Abstract
The thermoacidophilic archaeon Sulfolobus solfataricus P2 encodes three hypothetic endo-β-glucanases, SSO1354, SSO1949 and SSO2534. We cloned and expressed the gene sso1949 encoding the 334 amino acids containing protein SSO1949, which can be classified as a member of glycoside hydrolase family 12. The purified recombinant enzyme hydrolyses carboxymethylcellulose as well as cello-oligomers, with cellobiose and cellotriose as main reaction products. By following the hydrolysis of a fluorescently labelled cellohexaoside under a wide variety of conditions, we show that SSO1949 is a unique extremophilic enzyme. This archaeal enzyme has a pH optimum of approx. pH 1.8 and a temperature optimum of approx. 80 °C. Furthermore, the enzyme is thermostable, with a half-life of approx. 8 h at 80 °C and pH 1.8. The thermostability is strongly pH-dependent. At neutral pH, the thermal inactivation rate is nearly two orders of magnitude higher than at pH 1.8. Homology modelling suggests that the catalytic domain of SSO1949 has a similar fold to other mesophilic, acidophilic and neutral cellulases. The presence of a signal peptide indicates that SSO1949 is a secreted protein, which enables S. solfataricus to use cellulose as an external carbon source. It appears that SSO1949 is perfectly adapted to the extreme environment in solfataric pools. A cellulolytic enzyme with such a combination of stability and activity at high temperatures and low pH has not been described so far and could be a valuable tool for the large-scale hydrolysis of cellulose under acidic conditions.
Keywords: cellulase, endo-β-glucanase, extremophile, glycoside hydrolase, Sulfolobus
Abbreviations: CMC, carboxymethylcellulose; FRET, fluorescence resonance energy transfer; GH, glycoside hydrolase
INTRODUCTION
Cellulose, a main component of plant cell walls, is the most abundant biopolymer in the world and is considered to be an important alternative source of renewable energy [1]. Cellulose is a linear biopolymer of D-glucose, linked by β-1,4-glucosyl linkages. The high degree of intermolecular hydrogen bonding in crystalline cellulose explains its exceptional stability. High temperatures combined with acids are required to hydrolyse cellulose chemically [2]. Cellulose can also be hydrolysed enzymically under milder reaction conditions. A cellulosic enzyme (cellulase) system consists of three major components: endo-β-glucanase (EC 3.2.1.4), cellobiohydrolase (EC 3.2.1.91) and β-glucosidase (EC 3.2.1.21). Endo-β-glucanases randomly hydrolyse the internal glycosidic bond to decrease the length of the cellulose chains. Cellobiohydrolases are exo- or endo-processive enzymes that split off cellobiose. Cellobiose is subsequently hydrolysed by β-glucosidases to glucose [3]. The cellulolytic enzymes are members of a superfamily of GHs (glycoside hydrolases), with more than 2200 known protein sequences. To date, the GHs have been classified into more than 80 different GH families based on their amino acid sequence similarities [4,5]. Cellulases are found in families 5–12, 26, 44, 45 and 48 [6]. Family 12 comprises endoglucanases from mesophilic and thermophilic bacteria, fungi and archaea. Currently, the X-ray structures of five enzymes of GH family 12 have been solved [7–11]. Unlike other cellulase families, the cellulases of family 12 do not contain a cellulose-binding module, which explains the low activity on crystalline cellulose of these enzymes. As catalytic residues, two highly conserved glutamic residues have been identified in the cellulases, which are supposed to act as a nucleophile and acid/base catalyst [12].
The hyperthermophilic archaea have received considerable attention because of the high thermostability of their enzymes. The crenarchaeote Sulfolobus solfataricus, which thrives in acidic volcanic hot springs, is a thermoacidophile growing optimally at approx. 80 °C and pH 2– 4. The genome of S. solfataricus has been sequenced, and three genes (sso1354, sso1949 and sso2534) encoding potentially secreted endo-β-glucanases of GH family 12 are found in the genome [13]. The protein SSO2534 has been characterized and is active at approx. pH 5.8 [14].
In the present study, we report the cloning, expression, purification and characterization of the endo-β-glucanase encoded by gene sso1949 from the thermoacidophilic archaeon S. solfataricus P2. The enzyme shows an exceptional activity at extremely low pH and is thermostable. To our knowledge, a similar combination of acid and heat stability has not yet been reported for other GHs.
EXPERIMENTAL
Cloning, expression and purification of SSO1949
Genomic DNA of S. solfataricus was isolated from a 5 ml culture of S. solfataricus by CTAB (hexadecyltrimethylammonium bromide) extraction. The region of the SSO1949 gene encoding the hypothetic extracellular cellulase SSO1949 was amplified by PCR with primers sso1949-for (5′-GCTAGCGCTATTTACCTACACC-3′) and sso1949-rev (5′-AAGCTTAGAGGAGAGTTTCAGAAAAGTTGG-3′). These primers create an NheI and a HindIII (bold letters) cleavage site upstream and downstream of the sso1949 gene respectively. The PCR product was sequenced and cloned into pET-28c (Novagen, Madison, WI, U.S.A.) to yield the plasmid pET-28c-1949Nhis. In this construct, SSO1949 carries a fusion of six histidine residues at its N-terminus. The native signal peptide was deleted by this cloning strategy. The N-terminally truncated SSO1949 lacking the serine- and threonine-rich region (amino acids 25–85) was cloned as follows: a PCR fragment obtained with the forward primer 5′-GCTAGCTTTTATCTTGAAGTGAACATGTGG-3′ and primer sso1949-rev was cut with NheI and HindIII and ligated into the vector pET-28c. The expression plasmids were used to transform Escherichia coli BL21 (codon+) cells (Novagen).
