Abstract
To examine the roles of active hypusinated eIF5A (eukaryotic translation initiation factor 5A) and polyamines in cell proliferation, mouse mammary carcinoma FM3A cells were treated with an inhibitor of deoxyhypusine synthase, GC7 (N1-guanyl-1, 7-diaminoheptane), or with an inhibitor of ornithine decarboxylase, DFMO (α-difluoromethylornithine), or with DFMO plus an inhibitor of spermine synthase, APCHA [N1-(3-aminopropyl)-cyclohexylamine]. Treatment with GC7 decreased the level of active eIF5A on day 1 without affecting cellular polyamine content, and inhibition of cell growth occurred from day 2. This delay reflects the fact that eIF5A was present in excess and was very stable in these cells. Treatment with DFMO or with DFMO plus APCHA inhibited cell growth on day 1. DFMO considerably decreased the levels of putrescine and spermidine, and the formation of active eIF5A began to decrease when the level of spermidine fell below 8 nmol/mg of protein after 12 h of incubation with DFMO. The combination of DFMO and APCHA markedly decreased the levels of putrescine and spermine and significantly decreased the level of spermidine, but did not affect the level of active eIF5A until day 3 when spermidine level decreased to 7 nmol/mg of protein. The results show that a decrease in either active eIF5A or polyamines inhibits cell growth, indicating that eIF5A and polyamines are independently involved in cell growth.
Keywords: biosynthetic inhibitor, cell proliferation, deoxyhypusine synthase, eukaryotic translation initiation factor 5A (eIF5A), hypusine, polyamine
Abbreviations: APCHA, N1-(3-aminopropyl)cyclohexylamine; DFMO, α-difluoromethylornithine; eIF5A, eukaryotic translation initiation factor 5A; FUT-175, 6-amino-2-naphthyl-4-guanidinobenzoate dihydrochloride; GC7, N1-guanyl-1,7-diaminoheptane
INTRODUCTION
Polyamines (putrescine, spermidine and spermine) are necessary for cell growth [1,2]. Since polyamines exist mainly as polyamine–RNA complexes in both Escherichia coli and animal cells [3,4], their proliferative effects are presumed to be primarily due to the stimulation of protein synthesis [2]. Indeed, we have found that protein synthesis in general is increased in the presence of polyamines through the stimulation of the maturation of 30 S ribosomal subunits [5,6], and that synthesis of several proteins, important for cell growth and viability, is stimulated by polyamines at the translation level in E. coli [7–9]. Regarding the proliferative effects of polyamines in mammalian cells, it is supposed that both the stimulation of protein synthesis by polyamines [10,11] and the function of active eIF5A (eukaryotic translation initiation factor 5A) [12–15] are involved in the cell proliferation; eIF5A contains hypusine [Nε-(4-amino-2-hydroxybutyl)lysine] derived from spermidine. The essential nature of spermidine for hypusine modification of eIF5A and thus for cell proliferation is well established [16]. Synthesis of hypusine in this protein occurs by conjugation of the aminobutyl moiety of spermidine to a specific lysine residue by deoxyhypusine synthase, followed by deoxyhypusine hydoroxylase-mediated hydroxylation. This posttranslational modification converts an inactive eIF5A precursor into an active eIF5A (hypusine-containing form).
Extensive studies to clarify the role of eIF5A and polyamines in cell proliferation have been undertaken using inhibitors of the polyamine biosynthetic enzymes. Inhibitors of S-adenosylmethionine decarboxylase, e.g. 5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxyadenosine (AbeAdo), caused the depletion of spermidine and spermine with a compensatory increase in putrescine, leading to a delayed cytostasis in L1210 cells [17]. This growth arrest of L1210 cells by AbeAdo was supposed to be attributed mainly to the deprivation of hypusine in eIF5A. Inhibition of ornithine decarboxylase by DFMO (α-difluoromethylornithine) caused an effective depletion of putrescine and spermidine, and this was accompanied by the inhibition of cell proliferation in rat HTC (hepatoma tissue culture) cells [18] and other mammalian cells [19–21]. Since only a very small portion of cellular spermidine was used for hypusine synthesis, it was not clear whether the growth inhibition, in the case of DFMO treatment, was due to the depletion of hypusine-containing eIF5A or a decrease in total polyamines. We had reported previously that an inhibitor of spermine synthase, APCHA [N1-(3-aminopropyl)cyclohexylamine], inhibited growth of FM3A cells without a significant reduction of cellular spermidine [22]. It had been also reported that GC7 (N1-guanyl-1,7-diaminoheptane), a potent inhibitor of deoxyhypusine synthase, selectively inhibits the formation of active eIF5A [13].
In the present study, we compared the effects of GC7 (inhibition of hypusine synthesis without decrease in polyamines), DFMO (decrease in both total polyamines and hypusine synthesis) and DFMO plus APCHA (reduction of total polyamines with little or no reduction of hypusine synthesis) on the growth of FM3A cells. Our results demonstrate that both eIF5A and polyamines influence cell growth, but in an independent manner.
MATERIALS AND METHODS
Materials
Antibodies against human recombinant eIF5A precursor and rat deoxyhypusine synthase were prepared as described previously [15,23]. GC7, an inhibitor of deoxyhypusine synthase was prepared by the method of Jakus et al. [13]. APCHA, an inhibitor of spermine synthase, was synthesized as described previously [24]. DFMO, an irreversible inhibitor of ornithine decarboxylase, was donated by Aventis Pharma (Tokyo, Japan). FUT-175 (6-amino-2-naphthyl-4-guanidinobenzoate dihydrochloride), an inhibitor of serine protease [25], was a gift from Torii Pharmaceutical (Tokyo, Japan).
