Abstract
Two β-agarase genes, agaA and agaB, were functionally cloned from the marine bacterium Zobellia galactanivorans. The agaA and agaB genes encode proteins of 539 and 353 amino acids respectively, with theoretical masses of 60 and 40 kDa. These two β-agarases feature homologous catalytic domains belonging to family GH-16. However, AgaA displays a modular architecture, consisting of the catalytic domain (AgaAc) and two C-terminal domains of unknown function which are processed during secretion of the enzyme. In contrast, AgaB is composed of the catalytic module and a signal peptide similar to the N-terminal signature of prokaryotic lipoproteins, suggesting that this protein is anchored in the cytoplasmic membrane. Gel filtration and electrospray MS experiments demonstrate that AgaB is a dimer in solution, while AgaAc is a monomeric protein. AgaAc and AgaB were overexpressed in Escherichia coli and purified to homogeneity. Both enzymes cleave the β-(1→4) linkages of agarose in a random manner and with retention of the anomeric configuration. Although they behave similarly towards liquid agarose, AgaAc is more efficient than AgaB in the degradation of agarose gels. Given these organizational and catalytic differences, we propose that, reminiscent of the agarolytic system of Pseudoalteromonas atlantica, AgaA is specialized in the initial attack on solid-phase agarose, while AgaB is involved with the degradation of agarose fragments.
Keywords: β-agarase, agarose, CFB group bacteria, family GH-16, marine bacterium, Zobellia galactanivorans
Abbreviations: AgaAc, catalytic domain of AgaA; DQF-COSY, double-quantum-filtered correlation spectroscopy; ESI, electrospray ionization; HMBC, heteronuclear mutliple bond correlation; HMQC, heteronuclear single quantum correlation; HPAEC, high-performance anion-exchange chromatography; HPAEC-PAD, HPAEC with pulsed amperometric detection; ORF, open reading frame; Q-TOF, quadrupole–time-of-flight
INTRODUCTION
Agars, the main matrix polysaccharides of Ahnfeltiales, Gelidiales and Gracilariales (Rhodophyta), consist of a linear backbone of alternating L- and D-galactose residues linked by α-(1→3) and β-(1→4) linkages respectively, with various substituents such as ester-sulphonic groups, methyl ethers and pyruvic acid [1]. In agarose, the neutral fraction of agar, the α-(1→4)-linked units exhibit 3,6-anhydro bridges which, upon the disorder–order conformational transition, stabilize the molecules into double helices, leading to the formation of pseudo-crystalline fibres made of aggregates of double-stranded agarose chains [2]. Carrageenans, the other category of red algal gel-forming polysaccharides from the orders Gigartinales and Solieriales, differ from agars in the D-configuration of the (1→4)-linked galactose residues and in the density of ester-sulphate substituents per digalactose repeating unit (one in κ-carrageenan, two in ι-carrageenan and three in λ-carrageenan).
A number of micro-organisms have been reported to degrade agarose, including marine bacteria from the genera Pseudomonas or Alteromonas [3–6], Vibrio [7,8] and Cytophaga [9], as well as the land bacterium Streptomyces coelicolor [10]. With the exception of the enzyme from Alteromonas agarilytica [11], they all attack agarose by cleavage of the β-(1→4) linkages, yielding agaro-oligosaccharides in the series related to neoagarobiose [O-3,6-anhydro-α-L-galactopyranosyl-(1→3)-D-galactose]. The agarolytic system of Pseudoalteromonas atlantica first involves the extracellular endo-β-agarase I, which hydrolyses agarose to neoagarotetraose. The end-products of this enzyme are hydrolysed further by two periplasmic enzymes, neoagarotetraose hydrolase (also known as β-agarase II) and neoagarobiose hydrolase, leading eventually to the release of 3,6-anhydro-L-galactose and D-galactose for use as a carbon source [5,12]. A similar mode of agar degradation was reported for Pseudomonas elongata [13] and Cytophaga flavensis [14]. In contrast, Vibrio sp. (strain JT0107) features two extracellular β-agarases, referred to as AgaA and AgaB, which hydrolyse agarose to neoagarobiose and neoagarotetraose respectively [8,15], as well as a periplasmic α-L-galactosidase which hydrolyses the α-(1→3) linkages from the non-reducing ends of neoagaro-oligosaccharides smaller than the hexamer [16].
Protein–polysaccharide interactions have been investigated extensively in the case of neutral glycans such as amylose and cellulose. Far less is known for the case of acidic polysaccharides. In this light, agarases and carrageenases provide an interesting model system to investigate the molecular bases of the solid-phase degradation of sulphated polysaccharides, and we have undertaken to unravel the structural biology of these hydrolases. Five genes encoding proteins with β-agarase activity (EC 3.2.1.81) have been reported so far, falling into three distinct structural families of glycoside hydrolases (http://afmb.cnrs-mrs.fr/CAZY) [17]: the families GH-16 [10,18], GH-50 [15,19] and GH-86 [20]. While families GH-50 and GH-86 encompass only β-agarases, family GH-16 includes glycoside hydrolases with various substrate specificities, namely κ-carrageenases, lichenases (β-1,3–1,4-glucanases), laminarinases (β-1,3-glucanases), xylo-endo-β-1,4-glucan transferases and endo-β-galactosidases [21]. Within this family, a first crystal structure (a curved-β-sandwich) was solved for Bacillus macerans lichenase [22]. We then reported the crystal structure of Pseudoalteromonas carrageenovora κ-carrageenase, which, unlike the lichenases, exhibits a tunnel-shaped active-site cavity. This conspicuous structural feature probably accounts for the processivity of this enzyme [23]. While κ-carrageenases are structurally related to the family GH-16 β-agarases, ι-carrageenases unexpectedly constitute a distinct structural family, GH-82 [24]. These processive glycoside hydrolases fold into a right-handed parallel β-helix [25], which again adopts a tunnelled conformation in the presence of ι-carrageenan [26].
Zobellia galactanivorans, a marine aerobic Gram-negative gliding bacterium isolated from the red alga Delesseria sanguinea and belonging to the family Flavobacteriacea (Bacteroidetes phylum; CFB group) produces various hydrolases specific for the red algal matrix polysaccharides, such as κ- and ι-carrageenases as well as β-agarases [27]. The genes for the κ-carrageenase and ι-carrageenase from Z. galactanivorans have been described previously [21,24]. We report here the functional cloning, overexpression, purification and biochemical characterization of two β-agarases from Z. galactanivorans, referred to as AgaA and AgaB. Sequence analyses indicate that these two β-agarases feature homologous catalytic domains belonging to the family GH-16. This result was recently confirmed by the crystal structures of the recombinant catalytic domain of Z. galactanivorans AgaA and AgaB [28,29]. However, these paralogous proteins behave differently with regard to their interaction with solid-phase agarose, and differ in their overall molecular organization. AgaA is an extracellular, monomeric protein displaying a modular architecture with two additional C-terminal domains of unknown function. In constrast, AgaB is composed of only one, possibly membrane-anchored, domain, and forms a dimeric protein in solution. Given these organizational and catalytic differences, we propose that, reminiscent of the agarolytic system of Pseudoalteromonas atlantica, AgaA is specialized in the initial attack of solid-phase agarose, while AgaB is involved in the degradation of agarose fragments.
