Abstract
The clinical manifestation of leishmaniasis has historically been determined by the Leishmania species involved. However, recent emergence of novel Leishmania lineages has caused atypical pathologies. We isolated and characterized 2 new Leishmania donovani parasites causing cutaneous leishmaniasis in Himachal Pradesh, India.
Keywords: leishmaniasis, Leishmania donovani, cutaneous leishmaniasis, hybrid parasites, whole-genome sequencing, Himachal Pradesh, surveillance, parasites, India
Leishmaniasis is a neglected tropical disease caused by the protozoan parasite Leishmania. The manifestation of the disease has historically been species-specific: Leishmania donovani and Leishmania infantum cause visceral leishmaniasis (VL), also called kala-azar, and many species such as Leishmania tropica and Leishmania major cause cutaneous leishmaniasis (CL) (1). In recent years, however, the existence of interspecies and intraspecies hybrids has emerged, and hybridization has been associated with a potential cause of CL in Sri Lanka (2) and Himachal Pradesh, India (3). In Sri Lanka, CL is mostly caused by an atypical L. donovani (4–6) and CL cases were recently observed to be associated with L. donovani/L. major hybrids or L. donovani/L. tropica hybrids (2). CL is an emerging disease in Himachal Pradesh, where a recently identified L. donovani intraspecies hybrid isolated from a CL patient belonged to the Indian subcontinent 1 (ISC1) Yeti clade (3). Further, the recent discovery of ISC1 Leishmania parasites in the neighboring region of West-Nepal supports the establishment of the ISC1 clade in the area (7). Therefore, continued monitoring for emergence of CL in Himachal Pradesh is necessary to identify new L. donovani lineages associated with cutaneous disease outcomes. We report 2 new cases of CL in Himachal Pradesh caused by L. donovani belonging to the ISC1 Yeti clade that are not hybrid parasites previously identified from this region (GenBank BioProject no. PRJNA701770) (Appendix Figure 1) (3). Genome surveillance of CL parasites coming from Himachal Pradesh can help identify gene sequences associated with CL disease outcomes and identify the origin and transmission of emerging L. donovani parasites.
We performed a phylogenetic analysis by adding LdHPCL71 and LdHPCL76 to a previously generated tree containing 685 whole-genome L. donovani isolates (3). As shown previously, the topology of the tree matches the geographic origin of the samples used (2). Consistent with the previous report from Himachal Pradesh (3), phylogenetic analysis of the new LdHPCL71 and LdHPCL76 CL lineages revealed that they also clustered within the ISC1 Yeti clade of L. donovani (Figure). Because the previously identified cutaneous lineage LdHPCL66 from Himachal Pradesh was an intraspecific hybrid (3), we next investigated whether the LdHPCL71 and LdHPCL76 parasites were also hybrids. We used VarScan (https://varscan.sourceforge.net) to identify all single-nucleotide polymorphisms (SNPs) by using the Sri Lanka CL L. donovani reference strain (6) and compared the SNP frequencies with previous data on hybrid parasites (Appendix Figure 2, panel A). We plotted the full genomic representation for all 36 chromosomes for each parasite by using Circos (https://circos.ca) (Appendix Figure 2, panel B).
Figure.
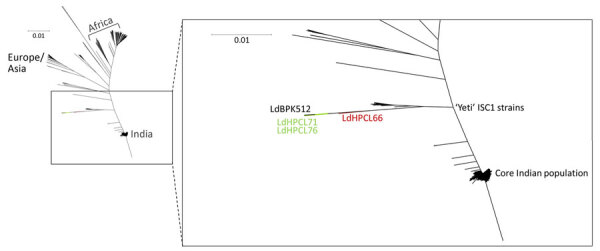
Phylogeny of Leishmania donovani lineages associated with cutaneous leishmaniasis, Himachal Pradesh, India, 2023. Novel L. donovani from this study (green) are compared with the global population of the 684 parasites previously reported in the L. donovani complex, including reference strains L. donovani LV9, L. donovani BHU 1220, and L. donovani BPK282A1. The previously isolated interspecific hybrid LdHPCL66 (red) falls halfway between the unique LDBPK512 and Yeti-ISC1 lineages. The novel LdHPCL71 and LdHPCL76 (green) nonhybrid parasites are more closely related to the unique LdBPK512 parasite. Scale bar indicates the modified Euclidian distance as calculated by TASSEL (https://tassel.bitbucket.io).
The heterozygosity index of LdHPCL71 was 0.168 and of LdHPCL76 was 0.158, suggesting that they are not hybrids because the known hybrids have been shown to contain a large portion of heterozygous SNPs (2,3). In addition, the many SNPs seen (Appendix Figure 2, panel B) are homozygous, indicating the lineages are distant from the core L. donovani parasite population in India but neither parasite seems to be a hybrid. We compared the SNPs from each of the newly identified parasites with the SNPs from the previously isolated LdHPCL66 hybrids from the Yeti ISC1 group to determine if a subset of SNPs is common among the 3 lineages (Appendix Table). We found too many SNPs in common between the parasites to specifically associate any of them with the CL manifestations in patients because they are from 3 divergent lineages in an undersampled population.
