Abstract
HnRNP K (heterogeneous nuclear ribonucleoprotein K) was biochemically purified from a screen of proteins co-purifying with binding activity to the osteocalcin promoter. We identify hnRNP K as a novel repressor of osteocalcin gene transcription. Overexpression of hnRNP K lowers the expression of osteocalcin mRNA by 5-fold. Furthermore, luciferase reporter assays demonstrate that overexpression of hnRNP K represses osteocalcin transcription from a CT (cytosine/thymidine)-rich element in the proximal promoter. Electrophoretic mobility-shift analysis reveals that recombinant hnRNP K binds to the CT-rich element, but binds ss (single-stranded), rather than ds (double-stranded) oligonucleotide probes. Accordingly, hnRNP K antibody can supershift a binding activity present in nuclear extracts using ss sense, but not antisense or ds oligonucleotides corresponding to the CT-rich −95 to −47 osteocalcin promoter. Importantly, addition of recombinant hnRNP K to ROS 17/2.8 nuclear extract disrupts formation of a DNA–protein complex on ds CT element oligonucleotides. This action is mutually exclusive with hnRNP K's ability to bind ss DNA. These results demonstrate that hnRNPK, although co-purified with a dsDNA-binding activity, does not itself bind dsDNA. Rather, hnRNP K represses osteocalcin gene transcription by inhibiting the formation of a transcriptional complex on the CT element of the osteocalcin promoter.
Keywords: cytosine/thymidine, heterogeneous nuclear ribonucleoprotein K, osteoblast, osteocalcin, transcription regulation
Abbreviations: CMV, cytomegalovirus; CT, cytosine/thymidine; ds, double-stranded; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; ERK, extracellular-signal-regulated kinase; GAL4BD, Gal4 DNA-binding domain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; hnRNP K, heterogeneous nuclear ribonucleoprotein K; MEM, minimal essential medium; Sp1, specificity protein 1; ss, single-stranded
INTRODUCTION
Osteocalcin is an osteoblast-specific protein secreted during the process of bone formation, where it modulates mineralization of the organic matrix [1,2]. Its expression is regulated by a variety of signals including hormones, growth factors, cytokines and mechanical stress required for normal bone homoeostasis. Because of its osteoblast-specific expression, the osteocalcin promoter has been used extensively to characterize tissue-specific transcriptional factors that are essential for osteogenic differentiation [3–7]. Despite the wealth of information produced by these studies, new regulatory elements and transcription factors that may be very important for osteogenic differentiation are still emerging. In particular, the proximal promoter region (−0.2 kb) of the rat osteocalcin gene contains several transcriptional elements that drive both basal and induced gene expression, including a Runx2-binding cognate [4], an FGF (fibroblast growth factor)-responsive element [8] and a cAMP-responsive region [9]. We have previously identified a CT (cytosine/thymidine)-rich region of the osteocalcin proximal promoter that binds two protein–DNA complexes and regulates osteocalcin gene transcription. We identified one of these complexes as containing the ubiquitous transcription factors, Sp1 (specificity protein 1) and Sp3, whose binding is modulated by gap junctional communication [10]. To define the second protein–DNA complex binding to this pyrimidine-rich element, we undertook a series of biochemical purifications of osteoblast nuclear extracts and identified the hnRNP K (heterogeneous ribonucleoprotein K) as a regulator of osteocalcin gene transcription.
The hnRNPs are a family of nuclear proteins that function in mRNA biogenesis, including pre-mRNA splicing [11], transport of mRNA from the nucleus to the cytosol [12] and translation [13]. HnRNP K is a unique member of this family, in that it preferentially binds ssDNA (single-stranded DNA), whereas other RNPs bind RNA [14]. Thus, hnRNP K is an intriguing, multifunctional molecule that can act both in the cytosol and in the nucleus (reviewed in [15,16]), and has been implicated in a vast number of cellular processes, including gene transcription [17,18] and chromatin remodelling [19], in addition to the more typical hnRNP functions of splicing and transport of mRNA to the cytoplasm [20]. In the present study, we report that hnRNP K is expressed by osteoblasts and regulates osteocalcin gene transcription by acting as a repressor. Although hnRNP K can bind the CT-rich element of the rat osteocalcin promoter, its repressive action appears to be mediated not by protein–DNA interactions, but rather by competitive removal of a transactivator from the CT element, resulting in diminished osteocalcin gene transcription.
MATERIALS AND METHODS
Cells and reagents
As previously described, the rat osteogenic sarcoma cells, ROS17/2.8 and the mouse osteoblast-like cells, MC3T3-E1, were cultured in MEM (minimal essential medium) containing 10% (v/v) fetal bovine serum and antibiotics [10,21]. Cell culture medium was obtained from Sigma Chemical (St Louis, MO, U.S.A.), fetal bovine serum was from Atlanta Biologicals (Norcross, GA, U.S.A.). Molecular biology reagents were from Promega (Madison, WI, U.S.A.), unless stated otherwise. Oligonucleotides were synthesized by the Washington University Protein and Nucleic Acid Core Laboratory. ECL® detection reagents, glutathione–Sepharose 4B and pGEX-5X-2 vectors were from Amersham Biosciences (Piscataway, NJ, U.S.A.). Novex 4–20% TBE (90 mM Tris/90 mM boric acid/0.2 mM EDTA) acrylamide gels, LIPOFECTAMINE™ and pcDNA3 were from Invitrogen (Carlsbad, CA, U.S.A.). SYBR Green I PCR master mix was purchased from Applied Biosystems (Foster City, CA, U.S.A.). Radioisotope was purchased from ICN (Costa Mesa, CA, U.S.A.). Rabbit anti-RNP K antibodies were raised against a peptide corresponding to amino acids 277–288 of human hnRNP K, conjugated to KLH (keyhole-limpet haemocyanin) through a cysteine residue (Polyquick antibody service; Zymed, South San Francisco, CA, U.S.A.). Affinity purification was performed using SulfoLink kit (Pierce, Rockford, IL, U.S.A.) following manufacturer's recommendations. Antibodies against α-Fos (K-25), α-Jun (D), ATF-1 and ATF-2 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.).
Plasmid constructs
The rat osteocalcin promoter-luciferase reporter construct (−92 OCLUC) contains the −92 to +32 5′-flanking sequence of the osteocalcin gene, relative to the transcriptional start site. This construct was cloned into the KpnI–MluI sites of pGL2Basic plasmid as described by Towler et al. [22]. The −92ΔCT OCLUC construct, in which the CT element was deleted from the −92 OCLUC reporter, was generated by amplifying the 5′- and 3′-flanking regions of the CT element respectively by PCR from the −92 OCLUC construct. The resulting PCR fragments were gel purified, blunt-end ligated to each other, and cloned into the KpnI–MluI sites of pGL2Basic. To generate the heterologous promoter constructs, CT-RSVLUC and mtCT-RSVLUC, oligonucleotides spanning the CT element (−74 to −57), CGATCTCCTGCCCCTCCTGCTGGAC and CAGCAGGAGGGGCAGGAGATCGGTAC or CGCATCTCCTGGGGGACCTGGTAC and CAGGTCCCCCAGGAGATGCGGTAC were annealed, phosphorylated with T4 DNA kinase and ligated into the KpnI site of a minimal RSVLUC promoter [9]. RNPK-pGEX and RNPK-pcDNA3 constructs were generated by PCR amplification of the coding sequence of the mouse hnRNP K gene from mouse cDNA. The amplified product was cloned in frame into the EcoRI–XhoI site of pGEX-5X-2 and pcDNA3 plasmids respectively. The RNPK-pcDNA3 contained an N-terminal myc tag. All constructs were sequenced to confirm the identity of the insert.
