. The D. gigas enzyme interacts with rubredoxin, whereas the corresponding E. coli enzyme, containing a C-terminal rubredoxin domain, is considered to be a fusion with its redox partner [22]. The E. coli enzyme was named FlRd (flavorubredoxin) and was also found to be the terminal oxygen reductase of a cytosolic electron transfer chain [22]. The rubredoxin domain of the E. coli FlRd enzyme receives electrons from a rubredoxin reductase named FlRd reductase [22]. Within E. coli FlRd electrons are then transferred through the FMN cofactor of the flavodoxin domain to the bi-iron site of the metallo-β-lactamase domain, where oxygen is reduced to water [22]. The crystal structure of D. gigas ROO was modelled with a dioxygen molecule in the vicinity of the bi-iron site [5]. ROO from D. gigas is a homodimer with the FMN cofactor of one subunit being approx. 6 Å from the bi-iron site of the second subunit [5]. Recently, it was discovered that E. coli FlRd also has nitric oxide reductase activity [25,26]. Its expression is up-regulated under anaerobic growth conditions, especially on increased nitrite levels [27].
The D. gigas ROO crystal structure revealed a unique metal-binding site within the metallo-β-lactamase superfamily. A common feature of hydrolytic enzymes with a metallo-β-lactamase fold is one zinc-binding site with three ligating histidine residues [4,6,28–30]. The second zinc-binding site is more versatile and is composed of ligating histidine and aspartate residues as well as ligating cysteine residues for the metallo-β-lactamases from B. cereus, B. fragilis and A. hydrophila (CphA enzyme) [4,28,31,32]. Hydrolytic proteins belonging to the metallo-β-lactamase family share the short sequence motif HXHXDH [2]. While the first three conserved amino acids are generally conserved metal ligands, the fourth amino acid participates in metal co-ordination only in the cases of glyoxalase II and the metallo-β-lactamases L1 and FEZ-1 of the B3 subclass [6,29,30,33]. For the bi-iron enzymes D. gigas ROO and E. coli FlRd, the conserved sequence motif exchanged to HXEXDH [22], with one histidine of the characteristic three histidine metal-binding site being replaced by a glutamate residue [5]. One iron ion is ligated by two histidine residues, one glutamate and one aspartate, whereas the second iron ion is co-ordinated by one histidine and two aspartate residues [5] (Figure 1). It has been speculated that this replacement of a histidine by an atypical glutamate plays a major role in the iron selectivity of ROO [34]. The metal ligands found in the ROO crystal structure are conserved in the E. coli FlRd sequence [34].
We aimed to explore the zinc affinity and selectivity of cytosolic metallo-β-lactamase domain proteins. While numerous studies were performed on the zinc-binding properties of extracellular metallo-β-lactamase domain proteins, the present study is the first comparative characterization of zinc binding to cytosolic metallo-β-lactamase domain proteins. We used E. coli ZiPD as a prototypical zinc-dependent hydrolase and the E. coli FlRd metallo-β-lactamase domain as a prototypical iron-selective enzyme. Zinc binding was structurally and functionally probed by means of X-ray absorption spectroscopy, activation of apo ZiPD and comparative isothermal zinc titration calorimetry.
The role of the atypical glutamate at the FlRd metal-binding site was investigated by mutation to a histidine ligand, restoring the typical three histidine metallo-β-lactamase zinc-binding site.
EXPERIMENTAL
Materials
Except when stated otherwise, all fine chemicals were purchased from Sigma (Deisenhofen, Germany). Restriction enzymes were from New England Biolabs (Frankfurt, Germany). Oligonucleotides were synthesized by Proligo (Paris, France). DNA sequencing was performed by MWG-Biotech (Ebersberg, Germany).
Expression plasmids
pET32a-ZiPD
The previously used petM11-ZiPD expression plasmid yielded ZiPD protein with an N-terminal His6 tag, which could not be removed despite the presence of a TEV (tobacco etch virus) protease cleavage site [11]. To obtain untagged ZiPD for ITC (isothermal titration calorimetry) measurements, the following construct was used. The ElaC gene was cloned from E. coli DH5α genomic DNA by PCR using the primers (restriction sites underlined) 5′-ATATGGATCCATGGAATTAATTTTTTTAGGTACTTCAGCC-3′ (5′-primer) and 5′-ATATTACTCGAGTTAAACGTTAAACACGGTGAAATCATTCGC-3′ (3′-primer). The PCR product was digested with BamHI and XhoI and cloned into the BamHI and XhoI sites of a pET32a vector (Novagen, Madison, WI, U.S.A.), yielding pET32a-ZiPD. The pET32a vector includes a thioredoxin tag, an S tag and a His6 tag at the N-terminus. All tags can be removed from the final gene product by an enterokinase cleavage site. The integrity of the cloning product was verified by DNA sequencing.
FlRd-lactamase domain
The FlRd-lactamase domain was cloned from E. coli DH5α genomic DNA by PCR using the primers (restriction sites underlined) 5′-TGAGGTTGCCATGGCTATTGTGGTGAA-3′ (5′-primer) and 5′-TCCTCGAGTTAATCAGCCGCCCATTTCAG-3′ (3′-primer, also introducing stop codon). The PCR product was digested with NcoI and XhoI and cloned into the NcoI and XhoI sites of a modified pET24d vector (Novagen), which contains an N-terminal His6 tag and a TEV protease cleavage site, yielding pETM11-FlRd-lactamase. DNA sequencing proved that the amplified sequence corresponds to the first 248 residues of the FlRd gene (accession no. U29579), hence representing the metallo-β-lactamase domain of the gene product [22].
FlRd-lactamase domain E81H (Glu81→His) mutant
Site-directed mutagenesis of the pETM11-FlRd-lactamase expression plasmid was performed essentially as described in [20]. pETM11-FlRd-lactamase was used as a template. The sequence of the forward primer was 5′-GTGATTAACCATGCACATGAGGACCACGCTGGG-3′ with the reverse primer being complementary. In addition to the E81H mutation (shown in boldface), silent mutations were introduced to delete an Ear1 restriction site of the wild-type gene, allowing for easy screening of the mutant vector. This was guided by the worldwide web-based program ‘The Primer Generator’ [35]. The resulting insert, coding for the E81H mutation of the FlRd-lactamase domain, was verified by DNA sequencing.
Protein expression and purification
ZiPD from pETM11-ZiPD
Expression and purification of ZiPD based on the petM11-ZiPD expression plasmid was described previously [11,20]. To obtain ‘as isolated’ protein, zinc was omitted from the lysis buffer.
