Abstract
Programmed cell death 1 (PD-1) is a premier cancer drug target for immune checkpoint blockade (ICB). Because PD-1 receptor inhibition activates tumor-specific T-cell immunity, research has predominantly focused on T-cell-PD-1 expression and its immunobiology. In contrast, cancer cell-intrinsic PD-1 functional regulation is not well understood. Here, we demonstrate induction of PD-1 in melanoma cells via type I interferon receptor (IFNAR) signaling and reversal of ICB efficacy through IFNAR pathway inhibition. Treatment of melanoma cells with IFN-α or IFN-β triggers IFNAR-mediated Janus kinase-signal transducer and activator of transcription (JAK/STAT) signaling, increases chromatin accessibility and resultant STAT1/2 and IFN regulatory factor 9 (IRF9) binding within a PD-1 gene enhancer, and leads to PD-1 induction. IFNAR1 or JAK/STAT inhibition suppresses melanoma-PD-1 expression and disrupts ICB efficacy in preclinical models. Our results uncover type I IFN-dependent regulation of cancer cell-PD-1 and provide mechanistic insight into the potential unintended ICB-neutralizing effects of widely used IFNAR1 and JAK inhibitors.
Subject terms: Melanoma, Cancer immunotherapy, Tumour immunology
Cancer cell-intrinsic PD-1 expression has been documented in multiple tumor types, including in melanoma. Here the authors identify a type I IFN-JAK/STAT signaling axis as a critical regulator of tumor cell-intrinsic PD-1 expression and targeting, with implications for cancer immunotherapy.
Introduction
Immune checkpoint therapies targeting the programmed cell death 1 (PD-1) pathway have revolutionized the clinical landscape for the treatment of advanced stage cancers1. To date, six therapeutic antibodies (abs) blocking the PD-1 receptor and three recognizing its major ligand, PD-L1, have been approved by the US Food and Drug Administration (FDA) for the treatment of patients with metastatic disease of various etiologies. In patients and preclinical tumor models, PD-1:PD-L1 axis blockade stimulates antitumor immunity by increasing activation and proliferation of tumor-reactive T-cells1–3, including self-renewing stem-like CD8+ T-cells4. Consequently, most research investigations of PD-1 immunobiology in the cancer context have focused on T-cells5. Nevertheless, PD-1 is also critical for maintaining T-cell homeostasis and tolerance to prevent autoimmunity and dampen immune hyperactivation6.
Cytokines and growth factor networks are crucial regulators of immune surveillance and crosstalk in these various scenarios7, in part through regulation of T-cell-PD-1 expression8. Indeed, the T-cell-PD-1 axis can be induced or modulated by multiple members of the interleukin (IL) family of cytokines9,10. Several growth factors, such as transforming growth factor (TGF)-β, tumor necrosis factor (TNF)-α, and vascular endothelial growth factor (VEGF) also regulate PD-1 pathway member expression on T-cells11–13.
The interferon (IFN) cytokine family plays a crucial role in regulating T-cell-PD-1 expression in concert with T-cell receptor (TCR) stimulation14. It also induces PD-L1 and PD-L2 in cancer cells15,16, macrophages17, and other immune cell types within the tumor microenvironment (TME)18 to orchestrate an immunoevasive program. Consistently, intratumoral IFN signatures and downstream loss-of-function mutations have been associated with resistance to PD-1 immune checkpoint blockade (ICB)19–21. Conversely, IFNs can stimulate antitumor T-cell responses and are thus also correlates of cancer cell immunosurveillance and clinical ICB benefit22–24. Two main classes of IFNs exist, type I and type II25. The type I IFN family comprises multiple subtypes, including IFN-α and IFN-β, whereas type II IFN consists only of IFN-γ. Type I IFNs signal through the IFN-α receptor (IFNAR) 1 and 2 heterodimeric complex, while type II IFN binds to the IFN-γ receptor (IFNGR) 1 and 2 ternary complex25. Type I and type II IFNs activate overlapping, yet partly distinct, Janus kinase (JAK) signal transducer and activator of transcription (STAT) effectors to regulate the expression of diverse target genes, including PD-1 pathway members26.
In a broad range of autoimmune and inflammatory disorders characterized by aberrant IFN pathway activation, inhibitors of the IFN signaling axis have produced remarkable clinical benefit27. These include IFNAR1 blocking abs, such as anifrolumab, which received FDA approval for the treatment of systemic lupus erythematosus (SLE)28. Moreover, a panel of inhibitors targeting either JAK1/2, e.g. ruxolitinib or upadacitinib, or tyrosine kinase (TYK) 2, such as deucravacitinib, was also clinically approved for the treatment of vitiligo, psoriasis, atopic dermatitis, and other immunological conditions29–31. Despite their widespread clinical use and the elevated risk of developing cancer in patients with some of these pathologies32, potential IFN pathway inhibitor effects on T-cell-PD-1 expression and resultant PD-1 ICB outcomes have not been systematically analyzed.
There is growing recognition that PD-1 expression is not restricted to T-cells, but is also found on multiple additional TME cell types, including macrophages, B-cells, NK cells, and even cancer cells5,24,33–36. Tumor cell-intrinsic PD-1 expression was initially described in melanoma37–42 but has since also been found in hepatocellular43, non-small cell lung (NSCLC)44,45, and colorectal carcinomas46,47, amongst other malignancies36,48,49. Cancer cell-PD-1 exerts growth-inhibitory activities in NSCLC44,45 and colorectal carcinoma46,47 but promotes tumorigenesis in melanoma37,39,42 and other cancers36,43,48,49. Accordingly, inhibition of tumor cell-PD-1 and its downstream proproliferative signaling effectors in the latter cancers attenuates growth, including in T-cell-deficient mouse models37,39,43,48,49. While several molecular mechanisms governing PD-L1 induction in cancer cells have been defined15,16, regulation of tumor-intrinsic PD-1 receptor gene and protein expression and consequent impact on ICB remain poorly understood.
In this work, we examine cytokine and growth factor regulation of melanoma cell-intrinsic PD-1 expression and its impact on ICB efficacy, leveraging knowledge on established modulators of the T-cell-PD-1 receptor pathway. Analysis of a single cell (sc) RNA sequencing (seq) dataset50 of patient melanomas and assessment of gene expression in human and murine melanoma lines reveals preferential expression of IFNAR by PD-1+ versus PD-1- melanoma cell subsets. Treatment with IFN-α or IFN-β induces PD-1 gene (PDCD1/Pdcd1) and protein expression in multiple human and murine melanoma lines. ATAC-seq reveals type I IFN-mediated chromatin opening upstream of the PD-1 gene promoter in melanoma cells, at an enhancer cis-regulatory element (cCRE) containing annotated STAT1:STAT2 and IFN regulatory factor (IRF) binding sites. Physical interaction of phosphorylated (p) STAT1/2 and IRF9 at this PD-1 gene enhancer is confirmed by chromatin immunoprecipitation (ChIP)-qPCR. Inhibition of type I IFN signaling suppresses melanoma-PD-1 expression and eliminates PD-1 ICB-mediated tumor growth inhibition. Together, our findings identify a tumor cell-intrinsic type I IFN signaling axis as a pivotal regulator of melanoma-PD-1 expression and response to PD-1 ICB. Therefore, they raise concerns that clinically approved IFNAR1 and JAK inhibitors, currently used in patients with increased susceptibility to cancer32, might unintentionally disrupt PD-1 ICB benefit by suppressing PD-1 receptor targeting.
Results
The type I interferon heterodimeric receptor is highly expressed by PD-1+ melanoma cells
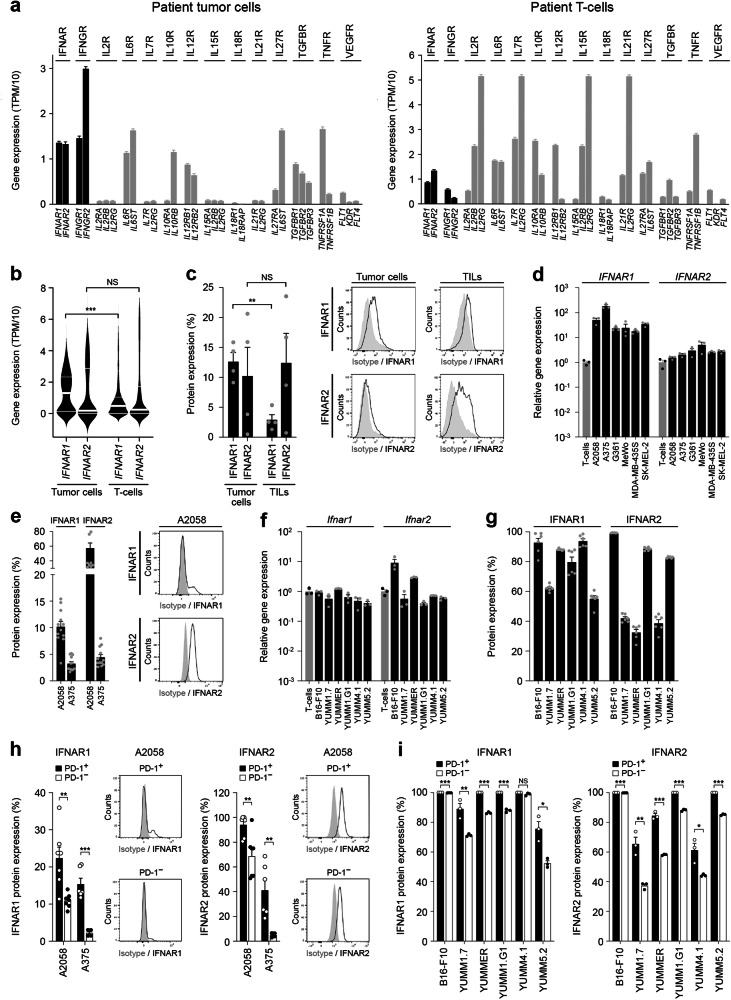
We analyzed a single-cell (sc) RNA-seq melanoma patient dataset50 for tumor cell expression of cytokine and growth factor receptors known to regulate the PD-1 pathway in T cells, including IFNAR, IFNGR, IL2R, IL6R, IL7R, IL10R, IL12R, IL15R, IL18R, IL21R, IL27R, TGFBR, TNFR, and VEGFR. Compared to melanoma-infiltrating T-cells, patient melanoma cells expressed significantly higher levels of IFNAR heterodimers (Fig. 1a, b), while most other receptors evaluated tended to be more prevalent on T-cells (Figs. 1a, S1a, b). Similarly, IFNGR heterodimers were overrepresented on patient melanoma cells vs. T-cells (Figs. 1a, S2a). FACS analysis revealed surface protein expression of IFNAR1 and IFNAR2 by patient tumor-infiltrating lymphocytes (TILs) and cancer cells, the latter of which expressed a significantly higher level of the rate-limiting subunit for the type I IFN receptor complex25, IFNAR1 (Figs. 1c, S3a). Similarly, RT-qPCR analysis revealed markedly greater gene expression levels of IFNAR1 and IFNAR2 (Fig. 1d) as well as IFNGR1 and IFNGR2 (Fig. S2b) in human melanoma cell lines, A2058, A375, G361, MeWo, MDA-MB-435S, and SK-MEL-2, compared to human T-cells. Flow cytometric analyses corroborated the expression of IFNAR (Figs. 1e, S3b) and IFNGR subunits (Fig. S2c) by human melanoma lines at the protein level. Similarly, Ifnar1/2 (Fig. 1f) and Ifngr1/2 heterodimer levels (Fig. S2d) by murine melanoma cells, B16-F10, YUMM1.7, YUMMER1.7D4, YUMM1.G1, YUMM4.1, and YUMM5.2 were equal to or exceeded those in murine T-cells. Protein expression of IFNAR and IFNGR subunits by murine melanoma lines was also substantial (Figs. 1g, S2e, S3c). In contrast, human or murine melanoma lines did not express or tended to express lower levels of IL2R/Il2r, IL7R/Il7r, IL10R/Il10r, IL12R/Il12r, IL15R/Il15r, IL18R/Il18r, IL21R/Il21r, IL27R/Il27r, or TNFR compared to T-cells, consistent with undetectable and/or minimal expression of at least one receptor heterodimer (Figs. S4a,b, S5a). Nevertheless, appreciable levels of melanoma-intrinsic IL6R/Il6r (Figs. S4a, S5a), TGFBR/Tgfbr, Tnfr, and VEGFR/Vegfr subunits (Figs. S4b, S5b) were detected. To determine if IFNAR and/or IFNGR might regulate PD-1 receptor levels on melanoma cells, we performed co-expression analyses by flow cytometry. Indeed, both IFNAR1 and IFNAR2 proteins were significantly enriched among native PD-1+ versus PD-1- human (Figs. 1h, S3d) and murine (Figs. 1i, S3e) melanoma cell subsets. Similarly, PD-1+ melanoma cell fractions co-expressed significantly higher levels of the IFNGR1 and IFNGR2 subunits compared to their PD-1- counterparts (Fig. S2f, g). Consistently, patient melanoma cells positive for the PD-1 gene (PDCD1) transcript co-express both IFNAR subunits in two independent scRNA-seq datasets50,51 (Fig. S6a, b), tended to be enriched in interferon-stimulated genes (ISG52, Fig. S6c), and in tumors of high versus low immunoscore53 (Fig. S6d). Together, these results demonstrate marked expression of type I and type II IFN receptors by melanoma cells at levels exceeding those in T-cells and preferential enrichment of IFNAR and IFNGR among PD-1+ melanoma subpopulations.
