Abstract
Dietary vegetable oils and fish oils rich in PUFA (polyunsaturated fatty acids) exert hypocholesterolaemic and hypotriglyceridaemic effects in rodents. The plasma cholesterol-lowering properties of PUFA are due partly to a diminution of cholesterol synthesis and of the activity of the rate-limiting enzyme HMG-CoA reductase (3-hydroxy-3-methylglutaryl-CoA reductase). To better understand the mechanisms involved, we examined how tuna fish oil and individual n−3 and n−6 PUFA affect the expression of hepatic FPP synthase (farnesyl diphosphate synthase), a SREBP (sterol regulatory element-binding protein) target enzyme that is subject to negative-feedback regulation by sterols, in co-ordination with HMG-CoA reductase. Feeding mice on a tuna fish oil diet for 2 weeks decreased serum cholesterol and triacylglycerol levels, by 50% and 60% respectively. Hepatic levels of FPP synthase and HMG-CoA reductase mRNAs were also decreased, by 70% and 40% respectively. Individual n−3 and n−6 PUFA lowered FPP synthase and HMG-CoA reductase mRNA levels in H4IIEC3 rat hepatoma cells to a greater extent than did stearate and oleate, with the largest inhibitory effects occurring with arachidonate, EPA (eicosapentaenoic acid) and DHA (docosahexaenoic acid). We observed a similar inhibitory effect on protein levels of FPP synthase. The suppressive effect of PUFA on the FPP synthase mRNA level was not due to a decrease in mRNA stability, but to transcription inhibition. Moreover, a lower nuclear availability of both SREBP-1 and SREBP-2 mature forms was observed in HepG2 human hepatoblastoma cells treated with arachidonate, EPA or DHA. Taken together, these data suggest that PUFA can down-regulate hepatic cholesterol synthesis through inhibition of HMG-CoA reductase and FPP synthase, at least in part through impairment of the SREBP pathway.
Keywords: farnesyl diphosphate synthase (FPP synthase), 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-CoA reductase), fish oil diet, hepatoma cell, mouse liver, sterol regulatory element-binding protein (SREBP)
Abbreviations: AOX, acyl-CoA oxidase; CMV, cytomegalovirus; DHA, docosahexaenoic acid; DMEM, Dulbecco's modified Eagle's medium; DRB, 5,6-dichlorobenzimidazole riboside; EPA, eicosapentaenoic acid; ER, endoplasmic reticulum; FPP synthase, farnesyl diphosphate synthase (geranyl-diphosphate geranyltranstransferase); HMG-CoA reductase, 3-hydroxy-3-methylglutaryl-CoA reductase; HSC70, heat-shock cognate 70 stress protein; LXR, liver X receptor; PPARα, peroxisome-proliferator-activated receptor α; PUFA, polyunsaturated fatty acids; RT-PCR, reverse transcription–PCR; SRE, sterol response element; SREBP, sterol regulatory element-binding protein; SCAP, SREBP cleavage-activating protein
INTRODUCTION
Epidemiological and clinical studies over the past 50 years have revealed numerous risk factors for atherosclerosis and related cardiovascular diseases. Environmental factors, such as the type of dietary fat, appear to be the most significant [1]. Ingestion of fish or fish oil rich in the n−3 PUFA (polyunsaturated fatty acids) EPA (eicosapentaenoic acid) and DHA (docosahexaenoic acid) may prevent the development of atherosclerosis [2,3]. A major effect of n−3 PUFA is to lower plasma triacylglycerols and lipoprotein concentrations, in normal as well as hypertriglyceridaemic subjects, and it is generally believed that PUFA exert their effects largely by suppression of the hepatic production of triacylglycerols and very-low-density lipoprotein [4,5], reflecting increased fatty acid oxidation and diminished fatty acid synthesis [6]. In contrast, dietary n−3 PUFA appear to have variable effects on levels of total cholesterol and low-density lipoprotein cholesterol, depending on several factors, in particular the dietary content of saturated and unsaturated fatty acids and the level of hyperlipidaemia [4,5]. However, under conditions of controlled dietary treatment, n−3 PUFA reduced total cholesterol in plasma to a lesser extent than triacylglycerols, and lowered low-density lipoprotein cholesterol in normolipidaemic individuals [7] and in hyperlipidaemic patients [8]. Like n−3 PUFA, n−6 PUFA that are abundant in vegetable oils (safflower oil and sunflower oil) appear to exert hypolipidaemic effects upon lowering plasma cholesterol [8,9]. Among experimental animals, some species, including the rat and the mouse, respond to the hypolipidaemic effects of fish oils and vegetable oils [10–12].
Dietary fish oils and vegetable oils have been shown to affect cholesterol synthesis. In humans, dietary safflower oil decreases the plasma lathosterol concentration, indicating a negative effect of n−6 PUFA on cholesterogenesis [13]. In rats, fish oil- and saf-flower oil-enriched diets were associated with lower cholesterol synthesis compared with an olive oil diet, and the level of inhibition depended on energy intake [14]. An inhibitory effect was obtained in hepatocytes from fish oil-fed rats as compared with hepatocytes from corn oil-fed rats [15]. In addition, hepatic HMG-CoA reductase (3-hydroxy-3-methylglutaryl-CoA reductase) activity was lower in rats fed diets rich in oils containing n−3 PUFA (fish, perilla and linseed oils) than in those fed n−6 PUFA-rich-oil diets (safflower and borage oils) [16,17]. The suppressive effect of fish oil on HMG-CoA reductase activity also occurred in rats with hereditary hypertriglyceridaemia [18]. From these data, it can be estimated that the n−6/n−3 and PUFA/saturated fatty acids ratios may control cholesterol synthesis.
Cholesterogenic genes are transactivated by the SREBPs (sterol regulatory element-binding proteins). The SREBPs are intrinsically bound to ER (endoplasmic reticulum) membranes and must be escorted by the trafficking protein SCAP (SREBP cleavage-activating protein) to the Golgi apparatus, where proteases liberate the N-terminal mature forms of SREBPs. Once released, mature SREBPs translocate to the nucleus and bind as dimers to the SRE (sterol response element) to activate the transcription of target genes, including genes involved in the synthesis of cholesterol, fatty acids and triacylglycerols [19–21]. Three SREBP isoforms (SREBP-1a, -1c and -2) are known to exist in animal tissues, and have different potencies for regulation of target gene expression. SREBP-1c preferentially activates lipogenesis genes, whereas SREBP-2 preferentially enhances the transcription of genes involved in cholesterogenesis. All SREBP-responsive genes are activated by SREBP-1a, supporting the view that SREBPs may act in complementary ways in response to cellular requirements for lipids. When overexpressed at supraphysiological levels, the three SREBPs activate all SREBP-responsive genes, by different magnitudes depending on their specificity [22,23]. The proteolytic processing of SREBPs is subject to negative-feedback regulation by cholesterol. When the cholesterol content rises in the ER membrane, retention of the SCAP–SREBP complex in the ER is favoured, which decreases the proteolytic cleavage of SREBP and hence target gene transcription, a form of regulation that occurs in cultured cells [19,21] and in the livers of rodents fed cholesterol-enriched diets [24]. Conversely, in sterol-depleted cells, proteolytic cleavage of SREBP occurs, thereby releasing mature forms of SREBPs [19].
