Abstract
Human Inborn Errors of Immunity (IEIs) encompass a clinically and genetically heterogeneous group of disorders, ranging from mild cases to severe, life-threatening types. Among these, Primary Immune Regulatory Disorders (PIRDs) constitute a subset of IEIs characterized by diverse clinical phenotypes, prominently featuring severe atopy, autoimmunity, lymphoproliferation, hyperinflammation, autoinflammation, and susceptibility to malignancies. According to the latest report from the International Union of Immunological Societies (IUIS), PIRDs arise from mutations in various genes including LYST, RAB27A, AP3B1, AP3D1, PRF1, UNC13D, STX11, STXBP2, FAAP24, SLC7A7, RASGRP1, CD70, CTPS1, RLTPR, ITK, MAGT1, PRKCD, TNFRSF9, SH2DIA, XIAP, CD27 (TNFRSF7), FAS (TNFRSF6), FASLG (TNFSF6), CASP10, CASP8, FADD, LRBA, STAT3, AIRE, ITCH, ZAP70, TPP2, JAK1, PEPD, FOXP3, IL2RA, CTLA4, BACH2, IL2RB, DEF6, FERMT1, IL10, IL10RA, IL10RB, NFAT5, TGFB1, and RIPK1 genes. We designed a targeted next-generation sequencing (TNGS) workflow using the Ion AmpliSeq™ Primary Immune Deficiency Research Panel to sequence 264 genes associated with IEIs on the Ion S5™ Sequencer. In this study, we report the identification of 38 disease-causing variants, including 16 novel ones, detected in 40 patients across 15 distinct PIRD genes. The application of next-generation sequencing enabled rapid and precise diagnosis of patients with PIRDs.
Keywords: Next-generation sequencing, PIRD, Novel mutation
Introduction
Inborn Errors of Immunity (IEIs) are a group of genetically and phenotypically heterogeneous inherited disorders. The identification of monogenic defects underlying IEIs has steadily increased over time [1]. In 2022, the count of known IEIs rose to 485, and the classification was updated to encompass more than 450 distinct gene defects [2]. Human IEIs are categorized into 10 groups based on shared pathogenesis and immune system components involved [2, 3]. As per the European Society for Immunodeficiencies (ESID), Primary Immune Regulatory Disorders (PIRDs) account for around 5.3% of IEIs, with 45 disease-causing genes currently categorized across four classes as follows: hemophagocytic lymphohistiocytosis (HLH), susceptibility to EBV, syndromes involving autoimmunity, and immune dysregulation with colitis [2–4] (refer to Table 1).
Table 1.
Diseases of immune dysregulation. A Hemophagocytic lymphohistiocytosis and EBV susceptibility. B Syndromes with autoimmunity and others
| A. Hemophagocytic lymphohistiocytosis and EBV susceptibility | |||
| HLH | Susceptibility to EBV | ||
| Hypopigmentation | Familial hemophagocytic lymphohistiocytosis syndromes | RASGRP1 deficiency RASGRP1 AR | EBV-associated HLH |
| Chediak Higashi syd. LYST AR | Perforin deficiency (FHL2) PRF1 AR | CD70 deficiency CD70 AR | XLP1 SH2DIA XL |
| Griscelli syd type 2 RAB27a AR | UNC13D/Munc13-4 deficiency (FHL3)UNC13D AR | CTPS1 deficiency CTPS1 AR | XLP2 XIAP XL |
| Hermansky Pudlak syd type 2 AP3B1 AR | Syntaxin 11 deficiency (FHL4) STX11 AR | RLTPR (CARMIL2) deficiency RLTPR AR | CD27 deficiency CD27(TNFRSF7) AR |
| Hermansky-Pudlak syd type 10 AP3D1 AR | STXBP2/Munc18-2 deficiency (FHL5) STXBP2 AR | ITK deficiency ITK AR | |
| FAAP24 deficiency FAAP24AR/AD | MAGT1 deficiency (XMEN) MAGT1 XL | ||
| SLC7A7 deficiencySLC7A7 AR | TET2 deficiency TET2 AR LOF | ||
| CDC42 deficiency (NOCARH syndrome) CDC42 AD | PRKCD deficiency PRKCD AR | ||
| RHOG deficiency AR | CD137 deficiencyTNFRSF9 AR | ||
| B. Syndromes with autoimmunity and others | |||
| Syndromes with autoimmunity | Immune dysregulation with colitis (IBD) | ||
| Increased CD4-CD8-TCR α/β double negative (DN) T cells? | IL-10 deficiency IL10 AR | ||
| Yes | No: regulatory T-cell defect? |
IL-10Ra deficiency IL10RA AR IL-10Rb deficiency IL10RB AR |
|
| No | Yes | ||
| ALPS | APECED (APS-1) AIRE AR/AD | IPEX FOXP3 XL | IL21 deficiency IL21AR |
| ALPS-FAS. TNFRSF6 AD | ITCH deficiency ITCH AR | CD25 deficiency IL2RA AR | NFAT5 haploinsufficiency NFAT5 AD |
| ALPS-FASLG TNFSF6 AR | ZAP-70 ZAP70 AR (LOF/GOF) | CTLA4 deficiency CTLA4 AD | TGFB1 deficiency TGFB1 AR |
| ALPS-Caspase10 CASP10 AD | Tripeptidyl Peptidase II deficiency TPP2 AR | BACH2 deficiencyBACH2 AD | RIPK1 deficiency RIPK1 AR |
| JAK1 GOF JAK1 AD | STAT3 GOF mutationsSTAT3 AR | ELF4 deficiency ELF4 XL | |
| Prolidase deficiency PEPD AR | CD122 deficiency IL2RB AR | ||
| Caspase 8 CASP8 AR | FERMT1 deficiency FERMT1 AR | ||
| FADD deficiency FADD AR | DEF6 deficiency DEF6 AR | ||
| SOCS1 deficiency SOCS1 AD | LRBA deficiency LRBA AR | ||
| IKAROS GOF IKZF1 AD | |||
ALPS autoimmune lymphoproliferative syndrome, HLH hemophagocytic lymphohistiocytosis, AD autosomal dominant transmission, XL X-linked transmission, AR autosomal recessive transmission, EBV Epstein–Barr virus, AD autosomal dominant transmission, XL X-linked transmission, AR autosomal recessive transmission, GOF gain of function, LOF loss of function
PIRDs constitute a growing collection of diseases arising from gene defects across various immune pathways, notably those affecting regulatory T-cell function. Unlike classical IEIs, PIRDs may present with autoimmune symptoms like cytopenia, enteropathy, and dermatitis as initial indications of the disorder [4–6].
