Abstract
NADH kinase (NADHK; ATP:NADH 2′-phosphotransferase; EC 2.7.1.86), an enzyme that preferentially utilizes NADH as the diphosphonicotinamide nucleotide donor, has been identified for the first time in plants. Low activity (0.4 nmol of NADPH produced/min per mg of protein) was observed in clarified protein extracts from Arabidopsis thaliana (thale cress) cell suspension cultures. However, unlike an NADHK from yeast (Saccharomyces cerevisiae) (POS5), the enzyme from Arabidopsis did not associate with the mitochondria. NADHK was cloned (gi:30699338) from Arabidopsis and studied as a recombinant protein following affinity purification from Escherichia coli. The enzyme had a pH optimum for activity of 7.9 and a subunit molecular mass of 35 kDa. Analytical gel filtration demonstrated that the recombinant enzyme exists as a dimer. Hyperbolic saturation kinetics were observed for the binding of NADH, ATP, free Mg2+ and NAD+, with respective Km values of 0.042, 0.062, 1.16, and 2.39 mM. While NADHK could phosphorylate NADH or NAD+, the specificity constant (Vmax/Km) for NADH was 100-fold greater than for NAD+. The enzyme could utilize UTP, GTP and CTP as alternative nucleotides, although ATP was the preferred substrate. PPi or poly-Pi could not substitute as phospho donors. PPi acted as a mixed inhibitor with respect to both NADH and ATP. NADHK was inactivated by thiol-modifying reagents, with inactivation being decreased in the presence of NADH or ATP, but not NAD+. This study suggests that, in Arabidopsis, NADP+/NADPH biosynthetic capacity could, under some circumstances, become uncoupled from the redox status of the diphosphonicotinamide nucleotide pool.
Keywords: Arabidopsis thaliana (thale cress), calcium/calmodulin, NAD+ kinase (NADK), NADH kinase (NADHK, ATP: NADH 2′-phosphotransferase, EC 2.7.1.86), nicotinamide (pyridine) nucleotide
Abbreviations: BLAST, basic local alignment sequence tool; CaM, calmodulin; pCMB, p-chloromercuribenzoate; DCPIP, 2,6-dichlorophenol-indophenol; DTT, dithiothreitol; NADK, NAD+ kinase; NADHK, NADH kinase (ATP:NADH 2′-phosphotransferase, EC 2.7.1.86); NCBI-CDD, National Center for Biotechnology Information conserved-domain database; PEP, phosphoenolpyruvate; ROS, reactive oxygen species
INTRODUCTION
The present study describes Arabidopsis thaliana (thale cress) NADHK (NADH kinase; ATP:NADH 2′-phosphotransferase, EC 2.7.1.86), a novel plant enzyme capable of synthesizing NADPH using NADH as the preferred diphosphonicotinamide (diphosphopyridine) nucleotide donor.
NAD+ and NADP+, both major carriers of cellular reducing equivalents, are of vital importance in diverse metabolic processes such as mitochondrial energy metabolism, and, depending on the context, production of, or protection from, ROS (reactive oxygen species) [1,2]. Efficient control and co-ordination of the reduction–oxidation (redox) status is therefore of crucial importance within the cell, where, for example, the NADP+/NAD+ ratio rapidly increases (over 10-fold) following pathogen attack [1,3]. In plants it is widely accepted that the only de novo mechanism of NADP+ biosynthesis is by NAD+ kinase (NADK, EC 2.7.1.23) using NAD+ and ATP as substrates [4,5]. The role of NADK has received much attention in plants because, in addition to the afore-mentioned importance in nicotinamide metabolism, isoenzymes have been characterized that are activated through interaction with Ca2+/CaM (calmodulin) [6].
Genes encoding NADK have been reported from bacterial [7,8], yeast (Saccharomyces cerevisiae) [9] and human sources [10] and, based on sequence homology searches, we identified NADK1 (At3g21070; gi30686057) and NADK2 (At1g21640; gi30687452), two candidate Arabidopsis NADK homologues [11]. Both Arabidopsis genes were subsequently cloned, expressed in Escherichia coli and verified as proteins with NADK activity, with one (NADK2) being shown to interact with Ca2+/CaM. Subsequent domain analysis using the National Center for Biotechnology Information conserved-domain database (NCBI-CDD; [12]), has identified NADK3 (At1g78590; gi30699338), a third gene product from Arabidopsis that contains a putative NADK-like domain. Binary sequence alignments of NADK3 revealed negligible similarity to either of the previously identified Arabidopsis NADKs. However, sequence analysis revealed that NADK3 possessed similarity to POS5 (YPL_188W) from yeast (S. cerevisiae), recently shown to be an NADHK [13,14]. This enzyme, which to date has only been described in yeast, is distinct from NADK [15,16] and localizes to the mitochondria [13,14,17]. Subsequent work with deletion mutants and recombinant protein expression has shown the POS5 enzyme to be of critical importance (non-redundant) in maintaining both mitochondrial redox balance and mitochondrial DNA stability, and in detoxification of ROS [13,14].
Owing to the importance of NADHK in yeast metabolism, and on the basis of database sequence analysis, we decided to study NADK3 and to determine whether higher plants also contain an NADHK. Below we present a kinetic characterization of recombinant NADK3, the first plant NADHK described, and show that the protein is distinct from several NADK enzymes thus far described in plants.
