Abstract
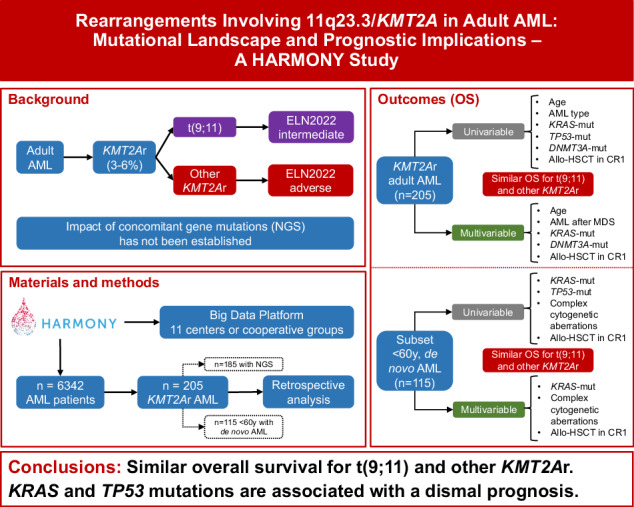
Balanced rearrangements involving the KMT2A gene (KMT2Ar) are recurrent genetic abnormalities in acute myeloid leukemia (AML), but there is lack of consensus regarding the prognostic impact of different fusion partners. Moreover, prognostic implications of gene mutations co-occurring with KMT2Ar are not established. From the HARMONY AML database 205 KMT2Ar adult patients were selected, 185 of whom had mutational information by a panel-based next-generation sequencing analysis. Overall survival (OS) was similar across the different translocations, including t(9;11)(p21.3;q23.3)/KMT2A::MLLT3 (p = 0.756). However, independent prognostic factors for OS in intensively treated patients were age >60 years (HR 2.1, p = 0.001), secondary AML (HR 2.2, p = 0.043), DNMT3A-mut (HR 2.1, p = 0.047) and KRAS-mut (HR 2.0, p = 0.005). In the subset of patients with de novo AML < 60 years, KRAS and TP53 were the prognostically most relevant mutated genes, as patients with a mutation of any of those two genes had a lower complete remission rate (50% vs 86%, p < 0.001) and inferior OS (median 7 vs 30 months, p < 0.001). Allogeneic hematopoietic stem cell transplantation in first complete remission was able to improve OS (p = 0.003). Our study highlights the importance of the mutational patterns in adult KMT2Ar AML and provides new insights into more accurate prognostic stratification of these patients.
Subject terms: Acute myeloid leukaemia, Acute myeloid leukaemia
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease in terms of clinical features and outcomes, in which the presence of cytogenetic aberrations and gene mutations provides crucial prognostic information that guides clinical decisions [1–5]. Balanced rearrangements involving the lysine methyltransferase 2a gene (KMT2A, previously known as MLL), located on chromosome band 11q23.3, have been described in 3–6% of adult patients with de novo AML [6–11].
Balanced chromosome rearrangements involving 11q23.3 and KMT2A (KMT2Ar) are very heterogeneous, as more that 100 fusion partners have already been described in leukemia patients [12]. Among them, the most frequent is the t(9;11)(p21.3;q23.3) resulting in a KMT2A::MLLT3 fusion, which will be referred to as t(9;11) hereafter. Other common translocations in adult AML are the t(6;11)(q27;q23.3)/KMT2A::AFDN [hereafter t(6;11)], t(11;19)(q23.3;p13.3)/KMT2A::MLLT1, t(11;19)(q23.3;p13.1) resulting in a KMT2A::ELL fusion, t/ins(10;11)(p12.3;q23.3)/KMT2A::MLLT10 [hereafter t(10;11)], and t(11;17)(q23.3;q25) resulting in a KMT2A::SEPTIN9 fusion [hereafter t(11;17)] [7, 13, 14].
There is a controversy in the literature regarding the prognostic impact of different KMT2Ar AML. Several studies have reported better outcomes in patients with t(9;11) when compared to the rest of KMT2Ar [7, 14–16], although some of them were restricted to specific populations (e.g., patients aged <60 years with de novo AML). On the other hand, these findings could not be confirmed by other studies [6, 17–19]. Moreover, heterogeneous results have also been reported regarding outcome in the post-allogeneic transplantation setting [20–22].
Additional cytogenetic aberrations have been identified in about 40% of KMT2Ar adult AML patients, but they were not associated with different patient outcomes [11, 16, 17]. While distinct gene mutations have proven to be excellent prognostic markers in AML [1–4, 23], KMT2Ar AML shows a lower incidence of co-occurring gene mutations, although a predominance of RAS mutations has been reported. The prognostic impact of these additional gene mutations in KMT2Ar adult AML has not been established yet [9, 14, 17, 19].
In order to address these open questions, we analyzed a large cohort of patients with KMT2Ar AML included in the Healthcare Alliance for Resourceful Medicine Offensive against Neoplasms in Hematology (HARMONY) AML multicenter database.
Methods
Patients
At the time of the analysis (December 2022) the HARMONY Alliance AML database contained 6342 AML patient data sets contributed by 11 European centers or cooperative groups (Supplementary Table S1). Conventional karyotype information was available for 6005 patients based on which a total of 205 KMT2Ar adult patients could be identified (incidence of KMT2Ar: 3.4%), who were diagnosed with AML between December 1996 and January 2020 (Supplementary Fig. S1). Some of the patients included in this analysis were also part of previously published AML cohorts [3, 9, 24–26].
For data upload into the HARMONY Big Data Platform, all patient data went through a robust double brokerage pseudonymization procedure in compliance with the General Data Protection Regulation. Next, data were harmonized and transformed using the Observational Medical Outcomes Partnership (OMOP) Common Data Model [27].
This study was performed in accordance with the Declaration of Helsinki and was approved by the HARMONY steering committee. The HARMONY research project was reviewed and approved by the Medicinal Research Ethics Committee of the University of Salamanca. For its studies, HARMONY provides an ethical and data-protection framework for the secondary use of data including a de facto anonymization. Written informed consent had been previously collected from all patients in the respective HARMONY partner institutions.
Cytogenetic and genetic analyses
Cytogenetic analyses were performed using standard G-banding with trypsin-Giemsa or trypsin-Wright staining in approved cytogenetic laboratories. The analysis and nomenclature of the chromosomes were based on International System for Human Cytogenetic Nomenclature (ISCN) [28]. Karyotypes were considered with clonal abnormalities if at least two metaphases showing the same abnormality were detected. The results of cytogenetic analysis of bone marrow were centrally reviewed by experienced cytogeneticists (JMHR and TGM) using standard ISCN-2020 criteria. Although t(11;19)(q23.3;p13.3) and t(11;19)(q23.3;p13.1) are known to be two distinct rearrangements [13], enough information to discriminate was only provided for around half of those patients, so we grouped them together as t(11;19)(q23;p13) [hereafter t(11;19)].
