Abstract
AGPAT (1-acyl-sn-glycerol 3-phosphate acyltransferase) exists in at least five isoforms in humans, termed as AGPAT1, AGPAT2, AGPAT3, AGPAT4 and AGPAT5. Although they catalyse the same biochemical reaction, their relative function, tissue expression and regulation are poorly understood. Linkage studies in humans have revealed that AGPAT2 contributes to glycerolipid synthesis and plays an important role in regulating lipid metabolism. We report the molecular cloning, tissue distribution, and enzyme characterization of mAGPATs (murine AGPATs) and regulation of cardiac mAGPATs by PPARα (peroxisome-proliferator-activated receptor α). mAGPATs demonstrated differential tissue expression profiles: mAGPAT1 and mAGPAT3 were ubiquitously expressed in most tissues, whereas mAGPAT2, mAGPAT4 and mAGPAT5 were expressed in a tissue-specific manner. mAGPAT2 expressed in in vitro transcription and translation reactions and in transfected COS-1 cells exhibited specificity for 1-acyl-sn-glycerol 3-phosphate. When amino acid sequences of five mAGPATs were compared, three highly conserved motifs were identified, including one novel motif/pattern KX2LX6GX12R. Cardiac mAGPAT activities were 25% lower (P<0.05) in PPARα null mice compared with wild-type. In addition, cardiac mAGPAT activities were 50% lower (P<0.05) in PPARα null mice fed clofibrate compared with clofibrate fed wild-type animals. This modulation of AGPAT activity was accompanied by significant enhancement/reduction of the mRNA levels of mAGPAT3/mAGPAT2 respectively. Finally, mRNA expression of cardiac mAGPAT3 appeared to be regulated by PPARα activation. We conclude that cardiac mAGPAT activity may be regulated by both the composition of mAGPAT isoforms and the levels of each isoform.
Keywords: 1-acyl-sn-glycerol 3-phosphate, acyltransferase, heart, murine, peroxisome-proliferator-activated receptor α (PPARα)
Abbreviations: AGPAT, 1-acyl-sn-glycerol 3-phosphate acyltransferase; hAGPAT, human AGPAT; mAGPAT, murine AGPAT; GPAT, glycerol-3-phosphate acyltransferase; LPCAT, lysophosphatidylcholine acyltransferase; LPEAT, lysophosphatidylethanolamine acyltransferase; LPGAT, lysophosphatidylglycerol acyltransferase; ORF, open reading frame; PA, phosphatidic acid; PPARα, peroxisome-proliferator-activated receptor α; RT, reverse transcriptase; WCE, whole cell extraction
INTRODUCTION
Mammalian AGPAT (1-acyl-sn-glycerol 3-phosphate acyltransferase; EC 2.3.1.51), also known as lysophosphatidic acid acyltransferase, catalyses the acylation of lysophosphatidic acid to form PA (phosphatidic acid), the precursor of all glycerolipids [1]. Lysophosphatidic acid and PA are both important lipid-signalling molecules involved in a variety of cellular processes, such as angiogenesis, atherosclerosis, inflammation and oncogenesis [2–8]. PA can also be hydrolysed by phosphatidic acid phosphohydrolase to yield diacylglycerol, which is a critical second messenger mediating intracellular signalling [9–13].
Mammalian organs contain several isoforms of AGPATs. In humans, at least 5 isoforms, namely hAGPAT1, hAGPAT2, hAGPAT3, hAGPAT4, hAGPAT5 (or hAGPAT-α, -β, -γ, -δ, -ε) have been described [14–17]. The physiological significance for the existence of more than one form of the enzyme, however, remains to be determined. AGPAT1 and AGPAT2 in human and AGPAT1 in mice have been cloned and characterized [15–19]. By linkage analysis, the mutations in hAGPAT2 have been proven to be one of the causes of congenital generalized lipodystrophy [20–23]. Recently, the involvement of hAGPAT2 in tumour proliferation and the specific inhibition of this enzyme leading to tumour cell apoptosis have also been demonstrated [24,25]. Thus the regulation of AGPATs appears to be critical in the generation of signalling molecules and in the development of human disorders, such as lipodystrophies, diabetes, atherosclerosis, obesity and cancer.
PPARs (peroxisome-proliferator-activated receptors) belong to the steroid/thyroid/retinoic acid receptor super family and act as ligand-dependent transcription factors [26–28]. The transcription of target genes by ligand-activated receptors depends on interactions between PPARs and their obligatory partner retinoid X receptor, RXR [28]. Three isotypes of PPAR have been identified: PPAR-α, -β and -γ. The importance of PPAR-α in lipid metabolism has been illustrated by the transcriptional regulation of lipid metabolizing enzymes [29–33]. Rodent heart contains PPAR-α, and the effect of the PPARα agonist, ethyl-p-chlorophenoxyisobutyrate (clofibrate), on cardiac fatty acid and phospholipid metabolism in rodent heart is well documented [31,34].
Recently, bioinformatic and molecular biological approaches have been shown to facilitate greatly the discovery and cloning of genes encoding AGPATs. However, a systemic study of all family members of cardiac AGPATs in rodents has not been reported. As the first step to elucidate the biological functions of each AGPAT isoform and their potential regulation, we have identified, cloned and characterized several mAGPAT (murine AGPAT) isoforms including mAGPAT2, mAGPAT3, mAGPAT4 and mAGPAT5, using bioinformatic data-mining, and molecular and biochemical approaches. We report the molecular cloning and functional expression of recombinant mAGPAT2 in vitro and in vivo. In addition, we characterized the tissue expression pattern of mAGPAT mRNAs, and the regulation of mAGPAT activity and mRNA expression in murine heart. Finally, we show that at least one of these isoforms, mAGPAT3, is regulated by PPARα activation.
MATERIALS AND METHODS
Materials
Dulbecco's modified Eagle's medium, PBS and other chemicals were obtained from Sigma (St. Louis, MO, U.S.A.). [35S]methionine (>1.0 Ci/mmol) at 10 mCi/ml and [1-14C]oleoyl-CoA (56 μCi/μl) in 0.001 M sodium acetate and [1-14C]oleic acid (52 μCi/μM) in ethanol were purchased from Amersham Biosciences (Bucks., U.K.). TLC plates (K6 Silica Gel 60 A) were products of Whatman International (Maidstone, Kent, U.K.).
Animals
Male C57BL/6N×Sv/129 mice were obtained from Central Animal Care Services (University of Manitoba, Winnipeg, Manitoba, Canada). PPARα null (–/–) mice were a gift from Dr F. Gonzalez (National Cancer Institute, NIH, Bethesda, MD, U.S.A.). Treatment of mice conformed to the guidelines of the Canadian Council on Animal Care. The mice were maintained on rat chow and water ad libitium. All animals were kept in identical housing units on a cycle of 12 h of light and 12 h of darkness. In the treatment group, mice were fed 0.5% clofibrate in a chow diet for 14 days.
Isolation of total RNA
Total RNA was extracted from murine organs using TRIzol® reagent (Invitrogen, Burlington, ON, Canada) [35]. Briefly, minced organs were homogenized with 1 ml TRIzol® reagent for 1 min. The homogenates were poured into a microcentrifuge tube and 0.2 ml of chloroform was added and shaken for 30 s. The upper aqueous phase containing RNA was obtained by centrifugation at 10000 g for 15 min at 4 °C and transferred to a sterile tube. Subsequently, an equal volume of ice-cold propan-2-ol was added into the mixture. The solution was mixed well and stored at room temperature (23 °C) for 5 min. The RNA pellets were collected by centrifugation at 10000 g for 10 min at 4 °C followed by washing with 1 ml of 75% ice-cold ethanol. The total RNA was resuspended in double-distilled water. The purity and the yield of isolated RNA were determined by monitoring the absorbance at A260 and A280. The integrity of the RNA was confirmed by performing denaturing agarose gel electrophoresis on the isolated RNA samples [36].