Cell culture and preparation of cell lysate
Mouse mammary carcinoma FM3A cells (Japan Health Science Foundation) were cultured in ES medium (Nissui Pharmaceutical, Tokyo, Japan), supplemented with 50 units/ml streptomycin, 100 units/ml penicillin G and 2% heat-inactivated fetal calf serum at 37 °C in an atmosphere of 5% CO2, according to the method described previously [26]. FM3A cells (2×106 cells) were suspended in 0.1 ml of a buffer containing 25 mM Hepes/KOH, pH 7.8, 0.1 mM EDTA, 6 mM 2-mercaptoethanol, 5% (v/v) glycerol and 20 μM FUT-175. The cells were lysed by repeated (three times) freezing and thawing with intermittent mechanical mixing. Cell lysate was obtained by centrifugation at 17000 g for 15 min. Protein content was determined by the method of Lowry et al. [27].
Measurement of polyamines
Polyamine contents in FM3A cells were determined according to the method described previously [28]. Cells were treated with 5% (w/v) trichloroacetic acid and centrifuged at 12000 g for 10 min. Polyamines in the supernatant were measured by using a TOSOH HPLC system (Tosoh, Tokyo, Japan) [28]. The precipitate was used to determine the protein content [27].
Measurement of hypusine formation and the half-life of eIF5A
To measure the hypusine formation in eIF5A, FM3A cells (10 ml) were cultured with 925 kBq [3H]spermidine (832.5 GBq/mmol; PerkinElmer Japan, Yokohama, Japan) in the presence of 1 mM aminoguanidine, an inhibitor of serum amine oxidase [29], for 12 or 24 h, and cell lysate was prepared as described above. Cell lysate (15 μg of protein) was separated by SDS/PAGE on a 13.5% gel as described by Laemmli [30], and fluorography was performed by the methods of Laskey and Mills [31]. Radioactivity was quantified using a Fujix Bas 2000II imaging analyser. Newly synthesized hypusine in active eIF5A was estimated based on the specific activity of [3H]spermidine in the cells, which was calculated from the radioactivity of incorporated [3H]spermidine for either 12 or 24 h and the spermidine content of the harvested cells. Cellular distribution of [3H]spermidine in cells should be same as that of non-labelled spermidine because cells were labelled with [3H]spermidine for 12 or 24 h.
To determine the half-life of eIF5A, cells were cultured in the presence of 2 μg/ml of cycloheximide (Sigma), an inhibitor of protein synthesis, at the designated time, and cell lysate was prepared as described above. Under these conditions, no significant cell growth was observed during the 36 h incubation. For Western blotting, cell lysate (15 μg of protein) was separated by SDS/PAGE, transferred on to Immobilon transfer membrane (Millipore), and eIF5A was detected by using an antibody against eIF5A [15], followed by ECL™ Western blotting detection reagents (Amersham Biosciences). The level of eIF5A was quantified by a LAS-1000 plus luminescent image analyser (Fuji Film). The half-life of active eIF5A was also estimated from the decay rate of 3H-labelled eIF5A. After cells were incubated with [3H]-spermidine for 24 h as described above, cells were washed with medium containing 1 mM spermidine and 1 mM aminoguanidine. Then, cells were incubated in the presence of 2 μg/ml cycloheximide and at the designated time the radioactivity remaining in the active eIF5A was estimated by fluorography.
Two-dimensional gel electrophoresis of proteins for detection of eIF5A precursors and the active protein
Cell lysate (200 μg of protein) for two-dimensional gel electrophoresis was prepared by ReadyPrep™ Sequential Extraction kit (Bio-Rad Laboratories) according to the recommended method. The first dimensional isoelectric focusing was performed in the PROTEAN® IEF Cell using 11 cm (pH range 4–7) ReadyStrip™ IPG Strip gel (Bio-Rad Laboratories). The proteins on the first dimensional Strip gel were further separated, in the second dimension, by SDS/PAGE on a 13.5% polyacrylamide gel, and transferred on to Immobilon transfer membrane (Millipore) for detection of the various forms of eIF5A. Western blotting was performed as described above using antibody raised against the recombinant eIF5A precursor. The position of active eIF5A was confirmed by fluorography using eIF5A labelled with [3H]spermidine.
Measurement of the level and the activity of deoxyhypusine synthase in FM3A cells
Cell lysate was prepared as described above. To determine the level of deoxyhypusine synthase, Western blotting was performed as described above using 30 μg protein of cell lysate and antibody against deoxyhypusine synthase [23]. The activity of deoxyhypusine synthase was measured as described previously [15]. The reaction mixture (50 μl) contained 250 mM glycine/NaOH, pH 9.3, 2 mM dithiothreitol, 10% glycerol, 0.5 mM NAD+, 150 mM KCl, 2 mM MgCl2, 10 μM FUT-175, 20 μM human eIF5A precursor prepared in E. coli, 30 μg protein of cell lysate and 15 μM [3H]spermidine (98.7 GBq/mmol). Cell lysate was dialysed before use against a buffer containing 50 mM Tris/HCl, pH 7.0, 1 mM dithiothreitol, 10% glycerol and 10 μM FUT-175. The reaction mixture, except [3H]spermidine, was preincubated at 37 °C for 10 min, and further incubation of the reaction mixture with [3H]spermidine was performed at 37 °C for 1 h. After incubation, 5% trichloroacetic acid-insoluble radioactivity of the reaction mixture (40 μl) was measured in a liquid-scintillation counter. The amino acid sequence of human eIF5A is exactly the same as that of mouse eIF5A [32].
