Abstract
The Pinna nobilis, a Mediterranean mollusc, has suffered population declines due to a massive mortality event associated with various factors including the parasite Haplosporidium pinnae. Some populations show resilience, possibly due to local environmental conditions. In this study, a molecular multiplex PCR method was developed using species‐specific primers targeting Internal Transcribed Spacer (ITS) regions of P. nobilis and P. rudis, allowing accurate species identification and hybrid detection. Samples from Mediterranean areas were analysed, including putative hybrids and individuals from five other bivalve species. DNA was isolated, ITS regions were amplified and sequenced, and phylogenetic analyses confirmed species differentiation and primer specificity. The multiplex‐PCR successfully identified P. nobilis, P. rudis, and their hybrids based on distinct amplicon patterns. This study highlights the value of molecular tools in species conservation, especially for monitoring and managing hybridization, supporting effective biodiversity conservation strategies.
Keywords: DNA barcode, endangered species, hybrid detection, molecular tools, multiplex PCR, ribosomal unit
The Pinna nobilis, a Mediterranean mollusc, has suffered population declines due to a mortality event linked to the parasite Haplosporidium pinnae, though some populations show resilience. This study developed a molecular multiplex‐PCR method using species‐specific primers to accurately identify P. nobilis, P. rudis, and their hybrids. The method's effectiveness was confirmed through analysis of samples from various Mediterranean areas, highlighting its value for species conservation and management.
1. INTRODUCTION
In recent years, the Mediterranean endemic bivalve Pinna nobilis, commonly known as the noble pen shell or fan mussel, has undergone a massive mortality event (MME). It has manifested as a rapid and widespread mortality of individuals throughout the Mediterranean Sea, raising concerns about the species' survival (Katsanevakis et al., 2022; Kersting et al., 2019). The exact cause of the MME in the Mediterranean Sea is still debated, and different hypotheses have been proposed to explain its origin. Multifactorial causes involving a combination of factors such as pathogens, parasites or environmental stressors have been also suggested (Box et al., 2020; Carella et al., 2019; Cižmek et al., 2020; Lattos et al., 2020; Scarpa et al., 2020). Nevertheless, one of the main pathogens suspected to be involved in the decline of Pinna nobilis populations is the protozoan parasite, Haplosporidium pinnae (Catanese et al., 2018; Darriba, 2017), which has been found in affected individuals and whose infection has been shown to play a major role in the onset of the mass mortality (Grau et al., 2022; Tiscar et al., 2022). More recently, an RNA virus has been also associated with disease and mortality in this species due to the virus's long‐term weakening effect on animals, increasing susceptibility and rendering them more vulnerable to pathogenic parasite attacks (Carella et al., 2023). Unfortunately, H. pinnae is now spreading even to previously presumed sanctuaries, areas that had remained H. pinnae‐free due to natural barriers such as river mouths, gulfs, and coastal marine lagoons (Donato et al., 2021, 2023; Foulquié et al., 2023; Labidi et al., 2023; Moro‐Martínez et al., 2023; Nebot‐Colomer et al., 2021; Prado et al., 2021). In some instances, summer water restrictions, like those observed in 2023 at the Delta Ebro region (Spain), may have disrupted the natural protective barrier of salinity gradients, potentially leading to an increased rate of parasite infections (CHEbro, 2023). In any case, H. pinnae seems to exhibit high species specificity, infecting solely P. nobilis and not its congeneric species Pinna rudis (Catanese et al., 2018; Vázquez‐Luis et al., 2017). Some populations of P. nobilis have currently remained alive apparently thanks to specific local abiotic conditions such as reduced temperature and extreme salinity (below or above typical Mediterranean Sea salinity), which hinder the spread of the pathogens (Acarlı et al., 2024; Cabanellas‐Reboredo et al., 2019; Çınar et al., 2021; Cortés‐Melendreras et al., 2022; Giménez‐Casalduero et al., 2020; Prado et al., 2021) or result in a greater host resilience to infections (Coupé et al., 2023; Karadurmuş et al., 2024; Papadakis et al., 2023; Salis et al., 2022). Such is the case of P. nobilis populations in the Sea of Marmara, where there is no documentation of pathogen infection, attributed to the low salinity of the region (Acarli et al., 2022; Karadurmuş et al., 2024). These populations can act as source of repopulation through larval dispersal, which could be transported by sea currents over large distances to other regions by sea currents, even to hundreds of kilometres away (Feria‐Rodríguez et al., 2024; Katsanevakis, 2016; Kersting et al., 2020; Kersting & García‐March, 2017; Papadakis et al., 2023; Peyran et al., 2022).
Scientists, environmentalists and government organizations are collaborating to implement a strategy for monitoring and safeguarding the species and to apply conservation and repopulation measures for surviving populations (García‐March et al., 2020). For instance, efforts to monitor reproductive events in the wild for juvenile recruitment have led to the deployment of collectors in various regions of the Mediterranean Sea (Cabanellas‐Reboredo et al., 2009; Kersting et al., 2020; Kersting & García‐March, 2017; Nebot‐Colomer et al., 2024). Through these methods, captured juveniles can also be bred under long‐term captivity conditions (Cortés‐Melendreras et al., 2022; García‐March et al., 2020; Hernandis et al., 2022, 2023; Prado et al., 2020) screened for pathogens, and then transferred to areas for repopulation initiatives of affected populations (Katsanevakis, 2016; Kersting et al., 2020; Kersting & García‐March, 2017).
Identifying juveniles of Pinna spp. based on specific shell features is feasible due to their small size, but distinguishing between different Pinna species can be challenging (Catanese, Tena‐Medialdea, et al., 2022). The congeneric species P. rudis is occasionally found in the Mediterranean, and it can be mainly found in waters from the Caribbean to the eastern Atlantic Ocean (Gvozdenović Nikolić et al., 2019; Huber, 2010). Recently it appears to have shown significant expansion into the Mediterranean Sea, as observed within Greek waters and documented through dedicated surveys and molecular confirmation. This expansion indicates an ecological phenomenon whereby P. rudis appears to be colonizing new habitats within the Mediterranean basin, some of which were previously occupied by P. nobilis (Kersting & Ballesteros, 2021; Oprandi et al., 2024; Zotou et al., 2023). During the diving surveys, some researcher observed individuals exhibiting morphological traits that seemed to be a mixture of the two Pinna species (Rubino et al., 2024; Vázquez‐Luis et al., 2021). Therefore, species sharing the same area may come into contact due to changes in habitat or environmental conditions, creating opportunities for hybridization. In fact, without spatial isolation, closely related species tend to hybridize more often (Abbott et al., 2013). Hybridization involves a complex interplay of ecological consequences. It can contribute to speciation by creating new hybrid taxa, and introgression of specific genetic loci may promote adaptive divergence, thus facilitating speciation (Abbott et al., 2013). Furthermore, hybridization can lead to the erosion of native genotypes and increase the invasive potential of certain species, enabling their successful establishment and expansion into new habitats (Seehausen, 2013). Hybridization can still occur in the absence of reproductive barriers between species, although it can be harmful and lead to non‐viable offspring (Abbott et al., 2013; Sedghifar et al., 2016).
In nature, the occurrence of natural hybridization among molluscs is quite common and it has been already described in many species including oysters (Huvet et al., 2004) and mussels (Beaumont et al., 2004; Rawson et al., 1996; Voroshilova et al., 2010).
Although rapid identification methods between P. nobilis and P. rudis have already been described, they are based on the sequencing or multiplex‐PCR amplification of mitochondrial DNA (mtDNA) fragments, which is normally transmitted through maternal inheritance (Catanese, Coupé, & Bunet, 2022; Catanese, Tena‐Medialdea, et al., 2022; Papadakis et al., 2023). Therefore, to identify juvenile or adult hybrids it is necessary to use additional genetic information present on the chromosomes from both parents. Each individual inherits half of its chromosomes from each parent during the breeding process and as a result, mtDNA versus nDNA alone cannot provide sufficiently detailed information to distinguish hybrids from pure lineage parental species.
Recently, morphological characteristics and molecular analyses were used to identify putative hybrids between congeneric P. nobilis and P. rudis (Vázquez‐Luis et al., 2021). However, employing a molecular method that relies on sequencing three fragments of the 28SrDNA gene to detect particular nucleotide positions, characterized by the presence of two distinct peaks of signal intensity in diagnostic sites, seems comfortless, costly, and susceptible to errors of interpretation.