KMT2Ar cases were also evaluated by fluorescence in situ hybridization (FISH) with a commercial break apart probe in order to confirm KMT2Ar. FISH results were also centrally reviewed and reported using standard ISCN-2020 nomenclature. For selected cases, the AF9::KMT2A fusion transcript was also validated by reverse transcription polymerase chain reaction (RT-PCR) using the generic PCR cycler program developed by the BIOMED-1 initiative for the standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia [29].
Next-generation sequencing (NGS) data using local custom diagnostic panels were available for 185 patients (90% of our KMT2Ar cohort). The different NGS panels overlapped with regard to 40 genes implicated in myeloid malignancies (Supplementary Table S2). Panel sequencing was performed and analyzed according to previous studies [3, 9, 24]. Gene variants were centrally reviewed, only pathogenic and likely pathogenic variants were considered, and data including variant allele frequency were harmonized.
Statistical analysis
Clinical endpoints were defined as recommended by international guidelines [4]. Composite complete remission (CRc) was defined as either complete remission (CR) or CR with incomplete hematologic recovery (CRi). Categorical variables were compared using Pearson’s Chi-squared test and Fisher’s exact test, while continuous variables were evaluated using Kruskal–Wallis test for multiple comparisons and Wilcoxon test for pairwise comparison. Overall survival (OS) and relapse-free survival (RFS) were estimated using the Kaplan–Meier method and differences between survival distributions were evaluated using the log-rank test. To estimate survival probabilities considering the effect of allogeneic hematopoietic stem cell transplantation (allo-HSCT) in first complete remission (CR1), Simon–Makuch method with clock-back correction was applied as previously reported [30, 31]. Cox proportional hazards model was used for multivariate survival analysis, including variables that were significant in the univariate analysis (age, AML type, KRAS-mut, DNMT3A-mut, TP53-mut and presence of complex cytogenetic aberrations), as well as KMT2A fusion partner. To examine the effect of allo-HSCT in CR1, multivariate Cox model with allo-HSCT as a time-depending intervening event was used. All reported p values are two-sided at the conventional 5% significance level. Data were analyzed as of December 2022 using R software (v3.6.3).
Results
Patient characteristics
The study population of 205 KMT2Ar adult AML patients included 54% females and median age at diagnosis was 48 years (range 18–86), while 75% of the patients were younger than 60 years. Most of the patients (73%) had de novo AML and 43% underwent allogeneic hematopoietic stem cell transplantation (allo-HSCT). Median follow-up was 4.9 years for those patients still alive. Patient baseline characteristics are summarized in Table 1 and Supplementary Table S3.
Table 1.
Baseline characteristics of KMT2Ar adult AML patients.
| n = 205 | |
|---|---|
| Female sex | 111 (54.1%) |
| Median age in years (range) | 48.1 (18.6–86.2) |
| Age ≥ 60 years | 51 (24.9%) |
| AML type | |
| De novo AML | 148 (72.2%) |
| Secondary AML | 57 (27.8%) |
| AML after MDS | 14 (6.8%) |
| Therapy-related AML | 43 (21%) |
| Hemoglobin (g/dL) | 9.5 [Q1 = 8.3, Q3 = 11.3] |
| WBC (×109/L) | 23.1 [Q1 = 4.7, Q3 = 58] |
| (>100 × 109/L) | 21 (10.2%) |
| Platelets (×109/L) | 51.5 [Q1 = 29.8, Q3 = 107.5] |
| Bone marrow % of blasts | 83 [Q1 = 70.2, Q3 = 90] |
| Chromosomal aberrations | |
| t(9;11) | 101 (49%) |
| t(11;19) | 33 (16%) |
| t(6;11) | 24 (12%) |
| t(10;11) | 10 (5%) |
| t(11;17) | 10 (5%) |
| Other KMT2Ar | 27 (13%) |
| Additional chromosomal aberrations | 85 (41.6%) |
| Complex aberrations (≥2 apart from KMT2Ar) | 38 (19%) |
| Trisomy 8 | 37 (18%) |
| Derivative 11 | 12 (6%) |
| ELN 2022 | |
| Favorable | – |
| Intermediate | 101 (49.3%) |
| Adverse | 104 (50.7%) |
| Treatment | |
| Intensive | 195 (95%) |
| Non-intensive | 10 (5%) |
| Composite complete response | 120 (63.8%) |
| Early death | |
| 30-day mortality | 28 (13.6%) |
| 60-day mortality | 36 (17.6%) |
| Allogeneic HSCT | 88 (42.9%) |
| In CR1 | 48 (23.4%) |
| In CR2 or later | 15 (7.3%) |
| Unknown response at allo-HSCT | 25 (12.2%) |
| Median survival in years (95% CI) | 1.4 (1.1–1.7) |
AML acute myeloid leukemia, MDS myelodysplastic syndrome, WBC white blood cell, ELN European LeukemiaNet, HSCT hematopoietic stem cell transplantation, CR1 first complete remission, CR2 second complete remission, CI confidence interval.
Frequency of specific 11q23.3/KMT2A rearrangements
Translocation t(9;11) was the most frequent KMT2Ar [n = 101, 49%], followed by t(11;19) [n = 33, 16%] and t(6;11) [n = 24, 12%]. Both t(10;11) and t(11;17) accounted for 5% of patients each (n = 10), while the less common translocations grouped together (hereafter “other KMT2Ar”) represented 13% of patients. A comparison of basic characteristics among different translocation partners showed differences in age distribution, with the youngest patients being in t(6;11) [median 41 years] and the oldest in the “other KMT2Ar” subgroup (median 55.9 years) (p = 0.004) (Supplementary Table S4). In addition, the proportion of therapy-related AML was highest in the t(9;11) [30.7%] subgroup, while none was found in the t(6;11) cases (p = 0.013). Allo-HSCT rates did not differ significantly among distinct KMT2Ar categories (p = 0.523).
Additional cytogenetic abnormalities
Additional cytogenetic abnormalities were present in 40% of patients. Complex aberrations (≥2 abnormalities in addition to the KMT2Ar) were the most frequent (19%), followed by trisomy 8 (18%), derivative 11 (6%) and trisomy 21 (5%). Isolated trisomy 8 as the only additional cytogenetic abnormality (apart from KMT2Ar) was found in 19 patients (9%) and was almost exclusively found in the t(9;11) subgroup (16 patients, p < 0.001). On the other hand, complex aberrations were more frequently associated with “other KMT2Ar” (p = 0.001).
