Abstract
The enhanced synthesis of fatty acids in the liver and adipose tissue in response to insulin is critically dependent on the transcription factor SREBP-1c (sterol-regulatory-element-binding protein 1c). Insulin increases the expression of the SREBP-1c gene in intact liver and in hepatocytes cultured in vitro. To learn the mechanism of this stimulation, we analysed the activation of the rat SREBP-1c promoter and its truncated or mutated congeners driving a luciferase reporter gene in transiently transfected rat hepatocytes. The rat SREBP-1c promoter contains binding sites for LXR (liver X receptor), Sp1, NF-Y (nuclear factor-Y) and SREBP itself. We have found that each of these sites is required for the full stimulatory response of the SREBP-1c promoter to insulin. Mutation of either the putative LXREs (LXR response elements) or the SRE (sterol response element) in the proximal SREBP-1c promoter reduced the stimulatory effect of insulin by about 50%. Insulin and the LXR agonist TO901317 increased the association of SREBP-1 with the SREBP-1c promoter. Ectopic expression of LXRα or SREBP-1c increased activity of the SREBP-1c promoter, and this effect is further enhanced by insulin. The Sp1 and NF-Y sites adjacent to the SRE are also required for full activation of the SREBP-1c promoter by insulin. We propose that the combined actions of the SRE, LXREs, Sp1 and NF-Y elements constitute an insulin-responsive cis-acting unit of the SREBP-1c gene in the liver.
Keywords: gene transcription, hepatocyte, insulin, promoter, sterol-regulatory-element-binding protein 1c (SREBP-1c)
Abbreviations: ChIP, chromatin immunoprecipitation; db-cAMP, dibutyryl cAMP; DTT, dithiothreitol; EMSA, electrophoretic mobility shift assay; ER, endoplasmic reticulum; ERK1/2, extracellular-signal-regulated kinase 1/2; FAS, fatty acid synthase; LDL, low-density lipoprotein; LXR, liver X receptor; LXRE, LXR response element; MAPK, mitogen-activated protein kinase; NF-Y, nuclear factor-Y; NP40, Nonidet P40; O-GlcNAc, O-linked N-acetyl-D-glucosamine; SRE, sterol response element; SREBP, sterol-regulatory-element-binding protein
INTRODUCTION
Insulin is a potent inducer of the enzymes catalysing de novo synthesis of fatty acids in liver and adipose tissue. The enhanced transcription of the FAS (fatty acid synthase) and acetyl-CoA carboxylase-1 genes produced by insulin is opposed by agents that raise intracellular concentrations of cAMP [1] and by polyunsaturated fatty acids [2]. Several lines of evidence point to SREBP-1c (sterol-regulatory-element-binding protein 1c) as a key transcription factor mediating the stimulatory effects of insulin [3].
Two separate genes encode SREBP-1 and SREBP-2. SREBP-2 primarily regulates genes associated with cholesterol synthesis or uptake, whereas SREBP-1 regulates genes that control lipogenesis as well. The use of alternative promoters generates SREBP-1a and SREBP-1c, which differ only in their first exon. In the human SREBP-1 gene, the alternative exons 1a and 1c are separated by about 14 kb [4]. SREBP-1c, the predominant isoform in liver and adipose tissue, has stronger effects on transcription of genes encoding lipogenic enzymes than those involved in cholesterol metabolism [5]. In contrast, SREBP-1a and SREBP-2 are the predominant isoforms in other cell types and in actively growing cell lines [5].
Nascent SREBP proteins are integrally inserted into the ER (endoplasmic reticulum) with the N- and C-terminal domains exposed to the cytoplasm. The ER-bound SREBP is associated with SCAP (SREBP-cleavage-activating protein) [6,7], a chaperone that accompanies SREBP from the ER to the Golgi in cholesterol-depleted states. Two successive cleavages liberate the N-terminal fragment of SREBP, which acts as a transcription factor in the nucleus. The levels of SREBP-1c mRNA change in response to insulin or cAMP. However, whether insulin also stimulates generation of the nuclear form of SREBP from its precursor is not known at present.
Hepatic levels of SREBP-1c mRNA and protein are reduced by fasting and increased by refeeding, whereas expression of SREBP-1a and SREBP-2 is only minimally affected under these conditions [8]. Levels of hepatic SREBP-1c mRNA also fall in experimental diabetes and return to normal after insulin treatment [9]. These physiological changes can be replicated in primary cultures of rat hepatocytes [10–12]. Insulin induces, whereas glucagon and db-cAMP (dibutyryl cAMP) suppress SREBP-1c gene transcription [12] and promoter activity [11].
Although the human [4] and mouse [13] SREBP-1 genes have been cloned, there is limited knowledge of the mechanisms by which hormones regulate transcription of SREBP-1c. A mouse SREBP-1c promoter and its response to recombinant SREBP-1c and to LXR (liver X receptor) agonists have been studied [14–16]. We cloned a 1.5 kb segment of the rat SREBP-1c promoter and found that it responds in a physiological manner, in transfected primary cultures of rat hepatocytes, to insulin, cAMP and polyunsaturated fatty acids [11]. The rat and mouse proximal SREBP-1c promoters are highly conserved, including similarly located Sp1 and NF-Y (nuclear factor-Y) elements, SRE (sterol response element) and LXRE (LXR response element). The present investigation was undertaken to determine the contribution of these elements to stimulation of the rat SREBP-1c promoter by insulin. In the present study we report that the full response of the proximal SREBP-1c promoter to insulin requires participation of LXR-, SREBP-, Sp1- and NF-Y-binding elements.
EXPERIMENTAL
Plasmids
Mutations in pSREBP(−1516/+40)-luc, described previously by Deng et al. [11], were created by site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA, U.S.A.). The sequences of forward primers used to generate site-directed mutations are as follows: mutSRE-luc, ctg att ggc cat gtg cgA GAG TAC gag ggg cgg ggc a; mutSp-luc, gct gat tgg cca tgt gcg ctc acc cga ggA AAT TTg cac ggg g; mutEbox-luc, ctg att ggc TGT ACG cgc tca ccc gag ggg cgg ggc acg g; mutNFY-luc, cgc ggc gcg gct gct gGt ACC cca tgt gcg ctc acc cga g; mutLXRE1-luc, ggg ctg gga cgg cag CCC GGG ATC CAC CAA TCG gcg cgc gct ggc; mutLXRE2-luc, cgg tta aag gcg gaC CCG GGA TCC ACC AAT CGg ccc cat tca gag. Successful mutations of the SRE complex in plasmid recovered from transformed cells were verified by sequencing.
Plasmids pCMX-hLXRα and pCyp7aLXRE(×3)-luc were a gift from Dr David J. Mangelsdorf (Department of Pharmacology, Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, TX, U.S.A.); pTarget-hLXRα was constructed by ligating the insert from pCMX-hLXRα into pTarget empty vector (Promega, Madison, WI, U.S.A.); pCMV-nSREBP-1a(1–460) was obtained from the A.T.C.C.; pSport-ADD1(1–403) was a gift from Dr Bruce M. Spiegelmann (Department of Cell Biology, Harvard Medical School, Boston, MA, U.S.A.); pCMV-Sp1 and pCMV-Sp3 were a gift from Dr Guntrum Suske (Institut für Molekularbiologie und Tumorforschung, Philipps-Universitaet Marburg, Marburg, Germany).
Plasmids used to prepare cDNA probes for measurement of SREBP-1, FAS and LXRα mRNAs were generously provided by Dr Bruce M. Spiegelman, Dr Stuart Smith (Children's Hospital Research Institute, Oakland, CA, U.S.A.), and Dr David J.Mangelsdorf. β-actin mRNA was measured using mouse β-actin DECA probe (Ambion, Austin, TX, U.S.A.).
EMSAs (electrophoretic mobility shift assays)
Double-stranded oligonucleotides corresponding to the SREBP-1c promoter sequence (−86/−47) were designed with XbaI overhangs and labelled with [α-32P]dCTP using the Klenow fragment of DNA polymerase. Radiolabelled probe (30000 dpm) and proteins were combined in buffer containing 10 mM Tris/HCl (pH 7.5), 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT (dithiothreitol), 50 mM NaCl, 1.0 μg of poly(dI-dC)·(dI-dC), and 5% glycerol in a total volume of 20 μl. Human SREBP-1a (1–460) was generated from pCMVnSREBP1a by in vitro translation using rabbit reticulocyte lysate (Proteinscript II, Ambion). In competition experiments, a 10-, 30- or 100-fold excess of unlabelled annealed oligonucleotides was added to the reaction mixture. In supershift experiments, antibodies were incubated with an in vitro translation mixture at 4 °C for 1 h before EMSA. Binding was carried out at room temperature (22 °C) for 20 min, and the protein–DNA complexes were then resolved in a non-denaturing 5% acrylamide gel in 20 mM Tris, 190 mM glycine and 0.1 mM EDTA. After electrophoresis (4 °C for 90 min at 80 V), the gels were dried and the radioactive bands were visualized by autoradiography.