Within eukaryotes, the Internal Transcribed Spacer (ITS) regions constitute non‐coding stretches situated within the ribosomal DNA repeats amidst the small subunit (18S rDNA) and large subunit (28S rDNA) ribosomal genes. They are subdivided into two primary segments: ITS1 and ITS2 with 5.8S rDNA genes intercalated between them. Both ITSs are used in characterizing eukaryotic organisms at the molecular level to discern species, evaluate biodiversity, and probe evolutionary relationships. Among species, they exhibit divergent lengths, sequence variabilities, and evolutionary dynamics, with applications in multiple domains including ecology, conservation biology, and forensics. The high sequence variation inherent in ITS regions can facilitate the discrimination of closely related taxa, using them as DNA barcodes in molecular biology and taxonomy for marine species identification (Aranishi, 2005; Moniz & Kaczmarska, 2010; Prasad et al., 2009; Schoch et al., 2012; Stern et al., 2012; Wiemers et al., 2009). Moreover, despite the substantial variability in these regions, specific segments flanking or included within them remain conserved across taxa. This conservation allows for the design of universal primers suitable for PCR amplification. The ITS1 region, known for its high variability and preferred use as a DNA barcode in molecular taxonomy, provides enhanced discriminatory power for closely related species, whereas ITS2, although shorter and more conserved than ITS1, features lower sequence variability but retains informative variations crucial for taxonomic and phylogenetic analyses, prompting researchers to often combine ITS1 and ITS2 sequences for comprehensive taxonomic and phylogenetic investigations. The rapid evolutionary rate of the ITS region significantly improves the accurate differentiation of closely related species, serving as an effective tool for plant species identification and as a supplementary marker alongside barcode for animal identification (Yao et al., 2010).
In this work, we developed a molecular multiplex‐PCR procedure using species‐specific primers targeting the ITS regions of P. nobilis and P. rudis. This approach enables the identification of these species based on the positive amplification of PCR fragments with distinct sizes. Additionally, our method allows for the detection of hybrids between these two species by observing positive amplification of two different PCR fragments.
2. MATERIALS AND METHODS
2.1. Sampling
A total of 41 biopsies (25 of P. nobilis and 16 of P. rudis) were collected between 2021 and 2023 in different areas of the Mediterranean Sea, the ratio P. nobilis/P. rudis in each location was as follows: Delta Ebro (5/0), Calpe (5/4), Balearic Islands (4/1) [Spain]; Messina (5/3) [Italy]; Crete (0/8) and Lesvos Island (6/0) [Greece]. Three putative hybrids were collected from the Balearic Islands (Spain). The biopsies consisted of a portion of the mantle of each individual that was excised and kept in absolute ethanol. All analysed Pinna individuals were adults. In addition, we also collected two samples of each species of the following molluscs: Pinctada sp., Spondylus gaederopus, Malleus sp., and Isognomon sp. from Messina (Italy) and Atrina pectinata from Calpe (Spain). The identification of all these species was mainly based on morphological features. Samplings were performed with the necessary permits of the competent authorities (national and/or local).
2.2. DNA isolation, amplification and sequencing
Total genomic DNA was isolated from 30 mg of tissue using the Tissue Genomic DNA Extraction Kit (Macherey‐Nagel), according to the manufacturer's instructions.
For the amplification of the ITS regions, primer pairs Pinna_ITSF (5’‐CCGTTGAACCTCCTTCGTGCT‐3′) and Pinna_ITSR (5’‐GGCTCTTCCCGCTTCACTC‐3′) were designed based on the partial sequences of 18S rDNA and 28S rDNA of Pinna available on GenBank. These primers were used in all the bivalve considered in this study, except for PCR amplification of Atrina species for which the primers ITS5 and ITS4, designed by White et al. (1990), were applied.
All PCR reactions were performed in a total volume of 20 μL containing: 10 μL of Kapa Taq Ready mix (Sigma‐Aldrich), 8.2 μL of sterile water, 0.4 μL of each primer (stock 20 Mmol) and 1 μL of DNA at 50 ng/μL. The following conditions of PCR were used: an initial denaturation step at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 50–60°C for 30 s and elongation at 72°C for 5 min. The PCR products were purified using a mi‐Gel Extraction Kit (Metabion) and bidirectionally sequenced using the Sanger's method at Secugen service (www.secugen.es; 15.02.2024). Species identification of Pinna sp. individuals based on mitochondrial DNA pattern was also carried out using the method described by Catanese, Tena‐Medialdea, et al. (2022).
2.3. Sequence comparison, genetic distance and phylogenetic analyses
Although, morphologically, the adult individuals of different bivalve families are possibly recognizable, we analysed the sequences of other species to be sure not to design primers that could give cross‐amplification. Therefore, to develop a multiplex PCR assay specific for P. nobilis and P. rudis identification a profound analysis of the nucleotide differences between the sequences of the considered species was carried out. The nucleotide sequences of ITS regions were edited and aligned using BioEdit (Hall, 1999) and MEGA 11 software (Tamura et al., 2021). The base composition, nucleotide differences and pairwise genetic distances were obtained using MEGA11.
The sequences were then analysed with JModelTest v2.1.7 (Posada, 2008) using the Akaike Information Criterion (AIC; Posada & Buckley, 2004) to select the appropriate model of evolution, as a criterion to determine the best model. A neighbour‐joining (NJ), from Tamura‐Nei genetic distances and a Maximum likelihood (ML) method was adopted to reconstruct the phylogenetic relationships using MEGA11, with 1000 bootstrap replicates. In addition to the obtained sequences for Pinna spp. and other molluscs, the following bivalve species ITS sequences described by other authors and retrieved from GenBank were used for sequence comparison: P. nobilis (KX101235), Atrina pectinata (NC020028), Spondylus gaederopus (KX257353) Mytilus edulis (AY695798), M. galloprovincialis (JX081670), and different species of the Haliotis family (from AF296851 to AF296869). The sequence of Ruditapes decussatus (HQ634139) was also included in the analysis as an outgroup.
2.4. Development of multiplex PCR assay for species identification and hybrid detection
For the development of the assay procedure for detection of hybrid between P. nobilis and P. rudis, amplicons with different size were considered in the primer design.
To lead it, we have designed species‐specific primers for each of the two species of the genus Pinna (Figure 1a,b). The specific primer pair for P. nobilis PCR amplification was PN_267 F (5′‐ACGCACGGAAAAAAAACGACAAAAAGTC‐3′), PN_267 R (5′‐TTGAACCTCGGCCCCACCC‐3′) while for P. rudis PCR amplification was PR_102 F (5′‐CCCCGTAGATTCGTTCAACACCAC‐3′), PN_102 R (5′‐CACCGACAACAACCCCCCA‐3′).
FIGURE 1.
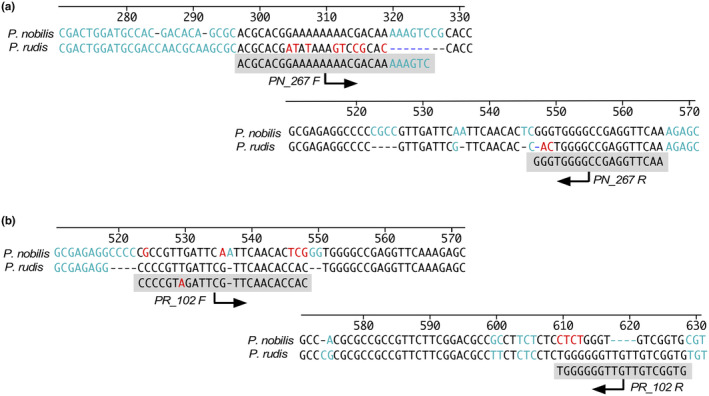
Primer positions in the ITS region for specific PCR amplifications: (a) Forward and reverse primers designed for the identification of P. nobilis; (b) Forward and reverse primers designed for the identification of P. rudis.
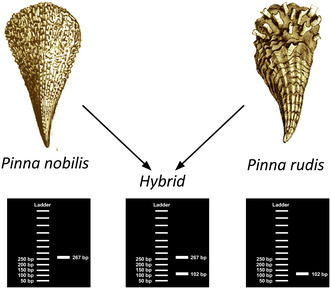
These primers have been designed in such a way that, after multiplex PCR amplification and subsequent visualization of the amplified products on an agarose gel, the different species can be differentiated based on the size of the specific amplified DNA fragments. In presence of a hybrid individual, two amplicons, one for each parental species, should be observed (Figure 2).
FIGURE 2.
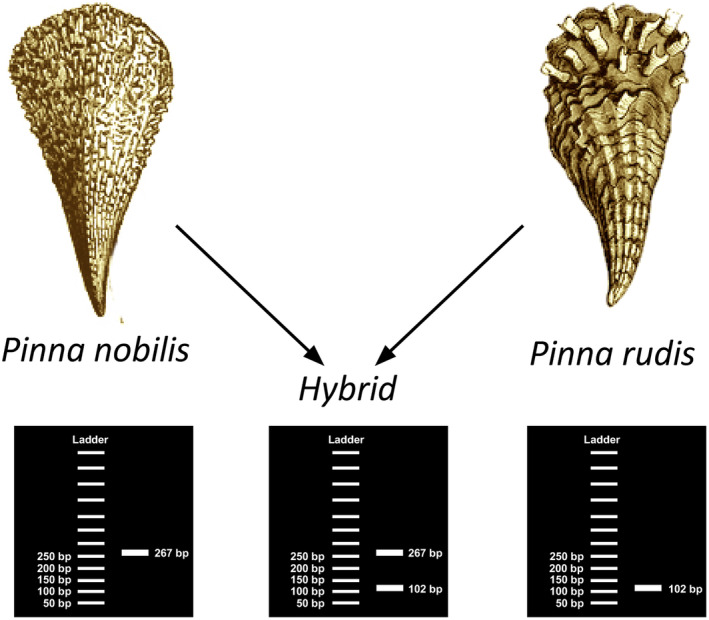
Overview of the developed PCR multiplex method for specific amplification and identification. This method targets P. nobilis, P. rudis, and their hybrids.
