Abstract
The actin-binding protein gelsolin is highly conserved in vertebrates and exists in two isoforms, a cytoplasmic and an extracellular variant, generated by alternative splicing. In mammals, these isoforms differ only by an N-terminal extension in plasma gelsolin, a short sequence of up to 25 amino acids. Cells and tissues may contain both variants, as plasma gelsolin is secreted by many cell types. The tertiary structure of equine plasma gelsolin has been elucidated, but without any information on the N-terminal extension. In this paper, we present topographical data on the N-terminal extension, derived using a biochemical and immunological approach. For this purpose, a monoclonal antibody was generated that exclusively recognizes cytoplasmic gelsolin but not the extracellular variant and thus allows isoform-specific immunodetection and quantification of cytoplasmic gelsolin in the presence of plasma gelsolin. Using limited proteolysis and pepscan analysis, we mapped the binding epitope and localized it within two regions in segment 1 of the cytoplasmic gelsolin sequence: Tyr34–Ile45 and Leu64–Ile78. In the tertiary structure of the cytoplasmic variant, these sequences are mutually adjacent and located in the proximity of the N-terminus. We therefore conclude that the binding site of the antibody is covered by the N-terminal extension in plasma gelsolin and thus sterically hinders antibody binding. Our results allow for a topological model of the N-terminal extension on the surface of the gelsolin molecule, which was unknown previously.
Keywords: actin-binding site, epitope mapping, gelsolin antibody, gelsolin structure, N-terminal extension, topology
INTRODUCTION
Gelsolin is a calcium ion- and phosphoinositide-regulated actin-binding protein that severs and caps actin filaments and also promotes nucleation of actin polymerization. A gelsolin molecule consists of six modular domains of approx. 14 kDa each, which are similar in sequence and structure (see [1–4] for reviews). Binding of Ca2+ to subdomain 4 in the C-terminal half induces a gross conformational change in the molecule [5–10], leading to the exposure of actin-binding sites. Subsequently, gelsolin can either bind to and sever actin filaments or bind two actin monomers to form a stable ternary complex [11,12] that acts as a potent nucleus in actin polymerization.
Gelsolin exists in two isoforms: cytoplasmic gelsolin and an extracellular variant termed plasma gelsolin. In mammals, both isoforms are splicing products of one gene [13]. The extracellular variant is abundant in blood plasma and interstitial fluids. Cells may contain both variants, since plasma gelsolin is secreted by many cell types [14] and is therefore present in both compartments of the secretory pathway and the interstitium. For the determination of gelsolin concentrations or its localization in cells and tissues, discrimination between the two variants is therefore essential.
The sequences of plasma and cytoplasmic gelsolins differ solely in an N-terminal extension, which is present only in the secreted form. The length of this extension is species-specific, i.e. 25 amino acids in human [13] and 9 amino acids in pig [15]. Although the structure of the gelsolin molecule common to both variants has been elucidated by X-ray crystallography [16], topological information is not available for the N-terminal extension.
In the present study, we have characterized a monoclonal antibody against gelsolin which recognizes the intracellular variant but not the secreted form (2E12). At first glance, this finding appears puzzling, since plasma gelsolin contains the complete sequence and therefore all potential epitopes of cytoplasmic gelsolin. A plausible explanation is that the epitope recognized by this antibody is not accessible in plasma gelsolin. In the present study, we present data from both immunochemical analysis and epitope mapping in support of this hypothesis and show that access to the epitope of 2E12 is sterically hindered by the N-terminal extension of plasma gelsolin. These findings allow for a topological model of the N-terminal extension on the surface of the gelsolin molecule.
MATERIALS AND METHODS
Preparation of proteins and tissue extracts
Cytoplasmic gelsolin was purified from pig stomach smooth muscles and human platelets; plasma gelsolin was purified from pig and human blood plasma by methods described earlier [17,18] and stored as an ammonium sulphate precipitate in liquid nitrogen. Before use, the precipitate was dissolved and dialysed against PSAM buffer (10 mM imidazole, 0.5 mM EGTA, 0.2 mM dithiothreitol and 2 mM NaN3, pH 7.0).
SDS/PAGE and immunoblotting
SDS/PAGE was performed using 15% (w/v) acrylamide and 0.1% bisacrylamide gels in the Laemmli buffer system [19]. To resolve cytoplasmic and plasma gelsolins from guinea-pig, the usual running time for the gels was increased from 2 to 6 h. Gels were either stained with Coomassie Brilliant Blue R-250 or used for polypeptide transfer on to nitrocellulose membranes for 2 h using a semi-dry type blotter [20]. After transfer, non-specific binding sites on the nitrocellulose membranes were blocked for 1 h with 3% (w/v) fish gelatin or non-fat milk powder in TTBS (Tris-buffered saline with 0.05% Tween 20) and incubated with the antibodies at room temperature (22 °C). Immunodetection of gelsolin was performed using either goat anti-mouse or goat antirabbit secondary antibodies (Sigma). Alkaline phosphatase-conjugated secondary antibodies were processed in 0.5% gelatin– TTBS, using BCIP (5-bromo-4-chloroindol-3-yl phosphate)/Nitro Blue Tetrazolium as the substrate or using peroxidase-conjugated antibodies in 0.5% non-fat milk powder–TTBS.
