Abstract
The DNMT3A (DNA methyltransferase 3A) and DNMT3B genes encode putative de novo methyltransferases and show complex transcriptional regulation in the presence of three and two different promoters respectively. All promoters of DNMT3A and DNMT3B lack typical TATA sequences adjacent to their transcription start sites and contain several Sp1-binding sites. The importance of these Sp1-binding sites was demonstrated by using a GC-rich DNA-binding protein inhibitor, mithramycin A, i.e. on the basis of decrease in the promoter activities and mRNA expression levels of DNMT3A and DNMT3B. Overexpression of Sp1 and Sp3 up-regulated the promoter activities of these two genes. The physical binding of Sp1 and Sp3 to DNMT3A and DNMT3B promoters was confirmed by a gel shift assay. Interestingly, Sp3 overexpression in HEK-293T cells (human embryonic kidney 293T cells) resulted in 3.3- and 4.0-fold increase in DNMT3A and DNMT3B mRNA expression levels respectively by quantitative reverse transcriptase–PCR, whereas Sp1 overexpression did not. Furthermore, an antisense oligonucleotide to Sp3 significantly decreased the mRNA levels of DNMT3A and DNMT3B. These results indicate the functional importance of Sp proteins, particularly Sp3, in the regulation of DNMT3A and DNMT3B gene expression.
Keywords: DNA methyltransferase (DNMT), gene regulation, mithramycin A, specificity protein 1 (Sp1), Sp3
Abbreviations: DNMT, DNA methyltransferase; EMSA, electrophoretic mobility-shift assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HEK-293T cells, human embryonic kidney 293T cells; RT, reverse transcriptase; Sp, specificity protein; TSP, transcription start site
INTRODUCTION
DNMTs (DNA methyltransferases) are the enzymes catalysing DNA methylation [1]. To date, three functional DNMTs have been identified. Dnmt1 has a 5–30-fold preference for hemimethylated substrates; therefore it has been designated as a maintenance methyltransferase [2]. Dnmt3A and Dnmt3B show equal preference for unmethylated and hemimethylated DNA in vitro [3]; therefore they are regarded as de novo methyltransferases.
The promoters of DNMT1 have already been characterized [4]. Increased DNMT1 mRNA expression has been reported for a number of human cancers [5]. In addition, it has been demonstrated that DNMT1 is the target of c-fos-transformed cells [6], shows up-regulated mRNA expression with an inflammatory cytokine interleukin-6 [7] and is controlled by Sp1 (specificity protein 1) and Sp3 [8].
On the other hand, to date, not much data concerning the transcriptional regulation of other methyltransferases have been reported. Dnmt3A and Dnmt3B are highly expressed in undifferentiated embryonic stem cells and embryonal carcinoma cells, whereas they are barely detectable in differentiated cells and adult tissues, except for some particular organs [3]. DNMT3A and DNMT3B mRNAs are reported to be overexpressed in tumours and cancer cell lines [5], and inhibition of DNMT3B by antisense oligonucleotide induces apoptosis in cancer cells but not in normal cells [9].
We previously identified the promoters of DNMT3A and DNMT3B [10]. DNMT3A has three alternative 1st exons named exons 1A, 1B and 1C, which are controlled by the 1st, 2nd and 3rd promoters respectively. DNMT3B has two alternative 1st exons, designated as exons 1A and 1B, from the 1st and 2nd promoters respectively. All promoters of DNMT3A and DNMT3B lack TATA sequences near their TSPs (transcription start sites). The 1st and 2nd promoters of DNMT3A and the 1st promoter of DNMT3B are CpG-rich and contain clusters of Sp1-binding sites in the proximal promoter region, whereas the 3rd promoter of DNMT3A and the 2nd promoter of DNMT3B are CpG-poor. However, the transcriptional regulation and major transcription factors that regulate these promoters have not yet been reported.
The Sp transcription factor family belongs to the conserved zinc finger DNA-binding domain proteins that recognize the putative DNA-binding motifs GC-box (GGGCGGG) and GT-box (GGTGTGGGG). They are important for the expression of many different housekeeping genes and genes that generally do not contain TATA- or CAAT-boxes in their proximal promoters, as well as tissue-specific genes [11]. Several Sp proteins (Sp1–Sp8) have been identified [11]. Sp1 and Sp3 are ubiquitously expressed [12], whereas the others show tissue-restricted expression patterns [11]. Sp1 is well known as a transcriptional activator, whereas Sp3 can be either a transcriptional activator [13] or repressor of Sp1-mediated transcription [14], depending on the promoter context and cell type.
In the present study, we focused on the transcriptional regulation of DNMT3A and DNMT3B promoters by the transcription factors Sp1 and Sp3. By means of various experimental approaches, we demonstrated that Sp proteins, particularly Sp3, were essential for the expression of DNMT3A and DNMT3B. Altogether, our results lead to the first basic understanding of the molecular transcriptional regulation of DNMT3A and DNMT3B.
MATERIALS AND METHODS
Plasmids
The reporter plasmids containing full-length human DNMT3A 1st+2nd promoters pGL3A-P1+2 (−2489/+640), DNMT3A 3rd promoter pGL3A-P3 (−3007/+1021), DNMT3B 1st promoter pGL3B-P1 (−2483/+309) and DNMT3B 2nd promoter pGL3B-P2 (−3531/+260) were described previously [10]. All deletion mutants were named according to the nucleotide numbers of their 5′- and 3′-ends relative to the TSPs of each exon (+1). The plasmid pCMV-Sp1 was a gift from S. Smale (University of California, Los Angeles, CA, U.S.A.). The plasmid pCMV4-Sp3/flu was obtained from J. M. Horowitz (North Carolina State University, Raleigh, NC, U.S.A.). Empty mammalian expression vector pRc/CMV (Invitrogen, Groningen, the Netherlands) was used as a negative control.