EXPERIMENTAL
Cloning of the agaA and agaB genes from the marine bacterium Zobellia galactanivorans
Genomic DNA was obtained as described previously [30] and partially digested by the restriction endonuclease NdeII. Fragments ranging from 4 to 10 kb in size were purified using a 5–40% (w/v) sucrose gradient in 10 mM Tris/HCl, pH 8, 10 mM NaCl and 5 mM EDTA after a centrifugation at 85000 g for 23 h. DNA fragments were ligated into the BamHI site of plasmid pAT153 and used to transform Escherichia coli competent cells (strain DH5α; recA1, endA1, gyrA96, thi1, hsdR17 [rK− mK+], supE44, relA1, LacZ ΔM15), yielding a genomic library of approx. 5000 recombinant clones. Agarase genes were identified by functional cloning as described previously for other gel-degrading enzymes [21,24]. Briefly, the transformed bacteria were picked and grown separately in 96-well plates at 37 °C in Luria–Bertani medium supplemented with ampicillin at 50 μg·ml−1, then plated at 22 °C on Zd medium [21], solidified with 1.5% (w/v) Difco agar. Those clones that had dug a hole in the substratum were considered as harbouring agarase activity. Plasmid DNA was isolated from the agarase-positive clones by the alkaline lysis method [31] and mapped with restriction endonucleases. The fragments were recovered from recombinant plasmids pAC5 and pAC6 using a Geneclean II kit (BIO 101) and subcloned into phagemid pBluescript II KS(−) (Stratagene). Sequencing was performed on both strands by the dideoxy-sequencing method [32]. The deduced amino acid sequences of the agarases from Z. galactanivorans were compared using GAP (Wisconsin Package, version 8; Genetics Computer Group, Madison, WI, U.S.A.), with a gap creation penalty of 3.0 and a gap extension penalty of 0.1. Galactan hydrolases were aligned using PILEUP (Wisconsin Package version 8), with a gap creation penalty of 3.0 and a gap extension penalty of 0.1. The sequences of the agaA and agaB genes were deposited in GenBank under accession numbers AF098954 and AF098955 respectively.
Production and purification of extracellular agarases from Z. galactanivorans
The marine bacterium Zobellia galactanivorans Dsij DSMZ 12802T and CIP 106680T [28] was grown for 18 h at 22 °C in ZoBell medium [33] (2.5 litres) supplemented with 2.50 g·l−1 agar to induce agarase activity. Agarase activity was monitored using a reducing sugar assay (see below). The culture was centrifuged at 10000 g for 20 min and the supernatant was reduced to 50 ml by tangential ultrafiltration (cut-off 10 kDa). Proteins were precipitated with ammonium sulphate and the pellet from 30–85% (w/v) ammonium sulphate saturation was resuspended in 9 ml of buffer I (50 mM Mes buffer, pH 6.2). Sepharose CL6B beads (2 ml) were added and the suspension was shaken gently for 2 h at 4 °C. After centrifugation at 200 g for 1 min, the supernatant was set apart and the beads were washed in 5 ml of buffer I. The supernatant and the wash buffer were pooled and tested for agarase activity. This protein pool was fractionated further on an anion-exchange column (Mono Q HR5; Amersham) with an elution gradient of NaCl (30 ml of buffer I with 0–500 mM NaCl). The protein fractions displaying agarase activity were analysed by SDS/PAGE and isoelectric focusing/PAGE on a 12% (w/v) polyacrylamide gel followed by staining with Coomassie Blue. A 5 ml column (IBF) was packed with the Sepharose beads washed by buffer I. Agaro-oligosaccharides were prepared by enzyme digestion of a 1% (w/v) agarose solution with Z. galactanivorans culture supernatant followed by ultrafiltration through a 10 kDa membrane. The Sepharose column was then eluted with 10 ml of buffer I containing agaro-oligosaccharides (5 mg·ml−1). The elution solution was tested for agarase activity and analysed by SDS/PAGE on 12% (w/v) polyacrylamide gels, followed by staining with Coomassie Blue. The partially purified agarases were separated by electrophoresis on acrylamide gels, stained with 0.003% Amido Black, and the major protein bands were microsequenced by Edman degradation (Pasteur Institute, Paris, France).
Overexpression and purification of recombinant agarases
Plasmids pACP5 and pASP5 were linearized with the restriction endonuclease XhoI. For subcloning of the agaA gene, the primers had the following sequences: 5′ CCCCGATATCGCACAGGACTGGAACGGAATT 3′ (forward primer; corresponding to the N-terminal end without the signal peptide) and 5′ CCCCCTCGAGACTTACCGCAACAGGCTTGTA 3′ (reverse primer; corresponding to the C-terminal end without the final 249 residues) (Figures 1A and 1C). For the agaB gene (Figure 1B), the primers had the following sequences: 5′ CCCCGATATCGGCGACAATTCAAAATTTGAT 3′ (forward primer; N-terminal end without the signal peptide) and 5′ CCCCCTCGAGTTTCTCTACAGGTTTATAGAT 3′ (reverse primer; C-terminal end). For both genes, forward and reverse primers harboured an EcoRV and XhoI site respectively at their N-terminal ends (underlined nucleotides). PCR was carried out in a volume of 100 μl containing 25 ng of template, 1 μM of each of the two specific primers, 250 μM of each dNTP, 2 mM MgSO4, 1×buffer purchased from the manufacturer, and 2 units of Deep Vent polymerase. Conditions were as follows: 10 min at 94 °C, then 35 cycles of 1 min at 94 °C, 1 min at 68 °C and 2.5 min at 72 °C, and finally a polymerization step of 10 min at 72 °C. PCR products were purified by gel electrophoresis and cloned in vector pCR2.1 TOPO (Invitrogen) after treatment with 5 units of Tfl DNA polymerase, 2.5 mM MgSO4 and 0.2 mM dATP for 15 min at 72 °C. ORFs (open reading frames) were purified by hydrolysing the recombinant plasmids with EcoRV and XhoI. The purified fragments were cloned in the pET20b plasmid (Novagen). After sequence validation, plasmids were transformed in E. coli strain Origami(DE3)pLysS {Δara-leu 7697, ΔlacX74, ΔphoAPvuII, phoR, araD139, galE, galK, rspL, F'[lac−(lacIq)pro], gor522::Tn10(TcR), trxB::kan, (DE3), pLysS (CmR)}.
Figure 1. Nucleotide and deduced amino acid sequences of the agaA (A) and agaB (B) genes from Zobellia galactanivorans.
Restriction sites are indicated above the nucleotide sequences. Putative −35 and −10 boxes in the promoter region are shown in bold. The potential ribosome-binding sites referred to as Ω and ε are underlined. Hairpin loops are indicated by arrows. Forward and reverse primers used to overexpress AgaAc and AgaB in E. coli are indicated and underlined. The proposed signal peptides are shown in italics, and the putative lipid-substituted cysteine residue in the signal peptide of AgaB is shown in bold. The underlined amino acids in the AgaA sequence were confirmed by internal peptide microsequencing. The catalytic glutamic acid residues are shown in bold. The general organization of AgaA and AgaB is shown in (C): SP, signal peptide; LP, lipoprotein signal peptide; UNK 1 and 2 refer to domains of unknown function. The amino acid numbers of each domain are indicated.
Recombinant bacteria were grown in a Bioflow 3000 fermentor (New Brunswick Scientific) at 37 °C to D600=0.8 in 5 litres of M9 medium supplemented with 2% (w/v) casaminoacids, 100 μg·ml−1 ampicillin, 35 μg·ml−1 chloramphenicol, 15 μg·ml−1 kanamycin and 15 μg·ml−1 tetracycline, then at 20 °C after addition of ampicillin (500 μg·ml−1). Bacteria were then allowed to grow in the presence of 0.5 mM isopropyl β-D-thiogalactoside for 18 h. The pH was maintained at 7.2 and the oxygen concentration was regulated at 70%. Cells were pelleted at 2000 g for 20 min and pellets stocked at −20 °C. Pellets (from approx. 800 ml of culture) were resuspended in 20 ml of buffer II (50 mM Tris/HCl, pH 8.0, 300 mM NaCl) supplemented with 800 μl of protease inhibitor cocktail (P8849; Sigma), and cells were disrupted using a French press (138 MPa; 20000 p.s.i.). After centrifugation at 20000 g for 1 h, supernatants were loaded on a 3 ml column of nickel–nitrilotriacetic acid (Superflow; Qiagen). After extensive washing with buffer II, the recombinant proteins AgaAc (the catalytic domain of AgaA) and AgaB were eluted in buffer II supplemented with 350 mM and 60 mM imidazole respectively. AgaB was purified further by size-exclusion chromatography with Sephacryl S200 in buffer II.
