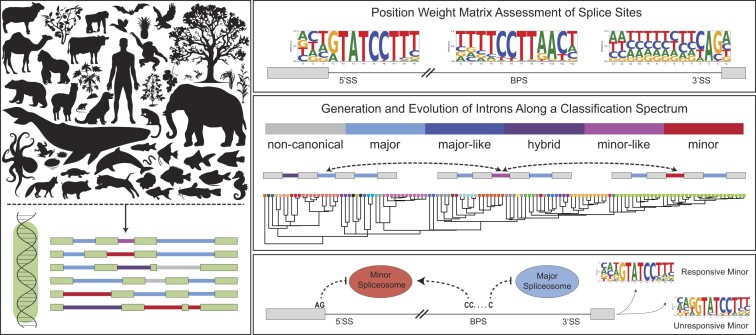
Abstract
Classification of introns, which is crucial to understanding their evolution and splicing, has historically been binary and has resulted in the naming of major and minor introns that are spliced by their namesake spliceosome. However, a broad range of intron consensus sequences exist, leading us to here reclassify introns as minor, minor-like, hybrid, major-like, major and non-canonical introns in 263 species across six eukaryotic supergroups. Through intron orthology analysis, we discovered that minor-like introns are a transitory node for intron conversion across evolution. Despite close resemblance of their consensus sequences to minor introns, these introns possess an AG dinucleotide at the –1 and –2 position of the 5′ splice site, a salient feature of major introns. Through combined analysis of CoLa-seq, CLIP-seq for major and minor spliceosome components, and RNAseq from samples in which the minor spliceosome is inhibited we found that minor-like introns are also an intermediate class from a splicing mechanism perspective. Importantly, this analysis has provided insight into the sequence elements that have evolved to make minor-like introns amenable to recognition by both minor and major spliceosome components. We hope that this revised intron classification provides a new framework to study intron evolution and splicing.
Graphical Abstract
Graphical Abstract.
Introduction
Introns are non-coding interruptions that fragment eukaryotic coding sequences. They occupy significantly more genomic space than coding exons and are therefore thought to act as a sponge for random mutations that would otherwise be detrimental to the organism if they occurred in exons. The higher degree of genetic drift in introns aligns with their gain and/or loss across evolution, which is reflected in the unequal intron density observed between eukaryotic lineages (1). Evolution of most eukaryotic lineages is thought to have involved the substantial loss of introns, exemplified by the presence of very few introns in many unicellular organisms (2). Nevertheless, eukaryotes such as yeast have maintained a small number of introns, as their presence provides adaptive advantage (3,4). At the same time, intron gain is thought to have accompanied major eukaryotic transitions, as observed in the intron-rich genomes of metazoa (2,5). In fact, organismal complexity observed in these lineages is in part owed to alternative splicing, which enables the production of a diverse proteome from a limited number of genes. As such, removal of introns by the spliceosome has become an important regulatory node for gene expression.
The spliceosome, which consists of five small nuclear RNAs (snRNAs) and associated proteins, is thought to have co-evolved with spliceosomal introns. Specifically, spliceosomal snRNAs are thought to have originated from the catalytic fragments of group II introns and are highly conserved across eukaryotic lineages (6–8). These spliceosome components are collectively essential for identifying the exon/intron boundaries at the 5′ and 3′ ends of the intron, referred to as 5′ and 3′ splice sites, as well as sequences within the intron body, such as the branch point sequence and polypyrimidine tract. As intron identification relies on the base pairing of snRNAs with these intronic sequence elements, these sequence motifs, despite the genetic drift in introns, are conserved. Since more than 99% of human introns contain sequence elements that are thought to be recognized by the snRNAs of the abundant, major spliceosome (consisting of U1, U2, U4, U5, U6 snRNP), these introns are referred to as major introns. However, there exists a small subset of introns (∼0.25% in human genome), called minor introns, which possesses sufficiently divergent consensus sequences such that they require a parallel spliceosome, called the minor spliceosome (consisting of U11, U12, U4atac, U6atac and U5 snRNP) (9,10).
This binary classification schema of introns and the implied obligate relationship with their splicing machinery, i.e. major versus minor, has proven invaluable to the study of their role in regulating eukaryotic gene expression. Nevertheless, data accrued from advancements in genomic and transcriptomic sequencing has suggested that the consensus sequences utilized for this binary classification show a high degree of variance. While divergence from the canonical major intron consensus sequences is well-documented, the consensus sequences of minor introns are more highly conserved (11). However, the identification of non-canonical minor introns in the slime mold Physarum polycephalum indicates that minor intron sequence elements may vary more than previously thought (12). Moreover, it suggests that the minor spliceosome, like the major spliceosome, may show flexibility in its function. This further underscores the need to revisit and refine the classification of introns beyond minor versus major. Now, with increased availability of annotated genomes across diverse eukaryotic lineages, we can re-evaluate the nuances observed in consensus sequences of splice sites to classify introns.
A refined classification of introns across a diverse group of species may also shed light on intron evolution. Both minor introns and minor spliceosome components have been identified in many eukaryotic lineages, although they are reportedly absent in genera such as yeast, green algae, Dictyostelium and Caenorhabditis (8,13,14). Paradoxically, the high conservation of minor intron splicing across eukaryotic supergroups, alongside the complete loss of minor intron splicing in other lineages, has raised questions as to their origin and evolution. Minor introns, while mostly ancient and likely present in the last eukaryotic common ancestor, are thought to have emerged after major introns (14,15). Nevertheless, there have also been reports of younger minor introns that have evolved more recently, with the massive minor intron gain in P. polycephalum providing an extreme example (12,16). The evolution of minor introns is particularly interesting as their loss and gain does not necessarily have to be the consequence of deletion or creation of a new intron. Instead, minor intron gain and loss might also be achieved through conversion from and to major introns, respectively (15,17). This switch between major and minor-type consensus sequences could be achieved relatively easily, as only a few sequential point mutations would be required (18). If conversion of minor introns to major introns and vice versa does occur, one would expect to detect introns that are in flux. These introns would possess degenerating consensus sequences that might not be recognized by either the major, or the minor spliceosome. Alternatively, the degenerate splice site motifs might facilitate recognition by both spliceosomes. Additionally, conversion of minor to major introns might result in the presence of introns with a minor-type 5′ splice site and major-type 3′ splice site, or vice versa. In fact, a few of these so-called ‘hybrid’ introns have previously been identified using position-weight matrices (19).
Here, we have designed a bioinformatics pipeline that leverages position weight matrices of splice sites and branch point sequences to classify introns as major or minor, and those with weak scores as minor-like, hybrid, major-like, and non-canonical. Identification of these six intron classes was performed across 263 genomes, representing six eukaryotic supergroups, and is accessible at http://midb.pnb.uconn.edu. We detected minor introns and minor spliceosome snRNAs in the genomes of species from all eukaryotic supergroups, supporting their ancient origin. The curated species provide an evolutionary snapshot, such that analysis of orthologous intron clusters revealed that minor-like introns may not only represent an intermediate evolutionary state in the conversion of minor introns to major introns, but also vice versa. Importantly, while the consensus sequences of minor-like introns share significant sequence similarity with minor introns, they also contain salient features of major introns, such as an AG at the -1 and -2 of the 5′ splice site. This suggests that minor-like introns might not only be an evolutionary transitory node, but might also be an intermediate intron class from a mechanistic perspective. By combining CLIP-seq, CoLa-seq and RNAseq data, we show that minor-like introns spliced by the minor spliceosome lack the A-2 G-1 at the 5′ splice site and are bound at a higher probability by U2AF1 than minor introns. In contrast, minor-like introns that were unaffected by inhibition of the minor spliceosome enriched for an A-2G-1 at the 5′ splice site. In all, our analysis has provided insight into the sequence elements that have made minor-like introns amenable to recognition by both minor and major spliceosome components. As such, we hope our intron classification schema provides a probability matrix that will be leveraged to study intron evolution and splicing mechanism of these various intron types. In all, we propose to decouple intron identity from the spliceosome that splices it and favor a sequence-based classification, which is more amenable for evolutionary studies.
Materials and methods
Phylogeny
The taxonomy of the 263 species analyzed in this manuscript was obtained from the NCBI Taxonomy Browser, and we used the criteria described in (20) to make changes to the classification of certain protists. Specifically, these changes consider Haptista and Cryptista as definitive supergroups, while the supergroup Excavata contains three clades with mutual relationships to other clades that remain uncertain. The updated taxonomy used in this study can be found in Supplementary Table S1. A phylogenetic tree with branch lengths that reflect time was obtained from timetree.org.
Classification of introns
The classification of introns was done using position weight matrices (PWMs), with several changes to previously described methods (21). Genome and intron data for all 263 species was extracted from FASTA and GTF files obtained from Ensembl (Supplementary Table S2). Introns were initially binned as ‘putative major’ (GT-AG and GC-AG), ‘putative minor’ (AT-AC), or ‘other’ based on their terminal dinucleotide sequence (Supplementary Figure S1A). For putative major introns, an initial PWM for the 5′ splice site was generated from the –2 to +6 nucleotides, as these are known to be important for base pairing with U1 and U6 snRNA during splicing (22,23) (Supplementary Figure S1B, S1Ci). For the construction of an initial major branch point sequence PWM, we employed a sliding window from the –44 to –18 position of all putative major introns to extract all potential seven nucleotide sequences with an adenine at the +6 position (Supplementary Figure S1Cii). This initial PWM was then utilized to score all putative branch point sequences generated by the sliding window. The highest scoring branch point sequence from each putative major intron (with a positive cumulative log-odds score) was extracted to generate a final putative major branch point sequence PWM (Supplementary Figure S1Cii). The initial PWM for the major polypyrimidine tract was then constructed from the –13 to –1 nucleotides of putative major introns (Supplementary Figure S1Ciii).
Analogously, an initial 5′ splice site PWM was constructed for putative minor introns using the +4 to +9 nucleotides, as U11 and U6atac are known to base pair with these nucleotides for splicing (24,25) (Supplementary Figure S2A, Supplementary Figure S2Bi). For the construction of an initial minor branch point sequence PWM, a sliding window from the –40 to –1 position was applied to all putative minor introns to extract all potential twelve nucleotide sequences with an adenine at the +9 or +10 position (Supplementary Figure S2B). This initial PWM was then utilized to score all putative branch point sequences generated by the sliding window. The highest scoring branch point sequence from each putative minor intron (with a positive cumulative log-odds score) was extracted to generate two initial minor branch point sequence PWM: one with adenine at the +9 position, and one with adenine at the +10 position (Supplementary Figure S2Bii-iii).
After generation of the initial major and minor PWM sets (Supplementary Figures S1C, S2B), all introns were scored against these initial PWMs (Supplementary Figure S3Ai). Based on these scores, introns were re-binned as putative major and minor introns (Supplementary Figure S3Aii). Specifically, introns that met the following three criteria: (i) score above 50 against the initial major 5′ splice site PWM, (ii) score higher against the initial major 5′ splice site PWM than the initial minor 5′ splice site PWM and (iii) score above 50 against the major polypyrimidine tract PWM, were classified as putative major introns (Supplementary Figure S3A-iia). In contrast, introns that met the criteria of: (a) score above 50 against the initial minor 5′ splice site PWM, (b) score higher against the initial minor 5′ splice site PWM than the initial major 5′ splice site PWM plus 22.5 and (c) score above 75 against the minor branch point sequence PWM, were classified as putative minor introns (Supplementary Figure S3A-iib). These two classes were then used to generate refined PWMs for the major and minor 5′ splice site, minor branch point sequence and major polypyrimidine tract (Supplementary Figure S3B–E). After rescoring all introns against these refined PWMs, introns were binned using the criteria outlined in Supplementary Figure S4. Notably, the major-type branch point sequence is relatively degenerate and unlike the minor-type branch point sequence, does not have a well-defined distance constraint from the 3′ splice site. As such, we have used the major-type polypyrimidine tract for intron classification rather than the major-type branch point sequence. The final intron classification for all 263 species is available at the Minor Intron Database, which can be accessed at http://midb.pnb.uconn.edu (21).
