Abstract
We performed a structure–function analysis of the plasma membrane-localized plant-specific barley (Hordeum vulgare) MLO (powdery-mildew-resistance gene o) protein. Invariant cysteine and proline residues, located either in extracellular loops or transmembrane domains that have been conserved in MLO proteins for more than 400 million years, were found to be essential for MLO functionality and/or stability. Similarly to many metazoan G-protein-coupled receptors known to function as homo- and hetero-oligomers, FRET (fluorescence resonance energy transfer) analysis revealed evidence for in planta MLO dimerization/oligomerization. Domain-swap experiments with closely related wheat and rice as well as diverged Arabidopsis MLO isoforms demonstrated that the identity of the C-terminal cytoplasmic tail contributes to MLO activity. Likewise, analysis of a progressive deletion series revealed that integrity of the C-terminus determines both MLO accumulation and functionality. A series of domain swaps of cytoplasmic loops with the wheat (Triticum aestivum) orthologue, TaMLO-B1, provided strong evidence for co-operative loop–loop interplay either within the protein or between MLO molecules. Our data indicate extensive intramolecular co-evolution of cytoplasmic domains in the evolutionary history of the MLO protein family.
Keywords: barley–powdery mildew interaction, fluorescence resonance energy transfer (FRET), intramolecular co-evolution, MLO, seven-transmembrane domain protein, structure–function analysis
Abbreviations: APB, acceptor photobleaching; BF, background fluorescence resonance energy transfer efficiency; Bgh, Blumeria graminis f. sp. hordei; CaM, calmodulin; CaMBD, calmodulin-binding domain; CFP, cyan fluorescent protein; CT, C-terminus; EF, fluorescence resonance energy transfer efficiency; EST, expressed sequence tag; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; GPCR, G-protein-coupled receptor; GUS, β-glucuronidase; MLO, powdery-mildew-resistance gene o; NOS, nopaline synthase; ROI, region of interest; SOE, splicing by overlap extension; YFP, yellow fluorescent protein
INTRODUCTION
The fully sequenced genomes of the dicotyledonous model plant Arabidopsis thaliana [1] and the monocotyledon Oryza sativa (rice) [2,3] provided for the first time a comprehensive overview of the gene content of two flowering plant species which diverged approx. 200 million years ago. The comparative analysis of these two genomes revealed important features: (i) approx. one third of the predicted Arabidopsis and rice genes appear to be plant-specific, since no homologous sequences could be identified in known non-plant genomes; (ii) approx. 30% of the predicted gene products in the rice or Arabidopsis genome cannot be assigned to any functional class and are currently considered to be ‘hypothetical’, ‘unknown’ or ‘putative’; and (iii) only a small proportion (as of December 2000: 9%) of the genes in the Arabidopsis genome have been characterized experimentally so far [1,2]. These findings emphasize the need for in-depth functional analysis of plant-specific genes and gene families encoding proteins with as yet unknown biochemical functions. It is evident that data for these protein families cannot be gained from the analysis of non-plant species. Structure–function analysis by site-directed mutagenesis or domain-swap experiments represents a means to reveal insights into the contribution of specific amino acids and/or domains to the function of a given protein.
Barley (Hordeum vulgare) MLO (powdery-mildew-resistance gene o) is the ‘founder’ of a family of plant-specific integral proteins with seven-transmembrane domains [4–6]. MLO-like proteins are encoded by medium-sized gene families present in all investigated land plant species, including mosses. This suggests an ancient origin of this protein family [6]. The Arabidopsis Mlo family comprises 15 members, whereas at least 11 distinct isoforms are encoded by the rice genome. Recessive loss-of-function alleles (mlo) of barley Mlo confer broad-spectrum resistance against the widespread powdery mildew fungus (Blumeria graminis f. sp. hordei; Bgh). Requirement of the wild-type protein for successful Bgh pathogenesis may indicate a role of MLO as endogenous plant defence modulator. Alternatively, MLO might be targeted by the fungal pathogen for suppression of host-defence pathways, possibly by interfering with a vesicle-associated and SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor)-protein-dependent resistance mechanism [7–9]. Apart from the role of barley MLO in powdery mildew pathogenesis, no function has been assigned to any other MLO isoform to date. However, closely related orthologues of barley Mlo in wheat and rice were shown to complement powdery mildew resistance of barley mlo mutants [10].
Since MLO proteins do not exhibit recognizable sequence similarity to other protein families and lack obvious protein motifs, the primary amino acid sequences of MLO isoforms do not provide clues with regard to biochemical activities. Topology studies revealed that barley MLO is a heptahelical integral membrane protein that accumulates in the plasma membrane, with the N-terminus positioned extracellularly, while the C-terminus faces the cytoplasm [5]. The proximal part of the cytoplasmic C-terminus of MLO proteins contains a binding site for the cytoplasmic calcium sensor, CaM (calmodulin) [11,12]. Ca2+-dependent binding of CaM to this binding domain (CaMBD) is required for full activity of barley MLO [12]. Although the global topological features, subcellular localization and the sequence diversification within the protein family are reminiscent of metazoan GPCRs (G-protein-coupled receptors), MLO proteins appear to function independently of heterotrimeric G-proteins [12]. Small G-proteins of the ROP (RHO of plants) family were shown to modulate MLO-dependent mildew susceptibility, but it is not known whether this involves a direct protein–protein interaction [13,14].
Particle-bombardment-mediated transient gene expression in single barley leaf epidermal cells and subsequent challenge with Bgh spores constitutes a convenient assay to assess quantitatively cell-autonomous MLO activity [15,16]. Recently, we developed a luciferase-reporter-protein-based assay that provides a quantitative estimate for in planta accumulation levels of MLO variants (J. Müller, P. Piffanelli, A. Devoto, M. Miklis, C. Elliott, B. Ortmann, P. Schulze-Lefert and R. Panstruga, unpublished work; for details, see the Materials and methods section). In the present study, we have combined both methods for a comprehensive structure–function analysis involving site-directed mutants of barley MLO and MLO chimaeras. This enabled us to discriminate between functional, non-functional/unstable and non-functional/stable variants. In addition, we used in planta FRET (fluorescence resonance energy transfer) analysis to study potential MLO oligomerization. Our data provide evidence for a contribution of the hypervariable extracellular domains and the distal cytoplasmic tail to MLO function. A series of domain-swap experiments between barley MLO and a wheat orthologue suggest co-operative interplay and co-evolution of cytoplasmic MLO domains. The results are discussed in the context of potential MLO dimerization/oligomerization.
MATERIALS AND METHODS
Full-size MLO sequences used for determining conserved amino acid sequences
Full-size MLO sequences used for the ClustalW alignment include A. thaliana AtMLO1–AtMLO15 (see http://www.arabidopsis.org/info/genefamily/mlo.html for details); Physcomitrella patens (moss) PpMLO1 (GenBank® accession number AY599871); Zea mays (maize) ZmMLO1–ZmMLO4 and ZmMLO6–ZmMLO8 (GenBank® accession numbers AY029312–AY029315 and AY029317–AY029319); H. vulgare (barley) MLO (GenBank® accession number Z83834), HvMLO2 (GenBank® accession number Z95496) and HvMLO3 (GenBank® accession number AY581255); Triticum aestivum (wheat) TaMLO-B1 (GenBank® accession number AF361932); and Oryza sativa (rice) OsMLO1–OsMLO11. For rice MLO sequences, mostly rice genomic sequences served as template for conceptual translation. Depending on coverage by genomic contigs, sequences derived from O. sativa ssp. indica [3] or japonica [2] were used: OsMLO1 (GenBank® accession number AAAA01004541), OsMLO2 (GenBank® accession number AF384030), OsMLO3 [TMRI (Torrey Mesa Research Institute) contig CL011892.313], OsMLO4 (GenBank® accession number AC073166), OsMLO5, (GenBank® accession number AP003431), OsMLO6 (TMRI contig CLB6565.21), OsMLO7 (TMRI contig CL018067.96), OsMLO8 (GenBank® accession number AAAA01001913), OsMLO9 (GenBank® accession number AAAA01000029), OsMLO10 (TMRI contig CLB6542.1) and OsMLO11 (TMRI contig CL035228.93). TMRI rice contigs of O. sativa ssp. japonica can be downloaded via the following webpage: http://www.tmri.org/en/partnership/access.aspx
Plant and fungal material
Barley cultivar BC (backcross Ingrid) mlo-5 was cultivated at 22 °C (16 h light/8 h darkness). The barley powdery mildew Bgh isolates A6 and K1 were propagated as described in [17].