The multiplex PCR reaction using DNA was carried out in a total volume of 20 μL (microlitres) containing: 10 μL (1.25 units) of KAPATaq DNA polymerase (Merck), 0.3 μL of each primer PN_267F and PN_267R (20 μM) (specific for P. nobilis) and 0.4 μL of each primer PR_102F and PR_102R (20 μM) (specific for P. rudis), 1 μL of template DNA from the sample (∼30–50 ng) and sterile distilled water until the total volume of 20 μL.
The PCR amplifications were performed with KapaTaq DNA polymerase under the following conditions: an initial denaturation step at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 62°C for 10 s and elongation at 72°C for 20 s.
The PCR products were loaded onto high‐resolution agarose gels containing Ultrapure Agarose (Invitrogen) with a minimum concentration of 2.5% and subsequently stained using GelRed (Biotium). Electrophoresis was conducted in 1x TBE buffer, and DNA fragments were visualized under ultraviolet light. The size of DNA fragments was determined using the standard 50 bp DNA Ladder (Norgen). This method facilitates the direct identification of each Pinna species through the observation of distinct PCR band sizes in agarose gel, eliminating the need for additional sequencing.
Then, we analysed samples collected from juveniles captured in collectors and samples of adults exhibiting morphological characteristics of both species. This analysis was conducted using the described specified method, in addition to the method reliant on species identification through mitochondrial DNA analysis (Catanese, Coupé, & Bunet, 2022; Catanese, Tena‐Medialdea, et al., 2022).
3. RESULTS
The different sequences of ITS regions were sent to GenBank and used for sequence alignment and phylogenetic tree. We characterized the ITS region of following bivalve species: P. nobilis (access. n. LC794141), P. rudis (LC794142); Pinctada sp (LC814847‐LC814850); Isognomon sp. (LC814845); Malleus sp. (LC814846) and we confirmed the already described sequences of Spondylus gaederopus (LC814851‐LC814852) and Atrina pectinata (LC814844).
The obtained sequences containing ITS regions varied in length from 1046 to 1377 bp approximately. The multiple sequence alignment revealed the presence of a more conserved region identified as 5.8S rRNA within the sequences from the studied species.
Considering only the comparison between the sequences obtained from P. nobilis and P. rudis, the alignment between them highlighted 107 polymorphic sites, 67 of which were indels and 40 nucleotide substitutions. No differences were observed among specimens of Pinna species from different geographic sampled areas sampled in the Mediterranean Sea.
The ITS regions of all these organisms proved to have similar base composition to those typical of the vast majority of other marine organisms with 51%–59% GC content (Chow et al., 2009).
3.1. Genetic distance and phylogenetic analysis
The genetic distance among the analysed sequences varied from 0.457 in the comparison between Malleus sp. and Isognomon sp. to 0.847 in the comparison between S. gaederopus and Haliotis iris (Table S1).
For phylogenetic analysis, the Akaike Information Criterion was applied using JModeltest software, indicating the general time reversible (GTR) as the best‐fit model of DNA sequence evolution, which was applied to build the ML tree. A neighbour‐joining, was also inferred using Tamura‐Nei genetic distances from the ITS sequences. The Phylogenetic trees resulting from the NJ and ML analyses were almost congruent with each other (Figure 3).
FIGURE 3.
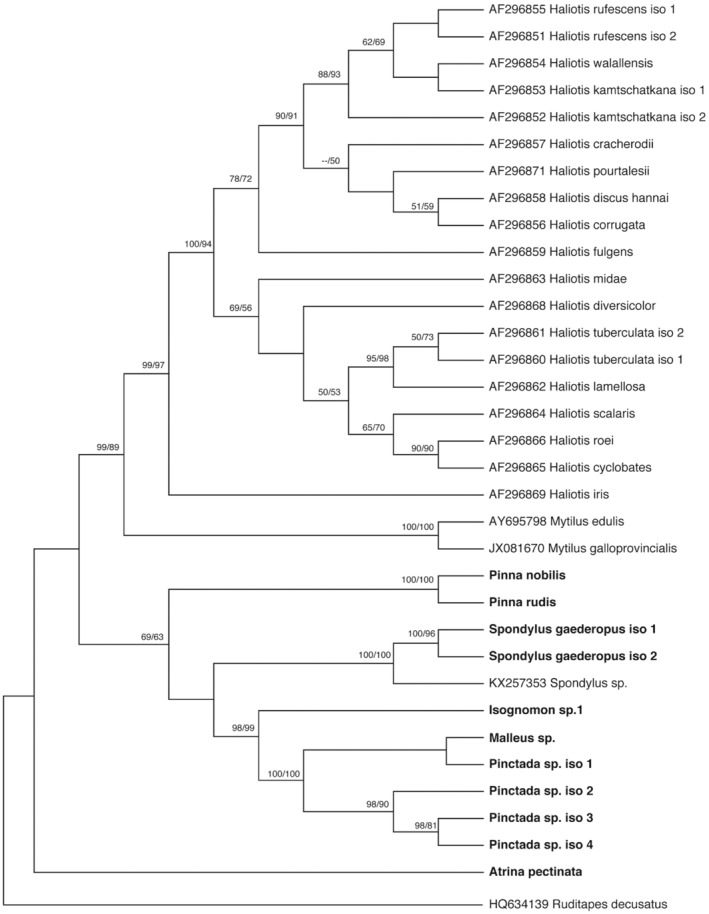
Phylogenetic relationships based on ITS sequences from various bivalve families. Bootstrap values from neighbour‐joining and maximum likelihood analyses greater than 50% are shown below the nodes, respectively.
The sequences of Pinna species were grouped in the same cluster, with high bootstrap (100%) and clearly separated from all other sequences.
3.2. Identification of Pinna sp. and hybrids
Analysing the alignment of ITS sequences between P. nobilis and P. rudis, to search specific primers for identification, the positions 320–326 and 546–549 were chosen as optimal to design the P. nobilis‐specific forward (PN_267 F) and reverse primers (PN_267 R) because the sequence permitted the detection of the nucleotide differences at the critical 3‐end between the two species (Figure 1a). The amplicon expected was 267 bp in length. This P. nobilis‐specific sequence stretch was completely conserved in all the sequences obtained in this work. Moreover, differences were observed in the comparison of the sequence of the primer regions in the other bivalves used in this study.
Similarly, the positions 545–547 and 609–612 of alignment were chosen as optimal for P. rudis‐specific forward (PR_102 F) and reverse primer (PR_102 R) which also showed nucleotide differences at the critical 3‐end (Figure 1b). The expected length of this specific amplicon was 102 bp. The other bivalves also showed greater differences in the sequence of the region of primers.
For the multiplex PCR, in addition to P. nobilis and P. rudis we tested different bivalve and three putative hybrid samples from specimens which showed indistinguishable morphological characteristics between the two species. No amplification was obtained for the other bivalve species (data not shown). In contrast, samples from P. nobilis and P. rudis exhibited amplification of the species‐specific fragment. Additionally, amplification of both PCR fragments was observed in the three putative hybrid samples, confirming the effectiveness of the developed method (Figure 4).
FIGURE 4.
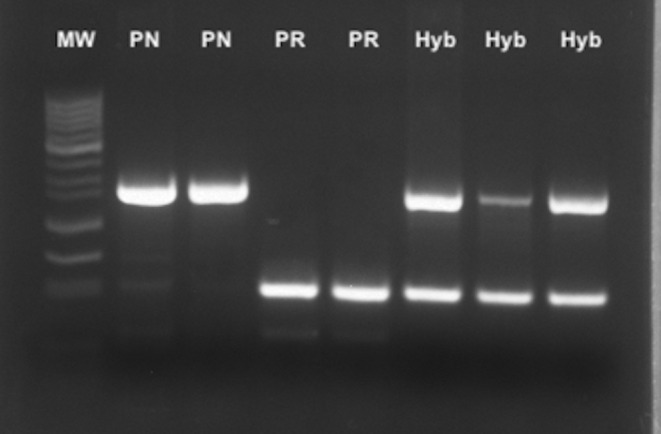
Electrophoresis in Agarose gel of multiplex‐PCR products from DNA of adult Pinna sp. individuals. Band sizes of the 50 bp DNA ladder (MW) are indicated on the left. PCR products are displayed as follows: Lanes 1–2: P. nobilis samples (PN); Lanes 3–4: P. rudis samples (PR); Lanes 5–7: Hybrid samples (Hyb).
The same samples indicated as putative hybrids were previously identified using the mitochondrial analysis method described by Catanese, Tena‐Medialdea, et al. (2022) as P. nobilis, which would confirm uniparental mitochondrial transmission in these species although a situation of hybridization or gene introgression between the two species is taking place.