Fig. 1. Expression of type I interferon receptors by melanoma cells.
Single-cell RNA-seq analysis of a type I interferon-α/β receptor (IFNAR) and type II interferon-γ receptor (IFNGR) subunit gene expression shown in units of transcript per million (TPM)/10 (black bars; mean ± SEM) in patient melanoma cells (left) versus tumor-infiltrating T-cells (right). Gene expression of additional cytokine and growth factor receptor subunits is also shown (gray bars). b Violin plots of IFNAR1 and IFNAR2 gene expression (TPM/10; median, bold white line; top and bottom quartiles, thin white lines) in patient melanoma (MM) versus tumor-infiltrating T-cells generated from the single-cell RNA-seq dataset above. c IFNAR1 and IFNAR2 surface protein expression (percent positivity, mean ± SEM, left) by patient tumor cells and tumor-infiltrating lymphocytes (TILs), with representative flow cytometric histograms shown (right). d Relative IFNAR1 and IFNAR2 gene expression (fold change, mean ± SEM) in human melanoma cell lines (black bars) versus human activated CD3+ T-cells (gray bars) as determined by RT-qPCR. e IFNAR1 and IFNAR2 surface protein expression (percent positivity, mean ± SEM, left) by human A2058 and A375 melanoma cells, with representative flow cytometric histograms shown for A2058 melanoma cells (right). f Relative Ifnar1 and Ifnar2 gene expression (fold change, mean ± SEM) in murine melanoma cell lines (black bars) versus murine activated CD3+ T-cells (gray bars) as determined by RT-qPCR. g IFNAR1 and IFNAR2 surface protein expression (percent positivity, mean ± SEM, left) by murine melanoma cells, with representative flow cytometric histograms shown for B16-F10 and YUMM1.7 melanoma cells (right). IFNAR1 (left) and IFNAR2 (right) surface protein expression (percent positivity, mean ± SEM) by PD-1+ versus PD-1- h, human A2058 and A375 and i, murine melanoma cell subsets, with representative flow cytometric histograms shown for A2058 cells. Results represent biologically independent samples of a, b n = 1252 MM cells (left panel) and 2040 T-cells (right panel), c n = 4, and biologically independent experiments of d, f, i n = 3, e n = 12, g n = 6, and h n = 7 (A2058) and n = 6 (A375). Statistical analyses included b Mann-Whitney test, two-sided, c paired t-test, one-sided, and h, i unpaired t-test, two-sided. *p < 0.05; **p < 0.01; ***p < 0.001; NS, not significant. See also Supplementary Figs. 1–7. Source data, including exact p-values, are provided as a Source Data file.
Type I interferons induce melanoma cell-intrinsic PD-1 expression
To directly examine whether IFNAR or IFNGR ligation functionally induces PD-1 expression by melanoma cells, we treated human melanoma cell lines with the type I IFNs, IFN-α or IFN-β, or the type II IFN, IFN-γ, and assessed PD-1 gene and protein expression. Both IFN-α and IFN-β induced PDCD1 gene expression up to 16-fold (Figs. 2a, S7a) and also significantly induced PD-1 protein expression (Fig. 2b) in human melanoma lines compared to controls. Addition of IFN-γ induced both PD-1 gene and protein expression only in A375, but not in A2058 human melanoma cells (Fig. 2a, b). Consistently, Pdcd1 gene and PD-1 protein levels were induced up to 42-fold and 5-fold, respectively, by IFN-α or IFN-β treatment in murine melanoma lines, B16-F10, YUMM1.7, YUMMER1.7D4, YUMM1.G1, YUMM4.1, and YUMM5.2, while IFN-γ stimulation variably induced PD-1 across these lines (Figs. 2c, d, S7b). Treatment of melanoma cells with IL-2, IL-6, IL-7, IL-10, IL-12, IL-15, IL-18, IL-21, IL-27, TGF-β, TNF-α, or VEGF did not induce substantial PD-1 gene (Fig. S8a) or protein expression (Fig. S8b), in agreement with generally lower overall cognate receptor expression on melanoma cells (Figs. 1, S1, 4, 5). Consistent with published reports15,16, human and murine PD-L1 gene (CD274/Cd274) and protein levels were elevated via IFN-α, IFN-β, or IFN-γ treatment across all melanoma lines examined (Fig. S9a-d), with type II IFN inducing PD-L1 protein to the greatest extent (Fig. S9d). These findings identify a melanoma cell-intrinsic type I IFN axis that not only functionally regulates PD-L1, as previously established15,16, but also PD-1 receptor expression.
Fig. 2. Type I interferons induce melanoma cell-intrinsic PD-1 expression.
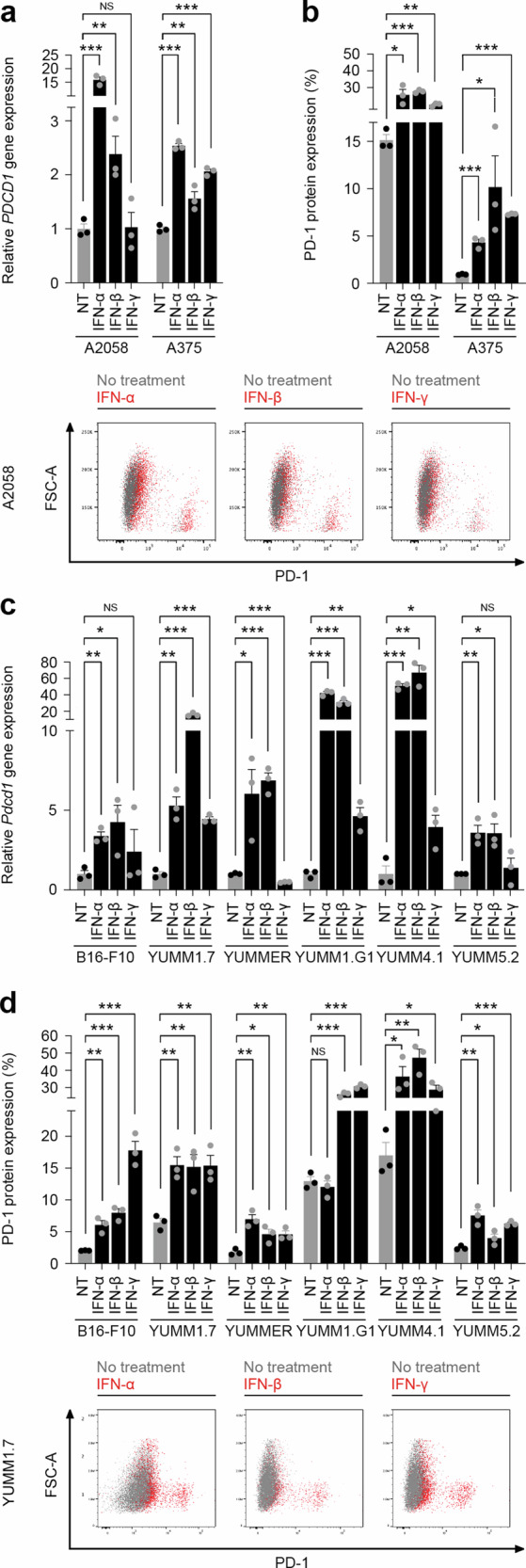
a Relative PDCD1 gene (fold change, mean ± SEM) and b PD-1 surface protein (percent positivity, mean ± SEM, top) expression by human A2058 and A375 melanoma cells treated (black bars) versus not treated (NT, gray bars) with human type I interferons, IFN-α or IFN-β, or type II interferon, IFN-γ, as determined by RT-qPCR and flow cytometry, respectively. Representative flow cytometric dot plot overlays of human PD-1 surface protein induction in IFN-α, IFN-β, or IFN-γ treated (red dots) versus no treatment control (gray dots) A2058 cells are shown (bottom). c Relative Pdcd1 gene (fold change, mean ± SEM) and d PD-1 surface protein (percent positivity, mean ± SEM) expression by murine melanoma cells treated (black bars) versus not treated (NT, gray bars) with murine IFN-α, IFN-β, or IFN-γ, as determined by RT-qPCR and flow cytometry, respectively. Representative flow cytometric dot plot overlays of murine PD-1 surface protein induction in IFN-α, IFN-β, or IFN-γ treated (red dots) versus no treatment control (gray dots) YUMM1.7 cells are shown (bottom). Results in (a-d) represent biologically independent experiments of n = 3. Statistical analyses included the (a, d) unpaired t-test, one-sided, and (b,c) unpaired t-test, two-sided. *p < 0.05; **p < 0.01; ***p < 0.001; NS, not significant. See also Supplementary Figs. 6, 8, 9. Source data, including exact p-values, are provided as a Source Data file.
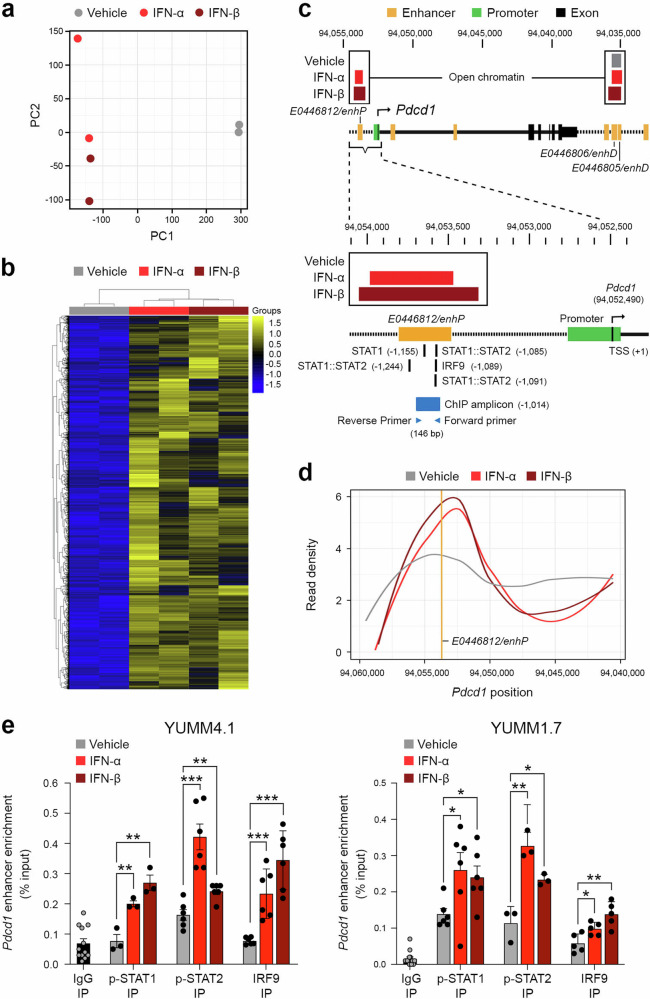
Type I interferons promote chromatin opening at a PD-1 gene enhancer region and consequent STAT-IRF transcription factor binding in melanoma cells
To dissect type I IFN-mediated PD-1 gene regulatory mechanisms in melanoma cells, we next performed unbiased ATAC-seq analysis of YUMM4.1 cells, because they demonstrated the greatest fold-induction of Pdcd1 transcription by IFN-α or IFN-β among all lines examined (Figs. 2c, S7b). Principal component (PC) analysis revealed similar global chromatin accessibility patterns between IFN-α and IFN-β treatment groups and substantial differences compared to vehicle control YUMM4.1 cohorts along PC1 (Fig. 3a). Pairwise comparisons of chromatin accessibility for IFN-α and IFN-β versus vehicle control showed strong consistency in open chromatin patterns between the type I IFNs (Fig. 3b, Data S1). Examination of ENCODE candidate cCREs for the Pdcd1 gene specifically uncovered selective chromatin opening at the E0446812/enhP enhancer located upstream of the Pdcd1 promoter in response to IFN-α or IFN-β versus vehicle control treatment (Fig. 3c, d). In contrast, constitutively open chromatin was observed in other Pdcd1 cCREs in both vehicle and type I IFN treatment conditions (e.g. enhancers E0446806/enhD or E0446805/enhD) (Fig. 3c). The E0446812/enhP Pdcd1 enhancer encodes predicted binding sequences for STAT1:STAT2 and IRF9 transcription factors (TFs, Fig. 3c). Consequently, we performed ChIP-qPCR analyses to directly assess binding of these TFs to the Pdcd1 enhancer element. IP for p-STAT1, p-STAT2, or IRF9 revealed significantly increased TF binding to the E0446812/enhP Pdcd1 enhancer in IFN-α or IFN-β compared to vehicle treated YUMM4.1 and YUMM1.7 melanoma cells, as quantified by qPCR (Fig. 3e). These results highlight type I IFN-dependent regulation of melanoma cell-intrinsic PD-1 gene expression via induction of chromatin opening within the E0446812/enhP Pdcd1 enhancer cCRE, thereby leading to increased binding of the IFN-stimulated gene factor 3 (ISGF3) complex components25, p-STAT1, p-STAT2, and IRF9.
