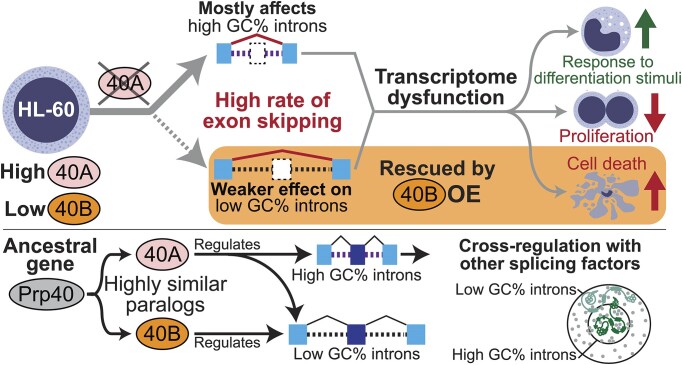
Abstract
We characterized the regulatory mechanisms and role in human myeloid cell survival and differentiation of PRPF40A, a splicing factor lacking a canonical RNA Binding Domain. Upon PRPF40A knockdown, HL-60 cells displayed increased cell death, decreased proliferation and slight differentiation phenotype with upregulation of immune activation genes. Suggestive of both redundant and specific functions, cell death but not proliferation was rescued by overexpression of its paralog PRPF40B. Transcriptomic analysis revealed the predominant role of PRPF40A as an activator of cassette exon inclusion of functionally relevant splicing events. Mechanistically, the exons exclusively upregulated by PRPF40A are flanked by short and GC-rich introns which tend to localize to nuclear speckles in the nucleus center. These PRPF40A regulatory features are shared with other splicing regulators such as SRRM2, SON, PCBP1/2, and to a lesser extent TRA2B and SRSF2, as a part of a functional network that regulates splicing partly via co-localization in the nucleus.
Graphical Abstract
Graphical Abstract.
Introduction
Alternative splicing plays crucial roles in human cell differentiation such as myelopoiesis, which is the generation of mature myeloid cells including monocytes, monocyte-derived macrophages and dendritic cells (DCs), as well as granulocytes (1). The main regulators of myelopoiesis are master transcription factors acting at different stages such as PU.1, C/EBPα and RUNX1 (2). Intron retention is a major alternative splicing event during myeloid-cell differentiation (3–6). Furthermore, alternative splicing modulates immune responses via toll-like receptors in myeloid cells (7–9). A few splicing factors have established roles in myeloid cell differentiation and function (10–13). In the pathological context, myeloid malignancies often exhibit splicing factor mutations, particularly in myelodysplastic syndromes (MDS) and to a lesser extent, acute myeloid leukemia (AML). Splicing factors mutations are reported at about 10% in de novo AML and up to 50% in secondary AML derived from myelodysplasia (14). These mutations are often associated with poor prognosis, making it important to understand their mode of action for targeted therapies. The top commonly mutated splicing factors in MDS and AML are SF3B1, SRSF2 and U2AF1 which are involved in recognition of the 3′ end of introns. Mutations of a few other splicing factors were also reported in MDS and AML patients, including PRPF40B in the original study (15), whose later transcriptomic analysis did not support a role in driving MDS (16). AML cells exhibit the largest number of aberrant splicing events, which adds up to approximately 29% of expressed genes (17), including oncogenes, tumour suppressors, as well as splicing factors (18). Beyond MDS/AML, the SRSF1 splicing factor is involved in drug resistance in Chronic Myeloid Leukemia (19). All these findings support the importance of characterizing alternative splicing in myeloid cells under both physiological and pathological conditions.
Alternative splicing affects the mRNA structure, stability, translatability, protein coding sequence and/or non-coding function. Apart from the essential cis-acting elements including the 5′/3′ splice sites (5′ss/3′ss) and the branch point sequence, alternative splicing is regulated by sequences that promote use of splice sites such as exonic or intronic splicing enhancers (ESEs and ISEs), and by other sequences that repress them, such as exonic or intronic splicing silencers (ESSs and ISSs) (20). These cis-acting elements are bound by trans-acting factors which include two major families, the Serine/Arginine-rich (SR) proteins and the heterogeneous nuclear ribonucleoproteins (hnRNPs).The typical trans-acting factors are RNA Binding Proteins with well-defined, canonical RNA Binding Domains (RBDs) including RNA recognition motifs (RRM), hnRNP K homology domains (KH) and zinc fingers (21). However, certain splicing factors like PRPF40A and PRPF40B lack a canonical RBD. Such splicing factors may not contact RNA at all and perform their splicing function via protein-protein interactions, or instead may utilize intrinsically disordered regions to mediate RNA binding (22).
The budding yeast ortholog of the human PRPF40A/B paralogs, namely the pre-mRNA processing protein Prp40, is an essential splicing factor in yeast (23). Consisting of two WW and four FF domains, yeast Prp40 is stably associated with the U1 snRNP and binds the branch-point binding protein (BBP)/Mud2p to bring the ends of the intron together (24). In humans, both PRPF40A and PRPF40B are composed of two WW and five FF domains which are conserved in vertebrates (23,25). Although the human PRPF40A/B proteins share 52% sequence similarity, available data hint at functional specialization of these two proteins (23,26). First, the WW domains in PRPF40A are essential for its localization to splicing factor-rich nuclear speckles, while the FF domains are sufficient for PRPF40B nuclear speckle localization (26,27). PRPF40A associates with the U2 snRNP while it is recruited to nascent transcripts during transcription (28). In addition, histone acetylation is necessary for PRPF40A to assist in the assembly of a functional spliceosome (29). On the other hand, PRPF40B interacts directly with SF1 mainly through its WW domains and more weakly through the FF domains, and it regulates alternative splicing associated with weak splice sites (16,26). Worm PRP-40 only regulates microexons, while knockdown of Prpf40A/B in mouse N2a cells affects microexons and not larger exons (30). PRPF40A but not PRPF40B plays other roles unlinked to splicing like cell migration (28). Finally, both PRPF40A/B paralogs might be involved in neurological disorders, because they bind huntingtin with expanded glutamine tracts as well as methyl-CpG-binding protein (MeCP2), the causative genes of Huntington's disease and Rett syndrome (31,32), respectively.
Recent studies suggest that PRPF40A might play a role in carcinogenesis, by either acting as a driver or as a suppressor. PRPF40A knockdown in a pancreatic cell line inhibited cell proliferation (33), and the levels of this factor correlate with hypoxia markers in non-small cell lung cancer (NSCLC) (33,34). In contrast, PRPF40A knockdown cells were enriched in a pooled in vivo RNAi screen for the identification of splicing factors with a role in AML, suggesting a leukemia suppressive role (35). Our previous transcriptomics of PRPF40B knockout (KO) in K562 myeloid cells revealed an upregulation of hypoxia-related pathways, suggesting that PRPF40B could inhibit hypoxia (16). TCGA data mining showed that PRPF40A is expressed highly in AML as compared to other cancers, while PRPF40B displayed the lowest expression in AML. Hence, we hypothesized that PRPF40A and PRPF40B might play a distinct role in myeloid cells with relevance to differentiation and leukemogenesis. Here we performed functional studies by knocking down PRPF40A/B in HL-60 myeloid cells, to uncover the PRPF40A role in myeloid cell proliferation, differentiation and immunity. Mechanistically, PRPF40A regulates splicing targets with short GC-rich introns, as part of a larger functional network regulating alternative splicing that is important for myeloid cell function.
Materials and methods
Cell lines and culture
We cultured the HL-60 cells (ATCC CCL-240) in RPMI-1640 (HyClone, GE Healthcare) that was supplemented with 20% fetal bovine serum (FBS) (Gibco, Thermo Fisher) and 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco, Thermo Fisher). We kept cells in a humidified incubator at 37ºC with 5% CO2.
Knockdown and Overexpression of PRPF40A or PRPF40B in HL-60
We performed the knockdowns using doxycycline-inducible EZ-Tet-pLKO-Puro, cloned with the respective shRNAs (target sequences in Supplementary Table S1). EZ-Tet-pLKO-Puro was a gift from Cindy Miranti (Addgene plasmid # 85966) (36). We added 50 ng/ml of doxycycline (Sigma) to 125 000 cells/ml in 4 ml of media to induce knockdown of target, and refreshed the doxycycline media every 48 h.
We overexpressed PRPF40B using pLV-EF1a-IRES-Blast using lentiviral transduction. The pLV-EF1a-IRES-Blast plasmid was a gift from Tobias Meyer (Addgene plasmid # 85133) (37), and the lentivirus packaging plasmids psPAX2 (Addgene plasmid # 12260) and pMD2.G (Addgene plasmid #12259) were gifts from Didier Trono. We generated the lentiviruses by transfecting HEK-293T cells with psPAX2, pMD2.G together with the transfer plasmid in a 1:1:2 molar ratio, using X-tremeGENE™ HP DNA Transfection Reagent (Roche Applied Science, Merck) in a 3:1 ratio of reagent to DNA. We harvested the lentiviral particles 48 h after transfection.
We transduced HL-60 cells with lentivirus by centrifugation at 800 × g for 30 min in the presence of 10 μg/ml polybrene (Sigma-Aldrich). We then selected the transduced cells with either 2.5 μg/ml of puromycin dihydrochloride (Gibco, Thermo Fischer) for 4 days or 10 μg/ml of blasticidin S HCl (Gibco, Thermo Fischer) for a week. We also introduced shRNA and overexpression plasmids into HEK-293T by transfection using X-tremeGENE™ 9 DNA Transfection Reagent (Roche Applied Science, Merck) in a 3:1 ratio of reagent to DNA.