Mutational landscape
NGS analysis was performed in 185 (90%) cases using a panel of up to 90 genes (Supplementary Table S2). A total of 263 mutations were found in 44 of the 90 analyzed genes. The median number of gene mutations per patient was 1 (range 0–6).
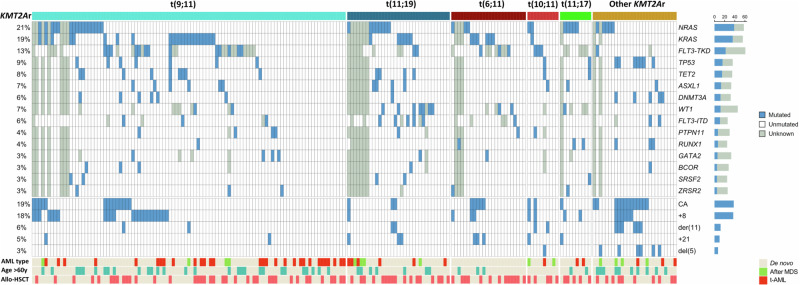
The most frequent gene mutations co-occurring with a KMT2Ar were NRAS (21%), KRAS (19.5%) and FLT3-TKD (13.3%), followed by TP53 (8.6%), TET2 (8.1%), ASXL1 (7%), WT1 (7%), DNTM3A (6.5%) and FLT3-ITD (5.8%). When grouped together, mutations in genes that comprise the RAS signaling pathway (NRAS, KRAS, PTPN11 and BRAF) were present in 42.1% of KMT2Ar AML. Of note, NRAS and KRAS mutations were not mutually exclusive in our cohort, as seven patients harbored mutations in both genes. The distribution of mutations within the different KMT2Ar subgroups is shown in Fig. 1. WT1 mutation was predominant in t(11;19) subgroup (p < 0.001), while TP53 mutation was recurrent in the “other KMT2Ar” subgroup (p = 0.002).
Fig. 1. Mutational landscape and additional cytogenetic aberrations in KMT2Ar adult AML by fusion partner.
Only mutations present in at least five patients and most frequent cytogenetic aberrations are shown. Allo-HSCT allogeneic hematopoietic stem cell transplantation; CA complex cytogenetic aberrations (≥2 apart from KMT2Ar); MDS myelodysplastic syndrome; t-AML therapy-related acute myeloid leukemia.
Patient outcomes
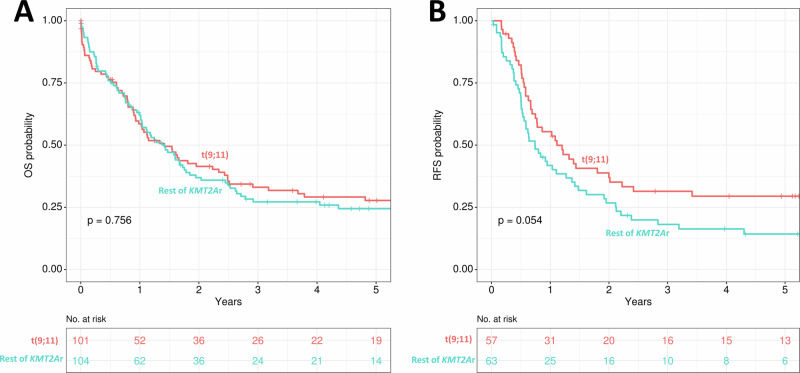
Median OS was 1.4 years for the whole cohort. Notably, OS was similar across the different KMT2Ar subgroups and we were not able to find any differences in OS between t(9;11) and the remainder of the KMT2Ar patients (p = 0.756, Fig. 2A). However, in t(9;11) patients there was a trend for better RFS (p = 0.054, Fig. 2B).
Fig. 2. Comparison of survival outcomes between t(9;11) and the rest of the KMT2Ar cohort.
A Overall survival. B Relapse-free survival.
Patients harboring concomitant cytogenetic aberrations had a higher rate of early deaths (first 60 days 23.2% vs 7.3%, p = 0.002), although those patients were older (median age 52 vs 46 years, p = 0.008) and median overall survival was similar to the rest of the cohort (12 vs 15 months, p = 0.529), as shown in Supplementary Fig. S2A. Complex aberrations (≥2 additional cytogenetic abnormalities) were associated with shorter OS, with median OS of 9 vs 18 months for the rest of the cohort (p = 0.046, Supplementary Fig. S2B). Otherwise, we did not find significant differences in OS between patients with trisomy 8 when compared to the remainder of patients (p = 0.920, Supplementary Fig. S2C). Patients with isolated trisomy 8 as the only additional cytogenetic aberration also presented with similar OS compared to the rest of the cohort (p = 0.402, Supplementary Fig. S2D).
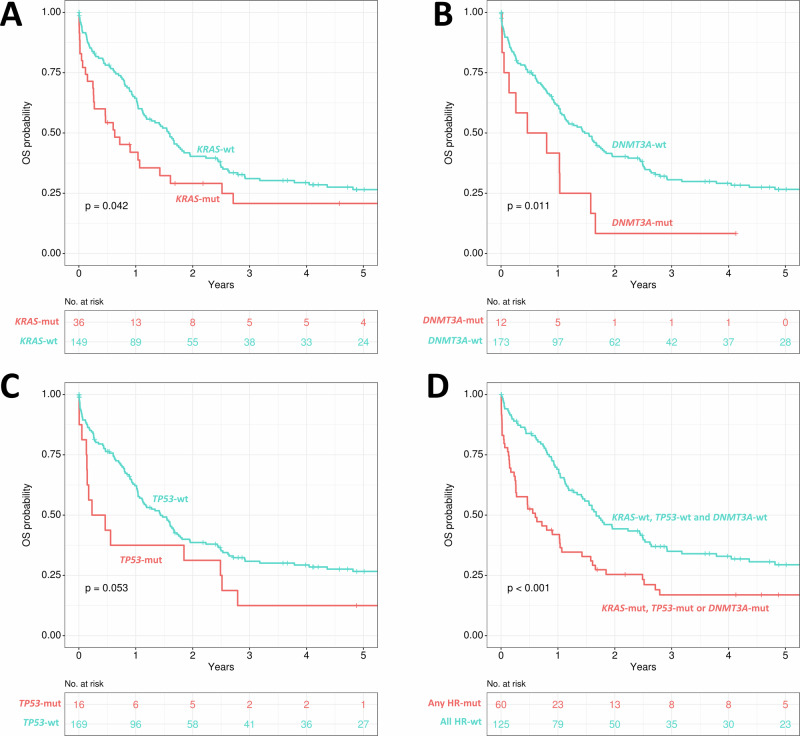
Given that NGS information on mutational landscape was available for the vast majority of the patients of the cohort (90%, n = 185), the impact of gene mutations on OS was further analyzed. KRAS mutations were present in 36 patients and were associated with shorter OS, with a median OS of 8 vs 19 months for KRAS-mut vs KRAS-wt patients (p = 0.042, Fig. 3A). NRAS-mut was the most frequent mutation (39 patients), but it was not associated with shorter OS (p = 0.883, Supplementary Fig. S3A) and only patients presenting both mutations (NRAS and KRAS, 7 patients) had reduced survival when compared to the rest of the cohort (p = 0.041, Supplementary Fig. S3B). Patients with DNMT3A-mut also presented with shorter OS with a median OS of 8 vs 18 months for DNMT3A-mut vs DNMT3A-wt patients (p = 0.011, Fig. 3B), and there was a trend for poor outcome in TP53-mut patients (p = 0.053, Fig. 3C).
