Abstract
Replicated genetic material must be partitioned equally between daughter cells during cell division. The precision with which this is accomplished depends critically on the proper functioning of the mitotic spindle. The assembly, orientation and attachment of the spindle to the kinetochores are therefore constantly monitored by a surveillance mechanism termed the SCP (spindle checkpoint). In the event of malfunction, the SCP not only prevents chromosome segregation, but also inhibits subsequent mitotic events, such as cyclin destruction (mitotic exit) and cytokinesis. This concerted action helps to maintain temporal co-ordination among mitotic events. It appears that the SCP is primarily activated by either a lack of occupancy or the absence of tension at kinetochores. Once triggered, the inhibitory circuit bifurcates, where one branch restrains the sister chromatid separation by inhibiting the E3 ligase APCCdc20 (anaphase-promoting complex activated by Cdc20) and the other impinges on the MEN (mitotic exit network). A large body of investigations has now led to the identification of the control elements, their targets and the functional coupling among them. Here we review the emerging regulatory network and discuss the remaining gaps in our understanding of this effective mechanochemical control system.
Keywords: cell cycle, checkpoint, kinetochore, mitotic exit, spindle
Abbreviations: APC/C, anaphase-promoting complex/cyclosome; Bub, budding uninhibited by benzimidazole; CBF3, centromere-binding factor 3; Cdc, cell division cycle; CDK, cyclin-dependent kinase; CENP-E, centromere protein-E; DAM1, Duo1- and monopolar-spindle-1-interacting; FEAR, Cdc fourteen early anaphase release; GAP, GTPase-activating protein; GEF, GTPase-exchange factor; Hct1, homologue of Cdc twenty; INCENP, inner centromere protein; Mad, mitotic arrest defective; MAP, microtubule-associated protein; MEN, mitotic exit network; Mps1, monopolar spindle 1; MT, microtubule; NPC, nuclear pore complex; PAK, p21Cdc42/Rac-activated kinase; PLK, Polo-like kinase; RENT, regulator of nucleolar silencing and telophase; SPB, spindle pole body; Scc, sister chromatid cohesion; SCF, Skp1–Cullin–F box E3 ligase complex; SCP, spindle checkpoint; Smc, structural maintenance of chromosome protein
INTRODUCTION
Almost a century ago, it was recognized that the segregation of chromosomes from mother to daughter cells constituted the basis of heredity and indeed, the very continuity of life [1,2]. The stunning precision that characterizes the partitioning of genomes between daughter cells reflects the remarkable fidelity of the regulatory mechanisms that govern this complex error-prone process. During cell division, transitions from one stage of the cell cycle to another are regulated by surveillance mechanisms called checkpoints that make one process dependent on another biochemically unrelated process or event. To ensure faithful transmission of chromosomes to progeny cells, cells are endowed with the SCP (spindle checkpoint) mechanism which is activated when cells encounter errors in spindle assembly or in the attachment of spindle MTs (microtubules) to chromosomes. This evolutionarily conserved checkpoint serves as an effective instrument for self diagnosis. Should problems arise in the attachment of chromosomes to the mitotic spindle, the SCP transduces an inhibitory signal to the cell cycle machinery and impinges on two major cell cycle transitions to ensure that (i) the onset of anaphase only occurs once all chromosomes have formed stable bipolar attachments to MTs, and (ii) mitotic exit is delayed until sister chromatids have segregated properly. In doing so, the SCP couples progression through and exit from mitosis to correct spindle assembly and function. The SCP is a mechanochemical system that comprises a highly sensitive error-detection mechanism, an intracellular signal transduction pathway and a target (effector) that work together to forestall the cell cycle and provide time for error correction.
The need to evolve such a proofreading mechanism is best appreciated by understanding why these two transitions are so crucial to cell division and survival. By initiating anaphase, a cell commits to the simultaneous and irreversible physical separation of all duplicated sister chromatids. Failure to execute this step perfectly can irremediably alter ploidy and has catastrophic consequences for the cell [3]. For instance, mutants in the SCP signalling pathway fail to arrest cell cycle progression when cells are treated with low doses of MT-disrupting agents, resulting in massive chromosome mis-segregation and eventually, cell death. When chromosome segregation is endangered, it also becomes imperative to block a much later event in mitosis, namely mitotic exit. Failure to do so would cause untimely inactivation of the mitotic kinase via B-type cyclin destruction, and re-initiation of a new cell cycle. Genome instability and aneuploidy caused by the malfunctioning of the SCP can lead to tumorigenesis [4]. In recent years, many excellent reviews describing how the SCP inhibits chromosome segregation have been published [5–7]. We will limit the scope of this review to mainly how the SCP, by inhibiting the onset of anaphase, simultaneously prevents mitotic exit. We will also highlight the gaps in our understanding of the intricate network of interactions that underlies this regulation. This review draws heavily from the budding yeast (Saccharomyces cerevisiae) system, since the study of this organism has contributed immensely to our understanding of the SCP mechanism. Where appropriate, parallels in other systems will also be highlighted. First, we will introduce the mitotic events that surround the SCP mechanism.