Size-exclusion chromatography
The apparent molecular masses of purified recombinant proteins AgaAc and AgaB were evaluated by size-exclusion chromatography using a Superdex 200 column with the Äkta system (Amersham Pharmacia Biotech). The column was equilibrated in 50 mM Tris/HCl (pH 8.0) buffer supplemented with 0, 10, 50 or 300 mM NaCl. Samples (50–100 μg) were eluted at a flow rate of 0.75 ml·min−1 with 1.5 column vol. The column was calibrated for each NaCl concentration with cytochrome oxidase (12.4 kDa), carbonic anhydrase (29 kDa), BSA (66 kDa), alcohol dehydrogenase (150 kDa) and β-amylase (200 kDa).
ESI (electrospray ionization)-MS
Prior to mass analysis, samples of AgaAc and AgaB were desalted at 4 °C by dialysis against 10 mM ammonium acetate (pH 6.8) for non-denaturing conditions and deionized water for denaturing conditions. ESI-MS measurements were performed on an ESI-Q-TOF (ESI-quadrupole–time-of-flight) mass spectrometer (Q-TOF II; Micromass, Altrincham, Cheshire, U.K.). Mass spectra were recorded at the exit of the TOF analyser, using the first quadrupole in the ‘RF only’ mode. The purity and homogeneity of each sample were estimated by mass analysis in denaturing conditions (resolution 10 p.p.m.); AgaAc and AgaB in water solution were diluted to 1 pmol·μl−1 in a mixture of water/acetonitrile/formic acid (50:50:0.2, by vol.). Mass spectra were recorded in the positive-ion mode in the mass range 500–2500 m/z, after calibration with horse heart myoglobin diluted to 200 fmol·μl−1 in water/acetonitrile/formic acid (50:50:0.2, by vol.). The accelerating voltage was set at 45 V, and the pressure in the interface of the mass spectrometer was 250 Pa (2.5 mbar). In non-denaturing conditions, the mass measurement of native AgaAc and AgaB was performed in ammonium acetate (pH 6.8). The concentration of the sample after desalting and dilution was estimated to be 2 μM for monomers. The sample was infused continuously into the ESI ion source at the rate of 5 μl·min−1. In order to preserve the integrity of the non-covalent assemblies and to enhance the sensitivity of detection, the pressure in the interface between the atmospheric source and the high vacuum region was increased to 650 Pa (6.5 mbar) by throttling the pumping line. The accelerating voltage applied on the sample cone ranged from 45 to 100 V, and the source and desolvation temperatures were 80 and 120 °C respectively. Data were accumulated over 3 min, and the accumulated spectra were treated with maximum entropy-based software (MaxEnt I) in order to find the approximated mass of each subassembly (monomer and oligomer) and hence the charge on each multiply charged peak. The mass spectra were obtained for a mass range of 35000 to 85000 Da with a resolution of 2 Da.
Measurement of agarase activity
A stock solution of 0.125% (w/v) agarose (Eurogentech) was prepared in 50 mM Mops, pH 7.5, and 300 mM NaCl. Aliquots (995 μl) were boiled and then maintained at 44 °C to prevent the polysaccharide from gelling. These solutions, referred to as melted agarose, were incubated in triplicate at 44 °C with 5 μl of agarase. The amount of reducing sugars released was assayed using a ferricyanide method adapted from that of Kidby and Davidson [34]. The ferricyanide reagent consisted of 300 mg of potassium hexacyanoferrate III and of 28 g of hydrated Na2CO3 dissolved in 1 litre of distilled water, to which 1 ml of 1 M aqueous NaOH was added. Aliquots (100 μl) of the incubation medium were mixed with 900 μl of ferricyanide reagent, and absorbances were recorded at 420 nm. Reducing sugar concentrations were calibrated using neoagarotetraose as standard. One enzyme unit is defined as the amount required to release 1 nM reducing sugar·min−1 under these conditions (1 ml of 0.125% melted agarose, 44 °C, pH 7.5).
The enzymic degradation of solid-phase agarose was also monitored, as follows. A solution of 0.125% (w/v) agarose, 50 mM Mops, pH 7.5, and 300 mM NaCl was first boiled to solubilize the powdered agarose, then left at room temperature to gel, and finally brought to 44 °C. Due to the hysteresis loop in the gel/sol phase transition, the agarose chains remain aggregated at this temperature and form a suspension of micro-gels. Purified recombinant AgaAc and AgaB were added to a final concentration of 4.66 nM to agarose micro-gels (2 ml) and incubated at 44 °C for 8 h. An aliquot (50 μl) of each reaction mixture was taken every 15 min and added to 950 μl of ferricyanide reagent to measure the release of reducing ends.
Kinetic characterization of AgaAc and AgaB
To determine their kinetic parameters, purified AgaAc and AgaB were used at a concentration of 4.66 nM and 7 nM respectively in 50 mM Mops, pH 7.5, and 300 mM NaCl. The protein concentrations were determined by Bradford assay and by absorbance at 280 nm using the molar absorption coefficient equation of Pace and co-workers [35]. The substrate concentrations tested were 0.015, 0.030, 0.060, 0.125, 0.250, 0.500 and 0.75% (w/v) melted agarose. The monomolar concentration of agarose was determined using the molecular mass of the repeating unit, neoagarobiose. Reaction mixtures were incubated at 44 °C for 24 min, and aliquots (100 μl) were taken every 3 min. Experiments were performed in quadruplicate. Values of Vmax, Km and kcat were determined from the Lineweaver–Burk plots.
HPAEC (high-performance anion-exchange chromatography) and 1H-NMR monitoring of the enzymic degradation of melted agarose
The enzymic degradation of melted agarose was monitored by HPAEC and 1H-NMR. Purified recombinant AgaAc or AgaB (100 μl, 100 nM) was added to a solution of melted agarose (9.9 ml; 0.125%, w/v) maintained at 44 °C. An aliquot of the reaction mixture (50 μl) was taken every 30 min, boiled to stop the reaction and filtered (pore size 0.22 μm). Aliquots were then analysed by HPAEC-PAD (HPAEC with pulsed amperometric detection), using a Dionex chromatograph DX 500 equipped with a 50 μl injection loop, a CarboPac PA100 column (4 mm×250 mm) and an electrochemical detector with a gold electrode. Analyses were performed according to a method adapted from Weinberger and co-workers as follows [36]. Elution was carried out at 1 ml·min−1 with a 30 min linear gradient of 0–300 mM sodium acetate in 150 mM NaOH. The column was calibrated with neoagarotetraose and neoagarohexaose standards (Dextra).
For 1H-NMR analyses, agarose powder (10 mg) was melted directly in the NMR tube in the presence of 2H2O and allowed to cool to 45 °C inside a 500 MHz Bruker NMR spectrometer (Service de RMN, Université de Bretagne Occidentale, Brest, France). After recording the spectrum of the substrate, 1 mg of lyophilized recombinant AgaAc or AgaB was then added rapidly to the NMR tube, which was immediately placed back in the spectrometer. 1H-NMR spectra were recorded every 5 min for 2 h, then at 24 h after the addition of the enzyme, i.e. when enzymic hydrolysis was complete and the mutarotation equilibrium had been reached. The NMR signals corresponding to purified agarose di-, tetra- and hexa-saccharides were fully assigned using DQF-COSY (double-quantum-filtered correlation spectroscopy), HMBC (heteronuclear mutliple bond correlation) and HMQC (heteronuclear single quantum correlation) pulse sequences. These data were used to assign the NMR signals of the kinetic spectra.