Fig. 3. Type I interferons promote chromatin opening and STAT-IRF transcription factor binding to a PD-1 gene enhancer in melanoma cells.
a Principal component analysis of chromatin accessibility as determined by ATAC-seq of YUMM4.1 cells treated with IFN-α, IFN-β, or vehicle control. b Heatmap showing a subset of peaks (~10%) found to be differentially bound between cells treated with IFN-α or IFN-β versus vehicle control. c, Schematic of open chromatin regions within the PD-1 gene (Pdcd1) locus of YUMM4.1 cells treated with IFN-α or IFN-β (red boxes) or vehicle control (gray box), as determined by ATAC-seq as above. Genomic locations for individual enhancer cis-regulatory elements (cCREs, yellow), E0446812/enhP (left), E0446806/enhD (middle), and E0446805/enhD (right) and gene promoter (green) relative to the Pdcd1 transcriptional start site (TSS, +1, black arrow) are shown. Annotated STAT1:STAT2 and IRF9 binding sites (black lines) within the E0446812/enhP enhancer motif as well as for primers used for subsequent ChIP-qPCR analysis (blue arrows) of the 146 base pair (bp) amplicon (blue box) are also illustrated. Both IFN-α and IFN-β treatment resulted in significantly increased chromatin opening at E0446812/enhP compared to vehicle control as shown above by the illustrated open chromatin and by d smoothed conditional means of read density of normalized counts in 100 base pair bin sizes. The E0446812/enhP Pdcd1 enhancer location is both represented and labeled as a vertical yellow line. e ChIP-qPCR analysis of E0446812/enhP Pdcd1 enhancer amplification (% input, mean ± SEM) in anti-p-STAT1, anti-p-STAT2, or anti-IRF9 versus control IgG immunoprecipitated (IP) lysates from YUMM4.1 (left) and YUMM1.7 (right) cells treated as above. Results represent biologically independent experiments of a–d n = 2 per treatment group and e n = 12 (IgG1), n = 3 (p-STAT1), n = 6 (p-STAT2), and n = 6 (IRF9) for YUMM4.1 and n = 14 (IgG1), n = 6 (p-STAT1), n = 3 (p-STAT2), and n = 5 (IRF9) for YUMM1.7. Statistical analyses in e included the unpaired t-test, two-sided. *p < 0.05; **p < 0.01; ***p < 0.001. See also Supplementary Data 1. Source data, including exact p-values, are provided as a Source Data file.
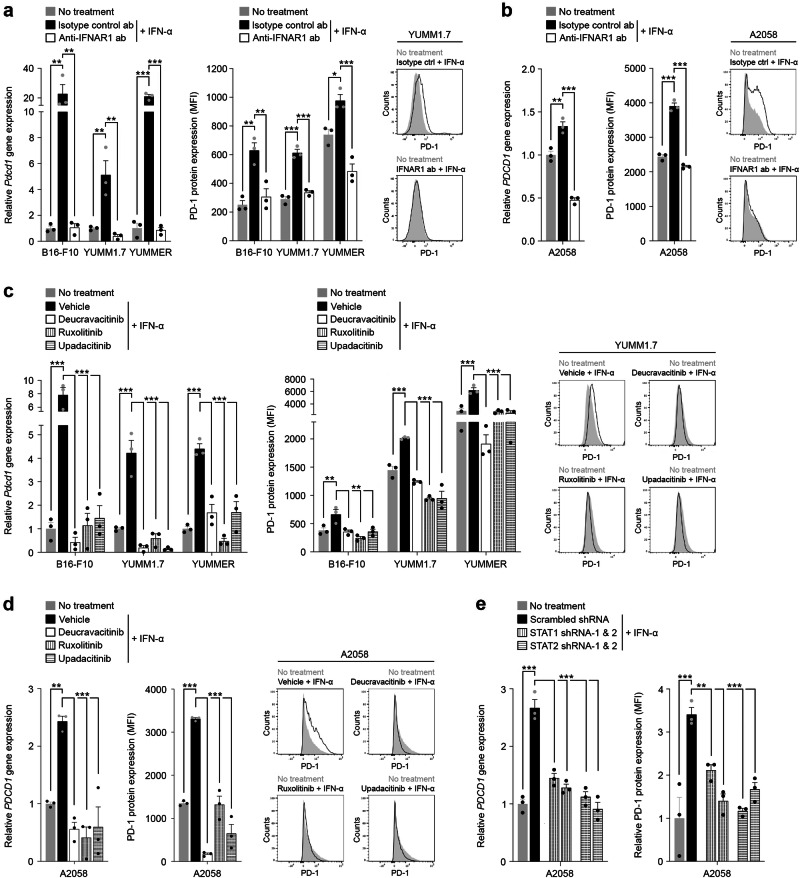
Inhibition of the IFNAR-JAK/STAT signaling axis reverses melanoma cell-PD-1 induction
To independently validate the functional involvement of specific IFNAR pathway members in regulating melanoma cell-intrinsic PD-1, we targeted several distinct signaling mediators using both experimental and clinically approved IFNAR1 blocking abs28, pharmacological antagonists of the IFNAR docking proteins25, JAK1 and TYK2, and downstream STAT1 and STAT2 gene knockdown approaches. Ab-mediated blockade of IFNAR1 on murine B16-F10, YUMM1.7, or YUMMER1.7D4 melanoma cells significantly reversed IFN-α-induced PD-1 gene and protein expression down to baseline levels (Fig. 4a). Blockade of human IFNAR1 with the FDA-approved inhibitor, anifrolumab28, also significantly suppressed IFN-α-dependent PD-1 expression by A2058 human melanoma cells (Fig. 4b), with significantly greater antagonism of PD-1 expression upon extended IFNAR1 ab treatment (Fig. S10a). Consistently, inhibition with the clinically approved pharmacologic antagonists targeting JAK1, ruxolitinib or upadacitinib29,31, or TYK2, deucravacitinib30, significantly attenuated the marked induction of PD-1 transcript and protein by IFN-α in both murine (Fig. 4c) and human (Figs. 4d, S10a) melanoma cell lines, as above. IFN-α or IFN-β-mediated activation of downstream STAT1 and STAT2 in melanoma cells was confirmed via intracellular flow cytometry with abs recognizing phosphorylated (p)-STATs (Fig. S10b). T-cells, on the other hand, showed selective phosphorylation of STAT1 but not STAT2 following type I IFN treatment (Fig. S10b). Consistent with basal PD-1 expression by unstimulated murine and human melanoma cells37,38, cell-intrinsic type I IFN effector molecules, as above, showed some degree of constitutive phosphorylation (Figs. S10c, d, S15) and were enriched among PD-1+ vs. PD-1- melanoma cell subsets (Fig. S10e, f). To address STAT functional dependence of IFN-mediated PD-1 regulation in melanoma cells, we generated stable STAT1 and STAT2 knockdown versus control A2058 tumor variant lines using two independent hairpins each. Silencing of STAT1 and STAT2 gene and protein (Figs. S10g, S16) expression was confirmed by RT-qPCR and immunoblotting, respectively. STAT1 and STAT2 shRNA-mediated knockdown significantly suppressed IFN-α-dependent PD-1 gene or protein induction compared to scrambled shRNA controls (Fig. 4e). Together, these findings identify a tumor-intrinsic type I IFN signaling axis, encompassing IFNAR1 and downstream JAK1, TYK2, STAT1, and STAT2 effector molecules, that functionally regulate melanoma cell-PD-1 receptor expression. They also reveal that clinically approved IFNAR1, JAK1, and TYK2 inhibitors, developed to antagonize IFN-dependent signaling in immune cells in patients with various inflammatory disorders, potently suppress PD-1 induction in melanoma cells. Widely used clinical IFNAR1 or JAK inhibitors might thus unintentionally disrupt PD-1 ICB efficacy, including at the level of the melanoma cell, by reducing PD-1 expression and its recognition by therapeutic PD-1 abs.
Fig. 4. Inhibition of the type I Interferon signaling axis reverses melanoma cell-PD-1 induction.
Relative PD-1 gene (fold change, mean ± SEM, left) and PD-1 surface protein (fluorescence intensity, mean ± SEM, right) expression by IFN-α stimulated a, murine and b, human melanoma cells pre-treated with either blocking anti-IFNAR1 (white bars) or isotype control ab (black bars), compared to no IFN-α treatment controls (gray bars), as determined by RT-qPCR and flow cytometry, respectively. Representative flow cytometric histograms for a YUMM1.7 and b A2058 cells are shown (right). Relative PD-1 gene (fold change, mean ± SEM, left) and PD-1 surface protein (fluorescence intensity, mean ± SEM, right) expression by IFN-α (white bars) versus vehicle control (black bars) treated c murine and d human melanoma cells pre-incubated with FDA-approved small molecule inhibitors targeting the type I interferon pathway mediators, TYK2 (deucravacitinib) or JAK1 (ruxolitinib or upadacitinib), compared to no IFN-α treatment controls (gray bars), as determined by RT-qPCR and flow cytometry, respectively. Representative flow cytometric histograms for c YUMM1.7 and d A2058 cells are shown (right). e, Relative PDCD1 gene (fold change, mean ± SEM, left) and PD-1 surface protein (fluorescence intensity, mean ± SEM, right) expression by IFN-α treated STAT1 or STAT2 knockdown (shRNA-1/-2, white patterned bars) versus scrambled control shRNA (black bars) melanoma A2058 cells, compared to no IFN-α treatment controls (gray bars), as determined by RT-qPCR and flow cytometry, respectively. Results in a–e represent biologically independent experiments of n = 3. Statistical analyses in a–e included the one-way ANOVA with Dunnett post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. See also Supplementary Fig. 10. Source data, including exact p-values, are provided as a Source Data file.
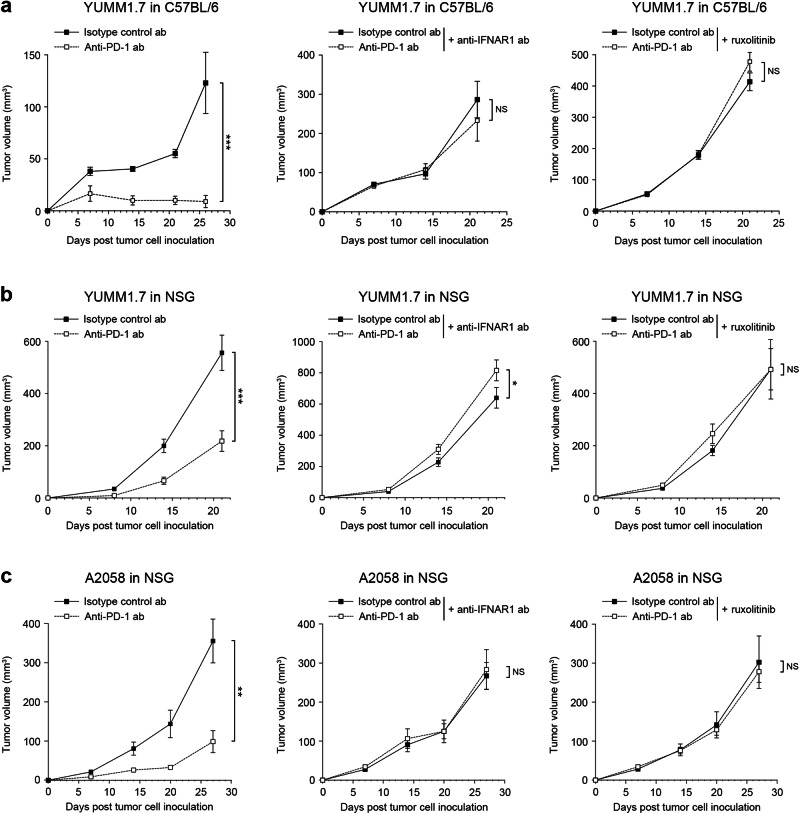
IFNAR-JAK/STAT pathway inhibition disrupts therapeutic efficacy of PD-1 checkpoint blockade
To directly address the above possibility, we treated melanoma-bearing mice with PD-1 blocking abs in the presence or absence of IFNAR1 neutralizing ab or the JAK1 inhibitor, ruxolitinib. Consistent with previous findings54, administration of anti-PD-1 ab significantly inhibited growth of YUMM1.7 tumors showing IFNAR1 expression and baseline STAT1 phosphorylation (Fig. S11) compared to isotype control treatment in immunocompetent C57BL/6 mice (Fig. 5a). Combination therapy with PD-1 ab and either IFNAR1 ab or ruxolitinib completely abrogated tumor growth suppression from PD-1 checkpoint blockade (Fig. 5a). Tumor volumes in both IFNAR1 and ruxolitinib treatment cohorts were significantly larger than those in respective isotype or vehicle control groups, consistent with the observed reduction in tumor-infiltrating T-cell frequency (Fig. S12a) and activation (Fig. S12b) within the former treatment groups. IFNAR1 ab or ruxolitinib administration significantly reduced PD-1 (Pdcd1) or PD-L1 (Cd274) gene (Fig. S12c), and/or PD-1 protein expression, as well as levels of ISGs, Cmpk2, Ifit2, Irf7, Isg20, and/or Oasl, but not p-STAT1 compared to control YUMM1.7 tumors in C57BL/6 mice (Fig. S12d, e). These results thus indicate divergent sensitivity to IFN-dependent PD-1 regulation among distinct TME cell types with varying levels of IFN-α and IFN-β expression (Figs. S11, S12f). In NSG mice devoid of adaptive immune cell-PD-1 and therefore comparatively enriched for cancer cell-PD-137, IFNAR1 inhibition or ruxolitinib treatment significantly suppressed PD-1 and/or PD-L1 transcript levels (Fig. S12g), PD-1 and p-STAT1 protein expression (Figs. S12h, S13a), as well as ISGs (Fig. S12i), indicative of tumor cell-intrinsic inhibition of type I IFN signaling and resultant PD-1 downregulation. Consistently, PD-1 monotherapy of YUMM1.7 tumors in NSG mice significantly reduced tumorigenesis compared to isotype control treatment, whereas administration of PD-1 ab in combination with melanoma-PD-1 downregulating agents (Figs. 4, S12c, d, g, h), IFNAR1 or JAK inhibitors, fully reversed PD-1 therapeutic benefit (Fig. 5b). Human A2058 melanoma growth in immunocompromised NSG mice was also significantly abrogated by PD-1 neutralizing ab, as above, whereas anifrolumab or ruxolitinib treatment reduced human tumor cell-intrinsic PD-1 and p-STAT1 protein expression (Figs. S13b, S14a, b) and thwarted PD-1 ICB efficacy (Fig. 5c). Murine IFNs did not induce PD-1 gene or protein expression by human A2058 melanoma cells (Fig. S14c). Nevertheless, A2058 cells produced marked in vivo amounts of IFN-α and IFN-β (Fig. S14d). Tumor-autocrine IFNA, IFNB, and ISG expression tended to correlate with PD-1 gene level (Fig. S14e, f), in agreement with IFNAR1 expression and baseline STAT1 phosphorylation in A2058 tumors (Fig. S13b). Together, these results demonstrate that IFNAR1 and JAK inhibitors commonly used in the clinic can unintentionally disrupt cancer therapeutic benefit when used in combination with PD-1 ICB regimens, by suppressing IFN pathway-dependent PD-1 receptor expression and targeting, including on melanoma cells.
