Abstract
Proteinase 3C of hepatitis A virus (HAV) plays a key role in the viral life cycle by generating mature viral proteins from the precursor polyprotein. In addition to its proteolytic activity, 3C binds to viral RNA, and thus influences viral genome replication. In order to investigate the interplay between proteolytic activity and RNA binding at the molecular level, we subjected HAV 3C and three variants carrying mutations of the cysteine residues [C24S (Cys-24→Ser), C172A and C24S/C172A] to proteolysis assays with peptide substrates, and to surface plasmon resonance binding studies with peptides and viral RNA. We report that the enzyme readily forms dimers via disulphide bridges involving Cys-24. Dissociation constants (KD) for peptides were in the millimolar range. The binding kinetics for the peptides were characterized by kon and koff values of the order of 102 M−1·s−1 and 10−2 to 10−1 s−1 respectively. In contrast, 3C binding to immobilized viral RNA, representing the structure of the 5′-terminal domain, followed fast binding kinetics with kon and koff values beyond the limits of the kinetic resolution of the technique. The affinity of viral RNA depended strongly on the dimerization status of 3C. Whereas monomeric 3C bound to the viral RNA with a KD in the millimolar range, dimeric 3C had a significantly increased binding affinity with KD values in the micromolar range. A model of the 3C dimer suggests that spatial proximity of the presumed RNA-binding motifs KFRDI is possible. 3C binding to RNA was also promoted in the presence of substrate peptides, indicating co-operativity between RNA binding and protease activity. The data imply that the dual functions of 3C are mutually dependent, and regulate protein and RNA synthesis during the viral life cycle.
Keywords: dimerization, hepatitis A virus, proteinase 3C, RNA binding, surface plasmon resonance (SPR), NMR
Abbreviations: Ac, N-acetyl; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; Fmoc, fluoren-9-ylmethoxycarbonyl; HAV, hepatitis A virus; pNA, p-nitroanilide; RT-PCR, reverse transcription PCR; RU, response units; SPR, surface plasmon resonance; STD NMR, saturation transfer difference NMR; C24S, Cys24→Ser; 3C, proteinase 3C
INTRODUCTION
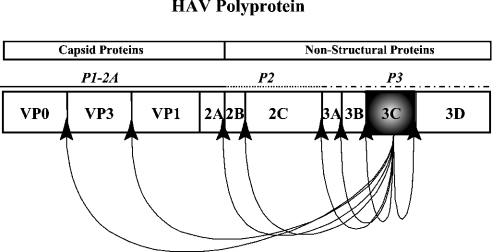
3C proteinases of picornaviruses (picornain 3C, EC 3.4.22.28) form a unique class of cysteine proteinases with a three-dimensional structure similar to that of chymotrypsin. As the major virus-encoded proteinase, 3C catalyses the cleavage of the polyprotein that is translated from the viral RNA. By its ability to liberate intermediate and mature viral proteins, 3C is a key player in the gene expression strategy of picornaviruses, and has been identified as a promising target for specific antiviral therapies of infections with picornaviruses, such as coxsackievirus, poliovirus, rhinovirus and hepatitis A virus (HAV) [1,2]. 3C maps to the P3 domain of the viral polyprotein, and is flanked by the RNA-dependent RNA polymerase 3D and the membrane-targeted primer protein 3AB. As an integral part of the polyprotein, proteinase 3C cleaves at eight specific sites in the viral polyprotein (Figure 1), and thus initiates both assembly of the viral capsid and the formation of the viral replication complex that is formed by proteins of the P2 and P3 domains and catalyses viral genome synthesis. In order to compensate for its restricted coding capacity, multiple functions are performed by individual portions of the picornaviral polyprotein. The activity and specificity of 3C proteinases is modulated by viral cofactors either covalently linked to 3C, e.g. 3AB or 3D, or specifically interacting with the proteinase [3,4].
Figure 1. 3C cleavage sites in the HAV polyprotein.
The three-dimensional structures of 3C of various picornaviruses have been determined by X-ray diffraction and found to be related to the chymotrypsin-like serine proteinases [5–11]. 3C proteinases that fold into a two-domain β-barrel structure display a thiol function of a cysteine residue as the active site nucleophile. Due to the shallow, and possibly more flexible, substrate-binding groove of HAV 3C, it might have a wider substrate specificity than, for example, poliovirus 3C [5–9]. Interestingly, it has been reported recently that HAV 3C differs from poliovirus 3C in its rapid degradation by the ubiquitin–proteasome system, which might explain its low concentration in HAV-infected cells [12].
In addition to its proteolytic activity, the ability to specifically bind viral RNA is unique to picornaviral proteinase 3C (and its precursors) and has attracted much attention for its role in viral RNA genome replication. For poliovirus 3C, the RNA-binding site has been mapped to the highly conserved amino acid motif KFRDI [13,14], which is also present in HAV 3C located on the side opposite to the active site of the enzyme. The secondary structures formed at the 5′-end of the genomes of poliovirus, rhinovirus and HAV were identified as specific RNA ligands [13–17]. Structural prerequisites for both viral RNA and 3C have been defined, and the role of the RNA–3C interaction in regulating viral RNA synthesis has been shown for poliovirus [13,14]. During the viral replication cycle, the positive-stranded picornaviral genome first serves as template for translation, before it is used as template for RNA synthesis. For poliovirus, it is now assumed that by binding to the 5′-end of the viral genome, 3C in its precursor form 3CD is involved in the switch from translation to RNA synthesis, two processes that use the same RNA template, yet in opposite directions [15]. Although 3C substrate specificity and cleavage activity, as well as the interaction with viral RNA, have been studied in some detail for 3C of various picornaviruses, including HAV, fundamental questions on the molecular requirements for both functions remain to be addressed [16–20]. It is an open question if, and to what extent, RNA binding and polyprotein cleavage by 3C are mutually modulating processes.
