Abstract
The present study provides functional characterization of alternative splicing of the NTPDase2 (ecto-nucleoside triphosphate diphosphohydrolase-2) involved in the regulation of extracellular nucleotide concentrations in a range of organ systems. A novel NTPDase2β isoform produced by alternative splicing of the rat NTPDase2 gene provides an extended intracellular C-terminus and distinguishes itself from NTPDase2α isoform in gaining several intracellular protein kinase CK2 (casein kinase 2) phosphorylation sites and losing the intracellular protein kinase C motif. The plasmids containing NTPDase2α or NTPDase2β cDNA were used to stably transfect Chinese-hamster ovary-S cells. Imaging studies showed that NTPDase2α was predominantly membrane-bound, whereas NTPDase2β had combined cell surface and intracellular localization. α and β isoforms showed variations in divalent cation dependence and substrate specificity for nucleoside-5′-triphosphates and nucleoside-5′-diphosphates. NTPDase2β exhibited reduced ATPase activity and no apparent ADPase activity. NTPDase2 isoforms demonstrated similar sensitivity to inhibitors such as suramin and pyridoxal phosphate-6-azophenyl-2′,4′-disulphonic acid, and differential regulation by protein kinases. NTPDase2β was up-regulated by intracellular protein kinase CK2 phosphorylation, whereas NTPDase2α activity was down-regulated by protein kinase C phosphorylation. The results demonstrate that alternative coding of the intracellular C-terminal domain contributes distinctive phenotypic variation with respect to extracellular nucleotide specificity, hydrolysis kinetics, protein kinase-dependent intracellular regulation and protein trafficking. These findings advance the molecular physiology of this enzyme system by characterizing the contribution of the C-terminal domain to many of the enzyme's signature properties.
Keywords: C-terminus; ecto-ATPase; ecto-nucleoside triphosphate diphosphohydrolase-2 (NTPDase2, CD39L1); protein kinase; splice variant
Abbreviations: CHO, Chinese-hamster ovary; CK2, casein kinase 2; CPA, cyclopiazonic acid; DAG, 1,2-diacylglycerol; DCF, divalent cation-free; E-NTPDase, ecto-nucleoside triphosphate diphosphohydrolase; ER, endoplasmic reticulum; GFP, green fluorescent protein; NGS, normal goat serum; OAG, 1-oleoyl-2-acetyl-sn-glycerol; PKC, protein kinase C; PPADS, pyridoxal phosphate-6-azophenyl-2′,4′-disulphonic acid; RP, reversed phase; RT, reverse transcriptase; TBB, 4,5,6,7-tetrabromobenzotriazole
INTRODUCTION
The proposed roles of nucleotides and nucleosides as extracellular signalling molecules have now gained widespread recognition [1]. Extracellular purines (ATP, ADP and adenosine) and pyrimidines (UTP and UDP) mediate diverse biological effects through cell surface P1 (adenosine) and P2 (nucleotide) receptors [2]. Two families of P2 receptors, the ligand-gated ion channels (P2X receptors) and the G-protein-coupled P2Y receptors [3–5], influence cellular function in most tissues. P2 receptor signalling appears to be involved in short- and long-term events such as neurotransmission, secretion, cell growth, proliferation and death [6].
Nucleotide signalling is regulated by ecto-nucleotidases, a diverse group of surface-located enzymes that hydrolyse extracellular nucleotides [7]. Several ecto-nucleotidase families with differential preference for nucleotide species have been identified and characterized in mammalian tissues. These include the E-NTPDase (ecto-nucleoside triphosphate diphosphohydrolase) family, E-NPP (ectonucleotide pyrophosphatase/phosphodiesterase) family, ecto-5′-nucleotidase and alkaline phosphatase [8,9]. Nucleoside 5′-tri- and diphosphates are hydrolysed by E-NTPDase, E-NPP and alkaline phosphatase activity. The complete dephosphorylation of nucleotides is achieved by ecto-5′-nucleotidase, which specifically hydrolyses nucleoside 5′-monophosphates [10].
Six members of the E-NTPDase family hydrolyse both purine and pyrimidine nucleotides and their activities strongly depend on the presence of Ca2+ and Mg2+ [7]. NTPDase1–3 have the topology of integral membrane proteins with short N- and C-terminal cytoplasmic domains and a large extracellular loop containing the five apyrase conserved regions [11] that play an important role in forming the catalytic site [12]. NTPDase5 (CD39L4; ER-UDPase, where ER stands for endoplasmic reticulum) and NTPDase6 (CD39L2; Golgi GDPase) lack the C-terminal transmembrane region and may be cleaved at the N-terminus to form a soluble protein [9]. Best characterized members of the E-NTPDase family are NTPDase1 (apyrase, CD39) that hydrolyses nucleoside 5′-tri- and diphosphates with similar rates, and NTPDase2 (ecto-ATPase, CD39L1) that has high preference for nucleoside 5′-triphosphates. These two enzymes probably have a different impact on P2 receptor signalling, based on their differential hydrolytic activity [13,14].
NTPDase2 has been cloned from chicken, human, mouse and rat tissues [15–17] and functionally characterized following expression in mammalian cell lines [13,18]. The open reading frame of NTPDase2 encodes 495 amino acid residues with a calculated protein molecular mass of 54.4 kDa [15,17,18]. The molecular mass of the glycosylated enzyme is 75 kDa [14]. The extracellular region of the protein contains seven potential N-glycosylation sites, 11 cysteine residues probably involved in disulphide bond formation [19], and several potential ecto-protein kinase phosphorylation sites. The intracellular C-terminus (12 amino acid residues) contains a putative PKC (protein kinase C) phosphorylation site [17]. Recent evidence suggests a role for NTPDase2 in platelet aggregation [14], cochlear function [20–22] and neuronal development [23].
The sequencing of a putative human NTPDase2 has revealed three alternatively spliced isoforms [24]. The full-length sequence [18,24] corresponds to the previously characterized rat NTPDase2 [17] and shares its functional properties. The other two isoforms, lacking 23 and 45 amino acid residues respectively in the extracellular domain, were produced by alternative splicing of human NTPDase2 and lack ectonucleotidase activity [24].
Alternative splicing has been also reported for the rat NTPDase2 gene [20]. A putative splice variant, designated as NTPDase2β, has an extended open reading frame providing the translation of an additional 50 amino acids at the C-terminus [20]. This isoform thus encodes 545 amino acid residues with a predicted molecular mass of 60 kDa. The extended C-terminal cytoplasmic tail (62 amino acid residues) contains three potential protein kinase CK2 (casein kinase 2) sites not present in NTPDase2α [17], whereas the putative PKC phosphorylation site has been spliced out. Both isoforms are expressed in a range of rat tissues with different abundances [20]. In the present study, we compare biochemical properties and regulation of these two rat NTPDase2 isoforms subsequent to stable expression in CHO (Chinese-hamster ovary) cells.