Site-directed mutagenesis was performed by a PCR-based approach. The Sp1-binding sites at −99/−87 of pGL3A-P3 (−334/+376) and −100/−92 of pGL3B-P2 (−469/+260) were replaced by an XbaI restriction site to generate pGL3A-P3 (−334/+376)-M and pGL3B-P2 (−469/+260)-M. As for pGL3B-P1 (−102/+309), the Sp1-binding sites at −99/−92 and −79/−70 were replaced by EcoRI and XbaI restriction sites respectively to generate pGL3B-P1 (−102/+309)-M. This mutant plasmid was further used as a template to generate pGL3B-P1 (−102/+309)-DM, in which the Sp1-binding site at −62/−49 (5′-GGGCGGGGGCGGGG-3′) was also mutated to 5′-GGGC-TTAAACGGGG-3′. All plasmids were sequenced before using them in transfection experiments.
Cell culture, transfection and luciferase assay
HEK-293T cells (human embryonic kidney 293T) cells, adenovirus-transformed HEK cells expressing simian-virus-40 large T antigen, and U-2OS human osteosarcoma cells were grown at 37 °C under a 5% CO2 and 95% air atmosphere in Dulbecco's modified Eagles's medium containing 10% (v/v) fetal bovine serum.
For the luciferase assay, HEK-293T and U-2OS cells were transfected in 24-well plates in duplicate with 50 ng of reporter plasmid and increasing amounts (100, 150 and 200 ng) of pCMV-Sp1, pCMV4-Sp3/flu or pRc/CMV, using TransIT-LT1 transfection reagent (Mirus, Madison, WI, U.S.A.) for 48 h. For the luciferase assay with a GC-rich DNA-binding protein inhibitor, mithramycin A (Sigma, St. Louis, MO, U.S.A.) was added to a final concentration of 100 nM for 24 h. Cells were lysed with Passive lysis buffer (Promega, Madison, WI, U.S.A.). The luciferase activity of the cell extract was measured with a Luminocounter 700 (Microtech Niti-On, Chiba, Japan) and normalized to the cellular protein concentration. Results were expressed as fold activation, i.e. the ratio of normalized luciferase activity of pCMV-Sp1 and pCMV4-Sp3/flu to that of pRc/CMV.
RNA extraction, cDNA synthesis and semiquantitative PCR analysis
The HEK-293T cells were transfected in 60 mm plates with 2 μg of pCMV-Sp1, pCMV4-Sp3/flu or pRc/CMV using TransIT-LT1 transfection reagent (Mirus) for 48 h. For the GC-rich DNA-binding protein inhibitor experiment, mithramycin A was added to a final concentration of 100 or 200 nM for 24 h. Total RNA was extracted by using an RNeasy Mini kit (Qiagen, Hilden, Germany). The isolated RNA (1 μg) was used for cDNA synthesis as described previously [15].
The synthesized cDNA was then amplified by the PCR method. The locations, sequences and annealing temperatures of primers are shown in Figure 1(A) and Table 1. PCRs (26–35 cycles) were used to cover the linear range of PCR amplification. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts were amplified as an internal control for 21 cycles. All RT (reverse transcriptase)–PCR products were ligated into the pGEM-T easy vector (Promega) and confirmed by direct sequencing.
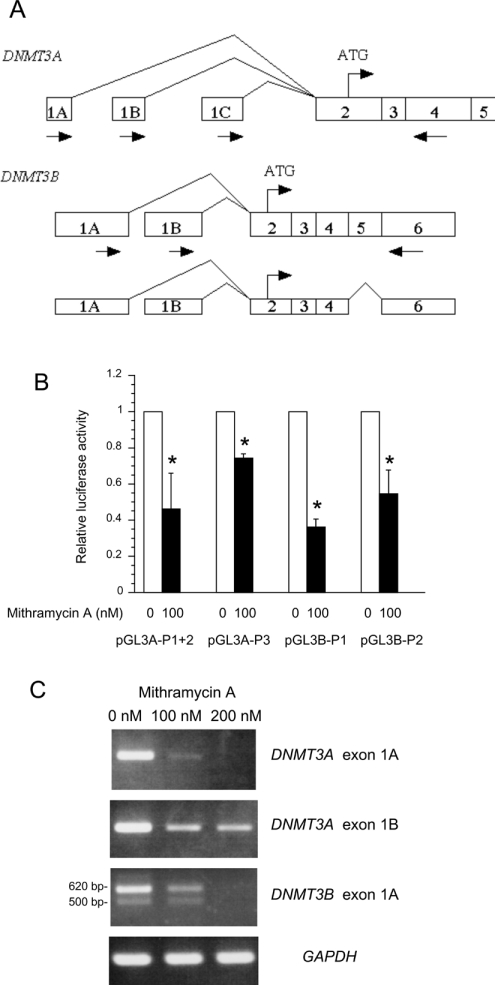
Figure 1. Mithramycin A inhibits DNMT3A and DNMT3B promoter activities and mRNA expression.
(A) Schematic structure of the 5′-region of the human DNMT3A and DNMT3B mRNAs. Boxed numbers indicate exons, and arrows indicate the positions of sense and antisense primers used for semiquantitative RT–PCR. Three distinct 1st exons of DNMT3A (1A, 1B and 1C) are driven by separate promoters (1st, 2nd and 3rd promoters respectively), and spliced to the common exon 2. The 5′-region of DNMT3B mRNA contains two alternative 1st exons (1A and 1B), which are spliced to a common exon 2. The structure of the novel alternative spliced variant of DNMT3B that lacks exon 5 is shown in the smaller Figure below. (B) The reporter construct containing DNMT3A promoters (1st and 2nd promoters, pGL3A-P1+2; 3rd promoter, pGL3A-P3) and DNMT3B promoters (1st promoter, pGL3B-P1; 2nd promoter, pGL3B-P2) were transfected into HEK-293T cells. Then, luciferase activity was determined in the absence or presence of 100 nM mithramycin A and expressed relative to the activity in the absence of mithramycin A. Results are expressed as the means±S.D. for three experiments. *P<0.05 with the paired t test. (C) Semiquantitative RT–PCR analysis of DNMT3A and DNMT3B 1st exon mRNA variants in HEK-293T cells after incubation with 100 and 200 nM mithramycin A.