RESULTS
Functional cloning of the agarase genes agaA and agaB from Zobellia galactanivorans
Within 2 months of plating the Z. galactanivorans genomic library at 22 °C on Zd agar broth, four independent colonies had made a hole in the substratum. The positive clones, referred to as pAC1–pAC4, contained inserts ranging from 6.6 to 9.7 kb. The physical maps of theses clones were not similar; the pAC1 and pAC3 plasmids shared a 5.0 kb SalI–PstI fragment, whereas pAC2 and pAC4 shared a 5.0 kb ClaI–PstI fragment. These two fragments were subcloned into pBluescript. Both subclones, referred to as pASP5 (SalI–PstI fragment) and pACP5 (ClaI–PstI fragment), displayed an agarase+ phenotype.
Plasmids pACP5 and pASP5 were used to determine, on both strands, the nucleotide sequences of the agarase genes of Z. galactanivorans (Figures 1A and 1B). The pACP5 insert was sequenced over 2.98 kbp. It contained a single ORF, 1617 bp in length, referred to as agaA (Figure 1A). Two hexamers, TaGAaA (nt 78–83) and TATAtT (nt 99–104), consistent with E. coli ‘−35’ and ‘−10’ consensus promoters [37], were found 84 nt and 63 nt respectively upstream of the putative start codon (ATG; nt 168–170). In the 3′ untranslated region, a stem–loop (nt 1808–1852) was present downstream of the TAA stop codon (nt 1785–1787), followed by three thymidine residues. Its free energy [ΔG=−96 kJ·mol−1 (−23 kcal·mol−1)] and the presence of thymidine residues suggested that this sequence can function as a site for rho-independent transcriptional termination [37].
The pASP5 insert was sequenced over 2.44 kbp. It contained a single complete ORF, 1059 bp in length, referred to as agaB (Figure 1B). Two putative −35 (TTGAgA; nt 125–130) and −10 (TATtcT; nt 148–153) boxes, separated by 17 nucleotides, were found upstream of the putative start codon of the agaB gene (ATG; nt 197–199). Like the κ- and ι-carrageenase genes of Z. galactanivorans [21,24], both the agaA and agaB genes lack a canonical Shine–Dalgarno sequence. However, upstream of their translation initiator codons, they feature AT-rich sequences, similar to the Ω sequences found in the untranslated 5′ leader region of tobacco mosaic virus [38] and functionally equivalent to Shine–Dalgarno sequences [39], as well as regions similar to the ε sequences of the 5′ untranslated regions of T7 phage gene 10 [40] and of E. coli atpE gene [41], and which are considered as translation enhancers [38]. In the 3′ untranslated region, a possible transcription terminator (nt 1326–1374) was identified downstream of the stop codon (TAA; nt 1256–1258).
The predicted product of the agaA gene is a protein of 539 amino acids, with a theoretical molecular mass of 60 kDa (AgaA). Upon hydropathy analysis, the N-terminus of the protein stands out as a domain with high hydrophobicity, suggesting that this domain is a signal peptide [42]. In accordance with the (−3, −1) rule [42], the most probable site of cleavage by signal peptidase I is between Ala-19 and Ala-20 (Figure 1A). The primary translation product of the agaB gene is a protein with a theoretical molecular mass of 40.68 kDa (AgaB). Its hydropathy profile indicates that the 20 N-terminal amino acids form a very hydrophobic segment. According to Nakai's expert system PSORT [43], this N-terminal hydrophobic segment is predicted to be uncleavable by signal peptidase I. Interestingly, however, this sequence is consistent with the N-terminal signature of prokaryotic lipoproteins. It harbours a consensual pattern for lipid attachment, Leu-Val-Phe-Cys-Cys-Ala-Leu-Leu-Leu-Gly-Cys (Figure 1B), with a probable site of cleavage by signal peptidase II between Gly-17 and Cys-18 (Prosite PS00013; http://www.expasy.org/prosite/). AgaB without its signal peptide (Gly-19–Lys-353) shares clear sequence similarity (44.5% identity) with the N-terminal part of AgaA (Ala-20–Ser-290).
Fractionation of extracellular Z. galactanivorans agarases
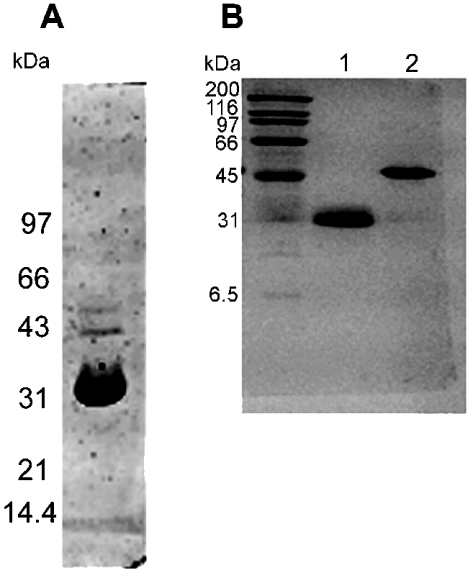
The presence of agar in the culture medium of Z. galactanivorans induced agarolytic activity, which was detected in the culture supernatant. The culture supernatant was fractionated on a column of Sepharose CL6B, i.e. beads of cross-linked agarose. An agarase activity was detected in the flow-through, while elution with neoagaro-oligosaccharides released another protein fraction with agarolytic activity. On SDS/PAGE analysis, the protein fraction that bound to the Sepharose matrix consisted of a major band with a mass of ∼31 kDa (Figure 2A), which was microsequenced by Edman degradation. As the N-terminal sequence was blocked, an internal oligopeptide, Ile-Gln-Tyr-Pro-Val-Tyr-Met-Glu-Ile-Lys, was identified following protein digestion with trypsin. This sequence corresponds to residues Ile-99–Lys-108 of the amino acid sequence deduced from agaA (Figure 1A). The unbound protein fraction was fractionated further on a Mono Q column into three fractions with agarase activity (f1, f2 and f3, eluting at 0, 50 and 200 mM NaCl respectively). On SDS/PAGE analysis, fractions f1 and f2 contained a single protein of ∼30 kDa, while fraction f3 featured a major band also at ∼30 kDa and several additional minor bands. By Edman degradation, the oligopeptide Ala-Thr-Tyr-Asp-Phe-Thr-Gly-Asn-Thr-Pro was identified as the N-terminus of the agarolytic protein present in fraction f2. This sequence matches neither the agaA gene nor the agaB gene. This protein is referred to as AgaC. No putative protein encoded by the agaB gene was identified in the supernatant of Z. galactanivorans.
Figure 2. SDS/PAGE analysis of native and recombinant β-agarases.
(A) SDS/PAGE analysis of the agarose-binding agarase fraction of Zobellia galactanivorans, after elution from a Sepharose CL6B affinity chromatography column. (B) SDS/PAGE analysis of the overexpressed β-agarases AgaAc (lane 1) and AgaB (lane 2) after purification on a nickel affinity chromatography column.
Production of AgaAc and AgaB recombinant agarases
A truncated form of AgaA (Ala-20–Ser-290), referred to as AgaAc, and the entire agaB gene product (Gly-19–Lys-353) were expressed without their own signal peptides in Escherichia coli as fusion proteins with a N-terminal signal peptide from E. coli (PelB) and a C-terminal histidine tag. The recombinant protein AgaAc corresponded to the N-terminal domain of AgaA homologous to the entire AgaB, and it included the internal oligopeptide Ile-Gln-Tyr-Pro-Val-Tyr-Met-Glu-Ile-Lys, microsequenced from the secreted mature protein. The two recombinant proteins were produced at 20 °C in soluble form, with production yields of 1 and 8 mg·l of culture medium−1 respectively. They were purified to electrophoretic homogeneity (Figure 2B) by metal affinity chromatography, followed by size-exclusion chromatography in the case of AgaB. They both clearly displayed agarase activity (specific activities of 160 and 100 units·μg−1 for AgaAc and AgaB respectively).
