To the Editor
Myeloproliferative neoplasms (MPN) are a group of clonal stem cell disorders with heterogeneous clinical presentation [1]. Due to the risk of severe thromboembolic complications and disease progression, the early recognition of an MPN prior to the appearance of clinical complications is clearly warranted to facilitate early pharmacologic intervention [2–4]. Detection of the somatic mutations by genotyping has become an essential part of the diagnostic work-up of suspected subjects, as well as of the risk stratification after the diagnosis of MPN has been confirmed [5]. However, in many parts of the world molecular testing is barely affordable.
We have established an immunofluorescence microscopy (IF)-based method for platelet phenotyping on the peripheral blood smear [6]. This method has been proven to be highly efficient in the diagnosis of diverse hereditary platelet disorders by recognizing disease-specific changes of cell structures, including alterations of leukocytes and red blood cells (RBC) [7, 8]. Major advantages of this approach are the need of small amounts of blood (<100 μL) and the possibility to send the blood films by regular mail even long distances.
It is well-known that morphology of peripheral blood cells is also often altered in MPN [9, 10]. However, due to different methods and the heterogeneity of the patients’ populations, results are difficult to compare.
In the present study, we aimed at assessing platelet phenotype using our IF method in a cohort of patients diagnosed with MPN. The study has been registered in the German Clinical Trials Register (DRKS-ID: DRKS00032588). Three German reference centers for diagnosis and treatment of MPN took part in the study: Internal Medicine C, University Medicine Greifswald; Internal Medicine 2, University Hospital Jena; and Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Germany. The study protocol was approved by the institutional review boards of all centers. Patients or their legal guardians signed written informed consent to the investigation, which was conducted according to the Declaration of Helsinki. Healthy controls were enrolled among blood donors at the Institute for Transfusion Medicine, University Medicine Greifswald, Germany.
Blood slides were prepared using fresh EDTA-anticoagulated blood and shipped by regular mail within 5 days to the Greifswald platelet laboratory, where the analysis was performed. One blood smear was stained using the May-Grünwald-Giemsa technique, and the others labeled with primary and secondary antibodies upon fixation as previously reported [7]. The panel of used antibodies is given in the Supplementary Table 1. In detail, we investigated 13 structures including platelet surface glycoprotein IIb/IIIa and Ib/IX, three components each for platelet alpha- (von Willebrand factor, P-selectin, and thrombospondin 1) and dense granules (LAMP 1, LAMP 2, and CD63), the cytoskeletal proteins non-muscular myosin IIA (NMMIIA), filamin A, β1- and α tubulin, and the stem cell antigen CD34.
Light- and standard IF microscopy were performed using an Olympus BX40 microscope (Olympus, Hamburg, Germany) equipped with an Olympus XC10 camera, and an UplanSApo 60x immersion objective lens and the following wave length filters: WIB 460–490 nm, and WG 510–550 nm. The microscopic assessment was performed by two independent observers, who were blinded for the clinical phenotype of the MPN patients. The morphologic changes were assigned to the specific cell structure, and reported by a semiquantitative grading system as previously described [8]. The interobserver concordance was high (91%).
We enrolled 135 MPN patients and 83 healthy controls. The demographic, clinical and molecular characteristics of the enrolled subjects are provided in Table 1 and Supplementary Table 2.
Table 1.
Clinical and morphologic features of the enrolled patients.