Antibodies
A polyclonal antibody was raised in rabbits using pig cytoplasmic gelsolin as antigen. Purified gelsolin (2 mg) was run on preparative SDS/PAGE and electrotransferred on to nitrocellulose. The bands were excised and, after pulverizing the nitrocellulose in a liquid N2-cooled mortar, used as antigen after resuspension in PBS and addition of adjuvant peptide [21]. Aliquots of 0.2 mg of gelsolin were administered by subcutaneous injection according to standard injection methods.
A monoclonal antibody (2E12) was generated by immunizing mice with porcine cytoplasmic gelsolin from smooth muscles by standard methods. Antibody-secreting hybridoma cells were grown either in serum-containing medium or in serum-free medium in the presence of 500 μg/ml serum albumin and 10 μg/ml transferrin. The culture supernatant containing 2E12 was concentrated by membrane filtration approx. 50-fold and stored freeze-dried at −80 °C. The antibody was classified as IgG1. Secondary antibodies were obtained from Sigma.
ELISA
For the quantification of gelsolin, a sandwich type of ELISA was used with a polyclonal gelsolin antibody as the first antibody for precoating the plates and the 2E12 antibody as detecting antibody. As a secondary antibody, a horseradish peroxidase-conjugated goat anti-mouse antibody (Sigma) was used. The reaction was developed using 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulphonic acid) as the colour reagent.
Samples were prepared either by diluting native or SDS-denatured purified gelsolins into sample buffer or by boiling homogenized tissues in 2% (w/v) SDS before dilution.
Pepscan analysis
For the identification of the 2E12 epitope, the sequence of the reactive region of gelsolin was spot-synthesized on cellulose membranes as 15-amino-acid-long peptides with an overlap of 12 amino acids [22]. 2E12 binding to these peptides was monitored as described previously [23,24].
Proteolytic cleavage
Purified porcine plasma gelsolin was cleaved sequentially with aminopeptidase M and with leucine aminopeptidase, both in the ratio 1:100 (w/w) and at 25 °C. The reaction with aminopeptidase M was terminated by the addition of 5 mM 1,10-phenanthroline before adding leucine aminopeptidase. Porcine cytoplasmic gelsolin was cleaved with chymotrypsin in the ratio 1:100 (w/w) and at 25 °C. The reaction was terminated by the addition of 1 mM PMSF followed by immediate boiling.
Molecular modelling
The crystal structure of equine plasma gelsolin [16] [Protein Data Bank no. 1D0N] allowed the 2E12 epitope to be localized structurally. Apart from different N-terminal extensions, porcine and equine gelsolins differ by a single amino acid in segment 1 (amino acid 55 is isoleucine in horse, aspartic acid in pig, cytoplasmic gelsolin numbering). Owing to the 25-amino-acid N-terminal extension of equine gelsolin [25], the amino acid numbering of the (porcine) peptide sequences nominally differs by 25 compared with the equine structure. Since the first 26 residues could not be located in the original crystal structure, the first residue included is Val27, which corresponds to Val2 of cytoplasmic porcine gelsolin. We have modelled a ten-residue N-terminal extension (17–26) on to the crystal structure of equine gelsolin to determine qualitatively whether such an extension would be sufficient to cover and thereby shield the 2E12 epitope in the plasma equine gelsolin. The position of the extension is purely hypothetical and is provided only to elucidate the findings of the present study in a structural sense.
RESULTS
2E12 specifically recognizes gelsolin without an N-terminal extension
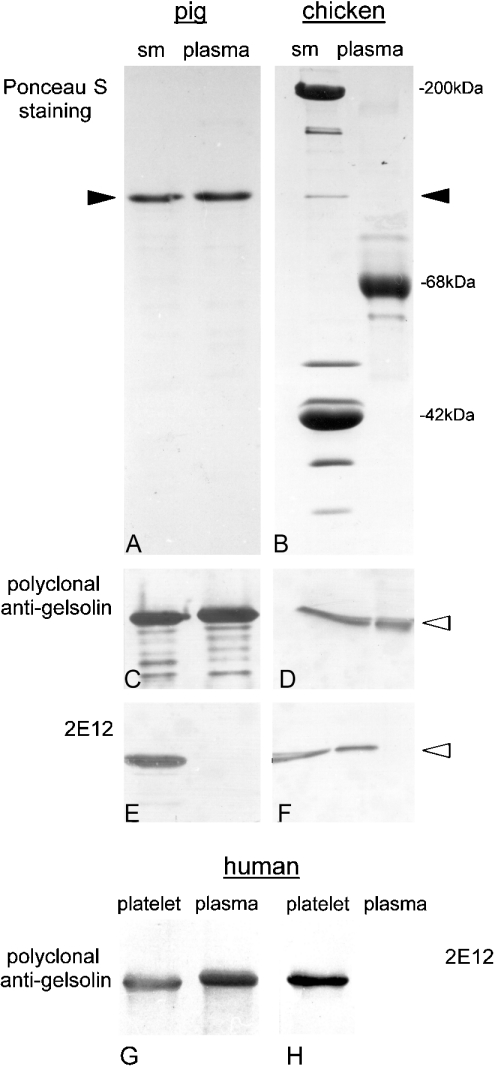
The reactivity of specific gelsolin antibodies with cytoplasmic and plasma gelsolins from pig and chicken respectively is shown in immunoblots in Figures 1(A)–1(F). A polyclonal antibody detected both isoforms from both species (Figures 1C and 1D), whereas the monoclonal antibody 2E12, raised against pig smooth-muscle gelsolin, reacted with the cytoplasmic gelsolin variant of both species and also with chicken plasma gelsolin, but not with pig plasma gelsolin (Figures 1E and 1F). In chicken, cytoplasmic and plasma gelsolins are identical in sequence and size [21,26], whereas pig plasma gelsolin carries a nine-amino-acid-long N-terminal extension [15]. Hence, we concluded that this extension interferes with the interaction between 2E12 and this gelsolin variant. This assumption was supported by the reactivity pattern obtained with human gelsolin. Figures 1(G) and 1(H) show that, whereas the cytoplasmic variant was recognized, plasma gelsolin, containing an N-terminal extension of 25 amino acids, was not labelled by 2E12. Again, the polyclonal antibody used as a reference detected both variants.