Fig. 3. Impact of different gene mutations on overall survival in KMT2Ar AML.
A KRAS, B DNMT3A, C TP53, D comparison of patients with either KRAS-mut, DNMT3A-mut or TP53-mut and the rest of the KMT2Ar AML cohort; OS overall survival; mut mutated; wt wildtype.
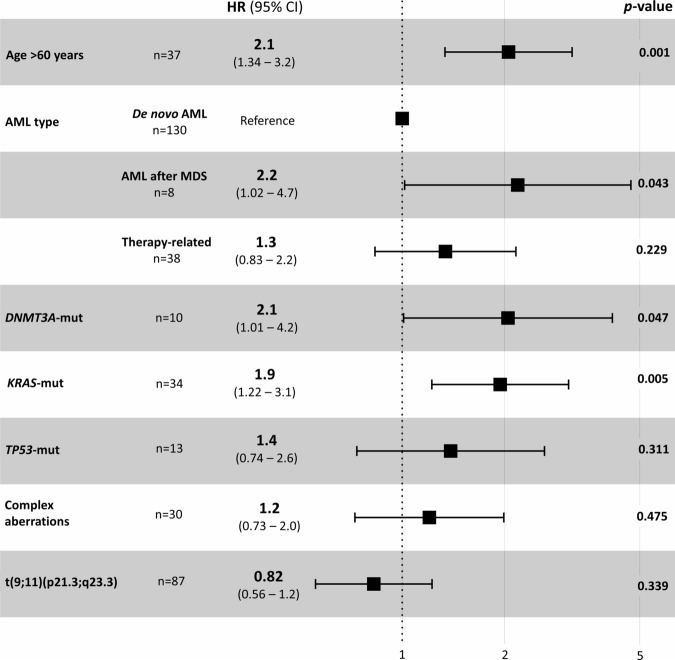
Patients harboring a mutation of any of the above-mentioned high-risk genes (either KRAS, n = 36; DNMT3A, n = 12 or TP53, n = 16) represented 32% of the cohort and had a median OS of 7 months, while the rest of the patients had a median OS of 20 months (p < 0.001, Fig. 3D). A multivariate Cox regression model for intensively treated patients identified the following independent pretreatment variables regarding OS: age > 60 years (HR 2.1, p = 0.001), secondary AML after MDS (HR 2.2, p = 0.043), DNMT3A-mut (HR 2.1, p = 0.047), KRAS-mut (HR 2.0, p = 0.005), as shown in Fig. 4. Of note, therapy-related AML, complex cytogenetic aberrations, t(9;11) or TP53-mut were not significant in this model.
Fig. 4. Multivariate Cox regression model for prediction of overall survival in intensively treated patients.
HR hazard ratio; CI confidence interval; AML acute myeloid leukemia; MDS myelodysplastic syndrome; mut mutated.
Effect of allo-HSCT in KMT2Ar AML
Allo-HSCT was performed in 88 patients (42.9%), at CR1 in 48 (23.4%) and CR2 or later in 15 (7.3%), while the response at allo-HSCT was not known in 25 (12.2%). The estimated survival probabilities using Simon–Makuch method with clock-back correction revealed a significant OS benefit of allo-HSCT in CR1 (p = 0.003, Supplementary Fig. S4A). Moreover, patients with KRAS-mut, TP53-mut or DNMT3A-mut AML also benefited from allo-HSCT in CR1 (p = 0.026, Supplementary Fig. S4B). Remarkably, from all patients who underwent allo-HSCT in CR1, OS for KRAS-mut, TP53-mut or DNMT3A-mut patients was similar to those patients lacking these mutations (p = 0.530, Supplementary Fig. S4C), as well as RFS (p = 0.884, Supplementary Fig. S4D). Allo-HSCT in CR1 was an independent prognostic variable for increased OS in a multivariate Cox regression model that included the same variables stated previously (p < 0.001, Supplementary Fig. S5).
Outcome in intensively treated younger (<60 years) KMT2Ar de novo AML patients
As the general cohort of 205 patients was heterogeneous in terms of age, origin of AML and treatment, an analysis in a subgroup of 115 patients aged <60 years at diagnosis, who received intensive chemotherapy regimens and who presented with de novo KMT2Ar AML (excluding both therapy-related and AML after MDS) was carried out. Median age of this subgroup was 42 years (range 19–59) and it included 51% females. Median follow-up for those patients alive was 5.1 year, CRc rate was 75.7%, allo-HSCT was performed in 53.9% and median OS was 1.7 years.
Notably, OS was still similar across the different translocation partners, without significant differences in OS and similar RFS between t(9;11) and the remainder of the patients (p = 0.916, Supplementary Fig. S6A, and p = 0.193, Supplementary Fig. S6B, respectively). Interestingly, there was a trend for longer OS for patients who harbored isolated trisomy 8 (n = 10) as the only additional cytogenetic abnormality (p = 0.072, Supplementary Fig. S7A). In fact, 80% of the patients achieving a CRc had long-lasting responses with a 2-year RFS of 75% vs 31% for the rest of cases (p = 0.016, Supplementary Fig. S7B).
