Abstract
There exist two SMN (survival motor neuron) genes in humans, the result of a 500 kb duplication in chromosome 5q13. Deletions/mutations in the SMN1 gene are responsible for childhood spinal muscular atrophy, an autosomal recessive neurodegenerative disorder. While the SMN1 and SMN2 genes are not functionally equivalent, up-regulation of the SMN2 gene represents an important therapeutic target. Consequently, we exploited in silico, in vitro and in vivo approaches to characterize the core human and mouse promoters in undifferentiated and differentiated P19 cells. Phylogenetic comparison revealed four highly conserved regions that contained a number of cis-elements, only some of which were shown to activate/repress SMN promoter activity. Interestingly, the effect of two Sp1 cis-elements varied depending on the state of P19 cells and was only observed in combination with a neighbouring Ets cis-element. Electrophoretic mobility-shift assay and in vivo DNA footprinting provided evidence for DNA–protein interactions involving Sp, NF-IL6 and Ets cis-elements, whereas transient transfection experiments revealed complex interactions involving these recognition sites. SMN promoter activity was strongly regulated by an NF-IL6 response element and this regulation was potentiated by a downstream Ets element. In vivo results suggested that the NF-IL6 response must function either via a protein-tethered transactivation mechanism or a transcription factor binding an upstream element. Our results provide strong evidence for complex combinatorial regulation and suggest that the composition or state of the basal transcription complex binding to the SMN promoter is different between undifferentiated and differentiated P19 cells.
Keywords: cellular differentiation, P19 cell, Sp1 and Ets cis-elements, spinal muscular atrophy, survival motor neuron gene, transcriptional regulation
Abbreviations: AhR, aromatic hydrocarbon receptor; C/EBP, CCAAT/enhancer-binding protein; CR, conserved region; DMS, dimethylsulphate; EC cells, embryonal carcinoma cells; EHMN, embryonic hybrid motor neuron; EMSA, electrophoretic mobility-shift assay; IL-6, interleukin-6; LMPCR, ligation-mediated PCR; NF1, nuclear factor-1; P19RA, P19 cells treated with retinoic acid; RA, retinoic acid; RLU, relative luciferase unit; SMA, spinal muscular atrophy; SMN, survival motor neuron; TF, transcription factor; TIS, transcription initiation site; UVC, ultraviolet C
INTRODUCTION
There exist two SMN (survival motor neuron) genes in humans, the result of a 500 kb segmental duplication in the q13 region of chromosome 5 [1]. The major functional difference between the SMN1 and SMN2 genes is a C→T transition in exon 7 [2], which first appeared in Homo sapiens [3] and which disrupts normal splicing by creating an exonic splicing silencer that blocks the inclusion of exon 7 in mRNA produced from the SMN2 gene [4]. Consequently, the SMN1 and SMN2 genes are not functionally equivalent, explaining why SMN2 cannot completely complement ablation/disruption of the SMN1 gene [5]. Mutations in the SMN1 gene cause childhood-onset SMA (spinal muscular atrophy), an autosomal recessive lower motor neuron disorder affecting approx. 1 in 10000 live newborns [6]. Deletions, most often associated with severe type I SMA, cause a drastic reduction in SMN protein, whereas gene conversions, often associated with milder type II and type III SMA, result in the overproduction of exon 7-lacking transcripts and variable amounts of functionally deficient SMN protein [7]. The strong correlation between SMN2 copy number and disease severity [8], coupled with the dose-dependent rescue of embryonic lethality in Smn knockout mice by the human SMN2 gene [5], has led to extensive efforts to identify compounds that can alter SMN splicing [9] or up-regulate SMN expression [10,11]. SMN is ubiquitously expressed but is most abundant in brain, spinal cord and kidney [12]. Furthermore, SMN expression is greatest during embryogenesis and is significantly down-regulated after birth [13]. SMN interacts with a number of proteins and the large, highly stable SMN complex can be found in the cytoplasm and nucleus. SMN is supposed to play an essential role in the assembly of ribonucleoprotein complexes, apoptosis and transcription [14]. More recently, the localization of SMN protein in axons, dendrites and the neuromuscular junction [15,16], coupled with observed arrested muscle maturation in human SMA muscle biopsies and data emerging from animal models, suggest that SMA is probably a developmental defect caused by dysregulation of naturally occurring cell death and/or neuromuscular maturation [17].
Transcriptional regulation of the human SMN gene has been investigated to compare the SMN1 and SMN2 promoters, dissect mechanisms governing temporal and spatial expression and identify potential targets for the treatment of SMA. The human promoters lie within 2 kb of exon 1 and their activities are indistinguishable [18–20]. Analysis of adult tissues or cells resulted in the identification of a TIS (transcription initiation site) approx. 162/163 bp upstream of the translation initiation site in coding exon 1 [18,20], designated the +1 TIS. A second TIS approx. 79 nt upstream of the +1 site has also been mapped and appears to be the main TIS used during fetal development [20]. The proximal promoter comprises 107 and 150 nt upstream and downstream of the +1 TIS respectively in a region that shares complete sequence identity between the SMN1 and SMN2 genes [20]. Interestingly, this promoter (designated m107p150) is less active after RA (retinoic acid)-induced differentiation of EC (embryonal carcinoma) P19 cells; thus this model can be used to dissect the temporal regulation of SMN expression. The mouse SMN promoter is contained within 455 nt upstream of exon 1 [21]. A number of cis-elements have been investigated, including one located at −65 that is responsive to β- and γ-interferons and requires the interferon regulatory factor-1 TF (transcription factor) [10]. Furthermore, enhanced SMN expression in interferon-treated SMA patient fibroblasts supported the expectation that up-regulation of SMN expression represents a potential therapeutic target. N-methyl-D-aspartate receptor stimulation [22] was also found to up-regulate Smn expression in differentiated EHMN (embryonic hybrid motor neuron) cells; however, the underlying mechanism responsible for this response is still not clear. In the present study, we have exploited in silico, in vitro and in vivo approaches to identify cis-elements required for expression of the human and mouse SMN genes in P19 EC cells and provide evidence for the combinatorial action of Sp and Ets family members on SMN promoter activity in undifferentiated and differentiated P19 cells.
EXPERIMENTAL
Preparation of SMN/Smn reporter gene constructs
The human SMN and mouse Smn promoter fragments were generated by PCR, using oligonucleotides (Alpha DNA, Montreal, QC, Canada) corresponding to the appropriate extremities, to generate the human SMN m107p156, m58p156, m107p127, m61p127, m61p117, m15p127, m15p117, m61p46, m15p93 and mouse Smn m105p125 and m46p125 fragments that were then purified and subcloned into the pGem®-T Easy vector (Promega, Madison, WI, U.S.A.). The EcoRI fragments were then subcloned into an EcoRI site that we had inserted between the BglII and HindIII recognition sites in the multiple cloning cassette upstream of the luciferase reporter gene in the promoterless pGL3-basic vector supplied by Promega. Each construct was sequenced in our institution's core facility to confirm correct orientation and to ensure that mutations had not been introduced during the amplification step. Mutant vectors were created using the Quik Change Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) according to the manufacturer's instructions and the following oligonucleotides (Alpha DNA). Oligonucleotides (5′ to 3′) gccacaaatgtgggattgcgataaccactcgtagaaaagcg (+11Sp1), ggcgataaccactcgtagaaagtttgagaagttactacaagcgg (+37AhR), gttactacaagcggtcctattggccaccgtactgttccg (+56Sp1), ggccaccgtactgtctcgcttcccagaagccccg (+79Ets), ccgtactgttccgcgacttgaagccccgggcggcgg (+86IL-6) and gccccgggcggcgagagtcgtcactctaagaag (+105Ets), and their complementary oligonucleotides (not listed) were used to mutate the human SMN promoter. Oligonucleotides (5′ to 3′) ggtctctggctgcctattggccaccgtactcttccg (+70Sp1), ccgtactcttccgggacttgaagccccatgacgg (+97IL-6), atgacgactctcgtcatggg (+115Ets), and their complementary oligonucleotides (not listed), were used to mutate the mouse Smn promoter. Mutant nucleotides are shown in boldface. Single mutations were introduced into the wild-type minimal promoter vector, whereas double and triple mutants were created sequentially using the appropriate mutant vector as template. All plasmids were prepared using the Qiagen Plasmid Midi kit (Qiagen, Mississauga, ON, Canada).