Table 1. Primer sequences for RT–PCR.
| Primer | Sequence | Tm (°C) |
|---|---|---|
| DNMT3A exon 1A sense | 5′-GATGCGGGGACCCAGCGCAGA-3′ | 62 |
| DNMT3A exon 1B sense | 5′-GCCCGACCCCACCGGCCATAC-3′ | 62 |
| DNMT3A exon 1C sense | 5′-GCCTGCAGCCTGAGCTCAGACC-3′ | 58 |
| DNMT3A exon 4 antisense | 5′-AGATCACCGCAGGGTCCTTTGGC-3′ | 62 |
| DNMT3A exon 17 sense | 5′-CATCACGGTGGGCATGGTGCG-3′ | 60 |
| DNMT3A exon 23 antisense | 5′-GTCTCTGCCTCGCCAAGC-3′ | 60 |
| DNMT3B exon 1A sense | 5′-AGTGGCCCAAGTAAACCTAGCTC-3′ | 62 |
| DNMT3B exon 1B sense | 5′-GACAGCCTGTCCACATGGAACC-3′ | 68 |
| DNMT3B exon 6 antisense | 5′-CGTTCCTGCCGATGCTGTTGC-3′ | 62 |
| DNMT3B exon 17 sense | 5′-CATTGCTGTTGGAACCGTGAA-3′ | 55 |
| DNMT3B exon 23 antisense | 5′-CTTCTGGCGGGCACCACG-3′ | 55 |
| GAPDH sense | 5′-GACCACAGTCCATGCCATCAC-3′ | 60 |
| GAPDH antisense | 5′-GTCCACCACCCTGTTGCTGTA-3′ | 60 |
LightCycler real-time PCR
The DNMT3A and DNMT3B mRNA levels in Sp1 or Sp3 transfected HEK-293T cells and Sp3-antisense- or mismatch oligonucleotides-treated cells were quantified with LightCycler™ system using LightCycler DNA Master SYBR Green I (Roche Molecular Biochemicals). As an internal control, GAPDH mRNA was also quantified with the same primers used for semiquantitative RT–PCR. The primers for DNMT3A were 5′-GACTCCATCACGGTGGGCATGG-3′ (sense) and 5′-TGTCCCTCTTGTCACTAACGCC-3′ (antisense). The primers for DNMT3B were 5′-GAGTCCATTGCTGTTGGAACCG-3′ (sense) and 5′-ATGTCCCTCTTGTCGCCAACCT-3′ (antisense). The amplification programmes were repeated for 40 cycles for DNMT3B and GAPDH or 45 cycles for DNMT3A. The specificity of PCR products was confirmed by melting curve analysis with the presence of single melting curve and reconfirmed by agarose-gelelectrophoresis. The standard curve was constructed with 5-fold serial dilutions of HEK-293T cDNA. The Fit Point method was performed for the determination of concentration using LightCycler software version 3.3 (Roche Molecular Biochemicals). The DNMT3A and DNMT3B mRNA levels were expressed relative to the levels of GAPDH mRNA in the same samples.
Western-blot analysis
Western blotting was performed essentially as described in [16]. Total protein (30 μg) from Sp1 or Sp3 transfected HEK-293T cells and Sp3-antisense- or mismatch oligonucleotide-treated cells were immunoblotted with anti-Sp1 (sc-59X; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) at a dilution of 1:2000, anti-Sp3 (sc-644X; Santa Cruz Biotechnology) at 1:6000 and anti-α-tubulin (Bio-Rad) at 1:8000.
EMSA (electrophoretic mobility-shift assay)
The methods for nuclear extract preparation and EMSA were described previously [15]. The sequences of wild-type and mutant-type oligonucleotides are available upon request. For gel supershifts, nuclear extracts were preincubated with 1 μl of the antibody against Sp1 (sc-59X) and/or Sp3 (sc-644X), according to the method described above.
Sp3 knockout with antisense oligonucleotide
The phosphorothioate-modified antisense oligonucleotide (5′-AGTAGCAGCACTTGGAATCTGGACT-3′) and mismatch oligonucleotide (5′-AGTACCAGGACTTCGAATGTGCACT-3′) of the Sp3 gene were synthesized and HPLC-purified (Qiagen). The HEK-293T cells were transfected with the antisense or mismatch oligonucleotides at a final concentration of 150 nM by using TransIT-Oligo transfection reagent (Mirus). Cells were incubated for 48 h and then harvested for semiquantitative RT–PCR, Light-Cycler real-time PCR and Western-blot analyses.
RESULTS
Mithramycin A represses DNMT3A and DNMT3B promoter activities and inhibits mRNA expression
Mithramycin A is an aureolic acid antibiotic that has been shown to inhibit gene expression selectively by displacing transcriptional activators which bind to GC-rich regions of promoters such as Sp1 [17]. Therefore we determined the effects of mithramycin A on the activities of DNMT3A and DNMT3B promoters. Plasmids containing promoters of DNMT3A 1st+2nd promoters pGL3A-P1+2, DNMT3A 3rd promoter pGL3A-P3, DNMT3B 1st promoter pGL3B-P1 and DNMT3B 2nd promoter pGL3B-P2 were transfected into HEK-293T cells and then mithramycin A was added to a concentration of 100 nM. As shown in Figure 1(B), the addition of mithramycin A significantly repressed all promoter activities of DNMT3A and DNMT3B in HEK-293T cells.
We next analysed the mRNA expression of DNMT3A and DNMT3B after mithramycin A treatment by the RT–PCR method. As shown in Figure 1(C), the parental HEK-293T cells expressed DNMT3A/1A and 1B (from DNMT3A 1st and 2nd promoters respectively) and DNMT3B/1A (from DNMT3B 1st promoter), whereas the expression of DNMT3A/1C (from DNMT3A 3rd promoter) and DNMT3B/1B (from DNMT3B 2nd promoter) were undetectable (results not shown). Mithramycin A treatment decreased the DNMT3A/1A, 1B and DNMT3B/1A mRNA levels in a dose-dependent manner (Figure 1C). With regard to DNMT3A/1C and DNMT3B/1B, expression levels remained undetectable after mithramycin A treatment (results not shown). Similar results were also obtained with U-2OS cells (results not shown).
Interestingly, although expressed at a much lower level, the RT–PCR product of DNMT3B/1A also gave a smaller band (approx. 500 bp) below the major band (620 bp). The direct sequencing result of the smaller product revealed a novel DNMT3B isoform that lacks exon 5 (126 bp). The structures of all the alternative spliced variants are shown in Figure 1(A).
