Abstract
Formin proteins, characterized by the FH2 domain, are critical in regulating actin-driven cellular processes and cytoskeletal dynamics during abiotic stress. However, no genome-wide analysis of the formin gene family has yet to be conducted in the economically significant plant potato (Solanum tuberosum L.). In this study, 26 formin genes were identified and characterized in the potato genome (named as StFH), each containing the typical FH2 domain and distributed across the ten chromosomes. The StFH was categorized into seven subgroups (A-G) and the gene structure and motif analysis demonstrated higher structural similarities within the subgroups. Besides, the StFH exhibited ancestry and functional similarities with Arabidopsis. The Ka/Ks ratio indicated that StFH gene pairs were evolving through purifying selection, with five gene pairs exhibiting segmental duplications and two pairs exhibiting tandem duplications. Subcellular localization analysis suggested that most of the StFH genes were located in the chloroplast and plasma membrane. Moreover, 54 cis-acting regulatory elements (CAREs) were identified in the promoter regions, some of which were associated with stress responses. According to gene ontology analysis, the majority of the StFH genes were involved in biological processes, with 63 out of 74 GO terms affecting actin polymerization. Six major transcription factor families, including bZIP, C2H2, ERF, GATA, LBD, NAC, and HSF, were identified that were involved in the regulation of StFH genes in various abiotic stresses, including drought. Further, the 60 unique microRNAs targeted 24 StFH by regulating gene expression in response to drought stress were identified. The expression of StFH genes in 14 different tissues, particularly in drought-responsive tissues such as root, stem, shoot apex, and leaf, underscores their significance in managing drought stress. RNA-seq analysis of the drought-resistant Qingshu No. 9 variety revealed the potential role of up-regulated genes, including StFH2, StFH10, StFH19, and StFH25, in alleviating drought stress. Overall, these findings provide crucial insights into the response to drought stress in potatoes and can be utilized in breeding programs to develop potato cultivars with enhanced drought-tolerant traits.
1.0 Introduction
Formins, a variety of multi-domain proteins, efficiently stimulate actin polymerization in living organisms, indicating their role as actin nucleators [1]. Formin proteins are comprised of multiple domains, prominently featuring FH1 (Formin Homology 1), FH2 (Formin Homology 2), and FH3 (Formin Homology 3) [2]. The FH1 domain contains polyproline tracks that bind to profilin [3]. Profilin transports actin subunits to the FH2 domain [4]. Additionally, profilin enhances the elongation rate of actin filaments associated with formins, often surpassing the speed of free actin filaments [5]. Plants have developed advanced strategies to deal with environmental stresses. The cytoskeleton, a dynamic network of actin filaments and microtubules within cells, is an integral aspect of this adaptation [6]. This actin filament structure plays a crucial role in facilitating rapid changes in the shape and structure of cells, enabling plants to adapt their organs and tissues for improved stress resilience [7]. Notably, cytoskeleton dynamics contribute to salt tolerance in Arabidopsis thaliana and enhance drought resistance in different plant species [8]. Although the precise mechanisms are still under investigation, the cytoskeleton plays a fundamental role in the plant stress response [9].
Plants generally rely on the cytoskeleton’s dynamic adaptation to various environmental stressors [10]. When stress challenges the cell, the cytoskeleton reorganizes its components, reinforcing weak points to ensure resilience [11]. Specialized proteins such as tubulins and actins are crucial in the cytoskeleton’s stress response [12]. These proteins maintain structural integrity, facilitate movements, and support cellular communication for collective stress response [8]. Another aspect of the cell’s internal structure, microtubules (MT), act as the cell’s internal support system. When the cell faces stress, especially from salt, these microtubules help by both building up and breaking down. Signal transduction mechanisms such as abscisic acid (ABA) regulation, cytosolic calcium ions, and reactive oxygen species (ROS) are crucial in controlling stress in plants. High levels of cytosolic calcium ions and ABA in the root ensure that the structure of cortical MTs breaks down to depolymerize. Conversely, ROS in plants rebuilds these microtubules by rearranging α-tubulin and β-tubulin to encounter the drought stress conditions [8, 13, 14].
Potato (Solanum tuberosum L.), commonly known as white potato or Irish potato, is cultivated on 19.3 million hectares globally and yields almost 400 million metric tons annually, ranking only behind rice, wheat, and maize [15]. Stress-tolerant potato cultivars are necessary to maintain the food supply despite climate change and protect global populations [16]. Although potatoes are susceptible to common stressors such as drought and late blight, leading to significant reductions in yield, they demonstrate inherent adaptability through several stress-response mechanisms, such as salinity adaptation and antioxidant production. The formin gene family members have been identified in various economically valuable plant species, such as 25 and 34 in wheat (Triticum aestivum L.) [17] and soybean (Glycine max. L.) [18], respectively. However, formin genes have not been investigated in the genome of potato (S. tuberosum L.).
This study characterized 26 StFH genes, revealing their physical and chemical properties, phylogenetic comparison, genomic evolution, gene structure, conserved domain, motifs, gene duplication, chromosome mapping, subcellular localization, cis-acting regulatory elements, tissue-specific expression, and RNA-seq data analysis under drought stress condition. The findings from this study will build the basis for functional investigations on the StFH genes and offer excellent opportunities to improve this crop species with drought-tolerant traits in future breeding programs.
2.0 Materials and methods
2.1 Database searching and retrieval of formin proteins in the potato genome
The A. thaliana formin DNA-binding domains were used to retrieve gene-encoded formin proteins in the potato (S. tuberosum) genome using the BlastP version 2.15.0 (Protein-basic local alignment search tool) program at Phytozome v13 (https://phytozome-next.jgi.doe.gov/) [19] (S1 Data). The comparison matrix (BLOSUM62), an expected (E) threshold value of -1, and other default parameters were employed. The Pfam database as Hidden Markov Models (HMMER) [20], Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) [21], and NCBI Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) [22] were used to analyze the presence of the FH2 conserved domain with default parameters. StFH proteins containing FH2 domains were renamed according to their chromosomal positions.
2.2 Determination of physio-chemical properties for StFH
The physiochemical characteristics of StFH proteins, including the number of amino acids (A.A.), molecular weight, isoelectric points (pI), and grand average hydrophilicity (GRAVY), were determined in the ProtParam online program (http://web.expasy.org/protparam/) [23].
2.3 Phylogenetic relationship analysis between ZmFH, OsFH, AtFH, MtFH, LjFH, and StFH
The evolutionary phylogenetic tree of FH2 proteins between Zea mays, Oryza sativa, Arabidopsis thaliana, Medicago truncatula, Lotus japonicas, and Solanum tuberosum was constructed (S2 Data). The sequences were aligned using the MEGA version 11.0.10 software (https://www.megasoftware.net/) [24]. The ClustalW [25] program was employed to align amino acid sequences. The Maximum Likelihood (ML) method was used with default parameters and a significant 1000 bootstrap value to strengthen branch values [26]. The FASTA format of the phylogenetic tree was further processed using IQ Tree version 1.6.12 with default parameters (http://www.iqtree.org/) [27]. The finalized phylogenetic tree was uploaded to iTOL v6.7.4 (https://itol.embl.de/) for an attractive visual representation [28].
2.4 Gene structure analysis of StFH
CDS (coding sequence) and genomic DNA sequences were extracted in the FASTA format from Phytozome v13.0 to elucidate the structure of the StFH genes using the Gene Structure Display Server (GSDS v2.0) (http://gsds.cbi.pku.edu.cn/) [29] (S3 and S4 Data).
2.5 Conserved domain and motif analysis of StFH
The DOG2.0 software was used to demonstrate the conserved domain of StFH [30]. The specific position details of the FH2 domain sequence were collected through MOTIF-searching protein sequence motifs (https://www.genome.jp/tools/motif/) [31]. To analyze the structural motifs of the StFH, Multiple EM for Motif Elicitation (MEME) tools (https://meme-suite.org/meme/tools/meme) was employed with a maximum of 20 motifs and other default parameters [32]. The structural pattern of motifs was visualized in TB tools v.2.010 [33].
2.6 The evolutionary Ka/Ks ratio analysis of StFH
The Ka (non-synonymous) and Ks (synonymous) substitution ratios of StFH were computed in the Ka/Ks calculation tool (https://services.cbu.uib.no/tools/kaks) [34]. Specifically, the MCScanX tool of TB tools v.2.010 was employed on the StFH CDS of duplicated genes. The Ka/Ks ratio was used to determine the rates of molecular evolution for each pair of paralogous genes, providing insights into the selective pressures acting on these duplicated sequences. Additionally, the duplication and time of divergence for the StFH genes were measured in million years ago (MYA) using the formula T = Ks/2x, where x was equal to 6.56×10−9 and MYA = 10−6 [35].
2.7 Collinearity and synteny analysis of StFH
The collinear relationships of StFH genes were analyzed based on gene duplication events. Additionally, the syntenic relationships between S. tuberosum and A. thaliana, Z. mays, and O. sativa were analyzed using MCScanX in TB tools v.2.010. Both analyses were visualized with TB tools v.2.010.
2.8 Chromosomal localization and duplication analysis of StFH
The details on chromosomes were extracted from Phytozome v13, and the distribution of StFH genes throughout the chromosomes was mapped using the MapGene2Chrom web v2 (MG2C) web server (http://mg2c.iask.in/mg2c_v2.0/) [36]. Segmental and tandem duplicated gene pairs were analyzed in the distributed chromosomes.