The comprehensive characterization of the multiple functions of 3C is of prime importance for the understanding of the finely tuned steps of protein and RNA synthesis in the viral life cycle. Our initial experiments were therefore aimed at characterizing HAV 3C binding to viral RNA and substrate peptides, and to investigate the interplay of proteolytic and RNA binding activity of HAV 3C at the molecular level. We have now assessed 3C binding to substrate peptides and to RNA using SPR (surface plasmon resonance). In this biosensor analysis, complex formation is monitored with one binding partner immobilized on the sensor chip, and the other component being passed over the chip surface in an aqueous buffer [21]. One advantage of this assay is that it allows the detection of low-affinity or transient interactions, with KD values in the millimolar range [22]. In addition, minimal amounts of the immobilized component are required. Wild-type HAV 3C and three mutants were used in combination with synthetic peptides representing the proteolytic cleavage sites, and synthetic RNA representing the 5′-end of the viral genome. Surprisingly, large differences were identified for the RNA-binding affinity of wild-type and mutant enzyme. Furthermore, the effect of peptide binding on the RNA-binding affinity of HAV 3C was assessed. 3C binding to RNA was significantly improved by substrate peptides. The potential biological implications of these observations are discussed.
EXPERIMENTAL
Expression and purification of recombinant 3C proteins
Wild-type HAV 3C proteinase and mutants carrying replacements of either one or both of the two cysteine residues at amino acid positions 24 and 172 [C24S (Cys-24→Ser), C172A and C24S/C172A] were kindly provided by Professor John C. Vederas (Department of Chemistry, University of Alberta, Edmonton, Alberta, Canada). Recombinant proteins were expressed in Escherichia coli XL-2 and purified as described previously [18,23,24]. The final protein concentration was 0.5 to 1 mg/ml. The proteins were stored at −70 °C in buffer containing 6 mM DTT (dithiothreitol). The presence of DTT was important for storage, however, for the SPR experiments it was necessary to transfer purified 3C to a DTT-free buffer prior to its immobilization on the CM5 chip. The purified protein was transferred into de-gassed HM buffer (10 mM Hepes, pH 7.4, 150 mM NaCl and 6 mM MgCl2), and, using a Vivaspin concentrator, 3C dimerization was checked by SDS/PAGE (12% gel) under reducing and non-reducing conditions.
SPR analysis to determine peptide binding to immobilized 3C
All experiments were performed at 25 °C on the Biacore 3000™. 3000–4000 RU (response units) of DTT-free preparations of recombinant 3C were immobilized on a CM5 chip (Biacore) at a flow rate of 5 μl/min after the dextran matrix had been activated for 20 min with EDC/NHS [N-(3-dimethyl-aminopropyl)-N-ethylcarbodi-imide/N-hydroxysuccinimide], according to the manufacturer's instructions. Peptide analytes Ac-ELRTQ-pNA and Ac-LRTQ-pNA (where Ac is N-acetyl, and pNA is p-nitroanilide) were diluted in HMD (HM buffer containing 6 mM DTT) at various concentrations (0.025 to 8 mM). Data were analysed and simulated using the BIAevaluation software, version 3.2.
Synthesis of peptides and 3C activity assay
Ac-ELRTQ-pNA was synthesized using an Fmoc (fluoren-9-ylmethoxycarbonyl)-based solid-phase synthesis strategy, as described previously [25]. The other peptides (Ac-LRTQ-amide, Ac-LAAQ-OH, Ac-ELRTQSFS-amide, Ac-ARTQSFSN-amide, Ac-LRTQFSFN-OH, Ac-WRTQFSFNL-OH, Ac-LRTQSGSNL-OH, Ac-ARTQSFSNL-OH and H2N-PGRAF-OH) were synthesized on a peptide synthesizer (Advanced ChemTech) by solid-phase synthesis using the Fmoc strategy. The proteolytic activity of the recombinant proteins was determined in a multiwell plate with Ac-ELRTQ-pNA as substrate. The amino acid sequence ELRTQ is part of the natural 3C cleavage sequence at the 2B/2C site in the P2 domain of the polyprotein (Figure 1 and [19]). The A405 was monitored over 6 min at room temperature (20 °C). For inhibition studies, 10 mM leupeptin (Bachem), a reversible inhibitor of cysteine proteinases, and 1 mM peptides (Ac-LRTQ-OH, Ac-LAAQ-OH, Ac-ELRTQSFS-NH2, Ac-ARTQSFSN-NH2 or H2N-PGRAF-OH) were added to a solution of 3C (3 μM) in assay buffer containing 6 mM DTT.
Preparation and infectivity of the full-length HAV RNA containing the C24S mutation
A cDNA construct containing the full-length HAV genome with the C24S mutation in 3C was prepared by two-primer PCR-based site-directed mutagenesis (QuikChange, Stratagene) using appropriate primers. The sequence of the mutated region was verified (AGOWA, Berlin, Germany). Synthetic in vitro transcripts of the wild-type construct and the 3C C24S mutant were transfected into Huh7 cells as described previously [4]. To determine the nucleotide sequence of the rescued virus, viral RNA obtained after the second cell-culture passage was reverse-transcribed and PCR-amplified with HAV 3C-specific primers. The RT-PCR (reverse transcription PCR) amplification product was used for nucleotide sequencing.
RNA in vitro transcripts, RNA 3′-biotinylation and interaction with 3C
Radioactively labelled and unlabelled run-off HAV RNA transcripts [RNA(1–148) and RNA(46–148)] were prepared from linearized plasmids as described previously [16]. RNA interaction of reduced and non-reduced HAV 3C was assessed by EMSAs (electrophoretic mobility-shift assays) with RNA(1–148) [16]. In vitro transcripts of HAV RNA(1–148) and RNA(46–148) were biotinylated at their 3′-end by periodate oxidation as described previously [26,27]. The amount and purity of biotinylated RNA was checked by gel-shift experiments on a 0.8% agarose gel using streptavidin as ligand. Between 20 and 50% of the RNA was found to be biotinylated. The final concentration of biotinylated RNA(1–148) and RNA(46–148) were 7 μM and 3 μM respectively.