Effects of the transcription factors Sp1 and Sp3 on DNMT3A and DNMT3B promoters
Sp1 and Sp3 are ubiquitously expressed and they are representative GC-rich DNA-binding proteins. Thus, to address the biological effects of Sp1 and Sp3 on DNMT3A and DNMT3B promoters, reporter plasmids containing the full-length promoters of DNMT3A and DNMT3B were co-transfected with increasing amounts of an expression vector encoding either Sp1 (pCMV-Sp1) or Sp3 (pCMV4-Sp3/flu) in HEK-293T cells. As shown in Table 2, co-transfection with either Sp1 or Sp3 resulted in the increase of pGL3A-P1+2 luciferase activity in a dose-dependent manner. Similar results were obtained from pGL3A-P3 and pGL3B-P2. With regard to pGL3B-P1, weaker activation by Sp1 and Sp3 was observed. Since transfection with this reporter vector alone has already produced very high basal luciferase activity in many cell lines tested compared with the other reporter vectors ([10]; results not shown), a weaker activation in the co-transfection experiments may result from the high cellular background of endogenous Sp proteins or other transcription factors. The results for other cells (U-2OS) revealed a similar tendency (results not shown).
Table 2. Effects of Sp1 and Sp3 on DNMT3A and DNMT3B promoters.
The reporter vectors containing full-length promoters of DNMT3A and DNMT3B were co-transfected with increasing amounts (100, 150 and 200 ng) of Sp1 expression vector pCMV-Sp1, Sp3 expression vector pCMV4-Sp3/flu or empty expression vector pRc/CMV. Luciferase activity was normalized to the protein concentration and expressed relative to the activity in the presence of pRc/CMV in each sample. The results represent the means±S.D. for two independent experiments.
| Expression vector… | pCMV-Sp1 | pCMV4-Sp3/flu | |||||
|---|---|---|---|---|---|---|---|
| Reporter vector | Amount (ng)… | 100 | 150 | 200 | 100 | 150 | 200 |
| pGL3A-P1+2 | 1.6±0.2 | 2.1±0.3 | 6.1±1.3 | 1.8±0.3 | 2.3±0.3 | 3.6±0.5 | |
| pGL3A-P3 | 2.4±0.5 | 4.6±0.4 | 13.7±2.3 | 3.1±0.5 | 4.6±0.6 | 6.6±0.1 | |
| pGL3B-P1 | 1.1±0.1 | 1.1±0.2 | 1.8±0.4 | 0.7±0.1 | 0.7±0.1 | 1.1±0.2 | |
| pGL3B-P2 | 1.1±0.5 | 1.9±0.5 | 10.5±1.3 | 0.7±0.2 | 0.9±0.3 | 2.9±0.3 | |
DNMT3A and DNMT3B expression levels in Sp1- and Sp3-overexpressing cells
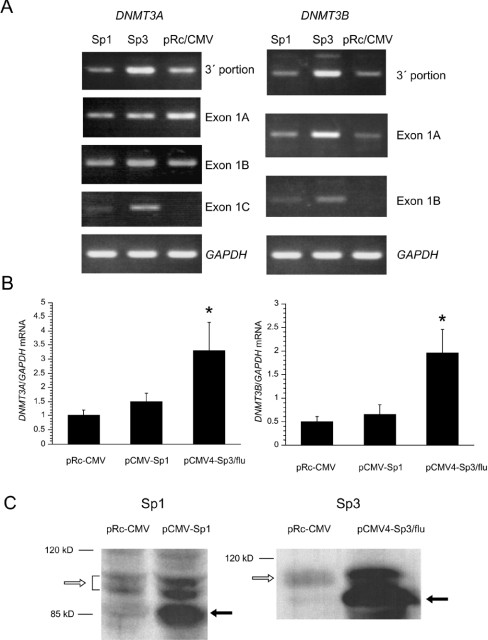
We next examined whether or not Sp1 and Sp3 overexpression further up-regulates the endogenous mRNA levels of DNMT3A and DNMT3B. The HEK-293T cells were transiently transfected with pCMV-Sp1, pCMV4-Sp3/flu or pRc/CMV as a negative control. Semiquantitative RT–PCR analysis with primers recognizing the 3′-portions of DNMT3A and DNMT3B cDNA showed that DNMT3A and DNMT3B mRNA expression was increased in HEK-293T cells transfected with the Sp3 vector compared with those transfected with the Sp1 vector and pRc/CMV (Figure 2A, top panels).
Figure 2. DNMT3A and DNMT3B expression levels in Sp1- or Sp3-overexpressing cells.
(A) Total RNA from HEK-293T cells transfected with Sp1, Sp3 or pRc/CMV were amplified by semiquantitative RT–PCR method using specific primers for the 3′-portions of DNMT3A and DNMT3B and all alternative 1st exons of each gene. The experiments were repeated three times using different cDNAs from three transfection experiments. (B) The DNMT3A and DNMT3B mRNA levels in the same cDNA preparation were quantified by using LightCycler real-time PCR. The DNMT3A (left) and DNMT3B (right) levels were expressed relative to GAPDH mRNA levels in the same samples. Results are expressed as the means±S.D. for three experiments. *P<0.05 with the paired t test. (C) Total protein from HEK-293T cells transfected with pCMV-Sp1, pCMV4-Sp3/flu or pRc/CMV was used for Western-blot analyses with anti-Sp1 and anti-Sp3 antibodies respectively. Open arrows indicate the endogenous Sp1 or Sp3 proteins in each Figure. Closed arrows indicate exogenous Sp1 or Sp3 proteins in each Figure.
To determine the promoters of DNMT3A from which the increase in mRNA expression is derived, we performed RT–PCR with primers specific to all alternative 1st exons of DNMT3A or DNMT3B. With regard to DNMT3A, DNMT3A/1A and 1B were expressed in the parental cells and no significant increase in mRNA expression was observed (Figure 2A). On the other hand, DNMT3A/1C, the expression of which was below the detectable level in the parental cells, was significantly up-regulated by Sp3 and to a lesser extent by Sp1. Concerning DNMT3B, DNMT3B/1A was up-regulated by Sp3. DNMT3B/1B was also up-regulated by Sp3 and to a lesser extent by Sp1. The experiments were repeated three times using different cDNAs from three transfection experiments and the same results were obtained.