2.9 Distribution of StFH on multiple sub-genomes of S. tuberosum
The chromosomal lengths, as well as the start, and end points for the three sub-genomes of S. tuberosum, including S. tuberosum cv. Otava (autotetraploid), S. tuberosum group Phureja DM 1–3516 R44 (doubled monoploid), and S. tuberosum group Tuberosum RH89-039-16 (heterozygous diploid), were obtained from the SpudDB database version 6.1 (http://spuddb.uga.edu/) [37]. The distribution of StFH genes in these sub-genomes was visualized using the MapGene2Chrom web v2 (MG2C) web server.
2.10 Subcellular localization analysis of StFH
The subcellular localization of StFH was analyzed in the Wolf PSORT online tool (https://wolfpsort.hgc.jp/) [38]. The predicted results were visualized using the RStudio 2023.06.1 version [39].
2.11 Cis-acting regulatory elements (CAREs) analysis of StFH
The 2000 bp from the 5ʹ untranslated region (5ʹ UTR) of the StFH gene was collected from the Phytozome v13 (S5 Data). The predictions of CAREs were conducted in the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [40]. The predicted CAREs were further classified and visualized as a heatmap in TB tools v.2.010.
2.12 Gene ontology (GO) analysis of StFH
GO analysis was performed by obtaining GO IDs from the Plant Transcriptional Regulatory Map database (PlantRegMap; https://plantregmap.gao-lab.org/go.php) using the p-value of 0.01 and other default parameters [41]. The ChiPlot online tool (https://www.chiplot.online/) was used to illustrate the categorization and functions of StFH genes [42].
2.13 Transcription factors (TF) analysis of StFH
The analysis of TF was conducted in the PlantRegMap database (https://plantregmap.gao-lab.org/binding_site_prediction.php) using the threshold p-value 1.0E-4 and other default parameters. RStudio version 2023.06.1 was used to visualize and categorize the TFs family.
2.14 Regulatory network between TFs and StFH
Cytoscape v3.10.0 was used to visualize interactions between major TF families and respective StFH genes [43].
2.15 Prediction of putative micro-RNAs (miRNAs) and networks targeting StFH
Potential regions targeted by miRNAs in StFH genes were identified using miRNA sequences of S. tuberosum from miRBase (https://mirbase.org/) [44]. The CDS of StFH genes were uploaded to the psRNATarget Server 18 (https://www.zhaolab.org/psRNATarget/analysis?function=2), using default parameters to predict potential miRNA interactions with StFH genes [45]. Cytoscape version 3.10.0 was used to construct and visualize the interaction network between the predicted miRNAs and the StFH genes targeted by miRNA.
2.16 Protein-protein interaction (PPI) of StFH
The web tool string version 12 (https://string-db.org/) was utilized to predict and construct the PPI network of StFH proteins based on homologous proteins in A. thaliana [46]. The parameters were as follows: network type-full STRING network, network edges evidence-meaning, active interaction source-text mining, experiments, databases, co‑expression, neighborhood, gene fusion, and co‑occurrence. The minimum required interaction score was defined as a medium confidence parameter (0.4). For the first shell, a maximum of 10 interactions were displayed, and the second shell was left blank. Network display options were enabled with a 3D bubble design.
2.17 Tissue-specific expression of StFH
The SpudDB database was used to extract the tissue-specific expression of the S. tuberosum group, Tuberosum RH89-039-16. The data consisted of 14 different tissues and were classified into four groups; floral (stamen and flower), leaf (leaf, petiole), stolon/tuber (stolon, tuber sprout, tuber peel, tuber pith, tuber cortex, young tuber, and mature tuber), and other tissues (stem, shoot apex, and root) [47]. Consequently, the PPM (parts per million) value greater than >0.2 is considered as an expressed gene. TB tools v.2.010 was used to illustrate the expression patterns.
2.18 Expression pattern of StFH under drought stress
The RNA-seq data was composed of two potato varieties; Atlantic (a drought-sensitive variety) and Qingshu No. 9 (a drought-resistant variety). The StFH genes were analyzed under drought stress in early flowering, full blooming, and flower falling states at three different time points (25, 50, and 75 days, respectively). The sequence read archive (SRA) at NCBI under the bio-project ID PRJNA541096 was extracted [48]. The trimmomatic package version 0.32 was utilized for quality control and trimming of the RNA-seq data [49]. Besides, RNA-seq data was aligned to the S. tuberosum reference genome using the STAR package version 2.7.11b [50]. Conversion of sequence alignment map (SAM) files to binary alignment map (BAM) files, sorting, and arrangement were performed using the samtools version 1.20 packages [51]. The fragments per kilobase million (FPKM) value was calculated using the RSEM package v1.1.17 [52]. The FPKM value greater than >0.2 was considered as an expressed gene. TB tools v.2.010 was employed for generating a heatmap to visually represent the expression patterns of StFH under drought stress.
3.0 Results
3.1 Determination of physio-chemical properties of StFH proteins
The physio-chemical properties of StFH indicated a significant variability in A.A., ranging from 79 A.A. (StFH14) to 1746 A.A. (StFH8), with an average length of 912.5 A.A. (Table 1). The encoded StFH proteins showed diverse molecular weights, ranging from 8881.02 kDa (StFH14) to 186282.23 kDa (StFH8). The pI measurements indicated a distinct acidic nature in 13 StFHs (StFH8, StFH9, StFH13, StFH14, StFH17, StFH19, StFH20, StFH21, StFH22, StFH23, StFH24, StFH25, and StFH26) with pI<7.0, whereas 13 others (StFH1, StFH2, StFH3, StFH4, StFH5, StFH6, StFH7, StFH10, StFH11, StFH12, StFH15, StFH16, and StFH18) exhibited an alkaline nature with pI>7.0. Moreover, 22 StFH proteins (StFH1, StFH2, StFH3, StFH4, StFH5, StFH6, StFH7, StFH8, StFH9, StFH10, StFH11, StFH12, StFH15, StFH16, StFH17, StFH18, StFH19, StFH20, StFH22, StFH24, StFH25, StFH26) with values exceeding instability index 40, indicating potential structural instability, whereas four proteins (StFH13, StFH14, StFH21, StFH23) exhibited lower instability index values less than 40.0. The aliphatic index exhibited significant diversity among the StFH genes, with StFH13 having the highest value of 107.03 and StFH26 having the lowest index of 66.79. The hydrophilic nature of all the StFH proteins was confirmed by the consistently negative GRAVY scores.
Table 1. List of 26 StFH proteins and their basic physio-chemical characterization.
| Gene name | Gene identifier | Size (A.A.) | Mass (kDa) | pI | Instability index | Aliphatic index | Grand average of hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|
| StFH1 | Soltu.DM.01G039360 | 900 | 99423.69 | 8.02 | 57.12 | 84.46 | -0.343 |
| StFH2 | Soltu.DM.02G027760 | 886 | 97885.23 | 9.11 | 55.73 | 75.15 | -0.606 |
| StFH3 | Soltu.DM.03G005470 | 881 | 97837.35 | 9.26 | 51.69 | 77.80 | -0.546 |
| StFH4 | Soltu.DM.03G027630 | 755 | 84258.78 | 8.13 | 53.26 | 75.92 | -0.583 |
| StFH5 | Soltu.DM.03G029790 | 1523 | 164989.25 | 7.94 | 66.52 | 66.79 | -0.556 |
| StFH6 | Soltu.DM.05G007060 | 800 | 87394.25 | 9.33 | 46.95 | 80.71 | -0.386 |
| StFH7 | Soltu.DM.05G018890 | 923 | 100495.71 | 8.57 | 45.06 | 80.92 | -0.453 |
| StFH8 | Soltu.DM.06G025740 | 1746 | 186282.23 | 6.88 | 78.20 | 68.14 | -0.538 |
| StFH9 | Soltu.DM.06G033180 | 944 | 103548.17 | 6.41 | 68.18 | 74.87 | -0.449 |
| StFH10 | Soltu.DM.07G000540 | 1470 | 160849.62 | 7.64 | 63.22 | 77.34 | -0.447 |
| StFH11 | Soltu.DM.07G018190 | 931 | 102161.23 | 8.95 | 60.06 | 79.88 | -0.441 |
| StFH12 | Soltu.DM.07G018220 | 107 | 12557.53 | 7.66 | 56.44 | 89.35 | -0.328 |
| StFH13 | Soltu.DM.07G018270 | 91 | 10140.41 | 4.64 | 37.20 | 107.03 | -0.226 |
| StFH14 | Soltu.DM.07G018320 | 79 | 8881.02 | 4.21 | 37.71 | 97.47 | -0.249 |
| StFH15 | Soltu.DM.07G018330 | 210 | 24194.69 | 8.20 | 52.41 | 86.38 | -0.280 |
| StFH16 | Soltu.DM.07G025400 | 880 | 96597.43 | 8.79 | 50.09 | 80.85 | -0.487 |
| StFH17 | Soltu.DM.08G006050 | 755 | 84664.51 | 6.22 | 55.20 | 86.54 | -0.296 |
| StFH18 | Soltu.DM.08G021110 | 1329 | 144466.43 | 8.17 | 66.32 | 75.38 | -0.458 |
| StFH19 | Soltu.DM.09G015150 | 983 | 107851.80 | 6.93 | 74.05 | 80.53 | -0.393 |
| StFH20 | Soltu.DM.10G001740 | 920 | 100485.36 | 6.46 | 48.47 | 79.75 | -0.432 |
| StFH21 | Soltu.DM.10G001770 | 50 | 54435.60 | 6.44 | 35.87 | 84.12 | -0.378 |
| StFH22 | Soltu.DM.10G001820 | 150 | 16612.90 | 6.74 | 41.54 | 81.33 | -0.538 |
| StFH23 | Soltu.DM.10G001850 | 382 | 42384.39 | 5.06 | 33.12 | 87.09 | -0.318 |
| StFH24 | Soltu.DM.10G001880 | 402 | 44744.22 | 5.38 | 51.21 | 86.37 | -0.301 |
| StFH25 | Soltu.DM.12G019570 | 944 | 103983.11 | 6.07 | 59.23 | 80.95 | -0.355 |
| StFH26 | Soltu.DM.12G027420 | 1324 | 147544.15 | 6.27 | 59.37 | 84.58 | -0.387 |
3.2 Phylogenetic relationship analysis between ZmFH, OsFH, AtFH, MtFH, LjFH, and StFH
The phylogenetic relationship analysis included a total of 117 formin proteins; 26 StFH, 21 AtFH, 20 ZmFH, 19 MtFH, 17 OsFH, and 14 LjFH, which were categorized into seven distinct groups: group A (19 proteins), group B (9 proteins), group C (14 proteins), group D (31 proteins), group E (7 proteins), group F (15 proteins), and group G (22 proteins) (Fig 1). The distribution of StFH proteins within these groups was as follows: group A (3 proteins)-StFH5, StFH8 and StFH18, group B (2 proteins)-StFH10, and StFH26, group C (2 proteins)-StFH4 and StFH6, group D (12 proteins)-StFH7, StFH11, STFH12, StFH13, StFH14, StFH15, StFH16, StFH20, StFH21, StFH22, StFH23, and StFH24, group E (1 protein)-StFH1, group F (2 proteins)-StFH2 and StFH3, and group G (4 proteins)-StFH9, StFH17, StFH19, and StFH25 (S6 Data).