AgaAc and AgaB cleave the β-(1→4) linkages of agarose in a random manner and with retention of the anomeric configuration
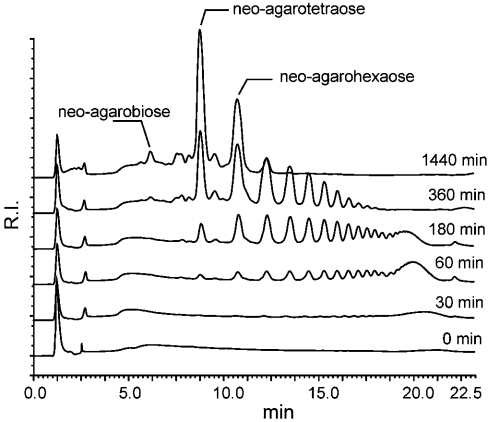
The enzymic mechanism of the recombinant agarases was first investigated using melted agarose maintained at 44 °C and HPAECPAD (Figure 3). AgaAc first produced high-molecular-mass oligosaccharides, which were converted progressively into smaller oligosaccharides, a digestion profile typical of random, endolytic glycoside hydrolases. As shown by 1H-NMR, the hydrolysis products belonged to the neoagarobiose series, indicating that AgaAc specifically cleaves the β-(1→4) linkages of agarose. This β-agarase essentially produced neoagarotetraose and neoagarohexaose as end-products (24 h). Neoagarobiose was also detected, but to a lesser extent. AgaB displayed a similar digestion profile, with its products also belonging to the neoagarobiose series, again indicating that this enzyme cleaves the β-(1→4) linkages of agarose in a random manner. After 24 h of hydrolysis, however, AgaB essentially produced neoagarotetraose and neoagarobiose. This latter oligosaccharide was produced to a greater extent (4-fold) than by AgaAc at the same protein concentration (results not shown).
Figure 3. HPAEC-PAD monitoring of the hydrolysis of agarose by the recombinant β-agarase AgaAc from Zobellia galactanivorans.
R.I., refractive index.
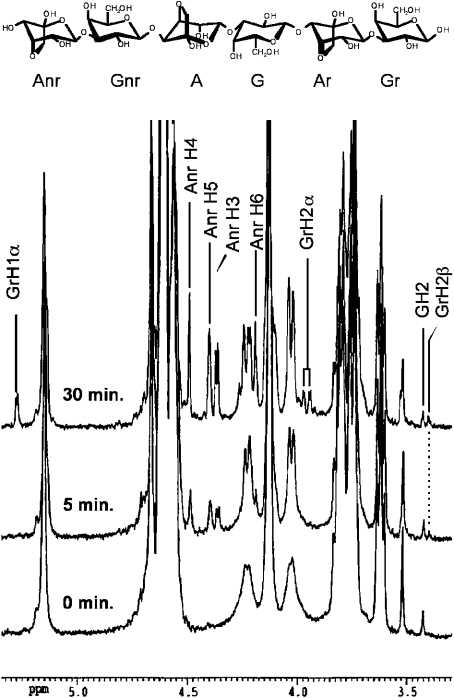
The hydrolysis of melted agarose by AgaAc and AgaB was also monitored by 1H-NMR spectroscopy at 500 MHz (Figure 4). The 1H-NMR signals were assigned on the basis of DQF-COSY, HMBC and HMQC spectra of neoagarotetraose. The 1H-NMR data were identical for both enzymes. Compared with the 1H-NMR spectrum of agarose (Figure 4), the spectra recorded from the reaction mixture (including as early as 5 min after the addition of the enzyme) showed several new resonances. These corresponded on the one hand to protons on the non-reducing ends released by hydrolysis (e.g. Anr H3−H6; Figure 4), and on the other hand to the release of β-anomers (GrH2β). Only after 30 min of hydrolysis was a signal corresponding to the presence of α-anomers (GrH1α) clearly visible (Figure 4). Therefore the β-agarases AgaAc and AgaB retain the anomeric bond configuration, producing β-anomers that give rise progressively to α-anomers when mutarotation takes place.
Figure 4. 1H-NMR monitoring of the hydrolysis of agarose by the recombinant β-agarase AgaAc from Zobellia galactanivorans.
Peak assignments are labelled according to the nomenclature defined in upper panel, where A and G refer to anhydrogalactose and galactose respectively, and nr and r refer to the non-reducing and reducing ends respectively of agarose oligomers.
Catalytic behaviour of AgaAc and AgaB towards liquid- and solid-phase agarose
The enzymic hydrolysis of agarose was also monitored by assaying the reducing sugars released from melted (i.e. liquid) agarose (44 °C). Based on this assay, the optimal pH values for AgaAc and AgaB were 6.0 and 7.0 respectively. Using liquid-phase agarose, both enzymes had degraded ∼100% of the substrate at completion (24 h), and their kinetic parameters were similar, as follows: AgaAc, Km=2±0.15 mM, kcat=150±10 s−1, kcat/Km=75±10 s−1·mM−1; AgaB, Km=1±0.13 mM, kcat=100±10 s−1, kcat/Km=105±20 s−1·mM−1. Moreover, the use of enzyme mixtures with various proportions of the two agarases did not affect significantly the rate or the extent of substrate degradation. However, the two enzymes differed markedly in their degradation of solid-phase agarose gels. Based on the reducing-sugar assay, AgaAc constantly degraded, at completion, 42% of a 0.125% (w/v) agarose gel, whereas AgaB was capable of degrading only 21% of the gel.
Molecular masses of recombinant AgaAc and AgaB agarases
The molecular masses of AgaAc and AgaB were evaluated by size-exclusion chromatography on a Superdex 200 column (Amersham), under various ionic strength conditions. Under all conditions, a single peak was always observed for each protein (results not shown), indicating that AgaAc and AgaB are monodisperse in solution. In the presence of 300 mM NaCl, their apparent molecular masses (AgaAc, 13.4 kDa; AgaB, 60 kDa) differed significantly from the theoretical values. In addition, under conditions of lower ionic strength, their apparent molecular masses increased markedly, to 15, 16 and 27 kDa for AgaAc and to 83, 98 and 120 kDa for AgaB at 50, 10 and 0 mM NaCl respectively.
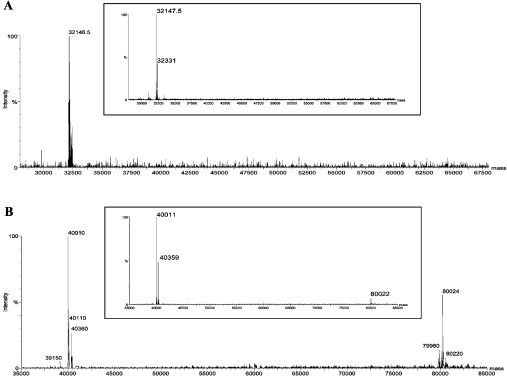
As determined by ESI-MS under denaturing conditions, the molecular masses of AgaAc and AgaB were 32148 Da and 40010 Da respectively, with a resolution better than 10 p.p.m. (Figure 5). These values correspond to the theoretical masses of the recombinant proteins without the PelB sequence, indicating that the E. coli signal peptide was cleaved correctly. AgaAc and AgaB were then analysed by ESI-MS under non-denaturing conditions (Figure 5). AgaAc was again seen as a monodisperse population, with a molecular mass of 32148±2 Da. In contrast, the mass spectrum of AgaB revealed two molecular species, one at 40010±2 Da (62.5% of the total population) and another at 80024±2 Da (37.5%).
Figure 5. ESI-MS spectra of the β-agarases AgaAc (A) and AgaB (B) under non-denaturing (main spectra) and denaturing (inset spectra) conditions.
