Abstract
PGHS-2 (prostaglandin H synthase-2) is induced in mammalian cells by pro-inflammatory cytokines in tandem with iNOS [high-output (‘inducible’) nitric oxide synthase], and is co-localized with iNOS and nitrotyrosine in human atheroma macrophages. Herein, murine J774.2 macrophages incubated with lipopolysaccharide and interferon γ showed induction of PGHS-2 and generated NO using iNOS that could be completely depleted by 12(S)-HPETE [12(S)-hydroperoxyeicosatetraenoic acid; 2.4 μM] or hydrogen peroxide (500 μM) (0.42±0.084 and 0.38±0.02 nmol·min−1·106 cells−1 for HPETE and H2O2 respectively). COS-7 cells transiently transfected with human PGHS-2 also showed HPETE- or H2O2-dependent NO decay (0.44±0.016 and 0.20±0.04 nmol·min−1·106 cells−1 for 2.4 μM HPETE and 500 μM H2O2 respectively). Finally, purified PGHS-2 consumed NO in the presence of HPETE or H2O2 (168 and 140 μM·min−1·μM enzyme−1 for HPETE and H2O2 respectively), in a haem-dependent manner, with 20 nM enzyme consuming up to 4 μM NO. Km (app) values for NO and 15(S)-HPETE were 1.7±0.2 and 0.45±0.16 μM respectively. These data indicate that PGHS-2 catalytically consumes NO during peroxidase turnover and that pro-inflammatory cytokines simultaneously upregulate NO synthesis and degradation pathways in murine macrophages. Catalytic NO consumption by PGHS-2 represents a novel interaction between NO and PGHS-2 that may impact on the biological effects of NO in vascular signalling and inflammation.
Keywords: cyclo-oxygenase, inflammation, nitric oxide, peroxidase, prostaglandin H synthase-2 (PGHS-2), reactive nitrogen species
Abbreviations: HETE, hydroxyeicosatetraenoic acid; HPETE, hydroperoxyeicosatetraenoic acid; IFNγ, interferon γ; iNOS, high-output (‘inducible’) nitric oxide synthase; LPS, lipopolysaccharide; L-NAME, NG-nitro-L-arginine methyl ester; NOS, nitric oxide synthase; PGG2, prostaglandin G2; PGHS, prostaglandin H synthase; hPGHS-2, human PGHS-2
INTRODUCTION
Nitric oxide plays essential roles in vascular homoeostasis through maintaining vascular tone and inhibiting leucocyte and platelet activation. While pathways of NO generation are well known, mechanisms of NO removal in specific cell types are only now being elucidated. We found previously that human PGHS (prostaglandin H synthase)-1 can consume NO via peroxidase turnover and that this attenuates its inhibitory effects on platelet function [1]. In contrast with PGHS-1, PGHS-2 is induced at high levels by pro-inflammatory cytokines in tandem with the high-output NOS (NO synthase) isoform, iNOS (‘inducible’ NOS). Indeed, in human atherosclerotic lesions PGHS-2 co-localizes with iNOS and nitrotyrosine, particularly in macrophages [2]. This makes it especially interesting as a candidate for controlling NO bioactivity, and several observations suggest that PGHS-2-dependent NO consumption may occur in cells and in vivo. Inhibition of PGHS-2 elevates iNOS-derived NO and promotes its toxicity in LPS (lipopolysaccharide)-dependent meningitis or LPS/IFNγ (interferon γ)-treated RAW 264.7 macrophages [3,4]. Also, administration of PGHS-2-selective inhibitors acutely increases NO bioactivity in hypertension or coronary artery disease, where PGHS-2 elevation is a consistent finding [5–9]. However, the mechanisms underlying these observations have not been characterized. This prompted us to examine the ability of both purified and cellular PGHS-2 to catalytically consume NO during turnover, particularly when generated intracellularly by iNOS in macrophages.
Herein, murine PGHS-2 and cells transfected with hPGHS-2 (human PGHS-2) consumed exogenously added NO via peroxidase turnover. Also, LPS/IFNγ-treated murine J774.2 macrophages rapidly consumed NO in the presence of HPETE (hydroperoxyeicosatetraenoic acid), H2O2 or arachidonate. The capacity of inflammation-activated J774.2 macrophages to consume NO far exceeded the synthetic capacity of iNOS. These data reveal a functional interplay between PGHS-2 and NO that leads to complete removal of endogenously synthesized NO in murine macrophages, and provide a potential mechanistic explanation for observations of improved NO bioactivity following administration of PGHS-2 inhibitors in inflammation and vascular disease.
EXPERIMENTAL
Materials
Recombinant murine PGHS-2 was purified as described in [10]. The apoenzyme was reconstituted with haematin (haematin/apoenzyme, 2:1) in 80 mM Tris, pH 7.8, for 20 min at 22 °C. 15(S)-HPETE, 12(S)-HPETE and 12(S)-HETE [12(S)-hydroxyeicosatetraenoic acid] were from Cayman Chemical (Ann Arbor, MI, U.S.A.). For enzyme assays, the sodium salt of HPETE was prepared in 10% (v/v) methanol/0.01 M NaOH.
Generation of hPGHS2pcDNA3 construct
HeLa OHIO cells were treated with 10 nM phorbol for 4 h to induce expression of hPGHS-2 [11]. Total RNA was extracted and purified using the Ultraspec™ RNA isolation system (Biotecx, Houston, TX, U.S.A.). First-strand cDNA synthesis was carried out using ImProm-II™ reverse transcriptase (Promega, Southampton, U.K.). The PGHS-2 coding region and flanking sequence were amplified by PCR using Elongase® (Invitrogen, Paisley, Scotland, U.K.) and the following sense and antisense primers, which contain restriction sites for BamHI and EcoRI respectively (in italics): sense, GGATCCAGACGCCCTCAGACAGCAAAGCCT; antisense, GAATTCCTCCGCAACAGGAGTACTGACTTCTG.
Amplified hPGHS-2 cDNA was desalted using the QIAquick PCR purification system (Qiagen, Crawley, Sussex, U.K.) and cloned into pGEM®-T (Promega). Positive clones were confirmed by sequencing using Big Dye chemistry (Applied Biosystems). The hPGHS-2 insert was excised by digestion with EcoRI and BamHI (Promega), purified by gel extraction and subcloned into the mammalian expression vector pcDNA3 (Promega). Recombinant pcDNA3 without the insert was also prepared for control transfections.
Expression of PGHS-2 in COS-7 cells
COS-7 cells were transiently transfected at 70% confluence with hPGHS2pcDNA3, using the ProFection® Mammalian Transfection System Calcium Phosphate (Promega). DNA (17 μg) in 125 mM CaCl2/Hepes-buffered saline was added dropwise to cells, then replaced by fresh medium after 5 h, and cells were harvested at 70 h and washed three times with PBS, pH 7.4.