ZiPD from pET32a-ZiPD
E. coli BL21(DE3) cells harbouring pET32-ZIPD were grown in Luria–Bertani medium to an absorbance A600 0.5 at 37 °C followed by an induction with 0.1 mM isopropyl β-D-thiogalactoside at 25 °C for 18 h. Cells were lysed by ultrasonication in 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 10% (v/v) glycerol, 0.2 mM DTT (dithiothreitol) and 70 μM zinc acetate. The recombinant protein was purified with Ni-NTA (Ni2+-nitrilotriacetate) agarose (Qiagen, Hilden, Germany). The eluted fraction was then dialysed against 50 mM Tris/HCl (pH 8.0), 0.5 mM DTT, and cleavage of the His6 tag was performed by adding 10 units of EnterokinaseMax (Invitrogen, Karlruhe, Germany) and incubating at 22 °C overnight. Ni-NTA agarose was used to remove the cleaved tag and uncleaved protein from the tag-free ZiPD. Purification of ZiPD from EnterokinaseMax and remaining contaminants was performed by gel filtration using a Superdex 200 16/60 column (Amersham Biosciences, Freiburg, Germany) equilibrated with 20 mM Tris/HCl (pH 7.4), 150 mM NaCl, 0.5 mM DTT and 70 μM zinc acetate. The recombinant protein eluted at a volume corresponding to a dimer of approx. 70 kDa.
FlRd-lactamase domain and its E81H mutant
For the expression of the FlRd-lactamase domain and its E81H mutant, BL21(DE3) cells harbouring pETM11-FlRd-lactamase or pETM11-FlRd-lactamase-E81H respectively were grown in Luria–Bertani medium to an A600 0.6, followed by induction with 0.1 mM isopropyl β-D-thiogalactoside at 25 °C for 16 h. Cells were lysed by ultrasonication in 50 mM Tris/HCl (pH 8.0), 150 mM NaCl and 10% glycerol. The recombinant protein was purified with Ni-NTA agarose (Qiagen). The buffer of the eluted fractions was changed to 50 mM Tris/HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, and the N-terminal His6 tag was removed by incubation with recombinant TEV protease (EMBL Protein Expression Core Facility, Heidelberg, Germany) at room temperature (22 °C) for 16 h. The TEV protease, which is itself produced as His6-tagged protein, as well as the remaining His-tagged recombinant protein was removed by a second Ni-NTA agarose (Qiagen) purification step, with the tag-free protein now being in the flow-through. This was followed by a gel filtration using a Superdex 75 16/60 column (Amersham Biosciences) equilibrated with 50 mM Tris/HCl (pH 7.4), 150 mM NaCl and 1 mM DTT. Both the FlRd-lactamase domain and its E81H mutant eluted at a volume corresponding to approx. 30 kDa.
Protein concentration
Protein concentrations were determined by measuring A280 values and using calculated molar absorption coefficients [36]. Absorbance values for a 0.1% (w/v) solution at 280 nm were: 0.67 for His-tagged ZiPD (molecular mass Mw 36.088 kDa), 0.62 for untagged ZiPD (Mw 33 kDa), 1.63 for FlRd metallo-β-lactamase domain (Mw 28.783 kDa) and 1.63 for the E81H mutant of the FlRd metallo-β-lactamase domain (Mw 28.791 kDa).
Metal analysis
Metal content of the protein was probed using PIXE (proton-induced X-ray emission) [37] at the LIPSION microprobe facility (Leipzig University, Leipzig, Germany) [38,39]. Protein samples of approx. 5 mg/ml in 100 mM Tris/acetic acid (pH 7.4) were deposited in 1 μl drops on a 0.9-μm-thick polyethylene terephthalate foil stretched on aluminium sample holders. A focused proton beam of 2.25 MeV and circular beam spot of approx. 2 μm diameter was used to scan the sample areas between 0.16 and 0.64 mm2 with currents between 200 and 400 pA. Simultaneous PIXE and RBS (Rutherford Back-scattering Spectroscopy) data were obtained; the RBS data were used to determine the sample thickness. X-rays were detected with a high-purity germanium, IGLET-X detector (EG&G Ortec) located at an angle of 135° with respect to the beam and protected from back-scattered particles by a 60 μm polyethylene filter. The back-scattered protons were detected by an annular Passivated Implanted Planar Silicon detector at an angle of 173° with respect to the beam, covering a solid angle of 75 msr. The number of metal atoms per protein was calculated by fitting the PIXE spectra using the analysis program GeoPIXE [40], taking into account X-ray emission yields and self-absorption of X-rays by the sample, especially the characteristic K lines from sulphur. For each sample, 5–10 scans were performed and analysed individually to ensure that no significant loss of sulphur or metal occurred.
Kinetic measurements
Phosphodiesterase activity of ZiPD was measured using the chromogenic substrate bpNPP [bis(p-nitrophenyl)phosphate] essentially as described in [11]. Reactions were conducted in 50 mM Tris/HCl (pH 7.4), 0.1% Triton X-100 and 2 mM 2-mercaptoethanol. For one particular series of experiments, the bpNPP solution also contained 10 mM EDTA (30 min preincubation at room temperature). One unit of activity corresponds to 1 μmol of p-nitrophenol liberated per min at 22 °C.
Activation of apo-ZiPD with zinc
Apo-ZiPD was produced as described previously [11]. The apo-enzyme was incubated for 16 h at 4 °C in solution containing zinc at concentrations ranging from 0 to 10 mM ZnCl2. Incubation buffer was 50 mM Tris/HCl (pH 7.4), 150 mM NaCl and 10% glycerol.
X-ray absorption spectroscopy
The FlRd-lactamase domain was incubated in 50 mM Tris/HCl (pH 7.4), 15% glycerol, 0.1 mM ZnCl2 at 4 °C for 16 h. Excess metal was removed by extensive dialysis against Chelex100 (Bio-Rad Laboratories, Hercules, CA, U.S.A.)-treated 50 mM Tris/HCl (pH 7.4)/15% glycerol. The protein was then concentrated to 1–2 mM, filled into plastic sample holders covered with polyimide windows, frozen in liquid nitrogen and kept at 30 K during the experiment. X-ray absorption spectra at the Zn K-edge for zinc-incubated FlRd-lactamase domain were recorded in fluorescence mode at the EMBL bending magnet beamline D2 (Deutsches Elektronen Synchrotron, Hamburg, Germany) equipped with a Si(111) double-crystal monochromator, a focusing mirror and a 13-element Ge solid-state fluorescence detector (Canberra, Meriden, CT, U.S.A.). Harmonic rejection was achieved by a focusing mirror cut-off at 21 keV and a monochromator detuning to 70% of peak intensity. Dead time correction was applied. Saturation was not observed because the dead time was always below 20%. The energy axis of each scan was calibrated by using the Bragg reflections of a static Si(220) crystal in back-reflection geometry [41]. Averaging of 20 scans and data reduction were performed with the EXPROG software package (C. Hermes and H. F. Nolting, EMBL, Hamburg, Germany) using the zinc absorption edge position E0,Zn=9660 eV. The EXAFS spectrum was analysed with EXCURV98 [42], applying constrained refinement. The R factor was used as a measure for the goodness of the fit [42]. The model for the zinc co-ordination of the FlRd-lactamase domain was based on the iron co-ordination found in the D. gigas ROO crystal structure [5]. The presence of two zinc ions results in an average co-ordination by 1.5 histidine and 2.0 aspartate residues as well as 1 water molecule. Histidine ligands are represented by an imidazole group as provided by EXCURV98, and aspartate residues by a carboxylate group with the adjacent carbon atom (Cβ for aspartic acid and Cγ for glutamic acid) as provided by EXCURV98. Owing to the limited resolution of EXAFS at Δk=3–14 Å−1, distances to first shell N and O atoms were refined collectively.
