Abstract
Background
Multidrug-resistant (MDR) bacteria are a significant cause of severe infections, particularly in human immunodeficiency virus (HIV)-positive individuals because of their weakened immunity. Since there was no previous pooled representative data regarding the MDR bacteria among HIV-positive individuals in Ethiopia, this systematic review and meta-analysis is required.
Methods
This study was conducted based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. A literature search was performed using PubMed, Medline, EMBASE, Google Scholar, Hinari, Web of Science, Science Direct, and African Journals Online databases. Data were extracted using Microsoft Excel 2019 and analyzed using STATA version 11.0 software. A random-effects model was used to estimate the pooled effect size of outcome variables across studies with a 95% confidence interval. The I2 statistic was used to check for heterogeneity. The presence of publication bias was determined using a funnel plot and Egger’s test with a p-value < 0.05 evidence of statistically significant bias.
Results
The pooled prevalence of MDR was 58.02% (95% CI: 46.32–69.73%) with high heterogeneity (I2 = 97.1%, (p < 0.001). In subgroup analysis, the highest multi-drug resistance was observed in the Oromia region (80.95%), patients with multiple infections (82.35%), and studies identified both Gram-positive and Gram-negative bacteria (61.45%). Furthermore, the pooled prevalence of MDR bacteria colonizing HIV-positive individuals was 48.76%. Regarding MDR species, Enterococci (77.41%) and Pseudomonas spp. (84.60%) were commonly identified in individuals with HIV infection.
Conclusion
Our study indicates a high burden of MDR among HIV-positive individuals in Ethiopia. The Oromia region, HIV patients with multiple infections, Pseudomonas spp., and Enterococci showed the highest MDR in the subgroup analysis. Therefore, regional hospitals should implement strategies to tackle MDR such as vaccination program, appropriate use of antibiotics, and further study on the associated factors of MDR bacteria in HIV are required.
Introduction
Starting from Alexander Fleming’s discovery of penicillin in 1928 till the present day, antimicrobial resistance (AMR) has been recognized as an ancient biological phenomenon, even as novel antimicrobial agents discovered and manufactured [1, 2]. The increased dissemination of AMR is threatening the world as it leads to serious illnesses and prolonged hospital admissions, an increase in healthcare costs, higher drug costs, and treatment failure [3]. It has been estimated that the burden of deaths by AMR may increase to 10 million each year by 2050, and it may claim around 2.4 million lives in Europe, North America, and Australia between 2015 and 2050 if effective action is not taken now [4].
Globally, drug-resistant infections contributed to 4.95 million deaths in 2019 and it is estimated that bacterial resistance was directly responsible for 1.27 million deaths [5]. In the World Health Organization (WHO) European region, an estimated 541,000 deaths are associated with bacterial AMR, and 133,000 deaths are attributable to bacterial AMR [6]. Above 1.05 million deaths were associated with AMR and 250,000 deaths were directly caused by bacterial resistance in the WHO African region [7]. In Ethiopia, 21,200 deaths were attributable to bacterial AMR and 85,300 deaths were associated with AMR in 2019 [8]. Lack of AMR surveillance, overuse of antimicrobials, clinical misuse of drugs, poor infection control practice, transmission of resistant pathogens in healthcare settings, prior antimicrobial use, poor healthcare contact, inadequate adherence to prescribed antibiotics, and the presence of underlying comorbid conditions are among the main factors that increase AMR [9, 10].
Multidrug-resistant (MDR) bacteria are defined as bacteria that are resistant to at least one antimicrobial agent from three or more antimicrobial classes [11]. Multi-drug resistance in bacteria is a significant concern in the healthcare industry, particularly in individuals infected with human immunodeficiency virus (HIV). The presence of MDR bacteria in HIV-positive individuals can be attributed to several factors, including a weakened immune system, frequent hospital admissions, clinic visits, and prolonged treatment with antimicrobials [12]. Overall, bacteria are one of the main causes of hospitalization and death in HIV-positive individuals; bacterial infection accounts for almost a third of all hospital admissions and a quarter of all deaths in these populations [13]. The most dangerous MDR bacteria that complicate the treatment of infections in HIV patients are extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa, Acinetobacter baumannii, Escherichia coli, Methicillin-resistant Staphylococcus aureus (MRSA), and Vancomycin-resistant Enterococci [14].
According to previous studies [15–36], the magnitude of infection or colonization of MDR bacteria related to HIV infection varies in different regions of Ethiopia. For instance, the prevalence of MDR ranged from 12% in Addis Ababa [33] to 88.4% in Gondar, Northwest Ethiopia [20]. To the best of our knowledge, there is no reported data on the national burden of MDR bacteria among HIV-positive individuals in Ethiopia, which is important for implementation of antimicrobial stewardship and infection prevention programs to minimize further complications of HIV. Therefore, this systematic review and meta-analysis provided a comprehensive understanding of the current status of MDR bacteria among HIV-positive individuals in Ethiopia.
Methods
Study design and protocol registration
This systematic review and meta-analysis were performed as per the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [37] (S1 Table). The protocol has been registered in the International Prospective Register of Systematic Reviews (PROSPERO) under the assigned number CRD42024498426.
Literature search strategy
This meta-analysis focused on MDR bacterial pathogens that cause infection or colonize individuals with HIV infection. A COCOPOP (Condition, Context, and Population) paradigm was used to determine the suitability of the included studies for this meta-analysis. The prevalence of MDR bacteria comprised the study’s condition (CO), HIV-positive individuals as the population (POP), and Ethiopia served as the context (CO). The search included studies published before January 2024 and the last search was performed between January 10 to 20/2024. Eight electronic databases (PubMed, Medline, EMBASE, Google Scholar, Hinari, Web of Science, Science Direct, and African Journals Online) were searched to identify articles reporting the prevalence of MDR bacteria in HIV-positive individuals in Ethiopia. We used search terms alone and in combination with Boolean operators such as "OR" or "AND". An example of a PubMed search strategy used was as follows: (((((bacteria) OR (infection)) OR (colonization)) AND ((((((antimicrobial resistance) OR (antibiotic resistance)) OR (multidrug-resistant)) OR (multi-drug resistance)) OR (MDR)) OR (antibiogram))) AND ((((human immunodeficiency virus) OR (immunocompromised)) OR (HIV/AIDS)) OR (HIV))) AND (Ethiopia). A manual search was conducted for relevant papers in the references of the included studies and other reviews. The articles retrieved were imported into EndNote X9 bibliographic software manager (Clarivate Analytics, Philadelphia, PA, USA).
Outcome of interest
The key outcome of interest in this study was the prevalence of MDR bacteria among HIV-positive individuals in Ethiopia, as described in the original study. Infection is described as the invasion and multiplication of pathogenic bacteria within a host organism [38]. Colonization refers to the establishment and persistence of bacteria in a host or a specific environment without necessarily causing harm or disease [39].
Studies eligibility
Three independent reviewers (MA, SB, and AA) screened the titles and abstracts of the identified studies to determine their eligibility. Full-text articles were then assessed for eligibility, and any disagreements between the reviewers were resolved through discussion. Articles published regarding MDR in the English language with a cross-sectional study design that included the prevalence of MDR bacteria for colonization or infection with HIV status in the results fulfilled the current definition of MDR, were conducted in Ethiopia and without limit on sample type, type of infection, and study period. The studies had to be original research articles and the data had to be presented in a format that allowed for meta-analysis. Studies not reporting MDR data, unclear results, not fulfilled current MDR definition, case reports, systematic reviews, meta-analysis studies, and conducted on tuberculosis were excluded from the study.