Induction of PGHS-2 and iNOS in J774.2 cells
Cells were incubated with 1 ng·ml−1 IFNγ and 10 ng·ml−1 LPS for 16 h before being harvested and washed four times with PBS. Western blotting (5–10 ng of protein) was performed using goat anti-hPGHS-2 polyclonal antibody (1:500; Santa Cruz Biotech) or rabbit anti-NOS2 polyclonal antibody (1:500; Santa Cruz Biotech) and visualized using ECL® (Amersham), following incubation with anti-goat or anti-rabbit IgG respectively conjugated to horseradish peroxidase [12].
Preparation and analysis of NO
Anaerobic solutions of NO were prepared by bubbling NO gas through argon-saturated deionized water. Contaminating NO2 and N2O3 were removed by flowing through 1 M NaOH. NO concentrations were measured electrochemically using an NO sensor (Harvard AmiNO100 or AmiNO700 with inNO meter; Harvard Apparatus, Edenbridge, Kent, U.K.). Where NO loss was not linear, rates are given as first-order rate constants (kobs). For all kobs values, the square of the Pearson product moment correlation coefficient (r) of the slope of the replotted data was greater than 0.9, confirming that the reaction was first order. Where NO consumption was linear, NO disappearance was determined and the background rate of NO loss subtracted. For measurement of NO consumption by purified PGHS-2, NO was added to 0.5 ml of 80 mM Tris, pH 7.8, at 37 °C with or without 15(S)-HPETE or H2O2 with stirring. Once the NO response had stabilized, haematin, apo-PGHS-2 or holo-PGHS-2 was added and NO decay rates were monitored. NO consumption by cells was measured following addition of H2O2 or HPETE to PBS containing (1–5)×106 cells·ml−1, 1 mM L-NAME (NG-nitro-L-arginine methyl ester), 1 mM CaCl2 and 0.95 μM NO at 37 °C with stirring. NO production by J774.2 cells was measured in the absence of L-NAME following addition of 1 mM L-arginine to PBS containing 1 mM CaCl2 and 107 cells·ml−1 at 37 °C with stirring.
Lipid extractions, absorbance spectroscopy of 15(S)-HPETE and calculation of Michaelis–Menten parameters
For analysis of conjugated dienes following PGHS-2 peroxidase turnover, 15(S)-HPETE (5 μM) was incubated in Tris, pH 7.8, for 5 min with or without the addition of 9.6 nM PGHS-2 and 7.6 μM NO, in triplicate. Following a 5 min incubation, samples were acidified and extracted twice using 2 vol. of diethyl ether. After drying under N2, samples were solubilized in methanol and scanned from 220 to 350 nm, using methanol as reference. Michaelis–Menten parameters were calculated using Enzfitter (Elsevier-Biosoft, Cambridge, U.K.), following determination of initial rates of NO consumption at various NO or 15(S)-HPETE concentrations, using 9.6 nM PGHS-2.
RESULTS
Characterization of NO loss in the electrode system
NO decay in aerobic buffer at 37 °C followed first-order kinetics, with kobs values of (1.81±0.28)×10−3 s−1 and (7.86±1.0)×10−3 s−1 (r2>0.9) for the AmiNO100 and AmiNO700 probes respectively (n=3, means±S.D.). This indicates that NO oxidation by the electrode and diffusion into the gas phase are the main causes of NO decay in these experiments. Using these kobs values, rates of background NO loss could be calculated and subtracted from all experimental values.
LPS/IFNγ treatment of murine monocytes stimulates lipohydroperoxidase-dependent NO consumption
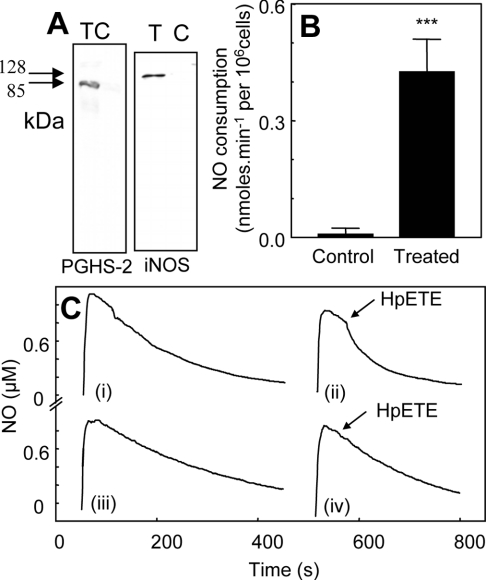
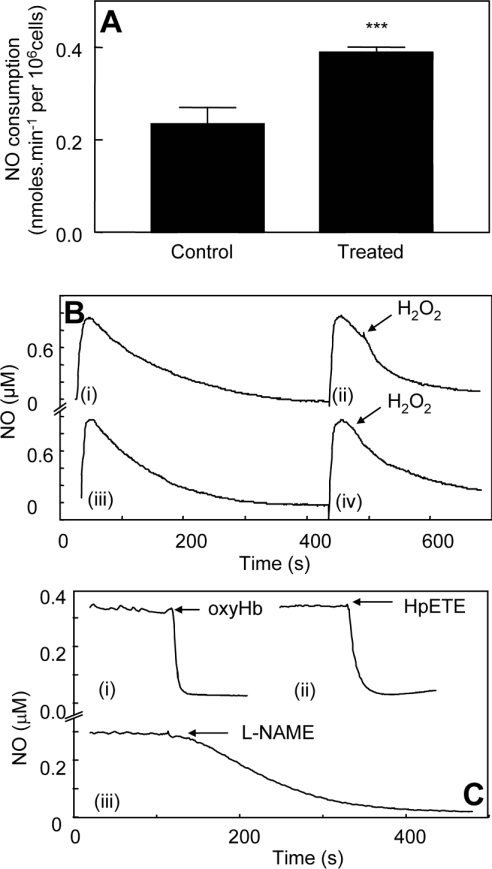
J774.2 cells were incubated with IFNγ/LPS for 16 h to induce PGHS-2, as described previously (Figure 1A, left panel) [13]. In experiments measuring exogenous NO decay for cytokine-treated cells, 1 mM L-NAME was routinely added to inhibit endogenous iNOS activity. Rates of NO decay in the presence of J774.2 cells (106 cells·ml−1) were increased over background [buffer alone, kobs=(1.5±0.1)×10−3 s−1], but were not significantly different for LPS/IFNγ-treated and control cells, and were still first order [kobs=(6.1±1.7)×10−3 s−1 and (6.7±0.6)×10−3 s−1 for treated and control cells respectively]. At 0.6 μM NO, this gave NO consumption rates of 0.16±0.06 and 0.19±0.02 nmol·min−1·106 cells−1 for LPS/IFNγ-treated and control cells respectively, indicating a low-level endogenous NO scavenging capacity of the cells that is unchanged by cytokine treatment. Addition of 2.4 μM 15(S)-HPETE to control cells did not increase NO decay. However, addition of 15(S)-HPETE to LPS/IFNγ-treated J774.2 cells immediately caused a significant increase in cell-dependent NO consumption (0.42±0.084 nmol·min−1·106 cells−1 after subtraction of cell-dependent rates) (Figures 1B and 1C). Using 500 μM H2O2 as substrate instead of 15(S)-HPETE, increased rates of NO decay were found for both cell populations; however, these were significantly higher for cytokine-induced cells (0.23±0.06 and 0.38±0.02 nmol·min−1·106 cells−1 for control and LPS/IFNγ-treated cells respectively, after subtraction of cell-dependent rates) (Figures 2A and 2B). Addition of H2O2 alone in the absence of cells did not accelerate NO decay (results not shown).