4. DISCUSSION
Natural hybridization plays a fundamental role in evolution due to its ability to influence genetic diversity, adaptation and speciation processes. This phenomenon involves genetic interaction between individuals of different species. At least 25% of plant species and 10% of animal species, mostly the youngest species, are involved in hybridization and potential introgression with other species (Mallet, 2005). Hybridization between marine species has been observed across various contexts. Examples include hybridization between the grey seal (Halichoerus grypus) and the common seal (Phoca vitulina) in the North Atlantic (Helyar et al., 2011), the bottlenose dolphin (Tursiops truncatus) and Risso's dolphin (Grampus griseus) in the Gulf of Mexico (Mazzoil et al., 2018), the American horseshoe crab (Limulus polyphemus) and Japanese horseshoe crab (Tachypleus tridentatus) (Shuster Jr et al., 2004), and different species of angelfish (Pterophyllum scalare and Pterophyllum altum) in South America (Le Bail & Keith, 2007).
In general, interbreeding between evolutionarily distinct lineages is termed “hybridization,” whereas “introgression” denotes gene flow between species resulting from this hybridization (Pfennig et al., 2016). Natural hybridization between species can introduce new alleles into populations, thus increasing genetic variability, and generating genetic combinations that confer adaptive advantages (Arnold, 1997). In fact, hybrid individuals may exhibit different phenotypes or combinations of genetic characteristics from the parental species, allowing them to adapt to environmental changes and new selective pressures. Hybridization can contribute to the generation of new hybrid species with unique characteristics.
However, even if the presence of hybrids can lead to an increase in genetic variability, genetic introgression can also compromise the survival of native populations. Although hybrid species could represent a resource for adapting to environmental changes by inheriting beneficial characteristics from their parents and favouring their survival in new environmental conditions, hybridization can threaten the genetic purity of the original species, especially if the parental species are at risk of extinction or are local endemics. If hybrid species show greater reproductive success or adaptation to the local environment, they could gradually displace parental species, leading to changes in the composition of marine communities (Todesco et al., 2016). Therefore, the fertility of hybrid species is an important aspect to consider in relation to their ecology and conservation. It can vary greatly depending on the species involved and the type of hybridization. Some hybrid species may be fertile and able to reproduce, while others may be sterile or have reduced fertility. Fertility is crucial to determining whether they can form a stable population in the long term. Indeed, they may contribute to a new gene pool that could evolve independently of the parental species.
In the case of Pinna hybrids, it is not known whether the first generation of offspring resulting from the crossbreeding of the two distinct parental lines (F1 generation) are fertile or infertile. If they were fertile and reproductively successful, they may require specific management in conservation.
Some authors emphasized the need for clear guidelines in conservation efforts to address hybridization issues effectively. Allendorf et al. (2001) proposed strategies for managing hybridization, including assessing the genetic and ecological impacts of hybrids and implementing appropriate management actions to conserve species integrity and diversity.
The study of hybridization events and the formation of sterile and non‐sterile hybrids can also provide information on the processes of speciation and diversification, the mechanisms that promote reproductive isolation between species and the different adaptability or diverse or fluctuating ecological niches. Furthermore, understanding the dynamics and consequences of hybridization is essential for biodiversity management and conservation.
However, even if the presence of hybrids can lead to an increase in genetic variability, it must be considered that hybrids could also indicate a problem since genetic introgression can also compromise the survival of native populations.
Hybrids often possess a broader range of ecological traits than their parent species. This broader ecological niche may include differences in habitat use and feeding behaviours. Greater ecological flexibility can reduce exposure to specific pathogens or minimize disease transmission within populations, thus contributing to overall resilience.
Considering that until now P. rudis has not shown massive mortality events, probably as a consequence of its greater adaptability to environmental changes and/or the presence of pathogens, hybrid individuals could also possess a greater adaptive potential to cope with emerging pathogens or changing environmental conditions. Rapid evolution through hybridization can lead to the development of new genetic combinations that confer resistance to prevalent pathogens.
Genome of the P. nobilis × P. rudis hybrids seems to be naturally resistant to the parasite (Vázquez‐Luis et al., 2021), suggesting that their innate immunity could be efficient enough to prevent from infection development and spreading.
Recently, RNA‐Seq comparative study revealed molecular effectors linked to the resistance of P. nobilis to H. pinnae parasite (Salis et al., 2022), while a preliminary research suggests a role of fast‐evolving immune gene TLR‐7 (toll‐like receptor) in P. nobilis resistance to H. pinnae, with possible TLR‐6 and TLR‐4 influence (Coupé et al., 2023). Although further studies are needed to clarify the role of TLRs in disease resistance, the presence of TLR genes from P. rudis in the genome of P. nobilis individuals resistant to H. pinnae, but not in those susceptible to the pathogen, suggests that resistance genes may have arisen through different ancestral evolutionary processes or have been introduced through adaptive hybridization over several generations.
However, our method was primarily designed to reliably identify first‐generation hybrids. Detecting individuals from older hybridization events, in the case of F1 hybrids are fertile, can be challenging. As F2 and subsequent generation hybrids may rapidly diverge both phenotypically and genotypically from F1 hybrids, accurate identification of these later‐generation hybrids could require the molecular analysis of multiple loci.
Anyway, from a conservation perspective, it is crucial to possess tools capable of detecting hybrids at the molecular level for several important reasons, particularly to ascertain the presence and scope of hybridization within natural populations. This information is vital for delineating conservation units and establishing conservation priorities, especially in scenarios where hybridization may pose risks or confer an advantage to endemic or endangered species.
Natural hybridization in molluscs is an intriguing phenomenon where individuals of different species can reproduce and produce viable offspring. Closely related mussel species engage in documented natural hybridization, especially in coastal areas with specific marine currents. In the specific case of mussels, it has been observed that they can hybridize in areas where their distributions overlap (Simonsen, 2001). For example, hybridization between the mussel species M. edulis and M. galloprovincialis and evidence of intragenic recombination in a genetic marker was found by Rawson et al. (1996) Furthermore, several researchers have investigated the genetic structure and distribution of hybrid mussel populations in various geographical regions (Coghlan & Gosling, 2007; Gosling et al., 2008; Inoue et al., 1997; Kartavtsev et al., 2014). In Pinna species, individuals featuring intermingled shell morphology and mantle coloration, with both P. nobilis and P. rudis traits have been observed. They were identified as putative hybrids by sequencing analysis detecting 9 diagnostic nucleotides of the 28S nuclear gene (Vázquez‐Luis et al., 2021). In our work, we used the same individuals for testing the developed method.
Several studies have incorporated phylogenetic analysis of the spacer regions of nuclear ribosomal genes. The ITS regions are particularly valuable in phylogenetic and taxonomic studies for inferring evolutionary relationships between species, owing to the conservation and co‐evolution of ribosomal unit sequences (Baldwin, 1992).
The ITS regions have emerged as a valuable tool for assessing plant, fungal, and algal lineages across various taxonomic levels due to their ease of amplification through PCR and sequenceability using conservative primers, proposing them as a potential barcode and for phylogenetic studies (Letsiou et al., 2024; Schoch et al., 2012; Stern et al., 2012; Yao et al., 2010). They provide high information content due to their variability between species and conservation within individuals of the same species, attributed to concerted evolution associated with the entire ribosomal unit. Concerted evolution refers to coordinated or simultaneous changes over time in DNA sequences encoding ribosomal subunits (rRNA) within an organism, ensuring the functional integrity and efficiency of the ribosomal system (Hillis et al., 1991).
The reconstruction of the phylogenetic history of some mollusc species has been attempted previously by different authors, using different gene loci and other aspects of DNA. In this study, the phylogenetic analyses were conducted on a partial scale rather than in‐depth, focusing solely on confirming significant molecular differences in ITS regions between species of taxonomically close families. Our goal was to ensure accurate primer design specific to Pinna species. Although the uniformity in sequence across specimens of Pinna species from various Mediterranean regions supports our results, further analysis of additional samples is needed to validate the observed low variability.
Finally, the choice between using a species‐specific PCR or sequencing a DNA fragment to detect hybrids between species is essential considering the practical and technical benefits of each approach. Species‐specific PCR offers a significant advantage in terms of sensitivity and specificity as well as in time and cost (Edwards & Gibbs, 1994) It allows highly accurate detection of sequences of interest, minimizing the possibility of cross‐amplification or false positives (Edwards & Gibbs, 1994). This precision is essential when searching for hybrids that may have genetic sequences similar but not identical to those of their parental species. The detection of hybrids between species can be indicated by successful amplification of the PCR product using primers designed for each parental species (Gross et al., 1996). In contrast, DNA sequencing can introduce ambiguities in interpretation when “double peak” patterns within DNA chromatograms are observed at nucleotide positions where parental sequences differ. The double peaks may complicate accurate identification of hybrids and could require additional bioinformatic analyses to confirm the presence of heterozygosity and hybridization.
Furthermore, species‐specific PCR requires less genetic material to carry out amplification than DNA sequencing, which is especially useful when working with limited or unpurified samples. The costs associated with species‐specific PCR are generally lower in terms of reagents and analysis time compared to DNA sequencing, making it a more accessible and practical option in many research contexts.