Identification of minor spliceosome snRNAs
All available sequences for U11 (RF00548), U12 (RF00007), U4atac (RF00618) and U6atac (RF00619) were downloaded from the Rfam database. For species that did not have any minor spliceosome snRNA recorded in the Rfam database, a blastn database was built from the genome FASTA file. We then performed blastn queries (word size = 7) on the Rfam fasta file with annotated snRNA sequences against these databases, to identify putative snRNA homologs in other species. All coordinates of hits with E value <10 were extracted and the sequences + flanking 200 nucleotides were identified. These putative snRNA homolog sequences (solely based on sequence similarity) were further analyzed using the Infernal package (26). To this end, the covariance models for U11, U12, U4atac and U6atac were downloaded from the Rfam database. This model was then run against the putative snRNA sequence fasta files using cmsearch. To identify high-confidence snRNA homologs, we first established score thresholds by comparing the covariance model for each snRNA that was downloaded from the Rfam database to the annotated snRNA sequences also hosted by the Rfam database. The score corresponding to the lowest scoring hit within the inclusion threshold was considered the threshold for identifying new snRNAs. These were: 37.3 for U11, 53.0 for U12, 53.2 for U4atac, and 22.9 for U6atac. All putative snRNA homologs above these thresholds were therefore considered reliable hits. If multiple hits were observed above this threshold, we recognized the potential for these to be gene copies or pseudogenes; if no hits were above this threshold but there were hits above the inclusion threshold set by cmsearch, they were separately analyzed.
Intron orthology
To determine gene, transcript and intron orthology, the genomic coordinates of all coding sequences (CDS) were extracted from the GTF of each species. The corresponding nucleotide sequence of individual CDS segments was extracted from the genome FASTA using BEDtools, and segments were merged to generate a model of the contiguous, spliced mRNA transcript (Supplementary Figure S5A). EMBOSS transeq v.6.6.0.0 was then used to translate all annotated protein-coding transcripts in silico, which were used as input to generate a DIAMOND database (DIAMOND v.0.09.10.111) (Supplementary Figure S5B). The protein sequences derived from transcripts of interest (i.e. transcripts of genes containing minor, minor-like or hybrid introns) were then blasted against the DIAMOND database of all 263 species using blastp (options: -max-target-sequence 1–e 10–10 –more-sensitive). A reciprocal best hit approach was then employed to identify orthologous transcripts (Supplementary Figure S5C). For this, the highest scoring protein in the queried species was blasted against the human DIAMOND database using the same parameters as described above to ensure the transcripts of interest were orthologous.
To identify orthologous introns, introns in orthologous transcripts were compared in an all-to-all approach (Supplementary Figure S5D). First, the position of all introns in orthologous transcript pairs were marked using custom scripts and the ten amino acids flanking each exon-intron boundary were extracted. For each intron in these transcripts, global pairwise alignment of the flanking twenty residues was then performed against all introns in the orthologous transcripts using the Biopython package pairwise2. The highest scoring intron with a match score ≥40% was then extracted as a putative intron ortholog. In case of tied match scores, the intron with the highest BLOSUM-62 score was extracted. A reciprocal best hit approach was employed to confirm that introns were orthologous.
Inferring ancestral intron classes
The ancestral intron class at vertebrate origin was determined for all human introns found in genes containing a minor, minor-like or hybrid intron. The ancestral state of each human intron was determined through analysis of the orthologous introns in protostomes (Annelida, Arthropoda, Mollusca, Nematoda, Rotifera and Brachipoda) and non-bilaterian species (Cnidaria, Porifera, Ctenophora, Placozoa). Human introns without orthologous introns in any of these non-vertebrate species were removed from the analysis. For the remaining introns, the presence of an orthologous minor intron would lead to the classification of an ancestral minor intron at vertebrate origin. Finally, we inferred a major origin for human introns that did not have any orthologous minor introns in non-vertebrate organisms.
CLIP-seq analysis
CLIP-seq for U2AF1 and ZRSR2 was obtained from (27). BedGraph files were downloaded from GSE203531 and converted to BED file format. The BED files were then converted to hg38 coordinates using the UCSC LiftOver tool, and used to re-generate BedGraph files. Peak calling was done as described in (27). Specifically, a minimum of eight reads spanning more than nine nucleotides was defined as a peak for ZRSR2, whereas a minimum of twenty reads spanning more than nine nucleotides was defined as a peak for U2AF1. Using BEDTools intersect, introns with a peak within 10 nucleotides of the 3′ intron-exon boundary were extracted and defined as CLIP-positive introns.
Branch point analysis
Experimentally validated branch point coordinates were obtained from the CoLa-seq analysis in (28). These experimentally validated branch point locations were then compared to the branch point nucleotide locations predicted by position weight matrices using BEDTools. In case the branch point adenosine predicted by the major and minor-type branch point sequence PWM was the same, and the location corresponded with the experimentally validated branch point, introns were binned as using both major and minor branch point. Similarly, if introns had multiple experimentally validated branch points, which corresponded to both the predicted major and minor-type branch point, these would be classified as using both major and minor branch point. If none of the experimentally validated branch points corresponded with the highest-scoring predicted branch points by PWM, introns were classified as having used a suboptimal branch point.
RNA-seq datasets
The splicing of introns was interrogated in a range of RNAseq datasets wherein components of the minor spliceosome was inhibited. For human, these included antisense morpholino oligonucleotides against U12, U4atac and U6atac snRNA (29) (N= 3 each), an auxin-inducible degron for CENATAC (30) (N= 3 per time-point), and peripheral blood mononuclear cells from individuals with Roifman syndrome (31) (mutation in U4atac snRNA; N= 3), microcephalic osteodysplastic primordial dwarfism type I (32) (mutation in U4atac snRNA; N= 3 amniotic; N= 5 fibroblast), and myelodysplastic syndrome (33) (mutation in ZRSR2; N= 8). For mouse, these included Emx1-mediated recombination of U11 snRNA (34) (N= 5 at E12, N= 2 at E14), and Prrx1-mediated recombination of U11 snRNA (35) (N= 3 for each limb for each timepoint). The zebrafish data was generated in this study and is described in detail below. For Drosophila melanogaster, analyzed RNAseq datasets included knockout larvae for smn, U12 and U6atac snRNA (36) (N= 2 each). Finally, for maize, data was obtained for roots and shoots of rgh3 knockouts (37) (N= 3 each) and roots of rbm48 mutants (38) (N= 3 for each point mutation). A full list with further details of the RNAseq datasets that were used in this study can be found in Supplementary Table S3, by accession number.
For zebrafish datasets, RNA from two independent rnpc3 mutant alleles and their respective wildtype controls (N= 3 for each genotype) were sequenced. Clbns846 identified in an ethylynitrosourea mutagenesis screen encodes a T to A transition in intron 13, creating a novel 3′SS 10 nucleotides upstream of the canonical 3′ splice site (39). This results in aberrant transcripts all containing premature stop codons with no correctly spliced exon 13–14 junction transcript detected. ClbnZM harbors a retroviral insertion in intron 1 of rnpc3; both alleles are functionally null (39). Total RNA was extracted from pools of genotyped 72-hour post fertilization larvae using TRIzol. Poly(A)-enriched RNA was used to generate cDNA libraries and sequenced using the Illumina HiSeq by the Australian Genomics Research Facility (AGRF), yielding 100 bp single end reads.
RNA-seq analysis for the identification of responsive introns
All introns classified as minor, minor-like or hybrid were used to generate BED files for splicing analysis. For the analysis of major and major-like intron splicing, a randomized list of introns were generated. For introns found in multiple transcripts, the longest transcript was considered as canonical, and used to extract the coordinates of the flanking exons. Reads were aligned to the Ensembl v.99 genome assembly of respective species using Hisat2 (40). Retention and alternative splicing analysis were then performed using custom scripts, as described previously (21,34). Significant differences in mis-splicing indices were determined by a one-tailed Welch's t-test using custom R scripts. Responsive introns were defined as those with a significant increase (P< 0.05) in mis-splicing index upon minor spliceosome inhibition. Additionally, we applied an expression filter to remove introns found in lowly expressed genes from the lists of responsive and unresponsive introns. To this end, read counts were determined using the R package featureCounts, and were then converted to TPM values. Genes expressed below 1TPM in experimental conditions were removed from further analysis.
Results
Sub-classification of introns on a spectrum of minor to major introns
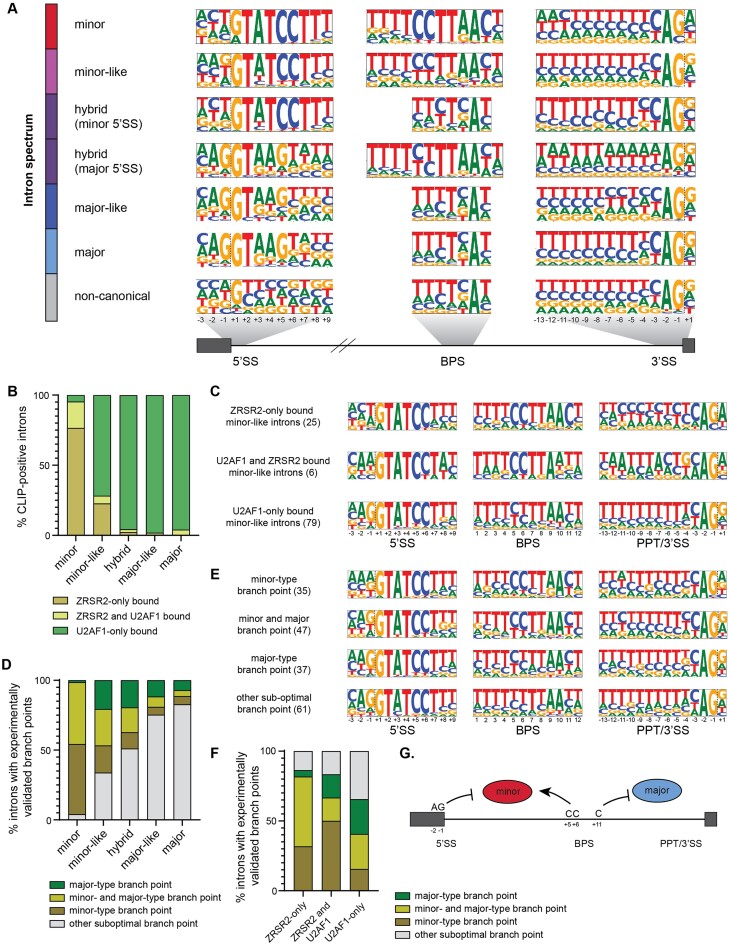
To date, eukaryotic introns have been classified as either major or minor based on the position weight matrix score of their 5′ splice site and branch point sequence. Nevertheless, there are introns with deviating splice sites and therefore reduced position weight matrix scores that confound a simple classification of these introns as entirely major or minor (41,42). These introns present a unique challenge to the commonly used binary classification system; in fact, we hypothesize that some of these introns might represent transitory intron states that are undergoing a conversion from minor to major, or vice versa. To gain insight into this process, we developed a new bioinformatics pipeline (Supplementary Figures S1-S4) to assess the prevalence of these introns in the genomes of 263 species, distributed over six of the seven defined eukaryotic supergroups (43). Specifically, we interrogated the genomes of 28 species in the TSAR supergroup, one species in Haptista, three species in Cryptista, 33 genomes in Archaeplastida, 192 genomes in the Amorphea supergroup, and six genomes in the Excavata supergroup (Supplementary Figure S6). Plotting the position weight matrix score of the major-type 5′ splice site against the minor-type 5′ splice site revealed that introns in all species were distributed along a continuum, rather than forming distinct clusters of major and minor introns. This distribution is incongruous with the assumption that major and minor introns are discrete populations with distinct sequence features (Supplementary Figure S7). This lack of distinction was even more apparent when looking at the distribution of position weight matrix scores for the major-type polypyrimidine tract against the minor-type branch point sequence (Supplementary Figure S7). This observation led us to deviate from the binary classification system of major and minor introns in favor of a sub-classification system that places introns into one of six categories (minor, minor-like, hybrid, major-like, major, and non-canonical introns) (see intron classification methods, Supplementary Figures S1–S4). Notably, we refer to non-canonical introns as those with a 5′ splice site position weight matrix score of less than 50, indicating the absence of any sequence motif that sufficiently resembles that of either major or minor introns (Supplementary Figures S4, S7). Using this method, we identified 850, 707, 684, 16 and 422 minor introns in the genomes of human, mouse, zebrafish, fruit fly and maize (Figure 1, Supplementary Figure S8). While these numbers are in keeping with previously published reports (Supplementary Figure S8), they are generally slightly higher. This is owed to the fact that we have classified all introns in the genome, rather than restricting our analysis to the introns of the longest isoform (19,44,45). In addition to minor introns, we also classified a small subset (0.5%) of introns in these species as minor-like and hybrid (Figure 1). Interestingly, in the human genome, a subset of introns with the same 5′ exon-intron boundary was classified as minor-like in one transcript, and as minor in another isoform. Thus, minor-like introns could be the consequence of alternative 3′ splice site usage in minor introns.