Cell proliferation assay with MTS
We used the MTS assay to determine cell survival upon PRPF40A/B knockdown. We incubated cells in a 96-well plate with CellTiter 96® AQueous One Solution Reagent (Promega) as recommended by the manufacturer. We recorded the absorbance at 490 nm with Tecan Infinite M200 PRO plate reader. We subtracted the absorbance readings with readings of culture medium with no cells and then normalized to untreated sample for comparison.
Giemsa stain
We used Giemsa stain to visualize cell morphology. We concentrated cells by centrifugation, spread them onto a glass slide, and performed fixation with methanol before staining with 10% Giemsa stain (Sigma-Aldrich, Merck) for 5 min. We washed off the stains with running water and air dried, and then viewed cells under the brightfield microscope.
Flow cytometry analysis
We analyzed cell death with an annexin V – FITC kit (Beckman Coulter), following the manufacturer's protocol. We pelleted cells at 300 × g for 10 min and washed them with PBS before staining with FITC-labelled annexin V and propidium iodide (PI). We then analyzed the stained cells using 5-laser LSRFortessa X-20 (BD Biosciences).
We used Hoechst 33342 stain for cell cycle analysis. We incubated ∼ 200000 cells with 1 μg/ml of Hoechst 33342 at 37°C for 30 min, then spun cells at 150 × g for 7 min, washed once with PBS before resuspension in PBS for analysis with 5-laser LSRFortessa X-20 (BD Biosciences).
For the analysis of cell surface markers, we harvested HL-60 cells by centrifugation at 150 x g for 7 min and resuspended them in staining buffer (5% FBS in PBS). We stained the harvested cells on ice and in the dark with anti-CD11b-PE (ICRF44, Biolegend), anti-CD14-APC (TÜK4, Miltenyi Biotec) and anti-CD86-PE (BU63, Biolegend) for 30 min, followed by a single wash with the staining buffer and finally resuspension in PBS for analysis. To label dead cells, in some experiments, we added 7-aminoactinomycin D (7-AAD) (Biolegend) to the resuspended cells and incubated them on ice in the dark for 10 min before analysis. We used appropriate isotypes as controls. We analyzed cells using a 5-laser LSRFortessa X-20 (BD Biosciences) to detect the surface expression of these differentiation markers.
HL-60 differentiation
We treated wildtype or transduced HL-60 cells with differentiation agents to promote differentiation. We used 100 nM of VD3 (Abcam) to differentiate cells into monocyte-like cells after 4 days. We also differentiated HL-60 into neutrophil-like cells using 2 μM all-trans retinoic acid (Abcam) and 1.25% dimethyl sulfoxide (Sigma-Aldrich) (ATRA/DMSO) for 4 days. We refreshed all media with respective differentiation agent after 48 h.
Validation of splicing changes with RT-PCR
We extracted total RNA from harvested cells with PureLink® RNA Mini Kit (Ambion, Thermo Fisher), followed by DNase treatment with RQ1-RNase-free DNase I (Promega). We then purified RNA with 100% ethanol and 3 M sodium acetate before using it for reverse transcription. We synthesized 1 μg of complementary DNA (cDNA) in 20 μl reactions by reverse transcription (RT) at 42°C for 1 h, using Moloney Murine Leukemia Virus reverse transcriptase (New England Biolabs) and oligo-dT (Integrated DNA Technologies) as primer.
We carried out PCR to analyze individual splicing events from cDNA samples. We prepared the reactions with 2X DreamTaq Green PCR Master Mix (Thermo Scientific) using 5 ng of cDNA plus 0.2 μM of forward and reverse primer each (targets and primers in Supplementary Table S2). The PCR program included an initial denaturation of 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 60 s, and a final elongation at 72°C for 5 min. We calculated the band intensities with Image Lab (Bio-Rad).
RNA Sequencing
We extracted total RNA from HL-60 cells after doxycycline induction, treated the RNA with DNAse, and then used it for preparation of RNA-seq libraries. We performed library preparation and sequencing at the Singapore Centre for Environmental Life Sciences Engineering (SCELSE) or NovogeneAIT Genomics Singapore. We prepared the RNA libraries using the TruSeq stranded mRNA library prep kit (Illumina), which were sequenced on an Illumina HiSeq 2500 sequencer for the PRPF40A knockdown datasets (SCELSE) and an Illumina NovaSeq 6000 sequencer for the PRPF40B-overexpression datasets (NovogeneAIT). We aligned the RNA-seq reads to the hg38 human genome and GENCODE V30 transcriptome using STAR (38). We performed differential alternative splicing analysis using rMATS (39) and differential transcript usage analysis using satuRn (40), as well as differential gene expression analysis using RSEM (41) and Limma (42). We filtered the DASEs detected by rMATS using cut-offs of FDR ≤ 0.05, |ΔPSI| ≥0.1 and a mean junction count ≥ 10 for each group of datasets. Similarly, we filtered the DEGs using cut-offs of adjusted P-value ≤ 0.05, |log2FC| ≥1 and mean TPM ≥ 10 across all samples. We performed gene enrichment analysis using Enrichr (43) for DEGs and NEASE (44) for DASEs. We performed motif discovery analysis using STREME (45) on sequences from the introns that flank differentially spliced cassette exons. We also analyzed published RNA-seq datasets using rMATS with the same cut-offs.
Immunofluorescence
We fixed cells with 4% paraformaldehyde and permeabilized them with 0.25% triton X-100, before blocking with 1% bovine serum albumin. We then incubated the cells with primary antibodies for α-PRPF40A (PA5-58319, Invitrogen), α-PRPF40B (16929-1-AP, Proteintech), α-SC-35 (ab11826, Abcam), or appropriate isotype controls. Following this, we washed and then stained the cells with fluorophore conjugated secondary antibodies, along with DAPI. We then visualized the samples using a Nikon Eclipse Ti fluorescence microscope and the NIS-Elements AR software, with image deconvolution done using the in-built Clarify.ai method. Subsequently, we processed the deconvoluted images using ImageJ, with DAPI to demarcate the nuclear boundaries of each cell and the immunofluorescent signals stacked as required. We acquired all images using the same exposure time, with the brightness and contrast left unaltered during the ImageJ processing.
Genomic loci visualization with Casilio
We cloned guide RNAs (Supplementary Table S3) into the pCR8-sgRNA-25xPBS9R backbone (Addgene plasmid #183217), with the gRNAs targeting sequences near exons of interest. We transduced HEK-293T cells to stably express dCas9. We seeded these cells at a density of 25 000 cells/well in glass bottom plates, before we transfected cells with the pMax-mRuby2-PUF9R (Addgene plasmid #183210) and gRNA plasmids 24 h later using XtremeGENE™ 360 DNA Transfection Reagent (Roche). We then visualized the transfected cells using a Nikon Eclipse Ti fluorescence microscope. The plasmids were a gift from Albert Cheng.
Results
PRPF40A promotes survival and proliferation of HL-60 cells
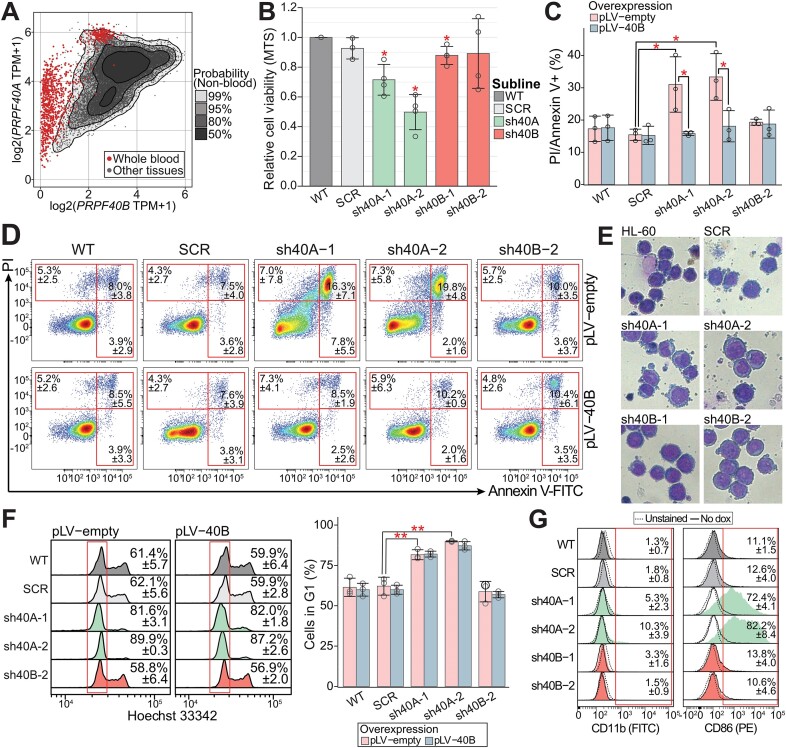
Exhaustive analysis of multiple databases converged that PRPF40A mRNA expression is high in blood and leukemia while PRPF40B expression is low (Figures 1A, S1A–C), as verified by our own expression profiling (Supplementary Figure S1D, E). To investigate the role of PRPF40A/B paralogs in leukemia or blood cells, we selected the myeloblastic HL-60 cell line because it is capable of chemical-induced terminal differentiation to diverse myeloid lineages (46–51). By lentiviral transduction, we generated HL-60 sublines with tetracycline/doxycycline inducible knockdown of either PRPF40A (PRPF40A-KD by sh40A-1/2) or PRPF40B (PRPF40B-KD by sh40B-1/2), plus a control (SCR). While PRPF40B shRNAs only showed a 50–80% reduction probably due to its low expression, we achieved >80% of PRPF40A depletion after 72 h (Supplementary Figure S2A). PRPF40B was also affected by the PRPF40A-KD, which was very unlikely caused by mispairing due to partial homology of the shRNA sequences (Supplementary Figure S2B). By overexpressing PRPF40B (PRPF40B-OE) in the knockdowns, we ensured that PRPF40B is not an off-target of PRPF40A shRNAs (Supplementary Figure S2C). Given the impossibility to overexpress PRPF40A in our system, we used PRPF40A-KD with or without PRPF40B overexpression for further phenotypic and splicing assays, to dissect the respective roles of these factors in myeloid cells.