Ease of implementation is also a point in favour of species‐specific PCR. This technique is well established in molecular biology laboratories and the necessary protocols and reagents are widely available. This facilitates its adoption and application in studies to detect hybrids between species, allowing researchers to obtain reliable results quickly and effectively. Moreover, this technique is highly recommended for both adult and juvenile individuals. It can be particularly effective when applied to fresh or frozen eDNA samples, eliminating the need for direct manipulation of individuals during sampling, similar to the mitochondrial assay used for Pinna identification.
In conclusion, species‐specific PCR emerges as a preferred strategy over DNA sequencing when seeking to detect hybrids between species. It offers sensitivity, specificity and efficiency, allowing clear and precise identification of genetic crosses between different species. This robust and practical technique is essential to advance our understanding of genetic diversity and evolution in contexts of interspecies hybridization.
The ongoing mortality event affecting Pinna nobilis in the Mediterranean Sea remains a critical concern for conservation efforts. Efforts to safeguard surviving populations of P. nobilis involve a multi‐pronged approach, including monitoring, captive breeding and employing collectors for juvenile recruitment. Captured juveniles are screened for pathogens and used in repopulation initiatives, in sites with specific local abiotic conditions such as extreme salinity or low temperature.
Hybridization events between P. nobilis and P. rudis could help conservation efforts due to their possible resilience to infection. Molecular techniques utilizing ITS regions enable the precise identification of hybrids and parental species, providing valuable insights into genetic interactions and potential introgression. Species‐specific PCR assays have proven effective in distinguishing hybrids, offering a practical and cost‐effective means of detecting genetic crosses.
The study underscores the significant implications of hybrids and the importance of using molecular tools for their identification. This is critical for effective biodiversity management and conservation strategies, ultimately ensuring the preservation of populations and deepening our understanding of the evolutionary consequences arising from genetic interactions between different species. Functional genomics research could reveal how hybridization affects physiology, behaviour, and ecological interactions, informing conservation plans. Additionally, studying the genetic resilience of Pinna species and their hybrids to climate change would turn out crucial for understanding adaptive capacities to rising sea temperatures and ocean acidification.
In conclusion, ongoing collaborative efforts involving scientists, environmentalists, and government organizations are essential for implementing effective monitoring, conservation, and repopulation strategies to safeguard P. nobilis populations against mortality events and emerging hybridization dynamics.
AUTHOR CONTRIBUTIONS
Gaetano Catanese: Conceptualization (lead); formal analysis (lead); funding acquisition (supporting); investigation (equal); methodology (lead); writing – original draft (lead); writing – review and editing (equal). Maite Vázquez‐Luis: Funding acquisition (supporting); investigation (equal); project administration (lead); resources (equal); writing – review and editing (equal). Salvatore Giacobbe: Investigation (equal); resources (equal); writing – review and editing (equal). José Rafael García‐March: Investigation (equal); project administration (equal); resources (equal); writing – review and editing (equal). Maria Zotou: Investigation (equal); resources (equal); writing – review and editing (equal). Prado Patricia: Investigation (equal); resources (equal); writing – review and editing (equal). Orestis Papadakis: Investigation (equal); resources (equal); writing – review and editing (equal). José Tena‐Medialdea: Investigation (equal); resources (equal); writing – review and editing (equal). Stelios Katsanevakis: Investigation (equal); resources (equal); writing – review and editing (equal). Amalia Grau: Funding acquisition (lead); investigation (equal); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
Data S1.
ACKNOWLEDGEMENTS
This research has been performed in the scope of the European Union's LIFE program through the project LIFE PINNARCA (NAT/ES/001265); BIODIV project: “Scientific and technical advice for the monitoring of marine biodiversity: protected marine areas and species of state competence (2022‐2025).” Project funded by the European Union – NextGenerationEU through the Recovery, Transformation and Resilience Plan and promoted by the Directorate General for Biodiversity, Forests and Desertification of the Ministry for Ecological Transition and the Demographic Challenge and CSIC, through the Spanish Institute of Oceanography (IEO‐CSIC); and the EsMarEs project (order IEO by Ministerio para la Transición Ecológica, Spanish Government). We thank the Regional Governments of local authorities (Conselleria de Agricultura, Pesca i Medi Natural – Govern de les Illes Balears) and national authorities (MITECO) for permits and support. We also thank the “Provincia Regionale di Messina” which strongly supported this research, both by granting permissions and providing facilities and services. We thank several colleagues for their help and fieldwork assistance.
Catanese, G. , Vázquez‐Luis, M. , Giacobbe, S. , García‐March, J. R. , Zotou, M. , Patricia, P. , Papadakis, O. , Tena‐Medialdea, J. , Katsanevakis, S. , & Grau, A. (2024). Internal transcribed spacer as effective molecular marker for the detection of natural hybridization between the bivalves Pinna nobilis and Pinna rudis . Ecology and Evolution, 14, e70227. 10.1002/ece3.70227
DATA AVAILABILITY STATEMENT
All experimental and unique haplotype data are available in the manuscript and deposited to NCBI Nucleotide Database (GenBank accession codes LC794141, LC794142 and from LC814844 to LC814852).
REFERENCES
- Abbott, R. , Albach, D. , Ansell, S. , Arntzen, J. W. , Baird, S. J. , Bierne, N. , Boughman, J. , Brelsford, A. , Buerkle, C. A. , Buggs, R. , Butlin, R. K. , Dieckmann, U. , Eroukhmanoff, F. , Grill, A. , Cahan, S. H. , Hermansen, J. S. , Hewitt, G. , Hudson, A. G. , Jiggins, C. , … Zinner, D. (2013). Hybridization and speciation. Journal of Evolutionary Biology, 26, 229–246. 10.1111/j.1420-9101.2012.02599.x [DOI] [PubMed] [Google Scholar]
- Acarli, D. , Acarli, S. , & Kale, S. (2022). The struggle for life: Pinna nobilis in the Marmara Sea (Turkey). Thalassas: An International Journal of Marine Sciences, 38, 1199–1212. 10.1007/s41208-022-00470-0 [DOI] [Google Scholar]
- Acarlı, S. , Acarlı, D. , & Kale, S. (2024). Distribution of the critically endangered fan mussel Pinna nobilis population in the Çanakkale Strait and Marmara Sea. Ege Journal of Fisheries and Aquatic Sciences, 41, 46–53. 10.12714/egejfas.41.1.07 [DOI] [Google Scholar]
- Allendorf, F. W. , Leary, R. F. , Spruell, P. , & Wenburg, J. K. (2001). The problems with hybrids: Setting conservation guidelines. Trends in Ecology & Evolution, 16, 613–622. 10.1016/S0169-5347(01)02290-X [DOI] [Google Scholar]
- Aranishi, F. (2005). PCR‐RFLP analysis of nuclear nontranscribed spacer for mackerel species identification. Journal of Agricultural and Food Chemistry, 53, 508–511. 10.1021/jf0484881 [DOI] [PubMed] [Google Scholar]
- Arnold, M. L. (1997). Natural hybridization and evolution. Oxford University Press. [Google Scholar]
- Baldwin, B. G. (1992). Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: An example from the Compositae. Molecular Phylogenetics and Evolution, 1, 3–16. [DOI] [PubMed] [Google Scholar]
- Beaumont, A. R. , Turner, G. , Wood, A. R. , & Skibinski, D. O. F. (2004). Hybridisations between Mytilus edulis and Mytilus galloprovincialis and performance of pure species and hybrid veliger larvae at different temperatures. Journal of Experimental Marine Biology and Ecology, 302, 177–188. 10.1016/j.jembe.2003.10.009 [DOI] [Google Scholar]