EMSA (electrophoretic mobility-shift assay)
EMSAs were performed as described previously, with minor modification [10]. Briefly, nuclear extracts were prepared according to the methods of Dignam et al. [23] with the addition of 1 mg/ml leupeptin, 1 mg/ml aprotinin, 1 mg/ml pepstatin, 1 mM NaVO4 and 1 mM PMSF. Extracts were dialysed against Dignam buffer D [20 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT (dithiothreitol)] and stored in 50 μl aliquots at −70 °C. Gel-purified oligonucleotides (10 pmol) were labelled with Klenow fragment and [α-32P]dCTP. Binding reactions were performed in the presence of 0.1 pmol of labelled oligonucleotide, 10 μg of acetylated BSA, 1 μg of double-stranded (ds) poly(dI/dC) and 2 μg of nuclear extract. Total reaction volume was adjusted to 15 μl with Dignam buffer D. For supershift experiments, 2 μg of antibody was added to the binding reaction. Binding reactions were performed for 20 min at 25 °C. The samples were electrophoresed on Novex 4–20% polyacrylamide gels in 0.375×TBE. Gels were dried and bands were visualized by autoradiography. The following oligonucleotides were used in this study (numeration is relative to the transcription start site of the rat osteocalcin promoter), sense CT (−74 to −56), CAGCATCTCCTGCCCCTCCTGCT; antisense CT (−74 to −56), GGAGCAGGAGGGGCAGGAGATGC; sense −95 to −47 osteocalcin, CCCCCAATTAGTCCTGGCAGCATCTCCTGCCCCTCCTGCTTGCATTAGG; antisense −95 to −47 osteocalcin, TAATGCAAGCAGGAGGGGCAGGAGATGCTGCCAGGACTAATAGGGGG.
Western blotting
Nuclear and cytoplasmic extract (20 μg), prepared by the method of Dignam et al. [23], were electrophoresed on SDS/PAGE (10% gels) and transferred to nitrocellulose membranes. Membranes were blocked in 5% (w/v) non-fat dry milk, probed with rabbit anti-hnRNP K antibodies (1:1000) and detected with horseradish peroxidase-conjugated anti-rabbit antibodies (1:5000) and ECL® detection system.
Southwestern blotting
The procedure described by Sambrook et al. [24] was followed to obtain concatemerized probes. Briefly, oligonucleotides (100 pmol) labelled using polynucleotide kinase (T4 kinase; Boehringer Mannheim, CT, U.S.A.) and [γ-32P]ATP (3000 Ci/mol; Amersham Biosciences) and subsequently concatemerized by overnight incubation with T4 ligase at 12–16 °C. Southwestern analysis was performed as described previously [25] with minor modifications. Nuclear extracts (30 μg) were electrophoresed on 10% (w/v) SDS/PAGE gel under reducing conditions and transferred to a nitrocellulose membrane. After air-drying, the proteins were denatured in 6 M guanidinium chloride in a buffer containing 25 mM Hepes (pH 7.6), 60 mM KCl, 1 mM EDTA and 0.5 mM DTT and renatured by a series of nine stepwise 1:2 dilutions in the same buffer at 4 °C with gentle shaking. Once the guanidinium chloride was diluted out from the buffer, the membrane was incubated for 1 h at room temperature (23 °C) in BLOTTO buffer (5% Carnation non-fat dry milk, 0.5 μg/ml sonicated calf thymus DNA, 1 mM DTT, 25 mM Hepes, pH 7.6, 60 mM KCl, 1 mM EDTA) with gentle shaking. After two 10 min washes with 0.5% non-fat dry milk, the filter was incubated with 1×106 Čerenkov c.p.m./ml of concatemerized 5′-labelled duplex probe in an identical buffer for 2 h at room temperature. After extensive washing, the filter was exposed to autoradiographic film at −70 °C with intensifying screens to visualize protein–DNA interactions.
Biochemical purification of hnRNP K
Large-scale cultures of ROS 17/2.8 cells (8×108 cells grown in roller bottles) were cultured in α-MEM containing 5% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin at the Washington University Tissue Culture and Media Center. Confluent cultures were washed in PBS, transferred to 50 ml conical tubes and mixed at 4 °C with 1 vol. of a solution containing 20 mM Hepes (pH 7.9), 0.3% Triton X-100, 1.5 mM MgCl2, 0.2 mM EDTA, 25% (v/v) glycerin, with 1.12% ammonium sulphate saturation, 1 mM DTT; 0.1 mM benzamidine, 1 mM PMSF, 6 μg/ml soya bean trypsin inhibitor and 1 μg/ml leupeptin (modified Dignam buffer C [23]). After the addition of prewashed glass beads (400–600 μm; Sigma) in Dignam buffer C, the cell extract was prepared by homogenization using the BioSpec Bead Beater (Bartlesville, OK, U.S.A.). After centrifugation at 3000 g for 15 min at 4 °C to clear the extract, ammonium sulphate fractionation was performed at 55% saturation. Proteins were dissolved in Dignam buffer D and dialysed using 5–8 kDa cutoff membrane (Spectrapor). Protein extract was subjected to batch mode DEAE-Sephacryl chromatography using DEAE-6CLB resin (Amersham Biosciences), and heparin chromatography using 1 ml Hi Trap Heparin columns (Amersham Biosciences) with 20 mM Hepes buffer (pH 7.9). Binding activity was eluted at 200 mM NaCl. A 5 ml Mono Q FPLC column (20 mM Tris buffered, pH 8.0; Amersham Biosciences) was used for anion exchange chromatography, and binding activity eluted at 500 mM NaCl. FPLC fractionation was achieved by a salt gradient from 0 to 400 mM, eluting at 1 ml/min flow rate with an increase in 13.8 mM NaCl/min in 0.1 mM EDTA and 1 mM 2-mercaptoethanol. The DNA-binding activity to the CT element was monitored in each fraction by EMSA and Southwestern blotting using a −95 to −47 rat osteocalcin promoter oligonucleotide. Peak activity was found in fraction 37 (274–311 mM NaCl), and this was further purified through a CM-Sepharose column, where the peak activity was collected in the flow-through from a 500 mM NaCl wash. Proteins were resolved by SDS/PAGE (10% gel) poured on virgin plastic plates. Fractionated proteins were visualized by staining in Coomassie Blue, briefly destained and excised with a sterile scalpel. Proteins were digested with 1:20 (w/w) of acetylated trypsin (Promega), and tryptic peptides were analysed by nanobore LC-MS/MS at the Harvard Microchemistry Facility (Harvard University, Boston, MA, U.S.A.) using a Finnigan ion trap mass spectrometer and identified as hnRNPK (four unique peptides).
Purification of recombinant hnRNP K
RNPK-pGEX constructs were transformed into Escherichia coli BL21 competent cells. Cultures were induced with 0.1 mM isopropyl β-D-thiogalactoside at 30 °C for 3 h. Recombinant proteins were isolated from extracts by affinity purification using glutathione–Sepharose 4B and eluted with 10 mM glutathione in 50 mM Tris (pH 8.0). Protein expression was monitored by SDS/PAGE and Coomassie Blue staining.
Transient transfection and luciferase reporter assays
ROS17/2.8 cells were seeded at high density (2×105 cells/well) in 24-well plates. The cells were transfected after 18 h with the appropriate plasmid using LIPOFECTAMINE™ reagent according to the manufacturer's instructions. Reporter plasmid (0.5 μg/well) and hnRNP K expression plasmid (0.25 μg/well) were used in each transfection. After 48 h, the cells were rinsed in PBS and lysed in 1 × passive lysis buffer. Luciferase activity was monitored using an Optocomp luminometer. Transfection efficiency was monitored by co-transfection with a β-galactosidase reporter. All experiments were performed in triplicate and repeated 2–5 times.