Figure 1. Cytokine induction of PGHS-2 in macrophages induces lipohydroperoxidase-dependent NO consumption.
(A) PGHS-2 or iNOS expression was examined by Western blotting following treatment of J774.2 macrophages with 1 ng·ml−1 IFNγ and 10 ng·ml−1 LPS for 16 h. T, cytokine-treated cells; C, control cells. Molecular-mass sizes are indicated in kDa. (B) 15(S)-HPETE (2.4 μM) was added to 1×106 J774.2 cells in the presence of 1 mM L-NAME, 1 mM CaCl2 and 0.8 μM NO in 1 ml of PBS (n=3; means±S.D.). Bars represent rates of NO decay for cells in the presence or absence of HPETE, as appropriate. ***P<0.001 compared with control (unpaired t test). (C) Representative traces from (B): (i) NO decay in 106 cytokine-treated J774 cells·ml−1, 1 mM L-NAME and 1 mM CaCl2 in PBS; (ii) 2.4 μM 15(S)-HPETE was added to cytokine-treated cells in (i); (iii) NO decay in 106 control J774 cells·ml−1, 1 mM L-NAME and 1 mM CaCl2 in PBS; (iv) 2.4 μM 15(S)-HPETE was added to control cells in (iii).
Figure 2. H2O2 induces NO consumption, and lipohydroperoxidase turnover depletes endogenously generated NO, in cytokine-treated macrophages.
(A) H2O2 (500 μM) was added to 2×106 cells, 1 mM L-NAME and 1 mM CaCl2 in 1 ml of PBS (n≥3; means±S.D.). Bars represent rates of NO decay for cells in the presence or absence of H2O2, as appropriate. ***P<0.05 compared with control (unpaired t test). (B) Representative traces from (A): (i) NO decay in PBS containing 2×106 cytokine-treated J774 cells·ml−1, 1 mM L-NAME and 1 mM CaCl2; (ii) 500 μM H2O2 was added to cytokinetreated cells in (i); (iii) NO decay in 2×106 control J774 cells·ml−1, 1 mM L-NAME and 1 mM CaCl2; (iv) 500 μM H2O2 was added to control cells in (iii). (C) OxyHb (3 μM) (i), 2.4 μM 15(S)-HPETE (ii) or 1 mM L-NAME (iii) was added to 107 cytokine-treated J774 cells·ml−1, 1 mM L-arginine and 1 mM CaCl2 in PBS.
Peroxidase turnover depletes endogenously synthesized NO in LPS/IFNγ-treated murine monocytes
Cytokine treatment of J774 cells causes iNOS to be induced in tandem with PGHS-2 (Figure 1A, right panel). In the absence of L-NAME, and with 1 mM L-arginine added as an iNOS substrate, LPS/IFNγ-treated J774.2 cells, but not untreated controls, produced NO, which reached a plateau of approx 0.3 μM after a few minutes and decreased slowly over several minutes thereafter (Figure 2C, and results not shown). Following addition of 2.4 μM 15(S)-HPETE, all endogenously synthesized NO was rapidly and totally consumed (Figure 2C). As controls, the NO was also effectively removed on addition of 2.5 μM oxyHb or 1 mM L-NAME (Figure 2C).
PGHS-2-expressing COS-7 cells consume NO
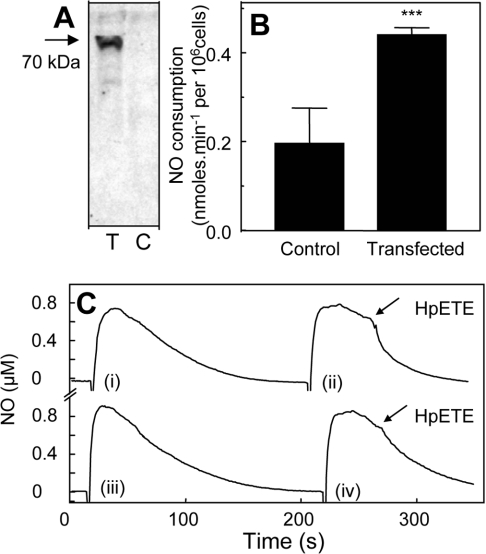
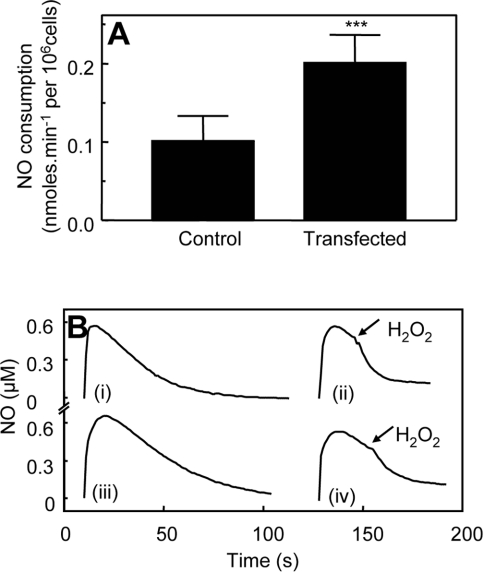
COS-7 cells transiently transfected with hPGHS-2 contained immunoreactive enzyme that was absent from cells transfected with empty vector (Figure 3A). NO disappearance in the presence of 3×106 COS-7 cells·ml−1 (either PGHS-2- or neo-transfected) was increased slightly over background (results not shown). However, rates were not significantly different for PGHS-2- and neo-transfected cells, indicating that this was a property intrinsic to the cell line itself (results not shown). Addition of 2.4 μM 15(S)-HPETE caused a small increase in NO consumption by control cells (0.20±0.079 nmol·min−1·106 cells−1 after subtraction of cell-dependent rates) (Figures 3B and 3C). In contrast, 15(S)-HPETE stimulated significantly higher rates of NO decay when added to hPGHS2pcDNA3-transfected cells (0.44±0.016 nmol·min−1·106 cells−1 after subtraction of cell-dependent rates) (Figures 3B and 3C). Similarly, 500 μM H2O2 increased NO decay to a significantly greater extent in hPGHS2pcDNA3-transfected COS-7 cells than in neo-transfected controls (0.20±0.04 and 0.10±0.03 nmol·min−1·106 cells−1 respectively, after subtraction of cell-dependent rates) (Figures 4A and 4B).