For expression, cells were grown overnight in 20 ml of Luria–Bertani medium with 50 μg/ml kanamycin and 50 μg/ml chloramphenicol at 37 °C. After inoculation of 4 litres of Luria–Bertani medium, the incubation was continued to an A600 of 0.6–0.8. Isopropyl β-D-thiogalactoside was then added to a final concentration of 1 mM and the culture was fermented for further 12 h at room temperature (25 °C). Cells were harvested by centrifugation. The cell pellet was resuspended in 50 ml of buffer A [100 mM Tris/HCl, pH 7.5, 1 M NaCl, 1 mM 2-mercaptoethanol, 100 μM PMSF, 5% (v/v) glycerol]. Cells were disrupted by a microfluidizer (Newton, MA, U.S.A.) and the cell debris was removed by centrifugation. The recombinant cellulase carrying a His6 tag was then purified by immobilized-metal- chelate affinity chromatography. SSO1949 was eluted at 50 mM imidazole from a Ni2+-nitrilotriacetate column. The protein pool was heat treated at 70 °C for 20 min, and denatured proteins were removed by centrifugation. The supernatant was dialysed against buffer B (20 mM potassium phosphate, pH 5.5) and further purified by anion-exchange chromatography on an EMD TMAE 650 (S) column. The protein was eluted at 500 mM NaCl. The active fractions were dialysed against the storage buffer (20 mM KPO4, pH 5.5, 1 mM 2-mercaptoethanol, 50% glycerol) and stored at −20 °C. The protein concentration was determined using the theoretical molar absorption coefficient of 97010 M−1·cm−1 at 280 nm.
Activity gel
The protein preparation was separated by SDS/PAGE in gels containing 0.2% (w/v) CMC (carboxymethylcellulose; Wolff Walsrode AG, Walsrode, Germany). The protein was then renatured by several wash steps, first with a mixture (4:1, v/v) of 20 mM potassium phosphate buffer (pH 1.8) and propan-2-ol for 30 min and then twice with 20 mM potassium phosphate buffer (pH 1.8) for 30 min. Finally, the gel was incubated in 10 mM potassium phosphate buffer (pH 1.8) at 75 °C for 10 h with two buffer exchanges. The gel was washed with 50 mM potassium phosphate buffer (pH 7) for 30 min and stained with 0.1% (w/v) Congo Red (Sigma, Deisenhofen, Germany) solution for 30 min and destained with 1 M NaCl.
Hydrolysis of CMC
Hydrolysis of CMC was measured by the decrease in viscosity and increase in reducing ends. For viscosity measurements, a 1% solution of CMC was incubated with SSO1949 at different pH and temperature values for 2 h. After adjustment of the samples to neutral pH, the apparent viscosity was measured by a cone-plate system (Thermo Haake Rheostress 6000 viscosimeter; Haake, Karslruhe, Germany) at 20 °C. For the reducing end determination, a 1% CMC solution was incubated with enzyme in 10 mM potassium phosphate. Aliquots of 50 μl were neutralized and the amount of reducing sugar ends was determined by the dinitrosalicyclic acid (DNS) method [15].
Analysis of degradation products by TLC
CMC (1%) and cello-oligosaccharides (8 mg/ml) (Fluka, Deisenhofen, Germany) were digested with 1.3 μM SSO1949 in 10 mM potassium phosphate buffer (pH 1.8) for 1 h at 80 °C. Aliquots (1–2 μl) were spotted on a silica 60 TLC plate (MachereyNagel, Düren, Germany), which was developed in ethyl acetate/acetic acid/water (2:1:1, by vol.) twice for 1 h [16]. Reducing sugars were stained with the thymol–sulphuric acid reagent (thymol/ethanol/sulphuric acid, 0.5:95:5, weight/vol./vol.) [17].
Fluorescent activity assay
The bifunctionalized cellohexaoside sodium N-{2-N-[(S-(4-deoxy-4-dimethylaminophenylazophenylthioureido-β-D-glucopyranosyl)-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl)-2-thioacetyl]aminoethyl}-1-naphthylamine-5-sulphonate was offered as a substrate [18]. The reaction was followed by monitoring EDANS {5-[(2-aminoethyl)-amino]naphthalene-1-sulphonic acid} fluorescence, which increases in the course of the enzymic reaction. The measurement was performed in 0.1 M potassium phosphate buffer at various pH and temperatures on a PerkinElmer LS50B spectrofluorimeter equipped with a thermostatically controlled cuvette holder. Excitation was at 340 nm and emission was observed at 470 nm. Initial rate constants were determined at several substrate concentrations in the presence of 0.1 μM SSO1949. The Michaelis–Menten constant Km was calculated using the formula V=Vmax[S/(S+Km)]. The maximal velocity Vmax was used to calculate the specific activity of 1 μmol·min−1·(mg of enzyme)−1. The pKa values were obtained by fitting the pH–activity profile data to the equation:
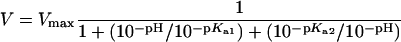 |
RESULTS
Sequence analysis, expression and purification of SSO1949
The gene sso1949 of the completely sequenced genome of S. solfataricus P2 has been predicted to encode an endo-β-glucanase [13]. SignalP analysis [19] indicates the presence of a signal peptide for amino acid positions 1–24, suggesting that SSO1949 is a secreted enzyme. Sequence comparison (Figure 1A) classifies this enzyme as a member of GH family 12 [5]. Sequence analysis does not predict the presence of a carbohydrate-binding domain in SSO1949.