To confirm the findings of semiquantitative RT–PCR, we performed LightCycler real-time PCR. As shown in Figure 2(B), Sp3 overexpression resulted in 3.3- and 4.0-fold increase in DNMT3A and DNMT3B levels compared with the levels in cells transfected with the empty vector respectively. As expected, slightly increased DNMT3A and DNMT3B levels were observed in Sp1-overexpressing cells.
We also confirmed the expression levels of Sp1 and Sp3 proteins in cells transfected with pCMV-Sp1 and pCMV4-Sp3/flu respectively using immunoblot analysis. In Figure 2(C), a significant increase in the exogenous Sp1 protein (approx. 85 kDa), which resulted from the lack of 85 amino acids in the N-terminus of Sp1 [18], was observed. The increase in endogenous Sp1 (upper bands) could be caused by autoregulation [19]. The doublet of Sp1 protein represented a different phosphorylated status. Similarly, the exogenous Sp3 protein, which resulted from the lack of approx. 70 amino acids in the N-terminus of Sp3 [13], was abundantly expressed.
Identification of the Sp1-binding sites responsible for minimal promoter activities in DNMT3A and DNMT3B promoters
On the basis of the findings for HEK-293T cells that mRNA expression from the DNMT3A 3rd promoter and DNMT3B 1st and 2nd promoters was up-regulated by Sp3 overexpression and to a lesser extent by Sp1 overexpression, we next examined the Sp1-binding sites responsible for these promoter activities. Previously, we identified the minimal promoters of the DNMT3A 3rd promoter pGL3A-P3 (−334/+376), the DNMT3B 1st promoter pGL3B-P1 (−604/+309) and the DNMT3B 2nd promoter pGL3B-P2 (−469/+260) in NT2 cells [10]. However, for the DNMT3B 1st promoter, pGL3B-P1 showed weak activation by Sp3 (Table 2). We next co-transfected the Sp3 expression vector with a series of 5′ deletion mutant reporter vectors of pGL3B-P1. Significant activation (approx. 2-fold) by Sp3 was observed in the deletion mutant pGL3B-P1 (−102/+309) (results not shown). Therefore we considered this region to be the minimal promoter of DNMT3B 1st promoter in this experiment. Alternatively, there may be suppressive sequences in the 5′-portion of DNMT3B 1st promoter, since the shorter reporter sequences exhibited a significant luciferase activity.
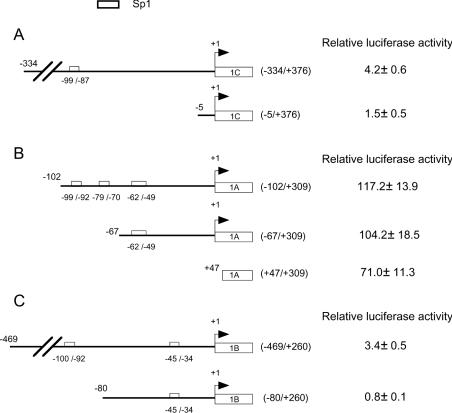
The minimal promoters, deletion mutants and the empty reporter vector pGL3-basic were transfected into HEK-293T cells (Figure 3). The results revealed that these minimal promoters still retained high basal activity compared with pGL3-basic. Further 5′ deletion of pGL3A-P3 (−334/+376) to (−5/+376) resulted in loss of basal activity (Figure 3A), indicating that crucial sites are located between −334 and −5. Deletion of pGL3B-P1 (−102/+309) to (−67/+309) resulted in a slight loss of the basal activity (Figure 3B). Further 5′ deletion to +47/+309 resulted in a 39.4% decrease in basal activity, suggesting that the region −67 to +47 is important. Further 5′ deletion of pGL3B-P2 (−469/+260) to (−80/+260) abrogated the basal activity (Figure 3C), which suggests that crucial sites are located between −469 and −80. We then checked the presence of Sp1-binding motifs in these regions with TFSEARCH (http://www.etl.go.jp/etl/ebrc/research/db/TFSEARCHJ.html). The results revealed one Sp1-binding site at −99 to −87 of the DNMT3A 3rd promoter, three Sp1-binding sites at −99 to −92, −79 to −70 and −62 to −49 of the DNMT3B 1st promoter and one Sp1-binding site at −100 to −92 of the DNMT3B 2nd promoter.
Figure 3. Identification of the Sp1-binding sites responsible for minimal promoter activities of DNMT3A and DNMT3B in HEK-293T cells.
Left panels: schematic structure of the minimal promoter of DNMT3A 3rd promoter pGL3A-P3 (−334/+376) (A), DNMT3B 1st promoter pGL3B-P1 (−102/+309) (B), DNMT3B 2nd promoter pGL3B-P2 (−469/+260) (C) and their deletion constructs are illustrated. The Sp1-binding sites are presented as open boxes and positions are shown relative to the TSP (+1) of each exon. Right panels: the plasmids containing minimal promoter of DNMT3A 3rd promoter, DNMT3B 1st and 2nd promoters and their deletion mutants were transfected into HEK-293T cells. Luciferase activity was expressed relative to the activity of empty reporter vector pGL3-basic. Results are expressed as the means±S.D. for three independent experiments.
Effect of site-directed mutagenesis of Sp1-binding sites on the DNMT3A and DNMT3B promoter activities
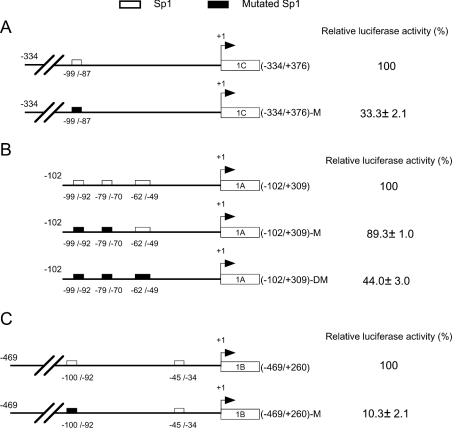
To investigate the role of each Sp1-binding site in the DNMT3A and DNMT3B minimal promoters, site-specific mutagenesis analyses for each Sp1-binding site were performed. As shown in Figure 4, mutations of the Sp1-binding sites at −99 to −87 of the DNMT3A 3rd promoter (Figure 4A) and −100 to −92 of DNMT3B 2nd promoter (Figure 4C) decreased the promoter activities to 33.3 and 10.3% respectively. Concerning the DNMT3B 1st promoter (Figure 4B), mutations of the Sp1-binding sites at −99 to −92 and −79 to −70 resulted in a slight loss of the promoter activity. However, when the binding site at −62 to −49 was also mutated, the promoter activity was reduced to 44.0%. The remaining promoter activity of pGL3B-P1 (−102/+309)-DM could be maintained by the existence of several Sp1-binding sites in the 1st exon [10]. However, these results indicated that these Sp1-binding sites are essential for DNMT3A and DNMT3B minimal promoter activities.