Though IEIs are rare globally, they exhibit higher prevalence in regions with significant consanguinity due to the prevalence of autosomal recessive IEIs [7]. The exact incidence of IEIs in Turkey remains uncertain due to the absence of comprehensive national disease registries and limited incidence studies. Nonetheless, given the elevated rates of consanguineous marriages in our country, the incidence of both IEIs and PIRDs is anticipated to surpass the reported Figs. [8–10].
The advent of Next-Generation Sequencing (NGS) has ushered in a new era in genomics research, offering unparalleled capabilities in terms of speed, cost-effectiveness, and throughput. In the context of PIRD, understanding molecular genetics is essential, and NGS stands as a powerful tool to unravel the complexities associated with these disorders. NGS significantly enhances diagnostic accuracy by providing a comprehensive genetic profile, minimizing the risk of misdiagnosis. This precision in diagnosis is paramount for effective patient management and timely interventions.
The substantial surge in recognized monogenic inborn errors of immunity disorders in recent years can be attributed to the wider adoption of next-generation sequencing (NGS) technology [11–14]. The application of multi-gene panels for targeted sequencing, especially for specific disease groups like PIRDs, has facilitated swift diagnoses through next-generation sequencing methods [14].
This study aims to examine the spectrum of PIRD gene mutations in Turkish patients, thereby extending our understanding of molecular genetics in this context.
Material method
Patients (n = 40) diagnosed with Primary Immune Regulatory Disorders (PIRDs) at the Medical Genetics Department between 2018 and 2021 were enrolled in this study. Comprehensive clinical and laboratory findings are summarized in Table 2. The study received approval from the ethics committee, and no external financial support was obtained. Written informed consent was secured from the parents of the participants.
Table 2.
Clinical classification features and disease-causing variants identified in PIRD patients
| Patient | Consanguinity | Gender | Age | Gene | Transcript | PIRD classification | Clinical presentation | Inheritance | Zygosity | cDNA | Protein | Novel/previously described | ACMG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Yes | F | 1 | LYST | NM_000081.3 | HLH | Recurrent cutaneous and systemic pyogenic infections | AR | Homozygous | c.6077_6078insA | p.Tyr2026Ter | Previously described | Likely pathogenic |
| 2 | Yes | F | 11 | LYST | NM_000081.3 | HLH | Septic sacroiliitis | AR | Homozygous | c.52C > T | p.Arg18Trp | Previously described | VUS (PM2,PM5) |
| 3 | Yes | M | 1 | LYST | NM_000081.3 | HLH | Recurrent infections | AR | Homozygous | c.10699C > T | p.Gln3567Ter | Novel | Pathogenic |
| 4 | Yes | F | 1 | LYST | NM_000081.3 | HLH | Hair hypopigmentation | AR | Homozygous | c.10423 T > C | p.Ser3475Pro | Novel | VUS (PM2) |
| 5 | Yes | F | 1 | RAB27A | NM_183235.3 | HLH | Hair hypopigmentation | AR | Homozygous | c.53_54delCT | p.Ser18fs | Previously described | Pathogenic |
| 6 | Yes | M | 20 | STXBP2 | NM_006949.4 | HLH | EBV infection | AR | Homozygous | c.1280-1G > C | - | Previously described | Pathogenic |
| 7 | No | M | 12 | MAGT1 | NM_001367916.1 | Susceptibility to EBV | Hodgkin lymphoma | XL | hemizygous | c.244C > T | p.Gln82Ter | Novel | Likely pathogenic |
| 8 | Yes | M | 23 | SH2D1A | NM_002351.4 | Susceptibility to EBV | ALPS? | XL | Hemizygous | c.164G > A | p.Arg55Gln | Previously described | Likely pathogenic |
| 9 | No | M | 28 | SH2D1A | NM_002351.4 | Susceptibility to EBV | Hyper IgM | XL | Hemizygous | c.163C > T | p.Arg55Ter | Previously described | Likely pathogenic |
| 10 | No | M | 2 | XIAP | NM_001167.3 | Susceptibility to EBV | Enteropathy, splenomegaly | XL | Hemizygous | c.482A > G | p.Tyr161Cys | Previously described | VUS (PM2,NBP4) |
| 11 | No | F | 8 | CASP10 | NM_032977.3 | Syndromes with autoimmunity | ALPS? | AD | Heterozygous | c.1297G > A | p.Glu433Lys | Previously described | VUS (PM2) |
| 12 | No | M | 4 | FAS | NM_032977.3 | Syndromes with autoimmunity | ALPS? | AD | Homozygous | c.869C > T | p.Ala290Val | Novel | VUS (PM2,PP2) |
| 13 | Yes | M | 5 | FAS | NM_032977.3 | Syndromes with autoimmunity | N/A | AD | Heterozygous | c.457A > T | p.Ile153Phe | Novel | VUS (PM2,PP2) |
| 14 | No | M | 2 | FAS | NM_000043.6 | Syndromes with autoimmunity | Neutropenia | AD | Heterozygous | c.452A > G | p.His151Arg | Novel | VUS (PM2,PP2) |
| 15 | Yes | M | 10 | LRBA | NM_001364905.1 | Syndromes with autoimmunity | ALPS? | AR | Homozygous | c.2496C > A | p.Cys832Ter | Novel | Likely pathogenic (PVS1,PM2) |
| 16 | Yes | F | 49 | LRBA | NM_001364905.1 | Syndromes with autoimmunity | Splenomegaly | AR | Homozygous | c.2165G > A | p.Arg722His | Previously described | VUS (PM2,PP3) |
| 17 | Yes | M | 15 | LRBA | NM_001364905.1 | Syndromes with autoimmunity | Autoimmune hemolytic anemia, splenomegaly | AR | Homozygous | c.5505delT | p.Ile1836fs | Previously described | Likely pathogenic |
| 18 | Yes | M | 15 | LRBA | NM_001364905.1 | Syndromes with autoimmunity | Hypogammaglobulinemia | AR | Homozygous | c.5123 T > G | p.Leu1708Arg | Novel | VUS (PM2) |