| Pat. no. | Clinical features | Morphologic features | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex/Agea | MPN subtypeb | Gene carrying the driver mutation | Additional somatic mutationsc/High-molecular risk statusd | Light microscopy | Immunofluorescence microscopy | ||||||
| Platelet anisocytosis | RBC anisopoikilocytosis | Platelet alpha granule defecte | Platelet dense granule defectf | Platelet cytoskeleton defectg | Platelet surface receptor defecth | RBC NMMIIA aggregatesi | |||||
| 1 | M/73 | PV | JAK2 | No | n.a. | n.a. | Yes | Yes | No | No | No |
| 2 | F/53 | MF | CALR | No | n.a. | n.a. | Yes | Yes | No | No | Yes |
| 3 | M/77 | PV | JAK2 | Yes/No | n.a. | n.a. | No | Yes | No | No | Yes |
| 4 | M/81 | PV | JAK2 | Yes/No | n.a. | n.a. | Yes | Yes | Yes | No | Yes |
| 5 | F/47 | ET | CALR | Yes/No | n.a. | n.a. | Yes | Yes | Yes | No | Yes |
| 6 | F/66 | ET | JAK2 | No | n.a. | n.a. | Yes | Yes | Yes | No | Yes |
| 7 | M/55 | ET | JAK2 | No | n.a. | n.a. | Yes | Yes | No | No | Yes |
| 8 | M/36 | MF | CALR | No | n.a. | n.a. | Yes | Yes | No | Yes | Yes |
| 9 | F/79 | PV | JAK2 | Yes/No | n.a. | n.a. | Yes | No | Yes | No | No |
| 10 | F/71 | MF | JAK2 | Yes/Yes | n.a. | n.a. | Yes | No | Yes | No | Yes |
| 11 | F/54 | MPN-U | JAK2 | No | n.a. | n.a. | No | No | No | No | Yes |
| 12 | M/44 | ET | JAK2 | No | n.a. | n.a. | No | Yes | Yes | No | Yes |
| 13 | F/79 | MF | CALR | Yes/No | n.a. | n.a. | Yes | Yes | No | No | Yes |
| 14 | F/39 | ET | CALR | No | n.a. | n.a. | No | No | No | No | No |
| 15 | F/61 | PV | JAK2 | Yes/No | n.a. | n.a. | Yes | No | No | No | No |
| 16 | M/59 | MF | JAK2 | No | n.a. | n.a. | Yes | Yes | Yes | No | Yes |
| 17 | M/80 | PV | JAK2 | Yes/No | n.a. | n.a. | Yes | No | Yes | No | Yes |
| 18 | M/63 | ET | JAK2 | Yes/No | n.a. | n.a. | Yes | No | No | Yes | Yes |
| 19 | M/40 | PV | JAK2 | No | n.a. | n.a. | No | No | No | No | Yes |
| 20 | F/63 | ET | JAK2 | Yes/No | n.a. | n.a. | Yes | Yes | No | No | Yes |
| 21 | F/69 | MF | JAK2 | No | n.a. | n.a. | Yes | No | No | No | Yes |
| 22 | M/67 | MF | CALR | Yes/Yes | n.a. | n.a. | No | No | No | No | No |
| 23 | F/62 | MF | CALR | No | n.a. | n.a. | No | Yes | No | No | Yes |
| 24 | M/78 | MF | CALR | No | n.a. | n.a. | No | Yes | No | No | Yes |
| 25 | F/57 | ET | CALR | Yes/No | n.a. | n.a. | No | No | No | Yes | Yes |
| 26 | M/58 | PV | JAK2 | Yes/No | n.a. | n.a. | No | No | No | No | Yes |
| 27 | F/43 | MF | JAK2 | No | n.a. | n.a. | No | Yes | No | No | No |
| 28 | M/70 | PV | JAK2 | Yes/No | n.a. | n.a. | Yes | No | No | No | Yes |
| 29 | M/79 | MF | CALR | No | n.a. | n.a. | No | No | No | No | No |
| 30 | M/69 | MF | JAK2 | Yes/Yes | n.a. | n.a. | No | Yes | No | No | Yes |
| 31 | M/69 | ET | JAK2 | Yes/No | n.a. | n.a. | No | No | Yes | No | No |