Quality assessment
After removing duplicated papers, all potentially eligible papers were reviewed. Full-text papers were retrieved for review and relevant information was extracted. The Joana Briggs Institute (JBI) critical appraisal checklist for simple prevalence was used to assess the quality of included studies [40]. This tool comprised nine questions. For each question, a score of 0 was assigned for ‘not reported’ or ‘not appropriate’ and 1 for ‘yes’. Then, the scores were summarized to obtain a total score ranging from 0 to 9. Based on the assigned points, articles were categorized as having a high (7–9), medium (5–7), or low (0–4) quality. Accordingly, articles with high (7–9) and medium (5–7) quality were included in the final analysis (S2 Table). Three independent authors (MA, SB, and AA) assessed the quality of the studies, and any disagreement was solved by discussing it with the fourth author (DB).
Data extraction
Data were extracted from each study using the designed tool in Microsoft Excel 2019 (Microsoft Corp., Redmond, WA, USA) by four independent authors (MA, SB, AA, and DB). Any ambiguity and difference during extraction were resolved through discussion. The data extracted from eligible studies were author name, year of publication, area in which the study was conducted, study design, type of sampling method, number of HIV-positive individuals, number of HIV-positive individuals on antiretroviral therapy (ART), sex (male), age group, type of infection or colonization, type of sample, number of MDR isolates, and type and number of bacterial species reported as MDR.
Statistical analysis
Eligible data were extracted into Microsoft Excel 2019 and then exported to the STATA version 11.0 software for analysis. The pooled prevalence of MDR and 95% confidence intervals were visually displayed using a forest plot. Subgroup analysis was performed based on regions in the country, sampling method, year of study, type of infection or colonization, and MDR bacterial group or species. The heterogeneity of the included studies was evaluated using an index of heterogeneity (I2 statistic) value of 0% = no heterogeneity, ≤ 25% = low, 25%– 50% = moderate, 50–75 = substantial, and ≥ 75% = high [41]. In all pooled analyses, heterogeneity resulting from differences in effects across different studies was accounted for using a random-effects model. A sensitivity analysis of each study’s impact on the overall prevalence was also conducted. Publication bias was statistically investigated using Egger’s test [42] and visual inspection of funnel plots. A p-value < 0.05 in Egger’s test was considered as evidence of statistically significant publication bias.
Results
Literature search results
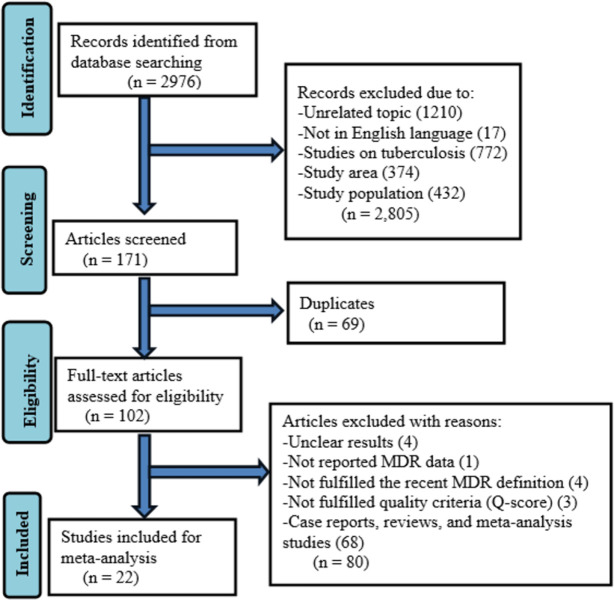
Initially, 2976 articles were identified in the literature search. The 171 were screened after removing articles for several reasons such as studies with unrelated topics, not written in the English language, studies on tuberculosis, study areas out of Ethiopia, and study populations other than HIV-infected individuals. Moreover, 69 duplicates were removed and 102 full-text articles were assessed for eligibility. Eighty full-text articles were excluded due to unavailable MDR data, not fulfilling the recent MDR definition, not fulfilling study quality criteria (Q-score), case reports, reviews, and meta-analysis studies. Finally, 22 studies were included in the meta-analysis (Fig 1).
Fig 1. Flow diagram describing the selection of studies for the systematic review and meta-analysis on the burden of MDR bacteria among HIV-positive individuals in Ethiopia.
Characteristics of the included studies
A total of 22 published articles [15–36] comprising 5636 HIV-positive individuals were included in this systematic review and meta-analysis. All of these studies used a cross-sectional study design. Regarding the sampling technique, 12 studies used systematic random sampling, 8 studies used convenient sampling, and the remaining 2 studies did not report their sampling techniques. Five regional states and one city administration in Ethiopia were represented by the studies: 10 from Amhara, 1 from Tigray, 7 from South Ethiopia, 1 from Sidama, 1 from Oromia, and 2 from Addis Ababa. The minimum and maximum numbers of study participants were 100 in Gondar [25] and 450 in Addis Ababa [21], respectively. Of the 5636 HIV-positive individuals, 3150 (55.9%) were females (Table 1).
Table 1. Descriptive summary of studies on the prevalence of MDR bacteria among HIV-positive individuals in Ethiopia.