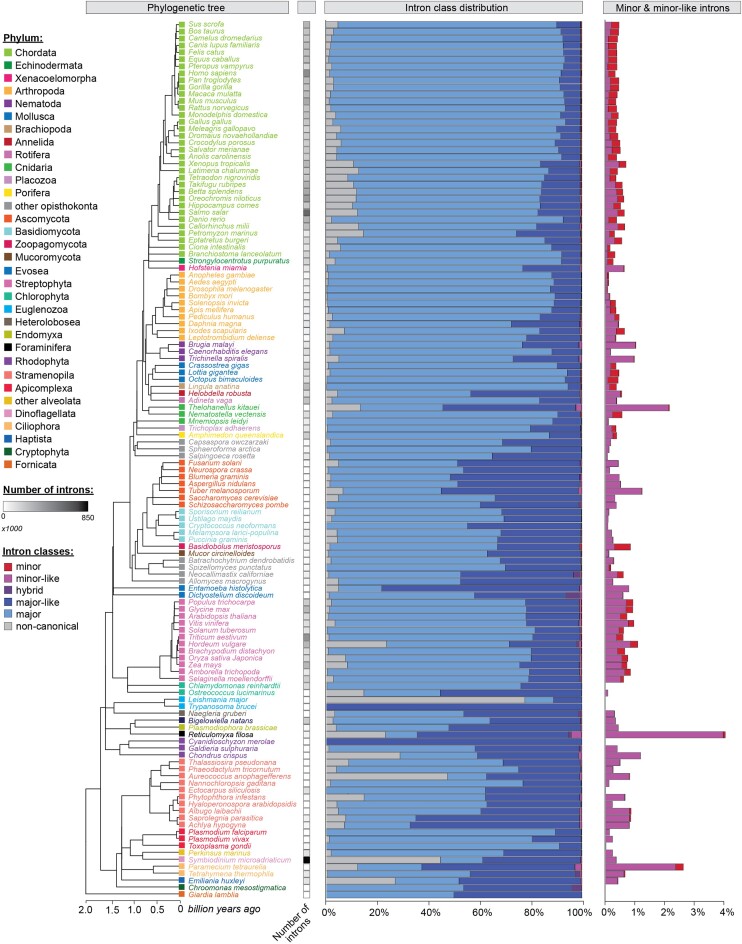
Figure 1.
Intron classification in diverse eukaryotic organisms. Phylogenetic tree (left) and bar graphs showing the distribution of the six intron classes (middle) in the genomes of a select set of eukaryotic organisms. A zoomed inset with the percentage of minor introns and minor-like introns can be seen on the right. Intron classes were defined using the criteria described in methods and Supplementary Figure S4. Species are color-coded by phylum. For a full list of intron numbers in all 263 eukaryotic organisms considered, see also Supplementary Table S4. See also Supplementary Figures S6–S10.
Presence of minor introns and minor spliceosome snRNAs across eukaryotic supergroups
We detected all six intron classes in the genomes of all investigated animals and streptophytes, including 27 land plants and the green algae Chara braunii, though not in red algae or green algae of Chlorophyta (Figure 1; Supplementary Table S4). Given that introns and snRNAs have co-evolved, we also explored the conservation of its unique snRNA components, i.e. U11, U12, U4atac and U6atac (9,10). The function of spliceosomal snRNAs depends not only on complementarity to the intronic sequence, but also their secondary structure. Therefore, we employed blastn, in combination with cmsearch, to identify putative snRNA genes. Using this approach, we identified genes encoding the minor spliceosome snRNAs in almost all metazoans (Figure 2). Moreover, we identified minor spliceosome snRNAs in all land plants but not in red algae and green algae of Chlorophyta, consistent with the presence and absence of minor introns in their respective genomes (Figures 1 and 2, Supplementary Figure S9, Supplementary Table S7). Given that minor introns have previously been reported as lost in nematodes, we were surprised to identify several in the genomes of Caenorhabditis elegans, Loa loa and Trichinella spiralis, among others (Figure 1; Supplementary Table S4). They comprised only ∼0.02% of all introns in these species and were therefore 10-fold less abundant than in chordates (Figure 1). In contrast, we found that the percentage of minor-like introns was almost 3-fold more in nematodes compared to chordates (Figure 1; Supplementary Table S4). Though we did not find minor spliceosome snRNAs in the genome of C. elegans, RNAseq analysis revealed the presence of reads spanning the correct exon-exon junctions (Supplementary Figure S10), indicating that the identified minor introns are spliced. While our approach did not detect minor spliceosome snRNAs, successful splicing of minor introns suggests the presence of a redundant mechanism that might leverage components of the major spliceosome, or an ancestral minor spliceosome-like machinery in C. elegans. The latter idea gains support from our discovery of genes encoding the U11, U12 and U6atac snRNA in the genome of the related species Trichinella spiralis, as well as evidence of U6atac snRNA for other nematodes (Figure 2, Supplementary Table S7). In all, nematodes might provide a snapshot of minor intron loss through conversion into minor-like introns, which could have been accompanied by degeneration of minor spliceosome snRNA genes, explaining the lack of detection in most of these species.
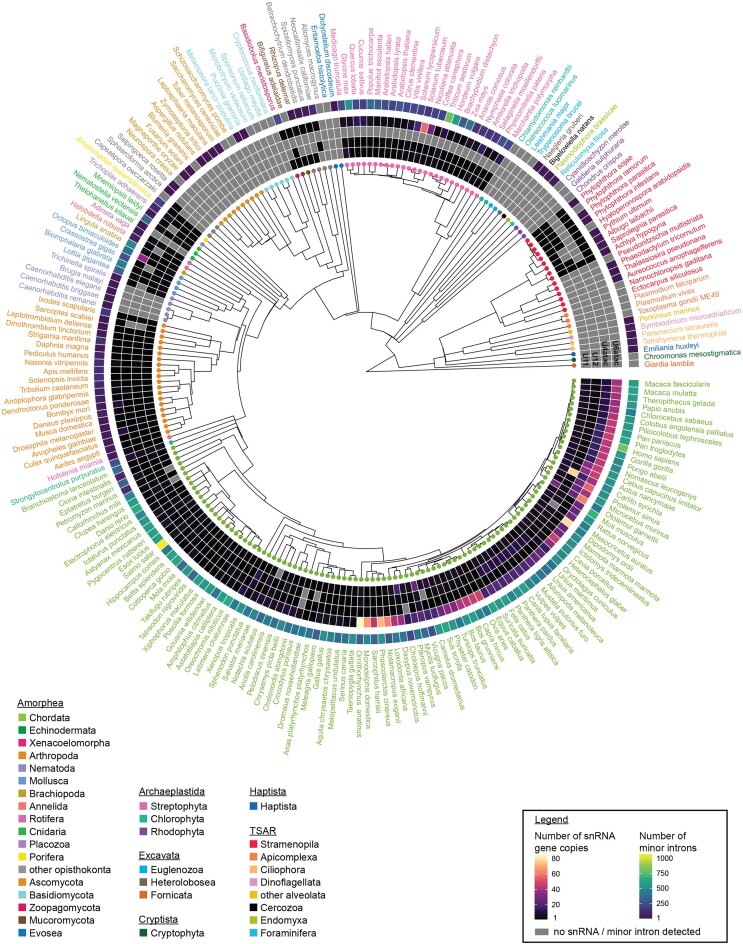
Figure 2.
Detection of minor spliceosome-specific snRNAs in a diverse set of eukaryotic organisms. Circos plot with heatmap of the number of detected gene copies for U11, U12, U4atac and U6atac snRNA in a select set of eukaryotic organisms. High confidence snRNAs were identified using a combination of blastn and cmsearch, according to inclusion thresholds described in the methods. The outer ring is a heatmap for the number of minor introns detected in each organism, as shown in Figure 1. Species are color-coded by phylum, as in Figure 1. For a full list of intron numbers in all 263 eukaryotic organisms considered, see also Supplementary Table S4. For a full list of both high and low-confidence snRNA gene copy counts, see also Supplementary Table S7. See also Supplementary Figures S24-S25.
Other organisms that have previously been reported to lack minor introns are the yeast Saccharomyces cerevisiae and other fungi (14,44,46). Accordingly, we detected few-to-none minor introns in the Ascomycota, Basidiomycota and Mucoromycota lineages (Figure 1; Supplementary Table S4). Interestingly, while we also did not identify minor spliceosome snRNAs for species in the fungal clades Ascomycota or Basidiomycota, we did detect U11, U12 and U6atac snRNA genes in the mucoromycote Bifiguratus adelaidae, suggesting fungi may once have possessed the minor spliceosome (Figure 2). Indeed, we found that other basal fungi also possess several minor spliceosome snRNA genes, in line with the relatively high minor intron densities observed in the zoopagomycote Basidiobolus meristosporus (237 minor introns; 0.5% of all introns) and neocallimastigomycote Neocallimastix californiae (129 minor introns; 0.2% of all introns) (Figures 1–2; Supplementary Table S4). Notably, the genome of the variosean amoeba Planoprotostelium fungivorum contained a substantially higher number of minor introns (0.16%) than other amoebozoa (0.01% on average) and possessed the U11, U12 and U6atac snRNA genes (Figures 1 and 2). Finally, in the slime mold Dictyostelium discoideum more than 5% of all introns were classified as hybrid, but no high confidence minor spliceosome snRNA genes were detected (Figures 1 and 2; Supplementary Table S4).
Relatively uncharacterized so far has been the distribution of minor introns in the supergroups Cryptista, Haptista, Excavata and TSAR. Our analysis revealed the presence of a few minor introns and a substantial number of minor-like introns in the haptophyte Emiliania huxleyi, as well as several gene copies of U6atac snRNA (Figures 1 and 2; Supplementary Table S4). Nonetheless, due to the lack of sequenced genomes in this supergroup, it remains unclear whether this is a general feature of this supergroup or specific to Emiliania huxleyi. Additionally, we identified minor introns in the genome of the cryptophyte Guillardia theta, which is more intron-rich than the other investigated cryptophyta (Figure 1; Supplementary Table S4). However, we did not detect evidence of minor spliceosome snRNA genes in this organism, thereby raising questions as to how these minor introns might be recognized and spliced. Similarly, we detected minor and minor-like introns in the genomes of multiple alveolates, but no minor spliceosome components (Figures 1 and 2, Supplementary Figure S9). Finally, several minor and minor-like introns were observed in the rhizaria and stramenopila lineages, such as the bygiria Blastocystis hominis (0.29% minor, 2.6% minor-like introns) and the foraminifera Reticulomyxa filosa (2.1% minor-like) (Figure 1; Supplementary Table S4). While we identified genes encoding U11, U12, and U6atac snRNA in the gyrista clade of stramenopiles, including for species lacking minor introns such as Phytophtora ramorum, we did not identify minor spliceosome components in other stramenopile clades such as diatoms, nor in rhizaria (Figure 2). Finally, we did not detect strong evidence for the presence of minor introns or minor spliceosome snRNA genes in any analyzed species of the Excavata supergroup.
