Dear Editor,
Considerable discussion surrounds the prognostic relevance of chromosome 1 aberrations in multiple myeloma (MM), from which most important are gains of 1q21 region and deletions of 1p32 locus [1]. Approximately 10–40% of MM patients develop extraosseous disease (EMM), where plasma cells outside of the bone marrow form tumors called plasmacytomas. Patients with EMM found at disease onset (primary EMM) represent a challenge due to a high risk of relapse and shorter survival. Patients developing plasmacytomas during therapy (secondary EMM) often experience an aggressive disease course, characterized by treatment resistance and early mortality. The exact mechanism of EMM development is not well known, but acquiring genetic alterations is one of the important hallmarks in clonal evolution, leading to EMM spread [2, 3]. Thus, we conducted a detailed evaluation of the distribution and clonal heterogeneity of chromosome 1 aberrations using paired samples from bone marrow and plasmacytoma tissue plasma cells. To assess the broader applicability of our findings, we performed a population-based cytogenetic analysis encompassing both EMM patients and a control cohort of MM patients without a history of EMM.
In twenty-two MM patients, who were diagnosed with EMM between 2022 and 2024 at the University Hospital Brno, a paired sampling of sorted bone marrow plasma cells (BMPCs) and plasmacytoma tissue plasma cells (TPCs) was performed. The interphase fluorescent in situ hybridization (I-FISH) analyses of commonly found aberrations were performed on sorted BMPCs or imprints of plasmacytoma tissue plasma cells (TPCs). Population cytogenetic data of EMM patients treated at the same site between 2015 and 2024 were retrospectively analyzed. During this period, a total of 1020 newly diagnosed multiple myeloma (NDMM) patients were identified at our institution. Among them, 16.3% (167/1020) of patients had a clear presence of EMM (primary EMM in 66.5% (111/167) and secondary EMM was identified in 33.5% of them (56/167)). For comparison, 243 NDMM patients without a history of EMM and with available cytogenetic data were used. Cytogenetics, clinical, and survival data of all primary EMM and control NDMM patients were evaluated from the time of diagnosis. For secondary EMM patients, data were evaluated from the onset of extraosseous plasmacytoma development. To present the cytogenetic findings, we organized them into two main categories reflecting key events in MM pathogenesis: primary events, which include hyperdiploidy (characterized by two or more trisomies of odd chromosomes) and translocations involving the IGH gene, and secondary events, which involve aberrations in chromosomes 1, 13, and 17 [4].
Description of patients‘ selection, methods, and statistical analysis is provided in a Supplementary data file.
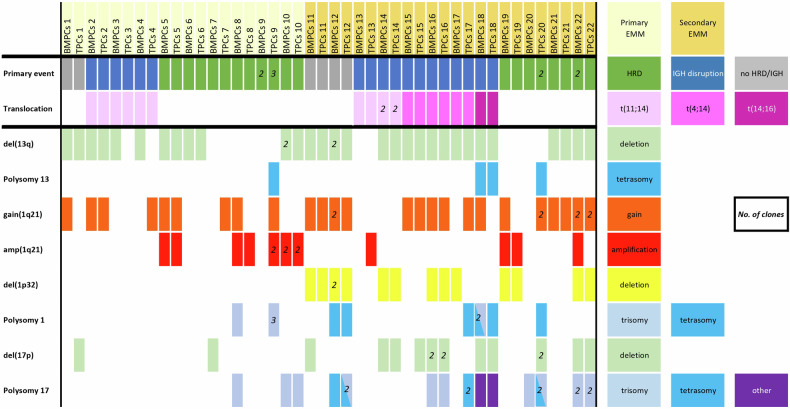
In total, we assessed paired BMPCs and TPCs samples from 10 primary EMM and 12 secondary EMM cases. In all patients, as expected, the primary events were always shared between BMPCs and TPCs. Del(13q14)/monosomy of 13 was found in 72.7% (16/22) of patients and shared in 87.5% (14/16) of cases. Del(17p)/monosomy of 17 was found in 40.9% (9/22) of patients and it was shared in 44.4% (4/9) of cases. We found chromosome 1 aberrations in 90.9% (20/22) of EMM patients, more often in TPCs than BMPCs (86.4% (19/22) vs 68.2% (15/22)). Only 35.0% (7/20) of patients shared the same 1q21 aberrations between BMPCs and TPCs. The 1q21 aberrations present only in TPCs were found in 30.0% (6/20) of patients. The del(1p32) was in 31.8% (7/22) of patients, in 85.7% (6/7) cases shared between BMPCs and TPCs. We found del(1p32) exclusively in samples from secondary EMM patients. In 71.4% (5/7) of cases, del(1p32) was associated with 1q21 gains. All chromosomal aberrations in patients‘ BMPCs and TPCs are shown in Fig. 1. Plasmacytoma sites, survival, and other clinical characteristic of those patients are presented in Supplementary Fig. 1 and Supplementary Table 1. Fluorescence microscopy images of BMPC and TPC sub-clones are presented in Supplementary Fig. 2.