Primary hepatocyte culture
Hepatocytes were obtained from livers of male Sprague–Dawley rats (approx. 300 g; Harlan Laboratories, Indianapolis, IN, U.S.A.) by collagenase perfusion, as described previously [17]. Cells were suspended in RPMI medium (Invitrogen, Carlsbad, CA, U.S.A.) containing 5% fetal bovine serum (Sigma, St. Louis, MO, U.S.A.), 10 mM glucose, 1 μM dexamethasone and 100 nM insulin. Each 60-mm diameter culture dish, coated with rat tail collagen (Collaborative Biochemical Products, Bedford, MA, U.S.A.), was seeded with 3×106 cells; after 4 h, non-adherent cells were removed, and adherent cells were incubated overnight in RPMI medium without serum or hormones. Incubation was continued for a further 24 h in fresh medium supplemented with 0.75% delipidated BSA, 20 mM glucose and 100 nM dexamethasone.
Measurement of steady-state levels of mRNAs by Northern hybridization
Total RNA was extracted with RNA Stat-60 (Tel-Test, Friendswood, TX, U.S.A.) and quantified at A260. Total RNA (20 μg) was loaded per lane of a formaldehyde/0.8% agarose gel, and electrophoresis was carried out in 1×Mops buffer, the gel was blotted on to Nytran Plus membranes (Schleicher and Schuell, Keane, NH, U.S.A.) and the UV cross-linked. rRNA bands were visualized by staining with ethidium bromide prior to the transfer step. Blots were prehybridized for 3 h at 42 °C in 50% formamide, 5×SSPE (0.15M NaCl/10 mM sodium phosphate (pH 7.4)/1 mM EDTA), 5×Denhardt's solution (5Prime-3Prime, Boulder, CO, U.S.A.), 7.5% dextran sulphate, 1.5% SDS and 100 μg/ml sheared salmon sperm DNA (Ambion). Overnight hybridization with the cDNA probes, 32P-labelled by the random primer method using a commercial kit (Invitrogen), was carried out at 42 °C. Unbound probe was removed by washing twice with 2×SSC (0.15 M NaCl/0.015 M sodium citrate) plus 0.1% SDS at room temperature and then twice with 0.1×SSC plus 0.1% SDS at 65 °C for 30 min each. Membranes were exposed to Bio-Max MS film (Eastman Kodak, Rochester, NY, U.S.A.); a digital image of the developed film was created and RNA bands quantified by densitometry (Alpha Innotech, San Leandro, CA, U.S.A.).
Measurement of LXR protein levels by Western blot analysis
Hepatocytes were washed twice and collected in ice-cold PBS. Centrifuged cells were resuspended in 1 ml of lysis buffer [20 mM Hepes (pH 7.9), 20% glycerol, 1% NP40 (Nonidet P40), 0.1% Triton X-100, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail (Sigma)]. After 15 min at 4 °C, lysates were centrifuged for 5 min at 500 g. The pelleted nuclei were resuspended in 1 ml of nuclear extraction buffer [20 mM Hepes (pH 7.9), 20% glycerol, 1% NP40, 0.1% Triton X-100, 420 mM NaCl, 100 mM KCl, 10 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail (Sigma)]. Samples were incubated on a rocking platform for 1 h and then centrifuged at 25000 g for 30 min, and the supernatant was stored at −80 °C until analysis. Samples containing 50 μg of nuclear protein were dissolved in Laemmli buffer, and the proteins were resolved by SDS/PAGE (7.5% gel), and transferred on to nitrocellulose membranes. Blots were blocked for 1 h with 5% non-fat dried milk in TBS-T [pH 8.0; 0.136 M NaCl, 20 mM Tris/HCl and 0.1% Tween 20]. Blots were probed for 1 h with 1 μg/ml goat polyclonal antiLXRα (Santa Cruz Biotechnology, Santa Cruz, CA) followed by 1 h with horseradish-peroxidase-linked anti-goat IgG (Santa Cruz Biotechnology). Blots were developed with SuperSignal reagent (Pierce Biotechnology, Rockford, IL, U.S.A.) and images captured with a digital imager (Alpha Innotech).
Transient transfection of primary cultures of rat hepatocytes
Freshly isolated rat hepatocytes were incubated for 18 h with 2 μg of pSREBP(−1516/+40)-luc or pSREBP(−109/+40)-luc plus 1 μg of p-RL-TK (Promega) dispersed with 8 μl of Lipofectin reagent (Invitrogen) in RPMI medium. Medium and vectors were then removed, and cells incubated for a further 24 h in RPMI medium containing 0.75% delipidated BSA, 20 mM glucose and 100 nM dexamethasone alone, or with the additional treatments as detailed. Cells were lysed and luciferase activity was quantified fluorimetrically (TD-20/20 Luminometer; Turner Designs, Sunnyvale, CA, U.S.A.) using the Dual-Luciferase Reporter Assay System (Promega). To normalize for variation in transfection efficiency, data were expressed as the ratio of Photinus to Renilla luciferase activity.
ChIP (chromatin immunoprecipitation) assay
Formaldehyde (final concentration 1%) was added to the culture medium and cells were incubated on a rocking platform for 10 min at room temperature. Cross-linking was stopped by addition of 125 mM glycine to each plate and cross-linked hepatocytes were washed with ice-cold PBS, and scraped into PBS containing protease inhibitors (1 mM PMSF, 1 μg of aprotinin and 1 μg/ml pepstatin A). Cells were concentrated by centrifugation and dispersed into SDS lysis buffer [1% SDS, 10 mM EDTA and 50 mM Tris/HCl (pH 8.1)]. The lysate was sonicated to shear the cross-linked DNA and an aliquot set aside for use as input DNA. The remaining DNA was diluted 10-fold into dilution buffer [1.2 mM EDTA, 16.7 mM Tris/HCl (pH 8.1) and 167 mM NaCl]. The samples were pre-cleared with 80 μl of salmon sperm DNA–Protein A agarose slurry (Upstate, Charlottesville, VA, U.S.A.) for 30 min at 4 °C. The samples were incubated overnight with 10 μg of rabbit polyclonal anti-SREBP-1 IgG (H-160; Santa Cruz Biotechnology) at 4 °C. Parallel samples were incubated with non-immune rabbit IgG or without antibody to serve as negative controls. IgG was adsorbed from the mixture with DNA–protein–Protein A agarose slurry (Upstate). The pelleted beads were washed once with low-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM, EDTA, 150 mM NaCl and 20 mM Tris/HCl (pH 8.1)], once with high-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris/HCl (pH 8.1)], once with LiCl wash buffer [250 mM LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris/HCl (pH 8.1)], and twice with TE buffer [1 mM EDTA and 10 mM Tris/HCl (pH 8.1)]. The adsorbed DNA was eluted with 1% SDS/100 mM NaHCO3 and protein cross-links were reversed by heating to 65 °C for 6 h. Protein was removed with proteinase K and DNA was recovered by chloroform/methanol extraction and ethanol precipitation, and was then used as template for PCR (30 cycles) to amplify a 260-bp segment of the rat SREBP-1c promoter between nt −267 and −8. The primers used were 5′-tggttgcctgtgcggcag-3′ (forward) and 5′-tcaggccccgccaggctttaa-3′ (reverse). Amplified products were resolved by electrophoresis on 2% agarose gel and visualized by ethidium bromide.
RESULTS
Rat SREBP-1c promoter is activated by the LXR agonist TO901317
The rat SREBP-1c promoter, which we have found previously to be stimulated by insulin in primary rat hepatocytes [11], contains two putative LXREs located at nucleotides −184/−178 and −234/−219. The mouse SREBP-1c promoter is stimulated by the synthetic LXR agonist TO901317 in transfected HEK-293 cells [15,16]. TO901317 also increases hepatic levels of SREBP-1c mRNA and stimulates lipogenesis in the livers of intact mice and hamsters [18]. The stimulatory effect of TO901317 on hepatic lipogenesis is sharply reduced in mice homozygous for an inactivating mutation in the SREBP-1c gene [19]. Thus, similar to insulin, the stimulatory effect of the LXR activator on lipogenesis is mediated by its ability to induce SREBP-1c. These results raise the possibility that stimulation of the SREBP-1c promoter by insulin is mediated in part by an effect on LXR activity. Insulin has been reported to increase levels of LXRα mRNA in rat hepatocytes and the ability of insulin to increase levels of hepatic SREBP-1 mRNA is blunted in mice genetically deficient in both LXRα and LXRβ [20], indicating that basal or stimulated expression of these receptors is required for the insulin effect in intact mouse liver.