Site-directed mutagenesis of barley Mlo cDNAs
The Mlo cDNA [4] was subcloned into a pBluescript II KS+ plasmid vector containing the NOS (nopaline synthase) terminator sequence and the resulting plasmid (pMloNOS) was used as a template for the introduction of point mutants or domain-swap variants using the method of SOE (splicing by overlap extension) [18]. Supplementary Table 1 (available at http://www.BiochemJ.org/bj/385/bj3850243add.htm) outlines the cloning strategy for all constructs made by SOE. Sequences of PCR primers used in the present study are listed in Supplementary Table 2 (available at http://www.BiochemJ.org/bj/385/bj3850243add.htm). Plasmids not made by SOE were constructed as follows. MLO E103N/R105S (Glu103→Asn/Arg105→Ser) was obtained by substitution of a HindIII/PstI fragment of pL2A [5] into pMloNOS. CT (C-terminus) swap was obtained by the substitution of a 215 bp BspM1/EcoRI fragment of pMloNOS into pTaMloB-1CT. All Mlo variants created were confirmed by sequencing and then subcloned into pUGLUM [a bifunctional plasmid containing both a GFP (green fluorescent protein) reporter plus a Mlo variant coding sequence each driven by a separate maize polyubiquitin promoter] [17], before ballistic delivery to barley leaf segments (see below). pUGN, the plasmid containing the GUS reporter gene (uidA β-glucuronidase gene of E. coli) contains a 1.9 kb fragment of the maize polyubiquitin 1 promoter followed by the Agrobacterium NOS terminator [19].
Single-cell complementation assay
Mlo variants were delivered ballistically to epidermal cells of detached barley leaf segments (resistant mlo-5 genotype) as described previously [10,15,17]. Leaf segments were subsequently inoculated with Bgh isolates A6 or K1 as described in [10]. When GUS was used as a reporter, leaves were vacuum-infiltrated with GUS-staining solution 48 h after inoculation according to the method of Schweizer et al. [20]. GUS-expressing epidermal cells were characterized for the presence of fungal haustoria (successful penetration) or fungal appressoria (aborted penetration attempt). When GFP was used as a reporter, leaves were observed 96 h after inoculation by UV light incident fluorescence microscopy (excitation filter, 450–490 nm; band pass filter, 515–565 nm). GFP-expressing cells challenged by the fungus were characterized for the presence or absence of colonies. Percent penetration success was calculated as the fraction of GUS- or GFP-expressing cells with a haustorium or colony of the total number of challenged GUS- or GFP-expressing cells. Susceptibility obtained with a wild-type Mlo construct was set as 100% relative penetration success in all experiments. Data for each construct are the means±S.D. obtained from at least three independent experiments.
Dual-luciferase-based in planta MLO accumulation assays
Plasmid K93 is a derivative of binary vector pAMPAT-MCS (GenBank® accession number AY436765) and carries two expression cassettes: one consisting of a doubled cauliflower mosaic virus 35 S promoter, an in-frame fusion of Mlo and Renilla luciferase cDNAs and a 35 S terminator, the second of a 35 S promoter, firefly luciferase and 35 S terminator. Derivatives of this plasmid expressing various Mlo variants or chimaeras as translational fusions with Renilla luciferase were generated by placement of suitable restriction fragments or cloning of respective SOE PCR products [18]. A. thaliana (ecotype Columbia 0, line At7) cells were propagated as reported by Trezzini et al. [21]. Protoplast preparations and transfections were carried out according to the protocol described by Sprenger-Haussels and Weisshaar [22] with a modified enzyme solution containing 12 units/ml cellulase Onuzoka R-10 (Merck, Darmstadt, Germany) and 1.5 units/ml Mazerozyme R-10 (Serva, Heidelberg, Germany). For dual-luciferase reporter assays (Promega, Madison, WI, U.S.A.), protoplast suspensions were diluted in 240 mM CaCl2, harvested by centrifugation (15000 g), shock-frozen in liquid nitrogen and extracted in the lysis buffer supplied with the dual-luciferase kit. Measurements were carried out according to the manufacturer's instructions. Data for each construct are the means±S.D. obtained from at least three independent experiments.
FRET analysis
FRET APB (acceptor photobleaching) experiments were performed essentially as described in [23]. Barley leaf epidermal cells were co-transformed ballistically with plasmid constructs encoding MLO–YFP (yellow fluorescent protein) and MLO–CFP (cyan fluorescent protein) or CaM–CFP. Cells exhibiting co-expression of both fluorescent proteins were bleached in the acceptor YFP channel by scanning an ROI (region of interest) using 40–50 pulses of the 514 nm argon laser line at 100% intensity (total bleach time 5–10 s, depending on the size of the ROI). Before and after APB, CFP intensity images were recorded to calculate changes in donor fluorescence.
FRET efficiency (EF) was calculated using the following formula EF=(I5−I4)×100/I5, where I5 is the CFP intensity after YFP photobleaching and I4 is the intensity just before photobleaching. This formula yields the increase in CFP fluorescence following a YFP bleach normalized by CFP fluorescence after the bleach [24]. In order to monitor potential changes in CFP fluorescence levels before the bleaching process, background FRET efficiency (BF) was calculated using the formula BF=(I4−I3)×100/I4, where I3 and I4 refer to the CFP intensity at time points 3 and 4 preceding the bleaching. In all cases, background FRET was insignificantly low, and thus background subtractions were not performed.
RESULTS
Relevance of invariant extracellular cysteine and transmembrane proline residues for MLO function
We aligned available deduced full-size MLO protein sequences using the ClustalW algorithm to identify highly conserved amino acid residues in MLO proteins (See Supplement 3 at http://www.BiochemJ.org/bj/385/bj3850243add.htm). This alignment includes a single sequence from the moss Physcomitrella patens, all 15 sequences from the dicotyledonous plant species A. thaliana (AtMlo1–AtMlo15) as well as one, three, seven and 11 sequences from the monocotyledonous species wheat (T. aestivum), barley (H. vulgare), maize (Z. mays) and rice (Oryza sativa) respectively (n=38 full-size MLO sequences; for details about sequences, see the Materials and methods section). In the present study, we focused on genomic and likely full-length cDNA sequences since single-pass ESTs (expressed sequence tags) do not provide the reliability required for the performed analysis. In total, 30 invariant amino acids (representing less than 10% of total MLO amino acids) were identified in this sample (Figure 1A). A further 17 amino acids are highly conserved, although not invariant among MLO family members. At each of these 17 positions, only one conservative amino acid replacement exists within the 38 tested MLO sequences. Of the 30 identified invariant residues, four (all cysteine residues) are located in extracellular loops 1 and 3, 12 in transmembrane domains (helices III, IV, V, VI and VII), whereas the majority (14 amino acids) reside in cytoplasmic regions with an apparent over-representation in the second intracellular loop (Figure 1A).
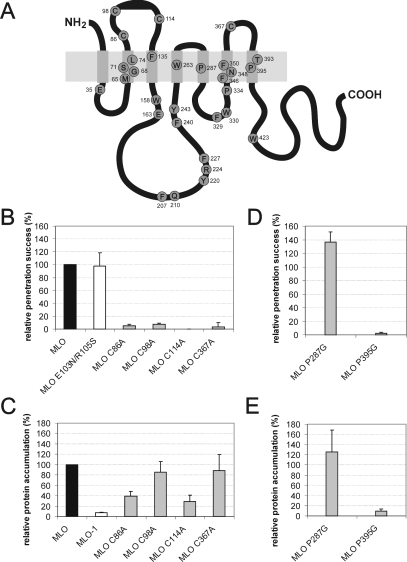
Figure 1. Invariant cysteine and proline residues are essential for MLO function and/or protein accumulation.