Previous studies have revealed that PUFA can regulate gene expression. Fish oil feeding drastically decreased mRNA levels for lipogenic enzymes in rodent liver, with a corresponding decrease in the mature form of SREBP-1 [25,26]. This suppressive effect could be attributed to a reduction in the content of SREBP-1c mRNA and to a decrease in the proteolytic release of mature SREBP-1c [26,27]. Similarly, in human embryonic HEK-293 kidney cells, SREBP-1c as well as SREBP-1a mRNA levels were markedly lowered by PUFA, and the proteolytic processing of SREBPs was also inhibited [28]. LXR (liver X receptor)-mediated regulation of SREBP-1c appears to be involved in the PUFA-mediated suppression of SREBP-1c transcription, as PUFA antagonize the activation of LXRs by their endogenous ligands [29].
Together, these findings led us to hypothesize that PUFA may control cholesterogenic gene expression through their effects on SREBP-dependent regulation. SREBP-responsive genes include those coding for the rate-limiting enzyme HMG-CoA reductase and for FPP synthase (farnesyl diphosphate synthase) [23]. Importantly, the product of FPP synthase activity, farnesyl diphosphate, is a precursor of both sterol and non-sterol isoprenoid compounds, which are required for the post-translational prenylation of several proteins. In addition, the molecular basis for the SREBP-regulated transcription of the FPP synthase gene has been well characterized [30]. Therefore FFP synthase appears to play key roles in several metabolic pathways that depend on SREBP proteins. Here we have investigated the regulation of FPP synthase by PUFA. Our results show that PUFA decreased FPP synthase mRNA and protein levels in rat hepatoma cells and in mouse liver, in correlation with their effects on HMG-CoA reductase. These effects were associated with decreased plasma levels of cholesterol and triacylglycerols in the animals.
MATERIALS AND METHODS
Cell culture and treatments
H4IIEC3 rat and HepG2 human hepatoma cells were maintained in DMEM (Dulbecco's modified Eagle's medium)/Ham F12 medium (Sigma) supplemented with 5 or 10% (v/v) foetal bovine serum (Sigma, St Quentin-Fallavier, France) respectively. After the growing period, the medium was changed to serum-free medium supplemented with 50, 100, 150 or 300 μM BSA (fatty acid-free; Sigma)-bound fatty acid for 18 h at a fatty acid/BSA ratio of 2:1. Untreated cells received an amount of BSA equal to that brought with the BSA–fatty-acid complex. For mRNA stability studies, DRB (5,6-dichlorobenzimidazole riboside) was used at a concentration of 100 μM for various periods with or without fatty acids.
Animals and experimental treatments
French guidelines for the use and care of laboratory animals were followed. C57BL/6 male mice (9–10 weeks old; 20±2 g) were purchased from IFFA CREDO. Animals were housed in a controlled environment (constant temperature and humidity; darkness from 20:00 to 08:00 hours). Mice were fed for 15 days ad libitum on a fat-free laboratory chow (UAR) supplemented with either 15% (w/w) olive oil (control group) or 15% (w/w) tuna fish oil (Clover Corporate Limited), corresponding to 30% of the energy in the diet. Olive oil contains 84% monounsaturated fatty acids, primarily oleic acid (74% of total fatty acids). Tuna fish oil has the highest proportion of n−3 fatty acids (36%) among fish oils, comprising mainly 25% DHA and 7.3% EPA. However, since fish oil is easily peroxidized to form hydroperoxides, which increase oxidative stress, the chows were supplemented or not with vitamin E [(+)-α-tocopherol acid succinate; Sigma] (2.24 mg/g of EPA and 2.68 mg/g of DHA). Importantly, tuna fish oil contains only 0.26% cholesterol (amounting to 0.04% of the diet). In addition, olive oil contains 0.18% phytosterols (which amounts to 0.03% of the diet), including 0.09% β-sitosterol, as determined by gas chromatography. Each treatment group contained four male mice. After the mice were killed, liver samples were obtained, snap frozen in liquid nitrogen and stored at −80 °C until RNAs were extracted. Cholesterol and triacylglycerol levels were assessed in arterial blood.
Plasmids
A 317 bp fragment of the FPP synthase gene promoter was amplified with primers FPPS 415 (sense; 5′-ACAAACAGGAAGTAGATAGCCGAATTCCCAG-3′) and FPPS 4 (antisense; 5′-TCTGTGCTTCAACGCGGATTC-3′) using rat genomic DNA. The product was subcloned into the cloning vector pGEM-T easy (Promega, Madison, WI, U.S.A.), and then into pGL3-basic (Promega). Sequencing of the plasmid pGL3-FPPS317 showed that the PCR-amplified promoter fragment exhibited 100% identity with the reported sequence [31]. The ADD1/SREBP-1c expression vector encoding the first 403 amino acids of rat ADD1/SREBP-1c was kindly provided by Dr I. Dugail (INSERM U465 Institut Biomedical des Cordeliers, Paris, France). pCMV-CSA, which encodes amino acids 1–490 of SREBP-1a, and pCMV-CS2, which encodes amino acids 1–481 of SREBP-2, were kindly provided by Dr T. Osborne (Department of Molecular Biology and Biochemistry, University of California, Irvine, CA, U.S.A.). The β-galactosidase plasmid (pCMVβ-gal; Clontech, Ozyme, France) used for transfection control consists of the LacZ gene driven by a CMV (cytomegalovirus) promoter.
Transfection and luciferase assays
H4IIEC3 and HepG2 cells were seeded into 24-well plates at 0.15×106 cells/well in DMEM/Ham F12 medium supplemented with 5% or 10% (v/v) foetal bovine serum respectively. After 24 h, cells were rinsed with serum-free medium, and transfected using Lipofectin® reagent (Invitrogen Life Technologies, Cergy Pontoise, France) as described by the manufacturer. In each well, 100 ng of pGL3-FFPS317 was co-transfected with 50 ng of pCMVβ-gal and with 20 ng of pCMV5 (Invitrogen Life Technologies) as control or with 20 ng of expression vector for each SREBP protein in the presence of 2 μl of Lipofectin® reagent. After 3–5 h, cells were washed with PBS and cultured in treatment medium, as indicated in the figure legends. After incubation for 24–36 h, cells were lysed with reporter lysis buffer (Promega) and luciferase activity was measured with a luminometer (Microlite TLX1; Dynex Technologies, Chantilly, VA, U.S.A.) with the luciferin reagent from Promega. The β-galactosidase activity was used to normalize for transfection efficiency. Each experimental point was performed in triplicate, and each experiment was repeated three or more times.