Reverse transcription and real-time PCR
Total RNA was isolated from confluent cultures of ROS 17/2.8 cells transfected with pcDNA or pcDNA-RNPK using TRIzol®. RNA (2 μg) was DNase I treated and then reverse transcribed using Superscript II reverse transcriptase and oligo(dT) primers. For real-time PCR analysis of the gene expression, 1/40 of this reaction was used with SYBR Green I dye chemistry. PCR product accumulation was monitored using a GeneAmp 5700 sequence detection system (Applied Biosystems). The mean cycle threshold value (Ct) from triplicate samples was used to calculate the gene expression. PCR products were normalized to levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Relative gene expression levels were determined as described in the User's Bulletin (P/N 4303859) from Applied Biosystems. The genespecific primers used were the following: OC-F, CCAGCGACTCTGAGTCTGACAA; OC-R, CCGGAGTCTATTCACCACCTTACT. GAPDH primers were from Applied Biosystems.
One-hybrid assay
The hnRNP K reading frame was PCR amplified from a mouse EST (expressed sequence tag) clone (Incyte Pharmaceuticals, St. Louis, MO, U.S.A.). The primers included BamHI and SacI sites for cloning into pFA-CMV (where CMV stands for cytomegalovirus; Stratagene). HnRNP K was fused in frame with the GAL4BD (GAL4 DNA-binding domain) to obtain pFA-GAL-4BD-RNPK, which drives the expression of the hnRNPK–GAL4BD fusion protein using the CMV promoter. The transcriptional activity of the hybrid pFA-GAL4BD-RNPK was assessed using pFR-LUC (Stratagene), which utilizes GAL4 response elements to drive a luciferase reporter (GAL4RE LUC). Experiments were performed by cotransfecting GAL4RE LUC with either pFA-GAL4BD-RNPK or vector alone (pFA-CMV) in MC3T3-E1 cells using LIPOFECTAMINE™ reagent, as described elsewhere [26]. Luciferase activity was monitored as described above.
Immunocytochemistry
A previously described procedure was followed [27]. Confluent ROS17/2.8 cells grown on glass coverslips were fixed in methanol/acetone (1:1) for 2 min at room temperature, washed, and incubated in PBS containing 2% (v/v) heat-inactivated goat serum and 0.5% Triton X-100 for 15 min. They were then incubated with anti-hnRNPK antibody (1:500 dilution) for 60 min at room temperature. After three washes in PBS, peroxidase-conjugated goat anti-rabbit IgG antibody (Roche, Indianapolis, IN, U.S.A.) was added (1:500 dilution) for 60 min at room temperature. Cells were subsequently washed three times in PBS, incubated with diaminobenzidine, counterstained with haematoxylin and visualized by optical microscopy. Negative controls were performed after omitting the primary antibody.
RESULTS
The −95 to −47 region of the osteocalcin promoter assembles unidentified protein/DNA activity
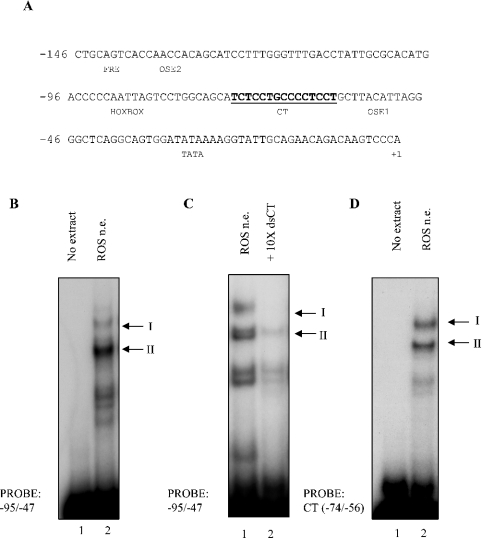
The proximal 146 bp of the rat osteocalcin promoter contain several regulatory elements, including a CT-rich stretch of DNA in the −74 to −56 region (Figure 1A). Confirming our previous results [10], two binding activities (denoted complex I and II respectively) were reproducibly observed to assemble on the −95 to −47 region of the osteocalcin promoter in EMSA using nuclear extract from ROS17/2.8 cells (Figure 1B). These two complexes assemble on the CT-rich region of the osteocalcin promoter (CT element) as demonstrated by competition of binding of both complexes in the presence of 10-fold molar excess of unlabelled oligonucleotides spanning this region (Figure 1C). By contrast, competition with unlabelled non-specific oligonucleotides did not appreciably reduce binding activity of either of the complexes (results not shown). Further, when radiolabelled CT element was used in EMSA, the two binding activities were reproduced as in the longer probe (Figure 1D), confirming that this element was responsible for assembling the two binding activities. The additional fast migrating bands present on the EMSA gels are probable proteolytic fragments and are not consistently observed.
Figure 1. Identification of binding activities on a CT-rich stretch on the −95 to −47 osteocalcin promoter.
(A) The −146 to +1 rat osteocalcin proximal promoter region is shown with the CT-rich DNA element shown in bold. Known transcription factor binding sites in the proximal promoter are indicated. Numbering is relative to the transcriptional start site. (B) EMSA was performed using ds −95 to −47 oligonucleotides incubated with (lane 2) or without (lane 1) nuclear extract (n.e.). Two binding activities were reproducibly observed and are denoted complex I and complex II, as indicated. (C) EMSA was performed with ds −95 to −47 osteocalcin probes in the presence (lane 2) or absence (lane 1) of a 10-fold molar excess of unlabelled, ds CT (−74 to −56) oligonucleotides. (D) EMSA was performed using the CT-rich −74 to −56 region of the osteocalcin promoter, incubated with (lane 2) or without (lane 1) nuclear extract.
Although we have previously identified that complex I contains the ubiquitous transcription factors, Sp1 and Sp3 [10], the nature of complex II remains unknown. To further map the core element within the CT sequence to which this faster migrating complex II binds, we used wild-type and a series of mutated CT cognate oligonucleotides in EMSA analysis. Complex II was unable to bind mut 6 and mut 9 oligonucleotides and could only weakly bind mut 15, whereas it could bind the mut 3 and mut 12 oligonucleotides (Figure 2A). On the basis of this mutation analysis, we define the minimal binding cognate as a TGCCCC motif located in the −68 to −62 region of the osteocalcin proximal promoter. No known transcription factor has been reported to bind specifically to this promoter sequence, and antibodies against transcription factors that have been proposed to bind the proximal osteocalcin promoter, c-jun, ATF-1, ATF-2 or c-fos, could not supershift complex II assembled on the -68 to -62 region of the promoter (Figure 2B).
Figure 2. Identification of the core binding cognate, but not the factor in the fast migrating complex.
(A) EMSA was performed using radiolabelled CT and mutated CT oligonucleotides (lanes 1–6). The identity of the various mutated CT oligonucleotides used as probes is listed below. Mutated bases are in lower case letters. (B) EMSA was performed using radiolabelled ds −95 to −47 oligonucleotides incubated with (lanes 2–6) or without (lane 1) nuclear extract. Extracts were incubated with anti-c-fos (lane 3), c-jun (lane 4), ATF-1 (lane 5) and ATF-2 (lane 6) antibodies.