SPR analysis with immobilized RNA
Biotinylated RNA was immobilized on a streptavidin-coated sensor chip (SA chip, Biacore). Before immobilization, the SA chip was treated 3 times for 60 s with 1 M NaCl in 50 mM NaOH at a flow rate of 5 μl/min. For immobilization, 14–30 nM solutions of biotinylated RNA in immobilization buffer (10 mM Hepes, pH 6.8, 600 mM NaCl and 1 mM EDTA) were injected until 800–1000 RU were obtained. Saturation of residual free streptavidin-binding sites was achieved with a 0.4% solution of biotin. Recombinant proteins (analytes) were prepared immediately before use by diluting them in HM or HMD buffers. Samples were injected for 300 s; the dissociation time was 300 s. Binding efficiency of the regenerated RNA chip was tested by reprobing with 3C. The RNA chip was used for about 100 injections. The data were analysed with the BIAevaluation software version 3.2.
Binding of 3C-peptide complexes to immobilized RNA
Binding of 3C (0.4 μM) to immobilized RNA(1–148) in the presence of up to a 500-fold molar excess of peptide (Ac-LRTQ-amide, Ac-ARTQSFSN-amide, Ac-LAAQ-OH, and H2N-PGRAF-OH as negative control) was compared with direct binding of the 1 to 2 mM peptide solution to the RNA chip.
Saturation transfer difference NMR experiments and molecular modelling
STD (saturation transfer difference) NMR spectra were performed as described at temperatures ranging from 278 to 310 K on a 500 and a 700 MHz Bruker DRX spectrometer, utilizing the C24S/C172A mutant as protein target [29,30]. Samples contained between 10 μM and 20 μM mutant 3C and peptide ligands at concentrations between 200 μM and 4 mM. A deuterated 20 mM Tris buffer, pH of 7.4, was used for all experiments. The following peptides were used as ligands at a 10 to 100-fold molar excess: Ac-LRTQpNA, Ac-LAAQ-OH, Ac-LRTQFSFN-OH, Ac-WRTQFSFNL-OH, Ac-LRTQSGSNL-OH and Ac-ARTQSFSNL-OH. For modelling, the structural data of HAV 3C obtained by X-ray analysis were used [7]. The crystal structure data (protein databank acquisition code 1QA7) displays four monomeric 3C molecules A–D in the asymmetric unit. Molecule D was chosen for further modelling studies because of its favoured temperature factors. Prior to docking experiments the protein was energy minimized using Sybyl molecular modeling software (version 6.7). Using the MUTATE function of the Sybyl program Cys-24 was replaced by a serine. The disulphide bridge was put either in the anti or in the gauche conformation. Docking of the peptide ligand LRTQFSF was achieved using the FLEXIDOCK and DOCK options of Sybyl. FLEXIDOCK simulations were performed with 60.000 generations. Flexibility was allowed for the amino acids Gln-15, His-44, Asn-124, Tyr-143, His-145, Lys-146, Cys-172, His-191 and Asn-196 in the binding pocket.
RESULTS
Characterization of recombinant 3C mutants
In order to study the proteolytic activity and the RNA-binding properties of wild-type HAV 3C and their potential interplay, we included three mutants in our experiments in addition to the wild-type enzyme. The catalytically inactive mutant C172A carries an alanine in place of the active site cysteine residue. The catalytically active single mutant C24S and the catalytically inactive double mutant C24S/C172A both lack the surface exposed thiol group in position 24 that is not conserved among the picornaviral proteinases. They allow a correlation of functional data with the three-dimensional structure of HAV 3C obtained from X-ray crystallography [6–8]. To assess the purity and the oligomeric status of the enzymes, recombinant 3C proteins purified by ion-exchange chromatography were first analysed by SDS/PAGE under reducing and non-reducing conditions, employing Coomassie or silver staining. Whereas all 3C variants migrated with the expected molecular mass of 24 kDa under reducing conditions (results not shown), the formation of dimers was observed under non-reducing conditions (Figure 2). Both forms of 3C lacking the surface-exposed cysteine residue (C24S and C24S/C172A) migrated as monomers under non-reducing conditions (Figure 2A, lanes 3 and 4). For the preparation of sensor chips, only freshly prepared proteins with minimal amounts of dimerized proteins were used (see below). After prolonged storage (Figure 2B), the proteolytically inactive 3C mutant (C172A) was predominantly in its dimeric form (Figure 2B, lane 2). After storage of wild-type 3C for 10 days at 4 °C, approx. 50% of the molecules were converted into dimers (Figure 2B, lane 1) with a concomitant, yet reversible, reduction of proteolytic activity (see below). Using a monospecific antibody directed against 3C [18], the identity of the higher molecular mass components was confirmed by immunoblotting (results not shown). In agreement with previous observations [6,24], these results show that Cys-24 is responsible for 3C dimerization. The process can be suppressed effectively by applying reducing conditions.
Figure 2. 3C dimerization after prolonged storage under non-reducing conditions.
Non-reducing SDS/PAGE analysis of HAV 3C and mutants freshly prepared (A, 2.5 μg of protein, Coomassie stain), or stored for approx. 10 days at 4 °C (B, 0.1 μg of protein, silver stain). Lanes 1 (in A and B), wild-type (wt) 3C; lanes 2, mutant C172A; lanes 3, mutant C24S; lanes 4, mutant C24S/C172A.
