Abstract
The human ribosomal protein L7a is a component of the major ribosomal subunit. We previously identified three nuclear-localization-competent domains within L7a, and demonstrated that the domain defined by aa (amino acids) 52–100 is necessary, although not sufficient, to target the L7a protein to the nucleoli. We now demonstrate that L7a interacts in vitro with a presumably G-rich RNA structure, which has yet to be defined. We also demonstrate that the L7a protein contains two RNA-binding domains: one encompassing aa 52–100 (RNAB1) and the other encompassing aa 101–161 (RNAB2). RNAB1 does not contain any known nucleic-acid-binding motif, and may thus represent a new class of such motifs. On the other hand, a specific region of RNAB2 is highly conserved in several other protein components of the ribonucleoprotein complex. We have investigated the topology of the L7a–RNA complex using a recombinant form of the protein domain that encompasses residues 101–161 and a 30mer poly(G) oligonucleotide. Limited proteolysis and cross-linking experiments, and mass spectral analyses of the recombinant protein domain and its complex with poly(G) revealed the RNA-binding region.
Keywords: RNA-binding motif, small nuclear RNA (snRNA), ribosomal protein, ribosome biogenesis, limited proteolysis analysis
Abbreviations: aa, amino acids; EMSA, electrophoretic mobility-shift assay; ESMS, electrospray MS; GST, glutathione S-transferase; IPTG, isopropyl β-D-thiogalactoside; LCMS, liquid chromatography MS; MALDI, matrix-assisted laser-desorption ionization; MALDI–TOF, MALDI–time-of-flight; RNP, ribonucleoprotein; r-protein, ribosomal protein; SECIS, selenocysteine insertion sequence; SBP2, SECIS-binding protein 2; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; snRNP, small nuclear RNP; TFA, trifluoroacetic acid
INTRODUCTION
The assembly of ribosomes takes place in the nucleolus and requires a number of co-ordinated events prior to nuclear export of the mature subunit to the cytoplasm [1]. These events include nucleolar transcription and processing of 18 S, 28 S and 5.8 S rRNA, nuclear import and nucleolar accumulation of ribosomal proteins (r-proteins), as well as nuclear transcription and nucleolar accumulation of 5 S rRNA. Thus the biogenesis of eukaryotic ribosomes requires complex intracellular trafficking to ensure that all the components necessary for efficient ribosome subunit assembly are present in the nucleoli. Import signals have been mapped in a number of r-proteins [2,3]; a common feature is their very basic nature and a greater complexity as compared with the classical nuclear localization sequence [4]. r-proteins enter the nucleus via a non-classical pathway [5]; once in the nucleus, they are sequestered in the nucleolus by different pathways, depending on a specific binding host in the nucleolus, as occurs for some non-ribosomal nucleolar proteins [6,7].
We previously showed that nucleolar accumulation of L7a in the nucleoli requires the presence of an essential domain that spans aa (amino acids) 52–100, and a positively charged stretch of amino acids that includes a nuclear localization sequence [3]. These results support the hypothesis that nucleolar targeting is mediated by interactions between one or more protein domains and macromolecules residing in the nucleolus [8,9].
The ribosome is an ancient protein–RNA complex, and r-proteins may have been among the first proteins to set up strategies to contact RNA. A putative RNA-binding motif (residues 129–160) has been identified in L7a on the basis of a predicted secondary structure similarity with an RNA-binding domain identified in some r-proteins and non-r-proteins [10]. Consequently, L7a belongs to a family of proteins (L7ae) that share a conserved region which is larger than the previously predicted RNA-binding domain, and that spans aa 135–189 in human L7a. This family includes Haloarcula marismortui L7ae, human r-protein S12, yeast r-protein L30, yeast protein NHP2 and the human orthologue 15.5 kDa tri-snRNP (small nuclear ribonucleoprotein) [11,12]. Most of these proteins are components of the RNP (ribonucleoprotein) complex. Furthermore, H. marismortui L7ae, yeast r-protein L30 and human 15.5 kDa tri-snRNP are known to bind a conserved RNA structural motif [13–15]. As a step toward a more detailed understanding of the mechanism of nucleolar accumulation of L7a, we have investigated the RNA-binding ability of L7a. Our results show that, in addition to the predicted RNA-binding domain (RNAB2), the domain previously shown to be essential for nucleolar accumulation of the human L7a r-protein [3] also exerts RNA-binding activity (RNAB1).
In the present study we report results leading to the definition of the L7a RNA-binding domains and the analysis of their specificities. A recombinant form of RNAB2, i.e. L7a(101–161), was expressed in Escherichia coli, purified and fully characterized. Topological investigation of this mutant protein and its complex with poly(G), carried out by limited proteolysis and cross-linking analysis, led to the definition of the RNA-binding region and provided experimental evidence of conformational changes induced by RNA–protein interactions.
EXPERIMENTAL
Plasmid constructs
To express recombinant r-protein L7a and L7a deletion mutants, we used the pRSET (Invitrogen) and pQE (Qiagen) vectors, both of which direct the production of histidine-tagged recombinant proteins. The pQE31/L7a construct encoding an L7a–His cDNA was generated by inserting an EcoR1–Pst1 fragment of L7a/pBTM116, containing the entire L7a coding region, into the SmaI–Pst1 sites of pQE31 after filling in the 3′-recessed end generated by EcoR1. To obtain cDNA constructs encoding different domains of the L7a protein, we used appropriate synthetic oligonucleotide primers and PCR amplification of the L7a cDNA template (pL7a) [3]. The primers used to amplify the cDNA coding aa 101–161 of L7a, i.e. construct L7a(101–161), were designed to carry the recognition sites for BamH1 and HindIII at the 5′- and 3′-termini respectively to allow cloning in the corresponding sites of the expression vector. The constructs L7a(1–40) (aa 1–40 of L7a) and L7a(1–161) (aa 1–161 of L7a) were generated by inserting the PCR products in the Pst1 and EcoR1 sites of pRSETC. The vector L7a(52–100) (aa 52–100 of L7a) was constructed by insertion of the PCR product in the BamH1–EcoR1 sites of pRSETC. For the expression of human 15.5 kDa protein, a cDNA was obtained by reverse transcription PCR on total RNA from HeLa cells. For the amplification step, oligonucleotide primers were designed using the cDNA sequence published in the GenBank® (accession number D50420). The resulting cDNA coding for the 15.5 kDa protein was inserted in the BamH1–Sal1 sites of a pGEX4T-3, which allows the production of a fusion protein GST (glutathione S-transferase)–15.5 kDa. All plasmid inserts were verified by sequencing.
Expression and purification of recombinant proteins
In order to produce the r-protein L7a and mutants thereof E. coli BL21(DE3)pLysS cells (Invitrogen) were transformed with each recombinant pRSET plasmid DNA. E. coli M15pREP cells (Qiagen) were transformed with each recombinant pQE plasmid. To produce the fusion protein GST–15.5 kDa, E. coli BL21(DE3) cells (Invitrogen) were transformed with the pGEX4T-3 derived recombinant plasmid. All recombinant proteins, with the exception of L7a(101–161), were expressed by growing cells to a D600 of 0.5 at 37 °C, and then inducing expression by exposing cultures for 3 h to 1 mM IPTG (isopropyl β-D-thiogalactoside) at 37 °C. L7a(101–161) was produced by growing cells at 30 °C to a D600 of 0.5 and inducing protein expression at 30 °C with 1 mM IPTG for 1 h. All His-tagged recombinant proteins were purified by affinity chromatography on Ni2+-nitrilotriacetate agarose beads (Qiagen), according to the manufacturer's instructions. After purification, the recombinant proteins used in filter-binding experiments [L7a–His and L7a(101–161)] were dialysed against 50 mM Tris (pH 7.4)/200 mM KCl. Purification of the GST and GST–15.5 kDa proteins was achieved by affinity chromatography on GSH–agarose beads (Pharmacia). The recombinant protein was eluted by using 10 mM GSH in 50 mM Tris/HCl, pH 8.0. After purification, the recombinant protein was dialysed against 50 mM Tris/HCl, pH 7.4, containing 10% glycerol.