Fig 1. Phylogenetic relationship between StFH, AtFH, ZmFH, MtFH, LjFH, and OsFH.
StFHs were classified into seven groups (A, B, C, D, E, F, and G). Different colors and shapes mark each gene family, with StFH labeled with red stars, AtFH with rectangular blue rectangles, ZmFH with dark yellow triangles, MtFH with green ellipses, OsFH with red-violet circles, and LjFH with purple rectangles.
3.3 Gene structure analysis of StFH
The structural diversity of StFH genes was analyzed by comparing introns and exons based on their distribution patterns. The group A members (StFH5, StFH8, and StFH18), each containing 17 exons, shared similar structural patterns (Fig 2 and S7 Data). Meanwhile, group B members (StFH10 and StFH26) exhibited structural similarity with group A, with StFH26 having the highest numbers of exons and introns (19 and 18, respectively). The group C members (StFH4 and StFH6) exhibited the lowest numbers of exons and introns but had similar structural patterns. Whereas, group D, containing 12 genes, exhibited the highest structural complexity with a total of 59 exons and 52 introns. But group E contained only StFH1, with four exons and three introns. Besides, group F exhibited distinct structures, with StFH2 having four exons and three introns, while StFH3 has five exons and four introns. Similarly, group G members (StFH9, StrFH17, StFH19, and StFH25) shared a consistent pattern of four exons and three introns.
Fig 2. The structure of StFH genes.
The grouping and colors of the StFH members were based on the phylogenetic tree. Black lines represent introns, deep blue represents exons, numbers 0, 1, and 2 represent intron phases, and deep yellow lines represent upstream/downstream.
3.4 Conserved domain and motif analysis of StFH
The analysis of conserved domains revealed the presence of the FH2 domain across all 26 StFH proteins (Fig 3). The motif analysis uncovered unique patterns across different StFH groups. The groups C (StFH4), G (StFH9, StFH19, and StFH25), and F (StFH3) each contained 11 motifs (Fig 4). While groups F (StFH2) and D (StFH11) both contained 12 motifs. Moreover, groups A members (StFH8, and StFH18) and B members (StFH10) each contained 14 motifs. StFH5 (group A) had 15 motifs, while StFH26 (group B) had 13 motifs. However, StFH1, the only member of group E, contained 13 motifs. Furthermore, other group D members displayed a variety of motifs, indicating structural diversity and genomic differences among them.
Fig 3. Feature domains of StFH.
Positions of the FH2 conserved domain and other domains are demonstrated in light blue color, whereas the entire protein sequence of the respective StFH is shown in purple.
Fig 4. The distribution of conserved motifs in StFH.
Each motif was illustrated by a color box aligned on the right side of the figure. Different colors indicate individual motifs within each StFH protein.
3.5 The evolutionary Ka/Ks ratio analysis of StFH
The estimated Ka values for the StFH genes ranged from 0.147052693 to 0.219661933, while the Ks values varied from 0.248696445 to 1.13504586 (Fig 5 and S8 Data). None of the StFH gene pairs were undergoing positive selection as their Ka/Ks ratio is less than 1. Gene pairs such as StFH5-StFH8 (0.219168631), StFH26-StFH10 (0.3238209), StFH2-StFH3 (0.234076034), StFH25-StFH17 (0.193526923), StFH9-StFH19 (0.25279716), StFH21-StFH24 (0.591293908), and StFH14-StFH12 (0.661554621) were all undergoing purifying selection.
Fig 5. Estimation of gene duplication time and Ka/Ks analysis of StFH.
The ratio of Ka to Ks is represented by Ka/Ks, with divergence time (measured in million years ago, MYA) also indicated. The color bar represents the data range.
3.6 Collinearity relationship analysis of StFH
A close relationship among the StFH genes was demonstrated through collinear analysis (Fig 6). Seven collinear pairs were identified within the StFH gene family. Chromosome 3 of StFH5 formed a pair with chromosome 6 of StFH8. Chromosome 2 of the StFH2 interacted with chromosome 3 of the StFH3. StFH10, located on chromosome 7, is paired with StFH26 on chromosome 12. StFH17 on chromosome 8 formed a collinear pair with StFH25 on chromosome 12. StFH9 on chromosome 6 interacted with StFH19 on chromosome 9. Additionally, StFH12 and StFH14 formed a pair on chromosome 7, and StFH21 and StFH24 were also observed forming collinear pairs on chromosome 10.
Fig 6. The collinearity analysis of the StFH gene family.
Different color rectangles represent chromosomes 1 to 12 in StFH. The dark blue lines linking chromosomes represent collinear relationships between them.
3.7 Synteny relationship analysis of StFH
The syntenic comparison of S. tuberosum (dicotyledon) was further examined with two monocotyledons, Z. mays and O. sativa, and one dicotyledon, A. thaliana (Fig 7). The results uncovered the close syntenic association of S. tuberosum (12 chromosomes) with A. thaliana (5 chromosomes). They shared 11 gene pairs on distinct chromosomes, indicating similarity in biological and physiological functions. On the contrary, among the monocotyledon species, S. tuberosum showed more similarity in evolutionary relations with 7 gene pairs with Z. mays (9 chromosomes) compared to 5 gene pairs with O. sativa (12 chromosomes). Thus, it contributed to understanding ancestry and genomic relations.
Fig 7. The synteny analysis of S. tuberosum with A. thaliana, O. sativa, and Z. mays.
The green, red, blue, and magenta-colored chromosome rectangles represent S. tuberosum, A. thaliana, O. sativa, and Z. mays. The dark blue color indicates a syntenic relationship between the species.
3.8 Chromosomal localization and duplication analysis of StFH
The 26 StFH genes were distributed on ten chromosomes (Fig 8). Single StFH genes (StFH1, StFH2, and StFH19) were located on chromosomes 1, 2, and 9, respectively. The remaining 23 StFH genes were located on chromosome 3 (StFH3, StFH4, and StFH5), chromosome 5 (StFH6 and StFH7), chromosome 6 (StFH8 and StFH9), chromosome 7 (StFH10, StFH11, StFH12, StFH13, StFH14, StFH15, and StFH16), chromosome 8 (StFH17 and StFH18), chromosome 10 (StFH20, StFH21, StFH23, and StFH24), and chromosome 12 (StFH25 and StFH26). However, chromosomes 4 and 11 did not contain any StFH genes. Meanwhile, two tandem and five segmental duplications were identified. The five identified segmental pairs were; StFH8-StFH5, StFH26-StFH10, StFH2-StFH3, StFH25-StFH17, and StFH9-StFH19. The two tandem pairs were StFH21-StFH24 and StFH12-StFH14.
Fig 8. The chromosomal locations and duplications of StFH.
The chromosome-scale is in millions of bases (Mb), showing the length of each chromosome on the left. The chromosomes are colored yellow. The blue lines indicate segmental duplications and the red stars represent tandem duplications.
3.9 Distribution of StFH on multiple sub-genomes of S. tuberosum
The StFH genes were distributed randomly across different chromosomes in S. tuberosum cv. Otava (autotetraploid), cv. Phureja DM 1–3516 R44 (doubled monoploid), and cv. Tuberosum RH89-039-16 (heterozygous diploid) (Fig 9). The cultivar Otava contained 22 StFH in 10 chromosomes, with chromosome 7 containing the highest number (8) of genes. Meanwhile, cultivar Phureja DM 1–3516 R44 had 23 StFH in 9 chromosomes, with a maximum number of 6 genes located in chromosome 7. The cultivar Tuberosum RH89-039-16 comprised 40 StFH distributed in 18 chromosomes. However, most of the three sub-genomic chromosomes in tuberosum contained one or two genes.
Fig 9. The distribution of StFH genes on multiple sub-genomes of S. tuberosum.
The Otava, Phureja DM 1–3516 R44, and Tuberosum RH89-039-16 chromosomes are purple, green, and red, respectively. The chromosome-scale is in millions of bases (Mb).