Figure 4. Effect of site-specific mutation of Sp1-binding sites on the DNMT3A and DNMT3B promoters.
Left panels: schematic representation of site-specific mutation of Sp1-binding sites on minimal promoters of DNMT3A 3rd promoter (A), DNMT3B 1st promoter (B) and DNMT3B 2nd promoter (C). The wild-type and mutant-type Sp1-binding sites are shown as open and filled boxes respectively. Right panels: luciferase activities (%) of the mutant reporter vectors were expressed relative to the activities of their wild-type counterparts. Results are expressed as the means±S.D. for three independent experiments.
Sp1 and Sp3 proteins bind to the DNMT3A and DNMT3B promoters
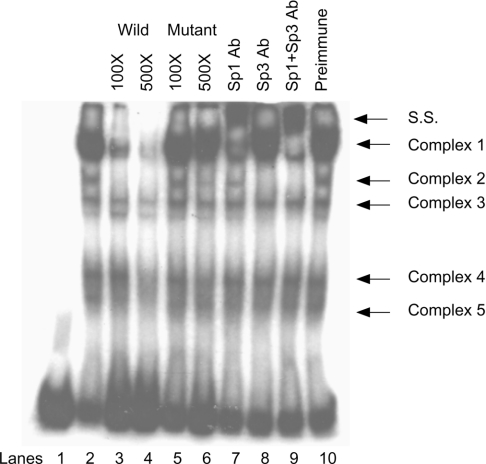
On the basis of the fact that the Sp1-binding sites at −99 to −87 of the DNMT3A 3rd promoter, −62 to −49 of the DNMT3B 1st promoter and −100 to −92 of the DNMT3B 2nd promoter are essential for the maintenance of the minimal promoter basal activity, we then examined the physical interaction between these binding sites and transcription factors. Oligonucleotide probes 3A-P3-W1 (from the DNMT3A 3rd promoter), 3B-P1-W1 (from the DNMT3B 1st promoter) and 3B-P2-W1 (from the DNMT3B 2nd promoter) were labelled and incubated with a nuclear extract of HEK-293T cells. The autoradiographs from all three probes were almost comparable. Representative data of the autoradiograph from probe 3B-P2-W1 is shown in Figure 5. The result revealed five major DNA–protein complexes (lane 2). Complexes 1 and 2 were completely competed for by a 500-fold excess of the unlabelled wild-type probe (lane 4), but were not competed for by a 500-fold excess of mutant-type probe (lane 6), indicating that they were specific complexes. To confirm the presence of Sp1 and Sp3 in these complexes, gel supershift was performed by using antibodies against Sp1 and Sp3. Complex 1 considerably decreased in the presence of Sp1 antibody (lane 7) and a combination of Sp1 and Sp3 antibodies further supershifted the remaining bands (lane 9), suggesting that Sp1 and Sp3 were components of complex 1. The addition of Sp3 antibody alone completely abolished complex 2 (lane 8), indicating that it was an Sp3–DNA complex. Since we detected a large Sp3 isoform (110–115 kDa) and small isoforms (70–80 kDa) in HEK-293T cells (Figure 6A), the upper and lower Sp3–DNA complexes may indicate the binding of large and small Sp3 isoforms respectively. No supershift complexes were observed in the presence of preimmune serum (lane 10). However, the last remaining bands of complex 1 (lane 9) could not be supershifted by either Sp1 and/or Sp3 antibodies; therefore their nature remains unknown. Altogether, these results indicated that Sp1 and Sp3 directly bind to the DNMT3A and DNMT3B promoters.
Figure 5. Sp1 and Sp3 bind to the 2nd promoter of DNMT3B.
The labelled synthetic oligonucleotide spanning nucleotides −119 to −73 relative to the TSP of the DNMT3B 2nd promoter (3B-P2-W1) was incubated with nuclear extract of the HEK-293T cells (lane 2) or in the presence of a 100 or 500 molar excess of the unlabelled wild-type probes (lanes 3 and 4) or the mutant-type probes in which the Sp1-binding site at −100/−92 (5′-GAGGCGGGG-3′) was mutated to 5′-ATATAGGGG-3′ (lanes 5 and 6). For the gel supershift assay, Sp1 and/or Sp3 antibodies were added to the reaction mixture (lanes 7–9). Preimmune serum was used as a control (lane 10). The supershifted bands (S. S.) are indicated.
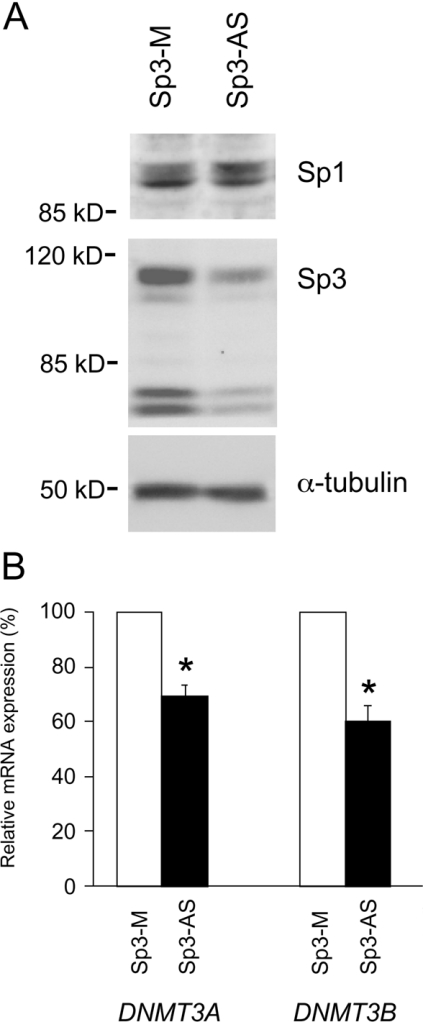
Figure 6. Down-regulation of DNMT3A and DNMT3B mRNAs with Sp3 antisense oligonucleotide.