EXPERIMENTAL
Materials
Nucleotides (ATP, GTP, UTP, ITP, CTP, ADP, GDP, UDP, IDP and CDP), suramin, PPADS (pyridoxal phosphate-6-azophenyl-2′,4′-disulphonic acid), OAG (1-oleoyl-2-acetyl-sn-glycerol), CPA (cyclopiazonic acid) and HPLC consumables were purchased from Sigma (Sydney, Australia). Restriction enzymes, pcDNA3.1 and pcDNA3.1/NT-GFP vectors, CHO-S-SFM II cells, cell culture reagents and LIPOFECTAMINE™ 2000 were obtained from Invitrogen (Groningen, The Netherlands). Alexa Fluor® 594 antibody was purchased from Molecular Probes (Eugene, OR, U.S.A.) and PKC inhibitor GF109203X was from Tocris (Tocris Cookson, Avonmouth, U.K.). Protein kinase CK2 inhibitor TBB (4,5,6,7-tetrabromobenzotriazole) was kindly provided by Professor L. A. Pinna (Dipartimento di Chimica Biologica, Università di Padova, Padova, Italy).
Construction of recombinant cDNA of NTPDase2 splice variant (NTPDase2β)
Recombinant NTPDase2β cDNA was produced by restriction digestion of cDNA encoding rat NTPDase2α (GenBank® accession no. Y11835) and inclusion of the cDNA fragment spanning the splice site. The digestion of cDNA encoding NTPDase2α with restriction enzymes BsaI and ApoI produced four fragments of 729, 598, 336 and 201 bp. Three fragments (729, 598 and 201 bp) were ligated with the 143 bp fragment produced by restriction digestion of partial NTPDase2β cDNA sequence (GenBank® accession no. AF129103) into the full-length NTPDase2β cDNA sequence (1.66 kb). The full-length cDNA con- struct was amplified directly from the ligation reaction by PCR using the primers corresponding to the 5′- and 3′-ends of NTPDase2α respectively (1–20 sense 5′-ATGGCTGGAAAGTTGGTGTC-3′ and 1836–1811 antisense 5′-TTTTCTAGGAAAAATCCTTTATTAC-3′). PCR with a 40 cycle profile was performed as follows: 94 °C denaturation (1 min), 55 °C annealing (1.5 min), 72 °C extension (2 min) steps using GeneAmp PCR system 2400 (PerkinElmer, Norwalk, CT, U.S.A.). The PCR product was excised from the gel and purified using the QIAEX II gel extraction kit (Qiagen, Clifton Hill, Australia). Recombinant cDNA was ligated in pCR2.1 vector and transformed into TOP10 competent cells (TA cloning kit; Invitrogen). After purification of plasmid DNA (QIAfilter Plasmid Midi; Qiagen), the identity of the construct was confirmed by restriction enzyme mapping and sequencing.
Amplification of NTPDase2α and NTPDase2β from rat tissues
To verify the expression of the full-length NTPDase2β isoform in tissues, mRNA was isolated from rat cochlea and thymus (Dynabeads mRNA DIRECT; Dynal A/S, Oslo, Norway). First-strand cDNA synthesis was performed in a 20 μl RT (reverse transcriptase) reaction using random primers (Invitrogen), dNTPs (Amersham Biosciences, Little Chalfont, Bucks., U.K.) and Superscript II RT (Invitrogen). NTPDase2-specific primers and PCR profile were the same as described above. PCR products were separated and the bands of predicted size excised from the gel, purified, cloned and sequenced.
Cell culturing and transfection of CHO cells
Serum-free medium-adapted CHO-S cells (Invitrogen) were cultured in Dulbecco's modified Eagle's medium supplemented with 0.1 mM non-essential amino acids, 10% (v/v) fetal calf serum, 50 units/ml penicillin and 50 μg/ml streptomycin and maintained at 37 °C in a humidified atmosphere with 5% CO2. The NTPDase2α cDNA and NTPDase2β cDNA were ligated into mammalian expression vector pcDNA3.1 (Invitrogen). CHO-S cells were transfected with plasmid DNA containing NTPDase2α or NTPDase2β using LIPOFECTAMINE™ 2000 (LF2000) reagent. Control cells were transfected with vector (pcDNA3.1) without insert. Transfection was performed in 24-well culture plates. The day before transfection, CHO-S cells were seeded on to culture plates at a density of 2×105 cells per well. For each well, a total of 0.8 μg of plasmid DNA in 50 μl of OPTI-MEM I reduced-serum medium was added to 2.5 μl of LF2000 reagent diluted in 50 μl of OPTI-MEM I. The diluted cDNA and LF2000 reagent were mixed and incubated for 20 min before adding to a 90% confluent monolayer of CHO-S cells. After the cells were incubated with the DNA-LF2000 complexes for 4.5 h, growth medium was replaced. Cells were diluted 24 hours after transfection, and cell populations stably expressing NTPDase2 isoforms were obtained by selection with 1.1 mg/ml of geneticin (G418 sulphate; Invitrogen). G418-resistant clones were selected after 2 weeks and subcultured. Clones with the highest expression of the NTPDase2α (wtC5) and NTPDase2β (svC1) were used for enzyme analysis.
Western blotting
The expression of NTPDase2 isoforms in CHO-S cells was confirmed by Western blotting and immunofluorescence using polyclonal anti-rat NTPDase2α antibody raised in rabbits by direct injection of the encoding cDNA in pcDNA3 [25]. CHO-S cells stably expressing NTPDase2α or NTPDase2β cDNA were stripped, washed with PBS and lysed in a buffer (150 mM NaCl, 5 mM EDTA, 10 mM Tris/HCl, pH 7.4, 1% Triton X-100) containing protease inhibitor cocktail (Roche, Mannheim, Germany). After centrifugation (at 16000 g for 10 min at 4 °C) to remove particulate materials, the protein concentration of the lysate was determined using the Bio-Rad DC assay kit (Bio-Rad Laboratories, Hercules, CA, U.S.A.). Approximately 60 μg of total protein in non-reducing sample buffer [1% SDS, 12.5% (v/v) glycerol, 0.1% Bromophenol Blue, 65 mM Tris/HCl, pH 6.8] was separated by SDS/PAGE on 10% polyacrylamide gel and electrophoretically transferred on to a PVDF membrane (Roche). After blocking with 5% (w/v) skimmed milk and 2% (v/v) NGS (normal goat serum) in TBS (20 mM Tris/HCl, pH 7.2, 137 mM NaCl, 0.1% Tween 20), the membrane was probed with NTPDase2 primary antibody (dilution 1:1000 in blocking buffer) for 3 h. The blotted membrane was incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (dilution 1:8000) before the bands were visualized by chemiluminescence (ECL® Western-blotting analysis system; Amersham Biosciences).