Figure 3. COS-7 cells transfected with hPGHS-2 consume NO via lipohydroperoxidase turnover.
(A) Western blot of COS-7 cells transfected with hPGHS-2 (T) or neo-transfected control cells (C). (B) 15(S)-HPETE (2.4 μM) was added to 1 ml of PBS containing 3×106 cells, 1 mM CaCl2 and 0.8 μM NO (n≥3; means±S.D.). Bars represent rates of NO decay for cells in the presence or absence of HPETE, as appropriate. ***P<0.01 compared with control (unpaired t test). (C) Representative traces from (B): (i) NO decay in PBS containing 3×106 PGHS-2-transfected COS-7 cells and 1 mM CaCl2; (ii) 2.4 μM 15(S)-HPETE was added to PGHS-2-transfected COS-7 cells in (i); (iii) NO decay in PBS containing 3×106 control COS-7 cells and 1 mM CaCl2; (iv) 2.4 μM 15(S)-HPETE was added to control COS-7 cells in (iii).
Figure 4. COS-7 cells transfected with hPGHS-2 consume NO following H2O2 addition faster than controls.
(A) NO consumption of COS-7 cells was determined as described in the Experimental section, following addition of 500 μM H2O2 to 1 ml of PBS containing 5×106 cells and 1 mM CaCl2 (n≥3; means±S.D.). Bars represent rates of NO decay for cells in the presence or absence of H2O2, as appropriate. ***P<0.05 compared with control (unpaired t test). (B) Representative traces from (A): (i) NO decay in PBS containing 5×106 PGHS-2-transfected COS-7 cells and 1 mM CaCl2; (ii) 500 μM H2O2 was added to PGHS-2-transfected COS-7 cells in (i); (iii) NO decay in PBS containing 5×106 control COS-7 cells and 1 mM CaCl2; (iv) 500 μM H2O2 was added to control COS-7 cells in (iii).
Purified recombinant PGHS-2 consumes NO during peroxidase turnover
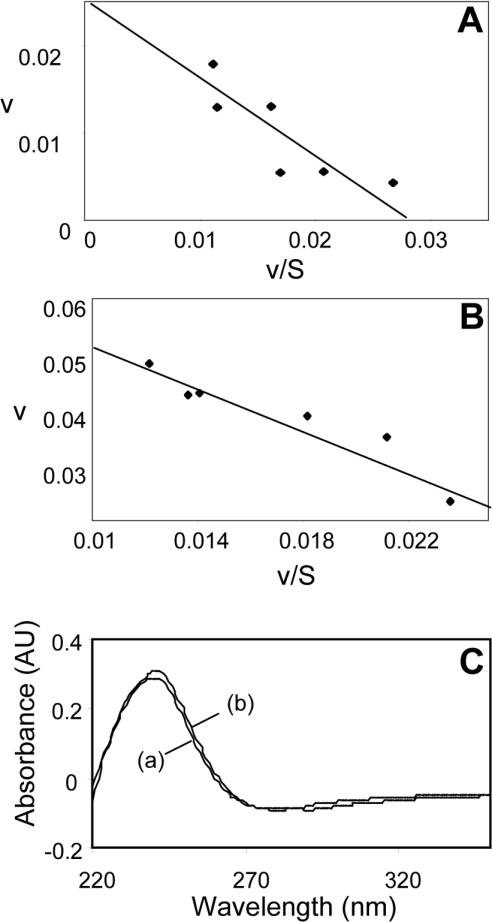
In the absence of peroxide substrate, haem-reconstituted PGHS-2 did not accelerate NO decay (Figure 5, Table 1). However, either 15(S)-HPETE or H2O2 stimulated PGHS-2-dependent NO disappearance significantly (168 and 140 μM·min−1·μM−1 PGHS-2 for 6.2 μM HPETE and 500 μM H2O2 respectively, after subtraction of background). Since amounts of PGHS-2 (up to 20 nM) added to the assay were far below amounts of NO consumed (up to 4 μM), peroxide-dependent NO consumption was a catalytic process. Hydroperoxide-stimulated NO consumption required haem reconstitution, indicating that NO was acting as a substrate for PGHS-2 peroxidase turnover (Figure 5, Table 1). Plotting initial rates of NO consumption at various substrate concentrations in the presence of 9.6 nM PGHS-2 gave Km (app) values for NO and 15(S)-HPETE of 1.7±0.2 μM and 0.45±0.16 μM respectively (Figures 6A and 6B). Finally, spectral analysis of 15(S)-HPETE before and after PGHS-2-dependent NO consumption revealed no loss of conjugated diene spectrum, with the absorbance maximum at 235 nm being retained (Figure 6C). This is consistent with NO acting as a reducing peroxidase substrate, stimulating reduction of HPETE to HETE.
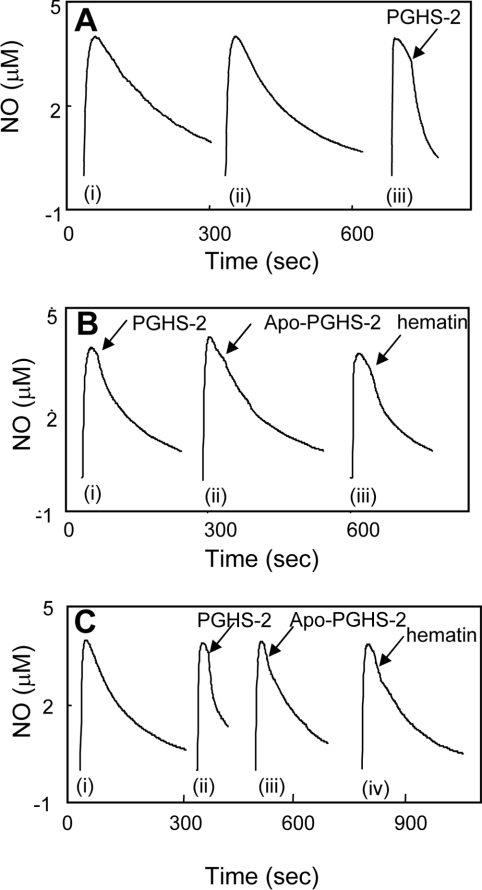
Figure 5. Purified murine PGHS-2 catalytically consumes NO via peroxidase turnover.