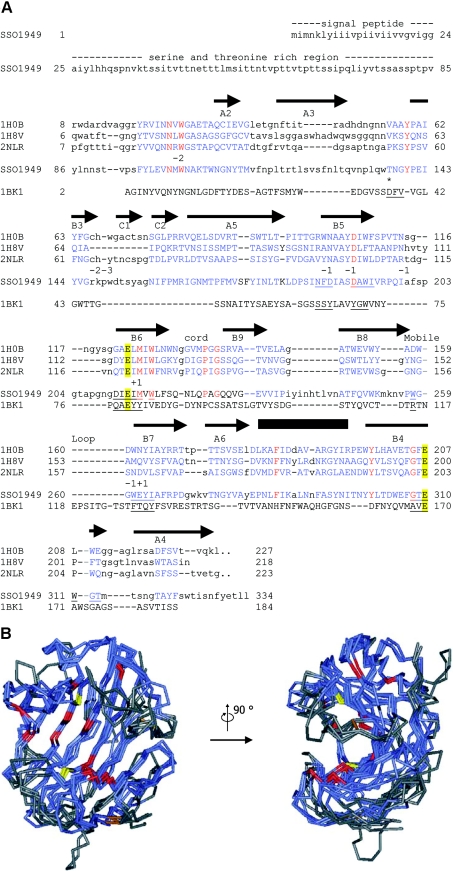
Figure 1. Sequence alignment and structural model of SSO1949.
(A) Aligned are the catalytic modules of the cellulases from Rhodothermus marinus (Protein Data Bank no. 1H0B) [9], T. reesei (PDB no. 1H8V) [8], Streptomyces lividans (PDB no. 2NLR) [7] and S. solfataricus SSO1949. The complete sequence is given only for SSO1949. The first 24 amino acids are predicted to be a signal sequence followed by a serine- and threonine-rich region (amino acids 25–85), which has no sequence similarity to other proteins. Regions of the GH domains which could be aligned are shown in blue capital letters. Identical residues are in red. The catalytic glutamate residues Glu-213 and Glu-310 are emphasized with yellow boxes. The arrows (β-strands) and the box (α-helix) above the sequence show the positions of the secondary-structure elements of the R. marinus cellulase. The elements are labelled according to Crennell et al. [9]. The numbers below the sequence 2NLR indicate the subsites of the active-site cleft. The alignment also includes the extremely acidophilic xylanase from A. kawachii (PDB no. 1BK1) [24]. This enzyme belongs to the GH family 11, which has a fold similar to the GH family 12 [4]. For the acidophilic enzymes from S. solfataricus and Aspergillus kawachii, residues within 0.5 nm of the catalytic glutamate residues are underlined. Thr-137 is homologous with Asp-37 of the A. kawachii xylanase, which is implicated in the low-pH optimum of this enzyme (*, see text for details). The alignment of the two acidophilic enzymes along with the neutral cellulases does not reveal any particular amino acid which could be responsible for the low pH optimum of the S. solfataricus enzyme. (B) The structure of SSO1949 was modelled with ESyPred3D [21] using the cellulase from R. marinus (PDB no. 1H0B) as template. Shown are the backbones of the model structure (amino acids 86–329) superimposed with the structures of the cellulases from R. marinus, T. reseii (PDB no. 1H8V) and S. lividans (PDB no. 2NLR). In blue are the parts of the protein which could be structurally aligned. Structural alignment was not possible for the regions in grey. The backbone trace is red for amino acids which are completely conserved within the four proteins. The catalytic glutamate residues are depicted in yellow. Disulphide bridges are displayed in brown. The region of the active site seems to be structurally well conserved and contains most of the completely conserved amino acids. The active-site cleft is best viewed from the side (right picture).
The catalytic domain of SSO1949 has significant amino acid sequence conservation compared with the thermostable cellulase Cel12A from Rhodothermus marinus (24.5% identity and 35.1% similarity), a GH 12 enzyme, whose crystal structure has been determined [20]. The sequence similarity allowed a homology modelling [21] of the catalytic domain (amino acids 86–329) of SSO1949 using the cellulase from R. marinus as template (Figure 1B). It was not possible to model the structure of the N-terminal amino acids 25–85. This part is Ser/Thr-rich and does not show homology to known sequences. From the sequence and structure-based alignment combined with the modelling, we infer that SSO1949 has a typical GH clan C fold [5] and an active site similar to that of other cellulases of family 12. The modelling places two putative catalytic glutamate residues, namely Glu-213 and Glu-310, to positions equivalent to that of catalytic glutamate residues of other known GH structures. On the basis of the known structures and well-characterized mechanisms of cellulases of GH family 12, Glu-213 should act as a nucleophile, whereas Glu-310 is a candidate for the acid–base function.
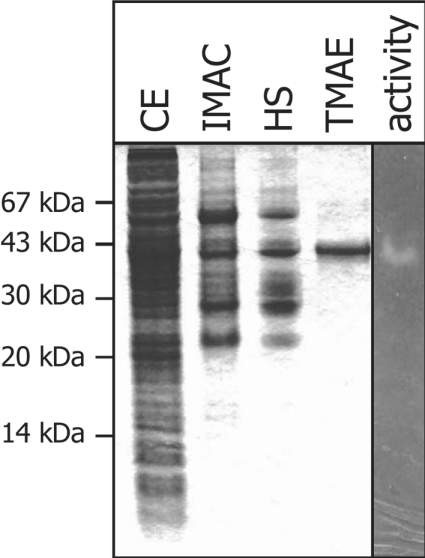
For expression, the putative signal peptide of 24 amino acids was substituted by an N-terminal His6 tag. The sso1949 gene contains 25 rare codons, and therefore the E. coli strain BL21 (codon+) was used for expression. The purification procedure included immobilized-metal-chelate affinity chromatography, heat denaturation and anion-exchange chromatography (Figure 2). The recombinant protein, purified to apparent homogeneity, consists of 333 amino acids and migrates in SDS gels with an apparent mass of 37 kDa, which is in agreement with the theoretical mass of 37.508 kDa. Total yield of a 4 litre culture was approx. 1 mg of purified SSO1949.