| 19 | Yes | M | 32 | LRBA | NM_001364905.1 | Syndromes with autoimmunity | Hypogammaglobulinemia | AR | Homozygous | c.6834delA | p.Glu2278fs | Novel | Likely pathogenic (PVS1,PM2) |
| 20 | Yes | F | 20 | LRBA | NM_001364905.1 | Syndromes with autoimmunity | Hepatosplenomegaly autoimmune cytopenia | AR | Homozygous | c.7943C > A | p.Ser2648Ter | Previously described | Likely pathogenic |
| 21 | No | M | 38 | CTLA4 | NM_005214.5 | Syndromes with autoimmunity | Autoimmune hemolytic anemia | AD | Heterozygous | c.436G > A | p.Gly146Arg | Previously described | VUS (PM2,PP5) |
| 22 | No | M | 23 | CTLA4 | NM_005214.5 | Syndromes with autoimmunity | ALPS | AD | Heterozygous | c.19C > T | p.Gln7Ter | Novel | Likely pathogenic (PVS1,PM2) |
| 23 | No | M | 25 | CTLA4 | NM_005214.5 | Syndromes with autoimmunity | Autoimmune polyglandular syndrome | AD | Heterozygous | c.495_496delinsAT | p.Trp165Ter | Novel | Likely pathogenic (PVS1,PM2) |
| 24 | No | M | 14 | CTLA4 | NM_005214.5 | Syndromes with autoimmunity | ALPS | AD | Heterozygous | c.60G > A | p.Trp20Ter | Previously described | Pathogenic |
| 25 | No | M | 19 | CTLA4 | NM_005214.5 | Syndromes with autoimmunity | Hepatosplenomegaly, enteropathy | AD | Heterozygous | c.518G > A | p.Gly173Glu | Previously described | VUS(PM2) |
| 26 | No | M | 30 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | Autoimmune hemolytic anemia | AD | Heterozygous | c.973C > T | p.Arg325Trp | Previously described | VUS (PM2,PM1,PP2,PP3) |
| 27 | No | F | 7 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | Autoimmune hemolytic anemia, hepatosplenomegaly | AD | Heterozygous | c.1981G > T | p.Asp661Tyr | Previously described | Likely pathogenic |
| 28 | No | F | 3 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | Hypereosinophilia | AD | Heterozygous | c.1295 T > C | p.Val432Ala | Novel | VUS (PM2,PM1,PP2,PP3) |
| 29 | No | F | 12 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | Hypereosinophilia | AD | Heterozygous | c.1144C > T | p.Arg382Trp | Previously described | Pathogenic |
| 30 | No | M | 15 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | ALPS | AD | Heterozygous | c.31G > A | p.Asp11Asn | Novel | VUS (PM2,PP1) |
| 31 | No | M | 45 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | Hepatosplenomegaly | AD | Heterozygous | c.1324G > C | p.Glu442Gln | Novel | VUS (PM2,PM1,PP2) |
| 32 | No | M | 18 | STAT3 | NM_139276.2 | Syndromes with autoimmunity | Agranulocytosis | AD | Heterozygous | c.454C > T | p.Arg152Trp | Previously described | Likely pathogenic |
| 33 | Yes | M | 3 | AIRE | NM_000383.4 | Syndromes with autoimmunity | Chronic mucocutaneous candidiasis | AR | Homozygous | c.769C > T | p.Arg257Ter | Previously described | Pathogenic |
| 34 | Yes | M | 30 | AIRE | NM_000383.4 | Syndromes with autoimmunity | Chronic mucocutaneous candidiasis | AR | Homozygous | c.769C > T | p.Arg257Ter | Previously described | Pathogenic |
| 35 | Yes | F | 14 | AIRE | NM_000383.4 | Syndromes with autoimmunity | Chronic mucocutaneous candidiasis | AR | Homozygous | c.769C > T | p.Arg257Ter | Previously described | Pathogenic |
| 36 | Yes | M | 2 | AIRE | NM_000383.4 | Syndromes with autoimmunity | Chronic mucocutaneous candidiasis | AR | Homozygous | Exon 2–3-4 deletion | Novel | Likely pathogenic | |
| 37 | Yes | M | 3 | ZAP70 | NM_001079.3 | Syndromes with autoimmunity | Recurrent CMV infection | AR | Homozygous | c.1448C > T | p.Ser483Phe | Novel | Likely pathogenic (PM2,PP3) |
| 38 | No | M | 6 | FOXP3 | NM_014009.3 | Syndromes with autoimmunity | Enteropathy | XL | Hemizygous | c.506G > A | p.Cys169Tyr | Previously described | VUS(PM2) |
| 39 | No | M | 3 | FOXP3 | NM_014009.3 | Syndromes with autoimmunity | N/A | XL | Hemizygous | c.1150G > A | p.Ala384Thr | Previously described | Pathogenic |
| 40 | No | F | 6 | IL10RB | NM_000628.5 | immune dysregulation with colitis (IBD) | Chronic diarrhea | AR | Homozygous | c.477G > A | p.Trp159Ter | Previously described | Pathogenic |
M male, F female, PIRD primary immune regulatory disorders, AD autosomal dominant, AR autosomal recessive, XL X-linked, HLH hemophagocytic lymphohistiocytosis, EBV Epstein–Barr virus ALPS autoimmune lymphoproliferative syndrome, N/A non available, ACMG American College of Medical Genetics, VUS variant of unknown significance
PVS1: Very strong evidence of pathogenicity Null variant (nonsense, frameshift, canonical ± 1 or 2 splice sites, initiation codon, single or multi-exon deletion) in a gene where loss of function (LOF) is a known mechanism of disease
PM2: Moderate evidence of pathogenicity, absent from controls (or at extremely low frequency if recessive) in exome sequencing project, 1000 genomes or ExAC
PM1: Moderate evidence of pathogenicity, located in a mutational hot spot and/or critical and well-established functional domain (e.g., active site of an enzyme) without benign variation
PP2: Missense variant in a gene that has a low rate of benign missense variation and where missense variants are a common mechanism of disease
PP3: Supporting evidence of pathogenicity, multiple lines of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.)