Identification of hnRNP K as a factor co-purifying with binding activity to the osteocalcin proximal promoter
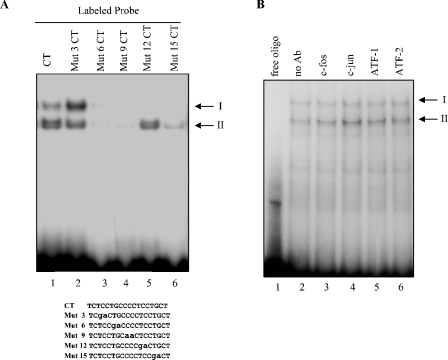
To characterize the unknown binding activity in complex II, proteins binding to the −95 to −47 osteocalcin proximal promoter were biochemically purified from ROS 17/2.8 nuclear extracts. The scheme is described in detail in the Materials and methods section. The DNA-binding activity, monitored by EMSA using the −95 to −47 osteocalcin promoter as a probe, was recovered in the eluate (fractions 36–38) of a NaCl gradient after FPLC through a Mono Q column (Figure 3A). Southwestern blotting using the −95 to −47 osteocalcin concatemerized probe revealed interaction of the probe with a protein of approx. 65 kDa after this purification step (Figure 3B). To further purify the protein(s) associated with DNA-binding activity, the eluate from the MonoQ column was then passed through a CM-Sepharose column. The DNA-binding activity remained almost exclusively in the flow-through from this column, and was enriched more than 10000-fold relative to the crude nuclear extract (Figure 3C, right panel). Although there were several bands of higher intensity by silver staining in the flow-through relative to the eluate, we focused on the bands migrating at approx. 65 kDa, which were only present in the flow-through (Figure 3C), and whose size corresponded to the bands observed in the Southwestern blot (Figure 3B). Thus the approx. 65 kDa band was excised and sequenced by MS. Four peptides corresponding to hnRNP K sequence were identified.
Figure 3. Elution profile of − 95 to − 47 binding activities and protein fractionation at various purification steps.
(A) EMSA of binding activities recovered from a NaCl gradient Mono Q FPLC fractionation, using the −95 to−47 region of the osteocalcin promoter as a probe. (B) Southwestern hybridizations were performed using concatamerized −95 to −47 oligonucleotide probes. (C) Left panel: silver stain of eluate from fraction 37 MonoQ column, flow through and eluate after further purification through a CM-Sepharose column. Black rectangle indicates the ∼65 kDa band submitted for microsequencing. Right panel: EMSA, using the flow-through and eluate fractions from the CM-Sepharose column and the starting nuclear extract of ROS 17/2.8 cells, were performed using the −95 to −47 probe.
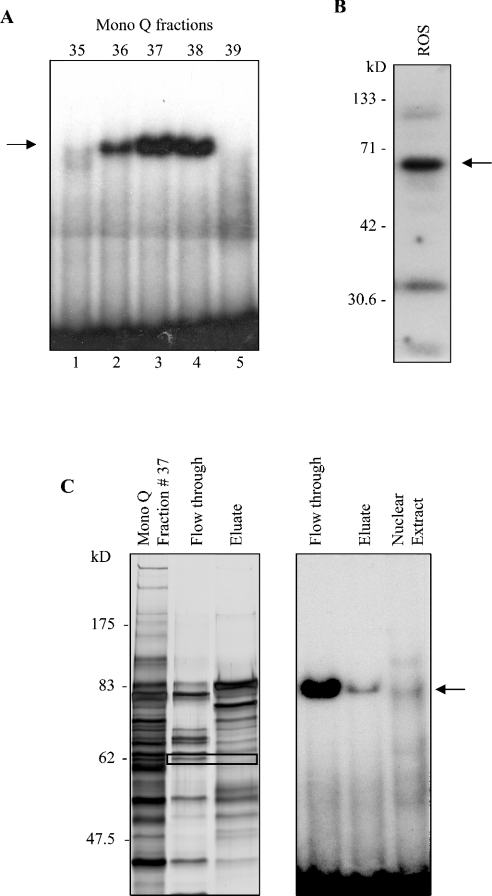
Expression of hnRNP K was confirmed in ROS 17/2.8 cells by immunoblot and immunocytochemistry using a polyclonal anti-hnRNP K antibody. Western-blot analysis revealed expression of hnRNP K protein most abundantly in the nuclear fraction of ROS 17/2.8 cells (Figure 4A). This conclusion is corroborated by expression of Sp3, a known nuclear transcription factor, almost exclusively in the nuclear fraction (Figure 4B). Likewise, hnRNP K protein was detected in nuclear extracts from primary murine calvarial osteoblasts and MC3T3-E1 cells (results not shown). Finally, immunocytochemistry revealed abundant hnRNP K-specific stain in the nuclei, with less intense stain in the cytoplasm (Figure 4C), a distribution pattern consistent with a possible role as a transcriptional regulator.
Figure 4. HnRNP K is expressed in osteoblastic cells.
(A) Cytosolic and nuclear extracts from ROS 17/2.8 cells were immunoblotted and probed with an anti-hnRNP K antibody, revealing expression primarily in the nucleus. (B) Immunoblotting with Sp3 reveals the specificity of the nuclear fraction. (C) Immunocytochemistry of ROS 17/2.8. Note that ROS17/2.8 cells show immunoreactivity (brown stain) for hnRNP K, which is more abundant in the nucleus than in the cytosol. In control cells, the primary antibody was omitted (×200 magnification).
HnRNP K represses osteocalcin gene transcription from the CT-element
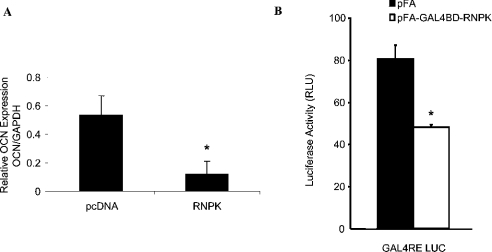
Previous studies have demonstrated hnRNP K as a bona fide transcription factor that can act through a CT-rich element in the c-myc promoter to activate gene transcription [18,28]. Therefore we tested whether hnRNPK is transcriptionally active on the osteocalcin promoter, and in particular on the identified CT element that contains the core-binding motif assembling complex II in EMSAs. Overexpression of hnRNP K in ROS 17/2.8 cells resulted in a 4.5-fold decrease in basal osteocalcin expression, assessed by quantitative real-time PCR, relative to cells transfected with empty vector (Figure 5A). To characterize the transcriptional activity of hnRNP K, we used the one-hybrid system, which allows detection of both transcriptional activation and repression, independently of other endogenous regulators. HnRNPK was fused in frame with the GAL4BD, using the pFA-CMV expression vector. Co-transfection of the GAL4BD-RNPK fusion plasmid with GAL4RE-LUC reporter construct in MC3T3-E1 cells resulted in a 1.6-fold decrease in luciferase activity compared with the empty vector (Figure 5B).
Figure 5. HnRNP K inhibits transcription of the osteocalcin gene.
(A) ROS17/2.8 cells were transiently transfected with RNPK-pcDNA3 or empty vector (pcDNA3). Total RNA was isolated from the cells 72 h post-transfection, and reverse transcribed. The cDNA was subjected to real-time PCR using osteocalcin-specific primers. Relative expression levels were determined by normalizing to GAPDH. Results (means±S.D.) are from a representative experiment performed in triplicate. Each experiment was repeated at least three times. *P<0.05 compared with control. (B) The function of hnRNP K was examined using a one-hybrid assay. Briefly, MC3T3-E1 cells were transiently co-transfected with GAL4RE-LUC (a GAL4 response element containing luciferase reporter) plasmid and GAL4BD–hnRNP K fusion protein expression plasmid (GAL4BD–RNPK). As a control, empty plasmid was used in a parallel experiment. Cells were analysed for luciferase activity 72 h post-transfection. Representative data (means±S.D.) are from an experiment performed in triplicate. Each experiment was repeated at least three times. *P<0.05 compared with control.