RNA extraction, and analysis by Northern blot and by RT-PCR (reverse transcription–PCR) experiments
RNA isolation and Northern blotting hybridization were performed as described previously [32]. The levels of mRNAs were quantified with Alpha Imager™ 1220 software (Alpha Innotech Corp.), and all mRNA expression levels were normalized to the intensity of 36B4 mRNA levels. Rat SREBP-1 cDNA was kindly provided by Dr I. Dugail. SREBP transcript levels were analysed by RT-PCR with specific primers. Portions of 2 μg of total RNA were reverse-transcribed using random hexamers as primers and MMLV (Moloney murine leukaemia virus) reverse transcriptase (Promega). Reverse transcription reactions (1/20 vol.) were used as templates for all PCR experiments. The PCR primers used in this study were: acidic ribosomal phosphoprotein (36B4), 5′-AATGTGGGCTCAAGCAGATG-3′ (sense) and 5′-CTGCTGAACATGCTGAACATCTC-3′ (antisense); SREBP-1a, 5′-ATGGACGAGCTGGCCTTCGGTGAGGCGGCT-3′ (sense) and 5′-CCAGAGAGGAACCCAGGGAAGCAG-3′ (antisense) [33]; SREBP-1c, 5′-GGAGCCATGGATTGCACATT-3′ (sense) and 5′-GGGTCCTCCCAGGAAGGCTTC-3′ (antisense) [34]; SREBP-2, 5′-GAGCTGACTCTCGGGGACAT-3′ (sense) and 5′-ACTGCCGCCACCACCTCCAG-3′ (antisense) [35].
Immunoblots
Nuclear extracts from HepG2 cells were purified as described previously [28], except that the cells were lysed with 1% (v/v) Nonidet P40. Portions of 25 μg of nuclear extracts were separated in SDS/8%-polyacrylamide gels and transferred on to Hybond-P membranes (Amersham Biosciences Europe GmbH). Immunoblot analysis was performed using the Western blotting luminol reagent kit (Santa Cruz Biotechnology, Tebu, France). Membrane sheets were first incubated with antibodies against human SREBP-1 (IgG 2A4; NeoMarkers, Fremont, CA, U.S.A.) or SREBP-2 (IgG 1C6; NeoMarkers) for 2 h at 22 °C, washed several times and incubated with peroxidase-conjugated affinity-purified goat anti-mouse IgG for 18 h at 4 °C.
Protein extraction from H4IIEC3 cells and mouse liver and FPP synthase protein level analysis were performed as described previously [36]. Antibody against HSC70 (heat-shock cognate 70 stress protein) (Santa Cruz Biotechnology) was used as a normalizing control.
RESULTS
A diet containing PUFA-rich tuna fish oil decreases FPP synthase mRNA levels in mouse liver
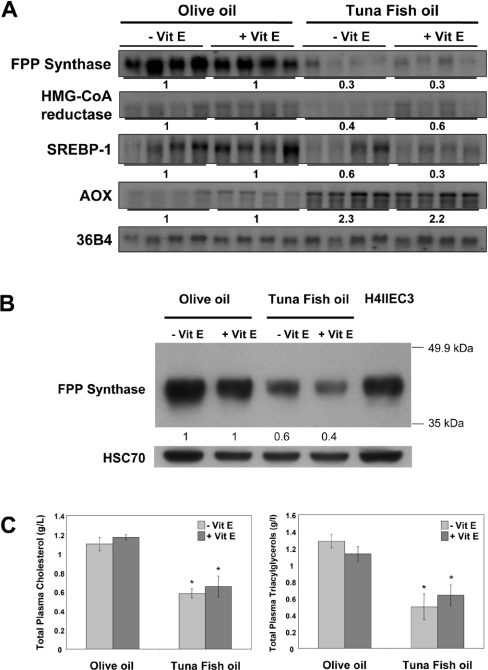
In order to investigate the effects of fatty acids on FPP synthase gene expression in vivo, we used mice fed for 2 weeks on a diet enriched in tuna fish oil, which is characterized by the highest proportion of n−3 fatty acids among fish oils, or with olive oil, used as a control (Figure 1A). After 2 weeks of the respective diets, the expression of genes involved in cholesterol synthesis had decreased markedly in the tuna fish oil group. FPP synthase mRNA levels decreased by 70%, and HMG-CoA reductase mRNA fell by 40–60%. SREBP-1 transcript levels were likewise markedly diminished by this diet, by 40–70%. Addition of vitamin E to the diet did not modify the expression of these genes (Figure 1A). RT-PCR analysis of individual SREBP mRNA levels showed that only SREBP-1c mRNA levels were decreased, and neither SREBP-1a nor SREBP-2 mRNA levels were markedly affected (results not shown). In order to determine if the negative effect of PUFA from the tuna fish oil diet on gene expression was a generalized inhibitory effect on transcription, we analysed the expression of AOX (acyl-CoA oxidase) mRNA, which is known to be induced by fatty acids through PPARα (peroxisome-proliferator-activated receptor α)-dependent transactivation. Indeed, AOX mRNA levels were induced by 2.2–2.3-fold (Figure 1A), indicating that the negative effect of PUFA on cholesterogenic gene expression was not a generalized suppressive effect.
Figure 1. Effects of a PUFA-rich diet on hepatic cholesterogenesis gene expression, FPP synthase protein levels and plasma cholesterol and triacylglycerol levels in mice.
(A) Northern blot analysis of FPP synthase, HMG-CoA reductase and SREBP-1 mRNA levels in liver. The AOX mRNA level was analysed as a marker of PPARα activation by PUFA. mRNA levels were normalized to those of 36B4 mRNA. mRNA levels in animals fed on a tuna fish oil-enriched diet (four male mice) relative to olive oil control treatment (determined by densitometry) are indicated. (B) Western blot analysis of FPP synthase protein levels in liver. Samples of 60 μg of total protein pooled from four male mice for each diet group were electrophoresed, and immunoblot analyses were performed with rabbit anti-(rat FPP synthase) IgG. Protein levels were normalized to amounts of HSC70 used as a control. Protein levels from tuna fish oil-fed mice relative to olive oil-fed control mice (determined by densitometry) are indicated. A H4IIEC3 protein sample was used as control of the immunoreactivity of anti-(rat FPP synthase) antibody. (C) Plasma cholesterol and triacylglycerol levels of mice fed on the olive oil or the tuna fish oil diet were assessed in arterial blood. Significance of differences: *P<0.05 compared with olive oil group (Student's t test). Vit E, vitamin E.
We next performed Western blot analyses to look at protein levels of FPP synthase in response to PUFA. As shown in Figure 1(B), the tuna fish oil diet markedly decreased FPP synthase protein levels, similar to the effect observed for mRNA levels.
Finally, the decrease in the expression of genes involved in cholesterol synthesis due to PUFA was associated with a 2-fold diminution of plasma cholesterol and triacylglycerol levels (Figure 1C).