MATERIALS AND METHODS
The vectors pBluescript II SK(+) and pET29a(+) were obtained from Stratagene and Novagen respectively. All other enzymes used during DNA manipulation and cloning were from MBI/Fermentas (Burlington, ON, Canada). Primers for PCR and DNA sequence analysis were synthesized by Cortec DNA Services (Kingston, ON, Canada). S-protein–agarose was from Novagen. Thrombin, Percoll, Sephadex PD-10 columns and native molecular-mass markers were from Amersham Biosciences. SDS/PAGE molecular-mass standards were from Bio-Rad. ATP, NAD+, NADH, glucose 6-phosphate and glucose-6-phosphate dehydrogenase (EC 1.1.1.49; from S. cerevisiae) were from Roche. All other chemicals were of analytical quality and obtained from either Sigma Chemical Co or Fisher Scientific. Arabidopsis (ecotype Columbia-O) liquid cell-suspension cultures were dark grown in Murashige and Skoog media (supplemented with 1% sucrose) on an orbital shaker (120 rev./min, 25 °C) with a 7-day culture cycle. Tissue was harvested by filtration, powdered under liquid nitrogen and stored at −80 °C until use. E. coli [BL21-CodonPlus(DE3)-RIL; Stratagene] were routinely cultured at 37 °C in Luria–Bertani media supplemented with appropriate antibiotic.
Amplification and cloning of NADK3
NADK3 was amplified by PCR (30 cycles, 55 °C annealing temperature) from an Arabidopsis inflorescence cDNA library (CD4-6; Arabidopsis Biological Resource Center stock number: 88580) using the following primer pair (based on At1g78590): 5′-GGA TCC ATG GCG ATT AGG AAG CTT TTG CTT-3′ and 5′-GAA TTC CT AGT ACC TTG ATC TGA TCT GAG ATA TGC-3′. The PCR product was cloned into pBluescript II SK(+) and subsequently into the pET29a(+) expression vector using BamHI and EcoRI restriction sites. This construct thus produced a 5′-(S-Tag·NADHK)-3′ fusion protein with an S-protein affinity tag. When required, the fusion protein was cleaved with thrombin protease according to the manufacturer's (Amersham Biosciences) protocol.
Expression and purification of recombinant NADK3
A 2 ml overnight culture of E. coli (pET29a(+):NADK3) was transfered into 100 ml of Luria–Bertani medium supplemented with 50 μg/ml kanamycin and cultured aerobically at 37 °C for 3 h. Recombinant protein expression was induced by the addition of 500 μM isopropyl β-D-thiogalactopyranoside and the cultures grown for a further 3 h at 30 °C. Cells were harvested by centrifugation (4000 g, 6 min), resuspended in 6 ml of STCB [S-Tag column buffer: 20 mM Tris/HCl (pH 7.9)/200 mM NaCl/0.2 mM dithiothreitol (DTT)/10% (v/v) glycerol/0.01% (v/v) Triton X-100] supplemented with 1 mM PMSF and 10 μM E-64 [trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane], and disrupted by a single pass through a French pressure cell (70 MPa). Cellular debris was removed by centrifugation (12000 g, 10 min), and the resulting supernatant incubated on a rocking platform with 2 ml of S-protein–agarose for 15 min at room temperature. The resin was packed into a column (0.8 cm×4 cm), washed with 20 column vol. of STCB and eluted with two 0.8 ml vol. of STCB supplemented with 3 M MgCl2. The column eluate was combined and desalted using a Sephadex PD-10 column that had previously been equilibrated in 20 mM Tris/HCl (pH 7.9)/200 mM NaCl/0.2 mM EDTA/0.2 mM DTT/10% (v/v) glycerol. Desalted fractions containing NADHK activity were pooled, frozen in aliquots under liquid nitrogen and stored at −80 °C. NADHK was stable for at least 3 months when stored frozen at −80 °C.
In planta analysis and subcellular fractionation of NADK3
Crude protein extracts were prepared by homogenizing (pestle and mortar) Arabidopsis suspension cells in 2 vol. of 50 mM Tris/HCl buffer, pH 7.9, containing 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 1 mM EDTA, 1 mM PMSF, 100 μM leupeptin, 10 μM E-64, 20% (v/v) glycerol, 2% (w/v) poly(ethylene glycol) 8000, 2% (w/v) polyvinylpolypyrrolidone and 0.05% (v/v) Nonidet P40 [poly(ethylene glycol) octaphenyl ether]. Extracts were clarified by centrifugation (12000 g, 10 min, 4 °C) and the supernatants desalted on Sephadex PD-10 columns (as for recombinantprotein purification). Mitochondria and plastids were isolated as described in [18]. Briefly, freshly harvested Arabidopsis suspension cells were disrupted in a buffer containing 450 mM sucrose as the osmoticum. Following collection of plastids by centrifugation (3500 g for 10 min), mitochondria were pelleted at 17000 g for 15 min and the resuspended pellet layered on to a Percoll discontinuous density gradient (18, 23, and 40%, v/v). The sample was centrifuged for 60 min at 75000 g and mitochondria collected from the 23% Percoll/40% Percoll interface.