ITC
ITC measurements were performed at 22 °C with a VP-ITC titration calorimeter (Microcal, Northampton, MA, U.S.A.). To obtain apo-protein, samples were dialysed for 40 h at 4 °C against 10 mM EDTA and 5 mM o-phenanthroline in 50 mM Tris/HCl (pH 7.4) 150 mM NaCl, 15% glycerol, followed by three dialysis steps in Chelex100 (Bio-Rad Laboratories)-treated 50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 15% glycerol to remove EDTA and phenanthroline. Apo-protein samples were analysed with a Superdex 75 10/30 gel filtration column to verify that no protein aggregation occurred. The dialysis buffer was used to prepare a 1.0 mM ZnCl2 solution used for titration. Protein concentrations in the sample cell were 18.2 μM for untagged ZiPD, 18.1 μM for FlRd-lactamase domain and 20.5 μM for the E81H mutant of the FlRd-lactamase domain. The titration was performed by a total of 60–80 injections of 4, 5 and 5 μl into ZiPD, FlRd and E81H solutions respectively until the protein sample was saturated with zinc. Blank titrations of the ZnCl2 solution into the dialysis buffer were performed to correct for the dilution heat of the zinc solution. Data reduction and analysis were performed with the Origin software (Microcal) applying the sequential binding sites model for two binding sites [43].
RESULTS
Zinc co-ordination in the FlRd binuclear active site
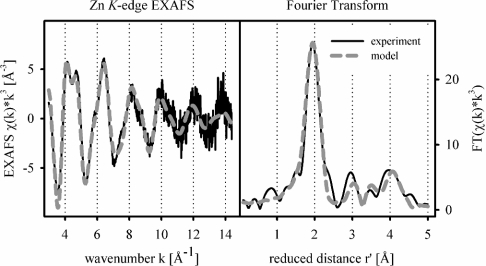
Both the FlRd-lactamase domain as well as its E81H mutant bound approx. 1.5 zinc ions per monomer on incubation of the apoenzyme with 75 μM zinc solution. To investigate further the zinc binding of the FlRd-lactamase domain, we applied EXAFS spectroscopy to characterize the zinc co-ordination environment. The Zn K-edge EXAFS spectrum shows typical features of protein-bound zinc, such as the double peak between k=4 and 5 Å−1 (Figure 2). EXAFS analysis provides evidence for a neighbouring zinc atom, obvious as an additional Fourier transform peak between 3 and 4 Å. The zinc–zinc contribution is of limited significance but a similar situation was observed for glyoxalase II, a metallo-β-lactamase domain protein with a bi-metal site. Gly-oxalase II also incorporated iron and manganese. While Fe and Mn K-edge EXAFS did not unambiguously prove the presence of a neighbouring metal, EPR spectroscopy clearly detected Fe3+Fe2+ and Mn2+Mn2+ centres [7]. Metal–metal contributions in X-ray absorption spectroscopy may be hidden due to phase cancellation by backscattering from light elements, e.g. belonging to imidazole groups [44–46]. In summary, the Zn K-edge EXAFS of the FlRd-lactamase domain is indicative of a bi-metal active site. The metal co-ordinating residues of D. gigas ROO are conserved in E. coli FlRd [22]. Given the presence of two zinc ions, the EXAFS spectrum is modelled with an average co-ordination by 1.5 histidine, 2.0 aspartate or glutamate residues and 1.0 water molecule or hydroxide ligands, all at 1.99±0.01 Å (Table 1). The zinc–zinc distance was refined to 3.29±0.02 Å. Zinc–ligand distances correspond to characteristic zinc co-ordination bond lengths found in small-molecule crystallography and high-resolution protein structures [47,48]. In essence, the FlRd metallo-β-lactamase domain is capable of accommodating zinc in its bi-nuclear active site.
Figure 2. Zn K-edge EXAFS and Fourier transform of FlRd-lactamase domain.
Zn K-edge EXAFS (left panel) and the corresponding Fourier transform (right panel) of FlRd-lactamase domain. Details of the model are stated in Table 1. χ(k), the EXAFS amplitude; r′, the metal–ligand distance corrected for first-shell phase shifts; FT, Fourier transform amplitude.
Table 1. Zinc co-ordination of FlRd-lactamase domain as derived by EXAFS analysis.
N is the co-ordination number, r the mean interatomic distance and σ2 the Debye–Waller factor. Numbers in parentheses represent the uncertainties of the last digit. The R factor of this model is 34.3% and the Fermi energy offset is −7.5 (3) eV. Quantitative EXAFS analysis determines co-ordination numbers with an accuracy of 20% as was found by a reference study on model compounds [62]. This intrinsic error is not explicitly stated. The fitting procedure is described in the Experimental section.
| Ligand | Atom* | N | r (Å) | σ2 (Å2) |
|---|---|---|---|---|
| His† | N1 | 1.5 | 1.99 (1) | 0.006 (1) |
| C2 | 3.03 | 0.012 (3) | ||
| C2 | 3.03 | 0.012 (3) | ||
| C3 | 4.14 | 0.003 (2)‡ | ||
| N3 | 4.17 | 0.003 (2)† | ||
| Asp/Glu† | O1 | 2.0 | 1.99 (1) | 0.006 (1) |
| C2 | 2.82 | 0.012 (3) | ||
| O2 | 3.03 | 0.012 (3) | ||
| C4 | 4.22 | 0.003 (2)‡ | ||
| OH−/water | O1 | 1.0 | 1.99 (1) | 0.006 (1) |
| Zn | 1.0 | 3.29 (2) | 0.011 (2) |
* For atoms with similar distances to the Zn centre, the Debye–Waller factors have been refined collectively. This is marked by identical subscripts.