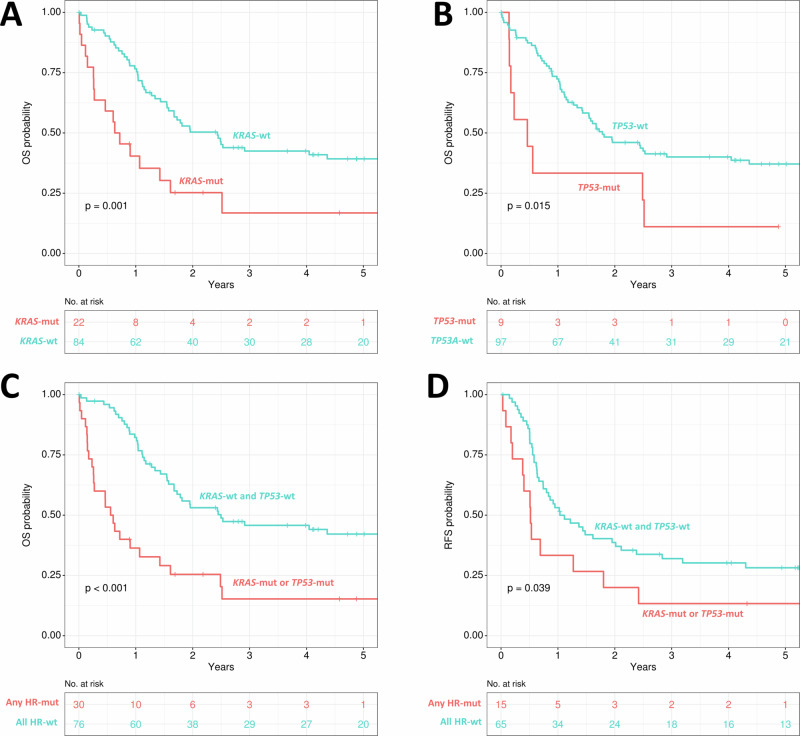
Regarding gene mutations, patients with KRAS-mut (n = 22) had a significantly shorter OS (p = 0.001, Fig. 5A) when compared to KRAS-wt (n = 84). Patients showing TP53-mut (n = 9) were also associated with shorter OS (p = 0.015, Fig. 5B), while DNMT3A-mut was not significant (p = 0.242), although there was also a low number of patients who presented with DNMT3A-mut (n = 5) in this subset of younger AML patients (<60 years of age).
Fig. 5. Impact of different gene mutations on overall survival in patients with de novo KMT2Ar AML and aged ≤60 years.
A KRAS, B TP53, C comparison of overall survival of patients with any high-risk genes mutated (either KRAS-mut, or TP53-mut) and the rest of patients aged ≤60 years with de novo AML, D comparison of relapse-free survival of patients with any high-risk mutated genes (either KRAS-mut or TP53-mut) and the rest of patients aged ≤60 years with de novo AML. OS overall survival; RFS relapse-free survival; mut mutated; wt wildtype; HR high-risk genes (KRAS, TP53).
Therefore, KRAS and TP53 were the most relevant genes linked to OS in this subgroup of patients. Grouped together (either KRAS-mut or TP53-mut) these mutations defined 28% (n = 30) of the cohort to have an adverse prognosis. These patients showed a significantly lower rate of CRc (50% vs 85.5%, p < 0.001). Median OS was 7 months and 5-year OS only 15%, while for the rest of patients median OS was 30 months and 5-year OS 42% (p < 0.001, Fig. 5C). Moreover, median RFS was 6 months for patients with KRAS-mut or TP53-mut, while median RFS was 13 months in all other younger KMT2Ar cases (p = 0.039, Fig. 5D).
A multivariate Cox regression in this subset of patients identified the following independent variables regarding OS: KRAS-mut (HR 2.6, p = 0.002), complex cytogenetic aberrations (HR 2.3, p = 0.019), and allo-HSCT in CR1 (HR 0.7, p = 0.003), while there was a trend for TP53-mut (HR 2.2, p = 0.077), as shown in Supplementary Fig. S8.
Historical comparison of outcomes in KMT2Ar AML
As the cohort was accrued over several decades, the impact of the year of AML diagnosis on OS was also analyzed. As we have reported improved patient outcomes for patients treated in the decade between 2007 and 2016 when compared to the decade between 1997 and 2006 in the HARMONY AML database [32] the same cut-off was used for this analysis. However, we could not find differences in OS when stratifying by the year of diagnosis (p = 0.458, Supplementary Fig. S9A). In the subset of de novo AML patients aged <60 years, early-death rates were similar with 10% for the 1996–2006 period and 6.2% for the 2007–2020 period (p = 0.508) and again without statistical differences in OS (median OS 19 vs 24 months respectively, p = 0.396, Supplementary Fig. S9B).
Discussion
KMT2Ar are recognized as recurrent genetic abnormalities in AML, although they are present only in a minority of adult AML patients [4, 5, 23]. In the HARMONY AML database, we found an incidence of 3.4% (205 out of 6005) for KMT2Ar AML, which is in line with previous studies [6, 7, 11, 15, 19]. This highlights the importance of large databases in order to obtain a representative cohort of KMT2Ar adult AML. However, based on conventional diagnostics the incidence of KMT2Ar AML might be slightly underestimated, as the development of molecular techniques has allowed the identification of cryptic KMT2A fusions, and thus the true prevalence might be higher [12]. While the incidence of KMT2Ar AML was recently shown to be significantly higher in patients <60 years, the median age of 48 years in our cohort was well within the range of previous reports [6, 7, 14, 15, 17, 19].
The prognostic impact of different translocation partners in KMT2Ar AML has been studied for several decades, with quite heterogeneous results [6, 7, 14–19]. Recently, Bill et al. [14] observed that the prognostic significance of t(9;11) was dependent on patient age, as it was associated with better outcomes only for patients aged <60 years with de novo AML. However, we could not corroborate that finding when we analyzed the same subset of patients. Interestingly, Bill et al. reported an uneven distribution of KRAS mutations among the different KMT2Ar, as 47% of their t(6;11) patients presented with KRAS-mut while only 3% of the t(9;11) had KRAS-mut. By contrast, in our cohort, KRAS-mut distribution was similar among the different translocations, with a prevalence of 19.5% for the global cohort and 24.2% for t(9;11) patients. Given the negative impact of KRAS-mut, this could explain the more favorable outcome of the t(9;11) cases reported in the Bill et al. study. In line with our observations, Grossman et al. [17] report a prevalence of 22.9% KRAS-mut in t(9;11) patients and they were also not able to find differences in OS for the distinct fusion partners. Therefore, the heterogeneous results reported in the literature with regard to a more favorable outcome of t(9;11) might be due to a potential bias in the distribution of KRAS mutations (rather than the fusion partner per se), although further studies will be required in order to confirm our hypothesis.