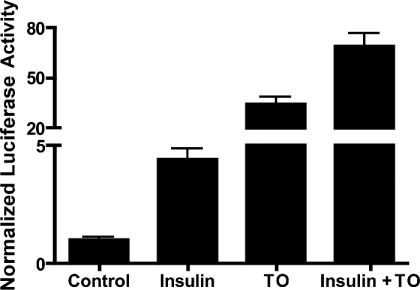
The stimulatory effect of LXR agonist (but not insulin) on the SREBP-1c promoter has been observed in transfected HEK-293 cells. As a first step in examining the contribution of LXR to transcriptional stimulation of the SREBP-1c gene by insulin, we determined whether the SREBP-1c promoter is also stimulated by activation of endogenous LXR in insulin-responsive rat hepatocytes. Cells were transfected with pSREBP(−1516/+40)-luc and incubated for 24 h with or without the synthetic LXR agonist TO901317 (10 μM). TO901317 increased luciferase expression 34.4±4.2-fold (Figure 1). In the present experiment, insulin alone (100 nM) increased luciferase expression 4.4±0.47-fold, and it enhanced expression in the presence of TO901317 to 68.9±8.0-fold. The endogenous agonists for LXR in the hepatocyte are oxysterols [21]. Our results suggest that intracellular levels oxysterol metabolites under basal conditions or in insulin-treated cells activate only a fraction of the receptors present, since high concentrations of a synthetic LXR agonist results in a much greater increase in SREBP-1c promoter activity than that provided by the physiological stimulus, insulin. Since LXRα and LXRβ are known to bind to LXREs as heterodimers with RXR (retinoic acid X receptor), we also treated transfected rat hepatocytes with the RXR agonist 9-cis retinoic acid (10 μM); in these experiments 9-cis retinoic acid did not significantly stimulate SREBP-1c promoter activity alone or in combination with TO901317 (results not shown).
Figure 1. SREBP-1c promoter is stimulated by the LXR agonist TO901317 and by insulin.
Rat hepatocytes were co-transfected with pSREBP (−1516/+40)-luc and with pRL-TK to determine efficiency of transfection. Cells were then incubated in medium with no additions or supplemented with insulin (100 nM), TO901317 (TO; 10 μM) or with insulin plus TO901317 (insulin+TO). Luciferase activity was measured 24 h after treatment of transfected cells. Normalized luciferase activities are expressed as the means±S.E.M. for four independent experiments.
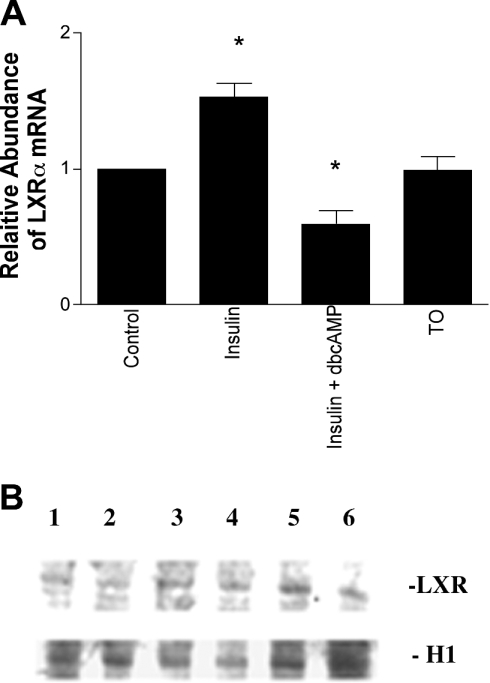
To test whether the synergy between insulin and TO901317 was mediated by altered expression of LXRα, we measured levels of LXRα mRNA in hepatocytes. As shown in Figure 2, insulin modestly increased the abundance of LXRα mRNA by 53±12%. In contrast, TO901317 did not affect LXRα mRNA levels. The human LXRα gene promoter contains LXREs, and treatment of human cell lines with TO901317 increases levels of LXRα mRNA [22]. In contrast, the mouse LXRα gene did not exhibit stimulation by TO901317 [22], which is similar to what we observed in rat hepatocytes (Figure 2), suggesting that the rat LXRα promoter also lacks the auto-regulatory element.
Figure 2. Insulin increases steady-state levels of LXRa mRNA, but not LXRa protein, in rat hepatocytes.
(A) Insulin increases steady-state levels of LXRα mRNA in rat hepatocytes. Cells were incubated for 24 h in medium with no additions or with insulin (100 nM), insulin (100 nM) plus db-cAMP (100 μM) or TO901317 (TO; 10 μM), and LXRα mRNA was measured by Northern analysis. Levels of β-actin mRNA were measured on the same blots to normalize the data. Measurements from four separate hepatocyte preparations are shown. *Statistically different compared with the control (P<0.05). (B) Steady-state levels of LXR protein in rat hepatocyte nuclear extracts. Cells were incubated for 24 h in medium with no additions (1,4) or with insulin (100 nM) (2,5), or db-cAMP (100 μM) (3,6) and LXR protein was measured by immunoblotting. Levels of histone H1 proteins were measured on the same blots to normalize the data. Data from two separate hepatocyte preparations are shown, which are representative of five separate experiments.
cAMP inhibits expression of lipogenic enzymes in the liver. We have previously found that db-cAMP reduces levels of SREBP-1 mRNA and SREBP-1c promoter activity in rat hepatocytes [11]. In the present experiments, db-cAMP (100 μM) eliminated insulin stimulation of the SREBP-1c promoter and reduced the stimulatory effect of TO901317 from 34.4±4.2-fold to 12.9±0.8-fold (n=4). Treatment with db-cAMP also reduced steady-state levels of LXRα mRNA to 59±0.06% of control, despite the presence of insulin (Figure 2A).
However, despite changes in LXRα mRNA produced by treatment of rat hepatocytes with insulin or db-cAMP, Western bot analysis failed to show corresponding changes in levels of LXR protein (Figure 2B). Although, the polyclonal antibody used in this study has significant cross-reactivity with LXRβ, and it is possible that changes in LXRα abundance were partially masked, these results suggest that the enhanced sensitivity of the SREBP-1c promoter to TO901317 in the presence of insulin and the inhibition of that response by db-cAMP are primarily due to increased transcriptional activity, rather than increased abundance of LXRα protein in rat hepatocytes.
Insulin increases transcriptional stimulation by LXRα in rat hepatocytes
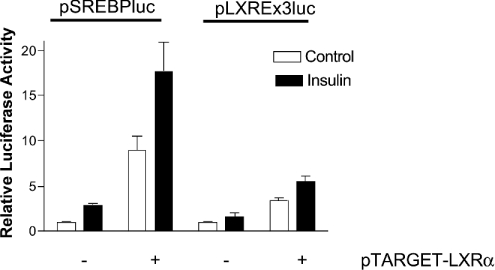
To determine whether insulin activates LXREs in rat hepatocytes, we compared the effect of insulin treatment on transcription of an artificial LXRE-driven reporter plasmid, pCyp7aLXRE(×3)-luc, which contains three LXREs ligated upstream from a luciferase reporter gene, and on pSREBP(−1516/+40)-luc. Each were co-transfected with the mammalian expression vector pTarget-hLXRα. SREBP-1c promoter activity was increased 8.9±1.6-fold by ectopic expression of LXRα, indicating that an increase in levels of LXRα within the hepatocyte results in increased activity of the SREBP-1c promoter. On the other hand, insulin treatment enhanced the SREBP-1c promoter activity resulting from ectopic expression of LXRα by a further 2-fold, suggesting that the stimulatory effect of insulin on the SREBP-1c promoter is not limited to increasing the expression of LXRα (Figure 3). As shown in Figure 3, the activation of pCyp7aLXRE(×3)-luc in the presence of insulin did not reach statistical significance. In contrast, co-transfection with pTarget-hLXRα stimulated activity of pCyp7aLXRE(×3)-luc by 3.46±0.33-fold. In the hepatocyte expressing ectopic LXRα, insulin stimulated activity of pCyp7aLXRE(×3)-luc by an additional 1.6-fold (Figure 3), confirming that LXRα exerts increased transcriptional effect in insulin-treated hepatocytes.
Figure 3. Exogenous expression of LXRα in rat hepatocytes stimulates SREBP-1c promoter activity.