Enzyme and protein assays
NAD(H)K activity was assayed (90 μl final volume) in microtitre plates essentially as described in [19]. Standard assay mixtures, unless otherwise specified, contained 50 mM Tris (pH 7.9), 4 mM ATP and 10 mM MgCl2. For the determination of NADHK or NADK activity, 0.8 mM NADH or 4 mM NAD+ was included in the reaction mixture respectively. Endogenous Arabidopsis NADK activity was measured in the presence of 1 mM CaCl2 and 300 nM CaM (prepared according to [20]). Reactions were initiated with up to 9 μl of protein extract and allowed to proceed for 30 min at 25 °C. NADP+ or NADPH formed was immediately detected by adding 20 μl of the NAD(H)K assay mixture to a cycling assay (250 μl final volume) containing 50 mM Tris/HCl, pH 7.9, 5 mM glucose 6-phosphate, 1 mM EGTA, 0.5 unit of glucose 6-phosphate dehydrogenase, 0.2 mg/ml DCPIP (2,6-dichlorophenol-indophenol), 0.1 mg/ml 5-methylphenazinium methyl sulphate. Reduction of DCPIP was monitored at 600 nm using a SpectroMax-Plus microplate reader (Molecular Devices) and the amount of NADP+ or NADPH quantified by comparison with a standard curve produced from genuine NADP+. When determining NADHK activity, the NADH concentration (0.8 mM or less) did not result in substantial non-enzymic DCPIP reduction in the subsequent cycling assay. In addition, no significant autoxidation of the reduced indophenol was observed. Therefore, carry-over of NADH into the cycling assay did not result in an overestimate of NADHK activity, and oxidation of excess substrate was unnecessary. All assays were linear with respect to time and concentration of enzyme assayed. One unit of NAD(H)K activity is defined as the amount of enzyme resulting in the production of 1 μmol of NADP+ or NADPH/min at 25 °C. Apparent Vmax and Km values were calculated from the Michaelis–Menten equation using a non-linear least-squares regression algorithm [21]. Competitive inhibition constants (Ki values) were determined using Dixon plots [22], whereas uncompetitive inhibition constants (Ki′ values) were obtained using Cornish-Bowden plots [23]. Kinetic parameters are the means of three or more independent determinations and are reproducible to within ±10% (S.E.M.) of the mean value. Free cation concentrations were calculated on the basis of their respective binding to organophosphates, nucleotides, EGTA and/or Cl− using a computer program that automatically corrects for temperature, pH and ionic strength [24]. To remove trace MgCl2 prior to cation cofactor analysis, NADHK was desalted for a second time on a Sephadex PD-10 column as described above. For kinetic studies of inactivation, NADHK was incubated at 37 °C with various concentrations of iodoacetic acid or p-chloromercuribenzoate (pCMB) in a total volume of 12 μl for up to 30 min. After incubation, residual NADHK activity was determined in the standard assay mixture. Inactivation of glucose-6-phosphate dehydrogenase was not apparent in the cycling assay. Substrate inactivation-protection was performed with either 1 mM ATP, NAD+ or NADH in the incubation mixture. ATP phosphatase, NADP+ phosphatase and alkaline pyrophosphatase activities were measured by monitoring phosphate release essentially as described in [25]. Assays were conducted in a final volume of 40 μl with either 50 mM Tris/HCl (pH 7.9) or 20 mM sodium acetate, pH 5.5, in the presence of 10 mM MgCl2 and contained either 10 mM ATP, 2 mM NADP+, 2 mM NADPH or 5 mM PPi. Citrate synthase (EC 2.3.3.1) was assayed according to [26] in a final volume of 200 μl. Protein concentration was determined by the dye-binding method of Bradford [27] as modified by Bollag et al. [28] with bovine γ-globulin as the protein standard.
Other methods
Determination of molecular mass by FPLC was performed with a Superose-6 HR10/30 column pre-equilibrated in 20 mM Tris/HCl (pH 7.9)/200 mM NaCl/5 mM MgCl2/0.5 mM EDTA/0.2 mM DTT/10% (v/v) glycerol. Native molecular masses were calculated from a plot of Kd (partition coefficient) against log (molecular mass) using the following protein standards: catalase (232 kDa), aldolase (158 kDa), BSA (67 kDa), ovalbumin (43 kDa) and carbonic anhydrase (29 kDa). SDS/PAGE was performed, using the method of Laemmli [29], according to the manufacturer's instructions (Bio-Rad) with 1.5-mm-thick slab gels. The final acrylamide concentrations of the resolving and stacking gel were 12 and 4% (w/v) respectively. Antiserum was prepared in rabbits using 0.2 mg of NADHK protein emulsified in Ribi adjuvant (Corixa Corporation, Seattle, WA, U.S.A.) according to the manufacturer's immunization schedule. Antiserum was immunopurified against pure recombinant NADHK immunoblotted on to nitrocellulose membrane. Immunoblotting was performed using standard protocols [28] using nitrocellulose membranes, non-fat skimmed milk as the blocking agent and goat anti-rabbit–alkaline phosphatase conjugate visualization.
RESULTS
Identification of NADK3 as an NADH kinase gene
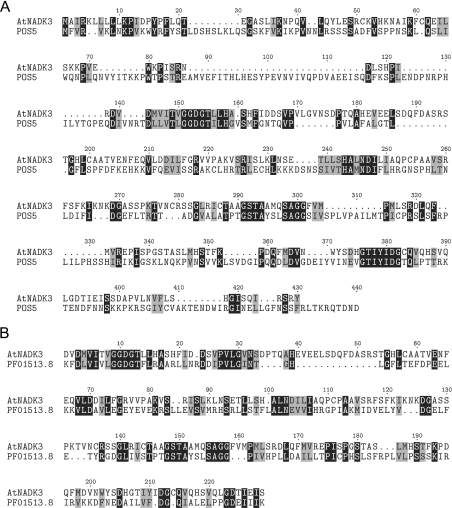
Currently the NCBI-CDD (v1.62; [12]) identifies three Arabidopsis genes that contain a putative NADK domain. We have previously cloned two of these genes (NADK1, NADK2) and confirmed they catalyse the synthesis of NADP+ from NAD+ [11]. Recently, the POS5-gene product from yeast was shown to encode an NADHK that preferentially used NADH as the diphosphonicotinamide nucleotide substrate [13,14]. NCBI BLAST (basic local alignment sequence tool) analysis [30] of POS5 revealed 25% identity (42% similarity) with NADK3, the third candidate NADK protein identified in the NCBI-CDD (Figure 1A). In addition, genomic and EST (expressed sequence tag) database sequence analysis detected highly conserved putative homologues of Arabidopsis NADK3 in various plants [e.g. barley (Hordeum vulgare), corn (Zea mays), rice (Oryza sativa), canola (Brassica napus oleifera) and potato (solanum tuberosum)], suggesting this enzyme is widespread among higher plants.