Dimerization affects the proteolytic activity of 3C
To assess the role of dimerization on the proteolytic activity of 3C, we developed a microassay using as substrate the chromogenic peptide Ac-ELRTQ-pNA that represents the 2B–2C cleavage site in the HAV polyprotein (see Figure 1). This peptide comprises the P1 to P5 positions on the N-terminal side of the scissile bond, and has been described as the most potent peptide substrate [19]. The measurement of initial reaction velocities as a function of the concentration of the substrate Ac-ELRTQ-pNA yielded the Michaelis–Menten constants KM of 24 and 27 mM for the wild-type 3C and mutant C24S respectively, corresponding to kcat/KM values of 2.07×103 M−1·s−1 and 1.94×103 M−1·s−1. Comparable small values have been reported elsewhere for 3C proteinases of other picornaviruses [19,31]. In the presence of competing peptides or leupeptin, the proteolytic activity was reduced by a factor of 2 to 5 (results not shown). No effect on the proteolytic activity of 3C was observed upon addition of the control peptide H2N-PGRAF-OH.
In order to estimate the effect of dimerization on the proteolytic activity, the microassay was performed with freshly prepared protein and after storage for various periods of time, applying reducing or non-reducing conditions. Freshly prepared proteinase was immediately subjected to SDS/PAGE, and to the microassay in order to correlate the dimerization status to the enzyme activity. Under these conditions the proteolytic activity was deliberately set at 100% as a reference. Upon transfer of wild-type 3C or the mutants to DTT-free buffer, they were again analysed by SDS/PAGE in the presence or absence of DTT, and their enzymic activity was assayed. As expected, the mutants C24S and C24S/C172A were monomeric (see above) and C24S was fully active. Although 10% of wild-type 3C was found as dimers in this experiment, the enzyme was still fully active (100%) when compared with the activity of the freshly prepared protein (Table 1). Addition of DTT completely restored the monomeric state of the enzymes. After protein storage in DTT-free buffer for 10 days at 4 °C, 50% of the wild-type enzyme was dimerized, and had lost 95% of its activity. Various kinds of modifications might be responsible for the loss of enzyme activity independent of dimerization [24]. The proteolytically inactive mutant C172A was almost completely dimerized. The mutant C24S was still in its monomeric state, but had lost 20% of its activity. This indicates that the active site cysteine residue at position 172 is less susceptible to reduction. Upon addition of DTT to these preparations, enzymic activity was largely recovered along with a complete reconstitution of the monomeric states. Taken together, the results suggest that prolonged 3C storage in DTT-free buffer resulted in 3C dimerization mostly via Cys-24, with a concomitant loss of proteolytic activity that may be recovered by reduction.
Table 1. HAV proteinase 3C dimerization and proteolytic activity.
NA, not applicable. C172A and C24S/C172A mutants are proteolytically inactive due to the active-site mutation.
| Freshly prepared 3C† | Stored 3C‡ | ||||
|---|---|---|---|---|---|
| C3 | DTT* | Activity (%) | Monomer (%) | Activity (%) | Monomer (%) |
| Wild-type | − | 100 | 90 | 5 | 50 |
| + | 100 | 100 | 70 | 100 | |
| C24S | − | 100 | 100 | 80 | 100 |
| + | 100 | 100 | 100 | 100 | |
| C172A | − | NA | 70 | NA | 1 |
| + | NA | 90 | NA | 100 | |
| C24S/C172A | − | NA | 100 | NA | 100 |
* 6 mM DTT was present or absent in the microassay.
† After purification in DTT-containing buffer, the proteins were directly subjected to the microassay or SDS/PAGE.
‡ Prior to the activity assay or SDS/PAGE, the proteins were transferred to DTT-free buffer and stored for approximately 10 days at 4 °C.
To determine the role of Cys-24 of 3C in HAV replication, this amino acid residue was replaced by a serine in an infectious full-length cDNA construct. After transfection of synthetic transcripts bearing the respective mutation, viral replication was assessed by the detection of viral antigen and infectious virus was rescued [4]. After RT-PCR amplification of the rescued viral RNA, nucleotide sequence analysis showed that the serine at amino acid position 24 of 3C was retained and that no second site mutation in 3C had occurred (results not shown). The specific infectivity of the infectious RNA carrying the 3C mutation was approximately reduced by 2 orders of magnitude. This indicates that 3C dimerization via Cys-24 is not a prerequisite for virus replication, but that it improves overall replication.
Binding affinity of a substrate peptide to HAV 3C and mutants
In order to determine the binding affinities of peptides Ac-ELRTQ-pNA and Ac-LRTQ-pNA to 3C and its mutants, we employed SPR experiments. The peptides were injected over a biosensor surface to which purified monomeric 3C had been immobilized. The ‘on’ and ‘off’ rates were at the detection limits of the SPR technique that allows to determine the on-rate constants, kon, between 102 to 107 M−1·s−1, and off-rate constants, koff, in the range between 10−1 and 10−6 s−1. For both peptides the equilibrium analysis yielded KD values in the millimolar range, albeit with large χ2 values (Table 2). In general, the closeness of the fit was better for the analyte Ac-LRTQ-pNA, and for data sets where the highest analyte concentration were omitted before the curve fitting.
Table 2. Binding of Ac-LRTQpNA and Ac-ELRTQpNA to HAV 3C.
Association rate constants kon, dissociation rate constants koff and dissociation constants KD were obtained from SPR experiments. For the kinetic analysis a Langmuir 1:1 binding model with corrections for a drifting baseline and mass transfer was applied using global fitting (experiments A and B) [35]. Analyte concentrations were: experiment A, 25, 50, 100, 250, 500, 1000 and 2000 μM; experiment B, 25, 50, 100, 250, 500, 2000 and 4000 μM. Data in experiments C and D resulted from an equilibrium analysis. The analyte concentrations were: *25, 50, 100, 250, 500, 2000, 4000, 6000 μM; †25, 50, 100, 250, 500, 2000 μM; ‡25, 50, 100, 250, 500, 2000, 4000, 8000 μM. 3000 to 4000 RU of ligand were immobilized in all cases. wt, wild-type.