Concentrations of recombinant proteins were estimated with the Bradford reagent (Bio-Rad protein assay) and checked by SDS/PAGE.
Nucleic acids
L7a mRNA, to be used in protein binding assays and in vitro translation experiments, was transcribed from a plasmid into which full-length human L7a cDNA [16] had been cloned adjacent to the phage SP6 RNA polymerase promoter (PGEM-4Z vector). RNA was synthesized in vitro by SP6 RNA polymerase according to the manufacturer's recommendations (Promega), and 50 μCi of [α-32P]CTP (Amersham) were included for the synthesis of radiolabelled RNA. The amount of RNA recovered was determined by measuring either the radioactivity present in the transcript or A260. For 5′-end labelling, gel-purified human 28 S RNA from HeLa cells was first dephosphorylated by using calf alkaline phosphatase (Biolabs) for 30 min at 55 °C. RNA was then extracted twice with phenol and 5′-end-labelled with phage T4 polynucleotide kinase (Roche) and 50 μCi of [γ-32P]ATP (5000 Ci/mmol, Amersham) for 1 h at 37 °C. Synthetic poly(G) (Eurogentec, Seraing, Belgium) and poly(C) (Sigma) were 5′-end-labelled by the same procedure without the dephosphorylation step. Total RNA was labelled in vivo by incubating HeLa cells with [32P]Pi (40 μCi/ml) for 2 h.
Cell-free translation
For cell-free translation, the Rabbit Reticulocyte Lysate System (Promega) was programmed with human L7a mRNA (10 μg) obtained by in vitro transcription. Translation reactions were performed using 17.5 μl of reticulocyte lysate and 20 μCi of [35S]-methionine (1000 Ci/mmol, Amersham). Translation was allowed to proceed for 90 min, according to the conditions indicated by the manufacturer (Promega). Aliquots of the translation product were used in the EMSAs (electrophoretic mobility-shift assays).
Northwestern experiments
Nitrocellulose filters carrying L7a–His protein or derived peptides and molecular mass protein markers (Gibco) as a control were prepared by electrophoretically transferring purified recombinant proteins resolved by SDS/PAGE (15% gel) on to a nitrocellulose membrane. Filters were incubated overnight at room temperature (25 °C) in a binding buffer (10 mM Tris/HCl, pH 7.4, 50 mM NaCl, 1 mM EDTA, 0.02% BSA, 0.02% Ficoll 400, 0.02% PVDF 150). The filters were then probed at room temperature for 1 h with labelled RNA (100000 cpm/ml) in the binding buffer containing 2 mg/ml heparin (porcine intestinal mucosa). Blots were washed three times for 15 min with binding buffer, air-dried and exposed to X-ray film for autoradiography.
Filter-binding assay
For filter-binding assays, 10 fmol of labelled RNA (L7a mRNA, human 28 S rRNA) were incubated at 60 °C for 15 min in 100 μl of TMK buffer (20 mM Tris/HCl, pH 7.4, 4 mM MgCl2, 200 mM KCl) and allowed to cool slowly to room temperature. L7a or derived peptides were added at the indicated concentrations in TMK buffer containing 20% glycerol, 1 mM dithiothreitol, 0.5 μg/ml tRNA and 4 μg/ml BSA. The protein/RNA mixtures were incubated for 30 min at room temperature and then filtered through a wet nitrocellulose filter (Schleicher and Schuell, BA85120) under gentle suction. The filter was washed twice with 300 μl of TMK buffer and dried at 80 °C. The percentage of 32P input retained on the filter was determined by Cerenkov counting in a Beckman scintillation counter. The dissociation constant (Kd) corresponds to the protein concentration at which half of the amount of RNA bound at saturation is retained on the filter [17]. All filter-binding experiments were performed at least four times and the results are expressed as the average amount of bound nucleic acid.
For competition experiments, unlabelled competitor RNA [ribo-homopolymer poly(A), poly(C) and poly(U) from Sigma, poly(G) from Eurogentec, yeast tRNA from Roche, poly(A)− RNA extracted from HeLa cells] or DNA (M13 single-strand genomic DNA, pGEM4Z DNA) was added to the binding reactions, which contained 10 fmol of labelled L7a mRNA, before the addition of L7a–His (500 nM).
EMSA
A U4 snRNA oligonucleotide (nucleotides 26-47, [12]) was obtained by Eurogentec, 5′-end labelled by using T4 polynucleotide kinase and [γ-32P]ATP (5000 Ci/mmol, Amersham), and gel-purified prior to use. Aliquots of the translation products of L7a mRNA, 14 μM recombinant GST or GST–15 kDa protein were incubated with 0.250 pmol of 32P-labelled U4 snRNA oligonucleotide in the presence of 20 μg of yeast tRNA (final volume of 20 μl in a buffer containing 20 mM Hepes, pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, pH 8, 0.1% Triton X-100) for 1 h at 4 °C [12]. The RNA–protein complexes were resolved by electrophoresis on a native 6% polyacrylamide gel in Tris/borate buffer (0.090 M Tris/borate, 0.002 M EDTA) and visualized by autoradiography.
Characterization of L7a(101–161)
Aliquots of L7a(101–161) were desalted on a Phenomenex Jupiter C4 reverse-phase column (250 mm×2.1 mm, 300 Å pore size) eluted at 0.2 ml/min. Solvent A was 0.1% TFA (trifluoroacetic acid) in water, solvent B was acetonitrile containing 5% water and 0.07% TFA. The protein was eluted by a linear gradient of solvent B from 5 to 95% in 15 min. Desalted proteins (approx. 1 nmol) were incubated with trypsin or endoprotease V8 in 50 mM ammonium bicarbonate, pH 8.5, at 37 °C overnight using an enzyme-to-substrate ratio of 1:50. Peptide mixtures were either directly analysed by MALDI (matrix-assisted laser-desorption ionization) MS or fractionated on a Phenomenex Jupiter C18 reverse-phase column using the solvent system described above with a linear gradient of solvent B from 5 to 65% in 60 min at a flow rate of 0.2 ml/min. Peptides were manually collected, vacuum dried and analysed by ESMS (electrospray MS).
Limited proteolysis experiments
Limited proteolysis experiments were carried out on 3 nmol of purified L7a(101–161) using trypsin, endoproteinase Lys-C, GluC protease, chymotrypsin and subtilisin as enzymic probes. All enzymic digestions were performed in 50 mM Tris/HCl buffer, pH 7.5, at 25 °C using enzyme-to-substrate ratios ranging from 1:100 to 1:5000 (see Table 1). The extent of digestion was monitored by taking samples of the reaction mixture at intervals from 15 to 60 min. The hydrolysis reaction was quenched by lowering the pH to about 2.0 with 0.1% TFA and the peptide mixture was fractionated by reverse-phase HPLC as described above. Individual fractions were collected and identified by ESMS or MALDI-MS. The L7a(101–161)–RNA complex was obtained by incubation of the protein and RNA in a molar ratio of 1:1.1 for 15 min at 25 °C in 50 mM Tris/HCl buffer, pH 7.5, containing 200 mM KCl. After limited proteolysis experiments on the complex, the products were analysed by LCMS (liquid chromatography MS). The elution was performed with a Phenomenex Jupiter C18 reverse-phase column equilibrated in 5% formic acid and 0.05% TFA in water (solvent A). The peptide mixtures generated during the proteolysis experiments were eluted by increasing the concentration of solvent B (95% acetonitrile containing 5% formic acid and 0.05% TFA) from 5 to 65% in 60 min.