Cell culture, transient transfections and enzymic assays
Mouse P19 EC cell lines (A.T.C.C. no. CRL-1825) and mouse EHMN cell lines (a gift from Dr A. Burghes, Medical Biochemistry Department, Ohio State University, Columbus, OH, U.S.A.) were maintained in α-minimal essential medium or Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum (Wisent, St. Bruno, QC, Canada) respectively, without antibiotics. Differentiation of P19 cells was accomplished using 4 μM RA (Sigma–Aldrich Canada, Oakville, ON, Canada) as described previously [16]. Transient transfections were conducted using 1.5×105 P19 cells and 2×105 P19RA cells (RA-treated P19 cells) plated on to 12-well tissue culture dishes. Each transfection was comprised of 0.35 μg of the vector containing the pGL3 luciferase reporter gene and 0.15 μg of the pUT535 vector containing the β-galactosidase gene under the control of the cytomegalovirus promoter. The latter vector was included to normalize the efficiency of each transfection. Vector DNAs were preincubated with 1.5 μl of FuGENE 6 (Roche Diagnostics Corp., Laval, QC, Canada) per reaction for 15 min at room temperature (20–23 °C). The DNA/FuGENE mixture was then added to the culture medium and transfections were allowed to proceed for 24 h at 37 °C. Each construct was transfected a minimum of six times. The pGL3-promoter vector containing the luciferase gene under the control of the simplex virus 40 promoter and the promoterless pGL3-basic vector served as positive and negative controls (Promega) respectively. At the end of each transfection, cells were rinsed in PBS, scraped in 160 μl of lysis buffer (0.1 M potassium phosphate buffer, pH 7.8, and 1% Triton X-100), transferred to an Eppendorf tube and vortex-mixed, the cellular debris was pelleted and the supernatant was recovered for protein, β-galactosidase and luciferase assays. Total protein was measured using the Bradford dye method and according to the instructions given by Bio-Rad (Mississauga, ON, Canada). To measure β-galactosidase activity, 20 μl of lysate was deposited into a 96-microwell plate, mixed with 20 μl of o-nitrophenyl β-D-galactopyranoside solution [40 mM NaH2PO4, 60 mM Na2HPO4, 1 M KCl, 0.1 M MgCl2, 50 mM 2-mercaptoethanol and 0.01 g/ml o-nitrophenyl β-D-galactopyranoside (Sigma–Aldrich Canada)], incubated for 30 min at 37 °C and the resultant activity measured using the Wallac #1420 luminometer (Fisher Scientific, Nepean, ON, Canada). Luciferase activity was measured using 30 μl of lysate mixed with 30 μl of luciferin solution [0.01 M tricine, 0.54 mM (MgCO3)4Mg(OH)2·5H2O, 1.34 mM MgSO4·7H2O, 0.05 mM EDTA, 0.13 mg/ml D-Luciferin (Roche Diagnostics Corp.), 0.5 mM ATP, 0.2 mg/ml CoA (Sigma–Aldrich Canada) and 0.5 mg/ml dithiothreito] and immediately read with a luminometer. Promoter activities are expressed as RLU (relative luciferase units) normalized with relative β-galactosidase units·(μg of transfected DNA)−1·(μg of protein)−1.
Preparation of nuclear extracts and EMSAs (electrophoretic mobility-shift assays)
Nuclear extracts were prepared from untreated P19 cells, RA-treated P19 cells and EHMN cells as described previously [23]. Total protein was measured as described above. Double-strand DNA probes (Alpha DNA) V (nt −3 to +47), VI (nt +37 to +87), VII (nt +78 to +127) and VIIa (nt +94 to +128) were 5′-end-labelled with [γ-32P]dCTP (Amersham Biosciences, Baie d'Urfé, QC, Canada) using T4 polynucleotide kinase (Invitrogen, Burlington, ON, Canada) and the unincorporated isotope removed using a Quick-spin G50 column (Amersham Biosciences). All the nucleotide positions are as given by Germain-Desprez et al. [20]. The nuclear extract (4 μg unless otherwise specified) was incubated with 100 fmol of labelled probe in a 20 μl volume of reaction buffer containing 3 μg of Poly[d(I-C)] (Roche Diagnostics Corp.), 100 mM Hepes (pH 7.9), 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (v/v) Tween 20 and 150 mM KCl. Where appropriate, unlabelled double-strand competitor oligonucleotides were added to the binding reaction. These were either wild-type (VIc nt +72 to +87; VIIa nt +94 to +128 and VIIb nt +78 to +93) or mutant SMN (VIIa* is the +105Ets oligonucleotide used for site-directed mutagenesis) probes synthesized for the present study or commercially available Sp1 and Ets-1/PEA3 consensus double-strand oligonucleotides (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). Binding reactions were performed at room temperature for 20 min and resolved on a 5% (w/v) non-denaturing polyacrylamide gel for 1 h 45 min at 8 V/cm. Gels were then dried and exposed to an X-ray film and two intensifying screens, overnight at −80 °C. For supershift assays, 0.4 μg of each antibody (Santa Cruz Biotechnology), directed against MAZ, Sp1, Elk-1 or Pea3 was added 20 min after mixing the nuclear extract with probe and the incubation was allowed to proceed for an additional 20 min at room temperature. EMSAs were repeated at least three times.
In silico analysis
Alignment of the promoter region from the human (SMN; GenBank® accession no. AF092925) and mouse genes (Smn; GenBank® accession no. AF027688) was accomplished using DALIGN (http://www.ch.embnet.org/cgibin/LALIGN/form.html). To identify potential consensus TF-binding sites in the human and mouse promoters, we used TESS (URL: http://www.cbil.upen.edu/tess) to query TRANSFAC v6.0 (URL: http://www.gene-regulation.com). Potential cis-elements investigated in the present study are annotated in Figure 2.
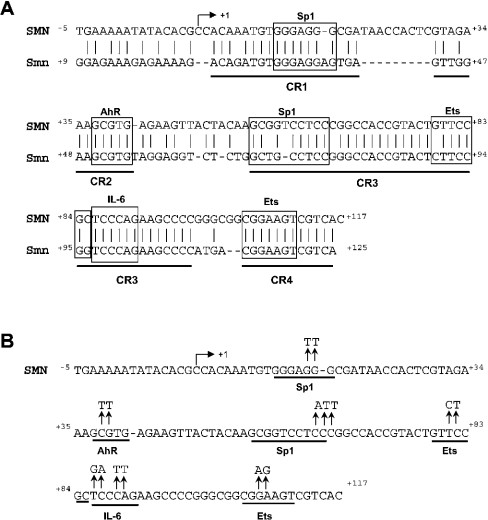
Figure 2. Phylogenetic footprinting.
(A) Alignment of the proximal human (SMN) and mouse (Smn) promoters; regions of conserved sequence identity are underlined (CR1–CR4). Vertical bars and boxes annotate sequence identity and putative cis-elements respectively. The IL-6 site corresponds to the NF-IL6 response element. (B) The human proximal promoter sequence and the mutations introduced by site-directed mutagenesis are shown above the putative Sp1, AhR, Ets and IL-6 cis-elements. The arrow ↱ corresponds to the +1 TIS.