(A) HEK-293T cells were incubated with either Sp3 antisense oligonucleotide (Sp3-AS) or mismatch oligonucleotide (Sp3-M). Total proteins from both groups were subjected to Western-blot analyses with anti-Sp1 and -Sp3 antibodies. (B) DNMT3A and DNMT3B mRNA levels from antisense and mismatch oligonucleotide-treated cells were quantified with Light-Cycler real-time PCR, then normalized to the GAPDH mRNA levels and expressed relative to the levels in mismatch oligonucleotide-treated cells. *P<0.05 by the t test. Results are expressed as the means±S.D. for three experiments.
Sp3-specific antisense oligonucleotide decreases DNMT3A and DNMT3B mRNA levels
To determine whether Sp1 and/or Sp3 regulate the DNMT3A and DNMT3B expression under the physiological conditions in parental HEK-293T cells, we specifically down-regulated Sp1 or Sp3 expression by using the antisense oligonucleotide. In Figure 6(A), Western-blot analysis showed that the Sp3 expression considerably decreased in antisense-treated cells, whereas the Sp1 level was unaffected. We further analysed the expression levels of DNMT3A and DNMT3B by LightCycler real-time PCR. As shown in Figure 6(B), Sp3 antisense oligonucleotide decreased the mRNA levels of DNMT3A and DNMT3B to 69.3 and 60.0% compared with the levels in mismatch oligonucleotide-treated cells respectively. Similar results were also obtained from semiquantitative RT–PCR analyses (results not shown). We inhibited Sp1 expression by an Sp1-specific antisense oligonucleotide but no significant decrease in DNMT3A and DNMT3B mRNA levels were observed (results not shown).
DISCUSSION
DNMTs are believed to play important roles in the development of cancer since they are essential for the maintenance of aberrant methylation in cancer cells [20] and for cancer cell survival [9]. However, there are a few studies of the mechanisms involved in the regulation of expression of DNMT3A and DNMT3B. In the present study, we indicated that Sp1 and Sp3 are essential for the regulation of DNMT3A and DNMT3B expression. Treatment with mithramycin A, a GC-rich DNA-binding protein inhibitor, repressed the promoter activities and the mRNA levels of DNMT3A and DNMT3B. The results from site-directed mutagenesis of Sp1-binding sites also suggested that the DNMT3A and DNMT3B promoter activities are largely dependent on these Sp1-binding sites and Sp proteins that interact with these binding sites. Whereas the results of some previous studies showed that Sp3 is a repressor of Sp1-mediated transcription [14,21], the results from our luciferase assay showed that both Sp1 and Sp3 were positive regulators of the DNMT3A and DNMT3B promoters not only in HEK-293T cells but also in several other cell lines (A. Jinawath, S. Miyake, Y. Yanagisawa, Y. Akiyama and Y. Yuasa, unpublished work). It has been reported that several promoters are also positively regulated by both Sp1 and Sp3 [8,22,23].
To determine whether the DNMT3A and DNMT3B genes are the downstream Sp1 and Sp3 target genes, overexpression and knockout experiments were performed. Overexpression of Sp3 in HEK-293T cells significantly increased DNMT3A and DNMT3B mRNA levels. Although we could not inhibit the Sp3 protein expression by using Sp3 antisense oligonucleotide completely, a significant decrease in DNMT3A and DNMT3B was found, supporting the view that Sp3 may be required for the expression of DNMT3A and DNMT3B in vivo. By a similar approach, the BACE1 (beta-amyloid precursor protein cleaving enzyme 1) gene was also shown to be regulated by Sp1, since the BACE1 mRNA level in Sp1 knockout cells was reduced to 58.6% [24], a reduction rate similar to ours. On the other hand, Sp1 overexpression did not induce significant up-regulation of DNMT3A and DNMT3B mRNA levels and treatment with an Sp1 antisense oligonucleotide did not significantly inhibit the expression of these two genes, although physical binding of Sp1 and Sp3 was found in the DNMT3A and DNMT3B promoters. These results suggest that Sp3 plays a more important role than Sp1 in the regulation of DNMT3A and DNMT3B in HEK-293T cells. Further experiments will be required to determine the more precise roles of Sp1 and Sp3 in the regulation of these promoters.
Sp3 functions as a repressor or weak activator when it binds to promoters containing multiple binding sites, whereas it functions as an activator when it binds to a promoter containing a single binding site [14]. The DNMT3A 3rd and DNMT3B 2nd minimal promoters contain one Sp1-binding site at −99 to −87 and −100 to −92 respectively; therefore these promoters may be up-regulated by Sp3 in this model. However, DNMT3B/1A, which is the major 5′ splicing variant of DNMT3B in all the cell lines examined (A. Jinawath, S. Miyake, Y. Yanagisawa, Y. Akiyama and Y. Yuasa, unpublished work), encoded from the DNMT3B 1st promoter containing a cluster of Sp1-binding sites, was also up-regulated by Sp3. Thus the responses of this promoter to Sp3 may depend on the context of a specific promoter. Promoters that contain multiple Sp1-binding sites and are positively regulated by Sp3 have also been reported [23,25].
Several DNMT3B isoforms resulting from alternative splicing of exons 10, 21 and/or 22 have been reported [5,26]. Here, we have shown a new splicing variant of the 5′-region of DNMT3B for the first time. This variant is associated with DNMT3B/1A and lacks exon 5, resulting in the loss of 42 amino acids from the N-terminal portion. We also found this splicing variant lacking exon 5 in association with DNMT3B/1B in other cell lines (A. Jinawath, S. Miyake, Y. Yanagisawa, Y. Akiyama and Y. Yuasa, unpublished work). The first 298 amino acids in the N-terminal portion of DNMT3B are essential for the physical binding with DNMT1 and DNMT3A, which may facilitate the rapid establishment of the de novo methylation pattern [27]. Therefore the loss of exon 5, which contains 42 amino acids (between amino acids 103 and 144), may impair the binding with other DNMTs. Determination of the functions of these 5′ splicing variants of DNMT3B and their association with 3′ alternative splicing of exons 10, 21 and/or 22 will require further experiments.