Sister chromatid cohesion and bi-orientation
For proper segregation of chromosomes, it is important that the structure of the spindle and its attachments to chromosomes are flawless. Following DNA replication, the duplicated sister chromatids establish amphitelic (bipolar) attachments (see Figure 1) to the spindle. Integral to this process is the detection and correction of inappropriate (syntelic or monopolar) attachments. Initially, sister chromatids are held together both at centromeres and at discrete sites along their arms [8,9] by multi-subunit cohesin complexes [subunits: Smc (structural maintenance of chromosome protein) 1, Smc3, Scc (sister chromatid cohesion) 1 and Scc3]. In mammalian cells, most of the cohesin is removed from chromosome arms in prophase via cohesin phosphorylation by the PLK (polo-like kinase). However, cohesin complexes persist at the centromeric region until the onset of anaphase. Cohesion between sister chromatids is essential for attainment of proper amphitelic attachment during metaphase. Amphitelic attachment, in turn, generates tension on sister kinetochores as the poleward force exerted on chromosomes by MTs is counteracted by cohesion between sister chromatids. The resulting tension is necessary for stabilizing the kinetochore–MT attachment. In the absence of proper bipolar attachment or sufficient tension across sister kinetochores, the SCP is triggered to inhibit the onset of anaphase.
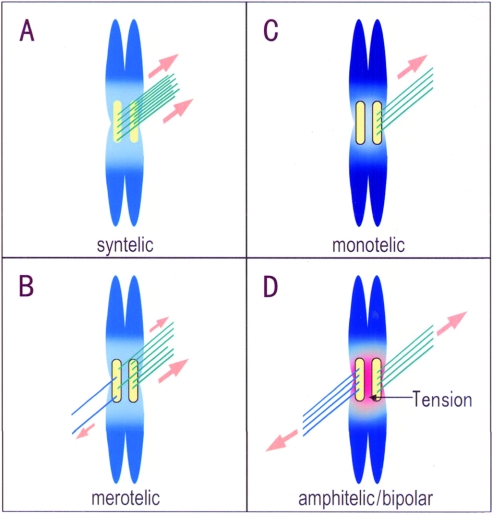
Figure 1. Attachment of spindle MTs to kinetochores.
The process by which chromosomes establish connections with MTs emanating from the two spindle poles is a highly stochastic one. Initially, different unstable modes of MT attachment may occur, for instance, syntelic attachment (A) or merotelic attachment (B). In such cases, action of Aurora B/Ipl1p disengages some of the MTs to promote monotelic attachment (C). Following this step, stable attachment is achieved when the sister kinetochore captures MTs originating from the spindle pole at the opposite end (amphitelic/bipolar attachment) (D). In such an arrangement, poleward forces generate tension between sister kinetochores.
A closer look at the chromosome–MT connection
Kinetochores are large protein complexes that assemble on centromeric DNA. Centromeres vary greatly in size and primary sequence: the budding yeast centromere is 125 bp long and is conserved among different chromosomes [10]. In contrast, metazoan centromeric DNA can be megabases in length and lacks easily recognizable consensus sequences [11]. Interestingly, centromeres of different species assemble similar multi-protein kinetochore complexes that mediate and monitor the connection between chromosomes and spindle MTs.
The kinetochore exhibits a layered organization of prohibitive complexity even in the simple eukaryote budding yeast (see Figure 2). About half the kinetochore components are encoded by essential genes, and many have close mammalian homologues [12], suggesting that important aspects of kinetochore architecture and function are likely to have been conserved. In the budding yeast, each kinetochore binds a single spindle MT. In vertebrates, a bundle of approx. 20 microtubules (a kinetochore fibre) attaches to the kinetochore [13]. The exact number varies between species, the stage of mitosis and the length of time for which the kinetochore has been attached to the pole.