Immunodetection
Transfected CHO-S cells were washed twice with sterile PBS (10.1 mM Na2HPO4, 1.8 mM KH2PO4, 136.9 mM NaCl, 2.7 mM KCl, pH 7.4). Non-specific binding of the antibody was blocked by incubating the cells for 20 min with 2.5% BSA and 1.5% NGS in PBS at room temperature (22 °C). Subsequently, NTPDase2 primary antibody (dilution 1:150 in 0.1 M PBS supplemented with 1% BSA and 1.5% NGS) was applied for 20 min at room temperature. CHO-S cells were rinsed with PBS and fixed in absolute methanol for 5 min at −20 °C. After washing the cells with PBS for 15 min, the secondary antibody (Alexa Fluor® 594 labelled goat anti-rabbit IgG) diluted in PBS/1.5% NGS (1:200) was applied for 20 min at room temperature. The cells were washed twice in PBS and visualized at 620 nm using an inverted epifluorescence microscope (DMIRB; Leica Lasertechnik GmbH, Heidelberg, Germany).
Cell imaging
To assess cellular distribution of NTPDase2α and NTPDase2β, chimaeras of the respective NTPDase2 isoforms with a GFP (green fluorescent protein) were generated at the N-terminus. CHO-S cells were transfected with GFP-NTPDase2α and GFP-NTPDase2β in pcDNA3.1 vector using LF2000 reagent as described previously. Control cells were transfected with an empty pcDNA3.1/NT-GFP vector. Transfection was performed in 24-well culture plates with a glass coverslip at the bottom of the wells. Transiently transfected CHO-S cells were visualized 24 h after plating using an inverted confocal microscope (Zeiss LSM 410). GFP fluorescence was excited using the 488 nm line of an argon laser and detected between 510 and 550 nm. Between 10 and 15 optical sections were acquired for each cell.
Measurement of NTPDase2α and NTPDase2β activity
NTPDase activity was measured in stably transfected CHO-S cells. Stable transfectants (wtC5 or svC1) were seeded in 24-well plates at a density of 5×104 cells per well. NTPDase2 activity was determined at 37 °C, 24 h after plating. The cells were washed twice with phosphate-free physiological saline solution (140 mM NaCl, 5 mM KCl, 10 mM glucose, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, pH 7.4) and subsequently incubated in 500 μl of substrate nucleotide solution. A sample of 60 μl collected from the cell culture was snap-frozen on dry ice and stored at −80 °C until further processing. Nucleotides and nucleosides were separated and quantified by RP (reversed phase) HPLC (Agilent 1100 series; Agilent Technologies, Palo Alto, CA, U.S.A.) using the Adsorbosphere® Nucleotide-Nucleoside column (Alltech; 7 μm, 250 mm×4.6 mm). The samples were eluted at 1 ml/min with the mobile phase consisting of buffer A (60 mM NH4H2PO4 and 5 mM tetrabutylammonium phosphate, pH 5.0) and buffer B (80% HPLC grade methanol with 5 mM tetrabutylammonium phosphate); gradient: 10–50% buffer B in buffer A in the first 10 min. The absorbance was detected at 254 nm, and the rate of hydrolysis was determined from the peak area in the chromatogram.
Time dependence of nucleotide hydrolysis and kinetic studies
Stably transfected CHO-S cells (wtC5, svC1 and control empty vector) were analysed for ATP (0.5 mM) and ADP (0.5 mM) hydrolysis at 37 °C in a time-dependent manner. A sample was collected at 0, 2, 5, 10, 20 and 30 min, stored at −80 °C and later analysed by RP-HPLC. For enzyme kinetic analysis, ATP at a concentration range from 10 to 5000 μM was incubated with CHO-S cells transfected with NTPDase2 isoforms. Enzyme kinetic parameters (Vmax and Km) were obtained using the non-linear least-squares fitting technique (EZ-Fit software; Perella Scientific, Amherst, NH, U.S.A.).
Divalent cation dependence and inhibitor studies
Divalent cation dependence was studied in Ca2+/Mg2+-free assay solution (140 mM NaCl, 5 mM KCl, 10 mM glucose, 10 mM Hepes, pH 7.4) containing 5 mM EDTA, and in solutions containing either added Ca2+ or Mg2+ (2 mM). Standard assay solution containing both cations served as control (100% activity). Putative inhibitors of NTPDase activity suramin (100 μM), PPADS (100 μM) and sodium azide (10 mM), were made up in phosphate-free physiological saline solution. The cells were preincubated with the inhibitors for 60 min before addition of the substrate (0.5 mM ATP was the final concentration in the reactions).
Substrate specificity
Nucleoside 5′-triphosphates (ATP, GTP, UTP, ITP and CTP) and nucleoside 5′-diphosphates (ADP, GDP, UDP, IDP and CDP) were prepared in phosphate-free physiological saline and applied at the final concentration of 0.5 mM. These substrates were incubated with transfected CHO-S cells for 5 min at 37 °C, and the hydrolysis rate was measured by RP-HPLC. Before these experiments, all commercial nucleoside triphosphates and diphosphates were analysed by HPLC to evaluate the level of contamination with nucleoside diphosphates and monophosphates. The contributions of GDP, UDP, CDP, IDP and ADP in the corresponding nucleoside triphosphate samples were 6.7, 7.6, 13.8, 9.6 and 4.1% respectively, and the contributions of GMP, UMP, CMP, IMP and AMP in the corresponding nucleoside diphosphate samples were 2.9, 5.6, 7.1, 5.7 and 4.8% respectively. These values and results of nucleotide hydrolysis obtained in the presence of mock-transfected cells were subtracted from nucleotide hydrolysis in the presence of transfected CHO-S cells.
Modulation of NTPDase2 activity by protein kinases
NTPDase2α and NTPDase2β have different putative intracellular phosphorylation sites (PKC and CK2 respectively). Hence, the regulation of enzyme activity by protein kinases was studied using PKC activator OAG (100 μM), PKC inhibitor GF109203X (10 μM) and CK2 inhibitor TBB (10 μM). The sarcoplasmic/endoplasmic reticulum Ca2+-ATPase-pump inhibitor CPA (10 μM) was used to determine Ca2+-dependence of intracellular phosphorylation. Transfected CHO-S cells were preincubated with OAG for 5 min, and with other compounds for 60 min before the addition of the substrate (ATP, 0.5 mM). The results were expressed as a percentage of ATPase activity in the absence of inhibitor/activator (100% activity).
RESULTS
NTPDase2β sequencing
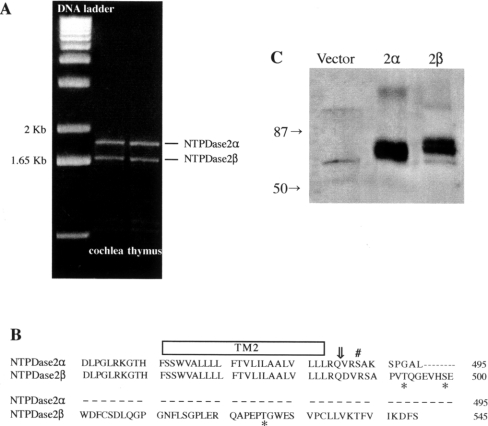
Two mRNA transcripts (1.85 and 1.66 kb), corresponding to full-length NTPDase2α [18] and NTPDase2β [20] respectively were detected by RT–PCR from rat cochlea and thymus (Figure 1A). DNA sequences of the recombinant NTPDase2β and the NTPDase2β amplified from the tissues were identical (DNA alignment not shown). Sequence analysis of NTPDase2β cDNA revealed a variance with rat brain NTPDase2α (GenBank® accession no. Y11835) at two sites (base 380, C-T conversion and base 382, T-C conversion), in addition to the alternatively spliced site shown in Figure 1(B). These two substitutions, reported previously for rat testicular NTPDase2α (GenBank® accession no. AF276940), altered the translation of the corresponding protein (proline to leucine at amino acid residue 127 and phenylalanine to leucine at residue 128).