Figure 1.
Phenotypic characterization of PRPF40A/B knockdown HL-60 cells. (A) PRPF40A and PRPF40B expression in blood cells and other tissues (the leukemia data in Supplementary Figure S1B). Data obtained from the GTEx database. (B) Cell viability MTS assay relative to wildtype HL-60 showed that PRPPF40A-KD HL-60 cells were less proliferative. The statistical significance was calculated against SCR using the Student's t-test and corrected with the Benjamini–Hochberg method. (C) Increased cell death in PRPF40A-KD HL-60 cells was also observed with AnnexinV/PI apoptosis assay. This was rescued with the overexpression of PRPF40B. The statistical significance was calculated between the knockdown samples (empty vector) against the scrambled control (empty vector) or the corresponding PRPF40B overexpression samples using the Welch's t-test and corrected with the Benjamini–Hochberg method. (D) Representative density plot of Annexin V (FITC) versus PI. (E) Giemsa stain of wildtype HL-60 and knockdown sublines after doxycycline induction. (F) Cell cycle analysis with Hoechst 33342 and flow cytometry likewise revealed an increased proportion of cells in the G1 phase upon PRPF40A-KD (highlighted in the red box), but PRPF40B overexpression did not significantly improve proliferation. The statistical significance was calculated between the knockdown samples (empty vector) against the scrambled control or the corresponding PRPF40B overexpression sample using the Welch's t-test and corrected with the Benjamini–Hochberg method. No statistical significance was found between corresponding empty vector controls and PRPF40B overexpression samples. (G) Flow cytometry analysis of myeloid cell markers showed increased CD86 expression in PRPF40A knockdown HL-60 cells. The statistical significance in all panels is presented as *P< 0.05, **P< 0.01.
Then we characterized the phenotypical changes of the sublines. MTS assay revealed a reduction in cell viability in PRPF40A-KD but not PRPF40B-KD cells after 72 h of induction (Figure 1B). An Annexin V and PI assay showed increased cell death upon PRPF40A-KD but not PRPF40B-KD (Figure 1C, D), and the fraction of dead cells upon PRPF40A-KD was reduced by half with PRPF40B-OE. Both HL-60 control and PRPF40B-KD cells showed a Giemsa stain pattern with large nuclei and small cytoplasm which is characteristic of a promyelocyte/myeloblast cell line (Figure 1E). In contrast, PRPF40A-KD cells exhibited signs of stress such as more blebbing at the cell membrane, consistent with increased cell death. Moreover, Hoechst 33342 staining revealed that cells with PRPF40A-KD were less proliferative with an increase in the G1 peak, and this phenotype was not rescued by PRPF40B-OE (Figure 1F). We conclude that PRPF40A is essential for the survival and proliferation of HL-60, and that only its protective role from apoptosis was rescued by PRPF40B-OE.
Next we measured few surface markers for signs of HL-60 differentiation, including CD11b, CD14 and CD86. PRPF40A-KD upregulated CD86 after 72 h of induction (Figure 1G), with the majority of cells with PRPF40A-KD as CD86 positive. Furthermore, PRPF40A-KD induced only a ∼10% increase in CD11b, which is upregulated early in myeloid-cell differentiation. This indicates that the CD86 upregulation in PRPF40A-KD HL-60 cells was not linked to the typical differentiation path of these cells. Overall, the surface markers suggest that PRPF40A-KD may sensitize these cells to differentiation agents, as characterized below.
PRPF40A depletion in HL-60 favors differentiation into monocyte-like cells
Analyses of a single-cell RNA-seq dataset with monocyte progenitors (52) (Supplementary Figure S2D), as well as neutrophil developmental stages from the BLUEPRINT database (53) (Supplementary Figure S2E) revealed that PRPF40A RNA levels are relatively stable during differentiation along these two lineages. However, PRPF40A protein levels decreased in HL-60 cells after treatment with all trans retinoic acid (ATRA) and DMSO (Supplementary Figure S2F), and a similar decrease was also observed in neutrophils based on mass spectrometry data from the ImmProt database (Supplementary Figure S2G). To further elucidate the PRPF40A function in myeloid differentiation, we used various differentiating agents to further elucidate the myeloid lineage that was induced upon PRPF40A-KD. We treated HL-60 cells with vitamin D3 (VD3) for 4 days to make monocyte-like cells (46), ATRA with DMSO for 4 days for neutrophil-like cells (48,54), phorbol 12-Myristate 13-acetate (PMA) for 3 days for macrophage-like cells (51), and calcium ionophore (CI) for 3 days for DC-like cells (50). The two last stimuli compromised cell viability which was aggravated by PRPF40A-KD, yielding too few viable cells for analysis. Therefore, the differentiation of PRPF40A-KD cells proceeded with only VD3 and ATRA/DMSO treatments, both confirmed by Giemsa (Supplementary Figure S2H).
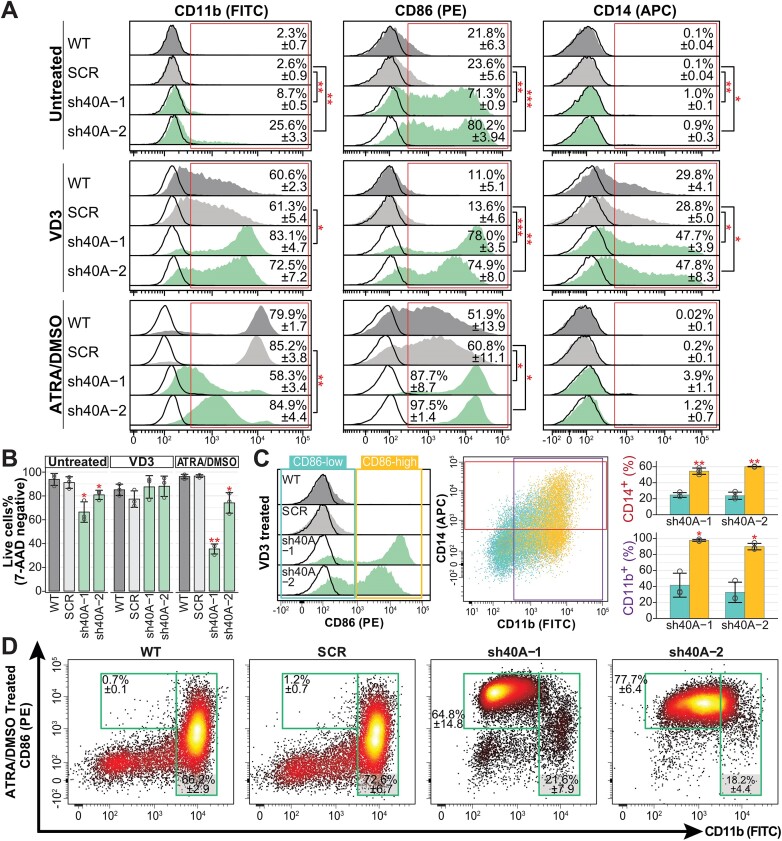
VD3 increased expression of CD11b and CD14 in HL-60, and these markers were further elevated in PRPF40A-KD cells (Figures 2A, S2I). Even though CD86 was not expressed in wildtype and SCR cells after VD3 treatment, its expression remained in PRPF40A-KD cells with VD3. In contrast to cells that were untreated or treated with ATRA/DMSO, VD3-treated cells did not exhibit an increased cell death phenotype upon PRPF40A depletion (Figure 2B).
Figure 2.
Phenotypic effects of the depletion of PRPF40A in HL-60 myeloid cells. (A) Flow cytometry data of surface markers for wildtype HL-60 and knockdown sublines upon doxycycline induction followed by differentiation with VD3 or ATRA/DMSO for 4 days. Representative histogram plots of CD11b (FITC), CD86 (PE) and CD14 (APC), with isotype controls represented by the black curves. The mean percentage of marker-positive cells is indicated in the histograms. (B) Cell viability obtained with 7-AAD staining. (C) CD11b and CD14 expression of CD86-low/high population after VD3 treatment. (D) Relative density plot of CD11b versus CD86 for ATRA/DMSO treated cells. The statistical significance depicted in all panels was calculated against the respective SCR controls using the Student's t-test and corrected with Benjamini–Hochberg method. The statistical significance in all panels is presented as *P< 0.05), **P< 0.01.