DISCUSSION
Based on their primary structures, β-agarases AgaA and AgaB share a common, family GH-16, catalytic domain
We show here that the Gram-negative marine bacterium Zobellia galactanivorans produces three different agarases, two of which are encoded by the agaA and agaB genes (Figures 1A and 1B) and one, referred to as AgaC, that features a N-terminal peptide different from those of AgaA and AgaB. These two latter glycoside hydrolases specifically cleave the β-(1→4) linkages of agarose, in a random manner and with retention of the anomeric configuration (Figures 3 and 4). AgaA and AgaC are extracellular, while AgaB is not secreted. This latter observation is consistent with the identification on AgaB of a signal for substitution at Cys-18 by a lipidic moiety (Figure 1C), which could function as a transmembrane anchor. The presence, three residues downstream of this cleavage site, of a negatively charged amino acid, Asp-20, further suggests that the protein is anchored to the plasma membrane [44].
The β-agarases AgaA and AgaB are, in their N-terminal domains, closely related proteins (Figure 1), with identity and similarity scores of 44.5% and 65.7% respectively. Upon BLAST analysis [45] using full-length AgaB as a query, 10 proteins were identified with significant sequence similarities to Z. galactanivorans β-agarases (E-values of 7e-92 to 2e-05). All of these proteins are glycoside hydrolases belonging to family GH-16. The functions of only two of them had been demonstrated previously, namely the β-agarases DagA from Streptomyces coelicolor [10] and AguB from an environmental sample [18]. Other family GH-16 enzymes, such as κ-carrageenases, endogalactosidases and laminarinases, are also structurally related to AgaA and AgaB, but with poor E-values and short sequences of similarity, essentially limited to the catalytic site of family GH-16 enzymes [21]. Therefore the catalytic domains of Z. galactanivorans β-agarases AgaA and AgaB belong to family GH-16 and, together with their 10 related proteins, constitute the GH-16 β-agarase subfamily, as defined by Allouch and co-workers [28].
β-Agarases AgaA and AgaB from Zobellia galactanivorans differ in their molecular organization
AgaA, a monomeric extracellular β-agarase with a modular architecture
As determined by MS, under both denaturing and non-denaturing conditions (Figure 5), the molecular mass of AgaAc was identical to the theoretical molecular mass of the recombinant protein, i.e. 32148 Da, indicating that the catalytic domain of AgaA is a monomeric polypeptide. Compared with the agaA gene product, however, the AgaA protein recovered from the culture medium of Z. galactanivorans (31 kDa) lacked the 245-amino-acid non-catalytic C-terminal domain. Therefore AgaA undergoes a two-step maturation process, involving cleavage of the signal peptide at its N-terminal end and removal of its C-terminal moiety. Such a C-terminal loss was also observed for the κ-carrageenases from Pseudoalteromonas carrageenovora and Z. galactanivorans (96 and 229 amino acids respectively), and we have proposed that this processing is involved in enzyme secretion [21,46]. However, it may seem surprising that as much as 50% of the protein is removed during secretion. The C-terminal domain of Pseudoalteromonas carrageenovora κ-carrageenase has since been identified as a bacterial immunoglobulin-like domain (Pfam family Big 2; http://www.sanger.ac.uk/cgi-bin/Pfam/). Such a module is usually involved in protein–protein interactions, and may locate the κ-carrageenase at the outer membrane surface of this Gram-negative marine bacterium. Similarly, a portion of the C-terminal sequence of Z. galactanivorans κ-carrageenase was recognized as a carbohydrate-binding module belonging to family CBM-16 (http://afmb.cnrs-mrs.fr/CAZY), although its substrate specificity has not been characterized. The search for related proteins using BLAST [45] indicates that the C-terminal sequence of AgaA is composed of two domains (Figure 1C). The first domain encompasses 132 residues (Asp-298–Val-429) and displays 28% identity and 46% similarity with various proteins of unknown function, i.e. BH3963, BH1116 and YesT from the alkalophilic bacteria Bacillus halodurans and YxiM from Bacillus subtilis. The second domain, 106 amino acids long (Asp-434–Gln-539), is 24% identical and 46% similar to a C-terminal domain of unknown function found in the family C10 peptidases PG21, PG57, PG91, PG97, PG99, PG102, HemR and PrtT from Porphyromonas gingivalis, in the putative β-agarases MS109, MS115, MS116 and MS130 from Microscilla spp., in the β-mannanase from Rhodothermus marinus and in the κ-carrageenase from Z. galactanivorans. It is noteworthy that all of these bacteria belong to the CFB phylum, suggesting that this module plays a functional role specific to this lineage of Gram-negative bacteria. These two domains do not belong to any CBM families (http://afmb.cnrs-mrs.fr/CAZY) or to known families of protein–protein interaction domains. Therefore the C-terminal domains of AgaA may represent a new family of protein- or carbohydrate-binding modules, rather than playing a role in the secretion of this enzyme. In this case, the truncated enzyme found in the bacterial culture medium would result from proteolysis at the level of the glycine-rich linker. To identify their function, the two C-terminal domains were cloned individually in the pET43 vector. Unfortunately, these recombinant proteins were expressed as inclusion bodies even at low temperature (results not shown).
AgaB, a dimeric β-agarase
Irrespective of the ionic strength conditions, the apparent molecular mass of AgaB as determined by gel filtration, i.e. 90 kDa on average, was far above its expected mass as a monomer, i.e. 40010 Da, suggesting that this protein is a dimer in solution. This was confirmed by MS measurements under non-denaturing conditions, showing that a significant proportion of the molecule remained dimeric, with a molecular mass of 80024±2 Da (Figure 5). The observation that a significant proportion (62.5%) of the molecule was monomeric under these ESI-MS analysis conditions was due to the fact that dimer dissociation was not completely prevented by the slower decreasing of the pressure gradient. Thus, in contrast with AgaAc, a monomeric protein, the β-agarase AgaB from Z. galactanivorans is a dimer in solution. This unusual situation for two paralogous proteins is not without precedent: the quinate/shikimate dehydrogenase YdiB from E coli was recently characterized as a dimeric protein, while its paralogue, the shikimate dehydrogenase AroE, is a monomer [47]. To our knowledge, AgaB is the first dimeric protein reported in the whole GH-B- clan (which encompasses families GH-7 and GH-16).
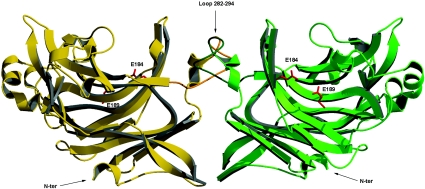
This result led us to reconsider the biological units present in the AgaB crystal [28]. This structure was solved by molecular replacement at 2.3 Å (0.23 nm) resolution using AgaAc as search model, and the asymmetric unit was depicted as containing four AgaB molecules. Analysis of the different protein–protein interfaces within the monoclinic crystals of AgaB shows that the largest contact surface area is between two molecules in the asymmetric unit, which thus contains two identical dimers (PDB ID code 1O4Z; http://www.rcsb.org/pdb/). Whereas each monomer subunit has a globular form, the dimer features a slightly elongated shape, with dimensions of 100 Å×58 Å×49 Å (Figure 6). The two AgaB molecules in one dimer are related by a quasi two-fold non-crystallographic symmetry axis, which is typical of biological dimers [48]. The positioning of the monomers in the dimer is such that the substrate-binding clefts of the two monomer are on the same side, tilted by approx. 45°. The dimerization of AgaB does not preclude its putative anchoring to the membrane, as the N-termini of both monomers are located on the same side, opposite to the active-site clefts (Figure 6).
Figure 6. Ribbon diagram of the dimer unit observed in the crystal structure of AgaB.