Figure 1. 2E12 reactivity is restricted to gelsolins without N-terminal extension.
Purified pig gelsolins from smooth muscles (sm) and blood plasma (A), as well as whole tissue homogenates of chicken gizzard (sm) and total extracts of chicken plasma (B) were subjected to SDS/PAGE, blotted on to nitrocellulose and stained with Ponceau S. Parallel blots were incubated with gelsolin antibodies as indicated (C–F). While the polyclonal antibody recognizes both cytoplasmic and plasma gelsolin (C, D), 2E12 is negative with pig plasma gelsolin (E) but shows a positive reaction with chicken plasma gelsolin (F; see text for explanation). Monoclonal antipig gelsolin 2E12 also discriminates between human cytoplasmic and plasma gelsolins (G, H). Immunoblots with polyclonal (G) and monoclonal (2E12; H) gelsolin antibodies using purified gelsolins from human plasma and human platelets as antigens were performed. Note the absence of 2E12 reactivity with human plasma gelsolin. Filled arrowheads represent expected position of gelsolin after SDS/PAGE and the empty arrowheads represent corresponding positions after immunoblotting.
2E12 can be used to quantify cytoplasmic gelsolin
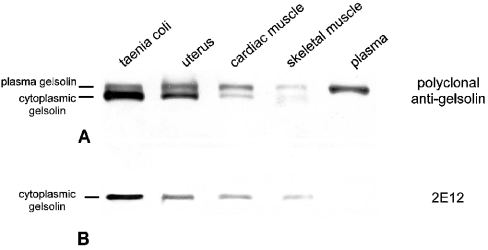
The following experiment was performed with guinea-pig tissue samples to show that the specific detection of cytoplasmic gelsolin by 2E12 is common among mammalian species. Since the molecular masses of intracellular and plasma gelsolins differ by only approx. 1 or 3 kDa in pig and human gelsolin respectively, the isoforms are not well separated by conventional SDS/PAGE. Hence, we applied specific ‘long-run’ conditions for electrophoresis of the guinea-pig tissues. Immunoblotting of these samples with the polyclonal anti-gelsolin revealed that tissues differed both in gelsolin content and in the ratio of cytoplasmic gelsolin to plasma gelsolin, as seen in Figure 2. Cross-striated muscles contained small amounts of total gelsolin and significantly more plasma gelsolin than cytoplasmic gelsolin. In contrast, smooth muscles contained high amounts of total gelsolin and a much higher share of the cytoplasmic variant (Figure 2A). 2E12 specifically detected only the lower band, comprising cytoplasmic gelsolin (Figure 2B). Therefore information obtained from immunoblots with 2E12 can be used to quantify the amounts of cytoplasmic gelsolin even under electrophoresis conditions where both variants are not well separated.
Figure 2. Tissues contain both variants of gelsolin in different amounts.
Immunoblots of homogenates from guinea-pig tissues (40 μg of protein in each lane), separated by SDS/PAGE and incubated with either the polyclonal (A) or monoclonal (2E12; B) anti-gelsolin. Note the double band recognized by the polyclonal antibody, showing different proportions of plasma and cytoplasmic gelsolins. The latter is identified by reactivity with 2E12.
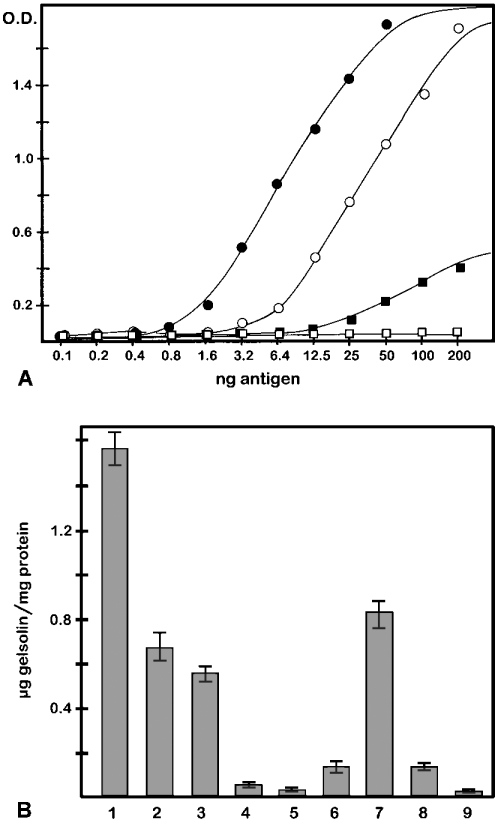
A sandwich ELISA with purified native gelsolins confirmed the high specificity of 2E12 for the cytoplasmic variant of gelsolin (Figure 3A). Plasma gelsolin yielded no signal even at high amounts (200 ng/well). Remarkably, even when gelsolin had been denatured by SDS, 2E12 was capable of discriminating between the two gelsolin variants with considerable specificity. The sensitivity of 2E12 for SDS-denatured cytoplasmic gelsolin was approx. 5-fold higher than that for the native protein, as can be deduced from the half-maximal values in the graphs shown. Denatured plasma gelsolin, at high amounts, showed a weak reaction with 2E12. Again, comparing the half-maximal values, the reactivity of 2E12 with SDS-treated plasma gelsolin was <5% of that seen with the SDS-denatured cytoplasmic protein.