3.10 Prediction of the subcellular localization of StFH
The subcellular localization analysis revealed that StFH protein signals were present in 11 different organelles, including the nucleus, mitochondria, chloroplast, cytoplasm, cytoskeleton, Golgi apparatus, peroxisome, vacuole, endoplasmic reticulum (ER), plasma membrane (PM), and extracellular membranes. StFH9 signals were detected at the highest number of sites, including all 11 sites of chloroplasts (Fig 10A). The protein signals were most abundant in the chloroplast (69.23%) and plasma membrane (57.69%), followed by the vacuole (53.84%) and nucleus (50%). Significant quantities of StFH protein signals were also found in the ER (46.15%), cytoplasm (42.3%), mitochondria (38.46%), extracellular cytoskeleton (30.76%), and Golgi body (26.92%). The peroxisome had the lowest concentration (3.84%) of StFH signals (Fig 10B). The bubble plot illustrated the redundancy of a particular StFH gene in specific organelles (S1 Fig).
Fig 10. Sub-cellular localization analysis of StFH.
(A) The heat map represents the sub-cellular localization analysis of StFH proteins. The names of each StFH protein are shown on the left side of the heat map, while the names of the respective cellular organelles are shown at the bottom. The intensity of the color indicates the presence of protein signals corresponding to the genes. The cellular organelles include nuclear, mitochondrial, cytoplasmic, chloroplast, cytoskeletal, peroxisomal, Golgi, vacuole, endoplasmic reticulum (ER), plasma membrane (PM), and extracellular locations. (B) The percentage distribution of StFH protein signals across various cellular organelles is represented by a bar diagram. The percentages of protein signals in different cellular organelles are shown on the left side of the diagram.
3.11 Cis-acting regulatory elements (CAREs) analysis in the promoters of StFH
The analysis of CAREs revealed 54 elements, with the most abundant category being Box 4, related to light responsiveness. CAREs were classified into four categories based on their functional regulation: light responsiveness, tissue-specific expression, phytohormone responsiveness, and stress responsiveness (Fig 11 and S9 Data). The largest category of CAREs related to light responsiveness included 26 elements such as 3-AF1 binding site, AAAC-motif, ACA-motif, ACE, AE-box, AT1-motif, ATC-motif, ATCT-motif, Box 4, Box II, CAG-motif, chs-CMA1a, chs-CMA2a, GA-motif, Gap-box, GATA-motif, G-Box, GT1-motif, GTGGC-motif, I-box, LAMP-element, MRE, Sp1, TCCC-motif, and TCT-motif. The tissue-specific expression CAREs included 13 elements, phytohormone responsiveness included 12 elements, and stress responsiveness included 4 elements.
Fig 11. A heatmap represents the distribution of putative CAREs on the 2.0 kb promoter region of StFH.
The names of the CAREs of each StFH gene are shown on the left side of the heat map. The number of putative CAREs for each StFH gene is displayed on the right side of the heat map and represented by different colors (black = 0, green = 1–5, yellow = 6–10). Functions associated with CAREs of the corresponding genes, such as light responsiveness, tissue-specific expression, phytohormone responsiveness, and stress responsiveness, are shown at the bottom of the heatmap and labeled green, red, blue, and magenta, respectively.
3.12 Gene ontology (GO) analysis of StFH
74 GO annotations for StFH genes, were classified into three categories: biological process, cellular component, and molecular function. Biological processes were the most predominant, with 63 GO terms identified, including a range of p-values from 2.4E-11 to 0.00561 (Fig 12 and S10 Data). The cellular component category encompassed six GO functions with p-values ranging from 5.50E-6 to 0.00775. The molecular function category included five GO functions with p-values between 4.40E-8 and 0.00029. The most abundant GO annotation in biological processes was "actin filament organization, polymerization, and regulation". In the context of cellular functions, the "membrane part" (GO:0044425; p-value: 0.00731) was strongly associated with StFH genes. The least number of GO annotations were linked to cellular function, specifically involving "macromolecular complex binding" (GO:0044877; p-value: 0.00029).
Fig 12. StFH gene function analysis through gene ontology.
The classification of the StFH gene function is shown on the right side of the circos plot. The number of genes involved under a specific GO ID, expected value, and rich factor are shown in distinctive colors. The scaling of the -log10 p-value is shown in three distinctive colors (red, yellow, and green).
3.13 Transcription factors (TFs) analysis of StFH
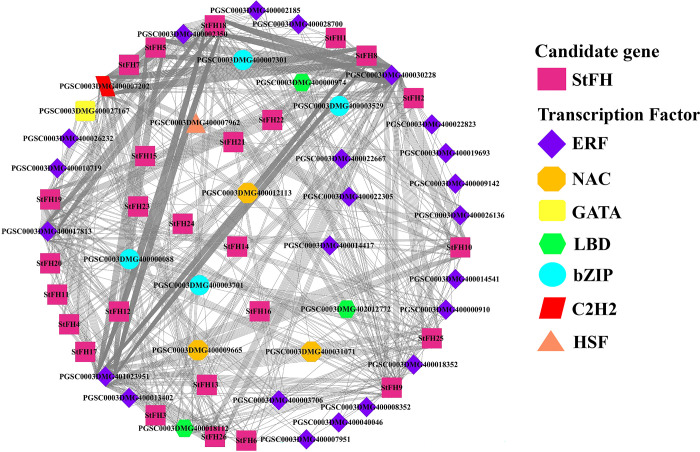
Thirty-six unique TFs regulating the StFH were identified and categorized into seven families: bZIP, C2H2, ERF, GATA, LBD, NAC, and HSF (Fig 13). The ERF family was the largest, comprising 26 TFs (PGSC0003DMG401023951, PGSC0003DMG400018352, PGSC0003DMG400000910, PGSC0003DMG400013402, PGSC0003DMG400009142, PGSC0003DMG400030228, PGSC0003DMG400017813, PGSC0003DMG400002350, PGSC0003DMG400026232, PGSC0003DMG400022305, PGSC0003DMG400014541, PGSC0003DMG400014417, PGSC0003DMG400010719, PGSC0003DMG400003706, PGSC0003DMG400022667, PGSC0003DMG400026136, PGSC0003DMG400007951, PGSC0003DMG400019693, PGSC0003DMG400002185, PGSC0003DMG400028700, PGSC0003DMG400022823, PGSC0003DMG400008352, PGSC0003DMG400040046). The bZIP family was the second largest. with four TFs (PGSC0003DMG400007301, PGSC0003DMG400003529, PGSC0003DMG400003701, PGSC0003DMG400000088). The LBD and NAC families each included three TFs (PGSC0003DMG400018112, PGSC0003DMG400000974, PGSC0003DMG402012772) and (PGSC0003DMG400012113, PGSC0003DMG400009665, PGSC0003DMG400031071), respectively, while the C2H2 and GATA families had one TF (PGSC0003DMG400007202 and PGSC0003DMG400027167, respectively).
Fig 13. A heatmap illustrates the TFs regulating StFH.
The StFH proteins are on the left, and TF names are at the bottom of the heat map. The color intensity on the right side of the heat map indicates the presence of TFs corresponding to the proteins. Distinctive colors represent the seven TF families: ERF (red), bZIP (orange), LBD (blueberry), NAC (magenta), C2H2 (light green), GATA (sky blue), and HSF (dark green).
3.14 Regulatory network between TF and StFH
All 36 TF members interact with both the 26 StFH genes and each other, revealing a complex network of interactions (Fig 14). The ERF family was prominent, interacting with 20 StFH genes, excluding StFH12, StFH13, StFH14, StFH15, StFH22, and StFH24. The bZIP family interacted only with the StFH25 gene. The C2H2 family interacted with 17 StFH genes, excluding several others. The HSF, NAC, LBD, and GATA families interacted with one (StFH15), four (StFH12, StFH13, StFH14, and StFH22), five (StFH5, StFH6, StFH18, StFH19, and StFH26), and fourteen StFH genes (StFH1, StFH3, StFH4, StFH5, StFH6, StFH8, StFH9, StFH10, StFH11, StFH17, StFH18, StFH19, StFH25, and StFH26), respectively.
Fig 14. The regulatory network between TF and StFH.
Different colors and shapes represent TFs and their interactions with StFH genes. The StFH genes are shown in magenta rectangles, and the TF families are represented by various shapes and colors: ERF (purple diamond), NAC (dark yellow hexagon), GATA (light yellow round rectangle), LBD (lime green hexagon), bZIP (bright aqua ellipse), C2H2 (red parallelogram), and HSF (pastel orange triangle).
3.15 Prediction of potential miRNAs targeting StFH
In this analysis, 170 putative miRNAs targeting 24 StFH genes were identified. The analysis revealed 60 unique miRNA sequences, with stu-miR156, stu-miR162, stu-miR167, and stu-miR172 being the most abundant (Fig 15A and 15B and S11 Data). These miRNAs targeted various StFH genes, with StFH8, StFH10, StFH17, StFH18, and StFH26 being the most frequently targeted.
Fig 15. Prediction of potential miRNAs targeting StFH.
(A) The network illustration of predicted miRNA targets shows StFH genes in blue-green and miRNAs in light pink ellipses. (B) The schematic diagram indicates the StFH genes targeted by miRNAs, with exons (purple), UTR (light green), introns (black lines), and miRNAs (red rectangles).