Rat hepatocytes were co-transfected with with pSREBP(−1516/+40)-luc (pSREBPluc) or pCyp7aLXRE(×3)-luc (pLXRE×3luc) and pTarget-hLXRα or empty vector. Co-transfection with pRL-CMV was included under all conditions to determine transfection efficiency. Photinus and Renilla luciferase activities were measured in cell extracts obtained after 24 h in medium with no additions (open bars) or supplemented with insulin (100 nM; closed bars). Normalized luciferase activities are expressed as the means±S.E.M. for five independent experiments.
We have not been able to compare insulin-mediated changes in the abundance of endogenous LXRα with those produced by co-transfection with the LXRα expression vector. However, since co-transfection with pTarget-hLXRα stimulated promoter activity of pCyp7aLXRE(×3)-luc and pSREBP(−1516/+40) to a greater extent than insulin treatment, we postulated that higher intracellular levels of LXRα were achieved by ectopic expression. Insulin treatment further enhanced the activity of pCyp7aLXRE(×3)-luc and pSREBP(−1516/+40)-luc in cells co-transfected with pTarget-hLXRα. On the basis of these observations, it is likely that insulin has additional actions on the intracellular concentrations of endogenous LXR agonists, on post-translational modification of LXRα or on the activity of other trans-acting factors with which LXR interacts.
In contrast with its effect on pSREBP(−1516/+40)-luc, insulin failed to stimulate pCyp7aLXRE(×3)-luc in the absence of exogenous LXRα. These results clearly indicate that insulin action is mediated by the combinatorial activity of LXRE and other cis-acting elements on the SREBP-1c promoter.
Ablation of LXREs in the SREBP-1c promoter blunts transcriptional stimulation by TO901317 and insulin
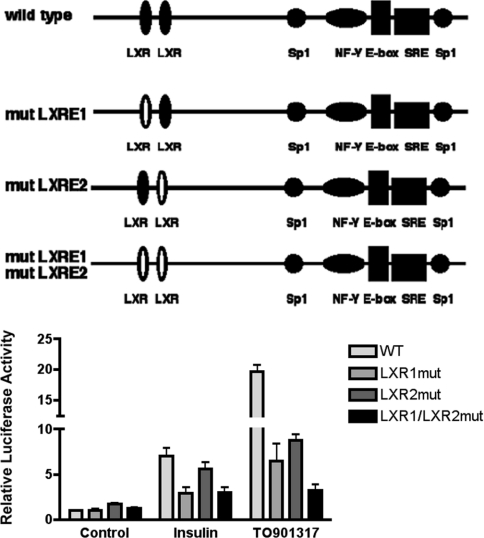
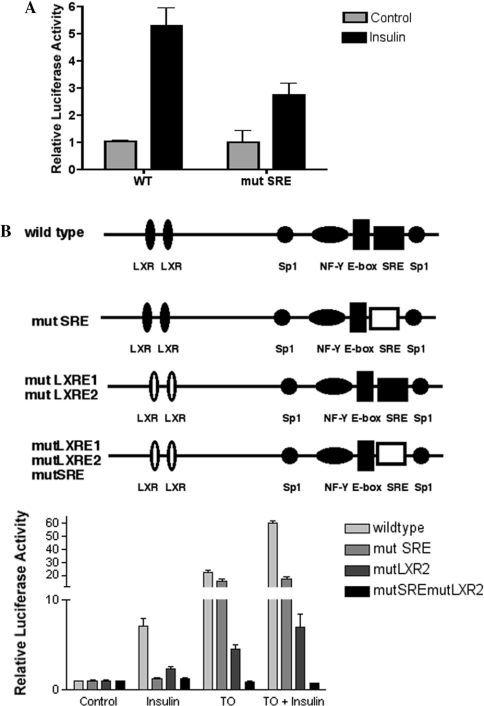
If insulin stimulates the SREBP-1c promoter through activation of LXR, ablation of the LXREs in the promoter should reduce its effect. To test this, we compared the response of wild-type pSREBP(−1516/+40)-luc with mutated versions of the promoter in which the LXREs at −234/−218 and at −184/−168 were inactivated by mutation [15]. In transiently transfected rat hepatocytes, mutation of the LXRE at −234/−218 resulted in 58% inhibition of the response to insulin and 67% inhibition of the response to TO901317 (Figure 4). Mutation of the LXRE at −184/−168 resulted in a 55% loss of response to TO901317, however, the 20% mean loss of response to insulin was not statistically significant. Simultaneous mutation of both LXREs resulted in an 84% loss of response to TO901317 and a 57% loss of response to insulin.
Figure 4. Mutations of the two LXREs in the proximal SREBP-1c promoter reduce its response to insulin and TO901317.
Rat hepatocytes were transfected with pSREBP(−1516/+40)-luc (wild-type/WT), pSREBP-(−1516/+40)-mutLXR1-luc, pSREBP(−1516/+40)-mutLXR2-luc or pSREBP(−1516/+40)-mutLXR1/mutLXR2-luc together with pRL-CMV. Photinus and Renilla luciferase activities were measured after 24 h incubation of transfected hepatocytes in medium with no additions or supplemented with insulin (100 nM) or TO901317 (10 μM). Normalized luciferase activities are expressed as the means±S.E.M. for four independent experiments.
It appears that the distal LXRE(−238/−218) is required for the full response of the SREBP-1c promoter to insulin, whereas loss of the proximal LXRE(−184/−168) had little effect. In contrast, loss of either LXRE inhibited the response to the synthetic LXR agonist to the approximately the same extent, and loss of both sites produced a greater loss in response. This confirms that both LXREs are functional in the context of the rat SREBP-1c promoter.
Our results indicate that that insulin stimulates SREBP-1c promoter activity in rat hepatocytes both by LXR-dependent and LXR-independent mechanisms. It is possible that the greater sensitivity of insulin response to the upstream LXRE results from selective interaction of LXR bound at that site with other factors bound to the SREBP-1c promoter.
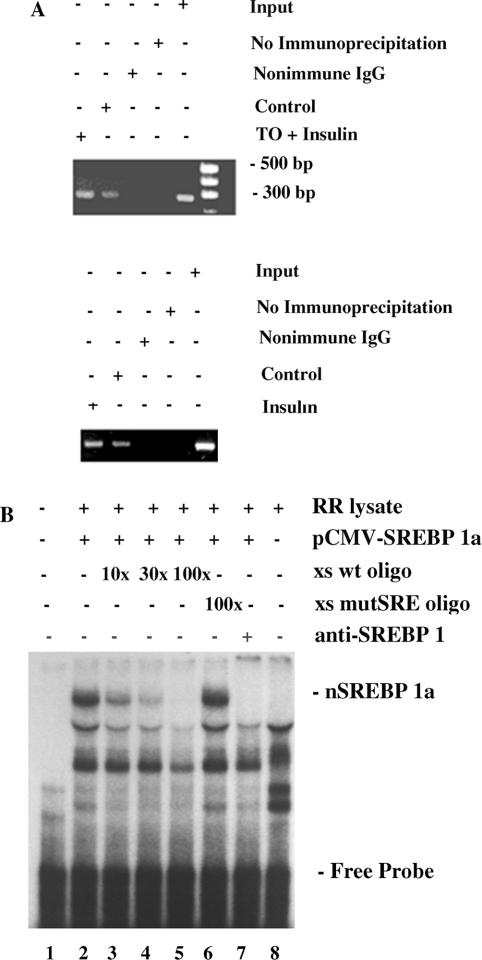
SREBP-1 associates with the SREBP-1c promoter in intact hepatocytes
The rat SREBP-1c gene contains an inverted CCAAT box, E-box, SRE and a GC box in its proximal promoter [11]. An analogous cluster of cis-acting elements in some promoters is thought to confer sensitivity to stimulation by SREBP [23–27], and the mouse SREBP-1c promoter is stimulated by ectopic expression of SREBP-1a or SREBP-1c in transfected HEK-293 cells [14]. To determine whether SREBP-1c associates with the SREBP-1c promoter in intact hepatocytes we conducted ChIP analysis using an antibody against rat SREBP-1 and primers designed to amplify a 260-bp segment of the proximal SREBP-1c promoter. SREBP-1 was found to be associated with the SREBP-1c promoter under basal conditions (Figure 5A; representative of three independent experiments). In the presence of insulin (100 nM) alone or combined with TO901317 (10 μM), conditions that potently stimulated SREBP-1c promoter activity (Figure 1), the extent of association of SREBP-1 with the SREBP-1c promoter was increased.
Figure 5. Rat SREBP-1c promoter contains a bona fide SRE that binds SREBP-1 in intact cells and in vitro.