DNA was extracted from whole blood utilizing the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). The amount of extracted DNA was quantified employing the Qubit™ dsDNA HS assay kit on the Qubit 2.0 fluorometer (Thermo Fisher Scientific), following the manufacturer’s guidelines. Library preparation was performed using the Ion Chef System (Thermo Fisher Scientific, San Francisco, CA, USA) as per the manufacturer’s protocols. Barcoded libraries were generated from 10 ng of DNA per sample through the Ion AmpliSeq Chef Solutions (Thermo Fisher Scientific) and the Ion AmpliSeq™ Primary Immune Deficiency Research Panel v2 (Thermo Fisher Scientific). This panel comprises 5241 amplicons within 264 genes. The resulting libraries were clonally amplified onto Ion Sphere Particles (ISP) using emulsion PCR within an Ion Chef System (Thermo Fisher Scientific), following the manufacturer’s guidelines. Enriched ISPs were loaded onto 530 chips accommodating 16 samples per sequencing run. Sequencing was executed using an Ion S5 Sequencer alongside an Ion 530 Chip and an Ion 530 kit–Chef Kit (all from Thermo Fisher Scientific). The sequences were aligned to the reference genome hg19, and base calling was carried out using the Torrent Suite software. The annotated variant-calling file (VCF) was subjected to filtration in Ion ReporterTM Software, displaying only the variants relevant to the 27 PIRD-related genes (LYST, RAB27A, PRF1, STX11, STXBP2, SLC7A7, CTPS1, ITK, MAGT1, PRKCD, SH2DIA, XIAP, FAS (TNFRSF6), FASLG (TNFSF6), CASP10, CASP8, FADD, LRBA, STAT3, AIRE, ITCH, ZAP70, FOXP3, CTLA4, IL10, IL10RA, IL10RB).
The clinical significance of the novel variants was assessed using the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) recommended standards and guidelines [15]. Minor Allele Frequencies were examined through databases including NCBI dbSNP build141, 1000 Genomes Project, Exome Aggregation Consortium (ExAC), and Genome Aggregation Database (gnomAD). Information specific to disease-related variants was obtained from ClinVar and OMIM. The impact of novel variants on protein structure was evaluated using prediction tools such as GERPP, Polyphen-2, and SIFT [16–18]. Variant pathogenicity was assessed in line with ACMG recommendations. All novel genetic variants were scrutinized for their pathogenicity, inheritance mode, and clinical manifestations. Missense variants affected conserved amino acids within functional protein domains. Notably, all variants were absent in gnomAD. Finally, candidate pathogenic variants identified through NGS were validated using Sanger sequencing on an ABI PRISM 3500 DNA analyzer (Applied Biosystems), followed by segregation analysis.
Results
The cohort consisted of 40 patients—14 pediatric and 26 adult individuals, with 28 males (70.0%) and 12 females (30%), resulting in a male-to-female ratio of 2.3 (28/12).
Allelic segregations for the identified gene variants were verified within the families. In total, 38 disease-causing variants were pinpointed across 15 distinct PIRD genes: STAT3 (n = 7), LRBA (n = 6), CTLA4 (n = 5), LYST (n = 4), AIRE (n = 4), FAS (n = 3), SH2D1A (n = 2), FOXP3 (n = 2), XIAP (n = 1), ZAP70 (n = 1), CASP10 (n = 1), RAB27A (n = 1), STXBP2 (n = 1), MAGT1 (n = 1), and IL10RB (n = 1). Our cohort encompassed HLH (n = 6), susceptibility to EBV (n = 4), syndromes featuring autoimmunity (n = 29), and one patient with immune dysregulation and colitis (IBD).
In this study, we unveil 16 novel variants, including three homozygous variants (c.2496C > A (p.Cys832Ter), c.5123 T > G (p.Leu1708Arg), c.6834delA (p.Glu2278fs)) in LRBA; two homozygous variants (c.10699C > T (p.Gln3567Ter), c.10423 T > C (p.Ser3475Pro)) in LYST; one homozygous deletion in AIRE spanning exons 2 to 3; one homozygous variant (c.1448C > T (p.Ser483Phe)) in ZAP70; one homozygous variant (c.869C > T (p.Ala290Val)) and two heterozygous variants (c.457A > T (p.Ile153Phe), c.452A > G (p.His151Arg)) in FAS; three heterozygous variants (c.1295 T > C (p.Val432Ala), c.31G > A (p.Asp11Asn), c.1324G > C (p.Glu442Gln)) in STAT3; two heterozygous variants (c.19C > T (p.Gln7Ter), c.495_496delinsAT (p.Trp165Ter) in CTLA4; and one hemizygous variant (c.244C > T (p.Gln82Ter)) in MAGT1. These novel variants expand the spectrum of clinical manifestations attributed to novel PIRD variants and broaden the genetic landscape of Human Inborn Errors of Immunity.