| Author, year | Study region | Study year | Sampling method | HIV-positive individuals (N) | HIV-positive individuals on ART (%) | Sex (M) | Age group (Years) | Infection/colonization | Sample source | Total isolates (N) | MDR (N) | MDR group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tilahun et al, 2023 [28] | Amhara | 2021 | Systematic random | 378 | 100 | 187 | ≥ 10 | Pneumonia | Sputum | 175 | 148 | GPB & GNB |
| Adhanom et al, 2019 [29] | Tigray | 2016 | NR | 252 | NR | 127 | ≥ 18 | Pneumonia | Sputum | 84 | 15 | GPB & GNB |
| Genetu& Zenebe, 2020 [32] | Amhara | 2019 | Convenient | 163 | 100 | 62 | ≥ 18 | Pneumonia | Sputum | 68 | 53 | GPB & GNB |
| Ayele et al, 2020 [23] | South Ethiopia | 2019 | Systematic random | 180 | NR | 84 | ≥ 15 | Diarrhea | Stool | 15 | 11 | GNB |
| Alebachew et al, 2016 [25] | Amhara | 2013 | Convenient | 100 | NR | 12 | all age | Bloodstream infection | Blood | 31 | 25 | GPB & GNB |
| Tilahun et al, 2023 [24] | Amhara | 2021 | Convenient | 384 | NR | 199 | all age | Multiple infections | Multiple samples | 34 | 28 | GPB |
| Tessema et al, 2020 [27] | Sidama | 2018 | Systematic random | 224 | NR | 93 | ≥ 18 | Urinary tract infection | Urine | 23 | 18 | GPB & GNB |
| Abebe et al, 2014 [20] | Amhara | 2013 | Systematic random | 113 | 50.7 | 53 | ≥ 11 | Colonization | Stool | 103 | 91 | GPB |
| Gebre et al, 2022 [33] | Addis Ababa | 2016–18 | NR | 183 | 81.4 | 90 | <15 | Colonization | Nasopharyngeal swab | 50 | 6 | GPB |
| Bayleyegn et al, 2021 [30] | Amhara | 2020 | Convenient | 161 | 100 | 77 | < 15 | Colonization | Stool | 186 | 71 | GNB |
| Jemal et al, 2020 [26] | Amhara | 2018 | Convenient | 384 | NR | 157 | all age | Bloodstream infection | Blood | 123 | 96 | GPB & GNB |
| Manilal et al, 2019 [16] | South Ethiopia | 2017 | Systematic random | 307 | NR | 131 | ≥ 18 | Colonization | Nasal swab | 64 | 12 | GPB |
| Dadi et al, 2021 [19] | South Ethiopia | 2020 | Systematic random | 200 | 100 | 100 | all age | Colonization | Stool | 123 | 61 | GPB |
| Fenta et al, 2016 [21] | Addis Ababa | 2015 | Convenient | 450 | 71.0 | 141 | ≥ 18 | Urinary tract infection | Urine | 51 | 32 | GPB & GNB |
| Simeneh et al, 2022 [22] | South Ethiopia | 2021 | Systematic random | 251 | NR | 114 | ≥ 18 | Urinary tract infection | Urine | 39 | 21 | GPB & GNB |
| Hantalo et al, 2020 [17] | South Ethiopia | 2018 | Systematic random | 205 | 82.4 | 81 | ≥ 18 | Urinary tract infection | Urine | 29 | 23 | GPB & GNB |
| Muhaba et al, 2022 [31] | Amhara | 2020 | Systematic random | 206 | 100 | 77 | all age | Colonization | Nasal swab | 127 | 70 | GPB |
| Seid et al, 2020 [18] | South Ethiopia | 2018 | Systematic random | 252 | 95.6 | 144 | ≥ 15 | Colonization | Nasal swab | 34 | 10 | GPB |
| Mulu et al, 2018 [15] | Amhara | 2016–17 | Convenient | 300 | NR | 153 | 6–15 | Colonization | Nasal and throat swab | 167 | 47 | GPB & GNB |
| Adisu et al, 2023 [34] | Oromia | 2021 | Convenient | 351 | 100 | 135 | ≥ 18 | Colonization | Nasal | 21 | 17 | GPB |
| Zike et al, 2024 [36] | Amhara | 2023 | Systematic random | 170 | NR | 65 | all age | Colonization | Stool | 95 | 83 | GPB |
| Mitiku et al, 2023 [35] | South Ethiopia | 2022 | Systematic random | 422 | NR | 204 | ≥ 18 | Diarrhea | Stool | 63 | 31 | GNB |
Note. HIV: human immunodeficiency virus, ART: antiretroviral therapy, NR: not reported, MDR: multi-drug resistance, GPB: Gram-positive bacteria, GNB: Gram-negative bacteria, M: male, N: number.
Prevalence of MDR among HIV-positive individuals
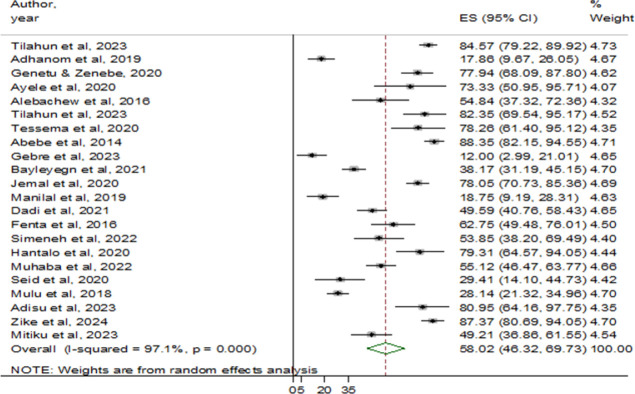
The minimum and maximum MDR prevalence reported by the studies was 12% (Addis Ababa) [33] and 88.4% (Gondar) [20], respectively. There were 1705 bacterial isolates identified from the eligible studies, of which 961 were MDR isolates (Table 1). Accordingly, the pooled prevalence of MDR was 58.02% (95% CI: 46.32–69.73), with high heterogeneity (I2 = 97.1%) and statistical significance (p < 0.001) (Fig 2).
Fig 2. The forest plot showed the pooled prevalence of MDR bacteria among HIV-positive individuals in Ethiopia.
Subgroup analysis
The prevalence of MDR among different types of infection or colonization, regions of the studies performed, MDR species, years of the study, and sampling method were analyzed by subgroup analysis. The pooled prevalence of MDR per year of study was as follows: 2013–15; 69.66% (95% CI: 47.26–92.06%), 2016–19; 48.83% (95% CI: 29.95–67.72%), and 2020–23; 64.53% (95% CI: 50.32–78.74%). According to sampling methods, the MDR prevalence was described as systematic random; 62.32% (95% CI: 48.57–76.07%), convenient sampling; 62.61% (95% CI: 45.51–79.72%), and studies have not reported the sampling method; 15.21% (95% CI: 9.15–21.27%). The combined prevalence of MDR among different regions showed that the highest MDR was reported in Oromia; 80.95% (95% CI: 64.16–97.75%), followed by Sidama; 78.26% (61.40–95.12%), and Amhara; 67.57% (95% CI: 52.70–82.43%) (Table 2).
Table 2. Subgroup analysis in the prevalence of MDR bacteria among HIV-positive individuals in Ethiopia.
| Characteristics | No. of Studies | Total isolates | MDR | Estimated pooled prevalence of MDR (95% CI) | Heterogeneity | Egger’s test | |
|---|---|---|---|---|---|---|---|
| I2 (%) | P-value | ||||||
| MDR in HIV | 22 | 1705 | 961 | 58.02 (46.32–69.73) | 97.1 | < 0.001 | 0.532 |
| Study year | |||||||
| 2013–15 | 3 | 185 | 140 | 69.66 (47.26–92.06) | 90.6 | < 0.001 | |
| 2016–19 | 10 | 657 | 287 | 48.83 (29.95–67.72) | 97.1 | < 0.001 | |
| 2020–23 | 9 | 824 | 509 | 64.53 (50.32–78.74) | 95.6 | < 0.001 | |
| Sampling method | |||||||
| Systematic random | 12 | 890 | 579 | 62.32 (48.57–76.07) | 96.0 | < 0.001 | |
| Convenient | 8 | 681 | 361 | 62.61 (45.51–79.72) | 95.9 | < 0.001 | |
| Not reported | 2 | 134 | 21 | 15.21 (9.15–21.27) | 0.0 | 0.346 | |
| Region | |||||||
| Amhara | 10 | 1109 | 704 | 67.57 (52.70–82.43) | 97.3 | < 0.001 | |
| Tigray | 1 | 84 | 15 | 17.86 (9.67–26.05) | NA | NA | |
| Addis Ababa | 2 | 101 | 38 | 37.13 (-12.60–86.86) | 97.4 | < 0.001 | |
| South Ethiopia | 7 | 367 | 169 | 49.70 (33.89–65.51) | 90.4 | < 0.001 | |
| Sidama | 1 | 23 | 18 | 78.26 (61.40–95.12) | NA | NA | |
| Oromia | 1 | 21 | 17 | 80.95 (64.16–97.75) | NA | NA | |
| Infection/colonization | |||||||
| Pneumonia | 3 | 327 | 216 | 60.15 (17.84–102.46) | 98.9 | < 0.001 | |
| Diarrhea | 2 | 78 | 42 | 59.39 (36.03–82.75) | 70.8 | 0.064 | |
| Urinary tract infection | 4 | 142 | 94 | 68.33 (56.48–80.18) | 59.6 | 0.059 | |
| Bloodstream infection | 2 | 154 | 113 | 67.86 (45.29–90.44) | 82.6 | 0.017 | |
| Multiple infections | 1 | 34 | 28 | 82.35 (69.54–95.17) | NA | NA | |
| Colonization | 10 | 970 | 489 | 48.76 (30.14–67.38) | 98.0 | < 0.001 | |
| MDR group | |||||||
| GPB & GNB | 10 | 790 | 470 | 61.45 (43.68–79.21) | 97.1 | < 0.001 | |
| GPB | 9 | 591 | 378 | 55.99 (35.28–76.70) | 97.8 | < 0.001 | |
| GNB | 3 | 264 | 113 | 50.53 (34.20–66.86) | 79.7 | 0.007 | |
Note. HIV: human immunodeficiency virus, NR: not applicable, MDR: multi-drug resistance, GPB: Gram-positive bacteria, GNB: Gram-negative bacteria, CI: confidence interval.