Next, we used luciferase reporter assays to examine the role and site of action of hnRNP K in regulating transcription in the context of the osteocalcin promoter. Expression of hnRNP K repressed transcription of a luciferase reporter driven by a single CT element in a heterologous Rous sarcoma virus minimal promoter (CT-RSVLUC) (Figure 6A). Though not quite as strong as was observed for the endogenous osteocalcin promoter (Figure 5A), this repression was approx. 3-fold (Figure 6A). Mutation of this CT-rich sequence (mtCT-RSVLUC) abrogated the sensitivity to hnRNP K repression (Figure 6A). These results indicate that the CT element is indeed an hnRNP K responsive element. Accordingly, hnRNP K repressed transcription from the −92 to +32 homologous osteocalcin promoter luciferase reporter construct (−92 OCLUC), which spans the CT element. Further, deletion of the CT element from the −92 OCLUC luciferase reporter (−92ΔCT OCLUC) relieves the repressive effects of hnRNP K overexpression (Figure 6B), indicating that the CT element is the sole hnRNP K responsive element in the osteocalcin proximal promoter (−92 to +32 region). No effect of hnRNP K was observed on either the promoterless pGL2 or RSV minimal promoter vector in the absence of the CT element (Figures 6A and 6B respectively).
Figure 6. Overexpression of hnRNP K decreases the transcriptional activity of the osteocalcin promoter through a CT-rich element.
(A) ROS17/2.8 cells were transiently co-transfected with CT-RSVLUC, mtCT-RSVLUC or ‘empty’ RSVLUC heterologous promoter reporter and pcDNA3 or mycRNPK-pcDNA3. Cells were analysed for luciferase activity 72 h post-transfection. Inset, overexpression of hnRNP K in the transfected cells was confirmed by immunoblotting using anti-hnRNP K and anti-myc antibodies. GAPDH antibodies were used as a control for loading. (B) ROS17/2.8 cells were transiently co-transfected with −92 OCLUC, −92ΔCT OCLUC homologous promoter reporter or ‘promoterless’ pGL2 basic and pcDNA3 or pcDNA3-mycRNPK. Expression of hnRNP K reduces reporter expression from the −92 osteocalcin promoter (−92 OCLUC). All data (means±S.D.) are from a representative experiment performed in triplicate. Each experiment was repeated at least three times. *P<0.05 compared with control.
HnRNP K binds ss, but not ds, CT element
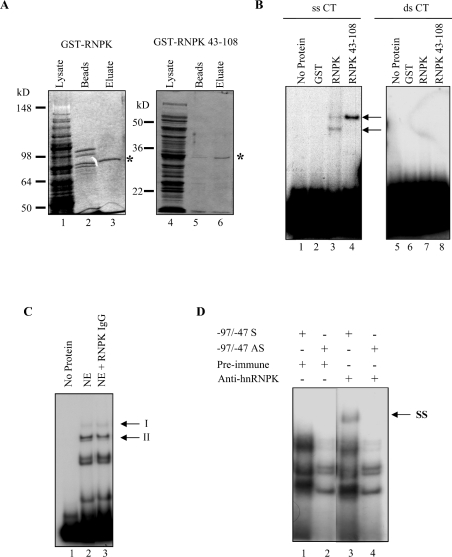
HnRNP K has been reported to act through ss CT-rich DNA in the c-myc promoter [18,28]. Therefore, recombinant GST-RNPK full-length protein as well as a minimal DNA-binding domain of the hnRNP K protein, including only the first two KH domains [GST (glutathione S-transferase)-RNPK 43–108], were bacterially expressed and purified. Coomassie Blue-stained SDS/PAGE gels reveal the relative purity of the recombinant proteins (Figure 7A). These recombinant proteins were then used in EMSA to test their DNA-binding activity. Both GST-RNPK and GST-RNPK 43–108 could bind radiolabelled, ss sense CT oligonucleotides (Figure 7B, lanes 3 and 4), whereas GST alone could not bind the CT probe (Figure 7B, lane 2). Surprisingly, these hnRNP K constructs were unable to bind ds CT oligonucleotides (Figure 7B, lanes 6–8). These results suggest that, although hnRNP K co-purifies with a ds binding factor, it is not itself the direct DNA-binding factor in complex II. Rather, it is probable that either hnRNP K acts in tandem by protein–protein interactions with a second DNA-binding factor to assemble in the transcription complex II, or that hnRNP K requires an endogenous co-factor to melt the dsDNA.
Figure 7. HnRNP K binds ss sense but not ds osteocalcin CT element.
(A) Coomassie Blue-stained SDS/PAGE of bacterially expressed GST-RNPK (left panel) and GST-RNPK 43–108 (right panel). A predominant approx. 90 kDa band corresponding to the predicted molecular mass of a GST-hnRNPK fusion protein is observed in the eluted fraction (lane 3, *). An approx. 33 kDa band corresponding to the predicted molecular mass of a GST-truncated hnRNPK fusion protein is observed in the eluted fraction (lane 6, *). Lanes 1 and 4, induced bacterial cell extract; lanes 2 and 5, glutathione–Sepharose bead fraction. Lanes 3 and 6, eluate fractions. (B) EMSA was performed using ss sense (lanes 1–4) or ds (lanes 5–8) CT element oligonucleotides. Bacterially expressed GST fusion proteins were analysed for DNA-binding activity. Lanes 1 and 5, no protein; lanes 2 and 6, 200 ng GST; lanes 3 and 7, 200 ng GST-RNPK; lanes 4 and 8, 200 ng GST-RNPK amino acids 43–108. Arrow denotes band of interest. (C) EMSA was performed using ds CT oligonucleotides (lanes 2 and 3). Anti-hnRNP K antibodies were co-incubated with nuclear extract (lane 3) to detect hnRNP K binding by supershift. Lane 1, no extract; NE, nuclear extract. (D) EMSA was performed using ss sense (lanes 1 and 3) or antisense (lanes 2 and 4) −95 to −47 oligonucleotides. Incubation of nuclear extract with an anti-hnRNP K antibody produced a supershift (SS) of binding using sense (lane 3), but not antisense (lane 4) oligonucleotides. Band migration pattern is compared with nuclear extract incubated with preimmune serum (lanes 1 and 2).
To test these hypotheses, supershift analysis was performed on hnRNP K using nuclear extract rather than purified protein. Presumably, the use of nuclear extract would provide necessary cofactors for hnRNP K action. However, as was observed using the recombinant hnRNP K, no hnRNP K was detected by supershift using a ds probe (Figure 7C). In contrast, incubation of anti-hnRNP K antibodies with ROS 17/2.8 nuclear extracts, followed by EMSA analysis produced a band of reduced mobility using sense but not antisense CT oligonucleotides (Figure 7D, compare lanes 1 versus 3 and lanes 2 versus 4 respectively). Thus hnRNP K genuinely binds DNA only in a ss conformation in osteoblastic cells. Consequently, DNA binding (either directly or indirectly) does not seem to explain the repressive activity of hnRNP K on the ds osteocalcin promoter.