(A) Schematic representation of MLO depicting strictly conserved residues within the MLO protein family. The light-grey box represents the lipid bilayer, whereas smaller dark-grey boxes symbolize the seven-transmembrane domains. Invariant amino acids of a sample of 38 full-size MLO sequences (see text) are shown as circles labelled using the one-letter amino acid code. Numbering of amino acids corresponds to barley MLO [4]. (B) Functional assay of single-amino-acid-substitution MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were bombarded with the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or a mutant version thereof (encoding GFP plus either the double-mutant MLO E104N/R106S or one of the four cysteine→alanine mutants). Leaves were then inoculated with Bgh A6 and GFP-fluorescent cells were inspected for fungal structures as described in the Materials and methods section. (C and E) Assessment of MLO protein accumulation. Relative accumulation of wild-type MLO and mutant versions MLO-1, C86A, C98A, C114A, C367A, P287G and P395G was determined by dual-luciferase assays of transfected Arabidopsis thaliana protoplasts as described in the Materials and methods section. (D) Functional assay of single-amino-acid-substitution MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were co-bombarded with a GUS reporter construct and the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or a mutant version thereof (encoding P287G or P395G single-amino-acid-substitution variants). Leaves were then inoculated with Bgh K1 and stained for GUS activity and fungal structures as described in the Materials and methods section.
Evolutionary invariant residues may be indicative of constraints, imposed by protein folding/conformation and/or function. Mutations affecting these residues are thus expected to often impair functionality and/or maturation. The latter frequently leads to altered steady-state protein accumulation. Previous analysis of powdery-mildew-resistant barley mlo mutants revealed DNA lesions in Mlo that affect in two cases invariant amino acid residues [Phe240 (mlo-12) and Pro334 (mlo-29); see Figure 1A] [4,25]. To examine the contribution of invariant amino acid residues for MLO function in a targeted manner, we performed site-directed mutagenesis of selected invariant residues. These included each of the four cysteine residues (Cys86, Cys98, Cys114 and Cys367) in extracellular loops 1 and 3 (Figure 1A), hypothesized to form two pairs of extracellular disulphide bridges, as well as two proline residues (Pro287 and Pro395) located in the centre of transmembrane domains V and VII (Figure 1A). Transmembrane helical proline residues are supposed to provide flexible kinks enabling intramolecular conformational changes (see the Discussion). We generated cysteine→alanine mutants for each of the four cysteine residues and proline→glycine mutants for two transmembrane helical proline residues. For the latter, we chose glycine instead of alanine to minimize potential steric perturbations of the α-helical structure of the respective transmembrane domains.
We assessed the functionality of the mutant MLO variants by transient gene expression in detached powdery-mildew-resistant (mlo genotype) barley leaves and subsequent powdery mildew challenge as described previously [10,15] (see also the Materials and methods section). In addition, we determined relative in planta accumulation of the mutant forms in comparison with wild-type MLO by dual-luciferase assays in A. thaliana protoplasts [12]. In this assay, translational fusions of MLO variants with the reporter protein Renilla luciferase are co-expressed with a second reporter protein, firefly luciferase, which serves as internal standard. Luminescence obtained from MLO–Renilla luciferase constructs normalized to firefly luciferase activity provides a relative measure for in planta accumulation of the respective MLO variant. Results from both the functional test and the protein-accumulation assay were expressed as relative values, with wild-type MLO activity/accumulation set as 100% (Figures 1B–1E).
We noted that all four cysteine→alanine mutants (C86A, C98A, C114A and C367A) lost the ability to complement the mlo mutant phenotype, while a control mutant, affecting two variable residues in extracellular loop 1 (E103N/R105S) [5] retained wild-type activity (Figure 1B). When tested for in planta accumulation, we found that two cysteine mutants (C98A and C367A) exhibited accumulation close to that of wild-type MLO (84% and 88% respectively), while the remaining two mutant variants (C86A and C114A) showed markedly reduced accumulation (39% and 29% respectively, P<0.05; Figure 1C). In this, and in the following experiments, we included the MLO-1 mutant (W162R) [4], which is a highly unstable MLO variant [12], as a control for severely impaired protein accumulation.
Next, we examined the effect of proline→glycine substitutions in MLO mutant variants P287G and P395G. We found that P395G failed to complement the mlo resistant phenotype, while the P287G variant exhibited enhanced (although statistically not significant at 95% confidence) functionality compared with wild-type MLO (Figure 1D). Assessment of in planta accumulation revealed that protein levels of the P395G mutant were very low (comparable with the unstable MLO-1 variant; see Figure 1C), while the P287G version accumulated to levels comparable with wild-type MLO (Figure 1E).
FRET analysis reveals evidence for MLO homo-oligomerization
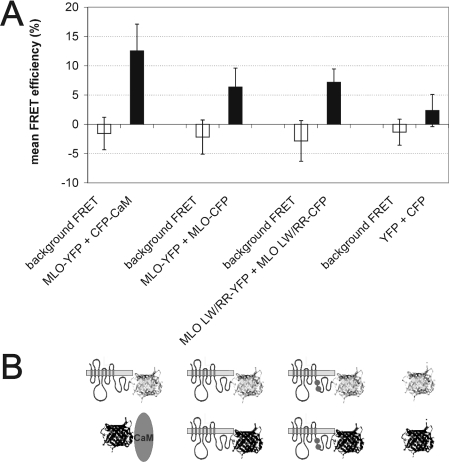
It is now commonly accepted that many members of the paradigm family of seven-transmembrane domain proteins, the metazoan GPCRs, form homo- and hetero-dimers/oligomers in vivo [26–28]. Conserved cysteine residues present in MLO proteins could thus either serve a role in the formation of intramolecular cysteine bridges or, alternatively, may participate in intermolecular bonds resulting in MLO dimers/oligomers. To test potential in planta dimerization/oligomerization of barley MLO, we applied FRET technology. We generated plasmids encoding fluorophore-tagged MLO variants by engineering C-terminal translational fusions of either YFP- or CFP-coding sequences to the cytoplasmic C-terminal tail of MLO. C-terminal fluorophore tagging was previously shown not to interfere with MLO function [15]. Upon transient gene expression in barley leaf epidermal cells, we determined FRET efficiencies by the APB method (for details, see the Materials and methods section). We included cytoplasmically expressed YFP and CFP as negative controls and fluorophore-tagged MLO and CaM, reported previously to interact in vivo [12], as a positive control in these experiments. In addition, we tested FRET between mutant variants of MLO unable to bind calmodulin (MLO L420R/W423R; [12]) to study whether CaM binding interferes with potential MLO dimerization/oligomerization. We found mean FRET efficiencies of 12.2±4.6% (MLO–YFP plus CaM–CFP; positive control; n=80), 6.4±3.3% (MLO–YFP plus MLO–CFP; n=26) and 7.7±3.3% (MLO L420R/W423R–YFP plus MLO L420R/W423R–CFP; n=40) respectively (Figures 2A and 2B). The latter two values are significantly different (Student's t test; P<10−7) from FRET between soluble cytoplasmic YFP and CFP (2.3±2.7%; negative control; n=80), thus indicating a basal level of MLO homodimerization/oligomerization for both wild-type MLO and the CaMBD mutant L420R/W423R. No significant difference was detected between wild-type MLO and the L420R/W423R CaMBD variant (P>0.2), suggesting that CaM binding contributes neither positively nor negatively to MLO dimerization/oligomerization.
Figure 2. FRET analysis reveals MLO homo-oligomerization.
(A) Quantitative FRET APB analysis. Background FRET (white bars) and FRET efficiencies (black bars) were calculated from barley leaf epidermal cells co-expressing the constructs shown below the graph (B). Results are means±S.D. for 26–80 cells each. (B) Schematic representation of constructs used for FRET analysis. MLO is depicted by its serpentine structure, CaM as an oval, and YFP and CFP fluorophores as light-grey (upper panel) and dark-grey (lower panel) ribbon models respectively. Two dots in the C-terminal tail of MLO symbolize the two amino acid substitutions in the L420R/W423R MLO CaMBD mutant variant.