Fig. 5. Antagonism of the type I interferon signaling pathway disrupts therapeutic efficacy of PD-1 checkpoint blockade.
Tumor growth kinetics (mean ± SEM) of murine YUMM1.7 cells in a C57BL/6 and b NSG mice, and of c, human A2058 cells in NSG mice treated with PD-1 blocking versus isotype-matched control abs, either in the absence (left) or presence of IFNAR1 blocking ab (middle) or the JAK1 inhibitor, ruxolitinib (right), at submaximal dosage (180 mg/kg/d, p.o.). Results represent biologically independent experiments involving a n = 18 mice (left, middle panels) and n = 28 (right panel), b n = 14 and 16 mice for anti-PD-1 ab and isotype control ab, respectively (left panel) and n = 14 mice (middle, right panels), c n = 6 mice (left, middle, and right panels). Statistical analyses in a–c included the two-way ANOVA with Bonferroni post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001; NS, not significant. See also Supplementary Figs. 11–14. Source data, including exact p-values and information on sex, are provided as a Source Data file.
Discussion
Blockade of the PD-1 receptor on tumor-infiltrating T-cells is a crucial requirement for eliciting robust clinical responses to PD-1 ICB in patients with cancer1,2. Multiple signaling networks regulate T-cell-PD-1 target expression in the TME5,9 and are thus important for predicting clinical benefit. The IFN receptor pathway stands out as a prominent inducer of PD-1 expression on TCR-engaged T-cells14,26 and has also been strongly associated with PD-1 ICB efficacy22–24. Additionally, type II, and to a lesser extent type I, IFNs upregulate PD-L1 expression on cancer cells15,16, macrophages17, and other TME cell types18 and similarly correlate with benefit from PD-1 checkpoint blockade22,23. Of note, these and several other TME cell lineages can also express PD-15,24,33–35,37,43,44,46,47, bind clinical PD-1 inhibitors44,46,47, and their TME prevalence has even been linked to ICB patient response24,34,44. Efficacy of PD-1 therapy might thus not only rely on targeting of T-cell-PD-1, but presumably also on antagonism of these additional PD-1-expressing cell types, the lineage-specific regulation of which is poorly understood. Consequently, dissection of the molecular mechanisms controlling PD-1 expression, not only in T-cells, but also in these other immune cell subsets and cancer cells could greatly enhance understanding of PD-1 ICB response or lack thereof and help optimize therapy.
Our studies demonstrate that PD-1 gene and protein expression in melanoma cells is robustly induced by type I IFN signaling through IFNAR, JAK1, TYK2, STAT1, STAT2, and IRF9 pathway effectors (Fig. 6). Melanoma cell-intrinsic PD-1 was also induced by type II IFN, albeit to a lesser extent and more inconsistently than type I IFNs across human and murine melanoma cell lines. In contrast, several other established regulators of T-cell-PD-19,12,13,26, including multiple ILs and growth factors, did not modulate melanoma-PD-1 expression. This is consistent with the more pronounced levels of IFN receptors and their STAT signaling mediators in comparison to other candidate PD-1-regulatory cytokine receptors in patient melanomas, established human and murine melanoma lines, relative to T-cells. These results highlight STAT1 and STAT2 as tumor-intrinsic targets of IFN-α and IFN-β activation, while IFN-γ only triggers STAT1 phosphorylation. Efficient stimulation of melanoma-PD-1 expression might thus require downstream signaling through both STATs. Indeed, stable knockdown of either STAT1 or STAT2 in melanoma cells independently repressed type I IFN-induced PD-1. In contrast, robust cancer cell-PD-L1 induction primarily relies on STAT1 only via type II IFN activation, as previously reported15,16, and consistent with our results. Accordingly, respective ratios of type I versus type II IFNs in the TME could differentially modulate the expression of distinct PD-1 pathway members.
Fig. 6. Significance of the melanoma cell-intrinsic type I interferon signaling axis in PD-1 immune checkpoint therapy.
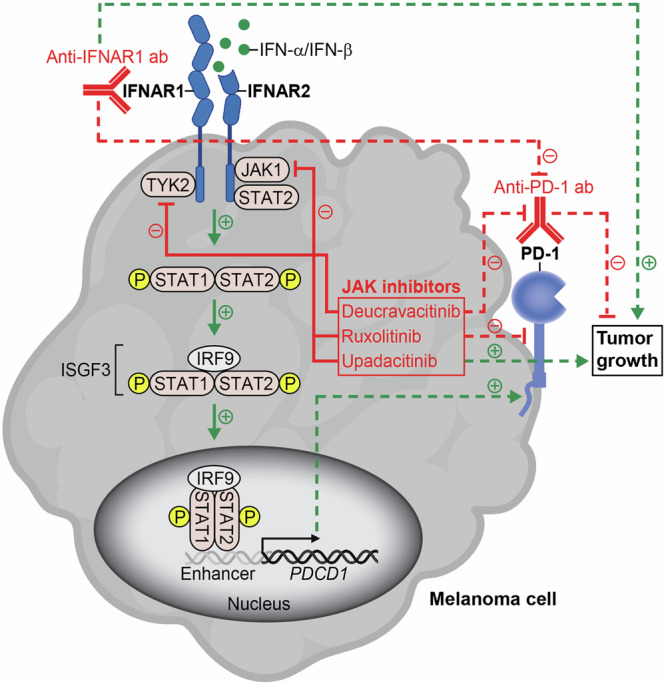
The melanoma cell-expressed IFNAR receptor binds type I interferons, IFN-α or IFN-β, thereby triggering downstream phosphorylation and activation of JAK/STAT pathway mediators (green arrows, solid lines). The protein complex containing phosphorylated (p)-STAT1, p-STAT2, and IRF9, commonly termed IFN-stimulated gene factor 3 (ISGF3), translocates to the nucleus and binds enhancer elements (gray) upstream of the PDCD1 promoter exposed upon type I interferon-induced chromatin remodeling. Consequently, both PD-1 gene (black arrow) and protein expression (green arrow, dashed line) are increased. Inhibition of the melanoma cell-IFNAR signaling axis with either blocking anti-IFNAR1 ab or pharmacological antagonists of JAK pathway effectors, TYK2 or JAK1 (red, solid lines), reduces PD-1 target receptor expression induced by type I interferons (red, dashed line) and thus unintentionally disrupts the therapeutic efficacy of anti-PD-1 immune checkpoint blockade (red, dashed lines) by promoting tumor growth (green arrows, dashed lines). Figure 6, created with BioRender.com, was released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License.
Unbiased ATAC-seq analysis revealed chromatin opening at an enhancer cCRE adjacent to the PD-1 promoter in melanoma cells in a type I IFN-inducible manner. This enhancer region contains annotated binding sites for canonical type I IFN signaling mediators, including STAT1 and STAT2 as well as IRF925. ChIP-qPCR assays confirmed direct binding of these type I IFN-associated ISGF3 complex members to this PD-1 gene enhancer element in IFN-α- and IFN-β-stimulated but not vehicle control-treated melanoma cells. Constitutive exposure of additional PD-1 proximal gene enhancer regions was even detected without IFN treatment, in agreement with basal JAK/STAT phosphorylation and PD-1 expression by untreated melanoma cells, as reported herein and previously37–40,42. IFN-α and IFN-β both recognize the same receptor, IFNAR25, and promoted similar chromatin accessibility patterns. However, IFN-β tended to more robustly augment melanoma-PD-1 expression, chromatin opening at the PD-1 promoter-proximal enhancer, and binding of p-STAT1 and IRF9 compared to IFN-α. These findings are consistent with higher binding affinity to IFNAR by IFN-β versus IFN-α variants25,55. Type I IFN treatment also stimulated chromatin opening at a PD-L1 gene enhancer region in melanoma cells, in line with the respective PD-L1 gene (CD274) and protein induction observed herein. In contrast, type II IFNs are known to trigger chromatin opening primarily at the CD274 promoter15,16 instead of enhancer regions, and resultant promoter binding of the IRF1 TF in cancer cells. Hence, regulation of tumor cell-intrinsic PD-1 pathway members is fine-tuned by composition of IFN-α, IFN-β, and IFN-γ in the TME, IFNAR and IFNGR expression, cell type-dependent activation of downstream effector molecules, and chromatin accessibility within specific PDCD1 or CD274 gene cCREs.
We leveraged several distinct inhibitory strategies targeting multiple IFNAR signaling effectors to rigorously validate functional dependence of melanoma cell-PD-1 regulation on type I IFNs, both in vitro and in vivo. In vivo administration of either IFNAR1 neutralizing ab or ruxolitinib fully eliminated inhibition of melanoma growth from PD-1 checkpoint blockade and also resulted in dramatically augmented tumor volumes independent of PD-1 ICB in T-cell-competent mice. Notably, both T-cell frequency and activation were markedly reduced in melanoma grafts in response to IFNAR1 or JAK inhibition compared to controls, thereby supporting an IFN-neutralizing effect on tumor-specific T-cell immunity, including in PD-1 ab-treated hosts. However, a complete reversal of PD-1 ICB efficacy also occurred in T-cell null mice grafted with either human or murine melanoma cells, thus also illuminating tumor cell-intrinsic, type I IFN-dependent regulation of PD-1 target expression and its relationship to PD-1 therapeutic response. In support, IFNAR1 blockade more robustly perturbed tumoral PD-1 and p-STAT1 levels in immunocompromised mice, wherein PD-1 expression is more relegated to tumor cells, but not in immunocompetent mice, in which PD-1 expression predominates on T-cells. Our results reveal that intact IFN signaling is required for efficient PD-1 ICB response, consistent with a prior report demonstrating that inactivating JAK mutations are enriched among melanoma patients exhibiting primary resistance to anti-PD-1 immunotherapy20. Loss-of-function alterations in JAK1 or JAK2 also arise through chronic T-cell pressure in response to prolonged PD-1 checkpoint blockade to cause acquired resistance21. Such disruptions of JAK/STAT signaling impaired tumor cell expression of PD-L1, as also found herein, as well as major histocompatibility complex (MHC)-I, thus leading to inefficiency of PD-1 therapy in patients20,21. In murine melanoma models, JAK antagonism by ruxolitinib attenuated tumoral PD-L1 levels, in agreement with our results, and partially reversed anti-PD-L1 ab-induced tumor growth suppression56. Our findings of complete eradication of PD-1 therapeutic efficacy as a consequence of IFNAR1 or JAK/STAT inhibition add an extra layer of complexity and mechanistic understanding to PD-1 ICB escape phenomena, by revealing PD-1 target downregulation at the level of the cancer cell (Fig. 6). Moreover, they raise concerns about widely used clinical JAK and IFNAR1 inhibitors in scenarios where patients might have undiagnosed cancers with active IFN signaling or are receiving PD-1 inhibitors for already diagnosed malignancies.