To understand whether expression of CD86 is associated with the upregulation of CD11b or CD14, we gated the VD3-treated cells into CD86-low and CD86-high (Figure 2C). Almost all CD86-high cells were CD11b positive while more than 50% were CD14 positive. CD86-high cells had significantly higher percentage of CD11b or CD14 positive cells compared to CD86-low cells, showing a strong correlation of CD11b and CD14 with CD86 expression. Last, most cells in all VD3-treated sublines showed indented nuclei as a sign of monocytic differentiation (Supplementary Figure S2H).
In turn, neutrophil-like ATRA/DMSO-treated HL-60 cells upregulated CD11b and CD86 but not CD14 (Figure 2A). ATRA/DMSO-treated PRPF40A-KD cells showed a further increase in CD86, but the CD86 positive population upon PRPF40A-KD expressed lower CD11b compared to controls (Figure 2D). In contrast to VD3, ATRA/DMSO-treated CD86 expression was not associated with CD11b, and ATRA/DMSO-treated PRPF40A-KD had increased cell death (Figure 2B).
All these data suggest that PRPF40A depletion promoted the differentiation into monocyte-like cells, consistent with the protein-expression changes during HL-60 and primary myeloid-cell differentiation. Next we performed transcriptomic analysis to elucidate the gene expression and splicing changes possibly accounting for these phenotypes.
PRPF40A acts as an activator of splicing with partially overlapping functions with PRPF40B
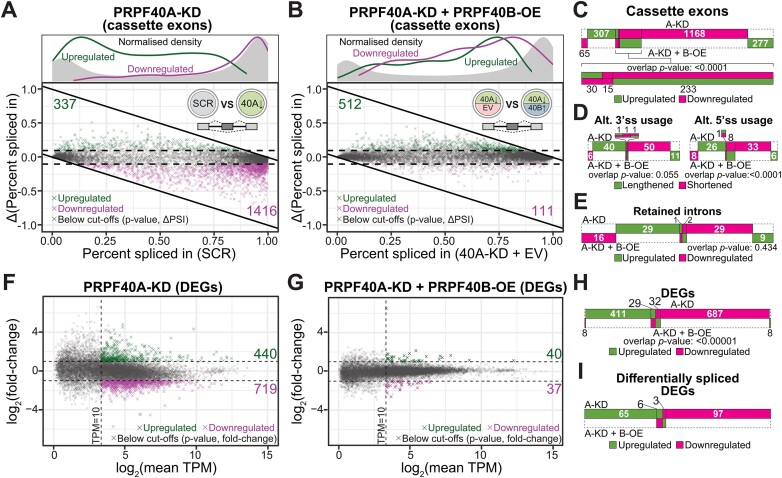
We performed RNA sequencing at 72 h post shRNA induction. We identified 1753 Differential Alternative Splicing Events (DASEs) upon PRPF40A-KD compared to SCR using rMATS (≥10% change in percent spliced in (ΔPSI), P ≤ 0.05 after false discovery rate (FDR), mean junction count ≥ 10) (Figure 3A, Supplementary Tables S4–S7). In turn, we found only 13 DASEs in the PRPF40B-KD cells (Supplementary Figure S3A) possibly because of the low knockdown rate, with seven DASEs in six genes shared with PRPF40A-KD, and six being regulated in the same direction. Cassette exons were predominantly more skipped by PRPF40A-KD, implying that PRPF40A mostly acts as a splicing activator (Figure 3A). We validated 21 out of 23 cassette exons using RT-PCR (Supplementary Figure S3B).
Figure 3.
Differential gene expression and splicing upon PRPF40A depletion without or with PRPF40B rescue. (A, B) Baseline percent spliced in (PSI) values of cassette exons plotted against the difference in PSI (ΔPSI) after PRPF40A-KD or rescue with PRPF40B-OE respectively. The density plots above indicate the distribution of points along the X-axis. (C–E) Distribution of differential cassette exons, alternate splice usage or intron retention between the two indicated datasets. The p-values for the overlap between the two datasets was calculated using the Fisher's exact test. (F, G) M–A plot (log-ratio versus mean expression) of differentially expressed genes for the indicated datasets. The dashed lines indicate thresholds. Only the statistically significant DEGs are highlighted in colour. (H) Distribution of DEGs between both datasets. The P-values for the overlap between the two datasets was calculated using the Fisher's exact test. (I) Distribution of DEGs which are also differentially spliced.
PRPF40B-OE in PRPF40A-KD cells predominantly led to cassette exon inclusion (Figures 3B, S3C, D). The PRPF40A DASEs corresponded to a majority of cassette exons followed by alternative 3′ss, alternative 5′ss and intron retention (Figure 3C–E). As many as 265 cassette exons altered upon PRPF40A-KD were rescued by PRPF40B-OE, which was a significant proportion (∼15%, P-value < 0.0001 by Fisher's exact test) (Figure 3C, Supplementary Table S4). Other PRPF40A-regulated alternative splicing events such as alternative 5′ss were also rescued by PRPF40B-OE, while no significant rescue was observed for retained introns and alternative 3′ss (Figure 3D, E, Supplementary Tables S5-S7). We also checked seven validated splicing events in PRPF40A-KD cells with PRPF40B-OE (Supplementary Figure S3E). Consistent with the RNA-seq data, PRPF40B-OE decreased the |ΔPSI| in PRPF40A-KD as compared to the empty plasmid counterpart in two of them (CHEK2-exon 10 and MYNN-exon 6).
Using RSEM (|log2FC| ≥ 1, P ≤ 0.05, mean TPM ≥ 10), we found 1159 Differentially Expressed Genes (DEGs) upon PRPF40A-KD, with 440 upregulated and 719 downregulated (Figure 3F, Supplementary Table S8). In contrast, PRPF40B-KD did not significantly alter the expression of any gene, including PRPF40B itself despite that qPCR and western blot showed > 50% knockdown. The second top upregulated gene in PRPF40A-KD was CD86, consistent with the observed increase in CD86 protein (Figure 2A). We tested 8 PRPF40A DEGs by qPCR and obtained a high overall correlation with the RNA-seq data (Supplementary Figure S3F, G). In contrast, PRPF40B-OE in PRPF40A-KD cells only affected 77 genes, among which 61 were rescued from the PRPF40A-KD (Figure 3G). Although this represented only 5.3% of the total PRPF40A-regulated DEGs, this overlap was highly significant (Figure 3H, Supplementary Table S8). As many as 171 of the DEGs were also differentially spliced (Figures 3I, S3H), indicating an enrichment of genes regulated by both expression and splicing. These DEGs/DASEs corresponded to an increase in non-coding transcript usage, especially for the downregulated DEGs (Supplementary Figure S3I, S3J). Nevertheless, the relatively small number of differentially spliced DEGs (despite the large number of DASEs) suggests that individual splicing perturbations are unlikely to directly drive the majority of gene-level expression changes.
The gene expression and splicing targets of PRPF40A are consistent with the HL-60 phenotypes
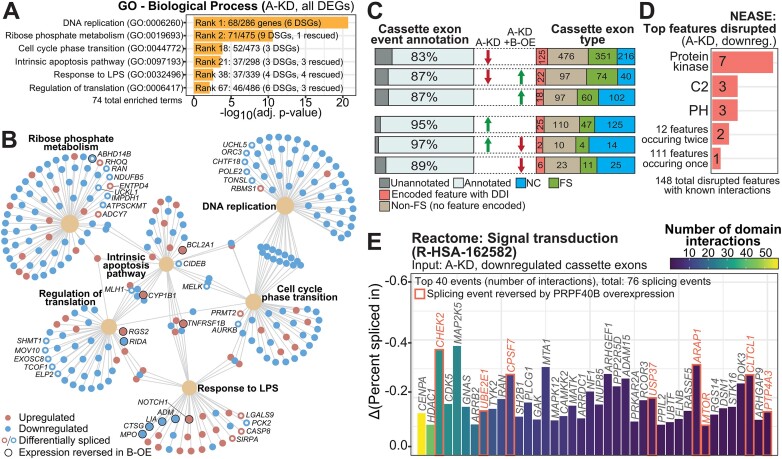
The top gene ontology category for the DEGs that are exclusively regulated by PRPF40A-KD corresponds to DNA replication (Figures 4A, B, S4A), which agrees with the phenotypic results above. Other enriched categories with little or no rescue by PRPF40B-OE are ribose phosphate metabolism, cell cycle and apoptosis. Importantly, these categories also include genes that undergo differential splicing upon PRPF40A-KD. Consistent with the differentiation phenotypes, the PRPF40A/B manipulations altered the expression of several monocytic markers obtained from the CellMarker 2.0 database (55) (Supplementary Figure S4B). Immune pathways like the response to lipopolysaccharide (LPS) were also an enriched DEG category.
Figure 4.
Enriched pathways and functions for the expression and splicing targets of PRPF40A. (A) Details of the Gene Ontology (GO) genesets selected for analysis of DEGs in the PRPF40A-KD (A-KD) dataset. DSGs: differentially spliced genes, DEGs that are also differentially spliced. The rescued genes refer to DEGs whose changes upon PRPF40A-KD were dampened by PRPF40B-OE. (B) Network plot of DEGs in the GO genesets indicated in Figure 4A. DEGs with concomitant differential splicing are indicated by hollow circles, while a black border indicates that the DEG was rescued by PRPF40B overexpression. (C) Distribution of cassette exon types by annotation for coding changes as in the key below, in addition to the percentage of differentially spliced cassette exons that are annotated as alternative exons in the GENCODE V43. (D) The three most numerous protein feature types suppressed by PRPF40A-KD, as reported by NEASE. PH: Pleckstrin homology domain. (E) The top 40 cassette exons with encoded protein features that are downregulated upon PRPF40A depletion, ranked by the number of known domain-domain interactions. Red borders indicate that the splicing event was rescuable by PRPF40B overexpression.