DNA modifications
Living cells and purified DNA (referred as in vivo and in vitro respectively) from undifferentiated and differentiated P19 cells were treated with one of the following probing agents: DMS (dimethylsulphate; Sigma–Aldrich Canada), UVC (ultraviolet C) irradiation (G15T8 germicidal lamp; Philips, Montreal, QC, Canada) and DNase I (Worthington Biochemical, Lakewood, NJ, U.S.A.). Specifically, cells were incubated in serum-free medium containing 0.2% DMS for 6 min at room temperature. After detaching cells with trypsin (Invitrogen), nuclei were isolated, and alkylated DNA was purified as described in [24]. Purified DNA (in vitro) was treated with DMS in a standard Maxam–Gilbert cleavage reaction. Hot piperidine (1 M, 80 °C for 30 min; Sigma–Aldrich Canada) was then used to chemically convert methylated guanines/adenines into single-strand DNA breaks. Alternatively, living cells and purified DNA were irradiated on ice with 254-nm germicidal lamps. The UVC dose and irradiation time were 1500 J/m and 32 s respectively. Cell lysis, sedimentation of nuclei and DNA purifications were performed as described in [24]. T4 endonuclease V (kindly provided by Dr R. S. Lloyd, The University of Texas Medical Branch, Galveston, TX, U.S.A.) was used to convert cyclobutane pyrimidine dimers, previously induced by UVC irradiation, into single-strand DNA breaks [25]. The resulting 5′-pyrimidine overhangs were then removed by photoreactivation using Escherichia coli photolyase (kindly provided by Dr T. R. O'Connor, Beckman Research Institute, City of Hope, Duarte, CA, U.S.A.) to generate ligatable ends. Finally, living cells were permeabilized with lysolecithin (Sigma–Aldrich Canada) as described in [24]. DNase I was added at a concentration of 7.5 μg/μl without removing the lysolecithin. After 8 min of a total incubation time of 20 min at 37 °C, cells were scraped, transferred to a conical 15 ml tube and returned to 37 °C for the remaining incubation time. After centrifugation for 1 min at 4500 g, the supernatant was removed and cells were resuspended in 1 ml of buffer B (150 mM NaCl and 5 mM EDTA, pH 7.8) and 1 ml of buffer C (20 mM Tris/HCl, pH 8.0, 20 mM NaCl, 20 mM EDTA and 1% SDS) containing 600 μg/ml proteinase K. The cells were incubated at 37 °C for 3 h followed by the addition of 200 μg/ml RNaseA and re-incubated for 1 h at 37 °C. DNA was then purified as described in [24]. To obtain in vitro DNA controls, 40 μg of purified DNA was digested with 5 ng/ml DNase I at room temperature for 20 min in 300 μl of water and 100 μl of solution II (150 mM sucrose, 80 mM KCl, 35 mM Hepes, pH 7.4, 5 mM MgCl2 and 2 mM CaCl2). The reaction was stopped by adding 400 μl of phenol, followed by extraction once with phenol–chloroform and once with chloroform. The DNA was then dissolved in water at a concentration of 0.5 μg/μl. The single-strand break frequencies were estimated after each treatment by alkaline gel electrophoresis.
LMPCR (ligation-mediated PCR) for in vivo footprinting
The LMPCR method has been previously described in detail [24,26]. Briefly, both strands of the approx. 170 bp murine SMN promoter (GenBank® accession no. AF027688), from nt −46 to +125 upstream and downstream of the TIS [21], were analysed using the following primer sets (5′ to 3′): gagaaaagacagatgtgggag (nt +16 to +36) and tgggaggagtgagttggaagcgtgtagg (nt +31 to +58) (set 5) were used to amplify the upper strand, and gcctgctaccacggac (nt +144 to +159) and ggacgctgccgggctcactcaatgacg (nt +147 to +121) (set 2), cctgcggatgtggc (nt +297 to +310) and caccgcctcggtctcgggcagcg (nt +265 to +287) (set 4), and cggaagagtacggtg (nt +81 to +95) and gccagagacctcctacacgcttccaac (nt +43 to +69) (set 6) were used to amplify the bottom strand. Primer positions are relative to the +1 TIS for the mouse Smn gene. A gene-specific primer was annealed to genomic fragments of variable sizes and then extended using cloned Pfu exo− DNA polymerase (Stratagene) to produce double-strand blunt ends. An asymmetric double-strand linker was then ligated to the phosphate terminal end of each fragment, providing a common sequence on the 5′-end of all fragments. Using Taq DNA polymerase (Roche Diagnostics Corp.) for primer sets SMN2, SMN4 and SMN5 or cloned Pfu exo− DNA polymerase for primer set SMN6, a linker-specific primer was used for a single round of linear amplification, followed by PCR amplification using the appropriate primer sets in conjunction with the linker primer. The desalted oligonucleotide primers were synthesized by Invitrogen. All primer extensions and PCR amplifications were performed on a T gradient thermocycler from Biometra (Kirkland, QC, Canada). The resulting products were subjected to PAGE (8% gel) alongside a Maxam and Gilbert sequencing ladder, followed by electrotransfer on to nylon membranes (Roche Diagnostics Corp.), hybridization to a 32P-labelled gene-specific probe and visualization by autoradiography. Each experimental condition was assayed in duplicate and then run on a screening sequencing gel using a portion of the DNA to ensure that there was no significant variation between samples. The duplicates were then pooled on a combined gel that served to analyse footprint patterns throughout the sequence, as described previously [26].
RESULTS
Delimiting the minimal human SMN and mouse Smn promoters
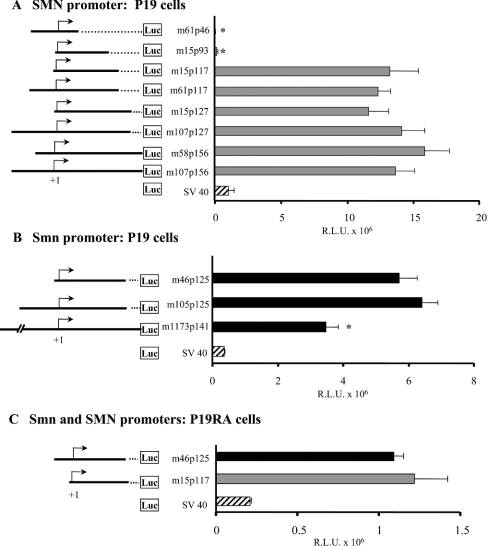
Our previous studies indicated that the human SMN promoter contained two TISs located 242 nt (−79 site) and 162 nt (+1 site) upstream of the initiating methionine in exon 1 [20]. Furthermore, the m107p156 construct (referred to as 107SMNCAT in [20]), containing 107 nt upstream and 156 nt downstream of the +1 TIS, was shown to have all of the sequences necessary for SMN promoter activity. To delimit further the human proximal promoter, a number of SMN promoters were prepared and cloned into pGL3-basic, a promoterless vector containing the luciferase reporter gene (Figure 1A). These constructs were transiently transfected into P19 cells and promoter activity was measured as RLU normalized against β-galactosidase activity as a measure of transfection efficiency. Five of the new constructs (SMN m58p156, m107p127, m15p127, m61p117 and m15p117) had the same promoter activity as described previously for SMN m107p156, namely 11.6–15.9×106 RLU. Thus the removal of 92 and 39 nt at the 5′- and 3′-end of m107p156 respectively did not have a significant effect on proximal promoter activity and all or most of the cis-elements required for this activity are contained within nt −15 to +117.
Figure 1. Characterization of the proximal SMN promoter.
A number of human SMN (A) and mouse Smn (B) promoter fragments were cloned upstream of the luciferase reporter gene. The names of different constructs are provided to the left of the bar graph and refer to the 5′ (m for minus) and 3′ (p for plus) nucleotide positions with respect to the +1 TIS (arrow). The vectors are depicted as solid lines, whereas dotted lines correspond to deleted segments. The human SMN-Luc (A) and mouse Smn-Luc (B) vectors were transiently transfected into undifferentiated (P19) and differentiated (P19RA) cells (C). The bar graph (grey for human, black for mouse and striped for simplex virus 40 constructs) depicts promoter activities (x-axis), expressed as RLU ×106 normalized against β-galactosidase activity as a measure of transfection efficiency. A Mann–Whitney U test was used to compare the promoter activities of deletion and m107p156 (human) or m105p125 (mouse) constructs; asterisks identify those with statistically significant differences (α≤0.05).
Comparison of the minimal proximal promoter m15p117 (13.2×106 RLU) with m15p93 (0.16×106 RLU) indicated an approx. 82-fold decrease in SMN promoter activity due to the removal of 24 nt between +93 and +117. Removing an additional 47 nt (nt +47 to +93) resulted in a further approx. 4.5-fold reduction in promoter activity. Taken together, these results suggested the presence of critical cis-elements in the +46 to +117 interval.
The mouse Smn promoter also possesses two TISs located 160 nt (+1 site) and 217 nt (−56 site) upstream of the initiating methionine (A. Semionov and L. R. Simard, unpublished work). The smallest promoter region (Smn m294p160) analysed to date contained 294 nt upstream and 160 nt downstream of the +1 TIS (referred to as Sal 1 −455/−1 in [21]). Having delimited the proximal human promoter, the mouse promoter constructs, Smn m105p125 (corresponding to SMN m107p117; results not shown) and Smn m46p125 (comparable with SMN m15p117) (Figure 1B), were transfected into P19 cells. As can be seen, m46p125 contained all of the cis-elements necessary to drive basal Smn promoter activity. Comparison of m105p125 and m46p125 (∼6.1×106 RLU) with the larger m1173p141 construct (3.5× 106 RLU) suggested the presence of a negative regulatory element(s) between −1173 and −105 of the mouse 5′-untranslated region.
Finally, we have previously demonstrated a 4-fold decrease in SMN promoter activity in differentiated P19 cells after RA treatment (P19RA) [20]. To determine if the minimal proximal promoters were also less active in differentiated cells, the human m15p117 and mouse m46p125 constructs were transfected into P19RA cells (Figure 1C). We observed an approx. 10- and 5-fold decrease in promoter activity respectively, indicating that the cis-element(s) mediating the observed decrease in promoter activity is conserved between species and is contained within the minimal proximal promoters.