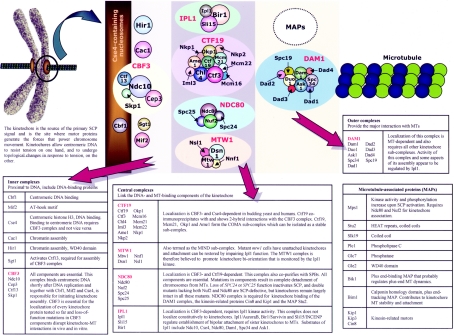
Figure 2. Schematic diagram of the organization of the budding yeast kinetochore.
In budding yeast, a spindle MT attaches to a single site on each chromatid (the centromere) where a complex multi-protein structure, termed the kinetochore, is formed. For purpose of simplicity, all kinetochore proteins are represented here as spheres and their relative sizes are only approximations of their true molecular masses. Interactions depicted between various components within each complex are purely schematic representations of the composition of each complex. The composition of the various complexes and some important details about them are summarized in brief in the accompanying Tables. Slk19, described herein as a MAP, is a kinetochore passenger protein and not much is known about its interaction with other kinetochore complexes. The budding yeast kinetochore is estimated to be at least 5 MDa, twice the size of a ribosome [141]. However, unlike ribosomes which self-assemble into a single complex, the kinetochore is composed of distinct subcomplexes. With the recent isolation of stable kinetochore subcomplexes, the COMA/C1 (Ctf19–Mcm21–Okp1–Ame1) subcomplex, the MIND/M1 (Dsn1–Nsl1–Nnf1–Mtw1) subcomplex and the Ndc80 complex, in association with the Cse4-containing nucleosome and the CBF3 complex, are proposed to form a linker platform for the further assembly of the yeast kinetochore [141,142]. The kinetochore is actually a dynamic structure whose composition and organization change during cell cycle progression: for instance, Ipl1, Sli15, Bir1, Ndc10 and Slk19 relocate from the kinetochores to the spindle mid-zone once anaphase is initiated. The kinesins Kip1 and Cin8 are degraded at the end of metaphase and during mitotic exit respectively [143,144]. Moreover, the assembly of the COMA subcomplex is cell-cycle-regulated [142].
In yeast, the kinetochore is composed of protein assemblies that can be broadly classified as inner, central or outer kinetochore complexes. The DAM1 [Duo1- and Mps1 (monopolar spindle 1)-interacting] complex, an outer kinetochore complex (see Figure 2), plays a crucial role in mediating the kinetochore–MT connection, and is regulated through phosphorylation by the Ipl1/Aurora B kinase. It interacts physically with proteins of the CBF3 (centromere-binding factor 3), CTF19 and Ndc80 kinetochore complexes and also binds MTs in vitro, although it is not clear if these interactions are constitutive or limited to specific stages of the cell cycle [14–16]. Like Dam1, Ipl1 and Sli15/INCENP (inner centromere protein) each bind MTs in vitro. Because of their association with both kinetochores and MTs, the Ipl1 and DAM1 complexes are believed to regulate various aspects of chromosome–spindle interaction [17]. The Ipl1 complex responds to the lack of tension in monotelic attachments, and acts to resolve these inappropriate attachments, probably through its substrates [15,18,19]. It has been shown that the interaction between Ndc80 and DAM1 proteins is essential for the association of the DAM1 complex with kinetochores, and that the DAM1–Ndc80 binding affinity is weakened by Ipl1-mediated phosphorylation [20]. In the absence of tension, Ipl1 causes an increased turnover of kinetochore–MT connections, perhaps by influencing the Ndc80–DAM1 interaction, thereby facilitating eventual bi-orientation of chromosomes [16,19,20]. In addition, Ipl1 may also regulate the assembly of the DAM1 complex itself [19]. Recent data suggest that the MTW1 complex promotes kinetochore bi-orientation, which is monitored by the Ipl1 kinase [21].
Experiments in budding yeast have demonstrated that the tension resulting from the physical connection between bi-oriented kinetochores, and the activity of Ipl1 (rather than any specific chromosomal architecture or kinetochore geometry) is sufficient for the proper alignment of sister chromatids [22]. In higher eukaryotes, where multiple MTs bind each kinetochore, Aurora B directs the co-ordinated disassembly of all the MTs that are bound to kinetochores in a syntelic manner [23]. However, it remains unclear how Aurora B/Ipl1 distinguishes deleterious syntelic and monotelic attachments from amphitelic ones, and selectively destabilizes the former. In mammalian cells, dynein and kinesin motors {e.g. CENP-E (centromere protein-E), XKCM1 (Xenopus kinesin catastrophe modulator 1)/MCAK (mitotic centromere-associated kinesin); [24]} as well as other MAPs (MT-associated proteins) are also likely to play important roles in mediating the connection between kinetochores and MTs.