Fig. 1. Chromosomal aberrations visualized in individual patient’s paired samples.
BMPCs—bone marrow plasma cells. TPCs—plasmacytoma tissue plasma cells. HRD—hyperdiploidy. Numbers in boxes indicate the number of clones found for individual aberration.
Evaluating the population cytogenetic data among patients with primary EMM, secondary EMM, and a control group of NDMM patients without EMM, we found a significant difference in the distribution of del(1p32). A higher proportion of del(1p32) was found in EMM patients, particularly among those with secondary EMM (20.7% (23/111) vs. 28.6% (16/56) vs. 11.1% (27/243); p = 0.002). Interestingly, in EMM patients, del(1p32) was significantly associated with 1q21 gains (12.6% (14/111) vs. 25.0% (14/56) vs. 6.2% (15/243); p < 0.001). The difference in the proportion of del(1p32) or del(1p32) with 1q21 gains was higher in secondary EMM but did not reach statistical significance (del(1p32) (28.6% (16/56) vs. 20.7% (23/111); p = 0.33) and del(1p32) + 1q21 gains (87.5% (14/16) vs. 60.9% (14/23); p = 0.086)). Furthermore, in secondary EMM patients, del(1p32) was more frequently observed in those with plasmacytoma found in soft tissues (extramedullary type, EMD) compared to those with plasmacytomas arising from the bone lesions (paraskeletal type, PS) (50.0% (8/16) vs. 20.0% (8/40); p = 0.046). Cytogenetics aberrations in all patients‘ groups are summarized in Table 1.
Table 1.
Cytogenetic data of patients‘ groups.
| Category | NDMM without plasmacytoma (N = 243) | Primary EMM (N = 111) | Secondary EMM (N = 56) | p-value a |
|---|---|---|---|---|
| t (4;14) | 9.1% (22/243) | 9.9% (11/111) | 14.3% (8/56) | 0.499 |
| t (14;16) | 2.9% (7/243) | 3.6% (4/111) | 1.8% (1/56) | 0.916 |
| Del(13q14)/monosomy 13 | 54.7% (133/243) | 56.8% (63/111) | 69.6% (39/56) | 0.124 |
| Gain (1q21) | 35.6% (84/243) | 36.0% (40/111) | 44.6% (25/56) | 0.477 |
| Amp (1q21) | 12.3% (30/243) | 12.6% (14/111) | 16.1% (9/56) | 0.477 |
| Del (1p32) | 11.1% (27/243) | 20.7% (23/111) | 28.6% (16/56) | 0.002 |
| Del (1p32) + 1q21 | 6.2% (15/243) | 12.6% (14/111) | 25.0% (14/56) | <0.001 |
| Chromosome 1 polysomy | 6.6% (16/243) | 7.2% (8/111) | 3.6% (2/56) | 0.640 |
| Del (17p)/monosomy 17 | 13.6% (33/243) | 18.9% (21/111) | 32.1% (18/56) | 0.004 |
| Chromosome 17 polysomy | 15.6% (38/243) | 18.9% (21/111) | 14.3% (8/56) | 0.653 |
aFisher exact test. Bold values indicate statistically significant results
Thus, we evaluated the survival of EMM patients in the context of del(1p32) and 1q21 gains. For primary EMM patients with both del(1p32) and 1q21 gains, the median PFS was significantly reduced to 7.9 months (95% CI: 3.4–NA), compared to 16.6 months (95% CI: 10.8–NA) in patients with only del(1p32), and 40.2 months (95% CI: 21.9–NA) in patients without either aberration (p = 0.003). The median OS for patients with both del(1p32) and 1q21 gains was also significantly reduced, to 10.6 months (95% CI: 8.3–NA), compared to 28.0 months (95% CI: 22.7–NA) and 69.4 months (95% CI: 45.5–NA), respectively (p = 0.03). In secondary EMM patients with both del(1p32) and 1q21 gains, the median PFS was significantly shorter at 2.0 months (95% CI: 1.5–5.3), compared to 0.5 months (95% CI: 0.4–NA) in patients with only del(1p32) and 6.6 months (95% CI: 4.8–17.2) in patients without either aberration (p < 0.001). Similarly, a significant reduction in median OS was observed in secondary EMM patients with both del(1p32) and 1q21 gains, namely 2.7 months (95% CI: 1.9–8.9), compared to 1.7 months (95% CI: 0.6–NA) and 22.2 months (95% CI: 10.6–NA), respectively (p < 0.001).