† The ∠Zn–N–C for the histidine ligand and ∠Zn–O–C for the carboxylate ligand were refined to allow for some structural flexibility of the ligands. ∠Zn–N–C=129±28°, ∠Zn–O–C=120±2°.
‡ The Debye–Waller factors of outer shell atoms at approx. 4 Å are considered to be too low. This is attributed to the correlation between the Debye–Waller factor and the co-ordination number. In the D. gigas ROO crystal structure a number of additional atoms are present at this distance, e.g. belonging to the non-ligating residue His-24 or water molecules.
Comparative zinc affinities of ZiPD, FlRd-lactamase domain and its E81H mutant
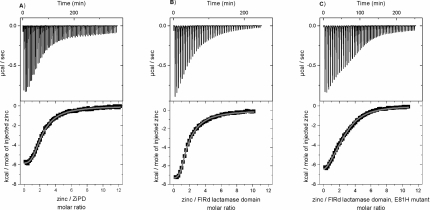
To determine and compare the zinc-binding affinities of ZiPD, FlRd-lactamase domain and its E81H mutant, we applied ITC. For all the three proteins, zinc binding heats were detected (Figure 3, Table 2). The integrated binding heats were indicative of two binding events, which agrees nicely with the number of predicted metal sites and the number of zinc ions per protein detected by metal analysis. The integrated binding heats were fitted to the sequential-binding site model with two binding sites. This model assumes interacting binding sites, which was the case for all previous studies on metal binding to metallo-β-lactamases [8,49,50]. ZiPD had a first dissociation constant (Kd1) of 2.2±0.2 μM and a second dissociation constant (Kd2) of 23.0±0.6 μM. Kd1=0.7±0.1 and Kd2=26.0±0.1 μM for FlRd-lactamase domain. ZiPD and the FlRd-lactamase domain, therefore, exhibit similar Kd1 and Kd2 values. Both enzymes displayed slightly negative co-operativity for zinc binding, since Kd2<Kd1. It was previously speculated that the atypical ligand E81 in the FlRd-lactamase domain functions to decrease the zinc affinity and enable selective iron binding [34]. Back-mutation of this atypical glutamate to histidine restores the typical three-histidine binding site of the metallo-β-lactamase domain. Surprisingly, the E81H mutant of the FlRd-lactamase domain displayed decreased zinc-binding affinity for the first binding event with Kd1=30±4 μM and Kd2=23±2 μM. Iron affinity was not probed by ITC due to the oxidation of ferrous iron to ferric iron. The use of reducing agents led to large dilution heats that made it impossible to determine the iron binding heat. It is of interest to determine whether a particular binding event can be assigned to a particular binding site (Figure 1). However, the present results do not resolve whether the sequence of binding events to particular binding sites remained unaltered for both the wild-type and E81H mutant.
Figure 3. ITC curves of zinc binding to ZiPD, FlRd-lactamase domain and its E81H mutant.
ITC measurements and data analysis were conducted as described in the Experimental section. (A) ZiPD, (B) FlRd metallo-β-lactamase domain and (C) E81H mutant of FlRd metallo-β-lactamase domain. Data were fitted to the sequential binding sites model with two binding sites yielding the thermodynamic parameters stated in Table 2 (continuous line). Upper panels, raw data for sequential injections of zinc solution into the protein solution. Lower panels, integrated-binding heats as a function of the Zn/protein ratio. 1 kcal=4.184 kJ.
Table 2. Zinc-binding properties of ZiPD, FlRd-lactamase domain and its E81H mutant as determined by ITC.
Isothermal titrations of zinc to ZiPD, FlRd-lactamase domain and its E81H mutant were performed as described in the Experimental section and as depicted in Figure 3. Isothermal titration curves were analysed applying the sequential binding sites model with two binding sites. Temperature was 22 °C. 1 cal=4.184 J.
| ZiPD | FlRd-lactamase domain | FlRd-lactamase domain, E81H | |
|---|---|---|---|
| Kd1 (μM) | 2.2±0.2 | 0.7±0.1 | 30±4 |
| ΔH1 (cal/mol) | −6511±90 | −7607±71 | −15990±1126 |
| ΔS1 [cal/(mol·deg)] | 3.9 | 2.5 | −33.5 |
| ΔG1 (cal/mol) | −7661 | −8345 | −6107 |
| Kd2 (μM) | 23.0±0.6 | 26.0±0.1 | 23±2 |
| ΔH2 (cal/mol) | −12090±158 | −12000±142 | −4008±1297 |
| ΔS2 [cal/(mol·deg)] | −19.8 | −19.7 | 7.6 |
| ΔG2 (cal/mol) | −6249 | −6189 | −6250 |
Activation of apo-ZiPD with zinc
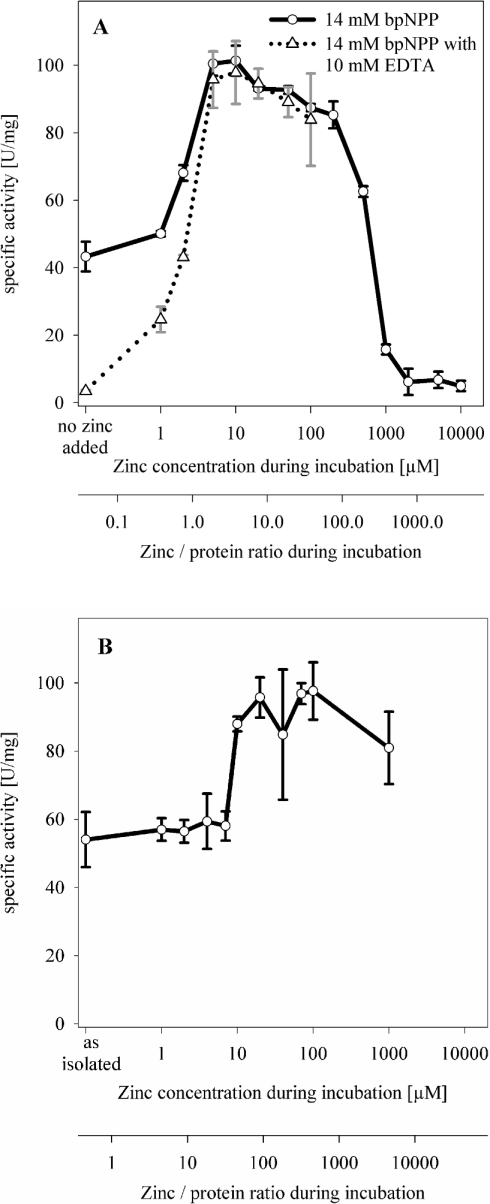
The phosphodiesterase activity of ZiPD depends on the presence of two zinc ions in its binuclear active site [11]. To determine the zinc concentration required by apo-ZiPD to gain full enzymic activity, we incubated apo-ZiPD sequentially with up to 10 mM ZnCl2 and determined the specific phosphodiesterase activity (Figures 4A and 4B). Apo-ZiPD with no added zinc seemed to have a specific activity of approx. 45 units/mg using the substrate bpNPP. Addition of a chelating agent (10 mM EDTA) to the substrate solution abolished the previously measured activity for apo-ZiPD, but had only a negligible effect on the maximum phosphodiesterase activity that was observed after the incubation of apo-ZiPD with 10 μM ZnCl2 (Figure 4A). We conclude that the previously observed residual activity of apo-ZiPD was due to traces of zinc in the substrate solution. At zinc concentrations ≪10 μM (preincubation), a zinc background of approx. 2 μM in the substrate solution led to partial activation of apo-ZiPD. We estimate the zinc background of the substrate solution to be approx. 