- Box, A. , Capó, X. , Tejada, S. , Catanese, G. , Grau, A. , Deudero, S. , Sureda, A. , & Valencia, J. M. (2020). Antioxidant response of the fan mussel Pinna nobilis related to the presence of Haplosporidium pinnae . Pathogens, 9, 932. 10.3390/pathogens9110932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanellas‐Reboredo, M. , Deudero, S. , Alós, J. , Valencia, J. M. , March, D. , Hendriks, I. E. , & Álvarez, E. (2009). Recruitment of Pinna nobilis (Mollusca: Bivalvia) on artificial structures. Marine Biodiversity Records, 2, e126. 10.1017/S1755267209001274 [DOI] [Google Scholar]
- Cabanellas‐Reboredo, M. , Vázquez‐Luis, M. , Mourre, B. , Álvarez, E. , Deudero, S. , Amores, A. , Addis, P. , Ballesteros, E. , Barrajón, A. , Coppa, S. , García‐March, J. R. , Giacobbe, S. , Giménez Casalduero, F. , Hadjioannou, L. , Jiménez‐Gutiérrez, S. V. , Katsanevakis, S. , Kersting, D. , Mačić, V. , Mavrič, B. , … Hendriks, I. E. (2019). Tracking a mass mortality outbreak of pen shell Pinna nobilis populations: A collaborative effort of scientists and citizens. Scientific Reports, 9, 13355. 10.1038/s41598-019-49808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella, F. , Aceto, S. , Pollaro, F. , Miccio, A. , Iaria, C. , Carrasco, N. , Prado, P. , & De Vico, G. (2019). A mycobacterial disease is associated with the silent mass mortality of the pen shell Pinna nobilis along the Tyrrhenian coastline of Italy. Scientific Reports, 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella, F. , Prado, P. , De Vico, G. , Palić, D. , Villari, G. , García‐March, J. R. , Tena‐Medialdea, J. , Cortés Melendreras, E. , Giménez‐Casalduero, F. , Sigovini, M. , & Aceto, S. (2023). A widespread picornavirus affects the hemocytes of the noble pen shell (Pinna nobilis), leading to its immunosuppression. Frontiers in Veterinary Science, 10, 1273521. 10.3389/fvets.2023.1273521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanese, G. , Coupé, S. , & Bunet, R. (2022). Mitogenome sequence comparison in the endangered congeneric Pinna nobilis and Pinna rudis bivalves. Molecular Biology Reports, 49, 3627–3635. 10.1007/s11033-022-07202-w [DOI] [PubMed] [Google Scholar]
- Catanese, G. , Grau, A. , Valencia, J. M. , García‐March, J. R. , Vázquez‐Luis, M. , Alvarez, E. , Deudero, S. , Darriba, S. , Carballal, M. J. , & Villalba, A. (2018). Haplosporidium pinnae sp. nov., a haplosporidan parasite associated with mass mortalities of the fan mussel, Pinna nobilis, in the Western Mediterranean Sea. Journal of Invertebrate Patholology, 157, 9–24. 10.1016/j.jip.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Catanese, G. , Tena‐Medialdea, J. , Bel Dajković, M. A. , Mičić, M. , & García‐March, J. R. (2022). An incubation water eDNA method for a non‐destructive rapid molecular identification of the two congeneric Pinna nobilis and Pinna rudis bivalve juveniles. MethodX, 9, 101708. 10.1016/j.mex.2022.101708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEbro (Confederación Hidrográfica del Ebro) . (2023). Plan de Sequía 2023. https://www.chebro.es/web/guest/plan‐de‐sequ%C3%ADa‐2023
- Chow, S. , Ueno, Y. , Toyokawa, M. , Oohara, I. , & Takeyama, H. (2009). Preliminary analysis of length and GC content variation in the ribosomal first internal transcribed spacer (ITS1) of marine animals. Marine Biotechnology, 11, 301–306. 10.1007/s10126-008-9153-2 [DOI] [PubMed] [Google Scholar]
- Çınar, M. , Bilecenoğlu, M. , Yokeş, M. B. , & Güçlüsoy, H. (2021). Pinna nobilis in the south Marmara Islands (sea of Marmara); it still remains uninfected by the epidemic and acts as egg laying substratum for an alien invader. Mediterranean Marine Science, 22, 161–168. 10.12681/mms.25289 [DOI] [Google Scholar]
- Cižmek, H. , Colíc, B. , Gracan, R. , Grau, A. , & Catanese, G. (2020). Emergency situation for pen shells in the Mediterranean: The Adriatic Sea, one of the last Pinna nobilis shelter, is now affected by the mass mortality event. Journal of Invertebrate Patholology, 173, 107388. 10.1016/j.jip.2020.107388 [DOI] [PubMed] [Google Scholar]
- Coghlan, B. , & Gosling, E. (2007). Genetic structure of hybrid mussel populations in the west of Ireland: Two hypotheses revisited. Marine Biology, 150, 841–852. 10.1007/s00227-006-0408-z [DOI] [Google Scholar]
- Cortés‐Melendreras, E. , Gomariz‐Castillo, F. , Alonso‐Sarría, F. , Giménez Martín, F. J. , Murcia, J. , Canales‐Cáceres, R. , Ramos Esplá, A. A. , & Barberá, C. (2022). The relict population of Pinna nobilis in the mar Menor is facing an uncertain future. Marine Pollution Bullettin, 18, 114376. 10.1016/j.marpolbul.2022.114376 [DOI] [PubMed] [Google Scholar]
- Coupé, S. , Giantsis, I. A. , Vázquez‐Luis, M. , Scarpa, F. , Foulquié, M. , Prévot, J.‐M. , Casu, M. , Lattos, A. , Michaelidis, B. , Sanna, D. , García‐March, J. R. , Tena‐Medialdea, J. , Vicente, N. , & Bunet, R. (2023). The characterization of toll‐like receptor repertoire in Pinna nobilis after mass mortality events suggests adaptive introgression. Ecology and Evolution, 13, e10383. 10.1002/ece3.10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba, S. (2017). First haplosporidan parasite reported infecting a member of the superfamily Pinnoidea (Pinna nobilis) during a mortality event in Alicante (Spain, Western Mediterranean). Journal of Invertebrate Pathology, 148, 14–19. [DOI] [PubMed] [Google Scholar]
- Donato, G. , Lunetta, A. , Spinelli, A. , Catanese, G. , & Giacobbe, S. (2023). Sanctuaries are not inviolable: Haplosporidium pinnae as responsible for the collapse of the Pinna nobilis population in Lake Faro (central Mediterranean). Journal of Invertebrate Pathology, 201, 108014. [DOI] [PubMed] [Google Scholar]
- Donato, G. , Vázquez‐Luis, M. , Nebot‐Colomer, E. , Lunetta, A. , & Giacobbe, S. (2021). Noble fan‐shell, Pinna nobilis, in Lake Faro (Sicily, Italy): Ineluctable decline or extreme opportunity? Estuarine, Coastal and Shelf Science, 261, 107536. [Google Scholar]
- Edwards, M. C. , & Gibbs, R. A. (1994). Multiplex PCR: Advantages, development and applications. PCR Methods and Applications, 3, S65–S75. [DOI] [PubMed] [Google Scholar]
- Feria‐Rodríguez, A. , March, D. , Mourre, B. , Hendriks, I. , & Vázquez‐Luis, M. (2024). Sink‐source connectivity for restocking of Pinna nobilis in the western Mediterranean Sea. Marine Environmental Research, 197, 106428. [DOI] [PubMed] [Google Scholar]
- Foulquié, M. , Coupé, S. , Vicente, N. , & Bunet, R. (2023). First detection of Pinna nobilis infection Haplosporidium pinnae in the sanctuary area of Thau lagoon, France. Mediterranean Marine Science, 24, 569–573. [Google Scholar]
- García‐March, J. R. , Tena, J. , Hernandis, S. , Vázquez‐Luis, M. , López, D. , Téllez, C. , Prado, P. , Navas, J. I. , Bernal, J. , Catanese, G. , Grau, A. , López‐Sanmartín, M. , Nebot‐Colomer, E. , Ortega, A. , Planes, S. , Kersting, D. , Jimenez, S. , Hendriks, I. , Moreno, D. , … Deudero, S. (2020). Can we save a marine species affected by a highly infective, highly lethal, waterborne disease from extinction? Biological Conservation, 243, 108498. 10.1016/j.biocon.2020.108498 [DOI] [Google Scholar]
- Giménez‐Casalduero, F. , Gomariz‐Castillo, F. , Alonso‐Sarría, F. , Cortés, E. , Izquierdo‐Muñoz, A. , & Ramos‐Esplá, A. (2020). Pinna nobilis in the Mar Menor coastal lagoon: A story of colonization and uncertainty. Marine Ecology Progress Series, 652, 77–94. 10.3354/meps13468 [DOI] [Google Scholar]
- Gosling, E. , Doherty, S. , & Howley, N. (2008). Genetic characterization of hybrid mussel (Mytilus) populations on Irish coasts. Journal of the Marine Biological Association of the UK, 88, 341–346. 10.1017/S0025315408000957 [DOI] [Google Scholar]
- Grau, A. , Villalba, A. , Navas, J. I. , Hansjosten, B. , Valencia, J. M. , García‐March, J. R. , Prado, P. , Follana‐Berná, G. , Morage, T. , Vázquez‐Luis, M. , Álvarez, E. , Katharios, P. , Pavloudi, C. , Nebot‐Colomer, E. , Tena‐Medialdea, J. , Lopez‐Sanmartín, M. , Peyran, C. , Čižmek, H. , Sarafidou, G. , … Catanese, G. (2022). Wide‐geographic and long‐term analysis of the role of pathogens in the decline of Pinna nobilis to critically endangered species. Frontiers in Marine Science, 9, 666640. 10.3389/fmars.2022.666640 [DOI] [Google Scholar]
- Gross, R. , Nilsson, J. , & Schmitz, M. (1996). A new species‐specific nuclear DNA marker for identification of hybrids between Atlantic salmon and brown trout. Journal of Fish Biology, 49, 537–540. [Google Scholar]