Figure 1. NTPDase2α and NTPDase2β expressions in rat tissues and CHO-S cells.
(A) NTPDase2α- and NTPDase2β-specific cDNAs generated by RT–PCR from rat cochlear and thymic mRNA (ethidium bromide-stained agarose gel). The molecular size of PCR products: NTPDase2α cDNA, 1.85 kb; NTPDase2β cDNA, 1.66 kb. (B) The alignment of C-terminal amino acid sequences of NTPDase2α (495 amino acid residues) and NTPDase2β (545 amino acid residues). The second transmembrane domain (TM2), splice site (⇓) and putative PKC (#) and CK2 (*) phosphorylation sites are shown. (C) Expression of rat NTPDase2 isoforms stably expressed in CHO-S cells (Western blotting). Molecular mass markers (kDa) are indicated on the left. Vector-transfected CHO-S cells were used as control.
Expression of NTPDase2 isoforms
Western-blot analysis revealed a small difference (∼6 kDa) in the molecular mass of the two NTPDase2 isoforms stably expressed in CHO-S cells (Figure 1C). This difference was predicted from their protein sequences [20]. Protein glycosylation apparently accounts for the larger molecular mass (75–82 kDa) when compared with that predicted from their amino acid backbones (54 and 60 kDa for NTPDase2α and NTPDase2β respectively).
Distribution of NTPDase2α and NTPDase2β in transfected CHO cells
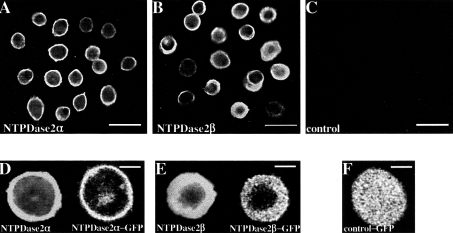
A polyclonal antibody raised against rat NTPDase2α was used to verify the expression of NTPDase2 isoforms in intact and viable CHO-S cells stably transfected with NTPDase2α and NTPDase2β isoforms (Figures 2A and 2B). Mock-transfected cells revealed no immunofluorescence (Figure 2C). Cellular distribution of NTPDase2α and NTPDase2β was demonstrated by intracellular immunofluorescence and NTPDase2-GFP fluorescence (Figures 2D–2F). NTPDase2α was predominantly located on the cell surface (Figure 2D), whereas NTPDase2β accumulated intracellularly in addition to the cell membrane localization (Figure 2E). A diffuse GFP fluorescence observed in cells transfected with an empty pcDNA3.1/NT-GFP vector indicated a lack of specific trafficking of GFP alone (Figure 2F).
Figure 2. The expression pattern of NTPDase2α and NTPDase2β isoforms in transfected CHO-S cells.
Immunofluorescence of both NTPDase2α and NTPDase2β was biased towards the cell membrane (A, B). No immunofluorescence was detected from mock-transfected CHO-S cells (C). Intracellular imaging using immunofluorescence and GFP-labelled constructs demonstrated cell-surface localization of NTPDase2α (D), whereas the NTPDase2β was not only expressed on the cell membrane but was also detected inside the cell (E). CHO-S cells transfected with GFP alone showed diffuse intracellular fluorescence (F). Scale bars, 20 μm (A–C), 5 μm (D–F).
Divalent cation dependence
NTPDase2α activity in stably transfected CHO-S cells was equally activated either by 2 mM Ca2+ or 2 mM Mg2+ (Table 1). In contrast, NTPDase2β activity was more dependent on the presence of Mg2+ than Ca2+ in the assay solution. Both enzyme isoforms were inhibited by 5 mM EDTA in the absence of Ca2+ or Mg2+, although NTPDase2β retained more activity in DCF (divalent cation-free) solution.
Table 1. Divalent cation dependence.
The effect of divalent cations on ATP hydrolysis by CHO-S cells stably transfected with NTPDase2α and NTPDase2β (means±S.D.; n=3 with duplicate sampling).
| ATPase activity (%) | |||
|---|---|---|---|
| Enzyme | Ca2+ only | Mg2+ only | DCF |
| NTPDase2α | 94.5±19.5 | 97.2±12.4 | 19.0±3.2 |
| NTPDase2β | 83.1±4.5 | 93.7±4.7 | 41.8±6.3 |
Nucleotide hydrolysis
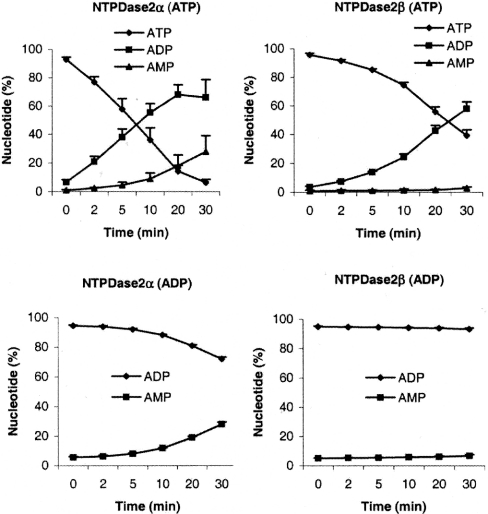
Hydrolysis of ATP was observed in CHO-S cells stably transfected with NTPDase2α or NTPDase2β (Figure 3). The pattern of ATP hydrolysis by NTPDase2α was characterized by fast accumulation of ADP and delayed appearance of AMP, similar to that described by Heine et al. [13]. The hydrolysis of ATP by NTPDase2β was slower, with gradual accumulation of ADP and negligible appearance of AMP in the assay medium. AMP was not hydrolysed by either of the two NTPDase2 isoforms. The degradation of ADP to AMP by NTPDase2α had a similar time course as hydrolysis of ATP to AMP (Figure 3). Significant accumulation of AMP was observed after the incubation of NTPDase2α-transfected CHO-S cells with ADP, whereas ADPase activity of NTPDase2β-transfected cells was negligible. CHO-S cells containing empty pcDNA3.1 vector did not hydrolyse ATP or ADP (results not shown), confirming low intrinsic ectonucleotidase activity of these cells [13].
Figure 3. Time dependence of nucleotide hydrolysis by CHO-S cells stably transfected with NTPDase2α and NTPDase2β isoforms.
ATP and ADP were used as substrates, and their dephosphorylation analysed by RP-HPLC. Each nucleotide is expressed as a percentage of the total nucleotide pool present in the sample, and data shown as means±S.D. (n=3, with duplicate determinations).