2 μM since apo-ZiPD incubated with 2 μM ZnCl2 had a specific activity of 43 units/mg when assayed in the presence of EDTA. This value corresponds nicely to the activity of apo-ZiPD with no added zinc when assayed in the absence of a chelating agent for the removal of the zinc background (45 units/mg). Notably, the duration of the kinetic measurement (2 min) was sufficiently short to prevent the removal of protein-bound zinc from ZiPD by the chelating agent EDTA, as indicated by similar maximum activities both in the presence and absence of EDTA. Since the chelating agent did not decrease maximum ZiPD activity, we assume that protein-bound zinc was not removed. Thus the low activity of apo-ZiPD in the presence of EDTA indicates that the zinc background of the apoenzyme solution was at least one order of magnitude lower than that in the substrate solution. In good agreement, zinc activation studies for the metalloprotease astacin yielded a zinc background of approx. 0.02 μM despite extensive measures to remove any metal contamination [60], whereas studies on the FEZ-1 metallo-β-lactamase revealed a zinc background of approx. 0.4 μM [61].
Figure 4. Activation of apo-ZiPD with zinc.
(A) Apo-ZiPD was incubated with zinc concentrations ranging from 0 to 10 mM as described in the Experimental section. Phosphodiesterase activity was measured with 14 mM bpNPP (solid line) in triplicate; data points designate the mean value, error bars designate the S.D. ZiPD concentration during the incubation was 0.093 mg/ml. In a second series (broken line), phosphodiesterase activity was measured with a 14 mM bpNPP solution containing 10 mM EDTA to remove free metal ions (30 min preincubation at room temperature). (B) Zinc activation of ZiPD as isolated, using a lower ZiPD concentration of 0.008 mg/ml. ZiPD is isolated with small, substoichiometric amounts of zinc (0.3 zinc/monomer [11]).
In good agreement with the Kd values measured by ITC, incubation with 10 μM zinc was required for the maximum activation of apo-ZiPD. Both independent methods, therefore, point towards a ZiPD zinc affinity in the low-μM range. Interestingly, mono-zinc ZiPD is the dominating fraction after incubation with 10 μM zinc, given the ITC dissociation constants of 2.2 and 23 μM. This observation suggests that ZiPD achieves maximum catalytic activity already with one bound zinc, similar to the metallo-β-lactamases from B. fragilis and B. cereus (strain 569/H/9) [9,10].
ZiPD had maximum phosphodiesterase activity after incubation with 10 μM ZnCl2 (Figure 4). On incubation with 1–10 μM ZnCl2 the specific activity of ZiPD considerably increased, and on incubation with 10 μM–0.2 mM ZnCl2 the ZiPD phosphodiesterase activity slightly decreased; for >0.2 mM ZnCl2, there was extensive decrease in activity. A similar zinc activation curve was obtained for a lower ZiPD concentration (Figure 4B). In the present study, the ZiPD catalytic activity considerably increased on incubation with 10 μM ZnCl2, which corresponds to a zinc/protein ratio of over 40. Additional slight activation was observed with 20 μM Zn, but higher zinc concentrations did not result in further significant activation. The zinc activation curve for the lower ZiPD concentration showed a specific activity of approx. 80 units/mg at 1 mM zinc (Figure 4B), whereas at the higher ZiPD concentration, the corresponding activity was only 16 units/mg (Figure 4A). Nevertheless, both ZiPD zinc activation curves clearly indicate the requirement of a zinc concentration over the range 10–20 μM for ZiPD activation. It is clear from Figure 4(B) that the zinc activation observed in Figure 4(A) was not due to a limiting zinc/protein ratio but reflected a zinc-binding affinity of ZiPD in the low-μM range.
DISCUSSION
FlRd and ZiPD from E. coli represent cases where strict metal selectivity is required for the activity. While both proteins share the metallo-β-lactamase fold, the redox enzyme FlRd has a biiron site, whereas the phosphodiesterase ZiPD depends on a bi-zinc site [11,22]. In the present study, zinc affinities of cytosolic metallo-β-lactamase domain proteins were determined for the first time. The metallo-β-lactamase fold was previously characterized to allow for a broad metal selectivity since recombinant glyoxalase II is functional with active site-bound iron, manganese and zinc [7]. Concerning ZiPD, the zinc dissociation constants measured by ITC (Kd1=2.2±0.2 μM and Kd2=25.6±0.6 μM) correspond well to a zinc concentration of approx. 10 μM required for the full activation of apo ZiPD. This zinc dissociation constant is increased in comparison with other metallo-β-lactamases. For the metallo-β-lactamase from B. cereus, the following dissociation constants were determined for zinc at pH 7.5: Kd1=0.62 nM and Kd2=1.5 μM [49]. A second study for the same enzyme yielded Kd1=1.8 nM and Kd2=1.8 μM as the zinc dissociation constants at pH 7.0 [50]. The metallo-β-lactamase from A. hydrophila, which is inhibited by a second zinc ion in the binuclear active site, has a dissociation constant for the first zinc ion of <20 nM at pH 6.5 [8]. The metallo-β-lactamase L1 from Stenotrophomonas maltophilia has a Kd1 of 2.6 nM and a Kd2 of 6.0 nM for zinc at pH 7.0 [50]. The increased Kd values of ZiPD are not due to the use of ITC since this method allows for the determination of zinc–protein dissociation constants as small as 0.8 pM [51]. Since all of the above metallo-β-lactamase proteins are extracellular enzymes, it will be of interest to compare the ZiPD zinc affinity with further cytosolic metallo-β-lactamase proteins that also require zinc for hydrolytic activity.