(A) Insulin and insulin plus TO901317 increase binding of SREBP-1 to the SREBP-1c promoter in intact rat hepatocytes. Rat hepatocytes were incubated for 24 h in control medium or in medium supplemented with insulin (100 nM) or with insulin (100 nM) plus TO901317 (TO; 10 μM). Formaldehyde cross-linked DNA was extracted and ChIP analysis performed, using PCR to amplify a 260 bp segment of the SREBP-1c promoter containing the SRE, as described in the Experimental section. The results shown are representative of data from three hepatocyte preparations. (B) Mutation of the SRE in the SREBP-1c promoter prevents binding of SREBP-1a. An SREBP-1c-specific double-stranded cDNA probe (nt −86/−47) was 32P-labelled using the Klenow fragment of DNA polymerase and subjected to EMSA, as described in the Experimental section. Lane 1, DNA probe only; lane 2, DNA probe incubated with in vitro-translated pCMVnSREBP-1a in rabbit reticulocyte lysate (RR lysate); lanes 3–5, same as lane 2, but with a 10-fold (lane 3), 30-fold (lane 4) or 100-fold (lane 5) excess of unlabelled wild-type double-stranded DNA (xs wt oligo); lane 6, same as lane 2, but with a 100-fold excess of unlabelled mutant SRE double-stranded DNA (xs mutSRE oligo); lane 7, same as lane 2, but with antibody against SREBP-1; lane 8, DNA probe incubated with rabbit reticulocyte lysate without pCMVnSREBP-1a. The DNA–protein band containing in vitro-translated SREBP-1a is eliminated by the antibody against SREBP-1 (lane 7) and by competition with unlabelled wild-type (lanes 3–5), but not mutant (lane 6), oligonucleotides.
Ectopic expression of SREBP-1c increased SREBP-1c promoter activity
Insulin increases transcription of SREBP-1c and association of SREBP-1 with the SREBP-1c promoter in rat hepatocytes. We therefore wished to determine whether the positive response of the SREBP-1c promoter to insulin could be mediated in part by a feed-forward amplification resulting from increased expression of endogenous SREBP-1c in insulin-treated cells. To avoid the additional complicating effects of insulin on LXRα expression or activity, we co-transfected truncated pSREBP(−109/+40)-luc (lacking the two LXREs) and pSVSport-ADD1(1–403), designed to express the transcriptionally active nuclear fragment of rat SREBP-1c [28].
Co-expression of pSVSport-ADD1(1–403) activated pSREBP(−109/+40)-luc by 6.82±0.66-fold, as seen in the results from four independent experiments. Under these conditions, insulin treatment further increased luciferase expression from 6.82±0.66- to 12.36±1.59-fold of the control value (n=4). Similar results were observed when hepatocytes were co-transfected with a vector expressing the nuclear form of SREBP-1a (results not shown). Although these results may reflect the additive effect of endogenous SREBP-1c induced by insulin treatment and ectopically expressed nuclear SREBP-1c, insulin may also activate SREBP-1c through post-translational modification. Evidence that insulin activates the N-terminal fragment of SREBP-1c was obtained in experiments in which activation of a FAS promoter vector by constitutive expression of nuclear SREBP-1c was increased 3-fold in rat-1 cells in the presence of 100 nM insulin [28]. Similar results have been reported in transfected MIN6 cells [29].
Mutation of the SRE of the proximal SREBP-1c promoter reduces its response to insulin
Since ectopic expression of SREBP-1c enhanced the activity of the pSREBP(−109/+40)-luc, we wished to assess the contribution of the SRE in the proximal SREBP-1c promoter to activation by insulin. The cis-acting sequence CTCACC of the SRE (−61 to −68) of full-length pSREBP(−1516/+40)-luc was mutated to CTCGTA. This mutation prevented binding of SREBP-1a to the proximal SREBP-1c promoter DNA, as determined by EMSA (Figure 5B), and reduced induction of the SREBP-1c promoter by insulin (Figure 6A). Mutation of the SRE element, however, had only a small effect on stimulation of the SREBP-1c promoter by TO901317 (Figure 6B). These results support the hypothesis that binding of SREBP-1c to the SRE within the SREBP-1c promoter is a component of the stimulatory effect of insulin on this promoter, and is critical to the additional stimulus provided by insulin in the face of maximum activation of LXRα by TO901317. Simultaneous mutation of both LXREs and the SRE eliminates all response of the SREBP-1c promoter to both insulin and TO-901317 (Figure 6B).
Figure 6. Mutating the SRE in the proximal SREBP-1c promoter reduces the stimulatory effect of insulin, but not TO901317.
(A) Rat hepatocytes were transfected with pSREBP(−1516/+40)-luc or pSREBP(−1516/+0)-mutSRE-luc and pRL-CMV. Photinus and Renilla luciferase activities were measured after incubation for 24 h in medium with no addition or supplemented with insulin (100 nM). The results are expressed as the means±S.E.M. for eight independent experiments. (B) Rat hepatocytes were transfected with pSREBP(−1516/+40)-luc, pSREBP (−1516/+0)-mutSRE-luc, pSREBP(1516/+40)-mutLXR1/mutLXR2-luc or pSREBP(1516/+40)-mutLXR1/mutLXR2/mutSRE-luc, together with pRL-CMV. Photinus and Renilla luciferase activities were measured after 24 h incubation in medium with no additions or supplemented with insulin (100 nM) or TO901317 (10 μM) or insulin plus TO901317.
SREBP isoforms can also bind to E-box elements in some gene promoters [30]. However, in contrast with the effect of ablation of the SRE, mutation of the adjacent E-box located at −73 to −78 from CATGTG to TGTACG had no effect on responsiveness of the SREBP-1c promoter to insulin, implying that the SRE, but not the E-box, interacts with SREBP-1c (Figure 7).
Figure 7. Proximal Sp1 site, but not the E-box, of the rat SREBP-1c promoter is activated by insulin and TO901317.
Rat hepatocytes were transfected with pSREBP(−1516/+40)-luc, pSREBP(−1516/+40)-mutSp1-luc, or pSREBP(−1516/+40)-mutE-luc, together with pRL-CMV. Photinus and Renilla luciferase activities were measured after 24 h of incubation with medium containing no additions, insulin (100 nM) or TO901317 (10 μM). The results are expressed as the means±S.E.M. for four independent experiments.
Stimulation of the SREBP-1c promoter by insulin is enhanced by binding of Sp1
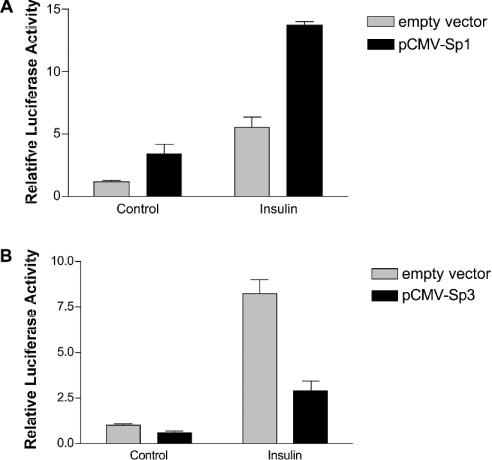
Our data indicate that an intact SREBP binding site is required for maximal stimulation of the SREBP-1c promoter by insulin. Trans-activation of the promoters of FAS, ATP-dependent citrate lyase and S14 by SREBP-1c requires simultaneous occupancy and activation of adjacent Sp1 and NF-Y sites [24,26,27]. The rat SREBP-1c promoter contains a binding site for Sp1 (−52/−60) adjacent to the SRE (−61/−68). To assess the role of Sp1 in stimulation of the SREBP-1c promoter by insulin, we mutated this site within pSREBP(−1516/+40)-luc from GGGCGGG to GAATTC. The insulin response of pSREBP(−1516/+40)-luc with the inactive Sp1 element was reduced from 8.2±0.8-fold to 3.2±1.0-fold (Figure 7). Interestingly, stimulation of the mutated SREBP promoter by TO901317 is also reduced by mutation of the proximal promoter Sp1 site (from 45±7.2-fold to 18±3.5-fold), indicating that binding of Sp1 to this site is needed for full transcriptional activation of the SREBP-1c promoter by both insulin and LXR agonist.
The levels of total and O-GlcNAc (O-linked N-acetyl-D-glucosamine)-modified Sp1 are increased by insulin in rat hepatoma cells [31]. To determine whether increased expression of Sp1 by insulin in rat hepatocytes would stimulate SREBP-1c promoter activity, rat hepatocytes were co-transfected with pSREBP(−1516/+40)-luc and pCMV-Sp1, expressing full-length human Sp1. Ectopic expression of Sp1 increased SREBP-1c promoter activity by 3.4±0.8-fold (Figure 8A), indicating that levels of Sp1 in rat hepatocytes under basal conditions are not saturating. In agreement with this, it has recently been reported that over-expression of protein tyrosine-phosphatase 1-B in rat hepatocytes increases levels of SREBP-1c mRNA by increasing Sp1 transcriptional activity [32].