Sp1 and Sp3 have been reported to participate in almost all biological aspects of cells including carcinogenesis. For example, overexpression of Sp3 contributes to a repression of TGF-β (transforming growth factor-β) receptor expression in breast cancer cells [21]. Sp1 and Sp3 are required for the induction of vascular endothelial growth factor-A by oxidative stress in gastric cancer cells [22] and constitutive expression of matrix metalloproteinase-2 in astrocytoma cells [28]. Our results indicate that Sp proteins, particularly Sp3, control both DNMT3A and DNMT3B expressions. Since DNMT3A and DNMT3B have been reported to be overexpressed in many tumours [5], it is tempting to speculate that the dysregulation of Sp3 and Sp1 contributes to DNMT3A and DNMT3B overexpression in cancer cells. However, the promoters of DNMT3A and DNMT3B also contain the binding sites for other transcription factors, including activator protein 1, nuclear factor-Y, E2F, Ets, SRY etc. [10]. To clarify further the mechanisms underlying the regulation of DNMT3A and DNMT3B expression in a cell- and tissue-specific manner, it is important to analyse other transcription factors as well as Sp proteins.
Acknowledgments
We thank S. Smale, J. M. Horowitz and T. Okamoto (Department of Molecular Genetics, Nagoya City University Medical School, Nagoya, Japan) for providing the plasmids. We also thank Yun-Qing Bai and Shoji Yamaoka for valuable technical advice and helpful discussions. This work was partially supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Baylin S. B., Herman J. G. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 2.Yoder J. A., Soman N. S., Verdine G. L., Bestor T. H. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 3.Okano M., Xie S., Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 4.Yoder J. A., Yen R. W., Vertino P. M., Bestor T. H., Baylin S. B. New 5′ regions of the murine and human genes for DNA (cytosine-5)-methyltransferase. J. Biol. Chem. 1996;271:31092–31097. doi: 10.1074/jbc.271.49.31092. [DOI] [PubMed] [Google Scholar]
- 5.Robertson K. D., Uzvolgyi E., Liang G., Talmadge C., Sumegi J., Gonzales F. A., Jones P. A. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakin A. V., Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science. 1999;283:387–390. doi: 10.1126/science.283.5400.387. [DOI] [PubMed] [Google Scholar]
- 7.Hodge D. R., Xiao W., Clausen P. A., Heidecker G., Szyf M., Farrar W. L. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J. Biol. Chem. 2001;276:39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- 8.Kishikawa S., Murata T., Kimura H., Shiota K., Yokoyama K. K. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur. J. Biochem. 2002;269:2961–2970. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 9.Beaulieu N., Morin S., Chute I. C., Robert M. F., Nguyen H., MacLeod A. R. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J. Biol. Chem. 2002;277:28176–28181. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- 10.Yanagisawa Y., Ito E., Yuasa Y., Maruyama K. The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents. Biochim. Biophys. Acta. 2002;1577:457–465. doi: 10.1016/s0167-4781(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 11.Bouwman P., Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 12.Hagen G., Muller S., Beato M., Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Udvadia A. J., Templeton D. J., Horowitz J. M. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majello B., De Luca P., Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J. Biol. Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 15.Bai Y. Q., Miyake S., Iwai T., Yuasa Y. CDX2, a homeobox transcription factor, upregulates transcription of the p21/WAF1/CIP1 gene. Oncogene. 2003;22:7942–7949. doi: 10.1038/sj.onc.1206634. [DOI] [PubMed] [Google Scholar]
- 16.Wen X. Z., Miyake S., Akiyama Y., Yuasa Y. BMP-2 modulates the proliferation and differentiation of normal and cancerous gastric cells. Biochem. Biophys. Res. Commun. 2004;316:100–106. doi: 10.1016/j.bbrc.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Chatterjee S., Zaman K., Ryu H., Conforto A., Ratan R. R. Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann. Neurol. 2001;49:345–354. [PubMed] [Google Scholar]
- 18.Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation. Cell (Cambridge, Mass.) 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas M., Noe V., Jensen K. B., Ciudad C. J. Cloning and characterization of the 5′-flanking region of the human transcription factor Sp1 gene. J. Biol. Chem. 2001;276:22126–22132. doi: 10.1074/jbc.M010740200. [DOI] [PubMed] [Google Scholar]
- 20.Rhee I., Bachman K. E., Park B. H., Jair K. W., Yen R. W., Schuebel K. E., Cui H., Feinberg A. P., Lengauer C., Kinzler K. W., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature (London) 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 21.Ammanamanchi S., Brattain M. G. Sp3 is a transcriptional repressor of transforming growth factor-beta. J. Biol. Chem. 2001;276:3348–3352. doi: 10.1074/jbc.M002462200. [DOI] [PubMed] [Google Scholar]
- 22.Schafer G., Cramer T., Suske G., Kemmner W., Wiedenmann B., Hocker M. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem. 2003;278:8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 23.Lee M. G., Pedersen P. L. Glucose metabolism in cancer: importance of transcription factor-DNA interactions within a short segment of the proximal region of the type II hexokinase promoter. J. Biol. Chem. 2003;278:41047–41058. doi: 10.1074/jbc.M307031200. [DOI] [PubMed] [Google Scholar]
- 24.Christensen M. A., Zhou W., Qing H., Lehman A., Philipsen S., Song W. Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1. Mol. Cell. Biol. 2004;24:865–874. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prowse D. M., Bolgan L., Molnar A., Dotto G. P. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 1997;272:1308–1314. doi: 10.1074/jbc.272.2.1308. [DOI] [PubMed] [Google Scholar]
- 26.Chen T., Ueda Y., Xie S., Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 27.Kim G. D., Ni J., Kelesoglu N., Roberts R. J., Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H., Sun Y., Benveniste E. N. The transcription factors Sp1, Sp3, and AP-2 are required for constitutive matrix metalloproteinase-2 gene expression in astroglioma cells. J. Biol. Chem. 1999;274:29130–29137. doi: 10.1074/jbc.274.41.29130. [DOI] [PubMed] [Google Scholar]