Baseline clinical characteristics and survival data of primary and secondary EMM patients in the context of chromosome 1 aberrations are shown in Supplementary Tables 2, 3A, 3B and Supplementary Figs. 3 and 4.
In conclusion, we found chromosome 1 aberrations in nearly all our paired samples with a slightly higher incidence in plasmacytoma tissue rather than bone marrow compartment. Moreover, we encountered very high intra-patient heterogeneity in the 1q21 region between BMPCs and TPCs samples. Our findings were in accord with other studies investigating plasmacytoma tissue samples, reporting the comparable prevalence and variability of these aberrations, with respect to the different detection limits of FISH analyses [5–8]. According to these findings, 1q21 gains appear to be a rather later event or aberration undergoing significant evolutionary changes within cancer cell lines [9]. Our findings regarding del(17p) revealed a shared pattern between BMPCs and TPCs in less than half of the samples bearing this aberration. Similar to 1q21 gains, del(17p) may represent a later event in EMM development [6–8].
The strong negative prognostic impact of del(1p32) in MM patients has been repeatedly confirmed [10, 11], and it is more pronounced in biallelic deletions or in combination with other high-risk aberrations, such as t(4;14), 1q21 gains or del(17p) [11]. Our results are consistent with these findings, highlighting the surprising association of this aberration with EMM patients. The highest incidence of del(1p32) along with 1q21 gains was observed in patients with secondary EMM, known for their poor prognosis [12]. The prognostic impact of these two aberrations combined was more severe across EMM patients than that of del(1p32) or 1q21 gains alone. Unfortunately, we were unable to detect biallelic deletions of 1p32 using I-FISH. These ultra-high-risk aberrations are rare, occurring in approximately 2% of cases. Other methods, such as single-nucleotide polymorphism arrays or next-generation sequencing, may be more effective in detecting them [11].
In the analysis of paired samples, deletions of 1p32 were found exclusively in secondary EMM patients, consistent with the population data mentioned above. Moreover, we observed this aberration more frequently in patients with EMD plasmacytomas. This coincidence may indicate a relation between del(1p32) and this aggressive entity [12]. The deletions of 1p32 were shared between bone marrow and plasmacytoma tissue in almost all cases. Shared deletions of 1p32 were also shown in the work of Liu Y et al. [7]. Compared to aberrations in the 1q21 region or TP53 deletions, our observation suggests that del(1p32) is likely a more conservative high-risk event in the clonal evolution and may play an important role in the initial stages of extraosseous disease development. Based on our results, we strongly recommend including the detection of del(1p32) alongside 1q21 gains in the standard prognostic assessment for MM patients.
Supplementary information
Acknowledgements
We acknowledge the CF Genomics CEITEC MU supported by the NCMG research infrastructure (LM2023067 funded by MEYS CR) for their support in obtaining scientific data presented in this paper. The authors would like to thank all the patients and their caregivers who participated in this study, and data managers from participating centers. We would like to thank Michael Elavsky for the grammatical revision of the text.
Author contributions
SM, OE, and PL designed the study. OE and BohM performed the FISH analysis, JM analyzed and evaluated the FISH data. SM, OE, BI, FD, AZ, KM, SV, KM, BorM, SS, JM, and PL co-wrote the paper. JZ, AT, KM, NV selected patients for tissue sampling and provided surgical biopsies. KZ and RL analyzed data. All authors critically reviewed and approved the manuscript.