Table 1. Summary of limited proteolysis analyses of L7a(101–161) and L7a(101–161)–RNA complex.
| Mass (Da) | |||||
|---|---|---|---|---|---|
| Protein | Enzyme | Enzyme/substrate ratio (w/w) | Peptide | Measured | Theoretical |
| L7a(101–161) | Trypsin | 1:5000 | 1–25 | 3117.7±0.4 | 3117.5 |
| 26–76 | 5429.6±0.7 | 5429.4 | |||
| 1–49 | 5671.0±0.9 | 5671.5 | |||
| 50–76 | 2875.1±0.3 | 2875.3 | |||
| LysC | 1:5000 | 1–22 | 2705.1±0.4 | 2705.0 | |
| 23–76 | 5842.1±1.1 | 5841.9 | |||
| GluC | 1:5000 | 1–21 | 2576.6±0.1 | 2576.8 | |
| 22–76 | 5970.0±0.2 | 5970.0 | |||
| 1–60 | 6756.3±0.8 | 6756.7 | |||
| 61–76 | 1789.8±0.3 | 1790.1 | |||
| Chymotrypsin | 1:10000 | 15–76 | 6838.6±0.6 | 6839.0 | |
| 1–27 | 3343.4±0.3 | 3343.8 | |||
| 28–76 | 5202.8±1.0 | 5203.1 | |||
| 1–58 | 6529.2±1.1 | 6528.5 | |||
| 59–76 | 2017.7±0.7 | 2018.4 | |||
| Subtilisin | 1:15000 | 1–58 | 6527.6±0.7 | 6528.5 | |
| 59–76 | 2017.7±0.8 | 2018.4 | |||
| 1–71 | 7958.8±0.3 | 7959.2 | |||
| L7a(101–161)– | Trypsin | 1:500 | 1–49 | 5670.9±0.9 | 5671.5 |
| RNA | 50–76 | 2875.1±0.3 | 2875.3 | ||
| 1–63 | 7127.5±0.8 | 7127.2 | |||
| 64–76 | 1419.5±0.2 | 1419.7 | |||
| LysC | 1:100 | 1–33 | 4027.9±0.6 | 4027.6 | |
| 34–76 | 4519.2±0.4 | 4519.3 | |||
| 1–63 | 7127.4±0.4 | 7127.2 | |||
| 64–76 | 1419.3±0.1 | 1419.7 | |||
| GluC | 1:2000 | 1–53 | 6012.6±0.7 | 6012.9 | |
| 54–76 | 2533.8±0.2 | 2534.0 | |||
| Chymotrypsin | 1:5000 | 15–76 | 6838.6±0.6 | 6839.0 | |
| 28–76 | 5202.8±1.1 | 5203.1 | |||
| Subtilisin | 1:12000 | 1–58 | 6529.0±1.0 | 6528.5 | |
| 59–76 | 2017.8±0.6 | 2018.4 | |||
| 1–53 | 6012.8±0.6 | 6012.9 | |||
| 54–76 | 2533.7±0.2 | 2534.0 | |||
Chemical cross-linking experiments
Cross-linking experiments were carried out by irradiating samples with a UV lamp (Spectronics Corporation) at 254 nm. In a typical experiment, an aliquot of the protein was combined with the polynucleotide in a 1:1.1 molar ratio in 50 mM Tris/HCl buffer, pH 7.5. The sample was then irradiated at 254 nm for 20 min at 25 °C at a distance of 7 cm. The cross-linked product was then digested with trypsin [enzyme/substrate ratio, 1:100 (w/w)] for 6 h at 25 °C in the same buffer, and enzymic hydrolysis was carried out with T1 ribonuclease (300 units at 37 °C for 30 min). The resulting peptide mixture was then directly analysed by MALDI-MS.
MS
Direct ESMS measurements and LCMS analyses were performed using a ZQ single quadruple instrument (Waters), equipped with an Alliance HPLC. For direct mass spectral analysis, samples were injected into the ion source by a Harward syringe pump at a flow rate of 3 μl/min. For LCMS analysis, the peptides were fractionated on a 2690 Alliance system using a Phenomenex Jupiter C18 reverse-phase column at a flow rate of 0.2 ml/min. Data were acquired and processed using Masslynx software (Waters-Micromass). Horse heart myoglobin was used to calibrate the instrument (average molecular mass 16951.5 Da); all masses are reported as average masses.
The MALDI–time-of-flight (MALDI–TOF) mass spectra were recorded with a Voyager DE-PRO instrument (Applied Biosystems). A mixture of analyte and matrix solution (10 mg/ml α-cyano-hydroxycinnamic acid in 66% acetonitrile, 0.1% TFA, in MilliQ water or a matrix, obtained by mixing in a 18:1:1 ratio 40 mg/ml picolinic acid in 66% acetonitrile, 3-hydroxypicolinic acid in 66% acetonitrile, ammonium citrate 40 mg/ml, in MilliQ water) was applied to the metallic sample plate and dried at room temperature. Mass calibration was performed using external peptide standards. Raw data were analysed using the computer software provided with the instrument and are reported as mono-isotopic masses.
RESULTS
Human r-protein L7a can bind RNA in vitro
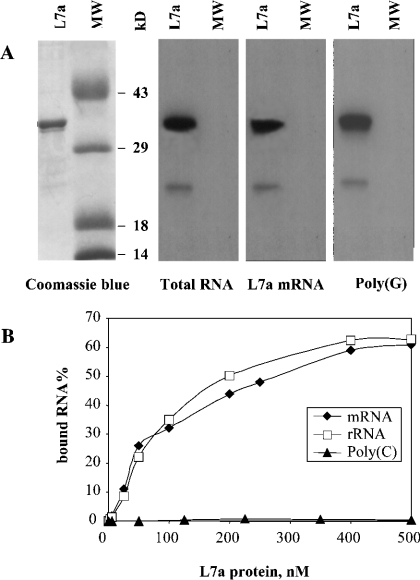
The RNA-binding ability of L7a was first tested in vitro by using an L7a protein produced by a cell-free translation system. With an assay widely used to test the RNA-binding activity of a protein whose RNA target is unknown [18], we evaluated the ability of L7a to bind poly(A), poly(G), poly(C) and poly(U) ribo-homopolymers or single-stranded DNA immobilized on agarose beads. High-affinity binding was observed only with poly(G), and was stable at NaCl concentrations up to 0.5 M (results not shown). The RNA-binding activity of L7a was confirmed by Northwestern (Figure 1A) and filter-binding assays (Figure 1B). For these experiments, the cDNA encoding L7a was expressed in E. coli as a fusion protein with an N-terminal His-tag, and purified by affinity chromatography on a Ni2+-nitrilotriacetic acid column. Figure 1(A) shows the results of a typical Northwestern experiment, demonstrating that the recombinant L7a protein is able to bind RNA from HeLa cells, 32P-labelled in vivo, in the presence of 1 mg/ml heparin (total RNA). In addition, the recombinant His–L7a protein was able to interact with 32P-labelled poly(G), and with radiolabelled L7a mRNA synthesized by an in vitro transcription system. The heparin-resistant binding and lack of binding to the molecular mass marker proteins (Figure 1A) demonstrate that the binding of L7a to RNA in stringent conditions was specific.