Figure 1. Alignment of deduced NADHK amino acid sequences.
(A) Alignment of Arabidopsis (At1g78590; gi 30699338) and yeast (POS5; YPL_188W) NADHK. Primary protein sequences were aligned using ClustalX and formatted using TeXshade (Biology Workbench, http://workbench.sdsc.edu). Black and grey boxes represent conserved and similar amino acids respectively. Dots indicate computer-generated gaps within the sequence. (B) Alignment of Arabidopsis NADHK (residues 75–296) and the NADK conserved domain (Pfam: PF01513.8). Primary sequence alignment and formatting were as detailed in (A).
NADK3 encodes a polypeptide of 317 amino acids with a predicted molecular mass of 35 kDa. Using the AGI (Arabidopsis Genome Initiative) protein data set at TAIR (The Arabidopsis Information Resource) [31], BLAST analysis of NADK3 did not detect close homology with any other Arabidopsis protein, with the two highest ranked alignments, NADK2 and NADK1, having low expect values of 0.019 and 0.096 respectively. However, when aligned to the NADK conserved domain at NCBI-CDD (substantially equivalent to Pfam PF01513.8 [32]), NADK3 aligned with over 81% of the 248-residue conserved domain with an expect value of 3×10−16 (Figure 1B). Conserved domain analysis and similarity to POS5 would strongly suggest that NADK3 encodes an NADHK. Therefore, as previously cloned Arabidopsis NADKs could not utilize NADH as a substrate [11] and NADHK activity has only been identified in yeast, we decided to clone and characterize NADK3.
Purification of recombinant Arabidopsis NADHK
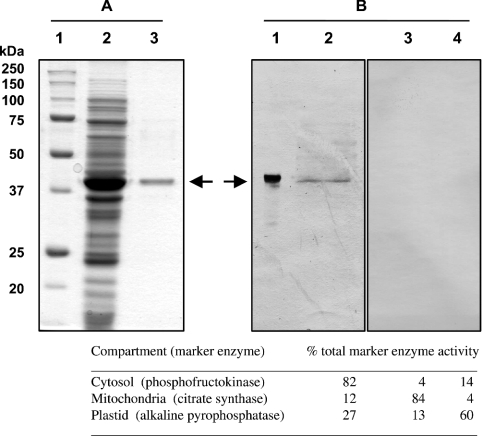
NADK3 was cloned and the recombinant protein affinity-purified from E. coli extracts using S-protein–agarose. The purified S-Tag fusion protein was homogeneous, as determined by staining with Coomassie Blue R-250 following SDS/PAGE, and had a molecular mass of 38 kDa, corresponding to that of the predicted NADHK polypeptide (Figure 2A). The native molecular mass of the protein, determined in triplicate by Superose-6 gel filtration FPLC, was 88±4 kDa, which suggests that NADHK is dimeric. To assess whether the S-protein affinity tag altered NADHK oligomerization, one Superose-6 gel-filtration experiment was conducted with thrombin-protease-cleaved NADK3. In this instance the molecular mass of NADHK was 78 kDa, which indicates the affinity tag does not alter the oligomeric state of the enzyme during gel-filtration chromatography. Immunoblotting of an Arabidopsis cell suspension culture extract revealed a single immunoreactive polypeptide when probed with anti-NADHK serum (Figure 2B). This demonstrates that NADK3 gene product is translated and the resulting protein is of the predicted molecular mass.
Figure 2. SDS/PAGE and immunoblot analysis of Arabidopsis NADHK.
(A) SDS/PAGE [12% (w/v) resolving gel] of recombinant NADHK purified from E. coli. Lane 1 contains 10 μg of various protein standards. Lane 2 contains 30 μg of a crude E. coli protein extract prepared after 3 h induction with 500 μM isopropyl β-D-thiogalactopyranoside. Lane 3 contains 2 μg of NADHK purified on S-protein–agarose. Protein staining was performed with Coomassie Blue R-250. (B) Immunoblot analysis performed using anti-NADHK serum. Lane 1 contains 0.05 μg of recombinant NADHK purified on S-protein–agarose. Lanes 2–4 each contain 50 μg of protein extracted from 7-day-old Arabidopsis cell suspension cultures enriched for various cellular compartments as follows (marker enzyme given in parentheses): lane 2, cytosol (phosphofructokinase); lane 3, mitochondria (citrate synthase); lane 4, plastid (alkaline pyrophosphatase). Antigenic polypeptides were visualized using alkaline phosphatase-tagged secondary antibody.
Characterization of the NAD+/NADH activity ratio of recombinant and native Arabidopsis NAD(H)Ks
Recombinant NAD(H)Ks were assayed for activity with either NAD+ (4 mM) or NADH (0.8 mM) as the diphosphonicotinamide nucleotide substrate and ATP as the nucleotide phospho donor. As shown in Table 1, NADK1 and NADK2, previously characterized as NADKs [11], both had an absolute requirement for NAD+, while NADHK could utilize NADH and NAD+, with a 1.8-fold preference for NADH. In addition, crude clarified extracts of Arabidopsis cell suspension cultures contained both NADHK and NADK activity (Table 1). However, taking recombinant NADHK substrate flexibility into account, the activity of NADK is approx. 17-fold higher than that of NADHK in Arabidopsis cell suspension culture extracts.
Table 1. Characterization of the NADHK NADH/NAD+ activity ratio and NADHK in Arabidopsis cell suspension cultures.