Based on GENCODE (release 43) isoform annotations, only 83% of exons downregulated upon PRPF40A-KD and not rescued by PRPF40B-OE were annotated cassette exons, in contrast to 87% and 95% for the rescued and upregulated events respectively (Figure 4C). PRPF40A-KD downregulated 147 frame-preserving exons (length multiple of 3) with encoded protein features suggestive of functional implications upon differential inclusion. In particular, NEASE analysis uncovered that protein kinase domains were the most frequently found protein features encoded in these exons (Figure 4D), which is not surprising because of the large number of human genes and exons that encode such domains (Supplementary Figure S4C). Among others, analysis of domain-domain interactions (DDI) for the PRPF40A-regulated cassette exons from the genes involved in signal transduction revealed CHEK2 exon 10 to have the third-most DDI annotations (Figure 4E). PRPF40A-KD increased exclusion of CHEK2 exon 10 whilst PRPF40B-OE increased its inclusion (Supplementary Figure S4D), as clear isoform switches without impact on overall abundance. In addition, IL17RA exon 11 (102 nt long) was more excluded upon PRPF40A-KD and rescued by PRPF40B-OE, as validated by RT-PCR (Supplementary Figure S3D, S4E). As exon 11 encodes the IL17RA transmembrane region, its skipping generates a soluble isoform (56) which is secreted into the extracellular space to bind and potentially inhibit its ligand IL-17A. Exon 2 of AURKB (aurora kinase B) is a putative first coding exon which encodes the KEN-box degron regulating AURKB protein turnover (57), so heightened exon 2 skipping upon PRPF40A-KD may result in the increase of isoforms that are resistant to degradation (Supplementary Figure S4F). This DASE is accompanied by an overall drop in AURKB mRNA levels, so it is regulated by both expression and splicing (Figure 3I). Overall, the aberrant splicing of these exons encoding domains may lead to altered protein function and influence downstream cellular events with many protein binding partners. Most likely the PRPF40A-induced phenotypes in these myeloid cells are specified by many DASEs with small effects rather than few with strong effects.
PRPF40A induces inclusion of cassette exons flanked by short GC-rich introns
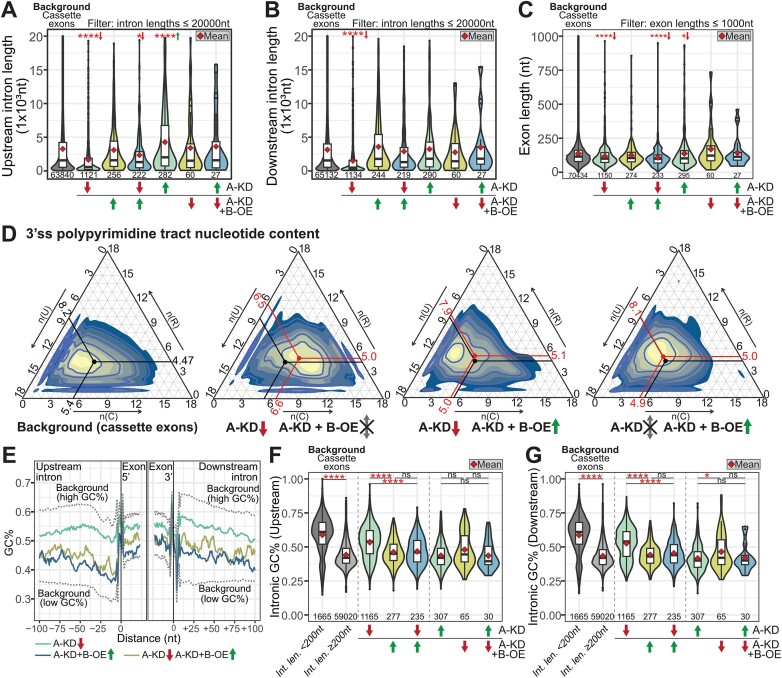
Next, we derived the features of the PRPF40A-regulated cassette exons. For the exons downregulated upon PRPF40A-KD and not rescued by PRPF40B-OE, both the upstream and downstream introns were significantly shorter than for the upregulated exons and all cassette exons (Figure 5A, B). Conversely, the upregulated exons in PRPF40A-KD had longer upstream introns compared to all cassette exons. The cassette exons downregulated upon PRPF40A-KD were also shorter than the controls (Figure 5C). We found that, in general, PRPF40A activates the inclusion of short cassette exons flanked by short introns, while it represses the inclusion of cassette exons preceded by long introns.
Figure 5.
Features of PRPF40A-regulated alternative splicing events in HL-60. (A–C) Distribution of intronic and exonic lengths of each differentially spliced cassette exon subset, with green and red arrows indicating up- and downregulation in the corresponding dataset. Mean values are indicated by the red diamond. The statistical significance between the background and each subset was calculated using the Wilcoxon's rank-sum test and corrected using the Benjamini–Hochberg method. The sample sizes are indicated below each violin plot. (D) Ternary plots of the nucleotide content of the 3′ss polypyrimidine tracts (18nt) for each indicated dataset. The black and red points indicate the mean values for the background and RNA-seq datasets, respectively. (E) Rolling window average (window of 5nt) plot of GC content for the indicated cassette exon subsets. The background values are indicated in dashed lines. (F, G) GC content of introns located up- and downstream of each cassette exon. The statistical significance within each group was calculated using the Kruskal–Wallis test within each subgroup (indicated by dashed lines) and corrected using the Benjamini–Hochberg method. The sample sizes are indicated below each violin plot. Int. len.: intronic length. The statistical significance in all panels is presented as *P< 0.05, ***P< 0.001, ****P< 0.0001. The arrows next to the asterisks indicate the direction of median change relative to background values.
Cassette exons regulated by PRPF40A-KD exhibited a slight decrease in 3′ss strength by the MaxEnt scores (Supplementary Figure S5A), while the 5′ss scores showed no difference (Supplementary Figure S5B). We observed a 3′ss signature with an enrichment of C and G instead of U in the polypyrimidine tract of exons that were downregulated by PRPF40A-KD and not rescued by PRPF40B-OE, while a different and weak trend was seen in upregulated and rescued exons (Figures 5D, S5C). Motif analysis showed that the 5′ss motifs for exons regulated by PRPF40A exhibit very small changes at some positions, with a slight enrichment in Gs at the intronic portion of the 5′ss of downregulated and not rescued exons (Supplementary Figure S5D). Although in PRPF40A-KD the presence of C or U respectively associates with either increased inclusion or exclusion of the cassette exons, the number of C or U does not correlate with the magnitude of splicing change or ΔPSI (not shown). Overall, the patterns of both splice-site motifs indicate that cassette exons with GC-rich splice sites are generally more excluded in PRPF40A-KD, while exons with AU-rich splice sites are more included.
By plotting the GC content of the upstream and downstream intron in a moving window of 5 nucleotides starting from either end of the cassette exon, we found that the high GC content for cassette exons downregulated upon PRPF40A-KD and not rescued by PRPF40B-OE extends into the introns (Figure 5E). Likewise, the total GC percentage in the introns flanking PRPF40A-KD downregulated and PRPF40B-OE-rescued exons was significantly higher as compared to the background Homo sapiens cassette exons (>50% versus 40–50% GC) (Figure 5F, G). Upregulated exons in PRPF40A-KD had an intronic GC content similar to that in the background exons.
Next we used STREME (45) to identify the motifs enriched in the cassette exons skipped upon PRPF40A-KD and not rescued by PRPF40B. Three of the top four motifs were respectively C-rich and a G-rich, as expected due to the high GC content of these introns (Supplementary Figure S5E). The other top motif resembled the binding site for SNRNP70 (Supplementary Figure S5F), with unknown significance.
In summary, most cassette exons downregulated by PRPF40A-KD are flanked by short introns with high GC content, and the upregulated exons had opposite features.
A common signature with short GC-rich introns regulated by a subset of splicing factors
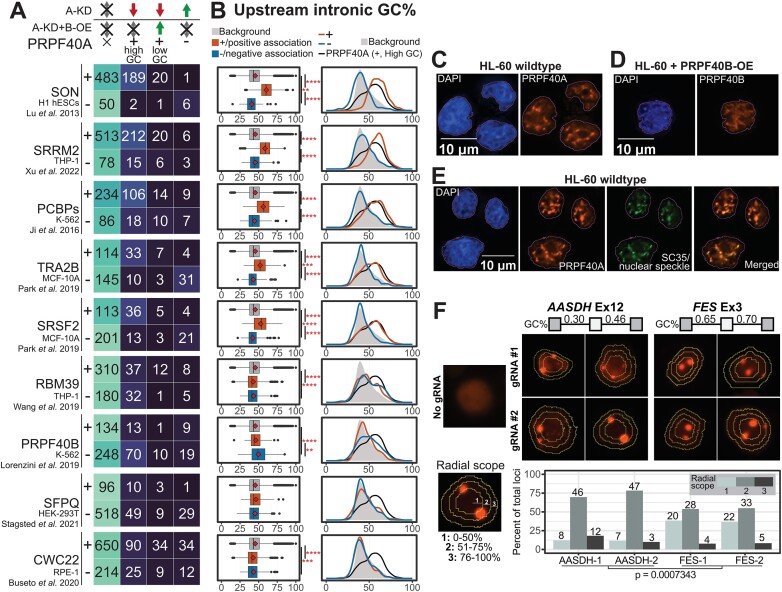
We compared the RNA-seq data of PRPF40A-regulated DASEs to the transcriptomes regulated by a wide variety of splicing factors with features that were shared or divergent with PRPF40A, with relation to homology, nuclear localization (nuclear speckles or radial localization), target composition (GC-content or others), or phenotype (AML) (details in Supplementary Figure S6 legend). The selected factors include PCBP1/2 knockdown in K562 (58), SRRM2 knockdown in the THP-1 AML cell line (13), SON knockdown in H1 human Embryonic Stem Cells (59), RBM39 knockout in THP-1 (60), PRPF40B knockout in K562 (16), SRSF2 P95H mutant in Human Erythroid Leukemia (HEL) cell line (61), SFPQ KD in HEK-293T (62), CWC22 knockdown in retinal cells (63), overexpression of SRSF2/4/6/9 and TRA2B in MCF10A breast cancer cells (64), and LUC7L knockdown in K562 (65). We classified the differential cassette exons for each dataset as being positively associated with RBP expression (with upregulated inclusion upon RBP overexpression or downregulated upon knockdown), or negatively associated with RBP expression (the reciprocal case).