PCR primers and RT (reverse transcriptase)–PCR conditions
The cDNA of each mAGPAT1, mAGPAT2, mAGPAT3, mAGPAT4 and mAGPAT5 was amplified with a pair of specific primers. The primers were designed using Vector NTI Suite software of InforMax (Vector NTI, version 7; Oxford, U.K.). All primers were synthesized by Invitrogen. Each pair of primers, the primer sites and the lengths of predicted PCR products and the GenBank® accession number for each primer design are summarized in Table 1.
Table 1. PCR primers.
| Primer name | Sequence | Product length (region) | GenBank® accession no. |
|---|---|---|---|
| mAGPAT1 (forward) | 5′-ACCAGAATGGAGCTGTGGCC-3′ | 846 bp (313–1158) | NM_018862 |
| mAGPAT1 (reverse) | 5′-CGCTCCCCCAGGCTTCTTCA-3′ | ||
| mAGPAT2 (forward) | 5′-CGCCGTCGGGGCTGGGGTGC-3′ | 861 bp (181–1041) | XM_130130 |
| mAGPAT2 (reverse) | 5′-CTGGGCTGGCAAGACCCCAG-3′ | ||
| mAGPAT3 (forward) | 5′-TGTTCTCAGTGAAGGACCGT-3′ | 1162 bp (70–1231) | NM_053014 |
| mAGPAT3 (reverse) | 5′-CTTAAGCTCTTGGTTGCCAT-3′ | ||
| mAGPAT4 (forward) | 5′-GATTTATCTCTTGAGAATCCCCACACC-3′ | 1161 bp (187–1347) | NM_026644 |
| mAGPAT4 (reverse) | 5′-GTCCGTTTGTTTCCGTTTGTTGTC-3′ | ||
| mAGPAT5 (forward) | 5′-AGAGGATGCTGCTGTCCCT-3′ | 1091 bp (721–1811) | XM_133988 |
| mAGPAT5 (reverse) | 5′-AACAAACCACAGGCAGCC-3′ | ||
| β-Actin (forward) | 5′-GTGGGGCGCCCCAGGCACCA-3′ | 540 bp | |
| β-Actin (reverse) | 5′-CTCCTTAATGTCACGCACGATTTG-3′ |
The first strand cDNA from 1 μg of total RNA was synthesized as described previously [31,36]. A typical reaction contained 150 units of MMLV (Moloney-murine-leukaemia virus) RT, 25 pmol of random hexamer primer, 20 units of RNase inhibitor, 1 mM dithiothreitol, and 10 pmol each of the four deoxynucleotides, in a total volume of 15 μl. The reaction was incubated at 37 °C for 1 h and terminated by boiling at 95 °C for 5 min. An aliquot of 1.0 μl of the resultant cDNA was used directly for each amplification reaction. PCR was performed in 20 μl of reaction mixture containing 8 pmol of each primer, 8 pmol of each deoxynucleotide and 0.4 unit Taq DNA polymerase. The mixture was overlaid with 20 μl of mineral oil to prevent evaporation. The reaction mixture was incubated in a PerkinElmer DNA Thermal Cycler and amplified for 25 cycles under the following conditions: AGPAT1 with denaturing for 1 min at 94 °C, annealing for 0.5 min at 62 °C and extension for 3 min at 72 °C; AGPAT2 with denaturing for 1 min at 94 °C and annealing and extension for 3 min at 72 °C; AGPAT3 and AGPAT4 with denaturing for 0.5 min at 94 °C, annealing for 0.5 min at 55 °C and extension for 2 min at 72 °C; AGPAT5 with denaturing for 1 min at 94 °C, annealing for 0.5 min at 58 °C and extension for 2 min at 72 °C; β-actin with denaturing for 1 min at 94 °C, annealing for 0.5 min at 55 °C and extension for 0.5 min at 72 °C.
Double-stranded DNA sequence analysis
The identity of each PCR product generated from RT–PCR using Taq or pfx DNA polymerase was verified by direct DNA sequencing. The gel fraction containing the DNA, including mAGPAT1, mAGPAT2, mAGPAT3, mAGPAT4 and mAGPAT5, was excised and purified using standard methods. Briefly, the agarose gel band with DNA fractions were cut under UV light, each fraction was placed into a Spin-X centrifuge tube equipped with a filter and 100 μl of water was added. The solution containing the DNA fraction was obtained by centrifugation at 11750 g for 5 min at 4 °C. The DNA in each fraction was extracted with phenol/chloroform, and the DNA in the aqueous fraction was allowed to precipitate in 1/10 vol. of 3 M sodium acetate (pH 5.5) and 2.5 vol. of 100% ethanol followed by a wash with 1.0 ml of 75% ethanol. The DNA was briefly air-dried and resuspended in water, and an aliquot was subjected to double-strained DNA sequencing analysis. The sequencing was performed using the PerkinElmer Applied Biosystems ABI310 Genetic Analyzer and the BigDye Terminator Cycle Sequencing Ready Reaction kit at the Institute of Cell Biology or Department of Microbiology, University of Manitoba, Winnipeg, Canada. The nucleotide sequences obtained were used for homologous search using BLAST to confirm the identity of the sequences.
Identification and construction of expression vectors for mAGPATs
Human AGPAT2 amino acid sequences were used to search against a non-redundant GenBank® BLASTP database. Several amino acid sequences were identified that showed significant similarity to hAGPAT2. One of those was mAGPAT2, which was later found to be identical with the sequence of mAGPAT2 (GenBank® accession no. XM_130130). The corresponding cDNA sequence was used to search against ResGen EST database, in which one of clones was identified to contain the mAGPAT2 coding sequence. The mAGPAT2 cDNA clone was then purchased from ResGen (Invitrogen). The double-stranded DNA sequencing revealed that the original clone (accession no. AA105947) contained the full-length ORF (open reading frame) of mAGPAT2, which was cloned into pBluescript SK −. Subsequently, the full-length ORF was subcloned into the BamHI and XhoI sites of pcDNA3.1 to construct the expression vector for mAGPAT2. The resulting fragment was then subcloned into pcDNA3.1 plasmid. The identity and orientation of the inserted sequence in pcDNA3.1 were confirmed by restriction enzyme digestion.
In vitro transcription and translation
The in vitro transcription and translation reactions were performed using a coupled transcription and translation system (TnT coupled Reticulocyte Lysate System; Promega). Reactions were performed according to the manufacturer's instructions as described previously [37,38]. TnT RNA polymerase T7 was used to set up a translation according to the promoter sequences available at the 5′-end of the inserted coding sequences. Two set-ups for the transcription and translation preparations were routinely used. These included [35S]methionine labelling to monitor the success of generation of enzyme protein and a non-radioactive reaction to generate enzyme protein for enzyme assay.
Cell culture and transfection
The COS-1 cell line, an SV40-transformed African green monkey cell line, was purchased from the A.T.C.C. (Rockville, MD, U.S.A.). These cells were maintained in culture flasks with Dulbecco's modified Eagle's medium containing 10% (v/v) fetal bovine serum, 100 units/ml penicillin G, 10 μg/ml streptomycin and 0.25 μg/ml amphotericin B.