Assays were conducted using the standard assay mixture (pH 7.9, 25 °C) with either 0.8 mM NADH or 4 mM NAD+ as the diphosphonicotinamide nucleotide substrate. The activity ratio given in parentheses represents the theoretical value calculated at saturating substrate concentrations. Abbreviation: n/d, not detected.
| Activity (μmol·min−1·mg−1) | |||
|---|---|---|---|
| Enzyme | NADH | NAD+ | Activity ratio [(NADH/NAD+)×100 (%)] |
| NADK3 | 40.1 | 15.6 | 257 (198) |
| Clarified extract | 4×10−4 | 62×10−4 | 6 (4) |
| NADK2 | n/d | 0.21 | − |
| NADK1 | n/d | 0.13 | − |
Yeast NADHK (POS5) has been localized to the mitochondria [13,14,17]. In Arabidopsis, attempts were made to monitor NADHK activity during mitochondrial enrichment on Percoll density gradients. However, the experimental conditions required for organelle isolation resulted in a total loss of NADHK activity and thus precluded activity as a means of determining the subcellular localization of the enzyme. Immunoblotting of fractions enriched for either mitochondria or plastids (based on marker enzyme activity; Figure 2B, lower panel) did not reveal the presence of an immunoreactive protein attributable to NADHK, with all of the detectable immunoreactive protein being associated with the cytosolic fraction (Figure 2B). This would indicate that NADHK is a cytosolic protein, although we cannot exclude the presence of NADHK in fragile subcellular compartments that were not recovered in the present study.
Substrate saturation kinetics and cation cofactor requirement of NADHK
The pH–activity profile was determined in standard NAD(H)K assay mixtures buffered with 20 mM Mes and 20 mM BisTris/propane in place of Tris/HCl. NADHK displayed a broad and symmetrical pH–activity profile with maximal activity at pH 7.9. No difference in the pH–activity profile was observed when either NAD+ or NADH were used as substrates, therefore all kinetic studies were performed at this pH. Table 2 summarizes the apparent Vmax and Km values for NAD+ and NADH. Both diphosphonicotinamide nucleotides exhibited hyperbolic saturation kinetics when assayed over the ranges 0.2–8 mM and 8–800 μM for NAD+ and NADH respectively. The specificity constant for NADH was approx. 100-fold higher than that obtained with NAD+ (Table 2), and therefore the protein was designated as an NADHK. The enzyme utilized alternative nucleoside triphosphates as substrates (Table 3). Saturation kinetics for ATP were hyperbolic, with no evidence of substrate inhibition (maximum ATP concentration 4 mM) as described for yeast NADHK [15,17]. NADHK activity showed an absolute requirement for a free bivalent-metal cofactor, with Mg2+ as the preferred cation, although Mn2+ could substitute with an approx. 12-fold lower catalytic efficiency (Table 4). NAD(H)K activity was not detected when Cu2+ or Co2+ were substituted in the reaction mixture. NADHK was incubated with an excess of either ATP or NADP+ for up to 1 h at 37 °C at pH 5.5 or pH 7.9. Enzymic phosphate release was not detected, indicating that the preparation could not function as an acid or alkaline ATP or NADP+ phosphatase (EC 3.1.3.–).
Table 2. Diphosphonicotinamide nucleotide substrate saturation kinetics of recombinant Arabidopsis NADHK.
Assays were conducted using the standard assay mixture (pH 7.9, 25 °C), except that the diphosphonicotinamide nucleotide concentration was varied.
| Nicotinamide nucleotide | Vmax (units·mg−1) | Km (mM) | Vmax/Km (units·mg−1·mM−1) |
|---|---|---|---|
| NADH | 41.2±0.9 | 0.042±0.001 | 981 |
| NAD+ | 23.2±0.6 | 2.39±0.05 | 10 |
Table 3. Use of alternative nucleotide triphosphates by recombinant Arabidopsis NADHK.
Assays were conducted using the standard assay mixture (pH 7.9, 25 °C), except that the NDP concentration was varied in the presence of either 0.8 mM NADH or 4 mM NAD+. Abbreviation: n/d, not detected.
| Vmax/Km (units·mg−1·mM−1) | |||||
|---|---|---|---|---|---|
| Nucleotide or phosphate | Nicotinamide nucleotide | Vmax (units·mg−1) | Km (mM) | NADH | NAD+ |
| ATP | NADH | 39.5±0.9 | 0.062±0.002 | 637 | − |
| NAD+ | 20.3±0.7 | 0.190±0.008 | − | 107 | |
| UTP | NADH | 22.6±0.1 | 0.156±0.005 | 145 | − |
| NAD+ | 11.5±0.1 | 0.885±0.020 | − | 13 | |
| GTP | NADH | 16.3±1.0 | 0.461±0.021 | 35 | − |
| NAD+ | 2.4±0.2 | − | − | − | |
| CTP | NADH | 13.8±0.6 | 0.759±0.01 | 18 | − |
| NAD+ | 2.0±0.1 | − | − | − | |
| PPi | NAD(H) | n/d | − | − | − |
| Poly- Pi | NAD(H) | n/d | − | − | − |
Table 4. Use of metal cation cofactors by recombinant Arabidopsis NADHK.
For the determination of relative activity, NADHK was assayed in the presence of 10 mM of the appropriate metal ion in the standard assay mixture (pH 7.9, 25 °C). NADHK activity was not detected in the absence of free metal ions. For kinetic analysis, assays were conducted using the standard assay mixture, except the free metal ion concentration was varied [24]. Prior to analysis, NADHK was desalted twice on Sephadex PD-10 columns to remove MgCl2.