Regarding infection or colonization, a higher combined prevalence of MDR bacteria was observed in HIV-positive individuals with multiple infections; 82.35% (95% CI: 69.54–95.17%), bloodstream infection (BSI); 67.86% (95% CI: 45.29–90.44%), pneumonia; 60.15% (95% CI: 17.84–102.46%), diarrhea; 59.39% (95% CI: 36.03–82.75%), and colonization; 48.76% (95% CI: 30.14–67.38%). Furthermore, the pooled prevalence of MDR in HIV-positive individuals per MDR group was 61.45% (95% CI: 43.68–79.21%) in studies identifying both Gram-positive and Gram-negative bacterial isolates, 55.99% (95% CI: 35.28–76.70%) in only Gram-positive bacteria, and 50.53% (95% CI: 34.20–66.86%) in Gram-negative bacteria (Table 2).
Pooled prevalence of MDR bacterial species
Of the included studies, the one conducted in Wolayta Sodo [17] did not report the type of bacterial species with MDR status was excluded. Bacterial species were included in the meta-analysis if reported by at least three studies. According to the type of bacterial species, four species of gram-positive bacteria and ten species of gram-negative bacteria were included in the meta-analysis. S. aureus, Enterococci, S. pneumoniae, and E. coli were the top four frequently identified isolates. The pooled proportion of MDR for each bacterial species was computed from the total isolates as summarized below (Tables 3 and 4).
Table 3. Summary on the estimate of MDR for gram-positive bacteria in HIV-positive individuals.
| Bacterial species | Estimate (95% CI) by infection and colonization | Overall pooled estimate (95% CI) | Heterogeneity | ||
|---|---|---|---|---|---|
| Infection | Colonization | I2 (%) | p-value | ||
| S. aureus | 64.41 (37.72–91.10) | 50.06 (13.10–87.03) | 57.72 (40.77–74.68) | 93.1 | < 0.001 |
| CoNS | 74.58 (49.05–100.11) | - | 74.58 (49.05–100.11) | 30.0 | 0.232 |
| Enterococci | 81.79 (68.22–95.36) | 75.95 (57.04–94.85) | 77.41 (63.24–91.58) | 87.2 | < 0.001 |
| S. pneumoniae | 84.43 (74.37–94.50) | 19.98 (-2.16–42.12) | 50.51 (14.12–86.90) | 86.5 | < 0.001 |
Note. CI: confidence interval, CoNS: Coagulase-negative Staphylococcus aureus.
Table 4. Summary on the estimate of MDR for gram-negative bacteria in HIV-positive individuals.
| Bacterial species | Estimate (95% CI) by infection and colonization | Overall pooled estimate (95% CI) | Heterogeneity | ||
|---|---|---|---|---|---|
| Infection | Colonization | I2 (%) | p-value | ||
| E. coli | 68.27 (52.33–84.22) | 41.44 (27.21–55.68) | 62.39 (45.30–79.49) | 69.7 | 0.003 |
| Klebsiella spp. | 65.11 (41.10–89.11) | 26.47 (-2.35–55.29) | 58.89 (34.40–83.39) | 88.3 | < 0.001 |
| Proteus spp. | 52.94 (5.41–100.48) | 75.00 (26.00–124.00) | 63.64 (29.52–97.75) | 0.0 | 0.727 |
| Citrobacter spp. | 66.47 (39.24–93.70) | 44.44 (-4.25–93.14) | 61.70 (38.36–85.04) | 0.0 | 0.447 |
| Acinetobacter spp. | 20.00 (-58.40–98.40) | - | 20.00 (-58.40–98.40) | NA | NA |
| Enterobacter spp. | 73.37 (47.26–99.49) | 16.67 (-34.98–68.32) | 57.11 (25.47–88.74) | 36.1 | 0.196 |
| Pseudomonas spp. | 84.60 (71.95–97.24) | - | 84.60 (71.95–97.24) | 0.0 | 0.584 |
| Salmonella spp. | 63.01 (40.07–85.95) | 57.14 (8.65–105.64) | 61.94 (41.20–82.68) | 0.0 | 0.971 |
| Shigella spp. | 38.71 (11.15–66.27) | 33.33 (-32.00–98.67) | 37.90 (12.51–63.29) | 0.0 | 0.882 |
| H. influenzae | 77.78 (46.98–108.58) | 33.33 (-32.00–98.67) | 65.28 (26.11–104.44) | 31.3 | 0.228 |
Note. CI: confidence interval, NA: not applicable.
Multi-drug resistance in Gram-positive bacteria
In this meta-analysis, bacterial isolates from three genera (Staphylococcus, Streptococcus, and Enterococci) have been included. Among the included species, the pooled proportion of MDR for S. aureus was found to be 64.41% (95% CI: 37.72–91.10%) for infection and 50.06% (95% CI: 13.10–87.03%) for colonization (Table 3). This study yielded an overall estimated MDR of 57.72% (95% CI: 40.77–74.68%) in S. aureus isolates. The estimate of MDR in Coagulase-negative Staphylococci (CoNS) was 74.58% (95% CI: 49.05–100.11%) for infection only. Concerning Enterococci, the pooled proportion of MDR was 81.79% (95% CI: 68.22–95.36%) for infection and 75.95% (95% CI: 57.04–94.85%) for colonization. The overall estimated MDR in Enterococci was 77.41% (95% CI: 63.24–91.58%). In the case of S. aureus, Enterococci, and S. pneumoniae, there was significant heterogeneity between the subgroups (infection and colonization) (Table 3) (S1 Fig.).
Multi-drug resistance in Gram-negative bacteria
Bacterial isolates belonging to the family Enterobacteriaceae, other gram-negative enteric bacteria, and genus Haemophilus have been included. The highest pooled MDR was observed in Pseudomonas spp. 84.60% (95% CI: 71.95–97.24%), followed by H. influenzae 65.28% (95% CI: 26.11–104.44%), Proteus spp. 63.64% (95% CI: 29.52–97.75%), and E. coli 62.39% (95% CI: 45.30–79.49%). In the case of E. coli and Klebsiella spp., there was higher heterogeneity between the subgroups (infection and colonization) (Table 4). The pooled proportion of MDR for each bacterial species and group differences are described in the forest plots (S1 Fig.).