(A) (i) Decay of 3.8 μM NO in Tris buffer, pH 7.8, at 37 °C; (ii) decay of NO in buffer with 6.2 μM 15(S)-HPETE; (iii) purified PGHS-2 (20 nM) was added to buffer containing 3.8 μM NO and 6.2 μM 15(S)-HPETE as shown. (B) (i) Purified murine PGHS-2 (20 nM) was added to buffer with 3.8 μM NO alone; (ii) apo-PGHS-2 (20 nM) was added to buffer containing 6.2 μM 15(S)-HPETE and 3.8 μM NO; (iii) haematin (40 nM) was added to buffer containing 6.2 μM 15(S)-HPETE and 3.8 μM NO. (C) (i) Decay of NO in buffer containing 500 μM H2O2; (ii) purified PGHS-2 (20 nM) was added to buffer containing 500 μM H2O2 and 3.8 μM NO; (iii) apo-PGHS-2 (20 nM) was added to buffer containing 500 μM H2O2 and 3.8 μM NO; (iv) haematin (40 nM) was added to buffer containing 500 μM H2O2 and 3.8 μM NO.
Table 1. NO uptake by purified PGHS-2.
NO (3.8 μM) was added to 80 mM Tris buffer, pH 7.8, at 37 °C containing H2O2 (500 μM) or 15(S)-HPETE (6.2 μM). Once the electrode response had stabilized, PGHS-2 (20 nM), apo-PGHS-2 (20 nM) or haematin (40 nM) was added, and rates of NO consumption were recorded. *For these samples, rates of NO loss at 3.0 μM were determined using the calculated first-order rate constant (kobs). For all other samples, initial linear rates of NO loss on addition of PGHS-2 were determined. Values are means±S.D.; n=3. For all kobs values, r2=0.99. ND, not determined.
| Sample | Rate (nmol·min−1) | kobs (s−1) |
|---|---|---|
| Background* | 0.7±0.09 | (7.86±1.0)×10−3 |
| +HPETE* | 0.6±0.03 | (6.93±0.36)×10−3 |
| +H2O2* | 0.77±0.1 | (8.6±1.2)×10−3 |
| +PGHS-2* | 0.75±0.02 | (8.5±0.2)×10−3 |
| +Apo-PGHS-2+HPETE* | 0.58±0.05 | (6.54±0.6)×10−3 |
| +Haematin+HPETE | 0.8±0.15 | ND |
| +PGHS-2+HPETE | 2.38±0.28 | ND |
| +Apo-PGHS-2+H2O2* | 0.81±0.07 | (9.06±0.8)×10−3 |
| +Haematin+H2O2* | 0.8±0.01 | (8.96±1.3)×10−3 |
| +PGHS-2+H2O2 | 2.1±0.3 | ND |
Figure 6. Calculation of kinetic parameters for HPETE-dependent NO consumption by PGHS-2.
(A) Km (app) was determined by measuring initial rates of NO consumption by 9.6 nM PGHS-2 at various 15(S)-HPETE concentrations. Michaelis–Menten parameters were calculated using Enzfitter (Elsevier–Biosoft). Rates of NO consumption are plotted as v against v/[S]. (B) Km (app) was determined as described in (A) but by varying the NO concentration. (C) HPETE (5 μM) was incubated for 5 min with or without 9.6 nM PGHS-2 and 7.2 μM NO in Tris buffer, pH 7.8, before being extracted using acidified diethyl ether, as described in the Experimental section, and reconstituted in methanol. Samples were scanned from 200 to 350 nm using methanol as reference: (a) control, (b) with PGHS-2 plus NO.
DISCUSSION
The present study shows that PGHS-2 peroxidase turnover can completely deplete endogenously synthesized NO in cytokine-activated J774.2 macrophages (Figures 1 and 2). Catalytic NO consumption represents a novel activity of PGHS-2 and provides a mechanistic explanation for previous observations of improved NO bioactivity following PGHS-2 inhibition in vivo and in vitro [3,4,8,9].
PGHS isoforms exhibit two catalytic activities, a cyclo-oxygenase activity that converts arachidonate into PGG2 (prostaglandin G2), and a haem-dependent peroxidase activity that reduces PGG2 to PGH2. In addition to PGG2, other hydroperoxides can serve as substrates, including lipid hydroperoxides (HPETEs), organic hydroperoxides and, to a lesser extent, H2O2. We showed previously that PGHS-1 could consume NO [1]. However, while PGHS-1 and PGHS-2 share many similarities, they are also both structurally and catalytically quite different. For example, they share only 60% identity at the protein level, and catalytic differences include: (i) differences in the threshold for hydroperoxidase activation of cyclo-oxygenase activity (10-fold lower for PGHS-2); (ii) negative allosteric regulation at low concentrations of arachidonic acid for PGHS-1 cyclo-oxygenase (at 500–1000 nM); (iii) depending on substrate concentration, PGHS-2 peroxidase catalyses different ratios of one- and two-electron reduction of HPETE; and (iv) PGHS-2 can oxidize additional substrates, such as arachidonyl ethanolamide, whereas PGHS-1 cannot [14–17]. Based on this, differences in kinetic parameters for the interaction of NO with PGHS-2 compared with PGHS-1 are expected. Finally, at a biological level, PGHS-2 is expressed in a very different manner (i.e. in response to pro-inflammatory cytokines) and in different cell types. Therefore the biological consequences of PGHS-2-dependent NO consumption will be very different from those of PGHS-1-dependent NO consumption. For these reasons, it is important to determine whether PGHS-2, independently of PGHS-1, can consume NO, and the potential impact of this on resulting cellular NO levels.