- Gvozdenović Nikolić, S. , Macic, V. , Pešić, V. , Nikolic, M. , Peraš, I. , & Mandic, M. (2019). Review on Pinna rudis (Linnaeus, 1758) (Bivalvia: Pinnidae) presence in the Mediterranean. Agriculture and Forestry, 65, 115–126. 10.17707/AgricultForest.65.4.10 [DOI] [Google Scholar]
- Hall, T. A. (1999). BioEdit: A user‐friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Helyar, S. J. , Hemmer‐Hansen, J. , Bekkevold, D. , Taylor, M. I. , Ogden, R. , Limborg, M. T. , Cariani, A. , Maes, G. E. , Diopere, E. , Carvalho, G. R. , & Nielsen, E. E. (2011). Application of SNPs for population genetics of nonmodel organisms: New opportunities and challenges. Molecular Ecology Resources, 11, 123–136. [DOI] [PubMed] [Google Scholar]
- Hernandis, S. , Ibarrola, I. , Tena‐Medialdea, J. , Vázquez‐Luis, M. , García‐March, J. R. , Prado, P. , & Albentosa, M. (2022). Scope for growth and dietary needs of Mediteranean Pinnids maintained in captivity. BMC Zoology, 27, 43. 10.1186/s40850-022-00141-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandis, S. , Prado, P. , García‐March, J. R. , Gairin, I. , Vázquez‐Luis, M. , & Tena‐Medialdea, J. (2023). Reproduction of the endangered fan mussel Pinna nobilis under natural and captivity conditions. Aquatic Conservation: Marine and Freshwater Ecosystems, 33, 1501–1513. 10.1002/aqc.4009 [DOI] [Google Scholar]
- Hillis, D. M. , Moritz, C. , Porter, C. A. , & Baker, R. J. (1991). Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science, 251, 308–310. [DOI] [PubMed] [Google Scholar]
- Huber, M. (2010). Compendium of bivalves. A full‐color guide to 3,300 of the world's marine bivalves. A status on Bivalvia after 250 years of research (p. 901). ConchBooks. [Google Scholar]
- Huvet, A. , Fabioux, C. , McCombie, H. , Lapègue, S. , & Boudry, P. (2004). Natural hybridization between genetically differentiated populations of Crassostrea gigas and C. Angulata highlighted by sequence variation in flanking regions of a microsatellite locus. Marine Ecology Progress Series, 272, 141–152. http://www.jstor.org/stable/24867455 [Google Scholar]
- Inoue, K. , Odo, S. , Noda, T. , Nakao, S. , Takeyama, S. , Yamaha, E. , Yamazaki, F. , & Harayama, S. (1997). A possible hybrid zone in the Mytilus edulis complex in Japan revealed by PCR markers. Marine Biology, 128, 91–95. [Google Scholar]
- Karadurmuş, U. , Benli, T. , & Sarı, M. (2024). Discovering new living Pinna nobilis populations in the sea of Marmara. Marine Biology, 171, 90. 10.1007/s00227-024-04416-3 [DOI] [Google Scholar]
- Kartavtsev, Y. P. , Katolikova, M. V. , Sharina, S. N. , Chichvarkhina, O. V. , & Masalkova, N. A. (2014). A population genetic study of the hybrid zone of Mytilus trossulus Gould, 1850 and an introduced species, M. Galloprovincialis Lamarck, 1819, (Bivalvia: Mytilidae) in peter the great bay in the sea of Japan. Russian Journal of Marine Biology, 40, 208–216. 10.1134/S1063074014030055 [DOI] [Google Scholar]
- Katsanevakis, S. (2016). Transplantation as a conservation action to protect the Mediterranean fan mussel Pinna nobilis . Marine Ecology Progress Series, 546, 113–122. 10.3354/meps11658 [DOI] [Google Scholar]
- Katsanevakis, S. , Carella, F. , Çinar, M. E. , Čižmek, H. , Jimenez, C. , Kersting, D. K. , Moreno, D. , Rabaoui, L. , & Vicente, N. (2022). The fan mussel Pinna nobilis in the brink of extinction in the Mediterranean. In DellaSala D. A. & Goldstein M. I. (Eds.), Imperiled: The encyclopedia of conservation (Vol. 2, pp. 700–709). Elsevier. 10.1016/B978-0-12-821139-7.00070-2 [DOI] [Google Scholar]
- Kersting, D. , & Ballesteros, E. (2021). Is the local extinction of Pinna nobilis facilitating Pinna rudis recruitment? Mediterranean Marine Science, 22, 623–626. 10.12681/mms.27809 [DOI] [Google Scholar]
- Kersting, D. , Benabdi, M. , Čižmek, H. , Grau, A. , Jimenez, C. , Katsanevakis, S. , Öztürk, B. , Tuncer, S. , Tunesi, L. , Vázquez‐Luis, M. , Vicente, N. , & Otero Villanueva, M. (2019). Pinna nobilis. The IUCN Red List of Threatened Species 2019: e.T160075998A160081499. 10.2305/IUCN.UK.2019-3.RLTS.T160075998A160081499.en Accessed on 26 April 2024. [DOI]
- Kersting, D. K. , & García‐March, J. R. (2017). Long‐term assessment of recruitment, early stages and population dynamics of the endangered Mediterranean fan mussel Pinna nobilis in the Columbretes Islands (NW Mediterranean). Marine Environmental Research, 130, 282–292. 10.1016/j.marenvres.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Kersting, D. K. , Vázquez‐Luis, M. , Mourre, B. , Belkhamssa, F. Z. , Álvarez, E. , Bakran‐Petricioli, T. , Barberá, C. , Barrajón, A. , Cortés, E. , Deudero, S. , García‐March, J. R. , Giacobbe, S. , Giménez‐Casalduero, F. , González, L. , Jiménez‐Gutiérrez, S. , Kipson, S. , Llorente, J. , Moreno, D. , Prado, P. , … Hendriks, I. E. (2020). Recruitment disruption and the role of unaffected populations for potential recovery after the Pinna nobilis mass mortality event. Frontiers in Marine Science, 7, 594378. 10.3389/fmars.2020.594378 [DOI] [Google Scholar]
- Labidi, S. , Vázquez‐Luis, M. , Catanese, G. , Grau, A. , Khammassi, M. , Ben Youssef, S. , & Sghaier, A. M. (2023). First detection of the invasive protozoan Haplosporidium pinnae in the critically endangered bivalve Pinna nobilis in south Mediterranean Sea (Bizerte lagoon, Tunis). Mediterranean Marine Science, 24, 470–481. [Google Scholar]
- Lattos, A. , Giantsis, I. A. , Karagiannis, D. , & Michaelidis, B. (2020). First detection of the invasive Haplosporidian and mycobacteria parasites hosting the endangered bivalve Pinna nobilis in Thermaikos gulf, North Greece. Marine Environmental Research, 155, 104889. 10.1016/j.marenvres.2020.104889 [DOI] [PubMed] [Google Scholar]
- Le Bail, P. Y. , & Keith, P. (2007). Confirmation of interspecific hybridization between Pterophyllum scalare and P. Altum (Cichlidae, Teleostei) through experimental crosses. Cybium, 31, 185–191. [Google Scholar]
- Letsiou, S. , Madesis, P. , Vasdekis, E. , Montemurro, C. , Grigoriou, M. E. , Skavdis, G. , Moussis, V. , Koutelidakis, A. E. , & Tzakos, A. G. (2024). DNA barcoding as a plant identification method. Applied Sciences, 14, 1415. 10.3390/app14041415 [DOI] [Google Scholar]
- Mallet, J. (2005). Hybridization as an invasion of the genome. Trends in Ecology & Evolution, 20, 229–237. 10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Mazzoil, M. , Shelden, K. E. W. , & Pierson, M. O. (2018). Evidence of hybridization between common bottlenose dolphins (Tursiops truncatus) and Risso's dolphins (Grampus griseus) in the northern Gulf of Mexico. Marine Mammal Science, 34, 476–488. [Google Scholar]
- Moniz, M. B. J. , & Kaczmarska, I. (2010). Barcoding of diatoms: Nuclear encoded ITS revisited. Protist, 161, 7–34. [DOI] [PubMed] [Google Scholar]
- Moro‐Martínez, I. , Vázquez‐Luis, M. , García‐March, J. R. , Prado, P. , Mičić, M. , & Catanese, G. (2023). Haplosporidium pinnae parasite detection in seawater samples. Microorganisms, 11, 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebot‐Colomer, E. , Álvarez, E. , Belando, M. D. , Deudero, S. , Catanese, G. , Bernardeau‐Esteller, J. , García‐Muñoz, R. , Ramos‐Segura, A. , Ruiz, J. M. , & Vázquez‐Luis, M. (2021). Living under threat: Will one of the last Pinna nobilis populations be able to survive? Aquatic Conservation: Marine and Freshwater Ecosystems, 32, 1–13. 10.1002/aqc.3738 [DOI] [Google Scholar]
- Nebot‐Colomer, E. , Hernandis, S. , Mourre, B. , Fraile‐Nuez, E. , Álvarez Pérez, E. , Deudero, S. , Albentosa, M. , & Vázquez‐Luis, M. (2024). No recruits for an ageing population: First signs of probable population extinction in one of the last reservoirs of the critically endangered species Pinna nobilis . Korean Journal of Nature Conservation, 79, 126600. 10.1016/j.jnc.2024.126600 [DOI] [Google Scholar]
- Oprandi, A. , Aicardi, S. , Azzola, A. , Benelli, F. , Bertolino, M. , Bianchi, C. N. , Chiantore, M. , Ferranti, M. P. , Mancini, I. , Molinari, A. , Morri, C. , & Montefalcone, M. (2024). A tale of two sisters: The southerner Pinna rudis is getting north after the regional extinction of the congeneric P. nobilis (Mollusca: Bivalvia). Diversity, 16, 120. 10.3390/d16020120 [DOI] [Google Scholar]