In silico phylogenetic analysis
Having defined the minimal promoter regions, the human and mouse sequences were compared using DALIGN and analysed with the TRANSFAC database to annotate CRs (conserved regions) and cis-elements potentially capable of binding known TFs. Alignment of the proximal promoters revealed four CRs (CR1–CR4), shown in Figure 2(A), corresponding to nt +3 to +20, +30 to +41, +56 to +98 and +105 to +116 relative to the human SMN promoter. Interrogation of the TRANSFAC database revealed numerous potential TF-binding sites; several of them, contained within the CRs, are annotated in Figure 2. The potential Sp1, Ets, AhR (aromatic hydrocarbon receptor) and IL-6 (interleukin-6) cis-elements were explored further.
EMSA
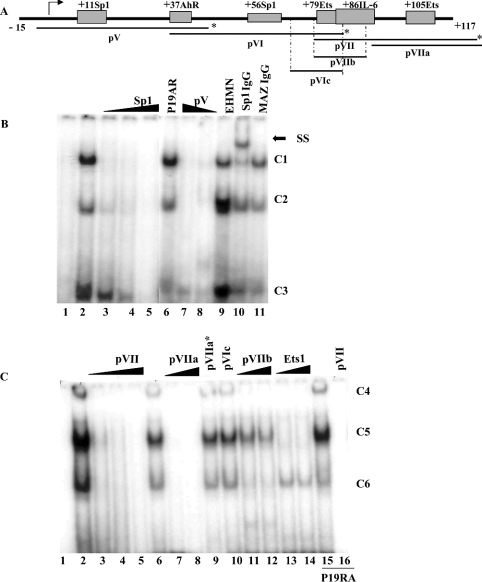
To analyse further the human promoter, a series of double-strand oligonucleotide probes, outlined in Figure 3(A), were end-labelled, incubated with nuclear extracts and mobility shifts of bound probe compared with unbound probe visualized on non-denaturing polyacrylamide gels. As can be seen in Figure 3(B), we detected three DNA–protein complexes (C1, C2 and C3) using probe V, which spans −3 to +47 nt and contains the putative +11Sp1 and +37AhR cis-elements in CR1 and CR2 respectively using nuclear extracts prepared from P19 (lanes 2–5, 7, 8, 10 and 11), P19RA (lane 6) and EHMN cells (lane 9). The specificity of these complexes was confirmed by competition with excess unlabelled probe V (lanes 7 and 8). C1 and C2 were easily displaced with 10-fold excess cold competitor consensus Sp1 oligonucleotides (lane 3) and, in the presence of anti-Sp1 antibody, the intensity of C1 (relative to C2 in P19 extracts) was diminished and is associated with the appearance of a supershift (SS) complex (lane 10). We did not detect a supershift when a polyclonal anti-MAZ antibody was added to the binding reaction (lane 11). The putative +37AhR cis-element was analysed by site-directed mutagenesis only (see below). The same three complexes were detected regardless of the nuclear extract employed; however, the relative intensity of these varied between P19 and EHMN cells.
Figure 3. EMSA analysis of the human SMN core promoter.
(A) Schematic representation of the proximal human SMN promoter (nt −15 to +117) and the putative cis-elements under investigation. Indicated below are the double-strand DNA probes used for EMSA studies, pV, pVI and pVII as well as competitor probes pVIc, pVIIa and pVIIb. The asterisk marks the extremity that was end-labelled with [γ-32P]dCTP. (B) EMSA analysis using end-labelled pV probe (lane 1) incubated with nuclear extract prepared from P19 (lanes 2–5, 7, 8, 10 and 11), P19RA (lane 6) or EHMN (lane 9) cells. A double-strand, unlabelled competitor oligonucleotide corresponding to the consensus Sp1 sequence was added in 10, 50 or 200 times excess in lanes 3–5. Double-strand pV competitor was added in 100 or 50 times excess in lanes 7 and 8 respectively. Binding reactions in the presence of 2 μl of anti-Sp1 or anti-MAZ antibody were run in lanes 10 and 11 respectively. The three DNA–protein complexes are designated as C1, C2 and C3 as shown to the right of the gel. A single supershift (SS) complex was observed in lane 10 as indicated by the arrow. (C) EMSA analysis using end-labelled pVII probe (lane 1) incubated with 4 μg of nuclear extract prepared from P19 (lanes 2–14) or P19RA (lanes 15 and 16). Lanes 2, 6 and 15 do not contain competitor probes. Double-strand, unlabelled competitor probes designated at the top of the gel were added in 10× (lane 3), 50× (lanes 4, 7, 11 and 13) or 200× excess (lanes 5, 8–10, 12, 14 and 16). The three DNA–protein complexes are designated as C4, C5 and C6 as shown to the right of the gel. The bottom of the gel was removed (B, C) so that the band corresponding to free probe is not shown. EMSAs were repeated at least three times.
Probe VI (+37 to +87) was used to study the +56Sp1 site and although we did detect a DNA–protein complex that was competed with excess cold Sp1 oligonucleotides, these EMSAs were difficult to reproduce, most probably due to weak DNA–protein interactions under the conditions employed (results not shown). This complex was not shifted in the presence of anti-Sp1 antibody (results not shown). Consequently, the +56Sp1 site was investigated by site-directed mutagenesis.
Probe VII (nt +78 to +127) was composed of the most highly conserved regions (CR3 and CR4) between the mouse and human promoters. A number of probes were used to analyse the putative +79Ets, +86IL-6, and +105Ets cis-elements; a representative result is provided in Figure 3(C). Once again, we detected three DNA–protein complexes (C4, C5 and C6) which were identical whether P19 (lanes 1–14), P19RA (lanes 15 and 16) or EHMN (results not shown) nuclear extracts were used in binding reactions. These complexes were sequence-specific as they were absent in the presence of as little as 10-fold excess unlabelled VII probe (lane 3). Addition of unlabelled VIIa that contained the +105Ets cis-element (lanes 7 and 8), VIIb that contained the +79Ets and +86IL-6 cis-elements (lanes 11 and 12) and the commercially available Ets1/Pea3 consensus (lanes 13 and 14) oligonucleotides efficiently competed the TF(s) present in C4. No competition was observed with probe VIIa* that contained a mutated +105Ets site (lane 9) or VIc that contained the putative +79Ets cis-element only (lane 10). These results suggested that C4 contained TFs interacting with the +86IL-6 and +105Ets cis-elements but not with +79Ets. Addition of unlabelled VIIa and Ets-1/Pea3 consensus oligonucleotides successfully competed for TF(s) present in C5, whereas unlabelled probes VIIa* and VIc did not. In contrast with C4, the unlabelled VIIb probe could not competitively remove a TF, suggesting that C5 contained an Ets-like but not an IL-6-like TF. Unlabelled VIIb, but not VIc or Ets-1/Pea3 probes, displaced TF(s) binding to C6, suggesting that this complex may contain an IL-6-like but not an Ets-like TF. Finally, the lack of competition by the Ets-1/Pea3 probe, despite efficient displacement by the pVIIa oligonucleotides, suggested the presence of an additional TF, which was most probably binding to the +94 to +105 or +116 to +128 interval. Probe pVIc was unable to displace a single complex, arguing against the hypothesis of a DNA–protein interaction within the conserved +71 to +86 region of the SMN promoter that contained the putative +79Ets cis-element. The addition of anti-Ets-1 or anti-Pea3 antibodies did not displace any of the complexes formed with probe pVII (results not shown). Results obtained by using probe VIIa (nt +94 to +128) as bait were consistent with the results obtained for probe VII (results not shown).