In the present study, recombinant murine PGHS-2 catalytically consumed NO in a haem-dependent manner that required hydroperoxide substrates, i.e. HPETE or H2O2 (Figure 5, Table 1). This confirms that NO can act as a reducing substrate for PGHS-2 peroxidase activity [1,18]. Far lower concentrations of HPETE than of H2O2 were required, consistent with previous reports of Km values for H2O2 being 50–100 times greater than for HPETEs [19–21]. Therefore HPETE will be the favoured substrate for PGHS-2-dependent NO consumption in cells. Kinetic analysis of NO consumption at various HPETE concentrations revealed a Km (app) for 15(S)-HPETE of 0.45±0.16 μM, which is almost 10-fold lower than that calculated for the hydroperoxidase-dependent consumption of NO by ovine PGHS-1 of 3.3±0.3 μM (Figure 6) [1]. Previous Km values for the reduction of lipid hydroperoxides by PGHS-1 are in the range 5–20 μM, similar to our calculated value for hydroperoxide-dependent NO consumption by PGHS-1 [19–21]. Although we were unable to find calculated Km values for lipid hydroperoxide reduction by PGHS-2, hydroperoxide initiator requirements for cyclo-oxygenase turnover, which are far below than those required for full peroxidase activity, are approx. 10-fold lower for PGHS-2 than for PGHS-1 (e.g. 2 nM and 20 nM respectively) [14]. This suggests that PGHS-2 will preferentially consume NO over PGHS-1 under conditions of low intracellular peroxide levels. Indeed, previous studies have shown that, in cells where HPETE is metabolized rapidly by glutathione peroxidase, PGHS-2 functions far more effectively than PGHS-1, due to its higher affinity for HPETEs. This enables PGHS-2 to compete effectively with glutathione peroxidases for HPETE reduction and function under conditions where PGHS-1 is catalytically inactive (Figure 6) [1,18,22,23].
On varying the NO concentration, a Km (app) for NO of 1.7±0.28 μM was determined (Figure 6). This is similar to the Km (app) for NO calculated for ovine PGHS-1 of 2.2±0.9 μM (V. B. O'Donnell, unpublished work). These concentrations of NO are expected to occur in vivo, particularly under inflammatory conditions following induction of iNOS [24]. Finally, following peroxidase-dependent NO decay, the conjugated diene spectrum of HPETE was retained (Figure 6C) [1]. While PGHS-1 catalyses predominantly the two-electron reduction of lipid hydroperoxides, PGHS-2 can catalyse either one- or two-electron reduction, with the proportion of one-electron reduction products increasing at higher HPETE concentrations. Thus at 2.5 μM HPETE the enzyme catalyses >97% two-electron reduction, with 60% reduction at 50 μM HPETE [15]. Our experiments, conducted at 5 μM HPETE, showed complete retention of the conjugated diene spectrum, consistent with reduction of HPETE to HETE, as found previously for PGHS-1 [1].
IFNγ/LPS-treated murine macrophages that express PGHS-2, but not untreated controls, consumed NO following addition of HPETE (Figure 1). Untreated control cells did not express detectable PGHS-1 or -2 (Figure 1 and results not shown). Using high concentrations of H2O2 as substrate, NO consumption was also observed; however, rates were lower and some H2O2-dependent NO consumption by control cells was also found (Figure 2). This may indicate activity of additional peroxidases that can reduce H2O2 but not HPETE, e.g. myeloperoxidase [25,26]. However, while previous studies showed that purified myeloperoxidase can consume NO, it has not been found to consume NO in intact cells, for example neutrophils [27], and it is not known to be expressed in J774 cells. Arachidonate also stimulated NO decay by LPS/IFNγ-treated cells which could be partially inhibited by aspirin or celecoxib (results not shown); however, rates were variable, and it was not possible to conclusively rule out a role for lipoxygenases that can (i) consume NO in the presence of arachidonate, and (ii) generate HPETE that may also be used as a peroxidase substrate for PGHS-2 [28,29]. In additional experiments, COS-7 cells transiently transfected with hPGHS-2 also exhibited HPETE- or H2O2-dependent NO consumption (Figures 3 and 4). Here, selective introduction of the gene encoding hPGHS-2 excludes NO consumption by additional peroxidases and confirms the ability of cellular PGHS-2 to consume NO during peroxidase turnover.
In vascular cells, iNOS and PGHS-2 are induced in tandem by the same pro-inflammatory cytokines. Previous studies have shown that these pathways interact at a number of levels, including both stimulation and suppression of PGHS-2-dependent prostaglandin generation by NO [30–33]. Herein, addition of L-arginine to cytokine-stimulated macrophages enabled NO generation by endogenous iNOS that could be completely depleted by HPETE addition (Figure 2C). Indeed, comparison of rates of NO consumption in our study (0.42±0.084 nmol·min−1·106 cells−1) with published estimates of NO generation by murine macrophages (0.1–0.2 nmol·min−1·106 cells−1) indicate further that PGHS-2 is expressed at levels that can impact significantly on NO levels under pro-inflammatory conditions in mammalian cells, and underscore its potential ability to modulate NO signalling in vivo [34,35].
Lipid hydroperoxides are generated by a number of pathways in vascular cells, including (i) PGHS-mediated oxidation of arachidonate to PGG2, which will be sensitive to aspirin, celecoxib or indomethacin, (ii) lipoxygenase-mediated oxidation of arachidonate to HPETE, and (iii) non-enzymic LDL peroxidation associated with macrophage-derived foam cells. All of these potential sources of PGHS substrate are elevated in vascular disease and inflammation, and could provide substrates for PGHS-2 peroxidase turnover. Indeed, numerous human studies have shown that PGHS-2-selective inhibitors or aspirin can acutely improve NO bioactivity in vivo in subjects with vascular disease [8,9,36–40].
In summary, PGHS-2-dependent NO consumption has been demonstrated by isolated enzyme, PGHS-2-transfected COS-7 cells and inflammation-activated murine macrophages, with turnover rates sufficient to fully deplete endogenous iNOS-derived NO. Catalytic NO scavenging by PGHS-2 provides a novel functional interaction between NO and PGHS-2 that may explain how PGHS-2 inhibition improves NO bioactivity in vitro in models of inflammation, and in human vascular disease in vivo [3,4,8,9].
Acknowledgments
We thank the British Heart Foundation (V. B. O'D., M. J. C., S. R. C.), the Wellcome Trust (V. B. O'D., P. B. A.) and the National Institutes of Health (CA89450 to L. J. M.) for financial support.