The transfection was performed using Effectene Transfection Reagent (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Briefly, cells were seeded in 10 cm dishes at (3–5)×105 cells per dish and cultured overnight until dishes were 40% confluent on the day of transfection. The transfection mixture was prepared according to the manufacturer's instructions, and then a portion of fresh medium was added. Finally, an aliquot of 3 ml per dish of the above mixture was added dropwise into the medium and dishes were gently swirled to ensure uniform distribution of the DNA–Effectene complexes.
Preparation of WCE (whole cell extraction) and protein determination
The transfected cells were maintained for 48 h and then harvested. The whole cell extracts were prepared by sonication of cell pellets on ice for three rounds (30 s/round) in a buffer containing 100 mM Tris/HCl (pH 7.4), 5 mM NaCl, 3 mM MgCl2 and 1 unit of protein inhibitors (proteinase inhibitor cocktail tablets were from Roche Diagnositcs GmbH). Subsequently, the cell lysate was centrifuged at 600 g for 10 min to remove cell debris and the resultant supernatant was designated as WCE. The protein concentration of WCE was determined by bicinchoninic acid assay (BCA Assay kit from Pierce) using BSA as a standard. An aliquot of 20–30 μg of protein was used to determine acyltransferase activity as described in the following section.
Western-blot analysis
Samples containing 30–50 μg of WCE protein from transfected COS-1 cells were subjected to SDS/PAGE (8% gel) with prestained protein markers as references as described previously [31]. Briefly, protein fractions in the gel were transferred on to PVDF membranes. The membrane was then incubated with the monoclonal anti-V5 antibodies (1:1000 dilution; Invitrogen). The membranes were then washed and incubated with peroxidase-labelled secondary antibodies. The protein bands were detected on film using a Western blotting detection reagent kit. The relative intensities of protein bands were acquired using Bio-Rad Multi-Imaging System (Bio-Rad Laboratories, Hercules, CA, U.S.A.).
Assay of acyltransferase activities
The activities of GPAT (glycerol-3-phosphate acyltransferase), AGPAT, LPCAT (lysophosphatidylcholine acyltransferase), LPEAT (lysophosphatidylethanolamine acyltransferase) and LPGAT (lysophosphatidylglycerol acyltransferase) were determined in reticulocyte lysates containing in vitro translated enzymes and in cell extracts from transient transfected cells with corresponding plasmid DNA. For in vitro translated enzymes, the assay mixture consisted of 100 mM Tris/HCl (pH 7.4), 3 mM MgCl2, 10 μl of 0.56 μCi [14C]oleoyl-CoA, 10 μl of reticulocyte lysate, 100 μM of either sn-glycerol 3-phosphate for GPAT or 1-oleoyl-sn-glycerol 3-phosphate for AGPAT or 1-oleoyl-sn-glycerol 3-phosphorylcholine for LPCAT or 1-palmitolyl-sn-glycerol 3-phosphorylethanolamine for LPEAT or 1-acyl-sn-glycerol 3-phosphorylglycerol for LPGAT, in a total volume of 100 μl. The reaction was incubated at 37 °C for 20 min. At the end of incubation, the reaction was terminated by the addition of 0.62 ml of methanol and 0.8 ml of chloroform. To facilitate extraction and phase separation, a solution containing 3 μl of saturated NaCl and 300 μl of Tris/HCl (pH 7.4) was added to the mixture. The labelled phospholipid in the lower phase was resolved by TLC in a solvent system consisting of chloroform/ethanol/acetic acid/water (25:10:3:1, by vol.). The lipid standard on the thin-layer chromatographic plate was visualized by iodine vapour and the density of radioactive bands were scanned and quantified by a Molecular Imager FX equipped with QuantityOne software (Bio-Rad Laboratories) or visualized by exposure to Kodak X-OMAT X-ray film.
Cardiac perfusion studies
Mice were fed ±0.5% clofibrate in a chow diet for 14 days and on the following day hearts were removed and perfused for 30 min in the Langendorff mode [39] with Krebs–Henseleit buffer [40] containing 0.1 mM [1-14C]oleic acid (1 μCi/heart) bound to albumin (1:1 molar ratio). The organic phase was isolated and radioactivity incorporated into PA was determined as described in [41].
Statistical analysis
The data were analysed by a two-tail independent Student's t test. For differences in the mean values among multiple groups, oneway ANOVA and Student–Newman–Keuls method were employed (SigmaStat for Windows version 1.0). In all cases, the level of significance was defined as P<0.05. Results are presented as the means±S.D.
RESULTS
AGPAT exists as at least five isoforms in humans, termed as AGPAT1, AGPAT2, AGPAT3, AGPAT4 and AGPAT5. In addition, AGPAT1 and AGPAT2 in human and AGPAT1 in mice have been cloned and characterized [15–19]. However, to date a full identification of mAGPAT2, its tissue expression and regulation have not been examined. In addition, the expression of mAGPAT3, mAGPAT4 and mAGPAT5 isoforms have not been examined.
The mAGPAT2 gene locus, mRNA and enzyme protein
The completed ORF of mAGPAT2 cDNA was amplified and cloned using mouse liver total RNA and RT–PCR. The PCR products were subsequently purified and sequenced. In parallel, a 1.8-kb cDNA clone corresponding to the single copy gene AGPAT2, located on murine chromosome 2, was purchased and sequenced. Sequences of these two sources of cDNAs were mapped on to the mAGAT2 locus on murine chromosome 2A3 (GenBank® accession no. 1914762; LocusID: 67512). The mAGPAT2 genomic sequence is comprised of six exons in the genomic region spanning 11.4 kb. Using translation tool http://ca.expasy.org/tools/dna.html/, this 1.8 kb cDNA was found to contain a single long ORF, which was predicted to encode a polypeptide having 278 amino acids with a calculated molecular mass of 31.6 kDa. During the process of our characterization of this putative enzyme, two sequence records appeared in GenBank® that assigned AGPAT as the mAGPAT2 gene product (GenBank® accession no. XM_130130; LocusID: 67512). Our cDNA sequencing data confirmed that the above gene assigned by the automatic genome annotation was indeed the murine gene encoding mAGPAT2.
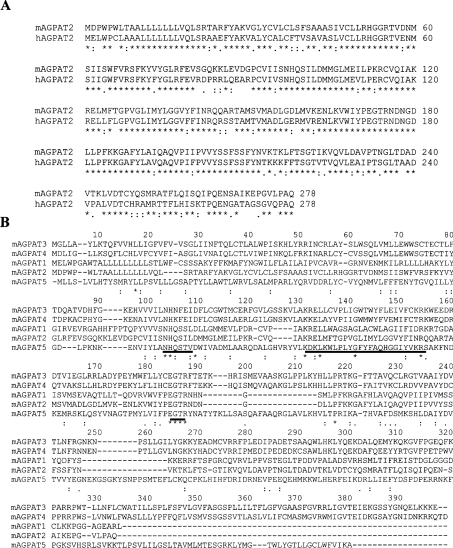
To examine the sequence similarities to the characterized human counterpart AGPAT2, protein sequence alignment was conducted using an on-line ClustalW alignment tool http://www.ebi.ac.uk/clustalw. The alignment revealed a significant sequence identity of 76% between the entire sequence of the mAGPAT2 and its human homologue (Figure 1A). The hydrophobicity plot of this 278-amino-acid polypeptide indicated the presence of four potential hydrophobic regions. Among these four regions, three were potential transmembrane helices: two at the N-terminus between positions 4 and 50 and one between positions 122 and 143 (http://www.cbs.dtu.dk/cgi-bin/, for the prediction of transmembrane helices). Subsequently, the subcellular localization of the encoded protein was analysed using protein subcellular localization prediction programs (PSORT II Server at http://psort.nibb.ac.jp/cgi-bin/ and TargetP server at http://www.cbs.dtu.dk/cgi-bin/). This protein was predicted to be localized in the endoplasmic reticulum. At its N-terminus, mAGPAT2 polypeptide contained a putative signal peptide with a predicted cleavage site between positions 45 and 46, which is consistent with hAGPAT2 described in a previous report [14] (http://www.cbs.dtu.dk/cgi-bin/, for a signal peptide and cleavage site prediction).