Monomer A in shown in yellow, and monomer B in green. The loops coloured dark green and dark yellow respectively are the main contributors to the inter-subunit interface. The active sites are marked by the red catalytic glutamic acid residues. This figure was prepared with Molscript [50].
Analysis of the dimer interface with the Protein–Protein Interaction Server (http://www.biochem.ucl.ac.uk/bsm/PP/server/) reveals typical characteristics of specific dimer interactions [48]. The surface area accessible to solvent (1.4 Å probe radius) in the isolated monomer A is 13070 Å2, while it is 12954 Å2 in monomer B. The surface area buried by the dimer interface is 1140 Å2 (≈570 Å2/monomer), which corresponds to 4% of the monomer solvent-exposed surface. Interestingly, it is one of the additional loops, comprising residues 282–294 in AgaB, that contributes primarily to the dimer interface. This large loop between α-helix 3 and β-strand 20 in monomer A (Figure 6) interacts with the corresponding loop in monomer B. Several hydrogen bonds are formed across the dimer interface, involving equivalent residue pairs: Tyr-155OH (A)–Trp-255NE1 (B), Tyr-155OH (A)–Arg-294O (B), Asp-292O (A)–Glu-300N (B), Asn-298ND2 (A)–Arg-294O (B), and Asn-298ND2 (A)–Thr-293O (B). Furthermore, hydrophobic contacts are formed between Lys-272 and Pro-254 from one monomer and Val-154 and Thr-283 respectively from the other monomer. These interactions are symmetrical and exist in both directions. No salt bridges are observed at the dimer interface. Thus 60% of the monomer contacts (closer than 4.0 Å) in the dimer interface involve hydrophobic interactions, while 40% involve polar interactions.
β-Agarases AgaA and AgaB from Zobellia galactanivorans differ in their behaviour towards solid agarose
In spite of the structural differences discussed above, and as indicated by their kcat/Km ratios when acting on melted agarose, AgaB was at least as efficient as AgaA in the degradation of agarose in the liquid phase. That AgaA and AgaB behave similarly in the presence of liquid agarose is substantiated further by their equal capacity to completely degrade the polysaccharide in solution, as well as by the absence of synergy between the two enzymes on melted agarose. However, and even though AgaA and AgaB display quasi-identical active sites, with eight subsites [28], AgaB has a greater efficiency to cleave neoagarohexaose into neoagarotetraose and neoagarobiose (present study and [28]).
It is also noteworthy that the proteins showed different behaviour in their interaction with solid-phase agarose. In gel filtration experiments involving Superdex 200, i.e. a chromatography matrix composed of dextran mixed with cross-linked agarose, AgaAc and AgaB displayed abnormally high retention times, suggesting that the proteins were retained by the matrix. The structural data on AgaAc and AgaB indicate that these proteins interact with agarose through hydrophobic interactions [28,29]. At low ionic strength, i.e. conditions where hydrophobic interactions are known to be weaker, the apparent molecular masses of AgaAc and AgaB did indeed increase. Interestingly, however, in those very ionic conditions (50 mM NaCl) where the estimation of the molecular mass of AgaB was correct, the apparent molecular mass of AgaA (15 kDa) was nevertheless substantially underestimated, indicating that this protein was bound to agarose more strongly than was AgaB. This is consistent with the retention of AgaA on Sepharose beads during fractionation of the bacterial culture medium, and it was further confirmed by our finding that AgaAc was twice as efficient as AgaB in the degradation of agarose gels. Altogether, AgaA appears more adapted than AgaB to bind and to degrade agarose in the solid phase.
The presence of a secondary agarose-binding site located at the outer, non-catalytic face of AgaAc [29] probably explains the higher efficiency of this enzyme in the degradation of agarose gels. Allouch and co-workers [29] suggested that this second, parallel, binding site may allow the enzyme to unwind the double-helical structure of agarose prior to catalytic cleavage. Since the agarose gel is composed of crystalline aggregates of double helices, the non-catalytic binding site is also likely to be involved in the dissociation of the agarose fibre, a crucial step before double-helix opening. Such a role for an additional carbohydrate-binding site was also proposed for the enzymic dissociation of ι-carrageenan and dermatan sulphate fibres by the β-helical ι-carrageenase and chondroitin B lyase respectively [26,49]. The residues involved in the surface agarose-binding site of AgaAc are not conserved in either AgaB or any other family GH-16 β-agarases [29]. Therefore the absence of a surface binding site in AgaB would explain its limited efficiency in degrading agarose gels. Altogether, considering their likely differential localization (extracellular versus membrane-anchored), AgaA and AgaB thus probably differ in their biological functions, with AgaA being specialized in the initial degradation of solid-phase agarose, while AgaB is involved in the subsequent degradation of agarose fragments.
Acknowledgments
A doctoral fellowship from the Conseil Régional de Bretagne to D.F. is gratefully acknowledged. We thank Dr Bernard Henrissat for helpful discussions, and Nelly Kervarec (UBO, Brest) for recording NMR spectra. We are grateful to Dr Frank Zal, who is in charge of the MS plate-form set-up in the Station Biologique de Roscoff. We also thank Waters Company for the special design of this instrument to allow the analysis of non-covalent complexes.
References
- 1.Craigie J. Cell walls. In: Cole K. D., Sheath R. G., editors. Biology of the Red Algae. Cambridge: Cambridge University Press; 1990. pp. 221–257. [Google Scholar]
- 2.Arnott S., Fulmer A., Scott W. E., Dea I. C., Moorhouse R., Rees D. A. The agarose double helix and its function in agarose gel structure. J. Mol. Biol. 1974;90:269–284. doi: 10.1016/0022-2836(74)90372-6. [DOI] [PubMed] [Google Scholar]
- 3.Day D. F., Yaphe W. Enzymatic hydrolysis of agar: purification and characterization of neoagarobiose hydrolase and p-nitrophenyl alpha-galactoside hydrolase. Can. J. Microbiol. 1975;21:1512–1518. doi: 10.1139/m75-223. [DOI] [PubMed] [Google Scholar]
- 4.Groleau D., Yaphe W. Enzymatic hydrolysis of agar: purification and characterization of beta-neoagarotetraose hydrolase from Pseudomonas atlantica. Can. J. Microbiol. 1977;23:672–679. doi: 10.1139/m77-100. [DOI] [PubMed] [Google Scholar]
- 5.Morrice L. M., McLean M. W., Williamson F. B., Long W. F. Beta-agarases I and II from Pseudomonas atlantica. Purifications and some properties. Eur. J. Biochem. 1983;135:553–558. doi: 10.1111/j.1432-1033.1983.tb07688.x. [DOI] [PubMed] [Google Scholar]
- 6.Vera J., Alvarez R., Murano E., Slebe J. C., Leon O. Identification of a marine agarolytic pseudoalteromonas isolate and characterization of its extracellular agarase. Appl. Environ. Microbiol. 1998;64:4378–4383. doi: 10.1128/aem.64.11.4378-4383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki T., Araki T., Kitamikado M. Purification and characterization of a novel beta-agarase from Vibrio sp. AP-2. Eur. J. Biochem. 1990;187:461–465. doi: 10.1111/j.1432-1033.1990.tb15326.x. [DOI] [PubMed] [Google Scholar]
- 8.Sugano Y., Terada I., Arita M., Noma M., Matsumoto T. Purification and characterization of a new agarase from a marine bacterium, Vibrio sp. strain JT0107. Appl. Environ. Microbiol. 1993;59:1549–1554. doi: 10.1128/aem.59.5.1549-1554.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Meulen H. J., Harder W. Production and characterization of the agarase of Cytoplaga flevensis. Antonie Van Leeuwenhoek. 1975;41:431–447. doi: 10.1007/BF02565087. [DOI] [PubMed] [Google Scholar]