Among these factors, the splicing targets of SON, SRRM2, PCBP1/2, TRA2B and SRSF2 tend to be positively associated with those of PRPF40A alone, while the overlap for the other splicing factors was much smaller (Figures 6A, S6A). However, the splicing events from the SON, SRRM2, PCBP1/2, TRA2B and SRSF2 datasets that overlapped the PRPF40A dataset were mostly different for each splicing factor (Supplementary Figure S6B). The splicing events regulated by these six splicing factors also had short introns (not shown) with high GC content, and while this trend was somehow weaker for TRA2B and SRSF2, it was completely erased in the other splicing factors (Figures 6B and S6C for upstream introns, S6D for downstream introns). Splice-site motif analysis revealed that the upregulated exons by SON, SRRM2 and PCBP1/2 had an enrichment of C in the 3′ss while 5′ss had an enrichment of G, with a similar yet stronger pattern than in PRPF40A (not shown). While TRA2B and SRSF2 showed a much weaker enrichment, the other splicing factors did not show any.
Figure 6.
Comparative meta-analysis reveals a subset of splicing factors with a common pattern of GC-rich splicing targets. (A) Overlap of differential cassette exons between each dataset and the PRPF40A/B target subsets, including exons downregulated by PRPF40A-KD and not rescued, downregulated exons rescued by PRPF40B-OE, and exons upregulated by PRPF40A-KD. The ‘+’ and ‘–’ indicate the exons that are positively and negatively associated with the expression of each splicing factor. The positively associated events are either upregulated in RBP overexpression datasets or downregulated in RBP knockdown datasets, while negatively associated events show the opposite trend. (B) Distribution of upstream intronic GC content for all human cassette exons (grey), positively correlated exons (+, orange) and negatively correlated exons (-, blue). The statistical significance within each group was calculated using the Wilcoxon's rank-sum test. **P< 0.01, ***P< 0.001, ****P< 0.0001. (C–E) Immunofluorescence assays for the localisation of the indicated splicing factors. The outlines indicate the DAPI-stained nuclear area. (F) Casilio-based tagging of genomic loci corresponding to exons flanked by lower GC content (AASDH Exon 12) and higher GC content (FES Exon 3). The concentric rings indicate the boundary of each radial scope, with the distribution of points across different cells indicated in the bar plot. For each gene, we summed up the loci counts across both gRNAs to get overall loci distributions, and applied the Kolmogorov–Smirnov test for the statistical differences between the AASDH and FES distributions (P-value as indicated).
Importantly, PRPF40B KO in K562 showed few overlapping cassette exons which tended to change in the opposite direction than upon PRPF40A-KD in HL-60 (Figure 6A) and devoid of a GC-rich signature (16), consistent with these paralogs regulating distinct sets of splicing events with distinct features. Our immunofluorescence labelling of PRPF40A and PRPF40B in HL-60 showed that PRPF40A formed clear puncta consistent with nuclear speckles by co-localization with SRSF2, while overexpressed PRPF40B had a more diffuse pattern (Figures 6C–E, S6E). As expected for a client protein, the depletion of PRPF40A does not perturb the integrity of the nuclear speckles (Supplementary Figure S6F).
Last, we used the Casilio genomic tagging system (66) to tag the exon loci in living cells. This system employs dCas9, mRuby2 fluorescent protein fused to a Pumilio/FBF (PUF) RBD, as well as a gRNA which targets a non-repetitive gene sequence and is appended with PUF-binding sites (PBS). The gRNAs used in this study were designed to label two different exon targets—FES and AASDH, which were downregulated or upregulated by PRPF40A-KD as well as of high or low GC content, respectively. We subsequently categorised the puncta based on their relative distance from the nuclear core to the periphery (Figure 6F). Previous studies reported that exons in high GC DNA regions tend to localise towards the center of the nucleus and share the same splicing regulation pattern (67,68). As expected, we observed that the FES target exon tended to be located closer to the nuclear center, while that of AASDH tended to be less central. Last, we examined the genomic loci radial scope data obtained in K562 myeloid cells (68) for the splicing events under regulation of each splicing factor, from 1 as the most central to 5 as the most peripheral score. As expected, the splicing events (full target genes and regulated exons only) upregulated by PRPF40A (and not rescued by PRPF40B-OE), SRRM2, SON and to a lesser extent by PCBP1/2, TRA2B and SRSF2, tended to be located closer to the nuclear core compared to the corresponding downregulated exons, while the other factors had low biases or regulated exons in more peripheral regions (Supplementary Figure S6G). For the nuclear subcompartment classification where A1 is more central while A2 is more peripheral, we see a clear enrichment for A1 in the upregulated genes and exons by SRRM2, SON, PRPF40A, PCBPs, TRA2B and SRSF2 compared to the downregulated exons for each factor, while the pattern is weaker or even reversed for the other factors (Supplementary Figure S6H).
These analyses indicate a functional connection between the splicing targets of PRPF40A and those of nuclear speckle scaffolds SRRM2 and SON (69) as well as PCBP1/2, TRA2B and SRSF2, in which their spatial localization within the nucleus likely plays an important role.
Discussion
Here, we characterized the role of PRPF40A in human myeloid cell differentiation via regulation of alternative splicing, and unveiled a common pattern of splicing regulation shared with other factors. We experimentally observed three main phenotypes upon PRPF40A-KD in HL-60, including increased cell death, loss of cell proliferation and myeloid cell differentiation, with the transcriptomic analysis correlating with these phenotypes. After many attempts, we could not achieve any PRPF40A overexpression either in wild-type or PRPF40A-KD HL-60 cells (not shown), likely because these myeloid cells tightly control its expression considering its effects in differentiation. Artificial PRPF40A downregulation in HL-60 also reduced PRPF40B levels unlikely via shRNA mispairing, which further highlights that the expression of these two factors is tightly regulated perhaps via indirect transcriptional control. As PRPF40A is primarily a splicing regulator, the effects of its manipulation on gene expression are likely indirect via altered splicing events that affect RNA levels. In any case, we cannot exclude that some of these phenotypes could be influenced by the PRPF40A role in actin polymerization and cell migration through binding formin (70) and N-WASP (27). PRPF40B-OE increased its levels beyond the normal expression in HL-60 which was very low, and importantly it rescued the cell viability but not proliferation (Figure 1D,F), indicating that cells might require one of the two paralogs to survive because of their fundamental role in splicing, while myeloid-cell division relies on PRPF40A because of its specific splicing regulation. The loss of proliferation upon PRPF40A-KD that was not rescued by PRPF40B-OE is consistent with the enriched DEG categories (Figure 4A, B), hence regulated at the RNA level.
The HL-60 acute myeloblastic leukemia cell line with maturation (FAB class M2) (71) is commonly used to study myeloid differentiation. The HL-60 cells possess differentiation blocks that can be alleviated by various chemical treatments, resulting in the generation of cells resembling mature myeloid cells (72,73). The co-treatment of PRPF40A shRNAs with VD3 or ATRA/DMSO in HL-60 showed that monocytic differentiation was favored upon PRPF40A depletion (Figure 2), hence supporting a role for PRPF40A in neutrophils or granulocytes, which in turn exhibit the lowest PRPF40B expression in all myeloid cells (Supplementary Figure S1B). PRPF40A-KD upregulates the mRNA and protein levels of the T-cell costimulatory molecule CD86 which is usually found on Antigen Presenting Cells (74). Other immune-relevant DEGs upon PRPF40A-KD include the top upregulated ADORA3 as a regulator of chemotaxis, phagocytosis and granule release in neutrophils (75). Among the downregulated genes upon PRPF40A-KD are four defensins (DEFA1, DEFA1B, DEFA3and DEFA4) which encode anti-microbial peptides in neutrophil granules (76). Together with the gene ontology analyses, these DEGs indicate that PRPF40A is important for the regulation of immune function in the HL-60 myeloid cells. Last, PRPF40A regulates several splicing events affecting certain protein regions important for kinases as well as PPIs (Figures 4D, E and S4C–E). Even if we expect that each PRPF40A-regulated DEG and/or DASE has a small contribution to these myeloid phenotypes, the functional dissection of these isoforms should be an important follow-up of this work.