Site-directed mutagenesis
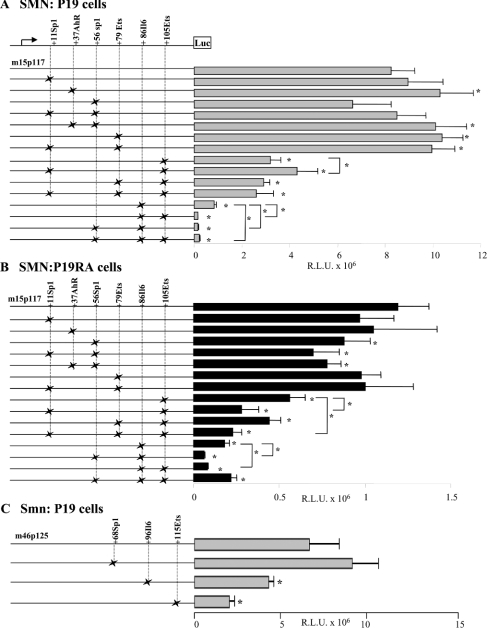
While EMSA provided evidence of clear DNA–protein interactions with the minimal SMN promoter and interaction with specific cis-elements could be inferred by competition assays, the functional relevance of these cis-elements was still not clear. Consequently, we created a number of SMN-promoter, luciferase-reporter gene constructs harbouring the mutations outlined in Figure 2(B), which were introduced by site-directed mutagenesis separately or in combination. These mutant constructs were transfected into P19 (Figure 4A) and P19RA (Figure 4B) cells and promoter activity compared with that observed for the minimal m15p117 promoter. SMN promoter activity generated by the mutant +11Sp1 construct was not significantly different from that observed for the wild-type construct in undifferentiated and differentiated P19 cells. Interestingly, mutations in the +11Sp1 cis-element did affect promoter activity when introduced in combination with the mutant +105Ets site (see below).
Figure 4. Transient transfection assays of wild-type and mutant prSMN-pGL3 constructs.
Mutations in the +11Sp1, +37AhR, +56Sp1, +79Ets, +86IL-6 and +105Ets were created singly or in combination as shown schematically to the left of the bar graphs. The × indicates the presence of a given mutation within the constructs shown along the Y-axis. SMN promoter activity is expressed as RLU×106 normalized against β-galactosidase activity as a measure of transfection efficiency (X-axis). Each construct was transiently transfected into undifferentiated (A) and differentiated (B) P19 cells. Note the different scale in (A, B) underscoring the lower SMN promoter activity in differentiated P19 cells. (C) The wild-type (m46p125) and mutant (+70Sp1, +97IL-6 and +115Ets cis-elements) mouse Smn promoter constructs schematically shown to the left of the bar graphs were transiently transfected into undifferentiated P19 cells. Asterisks identify those constructs that displayed SMN (A, B) or Smn promoter (C) activity significantly different from the corresponding wild-type minimal proximal SMN/Smn promoter constructs (m15p117 for A and B; or m46p125 for C) as determined using the Mann–Whitney U test. Asterisks identify statistically significant differences between mutant constructs (P≤0.05).
The +37AhR, +56Sp1 and +79Ets mutant constructs reacted differently in P19 cells compared with P19RA cells. We observed a small (1.2-fold) but significant increase in SMN promoter activity when the mutant +37AhR (P=0.009) and the +79Ets (P=0.002) constructs were introduced in P19 cells only (see Figure 4A). This same increase in activity was also observed with the +37AhR-+56Sp1 and +11Sp1-+79Ets double mutant constructs, indicating that the mutations introduced into the +11Sp1 and +56Sp1 sites had no effect on SMN promoter activity in P19 cells either alone or in combination with +37AhR or +79Ets mutations. In contrast, the mutation introduced into the putative +56Sp1 cis-element resulted in a 1.25-fold decrease in SMN promoter activity in P19RA cells only (P=0.04); an observation that was reproduced with the +11Sp1-+56Sp1 (P=0.01) and +37AhR-+56Sp1 (P=0.004) double mutant constructs. As there was no significant difference between these three mutant constructs, the observed reduction in SMN promoter activity in P19RA cells was attributed to the mutations introduced into the +56Sp1 cis-element alone. Thus the +37AhR and +79Ets cis-elements appear to be functional in P19 cells, whereas the +56Sp1 site appears to function in P19RA cells only. We did not find any evidence for an interaction between the two putative Sp1 cis-elements.
Mutations introduced into the putative +86IL-6 and/or +105Ets had the most significant effect on SMN promoter activity in P19 and P19RA cells (Figure 4). We observed a ≥2.5-fold decrease in SMN promoter activity when +105Ets, +79Ets-+105Ets and +11Sp1-+79Ets-+105Ets mutant constructs were transfected into P19 cells, suggesting that the observed decrease in promoter activity was the result of mutations introduced into the +105Ets cis-element. Interestingly, the effect of the +105Ets mutations was dampened from 2.5- to 1.9-fold in the presence of mutations in +11Sp1 (P=0.01). In contrast, mutation of the +105Ets cis-element also reduced SMN promoter activity by 2.1-fold in P19RA cells, this effect was further aggravated by mutations within the +11Sp1 site as the +11Sp1-+105Ets and +11Sp1-+79Ets-+105Ets constructs displayed a >4-fold decrease in SMN promoter activity (P=0.02). This observation suggested an antagonistic versus synergistic effect between TFs potentially binding to putative +11Sp1 and +105Ets cis-elements in P19 versus P19RA cells respectively. Disruption of the putative +86IL-6 cis-element resulted in an approx. 10- and 6-fold decrease in SMN promoter activity in P19 and P19RA cells respectively. This effect was further aggravated by mutating either the +105Ets or +56Sp1 cis-elements in which SMN promoter activity was decreased by >55-fold in P19 cells and >16-fold in P19RA cells. These results suggested a synergistic interaction between TFs binding to these two elements; however, the extent of this interaction was different in P19 and P19RA cells given that the double mutant had a 4-fold greater impact in undifferentiated (P19) compared with differentiated (P19RA) cells. We did not observe a significant difference in SMN promoter activity between the +105Ets and +79Ets-+105Ets constructs, arguing against an interaction between the two potential Ets cis-elements. Interestingly, in P19 cells only, the +56Sp1-+86IL-6-+105Ets triple mutant (∼38-fold decrease) was less detrimental compared with either double mutant, suggesting that these three sites combine in a complex manner to regulate SMN promoter activity in undifferentiated cells.
Conservation of cis-elements
The observed synergistic effect between the +56Sp1, +86IL-6 and +105Ets cis-elements within the core human promoter region led us to question whether these elements were also functional in the mouse promoter. Consequently, we introduced mutations into the putative +70Sp1, +97IL-6 and +115Ets cis-elements of the core mouse promoter. The mutant constructs were transfected into undifferentiated P19 cells and the resulting promoter activities are presented in Figure 4(C). As can be seen, inverting the guanine and adenine nucleotides in the +115Ets element resulted in a >3-fold decrease in Smn promoter activity, mimicking the effect observed with the human promoter. While the 4 nt substitution introduced in the +97IL-6 cis-element led to a 1.6-fold decrease in promoter activity, this effect was much less than the 10-fold decrease observed for the human SMN promoter. Mutations in the +70Sp1 cis-element did not affect Smn promoter activity in P19 cells (Figure 4C); however, when introduced into P19RA cells, we observed a 1.5-fold decrease in activity (results not shown), consistent with the results obtained for the +56Sp1 mutant human construct (Figure 4B). Taken together, these results suggest that these cis-elements are functionally conserved between species.
In vivo genomic footprinting
Having confined the proximal promoter activity to the m15p117 and m46p125 regions of the human SMN and mouse Smn promoters respectively, we employed in vivo genomic DNA footprinting, using LMPCR technology [24] to screen the minimal mouse Smn promoter. Specifically, we used DMS, UVC irradiation and DNase I as DNA-modifying agents to map single-strand DNA breaks, comparing in vitro (naked DNA) and in vivo (living cells) footprints. In vivo footprints corresponding to nt −46 to +125 encompassing the minimal core promoter are presented in Figure 5 and summarized in Figure 6.
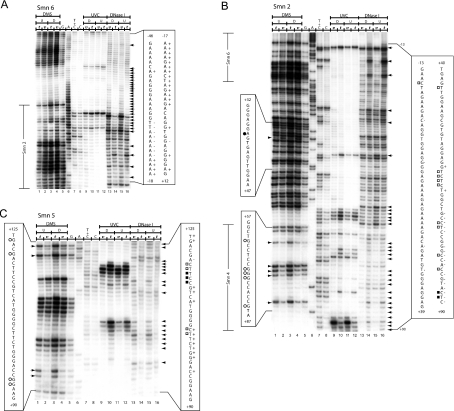
Figure 5. In vivo DNA footprint of the mouse Smn proximal promoter spanning nt −46 to +125.