Transcriptional repression by hnRNP K involves sequestering of an activator from the proximal osteocalcin promoter
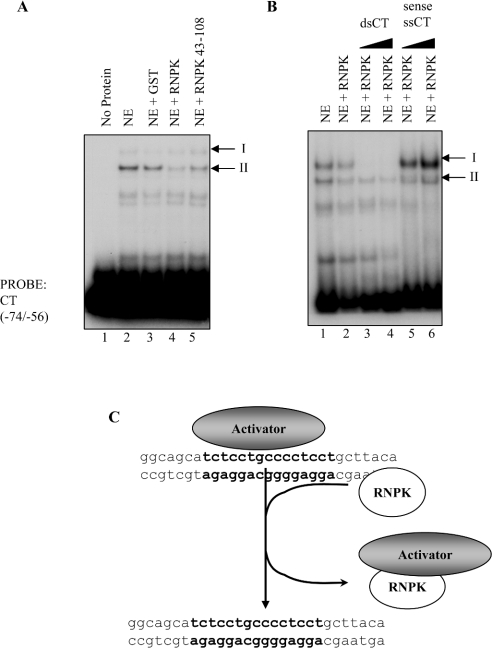
As an alternative mechanism of action, we hypothesized that hnRNP K may be acting indirectly, by a protein–protein interaction to repress gene transcription, perhaps by competing off an activator complex from the osteocalcin promoter. To test this hypothesis, recombinant hnRNP K was preincubated with nuclear extract before incubation with the ds CT element oligonucleotides (Figure 8A). Consistent with this hypothesis, recombinant hnRNP K (Figure 8A, lane 4), as well as the fragment including only the KH1 and KH2 domains, hnRNP K 43–108 (Figure 8A, lane 5), decreased the binding activity of the faster-migrating complex II assembling on the CT element compared with control (Figure 8A, lane 2), whereas the slower-migrating complex I remained unaffected. GST alone did not disrupt DNA binding of either complex I or II (Figure 8A, lane 3). Additionally, the ability of recombinant hnRNP K to disrupt the formation of the protein–DNA complex was examined following preincubation of hnRNP K with 50- or 100-fold molar excess of unlabelled ds and ss CT element (Figure 8B). As expected, preincubation of hnRNP K with unlabelled ds CT disrupted both complex I and II assembly on the labelled ds CT element (Figure 8B, lanes 3 and 4). In contrast, addition of excess unlabelled, ss CT element oligonucleotides to recombinant hnRNP K before incubation with nuclear extract abrogated the ability of hnRNP K to disrupt the protein–DNA complex II (Figure 8B, lanes 5 and 6). Complex I assembly was slightly enhanced under these conditions. These results suggest that hnRNP K disrupts formation of a protein–DNA complex (complex II) on the osteocalcin CT element without directly binding DNA (Figure 8C). Indeed, binding of hnRNP K to ss CT element suppresses its ability to disrupt the protein–DNA complex, indicating that the ability to compete off protein binding or bind ssDNA are mutually exclusive actions of hnRNP K.
Figure 8. HnRNP K disrupts DNA-binding activity on the CT element without directly binding DNA.
(A) EMSA was performed using ds CT element oligonucleotides. Crude nuclear extract was analysed for DNA-binding activity in the presence or absence of 200 ng recombinant GST, RNPK or RNPK amino acids 43–108. The binding activity of complex II is markedly diminished by the addition of recombinant RNPK (lane 4) or RNPK amino acids 43–108 (lane 5) to the nuclear extract, but not by GST (lane 3). Lane 1, contains no extract; lane 2, contains nuclear extract alone. (B) EMSAs were performed using radiolabelled ds CT element oligonucleotides. Lane 1, contains ROS17/2.8 nuclear extract. Incubation of nuclear extract with recombinant GST-RNPK disrupts formation of the faster migrating complex II (lane 2 versus lane 1). Preincubation of hnRNP K with 50- or 100-fold unlabelled ds CT oligonucleotides had greatly reduced the DNA-binding activities of complexes I and II, as expected (lanes 3 and 4). However, when GST-RNPK was preincubated with 50- or 100-fold unlabelled ss sense CT oligonucleotides (lanes 5 and 6 respectively), the ability of hnRNP K to disrupt complex II was attenuated. (C) HnRNP K can disrupt the formation of a protein–DNA complex on the osteocalcin CT element without directly binding to the DNA. In fact, the interaction between hnRNP K and ss DNA appears to be mutually exclusive to its ability to compete off a protein–DNA complex from the osteocalcin promoter. We hypothesize that the resulting repression of osteocalcin transcription caused by this interaction is probably due to the subtraction of an activator from the osteocalcin promoter.
DISCUSSION
We have identified hnRNP K as a new transcriptional regulator that can modulate transcription from a CT-rich element of the proximal osteocalcin promoter. HnRNP K has been shown to act at multiple levels of gene expression including transcription [18,29,30], translation [13,31] and RNA biogenesis [12,32]. The multifunctionality of hnRNP K is mediated by its numerous domains, including an N-terminal acidic domain, three DNA–RNA-binding KH domains (KH1, amino acids 46–98; KH2, amino acids 149–197; KH3, amino acids 391–439), a GRGG box (amino acids 236–273), an SH3-binding domain (amino acids 289–315) and a protein kinase-binding domain (amino acids 337–425) [16]. HnRNP K also interacts with numerous proteins in both the nucleus and cytosol, including signal transduction proteins, transcriptional activators and repressors. Thus, it has been postulated that hnRNP K may act as a docking platform or scaffold, shuttling from the cytoplasm to the nucleus [16]. Interestingly, we initially purified hnRNP K as a factor co-purifying with dsDNA-binding activity, under the assumption that we were purifying the components of complex II. However, our results clearly demonstrate that hnRNP K interacts only with ssDNA, indicating that hnRNP K co-purified with the dsDNA-binding activity but it was not the binding activity being tracked. Indeed, the result of the Southwestern blotting may reflect either denaturation of the dsDNA probe, or a protein other than hnRNP K, but part of the DNA-binding activity in complex II. Regardless, hnRNP K does interact with the fast migrating complex II binding the CT-element, and does have a bona fide effect on osteocalcin transcription.
It is this action as a transcriptional regulator that has recently attracted attention to hnRNP K. In fact, hnRNP K can activate the c-myc promoter by interaction with ssDNA on a CT-rich element [33,34], and can also activate c-fos gene transcription [35]. Although the c-myc and c-fos CT-rich elements are analogous to the one we have identified in the osteocalcin promoter, our results clearly demonstrate that hnRNP K acts as a repressor of osteocalcin transcription. Therefore hnRNP K may function as either a transactivator or transrepressor depending on the promoter context. Transcriptional repression by hnRNP K has been demonstrated in other promoters, including the acute phase response agp promoter [30], the human thymidine kinase promoter [36], and the β4 promoter of the neuronal nicotine acetylcholine receptor gene [37]. The ability of hnRNP K to act as an activator or a repressor is probably not due to variance in the cognate itself, as many of these promoters possess nearly identical primary sequence; rather it is quite probable that the presence of additional factors and their association with the DNA dictate hnRNP K functional activity.
There are at least two potential mechanisms by which transcriptional repression by hnRNP K may occur. The most likely scenario (proposed in Figure 8C) is that hnRNP K titrates off an unidentified transcription factor (complex II) that binds to the CT-rich region of the osteocalcin promoter resulting in the loss of an activator from the promoter and hence a net repression of gene transcription. This hypothesis is supported by the findings that neither recombinant nor endogenous hnRNP K is able to bind to the CT-rich region of the osteocalcin promoter in a ds conformation, and that either full-length hnRNP K or the KH1 and KH2 domains or hnRNP K 43–108 decrease binding activity of the fast migrating complex assembling on the −95 to −47 region of the osteocalcin promoter. Despite its inability to bind directly to this region, we were able to define a specific region of the osteocalcin promoter on which hnRNP K exerts its transcriptional repression activity, corresponding to the CT-rich TCTCCTGCCCCTCCT motif in the −74 to −56 region (core binding element underlined). Furthermore, the EMSA results suggest that the repressive effect of hnRNP K occurs only in the absence of DNA binding, supporting the idea that hnRNP K acts indirectly, possibly by interactions with other factors. There are many examples of transcriptional regulators that physically interact with hnRNP K, including the Y-box binding protein, YB1 [35], the transcriptional repressor Zik1 [38], the CCAAT-enhancer binding protein, C/EBPβ [30], Purα [39], as well as other hnRNPs [36]. In fact, protein–protein interactions between hnRNP K and other transcriptional regulators have been implicated as a mechanism of transcriptional repression of the thymidine kinase [36], neuronal nictonic acetylcholine receptor [37] and α-1 acid glycoprotein [30] promoters. Indeed, the interaction at the human thymidine kinase promoter is analogous to what was observed in our study, in which full-length GST-hnRNP K competitively inhibited binding of transcription factors without directly binding DNA [36].