Genotype–phenotype correlations of the patients with novel variations
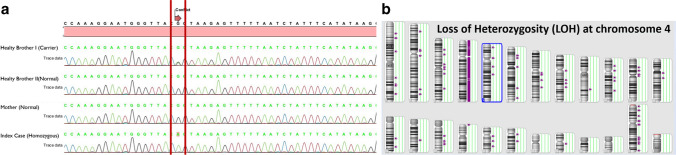
We present a 49-year-old female (case 16) diagnosed with common variable immune deficiency, recurrent respiratory tract infections, cough attacks, periodic hematuria, dysuria, and hypogammaglobulinemia (IgG < 34 mg/dl; IgA < 27 mg/dl; IgM < 17 mg/dl, IgE < 17.9 IU/ml). Over 11 years, she had asthma, while rheumatoid arthritis and hypothyroidism were diagnosed 3 years ago. Sequence analysis revealed a homozygous c.2165G > A (p.Arg722His) variation. Segregation testing was performed on the case’s mother and two healthy brothers, with one brother found to be heterozygous for c.2165G > A (p.Arg722His), while the mother and the other brother were non-carriers. The father’s DNA could not be sequenced due to his passing. Though we hypothetically considered the father as a carrier due to the autosomal recessive nature of LRBA deficiency, the circumstances remained unclear. This is inconsistent with the expected homozygous occurrence in our patient. To verify this, we confirmed genotypes via Sanger sequencing (Fig. 1a). Our findings suggested either a deletion encompassing the LRBA locus on the maternal chromosome 4 or the inheritance of two copies of the mutant maternal LRBA allele. Further analysis with an SNP array revealed loss of heterozygosity of the entire chromosome 4 (Fig. 1b).
Fig. 1.
a Segregation analysis of the family and b SNP array analysis of the index
Two male siblings with a novel LRBA variation presented with different initial symptoms: the younger one had chronic early-onset diarrhea, while the elder one had autoimmune hemolytic anemia. Both developed hypogammaglobulinemia, enteropathy, and lung involvement during long-term follow-up for the IPEX phenotype. Partial responses to immunosuppressive therapies were observed. Molecular diagnosis revealed a homozygous LRBA gene variation c.2496C > A (p.Cys832Ter), resulting in a premature stop codon. Subsequent treatment with abatacept, a target-specific molecule, showed promising results.
A novel hemizygous MAGT1 c.244C > T (p.Gln82Ter) variant was identified in a 13-year-old male patient presenting with swelling on the right side of the neck. Despite antibiotic treatment, neck lymphadenopathy persisted, leading to a referral for malignancy evaluation. B symptoms were absent during presentation, and physical examination revealed a 3 cm mobile rubbery lymphadenopathy in the right submandibular region. The patient’s parents were not consanguineous, and no family history of cancer was reported. Laboratory tests indicated normal complete blood count and biochemistry, with an LDH of 251 IU/l and an erythrocyte sedimentation rate of 9 mm/h. Excisional lymph node biopsy diagnosed the patient with classic Hodgkin’s lymphoma, nodular sclerosis type, and staging placed him in risk group TG1 under the GPOH-HD-2002 protocol. Following two cycles of OEPA treatment, a complete response was achieved, and treatment was concluded due to stage 1A and complete response attainment. Immunoglobulin levels at lymphoma diagnosis were IgA, 0.23 g/l; IgG, 6.74 g/l; and IgM, 0.23 g/l. No recurrent infections were documented, but intravenous immunoglobulin was administered during chemotherapy. Following chemotherapy, immunoglobulin levels remained low, prompting evaluation for potential immune deficiencies.
In our cohort, two novel heterozygous CTLA4 variants (c.19C > T (p.Gln7Ter) and c.495_496delinsAT (p.Trp165Ter)) were found in case 22 with cytopenia and case 23 with rheumatoid arthritis, diabetes mellitus, CVID, and alopecia totalis. These cases were treated with abatacept, yielding successful outcomes.
The preliminary diagnoses for cases with newly identified LYST variations c.10699C > T (p.Gln3567Ter) and c.10423 T > C (p.Ser3475Pro) were Chediak-Higashi syndrome.
Our cohort’s patients with novel STAT3 variants presented with c.1295 T > C (p.Val432Ala)-related hypereosinophilia, c.31G > A (p.Asp11Asn)- and c.1324G > C (p.Glu442Gln)-associated ALPS, and hepatosplenomegaly.
Discussion
Inborn Errors of Immunity (IEIs) encompass a group of genetically and phenotypically heterogeneous inherited disorders that impede the development and/or function of the human immune system [1]. Primary Immune Regulatory Disorders (PIRDs) are linked to autoimmunity, autoinflammation, and/or disruptions in lymphocyte homeostasis [4]. Commonly, defects in T cells and their tolerance induction, B cells, immunoglobulins, and class-switch recombination, as well as genes affecting multiple cellular subsets, constitute the prevalent issues predisposing IEI patients to autoimmunity [19]. The advent of NGS technologies has revealed an expanding list of monogenic defects underlying IEIs, with diagnostic yields ranging from 15 to 79% [13].