The identity and integrity of the C-terminus is critical for MLO function and accumulation
The C-terminal cytoplasmic tail of MLO proteins is highly variable both in length and amino acid sequence [6]. Extreme examples comprise AtMLO9 (31 amino acids) and OsMLO6 (172 amino acids). The proximal part of the C-terminus harbours the conserved CaMBD of MLO proteins (approximately amino acid residues 10–35; Figure 3A) [11,12]. The functional contribution of the hypervariable distal part, however, has remained unclear to date. We performed MLO domain-swap experiments, and generated a series of incremental truncations of the C-terminus to assess the functional role of this region.
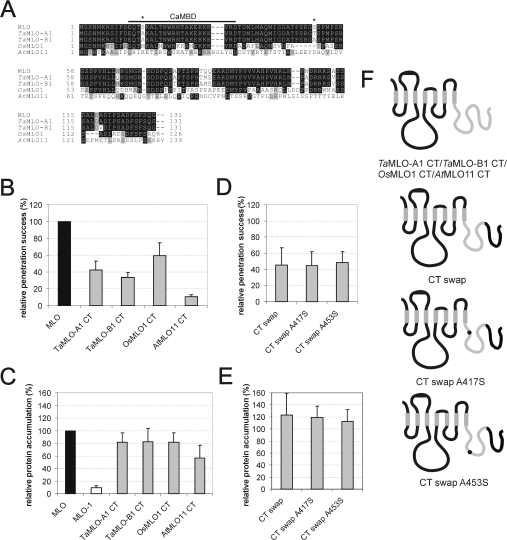
Figure 3. Identity of the C-terminus is critical for MLO functionality.
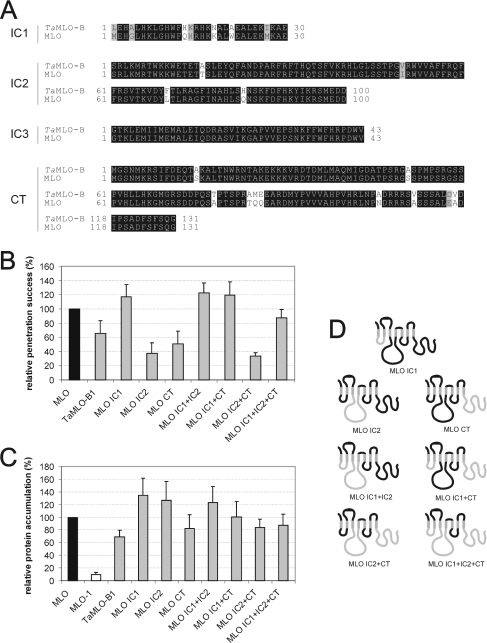
(A) Amino acid sequence alignment of the C-termini of barley (MLO), wheat (TaMLO-A1 and TaMLO-B1), rice (OsMLO1) and Arabidopsis (AtMLO11) MLO isoforms. The previously characterized CaMBD is indicated by a bar above the sequence. Two MLO serine residues (Ser417 and Ser453) identified to be crucial for full MLO function (see Figure 2D) are highlighted by an asterisk above the sequence. (B) Functional assay of MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were bombarded with the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or a mutant version thereof (encoding GFP plus TaMLO-A1 CT, TaMLOB1 CT, OsMLO1 CT or AtMLO11 CT). Leaves were then inoculated with Bgh A6 and GFP-fluorescent cells were inspected for fungal structures as described in the Materials and methods section. (C and E) Assessment of MLO protein accumulation. Relative accumulation of wild-type MLO, mutant version MLO-1, as well as chimaeras TaMLO-A1 CT, TaMLO-B1 CT, OsMLO1 CT, AtMLO11 CT, CT swap, CT swap A417S and CT swap A453S was determined by dual-luciferase assays of transfected A. thaliana protoplasts as described in the Materials and methods section. Note that the values obtained for MLO and MLO-1 result from a common experiment with the constructs shown in Figure 4(C). (D) Functional assay of single-amino-acid-substitution MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were co-bombarded with a GUS reporter construct and the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or mutant versions thereof (encoding variants CT swap, CT swap A417S or CT swap A453S). Leaves were then inoculated with Bgh K1 and were stained for GUS activity and fungal structures as described in the Materials and methods section. (F) Schematic representation of the domain-swap constructs TaMLO-A1 CT, TaMLOB1 CT, OsMLO1 CT, AtMLO11 CT, CT swap, CT swap A417S and CT swap A453S. Black bends illustrate barley MLO portions, whereas light-grey bends depict the respective heterologous fraction. Black dots symbolize single-amino-acid-replacements A417S or A453S.
We replaced the Mlo C-terminus with either the corresponding coding sequence of two wheat Mlo orthologues (TaMlo-A1 or TaMlo-B1 respectively), the C-terminus of the closely related rice OsMlo1 gene, or the similarly sized, but sequence diversified, Arabidopsis AtMlo11 C-terminus (Figures 3A and 3F). The corresponding identities at the amino acid level with barley MLO are 94% (TaMLO-A1), 92% (TaMLO-B1), 51% (OsMLO1) and 13% (AtMLO11; 42% similarity). We tested the functionality of the respective chimaeras by transient expression in detached barley leaves (mlo genotype). While the variants carrying C-termini of monocotyledonous MLO proteins TaMLO-A1, TaMLO-B1 or OsMLO1 partially complemented mlo resistance (43%, 33% and 60% respectively), the activity of the derivative containing the AtMLO11 C-terminus was drastically reduced (10%; Figure 3B). When tested for in planta protein accumulation, each of the hybrids bearing the monocotyledonous C-termini accumulated to levels close to wild-type MLO, whereas accumulation of the derivative with the AtMLO11 C-terminus was almost halved (56%; Figure 3C).
Since the C-termini of barley MLO and wheat TaMLO-B1 are 92% identical at the amino acid level (Figure 3A), we aimed to identify determinants that confer the observed 67% activity reduction on chimaera TaMLO-B1 CT (Figure 3B). We successively replaced the wheat sequence in TaMLO-B1 CT with the corresponding barley MLO sequence. We first replaced the distal half of the C-terminus in TaMLO-B1 CT with barley MLO by generating the construct ‘CT swap’ (Figure 3F). This chimaera differed by only two amino acids from barley MLO, yet exhibited less than 50% of wild-type MLO activity (Figure 3D), suggesting that the two respective amino acids (Ser417 and Ser453) are critical activity determinants in barley MLO.
Next, we individually substituted alanine residues for these serine residues in the CT swap construct, resulting in constructs CT swap A417S and CT swap A453S respectively (Figure 3F). Surprisingly, neither of these substitutions rescued MLO wild-type activity, indicating that either serine→alanine replacement is sufficient to halve MLO activity. Relative accumulation of variants CT swap, CT swap A417S and CT swap A453S was unaltered compared with wild-type MLO, indicating that protein instability does not account for the reduced complementation efficiency (Figure 3E).