| Metal-ion cofactor | Relative activity (%) | Vmax (units·mg−1) | Km (mM) | Vmax/Km (units·mg−1·mM−1) |
|---|---|---|---|---|
| Mg2+ | 100 | 40.3±1.2 | 1.16±0.02 | 35 |
| Mn2+ | 28 | 11.3±0.3 | 3.48±0.03 | 3 |
| Ca2+ | 8 | − | − | − |
| Zn2+ | 4 | − | − | − |
Activity effectors of recombinant NADHK
A variety of compounds were tested as possible effectors of NADHK at subsaturating concentrations of ATP (60 μM) and either NAD+ (2.0 mM) or NADH (40 μM). The following compounds had little or no influence (±15% control velocity) on activity: nicotinic acid, nicotinamide, nicotinic acid mononucleotide, quinolinic acid (0.2 mM each), 5 mM PEP (phosphoenolpyruvate), 200 mM sodium acetate and 1 mM glutathione. The kinetic parameters of yeast NADHK are altered (16- and 4-fold reduction in Km,ATP and Km,NADH respectively) by liposomes containing phosphatidylcholine and phosphatidylethanolamine [33]. In contrast, liposomes containing 50:50 (w/w) mixtures of phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine or phosphatidylinositol [34] did not influence the activity of NADHK. PPi was found to inhibit NADHK (Table 5A) and this was characterized further, with inhibition constants shown in Table 5(B). Mixed inhibition was observed with respect to either ATP or NADH, which suggests that PPi may act at an allosteric site. When PPi (5 mM) was substituted for ATP in the assay mixture, NAD(H)K activity was not observed (Table 3), indicating that PPi cannot substitute as the phospho donor. Whereas other plant NADKs are activated by reducing agents [4,5], these compounds had no effect on NADHK (Table 5A). However, similar to yeast NADHK [16] and plant NADK [4,5], the thiol-group-modifying agents pCMB or iodoacetic acid irreversibly inhibited NADHK activity after 30 min incubation at 37 °C (Table 5A). Incubation in the presence of either 1 mM ATP or NADH decreased pCMB inhibition by 21 and 43% respectively, whereas, with 1 mM NAD+, inhibition was not significantly altered. The effect of thiol-group-modifying reagents suggests that a cysteine residue is required for activity within the active site of NADHK.
Table 5. Effect of various substances on the activity of recombinant Arabidopsis NADHK.
(A) Assays were conducted using the standard assay mixture (pH 7.9, 25 °C) in the presence of subsaturating concentrations of NADH (40 μM) and ATP (60 μM) unless otherwise stated. (B) PPi inhibition pattern and enzyme inhibitor dissociation constants. The standard assay mixture was used with invariant substrate concentrations of 0.8 mM NADH or 4 mM ATP. The Ki and Ki′ values were determined as described in the Materials and methods section.
| (A) | |||
|---|---|---|---|
| Compound | Concentration | Relative activity (%) | |
| PPi | 1 mM | 48 | |
| PEP | 5 mM | 100 | |
| Ca2+/CaM | 1 mM/0.3 μM | 88 | |
| pCMB | 10 μM | 42 | |
| pCMB+1 mM NADH | 10 μM | 85 | |
| pCMB+1 mM NAD+ | 10 μM | 43 | |
| pCMB+1 mM ATP | 10 μM | 63 | |
| Iodoacetate | 1 mM | 64 | |
| Glutathione | 1 mM | 95 | |
| Sodium acetate | 200 mM | 106 | |
| (B) | |||
| Variable substrate | PPi inhibition pattern | Ki (mM) | Ki′ (mM) |
| ATP | Mixed | 0.23 | 1.21 |
| NADH | Mixed | 0.51 | 2.86 |
DISCUSSION
In the present paper we provide, to our knowledge, the first evidence that plants contain an NADHK (Tables 1 and 2) distinct from previously studied [4,5,35,37] NADK isoenzymes. The gene encoding NADHK, NADK3 (At1g78590), was identified by conserved-domain analysis [12] and, by subsequent similarity searching (BLAST), shown to be homologous to POS5 (Figure 1), a recently identified yeast NADHK [13,14]. In yeast, the POS5-gene product localized to the mitochondria [13,14,17]. However, in Arabidopsis, NADHK did not associate with a fraction enriched for mitochondria, as indicated by immunoblotting with an anti-NADHK serum (Figure 2B). This suggests [and this is supported by the lack of a predicted mitochondrial pre-sequence (Predotar; http://www.inra.fr/predotar; Version 0.5)] that, unlike the yeast enzyme, NADHK does not associate with the mitochondria. The difference in NADHK compartmentalization between the two systems therefore makes it difficult to speculate as to whether the plant enzyme has a similar physiological role in the protection from mitochondrial ROS that has been described in yeast [13,14]. However, it has been noted that NADP+ can have very different metabolic roles in plants and yeast [1,2] and, in contrast with yeast, NADP+ may be taken up by plant mitochondria [36]. In addition, Ca2+/CaM-dependent and -independent NADK isoenzymes have been localized to plant mitochondria in some species [37,38], which makes it difficult to assign a role for NADHK independently from that of NADK isoenzymes. Currently we are generating NAD(H)K transgenic Arabidopsis plants to allow a more comprehensive analysis of the subcellular compartmentalization of nicotinamide nucleotide metabolism and the respective roles of the corresponding NAD(H)K enzymes.