RESULTS
Effects of decreasing active eIF5A
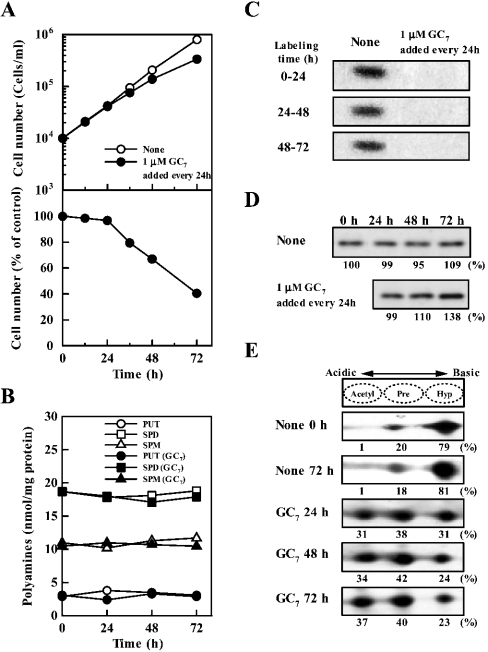
It has been reported that GC7 selectively inhibits the formation of active eIF5A (i.e. eIF5A containing hypusine) by inhibiting deoxyhypusine synthase [13]. We developed a paradigm in which cells were cultured for 72 h and 1 μM GC7 was added to the medium every 24 h. As shown in Figures 1(A) and 1(C), cell growth was not inhibited significantly on day 1 but it decreased by 30–60% on days 2–3, although formation of active eIF5A, measured by labelling of eIF5A with [3H]spermidine, was strongly inhibited even within the first 24 h. The degree of inhibition of active eIF5A formation was approx. 90% when the band detected by fluorography was quantified (Figure 1C). Under these conditions, there were no noticeable changes in the polyamine levels (Figure 1B). The level of total (active and inactive) eIF5A was then measured by Western-blot analysis using an antibody that recognizes both the active eIF5A and the inactive precursor proteins. In normal FM3A cells, the level of eIF5A was nearly equal during the 72 h cell culture (Figure 1D). However, in GC7 treated cells, there was actually an increase in the level of eIF5A (Figure 1D). It has been reported that there are three kinds of eIF5A: active eIF5A (hypusinated at Lys50), eIF5A precursor and acetylated eIF5A precursor (acetylated at Lys47) [33,34]. We measured active eIF5A, eIF5A precursor and acetylated eIF5A precursor by two-dimensional gel electrophoresis followed by Western blotting. As shown in Figure 1(E), approx. 80% was the active form when cells grew normally. The percentage of active eIF5A did not change significantly during the 72 h cell culture. By treating cells with GC7, the synthesis of active eIF5A was strongly inhibited. Thus the percentage of active eIF5A was decreased to 31% in cells cultured for 24 h, whereas the levels of both eIF5A precursor and acetylated eIF5A precursor increased. There was only a small further decrease in the percentage of active eIF5A between 24 and 72 h (Figure 1E).
Figure 1. Effect of 1 μM GC7 added every 24 h on cell growth (A), polyamine content (B), active eIF5A formation (C), total (active and inactive) level of eIF5A (D) and level of active eIF5A (E).
Cells were cultured for 72 h by adding 1 μM GC7 to the medium every 24 h. (A) The number of cells was counted in the presence of 0.05% Trypan Blue. Each value is the mean for triplicate determinations. S.D. was within ±10% for each point. (B) Polyamine contents in cells on days 1, 2 and 3 were measured by HPLC as described in the Materials and methods section. Each value is the mean for triplicate determinations. S.D. was within ±10% for each point. PUT, putrescine; SPD, spermidine; SPM, spermine. (C) Active eIF5A formation (hypusination of eIF5A) was measured as described in the Materials and methods section by labelling of eIF5A with [3H]spermidine for 24 h. For example, for 0–24 h data, cells were cultured in the presence of [3H]spermidine from 0 to 24 h. (D) Total (active and inactive) level of eIF5A was measured by Western blotting. (E) Acetylated eIF5A precursor, eIF5A precursor and active hypusinated eIF5A were separated by two-dimensional gel electrophoresis using 200 μg of protein from cell lysate. Intensity of each band for acetylated eIF5A precursor, eIF5A precursor and active eIF5A was measured using a LAS-1000 plus luminescent image analyser, and it was shown as percentage in total eIF5A.