Figure 3. Sandwich ELISA with gelsolin variants reveals high sensitivity and specificity of 2E12 for cytoplasmic gelsolin and can be used for its quantification in various tissues.
(A) Purified cytoplasmic (○, ●) and plasma (□, ■) gelsolins from pig – either as native proteins (○, □) or as SDS-denatured proteins (●, ■) – were applied to microwells precoated with polyclonal anti-gelsolin, then reacted with anti-gelsolin 2E12 and processed as described in the Material and methods section. 2E12 detects both native and SDS-denatured cytoplasmic gelsolins with high sensitivity, but shows only a weak reaction with SDS-denatured plasma gelsolin and no reaction at all with native plasma gelsolin. (B) Concentration of cytoplasmic gelsolin in various porcine tissues. Data were obtained from sandwich ELISAs as shown in (A), using whole tissue homogenates. Numbers refer to stomach smooth muscles (1), intestinal smooth muscles (2), uterus (3), cardiac muscles (4), skeletal muscles (5), brain (6), spleen (7), kidney (8) and erythrocytes (9). Note the high gelsolin concentrations in smooth muscles and spleen, and low to zero levels in striated muscle and erythrocytes, respectively. Values are means±S.E.M. for n=3–5 experiments.
The same assay was used for the quantitative determination of cytoplasmic gelsolin in a variety of tissues, as shown in Figure 3(B). Smooth-muscle tissues and spleen display high levels of cytoplasmic gelsolin: up to 0.1% of total protein. Since the specificity of 2E12 for cytoplasmic gelsolin is retained even in the presence of SDS (see above), SDS-denatured whole tissue homogenates can be used, rather than tissue extracts in buffers without SDS. This is advantageous, as significant amounts of gelsolin are not extracted under non-denaturing conditions (H. Hinssen, unpublished work). Therefore this assay allows the quantification of total cytoplasmic gelsolin in whole tissue homogenates. Furthermore, 2E12 can also be used for selective immunoprecipitation of cytoplasmic gelsolin (results not shown).
Limited proteolysis and pepscan analysis reveal the 2E12 epitope
The phenomenon that a monoclonal antibody exclusively reacts with one isoform of a protein when the non-reactive isoform contains all the sequence information of the reactive isoform is not immediately understood. From the data presented in Figure 1, it seemed plausible that steric hindrance caused by the N-terminal extension of plasma gelsolin is responsible for the observed discrimination between the two variants by 2E12. To challenge this assumption, we used limited proteolysis in two ways: first, to show directly whether the N-terminal extension is involved and, secondly, to assign the 2E12 epitope to one of the six gelsolin domains.
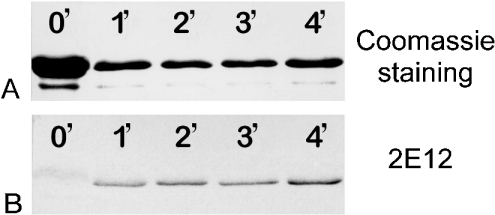
In the first approach, pig plasma gelsolin was incubated with aminopeptidase M, which specifically removes two amino acids from the N-terminal extension VSPMRPSTM and then stops at a proline residue. After inhibiting aminopeptidase M with 1,10-phenanthroline, the protein was incubated with leucine aminopeptidase, which removes two more amino acids and stops at arginine. Figure 4 shows a time sequence of the second reaction after SDS/PAGE. Whereas the Coomassie Blue-stained band remains unchanged within 4 min (Figure 4A), the corresponding immunoblot with 2E12 reveals the appearance of an antibody-reactive band after 1 min, which increases in intensity over time (Figure 4B). This indicates that the removal of no more than the first four amino acids of the N-terminal extension renders pig plasma gelsolin reactive to the 2E12 antibody.
Figure 4. Proteolytic removal of N-terminal amino acids from plasma gelsolin exposes the 2E12-binding site.
Purified pig plasma gelsolin was incubated sequentially with aminopeptidase M (removing the two terminal amino acids from the extension VSPMRPSTM) and leucine aminopeptidase (which removes two more amino acids). Numbers refer to exposure to the leucine aminopeptidase in min. Lane 0′, 6 μg; all other lanes, 2 μg. (A) Coomassie Blue-stained gelsolin band after SDS/PAGE. (B) Immunoblot with 2E12 of the same samples. Note the positive reaction with 2E12 even after a 1 min reaction period, whereas there was no reaction in the 0 min control even with a much higher load.