That NADK3 represents a bona fide NADHK was demonstrated by kinetic analysis of the recombinant protein expressed and affinity-purified from E. coli (Figure 2A). Table 2 shows the apparent Km value for NADH to be approx. 60-fold lower than that for NAD+, whereas the relative specificity constant (Vmax/Km) for NAD+ was only 1% of that for NADH. These values demonstrate that Arabidopsis NADHK has similar specificity for NADH compared with yeast [15,16] and contrasts with previously characterized plant NADK enzymes that do not utilize NADH as a substrate [11,35]. Therefore, NADK3 characterized in this work encodes an NADHK (as specified by EC 2.7.1.86, not EC 2.7.1.23) that will preferentially utilize NADH as the nicotinamide nucleotide phospho acceptor in vivo. The apparent and KmATP and KmNADH values for recombinant Arabidopsis NADHK are in good agreement with previous reports for yeast NADHK [33] and are within meaningful physiological ranges for these compounds [39,40]. Similar to partially purified NADHK from yeast [15], NADHK could utilize a range of nucleoside triphosphates as substrates (Table 3). However, as judged by the respective specificity constants (Table 3), ATP will be the preferred nucleotide substrate for the enzyme. PPi functions as an autonomous energy donor in many plant metabolic pathways [41] and is maintained in the approximate range of 0.2–0.3 mM in the cytosol and mitochondria [42]. PPi was not used as a substrate by Arabidopsis NADHK, but acted as a mixed inhibitor with respect to ATP and NADH (Table 5). As the inhibition constants of PPi for ATP are within the physiologically meaningful range [39,40], it is possible that, in vivo, the activity of NADHK is partly under the control of these respective compounds. In contrast with yeast NADHK [16,17], the plant enzyme was not inhibited by up to 5 mM PEP (phosphoenolpyruvate) at subsaturating concentrations (approx. Km) of ATP and NADH (Table 5). Many reports indicate that the primary control of plant glycolysis, unlike that in yeast and animals, is exerted at the level of PEP utilization [41]. As a consequence, differences in PEP inhibition may be attributable to differences in the primary mode of metabolic control between plants and yeast.
In the present study, Ca2+/CaM did not activate NADHK, but inhibited activity by 12% (Table 5). The enzyme is therefore similar to the Ca2+/CaM-independent NADK observed in some plants [5], but contrasts with the ubiquitous Ca2+/CaM-dependent NADK [4,35,38]. Clearly, within plants, there are three separate routes for the de novo biosynthesis of triphosphonicotinamide nucleotides. Thus control of nicotinamide nucleotide partitioning in plants is expected to differ substantially from that of yeast. This situation possibly reflects the different metabolic roles for NADP+ between the two systems as mentioned above [1,2]. Certainly it would appear that the presence of NAD+- and NADH-dependent NAD(H)K activities could uncouple the capacity to synthesize NADP+ from the redox status of the diphosphonicotinamide nucleotide pool, thus increasing metabolic flexibility of nicotinamide metabolism. Further work is needed to determine how control of the various NAD(H)K activities are integrated and how the nicotinamide nucleotide ratio is maintained at an optimal level in different subcellular compartments and under various environmental conditions. In addition, it will be important to determine the significance of a specific NADP+ phosphatase (EC 3.1.3.-) in the conversion of NADP+ into NAD+ and Pi [43]. Data from these studies could allow a model of nicotinamide nucleotide metabolism to be built based on the ‘kinase/phosphatase’ ratio, which will help explain how, for example, NAD+/NADP+ ratios can rapidly decrease by 10-fold in response to wounding or pathogen attack [1,3]. In addition, it is anticipated that a detailed understanding of these cellular responses could lead to the engineering of plants with enhanced abilities to respond to environmental stress without the need for de novo nicotinamide biosynthesis.
Acknowledgments
This work is supported by research and equipment grants from the Natural Sciences and Engineering Research Council of Canada (NSERC). We are also grateful to Dr William C. Plaxton of this Department for helpful discussions.
References
- 1.Moller I. M. Plant mitochondria and oxidative stress: electron transport, NADPH turnover and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- 2.Moller I. M., Rasmusson A. G. The role of NADP in the mitochondrial matrix. Trends Plant Sci. 1998;3:21–27. [Google Scholar]
- 3.Harding S. A., Oh S. H., Roberts D. M. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 1997;16:1137–1144. doi: 10.1093/emboj/16.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delumeau O., Renard M., Montrichard F. Characterization and possible redox regulation of the purified calmodulin-dependent NAD+ kinase from Lycopersicon pimpinellifolium. Plant Cell Environ. 2000;23:1267–1273. [Google Scholar]
- 5.Gallais S., Pou de Crescenzo M. A., Laval-Martin D. L. Characterization of soluble calcium calmodulin-dependent and -independent NAD+ kinases from Avena sativa seeds. Aust. J. Plant Physiol. 2001;28:363–371. [Google Scholar]
- 6.Snedden W. A., Fromm H. Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 2001;151:35–64. doi: 10.1046/j.1469-8137.2001.00154.x. [DOI] [PubMed] [Google Scholar]
- 7.Kawai S., Mori S., Mukai T., Hashimoto W., Murata K. Molecular characterization of Escherichia coli NAD+ kinase. Eur. J. Biochem. 2001;268:4359–4365. doi: 10.1046/j.1432-1327.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 8.Kawai S., Mori S., Mukai T., Suzuki S., Yamada T., Hashimoto W., Murata K. Inorganic polyphosphate/ATP-NAD+ kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 2000;276:57–63. doi: 10.1006/bbrc.2000.3433. [DOI] [PubMed] [Google Scholar]