As genomic features have been described as the most powerful predictors of OS in adult AML and the interaction of genomic aberrations has been linked to outcome [3], it is reasonable to think that additional cytogenetic aberrations or concomitant gene mutations might also play a role in KMT2Ar AML. In our study, 40% of KMT2Ar patients presented with additional cytogenetic abnormalities resulting in higher early-death rates, although poorer long-term outcome was only associated with cases with more complex genomic alterations. While both observations need to be confirmed in independent cohort, they definitely point to additional heterogeneity within KMT2Ar-driven leukemia biology that needs to be further understood.
In accordance, comprehensive molecular analysis of the HARMONY KMT2Ar adult AML cohort, which to the best of our knowledge is the largest cohort studied by panel sequencing to date, revealed additional heterogeneity on the molecular level. In line with previous studies [10, 14, 17, 19], we observed a relatively low incidence of gene mutations, with a median of only one mutation per patient. This is in contrast to the average amount of additional driver mutations seen in other AML subtypes [3], most likely due to the fact that KMT2A is one of the strongest oncogenic drivers and is sufficient to transform healthy hematopoietic stem cells in murine models as a single molecular lesion [33].
With regard to additional oncogenic drivers, NRAS was the most frequently mutated gene (21% of the patients). Overall, we found a high incidence (42.1%) of gene mutations involving the RAS signaling pathway (NRAS, KRAS, PTPN11 and BRAF), which is consistent with previous reports [14, 17, 34]. Furthermore, we found constitutive activation of FLT3 by mutations in 19.1% of cases, with TKD mutations being more frequent than ITD aberrations. This is in line with a previous report in childhood KMT2Ar leukemia that also reported more TKD than ITD mutations [35]. Of note, NPM1 mutations were detected only in 1% of the cohort. While this constitutes a different mutational landscape in KMT2Ar adult AML when compared to that described for the rest of adult AML [1–3], additional drivers do contribute to KMT2Ar leukemia pathogenesis and their impact has to be further unraveled.
With regard to outcome, we were able to demonstrate that adult patients with KMT2Ar AML have shorter OS when they present with a KRAS mutation. KRAS mutations have recently been described as an independent adverse prognostic factor in pediatric KMT2Ar AML by the Japanese Pediatric Leukemia/Lymphoma Study Group, which was validated using a smaller cohort of adult KMT2Ar patients [36]. However, several studies of KMT2Ar adult AML were not able to find differences in OS for KRAS-mut patients, which is most likely due to the relatively small sample size and a potential bias in the distribution of KRAS-mut [14, 17, 19]. While TP53-mut have been described as an adverse prognostic marker in a univariate OS analysis in one previous study [17], we also observed an unfavorable impact of this tumor suppressor gene mutation.
The importance of gene mutations was confirmed in a subset of patients aged <60 years at diagnosis receiving intensive chemotherapy regimens, with KRAS and TP53 being the most relevant genes for patient outcomes. Only 50% of the patients with either KRAS-mut or TP53-mut achieved CRc, and for those who did, median RFS was only 6 months, which resulted in a significantly shorter OS when compared to the rest of the patients of the subset. Previous studies have shown that KRAS mutations are likely subclonal and therefore relatively late events in KMT2Ar AML [9, 14, 17, 34]. However, they might promote disease progression and clonal expansion of KMT2Ar cells, as reported in a recent study using a retroviral mouse AML model [37], which could explain the low rate of treatment responses and early relapses that we observed in our patients.
On the other hand, allo-HSCT in CR might help overcome the poor prognostic impact of these additional driver mutations. The effect of allo-HSCT for KMT2Ar AML patients achieving CR has been explored in several studies, favoring the performance of allo-HSCT whenever feasible to improve patient outcomes [19, 20, 22]. We were able to confirm these findings in our cohort, showing the benefit of allo-HSCT in CR1. In fact, our results suggest that allo-HSCT could mitigate the deleterious effect of KRAS-mut, TP53-mut and DNMT3A-mut, as mutated patients that underwent allo-HSCT had similar OS and RFS when compared to unmutated patients.
However, the prognosis of adult KMT2Ar AML still remains poor, especially in patients who are not able to receive an allo-HSCT. While the development of menin inhibitors shows encouraging results in early-phase clinical trials and could change the treatment landscape of KMT2Ar AML in the future [38, 39], KRAS inhibitors that have been recently approved for advanced solid tumors [40] might also open novel possibilities of targeted AML therapy and might change the prognostic impact of the markers presented in this study.
In conclusion, our study reveals a different mutational landscape of KMT2Ar adult AML, which has important prognostic and therapeutic implications. Future studies will have to continue to better understand the disease biology underlying KMT2Ar leukemia, especially to better guide not only conventional but also novel targeted treatment approaches and further improve patient outcome.
Supplementary information
Acknowledgements
HARMONY and HARMONY PLUS are funded through the Innovative Medicines Initiative (IMI), Europe’s largest public-private initiative aiming to speed up the development of better and safer medicines for patients. Funding is received from the IMI 2 Joint Undertaking and is listed under grant agreement for HARMONY No. 116026 and grant agreement for HARMONY PLUS No. 945406. This Joint Undertaking receives support from the European Union’s Horizon 2020 Research and Innovation Program and the European Federation of Pharmaceutical Industries and Associations (EFPIA). This study presents results from the AML workgroup in HARMONY. AHS is supported by Contrato Río Hortega CM23/00101 (ISCIII). ATT received additional support by the Deutsche Forschungsgemeinschaft (DFG) grant FU 356/12-1 (ATT).
Author contributions
AHS, TG, JMHR, BH, GO, HD and LB designed the study. AHS, MS, TG, MA, PJMV, AVR, KHM, RA, JMT, JML, MP, JME, KIM, CT, GS, KD, MH, TH, ATT, DR and JMHR performed data collection and assembly of the data. AHS, TG, RS, GC, AVR, AB, JME, KD, ATT, JMHR, BH, GO, HD and LB performed data analysis and interpretation. AHS, JMHR and LB wrote the manuscript. BH, GO and HD critically reviewed the manuscript. LB supervised research and coordinated the HARMONY AML group. All authors had access to primary data, read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
After the publication of this article, data collected for this analysis and related documents will be made available to others upon reasonably justified request, which needs to be written and addressed to the attention of the corresponding author LB at the following e-mail address: lars.bullinger@charite.de. The HARMONY Alliance, via the corresponding author LB, is responsible to evaluate and eventually accept or refuse every request to disclose data and their related documents, in compliance with the ethical approval conditions, in compliance with applicable laws and regulations, and in conformance with the agreements in place with the involved subjects, the participating institutions, and all the other parties directly or indirectly involved in the participation, conduct, development, management and evaluation of this analysis.