3.16 Protein-protein interactions (PPI) of StFH
The PPI analysis uncovered a strong connection between the 26 StFH proteins and those from Arabidopsis, revealing meaningful associations in a biological context (Fig 16 and S12 Data). Specifically, among the StFH proteins, StFH11, StFH14, StFH15, StFH13, StFH16, StFH7, StFH24, StFH22, StFH23, StFH20, and StFH21 exhibited homology with AtFH5, which interacted with FH20, PRF1, PRF2, PRF3, PRF4, and PRF5. Notably, AtFH1 displayed homology with four StFH proteins, namely StFH12, StFH17, StFH25, and StFH9, and interacted with FH14, PRF1, PRF2, PRF3, PRF4, PRF5, and FH13. Furthermore, StFH10 and StFH26 demonstrated homology with AtFH13, which interacted with FH1, PRF2, PRF1, PRF4, and FH5. Meanwhile, AtFH6 exhibited homology with StFH2 and StFH3, and interacted with PRF2, PRF3, PRF4, and PRF5. Additionally, AtFH20 showed homology with StFH5 and StFH8, and AtFH4, AtFH8, AtFH11, and AtFH14 were found to be homologous with StFH6, StFH4, StFH1, and StFH18, respectively. The width or boldness, of the connecting lines between proteins, serves as an indicator of the strength or frequency of their interactions. A broader line connecting two proteins signified a higher interaction ratio, suggesting a more robust and frequent association between the respective proteins. This observation suggested that these StFH proteins played similar roles in biological processes through their interactions with Arabidopsis proteins, highlighting potential functional similarities or involvement in cellular activities.
Fig 16. Protein-protein interaction of StFH based on known Arabidopsis proteins.
The network nodes represent proteins, and the line colors indicate different data sources.
3.17 Tissue-specific expression of StFH
The tissue-specific expression analysis revealed similar expression levels in floral tissues. In the stamen, StFH7, StFH16, and StFH18 were highly expressed compared to other StFH genes (Fig 17). Additionally, StFH2, StFH7, StFH16, StFH18, and StFH19 showed higher expression in flowers (S13 Data). The expression levels in leaf tissues were relatively consistent, with StFH2, StFH17, StFH18, and StFH19 being actively expressed. In petiole tissue, StFH2, StFH18, StFH19, and StFH26 exhibited higher expression levels. The expression levels of StFH genes in tuber tissues were similar, with the highest expression observed in the stolon and tuber cortex (88%). Specifically, StFH1, StFH9, StFH18, StFH19, and StFH25 were highly expressed in the stolon. Moreover, StFH2, StFH9, StFH18, StFH19, StFH25, and StFH26 displayed similar expression patterns in six tuber tissues (tuber sprout, peel, pith, cortex, young, and mature tuber). StFH expression in other tissues such as stem, shoot apex, and root was also similar, with StFH2, StFH3, StFH9, StFH10, StFH18, StFH19, and StFH25 showing high levels of expression.
Fig 17. Tissue-specific expression pattern of StFH.
The respective StFH gene names are shown on the right side of the heat map. Various tissues are represented at the bottom of the heat map. The color gradient from green to red indicates the expression levels on the right side of the heat map.
3.18 Expression pattern of StFH under drought stress
The expression patterns of StFH genes were not similar in the drought-sensitive Atlantic variety and the drought-resistant Qingshu No. 9 variety. However, only nine StFH genes-StFH1, StFH2, StFH5, StFH10, StFH17, StFH19, StFH21, StFH25, and StFH26 were up-regulated, while most were down-regulated in Qingshu No. 9 at 25 days after early flowering (Fig 18). At 50 days after full flower blooming, StFH2, StFH5, StFH10, StFH17, StFH19, StFH21, and StFH25 were up-regulated (S14 Data). However, nine StFH genes showed decreased expression levels at 50 days in Qingshu No. 9, indicating down-regulation at the full blooming stage. Moreover, StFH2, StFH5, StFH10, StFH17, StFH19, StFH21, StFH25, and StFH26 were up-regulated in Qingshu No. 9 at 75 days after flower falling. In contrast, StFH9, StFH18, and StFH20 were down-regulated in Qingshu No. 9 but exhibited high levels of expression at all three stages in the Atlantic variety.
Fig 18. Expression pattern of StFH under drought stress.
The respective StFH gene names are shown on the right side of the heatmap. The stages of the plant in two varieties (Atlantic and Qingshu No. 9) are represented at the bottom of the heat map. The color gradient from white to red indicates the expression levels on the right side of the heat map.
4.0 Discussion
The formin family of proteins is widely distributed in plants and is essential to cellular growth and development [53]. The de novo production of actin filaments is regulated through actin cytoskeleton remodeling [54]. Recently, a consistent evolutionary framework has been performed within the Solanaceae family through phylogenetic analyses, which clearly define groups and relationships [55]. Genomic comparisons have revealed a significant expansion of the formin gene family in Solanaceae, particularly within the Solanum genus [54]. This expansion is driven by gene duplication events, resulting in a greater diversity of formin proteins with specialized functions. The physiochemical properties of StFH proteins exhibited significant variability in both size and mass, with their chemical nature indicating they were half acidic and half alkaline. Moreover, 22 StFH proteins are potentially unstable, as their instability index surpassed 40.0, suggesting their susceptibility to structural fluctuations [56]. Moreover, the aliphatic index indicated extensive diversity among StFH proteins. The negative GRAVY scores indicate a hydrophilic disposition and a preference for aqueous environments [57, 58]. Therefore, all the StFH proteins were hydrophilic.
The phylogenetic relationships play a crucial role in uncovering the molecular basis and evolutionary aspects of lineages, interactions, and diversity within various species [59]. This analysis revealed the patterns and rates of evolution for diverse species. The findings revealed significant clustering patterns, demonstrating that a substantial portion of StFH proteins formed clusters closely associated with AtFH and MtFH. The gene organization demonstrated that many introns allow for alternative splicing, which can be linked to specific and significant biological roles [60, 61]. Thus, StFH26 was involved in alternative splicing. Besides, genes with higher expression levels tend to possess longer introns, distinguishing them from low-expression genes, and reduced exon numbers were often marked as early-responsive genes [62, 63]. Therefore, StFH4, StFH6, and StFH13 genes exhibited reduced exon numbers, marking them as early response genes, aligning with their higher activation potential.
The FH2 domain predominantly plays a crucial role in cellular processes, particularly in the regulation of actin dynamics [64, 65]. Thus, the presence of the FH2 domain in all the StFH proteins suggested that they might have a role in cytoskeleton regulation. A conserved motif is a repeated and evolutionary preserved sequence pattern in proteins with vital biological functions [66]. The identified motifs in the StFH showed differences between groups while being relatively similar within the same group. For instance, the eleven motifs of StFH9 and StFH19 in group G were quite similar, suggesting their potential involvement in relevant biological processes. In contrast, other groups exhibited variations in these motif sets [67]. The Ka/Ks ratios among StFH gene pairs were less than 1, suggesting a purifying selection process within the gene family. Moreover, the evolutionary time divergence of StFH gene pairs revealed that they emerged between 18.96 to 86.51 MYA (million years ago) [68].
Chromosomal localization provides mapping of genes to specific locations on chromosomes. Moreover, OsFH genes were distributed on nine chromosomes [69]. Consequently, twenty-six StFH genes with the highest number of genes observed on chromosome 7, were mapped to 10 chromosomes. Besides, the presence of the highest number of genes on chromosome 7 in the three sub-genomes of S. tuberosum also highlighted its similarity. Thus, it sheds light on genome evolution, gene function, phenotypic variation, and improvement [70]. Besides, autotetraploid plants have shown higher drought tolerance and heterozygosity compared to diploids [71, 72]. The gene duplication mechanisms create genetic redundancy, allowing one copy of a gene to evolve new functions while the other maintains its original function. Tandem, whole genome, and segmental duplications are the main mechanisms for expanding the gene families in many plant species [73, 74]. The segmental and tandem duplications were pivotal in the expansion of the StZFP gene family [75]. Thus, five segmental and two tandem duplication events observed in StFH suggest that they persisted over time, most likely due to their significant functional importance. Furthermore, the syntenic comparison of S. tuberosum with three species (two monocotyledons and one dicotyledon) demonstrated a close evolutionary relationship and a higher degree of resemblance in genomic conservation with the dicotyledon, A. thaliana.
Formin proteins are critical actin filament builders and display various subcellular localization patterns based on their particular isoforms and cellular activities [76]. Additionally, some formins, like Fmn1 and mDia1, interact with microtubules and may coordinate cytoskeleton dynamics as well as regulate gene expression [77, 78]. The subcellular localization analysis revealed that StFH protein signals were most abundant in the chloroplast and plasma membrane, followed by the vacuole and nucleus.
Generally, GO is analyzed to distinguish the functions of individual genes into three categories; biological processes, cellular components, and molecular functions [79, 80]. This study identified most of the StFH genes engaged in biological processes regarding actin filament polymerization. CAREs are DNA sequences found in gene promoter regions, controlling gene expression and binding TFs [81]. They play crucial roles in biological processes like development, adaptation, and response to environmental stresses [82]. Most of the CARE motif in StFH was related to light response. However, some stress-responsive elements were also found. The stress responsiveness elements included LTR, MBS, TC-rich repeats, and WUN-motif, responsible for managing abiotic stress such as drought [83]. In Arabidopsis, AtMYB2 and AtMYB60 were involved in drought stress [84, 85]. The TFs play crucial roles in various biological functions, specifically regulating metabolism, promoting growth, facilitating progression, providing resistance against microbial infections, and responding to both biotic and abiotic stress [86, 87]. Among the seven TF families identified in StFH, the ERF (Ethylene Response Factor) TF in potatoes explores their role in negatively regulating specific processes [88]. Whereas, bZIP (basic Leucine Zipper) TF might confer resistance against abiotic stress such as dehydration, drought, and salt [67]. Moreover, drought-responsive elements, for instance, NAC TFs, are primarily identified as binding to drought-related transposons in citrus [89]. Furthermore, the up-regulation of HSF, GTE, DREB2B, bHLH, MYB, HD-ZIP, and ERF TF families during heat and cold stress in potatoes was observed [90]. Besides, the regulatory network between TFs and StFH interacts with each other, revealing a complex network of interactions.