Funding
This work was supported by the projects NU21-03–00076 and Conceptual development of research organization (FNBr 65269705) provided by the Ministry of Health of the Czech Republic, and National Institute for Cancer Research (Program EXCELLES, ID Project No. LX22NPO5102) funded by the European Union—Next-Generation EU.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee of the University Hospital Brno, Czech Republic (2016). Informed consent was obtained from all subjects involved in the study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Martin Stork, Eva Ondrouskova.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01131-6.
References
- 1.Neupane K, Fortuna GG, Dahal R, Schmidt TM, Fonseca R, Chakraborty R, et al. Alterations in chromosome 1q in multiple myeloma randomized clinical trials: a systematic review. Blood Cancer J. 2024;14:20–30. 10.1038/s41408-024-00985-0 10.1038/s41408-024-00985-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiménez-Segura R, Rosiñol L, Cibeira MT, Fernández de Larrea C, Tovar N, Rodríguez-Lobato LG, et al. Paraskeletal and extramedullary plasmacytomas in multiple myeloma at diagnosis and at first relapse: 50-years of experience from an academic institution. Blood Cancer J. 2022;12:135. 10.1038/s41408-022-00730-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sevcikova S, Minarik J, Stork M, Jelinek T, Pour L, Hajek R. Extramedullary disease in multiple myeloma - controversies and future directions. Blood Rev. 2019;36:32–9. 10.1016/j.blre.2019.04.002 10.1016/j.blre.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 4.Campo E, Jaffe ES, Cook JR, Quintanilla-Martinez L, Swerdlow SH, Anderson KC, et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A Report from the Clinical Advisory Committee. Blood. 2022;140:1229–53. 10.1182/blood.2022015851 10.1182/blood.2022015851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besse L, Sedlarikova L, Greslikova H, Kupska R, Almasi M, Penka M, et al. Cytogenetics in multiple myeloma patients progressing into extramedullary disease. Eur J Haematol 2016;97:93–100. 10.1111/ejh.12688 [DOI] [PubMed] [Google Scholar]
- 6.Xia Y, Shi Y, Chen Z, Zhang J, Zhu Y, Guo R, et al. Characteristics and prognostic value of extramedullary chromosomal abnormalities in extramedullary myeloma. Chin Med J. 2022;135:2500–2. 10.1097/CM9.0000000000002351 10.1097/CM9.0000000000002351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Jelloul F, Zhang Y, Bhavsar T, Ho C, Rao M, et al. Genetic basis of extramedullary plasmablastic transformation of multiple myeloma. Am J Surg Pathol 2020;44:838–48. 10.1097/PAS.0000000000001459 10.1097/PAS.0000000000001459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jelinek T, Zihala D, Sevcikova T, Anilkumar SA, Kapustova V, Sahinbegovic H, et al. Beyond the marrow: insights from comprehensive next-generation sequencing of extramedullary multiple myeloma tumors. Leukemia. 2024;38:1323–33. 10.1038/s41375-024-02206-w 10.1038/s41375-024-02206-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locher M, Steurer M, Jukic E, Keller MA, Fresser F, Ruepp C, et al. The prognostic value of additional copies of 1q21 in multiple myeloma depends on the primary genetic event. Am J Hematol 2020;95:1562–71. 10.1002/ajh.25994 10.1002/ajh.25994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebraud B, Leleu X, Lauwers-Cances V, Roussel M, Caillot D, Marit G, et al. Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: the IFM experience on 1195 patients. Leukemia. 2014;28:675–9. 10.1038/leu.2013.225 10.1038/leu.2013.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schavgoulidze A, Talbot A, Perrot A, Cazaubiel T, Leleu X, Manier S, et al. Biallelic deletion of 1p32 defines ultra-high-risk myeloma, but monoallelic del(1p32) remains a strong prognostic factor. Blood. 2023;141:1308–15. 10.1182/blood.2022017863 10.1182/blood.2022017863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pour L, Sevcikova S, Greslikova H, Kupska R, Majkova P, Zahradova L, et al. Soft-tissue extramedullary multiple myeloma prognosis is significantly worse in comparison to bone-related extramedullary relapse. Haematologica. 2014;99:360–4. 10.3324/haematol.2013.094409 10.3324/haematol.2013.094409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.