Competing interests
MS: honoraria from Novartis, Celgene, AOP Orphan, AbbVie. KHM: honoraria from AbbVie, Bristol Myers Squibb, Celgene, Janssen, Novartis, Pfizer, Otsuka; research funding from AbbVie. RA: honoraria from Astellas, Bristol Myers Squibb, Incyte, and Novartis. MP: honoraria from Novartis. CT: co-owner and CEO of AgenDix GmbH and has received lecture fees and/or participated in Ad-Boards from Novartis, Jazz Pharmaceuticals, Astellas, Janssen, Illumina; research funding from Novartis, Bayer. GS: honoraria from Takeda and has participated in Ad-Boards from Novartis, Celgene, Abbvie, Helsinn and Takeda. KD: honoraria from Novartis, Jazz Pharmaceuticals, Abbvie; has participated in Ad-Boards from Novartis, Bristol Myers Squibb, Jazz Pharmaceuticals, Abbvie; research funding from Novartis, Astellas, Agios, Bristol Myers Squibb, Kronos. MH: honoraria from Takeda, Novartis, Janssen, Jazz Pharmaceuticals, Eurocept, Abbvie and has participated in Ad-Boards from Kura Oncology, Glycostem, Daiichi Sankyo, Bristol Myers Squibb, Novartis, Jazz Pharmaceuticals, Abbvie, Pfizer, PinotBio, Roche, Tolremo; research funding from Glycostem, Daiichi Sankyo, Bristol Myers Squibb, Novartis, Jazz Pharmaceuticals, Abbvie, Pfizer, PinotBio, Roche, Astellas, Bayer, BergenBio, Loxo Oncology. TH: current employment at Munich Leukemia Laboratory, with part ownership. ATT: Consultancy for CSL Behring, Maat Pharma, Biomarin and Onkowissen; travel reimbursements from Neovii Biotech. DR: has participated in Ad-Boards from Bristol Myers Squibb, Novartis, Cerus, Medac, EUSA Pharma, Bluebird Bio; research funding from Novartis, BlueBird Bio. RSR: current employment at Bayer Pharma AG. MB: current employment at Abbvie. JMHR: honoraria from Bristol Myers Squibb, Pfizer, Amgen, Celgene, GSK, Novartis; advisory role for Bristol Myers Squibb, Pfizer, Amgen, Celgene, Novartis, Janssen, Roche, Abbvie, AstraZeneca, Beigene, Lilly, Gilead, Takeda, Jazz Pharmaceuticals, Rovi, Incyte; research funding from Bristol Myers Squibb, Celgene, Novartis. BH: honoraria from Pfizer, Bristol Myers Squibb, Novartis; research funding from AstraZeneca. GO: honoraria from Abbvie, Jazz Pharmaceuticals, Astellas, Gilead, Bristol Myers Squibb, Servier, Roche. HD: advisory role for AbbVie, Agios, Amgen, Astellas, AstraZeneca, Berlin-Chemie, Bristol Myers Squibb, Celgene, GEMoaB, Gilead Sciences, Janssen, Jazz Pharmaceuticals, Novartis, Syndax; research funding from AbbVie, Agios, Amgen, Astellas, Bristol Myers Squibb, Jazz Pharmaceuticals, Kronos-Bio, Novartis. LB: honoraria from AbbVie, Amgen, Astellas, Bristol Myers Squibb, Celgene, Daiichi Sankyo, Gilead, Hexal, Janssen, Jazz Pharmaceuticals, Menarini, Novartis, Pfizer, Roche and Sanofi; research funding from Bayer, Jazz Pharmaceuticals.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02333-4.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52. [DOI] [PubMed]
- 2.Bullinger L, Döhner K, Dohner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35:934–46. 10.1200/JCO.2016.71.2208 [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77. 10.1182/blood.2022016867 [DOI] [PubMed] [Google Scholar]
- 5.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization Classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19. 10.1038/s41375-022-01613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–402. 10.1182/blood-2003-02-0434 [DOI] [PubMed] [Google Scholar]
- 7.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. 10.1182/blood-2009-11-254441 [DOI] [PubMed] [Google Scholar]
- 8.Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–98. 10.1182/blood-2016-01-693879 [DOI] [PubMed] [Google Scholar]
- 10.Eisfeld AK, Mrózek K, Kohlschmidt J, Nicolet D, Orwick S, Walker CJ, et al. The mutational oncoprint of recurrent cytogenetic abnormalities in adult patients with de novo acute myeloid leukemia. Leukemia. 2017;31:2211–8. 10.1038/leu.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vetro C, Haferlach T, Meggendorfer M, Stengel A, Jeromin S, Kern W, et al. Cytogenetic and molecular genetic characterization of KMT2A-PTD positive acute myeloid leukemia in comparison to KMT2A-rearranged acute myeloid leukemia. Cancer Genet. 2020;240:15–22. 10.1016/j.cancergen.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 12.Meyer C, Larghero P, Almeida Lopes B, Burmeister T, Gröger D, Sutton R, et al. The KMT2A recombinome of acute leukemias in 2023. Leukemia. 2023;37:988–1005. 10.1038/s41375-023-01877-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatnagar B, Blachly JS, Kohlschmidt J, Eisfeld AK, Volinia S, Nicolet D, et al. Clinical features and gene-and microRNA-expression patterns in adult acute leukemia patients with t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3). Leukemia. 2016;30:1586–9. 10.1038/leu.2015.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bill M, Mrózek K, Kohlschmidt J, Eisfeld AK, Walker CJ, Nicolet D, et al. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proc Natl Acad Sci USA. 2020;117:26340–6. 10.1073/pnas.2014732117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mrózek K, Heinonen K, Lawrence D, Carroll AJ, Koduru PRK, Rao KW, et al. Adult patients with de novo acute myeloid leukemia and t(9; 11)(p22; q23) have a superior outcome to patients with other translocations involving band 11q23: a Cancer and Leukemia Group B Study. Blood. 1997;90:4532–8. [PubMed]
- 16.Krauter J, Wagner K, Schäfer I, Marschalek R, Meyer C, Heil G, et al. Prognostic factors in adult patients up to 60 years old with acute myeloid leukemia and translocations of chromosome band 11q23: individual patient data-based meta-analysis of the German acute myeloid leukemia intergroup. J Clin Oncol. 2009;27:3000–6. 10.1200/JCO.2008.16.7981 [DOI] [PubMed] [Google Scholar]
- 17.Grossmann V, Schnittger S, Poetzinger F, Kohlmann A, Stiel A, Eder C, et al. High incidence of RAS signalling pathway mutations in MLL-rearranged acute myeloid leukemia. Leukemia. 2013;27:1933–6. 10.1038/leu.2013.90 [DOI] [PubMed] [Google Scholar]