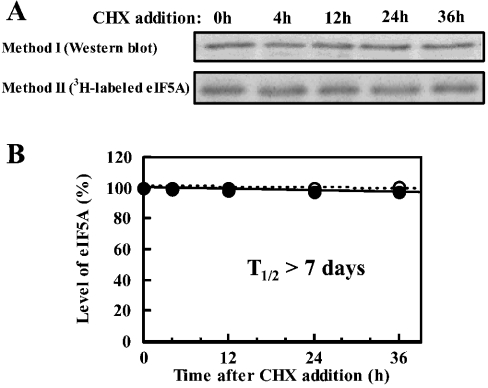
A possible reason for the weak inhibitory effect of GC7 on cell growth, in spite of the almost complete inhibition of new synthesis of active eIF5A, may be due to the stability of preformed eIF5A. We thus attempted to determine the stability of the eIF5A protein by measuring eIF5A by Western blotting or the decay of the labelled active eIF5A in the presence of cycloheximide. As shown in Figure 2, eIF5A was very stable, with a very long half-life (probably >7 days). A long half-life of eIF5A was reported in CHO and JURKAT T-cells [35,36]. Furthermore, the number of cells increased approx. 4-fold on day 1, but no significant effect of GC7 on cell growth was observed on day 1 although de novo formation of active eIF5A was strongly inhibited by GC7 at this time. These results suggest that active eIF5A is present in excess in FM3A cells. The correlation between the inhibition of cell growth and formation of active eIF5A by GC7 is summarized in Table 1. Inhibition of active eIF5A formation was calculated from the cell number (Figure 1A), total (active and inactive) eIF5A (Figure 1D) and the percentage of active eIF5A (Figure 1E). The percentage of the inhibition of active eIF5A formation on day 1, 2 and 3 was 82, 81 and 86% respectively, which is in good accordance with the value calculated from the labelling of eIF5A with [3H]spermidine (Figure 1C). The degree of inhibition of active eIF5A formation was always greater than the degree of inhibition of cell growth, supporting the idea that active eIF5A exists in excess in FM3A cells. The relative amount of active eIF5A in GC7 treated cells on day 1 was approx. 40% compared with that in normal cells, suggesting that the level of active eIF5A existing in untreated FM3A cells is at least 2.5 times more than the minimal level necessary for the cell growth.
Figure 2. Determination of half-life of eIF5A.
Cells (4×105/ml) were treated with 2 μg/ml cycloheximide (CCHX). Method I: at the designated time (0–36 h), the level of eIF5A was determined by Western blotting (A) and intensity of the band was estimated with a LAS-1000 plus luminescent image analyser (B). Method II: the level of active eIF5A prelabelled with [3H]spermidine was followed for 36 h by fluorography after treating cells with 2 μg/ml cycloheximide (A) and the intensity of the band was estimated with a Fujix Bas 2000II imaging analyser (B). Essentially the same results were obtained after treating cells with 10 μg/ml cycloheximide. (B) ●, eIF5A measured by Western blotting; ○, measurement of [3H]spermidine-labelled eIF5A. Each value is the mean for duplicate determinations.
Table 1. Correlation between inhibition of cell growth and of active eIF5A formation by GC7.
| Level of eIF5A (arbitrary units) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell growth | −GC7 | +GC7 | ||||||||||
| Number of cells/ml | Relative level of eIF5A/cell(%) | |||||||||||
| Day | −GC7 | +GC7 | Inhibition (%) | Total eIF5A/cell* | Active eIF5A/total cells† | Increased active eIF5A‡ | Total eIF5A/cell* | Active eIF5A/total cells† | Increased active eIF5A‡ | Inhibition by GC7 (%)§ | −GC7 | +GC7∥ |
| 0 | 1×104 | 1×104 | 0 | 1.00 | 79 | − | 1.00 | 79 | − | − | 100 | 100 |
| 1 | 4.26×104 | 4.12×104 | 3.3 | 0.99 | 334 | 255 | 0.99 | 126 | 47 | 82 | 99 | 39 |
| 2 | 20.7×104 | 13.9×104 | 32.9 | 0.95 | 1550 | 1470 | 1.10 | 367 | 288 | 81 | 95 | 36 |
| 3 | 80.0×104 | 33.7×104 | 57.9 | 1.09 | 6890 | 6810 | 1.38 | 1070 | 991 | 86 | 109 | 45 |
* Value from Figure 1(D).
† Percentage of active eIF5A (Figure 1E)×cell number×column*÷104.
‡ Value of column† on each day−the value of column† on day 0 (79).
§ Percentage inhibition of active eIF5A formation by GC7 was calculated by [column‡ (−GC7)−column† (+GC7)]÷column‡ (−GC7)×100.
∥ column*×percentage of active eIF5A (Figure 1E) divided by 79.