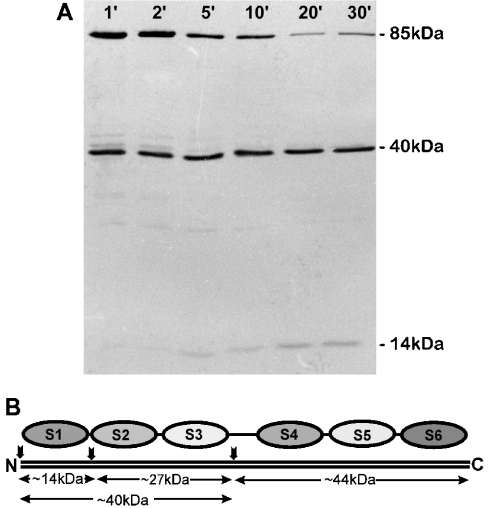
When purified cytoplasmic gelsolin is subjected to limited chymotryptic cleavage, the molecule is initially cleaved into two fragments of similar size, one with an apparent molecular mass of approx. 45 kDa comprising the C-terminal half of the molecule (S4–S6) and a second one of approx. 40 kDa containing the N-terminal part (S1–S3) [27]. With prolonged cleavage time, the 40 kDa fragment is further cleaved into an approx. 14 kDa part (S1) and an approx. 28 kDa part (S2+S3), whereas the C-terminal half of gelsolin is more resistant to chymotrypsin [27]. The immunoblot after SDS/PAGE of such a reaction is shown in Figure 5. A 2E12-reactive 40 kDa band appeared already within 1 min of cleavage and, subsequently, an approx. 14 kDa fragment was seen labelled by 2E12 and increasing in intensity with time, representing the gelsolin segment S1 [27]. These results assign the epitope to the N-terminal half of gelsolin, specifically to the N-terminal segment S1.
Figure 5. The epitope for 2E12 anti-gelsolin is located in the N-terminal approx. 14 kDa polypeptide.
(A) Immunoblot analysis with 2E12 of the chymotryptic cleavage pattern obtained with purified pig cytoplasmic gelsolin at various reaction times. 2E12 reacts with uncleaved gelsolin (85 kDa), a 40 kDa polypeptide and a 14 kDa polypeptide that appears after 5 min exposure to chymotrypsin and increases in intensity with time. (B) Schematic representation of the gelsolin segment structure, showing the 14 and 40 kDa immunoreactive bands to represent S1 and S1–S3 respectively. Note the absence of 2E12 reactivity with a 44 kDa chymotryptic fragment that would comprise S4–S6.
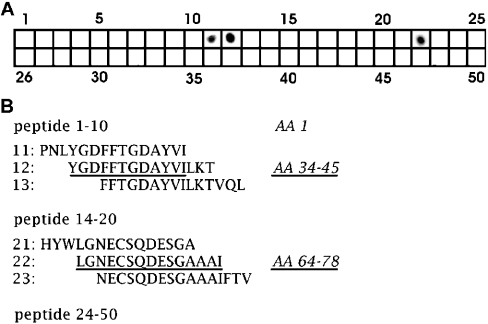
For more detailed information on the 2E12 epitope, we used pepscan analysis. Since the data shown in Figure 5 predicted the epitope location within the domain S1, amino acids 1–160 of cytoplasmic gelsolin were used to synthesize 50 peptides of 15 amino acids each, with an overlap of 12 amino acids. As seen in Figure 6, 2E12 reacted specifically with three of these: peptides 11, 12 and 22. By considering only those amino acids that are common to peptides 11 and 12, two regions in the sequence can be selected that should harbour the epitope for 2E12, amino acids 34–45 and 64–78, the latter being represented by peptide 22. Regarding the sequence, both regions are too far apart for the formation of a sequential epitope, suggesting that the binding epitope is conformational rather than sequential, with both sequences forming a continuous patch in the tertiary structure of gelsolin.
Figure 6. Pepscan analysis and mapping of the 2E12 epitope in cytoplasmic gelsolin.
(A) A dot array of 50 overlapping peptides from the porcine gelsolin sequence was spotsynthesized on cellulose, incubated with 2E12 and processed like an immunoblot. A positive reaction was observed with peptides 11, 12 and 22, covering amino acids 31–48 and 64–78. Since the reaction in both peptides 11 and 12 excludes the non-overlapping amino acids from the binding epitope, the potential binding motif in this part of the sequence is limited to amino acids 34–45. (B) Sequences of some of the peptides in the region where positive reaction was obtained. Taking into consideration the overlap with the non-reactive peptides, those sequence regions that are assumed to contribute to 2E12 binding are shown underlined.
2E12 recognizes a surface-exposed conformational epitope that is obscured by the N-terminal extension
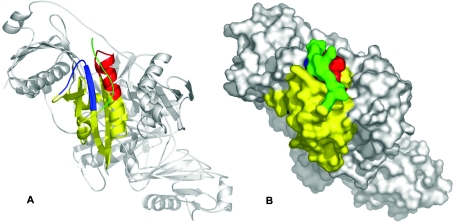
The crystal structure of equine gelsolin [16] indicates that the sequence motifs recognized in the pepscan analysis are adjacent to each other, allowing them to form a single epitope for antibody binding (Figure 7). Considering only surface-accessible amino acids further narrows the putative antibody-binding residues down to 6 and 13 amino acids respectively (results not shown). The 2E12 epitope thus appears to be located within amino acids 37–42 and 65–77 (or amino acids 62–67 and 90–102 for horse plasma gelsolin [16]), as shown in the ribbon model in Figure 7(A).