Figure 1. Amino acid sequences of mAGPAT2 and hAGPAT2, and alignment of predicted protein sequences of mAGPAT gene family members.
(A) Alignment of the amino acid residues from the complete amino acid sequences of mAGPAT2 and hAGPAT2 (GenBank® accession no. NM_006412). The amino acid sequence is shown in single-letter code. * indicates amino-acid identities of the aligned polypeptides. Numbering of amino acids begins with the first amino acid residue of predicted mAGPAT2. (B) The predicted protein sequences from five mAGPAT gene families were aligned with the ClustalW program (http://www.ebi.ac.uk). The conserved motifs are underlined and identical amino acid residues are indicated with an asterisk. GenBank® accession numbers are summarized in Table 1.
mAGPAT2 belongs to the AGPAT gene family
The amino acid sequence identity of the mAGPAT gene family members was investigated. Protein sequences of mAGPATs were aligned with the ClustalW alignment program (Figure 1B). The overall matches among mAGPATs ranged from 2 to 60%. While the overall matches between members varied, a core region containing conserved amino acid stretches was found within the catalytic domain. Two motifs that are critical for enzymic activities appeared to be conserved among all five mAGPATs [39,40]. These two motifs are NHX4D, which is involved in catalytic function and EGTR, which is involved in substrate recognition and binding. Interestingly, a novel conserved motif/pattern KX2LX6GX12R was apparent. This motif/pattern was found to lie between catalytic and substrate binding motifs. This conserved pattern might represent an unidentified motif for this gene family. The amino acid alignment scores of five members are shown in Table 2. According to their relative similarities, mAGPATs could be further subdivided into three subgroups. The alignment reveals that group I members (mAGPAT1 and mAGPAT2) and group II members (mAGPAT3 and mAGPAT4) share high percentage of sequence identities, whereas a group III member (mAGPAT5) remains as a singleton on the basis of a distinct sequence.
Table 2. Amino acid alignment scores of mAGPAT gene family members.
| Identity (%) | |||||
|---|---|---|---|---|---|
| Gene | mAGPAT1 | mAGPAT2 | mAGPAT3 | mAGPAT4 | mAGPAT5 |
| mAGPAT1 | − | 44 | 7 | 11 | 10 |
| mAGPAT2 | 44 | − | 9 | 9 | 2 |
| mAGPAT3 | 7 | 9 | − | 60 | 19 |
| mAGPAT4 | 11 | 9 | 60 | − | 17 |
| mAGPAT5 | 10 | 2 | 19 | 17 | − |
Functional expression of recombinant mAGPAT2 in vitro and in vivo
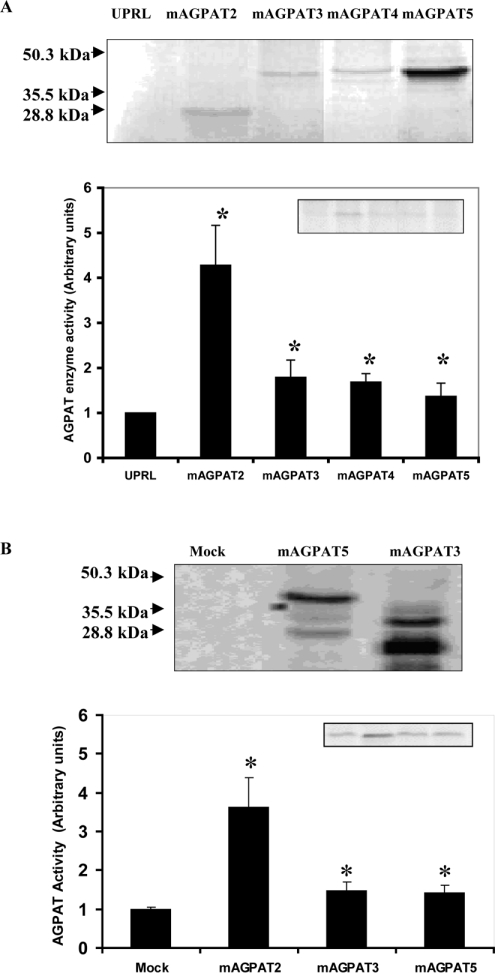
To examine whether our cloned mAGPAT2 cDNA encoded protein, mAGPAT proteins were expressed in two experimental systems. First, expression of the wild-type mAGPAT2 was tested in rabbit reticulocyte lysates using an in vitro transcription coupled translation system. Expression of in vitro translated mAGPAT2 enzyme protein was monitored through radiolabelling of newly synthesized protein with [35S]methionine. A major polypeptide of 31.6 kDa was present in the programmed lysates for mAGPAT2 (Figure 2A). Next, expression of the wild-type mAGPAT3, mAGPAT4 and mAGPAT5 were examined. A major polypeptide of 45.1 kDa was present in the programmed lysates for mAGPAT3, mAGPAT4 and mAGPAT5 (Figure 2A). As a control to monitor the expression of proteins, V5-His epitope tag fused to the C-terminus of mAGPAT3 and mAGPAT5 in the same expression vector pcDNA3.1 were produced and transfected into COS-1 cells. Cell lysates were then extracted and subjected to SDS/PAGE analysis. Specific tagged expressions of mAGPAT5 (Figure 2B, 45.1 kDa) and mAGPAT3 (Figure 2B, 35.5 and 28.8 kDa) were observed. Thus wild-type mAGPAT2 enzyme proteins could be produced under our experimental conditions.
Figure 2. Expression of mAGPAT in vitro and in vivo and enzyme activities of mAGPATs.
(A) Expression of mAGPAT2, mAGPAT3, mAGPAT4 and mAGPAT5 protein using the in vitro protein translation system. Reticulocyte lysates were prepared and proteins were [35S]methionine radiolabelled and visualized by exposure to X-ray film (upper panel). UPRL, unprocessed lysate. AGPAT enzyme activities were determined as described in the Materials and methods section (lower panel). (B) Expression of V5-His epitope tag fused to the C-terminus of mAGPAT3 and mAGPAT5 in COS-1 cells (upper panel). Appearance of the mAGPAT5 band is greater since the protein contains 13 methionine residues instead of 11 and 7 methionine residues in mAGPAT2 and mAGPAT3 respectively. Molecular mass markers are indicated on the left. An equal amount of protein from cell lysate was used to determine relative mAGPAT enzyme activitities as described in the Materials and methods section (lower panel).