References
- 1.O'Donnell V. B., Coles B., Lewis M. J., Crews B. C., Marnett L. J., Freeman B. A. Catalytic consumption of nitric oxide by prostaglandin H synthase-1 regulates platelet function. J. Biol. Chem. 2000;275:38239–38244. doi: 10.1074/jbc.M001802200. [DOI] [PubMed] [Google Scholar]
- 2.Baker C. S., Hall R. J., Evans T. J., Pomerance A., Maclouf J., Creminon C., Yacoub M. H., Polak J. M. Cyclooxygenase-2 is widely expressed in atherosclerotic lesions affecting native and transplanted human coronary arteries and colocalizes with inducible nitric oxide synthase and nitrotyrosine particularly in macrophages. Arterioscler. Thromb. Vasc. Biol. 1999;19:646–655. doi: 10.1161/01.atv.19.3.646. [DOI] [PubMed] [Google Scholar]
- 3.von Knethen A., Brune B. Cyclooxygenase-2: an essential regulator of NO-mediated apoptosis. FASEB J. 1997;11:887–895. [PubMed] [Google Scholar]
- 4.Boje K. M., Jaworowicz D., Jr, Raybon J. J. Neuroinflammatory role of prostaglandins during experimental meningitis: evidence suggestive of an in vivo relationship between nitric oxide and prostaglandins. J. Pharmacol. Exp. Ther. 2003;304:319–325. doi: 10.1124/jpet.102.041533. [DOI] [PubMed] [Google Scholar]
- 5.McAdam B. F., Catella-Lawson F., Mardini I. A., Kapoor S., Lawson J. A., FitzGerald G. A. Systemic biosynthesis of prostacyclin by cyclooxygenase (COX)-2: the human pharmacology of a selective inhibitor of COX-2. Proc. Natl. Acad. Sci. U.S.A. 1999;96:272–277. doi: 10.1073/pnas.96.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belton O., Byrne D., Kearney D., Leahy A., Fitzgerald D. J. Cyclooxygenase-1 and -2-dependent prostacyclin formation in patients with atherosclerosis. Circulation. 2000;102:840–845. doi: 10.1161/01.cir.102.8.840. [DOI] [PubMed] [Google Scholar]
- 7.FitzGerald G. A., Smith B., Pedersen K., Brash A. R. Increased prostacyclin biosynthesis in patients with severe atherosclerosis and platelet activation. N. Engl. J. Med. 1984;310:1065–1068. doi: 10.1056/NEJM198404263101701. [DOI] [PubMed] [Google Scholar]
- 8.Chenevard R., Hurlimann D., Bechir M., Enseleit F., Spieker L., Hermann M., Riesen W., Gay S., Gay R. E., Neidhart M., et al. Selective COX-2 inhibition improves endothelial function in coronary artery disease. Circulation. 2003;107:405–409. doi: 10.1161/01.cir.0000051361.69808.3a. [DOI] [PubMed] [Google Scholar]
- 9.Widlansky M. E., Price D. T., Gokce N., Eberhardt R. T., Duffy S. J., Holbrook M., Maxwell C., Palmisano J., Keaney J. F., Jr, Morrow J. D., Vita J. A. Short- and long-term COX-2 inhibition reverses endothelial dysfunction in patients with hypertension. Hypertension. 2003;42:310–315. doi: 10.1161/01.HYP.0000084603.93510.28. [DOI] [PubMed] [Google Scholar]
- 10.Rowlinson S. W., Crews B. C., Lanzo C. A., Marnett L. J. The binding of arachidonic acid in the cyclooxygenase active site of mouse prostaglandin endoperoxide synthase-2 (COX-2). A putative L-shaped binding conformation utilizing the top channel region. J. Biol. Chem. 1999;274:23305–23310. doi: 10.1074/jbc.274.33.23305. [DOI] [PubMed] [Google Scholar]
- 11.Dixon D. A., Kaplan C. D., McIntyre T. A., Zimmerman G. A., Prescott S. M. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J. Biol. Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 12.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swierkosz T. A., Mitchell J. A., Warner T. D., Botting R. M., Vane J. R. Co-induction of nitric oxide synthase and cyclo-oxygenase: interactions between nitric oxide and prostanoids. Br. J. Pharmacol. 1995;114:1335–1342. doi: 10.1111/j.1476-5381.1995.tb13353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulmacz R. J., Wang L. H. Comparison of hydroperoxide initiator requirements for the cyclooxygenase activities of prostaglandin H synthase-1 and -2. J. Biol. Chem. 1995;270:24019–24023. doi: 10.1074/jbc.270.41.24019. [DOI] [PubMed] [Google Scholar]
- 15.Landino L. M., Crews B. C., Gierse J. K., Hauser S. D., Marnett L. J. Mutational analysis of the role of the distal histidine and glutamine residues of prostaglandin-endoperoxide synthase-2 in peroxidase catalysis, hydroperoxide reduction, and cyclooxygenase activation. J. Biol. Chem. 1997;272:21565–21574. doi: 10.1074/jbc.272.34.21565. [DOI] [PubMed] [Google Scholar]
- 16.Swinney D. C., Mak A. Y., Barnett J., Ramesha C. S. Differential allosteric regulation of prostaglandin H synthase 1 and 2 by arachidonic acid. J. Biol. Chem. 1997;272:12393–12398. doi: 10.1074/jbc.272.19.12393. [DOI] [PubMed] [Google Scholar]
- 17.Kozak K. R., Crews B. C., Morrow J. D., Wang L. H., Ma Y. H., Weinander R., Jakobsson P. J., Marnett L. J. Metabolism of the endocannabinoids, 2-arachidonylglycerol and anandamide, into prostaglandin, thromboxane, and prostacyclin glycerol esters and ethanolamides. J. Biol. Chem. 2002;277:44877–44885. doi: 10.1074/jbc.M206788200. [DOI] [PubMed] [Google Scholar]
- 18.Curtis J. F., Reddy N. G., Mason R. P., Kalyanaraman B., Eling T. E. Nitric oxide: a prostaglandin H synthase 1 and 2 reducing cosubstrate that does not stimulate cyclooxygenase activity or prostaglandin H synthase expression in murine macrophages. Arch. Biochem. Biophys. 1996;335:369–376. doi: 10.1006/abbi.1996.0518. [DOI] [PubMed] [Google Scholar]
- 19.Marnett L. J., Bienkowski M. J., Pagels W. R. Oxygen 18 investigation of the prostaglandin synthetase-dependent co-oxidation of diphenylisobenzofuran. J. Biol. Chem. 1979;254:5077–5082. [PubMed] [Google Scholar]
- 20.Kulmacz R. J. Prostaglandin H synthase and hydroperoxides: peroxidase reaction and inactivation kinetics. Arch. Biochem. Biophys. 1986;249:273–285. doi: 10.1016/0003-9861(86)90003-2. [DOI] [PubMed] [Google Scholar]