Publication bias
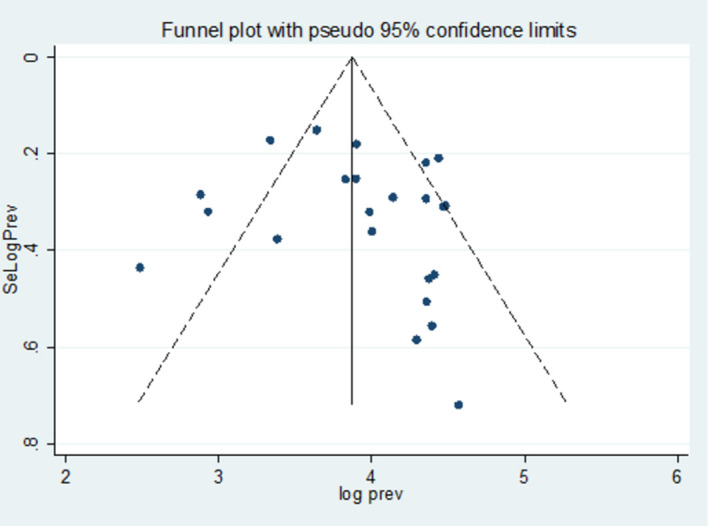
Studies were assessed for potential publication bias statistically using Egger’s test and funnel plot. The result of Egger’s test indicated no publication bias, -2.26 (95% CI: -9.68, 5.16, p-value = 0.532) (Table 5). This was depicted graphically by a funnel plot, which showed a symmetrical display of prevalence reported by the studies (Fig 3).
Table 5. Publication bias using Egger’s test.
| Egger’s test | |||||
|---|---|---|---|---|---|
| Std_Eff | Coef. | Std. Err. | t | p>t | [95% Conf. Interval] |
| Slope | 69.46217 | 16.6217 | 4.18 | 0.000 | 34.78991 104.1344 |
| bias | -2.261594 | 3.557294 | -0.64 | 0.532 | -9.681979 5.158791 |
Fig 3. The funnel plot indicated publication bias in the studies on the burden of MDR bacteria among HIV-positive individuals in Ethiopia.
Sensitivity analysis
A sensitivity analysis was performed using a random effects model because of the results’ extreme heterogeneity. Step-by-step removal of each study was used in a sensitivity analysis to determine how each study affected the pooled prevalence. The results showed that the omitted studies do not have a significant effect on the pooled prevalence of MDR among HIV-positive individuals (Table 6).
Table 6. Sensitivity analysis of the included studies to estimate the prevalence of MDR bacteria among HIV-positive individuals in Ethiopia.
| Study omitted | Estimate (95% confidence interval) | Heterogeneity | |
|---|---|---|---|
| I2 (%) | p-value | ||
| Tilahun et al, 2023 | 56.71 (44.74–68.68) | 96.8 | < 0.001 |
| Adhanom et al, 2019 | 59.98 (48.54–71.43) | 96.8 | < 0.001 |
| Genetu & Zenebe, 2020 | 57.06 (44.93–9.20) | 97.2 | < 0.001 |
| Ayele et al, 2020 | 57.34 (45.38–69.38) | 97.2 | < 0.001 |
| Alebachew et al, 2016 | 58.17 (46.11–70.23) | 97.2 | < 0.001 |
| Tilahun et al, 2023 | 56.88 (44.84–68.92) | 97.2 | < 0.001 |
| Tessema et al, 2020 | 57.12 (45.08–69.14) | 97.2 | < 0.001 |
| Abebe et al, 2014 | 56.52 (44.69–68.34) | 96.8 | < 0.001 |
| Gebre et al, 2022 | 60.25 (48.93–71.58) | 96.7 | < 0.001 |
| Bayleyegn et al, 2021 | 59.01 (46.84–71.18) | 97.1 | < 0.001 |
| Jemal et al, 2020 | 57.05 (44.81–69.23) | 97.1 | < 0.001 |
| Manilal et al, 2019 | 59.92 (48.28–71.57) | 96.9 | < 0.001 |
| Dadi et al, 2021 | 58.44 (46.17–70.72) | 97.2 | < 0.001 |
| Fenta et al, 2016 | 57.81 (45.68–69.94) | 97.2 | < 0.001 |
| Simeneh et al, 2022 | 58.22 (46.14–70.30) | 97.2 | < 0.001 |
| Hantalo et al, 2020 | 57.04 (44.99–69.08) | 97.2 | < 0.001 |
| Muhaba et al, 2022 | 58.18 (45.85–70.50) | 97.2 | < 0.001 |
| Seid et al, 2020 | 59.35 (47.38–71.31) | 97.2 | < 0.001 |
| Mulu et al, 2018 | 59.50 (47.74–71.24) | 96.8 | < 0.001 |
| Adisu et al, 2023 | 56.98 (44.97–69.00) | 97.2 | < 0.001 |
| Zike et al, 2024 | 56.57 (44.66–68.50) | 96.9 | < 0.001 |
| Mitiku et al, 2023 | 58.45 (46.32–70.58) | 97.2 | < 0.001 |
Discussion
In recent years, the prevalence of MDR bacteria among HIV-positive individuals has become a significant concern for healthcare providers and public health [13]. This systematic review and meta-analysis is the first to determine the pooled prevalence of MDR bacteria in HIV-positive individuals. The results included different types of bacterial infections and colonization in HIV-positive individuals in all age groups. Twenty-two studies based on 5636 HIV-positive individuals were included in this study. The results showed that the pooled prevalence of MDR bacteria among HIV-positive individuals was 58.02% (95% CI: 46.32–69.73%). Our finding is lower than systematic review and meta-analysis studies conducted in Ethiopia, which reported a 70.5% [43], 70.56% [44], 74.0% [45], and 74.2% [46] pooled prevalence of MDR bacteria. This might be due to the difference in types of infections, type of bacterial isolate, and study populations, which may increase the prevalence whereas our study was specific to HIV-positive individuals. Although there is no systematic review or meta-analysis conducted previously elsewhere focused on HIV-positive individuals, our finding indicated the higher burden of MDR. This might be due to the weakened immune system that leads to increased susceptibility to MDR bacterial infections, exposure to antibiotics, inadequate adherence to ART, and poor access to healthcare especially in resource-poor settings [47, 48]. Moreover, HIV infection alters the composition of the microbiome, which can create an environment conducive to the growth and persistence of MDR bacteria [49].
Our study indicates high heterogeneity (I2 = 97.1%) between the studies. Subgroup analysis was conducted to provide further details on the cause of this disparity. Accordingly, the prevalence of MDR in the different regions showed a statistically significant difference ranging from 80.95% to 17.86% with the highest and lowest pooled prevalence from Oromia and Tigray regions, respectively. The difference in number of studies between the regions could affect the MDR prevalence. This systematic review and meta-analysis included a single study from the Tigray [29], Sidama [27], and Oromia [34] regions based on our inclusion criteria. Most of the studies included were from Amhara, followed by South Ethiopia. This suggests that there are laboratory facilities for bacterial culture and susceptibility testing in these regions compared with others. In addition, the heterogeneity may be due to the few studies that were conducted on types of infection such as multiple infections [24], diarrhea [23, 35], and BSI [25, 26], a single study in the study period 2013–2015, and variations in the type and number of MDR bacteria identified between the studies.