Figure 1. Analysis of RNA-binding ability of human r-protein L7a.
Purified recombinant L7a–His protein (L7a) and molecular mass marker proteins (MW) were loaded on to an SDS gel. Upon electrophoresis, gel samples were stained with Coomassie Brilliant Blue (A, left-hand panel), or transferred on to a nitrocellulose membrane, probed with the indicated radiolabelled RNAs and subjected to autoradiography (A, right-hand panels; for an explanation for the lanes marked MW see the text). (B) Plot of binding activity of L7a–His to radiolabelled RNAs. For details of nucleic acid labelling and filter-binding assay, see the Experimental section.
We tested the RNA-binding capacity of the purified recombinant L7a protein also by filter-binding assay (Figure 1B). L7a mRNA, transcribed in vitro, and 28 S rRNA, electroluted from agarose gel, were used as ligands in the filter-binding analysis. L7a was able to bind, in a dose-dependent manner, its own mRNA and 28 S rRNA, whereas it failed to bind poly(C), thus confirming the results of the Northwestern assay. We determined from a graph an apparent Kd of 75 nM for L7a mRNA and of 85 nM for 28 S rRNA. The Kd values are in the range observed when assaying the RNA-binding activity of individual r-proteins in vitro; a co-operative effect mediated by interacting macromolecules ensures a greater affinity in vivo [19].
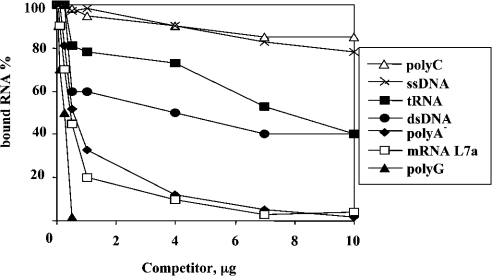
We also used the His–L7a protein to analyse the RNA-binding specificity of L7a in a filter-binding competition assay. Various nucleic acids served as competitors for the binding of 32P-labelled L7a mRNA to His–L7a. The L7a mRNA was chosen because it is able to bind L7a (see Figure 1), and it can be obtained as a pure single species by in vitro transcription. The binding activity of L7a to its own mRNA was considered to be 100%; as internal standard, we used unlabelled L7a mRNA as competitor. The results of this analysis are shown in Figure 2. RNA binding was specifically competed by low concentrations of unlabelled poly(G), but not by an excess of other ribo-homopolymers (as an example, the results obtained with poly(C) are reported in Figure 2), or single-stranded DNA.
Figure 2. RNA-binding specificity of human r-protein L7a.
Filter-binding assay of competition of the binding of L7a–His to radiolabelled L7a mRNA by increasing amounts of unlabelled nucleic acids. The binding reaction contained 10 fmol of labelled L7a mRNA, to which a competitor nucleic acid was added before the addition of 500 nM L7a–His. The assay was performed as described in the Experimental section. The bound RNA values were corrected for the radioactivity retained on the filter when no L7a protein was added to the labelled L7a mRNA. The RNA-binding activity of L7a protein to radiolabelled L7a mRNA in the absence of a competitor was considered as 100%.
Protein L7a carries two RNA-binding motifs
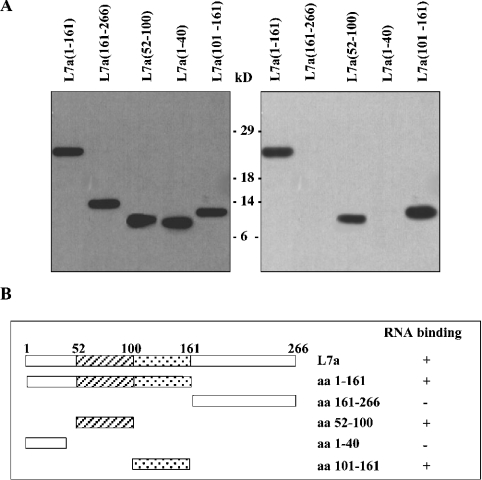
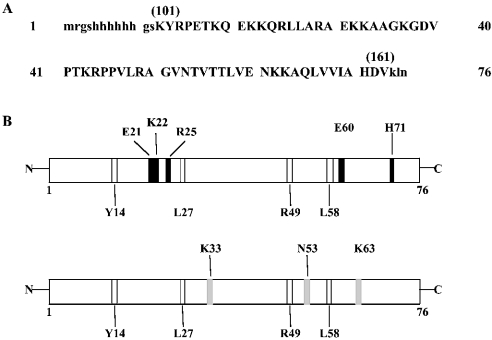
In a previous study, we dissected the L7a protein to identify domains that could mediate the nuclear import and nucleolar targeting of the protein [3], and demonstrated that a domain spanning aa 52–100 is essential for the nucleolar accumulation of L7a (Figure 3B). In an attempt to understand the relationship between RNA-binding activity and the subcellular localization of L7a, we expressed, as histidine-tagged proteins in E. coli, several L7a peptides corresponding to the previously identified functional domains and to other regions of the protein, and purified them by affinity chromatography on a Ni2+ column. The purified peptides were assayed in Northwestern experiments using radiolabelled L7a mRNA as a ligand. We found that a deletion mutant containing the first 161 aa of L7a [L7a(1–161) in Figure 3A] retained RNA-binding activity, whereas mutants lacking these sequences [L7a(161–266) in Figure 3A] failed to bind RNA. To identify the sequences within the 1–161 aa stretch that are responsible for RNA-binding activity, we generated three additional constructs (Figure 3A). Northwestern assay of these constructs using the L7a mRNA as a probe showed that the N-terminal region of the protein (aa 1–40) could be deleted without loss of RNA-binding activity. In fact, only mutants containing the domain essential for nucleolar targeting (aa 52–100), the predicted RNA-binding domain (aa 101–161), or both (aa 1–161), consistently retained RNA-binding activity (Figure 3B).
Figure 3. Definition of RNA-binding domains of the human r-protein L7a.
(A) Purified recombinant L7a-His deletion mutants were subjected to electrophoresis on an SDS/PAGE gel and then transferred on to a nitrocellulose membrane. The filter was probed with a commercially available antiserum against the His-tag (Santa Cruz Biotechnology) (left-hand panel). The RNA-binding activity of all deletion mutants was tested by Northwestern experiments using radiolabelled L7a mRNA (right-hand panel). (B) Summary of the RNA-binding ability of L7a deletion mutants. Striped boxes indicate the domain required for nucleolar targeting of the protein, and dotted regions indicate the putative RNA-binding motif [10].
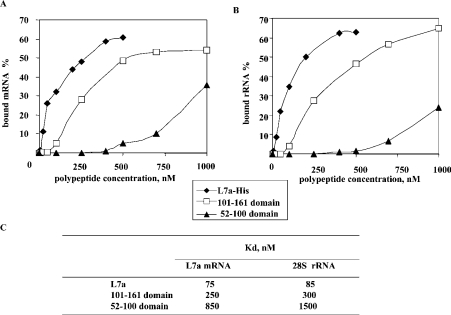
We next evaluated by filter-binding assay, the binding of these RNA-binding domains to L7a mRNA (Figure 4A) and human 28 S RNA (rRNA in Figure 4B). We also compared the RNA-binding activity of the isolated domains with that of the whole L7a protein. The dissociation constant (Kd, Figure 4C) of each peptide versus each RNA probe, graphically determined, was in the order of 0.1 μM.