Since both the PRPF40A/B paralogs lack a canonical RBD, their regulation on specific splicing targets might not rely on the direct binding to specific RNA motifs. The subset of splicing events altered upon PRPF40A-KD and rescued by PRPF40B-OE likely just need a Prp40-like protein irrespective of the paralog, to restore the cell viability. Importantly, most downregulated exons upon PRPF40A-KD were not rescued by PRPF40B-OE so they correspond to the PRPF40A specific targets, which exhibit the specific features of short exons and introns with a high GC content. This signature confers the selectivity for PRPF40A regulation because it disappears in the PRPF40B-OE rescued exons. As opposed to what was found in worms and mouse neuronal cells (30), we did not detect any changes in microexons likely because of the difference in cell types.
Since the motif analyses mostly reflected the GC-richness of the PRPF40A exclusive targets, we propose that the exon/intron architecture and GC content are the main determinants for the PRPF40A regulation, rather than directly or indirectly recognizing specific RNA sequences in their targets. Our comparative analysis showed a large overlap in the targets and features between PRPF40A and the nuclear speckle organizers SRRM2 and SON, as well as smaller overlaps with the splicing factors PCBP1/2, TRA2B and SRSF2. In fact, SRRM2 shares several features with PRPF40A: SRRM2 also lacks a canonical RBD, it is overexpressed in AML, it induces inclusion of exons flanked by short GC-rich introns, and its downregulation also compromises the viability and increases differentiation in myeloid cells, in this case THP-1 (13). SRRM2 is a predicted binder of PRPF40A based on a PPI network (33), and although it also binds CWC22 in the spliceosome (77), the target overlap between SRRM2 and CWC22 is small and devoid of the GC-rich pattern. In addition to SRRM2, SON is the other scaffold of nuclear speckles (69), hence its functional association with PRPF40A. For selected PRPF40A targets, we confirmed that their GC content and regulation were associated with the spatial localization of the DNA sequences, consistent with the recent evidence connecting GC content, nuclear spatial localization and splicing patterns (67,68). Our data strongly suggest that PRPF40A (and not its paralog PRPF40B) regulates its targets via increased localization to nuclear speckles in the nucleus center, as established by sequence composition (GC content) rather than by (in)direct target sequence recognition of specific RNA motifs. While this is the first mechanistic study to dissect the roles of the PRPF40A/B paralogs, their future mechanistic and biophysical characterization should shed light on to their disease roles, such as in neurodegenerative syndromes and potentially others affecting myeloid cell differentiation or function.
Supplementary Material
Acknowledgements
We thank all members of the Roca lab for fruitful discussions. We thank Cara Wee Ying Wong and Yi Lin Chua for help with the RNA-seq validations by RT-PCR, as well as Ruoqi Zhang, Chensi Lin and Zhaohan Xu for help in some computational analysis.
Notes
Present address: Xavier Roca, School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551 Singapore.
Contributor Information
Cheryl Weiqi Tan, School of Biological Sciences, Nanyang Technological University, 637551 Singapore.
Donald Yuhui Sim, School of Biological Sciences, Nanyang Technological University, 637551 Singapore.
Yashu Zhen, School of Biological Sciences, Nanyang Technological University, 637551 Singapore.
Haobo Tian, School of Biological Sciences, Nanyang Technological University, 637551 Singapore.
Jace Koh, School of Biological Sciences, Nanyang Technological University, 637551 Singapore.
Xavier Roca, School of Biological Sciences, Nanyang Technological University, 637551 Singapore.
Data availability
The RNA-seq data has been deposited in BioProject under accession number PRJNA1028250 (https://www.ncbi.nlm.nih.gov/bioproject).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
X.R. acknowledges funding from Singapore's National Research Foundation (NRF) [NRF2019-NRF-ISF003-3104]; Academic Research Fund Tier 1 RG24/22 from Singapore's Ministry of Education. The funders played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding for open access charge: Singapore’s Ministry of Education [RG24/22]; Singapore’s National Research Foundation (NRF) [NRF2019-NRF-ISF003-3104].
Conflict of interest statement. None declared.
References
- 1. Ng L.G., Liu Z., Kwok I., Ginhoux F.. Origin and heterogeneity of tissue myeloid cells: a focus on GMP-derived monocytes and neutrophils. Annu. Rev. Immunol. 2023; 41:375–404. [DOI] [PubMed] [Google Scholar]
- 2. Imperato M.R., Cauchy P., Obier N., Bonifer C.. The RUNX1-PU.1 axis in the control of hematopoiesis. Int. J. Hematol. 2015; 101:319–329. [DOI] [PubMed] [Google Scholar]
- 3. Green I.D., Pinello N., Song R., Lee Q., Halstead J.M., Kwok C.T., Wong A.C.H., Nair S.S., Clark S.J., Roediger B.et al.. Macrophage development and activation involve coordinated intron retention in key inflammatory regulators. Nucleic Acids Res. 2020; 48:6513–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song R., Tikoo S., Jain R., Pinello N., Au A.Y.M., Nagarajah R., Porse B., Rasko J.E.J., J J.L.W. Dynamic intron retention modulates gene expression in the monocytic differentiation pathway. Immunology. 2022; 165:274–286. [DOI] [PubMed] [Google Scholar]
- 5. Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K.et al.. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013; 154:583–595. [DOI] [PubMed] [Google Scholar]
- 6. Wong J.J., Schmitz U.. Intron retention: importance, challenges, and opportunities. Trends Genet. 2022; 38:789–792. [DOI] [PubMed] [Google Scholar]
- 7. Lee F.F.Y., Alper S.. Alternative pre-mRNA splicing as a mechanism for terminating Toll-like Receptor signaling. Front. Immunol. 2022; 13:1023567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Connor B.P., Danhorn T., De Arras L., Flatley B.R., Marcus R.A., Farias-Hesson E., Leach S.M., Alper S.. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 2015; 11:e1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pandya-Jones A., Bhatt D.M., Lin C.H., Tong A.J., Smale S.T., Black D.L.. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA. 2013; 19:811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu H., Lorenzini P.A., Zhang F., Xu S., Wong M.S.M., Zheng J., Roca X.. Alternative splicing analysis in human monocytes and macrophages reveals MBNL1 as major regulator. Nucleic Acids Res. 2018; 46:6069–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wagner A.R., Scott H.M., West K.O., Vail K.J., Fitzsimons T.C., Coleman A.K., Carter K.E., Watson R.O., Patrick K.L.. Global transcriptomics uncovers distinct contributions from splicing regulatory proteins to the macrophage innate immune response. Front. Immunol. 2021; 12:656885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. West K.O., Scott H.M., Torres-Odio S., West A.P., Patrick K.L., Watson R.O.. The splicing factor hnRNP M is a critical regulator of innate immune gene expression in macrophages. Cell Rep. 2019; 29:1594–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu S., Lai S.K., Sim D.Y., Ang W.S.L., Li H.Y., Roca X.. SRRM2 organizes splicing condensates to regulate alternative splicing. Nucleic Acids Res. 2022; 50:8599–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saez B., Walter M.J., Graubert T.A.. Splicing factor gene mutations in hematologic malignancies. Blood. 2017; 129:1260–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M.et al.. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011; 478:64–69. [DOI] [PubMed] [Google Scholar]
- 16. Lorenzini P.A., Chew R.S.E., Tan C.W., Yong J.Y., Zhang F., Zheng J., Roca X.. Human PRPF40B regulates hundreds of alternative splicing targets and represses a hypoxia expression signature. RNA. 2019; 25:905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dvinge H., Bradley R.K.. Widespread intron retention diversifies most cancer transcriptomes. Genome Medicine. 2015; 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou J., Chng W.J.. Aberrant RNA splicing and mutations in spliceosome complex in acute myeloid leukemia. Stem Cell Investigation. 2017; 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sinnakannu J.R., Lee K.L., Cheng S., Li J., Yu M., Tan S.P., Ong C.C.H., Li H., Than H., Anczukow-Camarda O.et al.. SRSF1 mediates cytokine-induced impaired imatinib sensitivity in chronic myeloid leukemia. Leukemia. 2020; 34:1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z., Burge C.B.. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008; 14:802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.M., Foehr S., Curk T., Krijgsveld J., Hentze M.W.. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016; 63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 23. Becerra S., Andres-Leon E., Prieto-Sanchez S., Hernandez-Munain C., Sune C.. Prp40 and early events in splice site definition. Wiley Interdiscip. Rev. RNA. 2016; 7:17–32. [DOI] [PubMed] [Google Scholar]
- 24. Abovich N., Rosbash M.. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997; 89:403–412. [DOI] [PubMed] [Google Scholar]
- 25. Katoh M. Identification and characterization of human FNBP3 gene in silico. Int. J. Mol. Med. 2003; 12:651–656. [PubMed] [Google Scholar]
- 26. Becerra S., Montes M., Hernandez-Munain C., Sune C.. Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo. RNA. 2015; 21:438–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizutani K., Suetsugu S., Takenawa T.. FBP11 regulates nuclear localization of N-WASP and inhibits N-WASP-dependent microspike formation. Biochem. Biophys. Res. Commun. 2004; 313:468–474. [DOI] [PubMed] [Google Scholar]