Figure 8. Sp1 and Sp3 transcription factors differentially regulate the rat SREBP-1c promoter.
(A) Ectopic expression of Sp1 augments SREBP-1c promoter activity in response to insulin. Rat hepatocytes were co-transfected with pSREBP(−1516/+40)-luc and pCMV-Sp1 or empty vector together with pRL-CMV. Cells were then incubated for 24 h in medium wth no additions or supplemented with 100 nM insulin. Photinus and Renilla luciferase activities were measured. The means±S.E.M. for four hepatocyte preparations are shown. (B) Ectopic expression of Sp3 reduces basal SREBP-1c promoter activity and blunts the effect of insulin. Rat hepatocytes were co-transfected with pSREBP(−1516/+40)-luc and pCMV-Sp3 or empty vector together with pRL-CMV. Cells were then incubated for 24 h in medium with no additions or supplemented with 100 nM insulin. Photinus and Renilla luciferase activities were measured. The means±S.E.M. for four hepatocyte preparations are shown.
Like Sp1, Sp3 binds to GC boxes; however, the actions of Sp1 and Sp3 are different [33]. To determine whether Sp3 regulates SREBP-1c promoter activity, rat hepatocytes were co-transfected with pSREBP(1516/+40)-luc and pCMV-Sp3. Ectopic expression of Sp3 reduced basal SREBP-1c promoter activity and severely blunted the insulin response (Figure 8B). These results suggest that Sp3 has an inhibitory effect on SREBP-1c transcription, in contrast to Sp1, which is required for the full stimulatory effect of insulin.
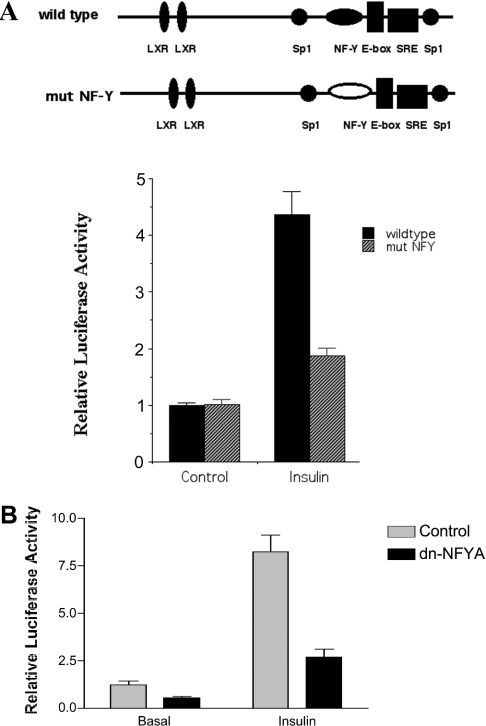
Stimulation of the SREBP-1c promoter by insulin is enhanced by binding of NF-Y
The SREBP-1c promoter contains an inverted CCAAT box (−80 to −85), a potential binding site for NF-Y, in close proximity to the SRE at −61 to −68. To assess the importance of the CCAAT box for activation of the SREBP-1c promoter by insulin, the putative NF-Y site was mutated to GACGC, and rat hepatocytes were transfected with wild-type or mutated pSREBP(−1516/+40)-luc reporter vector. Mutation of the putative NF-Y site in the SREBP-1c promoter blunts the ability of insulin to stimulate transcriptional activity (Figure 9A). Consistent with an important role for NF-Y in supporting activation of the SREBP-1c promoter, ectopic expression of a dominant-negative NF-YA reduces basal and insulin-stimulated SREBP-1c promoter activity (Figure 9B). Sp1 has been shown to interact with the NF-YA subunit in vitro and in vivo [34]. It is possible that the presence of both NF-Y and Sp1 is required for full transactivation by SREBP-1c bound to the SREBP-1c promoter. A similar dependence on the simultaneous presence of intact Sp1- and NF-Y-binding sites for a full stimulatory response to SREBP-1 has been reported for the 7-dehydrocholesterol reductase promoter [35].
Figure 9. Cis-acting NF-Y element is involved in insulin stimulation of the rat SREBP-1c promoter.
Rat hepatocytes were transfected with either pSREBP(−1516/+40)-luc (wild-type) or pSREBP(−1516/+40)-mut NFY-luc (mut NFY), together with pRL-CMV. Photinus and Renilla luciferase activities were measured after 24 h of incubation with medium containing either no additions or insulin (100 nM). The means±S.E.M. luciferase activities for four hepatocyte preparations are shown. (B) Ectopic expression of dominant-negative NF-YA affects both basal and insulin-stimulated SREBP-1c promoter activity by insulin. Rat hepatocytes were co-transfected with pSREBP(−1516/+40)-luc and a vector expressing dominant-negative NF-YA (dnNFYA) or empty vector, together with pRL-CMV. Cells were then incubated for 24 h in medium with no additions or supplemented with insulin (100 nM) and Photinus and Renilla luciferase activities were measured. The normalized luciferase activities (means±S.E.M.) for four independent experiments are shown.
DISCUSSION
The results of the present study suggest that activation of the SREBP-1c promoter in rat hepatocytes by insulin involves the combined action of several transcription factors, including LXRα, SREBP, Sp1 and NF-Y. Increased levels of LXRα protein is sufficient to elevate SREBP-1c promoter activity in rat hepatocytes, since ectopic expression of LXRα increased basal activity of pSREBP(−1516/+40)-luc. However, although insulin increases levels of LXRα mRNA in rat hepatocytes, we were unable to detect corresponding changes in levels of LXR protein. Moreover, the amount by which insulin stimulates SREBP-1c promoter activity (from 4-fold to 8-fold in separate experiments) is far greater than the increase in LXRα mRNA (1.6-fold), implying that other actions of insulin contribute to its stimulatory effect on this promoter.
It is of note that, whereas insulin may increase the activation rather than the abundance of LXRα, co-transfection with an LXRα expression vector had a stronger effect than insulin on SREBP-1c and pCyp7aLXRE(×3)-luc promoter activity. This implies that any increase in LXRα abundance produced by insulin is substantially lower than the constitutive exogenous expression provided by the LXR expression vector. Nonetheless, insulin further increased both pSREBP(−1516/+40)-luc and pCyp7aLXRE(×3)-luc promoter activity in the presence of ectopically expressed LXRα. These results suggest strongly that insulin increases the activity of LXR in rat hepatocytes by (i) increasing the concentration of endogenous LXR agonist, (ii) promoting covalent modification of LXRα, or (iii) modulating the interaction of LXRα with other factors bound to the SREBP-1c promoter. Of interest in this regard is the recent demonstration that LXRα associates with the co-repressor N-CoR (nuclear receptor co-repressor) and recruits N-CoR to the SREBC-1c promoter in a human hepatoma cell line [36]. Activation of LXRα with TO901317 reverses the association of LXRα with the co-repressor. Inactivating both LXREs by mutation reduced activation of the SREBP-1c promoter by insulin only by about 50%, indicating that insulin induction is not limited to these sites on the promoter.
The presence of an SREBP-1c-binding site in the SREBP-1c promoter itself suggests that changes in endogenous SREBP-1c expression may amplify transcriptional responses to insulin stimulation. This is supported by our finding that ablation of the SREBP-binding site in the SREBP-1c promoter reduces its response to insulin. This is consistent with the recent report that over-expression of Insig-1 in livers of transgenic mice, which inhibited migration of SREBP isoforms to the Golgi and liberation of the active N-terminal fragments, severely blunted the rise in hepatic levels of SREBP-1c mRNA resulting from re-feeding after a fast [37].
We have confirmed by ChIP methodology that SREBP-1 is associated with the SREBP-1c promoter in intact hepatocytes and that increasing the expression of SREBP-1c by incubation of the cells with insulin or with insulin plus TO901317 increases the degree of association. Hepatocytes differ in this regard from the CHO (Chinese-hamster ovary) cell line in which Bennett et al. [38] have reported that over-expression of nuclear SREBP-1c did not result in its association with the SRE in the FAS, HMG-CoA reductase or squalene synthase promoters, as demonstrated by ChIP assay. As insulin selectively increases expression of the SREBP-1c isoform in liver, and increased expression of SREBP-1c activates the FAS promoter in hepatocytes, Bennett et al. [38] speculate that hepatocytes may contain co-activators lacking in CHO cells that interact with SREBP-1c and open the nuclesosome structure to make the SRE accessible. The co-activators that mediate transcriptional stimulation by SREBP-1c have not been identified.