Figure 4. RNA-binding activity of L7a–His, and L7a-derived polypeptides with L7a mRNA (A) and 28 S RNA (B).
The filter-binding assays were carried out as described in the Experimental section. The resulting plots are the average of four independent experiments and constitute the basis for the calculation of the Kd values reported in (C).
Conservation of RNA-binding domains in L7a protein orthologues
Whereas there is no known eubacterial L7a orthologue, sequences are available for L7a orthologues from many eukaryotic organisms, and from some Archea prokaryotes [13]. Sequence alignment in Figure 5(A) shows that the RNA-binding domain defined by aa 52–100 in human L7a (RNA-binding domain 1, RNAB1) is well conserved among eukaryotic L7a, but totally absent in the Archea.
Figure 5. Evolutionary conservation of the RNA-binding domains of r-protein L7a.
(A) Alignment of the RNAB1 (aa 52–100) domain of the human L7a r-protein (RL7A_HUMAN; UniProt database accession number P11518) with the orthologue protein from chicken (RL7A_CHICK; P32429), Fugu rubripes (RL7A_FUGRU; O57592), Drosophila melanogaster (RL7A_DROME; P46223), Caenorhabditis elegans (RL7A_C. ELE; Q8T875), Arabidopsis thaliana (RL7A_ARATH; P49692), Oryza sativa (RL7A_ORYSA; P35685) and Saccharomyces cerevisiae (RL8B_YEAST; P29453). (B) Alignment of the RNAB2 (aa 101–161) domain of the human r-protein L7a with the eukaryotic orthologues shown in (A), and with the Archea H. marismortui (RL7A_HALMA; P12743) and P. abyssi (RL7A_PYRHO; O59165) orthologues. (C) Sequence alignment of the extended RNAB2 domain of human r-protein L7a with: Archea L7ae protein, human 15.5 kDa protein (NHPX_HUMAN; P55769), yeast L30 (RL30_YEAST; P14120), human SBP2 (SBP2_HUMAN; Q96T21). Identical residues are in black and conserved residues are grey boxes. The conserved residues are grouped DE, KRH, LIVAMPYFW and TSQCN.
The aa 101–161 region of the L7a r-protein (i.e. RNA-binding domain 2, RNAB2) is shown in Figure 5(B) aligned with the homologous region of some eukaryotic orthologues and with the L7ae protein from H. marismortui and Pyrococcus abyssi. Again, the RNAB2 domain is highly conserved among eukaryotes, whereas homology in the Archea is restricted to the RNA-binding motif postulated by Koonin et al. [10]. Although the aa 101–161 region exerts RNA-binding activity (see the 101–161 domain in Figure 4), the homology region extends further at the C-terminus (see Figure 5C), and is shared by L7ae proteins, by the 15.5 kDa/Snu13 protein [12], which is a component of the splicing machinery in eukaryotes, and by the SBP2 [SECIS (selenocysteine insertion sequence)-binding protein 2] [20]. In addition, the Archea L7ae protein is a functional homologue of the eukaryotic 15.5 kDa/Snu13 protein [21], and is thus both an r-protein and an snRNP core protein. The human 15.5 kDa protein bound to the U4 snRNA fragment containing the RNA target site has been crystallized [14]. This fragment, spanning nt 26–47 of U4 snRNA, folds in a specific asymmetric stem–loop structure named ‘kink-turn’ or ‘k-turn’ [15]. The kink-turn motif is also present in a region of the 23 S RNA target of protein L7ae [13].
RNA binding activity of human r-protein L7a has targets other than the kink-turn motif
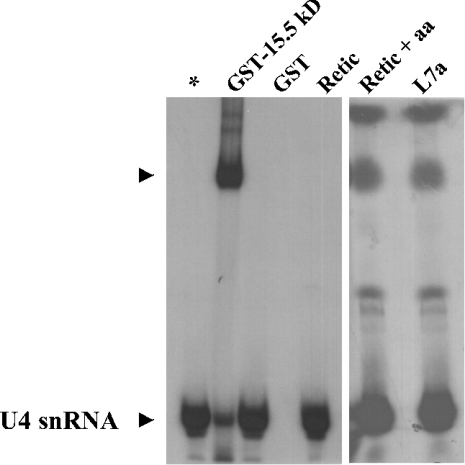
Because the activity of an RNA-binding domain shared by Archea rpL7ae, Eukarya 15.5 kDa/Snu13p, yeast rpL30 and human SBP2 [20–22] targets the same RNA kink-turn motif [15], we also asked whether the RNA-binding activity of the human rpL7a protein targets this structural motif. Figure 6 shows the results of EMSA experiments in which we compared the ability of the recombinant 15.5 kDa protein and the L7a protein to form a complex with an snU4 RNA fragment (nt 26–47, [12]). As expected, the U4 snRNA fragment showed a specific gel mobility shift when incubated with the recombinant 15.5 kDa protein (GST–15.5 kDa in Figure 6), which demonstrates the formation of an RNA–protein complex, whereas there was no shift when the fragment was incubated with the L7a protein produced by in vitro translation (L7a in Figure 6). Despite the background bands in the Retic+aa (unprogrammed rabbit reticulocyte lysate plus amino acids) and L7a lanes, we can exclude formation of the RNA–protein complex, because the U4 snRNA probe is clearly ‘unshifted’. Moreover, a filter-binding assay confirmed that L7a does not bind to the U4 snRNA fragment (results not shown). Interestingly, box C/D snoRNAs (small nucleolar RNAs) bind to the 15.5 kDa protein in vitro, and this is due to the fact that the box C/D motif folds into a structure almost identical to the kink-turn site of the U4 snRNA [23]. We carried out filter-binding experiments to assay the ability of protein L7a to bind to U14 snoRNA fragments [24], or to U24 snoRNA, a member of the C/D box snoRNA family encoded by an intron sequence in the L7a gene [25]. In no case did we observe any specific interaction (results not shown).
Figure 6. EMSA analysis of binding of proteins 15.5 kDa and L7a to U4 snRNA 5′-stem-loop oligonucleotide.
The recombinant GST–15.5 kDa fusion protein, or L7a protein produced by in vitro translation were incubated with the 5′-end labelled U4 snRNA oligonucleotide, and the protein–RNA complex was resolved by PAGE on a 6% native polyacrylamide gel. As a control, the EMSA was performed with the probe alone (*), with recombinant GST protein (GST), with unprogrammed rabbit reticulocyte lysate (Retic), and with unprogrammed rabbit reticulocyte lysate plus amino acids (Retic+aa).
Limited proteolysis on the native L7a(101–161) protein
Analysis by HPLC of recombinant L7a(101–161) showed the occurrence of a single major peak whose molecular mass was determined as 8528.9±0.1 Da by ESMS analyses. The measured mass value was in perfect agreement with the theoretical mass expected on the basis of the amino acid sequence (8528.9 Da), and the entire polypeptide sequence was verified by MALDI mapping. Limited proteolysis experiments were carried out with various proteases both on isolated L7a(101–161) and on the L7a(101–161)–RNA complex, according to a strategy described elsewhere [26–30]. The conditions for limited proteolysis were selected to maximize the stability of the protein or complex conformation and to increase the selectivity of proteolytic enzymes. The extent of enzymic digestion was monitored over time by sampling the incubation mixture at different intervals, and analysing the samples by HPLC. The fragments released from the protein were identified by ESMS, and assessed for preferential cleavage sites. Time-course analysis of the L7a(101–161)–RNA complex was monitored by direct LCMS procedures. After formation of the complex, the RNA shields the protein residues located within the interface region. Therefore, proteolytic patterns differed depending on whether digestions were carried out on the isolated protein or on the complex.