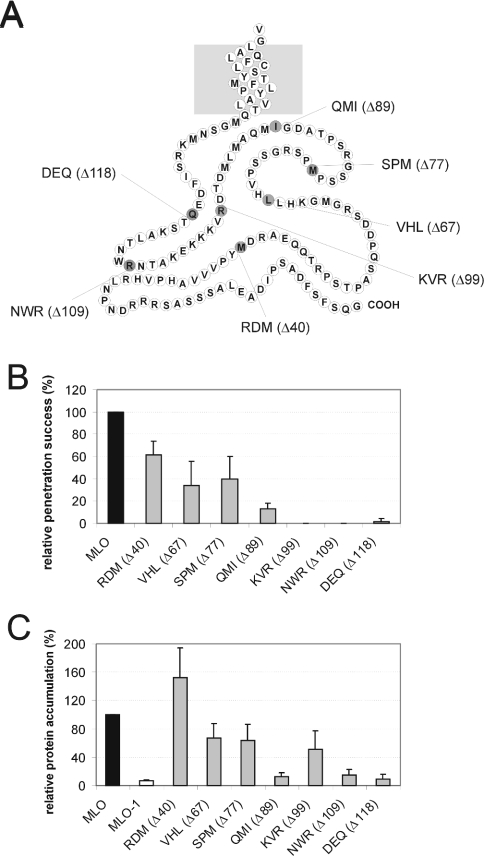
To test whether the length of the C-terminal tail contributes to MLO activity, we generated a series of successive truncations, resulting in constructs RDM (Δ40), VHL (Δ67), SPM (Δ77), QMI (Δ89), KVR (Δ99), NWR (Δ109) and DEQ (Δ118) lacking 40, 67, 77, 89, 99, 109 and 118 amino acids of the C-terminus respectively (Figure 4A). Complementation analysis by transient gene expression revealed that a truncation of the C-terminus by the distal 40 amino acids [construct RDM (Δ40)] resulted in a significant reduction in MLO functionality by approx. 40% (Figure 4B). This activity decrease was not due to diminished protein accumulation of the truncated derivative (Figure 4C). Constructs encoding MLO derivatives with increasingly shortened C-termini exhibited both progressively decreased activity and protein accumulation [VHL (Δ67), SPM (Δ77) and QMI (Δ89); Figures 4B and 4C]. Truncations of 99 amino acids or more in KVR (Δ99), NWR (Δ109) or DEQ (Δ118) resulted in complete loss of function (Figure 4B). Of these, the inactive variant KVR (Δ99) accumulated to a level comparable with variants VHL (Δ67) and SPM (Δ77) (Figure 4C). Since the latter two derivatives retained significant activity, complete loss of function of KVR (Δ99) is most likely due to missing essential amino acids residing distally of Arg434 (Figure 4A). Taken together, our data indicate that the length of the cytoplasmic C-terminal tail, as well as individual residues therein, are critical for MLO function and accumulation.
Figure 4. Integrity of the C-terminus is critical for MLO functionality.
(A) Schematic representation of the MLO C-terminus and indication of the derived C-terminal truncation variants. The light-grey box symbolizes the lipid bilayer. Individual amino acids are shown as circles labelled using the one-letter amino acid code. The last amino acid of the respective indicated constructs carrying C-terminal truncations is highlighted in dark grey. (B) Functional assay of MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were bombarded with the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or a mutant version thereof [encoding GFP plus either RDM (Δ40), VHL (Δ67), SPM (Δ77), QMI (Δ89), KVR (Δ99), NWR (Δ109) or DEQ (Δ118)]. Leaves were then inoculated with Bgh A6 and GFP-fluorescent cells were inspected for fungal structures as described in the Materials and methods section. (C) Assessment of MLO protein accumulation. Relative accumulation of wild-type MLO, mutant version MLO-1, as well as variants RDM (Δ40), VHL (Δ67), SPM (Δ77), QMI (Δ89), KVR (Δ99), NWR (Δ109) and DEQ (Δ118) carrying increasing C-terminal truncations was determined by dual-luciferase assays of transfected A. thaliana protoplasts as described in the Materials and methods section.
Cytoplasmic domain–domain interplay is required for MLO function
We demonstrated previously that wheat TaMlo-B1 is a functional orthologue of barley Mlo. Full-size TaMlo-B1 is capable of complementing a barley mlo mutant, although with slightly reduced efficiency (77% relative to barley Mlo) [10]. This seemed paradoxical in the context of our finding that replacement of the barley MLO C-terminus with the 92% sequence-identical C-terminus of TaMLO-B1 led to an approx. 65% reduction in MLO activity (see above, and Figures 3A and 3B). These data could be explained by assuming intra- or inter-molecular interactions between cytoplasmic domains of the heptahelical MLO protein that are critical for functionality.
To address this further, we generated a series of domain-swap constructs representing all possible permutations of different cytoplasmic MLO and TaMLO-B1 loops, as well as the C-termini. Since cytoplasmic loop 3 is identical between MLO and TaMLO-B1 (Figure 5A), the number of tested combinations was reduced to nine (Figure 5D). We examined the resulting constructs (MLO, TaMLO-B1, MLO IC1, MLO IC2, MLO CT, MLO IC1+IC2, MLO IC1+CT, MLO IC2+CT and MLO IC1+IC2+CT) for functionality as described above. Note that in each of these derivatives, extracellular domains and transmembrane helices are represented by barley MLO sequences. We confirmed reduced activity of the MLO CT variant in this independent set of experiments (Figure 5B; compare with construct TaMLO-B1 CT in Figure 3B) and found even greater impairment of functionality upon expression of MLO IC2 and MLO IC2+CT constructs. Interestingly, the activity of the MLO IC1 variant was slightly elevated compared with wild-type MLO (super-susceptibility; Figure 5B). Remarkably, adding wheat IC1 to either MLO IC2 or MLO CT rescued their impaired activity and generated slightly hyperactive proteins. Similarly, adding wheat IC1 to variant MLO IC2+CT rescued the impaired activity to above TaMLO-B1 wild-type levels. This suggests that the presence of wheat IC1 (hyper-)compensates for the penalty generated by introduction of either wheat IC2, CT, or a combination of both in barley MLO (Figure 5B). Although some variation was seen in the relative protein accumulation of these derivatives (Figure 5B), these differences cannot explain the observed activity rescue phenomenon (compare Figures 5B and 5C). Thus we conclude that the activity rescue is mediated by intra- and/or inter-molecular MLO domain interplay.
Figure 5. Domain-swap analysis with intracellular domains of barley MLO and TaMLO-B.
(A) Amino acid sequence alignment of the first to third intracellular loop (IC1–IC3), as well as the CT of MLO and TaMLO-B1. (B) Functional assay of single-amino-acid-substitution MLO variants. Leaf segments of the powdery-mildew-resistant barley cultivar BC Ingrid mlo-5 were co-bombarded with a GUS reporter construct and the bifunctional plasmid pUGLUM (encoding the GFP reporter plus wild-type MLO) or mutant versions thereof (encoding variants CT swap, CT swap A417S or CT swap A453S). Leaves were then inoculated with Bgh K1, and were stained for GUS activity and fungal structures as described in the Materials and methods section. (C) Assessment of MLO protein accumulation. Relative accumulation of wild-type MLO and mutant version MLO-1, as well as variants TaMLO-B1, MLO IC1, MLO IC2, MLO CT, MLO IC1+IC2, MLO IC1+CT, MLO IC2+CT and MLO IC1+IC2+CT was determined by dual-luciferase assays of transfected A. thaliana protoplasts as described in the Materials and methods section. Note that the values obtained for MLO and MLO-1 result from a common experiment with the constructs shown in Figures 2(C) and 2(E). (D) Schematic representation of the domain-swap constructs MLO IC1, MLO IC2, MLO CT, MLO IC1+IC2, MLO IC1+CT, MLO IC2+CT and MLO IC1+IC2+CT. Black bends illustrate MLO portions, whereas light-grey bends depict the respective TaMLO-B1 fraction.
DISCUSSION
Conservation of MLO proteins
Among MLO family members that belong to such distinct phyla as monocotyledons (barley, wheat, rice and maize), dicotyledons (A. thaliana) and mosses (Physcomitrella patens), we detected 30 invariant amino acids, representing less than 10% of the total protein. Unless these residues evolved by convergent evolution of the respective genes, the common ancestor of vascular plants (tracheophytes) and mosses (bryophytes) must have possessed Mlo gene(s)-encoding protein(s) bearing these invariant residues. Thus the 30 invariant residues within the MLO protein family were conserved during 400–450 million years of plant evolution. In a similar analysis of animal family 1 GPCRs, it was found that the extent of conservation varied greatly among individual subgroups: while the ADP-receptor-like GPR34 maintained only 17% of the amino acid residues during 450 million years of evolution, the retinal receptor rhodopsin preserved 43% of its residues over the same period [29]. This is thought to reflect a more stringent network of intramolecular constraints for the unique function of rhodopsin. Thus invariant amino acids must be interpreted in the context of a functional diversification of family members. Lacking knowledge about a potential functional specialization of MLO isoforms prohibits at present a more differentiated analysis of conserved residues.