Effects of simultaneously decreasing eIF5A and polyamines
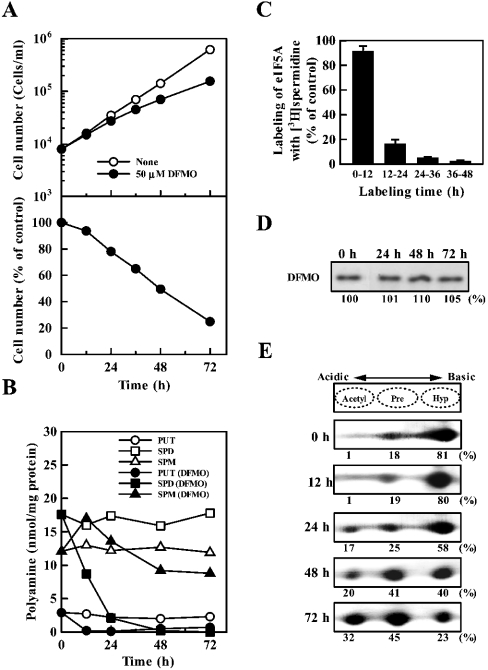
The effects of changing the levels of active eIF5A and polyamines together were examined. In these experiments, cells were treated with DFMO (an inhibitor of ornithine decarboxylase), which greatly decreases the levels of putrescine and spermidine. Under these conditions, there was inhibition of cell growth (Figure 3A), and a decrease in polyamine content and formation of active eIF5A (Figures 3B and 3C). Levels of putrescine and spermidine decreased significantly, but the decrease in spermine content was small (Figure 3B). The formation of active eIF5A started to decrease from 12 h after the onset of incubation (Figure 3C). At 12 h, spermidine content decreased from 18 to 8 nmol/mg of protein (Figure 3B). The percentage of active eIF5A on days 1, 2 and 3 decreased to 58, 40 and 23% respectively (Figure 3E). The level of total (active and inactive) eIF5A was nearly equal after DFMO treatment for 72 h (Figure 3D). The results indicate that the inhibition of cell growth by DFMO on day 1 was mainly due to a decrease in polyamine content, since inhibition of active eIF5A formation starts after 12 h of incubation and excess amounts of active eIF5A are present at the onset of incubation. On days 2 and 3, a decrease in both eIF5A and polyamines was probably involved in the inhibition of cell growth although the degree of contribution of each factor was not clear.
Figure 3. Effect of DFMO on cell growth (A), polyamine content (B), active eIF5A formation (C), total (active and inactive) level of eIF5A (D) and level of active eIF5A (E).
Cells were cultured in the presence of 50 μM DFMO for 72 h. Essentially, the same results were obtained in the presence of 150 μM DFMO. Experiments were performed as described in the legend of Figure 1, except that the labelling time of eIF5A with [3H]spermidine in control and DFMO-treated cells was for 12 h in (C). (A, B) Each value is the mean for triplicate determinations. S.D. was within ±10% for each point. (C) The values are expressed as means±S.D. for triplicate determinations.
Effects of reducing polyamine levels
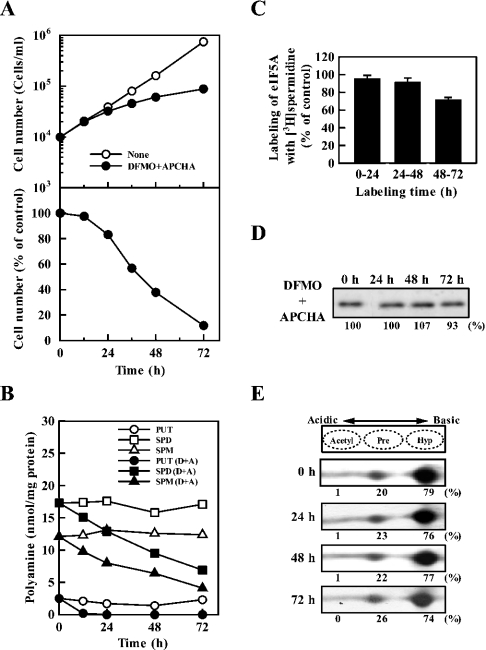
We then examined the effects of reducing polyamine levels under conditions where the formation of eIF5A was unaltered. For this purpose, cells were treated with DFMO and APCHA, an inhibitor of spermine synthase [24]. As shown in Figures 4(A) and 4(C), cell growth was inhibited at 24–72 h, and the formation of active eIF5A started to decrease on day 3. However, the level of total and active eIF5A was nearly equal for 72 h (Figures 4D and 4E). The percentage of active eIF5A on days 1, 2 and 3 was 76, 77 and 74% respectively. Under these conditions, the levels of putrescine, spermidine and spermine decreased, and spermidine content on day 3 was approx. 7 nmol/mg of protein compared with 18 nmol/mg of protein in untreated cells (Figure 4B). The results indicate that cell growth was strongly inhibited by decrease in polyamines without influencing the active eIF5A formation. During 48 to 72 h incubation, active eIF5A formation was inhibited by approx. 30%, and the level of spermidine decreased from 9 to 7 nmol/mg of protein. These results are in accordance with the previous results obtained with DFMO only: active eIF5A formation started to decrease when spermidine content decreased to 8 nmol/mg of protein. The results, taken together, indicate that both eIF5A and polyamines independently influence cell growth.
Figure 4. Effect of DFMO and APCHA on cell growth (A), polyamine content (B), active eIF5A formation (C), total (active and inactive) level of eIF5A (D) and level of active eIF5A (E).
Cells were cultured in the presence of 50 μM DFMO and 150 μM APCHA for 72 h as described previously [22]. Experiments were performed as described in the legend of Figure 1. (A, B) Each value is the mean for triplicate determinations. S.D. was within ±10% for each point. (C) Labelling time of eIF5A with [3H]spermidine in control and both DFMO- and APCHA-treated cells was for 24 h. The values are expressed as mean±S.D. for triplicate determinations.