| Analyte | Experiment | Ligand | kon (M−1·s−1) | koff (s−1) | KD×10−3 (M−1) | χ2 |
|---|---|---|---|---|---|---|
| Ac-LRTQ-pNA | A | |||||
| wt | 132 | 0.181 | 1.4 | 1.2 | ||
| C24S | 213 | 0.243 | 1.1 | 1.1 | ||
| C24S/C172A | 187 | 0.041 | 0.2 | 1.4 | ||
| Ac-ELRTQ-pNA | B | |||||
| wt | 35 | 0.014 | 0.4 | 2.8 | ||
| C24S | 18 | 0.035 | 1.9 | 2.8 | ||
| C24S/C172A | 37 | 0.015 | 0.4 | 2.7 | ||
| Ac-LRTQ-pNA | C | |||||
| wt* | 5.0 | 11 | ||||
| wt† | 1.8 | 3 | ||||
| C24S* | 3.9 | 7 | ||||
| C24S† | 1.8 | 2 | ||||
| C24S/C172A* | 6.4 | 11 | ||||
| C24S/C172A† | 2.6 | 5 | ||||
| Ac-ELRTQ-pNA | D | |||||
| wt‡ | 2.8 | 28 | ||||
| wt† | 1.3 | 11 | ||||
| C24S‡ | 4.3 | 54 | ||||
| C24S† | 1.0 | 18 | ||||
| C24S/C172A‡ | 5.7 | 136 | ||||
| C24S/C172A† | 0.5 | 30 |
We also performed a kinetic analysis of the data, although the binding kinetics of this system were at the limit of the SPR technique. These analyses utilized the global fitting features of the BIAevaluation software and revealed that association and dissociation reactions were both slow (Table 2, experiments A and B). At higher peptide concentrations a significant residual bulk effect was observed, especially for Ac-ELRTQ-pNA as the analyte. Applying corrections for this residual bulk effect, as well as for mass transfer and a drifting baseline, good fits were obtained, as reflected by the low χ2 values that were all well below 10. In an equilibrium analysis of the data, these corrections could be applied easily, and therefore rather poor fits were obtained (Table 2, experiments C and D).
STD NMR experiments
The peptides Ac-LRTQpNA, Ac-LAAQ-OH, Ac-LRTQFSFN-OH, Ac-WRTQFSFNL-OH, Ac-LRTQSGSNL-OH and Ac-ARTQSFSNL-OH were subjected to an NMR binding study, using STD NMR experiments [32] and the catalytically inactive mutant C24S/C172A as a receptor protein. The methodology that allows the study of complexes with dissociation constants KD of 1 nM to 10 mM requires that the dissociation rate constants koff are larger than approx. 1 s−1 for an efficient exchange between free and bound ligand molecules [29,30]. Under no experimental conditions could STD signals be detected for the peptides summarized above. Our failure to detect any STD effects strongly suggests slow dissociation rate constants koff for all peptides. In turn, this implies rather low association rate constants kon, given the millimolar dissociation constants.
Binding affinity of viral RNA to 3C is significantly increased upon dimerization of the enzymes
By genetic and gel-shift analyses, it was shown that a stretch of five highly conserved amino acid residues (KFRDI at HAV 3C amino acid positions 95–99) is involved in 3C binding to viral RNA structures formed by the 5′-terminal nucleotides of the viral RNA [13,16]. To estimate the quantitative parameters of 3C–RNA binding, we applied a Biacore analysis that is superior to traditional methods, such as gel-shift or filter-binding assays. The RNA fragments comprising the 5′ nucleotides 1–148 and 46–148 of the HAV genome were synthesized in vitro, biotinylated at the 3′-ends and bound to the sensor chip. This experimental setup was preferred over immobilization of 3C to the chip surface, since this assay format required only small amounts of RNA. Figure 3 shows sensorgrams at different concentrations of monomeric and dimeric wild-type 3C (left-hand panels), and the mutant C24S/C172A (right-hand panels), binding to immobilized RNA(1–148). It is immediately obvious from the sensorgrams that dimers of wild-type 3C present under non-reducing conditions produced a significantly higher SPR response than the monomeric protein (compare the upper and lower left-hand panels of Figure 3). For mutant C24S/C172A that is unable to dimerize, the sensorgrams were identical under reducing and non-reducing conditions (Figure 3, right-hand panels). The sensorgrams clearly display a binding kinetic with association and dissociation rate constants outside the kinetic resolution of the Biacore instrument. Therefore, we performed a steady-state equilibrium analysis using a 1:1 Langmuir binding model, yielding the data shown in Table 3. The equilibrium analysis under reducing conditions yielded KD values in the millimolar range for all 3C variants. Dimerization of wild-type 3C and of mutant C172A was associated with a significant decrease of the corresponding dissociation constants KD to values in the micromolar range. Binding of mutants C24S and C24S/C172A to RNA was unaffected by the presence or absence of reducing agent.
Figure 3. HAV 3C binding to RNA: sensorgrams from Biacore experiments.
Binding of wild-type (wt) 3C (left-hand panels) and mutant C24S/C172A (right-hand panels) to immobilized RNA(1–148). RNA(1–148) was biotinylated at its 3′-end and immobilized to a SA chip, resulting in 1000 RU. Protein concentration varied from 2 to 40 μM for each experiment. Binding reactions shown in the upper sensorgrams included DTT, the sensorgrams in the lower panels were obtained without DTT.
Table 3. Dissociation constants KD for the binding of wild-type and mutant proteinase 3C to viral RNAs.
KD values were calculated from SPR experiments applying a Langmuir 1:1 model.
| KD (M)/χ2 | ||||||
|---|---|---|---|---|---|---|
| Ligand | DTT | 3C… | Wild-type | C172A | C24S | C24S/C172A |
| RNA(1–148) | − | 8.8×10−6/12.6 | 7.5×10−6/42.4 | 1.3×10−4/0.9 | 1.5×10−3/0.5 | |
| + | 1.1×10−3/77.6 | 3.8×10−3/9.6 | 1.2×10−3/1.1 | 1.8×10−3/5.5 | ||
| RNA(46–148) | − | 1.7×10−5/4.6 | 2.3×10−5/22.7 | 6.1×10−3/2.3 | 2.2×10−3/2.4 | |
| + | 1.4×10−3/12.2 | 4.7×10−3/3.8 | 4.5×10−3/4.8 | 2.2×10−3/5.6 | ||
In order to further explore the specificity of the RNA–3C interaction, HAV RNA(46–148) was also immobilized on a sensor chip and subjected to a Biacore analysis. RNA(46–148) lacks the 5′-terminal stem-loop structure, and was shown previously by EMSA to interact less efficiently with 3C than RNA(1–148) [16]. Indeed, we found that RNA(46–148) formed complexes with 3C dimers with dissociation constants approx. 10-fold higher than observed for RNA(1–148) (Table 3).