The functional expression of mAGPAT isoform enzyme activities in both the in vitro and in vivo systems was then determined. In the programmed reticulocyte lysates, mAGPAT2 relative enzyme activity was >4-fold (4.28±0.88, P<0.05) over the control level of unprogrammed reticulocyte lysates, measured by the relative amount of PA produced by the enzyme reaction (Figure 2A, histogram). The other mAGPAT isoforms had lower activities but were increased (1.4- to 1.8-fold, P<0.05) when compared with control under identical experimental conditions. To verify the AGPAT activities of mAGPATs, the activities of the wild-type enzyme proteins were examined in COS-1 cells transfected with the mammalian expression vector pcDNA3.1 containing the ORF. As shown in Figure 2(B) (histogram), the AGPAT activities in the whole cell extracts from pcDNA3.1-mAGPAT2, -mAGPAT3 and -mAGPAT5 transfected cells were increased 2.5-fold (P<0.05), 48% (P<0.05) and 41% (P<0.05) respectively when compared with mock-transfected controls. mAGPAT4 activity was not observed under these conditions. Thus these mAGPAT isoforms had AGPAT activity.
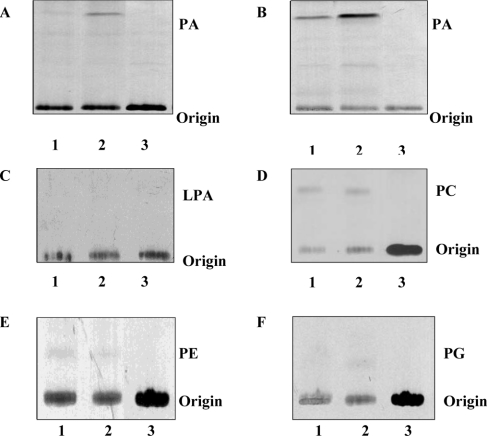
We next monitored enzyme activity and substrate specificity of mAGPAT2. mAGPAT enzyme activity was determined by the transfer of [1-14C]oleate from [1-14C]oleoyl-CoA to its unlabelled acceptor 1-oleoyl-sn-glycerol 3-phosphate to yield radioactive PA. Relative enzyme activity was determined in the in vitro transcription coupled translation system in reticulocyte lysates or in COS-1 cells transfected with mAGPAT2. The mAGPAT activity observed was greater in both in vitro and in vivo systems expressing mAGPAT2 (Figures 3A and 3B, lane 2) than the endogenous formation of radioactive PA (Figures 3A and 3B, lane 1). Thus the cloned mAGPAT2 exhibits functional AGPAT enzyme activity.
Figure 3. Specificity of mAGPAT2 activity.
AGPAT activities of mAGPAT2 from in vitro (A) and in vivo (B) sources. The film exposure time was 24 h for (A) and (B). (A) Lane 1, unprogrammed reticulocyte lysate; lane 2, reticulocyte lysate containing mAGPAT2; lane 3, [1-14C]oleoyl-CoA. (B) Lane 1, COS-1 cell lysate; lane 2, COS-1 cell lysate from mAGPAT2 transfected cells; lane 3, [1-14C]oleoyl-CoA. (C) GPAT activities in reticulocyte lysates. (D) LPCAT activities in reticulocyte lysates. (E) LPEAT activities in reticulocyte lysates. (F) LPGAT activities in reticulocyte lysates. Lane 1, unprogrammed reticulocyte lysate. Lane 2, reticulocyte lysate containing mAGPAT2. Lane 3, radiolabelled substrate [1-14C]oleoyl-CoA. The film was exposed for 72 h. Representative autoradiographs are depicted.
Substrate specificity of the cloned mAGPAT2 was then determined in the in vitro transcription coupled translation system in reticulocyte lysates using various lysophospholipids as substrates. GPAT, LPCAT, LPEAT and LPGAT activities were low in reticulocyte lysates indicating little endogenous acyltransferase activity (Figures 3C–3F, lanes 1). When lysates containing mAGPAT2 protein were used as enzyme sources, the acyltransferase activities for GPAT, LPCAT, LPEAT and LPGAT were not increased above endogenous activities indicating specificity of mAGPAT2 for 1-acyl-sn-glycerol 3-phosphate (Figures 3C–3F, lane 2 versus lane 1). Thus mAGPAT2 exhibits specificity for 1-acyl-sn-glycerol 3-phosphate consistent with the observed activities of hAGPAT2 [14–16].
Expression pattern of mAGPAT mRNAs in murine tissues
The validation of the quantitative measurement for each mAGPAT mRNA by RT–PCR was performed using an approach described previously [31]. Briefly, under the RT–PCR conditions used, the levels of the PCR products were dependent on the amount of templates used in the reaction. At least an 8–10-fold difference of cDNA levels of each mAGPAT was detected (results not shown). The size of each PCR product was consistent with the predicted size, and the identity of each PCR product was also confirmed by cloning and DNA sequencing analyses.
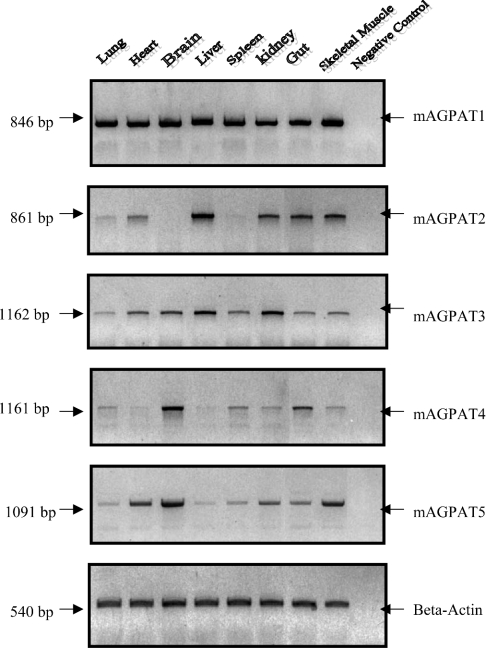
At least five isoforms have been identified and postulated as candidate members of AGPAT gene family. RT–PCR was used to detect transcripts of each candidate in various mouse tissues. To compare the relative abundance of each candidate, similar sets of primers (covering complete coding regions) and the same preparations of cDNAs were used for comparison between AGPAT isoform mRNA levels. mAGPAT1 was expressed ubiquitously at high levels in various tissues (Figure 4). Similarly, mAGPAT3 displayed a ubiquitous pattern with less abundance and some tissue variation. The pattern of expression of mAGPAT1 and mAGPAT3 differed from that of mAGPAT2, mAGPAT4 and mAGPAT5. mAGPAT2 was found to be expressed at high level in the liver, at intermediate levels in the kidney, gut, heart and skeletal muscles. However, mAGPAT2 was undetectable in brain and spleen. mAGPAT4 was expressed at a high level in the brain, at intermediate or low levels in skeletal muscles, gut, kidney, spleen and lung. mAGPAT4 was barely detectable in heart and liver. mAGPAT5 was expressed in all tissues at various levels.
Figure 4. mRNA expression profiles of mAGPAT isoforms in murine tissues.
Total RNAs were extracted and RT–PCR was performed as described in the Materials and methods section. Gel images obtained from RT–PCR showed the expression of AGPAT mRNAs in murine tissues. Results were from three independent RT–PCRs and representative gels are shown.