(A) The region shown was analysed with primer set Smn6 to reveal upper strand sequences from nt −46 to +12 relative to the +1 TIS. Lanes 1–4, LMPCR of DNA purified from undifferentiated (U) (lanes 1 and 2) and differentiated (D) (lanes 3 and 4) P19 cells, treated with DMS in vitro (t) (lanes 1 and 3) after DNA purification or in vivo (v) (lanes 2 and 4) before DNA purification. Lanes 5–8, Maxam–Gilbert sequencing reactions. Lanes 9–12, LMPCR of DNA purified from differentiated (D) (lanes 9 and 10) and undifferentiated (U) (lanes 11 and 12) P19 cells, irradiated with UVC in vivo (v) (lanes 9 and 11) before DNA purification or in vitro (t) (lanes 10 and 12) after DNA purification. Lanes 13–16, LMPCR of DNA purified from differentiated (D) (lanes 13 and 14) and undifferentiated (U) (lanes 15 and 16) P19 cells, treated with DNase I in vivo (v) (lanes 13 and 15) before DNA purification or in vitro (t) (lanes 14 and 16) after DNA purification. (B) The region shown was analysed with primer set Smn2 to reveal upper strand sequences from nt −13 to +90 relative to the +1 TIS. Lanes 1–16 and footprint characteristics for DMS, UVC and DNase I are as described in (A). Overlapping sequences detected with primer sets Smn6 and Smn4 are annotated to the left of the Figure. (C) The region shown was analysed with primer set Smn5 to reveal bottom strand sequences from nt +125 to +90 relative to the +1 TIS. Lanes 1–16 and footprint characteristics for DMS, UVC and DNase I are as described in (A). DMS protection and hyperreactivity is indicated by ○ and ● respectively (displayed on the left). UVC protection and hyperactivity is indicated by □ and ■ respectively (displayed on the right). DNase I protection and hyperactivity is indicated by ‘−’ and ‘+’ signs respectively (displayed on the right). As a reference, the corresponding portion of the Maxam–Gilbert-derived sequence is shown on both sides of the autoradiogram. Arrows indicate footprinted bands. Regions covered by superposing primer sets are indicated at the far left of the autoradiogram. The sequence overlapping with primer set Smn2 is annotated to the left of the Figure.
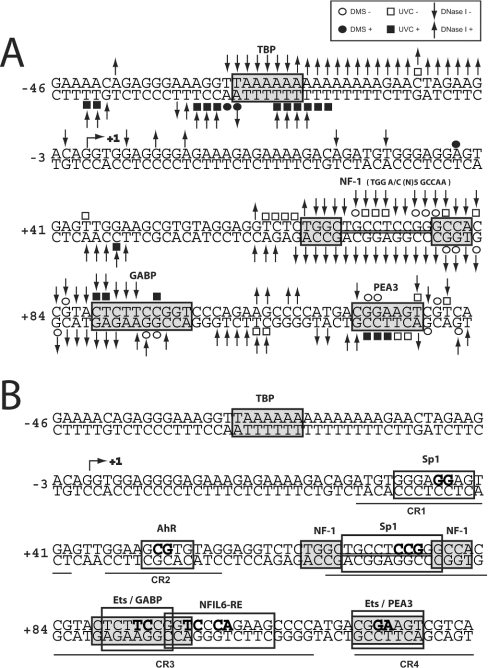
Figure 6. Summary of in vivo DMS, UVC and DNase I footprints of the mouse Smn proximal promoter.
(A) Nucleotides protected from DMS (○), UVC (□) or DNase I (↓) modification and DMS (●), UVC (■) or DNase I (↑) hypersensitive sites are indicated. The +1 TIS is annotated with an arrow ↱. (B) Regions of protected DNA corresponding to known cis-elements are highlighted by grey boxes. The putative TFs are annotated above their respective cis-elements. Regions of conserved sequence identity between the human and mouse promoters (CR1 to CR4) are underlined. The nucleotides targeted for site-directed mutagenesis are shown in boldface.
Within the core Smn promoter (m46p125), we observed a major DMS, UVC and DNase I hypersensitive region (−2 to −33) just upstream of the TIS, consistent with the fact that this promoter is active both in undifferentiated and differentiated P19 cells (Figures 5A and 5B). Embedded within this hypersensitive region is a ‘TTAAAAA’ element that was protected from DNase I digestion, suggesting that nt −29 to −23 of the upper non-transcribed strand interacts with a DNA-binding protein (Figure 5A). This sequence resembles a TATA box with respect to position and sequence relative to the TIS and may recruit TFIID to the promoter region. A similar atypical TATA box has been described for the mouse adenosine deaminase promoter [27]. We did not detect any significant footprints in the CR1 and CR2 segments, except for a hypersensitive region involving nt +44 to +48 of the lower transcribed strand just upstream of the putative AhR cis-element (Figure 5A). This result argues against possible binding of Sp1 or AhR family members within this interval in vivo. The largest stretch of DNA protected from all three probing agents was located between nt +62 to +94 (Figures 5B and 5C) with 26 nt of both strands (nt +65 to +90) resisting DNA modification. This region contains cis-elements recognized by Sp, NF1 (nuclear factor-1) and Ets family members and the presence of in vivo footprints suggested that TFs bind to DNA throughout CR3, except for the distal 12 nt that contains most of the potential IL-6 cis-element. In fact, the IL-6 cis-element was hypersensitive to UVC and DNase I treatment. In vivo footprinting also provided evidence for a protected region in CR4 involving nt +114 to +124 that included the +115 Ets cis-element. This footprint was immediately flanked by a hypersensitive zone spanning nt +125 to +137 (Figure 5B). We did not detect any difference in the DNA footprints when comparing P19 and P19RA cells.
DISCUSSION
A major challenge during the next phase of genomic research will be to understand how epigenetic changes and complex combinatorial events regulate protein-coding genes temporally and spatially. The importance of elucidating these events is underscored by the NIH ENCODE initiative to identify all functional elements in the human genome [28], the expected impact of such studies being the identification of pharmacological targets. To this end, we have employed in silico, in vitro and in vivo approaches to characterize the human and mouse SMN/Smn basal promoters because up-regulation of the human SMN2 gene is a recognized therapeutic target for SMA [1,29]. We have delimited the core human (−15 to +117) and mouse (−46 to +125) promoters, mapped critical cis-elements and have demonstrated that they are >5-fold less active in differentiated P19 cells. The latter suggests that the P19 cell system may be exploited to dissect the temporal down-regulation of SMN expression. Phylogenetic footprinting implicated a number of conserved cis-elements; however, only a subset of these was corroborated by in vivo DNA footprinting. Discordance between in vitro and in vivo binding assays underscores the necessity to establish in vivo evidence for DNA–protein interactions. Nonetheless, these studies have definitively implicated a number of cis-elements in the regulation of SMN/Smn promoters.
Ablating either the +86IL-6 or +105Ets cis-elements within the critical core promoter region had the greatest effect with the double mutant displaying more than 55-fold less activity compared with the wild-type SMN promoter. The biological relevance of the +105 Ets site was supported by the in vivo DNA footprint across nt +114 to +126 of the mouse Smn promoter corresponding to the conserved +105 to +116 interval of the human promoter containing the +105Ets cis-element. This result was consistent with EMSA studies. Ets TFs have a conserved winged helix–turn–helix DNA-binding motif and binding specificity depends on the GGAA/T core recognition site, the surrounding 9–15 nt and co-operative protein–protein interactions that combine to activate or repress a given promoter [30]. The more than 45 Ets proteins identified to date have a role in a number of physiological processes including cellular proliferation, differentiation, immune response and apoptosis. Interestingly, Ets1 expression is up-regulated by RA treatment [31], PEA3 is down-regulated [32] and Ets2 is ubiquitously expressed in undifferentiated and differentiated P19 cells [33]. EMSA studies so far have excluded Ets1 and Pea3 as potential candidates. In contrast, despite evidence of in vivo DNA binding across the +79Ets cis-element, EMSA studies did not corroborate Ets-binding to this site, suggesting that the TF(s) interacting with this region is not an Ets family member. Consistent with DNA–protein interactions detected in vivo, mutations involving the +79Ets cis-element caused a 1.25-fold increase in SMN promoter activity in P19 cells but not in P19RA cells. This small effect may reflect the fact that the most critical nucleotides have not yet been ablated. Re-inspection of in silico data revealed a bipartite TGGA/C(N)5GCCAA cis-element which overlaps the +56Sp1 site and recognizes NF1 [34]. Other examples of Sp1/NF1 composite sites do exist and NF1 has been shown to interfere with Sp1 binding to the rat poly(ADP-ribose) polymerase 1 promoter, thereby down-regulating Sp1 activity [35]. Further studies are underway to assess whether quantity and/or post-translational modification of these factors modulate SMN promoter activity.