- 10.Buttner M. J., Fernley I. M., Bibb M. The agarase gene (dagA) of Streptomyces coelicolor (A3)2: nucleotide sequence and transcriptional analysis. Mol. Gen. Genet. 1987;209:101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- 11.Potin P., Richard C., Rochas C., Kloareg B. Purification and characterization of the alpha-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur. J. Biochem. 1993;214:599–607. doi: 10.1111/j.1432-1033.1993.tb17959.x. [DOI] [PubMed] [Google Scholar]
- 12.Morrice L. M., McLean M. W., Long W. F., Williamson F. B. Beta-agarases I and II from Pseudomonas atlantica. Substrate specificities. Eur. J. Biochem. 1983;137:149–154. doi: 10.1111/j.1432-1033.1983.tb07808.x. [DOI] [PubMed] [Google Scholar]
- 13.Vattuone M. A., de Flores E. A., Sampietro A. R. Isolation of neoagarobiose and neoagarotetraose from agarose digested by Pseudomonas elongata. Carbohydr. Res. 1975;39:164–167. doi: 10.1016/s0008-6215(00)82653-1. [DOI] [PubMed] [Google Scholar]
- 14.Van der Meulen H. J., Harder W. Characterization of the neoagarotetra-ase and neoagarobiase of Cytophaga flevensis. Antonie Van Leeuwenhoek. 1976;42:81–94. doi: 10.1007/BF00399451. [DOI] [PubMed] [Google Scholar]
- 15.Sugano Y., Matsumoto T., Noma M. Sequence analysis of the agaB gene encoding a new beta-agarase from Vibrio sp. strain JT0107. Biochim. Biophys. Acta. 1994;1218:105–108. doi: 10.1016/0167-4781(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 16.Sugano Y., Kodama H., Terada I., Yamazaki Y., Noma M. Purification and characterization of a novel enzyme, alpha-neoagarooligosaccharide hydrolase (alpha-NAOS hydrolase), from a marine bacterium, Vibrio sp. strain JT0107. J. Bacteriol. 1994;176:6812–6818. doi: 10.1128/jb.176.22.6812-6818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coutinho P. M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G. J., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 18.Voget S., Leggewie C., Uesbeck A., Raasch C., Jaeger K. E., Streit W. R. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 2003;69:6235–6242. doi: 10.1128/AEM.69.10.6235-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugano Y., Matsumoto T., Kodama H., Noma M. Cloning and sequencing of agaA, a unique agarase 0107 gene from a marine bacterium, Vibrio sp. strain JT0107. Appl. Environ. Microbiol. 1993;59:3750–3756. doi: 10.1128/aem.59.11.3750-3756.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belas R. Sequence analysis of the agrA gene encoding beta-agarase from Pseudomonas atlantica. J. Bacteriol. 1989;171:602–605. doi: 10.1128/jb.171.1.602-605.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbeyron T., Gerard A., Potin P., Henrissat B., Kloareg B. The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 1998;15:528–537. doi: 10.1093/oxfordjournals.molbev.a025952. [DOI] [PubMed] [Google Scholar]
- 22.Keitel T., Simon O., Borriss R., Heinemann U. Molecular and active-site structure of a Bacillus 1,3-1,4-beta-glucanase. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5287–5291. doi: 10.1073/pnas.90.11.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel G., Chantalat L., Duee E., Barbeyron T., Henrissat B., Kloareg B., Dideberg O. The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of Clan-B glycoside hydrolases. Structure. 2001;9:513–525. doi: 10.1016/s0969-2126(01)00612-8. [DOI] [PubMed] [Google Scholar]
- 24.Barbeyron T., Michel G., Potin P., Henrissat B., Kloareg B. iota-Carrageenases constitute a novel family of glycoside hydrolases, unrelated to that of kappa-carrageenases. J. Biol. Chem. 2000;275:35499–35505. doi: 10.1074/jbc.M003404200. [DOI] [PubMed] [Google Scholar]
- 25.Michel G., Chantalat L., Fanchon E., Henrissat B., Kloareg B., Dideberg O. The iota-carrageenase of Alteromonas fortis. A beta-helix fold-containing enzyme for the degradation of a highly polyanionic polysaccharide. J. Biol. Chem. 2001;276:40202–40209. doi: 10.1074/jbc.M100670200. [DOI] [PubMed] [Google Scholar]
- 26.Michel G., Helbert W., Kahn R., Dideberg O., Kloareg B. The structural bases of the processive degradation of iota-carrageenan, a main cell wall polysaccharide of red algae. J. Mol. Biol. 2003;334:421–433. doi: 10.1016/j.jmb.2003.09.056. [DOI] [PubMed] [Google Scholar]
- 27.Barbeyron T., L'Haridon S., Corre E., Kloareg B., Potin P. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of. Int. J. Syst. Evol. Microbiol. 2001;51:985–997. doi: 10.1099/00207713-51-3-985. [DOI] [PubMed] [Google Scholar]
- 28.Allouch J., Jam M., Helbert W., Barbeyron T., Kloareg B., Henrissat B., Czjzek M. The three-dimensional structures of two beta-agarases. J. Biol. Chem. 2003;278:47171–47180. doi: 10.1074/jbc.M308313200. [DOI] [PubMed] [Google Scholar]
- 29.Allouch J., Helbert W., Henrissat B., Czjzek M. Parallel substrate binding sites in a beta-agarase suggest a novel mode of action on double-helical agarose. Structure. 2004;12:623–632. doi: 10.1016/j.str.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J. Bacteriol. 1984;160:586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ZoBell C. E. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 1941;4:41–75. [Google Scholar]
- 34.Kidby D. K., Davidson D. J. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal. Biochem. 1973;55:321–325. doi: 10.1016/0003-2697(73)90323-0. [DOI] [PubMed] [Google Scholar]
- 35.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberger F., Richard C., Kloareg B., Kashman Y., Hoppe H.-G., Friedlander M. Structure-activity relationships of oligoagar elicitors toward Gracilaria conferta (Rhodophyta) J. Phycol. 2001;37:418–426. [Google Scholar]
- 37.Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 38.Gallie D. R., Walbot V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanov I. G., Alexandrova R. A., Dragulev B. P., AbouHaidar M. G. A second putative mRNA binding site on the Escherichia coli ribosome. Gene. 1995;160:75–79. doi: 10.1016/0378-1119(95)00134-r. [DOI] [PubMed] [Google Scholar]
- 40.Olins P. O., Rangwala S. H. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. J. Biol. Chem. 1989;264:16973–16976. [PubMed] [Google Scholar]
- 41.McCarthy J. E., Schairer H. U., Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985;4:519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 43.Nakai K., Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 44.Gennity J. M., Inouye M. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 1991;266:16458–16464. [PubMed] [Google Scholar]
- 45.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potin P., Richard C., Barbeyron T., Henrissat B., Gey C., Petillot Y., Forest E., Dideberg O., Rochas C., Kloareg B. Processing and hydrolytic mechanism of the cgkA-encoded kappa-carrageenase of Alteromonas carrageenovora. Eur. J. Biochem. 1995;228:971–975. doi: 10.1111/j.1432-1033.1995.tb20348.x. [DOI] [PubMed] [Google Scholar]
- 47.Michel G., Roszak A. W., Sauve V., Maclean J., Matte A., Coggins J. R., Cygler M., Lapthorn A. J. Structures of shikimate dehydrogenase AroE and its paralog YdiB. A common structural framework for different activities. J. Biol. Chem. 2003;278:19463–19472. doi: 10.1074/jbc.M300794200. [DOI] [PubMed] [Google Scholar]
- 48.Lo Conte L., Chothia C., Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 49.Michel G., Pojasek K., Li Y., Sulea T., Linhardt R. J., Raman R., Prabhakar V., Sasisekharan R., Cygler M. The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J. Biol. Chem. 2004;279:32882–32896. doi: 10.1074/jbc.M403421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of proteins structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]