Enzyme kinetics of NTPDase2 isoforms
The initial rates of hydrolysis were measured over ATP concentrations ranging from 10 μM to 5 mM. The apparent Km and Vmax values for ATP hydrolysis were 567±92 μM and 137±7 nmol·min−1·(106 cells)−1 for NTPDase2α, and 525±34 μM and 55±1 nmol·min−1·(106 cells)−1 for NTPDase2β. Results are expressed as the means±S.E.M. for three independent experiments with duplicate determinations.
Substrate specificity
Both NTPDase2 isoforms hydrolysed effectively all nucleoside 5′-triphosphates (Table 2). The order of potency for NTPDase2α was ATP>UTP, CTP>GTP, ITP≫CDP, UDP≫ADP, GDP, IDP and for NTPDase2β it was ITP, ATP>CTP, GTP, UTP≫CDP, UDP≫ADP, GDP, IDP. All nucleoside 5′-diphosphates were hydrolysed at a rate lower than nucleoside 5′-triphosphates. Both NTPDase2 isoforms had higher preference for UDP and CDP compared with that for other nucleoside 5′-diphosphates. This was also reflected by the low UTP/UDP and CTP/CDP hydrolysis ratio.
Table 2. NTPDase2α and NTPDase2β substrate specificity.
Nucleotide hydrolysis by CHO-S cells stably transfected with NTPDase2α and NTPDase2β (means±S.D.; n=3 with duplicate sampling).
| Nucleotidase activity (%) | ||
|---|---|---|
| Substrates | NTPDase2α | NTPDase2β |
| ATP | 100 | 100 |
| GTP | 64.9±5.5 | 68.0±8.2 |
| UTP | 85.6±10.3 | 63.4±6.8 |
| ITP | 66.5±6.4 | 108.6±9.8 |
| CTP | 80.5±15.2 | 77.3±3.3 |
| ADP | 6.5±0.6 | 3.0±0.2 |
| GDP | 5.9±1.7 | 2.1±3.1 |
| UDP | 24.8±4.5 | 37.6±2.6 |
| IDP | 2.0±0.9 | 0 |
| CDP | 27.9±4 | 37.8±11.2 |
| Ratio of hydrolysis | ||
| ATP/ADP | 1:0.06 | 1:0.03 |
| GTP/GDP | 1:0.09 | 1:0.03 |
| UTP/UDP | 1:0.29 | 1:0.59 |
| ITP/IDP | 1:0.03 | 1:0 |
| CTP/CDP | 1:0.35 | 1:0.49 |
Enzyme inhibitors
The effect of putative NTPDase/P2 receptor inhibitors on NTPDase2 activity was tested and the results were expressed as a percentage of the ATPase activity in the absence of inhibitor (Table 3). Both NTPDase2 isoforms were inhibited by suramin (100 μM) and PPADS (100 μM), although the inhibitory effect was greater on NTPDase2β activity. Suramin was a more effective inhibitor than PPADS, and inhibited ATPase activity of NTPDase2α and NTPDase2β by 47.6 and 55.3% respectively (P<0.01, Student's t test). The NTPDase1 inhibitor sodium azide (10 mM) had no effect on the activity of NTPDase2 isoforms (Table 3).
Table 3. Effect of enzyme inhibitors on NTPDase2 isoforms.
Means±S.D.; n=3 with duplicate sampling; *P<0.05; **P<0.01; Student's t test.
| ATPase activity (%) | |||
|---|---|---|---|
| Enzyme | Suramin | PPADS | Sodium azide |
| NTPDase2α | 52.4±5.7** | 89.8±4.2* | 103.2±9.9 |
| NTPDase2β | 44.7±2.0** | 78.2±3.1** | 116.2±3.5 |
Effect of protein kinase modulators on NTPDase2 activity
Differential regulation of NTPDase2 isoforms through putative intracellular phosphorylation sites was tested using protein kinase inhibitors and activators (Table 4). Protein kinase CK2 inhibitor TBB [26] at a concentration of 10 μM selectively decreased NTPDase2β activity by 29.8% (P<0.01, Student's t test), whereas NTPDase2α activity was unaffected. The PKC inhibitor GF109203X (10 μM) increased NTPDase2α activity by 18.4% (P<0.05) and reduced the activity of NTPDase2β by 10.7% (P<0.05). In contrast, OAG (100 μM), a membrane permeable analogue of DAG (1,2-diacylglycerol) and PKC activator [27], decreased NTPDase2α activity by 32.2% and increased NTPDase2β activity by 26.4% (P<0.01). CPA, involved in receptor-independent mobilization of intracellular Ca2+ [28], did not have a significant effect on the activity of NTPDase2 isoforms.
Table 4. Effect of protein kinase inhibitors/activators on NTPDase2 activity.
Means±S.D.; n=3 with duplicate sampling; *P<0.05; **P<0.01; Student's t test.
| ATPase activity (%) | ||||
|---|---|---|---|---|
| Enzyme | TBB | GF109203X | OAG | CPA |
| NTPDase2α | 95.8±16.1 | 118.4±15.2* | 67.8±3.1** | 105.6±16.5 |
| NTPDase2β | 70.2±4.2** | 89.3±6.5* | 126.4±8.4** | 95.5±5.7 |
DISCUSSION
The present study provides the functional characterization of a novel NTPDase2β isoform produced by alternative splicing of the rat NTPDase2 gene [20]. For direct functional comparison with NTPDase2α previously isolated from rat brain [17], enzyme isoforms were stably expressed in CHO cells. These cells express endogenous P2X7 and P2Y2 receptor subunits [29,30], but have very low intrinsic ectonucleotidase activity [13] confirmed in the present study. Enzyme expression was verified by immunofluorescence using a polyclonal antibody specific for rat NTPDase2. The two enzyme isoforms demonstrated the functional similarities expected from their sequence homology, but also revealed differences in cellular distribution, catalytic properties and regulation by protein kinases, which are attributable to the variations in their C-terminal coding regions.
Biochemical properties
A significant residual NTPDase2β activity in DCF solution suggests that this isoform is less dependent on divalent cations when compared with the α isoform. Both isoforms had a high preference for nucleoside 5′-triphosphates over nucleoside 5′-diphosphates, whereas nucleoside 5′-monophosphates were not hydrolysed by either of the NTPDase2 isoforms. NTPDase2α had the highest preference for the hydrolysis of ATP, and NTPDase2β for ATP and ITP. CDPase and UDPase activities of both isoforms were significantly higher than previously reported for rat NTPDase2α [13].
NTPDase2 isoforms showed different patterns of ATP and ADP hydrolysis. The apparent Km values for ATP hydrolysis (567 μM NTPDase2α and 525 μM NTPDase2β) were consistent with previous kinetic studies of ecto-ATPases expressed in mammalian cell lines [18,31], but were significantly higher than values reported for purified enzyme preparations (10–20 μM) [9]. Km measurements for ATP hydrolysis by NTPDase2α and NTPDase2β are in a relevant range, given that extracellular nucleotide concentrations in cultured cells or tissues may reach millimolar levels in response to a variety of factors [32,33].