E. coli is reported to have a total zinc content of approx. 0.2 mM [52]. Most of the cellular zinc is bound to proteins or other molecules. The concentration of free or loosely bound zinc, metabolically available for incorporation into proteins or other biomolecules, is a matter of ongoing research. This quantity is difficult to determine and the results depend on the methods used, as described in [53]. The E. coli proteome, together with other biomolecules such as nucleic acids, is suggested to have an overcapacity for zinc binding without any free zinc in the cytoplasm [52]. However, other reports state a free zinc concentration between the pmol and the nmol range for E. coli [54]. For mammalian cells, the free zinc concentration was estimated at 20 μM [55]. Interestingly, the zinc export transporter ZntA of E. coli has an apparent Km of approx. 10 μM for Zn(II) [56]. Disruption of the E. coli ZntA zinc export protein lowers the zinc tolerance from mmol to μmol concentrations [56]. These findings originally led to the assumption of a free intracellular zinc concentration in E. coli over the range of 10 μM. Rapid purification and metal analysis of native ZiPD from its natural host may elucidate to what extent this enzyme is loaded with zinc in vivo. However, ZiPD was found to be expressed at comparably basal levels in E. coli [57].
For the FlRd-lactamase domain, X-ray absorption spectroscopy determined the binding of zinc to the bi-nuclear metal-binding site on zinc incubation of the apoenzyme. This EXAFS study represents the first structural characterization of the FlRd bi-metal site, proving that it is similar to ROO from D. gigas. It was previously shown that the ROO metal ligands are conserved in the FlRd sequence [22]. Surprisingly, the zinc dissociation constants of the FlRd-lactamase domain are in the range of the values observed for ZiPD. The metal-binding sites of both enzymes, although being structurally different, share common features, such as a bridging aspartate and a co-ordination environment solely composed of N/O ligands (Figure 1). Within this context, it is of interest to note that ZiPD and glyoxalase II share similar metal-binding sites [20]. Yet, recombinant glyoxalase II is isolated with various amounts of iron, manganese and zinc, whereas recombinant ZiPD has only substoichiometric metal content [7,11]. Whereas ZiPD is exclusively activated by zinc, plant glyoxalase II achieves comparable kcat/Km values with different ratios of protein-bound iron, manganese and zinc [7,11]. Taken together, these findings strengthen previous doubts concerning the exclusive role of first-shell metal ligands in metal selectivity.
Recently, metal contents were reported for a recombinant ROO homologue protein from Moorella thermoactica overexpressed in E. coli [58]. The enzyme contained two iron ions per monomer when produced in minimal medium supplemented with FeSO4. However, two zinc ions per monomer were found on replacement of FeSO4 with ZnSO4. This confirms our finding that the E. coli FlRd metallo-β-lactamase domain is not strictly iron-selective but maintains a zinc affinity comparable with zinc-dependent metallo-β-lactamase proteins. Mutation of the atypical E81 to histidine, which restores the typical three-histidine metallo-β-lactamase zinc-binding site, unexpectedly resulted in a decreased zinc affinity for the first binding event. This experimental finding challenges the previous theoretical assumption that the atypical glutamate residue plays a major role in the metal selectivity of iron-dependent metallo-β-lactamase domains [34]. For ZiPD, mutation of the corresponding histidine to a glutamate yielded a catalytically inactive enzyme, whereas alanine mutations of metal ligands, including the particular histidine under study, retained partial catalytic activity [20]. We therefore conclude that the atypical glutamate does not trigger metal selectivity but suppresses possible hydrolytic cross-reactivity of the redox enzyme.
It is worth noting that the E81H mutation resulted in a slightly positive co-operativity of the zinc binding in contrast with the wild-type enzyme. However, the results do not allow to assign binding events to binding sites. The present results do not distinguish whether the sequence of binding events to particular binding sites is conserved or inverted by the E81H mutation.
The crystal structure of D. gigas ROO characterized the enzyme as a homodimer with the FMN cofactor of one monomer being in the vicinity of the bi-iron site of the second monomer. Given our findings about the role of the atypical glutamate, it is possible that this structural arrangement partially determines the required metal selectivity of ROO-like proteins. This is in contrast with the in vivo zinc binding to recombinant ROO from M. thermoactica. However, this enzyme was heterologously overexpressed in E. coli, whereas native ROO from its natural host D. gigas contains 1.4±0.3 iron/protein molecule [59].
Acknowledgments
We acknowledge W. Stanley (EMBL, Hamburg, Germany) for support with the ITC.
References
- 1.Vetting M. W., Wackett L. P., Que L., Jr, Lipscomb J. D., Ohlendorf D. H. Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases. J. Bacteriol. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L. An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol. 1999;1:69–91. [PubMed] [Google Scholar]
- 3.Carfi A., Pares S., Duee E., Galleni M., Duez C., Frère J. M., Dideberg O. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Concha N. O., Rasmussen B. A., Bush K., Herzberg O. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 5.Frazao C., Silva G., Gomes C. M., Matias P., Coelho R., Sieker L., Macedo S., Liu M. Y., Oliveira S., Teixeira M., et al. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 2000;7:1041–1045. doi: 10.1038/80961. [DOI] [PubMed] [Google Scholar]
- 6.Cameron A. D., Ridderstrom M., Olin B., Mannervik B. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure Fold. Des. 1999;7:1067–1078. doi: 10.1016/s0969-2126(99)80174-9. [DOI] [PubMed] [Google Scholar]