Regulation of mAGPAT in murine heart through PPARα activation
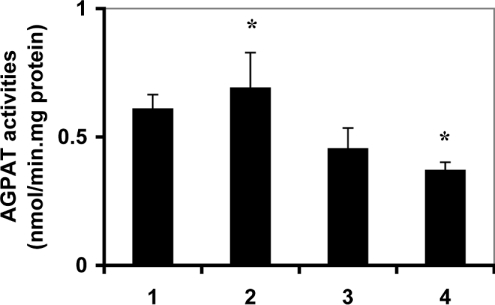
It is well documented that cardiac phospholipid biosynthetic pathways may be regulated by PPARα activation [30]. Hence, regulation of cardiac mAGPATs by PPARα activation was investigated. mAGPAT enzyme activities were examined in hearts prepared from wild-type and PPARα null mice. Cardiac mAGPAT activities were 25% (P<0.05) lower in PPARα null mice compared with wild-type animals (Figure 5, column 1 versus column 3). Mice were then fed either regular chow or chow containing 0.5% clofibrate for 14 consecutive days and cardiac mAGPAT activities were determined. Clofibrate feeding did not affect mAGPAT activities in wild-type mice when compared with controls (Figure 5, column 1 versus column 2). In addition, clofibrate feeding did not affect cardiac mAGPAT activities in PPARα null mice compared with controls (Figure 5, column 3 versus column 4). Interestingly, mAGPAT activity was 50% (P<0.05) lower in hearts prepared from PPARα null mice fed clofibrate compared with wild-type animals fed clofibrate (Figure 5, column 2 versus column 4). These data provided indirect evidence that cardiac mAGPAT activity could be regulated by PPARα activation.
Figure 5. AGPAT activity in wild-type and PPARα null mice treated with or without clofibrate.
Mice were fed 0.5% clofibrate or normal chow consecutively for 14 days and hearts were collected. Whole cell extracts were prepared and AGPAT activity was determined as described. Lane 1, wild-type control; lane 2, wild-type fed clofibrate; lane 3, PPARα null control; lane 4, PPARα null fed clofibrate. Values represent the means±S.D. (n=5), *P<0.05.
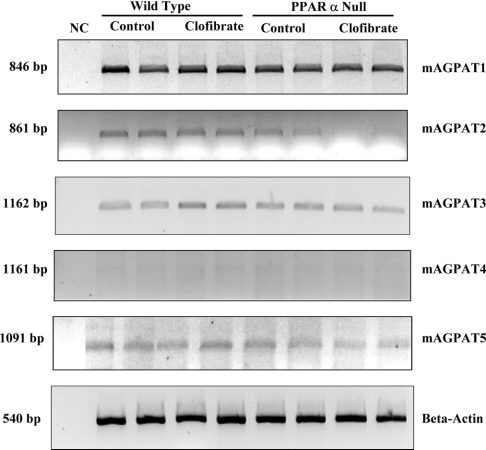
To confirm that cardiac mAGPAT activity could be regulated by PPARα activation, the mRNA levels of the five cardiac isoforms of the enzyme were examined in wild-type and PPARα null mice fed clofibrate. Consistent with the tissue distribution data of mAGPATs expression in heart (Figure 4, lane 2) cardiac mAGPAT4 mRNA was not detected (Figure 6). The mRNA levels of mAGPAT1 and mAGPAT5 were unaltered in both wild-type mice or PPARα null mice fed clofibrate when compared with controls (Figure 6). In contrast, cardiac mAGPAT3 mRNA expression was increased by 51% (P<0.05) in wild-type mice fed with clofibrate compared with controls. Thus it can be concluded that cardiac mAGPAT3 mRNA expression may be regulated by PPARα activation. It is interesting to note that the magnitude of change seen in the cardiac mAGPAT3 mRNA expression in wild-type and PPARα null mice was similar to that found for the mitochondrial fatty acid β-oxidation enzymes [34].
Figure 6. Expression of mAGPAT mRNAs in wild-type and PPARα null mice treated with or without clofibrate.
The collected mouse heart samples were used to determine mRNA levels of mAGPATs by RT–PCR as described in the Materials and methods section. The representative blots are shown. The densitometry of each band was normalized to the signal generated from β-actin (n=4 in each group).
Intriguingly, cardiac mAGPAT2 mRNA expression was decreased by 42% (P<0.05) in PPARα null mice compared with that of wild-type mice (Figure 6). In addition, cardiac mAGPAT2 mRNA expression was decreased by 70% (P<0.05) in PPARα null mice fed with clofibrate compared with that of wild-type mice fed with clofibrate. Thus the presence of PPARα appears to be important for the expression of the mAGPAT2. This might explain why there was a lower cardiac AGPAT activity in PPARα null mice compared with wild-type (Figure 5). In summary, these data suggest that the different cardiac mAGPAT isoforms may be differentially regulated by PPARα.
DISCUSSION
In mammalian cells, AGPAT exists in at least five isoforms. Previously, it has been found through Northern-blot analysis that AGPAT1 mRNA is ubiquitously expressed at high level in both human and mouse tissues [18,19]. The expression level of hAGPAT2 mRNA, however, varies in different tissues: with a large amount in the liver, a lower amount in brain and placenta, and an intermediate amount in the heart, skeletal muscles and small intestine [14]. The expression patterns for the other isoforms, AGPAT3, AGPAT4 and AGPAT5 were lacking. In the present study, we observed a similar expression pattern of both AGPAT1 and AGPAT2 mRNAs in various murine tissues, confirming the previous observation [15,16,18,19]. In addition, we analysed the mRNA expression pattern of the three other isoforms, mAGPAT3, mAGPAT4 and mAGPAT5. The amount of mAGPAT4 mRNA was expressed at high level in the brain, at intermediate or low levels in skeletal muscles, gut, kidney, spleen and lung. mAGPAT4 was barely detectable in heart and liver and its expression level varied in other tissues. mAGPAT3 and mAGPAT5 were expressed in all tissues at various levels. This is the first report to address the tissue distribution of multiple AGPAT isoforms in multiple tissues within one species. To address the sequence similarities among mAGPAT members, multiple sequence alignments were performed with complete amino acid sequences using ClustalW. The alignment reveals that mAGPAT1 and mAGPAT2 share high percentage of sequence identities, whereas mAGPAT3 and mAGPAT4 are more alike. However, mAGPAT5 remains as a singleton on the basis of distinct sequences. Thus these five members can be further divided into three subgroups according to their sequence identities. Accordingly, each subgroup of mAGPAT isoforms tends to exhibit a complementary expression pattern. For example, in group I, mAGPAT1 was ubiquitously expressed in most tissues, whereas mAGPAT2 displayed a tissue-specific expression pattern. In group II, mAGPAT3 appears to be ubiquitously expressed, whereas mAGPAT4 displays a tissue-specific expression pattern. Although the group III enzyme mAGPAT5 remains as a singleton and exhibits a ubiquitous expression pattern with some tissue variations, it is possible that a complementary isoform has yet to be discovered.
Surprisingly, two critical motifs for AGPATs were found to be 100% conserved among these five members. These included the catalytic motif NHX4D and substrate binding motif EGTR. The results support the original observation that these two motifs are conserved among the glycerophospholipid acyltransferase gene family [42,43]. Interestingly, when the amino acid sequences of the five mAGPATs are compared, a novel motif/pattern KX2LX6GX12R was apparent. This motif lies between the two conserved motifs within the catalytic region. This conserved motif is essentially an extension of motif II previously identified by site-directed mutagenesis and is conserved in some members of sn-glycerol 3-phosphate acyltransferases [42]. However, the motif identified in the present study is distinct from motif II in two respects: (i) the pattern and the composition of conserved amino acids are different; (ii) the KX2LX6GX12R motif is much longer than the motif II. Surprisingly, the amino acid conservation within these three motifs constitutes 11 out of 13 amino acid identities aligned along the entire sequences. These observations support the notion that AGPATs contain common structural motifs that are evolutionarily conserved within this gene family.