Figure 2. Purification of recombinant SSO1949.
Protein fractions were analysed by SDS/PAGE followed by Coomassie Blue staining: crude extract (lane CE), the pool after Ni2+-nitrilotriacetate chromatography (lane IMAC), supernatant of heat denaturation (lane HS) and the pool after anion-exchange chromatography (lane TMAE). A sample of the purified protein was also loaded on a CMC-containing activity gel (lane activity). After renaturation (see the Experimental section) cellulase activity was revealed by staining with Congo Red.
In an attempt to improve overexpression, an N-terminal deletion mutant lacking the serine- and threonine-rich region of amino acids 25–85 was expressed and purified. However, this truncated protein was inactive in the cellulose assays (results not shown).
Characterization of GH activity
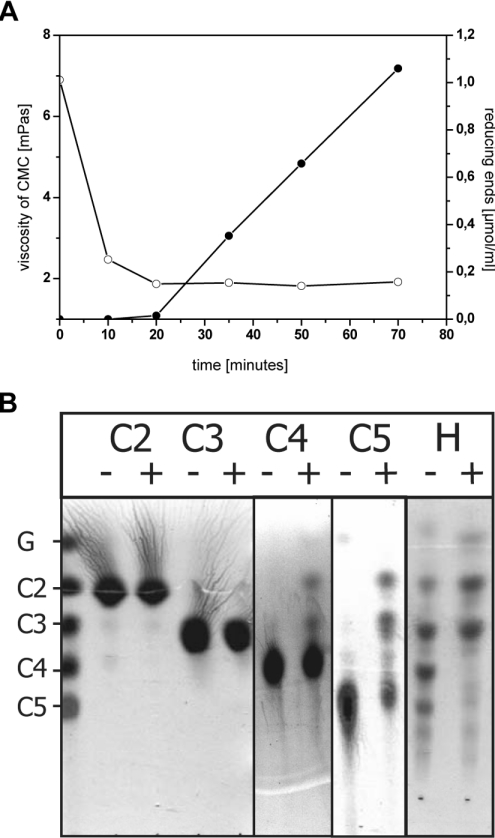
The cellulase activity of SSO1949 is readily detected by the release of reducing sugar ends from CMC and by viscosity measurements. These experiments show a rapid decrease in viscosity at the beginning of hydrolysis that is not accompanied by a large increase in reducing sugar ends. However, at longer reaction times viscosity stays nearly constant, whereas a strong increase in reducing sugar ends was observed (Figure 3A). The initial decrease in viscosity indicates an endoglycolytic action of the enzyme, which leads to a fast decrease in chain length.
Figure 3. SSO1949 degrades CMC and cello-oligomers.
(A) A 1% CMC solution was incubated with 1.1 μM SSO1949 at pH 1.8 and 80 °C. Samples were withdrawn at several time points, and the viscosity (○) and the amount of reducing ends (●) were measured. (B) Cellobiose (C2), cellotriose (C3), cellotetraose (C4), cellopentose (C5) and a hydrolysate of cellulose (H), each 8 mg/ml, were incubated with 1.3 μM SSO1949 in 20 mM potassium phosphate (pH 1.8) for 60 min at 80 °C. Reactions with (+) and without (−) enzyme were spotted on silica 60 plates and developed with a mixture of ethyl acetate/acetic acid/water (2:1:1, by vol.). Sugars were visualized with thymol sulphuric acid. Only oligomers larger than three units are degraded. Cellobiose and cellotriose are the main products of enzymic hydrolysis by SSO1949.
Next, we characterized the enzymic activity of SSO1949 by degradation of cello-oligomers followed by TLC to identify the reaction products. As shown in Figure 3(B), SSO1949 is active towards cellotetraose, whereas cellotriose and cellobiose are not hydrolysed under these assay conditions. When cellopentose or a mixture of higher oligosaccharides produced by HCl treatment of crystalline cellulose (Figure 3B) is offered, mainly cellotriose and cellobiose are obtained as degradation products. Glucose is found only in minor amounts. These observations also indicate that SSO1949 is an endoglucanase, which requires a β-linked cellulose tetramer as minimal substrate length and mainly forms cellotriose and cellobiose as reaction products. Furthermore, SSO1949 does not hydrolyse p-nitrophenyl β-D-cellobioside and p-nitrophenyl β-D-cellotrioside (results not shown). These compounds are used as model substrates for cellobiohydrolases, and the lack of activity towards these substrates further underscores that SSO1949 is not such an enzyme.
Alternative substrates of β-glucanases, such as xylan, laminarin and lichenan, appear not to be substrates for SSO1949. At low temperatures and at neutral pH, SSO1949 does not show activity towards these substrates. At the optimal reaction condition of SSO1949 (high temperature and low pH, see below), these polysaccharides show a high rate of spontaneous hydrolysis and the activity could not be measured reliably.
No activity could be detected with crystalline cellulose at both acidic and neutral pH values. In the sequence of SSO1949, a cellulose-binding module cannot be identified and the failure to detect activity towards crystalline cellulose is ascribed to the lack of this domain.