Anaphase onset and regulation of the APC/C (anaphase-promoting complex/cyclosome) function
Once chromosomes have successfully bi-oriented, cells utilize an evolutionarily conserved machinery to initiate anaphase. Rapid disjunction of sister chromatids at anaphase requires cleavage of cohesin subunit Scc1 by a cysteine protease called separase (Esp1 in budding yeast, Cut1 in Schizosaccharomyces pombe) [25,26]. Owing to the irreversible nature of Scc1 cleavage, this step is very tightly controlled. The separase is therefore kept inactive through most of the cell cycle by its association with an inhibitor, securin [26,27]. The metaphase-to-anaphase transition is triggered when securin [Pds1 in budding yeast, Cut2 in Schiz. pombe, PTTG (pituitary tumour transforming gene) in humans] is degraded by the proteasome following ubiquitination by the multi-component E3 ubiquitin ligase known as the APC/C. APC/C function is regulated by (i) phosphorylation, and (ii) association of activator proteins such as Cdc (cell-division cycle) 20 and its homologue Hct1 (homologue of Cdc twenty)/Cdh1, which modulate the affinity of APC/C for different substrates [28]. The activation of the SCP inhibits the ubiquitin-dependent proteolysis of securin by the APCCdc20 (APC/C activated by Cdc20) [29,30]. In mammals, additional mechanisms that control separase activity, and therefore anaphase onset, must exist, since cell lines lacking securin are able to restrain anaphase initiation in response to spindle poisons [31].
Cdc20 has been implicated in the regulation of APC/C-dependent proteolysis and is essential for chromosome segregation [32,33]. Cdc20 expression is maximal at G2/M phase. At restrictive temperature, the budding yeast cdc20-1 temperature-sensitive mutant arrests with diploid DNA content, an undivided nucleus, undegraded securin and a short spindle, a phenotype strikingly similar to that of the APC/C mutants cdc16, cdc23 and cdc27 [32–35]. Cdc20-related proteins have also been identified in other organisms, from flies to humans [36,37]. In Xenopus egg extracts, Cdc20 function is inhibited during prophase by the binding of the F-box protein Emi1. Emi1 is degraded in mitosis following ubiquitination by another E3 ligase, SCF (Skp1–Cullin–F box E3 ligase complex) [38], thereby setting Cdc20 free. In addition to its role in promoting the ubiquitination of securin by APC/C, budding yeast Cdc20 also plays an essential role in mitotic exit (see below).
Cdh1/Hct1 plays a major role in late mitosis in proteolysis of the mitotic cyclin Clb2, but is not essential for viability [32,39]. Cdh1 is constitutively expressed, and its activity is regulated by phosphorylation. While phosphorylation of Cdh1 by the Cdc28-Clb (mitotic) kinase prevents it from activating APC/C [40,41], the dephosphorylation of Cdh1 in late mitosis by the phosphatase Cdc14 is the critical step in the activation of APCCdh1 (APC/C activated by Cdh1) in budding yeast and mammalian cell culture systems [42–44]. The activity of Cdc14 itself is under the control of the MEN (mitotic exit network) that includes Tem1, Net1 and Cdc15 proteins (see below).
Regulation of mitotic exit
The hallmark of mitotic exit is the inactivation of the mitotic kinase via APC/C-dependent proteolysis of B-type cyclins. In budding yeast, destruction of major mitotic cyclin Clb2 occurs in a biphasic manner: APCCdc20 is responsible for the first phase of Clb proteolysis that occurs when cellular levels of the Clbs are high. The resulting decrease in mitotic kinase activity tips the balance in favour of a net dephosphorylation of Cdh1 (by Cdc14) and is the decisive step in Cdh1 activation. Activated APCCdh1 then mediates the second phase of Clb destruction and triggers mitotic exit [45]. The extent of Clb2 proteolysis driven by APCCdc20 may itself be sufficient to permit mitotic exit even in the absence of Cdh1 and the CDK (cyclin-dependent kinase) inhibitor Sic1, and the main role of Cdh1 may be to maintain a low mitotic kinase state in G1 [46].