In these 40 index patients, a total of 38 disease-causing variants (16 of which were novel) were identified. Among these, 23 variants were deemed likely pathogenic or pathogenic, while 16 were classified as variants of unknown significance (VUS). Out of the 38 different PIRD mutations, 22 were missense, eight were nonsense, five were in/dels, and one represented a single-base substitution in an exon–intron junction sequence, likely affecting a splice site.
This study marks the first instance of using next-generation sequencing within our country to investigate the distribution of mutations in the PIRD patient population. Turkey has reported a notably high rate of consanguineous marriages [20]. This phenomenon has contributed to a heightened prevalence of autosomal recessive inherited diseases, including Inborn Errors of Immunity. Among our cohort, recessive PIRD genes constituted 45% of cases, while dominant variants accounted for 40%, and X-linked PIRD genes constituted 15%.
In a Dutch cohort, NGS-based assessment for IEIs showed the highest yields among pediatric patients, within the immune dysregulation cluster, most patients received a diagnosis of familial hemophagocytic lymphohistiocytosis (HLH), with additional cases including autoimmune lymphoproliferative syndrome (ALPS), primarily attributed to pathogenic FAS variants [14]. Conversely, our cohort is primarily clustered within the immune dysregulation category, with a diagnosis of syndromes characterized by autoimmunity.
In a study from India, diseases involving immune dysregulation were observed in 20 patients. Most frequently, most of the patients are diagnosed with FHL [21].
In another study from Egypt, genetic assessments were conducted for 39 patients exhibiting immune dysregulation disorders. Among them, 21 individuals from 15 distinct consanguineous families displayed variations in the LRBA gene. Other infrequent genetic diagnoses included variants in IL10RA, IL10RB, FOXP3, AIRE, DOCK8, SLC7A7, UNC13D, PRKCD, SH2D1A, RIPK1, and FAS variants [22].
Our immune gene panel differs from the Dutch, Iranian, and Egyptian cohorts, as it involves a distinct panel lacking certain HLH genes found in the Primary Immune Deficiency Research Panel v2. Our clinicians preferred the HLH panel over the immune panel for FHL, which resulted in relatively lower FHL findings when assessed with our immune panel.
LRBA deficiency is an autosomal recessive disorder arising from biallelic mutations in the LRBA gene (OMIM #614,700). Clinically, it is characterized by early-onset hypogammaglobulinemia, autoimmune manifestations, susceptibility to inflammatory bowel disease, and recurrent infections. While partial isodisomy-associated LRBA deficiency has been reported previously [23], our study reports the first instance of LRBA mutation becoming homozygous through whole chromosome uniparental disomy (UPD).
Homozygous AIRE mutations c.769C > T (Arg257Ter), c.415C > T (p.Arg139Ter), and c.254A > G (p.Tyr85Cys) exhibit a founder effect in the Finnish, Sardinian, and Iranian Jewish populations, respectively [24]. The c.769C > T (Arg257Ter) variant in exon 6 has been identified in 89% of Finnish APECED alleles but is also the most prevalent across other ethnic groups. A literature review involving 23 published Turkish APECED patients revealed that the Finnish major mutation, c.769C > T (Arg257Ter), is prevalent in the Turkish population [24]. Of significance, three out of four (75%) APECED cases in our study featured the c.769C > T (Arg257Ter) variation.
Currently, there are nine genes associated with IEIs in which mutations have been detected for both loss-of-function and gain-of-function mutations: CFB, C3, CARD11, STAT1, STAT3, WAS, JAK1, IFIH1, and ZAP70 [2]. Gain-of-function (GOF) mutations bestow a different function upon the mutant gene due to the mutation it undergoes, leading to unexpected protein production. This type of mutation increases the transcription of a gene, endowing it with heightened activity and mobility, often referred to as a hypermorphic gene. Conversely, loss-of-function (LOF) mutations render the gene product dysfunctional. A gene product completely devoid of function is termed a null allele or amorphous allele. If the mutant type retains partial function, it is referred to as a hypomorphic allele. For instance, while functional mutations in STAT3 may manifest with lymphoproliferation, including lymphadenopathy and hepatosplenomegaly, and early-onset multisystem autoimmunity, STAT3 loss-of-function mutations underlie hyperimmunoglobulin E syndrome (Job’s syndrome). This syndrome is characterized by recurrent infections, eczema-like skin rashes, and vulnerability to severe lung infections. While both LOF and GOF of STAT3 cause immune deficiency, GOF leads to infections distinct from those observed with LOF, accompanied by more common connective tissue abnormalities [25, 26]. Moreover, a 5-year-old male with a novel ZAP70 c.1448C > T (p.Ser483Phe) variant underwent hematopoietic stem cell transplantation (HSCT), yielding successful clinical and immunologic outcomes.
CTLA4 deficiency is a rare disorder profoundly disrupting immune system regulation, leading to conditions such as intestinal disease, respiratory infections, autoimmune issues, and enlarged lymph nodes, liver, and spleen. Abatacept offers a potentially effective treatment for patients with documented CTLA-4 deficiency, inducing and sustaining remission of enteropathy [27].
In summary, PIRDs can manifest similar clinical profiles despite distinct genetic defects, and conversely, the same genetic defect can result in diverse clinical presentations. Beyond clinical diagnosis, identifying the molecular defect causing the disease is pivotal for prognosis prediction, treatment planning (e.g., abatacept, HSCT), preimplantation genetics, prenatal diagnosis, and carrier identification. Despite the genetic diversity underpinning PIRDs, genetic counseling assumes a crucial role in managing and shaping future decisions for affected families.