Several control experiments were performed to ensure that the cellulase activity was due to the recombinant SSO1949. When an empty vector was used for the purification, no cellulose activity could be measured (results not shown). Furthermore, the cellulase activity is abolished in both the neutral and acidic pH range on pretreatment of the enzyme with proteinase K (results not shown).
pH and temperature dependence of SSO1949 activity
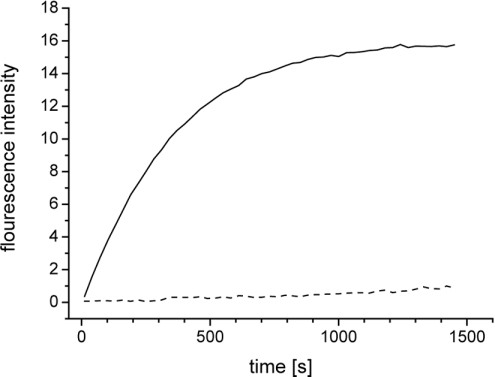
For a more detailed characterization of the cellulase activity of SSO1949, a FRET (fluorescence resonance energy transfer)-based assay was used. In this assay, a cellohexaoside is offered as substrate which carries the donor fluorophore EDANS at one end and the chromophore 4-(4′-dimethylaminobenzeneazo)benzene as acceptor chromophore at the other end [18]. Incubation of SSO1949 with the fluorescent hexaoside leads to an increase in fluorescence at 490 nm, indicating cleavage of this substrate (Figure 4). The FRET assay proved to be much more sensitive than the viscosity and reducing-sugar-end assays. The fluorescent cellohexaoside is very stable under the extreme pH and temperature conditions and allows precise measurements of cellulase activity over a wide range of conditions.
Figure 4. SSO1949 hydrolyses a fluorescent cellohexaoside.
Fluorescent hexaoside (3 μM) [18] was incubated with 0.1 μM SSO1949 in 100 mM potassium phosphate (pH 1.8) at 75 °C. Excitation was at 340 nm and fluorescence increase due to the hydrolysis of the hexaoside was monitored at 470 nm. The fluorescence intensity was sampled every 30 s. Shown are a progress curve with enzyme (—) and a control reaction without enzyme (---). Spontaneous degradation of the substrate even at these extreme assay conditions is very low, with a half-life of several hours.
Measurements of the initial kinetics at various substrate concentrations yielded a Km value of 2 μM for the fluorescent cellohexaoside at pH 1.8 and 80 °C. The specific activity of the purified SSO1949 was 1.2 μmol·min−1·mg−1 as measured by the reducing sugar method and 1.0 μmol·min−1·mg−1 as measured by the fluorescence method.
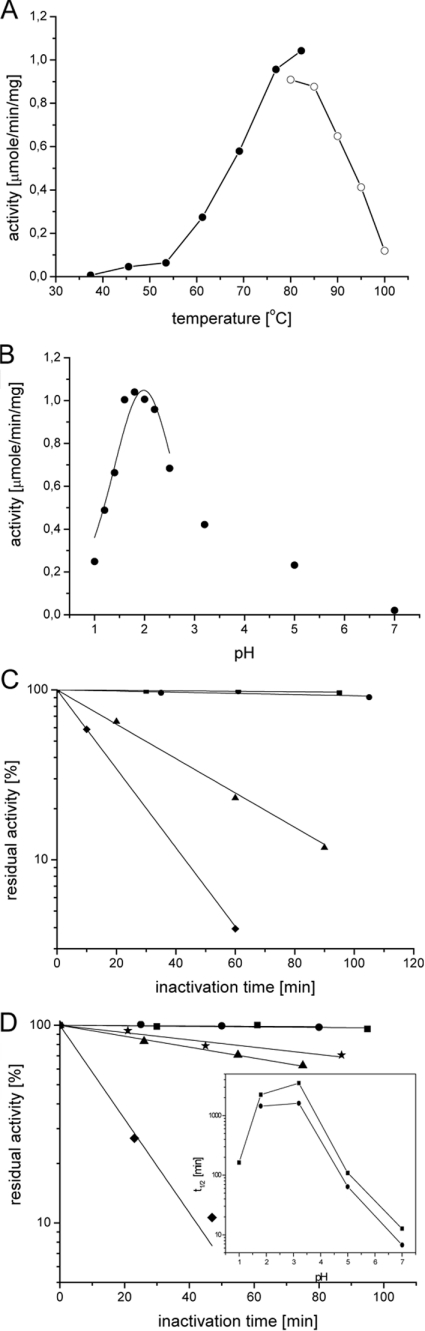
The most intriguing property of SSO1949 is its optimal activity at acidic pH and high temperatures. FRET measurements of SSO1949 activity at various pH values reveal a bell-shaped pH– activity profile with an optimum in the strongly acidic range. At 80 °C, SSO1949 shows the highest activity at pH 1.8 and a rather low activity at neutral pH values (Figure 5B). Most remarkably, the residual activity of the enzyme can still be detected at pH 1. The same pH–activity profile was obtained from measurements with CMC as a substrate and from viscosity measurements, although these measurements showed a high background at the strongly acidic pH values.
Figure 5. Activity and stability profile of SSO1949.