| 32 | F/67 | MF | JAK2 | Yes/Yes | n.a. | n.a. | Yes | No | No | No | Yes |
| 33 | M/62 | MF | JAK2 | No | n.a. | n.a. | Yes | No | No | No | Yes |
| 34 | F/69 | ET | JAK2 | Yes/No | n.a. | n.a. | No | No | No | No | No |
| 35 | M/55 | MF | CALR | n.a./n.a. | n.a. | n.a. | No | No | Yes | No | Yes |
| 36 | M/52 | MF | CALR | No | n.a. | n.a. | No | No | No | No | Yes |
| 37 | F/68 | MPN-U | TN | Yes/- | n.a. | n.a. | No | No | No | No | No |
| 38 | F/27 | ET | JAK2 | No | n.a. | n.a. | No | No | No | Yes | No |
| 39 | F/34 | ET | JAK2 | No | n.a. | n.a. | No | Yes | No | No | No |
| 40 | F/52 | PV | JAK2 | Yes/No | Yes | No | Yes | Yes | No | No | Yes |
| 41 | F/48 | MF | JAK2 | No | Yes | No | Yes | Yes | Yes | No | Yes |
| 42 | M/68 | MF | JAK2 | Yes/Yes | Yes | Yes | Yes | Yes | No | No | Yes |
| 43 | F/51 | PV | JAK2 | No | No | No | Yes | No | No | No | No |
| 44 | F/22 | MF | JAK2 | No | Yes | No | Yes | No | No | No | Yes |
| 45 | M/58 | MF | JAK2/MPL | Yes/Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| 46 | M/82 | MF | JAK2 | Yes/No | Yes | No | Yes | No | No | No | Yes |
| 47 | M/71 | PV | JAK2 | No | n.a. | n.a. | Yes | Yes | No | No | No |
| 48 | M/67 | MF | MPL | Yes/No | Yes | Yes | Yes | No | No | No | Yes |
| 49 | M/69 | PV | JAK2 | Yes/No | Yes | No | Yes | Yes | No | No | No |
| 50 | F/64 | PV | JAK2 | No | Yes | No | No | Yes | No | No | No |
| 51 | F/52 | MF | JAK2 | No | Yes | No | Yes | No | No | No | No |
| 52 | F/55 | MF | JAK2 | Yes/No | No | Yes | Yes | No | No | No | Yes |
| 53 | M/81 | PV | JAK2 | No | n.a. | n.a. | Yes | No | No | No | Yes |
| 54 | M/75 | MF | JAK2 | No | Yes | No | No | No | No | No | Yes |
| 55 | F/68 | MF | JAK2 | Yes/No | No | No | No | No | No | No | No |
| 56 | M/48 | ET | JAK2 | Yes/No | Yes | No | Yes | No | No | No | Yes |
| 57 | M/80 | MF | JAK2 | No | Yes | Yes | Yes | Yes | No | No | Yes |
| 58 | F/62 | MF | JAK2 | Yes/Yes | Yes | Yes | Yes | No | No | No | Yes |
| 59 | F/68 | MF | CALR | No | Yes | Yes | No | No | No | No | Yes |
| 60 | M/64 | MF | CALR | Yes/Yes | Yes | Yes | No | No | No | No | Yes |
| 61 | F/62 | PV | JAK2 | No | Yes | No | No | No | No | No | No |
| 62 | F/59 | ET | JAK2 | No | Yes | No | No | No | No | No | No |
| 63 | F/39 | MF | MPL | No | Yes | Yes | Yes | No | No | No | Yes |
| 64 | F/74 | ET | JAK2 | Yes/No | Yes | No | No | No | No | No | Yes |
| 65 | M/77 | MPN-U | JAK2 | Yes/- | Yes | No | Yes | Yes | No | No | Yes |
| 66 | F/53 | PV | JAK2 | Yes/No | Yes | No | No | No | No | No | Yes |
| 67 | M/77 | ET | MPL | Yes/No | Yes | Yes | Yes | Yes | No | No | Yes |
| 68 | F/63 | MF | JAK2 | No | Yes | Yes | Yes | No | No | No | Yes |
| 69 | F/73 | PV | JAK2 | Yes/No | No | No | No | Yes | No | No | Yes |