Figure 7. Cytoplasmic gelsolin, the 2E12-binding epitope and the modelled N-terminal extension.
Based on the crystal structure of horse gelsolin ([16]; PDB code 1D0N), a qualitative model of the N-terminal extension (green) was prepared, indicating that it is structurally feasible for the extension to cover the 2E12 epitope. (A) Ribbon diagram of the entire equine gelsolin molecule including domain S1 (yellow) as well as the 2E12-binding regions [amino acids 34–45 (blue) and amino acids 64–78 (red)] and the modelled N-terminal extension (green). (B) Space-filling model of gelsolin with the inferred position of the N-terminal extension (green). Note that both 2E12-binding regions are mutually adjacent. Images were prepared using Pymol (www.pymol.org).
The 25 N-terminal residues of horse plasma gelsolin were not resolved in the electron density map and thus not included in the published three-dimensional structure of the molecule [16]. Structural models of a complex of human gelsolin segment I with actin also do not include the N-terminal extension [28,29]. However, the resulting N-terminus is located in close proximity to the described antibody-binding patch (Figure 7A). Since the N-terminus of the published molecule starts at Val27 (Val2 of porcine cytoplasmic gelsolin), this structure is identical with the cytoplasmic isoform. We therefore conclude that the N-terminal extension of plasma gelsolin occupies a defined position on the molecular surface close to or overlapping the binding epitope of 2E12 in such a way that it interferes with antibody binding. This would explain all the data reported here. Correspondingly, we have extended the N-terminus by ten amino acids (Program O, [30]) to provide a qualitative structural model (Figures 7A and 7B) that graphically depicts the topological relationship between the N-terminal extension and the published structure [16].
DISCUSSION
In the present study, we describe the properties of a monoclonal antibody directed against cytoplasmic gelsolin. This antibody discriminates between plasma gelsolin and the intracellular gelsolin variant despite the fact that both forms differ only in the N-terminal extension present in plasma gelsolin. This is surprising, as an isoform specificity of antibodies might only be expected for the plasma variant, provided the N-terminal extension served as the immunogenic peptide.
Because of its high specificity for cytoplasmic gelsolin, the antibody is a valuable tool for the quantification and localization of gelsolin in cells and tissues. In contrast with monoclonal antibodies described earlier [31], it permits the monitoring of cytoplasmic gelsolin independent of contaminating or endogenously present plasma gelsolin. It has already been used successfully for immunofluorescence localization of gelsolin in previous studies [32,33], where the presence of plasma gelsolin both in interstitial space and in the cytoplasm presented a problem. A specific detection and quantification of the cytoplasmic variant of gelsolin has been shown earlier at the nucleic acid level [14], but never before at the protein level. Earlier publications have pointed out several problems with gelsolin localization by antibodies. For example, plasma gelsolin present in unfractionated gelsolin antiserum may bind to actin filaments in cells or tissue sections, thus leading to a misdirection of the antibody and incorrect conclusions on the localization of cytoplasmic gelsolin [34].
Two possibilities may explain an exclusive recognition of cytoplasmic gelsolin, as observed here: first, conformational differences may exist between the cytoplasmic variant and the secreted variant and, secondly, the binding epitope may sterically interfere with the N-terminal extension of plasma gelsolin. In fact, conformational differences between both variants do exist due to a disulphide bridge formed between Cys180 and Cys200 in plasma gelsolin. This modification also seems to affect functional properties [35,36]. However, this conformational difference cannot account for the selective reactivity of 2E12, as it is located in subdomain 2. No other conformational differences between the two gelsolin variants have been observed. Therefore it is quite probable that the N-terminal extension is responsible for the inhibition of interaction of 2E12 with plasma gelsolin. This extension differs between species not only in length (e.g. 9 amino acids in pig and 25 amino acids in human) but also in sequence. For example, when compared with the pig plasma gelsolin N-terminal sequence VSPMRPSTM [15], the respective part in mouse plasma gelsolin is VSEARPSTM [37] and in the human counterpart VPEARPNSM [13], with only four (V in position 1, R and P in positions 5 and 6 and M in position 9) out of nine amino acids conserved in all three sequences. It is tempting to speculate that some of these conserved residues are involved in the binding of the N-terminal extension to the core of the molecule. Notably, we have shown that the specificity of the 2E12 antibody for cytoplasmic gelsolin is conserved in samples of human (Figure 1), guinea-pig (see Figure 2) and mouse (results not shown) origin and is therefore not dependent on a specific sequence.