To confirm further the effects of dimerization on the RNA–3C interaction, we performed EMSA as described previously [16]. Figure 4 shows the altered electrophoretic mobility of radiolabelled RNA(1–148) after incubation with 3C monomers or dimers. Under reducing conditions when most 3C molecules were present in their monomeric state, only small portions of RNA migrated with retarded mobility (Figure 4B, lanes 2–5). The majority of RNA molecules migrated as free RNA, confirming that the interaction of RNA with monomeric wild-type 3C or mutant C172A was weak (Figure 4B, lanes 2 and 3). In the absence of reducing agent, RNA mobility was completely unaffected by monomeric mutants C24S or C24S/C172A (Figure 4A, lanes 4 and 5) when compared with free RNA (Figure 4A, lane 1). However, RNA interacting with dimeric proteins (wild-type 3C and mutant C172A) that were generated under non-reducing conditions resulted in a clear band shift (Figure 4A, lanes 2 and 3). The mutant C172A shifted the mobility of RNA(1–148) to a slower migrating position than wild-type 3C, suggesting that the mutant C172A formed more stable RNA complexes. Our findings, based on the SPR interaction studies and EMSA, clearly indicate that 3C binds to viral RNA more efficiently in its dimeric form than in its monomeric state.
Figure 4. Effect of 3C dimerization on RNA binding as determined by EMSA.
Wild-type (wt) 3C and mutants were incubated with radiolabelled RNA(1–148) in the presence (B) and absence of DTT (A). Complexes were run on a 5% gel containing DTT (B) or no DTT (A). The position of free and protein-complexed RNA is indicated on the left-hand side.
Substrate peptides modulate HAV 3C binding to viral RNA
The dual function of HAV 3C in polyprotein processing and RNA binding has so far only been studied in independent experimental systems [16,18,19,23]. Due to experimental limitations, possible mutual effects were not shown. We initially assayed the cleavage of the chromogenic substrate in the presence of up to a 15-fold molar excess of viral and control RNAs (results not shown). Owing to the low binding constant of wild-type 3C to RNA(1–148) (see Table 3) it can be estimated that under these conditions only 4 to 10% of the total 3C was bound to RNA.
Next we utilized Biacore experiments to test for the effect of peptide binding on the 3C–RNA interaction. Purified wild-type 3C was incubated with increasing amounts of peptides Ac-LRTQ-OH, Ac-LAAQ-OH or Ac-ELRTQSFS-NH2 before injection of the peptide–proteinase complex on to the SA chip carrying viral RNA(1–148). As seen from the data presented in Figure 5 (upper panel), the binding of LRTQ enhanced the 3C binding to viral RNA(1–148) in a concentration-dependent manner. The same results were obtained for Ac-LAAQ-OH and Ac-ELRTQSFS-NH2 (results not shown). At a 125-fold molar excess of peptide over 3C corresponding to approximately all of the peptide-binding pockets being occupied, a significant increase in RNA-binding was observed in all instances. Under the same experimental conditions, no effect on RNA binding was observed with the control peptide PGRAF (Figure 5, lower panel). The peptides alone did not interact with viral RNA (Figure 5). Equivalent results were obtained for the mutant C172A, but no effect of enhanced binding upon addition of peptide ligands was observed for the mutants C24S and C24S/C172A. To rule out the possibility that the increased response in the Biacore experiment was due to dimerization of the enzyme via disulphide bridges, we tested the dimerization status of the proteins at the time of injection of the sample. There was no indication that the addition of peptides induced an accelerated dimerization of the protein. Therefore, these results indicate that binding of HAV 3C to viral RNA is enhanced at elevated substrate concentrations, and they suggest that RNA and substrate bind co-operatively to 3C.
Figure 5. Sensorgrams for the binding of wild-type HAV 3C to RNA(1–148) in the presence of varying concentrations of peptides.
The biotinylated RNA(1–148) (800 RU) was immobilized on a SA chip. The bottom lines (·-·-·) of the sensorgrams were obtained with the pure peptides without 3C present (upper panel, 2 mM LRTQ; lower panel, 1 mM PGRAF). The injection time was 300 s. Upper panel: effect of peptide LRTQ on 3C binding to RNA(1–148). The sensorgram with the lowest solid line reflects the binding of the wild-type 3C (4 μM) without substrate peptide present. The sensorgram for the addition of peptide at a 10-fold excess (40 μM) is shown (······). In this case the injection time was 230 s. The other solid line sensorgrams reflect 3C binding in the presence of increasing concentrations of LRTQ (0.5 mM, 1:125; 1 mM, 1:250; 2 mM, 1:500). Lower panel: effect of the control peptide PGRAF on 3C binding to RNA(1–148). The sensorgram of wild-type 3C without PGRAF present is shown (······). The two solid-line sensorgrams were obtained in the presence of 1 and 2 mM peptide.
DISCUSSION
Viral proteinase 3C catalyses cleavage at defined sites in the viral polyprotein, and binds specifically to viral RNA structures formed at the 5′- and 3′-termini of the viral genome. By genetic analyses, both functions were mapped and their biological role in the polioviral life cycle was directly shown [14]. In all previous investigations the mutual effect of RNA binding and proteolytic activities was not studied in detail. To address this functional duality of HAV 3C on the molecular level, we applied SPR (Biacore) experiments. This method analyses biomolecular interactions in real time, and also allows monitoring of low-affinity or transient interactions. Moreover, this technique is useful to determine heterologous binding of complex ligands to a solid-phase partner [21,22].