Effect of decreasing active eIF5A and polyamines on deoxyhypusine synthase
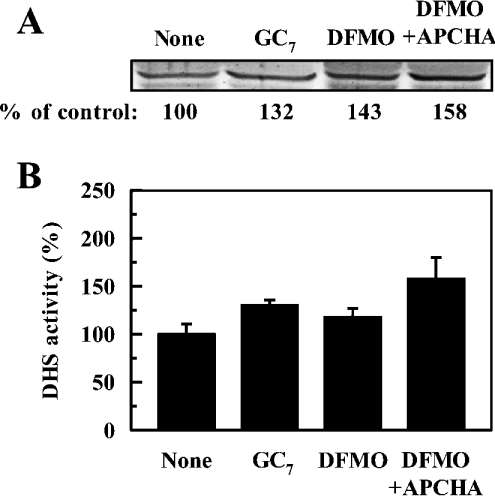
Since deoxyhypusine synthase appears to be a limiting factor for the formation of active eIF5A in FM3A cells, the level and the activity of deoxyhypusine synthase were measured under various conditions. As shown in Figure 5, both the level and the activity of deoxyhypusine synthase were slightly increased by the treatment with GC7, DFMO and the combination of DFMO plus APCHA. Thus the activity of deoxyhypusine synthase does not seem to be regulated by the cellular level of active eIF5A or polyamines. The results indicate that the inhibition of hypusine formation in DFMO-treated cells is due to the depletion of the substrate, spermidine, rather than due to a decrease in the level and the activity of deoxyhypusine synthase.
Figure 5. Effect of GC7, DFMO and DFMO plus APCHA on deoxyhypusine synthase.
Cells were cultured for 72 h in the absence and presence of 1 μM GC7 added every 24 h, 50 μM DFMO or 50 μM DFMO plus 150 μM APCHA. The level (A) of deoxyhypusine synthase (DHS) was measured by Western-blot analysis and the activity (B) was measured at pH 9.3 using dialysed cell lysate. The 100% activity of deoxyhypusine synthase corresponds to 4.26±0.53 pmol·min−1·(mg of protein)−1. The values are means±S.D. for triplicate determinations.
DISCUSSION
We have studied the properties of active eIF5A, its relationship with levels of polyamines and its role in cell proliferation. We confirmed that eIF5A is a very stable protein (T1/2>7 days). During the first 24 h exposure to GC7, the number of FM3A cells increased 4-fold and the formation of active eIF5A was inhibited by approx. 90% but the cell growth was not affected. Our results suggest that there is at least a 2.5-fold excess of active eIF5A normally present in FM3A cells. The synthesis of eIF5A precursor and the activity of deoxyhypusine synthase were not influenced significantly by the level of polyamines. Thus the level of active eIF5A in cells is strongly dependent on the level of spermidine. We measured the Km value of spermidine for human deoxyhypusine synthase in the presence of 150 mM KCl and 2 mM Mg2+ at pH 9.3, and it was estimated to be 21 μM. The Km value was higher than the value (7.2 μM) measured in the absence of KCl and MgCl2 [37], and it increased significantly when the assay was performed at neutral pH (results not shown). The formation of active eIF5A started to decrease when spermidine concentration in cells decreases from 18 to 8 nmol/mg of protein. The cellular distribution of spermidine was determined by the method of Watanabe et al. [3], and it was estimated that 8 nmol spermidine/mg of protein corresponds to 63 μM free spermidine. If the Km value were <10 μM, a decrease in the formation of active eIF5A would start when spermidine content decreased to 4 nmol/mg of protein, which corresponds to approx. 30 μM free spermidine. Thus we expect the Km value of spermidine for deoxyhypusine synthase in FM3A cells to be approx. 20–30 μM. The spermidine content in mammary carcinoma FM3A cells is high compared with normal cells that make up various organs and tissues (7–10 nmol/mg of protein; [3]). So, the level of eIF5A is more closely related to spermidine content in normal cells. If polyamine levels begin to decrease, the level of eIF5A would decrease in parallel.
We confirmed that the inhibition of hypusine synthesis, either by inhibition of deoxyhypusine synthase with GC7 or by depletion of the substrate spermidine in DFMO-treated cells, leads to arrest in cell proliferation. Our results indicate that in addition to the indispensable role of spermidine for hypusine modification in eIF5A, polyamines are also required for optimal growth of mammalian cells. This is the first report that clearly shows the critical requirement of polyamines, independent of hypusine synthesis, in mammalian cell proliferation. We previously reported that the role of polyamines in cell proliferation is mainly at the translation level, analogous to their role in E. coli [10,11]. Since eIF5A is a protein with a long half-life and active eIF5A exists approximately at a 2.5-fold excess in FM3A cells, the growth inhibition observed in DFMO-treated FM3A cells during the 48 h (Figure 3) may be largely due to a decrease in total polyamines rather than reduction of active eIF5A. However, the sensitivity or response to inhibitors of polyamine biosynthesis may vary in different mammalian cells, since the levels of polyamines and eIF5A are different in each cell type. Thus the participation of active eIF5A and polyamines in cell proliferation has to be analysed carefully in each cell type.
Our results indicate that polyamines and eIF5A can independently affect cell growth. However, the physiological functions of eIF5A are still not well understood. It was first identified as a stimulator of the synthesis of methionylpuromycin by Kemper et al. [38]. In addition, eIF5A directly influences the nuclear export of Rev, a nucleolar protein, which shuttles constantly between the nucleus and cytoplasm [39]. Then, Zuk and Jacobson [40] reported that decapped mRNA accumulates at the non-permissive temperature using yeast eIF5A temperature-sensitive mutant. We confirmed that decapped mRNA accumulates in mouse FM3A cells in which the formation of active eIF5A is inhibited by deoxyspergualin [15]. We also observed that decapped mRNA was accumulated in the cells treated with GC7 (K. Nishimura and K. Igarashi, unpublished work). Thus we think that one important function of eIF5A is at the level of mRNA turnover, probably acting downstream of decapping. If specific kinds of mRNAs are decapped and accumulated after the treatment of cells with GC7, the identification of these mRNAs would help to clarify the physiological functions of eIF5A.