The pivotal role of PPIs in shaping the evolution and functionality of living organisms has been well established [91]. It provides insights into species diversification and the complex regulation of cellular functions, which allow for the coordination and communication of diverse cellular processes [92]. The StFH showed homology with Arabidopsis and interacted with the FH family (FH1, FH3, FH4, FH5, FH6, FH8, FH11, FH13, FH14, and FH20) and PRF (Profilin) family (PRF1, PRF2, PRF3, PRF4, and PRF5). The FH family proteins are primarily recognized as cytoskeletal dynamics regulators, emerging as potential actin nucleation agents. Whereas PRF, a low-molecular-weight actin-binding protein, plays a crucial role in plant development during cell elongation and division [93–95]. The role of miRNA is not confined to cellular signaling, as it has potential involvement in various abiotic stresses such as heat, salinity, low temperature, drought, and biotic stresses like viral and bacterial attack [96]. Further, 60 unique miRNAs were identified to control essential biological processes in StFH, particularly their potential role in abiotic stress management. The crucial functions of stu-miR156 in the biological processes of potatoes, such as tuberization, can lead to the formation of aerial tubers under specific conditions (Table 2). Moreover, miR156 plays a vital role in controlling different aspects, notably influencing the development of lateral roots in potatoes [97]. Besides, miR156 is induced by drought stress [98]. Besides, Stu-miR172, a phloem-mobile miRNA, plays a crucial role in sugar-dependent signal transduction pathways, influencing flower and tuber induction [99]. However, the most pivotal role of miR172 in potatoes is managing drought stress [100]. Meanwhile, stu-miR162 in potatoes regulates miRNA biogenesis, plant development, and abiotic stress responses [101]. Stu-miR167 in potatoes is involved in positively regulating nuclear factor Y subunit A (NF-YA) and flavin-binding monooxygenase family protein (YUC2), activating the auxin signaling pathway [102].
Table 2. Information about abundant miRNA ID, functions, and their targeted StFH genes.
| miRNA ID | Functions | Targeted genes |
|---|---|---|
| stu-miR156 | It influences the development of lateral roots in potatoes as well as links to the regulatory mechanisms involved in tuberization. It is enhanced by drought stress. | StFH5, StFH10, StFH11, StFH26 |
| stu-miR162 | miRNA biogenesis, plant development, and abiotic stress responses. | StFH10, StFH17, StFH26 |
| stu-miR167 | Its positively regulates nuclear factor Y subunit A (NF-YA) and flavin-binding monooxygenase family protein (YUC2), activating the auxin signaling pathway. | StFH3, StFH8, StFH10 |
| stu-miR172 | It is a sugar-dependent signal transduction pathway and influences flower and tuber induction. It also regulates the transitions between developmental stages and specifies floral organ identity. It activates under drought conditions. | StFH1, StFH5, StFH11, StFH17, StFH25 |
Tissue-specific gene expression is a core biological mechanism that allows plants to respond more efficiently to stress by stress-responsive genes in specific cells or tissues, such as roots or leaves [103, 104]. For example, certain tissues like the root (which functions as a water and nutrient uptake system and can produce extensive lateral roots and deep root hair), the stem (which transports water, nutrients, and hormones), and the shoot apex (a center for auxin biosynthesis and growth regulation) are involved in sensing and initiating signaling cascades to counteract drought [105–108]. Similarly, the expression of StRFP2 in drought-responsive tissues (root, stem, and shoot apex) was observed [109]. Therefore, the expression of StFH2, StFH3, StFH9, StFH10, StFH18, StFH19, StFH25, and StFH26 in these drought-responsive tissues indicates a key role in drought tolerance. Moreover, leaf tissues contain stomata (which are centers for CO2 and O2 gas exchange) and guard cells (which regulate the opening and closing of stomata) to counteract drought stress [110]. In potatoes, the expression of StPIP1 in leaves was found to be involved in managing drought stress [111]. Consequently, the expression of StFH1, StFH18, and StFH19 in leaf tissue suggests their importance in enhancing drought tolerance. Other tissues, such as the stolon (responsible for forming tubers) and tuber (storehouse of carbohydrates), also play potential roles in plant growth, development, and metabolism under drought stress [112, 113]. The various expressions of StFH genes in these tissues highlighted their significance. The drought-resistant Qingshu No. 9, a high-yielding variety, further supports the significance of gene expression patterns in response to drought [114, 115]. The up-regulation of StFH2, StFH5, StFH10, StFH17, StFH19, StFH21, and StFH25 underscores their important roles in managing drought stress. Specifically, StFH2, StFH10, StFH19, and StFH25 exhibited significant expression in various tissues when responding to drought stress through up-regulation. In summary, this study emphasizes the crucial role of StFH genes in conferring drought tolerance and developing new strategies for breeding programs aimed at developing drought-resistant potato varieties.
5.0 Conclusion
In this study, 26 StFH genes were identified in the potato genome, distributed in ten chromosomes. These genes show ancestry and functional resemblance to the dicotyledon Arabidopsis, revealing similarities in their gene structures, typical domains, and motifs. The observation of both segmental and tandem duplications indicates the expansion of this gene family. The divergence time and evolutionary relationship analysis suggest that StFH genes have evolved through purifying selection, maintaining functional stability. Moreover, CARE analysis revealed the binding of major TFs such as ERF, bZIP, and C2H2 in the promoter region of StFH, indicating their role in regulating gene expression under various abiotic stresses, particularly drought. Most of the StFH genes were found to perform biological functions, and their expression was regulated by miRNAs under abiotic stress conditions such as drought. The expression of StFH2, StFH3, StFH9, StFH10, StFH18, StFH19, StFH25, and StFH26 in major drought-responsive tissues such as root, stem, and shoot apex indicated their involvement in stress response. The RNA-seq data confirmed the potential role of StFH2, StFH10, StFH19, and StFH25, as these genes showed significant up-regulation in the drought-resistant variety. Overall, the findings from this study provide valuable insights into candidate gene selection, gene function validation, stress mechanisms, and the development of stress-tolerant potato cultivars for future breeding programs.
Supporting information
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(TIFF)
Acknowledgments
The opportunity to conduct this research provided by the Laboratory of Functional Genomics and Proteomics, Department of Genetic Engineering and Biotechnology, Faculty of Biological Science and Technology, Jashore University of Science and Technology, Jashore 7408, Bangladesh, is gratefully acknowledged by the authors. Gratitude is extended to Mr. Tanzir Ahmed, Assistant Professor, Department of English, Faculty of Arts and Social Science, Jashore University of Science and Technology, Jashore 7408, Bangladesh for extensively editing the manuscript to avoid grammatical errors. The valuable comments and critical suggestions for improving the quality of this manuscript provided by the reviewers and members of the editorial panel are also acknowledged and appreciated.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Aspenström PJBeBA-MCR. Formin-binding proteins: modulators of formin-dependent actin polymerization. 2010;1803(2):174–82. doi: 10.1016/j.bbamcr.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 2.Blanchoin L, Staiger CJJBeBA-MCR. Plant formins: diverse isoforms and unique molecular mechanism. 2010;1803(2):201–6. doi: 10.1016/j.bbamcr.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 3.Campellone KG, Welch MDJNrMcb. A nucleator arms race: cellular control of actin assembly. 2010;11(4):237–51. doi: 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zweifel ME, Courtemanche NJJomb. Profilin’s affinity for formin regulates the availability of filament ends for actin monomer binding. 2020;432(24):166688. doi: 10.1016/j.jmb.2020.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez RJCrib, biology m. Actin filament nucleation and elongation factors–structure–function relationships. 2009;44(6):351–66. doi: 10.3109/10409230903277340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee S, Gardel ML, Schwarz USJArocmp. The actin cytoskeleton as an active adaptive material. 2020;11:421–39. doi: 10.1146/annurev-conmatphys-031218-013231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Lian N, Zhang Y, Man Y, Chen L, Yang H, et al. The cytoskeleton in plant immunity: Dynamics, regulation, and function. 2022;23(24):15553. doi: 10.3390/ijms232415553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun HJ, Baek D, Jin BJ, Cho HM, Park MS, Lee SH, et al. Microtubule dynamics plays a vital role in plant adaptation and tolerance to salt stress. 2021;22(11):5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Zhang L-J, Huang, behavior. Cytoskeleton and plant salt stress tolerance. 2011;6(1):29–31. doi: 10.4161/psb.6.1.14202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Mao TJCoipb. Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. 2019;52:86–96. doi: 10.1016/j.pbi.2019.08.002 [DOI] [PubMed] [Google Scholar]
- 11.Ma H, Liu MJMBR. The microtubule cytoskeleton acts as a sensor for stress response signaling in plants. 2019;46(5):5603–8. doi: 10.1007/s11033-019-04872-x [DOI] [PubMed] [Google Scholar]
- 12.Hohmann T, Dehghani FJC. The cytoskeleton—a complex interacting meshwork. 2019;8(4):362. doi: 10.3390/cells8040362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Li J, Yuan MJP, physiology c. Salt tolerance requires cortical microtubule reorganization in Arabidopsis. 2007;48(11):1534–47. doi: 10.1093/pcp/pcm123 [DOI] [PubMed] [Google Scholar]
- 14.Livanos P, Galatis B, Quader H, Apostolakos PJC. Disturbance of reactive oxygen species homeostasis induces atypical tubulin polymer formation and affects mitosis in root‐tip cells of Triticum turgidum and Arabidopsis thaliana. 2012;69(1):1–21. doi: 10.1002/cm.20538 [DOI] [PubMed] [Google Scholar]