| 70 | M/51 | MF | CALR | Yes/No | Yes | Yes | Yes | No | No | No | Yes |
| 71 | M/64 | MF | JAK2 | Yes/No | Yes | Yes | Yes | No | No | No | Yes |
| 72 | F/67 | MF | TN | Yes/Yes | Yes | Yes | No | Yes | No | No | Yes |
| 73 | M/47 | MF | MPL | Yes/Yes | Yes | No | Yes | Yes | No | No | No |
| 74 | F/63 | PV | JAK2 | No | No | No | Yes | Yes | No | No | No |
| 75 | F/33 | PV | JAK2 | No | Yes | No | No | No | No | No | No |
| 76 | M/46 | PV | JAK2 | Yes/No | No | Yes | No | No | No | No | Yes |
| 77 | F/59 | ET | JAK2 | No | Yes | Yes | Yes | No | No | No | No |
| 78 | F/75 | MF | JAK2 | Yes/No | Yes | No | Yes | No | Yes | No | Yes |
| 79 | M/54 | MF | MPL | No | No | No | No | No | No | No | Yes |
| 80 | F/78 | MF | CALR | Yes/No | Yes | No | Yes | No | Yes | No | Yes |
| 81 | F/80 | ET | MPL | No | Yes | No | Yes | No | Yes | No | Yes |
| 82 | F/31 | PV | JAK2 | No | No | No | No | No | Yes | No | Yes |
| 83 | M/60 | PV | JAK2 | Yes/No | Yes | No | No | No | Yes | No | Yes |
| 84 | M/44 | ET | CALR | Yes/No | No | No | Yes | No | No | No | No |
| 85 | F/43 | MF | JAK2 | No | Yes | Yes | Yes | Yes | Yes | No | Yes |
| 86 | F/45 | PV | JAK2 | Yes/No | No | No | Yes | Yes | No | No | No |
| 87 | M/65 | MF | JAK2 | Yes/No | Yes | Yes | No | Yes | No | No | Yes |
| 88 | F/39 | MF | JAK2 | Yes/No | No | No | No | No | No | No | Yes |
| 89 | F/64 | MPN-U | JAK2 | Yes/- | No | No | Yes | No | No | No | Yes |
| 90 | F/39 | ET | CALR | No | Yes | Yes | No | Yes | No | No | Yes |
| 91 | F/84 | PV | JAK2 | Yes/No | Yes | Yes | Yes | No | No | No | Yes |
| 92 | M/54 | MF | CALR | Yes/Yes | Yes | Yes | No | No | No | No | Yes |
| 93 | F/40 | ET | JAK2 | No | Yes | Yes | Yes | Yes | Yes | No | Yes |
| 94 | F/82 | MPN-U | JAK2 | Yes/- | Yes | Yes | Yes | Yes | No | No | Yes |
| 95 | F/58 | PV | JAK2 | No | Yes | No | Yes | No | No | No | Yes |
| 96 | M/60 | PV | JAK2 | No | Yes | Yes | Yes | Yes | No | No | Yes |
| 97 | M/58 | ET | JAK2 | No | Yes | No | No | No | No | No | Yes |
| 98 | M/79 | ET | MPL | Yes/No | Yes | Yes | Yes | Yes | No | No | Yes |
| 99 | F/76 | MPN-U | TN | Yes/- | No | No | No | No | No | No | No |
| 100 | M/31 | MF | TN | No | No | No | Yes | Yes | Yes | No | Yes |
| 101 | F/71 | MF | CALR | Yes/Yes | Yes | No | No | No | No | No | No |
| 102 | M/71 | ET | JAK2 | n.a./n.a. | Yes | Yes | Yes | Yes | Yes | No | Yes |
| 103 | F/60 | PV | JAK2 | No | Yes | No | No | No | No | No | No |
| 104 | F/78 | MF | JAK2 | Yes/Yes | Yes | Yes | Yes | No | No | No | Yes |
| 105 | F/80 | MF | JAK2 | Yes/No | Yes | Yes | Yes | Yes | No | No | Yes |
| 106 | M/73 | PV | JAK2 | Yes/No | Yes | Yes | Yes | Yes | No | No | Yes |
| 107 | F/74 | MF | CALR | No | Yes | No | Yes | Yes | No | No | Yes |
| 108 | F/43 | MPN-U | JAK2 | Yes/- | Yes | No | Yes | Yes | No | No | Yes |