We had previously demonstrated the interaction specificity for certain secondary structures formed by the viral RNA [16]. To compare binding of various forms of 3C and to assess the effect of peptide ligands on RNA binding, these studies were extended here using SPR technology. In order to preserve the RNA native folding, a capture technique with the 3′-end of the RNA immobilized to the chip was employed. Extending our previous observations, we show now that dimerization of HAV 3C or mutants has a significant effect on the binding affinity towards viral RNA. Upon dimerization, the binding affinity increased from milli- to micromolar KD values, which might be explained by the creation of an extended RNA-binding site comprising the KFRDI motif. On the basis of the crystal structure of HAV 3C proteinase [6,7], we obtained models for the HAV 3C dimer, as described in the Experimental section. The disulphide bond formed by the cysteine residue at position 24 was put either in an anti or in a gauche orientation, as shown in Figure 6. In both cases the KFRDI motifs may form an extended RNA-binding site. As an alternative explanation, the enhanced binding affinity might be due to bivalent binding of the enzyme to the immobilized RNA. Improved RNA binding of oligomeric proteins has been previously described for the HCV core protein and HIV Rev [28,33]. In contrast to our studies, for the HIV RNA–protein complex dissociation constants in the nanomolar range were observed, enabling a detailed kinetic analysis of the Biacore data, and indicating co-operativity of Rev binding [28].
Figure 6. Models for the HAV 3C dimer.
The disulphide bridge (located on the back of the molecules) was either anti- (right-hand panel) or gauche-oriented (left-hand panle). The KFRDI motifs in the adjacent monomers are highlighted. The two 3C molecules of the dimer are shaded differently.
The use of 3C variants mutated in their cysteine residues at amino acid positions 24 and 172 enabled us to control 3C dimerization in an easy way by changing the buffer conditions. As picornaviral 3C mostly resides in the cytoplasm of the infected cells, it is highly unlikely that 3C dimerization through disulphide bridges might occur under the reducing conditions of this cell compartment, and might thus not play a major role in vivo. Even though dimerization of mature 3C has not been directly proven in HAV or poliovirus infected cells, experimental evidence clearly suggests that 3C precursors are prone to form homodimers ([5,34], and V. Gauss-Müller and G. Morace, unpublished work). For HAV, precursor 3ABC binds viral RNA even more efficiently than 3C [20]. It is generally assumed that the regulated formation of precursor and mature proteins encoded by the P3 domain of the picornaviral genome (see Figure 1), their controlled enzymic activities and their half-lives are crucial in regulating the two major steps in the viral life cycle, namely viral protein and RNA synthesis. In both metabolic processes, 3C is a major player. We propose that our studies in vitro using biosensor technology reflect quite well the multiple interactions of precursor polypeptides that occur in infected cells.
Not only dimerization, but also binding of peptide substrates significantly improved 3C binding to RNA (see Figure 5). Since an unrelated peptide (PGRAF) that did not compete in the enzymic assay had no effect on 3C binding to RNA, our results provide direct evidence that substrate peptide binding to the active site of 3C enhances RNA affinity of wild-type 3C and the mutant with a wild-type Cys-24 residue. Although the critical importance of Cys-24 at first sight might suggest that the increased Biacore responses were due to peptide-mediated 3C dimerization, no increase in 3C dimers was observed after SDS/PAGE of the protein–peptide complexes. The significantly increased SPR responses in the presence of peptides that bind to the active site of wild-type 3C, or the C172A mutant may either be due to an allosteric interaction that rearranges the RNA-binding site, or to an aggregation reaction that takes place during the binding process, and that is triggered by peptides binding to the active site of the enzyme. At the moment we cannot decide which of these explanations is more likely. In the light of previous crystallographic studies [5], it is possible that peptide-enhanced 3C binding to RNA is due to conformational stabilization of the viral proteinase. Independent of its mechanism, it is unambiguous that the 3C interaction with viral RNA is modulated by the presence of peptides in a sequence-specific manner.
We assume that substrate-enhanced and dimerization-dependent interaction with RNA is a critical feature of 3C required to fulfill its multiple functions in the viral life cycle. On the basis of our observation that 3C dimerization is directly correlated with RNA binding and inversely related to its proteinase function, we propose that production of a critical mass of 3C (or its precursor) and its subsequent dimerization might reduce its proteolytic activity and promote RNA-binding function. Whether this conformational and functional change is the major regulatory step in the switch from translation to replication [15] needs to be addressed by further experiments.
Acknowledgments
We gratefully acknowledge the technical assistance of Andre Güllmar, Susanne Hengst, Thies Köhli and Wilfried Hellebrandt. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, grant PE-494). T.P. thanks the Fonds der Chemischen Industrie for generous financial support and the DFG for a grant for a 700 MHz NMR spectrometer (Grant Me1830). V.G.-M. and Y.Y.K. acknowledge the support from the Sonderforschungsbereich 367 (project B7). M.W. acknowledges generous financial support from Solvay Pharmaceuticals GmbH, Hannover. The helpful discussions with Dr U. Bierfreund (Biacore AB) are gratefully acknowledged.