Mutational analysis unravels a functional and/or structural role of invariant cysteine residues
In metazoan GPCRs, conserved extracellular cysteine residues in extracellular loops 1 and 2 have been shown to form disulphide bridges that are important for correct receptor folding and, as a consequence, for the establishment of high-affinity ligand-binding sites [30–35]. In addition, extracellular cysteine residues may serve a role in receptor oligomerization [32,36,37]. Likewise, the four invariant extracellular cysteine residues in MLO might either provide the structural basis for the formation of two intramolecular disulphide bonds bridging extracellular loops 1 and 3 (Figure 1A) [6] or contribute to MLO oligomerization (see below) by intermolecular disulphide links. In any case, the complete loss of function of the cysteine→alanine mutants is likely to reflect a specific role of the cysteine residues, since a double mutation of nearby glutamic acid (Glu103) and arginine (Arg105) residues in the first extracellular loop had no effect on MLO activity (Figure 2B).
It is noteworthy that the respective mutant variants accumulated in planta either comparably with wild-type MLO (C98A and C367A) or at moderately reduced levels (C86A and C114A). Because the majority of tested single-amino-acid replacements in MLO that lead to loss of protein function result in efficient removal by an ERAD (endoplasmic-reticulum-associated protein degradation)-like quality control mechanism [e.g. the W162R replacement in MLO-1 (J. Müller, P. Piffanelli, A. Devoto, M. Miklis, C. Elliott, B. Ortmann, P. Schulze-Lefert and R. Panstruga, unpublished work)], it seems unlikely that globally misfolded MLO proteins will pass such a rigid checkpoint. Therefore it is possible that the cysteine→alanine mutations either disrupt binding of a presumed agonist or impair subtle intra- or intermolecular interactions that are essential for MLO activity. Consistent with this, cysteine substitution mutants of the human mu opioid receptor were impaired in ligand binding, but were still targeted to the plasma membrane at reduced levels compared with that of wild-type [38].
Do conserved proline residues in MLO transmembrane domains function as molecular hinges?
Proline residues in transmembrane α-helices of integral membrane proteins have been suspected for a long time of playing a key role in helix packing and signal transduction by inducing regions of helix distortion and/or dynamic flexibility [39–42]. In this manner, helix kinks in transmembrane domains may act as hinges, swivels and/or switches, as suggested for ion channels and (G-protein coupled) receptors [40,43]. In the case of GPCRs, it is assumed that agonist binding induces a conformational change that is transduced across the membrane via conserved proline residues in transmembrane domains VI and VII, subsequently resulting in heterotrimeric G-protein activation at the cytoplasmic face of the lipid bilayer [40]. Recently, it has also been suggested that proline residues in transmembrane domains may be involved in helix–helix interactions of polytopic membrane proteins (‘helix packing’) [44]. Site-directed mutagenesis of invariant Pro287 and Pro395 residues in MLO had opposite effects on MLO function: mutant P287G was slightly hyperactive, whereas variant P395G was fully inactive (Figure 1D). Since the latter variant also exhibited drastically reduced steady-state protein accumulation (Figure 1E), we cannot yet distinguish whether the absence of MLO activity is attributable to a loss of function, severely impaired MLO levels, or a combination of both. Although the enhanced MLO activity of the P287G variant proved statistically not significant (P>0.05; Student's t test), all three replicate experiments provided evidence for hyperactivity, suggesting that P287G may represent a weak gain-of-function mutation. If so, this could reflect reduced transmembrane domain flexibility, possibly arresting cytoplasmic regions of MLO in a particular conformation.
Proline residues in proline-kinked transmembrane domains of GPCRs are frequently associated with accompanying serine or threonine residues. The presence of serine or threonine at position −1, −2 or +2 relative to proline is thought to increase the bending angle of the helix compared with a standard proline-kink, possibly by an additional hydrogen bond formed between the side chain of serine/threonine and the backbone carbonyl oxygen of proline [45]. In the MLO family, a threonine at position −2 relative to the invariant proline in transmembrane helix VII is also strictly conserved (Figure 1A), and its association with the invariant proline may be required to establish a functionally relevant structural conformation.
FRET analysis reveals potential MLO homo-oligomerization
Quantitative FRET analysis by APB revealed for the first time evidence for MLO homodimerization/oligomerization. Calculated FRET efficiencies for the MLO–YFP/MLO–CFP couple (6.4±3%) were significantly above the cytoplasmic YFP/CFP pair (2.3±2.7%), but significantly below the MLO–YFP/CaM–CFP positive control (12.2%; Figures 2A and 2B). Previously reported FRET efficiencies determined for GPCR oligomerization range from 11.5 to 18% [46–48]. It is unlikely that the moderate MLO–MLO FRET efficiency simply reflects reduced mobility of fluorophore-tagged proteins within a two-dimensional membrane compartment, since only little energy transfer was observed between two unrelated fluorophore-tagged GPCRs (2.9±1.9%; [48]), or a glucose transporter and a GPCR [46]. Instead, the modest MLO–YFP/MLO–CFP FRET efficiency may indicate that resonance energy transfer between donor and acceptor fluorophores is suboptimal, e.g. due to steric constraints or fluorophore spacing (FRET efficiency is inversely proportional to the sixth power of fluorophore distance). Alternatively, MLO dimerization/oligomerization may be very transient or might only occur within an MLO subpopulation. For metazoan GPCRs, oligomerization has been suggested to contribute to receptor maturation, intracellular trafficking, signal transduction, GPCR internalization or the expansion of receptor diversity [26–28]. It has been proposed that disulphide bridges, the C- or N-terminus, or transmembrane domains mediate GPCR dimerization [26,27,47–49]. While our experimental data rule out a contribution of CaM as a molecule linking two MLO proteins, it remains to be investigated by future experiments which MLO peptide domains may contribute to dimerization. It is likely that MLO only forms homo-oligomers or that at least only homo-oligomers functionally contribute to barley Bgh susceptibility, since mutational approaches revealed MLO as the sole barley powdery mildew ‘susceptibility’ locus. If MLO would also form functional hetero-oligomeric complexes with further barley MLO isoforms, then previous mutational analysis should have revealed the respective genes.
Both identity and integrity of the C-terminus are critical for MLO functionality
Replacement of the C-terminus with corresponding sequences encoded by highly similar monocotyledon Mlo genes (TaMLO-A1, TaMLO-B1 or OsMLO1) halved barley MLO activity, whereas substitution with the sequence-diversified dicotyledon AtMLO11 C-terminus resulted in a nearly inactive chimaera (Figure 3B). In a distinct set of experiments involving C-terminal truncations of MLO, constructs showing similar protein accumulation levels as AtMLO11 CT [e.g. VHL (Δ67) and SPM (Δ77)] were found to retain significant residual activity (Figures 4B and 4C). This provides clear evidence for a contribution of the C-terminus to MLO activity. It is counterintuitive that the OsMLO-1 C-terminus, exhibiting only 51% amino acid identity with barley MLO, was at least as active as the two chimaeras carrying the highly sequence-related wheat orthologue C-termini (94% and 92% identity with MLO; Figures 3A and 3B). This suggests that a three-dimensional conformation of the C-terminus and/or interactions with other intracellular domains rather than the amino acid identity itself determines MLO activity levels.
This interpretation may contradict the finding that replacement of either Ser417 or Ser453 in the C-terminal tail halved MLO activity (without recognizable alterations of protein accumulation; Figures 3D and 3E). In contrast with the strictly conserved cysteine and proline residues described above, the two serine residues occupy positions that are variable throughout the MLO protein family [5,6] (see Supplement 3 at http://www.BiochemJ.org/bj/385/bj3850243add.htm). Serine residues are potential targets for phosphorylation by specific serine/threonine protein kinases and have been shown to be important for receptor internalization [50,51]. Recently, a MS-based search for phosphorylated plasma-membrane-resident Arabidopsis proteins revealed phosphorylation of the C-termini of two distinct MLO isoforms [52,53]. Thus it is possible that isoform-specific phosphorylation patterns modulate MLO activity. The extent of MLO activity reduction in the mutants is reminiscent of activity impairment in MLO CaMBD mutants that carry amino acid substitutions of residues critical for CaM binding (e.g. L420R or W423R) [12]. Since Ser417 and Ser453 are either located within (Ser417) or adjacent to the CaMBD (Ser453; Figure 3A), a direct or indirect interplay between phosphorylation and in planta CaM binding appears plausible.