- Papadakis, O. , Mamoutos, I. , Ramfos, A. , Catanese, G. , Papadimitriou, J. A. , Theodorou, E. , Batargias, C. , Papaioannou, C. , Kamilari, M. , Tragou, E. , Zervakis, V. , & Katsanevakis, S. (2023). Status, distribution, and threats of the last surviving fan mussel populations in Greece. Mediterranean Marine Science, 24, 679–708. [Google Scholar]
- Peyran, C. , Boissin, E. , Morage, T. , Nebot‐Colomer, E. , Iwankow, G. , & Planes, S. (2022). Investigating population dynamics from parentage analysis in the highly endangered fan mussel Pinna nobilis . Ecology and Evolution, 12, e8482. 10.1002/ece3.8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig, K. S. , Kelly, A. L. , & Pierce, A. A. (2016). Hybridization as a facilitator of species range expansion. Proceedings of the Royal Society B: Biological Sciences, 283, 20161329. 10.1098/rspb.2016.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D. (2008). jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution, 25, 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Posada, D. , & Buckley, T. R. (2004). Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology, 53, 793–808. [DOI] [PubMed] [Google Scholar]
- Prado, P. , Cabanes, P. , Catanese, G. , Carella, F. , Carrasco, N. , Grau, A. , Hernandis, S. , García‐March, J. R. , Tena, J. , Caiola, N. , & Andree, K. A. (2020). Growth of juvenile Pinna nobilis in captivity conditions: Dietary and pathological constraints. Aquaculture, 522, 735167. 10.1016/j.aquaculture.2020.735167 [DOI] [Google Scholar]
- Prado, P. , Grau, A. , Catanese, G. , Cabanes, P. , Carella, F. , Fernández‐Tejedor, M. , Andree, K. , Añón, T. , Hernandis, S. , Tena, J. , & García‐March, J. R. (2021). Pinna nobilis in suboptimal environments are more resilient to disease but more vulnerable to catastrophic events. Marine Environmental Research, 163, 105220. [DOI] [PubMed] [Google Scholar]
- Prasad, P. K. , Tandon, V. , Biswal, D. K. , Goswami, L. M. , & Chatterjee, A. (2009). Phylogenetic reconstruction using secondary structures and sequence motifs of ITS2 rDNA of Paragonimus westermani (Kerbert, 1878) Braun, 1899 (Digenea: Paragonimidae) and related species. BMC Genomics, 10, 3S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson, P. D. , Secor, C. L. , & Hilbish, T. J. (1996). The effects of natural hybridization on the regulation of doubly uniparental mtDNA inheritance in blue mussels (Mytilus spp.). Genetics, 144, 241–248. 10.1093/genetics/144.1.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino, F. , Fanelli, G. , & Denti, G. (2024). The Queen is dead, long live the queen: The vanishing of Pinna nobilis and the onset of the Congeneric P. rudis (Mollusca: Bivalvia). Diversity, 16, 341. 10.3390/d16060341 [DOI] [Google Scholar]
- Salis, P. , Peyran, C. , Morage, T. , de Bernard, S. , Nourikyan, J. , Coupé, S. , Bunet, R. , & Planes, S. (2022). RNA‐Seq comparative study reveals molecular effectors linked to the resistance of Pinna nobilis to Haplosporidium pinnae parasite. Scientific Reports, 12, 21229. 10.1038/s41598-022-25555-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa, F. , Sanna, D. , Azzena, I. , Mugetti, D. , Cerruti, F. , Hosseini, S. , Cossu, P. , Pinna, S. , Grech, D. , Cabana, D. , Pasquini, V. , Esposito, G. , Cadoni, N. , Atzori, F. , Antuofermo, E. , Addis, P. , Sechi, L. A. , Prearo, M. , Peletto, S. , … Casu, M. (2020). Multiple non‐species‐specific pathogens possibly triggered the mass mortality in Pinna nobilis . Life, 10, 238. 10.3390/life10100238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch, C. L. , Seifert, K. A. , Huhndorf, S. , Robert, V. , Spouge, J. L. , Levesque, C. A. , Chen, W. , Fungal Barcoding Consortium, Fungal Barcoding Consortium Author List , Bolchacova, E. , Voigt, K. , Crous, P. W. , Miller, A. N. , Wingfield, M. J. , Aime, M. C. , An, K.‐D. , Bai, F.‐Y. , Barreto, R. W. , Begerow, D. , … Schindel, D. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. National Academy of Sciences of the United States of America, 109, 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedghifar, A. , Brandvain, Y. , & Ralph, P. (2016). Beyond clines: Lineages and haplotype blocks in hybrid zones. Molecular Ecology, 25, 2559–2576. [DOI] [PubMed] [Google Scholar]
- Seehausen, O. (2013). Conditions when hybridization might predispose populations for adaptive radiation. Journal of Experimental Biology, 26, 279–281. [DOI] [PubMed] [Google Scholar]
- Shuster, C. N., Jr. , Sekiguchi, K. , Brockmann, H. J. , & Jehle, R. (2004). The evolution of mating systems in horseshoe crabs: II. The application of molecular approaches to the study of Limulus polyphemus and Tachypleus tridentatus . Evolutionary Biology, 34(2), 140–175. [Google Scholar]
- Simonsen, V. (2001). Marine molluscs: Hybridization and introgression. Phuket Marine Biological Center Special Publication, 24, 315–322. [Google Scholar]
- Stern, R. F. , Andersen, R. A. , Jameson, I. , Kupper, F. C. , Coffroth, M.‐A. , Vaulot, D. , Le Gall, F. , Véron, B. , Brand, J. J. , Skelton, H. , Kasai, F. , Lilly, E. L. , & Keeling, P. J. (2012). Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One, 7, e42780. 10.1371/journal.pone.0042780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , & Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38, 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscar, P. G. , Rubino, F. , Paoletti, B. , Di Francesco, C. E. , Mosca, F. , Salda, L. D. , Hattab, J. , Smoglica, C. , Morelli, S. , & Fanelli, G. (2022). New insights about Haplosporidium pinnae and the pen shell Pinna nobilis mass mortality events. Journal of Invertebrate Pathology, 190, 107735. [DOI] [PubMed] [Google Scholar]
- Todesco, M. , Pascual, M. A. , Owens, G. L. , Ostevik, K. L. , Moyers, B. T. , Hübner, S. , Heredia, S. M. , Hahn, M. A. , Caseys, C. , Bock, D. G. , & Rieseberg, L. H. (2016). Hybridization and extinction. Evolutionary Applications, 22, 892–908. 10.1111/eva.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez‐Luis, M. , Álvarez, E. , Barrajón, A. , García‐March, J. R. , Grau, A. , Hendriks, I. E. , Jiménez, S. , Kersting, D. , Moreno, D. , Pérez, M. , Ruiz, J. M. , Sánchez, J. , Villalba, A. , & Deudero, S. (2017). S.O.S. Pinna nobilis: A mass mortality event in western Mediterranean Sea. Frontiers in Marine Science, 4, 220. 10.3389/fmars.2017.00220 [DOI] [Google Scholar]
- Vázquez‐Luis, M. , Nebot‐Colomer, E. , Deudero, S. , Planes, S. , & Boissin, E. (2021). Natural hybridization between pen shell species: Pinna rudis and the critically endangered Pinna nobilis may explain parasite resistance in P. Nobilis . Molecular Biology Reports, 48, 997–1004. 10.1007/s11033-020-06063-5 [DOI] [PubMed] [Google Scholar]
- Voroshilova, I. S. , Artamonova, V. S. , Makhrov, A. A. , & Slyn'ko, Y. V. (2010). Natural hybridization of two mussel species Dreissena polymorpha (Pallas, 1771) and Dreissena bugensis (Andrusov, 1897). Biology Bulletin of the Russian Academy of Sciences, 37, 542–547. 10.1134/S1062359010050158 [DOI] [PubMed] [Google Scholar]
- White, T. J. , Bruns, T. , Lee, S. , & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In Innis M. A., Gelfand D. H., Sninsky J. J., & White T. J. (Eds.), PCR protocols: A guide to methods and applications (pp. 315–322). Academic Press. [Google Scholar]
- Wiemers, M. , Keller, A. , & Wolf, M. (2009). ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evolutionary Biology, 9, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H. , Song, J. , Liu, C. , Luo, K. , Han, J. , Li, Y. , Pang, X. , Xu, H. , Zhu, Y. , Xiao, P. , & Chen, S. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One, 5, e13102. 10.1371/journal.pone.0013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotou, M. , Papadakis, O. , Catanese, G. , Stranga, Y. , Ragkousis, M. , Kampouris, T. E. , Naasan Aga‐Spyridopoulou, R. , Papadimitriou, E. , Koutsoubas, D. , & Katsanevakis, S. (2023). New kid in town: Pinna rudis spreads in the eastern Mediterranean. Mediterranean Marine Science, 24, 709–721. 10.12681/mms.35343 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
All experimental and unique haplotype data are available in the manuscript and deposited to NCBI Nucleotide Database (GenBank accession codes LC794141, LC794142 and from LC814844 to LC814852).