The biochemical data presented here clearly demonstrate that the 2E12 epitope is confined to segment 1 of the gelsolin molecule, despite the fact that the six segments are homologous and have significant sequence similarities [15]. Hence, we could restrict the search for the reactive epitope by pepscan analysis to only the first segment. The reactive peptides were found adjacent to each other in the tertiary structure, indicating that they form a conformational epitope for antibody binding. The topological vicinity of this epitope to the N-terminus of the molecule supports the assumption that the N-terminal extension in plasma gelsolin is responsible for its lack of reactivity with 2E12. This requires that the N-terminal extension of plasma gelsolin takes up a defined, fixed position on the surface of the molecule, rather than existing as a ‘floppy peptide’. Such an assumption is supported by the finding that 2E12 also discriminates between the two gelsolin variants in SDS-denatured samples, both in immunoblots and ELISA, indicating that this topological situation still prevails in denatured samples, either due to an incomplete unfolding of the proteins by SDS or by partial refolding by the antibody after transfer to nitrocellulose or on to the ELISA plates.
Attempts to define the position of the N-terminal extension on the gelsolin surface can lead to ambiguous results. If only the dimensions and geometry of the gelsolin structure and the length of the N-terminal extension are considered, the extension could interact with either segment S1 or S3. The interference with 2E12 antibody binding shown here clearly favours the first possibility, as depicted in our model (Figure 7). Notably, the proposed binding epitope resides within the region of G-actin binding that involves Asp25, Gln70, Gln82, Gln84, Gln85, Gln93 and Arg95 [6]. Of these residues, only Gln70 is shared between the identified 2E12 epitope and the actin-binding site, but there is no direct interference with our modelled N-terminal extension. It is at present unknown whether the N-terminal extension is of any significance for the function of gelsolin, and comparative studies with cytoplasmic and plasma gelsolins need to be performed to clarify whether the presence of the N-terminal extension has any effect on the interaction with actin.
Acknowledgments
We thank R. Frank (Gesellschaft für Biotechnologische Forschung, Braunschweig) for the spot synthesis of gelsolin peptides. This work was supported by the Deutsche Forschungsgemeinschaft (to B.M.J. and H.H.), the ‘Fonds der Chemischen Industrie’ (to B.M.J.) and by a Leopoldina Research Fellowship (BMBF-LPD 9901/8-74; to U.F.).
References
- 1.Burtnick L. D., Robinson R. C., Choe S. Structure and function of gelsolin. Results Probl. Cell Differ. 2001;32:201–211. doi: 10.1007/978-3-540-46560-7_14. [DOI] [PubMed] [Google Scholar]
- 2.Sun H. Q., Yamamoto M., Mejillano M., Yin H. L. Gelsolin, a multifunctional actin regulatory protein. J. Biol. Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 3.McGough A. M., Staiger C. J., Min J. K., Simonetti K. D. The gelsolin family of actin regulatory proteins: modular structures, versatile functions. FEBS Lett. 2003;552:75–81. doi: 10.1016/s0014-5793(03)00932-3. [DOI] [PubMed] [Google Scholar]
- 4.Kwiatkowski D. J. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr. Opin. Cell Biol. 1999;11:103–108. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 5.Hellweg T., Hinssen H., Eimer W. The Ca2+-induced conformational change of gelsolin is located in the carboxyl-terminal half of the molecule. Biophys. J. 1993;65:799–805. doi: 10.1016/S0006-3495(93)81121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson R. C., Mejillano M., Le V. P., Burtnick L. D., Yin H. L., Choe S. Domain movement in gelsolin: a calcium-activated switch. Science. 1999;286:1939–1942. doi: 10.1126/science.286.5446.1939. [DOI] [PubMed] [Google Scholar]
- 7.Lin K. M., Mejillano M., Yin H. L. Ca2+ regulation of gelsolin by its C-terminal tail. J. Biol. Chem. 2000;275:27746–27752. doi: 10.1074/jbc.M003732200. [DOI] [PubMed] [Google Scholar]
- 8.Kolappan S., Gooch J. T., Weeds A. G., McLaughlin P. J. Gelsolin domains 4-6 in active, actin-free conformation identifies sites of regulatory calcium ions. J. Mol. Biol. 2003;329:85–92. doi: 10.1016/s0022-2836(03)00383-8. [DOI] [PubMed] [Google Scholar]
- 9.Choe H., Burtnick L. D., Mejillano M., Yin H. L., Robinson R. C., Choe S. The calcium activation of gelsolin: insights from the 3A structure of the G4-G6/actin complex. J. Mol. Biol. 2002;324:691–702. doi: 10.1016/s0022-2836(02)01131-2. [DOI] [PubMed] [Google Scholar]
- 10.Narayan K., Chumnarnsilpa S., Choe H., Irobi E., Urosev D., Lindberg U., Schutt C. E., Burtnick L. D., Robinson R. C. Activation in isolation: exposure of the actin-binding site in the C-terminal half of gelsolin does not require actin. FEBS Lett. 2003;552:82–85. doi: 10.1016/s0014-5793(03)00933-5. [DOI] [PubMed] [Google Scholar]