Our data, combined with other reports of ISC1 spread (7), could support the theory that atypical L. donovani parasites are being increasingly encountered as a cause of CL in the Indian subcontinent. That hypothesis is further supported by a recent report on occurrence of CL cases in provinces in Nepal caused by L. donovani that are endemic and nonendemic for visceral disease (8). Of potential concern, this clade now includes both hybrid and nonhybrid parasites able to cause VL and CL. Emergence of such CL-causing L. donovani parasites highlights the urgent need for molecular surveillance as an integral part of the ongoing kala-azar elimination program in the Indian subcontinent. The ongoing regional strategic framework emphasizes the need to control post–kala-azar dermal leishmaniasis cases as a parasite reservoir to break the transmission cycle for sustaining the elimination program (9). On a similar note, CL cases caused by atypical L. donovani genotypes may also contribute to the transmission cycle of VL in some patients. Our observations support the argument that surveillance of atypical L. donovani lineages associated with CL should be included in VL elimination programs.
Additional information on emerging Leishmania donovani lineages associated with cutaneous leishmaniasis, Himachal Pradesh, India, 2023.
Acknowledgments
We thank Rentala Madhubala for generously gifting L. donovani and L. major standard cultures used as controls in initial species identification (data not shown).
Informed consent was obtained from the patients with due approval of the study by the Institutional Ethics Committee of Indira Gandhi Medical College, approval no. HFW (MS) G-5 (Ethics)/2014-10886.
G.M. and P.L. received support from the Canadian Institutes of Health Research (G.M. grant no. 153282, P.L. grant no. 187858). M.J. received a collaborative research grant from International Centre for Genetic Engineering and Biotechnology, Trieste, Italy (project no. CRP/IND19-01) under collaborative research project and Indian Council of Medical Research, ICMR (no. 6/9-7 272/KA/2021/ECD-II). Y.C. is supported by a fellowship from University Grant Commission, India. K.P. is supported by the Fonds de recherche du Québec (doctoral award).
Biography
Dr. Lypaczewski is a researcher with the Department of Microbiology & Immunology at McGill University. His research interests are parasitic diseases and all aspects of microbial genetics, including sample collection, DNA extraction, sequencing, and bioinformatics.
Footnotes
Suggested citation for this article: Lypaczewski P, Chauhan Y, Paulini K, Thakur L, Chauhan S, Roy EI, et al. Emerging Leishmania donovani lineages associated with cutaneous leishmaniasis, Himachal Pradesh, India, 2023. Emerg Infect Dis. 2024 Sep [date cited]. https://doi.org/10.3201/eid3009.231595
These first authors contributed equally to this article.
These authors were co–principal investigators.
References
- 1.Burza S, Croft SL, Boelaert M. Leishmaniasis. Lancet. 2018;392:951–70. 10.1016/S0140-6736(18)31204-2 [DOI] [PubMed] [Google Scholar]
- 2.Lypaczewski P, Matlashewski G. Leishmania donovani hybridisation and introgression in nature: a comparative genomic investigation. Lancet Microbe. 2021;2:e250–8. 10.1016/S2666-5247(21)00028-8 [DOI] [PubMed] [Google Scholar]
- 3.Lypaczewski P, Thakur L, Jain A, Kumari S, Paulini K, Matlashewski G, et al. An intraspecies Leishmania donovani hybrid from the Indian subcontinent is associated with an atypical phenotype of cutaneous disease. iScience. 2022;25:103802. 10.1016/j.isci.2022.103802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thakur L, Singh KK, Shanker V, Negi A, Jain A, Matlashewski G, et al. Atypical leishmaniasis: A global perspective with emphasis on the Indian subcontinent. PLoS Negl Trop Dis. 2018;12:e0006659. 10.1371/journal.pntd.0006659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang WW, Ramasamy G, McCall LI, Haydock A, Ranasinghe S, Abeygunasekara P, et al. Genetic analysis of Leishmania donovani tropism using a naturally attenuated cutaneous strain. PLoS Pathog. 2014;10:e1004244. 10.1371/journal.ppat.1004244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lypaczewski P, Hoshizaki J, Zhang WW, McCall LI, Torcivia-Rodriguez J, Simonyan V, et al. A complete Leishmania donovani reference genome identifies novel genetic variations associated with virulence. Sci Rep. 2018;8:16549. 10.1038/s41598-018-34812-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monsieurs P, Cloots K, Uranw S, Banjara MR, Ghimire P, Burza S, et al. Source tracing of Leishmania donovani in emerging foci of visceral leishmaniasis, Western Nepal. Emerg Infect Dis. 2024;30:611–3. 10.3201/eid3003.231160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rai T, Shrestha S, Prajapati S, Bastola A, Parajuli N, Ghimire PG, et al. Leishmania donovani persistence and circulation causing cutaneous leishmaniasis in unusual-foci of Nepal. Sci Rep. 2023;13:12329. 10.1038/s41598-023-37458-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A, Singh VK, Tiwari R, Madhukar P, Rajneesh, Kumar S, et al. Post kala-azar dermal leishmaniasis in the Indian sub-continent: challenges and strategies for elimination. Front Immunol. 2023;14:1236952. 10.3389/fimmu.2023.1236952 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information on emerging Leishmania donovani lineages associated with cutaneous leishmaniasis, Himachal Pradesh, India, 2023.