Our results further rationalize the possibility for improving PD-1 therapeutic efficacy by accentuating tumor cell-directed PD-1 receptor targeting via stimulation of the type I IFN pathway. In support, clinical trials of regimens involving PD-1 ICB in combination with IFN-α cytokine or IFN pathway activation, e.g. through stimulator of IFN genes (STING) or Toll-like receptor (TLR) agonists, have shown early success57–59. Such agonistic strategies might have multiple desired effects, including direct activation of cytotoxic T-cell (CTL) antitumor functions60 and, in light of our findings, induction of tumoral PD-1 target expression. Indeed, tumors with high autocrine expression of type I IFNs and ISGs52 are not only enriched in T-cells (immunologically hot)53 but also in melanoma cell-intrinsic PD-1 frequency. Type I IFN agonism additionally reverses repression caused by myeloid-PD-1:tumor cell-PD-L1 engagement found previously to impair CTL recruitment to the TME24. A potential caveat of sustained type I IFN stimulation is that it may facilitate acquired resistance to PD-1 blockade by enhancing intratumoral accumulation of immunosuppressive regulatory T-cells and myeloid lineages19. Together, these insights underscore the complexity of the IFN-PD-1 axis26,61 and thus highlight therapeutic approaches aimed at accentuating IFN signaling in a cell type-specific and temporally controlled fashion, including in cancer cells. This study represents a critical first step in this regard, by unraveling tumor cell-intrinsic IFN effects on PD-1 checkpoint regulation and function that might be of significance for improving ICB therapy.
Methods
Melanoma cell lines and clinical specimens, T-cell isolation, and culture methods
Authenticated, mycoplasma-free human A2058 (CRL-3601), A375 (CRL-1619), G361 (CRL-1424), MeWo (HTB-65), MDA-MB-435S (HTB-129), and SK-MEL-2 (HTB-68), and murine B16-F10 (CRL-6475), YUMM1.7 (CRL-3362), YUMMER1.7D4 (SCC243), YUMM1.G1 (CRL-3363), YUMM4.1 (CRL-3366), and YUMM5.2 (CRL-3367) melanoma cell lines were obtained from the American Type Culture Collection (ATCC, Gaithersburg, MD) or MilliporeSigma (St. Louis, MO). YUMM1.7 nuclear EGFP-expressing lines were generated as described62 and cultured as above. No commonly misidentified cell lines were used in the study. All cell lines were used at low passage, <70% confluency, and were cultured in RPMI-1640 medium (Life Technologies, Carlsbad, CA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, MilliporeSigma) and 1% (v/v) penicillin/streptomycin (Life Technologies) as described33,34,57. Single-cell suspensions were generated from patient tumor biospecimens by collagenase digestion, as described36,37.
Human peripheral blood mononuclear cells (PBMCs) were isolated from whole blood samples obtained from healthy donors by Ficoll-Paque PLUS (Cytiva, Marlborough, MA) density gradient centrifugation as described37,62. Human T-cells were activated with ImmunoCult Human CD3/CD28 T Cell Activator reagent (25 µL/mL, STEMCELL Technologies, Cambridge, MA) for 2–3 days in Advanced RPMI 1640 Medium (Life Technologies) supplemented with recombinant human IL-2 (20 ng/ml, BioLegend, San Diego, CA), 1% (v/v) GlutaMAX, 10 mM HEPES, 1% (v/v) penicillin/streptomycin (all from Life Technologies), and 10% (v/v) heat-inactivated FBS (MilliporeSigma) in 24-well plates (1–3 × 106 cells/ml) in tissue culture incubators at 37 °C and 5% CO2 as described37,62. Informed consent was obtained from all patients or volunteers, and all studies were conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (IRB) of Mass General Brigham, under protocol numbers 2022P002062, 2022P000827, and 2013P001014. Because only anonymous, randomized and de-identified human PBMC or tumor biospecimens were used without access to individual-level data, it is expected that such samples would be derived from female and male subjects across a range of ages.
Murine splenocytes were isolated by mechanical disruption of C57BL/6 mouse spleens as described38. Red blood cells were hypotonically lysed with the Ammonium-Chloride-Potassium (ACK) Lysing Buffer (Life Technologies) according to the manufacturer’s instructions. Splenocytes were resuspended at 1 × 106 cells/mL in Advanced RPMI 1640 Medium (Life Technologies) supplemented with 2 μg/mL soluble anti-mouse CD28 ab (clone 37.51, BD Biosciences, Franklin Lakes, NJ), 30 U/mL recombinant mouse IL-2 (BioLegend), 1% (v/v) GlutaMAX, 10 mM HEPES, 1% (v/v) penicillin/streptomycin (all from Life Technologies), and 10% (v/v) heat-inactivated FBS (MilliporeSigma), and then cultured in 24-well plates (Corning, Glendale, AZ) pre-coated with 10 μg/mL anti-mouse CD3 ab (clone 145-2C11, BD Biosciences) for five days as described38. All mice were at least 6 weeks of age and handled in accordance with the National Institutes of Animal Healthcare Guidelines under the Brigham and Women’s Hospital (BWH) Institutional Animal Care and Use Committee (IACUC)-approved experimental protocol 2016N000112.
Antibodies, recombinant cytokines, and biologic reagents
The following antibodies (abs) were used for flow cytometric analyses of human cells: Alexa Fluor 647-conjugated (BD Biosciences) or PerCP-eFluor 710-conjugated anti-human PD-1 (clone MIH4, 20 µg/mL, Thermo Fisher Scientific, Waltham, MA) and Alexa Fluor 647-conjugated or PerCP-eFluor 710-conjugated mouse IgG1 isotype controls (clone MOPC-31C, 20 µg/mL, BD Biosciences; clone P3.6.2.8.1, 20 µg/mL, Thermo Fisher Scientific), unconjugated anti-human PD-1 clinical ab, nivolumab (100 µg/mL, Bristol Myers Squibb, Cambridge, MA) was obtained from the BWH Pharmacy, and Ultra-LEAF unconjugated human IgG4 isotype control (clone QA16A15, 100 µg/mL, BioLegend), FITC-conjugated or PE-conjugated anti-human IgG4 (clone HP6023, 1:50, Abcam, Waltham, MA), PE-conjugated anti-human PD-L1 (clone 29E.2A3, 15 µg/mL, BioLegend) and PE-conjugated mouse IgG2b isotype control (clone MPC-11, 15 µg/mL, BioLegend), APC-conjugated anti-human PD-L2 (clone 24 F.10C12, 15 µg/mL, BioLegend) and APC-conjugated mouse IgG2a isotype control (clone MOPC-173, 15 µg/mL, BioLegend), PE-conjugated anti-human IFNAR1 (clone 85228, 20 µg/mL, Thermo Fisher Scientific) and PE-conjugated mouse IgG1 isotype control (clone P3.6.2.8.1, 20 µg/mL, Thermo Fisher Scientific) or PE-conjugated mouse IgG1 isotype control (clone MOPC-21, 20 µg/mL, BioLegend), Alexa Fluor 750-conjugated anti-human IFNAR1 (clone 85228, 20 µg/mL, R&D Systems, Minneapolis, MN) and Alexa Fluor 750-conjugated mouse IgG1 isotype control (clone 11711, 20 µg/mL, R&D Systems), APC-conjugated or APC-Vio770-conjugated anti-human IFNAR2 (clone REA124, 20 µg/mL, Miltenyi Biotec, Gaithersburg, MD) and APC-conjugated or APC-Vio770-conjugated human IgG1 isotype control (clone REA293, 20 µg/mL, Miltenyi Biotec), FITC-conjugated anti-human IFN-β (clone A1(IFNb)), 5 µg/mL, and FITC-conjugated mouse IgG1 (clone MOPC-21, 5 µg/mL, Thermo Fisher Scientific), PE-conjugated anti-human IFN-α[2b] (clone 7N4-1, 1.5 µg/mL) and PE-conjugated mouse IgG1 isotype control (clone MOPC-21, 1.5 µg/mL, BD Biosciences), FITC-conjugated anti-mouse Interferon alpha (clone RMMA-1, 50 µg/mL, PBL Assay Science, Piscataway, NJ) and FITC-conjugated rat IgG1 (clone eBRG1, 50 µg/mL, Thermo Fisher Scientific), anti-mouse Interferon-beta 1 (clone D2J1D, 75 ng/mL), rabbit IgG isotype control (clone DA1E, 75 ng/mL Cell Signaling Technology, Danvers, MA), and PE-conjugated donkey anti-rabbit IgG (clone Poly4064, 1.25 µg/mL BioLegend). The following abs were used to assess expression of JAK-STAT pathway proteins by intracellular flow cytometry: PE-conjugated anti-phospho (p)-STAT1 (Tyr701, clone 58D6, 1 µg/mL), PE-conjugated or Alexa Fluor 488-conjugated anti-p-STAT2 (Tyr690, clone D3P2P, 5 µg/mL), and PE-conjugated or Alexa-Fluor 488-conjugated rabbit IgG isotype control (clone DA1E, 1 and 5 µg/mL, respectively) (all from Cell Signaling Technology), APC-conjugated anti-p-JAK1 (Tyr1034 and Tyr1035, clone Jak1Y10221023-F11, 5 µg/mL, Abcam) and APC-conjugated rabbit IgG (clone EPR25A, 5 µg/mL, Abcam).
The following abs and/or reagents were used for flow cytometric analyses of murine cells: PE/Cyanine 7-conjugated or BV785-conjugated anti-mouse CD3 (clone 17A2, 10 µg/mL, BioLegend) and PE/Cyanine 7-conjugated or BV785-conjugated rat IgG2b isotype control (clone RTK4530, 10 µg/mL, BioLegend), Pacific Blue-conjugated or BV650-conjugated anti-mouse CD8a (clone 53-6.7, 10 µg/mL, BioLegend) and Pacific Blue-conjugated or BV650-conjugated rat IgG2a isotype control (clone RTK2758, 10 µg/mL, BioLegend), BV711-conjugated anti-mouse NK-1.1 (clone PK136, 10 µg/mL, BioLegend) and BV711-conjugated mouse IgG2a isotype control (clone MOPC-173, 10 µg/mL, BioLegend), BV570-conjugated anti-mouse CD11c (clone N418, 10 µg/mL, BioLegend) and BV570-conjugated armenian hamster IgG isotype control (clone HTK888, 10 µg/mL, BioLegend), BV605-conjugated anti-mouse CD45 (clone 30-F11, 10 µg/mL, BioLegend) and BV605-conjugated rat IgG2b isotype control (clone RTK4530, 10 µg/mL, BioLegend), Pacific Blue-conjugated anti-mouse CD31 (clone 390, 10 µg/mL, BioLegend) and Pacific Blue-conjugated rat IgG2a isotype control (clone RTK2758, 10 µg/mL, BioLegend), PE/Dazzle 594-conjugated anti-mouse CD140a (clone APA5, 10 µg/mL, BioLegend) and PE/Dazzle 594-conjugated rat IgG2a isotype control (clone RTK2758, 10 µg/mL, BioLegend), PE/Dazzle 594-conjugated or APC-eFluor 780-conjugated anti-mouse CD4 (clone GK1.5, 10 µg/mL, Thermo Fisher Scientific) and PE/Dazzle 594-conjugated or APC-eFluor 780-conjugated rat IgG2b isotype control (clone eB149/10H5, 10 µg/mL, Thermo Fisher Scientific), APC-eFluor 780-conjugated MHC Class I (H-2Kb, clone AF6-88.5.5.3, 5 µg/mL, Thermo Fisher Scientific), APC-eFluor 780-conjugated MHC Class I (H-2Kd, clone SF1-1.1.1, 5 µg/mL, Thermo Fisher Scientific) and APC-eFluor 780-conjugated mouse IgG2a isotype control (clone eBM2a, 5 µg/mL, Thermo Fisher Scientific), Alexa Fluor 700-conjugated anti-mouse CD19 (clone 1D3/CD19, 25 µg/mL, BioLegend) and Alexa Fluor 700-conjugated rat IgG2a isotype control (clone RTK2758, 25 µg/mL, BioLegend), RB705-conjugated rat anti-mouse CD11b (clone M1/70, 1.25 µg/mL, BD Biosciences) and RB705-conjugated rat IgG2b isotype control (clone R55-38, 1.25 µg/mL, BD Biosciences), PE/Cyanine 7-conjugated anti-mouse F4/80 (clone BM8, 10 µg/mL, BioLegend) and PE/Cyanine 7-conjugated rat IgG2a (clone RTK2758, 10 µg/mL, BioLegend), FITC-conjugated, APC-conjugated, PE-conjugated, PerCP/Cyanine 5.5-conjugated or Brilliant Violet 421-conjugated anti-mouse PD-1 (clone 29 F.1A12, 20 µg/mL, BioLegend) and FITC-conjugated, APC-conjugated, PE-conjugated, PerCP/Cyanine 5.5-conjugated or Brilliant Violet 421-conjugated rat IgG2a isotype control (clone RTK2758, 20 µg/mL, BioLegend), PE-conjugated anti-mouse PD-L1 (clone 10 F.9G2, 20 µg/mL, BioLegend) and PE-conjugated rat IgG2b isotype control (clone RTK4530, 20 µg/mL, BioLegend), PE-conjugated anti-mouse IFNAR1 (clone MAR1-5A3, 20 µg/mL, BioLegend) and PE-conjugated mouse IgG1 isotype control (clone MOPC-21, 20 µg/mL, BioLegend), PE-conjugated or APC-conjugated anti-mouse IFNAR2 (clone Q9D1R7, 20 µg/mL, R&D Systems) and PE-conjugated or APC-conjugated goat IgG isotype control (20 µg/mL, R&D Systems). Flow cytometric analyses additionally included the following reagents: Human TruStain FcX (Fc Receptor Blocking Solution, 1:20), TruStain FcX PLUS Antibody (anti-mouse CD16/32, clone S17011E, 2.5 µg/mL), Zombie NIR Fixable Viability Kit, Zombie Aqua Fixable Viability Kit, and Zombie Green Fixable Viability Kit (all from BioLegend, 1:1000).