Eukaryotic splicing, be it in concert across a single transcript or simultaneously at distinct transcripts, requires a high number of assembled spliceosomes. To meet this high demand for their expression, spliceosomal snRNAs are often encoded by multiple gene copies, as is the case for tRNAs (47). Indeed, multiple gene isotypes or variants of the major spliceosomal snRNAs have been previously described (8,48), but here, we found that mammals also contain several minor spliceosome snRNA variants. We found a particularly high number of U6atac variants in non-placental and placental mammals, and notably, a higher number of U12 snRNA variants in carnivores compared to other mammals (Figure 2). Finally, there was a clear enrichment of U4atac snRNA variants in the primate lineage (Figure 2, Supplementary Table S7). While we consistently detected U4atac snRNA genes across opisthokonts, we did not detect it in any species of other supergroups, with the exception of Achlya hypogyna, a facultative parasite of the stramenopile lineage (Figure 2, Supplementary Table S7). Notably, in some of these lineages U4atac genes were identified in a recent report by Larue et al, using a reduced threshold stringency (12). Therefore, experimental data will be essential in determining whether these more divergent U4atac variants are functional in the minor spliceosome.
In all, our findings suggest that the minor spliceosome and minor introns date back approximately 1.7 billion years. While minor introns are highly conserved in most metazoa and land plants, both the introns and the minor spliceosome machinery are absent from the genomes of many fungal, algae and protist lineages. Together, these facts suggest that minor introns have been both lost and gained throughout evolution.
Evolution of minor introns
To understand the trajectories of minor intron evolution, we identified the orthologous introns of human minor introns across all investigated species (Supplementary Figure S5). Most orthologs of human minor introns in chordates and mollusks were also classified as minor introns (Figure 3A, Supplementary Table S8). In contrast, only a few orthologous introns were classified as minor introns in land plants, even though their genomes contain a comparable number of minor introns (Figures 1, 3A). Instead, human minor introns were found to be orthologous to major-like/major introns in plants or to not have an orthologous intron in their corresponding gene, the latter of which could be indicative of intron loss or intron sliding (Figure 3A) (49,50). Similarly, orthologous introns of human minor introns were found to be either major introns or absent entirely in stramenopiles (Figure 3A). Finally, human minor introns often lack an ortholog in Ascomycota, whereas they are orthologous to major or major-like introns in other fungal lineages, pointing to distinct modes of minor intron loss within one kingdom (Figure 3A).
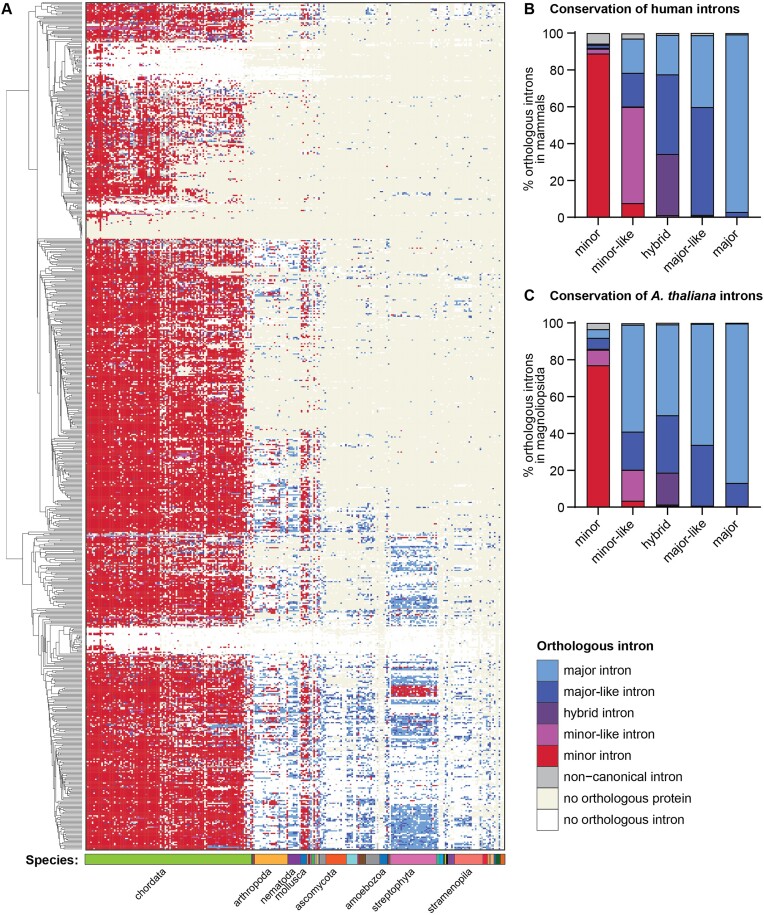
Figure 3.
Orthology of human minor introns in a diverse set of eukaryotic organisms. (A) Color-coded heatmap representing the identity of introns orthologous to human minor introns across all 263 eukaryotic organisms considered. Organisms are ordered by phylogeny as in Figures 1 and 2, such that primates are found on the left and TSAR/Haptista on the right. Introns were sorted for visualization purposes using hierarchical clustering of the intron classes. Ten clusters were identified using average linkage, using the nomclust package in R. (B) Conservation of human intron classes across mammalian genomes. (C) Conservation of intron classes in Arabidopsis thaliana across Magnoliopsidae genomes. Underlying data can be found in Supplementary Table S8. See also Supplementary Figure S5.
While minor intron loss through homologous recombination with a reverse transcribed, spliced mRNA has been documented previously (15,17,46,51), loss of minor introns through conversion to major introns has been relatively understudied. To gain more insight into the potential mechanisms of intron class switching, we took a closer look at orthologous intron clusters in mammals, which was the most densely sampled taxonomic clade in our dataset. Here, we found that human minor and major introns are highly conserved in other mammals as minor and major, respectively (Figure 3B). However, human major-like introns are orthologous to both major and major-like introns, suggesting an unsurprisingly high degree of similarity between these intron classes. In contrast, orthologs of human minor-like introns were classified as minor-like in only 50% of the mammals, suggesting they are less conserved than minor introns (Figure 3B). The remaining orthologous introns were either classified as major (∼20%), major-like (∼20%), or minor (∼10%). Given that minor-like introns are the only intron type with orthologs across all intron classes, we postulate that minor-like introns might act as a transitory node for conversion of minor to major introns and vice versa. This is bolstered by the observation that minor-like introns in Arabidopsis thaliana are not frequently conserved in other land plants as minor-like introns; rather, they are orthologous to minor, major-like or major introns (Figure 3C). In addition, minor introns in A. thaliana appear to undergo intron class switching to a higher extent in land plants than we observed for human minor introns in mammals (Figure 3B, C).
Major and minor-like introns both possess an AG at the –1 and –2 position of the 5′ splice site
To understand how minor-like introns might function as an intermediate during intron class conversion, we next investigated the sequence motifs associated with each intron class in mammals and land plants. Despite a reduced position weight matrix score, the consensus sequences of minor-like and major-like introns still resemble that of minor and major introns, respectively (Figure 4A, Supplementary Figure S11). Nevertheless, major-like introns generally possess a weaker polypyrimidine tract than major introns. Similarly, minor-like introns have a significant enrichment of a G at the –1 position of the 5′ splice site compared to minor introns (48% versus 11%; P< 0.0001; chi-squared test) (Figure 4A, Supplementary Figure S11). The G at the –1 position of the 5′ splice site is a hallmark feature of major introns and is normally used by U1 snRNA for base-pairing (52). Therefore, this raises the possibility that minor-like introns with a –1G at the 5′ splice site can be recognized by the major spliceosome, despite the overall resemblance of their consensus sequences to minor introns (Figure 4A, Supplementary Figure S11). In fact, a recent study noted a strong enrichment of not only –1G, but also of the A-2G-1 dinucleotide in major introns compared to minor introns (27). We therefore explored the enrichment of this dinucleotide in minor-like introns and found that this was also significant compared to minor introns (32% versus 3%; P< 0.0001; chi-squared test). Additionally, we observed a significantly reduced conservation of CC at the +5/+6 position of the branch point sequence (P< 0.0001; Chi-squared test) and enrichment of a G at the +1 position of the 3′ splice sites of minor-like introns, compared to minor introns (Figure 4A, Supplementary Figure S11). Together, these findings suggest that minor-like introns might not only be an intermediate intron class from a sequence and evolutionary perspective, but also raise the possibility that a subset might be recognized by components of both the major and minor spliceosome.
Figure 4.
Sequence elements informing recruitment of the major and minor spliceosome. (A) Frequency logos of consensus sequences for 5′ splice site, branch point sequence and 3′ splice site of the different intron classes in mammalian genomes. Dashed lines denote the exon-intron and intron-exon boundaries. (B) CLIP-seq analysis for U2AF1 and ZRSR2 across all human intron classes. (C) Frequency logos of consensus sequences for 5′ splice site, branch point sequence and 3′ splice site of human minor-like introns bound by either U2AF1, ZRSR2 or both. (D) CoLa-seq analysis for all human intron classes. The experimentally validated branch point coordinates were compared with the highest scoring branch points predicted by position weight matrices. (E) Frequency logos of consensus sequences for 5′ splice site, branch point sequence and 3′ splice site of human minor-like introns, separated by utilization of the branch point as determined by CoLa-seq. (F) Integration of the CoLa-seq data with the CLIP-seq data in human minor-like introns. (G) Simplified model with the sequence elements that inform recruitment of the major and minor spliceosome.
To gain a deeper understanding into the splicing mechanism of minor-like introns, we next analyzed recently published CLIP-seq data for the minor spliceosome component ZRSR2 and the major spliceosome-specific factor U2AF1 (27). ZRSR2 and U2AF1 were shown to function analogously by contacting the 3′ splice site of minor introns and major introns, respectively (53,54). As expected, 99% of all CLIP-positive major introns were bound by U2AF1, while ZRSR2 was recruited to the 3′ ends of 95% of the CLIP-positive minor introns (Figure 4B). It must be noted however, that 20% of the minor introns were also bound by U2AF1, in addition to ZRSR2 (Figure 4B), as has been reported previously (27). Thus, U2AF1 binding is not restricted to the 3′ ends of major introns. We next explored whether U2AF1 and/or ZRSR2 were recruited to the 3′ ends of minor-like introns. Interestingly, the majority of CLIP-positive minor-like introns were bound by U2AF1, while a quarter of all CLIP-positive minor-like introns were bound by ZRSR2 (Figure 4B). Analysis of the splice site sequences revealed a significantly reduced frequency of the branch point C+5C+6 of U2AF1-only bound minor-like introns compared to minor-like introns bound by ZRSR2 (23% versus 82%; P< 0.0001) (Figure 4C). This finding suggests that these nucleotides may facilitate recruitment of the minor spliceosome. Additionally, we observed that minor-like introns exclusively bound by ZRSR2 possess a cytosine at the +11 of the branch point sequence, while those minor-like introns that are bound by U2AF1 generally lacked this nucleotide (P= 0.0004) (Figure 4C). This finding suggests that the presence of a cytosine at the + 11 position of the branch point sequence might not favor recruitment of U2AF1. In contrast, we found that minor-like introns exclusively bound by U2AF1 contained a polypyrimidine tract and had a significant enrichment of an A-2G-1 at the 5′ splice site compared to those bound by ZRSR2 (P= 0.0005) (Figure 4C), in agreement with previous reports (27). Mutation of the –1 and –2 nucleotides of the 5′ splice site to A-2G-1 has recently been shown to prevent splicing of two minor introns by the minor spliceosome (27). Consistently, we observed enrichment of this AG dinucleotide in minor-like introns exclusively bound by U2AF1, but not in minor-like introns bound by both ZRSR2 and U2AF1. Together, these analyses revealed important sequence elements that can inform the recruitment of the major and minor spliceosome.