The subgroup analysis showed that the prevalence of MDR bacteria among HIV-positive individuals across all regions of Ethiopia was higher in Oromia, followed by Sidama, Amhara, South Ethiopia, Addis Ababa, and Tigray. This variability may be due to differences in the healthcare system, the burden of infectious disease in a community, and the characteristics of the study population. The prevalence of MDR among HIV-positive individuals shows a sudden increment across different categories of study years as it is 69.66% and 64.53% in 2013–15 and 2020–23, respectively. The emergence of drug-resistant organisms, new resistance mechanisms, and a decrease in the efficiency of antibiotics treating antibiotic-resistant bacteria change the MDR prevalence over time [50].
According to the type of infection, a higher prevalence of MDR bacteria among HIV-positive individuals was observed in multiple infections, followed by urinary tract infection (UTI), BSI, pneumonia, and diarrhea. The prevalence of MDR bacteria in UTI is higher because most study participants in our study were females, who have a high risk of drug drug-resistant bacteria than males because of the structure of the urethra and vagina, and sexual activity facilitating pathogen entry [51]. Higher MDR in BSI is due to co-infections and chronic inflammation, a common feature of HIV infection, which can contribute to the development of bacterial bloodstream infections by damaging the lining of blood vessels [52]. Bacterial infections such as diarrhea and pneumonia are also common problems among HIV-positive individuals due to the immune system’s inability to effectively combat bacterial infections allowing opportunistic pathogens, such as certain MDR bacteria, to thrive and cause infections in the body [53, 54].
The meta-analysis showed that the prevalence of MDR bacterial colonization in HIV-positive individuals is somehow higher, with a pooled prevalence of 48.76%, and Enterococci were identified as highly colonizing bacteria in the studies. A previous systematic review and meta-analysis reported that HIV-positive individuals had 2.12 higher odds for colonization and 1.90 higher odds for infection with MRSA [13]. Another study conducted on the global prevalence of MRSA colonization in HIV-positive individuals reported a pooled prevalence of 7.0% [55]. This highlights the importance of monitoring and managing MDR bacterial colonization in HIV-positive individuals, particularly those with advanced immunosuppression. The colonizing bacterial flora in HIV-positive individuals are intrinsic sources of opportunistic infections and antibiotic-resistance genes can be transferred from such commensals to pathogenic organisms [56].
In this study, the highest pooled proportion of MDR among Gram-positive bacteria was detected in Enterococci, followed by CoNS and S. aureus. In addition, Pseudomonas spp., followed by H. influenzae, Proteus spp., and E. coli were the top MDR gram-negative bacteria. However, there are certain pieces of evidence of the pooled prevalence of MDR bacterial species in Ethiopia. A systematic review and meta-analysis data showed that the common multidrug-resistant species of bacteria from human, animal, food, and environmental sources were S.aureus, CoNS, Pseudomonas spp., E.coli, Citrobacter spp., Klebsiella spp., Enterobacter spp., and Salmonella spp. [57]. Another study on Gram-negative bacteria in wound infections reported that the pooled estimate of MDR in Citrobacter spp., followed by K. pneumoniae, P. mirabilis, E. coli, Acinetobacter spp., and P. aeruginosa [58]. Moreover, a study on pregnant women with bacteriuria revealed the top four MDR bacteria such as E. coli, Klebsiella spp, S. aureus, and CoNS [59]. This variability of some bacterial species and MDR prevalence is due to the difference in study population, type of infection, source of specimen, and type of antibiotics tested. Several factors contribute to the existence of MDR bacterial species in individuals with HIV infection, including frequent hospital visits, prolonged use of prophylaxis, and the risk of MDR bacteria prevalent in hospital settings [60]. In addition, this MDR occurred among these bacterial species because of the transfer of resistance genes, less permeability of the lipopolysaccharide layer, expression of efflux pumps, secretion of degrading enzymes, alteration of the target site, extreme use of broad-spectrum antibiotics, and scarcity of target-oriented antibiotics [61].
Strengths
This systematic review and meta-analysis was the first report on the burden of MDR bacteria on HIV-positive individuals in Ethiopia. The strength of the study is its comprehensive literature search by three independent authors (MA, SB, and AA) to extract all available published articles. This study also focused on different types of bacterial infections and colonization in HIV-positive individuals.
Limitations
In this study, we included only Amhara, Tigray, Addis Ababa, Oromia, Sidama, and South Ethiopia regions suggesting that they may not accurately generalize the public health problem in the country. Due to the lack of information in the published articles, viral load, and CD4 count were not fully reported in our meta-analysis.
Conclusion and recommendations
This study highlights the alarming burden of MDR bacteria among HIV-positive individuals in Ethiopia with the common MDR species of Enterococci, CoNS, S. aureus, and Pseudomonas spp. A significant heterogeneity between the included studies and higher MDR prevalence in the Oromia, Sidama, and Amhara regions was observed. Therefore, regional hospitals should implement strategies to reduce antibiotic overuse and misuse, improve adherence to ART, optimize immune reconstitution, enhance access to healthcare services, further study on the complex relationship between HIV and the human microbiome, identifying risk factors of MDR in HIV, better access to diagnostic tools and antimicrobial stewardship, and early identification of opportunistic infections and colonization of bacteria in these regions with high MDR burden and all over the country is needed. Furthermore, the development of novel antimicrobial agents targeting MDR bacteria and vaccines for their prevention in HIV-positive individuals is crucial.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors thank the scientific researchers of the included studies in this systematic review and meta-analysis.
Abbreviations
- HIV
Human immunodeficiency virus
- MDR
multidrug-resistant/ce
- ART
Antiretroviral therapy
- MRSA
Methicillin-resistant Staphylococcus aureus
- BSI
Bloodstream infection
- UIT
Urinary tract infection
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- PROSPERO
International Prospective Register of Systematic Reviews
- WHO
World Health Organization.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Lobanovska M, Pilla G. Focus: drug development: Penicillin’s discovery and antibiotic resistance: lessons for the future? The Yale journal of biology medicine. 2017;90(1):135. [PMC free article] [PubMed] [Google Scholar]
- 2.Letek M. Alexander Fleming, The discoverer of the antibiotic effects of penicillin. Behind a great medical drug there is always a great scientist! 2023:64. [Google Scholar]
- 3.Dadgostar P. Antimicrobial resistance: implications and costs. Infection drug resistance. 2019:3903–10. doi: 10.2147/IDR.S234610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. Review on antimicrobial resistance. 2016. [Google Scholar]
- 5.Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55. [DOI] [PMC free article] [PubMed]
- 6.The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis. The Lancet Public health. 2022;7(11):e897-e913. [DOI] [PMC free article] [PubMed]
- 7.The burden of bacterial antimicrobial resistance in the WHO African region in 2019: a cross-country systematic analysis. The Lancet Global health. 2024;12(2):e201-e16. [DOI] [PMC free article] [PubMed]
- 8.Institute for Health Metrics and Evaluation. The burden of antimicrobial resistance (AMR) in Ethiopia [Available from: https://www.healthdata.org/sites/default/files/files/Projects/GRAM/Ethiopia_0.pdf.