The central event in the initiation of cyclin destruction is the release of Cdc14 from the RENT (regulator of nucleolar silencing and telophase) complex in the nucleolus where it is sequestered during most of the cell cycle (other components of the RENT complex are Cfi1/Net1 and Sir1). Once set free from the nucleolus, Cdc14 reverses CDK-dependent phosphorylation of Cdh1 [40,43]. Cdc14 release is controlled by two pathways: the FEAR (Cdc fourteen early anaphase release) network [47] and the MEN [40,48,49]. In budding yeast, separase (Esp1), the polo-like Cdc5 kinase, the kinetochore protein Slk19, Spo12 and its homologue Bns1 constitute the FEAR network and trigger partial release of Cdc14 from the nucleolus during early anaphase in a MEN-independent manner. Although the precise functional relationships between these components are not clear, some interactions among them have been reported: (i) Slk19, which localizes to kinetochores and the spindle mid-zone during anaphase, is a substrate of Esp1 [50,51], (ii) ESP1 functions upstream of SLK19, while SPO12 and BNS1 act in a parallel pathway [52], (iii) Esp1 and Slk19 may activate Cdc5 [53], and (iv) Cdc5 appears to induce MEN-independent phosphorylation of Cdc14 as well as MEN-dependent phosphorylation of Cfi1/Net1. Cdc14 release from the nucleolus occurs only when both Cdc14 and Net1 are phosphorylated [52]. In addition, the replication fork block protein Fob1, which resides within the nucleolus throughout the cell cycle, interacts physically with both Spo12 and Cfi1/Net1. It has been proposed that Fob1 prevents the dissociation of Cdc14 from Cfi1/Net1 before anaphase, and that Spo12 activation (perhaps via its phosphorylation by an unidentified kinase) during early anaphase promotes the release of Cdc14 by antagonizing Fob1 function [54]. Once released, Cdc14 localizes to SPBs (spindle pole bodies) and initially activates MEN signalling [55].
The MEN pathway, which is activated in late anaphase, is believed to promote and maintain Cdc14 in the released state [47], and also to facilitate its association with the SPB in mitosis [56]. The MEN pathway in budding yeast involves several genes, including TEM1, CDC15, CDC5, MOB1, DBF2, DBF20 and LTE1 [57], that display a wide array of genetic interactions among them/each other. Of these, Tem1 is a Ras-like GTPase, Cdc5, Cdc15, Dbf2 and Dbf20 are protein kinases [58–62], and Cdc15, Mob1, Lte1 and Dbf2 are phosphoproteins [63–65]. Temperature-sensitive mutations in the MEN pathway proteins arrest cells in telophase with high Cdc28-Clb activity. Tem1 is the most upstream element in this pathway, and Lte1 is its putative GEF (guanine nucleotide-exchange factor) [59]. Tem1 activity is downregulated by a heterodimeric GAP (GTPase-activating protein) complex composed of Bub (budding uninhibited by benzimidazole) 2 and Bfa1 [66]. GTP–Tem1 binds its target kinase Cdc15 at the SPB [67,68]; Cdc15 in turn directly phosphorylates and activates Mob1-bound Dbf2 [69]. Activated Dbf2 kinase presumably phosphorylates one or more substrates, and ultimately causes the release of more Cdc14 from the nucleolus. Cdc5 also acts downstream of Tem1 in the MEN to mediate full activation of Dbf2 [70] (see Figure 4). Cdc14, set free as a result of MEN signalling, performs several crucial tasks: first, it dephosphorylates and activates Cdh1, causing APCCdh1 to consign the Clbs for destruction, thereby triggering mitotic exit. Secondly, Cdc14 causes the accumulation of the CDK inhibitor Sic1 by up-regulation of its transcription via the dephosphorylation of transcription factor Swi5, and by stabilizing the Sic1 protein via down-regulation of its degradation. Although MEN-dependent release of Cdc14 from the nucleolus is essential for mitotic exit, the FEAR-network-dependent release of Cdc14 during early anaphase is not, though it may be required for timely exit from mitosis.
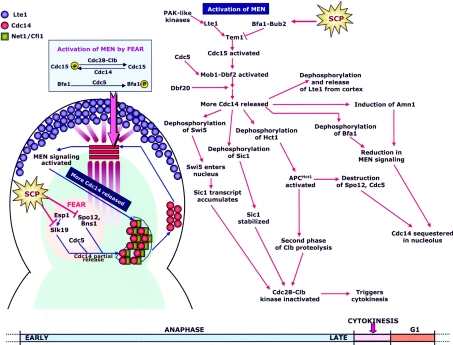
Figure 4. Pathways controlling onset of mitotic exit and their regulation by the SCP.