- 11.Bryan J. Gelsolin has three actin-binding sites. J. Cell Biol. 1988;106:1553–1562. doi: 10.1083/jcb.106.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khaitlina S., Hinssen H. Ca-dependent binding of actin to gelsolin. FEBS Lett. 2002;521:14–18. doi: 10.1016/s0014-5793(02)02657-1. [DOI] [PubMed] [Google Scholar]
- 13.Kwiatkowski D. J., Stossel T. P., Orkin S. H., Mole J. E., Colten H. R., Yin H. L. Plasma and cytoplasmic gelsolins are encoded by a single gene and contain a duplicated actin-binding domain. Nature (London) 1986;323:455–458. doi: 10.1038/323455a0. [DOI] [PubMed] [Google Scholar]
- 14.Kwiatkowski D. J., Mehl R., Izumo S., Nadal Ginard B., Yin H. L. Muscle is the major source of plasma gelsolin. J. Biol. Chem. 1988;263:8239–8242. [PubMed] [Google Scholar]
- 15.Way M., Weeds A. Nucleotide sequence of pig plasma gelsolin. Comparison of protein sequence with human gelsolin and other actin-severing proteins shows strong homologies and evidence for large internal repeats. J. Mol. Biol. 1988;203:1127–1133. doi: 10.1016/0022-2836(88)90132-5. [DOI] [PubMed] [Google Scholar]
- 16.Burtnick L. D., Koepf E. K., Grimes J., Jones E. Y., Stuart D. I., McLaughlin P., Robinson C. R. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell (Cambridge, Mass.) 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 17.Hinssen H., Small J. V., Sobieszek A. A Ca2+-dependent actin modulator from vertebrate smooth muscle. FEBS Lett. 1984;166:90–95. doi: 10.1016/0014-5793(84)80051-4. [DOI] [PubMed] [Google Scholar]
- 18.Pope B., Gooch J., Hinssen H., Weeds A. G. Loss of calcium sensitivity of plasma gelsolin is associated with the presence of calcium ions during preparation. FEBS Lett. 1989;259:185–188. doi: 10.1016/0014-5793(89)81524-8. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 21.Hinssen H., Vandekerckhove J., Lazarides E. Gelsolin is expressed in early erythroid progenitor cells and negatively regulated during erythropoiesis. J. Cell Biol. 1987;105:1425–1433. doi: 10.1083/jcb.105.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank R. Spot-synthesis. An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- 23.Mayboroda O., Schluter K., Jockusch B. M. Differential colocalization of profilin with microfilaments in PtK2 cells. Cell Motil. Cytoskeleton. 1997;37:166–177. doi: 10.1002/(SICI)1097-0169(1997)37:2<166::AID-CM9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Gonsior S. M., Platz S., Buchmeier S., Scheer U., Jockusch B. M., Hinssen H. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J. Cell Sci. 1999;112:797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- 25.Koepf E. K., Hewitt J., Vo H., Macgillivray R. T., Burtnick L. D. Equus caballus gelsolin – cDNA sequence and protein structural implications. Eur. J. Biochem. 1998;251:613–621. doi: 10.1046/j.1432-1327.1998.2510613.x. [DOI] [PubMed] [Google Scholar]
- 26.Nodes B. R., Shackelford J. E., Lebherz H. G. Synthesis and secretion of serum gelsolin by smooth muscle tissue. J. Biol. Chem. 1987;262:5422–5427. [PubMed] [Google Scholar]
- 27.Chaponnier C., Janmey P. A., Yin H. L. The actin filament-severing domain of plasma gelsolin. J. Cell Biol. 1986;103:1473–1481. doi: 10.1083/jcb.103.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaughlin P. J., Gooch J. T., Mannherz H. G., Weeds A. G. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature (London) 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 29.Irobi E., Burtnick L. D., Urosev D., Narayan K., Robinson R. C. From the first to the second domain of gelsolin: a common path on the surface of actin? FEBS Lett. 2003;552:86–90. doi: 10.1016/s0014-5793(03)00934-7. [DOI] [PubMed] [Google Scholar]
- 30.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Hiyoshi M., Nakao Y., Tatsumi N. Comparison of two monoclonal antibodies, H6B11 and GS2C4, against human plasma gelsolin. Biochem. Mol. Biol. Int. 1994;32:755–762. [PubMed] [Google Scholar]
- 32.Teubner A., Sobek-Klocke I., Hinssen H., Eichenlaub-Ritter U. Distribution of gelsolin in mouse ovary. Cell Tissue Res. 1994;276:535–544. doi: 10.1007/BF00343950. [DOI] [PubMed] [Google Scholar]
- 33.Dissmann E., Hinssen H. Immunocytochemical localization of gelsolin in fibroblasts, myogenic cells, and isolated myofibrils. Eur. J. Cell Biol. 1994;63:336–344. [PubMed] [Google Scholar]
- 34.Carron C. P., Hwo S. Y., Dingus J. U., Benson D. M., Meza I., Bryan J. A re-evaluation of cytoplasmic gelsolin localization. J. Cell Biol. 1986;102:237–245. doi: 10.1083/jcb.102.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen P. G. Functional consequences of disulfide bond formation in gelsolin. FEBS Lett. 1997;401:89–94. doi: 10.1016/s0014-5793(96)01439-1. [DOI] [PubMed] [Google Scholar]
- 36.Wen D., Corina K., Chow E. P., Miller S., Janmey P. A., Pepinsky R. B. The plasma and cytoplasmic forms of human gelsolin differ in disulfide structure. Biochemistry. 1996;35:9700–9709. doi: 10.1021/bi960920n. [DOI] [PubMed] [Google Scholar]
- 37.Dieffenbach C. W., SenGupta D. N., Krause D., Sawzak D., Silverman L. Cloning of murine gelsolin and its regulation during differentiation of embryonal carcinoma cells. J. Biol. Chem. 1989;264:13281–13289. [PubMed] [Google Scholar]