- 9.Castro-Sánchez E, Moore LS, Husson F, Holmes AH. What are the factors driving antimicrobial resistance? Perspectives from a public event in London, England. BMC Infect Dis. 2016;16:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulis G, Sayood S, Gandra S. Antimicrobial resistance in low-and middle-income countries: current status and future directions. Expert Rev Anti Infect Ther. 2022;20(2):147–60. doi: 10.1080/14787210.2021.1951705 [DOI] [PubMed] [Google Scholar]
- 11.Rafailidis PI, Kofteridis D. Proposed amendments regarding the definitions of multidrug-resistant and extensively drug-resistant bacteria. Expert Rev Anti Infect Ther. 2022;20(2):139–46. doi: 10.1080/14787210.2021.1945922 [DOI] [PubMed] [Google Scholar]
- 12.Henderson HI, Napravnik S, Kosorok MR, et al., editors. Predicting Risk of Multidrug-Resistant Enterobacterales Infections Among People With HIV. Open Forum Infectious Diseases; 2022: Oxford University Press US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olaru ID, Tacconelli E, Yeung S, et al. The association between antimicrobial resistance and HIV infection: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27(6):846–53. doi: 10.1016/j.cmi.2021.03.026 [DOI] [PubMed] [Google Scholar]
- 14.Rameshkumar MR, Arunagirinathan NJAiH, Control A. Drug-resistant bacterial infections in HIV patients. 2018;83. [Google Scholar]
- 15.Mulu W, Yizengaw E, Alemu M, et al. Pharyngeal colonization and drug resistance profiles of Morraxella catarrrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae among HIV infected children attending ART Clinic of Felegehiwot Referral Hospital, Ethiopia. PLoS One. 2018;13(5):e0196722. doi: 10.1371/journal.pone.0196722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manilal A, Shewangizaw M, Mama M, Gezmu T, Merdekios B. Methicillin-resistant Staphylococcus aureus colonization in HIV patients of Arba Minch province, Ethiopia: carriage rates, antibiotic resistance, and biofilm formation. Acta Microbiol Immunol Hung. 2019;66(4):469–83. doi: 10.1556/030.66.2019.014 [DOI] [PubMed] [Google Scholar]
- 17.Haile Hantalo A, Haile Taassaw K, Solomon Bisetegen F, Woldeamanuel Mulate Y. Isolation and Antibiotic Susceptibility Pattern of Bacterial Uropathogens and Associated Factors Among Adult People Living with HIV/AIDS Attending the HIV Center at Wolaita Sodo University Teaching Referral Hospital, South Ethiopia. HIV/AIDS-Research Palliative Care. 2020:799–808. doi: 10.2147/HIV.S244619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seid M, Beyene G, Alemu Y, et al. Does cotrimoxazole prophylaxis in HIV patients increase the drug resistance of pneumococci? A comparative cross-sectional study in southern Ethiopia. PLoS One. 2020;15(12):e0243054. doi: 10.1371/journal.pone.0243054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regasa Dadi B, Solomon Z, Tesfaye M. Vancomycin resistant Enterococci and its associated factors among HIV infected patients on anti-retroviral therapy in Ethiopia. PLoS One. 2021;16(6):e0251727. doi: 10.1371/journal.pone.0251727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abebe W, Endris M, Tiruneh M, Moges F. Prevalence of vancomycin resistant Enterococci and associated risk factors among clients with and without HIV in Northwest Ethiopia: a cross-sectional study. BMC Public Health. 2014;14(1):1–8. doi: 10.1186/1471-2458-14-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenta G, Legese M, Weldearegay G. Bacteriuria and their antibiotic susceptibility patterns among people living with HIV attending Tikur Anbessa Specialized and Zewditu Memorial Hospital ART Clinics, Addis Ababa, Ethiopia. Bacteriol Parasitol. 2016;7(05). [Google Scholar]
- 22.Simeneh E, Gezimu T, Woldemariam M, Alelign D. Magnitude of multidrug-resistant bacterial uropathogens and associated factors in urinary tract infection suspected adult HIV-positive patients in southern Ethiopia. The Open Microbiology Journal. 2022;16(1). [Google Scholar]
- 23.Ayele A, Tadesse D, Manilal A, Yohanes T, Seid M, Mekuria MS. Prevalence of enteric bacteria and enteroparasites in human immunodeficiency virus-infected individuals with diarrhoea attending antiretroviral treatment clinic, Arba Minch General Hospital, southern Ethiopia. New Microbes New Infections. 2020;38:100789. doi: 10.1016/j.nmni.2020.100789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tilahun M, Gedefie A, Sahle Z. Asymptomatic Carriage Rate, Multidrug Resistance Level, and Associated Risk Factors of Enterococcus in Clinical Samples among HIV-Positive Patients Attending at Debre Birhan Comprehensive Specialized Hospital, North Showa, Ethiopia. BioMed Research International. 2023;2023. doi: 10.1155/2023/7310856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alebachew G, Teka B, Endris M, Shiferaw Y, Tessema B. Etiologic agents of bacterial sepsis and their antibiotic susceptibility patterns among patients living with human immunodeficiency virus at Gondar University Teaching Hospital, Northwest Ethiopia. BioMed Research International. 2016;2016. doi: 10.1155/2016/5371875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jemal M, Deress T, Belachew T, Adem Y. Antimicrobial resistance patterns of bacterial isolates from blood culture among HIV/AIDS patients at Felege Hiwot Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2020;2020. doi: 10.1155/2020/8893266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tessema NN, Ali MM, Zenebe MH. Bacterial associated urinary tract infection, risk factors, and drug susceptibility profile among adult people living with HIV at Haswassa University Comprehensive Specialized Hospital, Hawassa, Southern Esthiopia. Sci Rep. 2020;10(1):10790. doi: 10.1038/s41598-020-67840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilahun M, Gebretsadik D, Seid A, et al. Bacteriology of community-acquired pneumonia, antimicrobial susceptibility pattern and associated risk factors among HIV patients, Northeast Ethiopia: cross-sectional study. SAGE open medicine. 2023;11:20503121221145569. doi: 10.1177/20503121221145569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adhanom G, Gebreegziabiher D, Weldu Y, et al. Species, risk factors, and antimicrobial susceptibility profiles of bacterial isolates from HIV-infected patients suspected to have pneumonia in Mekelle zone, Tigray, northern Ethiopia. BioMed research international. 2019;2019. doi: 10.1155/2019/8768439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayleyegn B, Fisaha R, Kasew D. Fecal carriage of extended spectrum beta-lactamase producing Enterobacteriaceae among HIV infected children at the University of Gondar Comprehensive Specialized Hospital Gondar, Ethiopia. AIDS Research Therapy. 2021;18(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhaba H, Fenta GM, Gebretsadik D. Methicillin resistance Staphylococcus aureus nasal carriage and its associated factors among HIV patients attending art clinic at Dessie comprehensive specialized hospital, Dessie, North East Ethiopia. PLOS Global Public Health. 2022;2(9):e0000838. doi: 10.1371/journal.pgph.0000838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genetu DE, Zenebe Y. Bacterial Profile and Their Antibiotic Resistance Pattern Among HIV Patients Diagnosed with Pneumonia in Felege-Hiwot Referral Hospital, Bahir Dar, Northwest Ethiopia. 2020. [Google Scholar]
- 33.Gebre HA, Wami AA, Kebede ES, Yidnekachew M, Gebre M, Negash AA. Nasopharyngeal Staphylococcus aureus colonization among HIV-infected children in Addis Ababa, Ethiopia: antimicrobial susceptibility pattern and association with Streptococcus pneumoniae colonization. Access Microbiology. 2023;5(8):000557. v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adisu M, Amankwah S, Tamrat R, Alemu Y, Akeba T, Tadesse M. Colonization and Associated Risk Factors of Methicillin-Resistant Staphylococcus aureus Among People Living with HIV at Jimma Medical Center, Southwest Ethiopia. [Google Scholar]
- 35.Mitiku A, Solomon Z, Gidisa B, et al. Prevalence, Antibiotic Susceptibility Pattern, and Associated Factors of Enteric Bacterial Pathogens Among HIV Infected Patients with Diarrhea Attending the ART Clinic of Dilla University Referral Hospital, Southern Ethiopia. Infection and Rrug Resistance. 2023:4227–36. doi: 10.2147/IDR.S410759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zike M, Ahmed AM, Hailu A, Hussien B. Vancomycin Resistant Enterococci Prevalence, Antibiotic Susceptibility Patterns and Colonization Risk Factors Among HIV-Positive Patients in Health-Care Facilities in Debre Berhan Town, Ethiopia. Infection and Drug Resistance. 2024:17–29. doi: 10.2147/IDR.S440479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. International journal of surgery. 2010;8(5):336–41. doi: 10.1016/j.ijsu.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 38.Thomson PD, Smith DJ Jr. What is infection? The American journal of surgery. 1994;167(1):S7–S11. [DOI] [PubMed] [Google Scholar]
- 39.Pirofski L-a, Casadevall A. The meaning of microbial exposure, infection, colonisation, and disease in clinical practice. The Lancet Infectious Diseases. 2002;2(10):628–35. doi: 10.1016/s1473-3099(02)00398-5 [DOI] [PubMed] [Google Scholar]
- 40.JB. I. The Joanna Briggs Institute Critical Appraisal Tools for Use in JBI Systematic Reviews Checklist for Analytical Cross Sectional Studies. North Adelaide, Australia. The Joanna Briggs Institute. 2017.