The ability of insulin to enhance responses of the SREBP-1c promoter to ectopically expressed SREBP-1c may reflect actions at other sites on the promoter that augment its response to SREBP. However, it is also possible that insulin acts directly on the nuclear form of SREBP to increase its transcriptional activity or to increase its concentration in the nucleus. The N-terminal fragments of SREBP-1c and SREBP-1a can be phosphorylated in vitro by MAPK (mitogen-activated protein kinase) isoforms, including ERK1/2 (extracellular-signal-regulated kinase 1/2), p38 MAPK and JNK (c-Jun N-terminal kinase) [39,40]. The major site of phosphorylation of human SREBP-1a by ERK1/2 has been identified as Ser117 [41], a sequence shared by SREBP-1c. Mutation of this site prevents insulin from stimulating the LDL (low-density lipoprotein) receptor promoter through ectopically expressed SREBP-1a in transfected HepG2 cells (in which endogenous SREBP-1 levels are not increased by insulin). Activation of MAPK in other transformed cells can lead to up-regulation of SREBP-1c expression and lipogenesis (for example, [42,43]). Whether changes in the phosphorylation state of SREBP occur in rat hepatocytes in response to insulin treatment is not known at present.
Nuclear SREBP is subject to proteasomal degradation following ubiquitination [44]. Acetylation of the nuclear forms of SREBP-1a and SREBP-2 by the co-activator p300 prevents ubiquitination and increases the nuclear content of these transcription factors [45]. If SREBP-1c is stabilized in a similar fashion by p300 or another co-activator with intrinsic acetyl transferase activity, insulin might increase its effective nuclear concentration by increasing association with co-activators at the SREBP-1c promoter.
Insulin may also promote association of SREBP with other transcription factors bound to adjacent sites on the SREBP-1c promoter. SREBP-1 and Sp1 are able to interact in vitro, and SREBP recruits Sp1 to its DNA-binding site in the LDL receptor promoter [46,47]. In the present study, mutation of the Sp1 site adjacent to the SRE reduced activation of the SREBP-1c promoter by insulin, demonstrating the importance of this site for expression of the full effect of the hormone. In addition, ectopic expression of Sp1 stimulated SREBP-1c promoter activity, showing that changes in Sp1 abundance or activity in the rat hepatocyte can modulate SREBP-1c gene expression.
Sp1 is subject to phosphorylation by a number of protein kinases, with uncertain consequences for its transcriptional activity (reviewed in [33,48]). Sp1 is also subject to O-GlcNAc modification at sites within the activation domain [33]. O-GlcNAc modification may assist the nuclear localization [49] or reduce the proteasomal degradation [50,51] of a number of proteins, including Sp1. Insulin has been reported to increase O-glycosylation and nuclear content of Sp1 and to increase Sp1-dependent activity of the calmodulin gene promoter in H4IIE hepatoma cells; accumulation of Sp1 and induction of calmodulin were blocked when O-glycosylation was inhibited [52]. Insulin also promotes O-glycosylation of Sp1 in L1 myotubes [53]. O-GlcNAc modification of Sp1 in aortic endothelial cells and in HeLa cells increases its transcriptional activity [54,55]. A similar effect of insulin in rat hepatocytes could contribute to activation of the SREBP-1c promoter.
While the present study was under review, some similar results from transfection experiments using the mouse SREBP-1c promoter were reported by Chen et al. [56]. In contrast to the results reported here, Chen et al. [56] found that mutation of the LXREs effectively eliminated the response of the mouse promoter to insulin, but that mutation of the SRE had relatively little effect on insulin response. At present we do not know whether this represents a species difference in the behaviour of the SREBP-1c promoter or reflects differences in the experimental protocols. The results presented here support a significant contribution of the SRE to the stimulatory effect of insulin on the rat SREBP-1c, and indicate that the full complement of LXRE, SRE, Sp1 and NF-Y sites is required for the maximum effect of insulin to be exerted.
Acknowledgments
We thank Poonam Kumar for expert technical assistance. This work was supported in part by grants from the Office of Research and Development, Department of Veterans Affairs (DVA), from the American Heart Association, Southeast Affiliate, the University of Tennessee Vascular Biology Center of Excellence and by a grant from the National Institutes of Health (grant number DK-059368 to E. A. P.). X. D. is a recipient of a Postdoctoral Fellowship Award from the American Heart Association, Southeast Affiliate. R. R. is a senior research scientist for the DVA.
References
- 1.Rangan V. S., Oskouian B., Smith S. Identification of an inverted CCAAT box motif in the fatty acid synthase gene as an essential element for the modification of transcriptional regulation by cAMP. J. Biol. Chem. 1996;271:2307–2312. doi: 10.1074/jbc.271.4.2307. [DOI] [PubMed] [Google Scholar]
- 2.Clarke S. D. Polyunsaturated fatty acid regulation of gene transcription: a molecular mechanism to improve the metabolic syndrome. J. Nutr. 2001;131:1129–1132. doi: 10.1093/jn/131.4.1129. [DOI] [PubMed] [Google Scholar]
- 3.Osborne T. F. Sterol regulatory element-binding poteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 2000;276:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 4.Hua X., Wu J., Goldstein J. L., Brown M. S., Hobbs H. H. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- 5.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown M. S., Goldstein J. L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Soc. Natl. Acad. Sci. U.S.A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards P. A., Tabor D., Kast H. R., Venkateswaran A. Regulation of gene expression by SREBP and SCAP. Biochim. Biophys. Acta. 2000;1529:103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 8.Horton J. D., Bashmakov Y., Shimomura I., Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura I., Bashmakov Y., Ikemoto S., Horton J. D., Brown M., Goldstein J. L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozatocin-induced diabetes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azout-Marniche D., Bécard D., Guichard C., Foretz M., Ferré P., Foufelle F. Insulin effects on sterol regulatory element binding protein 1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem. J. 2000;350:389–393. [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X., Cagen L. M., Wilcox H. G., Park E. A., Raghow R., Elam M. B. Regulation of the rat SREBP-1c promoter in primary rat hepatocytes. Biochem. Biophys. Res. Commun. 2002;290:256–262. doi: 10.1006/bbrc.2001.6148. [DOI] [PubMed] [Google Scholar]
- 12.Foretz M., Guichard C., Ferré P., Foufelle F. Sterol regulatory element binding protein 1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimano H., Horton J. D., Shimomura I., Hammer R. E., Brown M. S., Goldstein J. L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amemiya-Kudo M., Shimano H., Yoshikawa T., Yahagi N., Hasty A. H., Okazaki H., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K., et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 15.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J.-M. A., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. Regulation of mouse sterol regulatory element binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshikawa T., Shimano T., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. Identification of liver X receptor as an activator of the sterol regulatory element binding protein 1c gene promoter. Mol. Cell. Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorngate F. E., Raghow R., Wilcox H. G., Werner C. S., Elam M. B. Insulin promotes the biosynthesis and secretion of apolipoprotein B-48 by dramatically altering apolipoprotein B mRNA editing. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5392–5396. doi: 10.1073/pnas.91.12.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultz J. R., Hua T., Luk A., Repa J. J., Medina J. C., Li L., Schwender S., Wang S., Thoolen M., Mangelsdorf D. J., et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang G., Yang J., Horton J. D., Hammer R. E., Goldstein J. L., Brown M. S. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 20.Tobin K. A., Ulven S. M., Schuster G. U., Steineger H. H., Andresen S. M., Gustaffson J.-A., Nebb H. I. LXRs as insulin mediating factors in fatty acid and cholesterol biosynthesis. J. Biol. Chem. 2002;277:10691–10697. doi: 10.1074/jbc.M109771200. [DOI] [PubMed] [Google Scholar]