| 109 | M/68 | MF | MPL | Yes/Yes | Yes | Yes | Yes | Yes | No | No | Yes |
| 110 | M/80 | PV | JAK2 | Yes/No | No | Yes | Yes | Yes | No | No | Yes |
| 111 | M/45 | PV | JAK2 | No | Yes | No | Yes | Yes | No | No | Yes |
| 112 | M/79 | MF | JAK2 | Yes/No | n.a. | n.a. | Yes | Yes | No | No | Yes |
| 113 | M/74 | MF | JAK2 | Yes/No | Yes | No | No | Yes | No | No | No |
| 114 | F/39 | ET | JAK2 | No | n.a. | n.a. | No | Yes | No | No | No |
| 115 | F/80 | MF | JAK2 | No | Yes | Yes | Yes | Yes | No | No | Yes |
| 116 | F/67 | PV | JAK2 | No | No | No | Yes | Yes | No | No | Yes |
| 117 | F/68 | ET | JAK2 | Yes/Yes | No | Yes | Yes | Yes | No | No | Yes |
| 118 | M/50 | ET | JAK2 | Yes/No | Yes | Yes | No | Yes | No | No | Yes |
| 119 | F/71 | MF | JAK2 | Yes/Yes | No | Yes | Yes | No | No | No | Yes |
| 120 | M/75 | MF | JAK2 | No | Yes | Yes | Yes | No | Yes | No | Yes |
| 121 | F/62 | MF | CALR | Yes/No | Yes | Yes | No | No | No | No | Yes |
| 122 | M/67 | MF | JAK2 | Yes/Yes | Yes | No | Yes | Yes | No | No | Yes |
| 123 | M/55 | MF | CALR | No | Yes | Yes | Yes | No | Yes | No | Yes |
| 124 | F/45 | PV | JAK2 | Yes/No | Yes | No | Yes | Yes | No | No | Yes |
| 125 | F/61 | MF | JAK2 | Yes/No | Yes | No | No | No | No | No | No |
| 126 | M/28 | MF | CALR | No | Yes | No | No | No | No | No | No |
| 127 | F/79 | PV | JAK2 | No | Yes | No | Yes | Yes | No | No | Yes |
| 128 | F/48 | ET | JAK2 | Yes/No | Yes | No | Yes | No | No | No | No |
| 129 | M/87 | MPN-U | JAK2 | Yes/- | Yes | No | Yes | No | No | No | Yes |
| 130 | F/60 | MF | JAK2 | Yes/No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 131 | F/47 | ET | JAK2 | Yes/No | Yes | No | No | Yes | No | No | No |
| 132 | F/61 | ET | CALR | Yes/No | Yes | Yes | Yes | No | No | No | No |
| 133 | M/62 | ET | CALR | Yes/No | Yes | No | No | No | No | No | Yes |
| 134 | F/44 | PV | JAK2 | Yes/No | No | No | No | No | No | No | No |
| 135 | M/70 | PV | JAK2 | n.a./n.a. | No | Yes | Yes | Yes | No | No | Yes |
Pat. patient, No. number, F female, M male, MPN myeloproliferative neoplasm, PV polycythemia vera, ET essential thrombocythemia, MF primary or secondary myelofibrosis, MPN-U unclassifiable MPN, TN triple-negative, i.e., absence of mutations hitting either JAK2 or CALR or MPL; n.a. not available, RBC red blood cell, NMMIIA non-muscular myosin IIA.
aAt time of investigation.
bAccording to the WHO 2016 classification [13].
c≥1 additional somatic mutation detected by NGS analysis of 33 genes (ASXL1, BCOR, CBL, CEBPA, CUX1, DNMT3A, EZH2, GATA2, GNAS, GNB1, IDH1, IDH2, NF1, PHF6, PHIP, PPM1D, PRPF8, PTPN11, RAD21, RAS, RB1, RUNX1, SETBP1, SF3B1, SH2B3, SMC1A, SMC4, SRSF2, STAG2, TET2, TP53, U2AF1, ZRSR2) with a variant allele frequency ≥ 2%.