Acknowledgments
We thank Dr K. Williams and Dr A. J. Michael for their help in preparing this manuscript. We also thank Aventis Pharma and Torii Pharmaceutical for providing DFMO and FUT-175. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Cohen S. S. Oxford: Oxford University Press; 1998. A Guide to the Polyamines; pp. 1–595. [Google Scholar]
- 2.Igarashi K., Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe S., Kusama-Eguchi K., Kobayashi H., Igarashi K. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem. 1991;266:20803–20809. [PubMed] [Google Scholar]
- 4.Miyamoto S., Kashiwagi K., Ito K., Watanabe S., Igarashi K. Estimation of polyamine distribution and polyamine stimulation of protein synthesis in Escherichia coli. Arch. Biochem. Biophys. 1993;300:63–68. doi: 10.1006/abbi.1993.1009. [DOI] [PubMed] [Google Scholar]
- 5.Igarashi K., Kishida K., Kashiwagi K., Tatokoro I., Kakegawa T., Hirose S. Relationship between methylation of adenine near the 3′ end of 16-S ribosomal RNA and the activity of 30-S ribosomal subunits. Eur. J. Biochem. 1981;113:587–593. doi: 10.1111/j.1432-1033.1981.tb05103.x. [DOI] [PubMed] [Google Scholar]
- 6.Igarashi K., Kashiwagi K., Kishida K., Kakegawa T., Hirose S. Decrease in the S1 protein of 30-S ribosomal subunits in polyamine-requiring mutants of Escherichia coli grown in the absence of polyamines. Eur. J. Biochem. 1981;114:127–131. doi: 10.1111/j.1432-1033.1981.tb06182.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida M., Meksuriyen D., Kashiwagi K., Kawai G., Igarashi K. Polyamine stimulation of the synthesis of oligopeptide-binding protein (OppA). Involvement of a structural change of the Shine-Dalgarno sequence and the initiation codon AUG in OppA mRNA. J. Biol. Chem. 1999;274:22723–22728. doi: 10.1074/jbc.274.32.22723. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida M., Kashiwagi K., Kawai G., Ishihama A., Igarashi K. Polyamine enhancement of the synthesis of adenylate cyclase at the translational level and the consequential stimulation of the synthesis of the RNA polymerase σ28 subunit. J. Biol. Chem. 2001;276:16289–16295. doi: 10.1074/jbc.M011059200. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida M., Kashiwagi K., Kawai G., Ishihama A., Igarashi K. Polyamines enhance synthesis of the RNA polymerase σ38 subunit by suppression of an amber termination codon in the open reading frame. J. Biol. Chem. 2002;277:37139–37146. doi: 10.1074/jbc.M206668200. [DOI] [PubMed] [Google Scholar]
- 10.Ogasawara T., Ito K., Igarashi K. Effect of polyamines on globin synthesis in a rabbit reticulocyte polyamine-free protein synthetic system. J. Biochem. (Tokyo) 1989;105:164–167. doi: 10.1093/oxfordjournals.jbchem.a122633. [DOI] [PubMed] [Google Scholar]
- 11.Shimogori T., Kashiwagi K., Igarashi K. Spermidine regulation of protein synthesis at the level of initiation complex formation of Met-tRNAi, mRNA and ribosomes. Biochem. Biophys. Res. Commun. 1996;223:544–548. doi: 10.1006/bbrc.1996.0931. [DOI] [PubMed] [Google Scholar]
- 12.Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., Hershey J. W. B. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakus J., Wolff E. C., Park M. H., Folk J. E. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines. J. Biol. Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- 14.Park M. H., Joe Y. A., Kang K. R. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:1677–1683. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura K., Ohki Y., Fukuchi-Shimogori T., Sakata K., Saiga K., Beppu T., Shirahata A., Kashiwagi K., Igarashi K. Inhibition of cell growth through inactivation of eukaryotic translation initiation factor 5A (eIF5A) by deoxyspergualin. Biochem. J. 2002;363:761–768. doi: 10.1042/0264-6021:3630761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park M. H., Lee Y. B., Joe Y. A. Hypusine is essential for eukaryotic cell proliferation. Biol. Signals. 1997;6:115–123. doi: 10.1159/000109117. [DOI] [PubMed] [Google Scholar]
- 17.Byers T. L., Ganem B., Pegg A. E. Cytostasis induced in L1210 murine leukemia cells by the S-adenosyl-L-methionine decarboxylase inhibitor 5′-{[(z)-4-amino-2-butenyl]methylamino}-5′-deoxyadenosine may be due to hypusine depletion. Biochem. J. 1992;287:717–724. doi: 10.1042/bj2870717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mamont P. S., Duchesne M.-C., Grove J., Bey P. Anti-proliferate properties of DL-α-difuloromethylornithine in cultured cells. A consequence of the irreversible inhibition of ornithine decarboxylase. Biochem. Biophys. Res. Commun. 1978;81:58–66. doi: 10.1016/0006-291x(78)91630-3. [DOI] [PubMed] [Google Scholar]