Our reults also demonstrate that amino acids 43–108 spanning the KH1 and KH2 domains of hnRNP K are sufficient to mediate the effects of hnRNP K on protein–DNA complex disruption. In fact, interaction with ssDNA and ability to compete off the unidentified transcription factor are mutually exclusive functions of hnRNP K, suggesting the involvement of a common domain, and in particular the two KH domains, for both protein–protein and protein–ssDNA interactions. Previously, hnRNP K has been demonstrated to interact with other proteins by an SH3-binding domain at amino acids 289–315, which binds Src [40], Fyn, Lyn and Vav [41–43], or a repressor binding domain implicated in Zik1 and K protein kinase binding (amino acids 209–337) [38]. Thus to our knowledge this is the first observation of a protein–protein interaction mediated by the KH1 and KH2 domains of hnRNP K, although we have not systematically ruled out participation of other domains in the intervening sequence between KH1 and KH2.
A second hypothesis is that hnRNP K in the context of in vivo chromatin-bound DNA acts differently compared with what is seen in in vitro experiments. HnRNP K has been shown to bind ssDNA in partnership with cellular nucleic acid binding protein, CNBP, which helps to stabilize the ss structure [44]. The failure in EMSA experiments to supershift hnRNP K bound to dsDNA suggests that hnRNP K, even when binding partners are provided in the form of nuclear extract, is unable to form stabilized ssDNA on oligonucleotide probes. However, in the context of nucleosome occupied DNA, an ss intermediate may be more stable, allowing CNBP and hnRNP K to bind. In fact, the presence of ss CT-rich, hnRNP K binding elements in the c-myc promoter is associated with activated chromatin [45]. However, this possibility remains speculative at this stage. Theoretically, the requirement of an organized chromatin structure may explain the slightly greater repressive effect of hnRNP K on the endogenous promoter, relative to the single CT element. However, we favour the notion that hnRNP K may act at additional poly-pyrimidine sites, for example those present in the intronic sequence of the osteocalcin gene [46], resulting in a repression stronger than what is observed using the −92 to +32 region of the osteocalcin promoter. Further, the ability of hnRNP K to repress transcription in the luciferase outputs is clearly not a chromatin-mediated event, thus supporting our initial hypothesis.
Interestingly, hnRNP K inhibits Sp1 but not Sp3 binding to CT-rich cognates, preventing the formation of Sp1–DNA but not Sp3–DNA complexes [37]. We have previously demonstrated that the CT-rich region of the osteocalcin promoter is capable of binding both Sp1 and Sp3 [10]. However, in contrast with the Sp1-inhibitory effects observed on the nicotinic acetylcholine receptor promoter [37], the effects of hnRNP K do not involve disruption of Sp1 binding, since this factor is part of the slower-migrating complex I binding to the CT-rich element in the osteocalcin promoter, a complex unaffected by hnRNP K.
HnRNP K has been described as a docking platform that bridges signal-transduction pathways in the cytosol to nuclear function. In fact, hnRNP K is phosphorylated by Src and Lck [47] and by ERK (extracellular-signal-regulated kinase) [48]. ERK phosphorylation has been shown to drive the cytoplasmic accumulation of hnRNP K [48], while tyrosine phosphorylation of hnRNP K by the Src family of protein kinases has been shown to alter hnRNP K protein–protein and protein–nucleic acid binding activity and binding partners [47]. Of note, ERK activation stimulates osteocalcin transcription [49,50], whereas Src activity has been shown to regulate negatively osteocalcin gene transcription [51]. It is tempting to speculate that phosphorylation of hnRNP K by the Src/ERK pathways may alter the molecular interactions of hnRNP K and drive it from the nucleus to the cytosol, resulting in a de-repression of osteocalcin transcription. In support of this notion, we have demonstrated that ERK regulates transcription from this region of the osteocalcin promoter (J. P. Stains and R. Civitelli, unpublished work).
In summary, we have demonstrated that hnRNP K co-purifies with binding activity to the osteocalcin proximal promoter and regulates osteocalcin gene transcription. We further demonstrated that although hnRNP K does not interact with the osteocalcin proximal promoter directly, it functions as a transcriptional repressor by displacing an unidentified transcription factor complex (complex II) from a CT-rich region of the osteocalcin promoter. Since hnRNP K can shuttle between the cytosol and the nucleus and be a target of many signal transduction pathways, this multifunctional factor may represent an important convergence point between cell signalling and gene transcription.
Acknowledgments
We thank the members of the Civitelli and Towler laboratories for useful discussions and insights. We also thank W. Lane (Harvard Microchemistry Facility) for assistance in the characterization of purified DNA-binding protein by MS. This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health (R01 AR41255 to R. C. and T32 AR07033 to J. P. S.), and by funds from the Barnes-Jewish Hospital Foundation (St. Louis, MO, U.S.A.). F. L. was a Fellow from Ramón y Cajal Program, and was supported by funds from ‘UTE project CIMA’ agreement, the Ministry of Science and Technology (Plan Nacional I+D+I, Ref. SAF2001-1220), funds from the Spanish Ministry of Health (Fondo de Investigaciones Sanitarias, RTIC C10/03) and Government of Navarra (Ortiz de Landázuri Scholarship).
References
- 1.Romberg R. W., Werness P. G., Riggs B. L., Mann K. G. Inhibition of hydroxyapatite crystal growth by bone-specific and other calcium-binding proteins. Biochemistry. 1986;25:1176–1180. doi: 10.1021/bi00353a035. [DOI] [PubMed] [Google Scholar]
- 2.Hoang Q. Q., Sicheri F., Howard A. J., Yang D. S. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature (London) 2003;425:977–980. doi: 10.1038/nature02079. [DOI] [PubMed] [Google Scholar]
- 3.Ducy P., Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell (Cambridge, Mass.) 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 5.Towler D. A., Rutledge S.-J. C., Rodan G. A. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol. Endocrinol. 1994;8:1484–1493. doi: 10.1210/mend.8.11.7877617. [DOI] [PubMed] [Google Scholar]
- 6.Bortell R., Owen T. A., Bidwell J. P., Gavazzo P., Breen E., Van Wijnen A. J., DeLuca H. F., Stein J. A., Lian J. B., Stein G. S. Vitamin D-responsive protein–DNA interactions at multiple promoter regulatory elements that contribute to the level of rat osctocalcin gene expression. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6119–6123. doi: 10.1073/pnas.89.13.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lian J. B., Stein G. S., Stein J. L., van Wijnen A. J. Osteocalcin gene promoter: unlocking the secrets for regulation of osteoblast growth and differentiation. J. Cell. Biochem. 1998;30–31(Suppl.):62–72. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<62::AID-JCB10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Willis D. M., Loewy A. P., Charlton-Kachigian N., Shao J. S., Ornitz D. M., Towler D. A. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J. Biol. Chem. 2002;277:37280–37291. doi: 10.1074/jbc.M206482200. [DOI] [PubMed] [Google Scholar]
- 9.Towler D. A., Rodan G. A. Identification of a rat osteocalcin promoter 3′,5′-cyclic adenosine monophosphate response region containing two PuGGTCA steroid hormone receptor binding motifs. Endocrinology. 1995;136:1089–1096. doi: 10.1210/endo.136.3.7867563. [DOI] [PubMed] [Google Scholar]
- 10.Stains J. P., Lecanda F., Screen J., Towler D. A., Civitelli R. Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J. Biol. Chem. 2003;278:24377–24387. doi: 10.1074/jbc.M212554200. [DOI] [PubMed] [Google Scholar]