- 28. Makarov E.M., Owen N., Bottrill A., Makarova O.V.. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res. 2012; 40:2639–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhn A.N., van Santen M.A., Schwienhorst A., Urlaub H., Luhrmann R.. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA. 2009; 15:153–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choudhary B., Marx O., Norris A.D.. Spliceosomal component PRP-40 is a central regulator of microexon splicing. Cell Rep. 2021; 36:109464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buschdorf J.P., Stratling W.H.. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J Mol. Med. (Berl). 2004; 82:135–143. [DOI] [PubMed] [Google Scholar]
- 32. Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E.. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998; 7:1463–1474. [DOI] [PubMed] [Google Scholar]
- 33. Huo Z., Zhai S., Weng Y., Qian H., Tang X., Shi Y., Deng X., Wang Y., Shen B.. PRPF40A as a potential diagnostic and prognostic marker is upregulated in pancreatic cancer tissues and cell lines: an integrated bioinformatics data analysis. OncoTargets Ther. 2019; 12:5037–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oleksiewicz U., Liloglou T., Tasopoulou K.M., Daskoulidou N., Gosney J.R., Field J.K., Xinarianos G.. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2017; 143:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ge Y., Schuster M.B., Pundhir S., Rapin N., Bagger F.O., Sidiropoulos N., Hashem N., Porse B.T.. The splicing factor RBM25 controls MYC activity in acute myeloid leukemia. Nat. Commun. 2019; 10:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frank S.B., Schulz V.V., Miranti C.K.. A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector. BMC Biotechnol. 2017; 17:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hayer A., Shao L., Chung M., Joubert L.M., Yang H.W., Tsai F.C., Bisaria A., Betzig E., Meyer T.. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol. 2016; 18:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gilis J., Vitting-Seerup K., Van den Berge K., Clement L.. satuRn: Scalable analysis of differential transcript usage for bulk and single-cell RNA-sequencing applications. F1000Res. 2021; 10:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A.et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Louadi Z., Elkjaer M.L., Klug M., Lio C.T., Fenn A., Illes Z., Bongiovanni D., Baumbach J., Kacprowski T., List M.et al.. Functional enrichment of alternative splicing events with NEASE reveals insights into tissue identity and diseases. Genome Biol. 2021; 22:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bailey T.L. STREME: accurate and versatile sequence motif discovery. Bioinformatics. 2021; 37:2834–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bar-Shavit Z., Teitelbaum S.L., Reitsma P., Hall A., Pegg L.E., Trial J., Kahn A.J.. Induction of monocytic differentiation and bone resorption by 1,25-dihydroxyvitamin D3. Proc. Nat. Acad. Sci. U.S.A. 1983; 80:5907–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barber N., Belov L., Christopherson R.I.. All-trans retinoic acid induces different immunophenotypic changes on human HL60 and NB4 myeloid leukaemias. Leuk. Res. 2008; 32:315–322. [DOI] [PubMed] [Google Scholar]
- 48. Collins S.J., Ruscetti F.W., Gallagher R.E., Gallo R.C.. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc. Nat. Acad. Sci. U.S.A. 1978; 75:2458–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fischkoff S.A., Pollak A., Gleich G.J., Testa J.R., Misawa S., Reber T.J.. Eosinophilic differentiation of the human promyelocytic leukemia cell line, HL-60. J. Exp. Med. 1984; 160:179–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koski G.K., Schwartz G.N., Weng D.E., Gress R.E., Engels F.H., Tsokos M., Czerniecki B.J., Cohen P.A.. Calcium ionophore-treated myeloid cells acquire many dendritic cell characteristics independent of prior differentiation state, transformation status, or sensitivity to biologic agents. Blood. 1999; 94:1359–1371. [PubMed] [Google Scholar]
- 51. Rovera G., Santoli D., Damsky C.. Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc. Nat. Acad. Sci. U.S.A. 1979; 76:2779–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie X., Liu M., Zhang Y., Wang B., Zhu C., Wang C., Li Q., Huo Y., Guo J., Xu C.et al.. Single-cell transcriptomic landscape of human blood cells. Natl. Sci. Rev. 2021; 8:nwaa180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grassi L., Pourfarzad F., Ullrich S., Merkel A., Were F., Carrillo-de-Santa-Pau E., Yi G., Hiemstra I.H., Tool A.T.J., Mul E.et al.. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 2018; 24:2784–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Breitman T.R., Selonick S.E., Collins S.J.. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc. Nat. Acad. Sci. U.S.A. 1980; 77:2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hu C., Li T., Xu Y., Zhang X., Li F., Bai J., Chen J., Jiang W., Yang K., Ou Q.et al.. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023; 51:D870–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sohda M., Misumi Y., Tashiro K., Yamazaki M., Saku T., Oda K.. Identification of a soluble isoform of human IL-17RA generated by alternative splicing. Cytokine. 2013; 64:642–645. [DOI] [PubMed] [Google Scholar]
- 57. Nguyen H.G., Chinnappan D., Urano T., Ravid K.. Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property. Mol. Cell. Biol. 2005; 25:4977–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ji X., Park J.W., Bahrami-Samani E., Lin L., Duncan-Lewis C., Pherribo G., Xing Y., Liebhaber S.A.. αCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res. 2016; 44:2283–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu X., Goke J., Sachs F., Jacques P.E., Liang H., Feng B., Bourque G., Bubulya P.A., Ng H.H.. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 2013; 15:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang E., Lu S.X., Pastore A., Chen X., Imig J., Chun-Wei Lee S., Hockemeyer K., Ghebrechristos Y.E., Yoshimi A., Inoue D.et al.. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell. 2019; 35:369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liang Y., Tebaldi T., Rejeski K., Joshi P., Stefani G., Taylor A., Song Y., Vasic R., Maziarz J., Balasubramanian K.et al.. SRSF2 mutations drive oncogenesis by activating a global program of aberrant alternative splicing in hematopoietic cells. Leukemia. 2018; 32:2659–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stagsted L.V.W., O’Leary E.T., Ebbesen K.K., Hansen T.B.. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. eLife. 2021; 10:e63088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Busetto V., Barbosa I., Basquin J., Marquenet E., Hocq R., Hennion M., Paternina J.A., Namane A., Conti E., Bensaude O.et al.. Structural and functional insights into CWC27/CWC22 heterodimer linking the exon junction complex to spliceosomes. Nucleic Acids Res. 2020; 48:5670–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Park S., Brugiolo M., Akerman M., Das S., Urbanski L., Geier A., Kesarwani A.K., Fan M., Leclair N., Lin K.T.et al.. Differential functions of splicing factors in mammary transformation and breast cancer metastasis. Cell Rep. 2019; 29:2672–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Daniels N.J., Hershberger C.E., Gu X., Schueger C., DiPasquale W.M., Brick J., Saunthararajah Y., Maciejewski J.P., Padgett R.A.. Functional analyses of human LUC7-like proteins involved in splicing regulation and myeloid neoplasms. Cell Rep. 2021; 35:108989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Clow P.A., Du M., Jillette N., Taghbalout A., Zhu J.J., Cheng A.W.. CRISPR-mediated multiplexed live cell imaging of nonrepetitive genomic loci with one guide RNA per locus. Nat. Commun. 2022; 13:1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barutcu A.R., Wu M., Braunschweig U., Dyakov B.J.A., Luo Z., Turner K.M., Durbic T., Lin Z.Y., Weatheritt R.J., Maass P.G.et al.. Systematic mapping of nuclear domain-associated transcripts reveals speckles and lamina as hubs of functionally distinct retained introns. Mol. Cell. 2022; 82:1035–1052. [DOI] [PubMed] [Google Scholar]
- 68. Tammer L., Hameiri O., Keydar I., Roy V.R., Ashkenazy-Titelman A., Custodio N., Sason I., Shayevitch R., Rodriguez-Vaello V., Rino J.et al.. Gene architecture directs splicing outcome in separate nuclear spatial regions. Mol. Cell. 2022; 82:1021–1034. [DOI] [PubMed] [Google Scholar]
- 69. Ilik I.A., Malszycki M., Lubke A.K., Schade C., Meierhofer D., Aktas T.. SON and SRRM2 are essential for nuclear speckle formation. eLife. 2020; 9:e60579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chan D.C., Bedford M.T., Leder P.. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996; 15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 71. Dalton W.T. Jr, Ahearn M.J., McCredie K.B., Freireich E.J., Stass S.A., Trujillo J.M. HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood. 1988; 71:242–247. [PubMed] [Google Scholar]
- 72. Birnie G.D. The HL60 cell line: A model system for studying human myeloid cell differentiation. Br. J. Cancer Suppl. 1988; 9:41–45. [PMC free article] [PubMed] [Google Scholar]
- 73. Collins S.J. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood. 1987; 70:1233–1244. [PubMed] [Google Scholar]
- 74. Azuma M., Ito D., Yagita H., Okumura K., Phillips J.H., Lanier L.L., Somoza C.. B70 antigen is a second ligand for CTLA-4 and CD28. Nature. 1993; 366:76–79. [DOI] [PubMed] [Google Scholar]
- 75. Barletta K.E., Ley K., Mehrad B.. Regulation of neutrophil function by adenosine. Arterioscler. Thromb. Vasc. Biol. 2012; 32:856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bowdish D.M., Davidson D.J., Hancock R.E.. Immunomodulatory properties of defensins and cathelicidins. Curr. Top. Microbiol. Immunol. 2006; 306:27–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wan R., Bai R., Zhan X., Shi Y.. How is precursor messenger RNA spliced by the spliceosome?. Annu. Rev. Biochem. 2020; 89:333–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data has been deposited in BioProject under accession number PRJNA1028250 (https://www.ncbi.nlm.nih.gov/bioproject).