In conclusion, the integration of NGS into the study of PIRD molecular genetics provides a comprehensive and nuanced understanding of the disorder. From identifying rare variants to uncovering novel mutations, NGS emerges as a cornerstone technology, offering unprecedented insights that pave the way for personalized medicine and improved patient outcomes in the realm of Inborn Errors of Immunity. A key conclusion from this multicenter study: This report represents the inaugural utilization of NGS for diagnosing the Turkish PIRD cohort, offering novel insights that expand the spectrum of clinical manifestations attributed to various PIRD-related mutations.
Author contributions
A. A. and A.D, wrote the main manuscript text and N. K., N. G., F.G., F. C. M. T. C. M. A., D.C. F. E. C, A. K., A. Y., K.U, S.S.K. prepared tables and provided clinical information,G. A, O.A and N. K reviewed the manuscript.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Data availability
There is no applicable data available for this study.
Compliance with ethical standards
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The protocol of this study was reviewed and approved by the local ethics committees of the participating centers. Informed consent was obtained from study participants and/or their legal guardians in accordance with the requirements of the local ethics committees.
Consent for publication
The authors confirm that human research participants provided informed consent for the publication of their individual details.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Notarangelo LD. Primary immunodeficiencies. J Allergy Clin Immunol. 2010;125(2):S182–94. 10.1016/j.jaci.2009.07.053 [DOI] [PubMed] [Google Scholar]
- 2.Bousfiha Aziz, Moundir Abderrahmane, Tangye Stuart G, Picard Capucine, Jeddane Leïla, Al-Herz Waleed, Rundles Charlotte C, Franco Jose Luis, Holland Steven M, Klein Christoph, Morio Tomohiro, Oksenhendler Eric, Puel Anne, Puck Jennifer, Seppänen Mikko R J, Somech Raz, Su Helen C, Sullivan Kathleen E, Torgerson Troy R, Meyts Isabelle. The 2022 Update of IUIS phenotypical classification for human inborn errors of immunity. J Clin Immunol. 2022;42(7):1508–20. 10.1007/s10875-022-01352-z [DOI] [PubMed] [Google Scholar]
- 3.Tangye Stuart G, Al-Herz Waleed, Bousfiha Aziz, Cunningham-Rundles Charlotte, Franco Jose Luis, Holland Steven M, Klein Christoph, Morio Tomohiro, Oksenhendler Eric, Puel Capucine Picard Anne, Puck Jennifer, Seppänen Mikko R J, Somech Raz, Su Helen C, Sullivan Kathleen E, Torgerson Troy R, Meyts Isabelle. Human inborn errors of immunity: 2022 update on the classification from the international union of immunological societies expert committee. J Clin Immunol. 2022;42(7):1473–507. 10.1007/s10875-022-01289-3. 10.1007/s10875-022-01289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.López-Nevado Marta, González-Granado Luis I, Ruiz-García Raquel, Pleguezuelo Daniel, Cabrera-Marante Oscar, Salmón Nerea, Blanco-Lobo Pilar, Domínguez-Pinilla Nerea, Rodríguez-Pena Rebeca, Sebastián Elena, Cruz-Rojo Jaime, Olbrich Peter, Ruiz-Contreras Jesús, Paz-Artal Estela, Neth Olaf, Allende Luis M. Primary immune regulatory disorders with an autoimmune lymphoproliferative syndrome-like phenotype: immunologic evaluation, early diagnosis and management. Front Immunol. 2021;12:671755. 10.3389/fimmu.2021.671755. (eCollection 2021). 10.3389/fimmu.2021.671755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walter JE, Ayala IA, Milojevic D. Autoimmunity as a continuum in primary immunodeficiency. Curr Opin Pediatr. 2019;31:851–62. 10.1097/MOP.0000000000000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Notarangelo LD, Bacchetta R, Casanova JL, Su HC. Human inborn errors of immunity: an expanding universe. Sci Immunol. 2020;5:eabb1662. 10.1126/sciimmunol.abb1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abolhassani H, Azizi G, Sharifi L, Yazdani R, Mohsenzadegan M, Delavari S, Sohani M, Shirmast P, Chavoshzadeh Z, Mahdaviani SA, Kalantari A, Tavakol M, Jabbari-Azad F, Ahanchian H, Momen T, Sherkat R, Sadeghi-Shabestari M, Aleyasin S, Esmaeilzadeh H, Al-Herz W, Bousfiha AA, Condino-Neto A, Seppänen M, Sullivan KE, Hammarström L, Modell V, Modell F, Quinn J, Orange JS, Aghamohammadi A. Global systematic review of primary immunodeficiency registries. Expert Rev Clin Immunol. 2020;16(7):717–32. 10.1080/1744666X.2020.1801422. 10.1080/1744666X.2020.1801422 [DOI] [PubMed] [Google Scholar]
- 8.Sanal O, Tezcan I. Thirty years of primary immunodeficiencies in Turkey. Ann N Y Acad Sci. 2011;1238:15–23. 10.1111/j.1749-6632.2011.06242.x [DOI] [PubMed] [Google Scholar]
- 9.Kolukısa B, Barış S. Primary immune regulatory disorders and targeted therapies. Turk J Haematol. 2021;38(1):1–14. 10.4274/tjh.galenos.2021.2020.0724. 10.4274/tjh.galenos.2021.2020.0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilic SS, Ozel M, Hafizoglu D, Karaca NE, Aksu G, Kutukculer N. The prevalances and patient characteristics of primary immunodeficiency diseases in Turkey two centers study. J Clin Immunol. 2013;33:74–83. 10.1007/s10875-012-9763-3 [DOI] [PubMed] [Google Scholar]