References
- 1.Hayden F. G., Turner R. B., Gwaltney J. M., Chi-Burris K., Gersten M., Hsyu P., Patick A. K., Smith G. J., 3rd, Zalman L. S. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patick A. K., Potts K. E. Protease inhibitors as antiviral agents. Clin. Microbiol. Rev. 1998;11:614–627. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Probst C., Jecht M., Gauss-Muller V. Processing of proteinase precursors and their effect on hepatitis A virus particle formation. J. Virol. 1998;72:8013–8020. doi: 10.1128/jvi.72.10.8013-8020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusov Y., Gauss-Muller V. Improving proteolytic cleavage at the 3A/3B site of the hepatitis A virus polyprotein impairs processing and particle formation, and the impairment can be complemented in trans by 3AB and 3ABC. J. Virol. 1999;73:9867–9878. doi: 10.1128/jvi.73.12.9867-9878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosimann S. C., Cherney M. M., Sia S., Plotch S., James M. N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann E. M., Mosimann S. C., Chernaia M. M., Malcolm B. A., James M. N. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann E. M., Cherney M. M., Mckendrick J., Frormann S., Luo C., Malcolm B. A., Vederas J. C., James M. N. Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV. Virology. 1999;265:153–163. doi: 10.1006/viro.1999.9968. [DOI] [PubMed] [Google Scholar]
- 8.Allaire M., Chernaia M. M., Malcolm B. A., James M. N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature (London) 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 9.Malcolm B. A. The picornaviral 3C proteinases: cysteine nucleophiles in serine proteinase folds. Protein Sci. 1995;4:1439–1445. doi: 10.1002/pro.5560040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews D. A., Smith W. W., Ferre R. A., Condon B., Budahazi G., Sisson W., Villafranca J. E., Janson C. A., McElroy H. E., Gribskov C. L., et al. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 11.Ryan M. D., Flint M. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 1997;78:699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- 12.Losick V. P., Schlax P. E., Emmons R. A., Lawson T. G. Signals in hepatitis A virus P3 region proteins recognized by the ubiquitin-mediated proteolytic system. Virology. 2003;309:306–319. doi: 10.1016/s0042-6822(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 13.Andino R., Rieckhof G. E., Achacoso P. L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 15.Gamarnik A. V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusov Y. Y., Gauss-Muller V. In vitro RNA binding of the hepatitis A virus proteinase 3C (HAV 3Cpro) to secondary structure elements within the 5′ terminus of the HAV genome. RNA. 1997;3:291–302. [PMC free article] [PubMed] [Google Scholar]
- 17.Walker P. A., Leong L. E., Porter A. G. Sequence and structural determinants of the interaction between the 5′-noncoding region of picornavirus RNA and rhinovirus protease 3C. J. Biol. Chem. 1995;270:14510–14516. doi: 10.1074/jbc.270.24.14510. [DOI] [PubMed] [Google Scholar]
- 18.Schultheiss T., Sommergruber W., Kusov Y., Gauss-Muller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J. Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jewell D. A., Swietnicki W., Dunn B. M., Malcolm B. A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 20.Kusov Y. Y., Morace G., Probst C., Gauss-Muller V. Interaction of hepatitis A virus (HAV) precursor proteins 3AB and 3ABC with the 5′ and 3′ termini of the HAV RNA. Virus Res. 1997;51:151–157. doi: 10.1016/s0168-1702(97)00089-0. [DOI] [PubMed] [Google Scholar]
- 21.Rich R. L., Myszka D. G. Advances in surface plasmon resonance biosensor analysis. Curr. Opin. Biotechnol. 2000;11:54–61. doi: 10.1016/s0958-1669(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 22.Morton T. A., Myszka D. G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- 23.Malcolm B. A., Chin S. M., Jewell D. A., Stratton-Thomas J. R., Thudium K. B., Ralston R., Rosenberg S. Expression and characterization of recombinant hepatitis A virus 3C proteinase. Biochemistry. 1992;31:3358–3363. doi: 10.1021/bi00128a008. [DOI] [PubMed] [Google Scholar]
- 24.Chernaia M. M., Malcolm B. A., Allaire M., James M. N. Hepatitis A virus 3C proteinase: some properties, crystallization and preliminary crystallographic characterization. J. Mol. Biol. 1993;234:890–893. doi: 10.1006/jmbi.1993.1636. [DOI] [PubMed] [Google Scholar]
- 25.Kaspari A., Schierhorn A., Schutkowski M. Solid-phase synthesis of peptide-4-nitroanilides. Int. J. Pept. Protein Res. 1996;48:486–494. doi: 10.1111/j.1399-3011.1996.tb00867.x. [DOI] [PubMed] [Google Scholar]
- 26.Hansske F., Cramer F. Modification of the 3′ terminus of tRNA by periodate oxidation and subsequent reaction with hydrazides. Methods Enzymol. 1979;59:172–181. doi: 10.1016/0076-6879(79)59079-x. [DOI] [PubMed] [Google Scholar]
- 27.von Ahsen U., Noller H. F. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- 28.Van Ryk D. I., Venkatesan S. Real-time kinetics of HIV-1 Rev-Rev response element interactions. Definition of minimal binding sites on RNA and protein and stoichiometric analysis. J. Biol. Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 29.Mayer M., Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 30.Meyer B., Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chemie Int. Ed. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q. M., Johnson R. B., Cox G. A., Villarreal E. C., Loncharich R. J. A continuous colorimetric assay for rhinovirus-14 3C protease using peptide p-nitroanilides as substrates. Anal. Biochem. 1997;252:238–245. doi: 10.1006/abio.1997.2315. [DOI] [PubMed] [Google Scholar]
- 32.Müller K. M., Arndt K. M., Plückthun A. Model and simulation of multivalent binding to fixed ligands. Anal. Biochem. 1998;261:149–158. doi: 10.1006/abio.1998.2725. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka Y., Shimoike T., Ishii K., Suzuki R., Suzuki T., Ushijima H., Matsuura Y., Miyamura T. Selective binding of hepatitis C virus core protein to synthetic oligonucleotides corresponding to the 5′ untranslated region of the viral genome. Virology. 2000;270:229–236. doi: 10.1006/viro.2000.0252. [DOI] [PubMed] [Google Scholar]
- 34.Xiang W., Cuconati A., Hope D., Kirkegaard K., Wimmer E. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 1998;72:6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weimar T., Haase B., Köhli T. Low affinity carbohydrate lectin interactions examined with surface plasmon resonance. J. Carbohydr. Chem. 2000;19:1083–1089. [Google Scholar]