We used five different proteases in the limited proteolysis experiments in an attempt to create conditions in which the selectivity of the cleavages was not related to, or limited by, the specificity of the enzyme. For each protease, the appropriate enzyme/protein or enzyme/complex ratio was determined, so as to generate a limited number of proteolytic events and to direct protease action towards the most flexible and solvent-exposed sites. Preferential cleavage sites were assigned by the identification of the two complementary peptides generated by a single proteolytic event occurring on the intact protein or complex molecule, as described elsewhere [26].
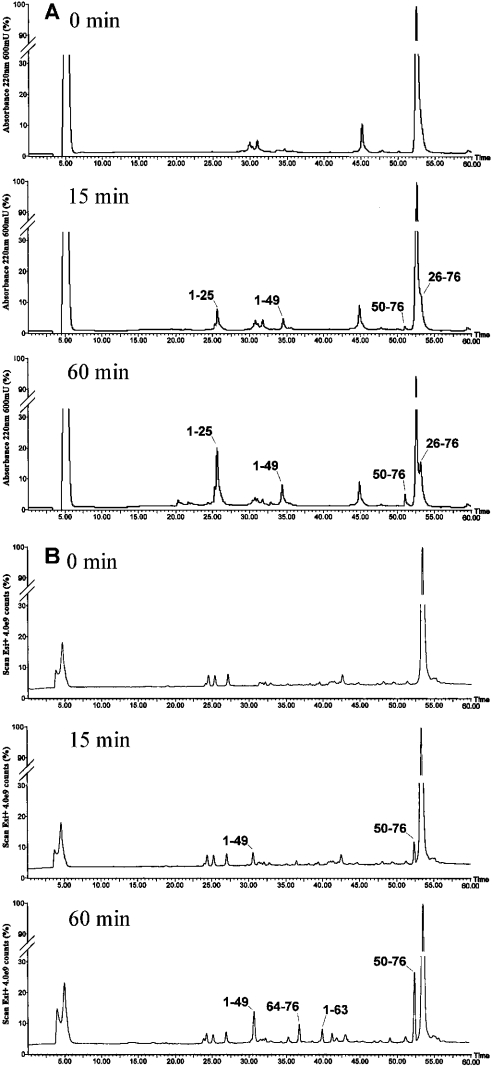
As an example, Figure 7(A) shows the HPLCs of samples taken after 15 min, 30 min and 60 min of trypsin digestion. The protein was immediately cleaved at Arg25 and Arg49, thereby releasing the complementary fragments 1–25 (3117.7±0.4 Da) and 26–76 (5429.6±0.7 Da), and 1–49 (5671.0±0.9 Da) and 50–76 (2875.1±0.3 Da). Similar results were obtained with the endoproteases LysC and GluC. The preferential proteolytic sites were identified at Lys22, Glu21 and Glu60.
Figure 7. Limited proteolysis of native L7a(101–161) (A), and the L7a(101–161)-poly(G) complex (B) with trypsin.
HPLC analysis of the native L7a(101–161) and of the L7a(101–161)–poly(G) complex after 0 min, 15 min and 60 min of incubation with trypsin. The indicated fractions were manually collected and the eluted peptides were identified by ESMS in the case of the isolated protein, and directly by the LCMS procedure in the case of the complex.
In other limited proteolysis experiments we incubated L7a-(101–161) with proteases chymotrypsin and subtilisin, which have a broader specificity. The HPLC time-course analysis showed the complementary peptide pair 1–58 and 59–76 (molecular mass values 6529.2±1.1 and 2017.7±0.7 Da respectively), indicating the occurrence of a single proteolytic event at Leu58. A second proteolytic cleavage site was identified at Leu27, as inferred by the presence of peptides 1–27 and 28–76. Finally, the presence of peptide 15–76 eluted under the protein peak indicated a preferential proteolytic site at Tyr14.
The results from the limited proteolysis experiments are summarized in Figure 8(B) and Table 1. Preferential proteolytic sites were clustered within two regions of the protein: the N-terminal segment encompassing residues 21–27 and the C-terminal region 58–71 residues. Tyr14 and Arg49 were isolated cleavage sites.
Figure 8. Location of proteolytic sites in the native L7a(101–161) protein and in the L7a(101–161)–poly(G) complex.
(A) Sequence of the L7a(101–161) recombinant polypeptide. Capital letters indicate the amino acid residues of L7a; the corresponding position in the protein is indicated in parentheses. Lower case letters indicate the amino acid residues added by the cloning strategy. (B) Schematic representation of the results obtained from limited proteolysis experiments on L7a(101–161) in the absence (upper panel) and in the presence of poly(G) (lower panel). Preferential proteolytic sites occurring only in the native protein are shown in black; those recognized by proteases in the complex are shown in grey, and those identified in all conditions are shown in white.
Limited proteolysis on the L7a(101–161)–RNA complex
The complex of L7a(101–161) with RNA was formed by incubating the protein with an equimolar excess of RNA solution in water for 15 min at 25 °C in Tris/HCl buffer 50 mM, pH 7.5. The complex was then submitted to limited proteolysis experiments as described above. Controlled trypsin hydrolysis showed that Arg49 and Lys63 were the preferential cleavage sites (Figure 7B). ESMS analysis of individual tryptic peptides released from the complex revealed the complementary peptide pair 1–49 (5670.9±0.9 Da) and 50–76 (2875.1±0.3 Da). These results demonstrate that a proteolytic event occurred at the level of Arg49. At later stages of incubation, a secondary cleavage site at Lys63 was inferred by the presence of species 1–63 and 64–76. Similar analyses with endoproteases LysC and GluC showed the occurrence of preferential cleavages at Lys33, and Lys63 and Asn53 respectively, as revealed by the release of the complementary species 1–33/34–76 and 1–63/64–76 and 1–53/54–76. The chymotryptic experiment identified fragments 15–76 and 28–76, thereby indicating Tyr14 and Leu27 as preferential cleavage sites. Finally, the limited proteolysis experiments with subtilisin identified Leu58 and Asn53 (fragments 1–58 and 59–76 and 1–53 and 54–76) as primary sites of hydrolysis.
A comparison of the results of the limited proteolysis experiments carried out with the L7a(101–161)–RNA complex (Figure 8B and Table 1) versus results obtained with the isolated L7a(101–161) protein clearly show that the poly(G) molecule had a specific shielding effect. The N-terminal region encompassing residues 21–32, easily accessible to proteases in L7a(101–161), became completely protected in the complex. On the contrary, the preferential cleavage sites were similarly distributed in the C-terminal part of the protein; the small differences found could be due to conformational changes consequent to formation of the complex. These limited proteolysis data indicate that the N-terminal segment 21–32 of L7a(101–161) specifically interacted with a poly(G) molecule.