- 11.Heimall JR, Hagin D, Hajjar J, Henrickson SE, Hernandez-Trujillo HS, Tan Y, et al. Use of genetic testing for primary immunodeficiency patients. J Clin Immunol. 2018;38:320–9. 10.1007/s10875-018-0489-8. 10.1007/s10875-018-0489-8 [DOI] [PubMed] [Google Scholar]
- 12.Seleman Michael, Hoyos-Bachiloglu Rodrigo, Geha Raif S, Chou Janet. Uses of next-generation sequencing technologies for the diagnosis of primary immunodeficiencies. Front Immunol. 2017;8:847. 10.3389/fimmu.2017.00847. 10.3389/fimmu.2017.00847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yska HA, Elsink K, Kuijpers TW, Frederix GW, van Gijn ME, van Montfrans JM. Diagnostic yield of next generation sequencing in genetically undiagnosed patients with primary immunodeficiencies: a systematic review. J Clin Immunol. 2019;39(6):577–591. 10.1007/s10875-019-00656-x. [DOI] [PMC free article] [PubMed]
- 14.Elsink K, Huibers MMH, Hollink IHIM, Simons A, Zonneveld-Huijssoon E, van der Veken LT, Leavis HL, Henriet SSV, van Deuren M, van de Veerdonk FL, Potjewijd J, Berghuis D, Dalm VASH, Vermont CL, van de Ven AAJM, Lambeck AJA, Abbott KM, van Hagen PM, de Bree GJ, Kuijpers TW, Frederix GWJ, van Gijn ME, van Montfrans JM, the Genetics First for Primary Immunodeficiency Disorders Consortium. Implementation of early next-generation sequencing for inborn errors of immunity: a prospective observational cohort study of diagnostic yield and clinical implications in Dutch genome diagnostic centers. Front Immunol. 2021;12:780134. 10.3389/fimmu.2021.780134. [DOI] [PMC free article] [PubMed]
- 15.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. 10.1038/gim.2015.30. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Computational Biol. 2010;6(12):e1001025. 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013, Chapter 7:Unit7.20. 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed]
- 18.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. 10.1038/nprot.2009.86. [DOI] [PubMed]
- 19.Azizi G, Pouyani MR, Abolhassani H, Sharifi L, Dizaji MZ, Mohammadi J, Mirshafiey A, Aghamohammadi A. Cellular and molecular mechanisms of immune dysregulation and autoimmunity. Cell Immunol. 2016;310:14–26. 10.1016/j.cellimm.2016.08.012. [DOI] [PubMed]
- 20.Tüik 2016. In: newsletter, no: 24646. http://www.tuik.gov.tr. Accessed 10 May 2017.
- 21.Arunachalam Arun Kumar, Maddali Madhavi, Aboobacker Fouzia N, Korula Anu, George Biju, Mathews Vikram, Edison Eunice Sindhuvi. Primary immunodeficiencies in India: molecular diagnosis and the role of next-generation sequencing. J Clin Immunol. 2021;41(2):393–413. 10.1007/s10875-020-00923-2. 10.1007/s10875-020-00923-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabab E El Hawary, Safa S Meshaal, Dalia S Abd Elaziz, Radwa Alkady, Sohilla Lotfy, Alia Eldash, Aya Erfan, Engy A Chohayeb, Mai M Saad, Rania K Darwish, Jeannette A Boutros, Nermeen M Galal, Aisha M Elmarsafy Genetic testing in Egyptian patients with inborn errors of immunity: a single-center experience. J Clin Immunol. 2022; 42(5):1051-1070. 10.1007/s10875-022-01272-y [DOI] [PMC free article] [PubMed]
- 23.Soler-Palacín P, Garcia-Prat M, Martín-Nalda A, Franco-Jarava C, Rivière JG, Plaja A, Bezdan D, Bosio M, Martínez-Gallo M, Ossowski S, Colobran R. LRBA Deficiency in a patient with a novel homozygous mutation due to chromosome 4 segmental uniparental isodisomy. Front Immunol. 2018;16(9):2397. 10.3389/fimmu.2018.02397.eCollection2018. 10.3389/fimmu.2018.02397.eCollection2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahonen P, Myllärniemi S, Sipilä I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322(26):1829–36. 10.1056/NEJM199006283222601. 10.1056/NEJM199006283222601 [DOI] [PubMed] [Google Scholar]
- 25.Fierabracci A, Pellegrino M, Frasca F. Sara Sebnem Kilic, Corrado Betterle APECED in Turkey: a case report and insights on genetic and phenotypic variability. Clin Immunol. 2018;194:60–6. 10.1016/j.clim.2018.06.012. 10.1016/j.clim.2018.06.012 [DOI] [PubMed] [Google Scholar]
- 26.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, Lyons JJ, Engelhardt KR, Zhang Y, Topcagic N, Roberson EDO, Matthews H, Verbsky JW, Dasu T, Vargas-Hernandez A, Varghese N, McClain KL, Karam LB, Nahmod K, Makedonas G, Mace EM, Sorte HS, Perminow G, Koneti Rao V, O’Connell MP, Price S, Su HC, Butrick M, McElwee J, Hughes JD, Willet J, Swan D, Xu Y, Santibanez-Koref M, Slowik V, Dinwiddie DL, Ciaccio CE, Saunders CJ, Septer S, Kingsmore SF, White AJ, Cant AJ, Hambleton S, Cooper MA. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125(4):591–9. 10.1182/blood-2014-09-602763 [DOI] [PMC free article] [PubMed]
- 27.Dhunputh C, Ducassou S, Fernandes H, Picard Capucine, FrédéricRieux-Laucat J-F, Viallard E Lazaro, Hermine O, Jouvray M, Machelard I, Lambilliotte A, Malphettes M, Moshous D, Neven B, Gauthier A, Garnier N, Leblanc T, Landman-Parker J, Leverger G, Aladjidi N. Abatacept is useful in autoimmune cytopenia with immunopathologic manifestations caused by CTLA-4 defects. Blood. 2022;139(2):300–4. 10.1182/blood.2021013496. 10.1182/blood.2021013496 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no applicable data available for this study.