- 15.Jia W, Xiong Y, Li M, Zhang S, Han Z, Li KJFiG. Genome-wide identification, characterization, evolution and expression analysis of the DIR gene family in potato (Solanum tuberosum). 2023;14:1224015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahal K, Li X-Q, Tai H, Creelman A, Bizimungu BJFips. Improving potato stress tolerance and tuber yield under a climate change scenario–a current overview. 2019;10:563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan W-j, Liu Z-h, Bai J-f, Yuan S-h, Li Y-m, Lu F-k, et al. Comprehensive analysis of formin gene family highlights candidate genes related to pollen cytoskeleton and male fertility in wheat (Triticum aestivum L.). 2021;22:1–16. doi: 10.1186/s12864-021-07878-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Zhang Z, Shan M, Amjad Z, Xue J, Zhang Z, et al. Genome-Wide Studies of FH Family Members in Soybean (Glycine max) and Their Responses under Abiotic Stresses. 2024;13(2):276. doi: 10.3390/plants13020276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. 2012;40(D1):D1178–D86. doi: 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. 2016;44(D1):D279–D85. doi: 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I, Khedkar S, Bork PJNar. SMART: recent updates, new developments and status in 2020. 2021;49(D1):D458–D60. doi: 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. 2020;48(D1):D265–D8. doi: 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasteiger E, Hoogland C, Gattiker A, Duvaud Se, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server: Springer; 2005. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Stecher G, Kumar SJMb, evolution. MEGA11: molecular evolutionary genetics analysis version 11. 2021;38(7):3022–7. doi: 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJJNar. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZJMb, evolution. PAML 4: phylogenetic analysis by maximum likelihood. 2007;24(8):1586–91. doi: 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 27.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular biology and evolution. 2015;32(1):268–74. Epub 2014/11/06. doi: 10.1093/molbev/msu300 ; PubMed Central PMCID: PMC4271533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork PJNar. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. 2021;49(W1):W293–W6. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao GJB. GSDS 2.0: an upgraded gene feature visualization server. 2015;31(8):1296–7. doi: 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao XJCr. DOG 1.0: illustrator of protein domain structures. 2009;19(2):271–3. doi: 10.1038/cr.2009.6 [DOI] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Kawashima S, Nakaya AJNar. The KEGG databases at GenomeNet. 2002;30(1):42–6. doi: 10.1093/nar/30.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Johnson J, Grant CJTMs. Noble WSJNar. 2015;43(W1):W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molecular plant. 2020;13(8):1194–202. Epub 2020/06/26. doi: 10.1016/j.molp.2020.06.009 . [DOI] [PubMed] [Google Scholar]
- 34.Lynch M, Conery JSJs. The evolutionary fate and consequences of duplicate genes. 2000;290(5494):1151–5. doi: 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- 35.Mushtaq N, Munir F, Gul A, Amir R, Zafar Paracha R. Genome-wide analysis, identification, evolution and genomic organization of dehydration responsive element-binding (DREB) gene family in Solanum tuberosum. PeerJ. 2021;9:e11647. Epub 2021/07/06. doi: 10.7717/peerj.11647 ; PubMed Central PMCID: PMC8236231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao J, Li Z, Sun Y, Aluko OO, Wu X, Wang Q, et al. MG2C: A user-friendly online tool for drawing genetic maps. 2021;1(1):1–4. doi: 10.1186/s43897-021-00020-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirsch CD, Hamilton JP, Childs KL, Cepela J, Crisovan E, Vaillancourt B, et al. Spud DB: A resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. 2014;7(1):plantgenome2013.12.0042. [Google Scholar]
- 38.Horton P,Park K-J, Obayashi T, Nakai K, editors. Protein subcellular localization prediction with WoLF PSORT. Proceedings of the 4th Asia-Pacific bioinformatics conference; 2006: World Scientific.
- 39.Team RCJC. RA language and environment for statistical computing, R Foundation for Statistical. 2020. [Google Scholar]
- 40.Rombauts S, Déhais P, Van Montagu M, Rouzé PJNar. PlantCARE, a plant cis-acting regulatory element database. 1999;27(1):295–6. doi: 10.1093/nar/27.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian F, Yang D-C, Meng Y-Q, Jin J, Gao GJNar. PlantRegMap: charting functional regulatory maps in plants. 2020;48(D1):D1104–D13. doi: 10.1093/nar/gkz1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie J, Chen Y, Cai G, Cai R, Hu Z, Wang HJNar. Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. 2023;51(W1):W587–W92. doi: 10.1093/nar/gkad359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A, Birgaoanu M, Griffiths-Jones SJNar. miRBase: from microRNA sequences to function. 2019;47(D1):D155–D62. doi: 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai X, Zhao PXJNar. psRNATarget: a plant small RNA target analysis server. 2011;39(suppl_2):W155-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. 2019;47(D1):D607–D13. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, Buell CRJPo. The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. 2011;6(10):e26801. doi: 10.1371/journal.pone.0026801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao S, Liu Z, Kang Y, Zhang R, Wang Y, Zhang J, et al. Identification of the GAox gene family in potato (Solanum tuberosum L.) and its expression analysis in response to drought stress. 2024;11(1):1–14. [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel BJB. Trimmomatic: a flexible trimmer for Illumina sequence data. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 2013;29(1):15–21. Epub 2012/10/30. doi: 10.1093/bioinformatics/bts635 ; PubMed Central PMCID: PMC3530905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Dewey CNJBb. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. 2011;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei MJMb, evolution. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. 2008;25(12):2717–33. doi: 10.1093/molbev/msn215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo C, Ren HJCSB. Formins: bringing new insights to the organization of actin cytoskeleton. 2006;51:2937–43. [Google Scholar]
- 55.Särkinen T, Bohs L, Olmstead RG, Knapp SJBeb. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. 2013;13:1–15. doi: 10.1186/1471-2148-13-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guruprasad K, Reddy BB, Pandit MWJPE, Design, Selection. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. 1990;4(2):155–61. [DOI] [PubMed] [Google Scholar]
- 57.Kyte J, Doolittle RFJJomb. A simple method for displaying the hydropathic character of a protein. 1982;157(1):105–32. doi: 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Zhong H, Gao C, Zang J, Yang DJIJoMS. The distinct properties of the consecutive disordered regions inside or outside protein domains and their functional significance. 2021;22(19):10677. doi: 10.3390/ijms221910677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soltis DE, Soltis PSJPp. The role of phylogenetics in comparative genetics. 2003;132(4):1790–800. doi: 10.1104/pp.103.022509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bondarenko VS, Gelfand MSJPO Evolution of the exon-intron structure in ciliate genomes. 2016;11(9):e0161476. doi: 10.1371/journal.pone.0161476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koralewski TE, Krutovsky KVJPo. Evolution of exon-intron structure and alternative splicing. 2011;6(3):e18055. doi: 10.1371/journal.pone.0018055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heidari P, Puresmaeli F, Mora-Poblete FJA. Genome-wide identification and molecular evolution of the magnesium transporter (MGT) gene family in Citrullus lanatus and Cucumis sativus. 2022;12(10):2253. [Google Scholar]
- 63.McKee AE, Neretti N, Carvalho LE, Meyer CA, Fox EA, Brodsky AS, et al. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome biology. 2007;8(8):R159. Epub 2007/08/09. doi: 10.1186/gb-2007-8-8-r159 ; PubMed Central PMCID: PMC2374990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu J, Meng W, Poy F, Maiti S, Goode BL, Eck MJJJomb. Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. 2007;369(5):1258–69. doi: 10.1016/j.jmb.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MKJN. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. 2005;433(7025):488–94. doi: 10.1038/nature03251 [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Goikoetxea M, Lupas ANJPO. A conserved motif suggests a common origin for a group of proteins involved in the cell division of Gram-positive bacteria. 2023;18(1):e0273136. doi: 10.1371/journal.pone.0273136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar P, Kumar P, Sharma D, Verma SK, Halterman D, Kumar A. Genome-wide identification and expression profiling of basic leucine zipper transcription factors following abiotic stresses in potato (Solanum tuberosum L.). PloS one. 2021;16(3):e0247864. Epub 2021/03/13. doi: 10.1371/journal.pone.0247864 ; PubMed Central PMCID: PMC7954325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hurst LDJTiG. The Ka/Ks ratio: diagnosing the form of sequence evolution. 2002;18(9):486–7. [DOI] [PubMed] [Google Scholar]
- 69.Li B, Du Z, Jiang N, He S, Shi Y, Xiao K, et al. Genome-Wide Identification and Expression Profiling of the FORMIN Gene Family Implies Their Potential Functions in Abiotic Stress Tolerance in Rice (Oryza sativa). Plant Molecular Biology Reporter. 2023;41(4):573–86. doi: 10.1007/s11105-023-01387-5 [DOI] [Google Scholar]
- 70.Manrique-Carpintero NC, Coombs JJ, Pham GM, Laimbeer FPE, Braz GT, Jiang J, et al. Genome reduction in tetraploid potato reveals genetic load, haplotype variation, and loci associated with agronomic traits. 2018;9:944. doi: 10.3389/fpls.2018.00944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou K, Liu B, Wang Y, Zhang X, Sun GJE, botany e. Evolutionary mechanism of genome duplication enhancing natural autotetraploid sea barley adaptability to drought stress. 2019;159:44–54. [Google Scholar]
- 72.Wang F, Xia Z, Zou M, Zhao L, Jiang S, Zhou Y, et al. The autotetraploid potato genome provides insights into highly heterozygous species. 2022;20(10):1996–2005. doi: 10.1111/pbi.13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang S-Y, Ramachandran SJGB, Evolution. Expansion mechanisms and evolutionary history on genes encoding DNA glycosylases and their involvement in stress and hormone signaling. 2016;8(4):1165–84. doi: 10.1093/gbe/evw067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morales-Cruz A, Amrine KC, Blanco-Ulate B, Lawrence DP, Travadon R, Rolshausen PE, et al. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. 2015;16(1):1–22. doi: 10.1186/s12864-015-1624-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Z, Coulter JA, Li Y, Zhang X, Meng J, Zhang J, et al. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). 2020;153:327–40. doi: 10.1016/j.ijbiomac.2020.03.022 [DOI] [PubMed] [Google Scholar]
- 76.Glory E, Murphy RFJDc. Automated subcellular location determination and high-throughput microscopy. 2007;12(1):7–16. doi: 10.1016/j.devcel.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 77.Kollárová E, Baquero Forero A, Stillerová L, Přerostová S, Cvrčková FJIJoMS. Arabidopsis class II formins AtFH13 and AtFH14 can form heterodimers but exhibit distinct patterns of cellular localization. 2020;21(1):348. doi: 10.3390/ijms21010348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Otomo T, Tomchick DR, Otomo C, Machius M, Rosen MKJPO. Crystal structure of the Formin mDia1 in autoinhibited conformation. 2010;5(9):e12896. doi: 10.1371/journal.pone.0012896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.research GOCJG. Creating the gene ontology resource: design and implementation. 2001;11(8):1425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masseroli M, Tagliasacchi M, Chicco D, editors. Semantically improved genome-wide prediction of Gene Ontology annotations. 2011 11th International Conference on Intelligent Systems Design and Applications; 2011: IEEE.