- 41.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 42.Egger M, Smith GD, Altman D. Systematic reviews in health care: meta-analysis in context: John Wiley & Sons; 2008. [Google Scholar]
- 43.Alemayehu T. Prevalence of multidrug-resistant bacteria in Ethiopia: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2021;26:133–9. doi: 10.1016/j.jgar.2021.05.017 [DOI] [PubMed] [Google Scholar]
- 44.Beyene AM, Gezachew M, Mengesha D, Yousef A, Gelaw B. Prevalence and drug resistance patterns of Gram-negative enteric bacterial pathogens from diarrheic patients in Ethiopia: A systematic review and meta-analysis. PLoS One. 2022;17(3):e0265271. doi: 10.1371/journal.pone.0265271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gemeda BA, Assefa A, Jaleta MB, Amenu K, Wieland B. Antimicrobial resistance in Ethiopia: A systematic review and meta-analysis of prevalence in foods, food handlers, animals, and the environment. One health (Amsterdam, Netherlands). 2021;13:100286. doi: 10.1016/j.onehlt.2021.100286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abayneh M, HaileMariam S, Asnake MJJoLM. Bacterial profile and multi-drug resistance pattern of bacterial isolates among septicemia suspected cases: a meta-analysis report in Ethiopia. 2021;45(3):167–78. [Google Scholar]
- 47.Ngowi BN, Sunguya B, Herman A, et al. Prevalence of multidrug resistant UTI among people living with HIV in northern Tanzania. Infection drug resistance. 2021:1623–33. doi: 10.2147/IDR.S299776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rameshkumar MR, Arunagirinathan N. Drug-resistant bacterial infections in HIV patients. Advances in HIV AIDS Control. 2018;83. [Google Scholar]
- 49.Williams B, Landay A, Presti RM. Microbiome alterations in HIV infection a review. Cell Microbiol. 2016;18(5):645–51. doi: 10.1111/cmi.12588 [DOI] [PubMed] [Google Scholar]
- 50.Rodríguez-Villodres Á, Martín-Gandul C, Peñalva G, et al. Prevalence and Risk Factors for Multidrug-Resistant Organisms Colonization in Long-Term Care Facilities Around the World: A Review. Antibiotics (Basel, Switzerland). 2021;10(6). doi: 10.3390/antibiotics10060680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onyango HA. Prevalence, Antimicrobial Susceptibility Profiles and Genotypic Characterization of Isolates obtained from Urine Samples of Pregnant Women Attending Antenatal Clinic at Pumwani Maternity Hospital, Kenya: JKUAT-COHES; 2018.
- 52.Kwiecinski JM, Horswill AR. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr Opin Microbiol. 2020;53:51–60. doi: 10.1016/j.mib.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belay A, Ashagrie M, Seyoum B, Alemu M, Tsegaye A. Prevalence of enteric pathogens, intestinal parasites and resistance profile of bacterial isolates among HIV infected and non-infected diarrheic patients in Dessie Town, Northeast Ethiopia. PLoS One. 2020;15(12):e0243479. doi: 10.1371/journal.pone.0243479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seidenberg P, Mwananyanda L, Chipeta J, et al. The etiology of pneumonia in HIV-infected Zambian children: findings from the Pneumonia Etiology Research for Child Health (PERCH) Study. The Pediatric infectious disease journal. 2021;40(9):S50. doi: 10.1097/INF.0000000000002649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabbagh P, Riahi SM, Gamble HR, Rostami A. The global and regional prevalence, burden, and risk factors for methicillin-resistant Staphylococcus aureus colonization in HIV-infected people: A systematic review and meta-analysis. Am J Infect Control. 2019;47(3):323–33. doi: 10.1016/j.ajic.2018.06.023 [DOI] [PubMed] [Google Scholar]
- 56.Sampane-Donkor E, Badoe EV, Annan JA, Nii-Trebi N. Colonisation of antibiotic resistant bacteria in a cohort of HIV infected children in Ghana. The Pan African Medical Journal. 2017;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tweldemedhin M, Muthupandian S, Gebremeskel TK, et al. Multidrug resistance from a one health perspective in Ethiopia: A systematic review and meta-analysis of literature (2015–2020). One health (Amsterdam, Netherlands). 2022;14:100390. doi: 10.1016/j.onehlt.2022.100390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chelkeba L, Melaku T, Mega TA. Gram-Negative Bacteria Isolates and Their Antibiotic-Resistance Patterns in Patients with Wound Infection in Ethiopia: A Systematic Review and Meta-Analysis. Infect Drug Resist. 2021;14:277–302. doi: 10.2147/IDR.S289687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chelkeba L, Fanta K, Mulugeta T, Melaku T. Bacterial profile and antimicrobial resistance patterns of common bacteria among pregnant women with bacteriuria in Ethiopia: a systematic review and meta-analysis. Arch Gynecol Obstet. 2022;306(3):663–86. doi: 10.1007/s00404-021-06365-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blake KS, Choi J, Dantas G. Approaches for characterizing and tracking hospital-associated multidrug-resistant bacteria. Cellurar Molecular Life Sciences. 2021;78(6):2585–606. doi: 10.1007/s00018-020-03717-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bharadwaj A, Rastogi A, Pandey S, Gupta S, Sohal JS. Multidrug-Resistant Bacteria: Their mechanism of action and prophylaxis. BioMed research international. 2022;2022. doi: 10.1155/2022/5419874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.