Unsaturated fatty acids decrease FPP synthase gene expression in H4IIEC3 cells
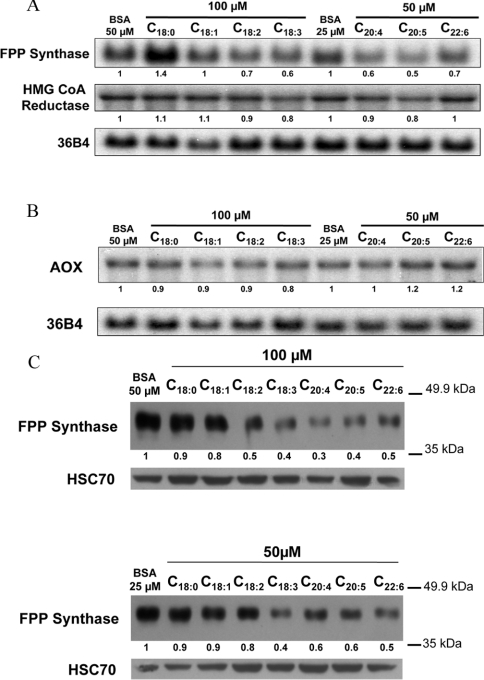
The effects of individual fatty acids on FPP synthase and HMG-CoA reductase gene expression were evaluated in H4IIEC3 cells, a well-differentiated rat hepatoma cell line. The fatty acids used were stearate (C18:0), oleate (C18:1), n−6 PUFA [linoleate (C18:2) and arachidonate (C20:4)] and n−3 PUFA [α-linolenate (C18:3), EPA (C20:5) and DHA (C22:6)]. The cells were cultured in the presence of fatty acid bound to fatty acid-free BSA for 18 h, in the absence of foetal bovine serum in order to control the exogenous supply of fatty acids. Pilot experiments were first performed to determine the optimal concentrations of PUFA. Cells were treated with 25, 50 or 100 μM PUFA for 18 h. Concentrations of 50 or 100 μM, depending on the fatty acid, were chosen as those provoking the most marked effects in the absence of toxicity, as determined by measurement of lactate dehydrogenase activity in the culture medium (results not shown). The FPP synthase mRNA level was induced by stearate, the only saturated fatty acid used in the present study, but no induction was found for HMG-CoA reductase mRNA, as compared with fatty acid-free controls (Figure 2A). By contrast, most unsaturated fatty acids, including linoleate, linolenate, arachidonate, EPA and DHA, decreased the FPP synthase mRNA level by up to 50%, whereas expression of the HMG-CoA reductase gene was only marginally decreased. As a control of PPARα-dependent activation and induction of peroxisomal β-oxidation by fatty acids in these cells, AOX mRNA levels were determined by Northern blot analysis (Figure 2B). Among the fatty acids used, only EPA and DHA had a weak inducing effect on AOX gene expression.
Figure 2. Effects of fatty acids on the expression of genes involved in cholesterol synthesis and in peroxisomal fatty acid β-oxidation, and on FPP synthase protein expression, in H4IIEC3 rat hepatoma cells.
Northern blot analysis of the expression of FPP synthase, HMG-CoA reductase (A) and AOX (B) after fatty acid treatment for 18 h. mRNA levels were normalized to those of 36B4 mRNA. mRNA levels relative to BSA control treatments (determined by densitometry) are indicated. One Northern blot analysis representative of five independent experiments is shown. (C) Western blot analysis of FPP synthase protein levels after treatment with 50 μM or 100 μM fatty acid for 18 h, normalized to HSC70 protein levels. Protein levels relative to BSA control treatment (determined by densitometry) are indicated (data from one experiment representative of two independent experiments).
As shown in Figure 2(C), all PUFA (but not stearate or oleate) triggered a drop in FPP synthase protein levels, similar to their effects on mRNA levels.
Fatty acids decrease SREBP-1c mRNA levels in H4IIEC3 cells
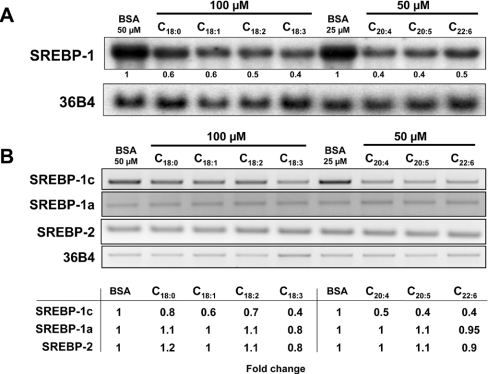
Since the FPP synthase gene is under the control of SREBP transcription factors, we analysed the effects of fatty acids on SREBP-1 mRNA levels. All of the fatty acids used decreased SREBP-1 mRNA levels by up to 60% compared with controls without fatty acids (Figure 3A). Linoleate, linolenate, arachidonate, EPA and DHA had the strongest negative effects on SREBP-1 gene expression.
Figure 3. Effects of fatty acids on SREBP gene expression in H4IIEC3 rat hepatoma cells.
(A) Northern blot analysis of SREBP-1 gene expression in H4IIEC3 cells after fatty acid treatment for 18 h. (B) RT-PCR analysis of SREBP-1a, -1c and -2 mRNA levels after fatty acid treatment for 18 h. mRNA levels relative to BSA control treatments (determined by densitometry) are indicated under each lane in (A) and in the table for (B). The data are representative of three independent experiments.
In order to discriminate between the highly similar SREBP-1a and -1c transcripts, the levels of each SREBP isoform were analysed by RT-PCR with specific primers. As shown in Figure 3(B), only SREBP-1c levels were decreased.
Unsaturated fatty acids suppress FPP synthase promoter activity
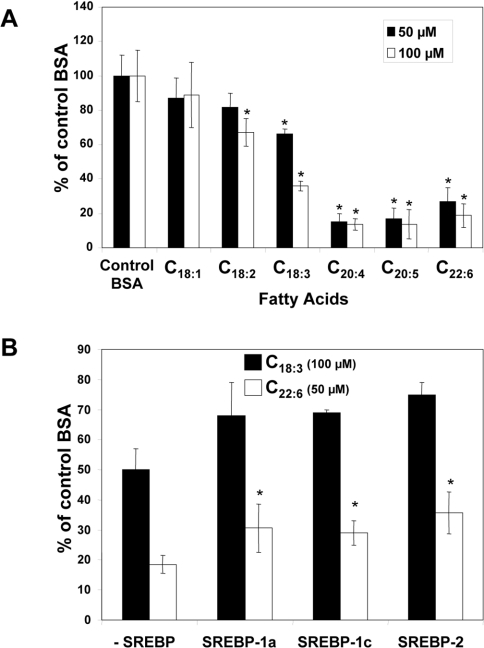
To determine the effects of fatty acids on FPP synthase promoter activity, H4IIEC3 cells were transiently transfected with pGL3-FPPS317, containing the luciferase reporter gene under the control of a FPP synthase promoter fragment. Transfected cells were incubated with 50 and 100 μM fatty acids of increasing chain length and unsaturation (Figure 4A). The results show that oleate had a weak inhibitory effect on luciferase gene expression. In contrast, all PUFA caused marked down-regulation.
Figure 4. Transcriptional analysis in H4IIEC3 cells.
(A) Cells were transiently transfected with pGL3-FFPS317 plus pCMVβ-gal with Lipofectin® for 4 h. Cells were then incubated for 18 h with BSA alone or with 50 or 100 μM fatty acid. Luciferase and β-galactosidase activities were determined as described in the Materials and methods section. Data are expressed as percentages of values following control BSA treatment, and represent results from at least three experiments performed in triplicate. In the treatments with 50 μM fatty acid, the BSA concentration was 25 μM, while it was 50 μM in the presence of 100 μM fatty acid.*P<0.05, statistically different from control (Student's t test). (B) Cells were transiently transfected with pGL3-FPPS317, pCMVβ-gal and an expression vector for SREBP-1a, -1c or -2 (or empty vector as a control). After transfection, cells were incubated with BSA alone or with 100 μM linolenate (C18:3) or 50 μM DHA (C22:6) for 18 h. Activities were measured as in (A). Data are expressed as percentages of values following control BSA treatment, and represent results from at least three experiments performed in triplicate. *P<0.05, statistically different from control (Student's t test).