Following the observation of U2AF1-bound minor-like introns, we asked whether the minor-type branch point sequence identified in these introns was still being utilized for splicing, or whether alternative, suboptimal branch point sequences were employed instead. After all, the major branch point sequence is sufficiently degenerate (i.e. yUnAy (55)) that it remains plausible that the U2 snRNA could also base pair with a degenerate minor-type branch point sequence. Towards addressing this, we analyzed CoLa-seq data containing >150 000 experimentally validated branch points, which were identified using intron lariat sequencing (28). This revealed branch points for 563 minor, 180 minor-like and 129 hybrid introns in K562 cells. For 94% of the minor introns the branch point(s) utilized was the same as the one predicted by our position weight matrix scoring (Figure 4D), suggesting that a strong minor-type branch point sequence is sufficient for recruiting the minor spliceosome. However, for only 46% of minor-like introns did the experimentally validated branch point(s) correspond with the bioinformatically predicted minor-type branch point sequence (Figure 4D). Examination of these minor-like introns where the minor-type branch point was utilized again revealed the presence of C+5C+6 at the branch point sequence, which was significantly reduced in minor-like introns without evidence of minor-type branch point utilization (18% versus 52%; P< 0.0001) (Figure 4E). This finding suggests that a strong minor-type branch point sequence, characterized by two cytosines at the +5/+6 position is an important feature to recruit the minor spliceosome.
Intersection of the CoLa-seq and CLIP-seq data revealed that minor-like introns that were exclusively bound by ZRSR2 predominately utilized the highest-scoring minor-type branch point (Figure 4F). Only a small percentage of the ZRSR2-only bound minor-like introns were not spliced using the minor-type branch point, indicating the presence of other cis-regulatory elements and/or trans-acting factors that can inform splice site usage in these introns. Finally, in the case whereby both U2AF1 and ZRSR2, or exclusively U2AF1, were recruited to a minor-like intron, the branch point that was most frequently used was either the highest-scoring major-type or another, suboptimal branch point (Fisher exact, P< 0.01) (Figure 4F). In all, our findings suggest that minor-like introns might break free from the obligatory relationship with the minor or major spliceosome for their removal.
Investigating the effect of minor spliceosome inhibition on novel intron classes
The CLIP-seq analysis revealed the recruitment of ZRSR2 to a subset of minor-like introns (Figure 4B). To test whether these minor-like introns are real targets of the minor spliceosome, we analyzed RNA sequencing data from various model organisms in which core subunits of the minor spliceosome were inhibited (Supplementary Table S3). These included data from patients with mutations in U4atac snRNA and ZRSR2, as well as experimental data from different loss-of-function models for the minor spliceosome U11, U12, U4atac and U6atac snRNAs, and minor spliceosome proteins CENATAC, RBM48 and rgh3 (ortholog of ZRSR2) (29–35,37,38). Moreover, we performed RNAseq of zebrafish larvae with two different mutations in the same gene, rnpc3, which encodes a critical minor spliceosome-specific protein (39). Collectively, analysis of these datasets was designed to identify ‘responsive’ introns, i.e. introns in expressed genes that are retained or alternatively spliced at significantly higher levels following minor spliceosome inhibition.
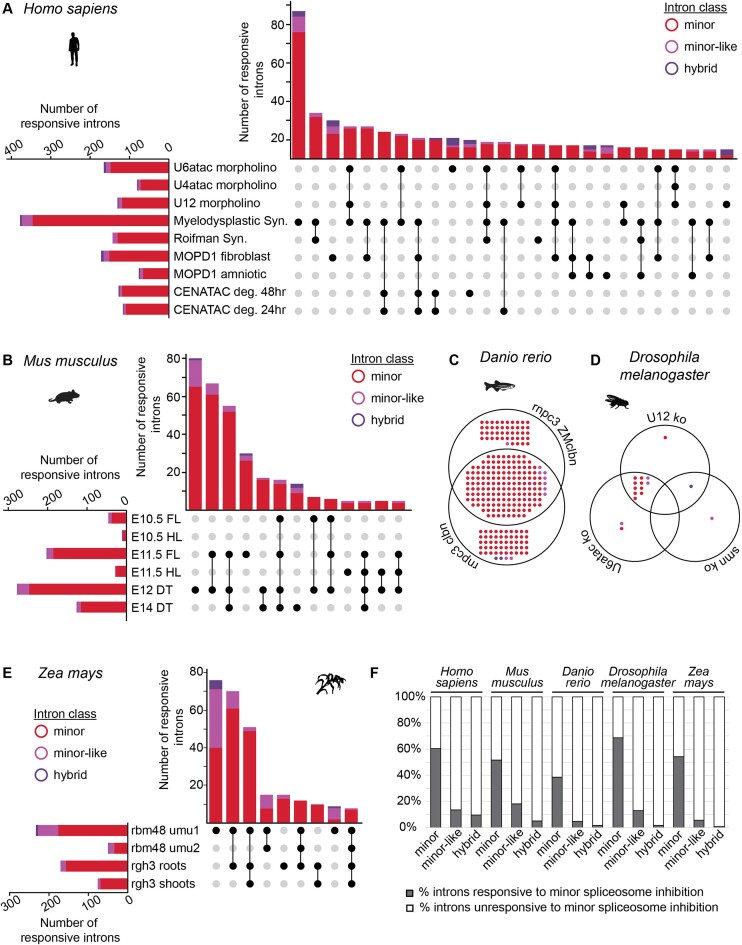
As expected, bioinformatically classified minor introns were retained at higher levels following minor spliceosome inhibition, reflected by a positive difference in mis-splicing index between the different experimental and respective control conditions (Supplementary Figures S12-S16). Using our bioinformatics pipeline, we detected the highest number of responsive minor introns in peripheral blood mononuclear cells of individuals with ZRSR2-linked myelodysplastic syndrome (59%) (Figure 5A), which is a similar percentage compared to previous studies, despite differences in bioinformatics pipelines (Supplementary Table S5) (30,33,56). All analyzed datasets showed evidence of minor intron mis-splicing, though the extent of mis-splicing varied considerably between datasets. For example, we detected less minor intron mis-splicing in amniotic fluid (10%) compared to fibroblasts (25%) from individuals with microcephalic osteodysplastic primordial dwarfism type 1, which agrees with the cell type-specific expression and splicing of minor intron-containing genes that has been reported previously (Figure 5A, Supplementary Table S6) (21,32). Moreover, we observed little mis-splicing of minor introns in the limbs of U11 conditional knockout embryos at E10.5 (∼5%), while > 45% of expressed minor introns were responsive in the developing cortex of E12 U11-null embryos (Figure 5B, Supplementary Table S6). Together, these findings underscore the importance of integrating multiple datasets towards understanding the dynamic relationship between the minor spliceosome and its intron targets. Moreover, they reveal the importance of trans-acting factors that might function in a cell-type and context-specific manner. Intersection of the responsive minor introns revealed great overlap between some datasets, such as the two zebrafish models with rnpc3 loss-of-function (62%) or the U12 and U6atac knockout Drosophila melanogaster larvae (82%) (Figure 5C, D, Supplementary Table S6). However, this overlap was significantly reduced in human, mouse and maize, for which we analyzed a larger number of datasets. While almost 80% of all minor introns affected upon CENATAC depletion were also mis-spliced in individuals with myelodysplastic syndrome, only 30% of the CENATAC-sensitive minor introns were also affected in Roifman syndrome (Supplementary Figure S17, Supplementary Table S6). Only the minor intron in NAA60 was mis-spliced in all human datasets (Supplementary Table S6). Similarly, only seven minor introns were mis-spliced in all analyzed maize datasets, and none were commonly identified in mouse (Figure 5B, E, Supplementary Table S6). In all, we found that approximately 60% of all expressed minor introns were affected by minor spliceosome inhibition in at least one human dataset (Figure 5F, Supplementary Table S6). Similar numbers of responsive minor introns were observed for mouse, zebrafish, fruit fly and maize (Figure 5F, Supplementary Table S6).
Figure 5.
Identification of introns responsive to minor spliceosome inhibition. (A, B) Upset plot for mis-spliced introns in different (A) human and (B) mouse datasets in which the minor spliceosome is inhibited. Intersections with fewer than five introns have been omitted. (C, D) Venn diagram for mis-spliced introns in different (C) zebrafish and (D) fruit fly datasets in which the minor spliceosome is inhibited. (E) Upset plot for mis-spliced introns in different maize datasets in which the minor spliceosome is inhibited. Intersections with fewer than five introns have been omitted. Color-coding for intron classes in Figure 5 is the same as in Figure 1. Significant retention and/or alternative splicing of introns was identified using a one-tailed Welch's t-test. Responsive introns were defined as those found in genes expressed above 1 TPM, with a significantly increased mis-splicing index (P< 0.05) in minor spliceosome loss-of-function conditions. (F) Bar graphs with total number of responsive minor, minor-like and hybrid introns in the different model organisms. For more information on the analyzed RNAseq datasets (including experimental conditions and N-value), see also Supplementary Table S3. See also Supplementary Figure S12-S18 and Supplementary Table S6. Deg = degron; syn = syndrome; FL = forelimb; HL = hindlimb; DT = dorsal telencephalon.
Unlike minor introns, which were expected to be affected upon minor spliceosome inhibition, the effect of minor spliceosome inhibition on minor-like and hybrid introns was less apparent. Our analysis showed that in all human datasets, a low number of minor-like and hybrid introns were mis-spliced (Figure 5A, Supplementary Table S6). Moreover, only a few responsive minor-like and hybrid introns were affected in more than one or two datasets, pointing to a variable dependence of these introns on individual minor spliceosome components. Similarly, we observed that 5–10% of responsive introns in species such as mouse, zebrafish and fruit fly were minor-like or hybrid (Figure 5B–D, Supplementary Table S6). This was much higher in the rbm48 maize mutants, where approximately 25% of all responsive introns were minor-like (Figure 5E, Supplementary Table S6). Finally, no more than 1–2% of major and major-like introns were responsive in any of the datasets, and these are likely due to secondary effects of disrupting minor-intron containing genes that themselves are RNA processing factors. In all, we found that approximately 55% of all expressed minor introns, 10% of all expressed minor-like introns, and 5% of all expressed hybrid introns were affected across the five model organisms (Figure 5F, Supplementary Table S6).
Inherent to the variation in the sampling procedures, sample sizes, and methods across the datasets used in our analyses, is the differing levels of power to identify responsive introns. To ensure that the ∼45% of minor introns classified as unresponsive were not the result of insufficient power to correctly classify them as responsive (i.e. false negatives), we conducted a sensitivity analysis to evaluate the effect of alpha on the percentage of introns identified as responsive to inhibition of the minor spliceosome (Supplementary Figure S18). As expected, increasing alpha beyond 0.05 led to an increase in the proportion of minor introns that were identified as responsive. However, increasing alpha concomitantly increased the proportion of responsive introns for all other intron classes as well. When combining all human datasets (Supplementary Figure S18A), we found that increasing the alpha value affected the percentage of responsive major introns more drastically than that of responsive minor introns. Given that inhibition of minor spliceosome components is not thought to affect major intron splicing directly, a high percentage of responsive major introns is likely a sign of having inflated type I errors (false positives). We therefore conclude that the effect of false negatives on our responsive versus unresponsive classification is limited upon integration of the individual datasets. Though we anticipate the identification of additional responsive minor introns with the generation of more knockout models and deeper transcriptomic data, the current list provides insight into the introns that are highly dependent on minor spliceosome function for their splicing in these systems.