- 11.Expert-Bezancon A., Le Caer J. P., Marie J. Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken beta-tropomyosin pre-mRNA. J. Biol. Chem. 2002;277:16614–16623. doi: 10.1074/jbc.M201083200. [DOI] [PubMed] [Google Scholar]
- 12.Michael W. M., Eder P. S., Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostareck D. H., Ostareck-Lederer A., Wilm M., Thiele B. J., Mann M., Hentze M. W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell (Cambridge, Mass.) 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 14.Siomi H., Choi M., Siomi M. C., Nussbaum R. L., Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell (Cambridge, Mass.) 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 15.Krecic A. M., Swanson M. S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 16.Bomsztyk K., Van S. I., Suzuki H., Denisenko O., Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403:113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 17.Tomonaga T., Levens D. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelotti E. F., Michelotti G. A., Aronsohn A. I., Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denisenko O., Bomsztyk K. Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length. Mol. Cell. Biol. 2002;22:286–297. doi: 10.1128/MCB.22.1.286-297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreyfuss G., Matunis M. J., Pinol-Roma S., Burd C. G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 21.Lecanda F., Towler D. A., Ziambaras K., Cheng S.-L., Koval M., Steinberg T. H., Civitelli R. Gap junctional communication modulates gene expression in osteoblastic cells. Mol. Biol. Cell. 1998;9:2249–2258. doi: 10.1091/mbc.9.8.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Towler D. A., Bennett C. D., Rodan G. A. Activity of the rat osteocalcin basal promoter in osteoblastic cells is dependent upon homeodomain and CP-1 binding motifs. Mol. Endocrinol. 1994;8:614–624. doi: 10.1210/mend.8.5.7914673. [DOI] [PubMed] [Google Scholar]
- 23.Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J., Fritsch E. F., Maniatis T. Planview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning – A Laboratory Manual. [Google Scholar]
- 25.Michael L. F., Alcorn J. L., Gao E., Mendelson C. R. Characterization of the cyclic adenosine 3′,5′-monophosphate response element of the rabbit surfactant protein-A gene: evidence for transactivators distinct from CREB/ATF family members. Mol. Endocrinol. 1996;10:159–170. doi: 10.1210/mend.10.2.8825556. [DOI] [PubMed] [Google Scholar]
- 26.Bidder M., Shao J. S., Charlton-Kachigian N., Loewy A. P., Semenkovich C. F., Towler D. A. Osteopontin transcription in aortic vascular smooth muscle cells is controlled by glucose-regulated upstream stimulatory factor and activator protein-1 activities. J. Biol. Chem. 2002;277:44485–44496. doi: 10.1074/jbc.M206235200. [DOI] [PubMed] [Google Scholar]
- 27.Ziambaras K., Lecanda F., Steinberg T. H., Civitelli R. Cyclic stretch enhances gap junctional communication between osteoblastic cells. J. Bone Miner. Res. 1998;13:218–228. doi: 10.1359/jbmr.1998.13.2.218. [DOI] [PubMed] [Google Scholar]
- 28.Tomonaga T., Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J. Biol. Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 29.Du Q., Melnikova I. N., Gardner P. D. Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J. Biol. Chem. 1998;273:19877–19883. doi: 10.1074/jbc.273.31.19877. [DOI] [PubMed] [Google Scholar]
- 30.Miau L. H., Chang C. J., Shen B. J., Tsai W. H., Lee S. C. Identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a repressor of C/EBPbeta-mediated gene activation. J. Biol. Chem. 1998;273:10784–10791. doi: 10.1074/jbc.273.17.10784. [DOI] [PubMed] [Google Scholar]
- 31.Collier B., Goobar-Larsson L., Sokolowski M., Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 1998;273:22648–22656. doi: 10.1074/jbc.273.35.22648. [DOI] [PubMed] [Google Scholar]
- 32.Ostareck-Lederer A., Ostareck D. H., Hentze M. W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 33.Michelotti E. F., Michelotti G. A., Aronsohn A. I., Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takimoto M., Tomonaga T., Matunis M., Avigan M., Krutzsch H., Dreyfuss G., Levens D. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 1993;268:18249–18258. [PubMed] [Google Scholar]
- 35.Shnyreva M., Schullery D. S., Suzuki H., Higaki Y., Bomsztyk K. Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J. Biol. Chem. 2000;275:15498–15503. doi: 10.1074/jbc.275.20.15498. [DOI] [PubMed] [Google Scholar]
- 36.Lau J. S., Baumeister P., Kim E., Roy B., Hsieh T. Y., Lai M., Lee A. S. Heterogeneous nuclear ribonucleoproteins as regulators of gene expression through interactions with the human thymidine kinase promoter. J. Cell. Biochem. 2000;79:395–406. doi: 10.1002/1097-4644(20001201)79:3<395::aid-jcb50>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 37.Du Q., Melnikova I. N., Gardner P. D. Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J. Biol. Chem. 1998;273:19877–19883. doi: 10.1074/jbc.273.31.19877. [DOI] [PubMed] [Google Scholar]
- 38.Denisenko O. N., O'Neill B., Ostrowski J., Van S. I., Bomsztyk K. Zik1, a transcriptional repressor that interacts with the heterogeneous nuclear ribonucleoprotein particle K protein. J. Biol. Chem. 1996;271:27701–27706. doi: 10.1074/jbc.271.44.27701. [DOI] [PubMed] [Google Scholar]
- 39.Da Silva N., Bharti A., Shelley C. S. hnRNP-K and Pur(alpha) act together to repress the transcriptional activity of the CD43 gene promoter. Blood. 2002;100:3536–3544. doi: 10.1182/blood.V100.10.3536. [DOI] [PubMed] [Google Scholar]
- 40.Taylor S. J., Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature (London) 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 41.Weng Z., Thomas S. M., Rickles R. J., Taylor J. A., Brauer A. W., Seidel-Dugan C., Michael W. M., Dreyfuss G., Brugge J. S. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol. Cell. Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hobert O., Jallal B., Schlessinger J., Ullrich A. Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J. Biol. Chem. 1994;269:20225–20228. [PubMed] [Google Scholar]
- 43.Bustelo X. R., Suen K. L., Michael W. M., Dreyfuss G., Barbacid M. Association of the vav proto-oncogene product with poly(rC)-specific RNA-binding proteins. Mol. Cell. Biol. 1995;15:1324–1332. doi: 10.1128/mcb.15.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michelotti E. F., Tomonaga T., Krutzsch H., Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 45.Michelotti G. A., Michelotti E. F., Pullner A., Duncan R. C., Eick D., Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kearns A. E., Goto K., Gianakakos G., Lippmann W., Demay M. B. Transcriptional repression of the rat osteocalcin gene: role of two intronic CCTCCT motifs. Endocrinology. 1999;140:4120–4126. doi: 10.1210/endo.140.9.6963. [DOI] [PubMed] [Google Scholar]
- 47.Ostrowski J., Schullery D. S., Denisenko O. N., Higaki Y., Watts J., Aebersold R., Stempka L., Gschwendt M., Bomsztyk K. Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J. Biol. Chem. 2000;275:3619–3628. doi: 10.1074/jbc.275.5.3619. [DOI] [PubMed] [Google Scholar]
- 48.Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A. L., Shokat K. M., Hentze M. W., Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 49.Xiao G., Gopalakrishnan R., Jiang D., Reith E., Benson M. D., Franceschi R. T. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J. Bone Miner. Res. 2002;17:101–110. doi: 10.1359/jbmr.2002.17.1.101. [DOI] [PubMed] [Google Scholar]
- 50.Zhao M., Xiao G., Berry J. E., Franceschi R. T., Reddi A., Somerman M. J. Bone morphogenetic protein 2 induces dental follicle cells to differentiate toward a cementoblast/osteoblast phenotype. J. Bone Miner. Res. 2002;17:1441–1451. doi: 10.1359/jbmr.2002.17.8.1441. [DOI] [PubMed] [Google Scholar]
- 51.Marzia M., Sims N. A., Voit S., Migliaccio S., Taranta A., Bernardini S., Faraggiana T., Yoneda T., Mundy G. R., Boyce B. F., et al. Decreased c-Src expression enhances osteoblast differentiation and bone formation. J. Cell Biol. 2000;151:311–320. doi: 10.1083/jcb.151.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]