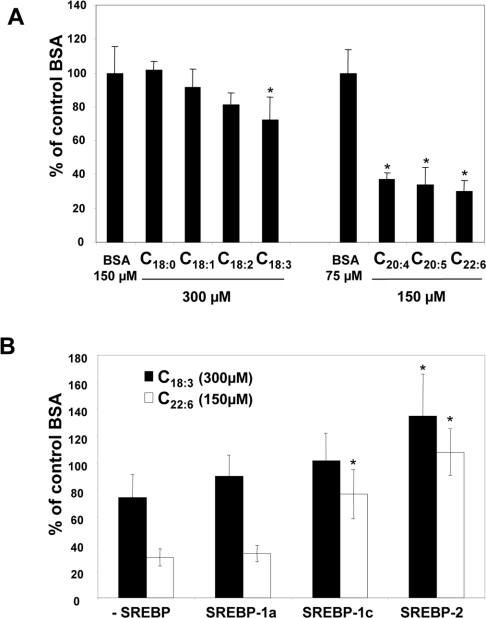
Since SREBPs are known to activate transcription of the FPP synthase gene, we tested the ability of SREBPs to compensate for the inhibitory effect of fatty acids on FPP synthase gene expression. To this end, we used linolenate and DHA, which had marked inhibitory effects on FPP synthase promoter activity, with cells co-transfected with the FPP synthase gene promoter and SREBP expression plasmids (Figure 4B). The three SREBP isoforms had only a weak compensatory effect on the inhibition of FPP synthase promoter activity brought about by linolenate. In contrast, the inhibitory effect of DHA could be partially compensated by expression of the three SREBP isoforms.
Northern blotting experiments were then performed with RNA from HepG2 human hepatoma cells. We first confirmed that PUFA treatment diminished FPP synthase mRNA levels in HepG2 cells as well (results not shown). Higher doses of PUFA were required compared with H4IIEC3 cells, but no toxicity was observed with the doses used. As shown in Figure 5(A), linolenate, arachidonate, EPA and DHA decreased FPP synthase promoter activity in HepG2 cells as they did in H4IIEC3 cells. These results indicate that the primary effect of these fatty acids is to reduce FPP synthase gene transcription. The ability of SREBP isoforms to prevent inhibition of FPP synthase promoter activity by PUFA was also analysed in HepG2 cells (Figure 5B). SREBP-1a provided only low compensation for the negative effects of linolenate and DHA. However, SREBP-1c and SREBP-2 had a significant compensatory effect.
Figure 5. Transcriptional analysis in HepG2 cells.
(A) Cells were transiently transfected with pGL3-FFPS317 plus pCMVβ-gal with Lipofectin® for 4 h. Cells were then incubated for 18 h with BSA alone or with 150 or 300 μM fatty acid. Luciferase and β-galactosidase activities were determined as described in the Materials and methods section. Data are expressed as percentages of values following control BSA treatment, and are from a representative experiment performed in triplicate. *P<0.05, Statistically different from control (Student's t test). (B) Cells were transiently transfected with pGL3-FPPS317, pCMVβ-gal and an expression vector for SREBP-1a, -1c or -2 (or with an empty vector as control). After transfection, cells were incubated with BSA alone or with 300 μM linolenate (C18:3) or 150 μM DHA (C22:6) for 18 h. Activities were measured as in (A). Data are expressed as percentages of values following control BSA treatment, and represent results from at least three experiments performed in triplicate. *P<0.05, Statistically different from control (Student's t test).
Linolenate and DHA do not modify FPP synthase mRNA stability
In order to determine if the decrease in FPP synthase mRNA levels in linolenate-and DHA-treated cells was due in part to a decrease in mRNA stability, we treated H4IIEC3 cells with DRB, an inhibitor of RNA polymerase II. The rate of decay of FPP synthase mRNA in the presence of DRB was not affected by either fatty acid (Figure 6). The efficiency of linolenate and DHA treatments in these experiments was confirmed by the analysis of FPP synthase mRNA levels, which had decreased by 50% after 12 h with linolenate and after 8 h with DHA.
Figure 6. Effects of linolenate and DHA on the turnover of FPP synthase mRNA in H4IIEC3 cells.
Cells were treated with DRB (100 μM) in the absence or the presence of 100 μM linolenate (C18:3) or 50 μM DHA (C22:6). Total RNA isolated from cells 0, 4, 8, 12, 16, 20 or 24 h (A) or 0, 4, 8, 14, 18 or 24 h (B) following DRB treatment was analysed by Northern blot with an FPP synthase probe, as described in the Materials and methods section. The FPP synthase mRNA level was standardized to 18 S rRNA levels. The data are from one representative experiment of three independent experiments for linolenate and two independent experiments for DHA. The FPP synthase mRNA decay equations for BSA and linolenate are y=−0.0335x+1.0138 and y=−0.0314x+1.002 respectively. The FPP synthase mRNA decay equations for BSA and DHA are y=−0.0369x+0.8267 and y=−0.0377x+0.8128 respectively.
Influence of fatty acids on nuclear levels of SREBP isoforms in HepG2 cells
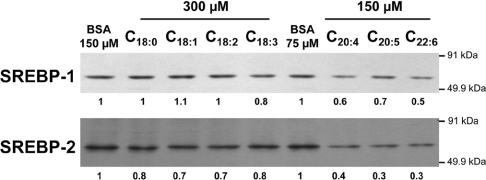
In order to determine the levels of nuclear SREBP isoforms following fatty acid treatment, immunoblot experiments were performed with HepG2 cell nuclear extracts (Figure 7). Whereas weak effects were observed for stearate, oleate, linoleate and linolenate, marked decreases were observed in nuclear levels of both SREBP-1 and SREBP-2 in cells treated with arachidonate, EPA or DHA, which also had the strongest negative effects on FPP synthase mRNA levels (Figure 2A) and promoter activity (Figures 4A and 5A).
Figure 7. SREBP immunoblot analysis of nuclear extracts of HepG2 cells.
Cells were incubated with control BSA medium or medium containing BSA-bound fatty acids (300 or 150 μM) for 18 h. Aliquots of each sample (25 μg of nuclear protein) were electrophoresed on SDS/10%-polyacrylamide gels and immunoblot analyses were performed with mouse monoclonal antibody IgG 2A4 against amino acids 301–407 of human SREBP-1, or with mouse monoclonal antibody IgG 1C6 against amino acids 833–1141 of human SREBP-2. Protein levels relative to BSA control treatments (determined by densitometry) are indicated under each lane. One representative Western blot from two independent experiments is shown.