Minor-like introns as an intermediate intron class between minor and major introns
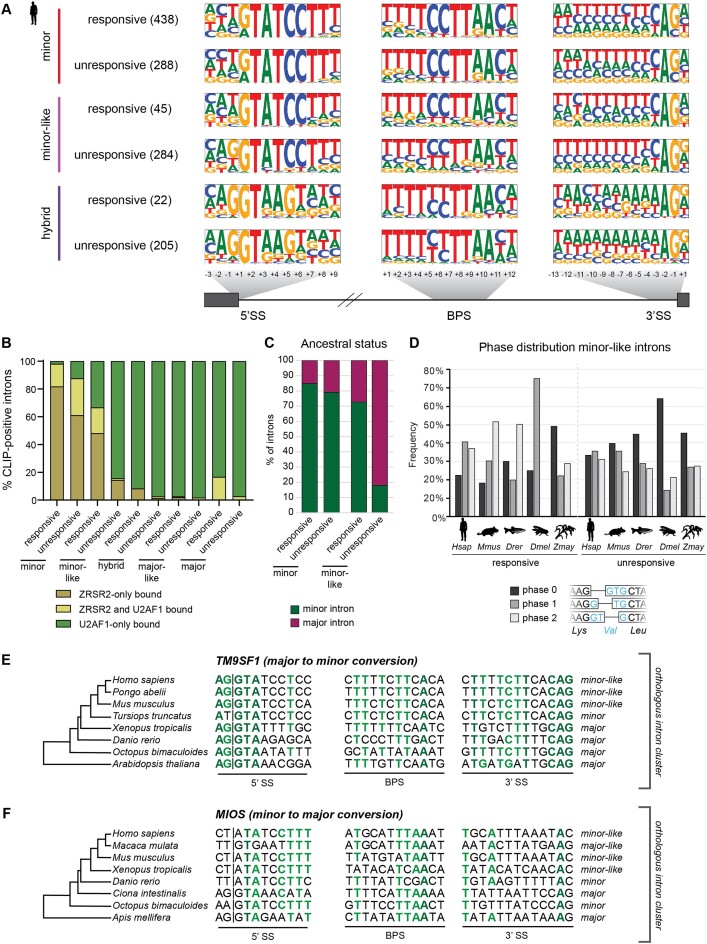
Our transcriptomic analyses showed that the splicing of some, but not all, minor, minor-like and hybrid introns was affected upon minor spliceosome inhibition. We therefore sought to identify which features contributed to a ‘responsive’ versus ‘unresponsive’ characterization of introns within the framework of our bioinformatics pipeline. Analysis of the consensus sequences from responsive and unresponsive minor introns did not reveal a significant difference in splice site motifs (Figure 6A). However, we did note a difference in the 5′ splice site consensus sequence of responsive and unresponsive minor-like introns in humans. Specifically, there was a bias against the G at the –1 position for minor-like introns affected by minor spliceosome inhibition (P< 0.001; chi-squared test) (Figure 6A). Similarly, we noted a bias against the G at the +1 position in the 3′ splice site of responsive minor-like introns (P< 0.05; Chi-squared test) (Figure 6A). Notably, this motif remained even when increasing alpha beyond 0.05 in our responsive vs. unresponsive intron classification and is consistent with the findings obtained through CLIP-seq analysis (Figure 4C; Supplementary Figure S19). The close resemblance of responsive minor-like introns to consensus sequences of minor introns suggests that the presence of a guanine at the exon-intron boundaries may reduce their reliance on the minor spliceosome through compensatory recognition by the major spliceosome.
Figure 6.
Features of responsive minor-like introns resemble that of minor introns. (A) Frequency logos of consensus sequences for 5′ splice site, branch point sequence and 3′ splice site of human minor, minor-like and hybrid introns whose splicing is either responsive (affected) or unresponsive (unaffected) to minor spliceosome inhibition. Introns found in genes that were expressed below 1 TPM were excluded from all analyses. The number in parentheses denotes the number of introns in each group used to create frequency logos. Dashed lines denote the exon-intron and intron-exon boundaries. (B) Bargraph showing the distribution of human introns bound by ZRSR2 and/or U2AF1. (C) Ancestral status of minor and minor-like introns at vertebrate origin. Ancestral status was determined by a joint call between protostomes and non-bilaterians. (D) Phase distribution for responsive (left) and unresponsive (right) minor-like introns in Homo sapiens (Hsap), Mus musculus (Mmus), Danio rerio (Drer), Drosophila melanogaster (Dmel), Zea mays (Zmay). An example of the different intron phases is shown below. (E, F) Gene schematics and splice site sequences for an orthologous intron cluster in TM9SF1 and MIOS, containing an unresponsive (E) and responsive (F) minor-like intron in Homo sapiens, respectively. Nucleotides in dark green are 100% conserved between orthologous introns of the cluster, while nucleotides in light green are 75% conserved. See also Supplementary Figures S19–S23.
Further, we found that the distance of the branch point to the 3′ end of responsive minor-like introns was shorter than that of unresponsive minor-like introns (Supplementary Figure S20). The distribution of these branch point distances also resembled that of minor introns, while the distribution for unresponsive minor-like introns was more uniform (Supplementary Figure S21). These findings may lead one to think that the minor-like intron class, despite close sequence resemblance to minor introns, is merely a mixed population consisting of both minor (responsive) and major (unresponsive) introns. To test whether minor-like introns represented a mixed population of minor and major introns, rather than a mechanistically intermediate class that can be recognized by components of both spliceosomes, we returned to the CLIPseq data. The rationale is that unlike responsive minor introns, responsive minor-like introns would show a higher probability of U2AF1 binding, thereby indicating that minor-like introns are a mechanistically intermediate class. Indeed, analysis of the CLIP-seq data revealed that a significantly higher proportion of responsive minor-like introns are bound by U2AF1, compared to responsive minor introns (Figure 6B). Thus, we propose that minor-like introns are a unique, mechanistically intermediate intron class.
The reduced dependency of minor-like introns on the minor spliceosome compared to minor introns is in keeping with the idea that minor-like introns could be an intermediate in the conversion of minor introns to major introns. To gain further insight into the conversion path of introns, we inferred the ancestral vertebrate intron class for human introns (Figure 6C). This revealed a high ancestral minor origin for both responsive and unresponsive minor introns, as expected. Similarly, most responsive minor-like introns have an inferred ancestral minor origin (Figure 6C). In contrast, the majority of unresponsive minor-like introns have an inferred major status at vertebrate origin (Figure 6C). This would suggest that while most responsive minor-like introns originate from a minor intron, the unresponsive minor-like introns predominately originate from major introns. As such, one could speculate that responsive minor-like introns represent an intermediate state in the conversion of minor introns to major introns, whereas unresponsive minor-like introns could be an intermediate in the conversion of major introns to minor introns. Indeed, we found that 15–20% of human minor introns had an inferred ancestral major origin, supporting the notion of occasional minor intron gain (Figure 6C). Consistent with the idea that unresponsive minor-like introns might represent an intermediate in the conversion of major introns to minor introns is the enrichment of the AG dinucleotide at the 5′ splice site of these introns, which is a feature observed in major introns (Figures 4A, 6A). Further support for this hypothesis stems from the phase distribution of the different intron classes. Introns are not positioned randomly in the genome; rather, it has long been known that an excess of major introns are found in phase 0 (i.e. positioned between two codons), while minor introns are predominately found in phase 1 (i.e. after the first nucleotide of a codon) or phase 2 (i.e. after the second nucleotide of a codon) (15,57). While the reason for this positional bias remains unresolved, it has been proposed that minor introns in phase 0 may be more readily converted to a phase 0 major intron (44). Indeed, we found that responsive minor-like introns in vertebrates were biased against phase 0, like minor introns (P< 0.01; Chi-squared test). In contrast, unresponsive minor-like introns were more equally distributed between the phases (Figure 6D, Supplementary Figures S22-S23). In all, our findings are compatible with a model wherein minor-like introns represent a transitory node in the conversion between minor and major introns.
Discussion
Classification of introns along a spectrum
The binary classification of minor versus major introns has served well in the study of introns and their regulation (1,12,19,21,29,44). However, inherent to this dichotomous classification remains the forced binning of introns with ambiguous features in either minor or major categories. Given that splicing is a biochemical process, reliant on RNA and protein binding affinities that are rarely binary, imposing a binary classification system is inherently flawed. Therefore, we have here placed introns along a spectrum where minor and major introns are the bookends, with other intron types in between. We have limited our classification to six subcategories so that the bins contain enough introns to perform statistical analyses. However, it would not be surprising for further experimental evidence to refine our understanding of intron subclassification. For example, here we report hybrid introns as one intron class, though they may be further distinguished as minor-hybrid or major-hybrid based on the 5′ splice site (Supplementary Figure S4). Moreover, our classification system revealed several extant organisms with a rather large proportion of non-canonical introns, such as Emiliana huxleyi, Thecamonas trahens, Symbiodinium microadriaticum and Aureococcus anophagefferens (Figure 1; Supplementary Table S4). While this might merely be the consequence of poor annotation of exon-intron boundaries in their genomes, an alternative biological explanation might be that these species have more flexibility in the splicing mechanism of these introns.
The possibility of a non-canonical spliceosome
The energetic burden associated with maintaining a spliceosome has likely led to an evolutionary pressure to streamline its composition. Indeed, mass spectrometry analysis of the major spliceosome B and C complexes in human and yeast has revealed that yeast spliceosomes contain drastically fewer proteins than those in humans (58). Attempts at bioinformatically identifying orthologs of these ∼60 core spliceosome proteins in more extant species such as Giardia lamblia and Encephalitozoon cuniculi, revealed the existence of only 30–35 proteins (59,60). Further evidence of the dramatic reduction of spliceosome size in ancient species stems from Cyanidioschyzon merolae, which lacks both U1 snRNA and all associated proteins (61). Despite the absence of many spliceosome components, introns in these species are spliced, thereby underscoring the extreme diversity and flexibility that exists in the splicing reaction. Here, we have detected minor introns in the genomes of several species that lack either specific or all minor spliceosome snRNAs (Supplementary Figure S24). Most notably, we did not detect any high confidence snRNAs in C. elegans and E. huxleyi. This might be explained by the fact that the genomes of extant species such as E. huxleyi are frequently incomplete and contain mistakes. However, this rationale does not extend to the well-characterized model organism C. elegans, where we confirmed that the identified minor introns are successfully spliced (Supplementary Figure S10). We can speculate on several different explanations for the splicing of minor introns in this organism. For one, the sequence and structure of spliceosomal snRNAs might have diverged so significantly that they fail to be detected using our computational approach. Indeed, a closer look at the U12/U6atac snRNA duplex that forms the catalytic center of the activated spliceosome for T. spiralis, a closely related species of C. elegans, revealed significant divergence of the nucleotides around the GAGA box and helix III (Supplementary Figure S24). Notably, the importance of helix III is unresolved, as it is also not conserved in Arabidopsis thaliana (62). Thus, it might be possible that some species possess highly divergent minor spliceosome snRNAs that can splice the minor introns we detected. An alternative explanation could be that major spliceosome snRNAs are not only capable of recognizing major, but also minor introns in C. elegans. Indeed, the major spliceosome has previously been shown to be capable of splicing major introns with AT-AC terminal dinucleotides in humans and generally uses fewer nucleotides for base pairing compared to the minor spliceosome. While experimental data would be needed to verify such a hypothesis, splicing of minor introns by major spliceosome components would provide another example of the enormous flexibility that can exist in the splicing reaction and supports a decoupling of intron identity from the spliceosome it splices.