The series of C-terminal deletion constructs revealed the importance of the full-length cytosolic C-terminus for MLO function (Figure 4B). This finding was unexpected, since the C-termini of MLO proteins are highly divergent both in length and sequence [6]. The distal C-terminal portion of some GPCRs has been shown to be dispensable for G-protein signalling [50,54]. More recently, however, the C-terminus of some GPCRs was shown to provide an interaction interface for a plethora of proteins that are either responsible for GPCR trafficking, modulating their clustering with various effectors, or regulating GPCR functions in an allosteric manner [55]. For this reason, it is conceivable that MLO C-termini may anchor, along with CaM, other interacting proteins by as yet unknown interaction interfaces.
Functional interplay and co-evolution of cytoplasmic domains
Functional analysis of chimaeras carrying all possible permutations of cytoplasmic domains derived from barley MLO and the wheat orthologue TaMLO-B1 provided strong evidence for cytoplasmic domain interactions (Figure 5B). If this were true, one would predict that co-evolution occurred between the intracellular regions to maintain the interplay over time. Consequently, any deleterious changes of amino acids in one domain must either be selected against, or counteracted by a complementary change in an interacting domain [56,57]. Previous computational analysis of the MLO protein family revealed indirect evidence for intramolecular co-evolution of cytoplasmic MLO domains [6]. Interdomain correlation analysis detected a particular likelihood for co-evolution of IC2, IC3 and the C-terminus [6]. Our experimental data obtained in the present study are fully consistent with the computational analysis. It has been shown for mammalian GPCRs that IC2, IC3 and often the proximal part of the C-terminus are collectively involved in G-protein coupling [58–60]. Likewise, it is possible that cytoplasmic domains of one or several MLO molecules (within an oligomeric complex) provide a co-operative interface for protein–protein communication with as yet unknown downstream signalling components.
Online data
Acknowledgments
We thank Professor Ralf Reski (University of Freiburg, Germany), and Markus Frank and colleagues at BASF (Ludwigshafen, Germany) for providing the full-length Physcomitrella Mlo cDNA sequence as well as Patrick Schweizer (IPK Gatersleben, Germany) for making available barley EST clone HO15N10S (GenBank® accession number CD057673). We acknowledge excellent technical assistance in molecular cloning and particle bombardment by Anja Reinstädler and Brigitte Koop. We are grateful to Jochen Kleemann for helping in the microscopic analysis of ballistically transformed barley leaf specimens.
References
- 1.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.Goff S. A., Ricke D., Lan T. H., Presting G., Wang R. L., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Hu S. N., Wang J., Wong G. K. S., Li S. G., Liu B., Deng Y. J., Dai L., Zhou Y., Zhang X. Q., et al. A draft sequence of the rice genome (Oryza sativa L. ssp indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 4.Büschges R., Hollricher K., Panstruga R., Simons G., Wolter M., Frijters A., van Daelen R., van der Lee T., Diergaarde P., Groenendijk J., et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 5.Devoto A., Piffanelli P., Nilsson I., Wallin E., Panstruga R., von Heijne G., Schulze-Lefert P. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 6.Devoto A., Hartmann H. A., Piffanelli P., Elliott C., Simmons C., Taramino G., Goh C. S., Cohen F. E., Emerson B. C., Schulze-Lefert P., Panstruga R. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 2003;56:77–88. doi: 10.1007/s00239-002-2382-5. [DOI] [PubMed] [Google Scholar]
- 7.Panstruga R., Schulze-Lefert P. Corruption of host seven-transmembrane proteins by pathogenic microbes: a common theme in animals and plants? Microbes Infect. 2003;5:429–437. doi: 10.1016/s1286-4579(03)00053-4. [DOI] [PubMed] [Google Scholar]
- 8.Schulze-Lefert P., Panstruga R. Establishment of biotrophy by parasitic fungi and reprogramming of host cells for disease resistance. Annu. Rev. Phytopathol. 2003;41:641–667. doi: 10.1146/annurev.phyto.41.061002.083300. [DOI] [PubMed] [Google Scholar]
- 9.Collins N. C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J. L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S. C., Schulze-Lefert P. SNARE-protein-mediated disease resistance at the plant cell wall. Nature (London) 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 10.Elliott C., Zhou F. S., Spielmeyer W., Panstruga R., Schulze-Lefert P. Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol. Plant Microbe Interact. 2002;15:1069–1077. doi: 10.1094/MPMI.2002.15.10.1069. [DOI] [PubMed] [Google Scholar]
- 11.Kim M. C., Lee S. H., Kim J. K., Chun H. J., Choi M. S., Chung W. S., Moon B. C., Kang C. H., Park C. Y., Yoo J. H., et al. Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein: isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 2002;277:19304–19314. doi: 10.1074/jbc.M108478200. [DOI] [PubMed] [Google Scholar]
- 12.Kim M. C., Panstruga R., Elliott C., Müller J., Devoto A., Yoon H. W., Park H. C., Cho M. J., Schulze-Lefert P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature (London) 2002;416:447–450. doi: 10.1038/416447a. [DOI] [PubMed] [Google Scholar]
- 13.Schultheiss H., Dechert C., Kogel K. H., Hückelhoven R. A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 2002;128:1447–1454. doi: 10.1104/pp.010805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultheiss H., Dechert C., Kogel K. H., Hückelhoven R. Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J. 2003;36:589–601. doi: 10.1046/j.1365-313x.2003.01905.x. [DOI] [PubMed] [Google Scholar]
- 15.Shirasu K., Nielsen K., Piffanelli P., Oliver R., Schulze-Lefert P. Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 1999;17:293–299. [Google Scholar]
- 16.Panstruga R. A golden shot: how ballistic single cell transformation boosts the molecular analysis of cereal–mildew interactions. Mol. Plant Pathol. 2004;5:141–148. doi: 10.1111/j.1364-3703.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou F. S., Kurth J. C., Wei F. S., Elliott C., Vale G., Yahiaoui N., Keller B., Somerville S., Wise R., Schulze-Lefert P. Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell. 2001;13:337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. Engineering hybrid genes without the use of restriction enzymes: gene-splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen K., Olsen O., Oliver R. A transient expression system to assay putative antifungal genes on powdery mildew infected barley leaves. Physiol. Mol. Plant Pathol. 1999;54:1–12. [Google Scholar]
- 20.Schweizer P., Pokorny J., Abderhalden O., Dudler R. A transient assay system for the functional assessment of defense-related genes in wheat. Mol. Plant Microbe Interact. 1999;12:647–654. [Google Scholar]
- 21.Trezzini G. F., Horrichs A., Somssich I. E. Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Mol. Biol. 1993;21:385–389. doi: 10.1007/BF00019954. [DOI] [PubMed] [Google Scholar]
- 22.Sprenger-Haussels M., Weisshaar B. Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 2000;22:1–8. doi: 10.1046/j.1365-313x.2000.00687.x. [DOI] [PubMed] [Google Scholar]
- 23.Bhat R. A., Borst J. W., Riehl M., Thompson R. D. Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 2004 doi: 10.1007/s11103-004-0553-z. in the press. [DOI] [PubMed] [Google Scholar]