DISCUSSION
Fish oils and vegetable oils exert hypocholesterolaemic effects in rodents [10–12], presumably in part by triggering a decrease in cholesterol synthesis [14,15]. The present study was initiated in order to determine the effects of PUFA on FFP synthase gene expression in liver cells and in mouse liver, in comparison with those of the key enzyme HMG-CoA reductase. Our results demonstrate that n−3 and n−6 PUFA decreased FPP synthase mRNA levels, and those of HMG-CoA reductase (albeit less strongly), in hepatoma cells. These effects are likely to occur at the transcriptional level, as indicated from the results of FPP synthase mRNA half-life determination in the case of linolenate and DHA, and following transient transfection of the SRE-containing FPP synthase promoter. Such a negative effect on FPP synthase and HMG-CoA reductase levels was also observed in the livers of mice fed fish oil, which naturally contains high levels of PUFA. As a control for the effect of this regimen, plasma cholesterol and triacylglycerol levels were reduced by 40–60% in these mice. This decrease was paralleled by down-regulation of SREBP-1 mRNA and of the nuclear mature forms of both SREBP-1 and SREBP-2 proteins following treatment of human hepatoma cells with arachidonate, EPA or DHA.
Our findings agree with the general consensus that the physiological activity of dietary fish oil is to decrease plasma lipid levels by virtue of its high content of n−3 PUFA, i.e. DHA and EPA. We can exclude the possibility that part of the decrease in FPP synthase expression in the tuna fish oil feeding experiment was due to feedback inhibition by dietary cholesterol, since the change in dietary cholesterol levels due to the fish oil diet is much lower (0.04%) than the concentration (5%) reported previously to decrease FPP synthase expression [37]. Our results also agree with the observation that dietary fish oil is more effective as a hypocholesterolaemic trigger than vegetable oil in mice [26] or rats [38]. Accordingly, we chose to use olive oil as a control. Indeed, oleate, the predominant fatty acid in olive oil (74%), did not change FPP synthase mRNA levels in H4IIEC3 cells (the present work), and dietary olive oil was reported to have no effect on HMG-CoA reductase levels [39]. In addition, dietary oleate, i.e. triolein, had no inhibitory effect on the expression of the SREBP-1 and fatty acid synthase genes in rat liver, whereas dietary safflower oil or fish oil suppressed the liver content of the precursor and mature forms of SREBP-1 [27].
One of the major problems associated with the use of highly unsaturated fatty acids such as arachidonate, EPA and DHA is their susceptibility to peroxidation, forming lipid radicals and lipid hydroperoxides, which can cause cell damage. Using vitamin E as an antioxidant in the fish oil diet, we found, however, that the inhibitory effects on FPP synthase and HMG-CoA reductase expression were not linked to the capacity of PUFA to undergo peroxidation.
In the present study, the fish oil diet (in which the fish oil comprised 30% of the energy content of the diet) decreased SREBP-1 mRNA levels in the liver, probably corresponding mostly to SREBP-1c, since the SREBP-1c/SREBP-1a transcript ratio is 9:1 in mouse liver [40]. A similar suppressive effect on the SREBP-1c mRNA level was induced in H4IIEC3 cells by linoleate, linolenate, arachidonate, EPA and DHA, while SREBP-1a and SREBP-2 mRNA levels remained unchanged, as observed on RT-PCR analyses. In addition, the amounts of mature SREBP-1 and SREBP-2 proteins were decreased, with the greatest effects being observed for arachidonate, EPA and DHA. Previous studies reported similar profiles of SREBP-1 expression in vivo and in vitro. In rats, fish oil and safflower oil diets (20% of energy content) reduced the hepatic level of SREBP-1 mRNA and of precursor and mature SREBP-1, compared with a fat-free diet or one containing triolein [27]. In mice, a high fish oil diet (40% or 60% of energy content) induced similar decreases in SREBP-1 expression [25,26]. In Caco-2 human intestinal cells, PUFA decreased both SREBP-1 gene and protein expression [41]. With respect to the transcriptional inhibition of SREBP-1c mRNA synthesis, it has been shown that transcription of SREBP-1c is under the positive control of the LXR [42]. The negative effect of PUFA is believed to depend on a negative interference with the activation of LXRs by their endogenous ligands [29]. In addition, PUFA have been reported to lower SREBP-1c levels by accelerating the degradation of its mRNA [43].
With regard to nuclear mature SREBP, feeding with low concentrations of fish oil inhibited the production of mature SREBP-1 in mouse liver, while a large decrease in the SREBP-1 mRNA level was obtained with higher concentrations [44]. Although the main effect of PUFA on SREBP is to decrease the SREBP-1 isoform, some studies have shown that levels of nuclear mature SREBP-2 could be slightly lowered by a sardine oil-rich diet [25] or by arachidonate in rat hepatoma cells [29]. High fish oil feeding also reduced SREBP-2 mRNA and protein levels in mouse liver [26]. However, the effect of PUFA on mature SREBP-2 protein levels was less than that on SREBP-1. In the present study, we do not know whether the observed decrease in the nuclear levels of SREBP-2 was due to a decrease in the amounts of precursor proteins, to decreased maturation, or to a combination of the two events. However, the fall in the level of the nuclear mature form of SREBP-1 is likely to depend on a decrease in the amount of the precursor form, since the level of SREBP-1c mRNA was strongly diminished.
Transient transfection analyses showed that the negative effects of PUFA occurred at the transcriptional level to reduce FPP synthase promoter activity in both H4EIIC3 and HepG2 cells. The inhibition appears to be correlated with the degree of unsaturation and chain length of PUFA, and was attenuated by co-transfection of SREBP-2 expression plasmids. Because SREBP-2 is a selective activator of cholesterogenic gene expression, whereas SREBP-1c is a weaker activator, the more plausible regulatory mechanism for the PUFA-mediated suppression of FPP synthase gene expression may involve mainly a decrease in mature SREBP-2. However, as postulated by Kim et al. [26], basal expression of cholesterogenic genes may also depend on SREBP-1c expression, since the diminution of its mature form in the nucleus was shown to lead to a decrease in the expression of lipogenic and cholesterogenic genes.
The regulation of the level of nuclear SREBP is closely dependent on the ability of cholesterol to block proteolytic activation into its mature form when cholesterol is in excess in cells [19,21]. It is plausible that dietary PUFA may modify the cholesterol content in hepatocytes, since they induce alterations in many cholesterol metabolic pathways. The cholesterol content may be lowered by a decrease in cholesterol synthesis and by stimulation of biliary cholesterol secretion [45]. In contrast, it may be increased as a consequence of depressed intracellular cholesteryl ester levels [46] and very-low-density lipoprotein secretion [47], or of stimulated low-density lipoprotein receptor activity [48] and reverse cholesterol transport [49]. Thus it is difficult to evaluate the net change in cellular cholesterol content or subcellular localization in response to dietary PUFA. However, by increasing the levels of PUFA in hepatocyte membrane phospholipids [16], a PUFA-rich diet may modify membrane fluidity, which could be followed by enrichment in cholesterol in order to preserve membrane stability. This change in membrane composition could inhibit the transfer of the SCAP–SREBP complex from the ER to the Golgi apparatus, where the proteolytic cleavage takes place. Alternatively, the PUFA enrichment of membranes could alter the microenvironment of membrane-associated SREBP and thereby suppress the maturation process. The differences observed in PUFA inhibition of cleavage between SREBP-1 and SREBP-2 could be due to the fact that SCAP or another ‘lipid sensor’ may be more sensitive to the PUFA content of ER membranes when it is associated with SREBP-1 rather than with SREBP-2. In addition, a recent study showed that the FPP synthase gene is a target for LXR in neuronal cells [50]. Since PUFA may antagonize LXR function, it is possible that the decrease in FPP synthase gene expression may reflect a decreased rate of LXR-dependent regulation. This hypothesis would, however, require further investigation.