- 19.McConlogue L., Coffino P. A mouse lymphoma cell mutant whose major protein product is ornithine decarboxylase. J. Biol. Chem. 1983;258:12083–12086. [PubMed] [Google Scholar]
- 20.Fukuchi J., Kashiwagi K., Kusama-Eguchi K., Terao K., Shirahata A., Igarashi K. Mechanism of the inhibition of cell growth by N1, N12-bis(ethyl)spermine. Eur. J. Biochem. 1992;209:689–696. doi: 10.1111/j.1432-1033.1992.tb17337.x. [DOI] [PubMed] [Google Scholar]
- 21.Korhonen V. P., Niiranen K., Halmekyto M., Pietila M., Diegelman P., Parkkinen J. J., Eloranta T., Porter C. W., Alhonen L., Jänne J. Spermine deficiency resulting from targeted disruption of the spermine synthase gene in embryonic stem cells leads to enhanced sensitivity to antiproliferative drugs. Mol. Pharmacol. 2001;59:231–238. doi: 10.1124/mol.59.2.231. [DOI] [PubMed] [Google Scholar]
- 22.He Y., Shimogori T., Kashiwagi K., Shirahata A., Igarashi K. Inhibition of cell growth by combination of α-difluoromethylornithine and inhibitor of spermine synthase. J. Biochem. (Tokyo) 1995;117:824–829. doi: 10.1093/oxfordjournals.jbchem.a124782. [DOI] [PubMed] [Google Scholar]
- 23.Wolff E. C., Lee Y. B., Chung S. I., Folk J. E., Park M. H. Deoxyhypusine synthase from rat testis: purification and characterization. J. Biol. Chem. 1995;270:8660–8666. doi: 10.1074/jbc.270.15.8660. [DOI] [PubMed] [Google Scholar]
- 24.Shirahata A., Takahashi N., Beppu T., Hosoda H., Samejima K. Effects of inhibitors of spermidine synthase and spermine synthase on polyamine biosynthesis in rat tissues. Biochem. Pharmacol. 1993;45:1897–1903. doi: 10.1016/0006-2952(93)90449-7. [DOI] [PubMed] [Google Scholar]
- 25.Fujii S., Hitomi Y. New synthetic inhibitors of C1r, C1 esterase, thrombin, plasmin, kallikrein and trypsin. Biochim. Biophys. Acta. 1981;661:342–345. doi: 10.1016/0005-2744(81)90023-1. [DOI] [PubMed] [Google Scholar]
- 26.Ayusawa D., Iwata K., Seno T. Alteration of ribonucleotide reductase in aphidicolin-resistant mutants of mouse FM3A cells with associated resistance to arabinosyladenine and arabinosylcytosine. Somat. Cell Genet. 1981;7:27–42. doi: 10.1007/BF01544746. [DOI] [PubMed] [Google Scholar]
- 27.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 28.Igarashi K., Kashiwagi K., Hamasaki H., Miura A., Kakegawa T., Hirose S., Matsuzaki S. Formation of a compensatory polyamine by Escherichia coli polyamine requiring mutants during growth in the absence of polyamines. J. Bacteriol. 1986;166:128–134. doi: 10.1128/jb.166.1.128-134.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shore P. A., Cohn V. H., Jr Comparative effects of monoamine oxidase inhibitors on monoamine oxidase and diamine oxidase. Biochem. Pharmacol. 1960;5:91–95. doi: 10.1016/0006-2952(60)90012-5. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur. J. Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins Z. A., Hååg P. G., Johansson H. E. Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics. 2001;71:101–109. doi: 10.1006/geno.2000.6418. [DOI] [PubMed] [Google Scholar]
- 33.Park M. H. The identification of an eukaryotic initiation factor 4D precursor in spermidine deleted Chinese hamster ovary cells. J. Biol. Chem. 1988;263:7447–7449. [PubMed] [Google Scholar]
- 34.Klier H., Csonga R., Joao H. C., Eckerskorn C., Auer M., Lottspeich F., Eder J. Isolation and structural characterization of different isoforms of the hypusine-containing protein eIF-5A from HeLa cells. Biochemistry. 1995;34:14693–14702. doi: 10.1021/bi00045a010. [DOI] [PubMed] [Google Scholar]
- 35.Torrelio B. M., Paz M. A., Gallop P. M. The formation and stability of the hypusine containing protein in Chinese hamster ovary cells. Biochem. Biophys. Res. Commun. 1987;145:1335–1341. doi: 10.1016/0006-291x(87)91584-1. [DOI] [PubMed] [Google Scholar]
- 36.Bergeron R. J., Weimer W. R., Müller R., Zimmerman C. O., McCosar B. H., Yao H., Smith R. E. Effect of polyamine analogues on hypusine content in JURKAT T-cells. J. Med. Chem. 1998;41:3901–3908. doi: 10.1021/jm980390o. [DOI] [PubMed] [Google Scholar]
- 37.Joe Y. A., Wolff E. C., Park M. H. Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins. J. Biol. Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- 38.Kemper W. M., Berry K. W., Merrick W. C. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- 39.Bevec D., Jaksche H., Oft M., Wöhl T., Himmelspach M., Pacher A., Schebesta M., Koettnitz K., Dobrovnik M., Csonga R., et al. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 40.Zuk D., Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]