(A) The cellulolytic activity of SSO1949 was assayed at various temperatures. SSO1949 (0.1 μM) was incubated with 3 μM fluorescently labelled hexaoside (Km=2 μM) in 100 mM potassium phosphate buffer (pH 1.8). The initial rates of hydrolysis (●) were calculated after correcting for the temperature-dependent fluorescence intensity. SSO1949 works best at approx. 80 °C. At temperatures above 85 °C the enzymic hydrolysis was determined with a single end-point determination after 5 min (○). The ascending limb of the temperature profile yielded an activation energy of 59 kJ/mol after Arrhenius analysis. (B) The pH optimum of SSO1949 was determined at 80 °C. The pH profile displays a bell-shaped behaviour typical for active sites with protonable groups. The data obtained for the range of pH values from 1 to 3.2 were fitted assuming two protonable groups and yielded the apparent pKa values of 1.7 and 2.3. (C) SSO1949 was preincubated for up to 2 h at various temperatures before the activity was determined at the optimal conditions (pH 1.8, 80 °C). SSO1949 resists a 2 h incubation at pH 1.8 at 80 °C (■) and 85 °C (●). At 90 °C (▲) and 95 °C (◆) the half-life of the enzyme decreases to 30 and 13 min respectively. (D) SSO1949 was preincubated at 80 °C at different pH before the activity was determined. SSO1949 is stable at pH 1.8 (■) and 3.2 (●). At the extreme pH of 1 (∗) SSO1949 is less stable, with a half-life of 160 min. At pH 5 (▲) and 7 (◆) SSO1949 is inactivated, with half-lives of 110 and 13 min respectively. Inset, the half-lives of SSO1949 are plotted as a function of pH at 80 °C (■) and 85 °C (●). SSO1949 is most stable at approx. pH 2.
SSO1949 is both an extremely acidophilic and thermophilic enzyme. Figure 5(A) shows the temperature profile of SSO1949 activity at pH 1.8. The enzyme has the highest turnover in the range from 75 to 85 °C and only low activity at 30–50 °C. For technical reasons, the enzymic activity above 85 °C was measured by an end-point determination (see the legend for Figure 5 for details).
Stability of SSO1949
In view of the unusual activity profile of SSO1949, it was important to investigate its stability at the extreme conditions of its pH and temperature optimum. For these experiments, we incubated the enzyme at various temperatures and pH values for prolonged times and then measured the residual activity by the FRET assay at pH 1.8 and 80 °C.
The data in Figure 5(C) show that SSO1949 retains its activity on prolonged incubation at high temperatures and acidic pH. Preincubation at pH 1.8 and 80 °C for 2 h does not lead to a significant decrease in cellulase activity. To induce inactivation of the enzyme, preincubation temperatures higher than 85 °C are required. When incubated at 95 °C and pH 1.8, SSO1949 is rapidly inactivated.
Interestingly, the thermal denaturation of SSO1949 is strongly pH-dependent. Preincubation at high temperatures and neutral pH leads to a fast inactivation of the enzyme, e.g. when the enzyme is incubated at 80 °C and pH 7, SSO1949 rapidly loses its activity (Figure 5D). On the other hand, SSO1949 is remarkably stable at pH 1 and 80 °C. The enzyme has a half-life of 160 min under these conditions. The half-lives determined from these experiments are summarized in the inset of Figure 5(D). At pH 7, the thermal inactivation rate at 80 and 85 °C is nearly 100 times higher when compared with pH 2.
We do not know of other enzymes with a similar combination of temperature and pH-dependence of activity and such a pronounced preferential stabilization at low pH. Our data identify SSO1949 as an enzyme with extraordinary properties that has evolved to work optimally both at high temperatures and at acidic pH. SSO1949 thus provides an outstanding example for the evolutionary adaptation of enzymic activity and stability under extreme conditions.
DISCUSSION
The gene sso1949 from the crenarchaeote S. solfataricus encodes a thermoacidophilic endo-β-glucanase, which is remarkably active and stable at acidic pH and high temperatures. Sequence comparison indicates significant homology to cellulases of GH family 12. Sequence similarity is highest with the endo-β-glucanase EglA from the hyperthermophilic archaeon Pyrococcus furiosus. This enzyme has a high temperature optimum at 100 °C, but in contrast with the enzyme from S. solfataricus it is most active in the neutral pH range, i.e. at pH 6.0 [22]. The preferred substrates of SSO1949 are higher cello-oligomers. The main products of the reaction are cellobiose and cellotriose. Probably due to the lack of a cellulose-binding domain, SSO1949 is inactive towards crystalline cellulose.
The unique property of SSO1949 is its ability to degrade CMC and higher cello-oligosaccharides, with a pH optimum of approx. 1.8 and a temperature optimum of approx. 80 °C. We know of only one other enzyme for which a similar combination of acid and heat activity has been reported, namely the protease thermopsin from S. acidocaldarius [23]. For this enzyme, the heat denaturation has not been studied at different pH values. Within GHs, SSO1949 is the first member which is active at high temperatures and low pH. Other acidophilic GHs are not thermophilic, e.g. the related xylanase C from Aspergillus kawachii has a pH optimum of 2.0 but is a mesophilic enzyme [24].