We did not detect an in vivo footprint across the +86IL-6 cis-element despite the fact that mutations in this site had the greatest effect on SMN promoter activity either alone or combined with mutant +56Sp1 or +105Ets cis-elements. These effects were greater in P19 compared with P19RA cells, suggesting that these elements are less active in differentiated P19 cells (see Figure 7). Thus nt +86, 87, 89 and 90 found within the IL-6 consensus element are critical for activating SMN expression. While in vivo footprinting argues against a role for an IL-6 TF in regulating the SMN promoter, other examples where transcriptional regulation occurs without DNA binding to its cognate element exist [36]. NF-IL6, also known as C/EBPβ, is a member of the leucine zipper C/EBP (CCAAT/enhancer-binding protein) family of TFs that have been implicated in a variety of physiological processes including cellular proliferation and differentiation [37]. IL-6 is neuroprotective against lethal viral infection [38], can induce neurite outgrowth [39] and overexpression in mice causes neurological symptoms [40]. Interestingly, ETS and bZIP cis-elements are frequently found in composite sites and members of these two families of TFs often co-ordinate transcriptional regulation [36]. In one instance, a protein-tethered transactivation mechanism was proposed because activation of the il-1b core promoter necessitated NF-IL6's transactivation domain, binding between NF-IL6 and Spi-1 (an ETS family member), but did not depend on NF-IL6 binding to its cognate element [36]. A similar mechanism may be involved in our observed Ets-IL6 transactivation of the SMN promoter. Alternatively, the TF binding affinity may be weak, the proportion of cells with bound protein too low to detect an in vivo footprint or the mutations affect the binding of nearby proteins within the +48 to +83 footprint. These possibilities are currently under investigation.
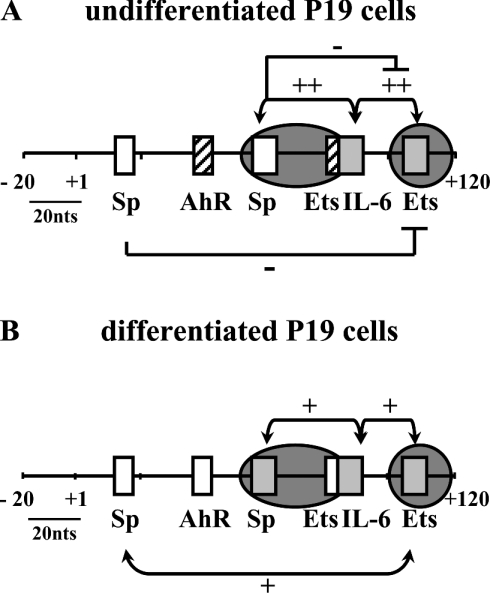
Figure 7. Summary of in silico, in vitro and in vivo analyses of transcriptional activation of the SMN proximal promoter in undifferentiated (A) and differentiated (B) P19 cells.
Schematic representation of the −20 to +120 bp region of the SMN promoter region summarizing the role of putative Sp, AhR, Ets and IL-6 cis-elements in regulating SMN expression. Cis-elements that had no effect (open boxes), activate (grey boxes) or repress (striped boxes) SMN promoter activity are shown and the results of single-site mutagenesis is summarized. Combinatorial effects are shown above and below the promoter region with the ‘+’ and ‘−’ signs designating synergistic and antagonistic effects respectively. Regions protected from DMS, UV or DNase I treatment in vivo are shown by the presence of dark grey circles depicting regions of DNA–protein interactions.
Binding-perturbing mutations revealed complex combinatorial effects involving the Sp, Ets and IL-6 cis-elements. We did not find any in vivo evidence implicating the putative +11Sp1 and +37AhR cis-elements identified by phylogenetic footprinting and deleting these sites had no effect (see Figure 7). The AhR cis-element was of interest since AhR expression is suppressed by RA [41]. Mutating the +37AhR cis-element had no effect on SMN promoter activity in P19RA cells, although the +11Sp1 and +105Ets cis-elements displayed a synergistic effect in P19RA cells. We detected an antagonistic effect of +56Sp1 mutations in conjunction with the +86IL-6-+105Ets double mutant, as the triple mutant recovered some activity in P19 cells only, despite the fact that mutating +56Sp1 alone had no effect on SMN promoter activity. Thus the +11Sp1, +37AhR and +56Sp1 cis-elements do not appear to be involved in the observed decrease in SMN promoter activity in P19RA cells.
Taken together, we provide evidence for combinatorial regulation of the SMN promoter, implicating at least Sp and Ets and perhaps also C/EBP family member(s). Our results indicated that the composition of the transcription complex binding to the SMN core promoter is most probably different in P19 and P19RA cells and that Sp family members may play a key role in modulating SMN promoter activity. This hypothesis is corroborated by the fact that binding-perturbing mutations, especially those involving Sp1 cis-elements, did not have the same effect in undifferentiated cells as in differentiated cells, whereas no differences in DNA–protein complexes were detected either in vitro or in vivo. Ubiquitously expressed Sp family proteins bind GC and GT boxes via C-terminal zinc finger domains and can interact with different cofactors to activate or repress a wide variety of genes [42,43]. Sp proteins can be modified post-translationally [44], can compete with each other [42] and can interact with other TFs, including Ets [45] and C/EBPβ [46] proteins. Direct interaction between Sp1 and RA receptors can potentiate Sp1 DNA binding in the absence of an RA response element [47]. Of particular interest to the SMN gene, both Ets [48] and Sp [43] proteins interact with the sin3/HDAC system, thereby mediating changes in chromatin structure associated with acetylation/deacetylation of core histones and/or cofactors. Consistent with the possible involvement of chromatin remodelling in regulating SMN expression, treatment of human fibroblasts or rat hippocampal slice cultures [49,50] with valproic acid, a histone-deacetylase inhibitor, activated SMN expression. However, it is unlikely that Sp1 alone is responsible for the 10-fold decrease in SMN promoter activity in differentiated cells, especially in the light of the antagonist effect of the +11Sp1 or +56Sp1 sites on more distal cis-elements in undifferentiated P19 cells. Further in vivo studies are underway to identify the specific TFs and cofactors contained within the multiprotein complex binding to the SMN promoter, compare and contrast this complex in P19 versus P19RA cells and determine how it interacts with regulatory elements outside the core promoter region.
Acknowledgments
We thank S. Morissette and L. Gallant for technical and secretarial assistance. We are grateful to Dr R. S. Lloyd and Dr T. R. O'Connor for supplying T4 endonuclease V and photolyase respectively and Dr A. Burghes for the EHMN cell line. This work was funded by the Canadian Institutes of Health Research (MOP-G-90071-21613) in partnership with Muscular Dystrophy Canada and the Amyotrophic Lateral Sclerosis Society of Canada, and partially supported by the Hospital for Sick Children Foundation. Research in R.D.'s laboratory was partly funded by the Canada Research Chairs Program and by the Canadian Genetic Diseases Network (MRC/NSERC NCE Program). R.D. holds the Canada Research Chair in Genetics, Mutagenesis and Cancer.