Cross-linking experiments
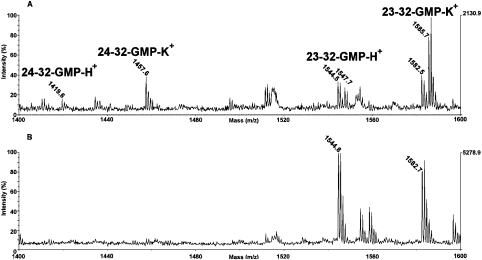
A complementary approach with which to investigate the formation of a complex between L7a(101–161) and RNA is to determine the formation of photo-chemical cross-links between the protein and the ligand. Identification of the cross-linked residues would indicate amino acid residues that specifically interact with the RNA molecule in the complex. We carried out cross-linking experiments at room temperature and at pH 7.5 by incubating the protein with an equimolar amount of RNA. The sample was then irradiated with UV light at 254 nm and the cross-linked products were digested with trypsin. The peptide mixture was submitted to enzymic hydrolysis with T1 ribonuclease to digest the polynucleotide chain. The resulting peptide mixture was directly analysed by MALDI-MS to identify the amino acid residues involved in the covalent linkages. Figure 9 shows the MALDI spectra obtained. Most of the signals were assigned to L7a fragments on the basis of their molecular mass and the specificity of the enzyme. However, the mass signals at m/z 1419.6 and 1457.6 could not be assigned to any fragment within the protein sequence and were thus candidates for intermolecularly cross-linked fragments. This hypothesis was supported by the observation that all these signals were accompanied by 38-Da higher satellite peaks, which correspond to a potassium adduct characteristic of nucleotide-containing samples. On the basis of the L7a(101–161) sequence and ribonuclease T1 digestions, the two signals were identified as peptides 24–32 and 23–32 linked to a GMP moiety. These results confirm the limited proteolysis data on the L7(101–161)–poly(G) complex in that the protein displays a specific binding site for the polynucleotide located within the N-terminal segment 23–32.
Figure 9. Cross-linking experiment on the L7a(101–161)–poly(G) complex.
Partial MALDI-MS spectrum of the cross-linked product (A) and of the control isolated protein (B) digested with trypsin and T1 ribonuclease. The attribution of the cross-linked peptides is shown.
DISCUSSION
In a first set of experiments we established that human rpL7a, when tested in vitro for RNA binding to synthetic ribo-homopolymers, specifically bound poly(G). Like other RNA-binding proteins in vitro, poly(G) presumably mimics RNA secondary structures that are sufficiently adaptable for specific binding [18]. The RNA-binding ability of L7a was confirmed by results showing the binding of L7a to rRNA and to the mRNA of L7a itself (Figures 1 and 2). The apparent Kds were 85 nM for 28 S RNA and 75 nM for L7a mRNA, which are in the range observed when assaying the RNA-binding activity of individual r-proteins in vitro. The RNA-binding activity is much greater in vivo because of co-operative interactions with other macromolecules [19]. When the binding of L7a to its own mRNA (100% in Figure 2) was challenged by increasing amounts of various nucleic acids as competitors, the specificity results were: poly(G)>L7a mRNA>poly(A)− RNA>dsDNA>tRNA>ssDNA>poly(C). Poly(G) proved to be a better competitor than L7a mRNA itself. Binding was also affected by competition with unfractionated rRNA [poly(A)− RNA], whereas tRNA and dsDNA, although highly folded molecules, competed poorly for binding. These results, although not identifying a specific target, indicate that complex folded molecules, such as mRNA and rRNA, carry structures preferentially recognized by protein L7a.
We previously demonstrated that a domain of protein L7a spanning aa 52–100 is essential for the nucleolar accumulation of L7a ([3], and see also Figure 3B). This observation suggested that the nucleolar targeting of L7a might occur through interactions of the 52–100 aa domain with macromolecules (RNA or protein) residing in the nucleolus. Koonin et al. [10] suggested that r-protein L7a exerts RNA-binding activity, because it contains a signature domain (spanning aa 130–160 in L7a) shared by a number of RNA-binding proteins. Subsequently, the predicted RNA-binding motif was shown to be functional in the L7ae r-protein from H. marismortui [13], in the yeast rpL30 protein [22], in the 15.5 kDa protein of the splicing apparatus [21], and in the human SBP2 protein [20]. In addition, a relevant amount of structural information became available when a crystal structure of the 15.5 kDa protein bound to the U4 snRNA target site was elucidated [14], and when the structure of the protein–RNA complex involving yeast rpL30 and its target mRNA was resolved by NMR [22]. This large body of data prompted us to analyse the RNA-binding ability of some L7a domains whose function we had characterized [3].
Given the lack of natural targets of L7a, we used L7a mRNA and human 28 S rRNA in the RNA-binding assays, because the whole L7a protein specifically binds to these RNA species (Figures 1 and 2). The results of the analysis of RNA binding activity of several L7a-derived polypeptides (Figure 3), among which a polypeptide defining the domain competent for accumulation of L7a in the nucleoli, revealed two distinct RNA-binding domains in L7a: RNAB1 (aa 52–100) and RNAB2 (aa 101–161). The Kd, determined graphically (Figure 4C), for each peptide versus each RNA probe was in the order of 0.1 μM. Thus the binding affinity was low. This is not surprising, because in vitro experiments of r-protein binding to RNA do not reproduce the cooperative interactions that make the RNA–protein binding in the ribosome subunit irreversible [19], and that could be required for efficient regulation of L7a binding to some non-ribosomal target RNAs. The effect of lack of co-operation plus a spurious RNA target might have been even more dramatic, and could have caused the weaker RNA binding obtained with the isolated protein domains (Figure 4, and see also [31]).
A database search did not show any significant similarity between the RNAB1 domain of L7a and other known RNA-binding motifs. However, the RNAB1 domain appeared to be strikingly conserved in Eukarya L7a, but lacking in the Archea L7a orthologues (Figure 5A). In the light of these observations, and of the data demonstrating that RNAB1 is essential for nucleolar targeting of human r-protein L7a [3], RNAB1 might represent a new RNA-binding motif that is committed to a special eukaryotic function.
The calculated Kds of RNAB1 for these L7a mRNA and rRNA are in the micromolar range (0.8 μM and 1.5 μM, respectively). The low affinity of the RNAB1 domain for these RNA species may be because we used non-specific targets, i.e. L7a mRNA and rRNA, or because of a lack of co-operative interactions with other macromolecules that regulate the RNA-binding efficiency in vivo. The identification of a specific physiological RNA target of RNAB1 will clarify the function of this L7a protein domain.
The RNAB2 domain is highly conserved among Eukarya L7a orthologues (Figure 5B), whereas the similarity with the Archea L7ae protein is restricted to the region that includes the RNA-binding motif predicted by Koonin et al. [10]. However, the similarity extends further at the C-terminus in proteins that recognize the secondary structure motif designated kink-turn [15] (Figure 5C). Since binding to a kink-turn seemed to be a common functional feature of L7ae proteins, we carried out experiments to verify whether human r-protein L7a would bind to an oligonucleotide fragment of spliceosomal U4 snRNA, which had been crystallized bound to the 15.5 kDa protein [14]. To our surprise, neither the whole protein L7a (see Figure 6) nor its isolated RNA-binding domains bound to the U4 snRNA oligonucleotide. Archea L7a was recently found to bind in vitro to Archea sRNAs of the C/D box family, which contain a kink-turn RNA motif [32]. By filter-binding assay, we did not detect any specific interaction of human r-protein L7a with U14 or U24 snoRNA, both of which belong to the C/D box family of Eukarya snoRNAs.