- 7.Schilling O., Wenzel N., Naylor M., Vogel A., Crowder M., Makaroff C., Meyer-Klaucke W. Flexible metal binding of the metallo-β-lactamase domain: glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry. 2003;42:11777–11786. doi: 10.1021/bi034672o. [DOI] [PubMed] [Google Scholar]
- 8.Valladares M. H., Felici A., Weber G., Adolph H. W., Zeppezauer M., Rossolini G. M., Amicosante G., Frère J. M., Galleni M. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry. 1997;36:11534–11541. doi: 10.1021/bi971056h. [DOI] [PubMed] [Google Scholar]
- 9.Paul-Soto R., Bauer R., Frère J. M., Galleni M., Meyer-Klaucke W., Nolting H., Rossolini G. M., de Seny D., Hernandez-Valladares M., Zeppezauer M., et al. Mono- and binuclear Zn2+-β-lactamase: role of the conserved cysteine in the catalytic mechanism. J. Biol. Chem. 1999;274:13242–13249. doi: 10.1074/jbc.274.19.13242. [DOI] [PubMed] [Google Scholar]
- 10.Paul-Soto R., Hernandez-Valladares M., Galleni M., Bauer R., Zeppezauer M., Frère J. M., Adolph H. W. Mono- and binuclear Zn-β-lactamase from Bacteroides fragilis: catalytic and structural roles of the zinc ions. FEBS Lett. 1998;438:137–140. doi: 10.1016/s0014-5793(98)01289-7. [DOI] [PubMed] [Google Scholar]
- 11.Vogel A., Schilling O., Niecke M., Bettmer J., Meyer-Klaucke W. ElaC encodes a novel binuclear zinc phosphodiesterase. J. Biol. Chem. 2002;277:29078–29085. doi: 10.1074/jbc.M112047200. [DOI] [PubMed] [Google Scholar]
- 12.Tavtigian S. V., Simard J., Teng D. H., Abtin V., Baumgard M., Beck A., Camp N. J., Carillo A. R., Chen Y., Dayananth P., et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 13.Smith M. S., Levitan D. J. The Caenorhabditis elegans homolog of the putative prostate cancer susceptibility gene ELAC2, hoe-1, plays a role in germline proliferation. Dev. Biol. 2004;266:151–160. doi: 10.1016/j.ydbio.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Schiffer S., Rösch S., Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 2002;21:2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takaku H., Minagawa A., Takagi M., Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrovsky E. B., Dubrovskaya V. A., Levinger L., Schiffer S., Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minagawa A., Takaku H., Takagi M., Nashimoto M. A novel endonucleolytic mechanism to generate the CCA 3′-termini of tRNA molecules in Thermotoga maritima. J. Biol. Chem. 2004;279:15688–15697. doi: 10.1074/jbc.M313951200. [DOI] [PubMed] [Google Scholar]
- 18.Schierling K., Rösch S., Rupprecht R., Schiffer S., Marchfelder A. tRNA 3′ end maturation in archaea has eukaryotic features: the RNase Z from Haloferax volcanii. J. Mol. Biol. 2002;316:895–902. doi: 10.1006/jmbi.2001.5395. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrini O., Nezzar J., Marchfelder A., Putzer H., Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogel A., Schilling O., Meyer-Klaucke W. Identification of metal binding residues for the binuclear zinc phosphodiesterase reveals identical coordination as glyoxalase II. Biochemistry. 2004;43:10379–10386. doi: 10.1021/bi049703+. [DOI] [PubMed] [Google Scholar]
- 21.Wasserfallen A., Ragttli S., Jouanneau Y., Leisinger T. A family of flavoproteins in the domains archaea and bacteria. Eur. J. Biochem. 1998;254:325–332. doi: 10.1046/j.1432-1327.1998.2540325.x. [DOI] [PubMed] [Google Scholar]
- 22.Gomes C. M., Vicente J. B., Wasserfallen A., Teixeira M. Spectroscopic studies and characterization of a novel electron-transfer chain from Escherichia coli involving a flavorubredoxin and its flavoprotein reductase partner. Biochemistry. 2000;39:16230–16237. doi: 10.1021/bi001844y. [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Liu M. Y., LeGall J., Fareleira P., Santos H., Xavier A. V. Rubredoxin oxidase, a new flavo-hemo-protein, is the site of oxygen reduction to water by the ‘strict anaerobe’ Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 1993;193:100–105. doi: 10.1006/bbrc.1993.1595. [DOI] [PubMed] [Google Scholar]
- 24.Gomes C. M., Silva G., Oliveira S., LeGall J., Liu M. Y., Xavier A. V., Rodrigues-Pousada C., Teixeira M. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J. Biol. Chem. 1997;272:22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- 25.Gardner A. M., Helmick R. A., Gardner P. R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 26.Gomes C. M., Giuffrè A., Forte E., Vicente J. B., Saraiva L. M., Brunori M., Teixeira M. A novel type of nitric-oxide reductase. J. Biol. Chem. 2002;277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 27.da Costa P. N., Teixeira M., Saraiva L. M. Regulation of the flavorubredoxin nitric oxide reductase gene in Escherichia coli: nitrate repression, nitrite induction and possible post-transcription control. FEMS Microbiol. Lett. 2003;218:385–393. doi: 10.1016/S0378-1097(02)01186-2. [DOI] [PubMed] [Google Scholar]
- 28.Carfi A., Duee E., Galleni M., Frère J. M., Dideberg O. 1.85 Å resolution structure of the zinc (II) β-lactamase from Bacillus cereus. Acta Crystallogr. D. 1998;54:313–323. doi: 10.1107/s0907444997010627. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Saez I., Mercuri P. S., Papamicael C., Kahn R., Frère J. M., Galleni M., Rossolini G. M., Dideberg O. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-β-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril. J. Mol. Biol. 2003;325:651–660. doi: 10.1016/s0022-2836(02)01271-8. [DOI] [PubMed] [Google Scholar]
- 30.Ullah J. H., Walsh T. R., Taylor I. A., Emery D. C., Verma C. S., Gamblin S. J., Spencer J. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J. Mol. Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez Valladares M., Kiefer M., Heinz U., Soto R. P., Meyer-Klaucke W., Nolting H. F., Zeppezauer M., Galleni M., Frere J. M., Rossolini G. M., et al. Kinetic and spectroscopic characterization of native and metal-substituted beta-lactamase from Aeromonas hydrophila AE036. FEBS Lett. 2000;467:221–225. doi: 10.1016/s0014-5793(00)01102-9. [DOI] [PubMed] [Google Scholar]
- 32.Heinz U., Bauer R., Wommer S., Meyer-Klaucke W., Papamichaels C., Bateson J., Adolph H. W. Coordination geometries of metal ions in d- or l-captopril-inhibited metallo-beta-lactamases. J. Biol. Chem. 2003;278:20659–20666. doi: 10.1074/jbc.M212581200. [DOI] [PubMed] [Google Scholar]