The pH–activity profiles of GHs are bell shaped, which is caused by the ionization states of the two catalytic acidic residues. In the reaction mechanism of glycoside hydrolysis, two carboxylate side chains co-operate; one of them acts as a nucleophile and is required to be in the deprotonated state, whereas the other donates a proton and therefore is required to be in the protonated state. The bell-shaped pH-activity profile of SSO1949 resembles that of other acidophilic GHs like xylanase C from A. kawachii and xylanase I from Trichoderma reesei, except that the activity profile of SSO1949 is sharper and is shifted to the acidic range. The pH–activity profile of SSO1949 indicates two ionization steps of apparent pKa values of 1.7 and 2.3 to be involved in catalysis. Our modelling of SSO1949 proposes a similar overall structure of the catalytic site in comparison with other mesophilic cellulases, and it suggests Glu-213 and Glu-310 to be involved in these ionization steps. When assayed at pH 1 and 80 °C, the enzyme shows 20% of its maximal activity. The decreased activity, however, cannot be ascribed to an inactivation of the enzyme. On preincubation at pH 1 for 1 h at 80 °C, the enzyme retains approx. 80% of its initial activity. Therefore we suggest that the decreased activity below pH 1.8 is due to protonation of a catalytically essential group. According to the homology modelling, a candidate for this group is the nucleophile Glu-213, whose pKa value might be lowered to the strongly acidic range by H-bonding with surrounding residues or by electrostatic effects. There are several examples of lowering pKa values of acidic amino acids by H-bond formation. In RNase T1, an aspartate residue has the extremely low pKa value of 0.5. The large shift of the acid ionization constant of this amino acid is explained by the stabilization of the charged side chain through four hydrogen bonds [25]. The constellation of a hydrogen-bonded glutamate–aspartate pair could be shown for xylanase C from A. kawachii to be responsible for the activity at low pH [24]. In this system, substitution of the aspartic acid 37 to asparagine shifts the pH optimum from 2.0 to 5.0. A similar observation has been reported for the xylanase from Bacillus circulans, which has an asparagine residue at the homologous position and has a pH optimum of 5.7. In the mutant protein N35D, a hydrogen bond between the aspartate and the catalytic glutamate residue is observed and the pH optimum is lowered to 4.6 [26]. The mechanistic basis for the change in pH optimum is, however, not completely understood in both the cases. Aspartate probably stabilizes the deprotonated catalytic glutamic acid by a hydrogen bond, thus lowering the pKa value of the glutamate. Comparison of the amino acid sequence of SSO1949 with the acidophilic xylanase from A. kawachii (Figure 1A) and the inspection of the active site of the modelled structure of SSO1949 did not allow us to identify a clear candidate for the low pH optimum of SSO1949. Thr-137, which is homologous with Asp-37 of the A. kawachii xylanase (see Figure 1A), probably could hydrogen bond with the acid/base catalyst Glu-310 and lower its pKa value.
Reasons for the extraordinary stability of SSO1949 at high temperature and acidic pH are also not known. The stability of two well characterized acidophilic enzymes, porcine pepsin and xylanase C, has been interpreted in terms of an excess of acidic residues on the surface and a low pI. The structure of the extremely acidophilic and acid-stable xylanase C, which has a pI of approx. 3.5, shows a clustering of acidic residues in coiled regions and has a negatively charged surface. Xylanases which are not acid-stable lack a similar negatively charged surface [24]. Porcine pepsin, the best studied example of an extremely acid-stable enzyme, carries a large excess of negatively charged groups over positively charged groups and it has an extremely low pI of approx. 1. It is rapidly denatured at pH>6.5, and this instability has been ascribed to a repulsion of the excess negative charges at high pH [27].
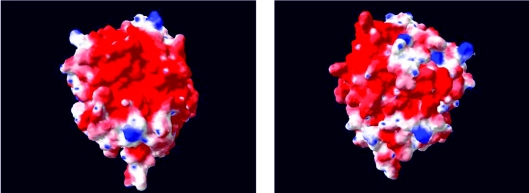
The modelling of the surface charges of SSO1949 at pH 7 also yields a mostly negatively charged surface (Figure 6), however, with some positive charges interspersed. Other β-glucanases, which are active in the neutral or slightly acidic pH-range, have a similar predicted pI value as SSO1949. Therefore the net charge of SSO1949 seems not to be solely responsible for its extreme acidic stability.
Figure 6. Charge distribution on the surface of SSO1949.
Charges on the surface were calculated at pH 7.0 under the assumption that pKa values of the side chains do not deviate from pKa values in solution. Positive charges are depicted in blue and negative charges in red. Left panel, the reader views in the substrate cleft with the active site. Right panel, the ‘back’ of the protein is shown.
The complete loss of activity observed on deletion of amino acids 25–85 indicates an essential function of this region for SSO1949 activity. The N-terminus, which is Ser/Thr rich, appears to stabilize the active conformation of SSO1949 at the extreme conditions.
Overall, SSO1949 is an enzyme that appears to be optimally adapted to work under acidic conditions and at high temperatures. Owing to these properties, SSO1949 has the potential to become an economically important enzyme. SSO1949 might be used as a key enzyme in the production of bioethanol from cellulose because of its unique combination of temperature and pH stability. For the production of bioethanol, cellulose is hydrolysed by acids at high temperatures. In the subsequent step of enzymic hydrolysis, an acid- and heat-tolerant cellulase such as SSO1949 could simplify the reaction process and reduce production costs.
S. solfataricus is a thermoacidophile growing optimally at approx. 80 °C and pH 2– 4. Under these conditions, spontaneous degradation of cellulose takes place and cello-oligomers will form. Probably, the physiological role of secreted SSO1949 is to catalyse the hydrolysis of cello-oligomers to cellobiose, cellotriose and glucose in the surrounding medium. These reaction products may then be imported into the cells and hydrolysed further. By this mechanism, the extracellular SSO1949 might enable S. solfataricus to use cellulose as carbon and energy source. Our results show that SSO1949 is perfectly adapted to work at high temperature and low pH, its physiological environment. The architecture of the active site seems to be unchanged with respect to ‘neutral’ and ‘alkaline’ cellulases of GH family 12 underscoring the potential of a 20-amino-acid protein world for functional adaptation to extremely harsh conditions.
Acknowledgments
This work was supported by High Tech Offensive Bayern.