- 9.Kawai S., Suzuki S., Mori S., Murata K. Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD+ kinase. FEMS Microbiol. Lett. 2001;200:181–184. doi: 10.1111/j.1574-6968.2001.tb10712.x. [DOI] [PubMed] [Google Scholar]
- 10.Lerner F., Niere M., Ludwig A., Ziegler M. Structural and functional characterization of human NAD+ kinase. Biochem. Biophys. Res. Commun. 2001;288:69–74. doi: 10.1006/bbrc.2001.5735. [DOI] [PubMed] [Google Scholar]
- 11.Turner W. L., Waller J. C., Snedden W. A. Cloning and characterization of two NAD+ kinases from Arabidopsis. Identification of a calmodulin binding isoform. Plant Physiol. 2004;135:1243–1245. doi: 10.1104/pp.104.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchler-Bauer A., Panchenko A. R., Shoemaker B. A., Thiessen P. A., Geer L. Y., Bryant S. H. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Outten C. E., Culotta V. C. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2003;22:2015–2024. doi: 10.1093/emboj/cdg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryotic Cell. 2003;2:809–820. doi: 10.1128/EC.2.4.809-820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths M. M., Bernofsky C. Purification and properties of reduced diphosphopyridine nucleotide kinase from yeast mitochondria. J. Biol. Chem. 1972;247:1473–1478. [PubMed] [Google Scholar]
- 16.Iwahashi Y., Hitoshio A., Tajima N., Nakamura T. Characterization of NADH kinase from Saccharomyces cerevisiae. J. Biochem. (Tokyo) 1989;105:588–593. doi: 10.1093/oxfordjournals.jbchem.a122709. [DOI] [PubMed] [Google Scholar]
- 17.Iwahashi Y., Nakamura T. Localization of the NADH kinase in the inner membrane of yeast mitochondria. J. Biochem. (Tokyo) 1989;105:916–921. doi: 10.1093/oxfordjournals.jbchem.a122779. [DOI] [PubMed] [Google Scholar]
- 18.Werhahn W., Niemeyer A., Jansch L., Kruft V., Schmitz U. K., Braun H. Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol. 2001;125:943–954. doi: 10.1104/pp.125.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmon A. C., Jarrett H. W., Cormier M. J. An enzymatic assay for calmodulins based on plant NAD+ kinase activity. Anal. Biochem. 1984;141:168–178. doi: 10.1016/0003-2697(84)90441-x. [DOI] [PubMed] [Google Scholar]
- 20.Fromm H., Chua N.-H. Cloning of plant cDNAs encoding calmodulin-binding proteins using 35S-labeled recombinant calmodulin as a probe. Plant Mol. Biol. Rep. 1992;10:199–206. [Google Scholar]
- 21.Brooks S. P. A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 22.Dixon M., Webb W. C., Thorne C. J. R., Tipton K. F. London: Academic Press; 1979. Enzymes. [Google Scholar]
- 23.Cornish-Bowden A. London: Butterworth; 1979. Fundamentals of Enzyme Kinetics. [Google Scholar]
- 24.Brooks S. P., Storey K. B. Bound and determined: a computer program for making buffers of defined ion concentrations. Anal. Biochem. 1992;201:119–126. doi: 10.1016/0003-2697(92)90183-8. [DOI] [PubMed] [Google Scholar]
- 25.Turner W. L., Plaxton W. C. Purification and characterization of banana fruit acid phosphatase. Planta. 2001;214:243–249. doi: 10.1007/s004250100607. [DOI] [PubMed] [Google Scholar]
- 26.Srere P. A. Citrate synthase. Methods Enzymol. 1967;13:3–11. [Google Scholar]
- 27.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Bollag D. M., Rozycki M. D., Edelstein S. J. New York: Wiley–Liss; 1996. Protein Methods. [Google Scholar]
- 29.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Huala E., Dickerman A. W., Garcia-Hernandez M., Weems D., Reiser L., LaFond F., Hanley D., Kiphart D., Zhuang M., Huang W., et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R., Griffiths-Jones S., Howe K. L., Marshall M., Sonnhammer E. L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwahashi Y., Nakamura T. Orientation and reactivity of NADH kinase in proteoliposomes. J. Biochem. (Tokyo) 1989;105:922–926. doi: 10.1093/oxfordjournals.jbchem.a122780. [DOI] [PubMed] [Google Scholar]
- 34.Dabrowska A., Terlecki G., Czapinska E., Gutowicz J. Interaction of bovine heart pyruvate kinase with phospholipids. Biochim. Biophys. Acta. 1995;1236:299–305. doi: 10.1016/0005-2736(95)00061-7. [DOI] [PubMed] [Google Scholar]
- 35.Muto S. Kinetic nature of calmodulin-dependent NAD+ kinase from pea seedlings. Z. Pflanzenphysiol. 1983;109:385–393. [Google Scholar]
- 36.Bykova N. V., Moller I. M. Involvement of matrix NADP+ turnover in the oxidation of NAD+-linked substrates by pea leaf mitochondria. Physiol. Plant. 2001;111:448–456. doi: 10.1034/j.1399-3054.2001.1110404.x. [DOI] [PubMed] [Google Scholar]
- 37.Pou de Crescenzo M. A., Gallais S., Leon A., Laval-Martin D. L. Tween-20 activates and solubilizes the mitochondrial membrane-bound, calmodulin dependent NAD+ kinase of Avena sativa. L. J. Membr. Biol. 2001;182:135–146. doi: 10.1007/s0023201-0039-8. [DOI] [PubMed] [Google Scholar]
- 38.Sauer A., Robinson D. G. Calmodulin dependent NAD+-kinase is associated with both the outer and inner mitochondrial membranes in maize roots. Planta. 1985;166:227–233. doi: 10.1007/BF00397353. [DOI] [PubMed] [Google Scholar]
- 39.Gallais S., Pou de Crescenzo M. A., Laval-Martin D. L. Pyridine nucleotides and redox charges during germination of non-dormant and dormant caryopses of Avena sativa. L. J. Plant Physiol. 2001;153:664–669. [Google Scholar]
- 40.Stitt M., Lillet R. M., Heldt H. W. Adenine nucleotide levels in the cytosol, chloroplasts and mitochondria of wheat leaf protoplasts. Plant Physiol. 1982;70:977–983. doi: 10.1104/pp.70.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plaxton W. C. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- 42.Casolo V., Micolini S., Macri F., Vianello A. Pyrophosphate import and synthesis by plant mitochondria. Physiol. Plant. 2002;114:516–523. doi: 10.1034/j.1399-3054.2002.1140403.x. [DOI] [PubMed] [Google Scholar]
- 43.Gallais S., Pou de Crescenzo M. A., Laval-Martin D. L. Evidence of active NADP+ phosphatase in dormant seeds of Avena sativa. L. J. Exp. Bot. 2000;51:1389–1394. [PubMed] [Google Scholar]