- 21.Peet D. J., Janowski B. A., Mangelsdorf D. J. The LXRs: a new class of oxysterol receptors. Curr. Opin. Genet. Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Bolten C., Bhat G., Woodring-Dietz J., Li S., Prayaga S. K., Xia C., Lala D. S. Induction of human liver X receptor a gene expression via an autoregulatory loop mechanism. Mol. Endocrinol. 2002;16:506–514. doi: 10.1210/mend.16.3.0789. [DOI] [PubMed] [Google Scholar]
- 23.Jackson S. M., Ericsson J., Mantovani R., Edwards P. A. Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein. J. Lipid Res. 1998;39:767–776. [PubMed] [Google Scholar]
- 24.Jump D. B., Thelen A. P., Mater M. K. Functional interaction between SREBP-1c, NF-Y, and T3 nuclear receptors. J. Biol. Chem. 2001;276:34419–34427. doi: 10.1074/jbc.M105471200. [DOI] [PubMed] [Google Scholar]
- 25.Magaña M. M., Lin S. S., Dooley K. A., Osborne T. J. Sterol regulation of acetyl coenzyme A carboxylase promoter requires two interdependent binding sites for sterol regulatory element binding proteins. J. Lipid Res. 1997;38:1630–1638. [PubMed] [Google Scholar]
- 26.Magaña M. M., Koo S.-H., Towle H. C., Osborne T. F. Different sterol regulatory element-binding protein isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J. Biol. Chem. 2000;275:4726–4733. doi: 10.1074/jbc.275.7.4726. [DOI] [PubMed] [Google Scholar]
- 27.Moon Y.-A., Lee J.-J., Park S.-W., Ahn Y.-H., Kim K.-S. The roles of sterol regulatory element binding proteins in the transactivation of the rat ATP citrate-lyase promoter. J. Biol. Chem. 2000;275:30280–30286. doi: 10.1074/jbc.M001066200. [DOI] [PubMed] [Google Scholar]
- 28.Kim J. B., Sarraf P., Wright M., Yao K. M., Mueller E., Solanes G., Lowell B. B., Spiegelman B. M. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreolas C., Xavier G., Diraison F., Zhao C., Varadi A., Lopez-Casillas F., Ferré P., Foufelle F., Rutter G. A. Stimulation of acetyl-CoA carboxylase gene expression by glucose requires insulin release and sterol regulatory element binding protein 1c in pancreatic Min6 cells. Diabetes. 2002;51:2536–2545. doi: 10.2337/diabetes.51.8.2536. [DOI] [PubMed] [Google Scholar]
- 30.Kim J. B., Spotts G. D., Halvorsen D., Shih H. M., Ellenberger T., Towle H. C., Spiegelman B. M. Dual DNA binding specificity of ADD1/SREBP-1 is controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan X., Solomon S. S., Borromeo D. M., Martinez-Hernandez A., Raghow R. Insulin deprivation leads to deficiency of Sp1 transcription factor in H4IIE hepatoma cells and in streptozotocin-induced diabetic ketoacidosis in the rat. Endocrinology. 2001;142:1635–1641. doi: 10.1210/endo.142.4.8083. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu S., Ugi S., Maegawa H., Egawa K., Nishio Y., Yoshizaki T., Shi K., Nagai Y., Morino K., Nemoto K., et al. Protein–tyrosine phosphatase 1B as a new activator fro hepatic lipogenesis via sterol regulatory element-binding protein gene expression. J. Biol. Chem. 2003;278:43095–43101. doi: 10.1074/jbc.M306880200. [DOI] [PubMed] [Google Scholar]
- 33.Bouwman P., Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:37–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 34.Roder K., Wolf S. S., Larkin K. J., Schweizer M. Interaction between the two ubiquitously expressed transcription factors NF-Y and Sp1. Gene. 1999;23:61–69. doi: 10.1016/s0378-1119(99)00180-8. [DOI] [PubMed] [Google Scholar]
- 35.Kim J.-H., Lee J. N., Paik Y.-K. Cholesterol biosynthesis from lanosterol. J. Biol. Chem. 2001;276:18153–18160. doi: 10.1074/jbc.M101661200. [DOI] [PubMed] [Google Scholar]
- 36.Hu X., Li S., Wu J., Chunsheng S., Lala D. S. Liver X receptors interact with corepressors to regulate gene expression. Mol. Endocrinol. 2003;17:1019–1026. doi: 10.1210/me.2002-0399. [DOI] [PubMed] [Google Scholar]
- 37.Engelking L. J., Kuriyama H., Hammer R. E., Horton J. D., Brown M. S., Goldstein J. L., Liang G. Overexpression of Insig-1 in livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 2004;113:1168–1175. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett M. K., Toth J. L., Osborne T. F. Selective association of sterol regulatory element binding protein isoforms with target promoters in vivo. J. Biol. Chem. 2004;279:37360–37367. doi: 10.1074/jbc.M404693200. [DOI] [PubMed] [Google Scholar]
- 39.Kotzka J., Müller-Wielande D., Koponen A., Njamen D., Kremer L., Roth G., Munck M., Knebel B., Krone W. ADD1/SREBP-1c mediates insulin-induced gene expression linked to the MAP kinase pathway. Biochem. Biophys. Res. Commun. 1998;249:375–379. doi: 10.1006/bbrc.1998.9161. [DOI] [PubMed] [Google Scholar]
- 40.Kotzka J., Müller-Wieland D., Roth G., Kremer L., Munck M., Schürmann S., Knebel B., Krone W. Sterol regulatory element binding proteins (SREBP)-1a and SREBP-2 are linked to the MAP kinase cascade. J. Lipid Res. 2000;41:99–108. [PubMed] [Google Scholar]
- 41.Roth G., Kotzka J., Kremer L., Lehr S., Lohaus C., Meyer H. E., Krone W., Müller-Wieland D. MAP kinases ERK1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J. Biol. Chem. 2000;275:33302–33307. doi: 10.1074/jbc.M005425200. [DOI] [PubMed] [Google Scholar]
- 42.Swinnen J. V., Heemer H., Deboel L., Foufelle F., Heyns W., Verhoeven G. Stimulation of tumor-associated fatty acid synthase expression by growth factor activation of the sterol regulatory element-binding protein pathway. Oncogene. 2000;19:5173–5181. doi: 10.1038/sj.onc.1203889. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y. A., Han W. F., Morin P. J., Chrest F. J., Pizer E. S. Activation of fatty acid synthesis during neoplastic transformation; role of mitogen-activated protein kinase and phosphatidylinositol 3-kinase. Exp. Cell Res. 2002;279:80–90. doi: 10.1006/excr.2002.5600. [DOI] [PubMed] [Google Scholar]
- 44.Hirano Y., Yoshida M., Shimizu M., Sato R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding protein by the ubiquitin-proteasome pathway. J. Biol. Chem. 2001;276:36431–36437. doi: 10.1074/jbc.M105200200. [DOI] [PubMed] [Google Scholar]
- 45.Giandomenico V., Simonsson M., Grönroos E., Ericsson J. Coactivator dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell. Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Athanikar J. N., Sanchez H. B., Osborne T. F. Promoter selective transcriptional synergy mediated by sterol regulatory element binding protein and Sp1: a critical role for the Bd domain of Sp1. Mol. Cell. Biol. 1997;17:5193–5200. doi: 10.1128/mcb.17.9.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yieh L., Sanchez H. B., Osborne T. F. Domains of transcription factor Sp1 required for synergistic activation with sterol regulatory element binding protein 1 of low density lipoprotein promoter. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6102–6106. doi: 10.1073/pnas.92.13.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samson S. L., Wong N. C. Role of Sp1 in insulin regulation of gene expression. J. Mol. Endocrinol. 2002;29:265–279. doi: 10.1677/jme.0.0290265. [DOI] [PubMed] [Google Scholar]
- 49.Zachara N. E., Hart G. W. The emerging significance of O-GlcNAc in cellular regulation. Chem. Rev. 2002;102:431–438. doi: 10.1021/cr000406u. [DOI] [PubMed] [Google Scholar]
- 50.Han I., Kudlow J. Reduced O-glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F., Su K., Yang X., Bowe D. B., Paterson A. J., Kudlow J. E. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 52.Majumdar G., Harmon A., Candelaria Martinez-Hernandez A., Raghow R., Solomon S. S. O-glycosylation of Sp1 and transcriptional regulation of the calmodulin gene by insulin and glucagon. Am. J. Physiol. 2003;285:E584–E591. doi: 10.1152/ajpendo.00140.2003. [DOI] [PubMed] [Google Scholar]
- 53.Walgren J. L. E., Vincent T. S., Schey K. L., Buse M. G. High glucose and insulin promote O-GlcNAc modification of proteins, including a-tubulin. Am. J. Physiol. 2002;284:E424–E434. doi: 10.1152/ajpendo.00382.2002. [DOI] [PubMed] [Google Scholar]
- 54.Du X.-L., Edelstein D., Rosetti L., Fantus I. G., Goldberg H., Ziyadeh F., Wu J., Brownlee M. Hyperglycemia-induced superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12222–12226. doi: 10.1073/pnas.97.22.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang H. T., Ju J. W., Cho J. W., Hwang E. S. Down-regulation of Sp1 activity through modulation of O-glycosylation by treatment with a low glucose-mimetic, 2-deoxyglucose. J. Biol. Chem. 2003;278:51223–51231. doi: 10.1074/jbc.M307332200. [DOI] [PubMed] [Google Scholar]
- 56.Chen G., Liang G., Ou J., Goldstein J. L., Brown M. S. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]