Linkage studies in humans have revealed that the AGPAT2 plays an important role in the regulation of lipid metabolism. mAGPAT2 expressed by in vitro transcription and translation reactions and in transfected COS-1 cells exhibited specificity for 1-acyl-sn-glycerol 3-phosphate. Cardiac mAGPAT activities were 25% lower (P<0.05) in PPARα null mice when compared with wild-type. In addition, cardiac mAGPAT activities were 50% lower (P<0.05) in PPARα null mice fed with clofibrate compared with clofibrate fed wild-type animals. This modulation of AGPAT activity was accompanied by significant enhancement/reduction of the mRNA levels of mAGPAT3/mAGPAT2 respectively. Interestingly, perfusion of hearts from wild-type mice with clofibrate increased incorporation of [1-14C]oleic acid into PA (1119 d.p.m./mg of protein, average of two hearts) compared with controls (342 d.p.m./mg of protein, average of two hearts) implying increased PA metabolism which might correspond to alterations in mAGPAT expression. However, caution should be used in the interpretation of this data as clofibrate mediates the expression of a plethora of lipid metabolic enzymes and alteration in the expression of these enzymes could account for the increase in [1-14C]oleic acid incorporated into PA [30,31,34].
The biological significance that arises from multiple isoforms of mAGPATs remains to be determined. One obvious explanation is to provide a mechanism to compensate the potential defects derived from one gene product. Congenital lipodystrophy occurs as a result of mutations in hAGPAT2 [20–23]. In this situation, the metabolic defect is not restored by compensatory mechanisms. These findings lead to the hypothesis that various AGPAT isoforms may perform different biological functions. This view is supported by the fact that, (i) mAGPATs are not identical enzyme proteins and their level of activities are different: mAGPAT2 appears to have higher AGPAT activity when compared with that of mAGPAT3 or mAGPAT5; (ii) enhancement/inhibition of AGPAT activities in different cell types may lead to different biological effects, such as enhancement of immunoresponse [17] and induction of apoptosis in tumour cells [24], and are compartmentalized within different subcellular locations; and (iii) cardiac mAGPAT activities and their mRNA levels may be differentially regulated through PPARα.
Acknowledgments
We thank Dr R.A. Coleman for helpful discussions. This work was supported by operating grants from the Canadian Institutes of Health Research (G.M.H. and P.C.C.) and the Heart and Stroke Foundation of Manitoba (G.M.H.).
References
- 1.Vance D. E. Phospholipid biosynthesis in eukaryotes. In: Vance D. E., Vance J., editors. Biochemistry of Lipids, Lipoproteins and Membranes. 4th edn. Amsterdam: Elsevier Science; 2002. pp. 205–231. [Google Scholar]
- 2.Bursten S. L., Harris W. E., Bomsztyk K., Lovett D. Interleukin-1 rapidly stimulates lysophosphatidate acyltransferase and phosphatidate phosphohydrolase activities in human mesangial cells. J. Biol. Chem. 1991;266:20732–20743. [PubMed] [Google Scholar]
- 3.English D., Garcia J. G., Brindley D. N. Platelet-released phospholipids link haemostasis and angiogenesis. Cardiovasc. Res. 2001;49:588–599. doi: 10.1016/s0008-6363(00)00230-3. [DOI] [PubMed] [Google Scholar]
- 4.Hla T., Lee M. J., Ancellin N., Paik J. H., Kluk M. J. Lysophospholipids–receptor revelations. Science. 2001;294:1875–1878. doi: 10.1126/science.1065323. [DOI] [PubMed] [Google Scholar]
- 5.McIntyre T. M., Pontsler A. V., Silva A. R., St Hilire A., Su Y., Hinshow J. C., Zimmerman G. A., Hama K., Aoki J., Arai H., et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. U.S.A. 2003;100:131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang X., Yu S., Bast R. C., Liu S., Xu H. J., Hu S. X., LaPushin R., Claret F. X., Aggarwal B. B., Lu Y., et al. Mechanisms for lysophosphatidic acid-induced cytokine production in ovarian cancer cells. J. Biol. Chem. 2004;279:9653–9661. doi: 10.1074/jbc.M306662200. [DOI] [PubMed] [Google Scholar]
- 7.Segura B. J., Zhang W., Cowles R. A., Xiao L., Lin T. R., Logsdon C., Mulholland M. W. Lysophosphatidic acid stimulates calcium transients in enteric glia. Neuroscience. 2004;123:687–693. doi: 10.1016/j.neuroscience.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Yamada T., Sato K., Komachi M., Malchinkhuu E., Tobo M., Kimura T., Kuwabara A., Yanagita Y., Ikeya T., Tanahashi Y., et al. Lysophosphatidic acid (LPA) in malignant ascites stimulates motility of human pancreatic cancer cells through LPA1. J. Biol. Chem. 2004;279:6595–6605. doi: 10.1074/jbc.M308133200. [DOI] [PubMed] [Google Scholar]
- 9.Akita Y., Altman A., Villalba M., Kikkawa U., Matsuzaki H., Yamamoto T. Protein kinase C-epsilon (PKC-epsilon): its unique structure and function. J. Biochem. (Tokyo) 2002;132:847–852. doi: 10.1093/oxfordjournals.jbchem.a003296. [DOI] [PubMed] [Google Scholar]
- 10.Altman A., Villalba M. Protein kinase C-theta (PKC theta): a key enzyme in T cell life and death. J. Biochem. (Tokyo) 2002;132:41–46. doi: 10.1093/oxfordjournals.jbchem.a003295. [DOI] [PubMed] [Google Scholar]
- 11.Kikkawa U., Matsuzaki H., Yamamoto T. Protein kinase C delta (PKC delta): activation mechanisms and functions. J. Biochem. (Tokyo) 2002;132:831–839. doi: 10.1093/oxfordjournals.jbchem.a003294. [DOI] [PubMed] [Google Scholar]
- 12.Saito N., Shirai Y. Protein kinase C gamma (PKC gamma): function of neuron specific isotype. J. Biochem. (Tokyo) 2002;132:683–687. doi: 10.1093/oxfordjournals.jbchem.a003274. [DOI] [PubMed] [Google Scholar]
- 13.Shirai Y., Saito N. Activation mechanisms of protein kinase C: maturation, catalytic activation, and targeting. J. Biochem. (Tokyo) 2002;132:663–668. doi: 10.1093/oxfordjournals.jbchem.a003271. [DOI] [PubMed] [Google Scholar]
- 14.Eberhardt C., Gray P. W., Tjoelker L. W. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J. Biol. Chem. 1997;272:20299–20305. doi: 10.1074/jbc.272.32.20299. [DOI] [PubMed] [Google Scholar]
- 15.Eberhardt C., Gray P. W., Tjoelker L. W. cDNA cloning, expression and chromosomal localization of two human lysophosphatidic acid acyltransferases. Adv. Exp. Med. Biol. 1999;469:351–356. doi: 10.1007/978-1-4615-4793-8_51. [DOI] [PubMed] [Google Scholar]