In the absence of a natural RNA target for the human L7a r-protein, we carried out a structural analysis of the complex formed by the RNAB2 domain of r-protein L7a and poly(G). Our aim was to obtain information about the structural modifications undergone by the r-protein after its binding to the surrogate RNA target. We used the limited proteolysis technique followed by ESMS analysis of the products (Figures 7 and 8, and Table 1), and cross-linking experiments followed by MALDI-MS to identify amino acid residues involved in covalent linkages (Figure 9). The results obtained with the two methodological approaches led to the identification of a region (KQRLLARAEK) that was shielded from proteases consequent to binding to poly(G) and that was involved in covalent linkages after UV cross-linking. Allmang et al. [20] addressed the interesting issue of how the various L7ae proteins specifically recognize their cognate RNA, despite a high degree of similarity in their RNA-binding domain by comparing the amino acid residues relevant for contact with the cognate RNA in human SBP2 and 15.5 kDa proteins [20]. They identified 12 aa residues that are important for SECIS RNA binding: most contact residues were predicted from the sequence similarity between SBP2 and 15.5 kDa RNA-binding domains, albeit their contribution to binding efficiency was different. The contact role played by three residues could not be predicted from sequence similarity, which suggested that non-conserved amino acids could be instrumental in determining the specificity of interaction. Our findings lend weight to this hypothesis. In addition, they raise the possibility that a region flanking the RNA-binding domain might contribute to the specificity of binding of a protein to its cognate RNA.
Acknowledgments
This work has been supported by MIUR, Progetti di Ricerca di Interesse Nazionale (PRIN 2003) and Fondo Investimenti Ricerca di Base (FIRB). We thank Jean A. Gilder for editing the text.
References
- 1.Hernandez-Verdun D., Roussel P., Gebrane-Younes J. Emerging concepts of nucleolar assembly. J. Cell Sci. 2002;115:2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- 2.Rudt F., Pieler T. Cytosolic import factor- and Ran-independent nuclear transport of ribosomal protein L5. Eur. J. Cell Biol. 2001;80:661–668. doi: 10.1078/0171-9335-00212. [DOI] [PubMed] [Google Scholar]
- 3.Russo G., Ricciardelli G., Pietropaolo C. Different domains cooperate to target the human ribosomal L7a protein to the nucleus and to the nucleoli. J. Biol. Chem. 1997;272:5229–5235. doi: 10.1074/jbc.272.8.5229. [DOI] [PubMed] [Google Scholar]
- 4.Damelin M., Silver P. A., Corbett A. H. Nuclear protein transport. Methods Enzymol. 2002;351:587–607. doi: 10.1016/s0076-6879(02)51870-x. [DOI] [PubMed] [Google Scholar]
- 5.Jakel S., Mingot J. M., Schwarzmaier P., Hartmann E., Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosorius O., Fries B., Stauber R. H., Hirschmann N., Bevec D., Hauber J. Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem. 2000;275:12061–12068. doi: 10.1074/jbc.275.16.12061. [DOI] [PubMed] [Google Scholar]
- 7.Etheridge K. T., Banik S. S., Armbruster B. N., Zhu Y., Terns R. M., Terns M. P., Counter C. M. The nucleolar localization domain of the catalytic subunit of human telomerase. J. Biol. Chem. 2002;277:24764–24770. doi: 10.1074/jbc.M201227200. [DOI] [PubMed] [Google Scholar]
- 8.Lee C. H., Chang S. C., Chen C. J., Chang M. F. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 1998;273:7650–7656. doi: 10.1074/jbc.273.13.7650. [DOI] [PubMed] [Google Scholar]
- 9.Lixin R., Efthymiadis A., Henderson B., Jans D. A. Novel properties of the nucleolar targeting signal of human angiogenin. Biochem. Biophys. Res. Commun. 2001;284:185–193. doi: 10.1006/bbrc.2001.4953. [DOI] [PubMed] [Google Scholar]
- 10.Koonin E. V., Bork P., Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henras A., Henry Y., Bousquet-Antonelli C., Noaillac-Depeyre J., Gelugne J. P., Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nottrott S., Hartmuth K., Fabrizio P., Urlaub H., Vidovic I., Ficner R., Luhrmann R. Functional interaction of a novel 15.5 kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 14.Vidovic I., Nottrott S., Hartmuth K., Luhrmann R., Ficner R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 15.Klein D. J., Schmeing T. M., Moore P. B., Steitz T. A. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Falco S., Russo G., Angiolillo A., Pietropaolo C. Human L7a ribosomal protein: sequence, structural organization, and expression of a functional gene. Gene. 1993;126:227–235. doi: 10.1016/0378-1119(93)90371-9. [DOI] [PubMed] [Google Scholar]
- 17.Carey J., Uhlenbeck O. C. Kinetic and thermodynamic characterization of the R17 coat protein–ribonucleic acid interaction. Biochemistry. 1983;22:2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- 18.Fabre E., Boelens W. C., Wimmer C., Mattaj I. W., Hurt E. C. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 19.Draper D. E. RNA–protein interactions in ribosomes. In: Hames B. D., Glover D. M., editors. Frontiers in Molecular Biology, vol. 6. Oxford University Press; 1994. pp. 82–102. [Google Scholar]
- 20.Allmang C., Carbon P., Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn J. F., Tran E. J., Maxwell E. S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao H., White S. A., Williamson J. R. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- 23.Watkins N. J., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 24.Watkins N. J., Newman D. R., Kuhn J. F., Maxwell E. S. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA. 1998;4:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu L. H., Henry Y., Nicoloso M., Michot B., Azum M. C., Renalier M. H., Caizergues-Ferrer M., Bachellerie J. P. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28 S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zappacosta F., Pessi A., Bianchi E., Venturini S., Sollazzo M., Tramontano A., Marino G., Pucci P. Probing the tertiary structure of proteins by limited proteolysis and mass spectrometry: the case of Minibody. Protein Sci. 1996;5:802–813. doi: 10.1002/pro.5560050502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zappacosta F., Ingallinella P., Scaloni A., Pessi A., Bianchi E., Sollazzo M., Tramontano A., Marino G., Pucci P. Surface topology of Minibody by selective chemical modifications and mass spectrometry. Protein Sci. 1997;6:1901–1909. doi: 10.1002/pro.5560060911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scaloni A., Miraglia N., Orru S., Amodeo P., Motta A., Marino G., Pucci P. Topology of the calmodulin–melittin complex. J. Mol. Biol. 1998;277:945–958. doi: 10.1006/jmbi.1998.1629. [DOI] [PubMed] [Google Scholar]
- 29.Orru S., Dal Piaz F., Casbarra A., Biasiol G., De Francesco R., Steinkuhler C., Pucci P. Conformational changes in the NS3 protease from hepatitis C virus strain Bk monitored by limited proteolysis and mass spectrometry. Protein Sci. 1999;8:1445–1454. doi: 10.1110/ps.8.7.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monti M., Principe S., Giorgetti S., Mangione P., Merlini G., Clark A., Bellotti V., Amoresano A., Pucci P. Topological investigation of amyloid fibrils obtained from beta2-microglobulin. Protein Sci. 2002;11:2362–2369. doi: 10.1110/ps.0206902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Mikecz A., Neu E., Krawinkel U., Hemmerich P. Human ribosomal protein L7 carries two nucleic acid-binding domains with distinct specificities. Biochem. Biophys. Res. Commun. 1999;258:530–536. doi: 10.1006/bbrc.1999.0682. [DOI] [PubMed] [Google Scholar]
- 32.Rozhdestvensky T. S., Tang T. H., Tchirkova I. V., Brosius J., Bachellerie J. P., Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]