Enzyme inhibitors were used to further characterize NTPDase2β activity. Even though a large number of compounds had been investigated as potential E-NTPDase inhibitors [9], very few appear to affect selectively ATP hydrolysis without inhibiting P2 receptors. Non-selective P2 receptor inhibitors suramin and PPADS were used in our study, as their inhibitory effect on NTPDase2α activity had been previously reported [13]. The inhibitory effect of suramin was greater than PPADS on both NTPDase2 isoforms, but NTPDase2β showed slightly higher sensitivity to inhibition when compared with NTPDase2α. As predicted, the NTPDase1 inhibitor sodium azide [34] did not affect the activity of NTPDase2 isoforms.
NTPDase2 regulation
E-NTPDase activity can be modulated in different physiological and pathological situations [35,36], suggesting the existence of regulatory mechanisms. Protein phosphorylation, a universal mechanism of cellular regulation and a final step in a signal transduction pathway [37], appears to be a probable candidate for E-NTPDase regulation [38]. NTPDase2 has potential intracellular and extracellular phosphorylation sites [15,17,18] that may be involved in cross-talk between extracellular nucleotides and intracellular signalling mechanisms [20]. Whereas NTPDase2α contains a potential PKC phosphorylation site in the C-terminus (Ser-488), the C-terminal cytoplasmic tail of NTPDase2β contains instead three potential protein kinase CK2 phosphorylation sites in amino acid positions 493, 499 and 526. In the present study, we investigated the regulation of NTPDase2 isoforms by using the compounds that selectively affect PKC or CK2 activity.
Protein kinase CK2 is a multifunctional Ser/Thr-specific protein kinase apparently resistant to many regulatory mechanisms known for other protein kinases [39]. A recently derived ATP-site-directed cell-permeant inhibitor of CK2, TBB, has provided a selective new tool for assessing the role of CK2 in cellular function [26]. In the present study, TBB selectively inhibited NTPDase2β activity, whereas NTPDase2α activity was unaffected. These results suggest that the NTPDase2β splice variant is indeed regulated by protein kinase CK2, as predicted from its molecular structure [20]. The lack of inhibition of NTPDase2α isoform suggests that the putative extracellular CK2 phosphorylation sites present in this isoform [17,18] are not involved in NTPDase2 regulation.
PKC consists of a family of 11 different isoforms, which have individual enzymological characteristics and specific tissue distribution [40]. GF109203X, a strong PKCα inhibitor and a weaker inhibitor of PKC δ and μ [41,42], had a dual effect on NTPDase2 isoforms. It stimulated NTPDase2α activity, and reduced the activity of NTPDase2β. The PKC activator OAG had an opposite effect, suggesting the involvement of DAG-responsive PKC isoforms. CPA did not have significant effect on the activity of NTPDase2 isoforms, suggesting that PKC activity was independent of Ca2+ mobilization. These results are therefore consistent with Ca2+-independent, DAG-activated nPKCs (δ and μ). Our results also implicate ecto-PKC involvement in the regulation of the NTPDase2β isoform, which lacks intracellular PKC phosphorylation sites, but has potential extracellular sites at positions 136, 365 and 419. Ecto-protein kinases are expressed on the outer cell surface of a variety of cells, and phosphorylate cell-surface proteins and soluble extracellular substrates primarily on ecto-domains [43]. Our results imply that extracellular and intracellular phosphorylations of NTPDase2 isoforms were mediated by different PKC subtypes, as the compounds that modulate PKC activity had opposite effects on the regulation of NTPDase2α and NTPDase2β isoforms. The differential effects of CK2 and PKC inhibitors on CHO cells transfected with either α or β NTPDase2 isoforms which showed variation only in the C-terminal region suggest that the effect of inhibitors was due to differential phosphorylation rather than non-specific pharmacological action, which would have produced comparable effect in both isoforms.
Functional significance
The functional characterization of the NTPDase2β splice variant stably expressed in CHO cells revealed unusual biochemical properties and cellular distribution. The differences in cellular distribution may have a considerable impact on catalytic properties of the alternatively spliced NTPDase2 isoforms. The stretch of 10 C-terminal amino acid residues (486–495) of the NTPDase2α isoform, substituted by a different coding region in NTPDase2β (60 amino acid residues), may have an important role in enzyme transport from the intracellular stores (ER and Golgi apparatus) to the cell surface. Impaired trafficking of NTPDase2β to the plasma membrane may arise from folding alterations that lead to partial intracellular retention, as described for human NTPDase2 splice variants [24]. The C-terminal domain of NTPDase2β contains a di-leucine motif (LL534-535), which serves as a checkpoint preventing inappropriate surface expression of misfolded polypeptides or unassembled subunits of oligomeric proteins. This motif mediates the internalization of surface-expressed mutant proteins and their subsequent relocation to the intracellular compartments [44]. This suggests a dynamic NTPDase2β expression on the cell membrane and the possibility that the enzyme may undergo degradation on endocytosis. Protein kinases may influence intracellular protein trafficking by suppressing ER retention [45] and thus modify the relative abundance of the alternatively spliced isoforms on the cell surface with subsequent impact on the phenotype of extracellular nucleotide hydrolysis. The role of protein kinases PKC and CK2 in trafficking of NTPDase2 isoforms is under investigation.
It is evident from the present study that the modification of the intracellular C-terminal coding region directly influences the selectivity and kinetics of the extracellular nucleotide hydrolysis, presumably arising from steric influences on the extracellular catalytic site associated with the apyrase conserved regions [12]. The long cytoplasmic C-terminus of NTPDase2β, not present in the NTPDase2α isoform, may play a significant role in intracellular events, including enzyme regulation by protein phosphorylation. Different signal transduction pathways linking P2 receptors and intracellular phosphorylation sites may be involved in the regulation of NTPDase2 isoforms [20]. The present study supports our previously proposed model of NTPDase2-P2 receptor feedback regulation associated with G-protein-coupled P2Y receptors that stimulate PKC activation through Gq protein–phospholipase C-β–DAG signal transduction pathway [20]. However, it does not support the feedback mechanism based on increase in intracellular Ca2+ induced by the activation of ATP-gated ion channels (P2X receptors), due to the lack of Ca2+-sensitive intracellular phosphorylation sites in NTPDase2 isoforms.
The physiological significance of multiple transcripts for the same enzyme is emerging, as the alternative splicing appears to be a common mechanism for the regulation of expression of ecto-ATPases and P2 receptors [20,24,46]. NTPDase2α is the dominant isoform at the cell surface, and it may provide the regulation of purinergic signalling both under quiescent conditions and during enhanced nucleotide release. However, the activation of P2Y receptors may induce PKC-mediated down-regulation of this enzyme isoform, raising the possibility that NTPDase2β, which is up-regulated by PKC and CK2 phosphorylation, may be the major regulator of P2Y receptor signalling. NTPDase2 is closely associated with P2Y receptors in different cells and tissues, including astrocytes, glial cells of the peripheral ganglia, Schwann cells and secretory tissues of the cochlea [21,23,47]. In addition to its role in the termination of P2 receptor signalling in these tissues, NTPDase2 may allow preferential activation of specific subsets of P2 receptors sensitive to ADP and UDP (P2Y1, P2Y6, P2Y12, P2Y13). Development of isoform-specific antibodies and co-localization studies with P2X and P2Y receptors will probably elucidate the role of NTPDase2 isoforms in the regulation of P2 receptor signalling.