- 16.Stamps A. C., Elmore M. A., Hill M. E., Kelly K., Makda A. A., Finnen M. J. A human cDNA sequence with homology to non-mammalian lysophosphatidic acid acyltransferases. Biochem. J. 1997;326:455–461. doi: 10.1042/bj3260455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West J., Tompkins C. K., Balantac N., Nudelman E., Meengs B., White T., Bursten S., Coleman J., Kumar A., Singer J. W., et al. Cloning and expression of two human lysophosphatidic acid acyltransferase cDNAs that enhance cytokine-induced signaling responses in cells. DNA Cell Biol. 1997;16:691–701. doi: 10.1089/dna.1997.16.691. [DOI] [PubMed] [Google Scholar]
- 18.Aguado B., Campbell R. D. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J. Biol. Chem. 1998;273:4096–4105. doi: 10.1074/jbc.273.7.4096. [DOI] [PubMed] [Google Scholar]
- 19.Kume K., Shimizu T. cDNA cloning and expression of murine 1-acyl-sn-glycerol-3-phosphate acyltransferase. Biochem. Biophys. Res. Commun. 1997;237:663–666. doi: 10.1006/bbrc.1997.7214. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal A. K., Arioglu E., De Almeida S., Akkoc N., Taylor S. I., Bowcock A. M., Barnes R. I., Garg A. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 2002;31:21–23. doi: 10.1038/ng880. [DOI] [PubMed] [Google Scholar]
- 21.Magre J., Delepine M., Van Maldergem L., Rpvert J. J., Maassen J. A., Meier M., Panz V. R., Kim C. A., Tubiana-Rufi N., Czernichow P., et al. Prevalence of mutations in AGPAT2 among human lipodystrophies. Diabetes. 2003;52:1573–1578. doi: 10.2337/diabetes.52.6.1573. [DOI] [PubMed] [Google Scholar]
- 22.Simha V., Garg A. Phenotypic heterogeneity in body fat distribution in patients with congenital generalized lipodystrophy caused by mutations in the AGPAT2 or seipin genes. J. Clin. Endocrinol. Metab. 2003;88:5433–5437. doi: 10.1210/jc.2003-030835. [DOI] [PubMed] [Google Scholar]
- 23.Gomes K. B., Fernandes A. P., Ferreira A. C., Pardini H., Garg A., Magre J., Pardini V. C. Mutations in the seipin and AGPAT2 genes clustering in consanguineous families with Berardinelli-Seip congenital lipodystrophy from two separate geographical regions of Brazil. J. Clin. Endocrinol. Metab. 2004;89:357–361. doi: 10.1210/jc.2003-030415. [DOI] [PubMed] [Google Scholar]
- 24.Bonham L., Leung D. W., White T., Hollenback D., Klein P., Tulinsky J., Coon M., de Vries P., Singer J. W. Lysophosphatidic acid acyltransferase-beta: a novel target for induction of tumour cell apoptosis. Expert. Opin. Ther. Targets. 2003;7:643–661. doi: 10.1517/14728222.7.5.643. [DOI] [PubMed] [Google Scholar]
- 25.Hideshima T., Chauhan D., Hayashi T., Podar K., Akiyama M., Mitsiades C., Mitsiades N., Gong B., Bonham L., de Vries P., et al. Antitumor activity of lysophosphatidic acid acyltransferase-beta inhibitors, a novel class of agents, in multiple myeloma. Cancer Res. 2003;63:8428–8436. [PubMed] [Google Scholar]
- 26.Evans R. M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata H., Spencer T. E., Onate S. A., Jenster G., Tsai S. Y., O'Malley B. W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent. Prog. Horm. Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 28.McKenna N. J., Lanz R. B., O'Malley B. W. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 29.Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y. J., Hatch G. M., Mymin D., Dembinski T., Kroeger E. A., Choy P. C. Modulation of cytosolic phospholipase A2 by PPAR activators in human preadipocytes. J. Lipid. Res. 2001;42:716–724. [PubMed] [Google Scholar]
- 31.Jiang Y. J., Lu B., Xu F. Y., Gartshore J., Taylor W. A., Halayko A. J., Gonzalez F. J., Takasaki J., Choy P. C., Hatch G. M. Stimulation of cardiac cardiolipin biosynthesis by PPARalpha activation. J. Lipid. Res. 2004;45:244–252. doi: 10.1194/jlr.M300314-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Cheung C., Akiyama T. E., Ward J. M., Nicol C. J., Feigenbaum L., Vinson C., Gonzalez F. J. Diminished hepatocellular proliferation in mice humanized for the nuclear receptor peroxisome proliferator-activated receptor alpha. Cancer Res. 2004;64:3849–3854. doi: 10.1158/0008-5472.CAN-04-0322. [DOI] [PubMed] [Google Scholar]
- 33.Desvergne B., Michalik L., Wahli W. Be fit or be sick: peroxisome proliferator-activated receptors are down the road. Mol. Endocrinol. 2004;18:1321–1332. doi: 10.1210/me.2004-0088. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe K., Fujii H., Takahashi T., Kodama M., Aizawa Y., Ohta Y., Ono T., Hasegawa G., Naito M., Nakajima T., et al. Constitutive regulation of cardiac fatty acid metabolism through peroxisome proliferator-activated receptor alpha associated with age-dependent cardiac toxicity. J. Biol. Chem. 2000;275:22293–22299. doi: 10.1074/jbc.M000248200. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y. J., Xu T. R., Lu B., Mymin D., Kroeger E. A., Dembinski T., Yang X., Hatch G. M., Choy P. C. Cyclooxygenase expression is elevated in retinoic acid-differentiated U937 cells. Biochim. Biophys. Acta. 2003;1633:51–60. doi: 10.1016/s1388-1981(03)00072-6. [DOI] [PubMed] [Google Scholar]
- 36.Lu B., Leygue E., Dotzlaw H., Murphy L. J., Murphy L. C., Watson P. H. Estrogen receptor-beta mRNA variants in human and murine tissues. Mol. Cell. Endocrinol. 1998;138:199–203. doi: 10.1016/s0303-7207(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 37.Lu B., Leygue E., Dotzlaw H., Murphy L. J., Murphy L. C. Functional characteristics of a novel murine estrogen receptor-beta isoform, estrogen receptor-beta 2. J. Mol. Endocrinol. 2000;25:229–242. doi: 10.1677/jme.0.0250229. [DOI] [PubMed] [Google Scholar]
- 38.Peng B., Lu B., Leygue E., Murphy L. C. Putative functional characteristics of human estrogen receptor-beta isoforms. J. Mol. Endocrinol. 2003;30:13–29. doi: 10.1677/jme.0.0300013. [DOI] [PubMed] [Google Scholar]
- 39.Langendorff O. Untersuchungen am uberlebenden saugetierberzen. Pfluegers Arch. 1895;61:291–332. [Google Scholar]
- 40.Krebs H. A., Henseleit K. Untersuchungen uber die Harstoffibildung in Tierkorper. Hoppe-Seyler's Z. Physiol. Chem. 1932;210:22–66. [Google Scholar]
- 41.Poorthuis B. J., Yazaki P. J., Hostetler K. Y. An improved two dimensional thin-layer chromatography system for the separation of phosphatidylglycerol and its derivatives. J. Lipid Res. 1976;17:433–437. [PubMed] [Google Scholar]
- 42.Lewin T. M., Wang P., Coleman R. A. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 43.Leung D. W. The structure and functions of human lysophosphatidic acid acyltransferases. Front. Biosci. 2001;6:D944–D953. doi: 10.2741/leung. [DOI] [PubMed] [Google Scholar]