References
- 1.Bayer E. A., Chanzy H., Lamed R., Shoham Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 2.Kim J. S., Lee Y. Y., Torget R. W. Cellulose hydrolysis under extremely low sulfuric acid and high-temperature conditions. Appl. Biochem. Biotechnol. 2001;91–93:331–340. [PubMed] [Google Scholar]
- 3.Wood T. M., Garcia-Campayo V. Biodegradation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1990. Enzymology of cellulose degradation; pp. 147–161. [Google Scholar]
- 4.Henrissat B., Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 5.Bourne Y., Henrissat B. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 2001;11:593–600. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- 6.Coutinho P. M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 7.Sulzenbacher G., Mackenzie L. F., Wilson K. S., Withers S. G., Dupont C., Davies G. J. The crystal structure of a 2-fluorocellotriosyl complex of the Streptomyces lividans endoglucanase CelB2 at 1.2 Å resolution. Biochemistry. 1999;38:4826–4833. doi: 10.1021/bi982648i. [DOI] [PubMed] [Google Scholar]
- 8.Sandgren M., Shaw A., Ropp T. H., Wu S., Bott R., Cameron A. D., Stahlberg J., Mitchinson C., Jones T. A. The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 Å resolution. J. Mol. Biol. 2001;308:295–310. doi: 10.1006/jmbi.2001.4583. [DOI] [PubMed] [Google Scholar]
- 9.Crennell S. J., Hreggvidsson G. O., Nordberg K. E. The structure of Rhodothermus marinus Cel12A, a highly thermostable family 12 endoglucanase, at 1.8 Å resolution. J. Mol. Biol. 2002;320:883–897. doi: 10.1016/s0022-2836(02)00446-1. [DOI] [PubMed] [Google Scholar]
- 10.Khademi S., Zhang D., Swanson S. M., Wartenberg A., Witte K., Meyer E. F. Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride. Acta Crystallogr. D Biol. Crystallogr. 2002;58:660–667. doi: 10.1107/s0907444902003360. [DOI] [PubMed] [Google Scholar]
- 11.Sandgren M., Gualfetti P. J., Shaw A., Gross L. S., Saldajeno M., Day A. G., Jones T. A., Mitchinson C. Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Sci. 2003;12:848–860. doi: 10.1110/ps.0237703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White A., Rose D. R. Mechanism of catalysis by retaining β-glycosyl hydrolases. Curr. Opin. Struct. Biol. 1997;7:645–651. doi: 10.1016/s0959-440x(97)80073-5. [DOI] [PubMed] [Google Scholar]
- 13.She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C., Clausen I. G., Curtis B. A., De Moors A., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limauro D., Cannio R., Fiorentino G., Rossi M., Bartolucci S. Identification and molecular characterization of an endoglucanase gene, celS, from the extremely thermophilic archaeon Sulfolobus solfataricus. Extremophiles. 2001;5:213–219. doi: 10.1007/s007920100200. [DOI] [PubMed] [Google Scholar]
- 15.Miller G. L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426. [Google Scholar]
- 16.Eckert K., Zielinski F., Lo L. L., Schneider E. Gene cloning, sequencing, and characterization of a family 9 endoglucanase (CelA) with an unusual pattern of activity from the thermoacidophile Alicyclobacillus acidocaldarius ATCC27009. Appl. Microbiol. Biotechnol. 2002;60:428–436. doi: 10.1007/s00253-002-1131-4. [DOI] [PubMed] [Google Scholar]
- 17.Jork H., Funk W., Fischer W., Wimmer H. Weinheim: VCH; 1994. Thin-Layer Chromatography. [Google Scholar]
- 18.Boyer V., Fort S., Frandsen T. P., Schulein M., Cottaz S., Driguez H. Chemoenzymatic synthesis of a bifunctionalized cellohexaoside as a specific substrate for the sensitive assay of cellulase by fluorescence quenching. Chem. Eur. J. 2002;8:1389–1394. doi: 10.1002/1521-3765(20020315)8:6<1389::aid-chem1389>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen H., Brunak S., von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 20.Halldorsdottir S., Thorolfsdottir E. T., Spilliaert R., Johansson M., Thorbjarnardottir S. H., Palsdottir A., Hreggvidsson G. O., Kristjansson J. K., Holst O., Eggertsson G. Cloning, sequencing and overexpression of a Rhodothermus marinus gene encoding a thermostable cellulase of glycosyl hydrolase family 12. Appl. Microbiol. Biotechnol. 1998;49:277–284. doi: 10.1007/s002530051169. [DOI] [PubMed] [Google Scholar]
- 21.Lambert C., Leonard N., De Bolle X., Depiereux E. ESyPred3D: prediction of protein 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 22.Bauer M. W., Driskill L. E., Callen W., Snead M. A., Mathur E. J., Kelly R. M. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes beta-1,4 bonds in mixed-linkage (1→3),(1→4)-beta-D-glucans and cellulose. J. Bacteriol. 1999;181:284–290. doi: 10.1128/jb.181.1.284-290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fusek M., Lin X. L., Tang J. Enzymic properties of thermopsin. J. Biol. Chem. 1990;265:1496–1501. [PubMed] [Google Scholar]
- 24.Fushinobu S., Ito K., Konno M., Wakagi T., Matsuzawa H. Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: biased distribution of acidic residues and importance of Asp37 for catalysis at low pH. Protein Eng. 1998;11:1121–1128. doi: 10.1093/protein/11.12.1121. [DOI] [PubMed] [Google Scholar]
- 25.Giletto A., Pace C. N. Buried, charged, non-ion-paired aspartic acid 76 contributes favorably to the conformational stability of ribonuclease T1. Biochemistry. 1999;38:13379–13384. doi: 10.1021/bi991422s. [DOI] [PubMed] [Google Scholar]
- 26.Joshi M. D., Sidhu G., Pot I., Brayer G. D., Withers S. G., McIntosh L. P. Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 2000;299:255–279. doi: 10.1006/jmbi.2000.3722. [DOI] [PubMed] [Google Scholar]
- 27.Sielecki A. R., Fedorov A. A., Boodhoo A., Andreeva N. S., James M. N. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 Å resolution. J. Mol. Biol. 1990;214:143–170. doi: 10.1016/0022-2836(90)90153-D. [DOI] [PubMed] [Google Scholar]