d≥1 high-molecular risk mutation according to the prognostic panels of PV, ET and PMF [11, 12] - where applicable.
eDefined as reduced expression of at least two out of the three investigated markers of alpha granules (von Willebrand factor, P-selectin, thrombospondin 1) compared to control, as reported [8].
fDefined as reduced or altered expression of at least two out of the three investigated markers of lysosomes and dense granules (LAMP-1, LAMP-2, CD63) compared to control, as reported [7].
gDefined as altered expression of at least three out of the four investigated cytoskeletal proteins (filamin A, NMMIIA, α-tubulin, β1-tubulin) compared to control, as reported [7].
hDefined as reduced expression of the surface glycoprotein Ib/IX or IIb/IIIa compared to control, as reported [7];
iDetection of NMMIIA aggregates in RBC, as reported [8]. Pat. = patient; No. = number; F = female; M = male; MPN = myeloproliferative neoplasm; PV = polycythemia vera; ET = essential thrombocythemia; MF = primary or secondary myelofibrosis; MPN-U = unclassifiable MPN; TN = triple-negative, i.e., absence of mutations hitting either JAK2 or CALR or MPL; n.a. = not available; RBC = red blood cell; NMMIIA = non-muscular myosin IIA.
Sixty-one of the 135 MPN patients (45%) were male, and 74 (55%) females. The median age was 63 years (range: 22–87). Thirty-six subjects (27%) had received a diagnosis of polycythemia vera; 31 (23%) of essential thrombocythemia; 60 (44%) of primary or secondary myelofibrosis; and eight (6%) of unclassifiable MPN.
Forty-two of the 83 healthy controls (51%) were male, and 41(49%) females. The median age was 36 years (range: 18–64).
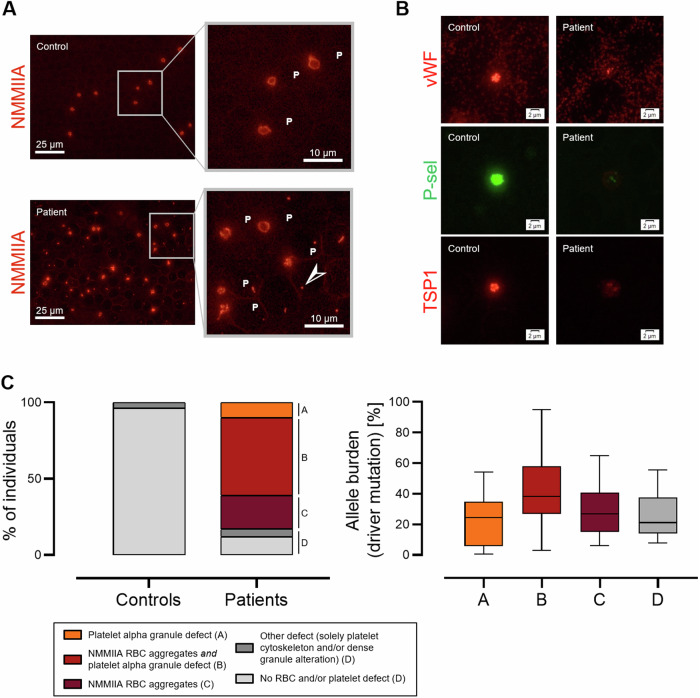
By IF microscopy, we identified two frequently altered structures in MPN patients, i.e., aggregates consisting of NMMIIA in the RBC and altered expression of platelet alpha granules. RBC aggregates of NMMIIA were found in 98 (73%) of the MPN patients (Fig. 1A). In 68 of these subjects (69%), this finding was accompanied by reduced expression of at least two out of the three investigated platelet alpha granule markers (Fig. 1B). Fourteen (10%) MPN subjects showed solely reduced expression of at least two alpha granule markers. Only 16 (12%) MPN patients displayed normal findings. In strong contrast, none of the controls showed either NMMIIA aggregates in the RBC or alterations of more than one platelet alpha granule marker. The other investigated structures were substantially non-informative as only 7 MPN patients (5%) displayed alterations assignable to platelet cytoskeleton and/or dense granules. Of the MPN patients with altered phenotype by IF microscopy, 41 (52%) and 62 (78%) of the 79 evaluable subjects displayed by light microscopy a remarkable RBC anisopoikilocytosis and platelet anisocytosis, respectively. Except for a leuko-erythroblastic picture, which was detectable in in 25/79 subjects (32%), no major alterations of the RBC or white blood cells were apparent.