The following abs were used for Western blotting: anti-phospho STAT1 (Tyr701, clone 58D6, 1:1000), anti-STAT1 (clone D1K9Y, 1:1000), anti-STAT1 (clone D4Y6Z, 1:1000), anti-phospho STAT2 (Tyr690, clone D3P2P, 1:1000), anti-STAT2 (clone D9J7L, 1:1000), anti-phospho JAK1 (Tyr1034/Tyr1035, clone D7N4Z, 1:1000), anti-JAK1 (clone 6G4, 1:1000), anti-phospho Tyk2 (Tyr1054/Tyr1055, clone D7T8A, 1:1000), anti-Tyk2 (clone D4I5T, 1:1000) (all from Cell Signaling Technology) anti-β-actin (clone C4, 1:10000, BD Biosciences), and horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse (Cell Signaling Technology). The following abs from Cell Signaling Technology were used for chromatin immunoprecipitation and quantitative polymerase chain reaction (ChIP-qPCR): anti-p-STAT1 (Tyr701, clone 58D6, 0.59 µg), STAT2 (clone D9J7L, 0.77 µg) and anti-IRF9 (clone D9I5H, 1.33 µg).
The following abs and/or pharmacologic reagents were used for in vitro and/or in vivo inhibition experiments (concentrations shown under ‘Recombinant cytokine treatment’ and/or ‘In vivo tumorigenicity studies’, respectively): InVivoMAb anti-human PD-1 (clone J116, Bio X Cell, Lebanon, NH) and InVivoMAb mouse IgG1 isotype control (clone MOPC-21, Bio X Cell), anti-human IFNAR1 clinical ab, anifrolumab (MedChemExpress, Monmouth Junction, NJ), and Ultra-LEAF Purified human IgG1 isotype control (clone QA16A12, BioLegend), Ultra-LEAF Purified anti-mouse PD-1 (clone 29 F.1A12, BioLegend) and Ultra-LEAF Purified rat IgG2a isotype control (clone RTK2758, BioLegend), Ultra-LEAF Purified anti-mouse IFNAR1 (clone MAR1-5A3, BioLegend) and Ultra-LEAF Purified mouse IgG1 isotype control (clone MOPC-21, BioLegend). Pharmacologic inhibitors of JAK1/2, ruxolitinib (INCB18424), and JAK1, upadacitinib (ABT-494), were from MedChemExpress, and the TYK2 inhibitor, deucravacitinib (BMS-986165), was from Selleckchem (Houston, TX).
The following recombinant human proteins were used to treat human melanoma cells in vitro: human type I interferons (IFN), IFN-α2 (IFN-α) and IFN-β (both at 60 ng/mL and from BioLegend or R&D Systems, Minneapolis, MN), human type II IFN, IFN-γ (5 ng/mL, R&D Systems), human interleukin (IL)-2 (BioLegend), human IL-6 (10 ng/mL, BioLegend), human IL-7 (10 ng/mL, BioLegend), human IL-10 (10 ng/mL, BioLegend), human IL-12 (10 ng/mL, BioLegend or Peprotech, Cranbury, NJ), human IL-15 (10 ng/mL, BioLegend), human IL-18 (10 ng/mL, BioLegend or R&D Systems), human IL-21 (10 ng/mL, BioLegend), human IL-27 (10 ng/mL, BioLegend), human transforming growth factor (TGF)-β1 (TGF-β, 10 ng/mL, BioLegend), human tumor necrosis factor (TNF)-α (10 ng/mL, BioLegend), and human vascular endothelial growth factor-165 (VEGFA, VEGF, 30 ng/mL, BioLegend). The following recombinant murine proteins were used to treat mouse melanoma cells in vitro: mouse IFN-α (60 ng/mL, BioLegend) or mouse IFN-α2 (IFN-α, 60 ng/mL, Thermo Fisher Scientific), mouse IFN-β1 (IFN-β, 60 ng/mL, BioLegend), and mouse IFN-γ (5 ng/mL, BioLegend).
RT-qPCR
Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen, Beverly, MA), according to the manufacturer’s protocol. RNA was subsequently converted to cDNA using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). Samples were assayed using the Fast SYBR Green Master Mix or TaqMan Fast Advanced Master Mix (both Thermo Fisher Scientific) with gene-specific primer sets recognizing human (Supplementary Tables 1 and 2) or mouse (Supplementary Tables 3 and 4) transcripts or TaqMan specific primers for Cd3 or Actb (Thermo Fisher Scientific), on a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). Thermal cycling was carried out as described35,36,57 at annealing temperatures shown in Supplementary Tables 1–4, followed by subsequent melt-curve validation. Data was normalized to human 18 s rRNA or human or murine β-actin. Relative transcript levels were calculated using the delta-delta-Ct method as described37,38,62.
Flow cytometry
Surface protein expression by established melanoma lines, cell suspensions derived from patient tumor biospecimens, or melanoma tumor xenografts was determined by multi-color flow cytometry as described37,38,62. Single-cell suspensions were generated from patient tumor biospecimens and melanoma tumor xenografts using collagenase digestion, as described36,37. Nonviable cells were excluded using the Zombie NIR Fixable Viability Kit (1:1000), Zombie Aqua Fixable Viability Kit (1:1000) or Zombie Green Fixable Viability Kit (1:1000), as per the manufacturer’s recommendations by staining for 10 min at room temperature (RT) in the dark. Cells were then blocked with Human TruStain FcX (Fc Receptor Blocking Solution, BioLegend) for 10 min at RT or TruStain FcX PLUS (anti-mouse CD16/32) Antibody (BioLegend) for 10 min at 4 °C. Cells were subsequently stained with fluorochrome-conjugated abs (20 μg/ml) or unconjugated abs, including nivolumab (100 μg/ml), in PBS + 2% (v/v) FBS, for 30 min at 4 °C, and then washed. Nivolumab binding was detected with FITC-conjugated or PE-conjugated anti-human IgG4 as described37,38,62. For analyses of p-STAT, p-JAK1 and p-Tyk2 proteins by intracellular flow cytometry, cell suspensions or human melanoma cells or PBMCs either not treated or stimulated with recombinant IFNs as described below were fixed in 4% (v/v) paraformaldehyde for 15 min at 37 °C, permeabilized with True-Phos Perm Buffer (BioLegend) and stained at RT with PE-conjugated anti-p-STAT abs as described above at the following concentrations: anti-p-STAT1 (1 μg/ml), anti-p-STAT2 (5 μg/ml), anti-p-JAK1 (5 μg/ml) and respective isotype control (1 or 5 μg/ml). For intracellular flow cytometric analyses of IFN production, cells incubated with Fc receptor blocking solution were fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences) and stained with anti-murine or anti-human IFNα and IFNβ abs for 30 min at 4 °C in the dark. In all flow cytometric experiments and analyses, isotype-matched control abs were employed and cell doublets excluded. Fluorescence emissions were acquired on an Aurora Spectral Analyzer (Cytek, Fremont, CA) and data analyzed with FlowJo software v10.10 (TreeStar, Ashland, OR).
Recombinant cytokine treatment
Melanoma lines were plated in RPMI-1640 medium containing 10% (v/v) heat-inactivated FBS and 1% (v/v) penicillin/streptomycin (complete media) in either 6-well plates, 60 mm cell culture dishes, or cell culture flasks at 37 °C, 5% CO2. For analyses of cytokine-dependent induction of either PD-1 gene expression by RT-qPCR, chromatin accessibility by ATAC-seq, or DNA-binding factors by ChIP-qPCR, cells were serum-starved for 6-12 hrs in RPMI-1640 medium containing 1% (v/v) penicillin/streptomycin and devoid of FBS and L-glutamine prior to addition of recombinant cytokines. For experiments of PD-1 surface protein expression, cells were first cultured in complete media for 3 days until reaching the desired confluency of approximately 70% and then serum-starved overnight as above. Serum-starved cells were treated with recombinant human or murine cytokines for 3, 6 or 12 hrs for gene expression studies by RT-qPCR, 3 hrs for ATAC-seq or ChIP-qPCR analyses, or 72 hrs or 14 days for protein expression assays, with respective cytokines freshly added each day. Recombinant cytokine concentrations were: IFN-α (60 ng/ml), IFN-β (60 ng/ml), and IFN-γ (5 ng/ml), with all other cytokines or growth factors used at 10-60 ng/ml. In experiments involving inhibitors, cells were pretreated prior to cytokine stimulation for 2 hrs with clinical anti-human IFNAR1 blocking ab (anifrolumab, 10 µg/ml), anti-mouse IFNAR1 blocking ab (clone MAR1-5A3, 10 µg/ml), or respective isotype-matched control abs (10 µg/ml), deucravacitinib (1 µM), ruxolitinib (1 µM), or upadacitinib (1 µM). For analyses of p-STAT protein levels by intracellular flow cytometry, serum-starved human melanoma cells at 70% confluency, or human PBMCs were treated for 15 min at 37 oC with IFN-α, IFN-β, or IFN-γ as above.
Generation of stable STAT1 and STAT2 knockdown melanoma cells
STAT1 and STAT2 knockdown (KD) A2058 melanoma variant cells were generated using lentiviral-based plasmids encoding short hairpin RNAs (shRNA) targeting human STAT1 or STAT2 (MilliporeSigma). A plasmid encoding a non-targeting scrambled shRNA (Addgene) was used as a control. The above shRNAs were packaged into lentiviral particles by HEK293 EBNA packaging cells co-transfected with the viral packaging plasmids pN8e-GagPolΔ8.1 and pN8e-VSV-G, viral supernatants were collected 48–72 hrs after transfection, and then filtered as described2. The target 21mers were (5’-3’): CTGGAAGATTTACAAGATGAA (shRNA-1) and CCCTGAAGTATCTGTATCCAA (shRNA-2) for human STAT1, and TGTCTTCTGCTTCCGATATAA (shRNA-1) and TAGGACTGAGGATCCATTATT (shRNA-2) for human STAT2. Human A2058 melanoma cells were infected with the respective lentiviral supernatants above and transductants selected in 1 μg/ml puromycin (Thermo Fisher Scientific) as described62. Knockdown of STATs was confirmed by RT-qPCR and immunoblotting.
Immunoblotting
Cells were lysed in ice-cold RIPA buffer supplemented with Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology) and vortexed for 30 min at 4 °C. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific), according to the manufacturer’s protocol. Lysates were resolved by SDS-PAGE gel electrophoresis and transferred to PVDF membranes (Bio-Rad) as described. Membranes were blocked in tris-buffered saline (TBS)/0.1% (v/v) Tween-20 (TBS-T, MilliporeSigma), containing 5% (w/v) bovine serum albumin (BSA, MilliporeSigma or Cell Signaling Technology) for at least 1 hr at RT. Membranes were incubated overnight at 4 °C with anti-phospho (p)-STAT1 (clone 58D6), anti-STAT1 (clone D4Y6Z), anti-STAT1 (clone D1K9Y), anti-p-STAT2 (clone D3P2P), anti-STAT2 (clone D9J7L), anti-p-JAK1 (clone D7N4Z), anti-JAK1 (clone 6G4), anti-p-TYK2 (clone D7T8A), anti-TYK2 (clone D4I5T), horseradish peroxidase (HRP)-conjugated anti-β-actin (clone D6A8) (all from Cell Signaling Technology, 1:1000), or anti-β-actin clone C4 (1:10,000, BD Biosciences) in TBS-T containing 5% (w/v) BSA or non-fat dry milk. Membranes were washed thrice in TBS-T and incubated with horseradish peroxidase (HRP)-conjugated secondary ab for 1 hr at RT. Antigens were visualized using the Lumi-Light Western Blotting Substrate (MilliporeSigma) on HyBlot CL Autoradiography Films (Thomas Scientific, Swedesboro, NJ) via a Kodak Min-R mammography processor (Kodak, Rochester, NY) or a ChemiDoc Imaging System (Bio-Rad).