- 81.Wittkopp PJ, Kalay GJNRG. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. 2012;13(1):59–69. [DOI] [PubMed] [Google Scholar]
- 82.Britten RJ, Davidson EHJS. Gene Regulation for Higher Cells: A Theory: New facts regarding the organization of the genome provide clues to the nature of gene regulation. 1969;165(3891):349–57. [DOI] [PubMed] [Google Scholar]
- 83.Ma R, Liu W, Li S, Zhu X, Yang J, Zhang N, et al. Genome-wide identification, characterization and expression analysis of the CIPK gene family in potato (Solanum tuberosum L.) and the role of StCIPK10 in response to drought and osmotic stress. 2021;22(24):13535. doi: 10.3390/ijms222413535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ESJG. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. 1998;149(2):479–90. doi: 10.1093/genetics/149.2.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. 2005;15(13):1196–200. doi: 10.1016/j.cub.2005.05.048 [DOI] [PubMed] [Google Scholar]
- 86.Khan S-A, Li M-Z, Wang S-M, Yin H-JJIJoMS. Revisiting the role of plant transcription factors in the battle against abiotic stress. 2018;19(6):1634. doi: 10.3390/ijms19061634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sasaki KJBs. Utilization of transcription factors for controlling floral morphogenesis in horticultural plants. 2018;68(1):88–98. doi: 10.1270/jsbbs.17114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian Z, He Q, Wang H, Liu Y, Zhang Y, Shao F, et al. The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. 2015;56(5):992–1005. doi: 10.1093/pcp/pcv025 [DOI] [PubMed] [Google Scholar]
- 89.de Oliveira TM, Cidade LC, Gesteira AS, Coelho Filho MA, Soares Filho WS, Costa MGJTg, et al. Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. 2011;7:1123–34. [Google Scholar]
- 90.TÜFEKÇİ ED, Behcet İJHTvGBD In silico analysis of drought responsive transposons and transcription factors in Solanum tuberosum L. 2019;23(2):189–95. [Google Scholar]
- 91.Rao VS, Srinivas K, Sujini G, Kumar GJIjop. Protein-protein interaction detection: methods and analysis. 2014;2014. doi: 10.1155/2014/147648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nooren IM, Thornton JMJTEj. Diversity of protein–protein interactions. 2003;22(14):3486–92. doi: 10.1093/emboj/cdg359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, Dong G, Wu L, Chen F, Yu Y, Ma DJEJoB. Identification and characterization of profilin gene family in rice. 2021;54:47–59. [Google Scholar]
- 94.Gupta CM, Ambaru B, Bajaj RJFiC, Biology D. Emerging functions of actins and actin binding proteins in trypanosomatids. 2020;8:587685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jockusch B, Murk K, Rothkegel MJRop, biochemistry, pharmacology. The profile of profilins. 2007:131–49. [DOI] [PubMed] [Google Scholar]
- 96.Deng K, Yin H, Xiong F, Feng L, Dong P, Ren MJP. Genome-wide miRNA expression profiling in potato (Solanum tuberosum L.) reveals TOR-dependent post-transcriptional gene regulatory networks in diverse metabolic pathway. 2021;9:e10704. doi: 10.7717/peerj.10704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jerome Jeyakumar JM, Ali A, Wang W-M, Thiruvengadam MJP. Characterizing the role of the miR156- SPL network in plant development and stress response. 2020;9(9):1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa AJPs. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. 2017;258:122–36. doi: 10.1016/j.plantsci.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 99.Garg V, Hackel A, Kühn. Expression level of mature miR172 in wild type and StSUT4-silenced plants of Solanum tuberosum is sucrose-dependent. 2021;22(3):1455. doi: 10.3390/ijms22031455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hwang E-W, Shin S-J, Park S-C, Jeong M-J, Kwon H-BJG, Genomics. Identification of miR172 family members and their putative targets responding to drought stress in Solanum tuberosum. 2011;33:105–10. [Google Scholar]
- 101.Yin Z, Xie F, Michalak K, Pawełkowicz M, Zhang B, Murawska Z, et al. Potato cultivar Etola exhibits hypersensitive resistance to PVYNTN and partial resistance to PVYZ‐NTN and PVYN‐Wi strains and strain‐specific alterations of certain host miRNAs might correlate with symptom severity. 2017;66(4):539–50. [Google Scholar]
- 102.Kaur G, Jain S, Bhushan S, Das N, Sharma M, Sharma DJPP, et al. Role of microRNAs and their putative mechanism in regulating potato (Solanum tuberosum L.) life cycle and response to various environmental stresses. 2024:108334. [DOI] [PubMed] [Google Scholar]
- 103.Yaschenko AE, Fenech M, Mazzoni-Putman S, Alonso JM, Stepanova AN. Deciphering the molecular basis of tissue-specific gene expression in plants: Can synthetic biology help? Current opinion in plant biology. 2022;68:102241. Epub 2022/06/15. doi: 10.1016/j.pbi.2022.102241 ; PubMed Central PMCID: PMC10605770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jopcik M, Bauer M, Moravcikova J, Boszoradova E, Matusikova I, Libantova, Tissue, et al. Plant tissue-specific promoters can drive gene expression in Escherichia coli. 2013;113:387–96. [Google Scholar]
- 105.Cai G, Ahmed MA. The role of root hairs in water uptake: recent advances and future perspectives. Journal of experimental botany. 2022;73(11):3330–8. Epub 2022/03/25. doi: 10.1093/jxb/erac114 . [DOI] [PubMed] [Google Scholar]
- 106.Ranjan A, Sinha R, Singla-Pareek SL, Pareek A, Singh AK. Shaping the root system architecture in plants for adaptation to drought stress. Physiologia plantarum. 2022;174(2):e13651. Epub 2022/02/18. doi: 10.1111/ppl.13651 . [DOI] [PubMed] [Google Scholar]
- 107.Shani E, Yanai O, Ori N. The role of hormones in shoot apical meristem function. Current opinion in plant biology. 2006;9(5):484–9. Epub 2006/08/01. doi: 10.1016/j.pbi.2006.07.008 . [DOI] [PubMed] [Google Scholar]
- 108.Sperry JSJIJoPS. Evolution of water transport and xylem structure. 2003;164(S3):S115–S27. [Google Scholar]
- 109.Qi X, Tang X, Liu W, Fu X, Luo H, Ghimire S, et al. A potato RING-finger protein gene StRFP2 is involved in drought tolerance. 2020;146:438–46. doi: 10.1016/j.plaphy.2019.11.042 [DOI] [PubMed] [Google Scholar]
- 110.Daszkowska-Golec A, Szarejko I. Open or close the gate—stomata action under the control of phytohormones in drought stress conditions. Frontiers in plant science. 2013;4:138. Epub 2013/05/30. doi: 10.3389/fpls.2013.00138 ; PubMed Central PMCID: PMC3652521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang L, Liu Y, Feng S, Yang J, Li D, Zhang JJFiPS. Roles of plasmalemma aquaporin gene StPIP1 in enhancing drought tolerance in potato. 2017;8:616. doi: 10.3389/fpls.2017.00616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fischer L, Lipavska H, Hausman JF, Opatrny Z. Morphological and molecular characterization of a spontaneously tuberizing potato mutant: an insight into the regulatory mechanisms of tuber induction. BMC plant biology. 2008;8:117. Epub 2008/11/26. doi: 10.1186/1471-2229-8-117 ; PubMed Central PMCID: PMC2613151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gong L, Zhang H, Gan X, Zhang L, Chen Y, Nie F, et al. Transcriptome Profiling of the Potato (Solanum tuberosum L.) Plant under Drought Stress and Water-Stimulus Conditions. PloS one. 2015;10(5):e0128041. Epub 2015/05/27. doi: 10.1371/journal.pone.0128041 ; PubMed Central PMCID: PMC4444143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qiu H, Sun C, Dormatey R, Bai J, Bi Z, Liu Y, et al. Thiamethoxam Application Improves Yield and Drought Resistance of Potatoes (Solanum tuberosum L.). Plants (Basel, Switzerland). 2024;13(4). Epub 2024/03/19. doi: 10.3390/plants13040477 ; PubMed Central PMCID: PMC10891895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen J, Wang Y, Huang C, Chen Q, Wei F, Han D, et al. Study on high-yield and high-efficiency cultivation techniques of high quality potato Qingshu 9. 2018;7(4):59–61. [Google Scholar]