- 33.Galleni M., Lamotte-Brasseur J., Rossolini G. M., Spencer J., Dideberg O., Frere J. M. Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2001;45:660–663. doi: 10.1128/AAC.45.3.660-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomes C. M., Frazao C., Xavier A. V., Legall J., Teixeira M. Functional control of the binuclear metal site in the metallo-β-lactamase-like fold by subtle amino acid replacements. Protein Sci. 2002;11:707–712. doi: 10.1110/ps.31202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turchin A., Lawler J. F., Jr The primer generator: a program that facilitates the selection of oligonucleotides for site-directed mutagenesis. Biotechniques. 1999;26:672–676. doi: 10.2144/99264st02. [DOI] [PubMed] [Google Scholar]
- 36.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 37.Garman E. Leaving no element of doubt: analysis of proteins using microPIXE. Structure Fold. Des. 1999;7:R291–R299. doi: 10.1016/s0969-2126(00)88335-5. [DOI] [PubMed] [Google Scholar]
- 38.Vogt J. G., Flagmeyer R. H., Heitmann J., Lehmann D., Reinert T., Jankuhn S., Spemann D., Tröger W., Butz T. Solid state analysis with the new Leipzig high-energy ion nanoprobe. Mikrochim. Acta. 2000;133:105–111. [Google Scholar]
- 39.Butz T., Flagmeyer R. H., Heitman J., Jamieson D. N., Legge G. J. F., Lehmann D., Reibetanz U., Reinert T., Saint A., Spemann D., et al. The Leipzig high-energy ion nanoprobe: a report on first results. Nucl. Instr. Methods. 2000;161:323–327. [Google Scholar]
- 40.Ryan C. G. Developments in dynamic analysis for quantitative PIXE true elemental imaging. Nucl. Instr. Methods. 2001;181:170–179. [Google Scholar]
- 41.Pettifer R. F., Hermes C. Absolute energy calibration of X-ray radiation from synchrotron sources. J. Appl. Crystallogr. 1985;18:404–412. [Google Scholar]
- 42.Binsted N., Strange R. W., Hasnain S. S. Constrained and restrained refinement in EXAFS data analysis with curved wave theory. Biochemistry. 1992;31:12117–12125. doi: 10.1021/bi00163a021. [DOI] [PubMed] [Google Scholar]
- 43.Microcal. Northampton, MA: 1998. ITC Data Analysis in Origin, Tutorial Guide, version 5.0. [Google Scholar]
- 44.Scott R. A. In: Physical Methods in Bioinorganic Chemistry: Spectroscopy and Magnetism, vol. 1. Que L. Jr, editor. Sausalito, CA: University Science Books; 2000. pp. 465–503. [Google Scholar]
- 45.Riggs-Gelasco P. J., Stemmler T. L., Penner-Hahn J. E. XAFS of dinuclear metal sites in proteins and model compounds. Coord. Chem. Rev. 1995;144:245–286. [Google Scholar]
- 46.Mijovilovich A., Meyer-Klaucke W. Determination of metal-metal distances: significance and accuracy. J. Synchrotron Radiat. 2001;8:692–694. doi: 10.1107/s0909049500014540. [DOI] [PubMed] [Google Scholar]
- 47.Harding M. M. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr. D. 1999;55:1432–1443. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 48.Harding M. M. Geometry of metal-ligand interactions in proteins. Acta Crystallogr. D. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 49.de Seny D., Heinz U., Wommer S., Kiefer M., Meyer-Klaucke W., Galleni M., Frère J. M., Bauer R., Adolph H. W. Metal ion binding and coordination geometry for wild type and mutants of metallo-β-lactamase from Bacillus cereus 569/H/9 (BcII): a combined thermodynamic, kinetic, and spectroscopic approach. J. Biol. Chem. 2001;276:45065–45078. doi: 10.1074/jbc.M106447200. [DOI] [PubMed] [Google Scholar]
- 50.Wommer S., Rival S., Heinz U., Galleni M., Frère J. M., Franceschini N., Amicosante G., Rasmussen B., Bauer R., Adolph H. W. Substrate-activated zinc binding of metallo-β-lactamases: physiological importance of the mononuclear enzymes. J. Biol. Chem. 2002;277:24142–24147. doi: 10.1074/jbc.M202467200. [DOI] [PubMed] [Google Scholar]
- 51.DiTusa C. A., Christensen T., McCall K. A., Fierke C. A., Toone E. J. Thermodynamics of metal ion binding: metal ion binding by wild-type carbonic anhydrase. Biochemistry. 2001;40:5338–5344. doi: 10.1021/bi001731e. [DOI] [PubMed] [Google Scholar]
- 52.Outten C. E., O'Halloran T. V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 53.Hantke K. Bacterial zinc transporters and regulators. Biometals. 2001;14:239–249. doi: 10.1023/a:1012984713391. [DOI] [PubMed] [Google Scholar]
- 54.Williams R. J. P., Fraústo da Silva J. J. R. The distribution of elements in cells. Coord. Chem. Rev. 2000;200–202:247–348. [Google Scholar]
- 55.Palmiter R. D., Findley S. D. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rensing C., Mitra B., Rosen B. P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schilling O., Rüggeberg S., Vogel A., Rittner N., Weichert S., Schmidt S., Doig S., Andrews S. C., Benes V., Franz T., et al. Characterization of an Escherichia coli elaC deletion mutant. Biochem. Biophys. Res. Commun. 2004;320:1365–1373. doi: 10.1016/j.bbrc.2004.05.227. [DOI] [PubMed] [Google Scholar]
- 58.Silaghi-Dumitrescu R., Coulter E. D., Das A., Ljungdahl L. G., Jameson G. N. L., Huynh B. H., Kurtz D. M. A flavodiiron protein and high molecular weight rubredoxin from Moorella thermoacetica with nitric oxide reductase activity. Biochemistry. 2003;42:2806–2815. doi: 10.1021/bi027253k. [DOI] [PubMed] [Google Scholar]
- 59.Chen L., Le Gall J., Xavier A. V. Purification, characterization and properties of an NADH oxidase from Desulfovibrio vulgaris (Hildenborough) and its coupling to adenylyl phosphosulfate reductase. Biochem. Biophys. Res. Commun. 1994;203:839–844. doi: 10.1006/bbrc.1994.2259. [DOI] [PubMed] [Google Scholar]
- 60.Stöcker W., Wolz R. L., Zwilling R. Astacus protease, a zinc metalloenzyme. Biochemistry. 1988;27:5026–5032. [Google Scholar]
- 61.Mercuri P. S., Garcia-Saez I., De Vriendt K., Thamm I., Devreese B., Van Beeumen J., Dideberg O., Rossolini G. M., Frere J. M., Galleni M. Probing the specificity of the subclass B3 FEZ-1 metallo-beta-lactamase by site-directed mutagenesis. J. Biol. Chem. 2004;279:33630–33638. doi: 10.1074/jbc.M403671200. [DOI] [PubMed] [Google Scholar]
- 62.Eggers-Borkenstein P., Priggemeyer S., Krebs B., Henkel G., Simonis U., Pettifer R. F., Nolting H. F., Hermes C. Extended X-ray absorption fine structure (EXAFS) investigations of model compounds for zinc enzymes. Eur. J. Biochem. 1989;186:667–675. doi: 10.1111/j.1432-1033.1989.tb15258.x. [DOI] [PubMed] [Google Scholar]