- 18.Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114:2489–96. 10.1182/blood-2009-04-215152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Issa GC, Zarka J, Sasaki K, Qiao W, Pak D, Ning J, et al. Predictors of outcomes in adults with acute myeloid leukemia and KMT2A rearrangements. Blood Cancer J. 2021;11:1–10. 10.1038/s41408-021-00557-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pigneux A, Labopin M, Maertens J, Cordonnier C, Volin L, Socié G, et al. Outcome of allogeneic hematopoietic stem-cell transplantation for adult patients with AML and 11q23/MLL rearrangement (MLL-r AML). Leukemia. 2015;29:2375–81. 10.1038/leu.2015.143 [DOI] [PubMed] [Google Scholar]
- 21.Menghrajani K, Gomez-arteaga A, Madero-Marroquin R, Zhang M, Bo-subait K, Sanchez J, et al. Risk classification at diagnosis predicts post-HCT outcomes in intermediate-, adverse-risk, and KMT2A-rearranged AML. Blood Adv. 2022;6:828–47. [DOI] [PMC free article] [PubMed]
- 22.Chen Y, Kantarjian H, Pierce S, Faderl S, O’Brien S, Qiao W, et al. Prognostic significance of 11q23 aberrations in adult acute myeloid leukemia and the role of allogeneic stem cell transplantation. Leukemia. 2013;27:836–42. 10.1038/leu.2012.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–28. 10.1182/blood.2022015850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, al Hinai A, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378:1189–99. 10.1056/NEJMoa1716863 [DOI] [PubMed] [Google Scholar]
- 25.Terwijn M, Kelder A, Huijgens PC, Dräger AM, Oussoren YJM, Scholten WJ, et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: Data from the HOVON/SAKK AML 42A study. J Clin Oncol. 2013;31:3889–97. 10.1200/JCO.2012.45.9628 [DOI] [PubMed] [Google Scholar]
- 26.Turki AT, Tsachakis-Muck N, Leserer S, Crivello P, Liebregts T, Betke L, et al. Impact of CMV reactivation on relapse of acute myeloid leukemia after HCT is dependent on disease stage and ATG. Blood Adv. 2022;6:28–36. 10.1182/bloodadvances.2021005509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belenkaya R, Gurley MJ, Golozar A, Dymshyts D, Miller RT, Williams AE, et al. Extending the OMOP common data model and standardized vocabularies to support observational cancer research. JCO Clin Cancer Inform. 2021;5:12–20. [DOI] [PMC free article] [PubMed]
- 28.McGowan-Jordan J, Hastings RJ, Moore S, editors. ISCN 2020: an international system for human cytogenomic nomenclature. S. Karger; 2020. [DOI] [PubMed]
- 29.Gabert J, Beillard E, van der Velden VHJ, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of ‘real time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – a Europe Against Cancer Program. Leukemia. 2003;17:2318–57. 10.1038/sj.leu.2403135 [DOI] [PubMed] [Google Scholar]
- 30.Bernasconi DP, Rebora P, Iacobelli S, Valsecchi MG, Antolini L. Survival probabilities with time-dependent treatment indicator: quantities and non-parametric estimators. Stat Med. 2016;35:1032–48. 10.1002/sim.6765 [DOI] [PubMed] [Google Scholar]
- 31.Döhner K, Thiede C, Jahn N, Panina E, Gambietz A, Larson RA, et al. Impact of NPM1/FLT3-ITD genotypes defined by the 2017 European LeukemiaNet in patients with acute myeloid leukemia. Blood. 2020;135:371–80. 10.1182/blood.2019002697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobas MA, Villaverde Ramiro A, Hernández Sánchez A, Martinez Elicegui J, González T, Azibeiro Melchor R, et al. Long-term follow-up of AML patients treated intensively before the era of targeted agents. A big data analysis from the Harmony Collaboration. Blood. 2022;140:6053–5. 10.1182/blood-2022-165088 [DOI] [Google Scholar]
- 33.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. 10.1038/nrc2253 [DOI] [PubMed] [Google Scholar]
- 34.Lavallée VP, Baccelli I, Krosl J, Wilhelm B, Barabé F, Gendron P, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47:1030–7. 10.1038/ng.3371 [DOI] [PubMed] [Google Scholar]
- 35.Fedders H, Alsadeq A, Schmäh J, Vogiatzi F, Zimmermann M, Möricke A, et al. The role of constitutive activation of FMS-related tyrosine kinase-3 and NRas/KRas mutational status in infants with KMT2A-rearranged acute lymphoblastic leukemia. Haematologica. 2017;102:e438–42. 10.3324/haematol.2017.169870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuo H, Yoshida K, Nakatani K, Harata Y, Higashitani M, Ito Y, et al. Fusion partner-specific mutation profiles and KRAS mutations as adverse prognostic factors in MLL-rearranged AML. Blood Adv. 2020;4:4623–31. 10.1182/bloodadvances.2020002457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyrenius-Wittsten A, Pilheden M, Sturesson H, Hansson J, Walsh MP, Song G, et al. De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat Commun. 2018;9:1770. 10.1038/s41467-018-04180-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Issa GC, Aldoss I, Dipersio J, Cuglievan B, Stone R, Arellano M, et al. The Menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature. 2023;615:920–4. 10.1038/s41586-023-05812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiskus W, Boettcher S, Daver N, Mill CP, Sasaki K, Birdwell CE, et al. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J. 2022;12:1–11. 10.1038/s41408-021-00603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383:1207–17. 10.1056/NEJMoa1917239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
After the publication of this article, data collected for this analysis and related documents will be made available to others upon reasonably justified request, which needs to be written and addressed to the attention of the corresponding author LB at the following e-mail address: lars.bullinger@charite.de. The HARMONY Alliance, via the corresponding author LB, is responsible to evaluate and eventually accept or refuse every request to disclose data and their related documents, in compliance with the ethical approval conditions, in compliance with applicable laws and regulations, and in conformance with the agreements in place with the involved subjects, the participating institutions, and all the other parties directly or indirectly involved in the participation, conduct, development, management and evaluation of this analysis.