- 21.Kulmacz R. J., Lands W. E. Requirements for hydroperoxide by the cyclooxygenase and peroxidase activities of prostaglandin H synthase. Prostaglandins. 1983;249:273–285. doi: 10.1016/0090-6980(83)90025-4. [DOI] [PubMed] [Google Scholar]
- 22.Capdevila J. H., Morrow J. D., Belosludtsev Y. Y., Beauchamp D. R., DuBois R. N., Falck J. R. The catalytic outcomes of the constitutive and the mitogen inducible isoforms of prostaglandin H2 synthase are markedly affected by glutathione and glutathione peroxidase(s) Biochemistry. 1995;34:3325–3337. doi: 10.1021/bi00010a023. [DOI] [PubMed] [Google Scholar]
- 23.Reddy S. T., Herschman H. R. Ligand-induced prostaglandin synthesis requires expression of the TIS10/PGS-2 prostaglandin synthase gene in murine fibroblasts and macrophages. J. Biol. Chem. 1994;269:15473–15480. [PubMed] [Google Scholar]
- 24.Brovkovych V., Stolarczyk E., Oman J., Tomboulian P., Malinski T. Direct electrochemical measurement of nitric oxide in vascular endothelium. J. Pharm. Biomed. Anal. 1999;19:135–143. doi: 10.1016/s0731-7085(98)00090-9. [DOI] [PubMed] [Google Scholar]
- 25.Abu-Soud H. M., Hazen S. L. Nitric oxide is a physiological substrate for mammalian peroxidases. J. Biol. Chem. 2000;275:37524–37532. doi: 10.1074/jbc.275.48.37524. [DOI] [PubMed] [Google Scholar]
- 26.Eiserich J. P., Baldus S., Brennan M. L., Ma W., Zhang C., Tousson A., Castro L., Lusis A. J., Nauseef W. M., White C. R., Freeman B. A. Myeloperoxidase, a leukocyte-derived vascular NO oxidase. Science. 2002;296:2391–2394. doi: 10.1126/science.1106830. [DOI] [PubMed] [Google Scholar]
- 27.Clark S. R., Coffey M. J., Collins P. W., Lewis P. W., Lewis M. J., Cross A. R., O'Donnell V. B. Characterization of nitric oxide metabolism pathways in normal, chronic granulomatous disease and myeloperoxidase-deficient human neutrophils. J. Immunol. 2002;169:5889–5896. doi: 10.4049/jimmunol.169.10.5889. [DOI] [PubMed] [Google Scholar]
- 28.O'Donnell V. B., Taylor K. B., Parthasarathy S., Kuhn H., Koesling D., Friebe A., Bloodsworth A., Darley-Usmar V. M., Freeman B. A. 15-Lipoxygenase catalytically consumes nitric oxide and impairs activation of guanylate cyclase. J. Biol. Chem. 1999;274:20083–20091. doi: 10.1074/jbc.274.29.20083. [DOI] [PubMed] [Google Scholar]
- 29.Coffey M. J., Natarajan R., Chumley P. H., Coles B., Thimmalapura P. R., Nowell M., Kuhn H., Lewis M. J., Freeman B. A., O'Donnell V. B. Catalytic consumption of nitric oxide by 12/15-lipoxygenase: inhibition of monocyte soluble guanylate cyclase activation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8006–8011. doi: 10.1073/pnas.141136098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salvemini D., Seibert K., Masferrer J. L., Misko T. P., Currie M. G., Needleman P. Endogenous nitric oxide enhances prostaglandin production in a model of renal inflammation. J. Clin. Invest. 1994;93:1940–1947. doi: 10.1172/JCI117185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minghetti L., Polazzi E., Nicolini A., Creminon C., Levi G. Interferon-gamma and nitric oxide down-regulate lipopolysaccharide-induced prostanoid production in cultured rat microglial cells by inhibiting cyclooxygenase-2 expression. J. Neurochem. 1996;66:1963–1970. doi: 10.1046/j.1471-4159.1996.66051963.x. [DOI] [PubMed] [Google Scholar]
- 32.Habib A., Bernard C., Lebret M., Creminon C., Esposito B., Tedgui A., Maclouf J. Regulation of the expression of cyclooxygenase-2 by nitric oxide in rat peritoneal macrophages. J. Immunol. 1997;158:3845–3851. [PubMed] [Google Scholar]
- 33.Marnett L. J., Wright T. L., Crews B. C., Tannenbaum S. R., Morrow J. D. Regulation of prostaglandin biosynthesis by nitric oxide is revealed by targeted deletion of inducible nitric-oxide synthase. J. Biol. Chem. 2000;275:13427–13430. doi: 10.1074/jbc.275.18.13427. [DOI] [PubMed] [Google Scholar]
- 34.Ischiropoulos H., Zhu L., Beckman J. S. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 1992;289:446–451. doi: 10.1016/0003-9861(92)90433-w. [DOI] [PubMed] [Google Scholar]
- 35.Lewis R. S., Tamir S., Tannenbaum S. R., Deen W. M. Kinetic analysis of the fate of nitric oxide synthesized by macrophages in vitro. J. Biol. Chem. 1995;270:29350–29355. doi: 10.1074/jbc.270.49.29350. [DOI] [PubMed] [Google Scholar]
- 36.Noon J. P., Walker B. R., Hand M. F., Webb D. J. Impairment of forearm vasodilatation to acetylcholine in hypercholesterolemia is reversed by aspirin. Cardiovasc. Res. 1998;38:480–484. doi: 10.1016/s0008-6363(98)00013-3. [DOI] [PubMed] [Google Scholar]
- 37.Mullen M. J., Kharbanda R. K., Cross J., Donald A. E., Taylor M., Vallance P., Deanfield J. E., MacAllister R. J. Heterogenous nature of flow-mediated dilatation in human conduit arteries in vivo: relevance to endothelial dysfunction in hypercholesterolemia. Circ. Res. 2001;88:145–151. doi: 10.1161/01.res.88.2.145. [DOI] [PubMed] [Google Scholar]
- 38.Taddei S., Virdis A., Ghiadoni L., Magagna A., Salvetti A. Cyclooxygenase inhibition restores nitric oxide activity in essential hypertension. Hypertension. 1997;29:274–279. doi: 10.1161/01.hyp.29.1.274. [DOI] [PubMed] [Google Scholar]
- 39.Husain S., Andrews N. P., Mulcahy D., Panza J. A., Quyyumi A. A. Aspirin improves endothelial dysfunction in atherosclerosis. Circulation. 1998;97:716–720. doi: 10.1161/01.cir.97.8.716. [DOI] [PubMed] [Google Scholar]
- 40.Graupera M., Garcia-Pagan J. C., Pares M., Abraldes J. G., Rosello J., Bosch J., Rodes J. Cyclooxygenase-1 inhibition corrects endothelial dysfunction in cirrhotic rat livers. J. Hepatol. 2003;39:515–521. doi: 10.1016/s0168-8278(03)00347-7. [DOI] [PubMed] [Google Scholar]