In conclusion, alternative splicing contributes to the diversity of nucleotide signalling, and may influence a number of physiological processes including thromboregulation, sound transduction, neural differentiation, and cross-talk between peripheral neurons and glia. The formation of functional hetero-oligomers by alternatively spliced isoforms may further diversify the regulation of nucleotide signalling. The NTPDase2β splice variant with a modified intracellular C-terminus clearly provides a distinctive extracellular nucleotide hydrolysis phenotype, which underpins P2 receptor signalling in a range of physiological systems.
Acknowledgments
This work was supported by the Auckland Medical Research Foundation, Maurice & Phyllis Paykel Trust and Health Research Council of New Zealand. N.B. and H.Z. were supported by DFG/SFB269/A4 and the Förderfonds der Chemischen Industrie. J.S. was supported by the Fonds de la Recherche en Santé du Québec and the Canadian Institutes of Health Research and S.C.R. by NIH (U.S.A.). We gratefully acknowledge Professor L.A. Pinna for providing CK2 inhibitor TBB.
References
- 1.Burnstock G. The past, present and future of purine nucleotides as signaling molecules. Neuropharmacology. 1997;36:1127–1139. doi: 10.1016/s0028-3908(97)00125-1. [DOI] [PubMed] [Google Scholar]
- 2.Ralevic V., Burnstock G. Receptors for purines and pyrimidines. Pharmacol. Rev. 1998;50:413–492. [PubMed] [Google Scholar]
- 3.Burnstock G., Kennedy C. Is there a basis for distinguishing two types of P2-purinoceptor? Gen. Pharmacol. 1985;16:433–440. doi: 10.1016/0306-3623(85)90001-1. [DOI] [PubMed] [Google Scholar]
- 4.Khakh B. S., Burnstock G., Kennedy C., King B. F., North R. A., Séguéla P., Voigt M., Humphrey P. P. A. International union of pharmacology. XXIV. Current status of the nomenclature and properties of P2X receptors and their subunits. Pharmacol. Rev. 2001;53:107–118. [PubMed] [Google Scholar]
- 5.Abbrachio M. P., Boeynaems J.-M., Barnard E. A., Boyer J. L., Kennedy C., Miras-Portugal M. T., King B. F., Gachet C., Jacobson K. A., Weisman G. A., et al. Characterisation of the UDP-glucose receptor (re-named here the P2Y14 receptor) adds diversity to the P2Y receptor family. Trends Pharmacol. Sci. 2003;24:52–55. doi: 10.1016/S0165-6147(02)00038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnstock G. Expanding field of purinergic signaling. Drug Dev. Res. 2001;52:1–10. [Google Scholar]
- 7.Plesner L. Ecto-ATPases: identities and functions. Int. Rev. Cytol. 1995;158:141–214. doi: 10.1016/s0074-7696(08)62487-0. [DOI] [PubMed] [Google Scholar]
- 8.Goding J. W. Ecto-enzymes: physiology meets pathology. J. Leukoc. Biol. 2000;67:285–311. doi: 10.1002/jlb.67.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann H. Ectonucleotidases: some recent developments and a note on nomenclature. Drug Dev. Res. 2001;52:44–56. [Google Scholar]
- 10.Zimmermann H. 5′-Nucleotidase: molecular structure and functional aspects. Biochem. J. 1992;285:345–365. doi: 10.1042/bj2850345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handa M., Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem. Biophys. Res. Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- 12.Smith T. M., Kirley T. L. Site-directed mutagenesis of a human brain ecto-apyrase: evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry. 1999;38:321–328. doi: 10.1021/bi9820457. [DOI] [PubMed] [Google Scholar]
- 13.Heine P., Braun N., Heilbronn A., Zimmermann H. Functional characterization of rat ecto-ATPase and ecto-ATP diphosphohydrolase after heterologous expression in CHO cells. Eur. J. Biochem. 1999;262:102–107. doi: 10.1046/j.1432-1327.1999.00347.x. [DOI] [PubMed] [Google Scholar]
- 14.Sévigny J., Sundberg C., Braun N., Guckelberger O., Csizmadia E., Qawi I., Imai M., Zimmermann H., Robson S. C. Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood. 2002;99:2801–2809. doi: 10.1182/blood.v99.8.2801. [DOI] [PubMed] [Google Scholar]
- 15.Kirley T. L. Complementary DNA cloning and sequencing of the chicken muscle ecto-ATPase. J. Biol. Chem. 1997;272:1076–1081. doi: 10.1074/jbc.272.2.1076. [DOI] [PubMed] [Google Scholar]
- 16.Chadwick B. P., Frischauf A.-M. Cloning and mapping of a human and mouse gene with homology to ecto-ATPase genes. Mamm. Genome. 1997;8:668–672. doi: 10.1007/s003359900534. [DOI] [PubMed] [Google Scholar]
- 17.Kegel B., Braun N., Heine P., Maliszewski C. R., Zimmermann H. An ecto-ATPase and an ecto-ATP diphosphohydrolase are expressed in rat brain. Neuropharmacology. 1997;36:1189–1200. doi: 10.1016/s0028-3908(97)00115-9. [DOI] [PubMed] [Google Scholar]
- 18.Mateo J., Harden T. K., Boyer J. L. Functional expression of a cDNA encoding a human ecto-ATPase. Br. J. Pharmacol. 1999;128:396–402. doi: 10.1038/sj.bjp.0702805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stout J. G., Kirley T. L. Control of cell membrane ecto-ATPase by oligomerization state: intermolecular cross-linking modulates ATPase activity. Biochemistry. 1996;35:8289–8298. doi: 10.1021/bi960563g. [DOI] [PubMed] [Google Scholar]
- 20.Vlajkovic S. M., Housley G. D., Greenwood D., Thorne P. R. Evidence for alternative splicing of ecto-ATPase associated with termination of purinergic transmission. Mol. Brain Res. 1999;73:85–92. doi: 10.1016/s0169-328x(99)00244-2. [DOI] [PubMed] [Google Scholar]
- 21.Vlajkovic S. M., Thorne P. R., Sévigny J., Robson S. C., Housley G. D. Distribution of ectonucleoside triphosphate diphosphohydrolases 1 and 2 in rat cochlea. Hear. Res. 2002;170:48–59. doi: 10.1016/s0378-5955(02)00460-4. [DOI] [PubMed] [Google Scholar]
- 22.Vlajkovic S. M., Thorne P. R., Sévigny J., Robson S. C., Housley G. D. NTPDase1 and NTPDase2 immunolocalisation in mouse cochlea: implications for regulation of P2 receptor signalling. J. Histochem. Cytochem. 2002;50:1435–1441. doi: 10.1177/002215540205001102. [DOI] [PubMed] [Google Scholar]