ATAC-seq
Serum-starved murine YUMM4.1 melanoma cells were treated with or without recombinant mouse IFN-α (60 ng/ml) or IFN-β (60 ng/ml) for 3 hrs as described above. Cells were harvested, washed twice in ice-cold PBS, centrifuged at 700 g for 5 min at 4 °C, and cell suspensions (~1 ×106, > 90% viability) cryopreserved according to GENEWIZ ATAC-seq sample preparation guidelines (Azenta Life Sciences, South Plainfield, NJ). ATAC-seq library preparation and DNA sequencing were conducted at Azenta Life Sciences as described63. In brief, cell samples were thawed, washed, and genomic DNA contamination was removed by DNase I (Thermo Fisher Scientific) treatment. Cell number and viability were quantified on a Countess Automated Cell Counter (Thermo Fisher Scientific). Cells were lysed, cytosolic fractions removed, nuclei treated with Tn5 transposase (Illumina, Hayward, CA) for 30 min at 37 °C, and tagmented DNA was purified using the MinElute PCR Purification Kit (Qiagen). Subsequently, tagmented DNA was barcoded using the Nextera Index Kit v2 (Illumina), PCR-amplified, and cleaned using solid-phase reversible immobilization (SPRI) beads to generate purified DNA libraries. Sequencing libraries were clustered on a flow cell, loaded onto an Illumina HiSeq instrument (model 4000 or equivalent) according to the manufacturer’s instructions and samples sequenced using a 2×150 bp Paired End (PE) configuration. Image analysis and base calling were conducted using Control Software (CS). Raw sequence bcl data files were converted to fastq files and de-multiplexed using bcl2fastq v2.17 conversion software (Illumina). One mismatch was allowed for index sequence identification.
Data was processed using the ATAC-seq pipeline implemented in bcbio-nextgen v1.2.9-8eb78b7. ATAC-seq data was evaluated for quality using FASTQC. Reads were filtered and trimmed with Atropos. High-quality reads were mapped to the Mus musculus genome (build mm10) using Bowtie2. After filtering reads from mitochondrial DNA, properly paired reads with high mapping quality (MAPQ score >10, non-duplicates, qualified reads) were retained using Sambamba for further analysis. The alignmentSieve function of Deeptools and the sort and index functions of Samtools were used to isolate fragments in nucleosome free regions (NFRs). Reads were shifted by 9 bp (+4 in positive and -5 in negative strand) to account for the dimeric binding of the Tn5 transposase that results in insertion of two adaptors separated by 9 bp. To call the peaks with unique reads, we used MACS2. ATAC-seq data quality was assessed using ataqv. CPM-normalized bigwig files (bin size=20) were visualized using IGV. Qualitative assessment of the data indicated that sample untreated 1 had fewer peaks called (15,939) compared to all other samples (range 91,542-103,254), higher proportions of mono- and di-nucleosomal fragments, and lower TSS enrichment. However, assessment of the peak overlap between the replicates untreated 1 and untreated 2 with ChIPpeakAnno v3.32.0 revealed that 69% of peaks called in untreated 1 overlapped with peaks called in untreated 2. Replicate data sets were thus merged using the bam files with Samtools v.1.14 to create pseudoreplicates using the (https://gist.github.com/brianhill11/7aeeeb6d94edfb868e5595aac04a0dd6) custom script. Peaks with unique reads were called using MACS2. Diffbind v3.4.11 was implemented in R 4.1.2 to generate a normalized count matrix for all samples. These count matrices were used in principal component analyses for all samples using the degPCA function from the DEGReport R package v.1.34.0. Differential accessibility was also assessed with Diffbind using DESeq2. Peaks were considered differentially enriched at FDR < 0.05. The initial count matrix was subset to only include differentially accessible regions and then a random subsample (3000 sites) was used to generate a heatmap with the pheatmap R package 1.0.12. Variability in read density of the Pdcd1 and Cd274 genes was examined and counts generated for 100 bp bins using the multiBamSummary function from deepTools. Counts were manually normalized by library size and smoothed conditional means plotted using the geom_smooth function from the R package ggplot2. Identical genomic regions were plotted using the gviz package implemented in R. Predicted STAT1, STAT2, and IRF9 binding sites were identified from the JASPAR database.
ChIP-qPCR
Chromatin immunoprecipitation and quantitative polymerase chain reaction (ChIP-qPCR) was performed as described64. Melanoma YUMM4.1 or YUMM1.7 cells (1 × 106) were seeded in 10-cm dishes (Corning) in normal growth media and either untreated or treated the following day with 60 or 180 ng/ml IFN-α or IFN-β (BioLegend) in media without serum for 3 hrs in a tissue culture incubator at 37 oC, 5% CO2. Samples were crosslinked with 1% (v/v) formaldehyde (MilliporeSigma) for 20 min at room temperature and quenched with 125 mM glycine (MilliporeSigma) for 5 min. Cells were washed twice in ice-cold PBS, pulse-centrifuged, and cell pellets resuspended in RIPA lysis buffer (200 mM Tris-HCl, pH 7.5, 130 mM NaCl, 10% (v/v) glycerol, 0.5% (w/v) sodium deoxycholate (DOC), 0.1% (w/v) SDS, 1% (v/v) NP-40) containing Complete ULTRA Protease Inhibitor tablets (Roche, Basel, Switzerland) and rotated for 1 hr at 4 °C. Chromatin was sonicated using the BioRuptor Sonicator (Diagenode, Denville, NJ) under the following settings: power = high, on interval = 30 s, off interval = 30 s, 7 cycles, 5 times. Cells were lysed in RIPA buffer for an additional 15 min at 4 °C. Chromatin was purified from cellular debris by centrifugation at 10,000 x g for 10 min at 4 °C. Supernatant fractions (10% of total) were removed and stored at −20 °C for subsequent use as input controls. Remaining supernatant fractions (90% of total) were incubated with pre-washed Dynabeads Protein A (Thermo Fisher Scientific) under constant rotation for 45 min at 4 °C. Samples were pulse-centrifuged, and abs (10 μl/sample, Cell Signaling Technology) recognizing p-STAT1 (clone 58D6, 0.59 μg), STAT2 (clone D9J7L, 0.77 μg), IRF9 (clone D9I5H, 1.33 μg), or Normal Rabbit IgG control (2729, input amount matched accordingly) were added to supernatants along with Dynabeads Protein A (30 μl). Samples were rotated overnight at 4 °C and then chromatin-ab complexes washed in the following order: first, ChIP digestion buffer (50 mM Tris pH 8.0, 25 mM EDTA, 1.25% SDS), second, TSE-150 (20 mM Tris pH 8.1, 150 mM NaCl, 2 mM EDTA, 0.10% SDS, 1% Triton X-100), third, TSE-500 (20 mM Tris pH 8.1, 500 mM NaCl, 2 mM EDTA, 0.20% SDS, 1% Triton X-100), fourth TSE-150, and fifth, ChIP digestion buffer. Samples were eluted in buffer consisting of 1% (w/v) SDS and 0.1 M NaHCO3 under continuous shaking for 15 min at room temperature, with the elution step repeated once. Samples were treated with 10 μg RNAse A (Thermo Fisher Scientific) and crosslinking reversed by overnight incubation on a desktop shaker set to 600 rpm in 5 mM NaCl at 65 °C. Samples were treated on a desktop shaker set to 800 rpm with 20 or 100 μg Proteinase K (New England Biolabs, Ipswich, MA) for 2.5 hrs at 55 °C. After centrifuging at 10,000 x g for 10 min at 4 °C, supernatants were transferred to fresh tubes, and the DNA purified using the ChIP DNA Clean & Concentrator kit (Zymo Research, Irvine, CA). Relative DNA levels were quantified by qPCR using the QuantStudio5 Real-Time PCR System via the following cycling program: 95 °C for 2 min, 40 cycles of (95 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s), and 72 °C for 2 min. Threshold cycle values were normalized to input control sample values. The following primer set was used for qPCR amplification of a 146 base pair fragment located 1014 bases upstream of the Pdcd1 gene (5’-3’, Fig. 3d): forward primer, TGGCTAGTCATTTCTGGGGC, and reverse primer, CGGGCTGCCTATTTTAGGGT.
In vivo tumorigenicity studies
Wildtype C57BL/6 and immunodeficient nonobese diabetic/severe combined immunodeficiency (NOD/SCID) IL-2Rγ(-/-) KO (NSG) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained at the BWH animal facility. Housing conditions for mice consisted of a dark/light cycle of light from 7 am to 7 pm, dark from 7 pm to 7 am, an ambient temperature range of 20–24 °C (68–75 °F), and a humidity level of 35-65%. All mice were female or male, at least 6 weeks of age, age-matched between experimental groups, and treated in accordance with the National Institutes of Animal Healthcare Guidelines under the BWH IACUC-approved experimental protocol 2016N000112. Human A2058 or nuclear EGFP-expressing62 murine YUMM1.7 melanoma lines were injected subcutaneously into the flanks of recipient mice at 5 × 105 cells/inoculum for A2058 cells in NSG mice, 2 × 105 cells/inoculum for YUMM1.7 in NSG mice, or 1 × 106 cells/inoculum for YUMM1.7 in wildtype C57BL/6 mice as described37,62. For PD-1 ab targeting studies, mice were injected intraperitoneally with 200 μg blocking InVivoMAb anti-human PD-1 (clone J116, Bio X Cell) or blocking Ultra-LEAF anti-mouse PD-1 (clone 29F.1A12, BioLegend) versus respective isotype-matched control ab as described above every three days starting on the day of tumor cell inoculation for the duration of the experiment. For IFNAR1 ab targeting studies, animals were injected intraperitoneally with 500 μg blocking anti-human IFNAR1 clinical ab (anifrolumab, MedChemExpress) or blocking Ultra-LEAF anti-mouse IFNAR1 ab (clone MAR1-5A3, BioLegend) versus respective isotype-matched control ab as described above on the day before, day of, and day after tumor cell inoculation, with abs (200 μg) subsequently administered every three days for the duration of the experiment. For JAK targeting studies, mice were fed a submaximal single daily dose (SD, 180 mg/kg/d, p.o.65) of the JAK inhibitor, ruxolitinib (MedChemExpress), or vehicle control incorporated into PicoLab Rodent Diet 20 5053 (Research Diets, New Brunswick, NJ) for the duration of the experiment. The SD was calculated per the following formula: DD = (SD x BW)/FI, where diet dose (DD) = 1285.714 mg compound/kg diet, body weight (BW) = 25 g BW/animal, and daily food intake (FI) = 3.5 g diet/day. Tumor formation/growth was measured once per week by determination of tumor volume (TV) according to the established formula [TV (mm3) = π/6 × 0.5 × length × (width)2] until either the experimental endpoint or excessive tumor burden or disease state required protocol-stipulated euthanasia as described37,62. Maximal tumor size/burden permitted by the BWH IACUC is 2 centimeters (cm) at the widest point. These limits were not exceeded in any experimental instance herein. Tumor ulceration or moribund condition also require euthanasia. These additional IACUC criteria were also met for all tumorigenicity experiments.
Statistics and reproducibility
Statistical differences between two experimental groups were determined using the paired or unpaired Student’s t-test or Mann-Whitney test, or in the case of three or more experimental groups, one-way ANOVA with Dunnett, Bonferroni, or Tukey post hoc test, two-way ANOVA with Bonferroni or Fisher’s Least Significance Difference (LSD) post hoc test, or Friedman test with Dunn’s post hoc test. In a few instances comparing control versus two experimental groups, a t-test was used to compare controls to either experimental condition when data was collected over different time periods. A one-sided or two-sided value of p < 0.05 was considered statistically significant. Pearson correlation coefficients were calculated to assess correlative trends between genes. All statistical analyses were performed using PRISM 10.2.3 software (GraphPad, San Diego, CA). Group and sample sizes were adjusted accordingly based on standard deviations and statistical assessments and/or prior studies conducted by our laboratories that yielded sufficient and reproducible power to detect statistically significant differences. Investigators were not blinded to group allocation during data collection or analysis because both experimental setups and analyses were generally performed by the same person(s).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by a Research Grant from the Dermatology Foundation, Milstein Research Scholar Award from the American Skin Association, Fund to Sustain Research Excellence from the Brigham Research Institute, Brigham and Women’s Hospital, NIH/NCI grants R01CA190838 (to T.S.), R01CA247957 and R01CA258637 (to S.R.B. and T.S.). Partial support was provided by a Developmental Project Grant from the Harvard Stem Cell Institute (to M.R.R. and T.S.), Klaus-Wolff Fellowships from the Austrian Society of Dermatology and Venereology (ÖGDV, to J.H.), Marietta Blau-Grant from Austria’s Agency for Education and Internationalisation (OeAD, to J.H.), Research Fellowship from the European Academy of Dermatology and Venereology (EADV, to A.D.), and a Walter Benjamin Scholarship from the German Research Foundation (DFG, to E.R.).
Author contributions
J.H., C.M, Z.K., A.D., S.R.B., and T.S. conceptualized the study. J.H., C.M., Z.K., A.D., E.R., L.M., P.S., E.S., M.R.R., S.R.B., and T.S. carried out experimental work. J.H., C.M., Z.K., A.D., E.R., L.M., P.S., E.S., J.L., K.B., J.S., W.H., E.L.B., S.H.S., M.R.R., S.R.B., and T.S. analyzed data. J.H., C.M., S.R.B., and T.S. wrote the paper. All authors discussed the results and commented on the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Murine ATAC-seq raw data have been deposited into the Gene Expression Omnibus (GEO) database under accession code GSE253179. The scRNA-seq publicly available data used in this study are also available in the GEO database under accession codes GSE72056 and GSE185386. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided as a Source Data file, which is also publicly available within the Harvard Dataverse under the following link: 10.7910/DVN/JRJUMR.