Fig. 1. Morphological changes of platelets and red blood cells (RBC) in 135 patients with myeloproliferative neoplasms (MPN) and 83 healthy controls by immunofluorescence microscopy on the blood smear.
A Representative picture of aggregates of non-muscular myosin IIA (NMMIIA) in RBC of a MPN patient in comparison to a healthy control. P indicates platelets, arrows indicate the RBC aggregates. B Reduced expression of the alpha granule markers von Willebrand factor (vWF), P-selectin (P-sel) and thrombospondin 1 (TSP1) in a MPN patient compared to a healthy control. C Left. Schematic representation of the spectrum of morphologic alterations found in patients and controls, and their prevalence. Right. Allele burden of mutations in JAK2, CALR or MPL gene within MPN patients grouped according to the found morphologic change(s), which are designated by capital letters at the stacked bar graph. Boxes represent the interquartile range, bars within the boxes designate the median values, and whiskers extend to the range of data (minimum and maximum).
We found morphologic changes of peripheral RBC and platelets, which clearly differentiated MPN patients from the healthy controls. Is interesting to note that the 68 MPN patients with concomitantly altered platelet- and RBC phenotype showed the highest median allele burden of their MPN driver mutations hitting JAK2-, CALR- or MPL gene (Table 1), whereas those with none of these morphological alterations had the lowest (Fig. 1C). The subgroup of patients with morphological changes in both platelets and RBC was also featured by the highest prevalence of individuals carrying additional, non-driver mutations overall (61%) as well as including at least one variant considered at-high-risk of progression (15%) [11, 12] (Supplementary Fig. 1). This suggests a possible correlation between the morphologic phenotype and other clinical features of MPN, which might be relevant also for the prognosis.
Aggregates of NMMIIA in the RBC are also typical for two hereditary platelet disorders associated with constitutional dyserythropoiesis due to germline mutations in the transcription regulator GATA1 or GFI1B [8], and might represent a novel marker of dyserythropoieis in the peripheral blood. This may link the pathogenesis of somatic mutations in MPN and germline mutations in hereditary platelet disorders [7, 10].
Further studies are required to investigate this aspect as well as to identify mechanisms leading to the morphologic changes found in platelets and RBC of MPN subjects.
In conclusion, blood cell phenotyping by IF on the peripheral blood smear seems to be a clinically useful and easy-to-apply additional diagnostic tool to identify patients with MPN. This approach might be particularly useful in low- and middle-income countries with limited access to second-level diagnostic tools to stratify patients who may benefit from further genetic testing, as well in high-income country as Germany as a screening tool prior to next generation sequencing.
Supplementary information
Acknowledgements
The authors acknowledge the patients and their families, and the colleagues referring the patients. The work of Ulrike Strobel, who processed the blood films in Greifswald laboratory, is highly appreciated.
Author contributions
CZ, AG, and FHH conceived the rationale of the study. FHH, CCC, and TMS collected and interpreted clinical and genetic data of the enrolled patients. LV, CZ, and LS performed and interpreted the microscopic assessment of the blood smears. LK performed the statistical analysis. JF, and CF performed the immunofluorescence staining. LV, and JW prepared the pictures. CZ, AG, and FHH wrote the manuscript. All the authors helped in data interpretation, critically reviewed and agreed to the final version of the manuscript.