- 24.Karpova T. S., Baumann C. T., He L., Wu X., Grammer A., Lipsky P., Hager G. L., McNally J. G. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. (Oxford) 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 25.Piffanelli P., Zhou F. S., Casais C., Orme J., Jarosch B., Schaffrath U., Collins N. C., Panstruga R., Schulze-Lefert P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 27.Rios C. D., Jordan B. A., Gomes I., Devi L. A. G-protein-coupled receptor dimerization: modulation of receptor function. Pharmacol. Ther. 2001;92:71–87. doi: 10.1016/s0163-7258(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 28.Terrillon S., Bouvier M. Roles of G-protein-coupled receptor dimerization: from ontogeny to signalling regulation. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulz A., Schöneberg T. The structural evolution of a P2Y-like G-protein-coupled receptor. J. Biol. Chem. 2003;278:35531–35541. doi: 10.1074/jbc.M303346200. [DOI] [PubMed] [Google Scholar]
- 30.Dohlman H. G., Caron M. G., Deblasi A., Frielle T., Lefkowitz R. J. Role of extracellular disulfide-bonded cysteines in the ligand-binding function of the β2-adrenergic receptor. Biochemistry. 1990;29:2335–2342. doi: 10.1021/bi00461a018. [DOI] [PubMed] [Google Scholar]
- 31.Savarese T. M., Wang C. D., Fraser C. M. Site-directed mutagenesis of the rat m1 muscarinic acetylcholine receptor: role of conserved cysteines in receptor function. J. Biol. Chem. 1992;267:11439–11448. [PubMed] [Google Scholar]
- 32.Noda K., Saad Y., Graham R. M., Karnik S. S. The high-affinity state of the β2-adrenergic receptor requires unique interaction between conserved and nonconserved extracellular loop cysteines. J. Biol. Chem. 1994;269:6743–6752. [PubMed] [Google Scholar]
- 33.Cook J. V. F., McGregor A., Lee T. W., Milligan G., Eidne K. A. A disulfide bonding interaction role for cysteines in the extracellular domain of the thyrotropin-releasing hormone receptor. Endocrinology. 1996;137:2851–2858. doi: 10.1210/endo.137.7.8770906. [DOI] [PubMed] [Google Scholar]
- 34.Cook J. V. F., Eidne K. A. An intramolecular disulfide bond between conserved extracellular cysteines in the gonadotropin-releasing hormone receptor is essential for binding and activation. Endocrinology. 1997;138:2800–2806. doi: 10.1210/endo.138.7.5233. [DOI] [PubMed] [Google Scholar]
- 35.Blanpain C., Lee B., Vakili J., Doranz B. J., Govaerts C., Migeotte I., Sharron M., Dupriez V., Vassart G., Doms R. W., Parmentier M. Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J. Biol. Chem. 1999;274:18902–18908. doi: 10.1074/jbc.274.27.18902. [DOI] [PubMed] [Google Scholar]
- 36.Kunishima N., Shimada Y., Tsuji Y., Sato T., Yamamoto M., Kumasaka T., Nakanishi S., Jingami H., Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature (London) 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 37.Giguère V., Gallant M. A., de Brum-Fernandes A. J., Parent J. L. Role of extracellular cysteine residues in dimerization/oligomerization of the human prostacyclin receptor. Eur. J. Pharmacol. 2004;494:11–22. doi: 10.1016/j.ejphar.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 38.Zhang P. S., Johnson P. S., Zollner C., Wang W. F., Wang Z. J., Montes A. E., Seidleck B. K., Blaschak C. J., Surratt C. K. Mutation of human mu opioid receptor extracellular “disulfide cysteine” residues alters ligand binding but does not prevent receptor targeting to the cell plasma membrane. Mol. Brain Res. 1999;72:195–204. doi: 10.1016/s0169-328x(99)00241-7. [DOI] [PubMed] [Google Scholar]
- 39.von Heijne G. Proline kinks in transmembrane α-helices. J. Mol. Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 40.Sansom M. S. P., Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane α-helices. Trends Pharmacol. Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 41.Cordes F. S., Bright J. N., Sansom M. S. P. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- 42.Bright J. N., Sansom M. S. P. The flexing/twirling helix: exploring the flexibility about molecular hinges formed by proline and glycine motifs in transmembrane helices. J. Phys. Chem. B. 2003;107:627–636. [Google Scholar]
- 43.Govaerts C., Blanpain C., Deupi X., Ballet S., Ballesteros J. A., Wodak S. J., Vassart G., Pardo L., Parmentier M. The TXP motif in the second transmembrane helix of CCR5: a structural determinant of chemokine-induced activation. J. Biol. Chem. 2001;276:13217–13225. doi: 10.1074/jbc.M011670200. [DOI] [PubMed] [Google Scholar]
- 44.Orzáez M., Salgado J., Gimenez-Giner A., Perez-Paya E., Mingarro I. Influence of proline residues in transmembrane helix packing. J. Mol. Biol. 2004;335:631–640. doi: 10.1016/j.jmb.2003.10.062. [DOI] [PubMed] [Google Scholar]
- 45.Deupi X., Olivella M., Govaerts C., Ballesteros J. A., Campillo M., Pardo L. Ser and Thr residues modulate the conformation of pro-kinked transmembrane α-helices. Biophys. J. 2004;86:105–115. doi: 10.1016/S0006-3495(04)74088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overton M. C., Blumer K. J. G-protein-coupled receptors function as oligomers in vivo. Curr. Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 47.Overton M. C., Blumer K. J. The extracellular N-terminal domain and transmembrane domains 1 and 2 mediate oligomerization of a yeast G protein-coupled receptor. J. Biol. Chem. 2002;277:41463–41472. doi: 10.1074/jbc.M205368200. [DOI] [PubMed] [Google Scholar]
- 48.Floyd D. H., Geva A., Bruinsma S. P., Overton M. C., Blumer K. J., Baranski T. J. C5a receptor oligomerization: II. Fluorescence resonance energy transfer studies of a human G protein-coupled receptor expressed in yeast. J. Biol. Chem. 2003;278:35354–35361. doi: 10.1074/jbc.M305607200. [DOI] [PubMed] [Google Scholar]
- 49.Klco J. M., Lassere T. B., Baranski T. J. C5a receptor oligomerization: I. Disulfide trapping reveals oligomers and potential contact surfaces in a G protein-coupled receptor. J. Biol. Chem. 2003;278:35345–35353. doi: 10.1074/jbc.M305606200. [DOI] [PubMed] [Google Scholar]
- 50.Wheeler M. B., Gelling R. W., Hinke S. A., Tu B., Pederson R. A., Lynn F., Ehses J., McIntosh C. H. S. Characterization of the carboxyl-terminal domain of the rat glucose-dependent insulinotropic polypeptide (GIP) receptor: a role for serines 426 and 427 in regulating the rate of internalization. J. Biol. Chem. 1999;274:24593–24601. doi: 10.1074/jbc.274.35.24593. [DOI] [PubMed] [Google Scholar]
- 51.Hanyaloglu A. C., Vrecl M., Kroeger K. M., Miles L. E. C., Qian H. W., Thomas W. G., Eidne K. A. Casein kinase II sites in the intracellular C-terminal domain of the thyrotropin-releasing hormone receptor and chimeric gonadotropin-releasing hormone receptors contribute to β-arrestin-dependent internalization. J. Biol. Chem. 2001;276:18066–18074. doi: 10.1074/jbc.M009275200. [DOI] [PubMed] [Google Scholar]
- 52.Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unson C. G., Cypess A. M., Kim H. N., Goldsmith P. K., Carruthers C. J. L., Merrifield R. B., Sakmar T. P. Characterization of deletion and truncation mutants of the rat glucagon receptor: seven transmembrane segments are necessary for receptor transport to the plasma membrane and glucagon binding. J. Biol. Chem. 1995;270:27720–27727. doi: 10.1074/jbc.270.46.27720. [DOI] [PubMed] [Google Scholar]
- 55.Bockaert J., Marin P., Dumuis A., Fagni L. The ‘magic tail’ of G protein-coupled receptors: an anchorage for functional protein networks. FEBS Lett. 2003;546:65–72. doi: 10.1016/s0014-5793(03)00453-8. [DOI] [PubMed] [Google Scholar]
- 56.Goh C. S., Bogan A. A., Joachimiak M., Walther D., Cohen F. E. Co-evolution of proteins with their interaction partners. J. Mol. Biol. 2000;299:283–293. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- 57.Pazos F., Valencia A. Similarity of phylogenetic trees as indicator of protein–protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- 58.Bourne H. R. How receptors talk to trimeric G proteins. Curr. Opin. Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 59.Cypess A. M., Unson C. G., Wu C. R., Sakmar T. P. Two cytoplasmic loops of the glucagon receptor are required to elevate cAMP or intracellular calcium. J. Biol. Chem. 1999;274:19455–19464. doi: 10.1074/jbc.274.27.19455. [DOI] [PubMed] [Google Scholar]
- 60.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.