For land plants, water molds in Gyrista, and several fungal species of the mucoromycote and zoopagomycote lineage, we identified high confidence gene variants for U11, U12 and U6atac snRNA, but not U4atac (Figure 2; Supplementary Table S7). Lowering the confidence threshold led to the identification of a putative divergent U4atac snRNA gene in Basidiobolus meristosporus and Neocallimastix neoformans (Supplementary Table S7). However, even by reducing our threshold, we did not observe divergent U4atac snRNAs in water molds of the Gyrista class. The fact that we can identify genes encoding U11, U12 and U6atac snRNA in these species, but never U4atac, suggests that this is unlikely due to incomplete genome annotation. Rather, this could mean that minor intron splicing in these non-metazoan species might occur without U4atac, that U4atac snRNA has diverged significantly, and/or that U4atac snRNA can be replaced by U4 snRNA. Unlike the U11, U12 and U6atac snRNA, which bind to proteins unique to the minor spliceosome, U4atac snRNA has no such protein interactions reported. While a few U4atac/U6atac-specific proteins, such as CENATAC, have been identified, they are all found in the mono-U6atac snRNP fraction and/or bound to U6atac snRNA (30,63). The importance of these minor spliceosome-specific proteins for the recruitment of the human U4atac/U6atac.U5 tri-snRNP to the human U11/U12 di-snRNP was recently demonstrated in the cryo-EM structure of the minor pre-B complex (64). This has revealed four distinct interfaces through which U11/U12 di-snRNP interacts with the tri-snRNP in humans, of which three are of sufficient quality to determine specific interactions. These include 1) the interaction of U11-25K and U11-59K with three nucleotides in the 3′ stem loop of U4atac snRNA, 2) the interaction of PRPF8 with U11 snRNA and associated proteins, and 3) the interaction of PRPF28 with U11 snRNP (64). Moreover, CENATAC was shown to interact with the cap of U4atac snRNA (64). While this study identifies several important nucleotides in U4atac snRNA for the formation of the minor pre-B complex in human (i.e. A88, G111, and G114), it must be noted that several of these nucleotides are not conserved across more distantly related animals such as D. melanogaster and A. queenslandica (Supplementary Figure S25A). This raises the possibility that non-metazoan organisms might be less dependent on interactions between U4atac snRNA and U11 snRNP for the formation of the minor pre-B complex. Similarly, CENATAC is not conserved across many fungal lineages, pointing to flexibility in the minor splicing reaction (30). Thus, the most critical features for a divergent U4atac or U4 snRNA to partake in the minor spliceosome remains its base pairing capacity with U6atac snRNA. Biochemical experiments have already revealed that the introduction of compensatory point mutations in U4 snRNA that maintain its base pairing capacity with U6atac snRNA is sufficient to activate minor intron splicing (65). Together, this opens the exciting possibility that non-metazoan species lacking U4atac snRNA can splice minor introns using a non-canonical, ‘hybrid’ spliceosome that consists of both major and minor spliceosome components. For example, in Phytopthora sojae U6atac snRNA has extensive sequence complementarity to U4 snRNA, such that they might be able to form a U4/U6atac di-snRNA (Supplementary Figure S25B).
Invoking a hybrid machinery for non-metazoan organism is not too far-fetched, considering that the minor spliceosome already uses many major spliceosome proteins, along with the shared U5 snRNA (66). In fact, the existence of such a hybrid spliceosome has been proposed for Giardia lamblia, which contains snRNAs possessing features of both major and minor spliceosome snRNAs. For instance, the U6 snRNA of G. lamblia is truncated at the 5′ end compared to the human U6 snRNA, and contains a 3′ stem loop that is characteristic of the human U6atac (67). This stem loop is important for minor intron splicing, as creation of a chimeric U6atac consisting of the human snRNA with the 3′ stem loop of Arabidopsis thaliana, Drosophila melanogaster, Trichinella spiralis, or Phytophtora infestans could only partially, or not at all, support the splicing of a minor intron in the P120 gene reporter (68).
Minor-like introns represent an intermediate in the conversion between minor and major introns
While it is generally thought that major introns emerged as a product of self-splicing group II introns, the positional bias of major introns at phase 0 in the genome suggests that these insertion events were unlikely to be random (1). One model proposed that major introns used proto-splice site sequences such as AG/G that are more amenable to their insertion (1). While these proto-splice sites are also overrepresented in phase 0, this unequal distribution is not sufficient to fully explain the phase bias observed for major introns (37,38). Further, the proto-splice site hypothesis fails to address the significant bias against phase 0 that is observed for minor introns (Supplementary Figure S22). Instead, it has been suggested that the conversion of minor to major introns through the accumulation of sequential point mutations at key nucleotides in minor intron splice sites has contributed to the underrepresentation of minor introns in phase 0 (29). In fact, acquisition of only three point mutations in the splice sites of the minor intron in P120 (now known as NOP2) is sufficient to change its dependency from the minor to major spliceosome (20). The identification of orthologous intron pairs in distant species, in which intron classification is major in one species yet minor in another, has previously been used in support of a minor-to-major intron conversion hypothesis (18,39). Support for the idea that minor-like introns represent an intermediate in this intron class conversion stems from the fact that these introns are the only ones found in orthologous intron clusters containing both minor, minor-like, major-like and major introns (Figure 3B, C). This is not to say that there is a specific path of precise point mutations that must occur in order to achieve intron class conversion. In fact, we propose that random genetic drift might result in sufficient point mutations in the consensus sequences of minor introns to ultimately reduce their position weight matrix score enough to result in a reclassification as minor-like intron. While the specific nucleotide changes that lead to such an intron class switch may vary, they are less likely to include the -1 and -2 nucleotide of the 5′ splice site, as these are coding nucleotides, and might therefore induce amino acid changes. For example, human minor-like introns originating from minor introns, such as the one in MIOS, will continue to be biased against the A-2G-1, and therefore still rely on the minor spliceosome for their splicing (Figure 6F). In contrast, minor-like introns representing an intermediate in the conversion of major introns to minor introns, such as that in TM9SF1, originate from major introns and therefore possess the A-2G-1 (Figure 6E). The presence of these nucleotides make this intron less reliant on the minor spliceosome for its splicing, even though the remainder of the splicing motifs may already resemble that of a minor intron. As such, minor-like introns originating from major introns remain unresponsive to minor spliceosome inhibition. In all, our findings suggest that minor-like introns represent a natural snapshot capturing the conversion of minor introns into major introns and vice versa.
The minor spliceosome regulates a subset of minor-like and hybrid introns
Our intron classification scheme, combined with CLIP-seq and transcriptome data, has provided insight into the genetic determinants of the major and minor splicing reaction. First, our transcriptomic analysis shows the existence of many minor and minor-like introns that are highly dependent on the proper function of the minor spliceosome, though a subset of each intron type remains unresponsive to minor spliceosome inhibition. We found that the splicing of many minor-like and hybrid introns is affected in individuals with mutation in ZRSR2, in addition to the previously reported minor intron retention (Figure 5A, Supplementary Table S6). Since minor-like and hybrid introns contain features that are amenable to recognition by either spliceosome, it is unsurprising that they are affected by loss-of-function of ZRSR2, which participates in both spliceosomes, albeit with different roles (53). Keeping with this idea, the splicing of many minor-like and hybrid introns was also affected upon inhibition of RBM48 in Zea mays, which, like ZRSR2, has protein interactions in both the major and minor spliceosome (Figure 5E) (38). Thus, our transcriptomic analysis points to a role for both spliceosomes in the mechanism of minor-like intron splicing.
Minor-like introns possess splice site sequence elements that are largely similar to those of minor introns (Figure 4A). However, the presence of a guanine at the –1 position of the 5′ splice site and +1 position of the 3′ splice site are characteristic features of major introns (Figure 4A). It has been proposed that guanines at these two positions can be leveraged by U5 snRNA to stabilize the pre-catalytic major spliceosome in the presence of a suboptimal intronic splice site sequence (69). This allows for the possibility that minor-like introns possessing a guanine at the –1 position can be spliced by the major spliceosome. In agreement with this hypothesis, we found that minor-like introns with a –1G were not responsive to minor spliceosome inhibition, while responsive minor-like introns had a significant bias against this –1G at the 5′ splice site (Figure 6A). In addition, we found that the majority of unresponsive minor-like introns were bound by major spliceosome component U2AF1 (Figure 6B). These findings suggests that the more abundant major spliceosome may normally compete for the splicing of minor-like introns with the less abundant minor spliceosome (70). Notwithstanding, in the absence of the –1G nucleotide there are insufficient Watson-Crick interactions to support splicing of the minor-like intron by the major spliceosome. Consequently, these minor-like introns would most likely depend on the minor spliceosome for their splicing.
In all, our intron classification strategy has provided insight into the evolution of minor introns through the identification of minor-like and hybrid introns. We have integrated this data into the Minor Intron Database (midb.pnb.uconn.edu) and hope that this resource will aid in expanding the study of splicing in non-model organisms. Moreover, we believe that the creation of a comprehensive list of the introns whose splicing is responsive to minor spliceosome inhibition will aid in the understanding of the molecular pathogenesis of spliceosomopathies (71).
Supplementary Material
Acknowledgements
We would like to acknowledge Dr Ion Mandoiu from the Computer Science Engineering Department of the University of Connecticut for creating the necessary infrastructure to perform all bioinformatics analysis. Moreover, we would like to thank Dr Bansal from the Computer Science Engineering Department and Dr Baumgartner from the Genetics department of Yale University for their valuable feedback on the evolution analysis. We would like to acknowledge the generosity of Dr Mazoyer from the Centre de Recherche en Neurosciences de Lyon, who shared the transcriptomic data obtained from MOPD1 patients (32). Finally, we would like to thank Emma Schwoerer and Nick Mcintosh for providing helpful feedback on the design and construction of MIDB.
Author contributions: Conceptualization – A.M.O., R.N.K, and C.F.S.; methodology – A.M.O., C.F.S. and R.N.K.; software – C.F.S., A.M.O., A.W.; investigation – A.M.O., C.F.S., I.A., K.N.G., S.M., J.K.H., A.C., J.B. and R.N.K.; resources – J.K.H., J.B., R.N.K.; statistics – T.E.M.; writing – A.M.O., K.N.G. and R.N.K.; supervision – R.N.K.; funding acquisition – R.N.K.
Contributor Information
Anouk M Olthof, Physiology and Neurobiology Department, University of Connecticut, Storrs, CT, USA.
Charles F Schwoerer, Physiology and Neurobiology Department, University of Connecticut, Storrs, CT, USA.
Kaitlin N Girardini, Physiology and Neurobiology Department, University of Connecticut, Storrs, CT, USA.
Audrey L Weber, Physiology and Neurobiology Department, University of Connecticut, Storrs, CT, USA.
Karen Doggett, Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia.
Stephen Mieruszynski, Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia.
Joan K Heath, Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia.
Timothy E Moore, Statistical Consulting Services, Center for Open Research Resources & Equipment, University of Connecticut, Storrs, CT, USA.
Jakob Biran, Department of Poultry and Aquaculture, Institute of Animal Science, Agricultural Research Organization, Rishon LeTsiyon, Israel.
Rahul N Kanadia, Physiology and Neurobiology Department, University of Connecticut, Storrs, CT, USA; Institute for Systems Genomics, University of Connecticut, Storrs, CT, USA.
Data availability
The intron classification for all 263 genomes can be found on the Minor Intron Database (MIDB), and can be accessed at midb.pnb.uconn.edu. Additionally, the database tables have been archived on FigShare at: https://figshare.com/projects/Taxonomy_of_introns_and_the_evolution_of_minor_introns/167411. The transcriptomic datasets generated and analyzed during this study are available in the Gene Expression Omnibus. Accession numbers for individual datasets can be found in Supplementary Table S3. Bioinformatics pipelines used to classify intron types, as well as evaluate their splicing, can be found on the Github repositories: https://github.com/amolthof/IntronClassification and https://github.com/amolthof/minor-intron-retention. The relevant code has beenarchived on Figshare and can be found at the following URL: https://figshare.com/account/home#/projects/167411 with the following permanent DOI: https://doi.org/10.6084/m9.figshare.26021926.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institute of Neurological Disorders and Stroke [R01NS102538 and R21NS096684 to R.N.K.]; Prostate Cancer Foundation [Igor Tulchinksy-Leerom Segal-PCF Challenge Award to R.N.K.]. Funding for open access charge: Igor Tulchinksy-Leerom Segal-PCF Challenge Award from the Prostate Cancer Foundation.
Conflict of interest statement. None declared.