Funding
CZ and AG research is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 374031971-TRR 240. FHH was supported in part by grants of the Deutsche Forschungsgemeinschaft (DFG): HE6233/15-1, project number 517204983 and HE6233/4-2, project number 320028127. Open Access funding enabled and organized by Projekt DEAL.
Data availability
Inquiries about data access should be made to the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andreas Greinacher, Florian H. Heidel.
Contributor Information
Carlo Zaninetti, Email: carlo.zaninetti@med.uni-greifswald.de.
Florian H. Heidel, Email: heidel.florian@mh-hannover.de
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02346-z.
References
- 1.Perner F, Perner C, Ernst T, Heidel FH. Roles of JAK2 in aging, inflammation, hematopoiesis and malignant transformation. Cells. 2019;8:854. 10.3390/cells8080854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palandri F, Breccia M, Tiribelli M, Bonifacio M, Benevolo G, Iurlo A, et al. Risk factors for progression to blast phase and outcome in 589 patients with myelofibrosis treated with ruxolitinib: real-world data. Hematol Oncol. 2020;38:372–80. 10.1002/hon.2737 [DOI] [PubMed] [Google Scholar]
- 3.Palandri F, Rossi E, Auteri G, Breccia M, Paglia S, Benevolo G, et al. Predictors of response to hydroxyurea and switch to Ruxolitinib in HU-Resistant Polycythaemia VERA Patients: a real-world PV-NET study. Cancers. 2023;15:3706. 10.3390/cancers15143706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verstovsek S, Krečak I, Heidel FH, De Stefano V, Bryan K, Zuurman MW, et al. Identifying patients with polycythemia vera at risk of thrombosis after hydroxyurea initiation: the polycythemia vera-advanced integrated models (PV-AIM) Project. Biomedicines. 2023;11:1925. 10.3390/biomedicines11071925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palandri F, Palumbo GA, Bonifacio M, Elli EM, Tiribelli M, Auteri G, et al. A prognostic model to predict Ruxolitinib discontinuation and death in patients with myelofibrosis. Cancers. 2023;15:5027. 10.3390/cancers15205027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greinacher A, Pecci A, Kunishima S, Althaus K, Nurden P, Balduini CL, et al. Diagnosis of inherited platelet disorders on a blood smear: a tool to facilitate worldwide diagnosis of platelet disorders. J Thromb Haemost. 2017;15:1511–21. 10.1111/jth.13729 [DOI] [PubMed] [Google Scholar]
- 7.Zaninetti C, Leinøe E, Lozano ML, Rossing M, Bastida JM, Zetterberg E, et al. Validation of immunofluorescence analysis of blood smears in patients with inherited platelet disorders. J Thromb Haemost. 2023;21:1010–9. 10.1016/j.jtha.2022.12.031 [DOI] [PubMed] [Google Scholar]
- 8.Zaninetti C, Rivera J, Vater L, Ohlenforst S, Leinøe E, Böckelmann D, et al. Aggregates of nonmuscular myosin IIA in erythrocytes associate with GATA1- and GFI1B-related thrombocytopenia. J Thromb Haemost. 2024;22:1179–86. 10.1016/j.jtha.2023.12.007 [DOI] [PubMed] [Google Scholar]
- 9.Ng ZY, Fuller KA, Mazza-Parton A, Erber WN. Morphology of myeloproliferative neoplasms. Int J Lab Hematol. 2023;45:59–70. 10.1111/ijlh.14086 [DOI] [PubMed] [Google Scholar]
- 10.Nann D, Fend F. Synoptic diagnostics of myeloproliferative neoplasms: morphology and molecular genetics. Cancers. 2021;13:3528. 10.3390/cancers13143528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tefferi A, Lasho TL, Guglielmelli P, Finke CM, Rotunno G, Elala Y, et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016;1:21–30. 10.1182/bloodadvances.2016000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guglielmelli P, Lasho TL, Rotunno G, Score J, Mannarelli C, Pancrazzi A, et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28:1804–10. 10.1038/leu.2014.76 [DOI] [PubMed] [Google Scholar]
- 13.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Inquiries about data access should be made to the corresponding author.