In conclusion, PUFA may affect cholesterogenesis by down-regulating expression of the FPP synthase and HMG-CoA reductase genes both in hepatoma cells and in mouse liver, and by suppressing fatty acyl synthase gene expression [26]. Furthermore, these effects are likely to occur as a result of SREBP gene down-regulation, as part of an integrated lipid sensor mechanism in the liver.
Acknowledgments
We thank the Regional Council of Burgundy and the French Ministry of Research and Technology for financial support. We are grateful to J. Gresti for technical assistance with chromatographic analysis of oil sterols, and to S. Tasic for help in the initial phases of this work. Thanks are due to Dr I. Dugail (INSERM U465 Institut Biomedical des Cordeliers, Paris, France) and Dr T. Osborne (Department of Molecular Biology and Biochemistry, University of California, Irvine, CA, U.S.A.) for the generous donation of the SREBP-1c and the SREBP-1a and SREBP-2 expression vectors respectively. Finally, we thank Dr L. Corcos for critical reading of the manuscript.
References
- 1.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature (London) 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 2.Hooper L., Summerbell C. D., Higgins J. P., Thompson R. L., Capps N. E., Smith G. D., Riemersma R. A., Ebrahim S. Dietary fat intake and prevention of cardiovascular disease: systematic review. Br. Med. J. 2001;322:757–763. doi: 10.1136/bmj.322.7289.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angerer P., von Schacky C. n-3 polyunsaturated fatty acids and the cardiovascular system. Curr. Opin. Lipidol. 2000;1:57–63. doi: 10.1097/00041433-200002000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Harris W. S. Fish oils and plasma lipid and lipoprotein metabolism in humans: a critical review. J. Lipid Res. 1989;6:785–807. [PubMed] [Google Scholar]
- 5.Nestel P. J. Effects of n-3 fatty acids on lipid metabolism. Annu. Rev. Nutr. 1990;10:149–167. doi: 10.1146/annurev.nu.10.070190.001053. [DOI] [PubMed] [Google Scholar]
- 6.Clarke S. D. Polyunsaturated fatty acid regulation of gene transcription: a molecular mechanism to improve the metabolic syndrome. J. Nutr. 2001;131:1129–1132. doi: 10.1093/jn/131.4.1129. [DOI] [PubMed] [Google Scholar]
- 7.Von Lossonczy T. O., Ruiter A., Bronsgeest-Schoute H. C., Van Gent C. M., Hermus R. J. The effect of a fish diet on serum lipids in healthy human subjects. Am. J. Clin. Nutr. 1978;8:1340–1346. doi: 10.1093/ajcn/31.8.1340. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann W., Biermann J., Kostner G. M. Comparison of effects of n-3 to n-6 fatty acids on serum level of lipoprotein(a) in patients with coronary artery disease. Am. J. Cardiol. 1995;76:459–462. doi: 10.1016/s0002-9149(99)80130-1. [DOI] [PubMed] [Google Scholar]
- 9.Friday K. E., Failor R. A., Childs M. T., Bierman E. L. Effects of n-3 and n-6 fatty acid-enriched diets on plasma lipoproteins and apolipoproteins in heterozygous familial hypercholesterolemia. Arterioscler. Thromb. 1991;11:47–54. doi: 10.1161/01.atv.11.1.47. [DOI] [PubMed] [Google Scholar]
- 10.Harris W. S. n-3 fatty acids and serum lipoprotein: animal studies. Am. J. Clin. Nutr. 1997;65:1611S–1616S. doi: 10.1093/ajcn/65.5.1611S. [DOI] [PubMed] [Google Scholar]
- 11.Lee J. H., Fukumoto M., Nishida H., Ikeda I., Sugano M. The interrelated effects of n-6/n-3 and polyunsaturated/saturated ratios of dietary fats on the regulation of lipid metabolism in rats. J. Nutr. 1989;119:1893–1899. doi: 10.1093/jn/119.12.1893. [DOI] [PubMed] [Google Scholar]
- 12.Dallongeville J., Bauge E., Tailleux A., Peters J. M., Gonzalez F. J., Fruchart J. C., Staels B. Peroxisome proliferator-activated receptor alpha is not rate-limiting for the lipoprotein-lowering action of fish oil. J. Biol. Chem. 2001;276:4634–4639. doi: 10.1074/jbc.M008809200. [DOI] [PubMed] [Google Scholar]
- 13.Cox C., Sutherland W., Mann J., De Jong S., Chisholm A., Skeaff M. Effects of dietary coconut oil, butter and safflower oil on plasma lipids, lipoproteins and lathosterol levels. Eur. J. Clin. Nutr. 1998;52:650–654. doi: 10.1038/sj.ejcn.1600621. [DOI] [PubMed] [Google Scholar]
- 14.Cha M. C., Jones P. J. Dietary fat type related changes in tissue cholesterol and fatty acid synthesis are influenced by energy intake level in rats. J. Am. Coll. Nutr. 1997;16:592–599. [PubMed] [Google Scholar]
- 15.Ribeiro A., Mangeney M., Cardot P., Loriette C., Rayssiguier Y., Chambaz J., Bereziat G. Effect of dietary fish oil and corn oil on lipid metabolism and apolipoprotein gene expression by rat liver. Eur. J. Biochem. 1991;196:499–507. doi: 10.1111/j.1432-1033.1991.tb15842.x. [DOI] [PubMed] [Google Scholar]
- 16.Choi Y. S., Goto S., Ikeda I., Sugano M. Effect of dietary n-3 polyunsaturated fatty acids on cholesterol synthesis and degradation in rats of different ages. Lipids. 1989;24:45–50. doi: 10.1007/BF02535263. [DOI] [PubMed] [Google Scholar]
- 17.Ihara-Watanabe M., Umekawa H., Takahashi T., Furuichi Y. Effects of dietary alpha- or gamma-linolenic acid on levels and fatty acid compositions of serum and hepatic lipids, and activity and mRNA abundance of 3-hydroxy-3-methylglutaryl CoA reductase in rats. Comp. Biochem. Physiol. Mol. Integr. Physiol. 1999;122:213–220. doi: 10.1016/s1095-6433(98)10176-9. [DOI] [PubMed] [Google Scholar]
- 18.Hromadova M., Sebokova E., Klimes I. HMG-CoA reductase activity in the liver of rats with hereditary hypertriglyceridemia: effect of dietary fish oil. Endocr. Regul. 1994;28:211–214. [PubMed] [Google Scholar]
- 19.Brown M. S., Goldstein J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 20.Brown M. S., Goldstein J. L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horton J. D., Goldstein J. L., Brown M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimano H., Horton J. D., Shimomura I., Hammer R. E., Brown M. S., Goldstein J. L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimomura I., Bashmakov Y., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yahagi N., Shimano H., Hasty A. H., Amemiya-Kudo M., Okazaki H., Tamura Y., Iizuka Y., Shionoiri F., Ohashi K., Osuga J., et al. A crucial role of sterol regulatory element-binding protein-1 in the regulation of lipogenic gene expression by polyunsaturated fatty acids. J. Biol. Chem. 1999;274:35840–35844. doi: 10.1074/jbc.274.50.35840. [DOI] [PubMed] [Google Scholar]