References
- 1. Rogozin I.B., Carmel L., Csuros M., Koonin E.V.. Origin and evolution of spliceosomal introns. Biol. Direct. 2012; 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Csuros M., Rogozin I.B., Koonin E.V.. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput. Biol. 2011; 7:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frumkin I., Yofe I., Bar-Ziv R., Gurvich Y., Lu Y.-Y., Voichek Y., Towers R., Schirman D., Krebber H., Pilpel Y.. Evolution of intron splicing towards optimized gene expression is based on various Cis- and trans-molecular mechanisms. PLoS Biol. 2019; 17:e3000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parenteau J., Durand M., Véronneau S., Lacombe A.-A., Morin G., Guérin V., Cecez B., Gervais-Bird J., Koh C.-S., Brunelle D.et al.. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol. Biol. Cell. 2008; 19:1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rogozin I.B., Wolf Y.I., Sorokin A.V., Mirkin B.G., Koonin E.V.. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003; 13:1512–1517. [DOI] [PubMed] [Google Scholar]
- 6. Vosseberg J., Snel B.. Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery. Biol. Direct. 2017; 12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marz M., Kirsten T., Stadler P.F.. Evolution of spliceosomal snRNA genes in metazoan animals. J. Mol. Evol. 2008; 67:594–607. [DOI] [PubMed] [Google Scholar]
- 8. López M.D., Alm Rosenblad M., Samuelsson T.. Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res. 2008; 36:3001–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tarn W.Y., Steitz J.A.. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996; 84:801–811. [DOI] [PubMed] [Google Scholar]
- 10. Tarn W.Y., Steitz J.A.. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996; 273:1824–1832. [DOI] [PubMed] [Google Scholar]
- 11. Patel A.A., Steitz J.A.. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003; 4:960–970. [DOI] [PubMed] [Google Scholar]
- 12. Larue G.E., Eliáš M., Roy S.W.. Expansion and transformation of the minor spliceosomal system in the slime mold physarum polycephalum. Curr. Biol. 2021; 31:3125–3131. [DOI] [PubMed] [Google Scholar]
- 13. Bartschat S., Samuelsson T.. U12 type introns were lost at multiple occasions during evolution. Bmc Genomics [Electronic Resource]. 2010; 11:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Russell A.G., Charette J.M., Spencer D.F., Gray M.W.. An early evolutionary origin for the minor spliceosome. Nature. 2006; 443:863–866. [DOI] [PubMed] [Google Scholar]
- 15. Burge C.B., Padgett R.A., Sharp P.A.. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998; 2:773–785. [DOI] [PubMed] [Google Scholar]
- 16. Baumgartner M., Drake K., Kanadia R.N.. An integrated model of Minor intron emergence and conservation. Front. Genet. 2019; 10:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coulombe-Huntington J., Majewski J.. Characterization of intron loss events in mammals. Genome Res. 2007; 17:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dietrich R.C., Incorvaia R., Padgett R.A.. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell. 1997; 1:151–160. [DOI] [PubMed] [Google Scholar]
- 19. Alioto T.S. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic. Acids. Res. 2007; 35:D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adl S.M., Bass D., Lane C.E., Lukeš J., Schoch C.L., Smirnov A., Agatha S., Berney C., Brown M.W., Burki F.et al.. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019; 66:4–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olthof A.M., Hyatt K.C., Kanadia R.N.. Minor intron splicing revisited: identification of new minor intron-containing genes and tissue-dependent retention and alternative splicing of minor introns. Bmc Genomics [Electronic Resource]. 2019; 20:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhuang Y., Weiner A.M.. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986; 46:827–835. [DOI] [PubMed] [Google Scholar]
- 23. Kandels-Lewis S., Séraphin B.. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993; 262:2035–2039. [DOI] [PubMed] [Google Scholar]
- 24. Kolossova I., Padgett R.A.. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA. 1997; 3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 25. Incorvaia R., Padgett R.A.. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA. 1998; 4:709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwon Y.-S., Jin S.W., Song H.. Global analysis of binding sites of U2AF1 and ZRSR2 reveals RNA elements required for mutually exclusive splicing by the U2- and U12-type spliceosome. Nucleic. Acids. Res. 2024; 52:1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeng Y., Fair B.J., Zeng H., Krishnamohan A., Hou Y., Hall J.M., Ruthenburg A.J., Li Y.I., Staley J.P.. Profiling lariat intermediates reveals genetic determinants of early and late co-transcriptional splicing. Mol. Cell. 2022; 82:4681–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olthof A.M., White A.K., Mieruszynski S., Doggett K., Lee M.F., Chakroun A., Abdel Aleem A.K., Rousseau J., Magnani C., Roifman C.M.et al.. Disruption of exon-bridging interactions between the minor and major spliceosomes results in alternative splicing around minor introns. Nucleic Acids Res. 2021; 49:3524–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Wolf B., Oghabian A., Akinyi M.V., Hanks S., Tromer E.C., van Hooff J.J.E., van Voorthuijsen L., van Rooijen L.E., Verbeeren J., Uijttewaal E.C.H.et al.. Chromosomal instability by mutations in the novel minor spliceosome component CENATAC. EMBO J. 2021; 40:e106536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merico D., Roifman M., Braunschweig U., Yuen R.K.C., Alexandrova R., Bates A., Reid B., Nalpathamkalam T., Wang Z., Thiruvahindrapuram B.et al.. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat. Commun. 2015; 6:8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cologne A., Benoit-Pilven C., Besson A., Putoux A., Campan-Fournier A., Bober M.B., De Die-Smulders C.E.M., Paulussen A.D.C., Pinson L., Toutain A.et al.. New insights into minor splicing-a transcriptomic analysis of cells derived from TALS patients. RNA. 2019; 25:1130–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y.et al.. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015; 6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baumgartner M., Olthof A.M., Aquino G.S., Hyatt K.C., Lemoine C., Drake K., Sturrock N., Nguyen N., Al Seesi S., Kanadia R.N. Minor spliceosome inactivation causes microcephaly, owing to cell cycle defects and death of self-amplifying radial glial cells. Development. 2018; 145:dev166322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drake K.D., Lemoine C., Aquino G.S., Vaeth A.M., Kanadia R.N.. Loss of U11 small nuclear RNA in the developing mouse limb results in micromelia. Development. 2020; 147:dev190967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L., Ding Z., Pang T.-L., Zhang B., Li C.-H., Liang A.-M., Wang Y.-R., Zhou Y., Fan Y.-J., Xu Y.-Z.. Defective minor spliceosomes induce SMA-associated phenotypes through sensitive intron-containing neural genes in Drosophila. Nat. Commun. 2020; 11:5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gault C.M., Martin F., Mei W., Bai F., Black J.B., Barbazuk W.B., Settles A.M.. Aberrant splicing in maize rough endosperm3 reveals a conserved role for U12 splicing in eukaryotic multicellular development. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2195–E2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bai F., Corll J., Shodja D.N., Davenport R., Feng G., Mudunkothge J., Brigolin C.J., Martin F., Spielbauer G., Tseung C.-W.et al.. RNA binding motif protein 48 is required for U12 splicing and maize endosperm differentiation. Plant Cell. 2019; 31:715–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Markmiller S., Cloonan N., Lardelli R.M., Doggett K., Keightley M.-C., Boglev Y., Trotter A.J., Ng A.Y., Wilkins S.J., Verkade H.et al.. Minor class splicing shapes the zebrafish transcriptome during development. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parada G.E., Munita R., Cerda C.A., Gysling K.. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 2014; 42:10564–10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R.. Comprehensive splice-site analysis using comparative genomics. Nucleic. Acids. Res. 2006; 34:3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burki F., Roger A.J., Brown M.W., Simpson A.G.B.. The new tree of eukaryotes. Trends Ecol. Evol. 2020; 35:43–55. [DOI] [PubMed] [Google Scholar]
- 44. Moyer D.C., Larue G.E., Hershberger C.E., Roy S.W., Padgett R.A.. Comprehensive database and evolutionary dynamics of U12-type introns. Nucleic Acids Res. 2020; 48:7066–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szcześniak M.W., Kabza M., Pokrzywa R., Gudyś A., Makałowska I.. ERISdb: a database of plant splice sites and splicing signals. Plant Cell Physiol. 2013; 54:e10. [DOI] [PubMed] [Google Scholar]
- 46. Larue G.E., Roy S.W.. Where the minor things are: a pan-eukaryotic survey suggests neutral processes may explain much of minor intron evolution. Nucleic Acids Res. 2023; 51:10884–10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iben J.R., Maraia R.J.. tRNA gene copy number variation in humans. Gene. 2014; 536:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mabin J.W., Lewis P.W., Brow D.A., Dvinge H.. Human spliceosomal snRNA sequence variants generate variant spliceosomes. RNA. 2021; 27:1186–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rogozin I.B., Lyons-Weiler J., Koonin E.V.. Intron sliding in conserved gene families. Trends Genet. 2000; 16:430–432. [DOI] [PubMed] [Google Scholar]
- 50. Roy S.W., Gilbert W.. The pattern of intron loss. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin C.-F., Mount S.M., Jarmołowski A., Makałowski W.. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol. Biol. 2010; 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mount S.M., Pettersson I., Hinterberger M., Karmas A., Steitz J.A.. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell. 1983; 33:509–518. [DOI] [PubMed] [Google Scholar]
- 53. Shen H., Zheng X., Luecke S., Green M.R.. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010; 24:2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gozani O., Potashkin J., Reed R.. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 1998; 18:4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gao K., Masuda A., Matsuura T., Ohno K.. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008; 36:2257–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oghabian A., Greco D., Frilander M.J.. IntEREst: intron-exon retention estimator. BMC Bioinf. 2018; 19:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fedorov A., Suboch G., Bujakov M., Fedorova L.. Analysis of nonuniformity in intron phase distribution. Nucleic Acids Res. 1992; 20:2553–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Lührmann R.. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009; 36:593–608. [DOI] [PubMed] [Google Scholar]
- 59. Collins L., Penny D.. Complex spliceosomal organization ancestral to extant eukaryotes. Mol. Biol. Evol. 2005; 22:1053–1066. [DOI] [PubMed] [Google Scholar]
- 60. Katinka M.D., Duprat S., Cornillot E., Méténier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P.et al.. Genome sequence and gene compaction of the eukaryote parasite encephalitozoon cuniculi. Nature. 2001; 414:450–453. [DOI] [PubMed] [Google Scholar]
- 61. Stark M.R., Dunn E.A., Dunn W.S.C., Grisdale C.J., Daniele A.R., Halstead M.R.G., Fast N.M., Rader S.D.. Dramatically reduced spliceosome in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1191–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shukla G.C., Padgett R.A.. Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA. 1999; 5:525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bai R., Wan R., Wang L., Xu K., Zhang Q., Lei J., Shi Y.. Structure of the activated human minor spliceosome. Science. 2021; 371:eabg0879. [DOI] [PubMed] [Google Scholar]
- 64. Bai R., Yuan M., Zhang P., Luo T., Shi Y., Wan R.. Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome. Science. 2024; 383:1245–1252. [DOI] [PubMed] [Google Scholar]
- 65. Shukla G.C., Padgett R.A.. U4 small nuclear RNA can function in both the major and minor spliceosomes. Proc. Natl. Acad. Sci. USA. 2004; 101:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Will C.L., Schneider C., Reed R., Lührmann R.. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999; 284:2003–2005. [DOI] [PubMed] [Google Scholar]
- 67. Hudson A.J., Moore A.N., Elniski D., Joseph J., Yee J., Russell A.G.. Evolutionarily divergent spliceosomal snRNAs and a conserved non-coding RNA processing motif in Giardia lamblia. Nucleic. Acids. Res. 2012; 40:10995–11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Singh J., Sikand K., Conrad H., Will C.L., Komar A.A., Shukla G.C.. U6atac snRNA stem-loop interacts with U12 p65 RNA binding protein and is functionally interchangeable with the U12 apical stem-loop III. Sci. Rep. 2016; 6:31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Artemyeva-Isman O.V., Porter A.C.G.. U5 snRNA interactions with exons ensure splicing precision. Front. Genet. 2021; 12:676971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Montzka K.A., Steitz J.A.. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. USA. 1988; 85:8885–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Olthof A.M., White A.K., Kanadia R.N.. The emerging significance of splicing in vertebrate development. Development. 2022; 149:dev200373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The intron classification for all 263 genomes can be found on the Minor Intron Database (MIDB), and can be accessed at midb.pnb.uconn.edu. Additionally, the database tables have been archived on FigShare at: https://figshare.com/projects/Taxonomy_of_introns_and_the_evolution_of_minor_introns/167411. The transcriptomic datasets generated and analyzed during this study are available in the Gene Expression Omnibus. Accession numbers for individual datasets can be found in Supplementary Table S3. Bioinformatics pipelines used to classify intron types, as well as evaluate their splicing, can be found on the Github repositories: https://github.com/amolthof/IntronClassification and https://github.com/amolthof/minor-intron-retention. The relevant code has beenarchived on Figshare and can be found at the following URL: https://figshare.com/account/home#/projects/167411 with the following permanent DOI: https://doi.org/10.6084/m9.figshare.26021926.