- 23.Braun N., Sévigny J., Mishra S. K., Robson S. C., Brath S. W., Gerstberger R., Hammer K., Zimmermann H. Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. Eur. J. Neurosci. 2003;17:1355–1364. doi: 10.1046/j.1460-9568.2003.02567.x. [DOI] [PubMed] [Google Scholar]
- 24.Mateo J., Kreda S., Henry C. E., Harden T. K., Boyer J. L. Requirement of Cys399 for processing of the human ecto-ATPase (NTPDase2) and its implications for determination of the activities of splice variants of the enzyme. J. Biol. Chem. 2003;278:39960–39968. doi: 10.1074/jbc.M307854200. [DOI] [PubMed] [Google Scholar]
- 25.Heine P., Braun N., Sévigny J., Robson S. C., Servos J., Zimmermann H. The C-terminal cysteine-rich region dictates specific catalytic properties in chimeras of the ectonucleotidases NTPDase1 and NTPDase2. Eur. J. Biochem. 2001;268:364–373. doi: 10.1046/j.1432-1033.2001.01896.x. [DOI] [PubMed] [Google Scholar]
- 26.Sarno S., Reddy H., Meggio F., Ruzzene M., Davies S. P., Donella-Deana A., Shugar D., Pinna L. A. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’) FEBS Lett. 2001;496:44–48. doi: 10.1016/s0014-5793(01)02404-8. [DOI] [PubMed] [Google Scholar]
- 27.Seibicke S., Strosznajder J. B., Haeffner E. W. Differential effect on inositol-phospholypid hydrolysis, cytosolic-free Ca2+ concentration, protein kinase C activity, and protein phosphorylation of 1-oleoyl-2-acetyl-sn-glycerol growth-stimulated ascites tumor cells. Eur. J. Cell Biol. 1988;46:403–410. [PubMed] [Google Scholar]
- 28.Seidler N. W., Jona I., Vegh M., Martonosi A. Cyclopiazonic acid is a specific inhibitor of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 1989;264:17816–17823. [PubMed] [Google Scholar]
- 29.Michel A. D., Chessell I. P., Hibell A. D., Simon J., Humphrey P. P. Identification and characterization of an endogenous P2X7 (P2Z) receptor in CHO-K1 cells. Br. J. Pharmacol. 1998;125:1194–1201. doi: 10.1038/sj.bjp.0702205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickenson J. M., Blank J. L., Hill S. J. Human adenosine A1 receptor and P2Y2-purinoceptor-mediated activation of the mitogen-activated protein kinase cascade in transfected CHO cells. Br. J. Pharmacol. 1998;124:1491–1499. doi: 10.1038/sj.bjp.0701977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vollmayer P., Koch M., Braun N., Heine P., Servos J., Israr E., Kegel B., Zimmermann H. Multiple ecto-nucleotidases in PC12 cells: identification and cellular distribution after heterologous expression. J. Neurochem. 2001;78:1019–1028. doi: 10.1046/j.1471-4159.2001.00480.x. [DOI] [PubMed] [Google Scholar]
- 32.Lazarowski E. R., Boucher R. C., Harden T. K. Interplay of consitutively released nucleotides, nucleotide metabolism and activity of P2Y receptors. Drug Dev. Res. 2001;53:66–71. [Google Scholar]
- 33.Lazarowski E. R., Boucher R. C., Harden T. K. Mechanisms of release of nucleotides and integration of their action as P2X- and P2Y-receptor activating molecules. Mol. Pharmacol. 2003;64:785–795. doi: 10.1124/mol.64.4.785. [DOI] [PubMed] [Google Scholar]
- 34.Knowles A. F., Nagy A. K. Inhibition of an ecto-ATP-diphosphohydrolase by azide. Eur. J. Biochem. 1999;262:349–357. doi: 10.1046/j.1432-1327.1999.00389.x. [DOI] [PubMed] [Google Scholar]
- 35.Braun N., Zhu Y., Krieglstein J., Culmsee C., Zimmermann H. Upregulation of the enzyme chain hydrolyzing extracellular ATP after transient forebrain ischemia in the rat. J. Neurosci. 1998;18:4891–4900. doi: 10.1523/JNEUROSCI.18-13-04891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonan C. D., Schetinger M. R. C., Battastini A. M. O., Sarkis J. J. F. Ectonucleotidases and synaptic plasticity: implications in physiological and pathological conditions. Drug Dev. Res. 2001;52:57–65. [Google Scholar]
- 37.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell (Cambridge, Mass.) 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 38.Wink M. R., Lenz G., Rodnight R., Sarkis J. J. F., Battastini A. M. O. Identification of brain ecto-apyrase as a phosphoprotein. Mol. Cell. Biochem. 2000;213:11–16. doi: 10.1023/a:1007194229587. [DOI] [PubMed] [Google Scholar]
- 39.Pinna L. A., Meggio F. Protein kinase CK2 (‘casein kinase-2’) and its implication in cell division and proliferation. Prog. Cell Cycle Res. 1997;3:77–97. doi: 10.1007/978-1-4615-5371-7_7. [DOI] [PubMed] [Google Scholar]
- 40.Gschwendt M. Protein kinase Cδ. Eur. J. Biochem. 1999;259:555–564. doi: 10.1046/j.1432-1327.1999.00120.x. [DOI] [PubMed] [Google Scholar]
- 41.Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marme D., Schächtele C. Selective inhibition of protein kinase C isoenzymes by the indolocarbazole Gö 6976. J. Biol. Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 42.Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H.-J., Johannes F.-J. Inhibition of protein kinase Cμ by various inhibitors. Differentiation from protein kinase C isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- 43.Redegeld F. A., Caldwell C. C., Sitkovsky M. V. Ecto-protein kinases: ecto-domain phosphorylation as a novel target for pharmacological manipulation? Trends Pharmacol. Sci. 1999;20:453–459. doi: 10.1016/s0165-6147(99)01399-1. [DOI] [PubMed] [Google Scholar]
- 44.Ren Z., Riley N. J., Garcia E. P., Sanders J. M., Swanson G. T., Marshall J. Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J. Neurosci. 2003;23:6608–6616. doi: 10.1523/JNEUROSCI.23-16-06608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Housley G. D., Greenwood D., Bennett T., Ryan A. F. Identification of a short form of the P2X2 purinoceptor subunit produced by alternative splicing in the pituitary and cochlea. Biochem. Biophys. Res. Commun. 1995;212:501–508. doi: 10.1006/bbrc.1995.1998. [DOI] [PubMed] [Google Scholar]
- 47.Braun N., Sévigny J., Robson S. C., Hammer K., Hanani M., Zimmermann H. Association of the ecto-ATPase NTPDase2 with glial cells of the peripheral nervous sytem. Glia. 2004;45:124–132. doi: 10.1002/glia.10309. [DOI] [PubMed] [Google Scholar]