- 26.Kim H. J., Takahashi M., Ezaki O. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. A possible mechanism for down-regulation of lipogenic enzyme mRNAs. J. Biol. Chem. 1999;274:25892–25898. doi: 10.1074/jbc.274.36.25892. [DOI] [PubMed] [Google Scholar]
- 27.Xu J., Nakamura M. T., Cho H. P., Clarke S. D. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. A mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J. Biol. Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 28.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. Unsaturated fatty acids down-regulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 29.Ou J., Tu H., Shan B., Luk A., DeBose-Boyd R. A., Bashmakov Y., Goldstein J. L., Brown M. S. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6027–6032. doi: 10.1073/pnas.111138698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ericsson J., Jackson S. M., Lee B. C., Edwards P. A. Sterol regulatory element binding protein binds to a cis element in the promoter of the farnesyl diphosphate synthase gene. Proc. Natl. Acad. Sci. U.S.A. 1996;93:945–950. doi: 10.1073/pnas.93.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spear D. H., Kutsunai S. Y., Correll C. C., Edwards P. A. Molecular cloning and promoter analysis of the rat liver farnesyl diphosphate synthase gene. J. Biol. Chem. 1992;267:14462–14469. [PubMed] [Google Scholar]
- 32.Le Jossic-Corcos C., Duclos S., Ramirez L. C., Zaghini I., Chevillard G., Martin P., Pineau T., Bournot P. Effects of peroxisome proliferator-activated receptor α (PPARα) activation on pathways contributing to cholesterol homeostasis in rat hepatocytes. Biochim. Biophys. Acta. 2004;1683:49–58. doi: 10.1016/j.bbalip.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Shimomura I., Bashmakov Y., Ikemoto S., Horton J. D., Brown M. S., Goldstein J. L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kakuma T., Lee Y., Higa M., Wang Z.-W., Pan W., Shimomura I., Unger R. H. Leptin, troglitazone, and the expression of sterol regulatory element binding proteins in liver and pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8536–8541. doi: 10.1073/pnas.97.15.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBose-Boyd R. A., Ou J., Goldstein J. L., Brown M. S. Expression of sterol regulatory element-binding protein1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta S. D., Mehan R. S., Tansey T. R., Chen H. T., Goping G., Goldberg I., Shechter I. Differential binding of proteins to peroxisomes in rat hepatoma cells: unique association of enzymes involved in isoprenoid metabolism. J. Lipid Res. 1999;40:1572–1584. [PubMed] [Google Scholar]
- 37.Ashby M. N., Edwards P. A. Identification and regulation of rat liver cDNA encoding farnesyl pyrophosphate synthetase. J. Biol. Chem. 1989;264:635–640. [PubMed] [Google Scholar]
- 38.Gaiva M. H., Couto R. C., Oyama L. M., Couto G. E. C., Silveira V. L. F., Ribeiro E. B., Nascimento C. M. O. Diets rich in polyunsaturated fatty acids: effects on hepatic metabolism in rats. Nutrition. 2003;19:144–149. doi: 10.1016/s0899-9007(02)00909-7. [DOI] [PubMed] [Google Scholar]
- 39.Salam W. H., Cagen L. M., Heimberg M. Regulation of hepatic cholesterol biosynthesis by fatty acids: effect of feeding olive oil on cytoplasmic acetoacetylcoenzyme A thiolase, beta-hydroxy-beta-methylglutaryl-CoA synthase, and acetoacetyl-coenzyme A ligase. Biochem. Biophys. Res. Commun. 1988;153:422–427. doi: 10.1016/s0006-291x(88)81241-5. [DOI] [PubMed] [Google Scholar]
- 40.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. Differential expression of exons 1a and 1c mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Field F. J., Born E., Murthy S., Mathur S. N. Polyunsaturated fatty acids decrease the expression of sterol regulatory element-binding protein-1 in CaCo-2 cells: effect on fatty acid synthesis and triacylglycerol transport. Biochem. J. 2002;368:855–864. doi: 10.1042/BJ20020731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J., Teran-Garcia M., Park J. H. Y., Nakamura M. T., Clarke S. D. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J. Biol. Chem. 2001;276:9800–9807. doi: 10.1074/jbc.M008973200. [DOI] [PubMed] [Google Scholar]
- 44.Nakatani T., Kim H. J., Kaburagi Y., Yasuda K., Ezaki O. A low fish oil inhibits SREBP-1 proteolytic cascade, while a high-fish-oil feeding decreases SREBP-1 mRNA in mice liver: relationship to anti-obesity. J. Lipid Res. 2003;44:369–379. doi: 10.1194/jlr.M200289-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Bravo E., Cantafora A., DeLuca V., Tripodi M., Avella M., Botham K. M. The mechanism underlying the hypocholesterolemic effect of chronic fish oil feeding in rats is not due to increased excretion of dietary cholesterol. Atherosclerosis. 1998;139:253–263. doi: 10.1016/s0021-9150(98)00075-6. [DOI] [PubMed] [Google Scholar]
- 46.Botham K. M., Maldonado E. N., Chico Y., Zheng X., Avella M., Ochoa B. The influence of chylomicron remnants on cholesteryl ester metabolism in cultured rat hepatocytes: comparison of the effects of particles enriched in n-3 or n-6 polyunsaturated fatty acids. Biochim. Biophys. Acta. 2001;1534:96–109. doi: 10.1016/s1388-1981(01)00177-9. [DOI] [PubMed] [Google Scholar]
- 47.Wong S. H., Nestel P. J., Trimble R. P., Storer G. B., Illman R. J., Topping D. L. The adaptive effects of dietary fish and safflower oil on lipid and lipoprotein metabolism in perfused rat liver. Biochim. Biophys. Acta. 1984;792:103–109. doi: 10.1016/0005-2760(84)90209-1. [DOI] [PubMed] [Google Scholar]
- 48.Spady D. K. Regulatory effects of individual n-6 and n-3 polyunsaturated fatty acids on LDL transport in the rat. J. Lipid Res. 1993;34:1337–1346. [PubMed] [Google Scholar]
- 49.Hatahet W., Cole L., Kudchodkar B. J., Fungwe T. V. Dietary fats differentially modulate the expression of lecithin: cholesterol acyltransferase, apoprotein-A1 and scavenger receptor b1 in rats. J. Nutr. 2003;133:689–694. doi: 10.1093/jn/133.3.689. [DOI] [PubMed] [Google Scholar]
- 50.Fukuchi J., Song C., Ko A. L., Liao S. Transcriptional regulation of farnesyl pyrophosphate synthase by liver X receptors. Steroids. 2003;68:685–691. doi: 10.1016/s0039-128x(03)00100-4. [DOI] [PubMed] [Google Scholar]