References
- 1.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell (Cambridge, Mass.) 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Monani U. R., Lorson C. L., Parsons D. W., Prior T. W., Androphy E. J., Burghes A. H., McPherson J. D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 3.Rochette C. F., Gilbert N., Simard L. R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 4.Kashima T., Manley J. L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–465. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 5.Monani U. R., Sendtner M., Coovert D. D., Parsons D. W., Andreassi C., Le T. T., Jablonka S., Schrank B., Rossol W., Prior T. W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(–/–) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 6.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 8.Feldkotter M., Schwarzer V., Wirth R., Wienker T. F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang J.-G., Hsieh-Li H.-M., Jong Y.-J., Wang N. M., Tsai C.-H., Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron-Delage S., Abadie A., Echaniz-Laguna A., Melki J., Beretta L. Interferons and IRF-1 induce expression of the survival motor neuron (SMN) genes. Mol. Med. 2000;6:957–968. [PMC free article] [PubMed] [Google Scholar]
- 11.Andreassi C., Jarecki J., Zhou J., Coovert D. D., Monani U. R., Chen X., Whitney M., Pollok B., Zhang M., Androphy E., et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum. Mol. Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- 12.Burlet P., Huber C., Bertrandy S., Ludosky M. A., Zwaenepoel I., Clermont O., Roume J., Delezoide A. L., Cartaud J., Munnich A., et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum. Mol. Genet. 1998;7:1927–1933. doi: 10.1093/hmg/7.12.1927. [DOI] [PubMed] [Google Scholar]
- 13.Battaglia G., Princivalle A., Forti F., Lizier C., Zeviani M. Expression of the SMN gene, the spinal muscular atrophy determining gene, in the mammalian central nervous system. Hum. Mol. Genet. 1997;6:1961–1971. doi: 10.1093/hmg/6.11.1961. [DOI] [PubMed] [Google Scholar]
- 14.Gubitz A. K., Feng W., Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 15.Pagliardini S., Giavazzi A., Setola V., Lizier C., Di Luca M., DeBiasi S., Battaglia G. Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum. Mol. Genet. 2000;9:47–56. doi: 10.1093/hmg/9.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Fan L., Simard L. R. Survival motor neuron (SMN) protein: role in neurite outgrowth and neuromuscular maturation during neuronal differentiation and development. Hum. Mol. Genet. 2002;11:1605–1614. doi: 10.1093/hmg/11.14.1605. [DOI] [PubMed] [Google Scholar]
- 17.Soler-Botija C., Ferrer I., Alvarez J. L., Baiget M., Tizzano E. F. Downregulation of Bcl-2 proteins in type I spinal muscular atrophy motor neurons during fetal development. J. Neuropathol. Exp. Neurol. 2003;62:420–426. doi: 10.1093/jnen/62.4.420. [DOI] [PubMed] [Google Scholar]
- 18.Echaniz-Laguna A., Miniou P., Bartholdi D., Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am. J. Hum. Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monani U. R., McPherson J. D., Burghes A. H. M. Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT) Biochim. Biophys. Acta. 1999;1445:330–336. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 20.Germain-Desprez D., Brun T., Rochette C., Semionov A., Rouget R., Simard L. R. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene. 2001;279:109–117. doi: 10.1016/s0378-1119(01)00758-2. [DOI] [PubMed] [Google Scholar]
- 21.DiDonato C. J., Brun T., Simard L. R. Complete nucleotide sequence, genomic organization, and promoter analysis of the murine survival motor neuron gene (Smn) Mamm. Genome. 1999;10:638–641. doi: 10.1007/s003359901061. [DOI] [PubMed] [Google Scholar]
- 22.Andreassi C., Patrizi A. L., Monani U. R., Burghes A. H. M., Brahe C., Eboli M. L. Expression of the survival of motor neuron (SMN) gene in primary neurons and increase in SMN levels by activation of the N-methyl-D-aspartate glutamate receptor. Neurogenetics. 2002;4:29–36. doi: 10.1007/s10048-001-0128-y. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber E., Tobler A., Malipiero U., Schaffner W., Fontana A. cDNA cloning of human N-Oct3, a nervous-system specific POU domain transcription factor binding to the octamer DNA motif. Nucleic Acids Res. 1993;21:253–258. doi: 10.1093/nar/21.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drouin R., Therrien J.-P., Angers M., Ouellet S. In vivo DNA analysis. In: Moss T., editor. DNA-protein Interactions, Principles and Protocols. Methods in Molecular Biology. 2nd edn. Totowa, NJ: Humana Press; 2001. pp. 175–219. [DOI] [PubMed] [Google Scholar]
- 25.Pfeifer G. P., Drouin R., Riggs A. D., Holmquist G. P. Binding of transcription factors creates hot spots for UV photoproducts in vivo. Mol. Cell. Biol. 1992;12:1798–1804. doi: 10.1128/mcb.12.4.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angers M., Cloutier J.-F., Castonguay A., Drouin R. Optimal conditions to use Pfu exo− DNA polymerase for highly efficient ligation-mediated polymerase chain reaction protocols. Nucleic Acids Res. 2001;29:E83. doi: 10.1093/nar/29.16.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Innis J. W., Moore D. J., Kash S. F., Ramamurthy V., Sawadogo M., Kellems R. E. The murine adenosine deaminase promoter requires an atypical TATA box which binds transcription factor IID and transcriptional activity is stimulated by multiple upstream Sp1 binding sites. J. Biol. Chem. 1991;266:21765–21772. [PubMed] [Google Scholar]
- 28.Collins F. S., Green E. D., Guttmacher A. E., Guyer M. S. A vision for the future of genomics research. Nature (London) 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 29.Burghes A. H. M. When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sementchenko V. I., Watson D. K. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 31.So E. N., Crowe D. L. Characterization of a retinoic acid responsive element in the human ets-1 promoter. Life. 2000;50:365–370. doi: 10.1080/713803742. [DOI] [PubMed] [Google Scholar]
- 32.Zin J. H., Cowie A., Lachance P., Hassell J. A. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 1992;6:481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]
- 33.Kola I., Brookes S., Green A. R., Garber R., Tymms M., Papas T. S., Seth A. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luciakova K., Barath P., Poliakova D., Persson A., Nelson B. D. Repression of the human adenine nucleotide translocase-2 gene in growth-arrested human diploid cells: the role of nuclear factor-1. J. Biol. Chem. 2003;278:30624–30633. doi: 10.1074/jbc.M303530200. [DOI] [PubMed] [Google Scholar]
- 35.Laniel M. A., Poirier G. G., Guerin S. L. Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity in vitro. J. Biol. Chem. 2001;276:20766–20773. doi: 10.1074/jbc.M010360200. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z., Wara-Aswapati N., Chen C., Tsukada J., Auron P. E. NF-IL6 (C/EBPbeta) vigorously activates il1b gene expression via a Spi-1 (PU.1) protein-protein tether. J. Biol. Chem. 2000;275:21272–21277. doi: 10.1074/jbc.M000145200. [DOI] [PubMed] [Google Scholar]
- 37.Lekstrom-Himes J., Xanthopoulos K. G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 38.Pavelko K. D., Howe C. L., Drescher K. M., Gamez J. D., Johnson A. J., Wei T., Ransohoff R. M., Rodriguez M. J. Interleukin-6 protects anterior horn neurons from lethal virus-induced injury. Neuroscience. 2003;23:481–492. doi: 10.1523/JNEUROSCI.23-02-00481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y. Y., Bradshaw R. A. Induction of neurite outgrowth by interleukin-6 is accompanied by activation of Stat3 signaling pathway in a variant PC12 cell (E2) line. J. Biol. Chem. 1996;271:13023–13032. doi: 10.1074/jbc.271.22.13023. [DOI] [PubMed] [Google Scholar]
- 40.Campbell I. L., Abraham C. R., Masliah E., Kemper P., Inglis J. D., Oldstone M. B., Mucke L. Neurologic disease induced in transgenic mice by cerebral overexpression of interleukin 6. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10061–10065. [Google Scholar]
- 41.Wanner R., Brömmer S., Czarnetzki B. M., Rosenbach T. The differentiation-related upregulation of aryl hydrocarbon receptor transcript levels is suppressed by retinoic acid. Biochem. Biophys. Res. Commun. 1995;209:706–711. doi: 10.1006/bbrc.1995.1556. [DOI] [PubMed] [Google Scholar]
- 42.Lania L., Majello B., De Luca P. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell. Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Dufau M. L. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 2002;277:33431–33438. doi: 10.1074/jbc.M204417200. [DOI] [PubMed] [Google Scholar]
- 44.Bonello M. R., Khachigian L. M. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J. Biol. Chem. 2004;279:2377–2382. doi: 10.1074/jbc.M308254200. [DOI] [PubMed] [Google Scholar]
- 45.Hadri L., Ozog A., Soncin F., Lompre A. M. Basal transcription of the mouse sarco(endo)plasmic reticulum Ca2+-ATPase type 3 gene in endothelial cells is controlled by Ets-1 and Sp1. J. Biol. Chem. 2002;277:36471–36478. doi: 10.1074/jbc.M204731200. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y. H., Williams S. C., Baer M., Sterneck E., Gonzalez F. J., Johnson P. F. The ability of C/EBP beta but not C/EBP alpha to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol. Cell. Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimada J., Suzuki Y., Kim S.-J., Wang P.-C., Matsumura M., Kojima S. Transactivation via RAR/RXR-Sp1 interaction: characterization of binding between Sp1 and GC box motif. Mol. Endocrinol. 2001;15:1677–1692. doi: 10.1210/mend.15.10.0707. [DOI] [PubMed] [Google Scholar]
- 48.Mavrothalassitis G., Ghysdael J. Proteins of the ETS family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- 49.Brichta L., Hofmann Y., Hahnen E., Siebzehnrubl F. A., Raschke H., Blumcke I., Eyupoglu I. Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 50.Sumner C. J., Huynh T. N., Markowitz J. A., Perhac J. S., Hill B., Coovert D. D., Schussler K., Chen X., Jarecki J., Burghes A. H., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]