Code availability
Code used for this manuscript has been made available in the Github repository under the following link: https://github.com/hbc/HBC04813_Schatton_mouse_ATACseq/tree/main. Methodological details on parameters used are available in the Methods section of this manuscript. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Julia Holzgruber, Christina Martins, Zsofi Kulcsar.
These authors jointly supervised this work: Steven R. Barthel, Tobias Schatton.
Contributor Information
Steven R. Barthel, Email: sbarthel@bwh.harvard.edu
Tobias Schatton, Email: tschatton@bwh.harvard.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51496-2.
References
- 1.Sharma, P. et al. The next decade of immune checkpoint therapy. Cancer Discov.11, 838–857 (2021). 10.1158/2159-8290.CD-20-1680 [DOI] [PubMed] [Google Scholar]
- 2.Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature515, 568–571 (2014). 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juneja, V. R. et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J. Exp. Med.214, 895–904 (2017). 10.1084/jem.20160801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill, A. L. et al. PD-1 blockade increases the self-renewal of stem-like CD8 T cells to compensate for their accelerated differentiation into effectors. Sci. Immunol.8, eadg0539 (2023). 10.1126/sciimmunol.adg0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patsoukis, N., Wang, Q., Strauss, L. & Boussiotis, V. A. Revisiting the PD-1 pathway. Sci. Adv.6, eabd2712 (2020). 10.1126/sciadv.abd2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharpe, A. H. & Pauken, K. E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol.18, 153–167 (2018). 10.1038/nri.2017.108 [DOI] [PubMed] [Google Scholar]
- 7.Schett, G., Elewaut, D., McInnes, I. B., Dayer, J.-M. & Neurath, M. F. How Cytokine Networks Fuel Inflammation: Toward a cytokine-based disease taxonomy. Nat. Med.19, 822–824 (2013). 10.1038/nm.3260 [DOI] [PubMed] [Google Scholar]
- 8.Boussiotis, V. A. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N. Engl. J. Med.375, 1767–1778 (2016). 10.1056/NEJMra1514296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinter, A. L. et al. The common gamma-chain cytokines IL-2, IL-7, IL-15, and IL-21 induce the expression of programmed death-1 and its ligands. J. Immunol.181, 6738–6746 (2008). 10.4049/jimmunol.181.10.6738 [DOI] [PubMed] [Google Scholar]
- 10.Hanna, B. S. et al. Interleukin-10 receptor signaling promotes the maintenance of a PD-1(int) TCF-1(+) CD8(+) T cell population that sustains anti-tumor immunity. Immunity54, 2825–2841.e2810 (2021). 10.1016/j.immuni.2021.11.004 [DOI] [PubMed] [Google Scholar]
- 11.Francisco, L. M., Sage, P. T. & Sharpe, A. H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev.236, 219–242 (2010). 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, B. V. et al. TGFβ1-mediated SMAD3 enhances PD-1 Expression on antigen-specific T cells in cancer. Cancer Discov.6, 1366–1381 (2016). 10.1158/2159-8290.CD-15-1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voron, T. et al. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J. Exp. Med.212, 139–148 (2015). 10.1084/jem.20140559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terawaki, S. et al. IFN-α directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J. Immunol.186, 2772–2779 (2011). 10.4049/jimmunol.1003208 [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Diaz, A. et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 Expression. Cell Rep.19, 1189–1201 (2017). 10.1016/j.celrep.2017.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, S. J. et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett.580, 755–762 (2006). 10.1016/j.febslet.2005.12.093 [DOI] [PubMed] [Google Scholar]
- 17.Loke, P. & Allison, J. P. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc. Natl Acad. Sci.100, 5336–5341 (2003). 10.1073/pnas.0931259100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng, Q. et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat. Commun.11, 4835 (2020). 10.1038/s41467-020-18570-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquelot, N. et al. Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res.29, 846–861 (2019). 10.1038/s41422-019-0224-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin, D. S. et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov.7, 188–201 (2017). 10.1158/2159-8290.CD-16-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaretsky, J. M. et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med.375, 819–829 (2016). 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayers, M. et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest.127, 2930–2940 (2017). 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grasso, C. S. et al. Conserved Interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell38, 500–515.e503 (2020). 10.1016/j.ccell.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klement, J. D. et al. Tumor PD-L1 engages myeloid PD-1 to suppress type I interferon to impair cytotoxic T lymphocyte recruitment. Cancer Cell41, 620–636.e629 (2023). 10.1016/j.ccell.2023.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platanias, L. C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol.5, 375–386 (2005). 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- 26.Saleiro, D. & Platanias, L. C. Interferon signaling in cancer. Non-canonical pathways and control of intracellular immune checkpoints. Semin. Immunol.43, 101299 (2019). 10.1016/j.smim.2019.101299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stebbing, J. & Lauschke, V. M. JAK inhibitors - more than just Glucocorticoids. N. Engl. J. Med.385, 463–465 (2021). 10.1056/NEJMe2108667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morand, E. F. et al. Trial of Anifrolumab in active systemic Lupus Erythematosus. N. Engl. J. Med.382, 211–221 (2020). 10.1056/NEJMoa1912196 [DOI] [PubMed] [Google Scholar]
- 29.Guttman-Yassky, E. et al. Once-daily upadacitinib versus placebo in adolescents and adults with moderate-to-severe atopic dermatitis (Measure Up 1 and Measure Up 2): results from two replicate double-blind, randomised controlled phase 3 trials. Lancet397, 2151–2168 (2021). 10.1016/S0140-6736(21)00588-2 [DOI] [PubMed] [Google Scholar]
- 30.Papp, K. et al. Phase 2 trial of selective Tyrosine Kinase 2 inhibition in Psoriasis. N. Engl. J. Med.379, 1313–1321 (2018). 10.1056/NEJMoa1806382 [DOI] [PubMed] [Google Scholar]
- 31.Rosmarin, D. et al. Two Phase 3, randomized, controlled trials of Ruxolitinib cream for Vitiligo. N. Engl. J. Med.387, 1445–1455 (2022). 10.1056/NEJMoa2118828 [DOI] [PubMed] [Google Scholar]
- 32.Vaengebjerg, S., Skov, L., Egeberg, A. & Loft, N. D. Prevalence, incidence, and risk of cancer in patients with Psoriasis and Psoriatic Arthritis: A systematic review and meta-analysis. JAMA Dermatol.156, 421–429 (2020). 10.1001/jamadermatol.2020.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon, S. R. et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature545, 495–499 (2017). 10.1038/nature22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helmink, B. A. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature577, 549–555 (2020). 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu, J. et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Invest.128, 4654–4668 (2018). 10.1172/JCI99317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martins, C. et al. Tumor cell-intrinsic PD-1 promotes Merkel cell carcinoma growth by activating downstream mTOR-mitochondrial ROS signaling. Sci. Adv.10, eadi2012 (2024). 10.1126/sciadv.adi2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleffel, S. et al. Melanoma cell-intrinsic PD-1 Receptor functions promote tumor growth. Cell162, 1242–1256 (2015). 10.1016/j.cell.2015.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martins, C. et al. Distinct antibody clones detect PD-1 checkpoint expression and block PD-L1 interactions on live murine melanoma cells. Sci. Rep.12, 12491 (2022). 10.1038/s41598-022-16776-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark, C. A. et al. Tumor-Intrinsic PD-L1 Signals Regulate Cell Growth, Pathogenesis, and Autophagy in Ovarian Cancer and Melanoma. Cancer Res.76, 6964–6974 (2016). 10.1158/0008-5472.CAN-16-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao, Z. et al. An unexpected role for p53 in regulating cancer cell-intrinsic PD-1 by acetylation. Sci. Adv.7, eabf4148 (2021). 10.1126/sciadv.abf4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanlorenzo, M. et al. BRAF and MEK inhibitors increase PD-1-positive Melanoma cells leading to a potential lymphocyte-independent synergism with anti-PD-1 antibody. Clin. Cancer Res24, 3377–3385 (2018). 10.1158/1078-0432.CCR-17-1914 [DOI] [PubMed] [Google Scholar]
- 42.Yang, S. et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun.10, 2782 (2019). 10.1038/s41467-019-10669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, H. et al. Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatol66, 1920–1933 (2017). 10.1002/hep.29360 [DOI] [PubMed] [Google Scholar]
- 44.Du, S. et al. Blockade of Tumor-Expressed PD-1 promotes lung cancer growth. Oncoimmunology7, e1408747 (2018). 10.1080/2162402X.2017.1408747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X. et al. Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc. Natl Acad. Sci.117, 6640–6650 (2020). 10.1073/pnas.1921445117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ierano, C. et al. In PD-1+ human colon cancer cells NIVOLUMAB promotes survival and could protect tumor cells from conventional therapies. J. Immunother. Cancer10, e004032 (2022). 10.1136/jitc-2021-004032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D’Alterio, C. et al. Targeting CXCR4 potentiates anti-PD-1 efficacy modifying the tumor microenvironment and inhibiting neoplastic PD-1. J. Exp. Clin. Cancer Res.38, 432 (2019). 10.1186/s13046-019-1420-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao, M. et al. Direct therapeutic targeting of immune checkpoint PD-1 in pancreatic cancer. Br. J. Cancer120, 88–96 (2019). 10.1038/s41416-018-0298-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirzaei, R. et al. PD-1 independent of PD-L1 ligation promotes glioblastoma growth through the NFκB pathway. Sci. Adv.7, eabh2148 (2021). 10.1126/sciadv.abh2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science352, 189–196 (2016). 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biermann, J. et al. Dissecting the treatment-naive ecosystem of human melanoma brain metastasis. Cell185, 2591–2608.e2530 (2022). 10.1016/j.cell.2022.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu, H. et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med25, 95–102 (2019). 10.1038/s41591-018-0302-5 [DOI] [PubMed] [Google Scholar]
- 53.Wang, H. et al. Tumor immunological phenotype signature-based high-throughput screening for the discovery of combination immunotherapy compounds. Sci. Adv.7, eabd7851 (2021). 10.1126/sciadv.abd7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, N. et al. Supplementation with alpha-ketoglutarate improved the efficacy of anti-PD1 melanoma treatment through epigenetic modulation of PD-L1. Cell Death Dis.14, 170 (2023). 10.1038/s41419-023-05692-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zemek, R. M. et al. Temporally restricted activation of IFNbeta signaling underlies response to immune checkpoint therapy in mice. Nat. Commun.13, 4895 (2022). 10.1038/s41467-022-32567-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo, N. et al. Melanoma response to anti-PD-L1 immunotherapy requires JAK1 signaling, but not JAK2. Oncoimmunology7, e1438106 (2018). 10.1080/2162402X.2018.1438106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davar, D. et al. Phase Ib/II Study of Pembrolizumab and Pegylated-Interferon Alfa-2b in Advanced Melanoma. J. Clin. Oncol.36, JCO1800632 (2018). 10.1200/JCO.18.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu, J. et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med.7, 283ra252 (2015). 10.1126/scitranslmed.aaa4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Razaghi, A., Durand-Dubief, M., Brusselaers, N. & Bjornstedt, M. Combining PD-1/PD-L1 blockade with type I interferon in cancer therapy. Front. Immunol.14, 1249330 (2023). 10.3389/fimmu.2023.1249330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu, C. et al. Type I interferon suppresses tumor growth through activating the STAT3-granzyme B pathway in tumor-infiltrating cytotoxic T lymphocytes. J. Immunother. Cancer7, 157 (2019). 10.1186/s40425-019-0635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boukhaled, G. M., Harding, S. & Brooks, D. G. Opposing roles of Type I Interferons in cancer immunity. Annu. Rev. Pathol.16, 167–198 (2021). 10.1146/annurev-pathol-031920-093932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schatton, T. et al. Inhibition of melanoma cell-intrinsic Tim-3 stimulates MAPK-dependent Tumorigenesis. Cancer Res.82, 3774–3784 (2022). 10.1158/0008-5472.CAN-22-0970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol.109, 21 29 21–21 29 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramsey, M. R., He, L., Forster, N., Ory, B. & Ellisen, L. W. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res.71, 4373–4379 (2011). 10.1158/0008-5472.CAN-11-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen, H. et al. Selective suppression of melanoma lacking IFN-gamma pathway by JAK inhibition depends on T cells and host TNF signaling. Nat. Commun.13, 5013 (2022). 10.1038/s41467-022-32754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Murine ATAC-seq raw data have been deposited into the Gene Expression Omnibus (GEO) database under accession code GSE253179. The scRNA-seq publicly available data used in this study are also available in the GEO database under accession codes GSE72056 and GSE185386. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided as a Source Data file, which is also publicly available within the Harvard Dataverse under the following link: 10.7910/DVN/JRJUMR.
Code used for this manuscript has been made available in the Github repository under the following link: https://github.com/hbc/HBC04813_Schatton_mouse_ATACseq/tree/main. Methodological details on parameters used are available in the Methods section of this manuscript. Source data are provided with this paper.