The proteolysis of securin at the metaphase-to-anaphase transition liberates separase, which together with the other components of the FEAR pathway, triggers the partial release of Cdc14 from the RENT complex in early anaphase. Cdc14 activates the Cdc15 kinase (component of the MEN pathway) at the SPB, while Cdc5 inactivates Bfa1 of the Tem1 GAP complex, contributing to Tem1 activation. Moreover, the translocation of the daughter SPB into the bud brings the Tem1 GTPase in contact with Lte1, its putative exchange factor, which is also believed to lead to Tem1 activation. The PAK-like kinases Cla4 and Ste20 also contribute to full Tem1 activation by promoting the localization of Lte1 to the cortex. The result is the activation of MEN signalling and the initiation of a positive-feedback loop causing further release of Cdc14 from the nucleolus. Activation of MEN signalling and the concomitant release of Cdc14 in late anaphase is essential for activation of APCHct1 (APC/C activated by Hct1) and the commencement of the second phase of Clb proteolysis (the first phase of Clb proteolysis occurs as cells enter anaphase and is triggered by APCCdc20), leading to mitotic exit, and also cytokinesis. By triggering the degradation of FEAR proteins Spo12 and Cdc5, by dephosphorylating and promoting the GAP activity of Bfa1 and by causing induction of the Amn1 protein, Cdc14 itself turns on the switch that down-regulates MEN signalling and eventually leads to the re-sequestration of Cdc14 in the nucleolus. The main targets of regulation by the SCP are highlighted in the Figure.
Tying mitotic exit to chromosome segregation
How do cells ensure that mitotic exit follows anaphase? It has been proposed that a spatial strategy is utilized by the cell to ensure this co-ordination. Tem1p is synthesized and asymmetrically localized to the SPB destined to migrate into the bud. Although Tem1 associates with the cytoplasmic face of the SPB, its GEF Lte1 is concentrated away from it at the bud cortex. Tem1 presumably becomes activated when the SPB is translocated into the bud during anaphase, which brings it in proximity to Lte1 [63]. However, the co-ordination of chromosome segregation and mitotic exit remains largely unperturbed in the absence of Lte1, suggesting that spatial segregation of Tem1 and Lte1 until anaphase is not a critical element in this co-ordination.
Another reason why the initiation of mitotic exit is contingent on anaphase onset could be that liberation of separase is necessary for the FEAR-network-dependent early release of Cdc14. Although Slk19 is a substrate of Esp1, the early release of Cdc14 does not require the cleavage of Slk19 by Esp1 [47,53]. The separase Esp1 promotes phosphorylation of Net1 by Cdc5, to facilitate Cdc14 release. This novel separase function is co-regulated with its proteolytic activity by securin. After securin degradation at the onset of anaphase, separase is set free to cleave Scc1 (thereby triggering chromosome segregation) and then to initiate mitotic exit via a non-proteolytic mechanism [53]. We will see in later sections that, in addition to the mechanisms described above, regulation by the SCP also ensures that mitotic exit is not initiated until chromosomes have partitioned successfully.
Closing the cell cycle
At the end of mitosis, daughter cells of the budding yeast activate an AMEN (antagonist of MEN) pathway in part through induction of the Amn1 protein in a Cdc14-dependent manner. Amn1 binds directly to Tem1 and prevents Tem1's association with Cdc15. Amn1 is thus part of a daughter-specific switch that helps cells to terminate the division cycle and reset the biochemical state of the cell to initiate a new cycle [71]. Cdc14 also reactivates Bfa1, promoting GTP hydrolysis on Tem1 and thus down-regulating MEN signalling. Furthermore, the FEAR proteins Spo12 and Cdc5 are targeted for destruction by APCCdh1, thereby reducing FEAR activity [72]. Termination of FEAR and MEN signalling presumably causes the re-sequestration of Cdc14 in the nucleolus (see Figure 4), although the detailed mechanism remains unknown. Early in the following cycle, Amn1 protein is phosphorylated by the Cdc28-Cln (G1) kinase, ubiquitinated by the SCF complex, and degraded. Cdh1 activity is then extinguished in S phase due to Cdh1 phosphorylation by Cdc28 in association with the early expressed Clbs [73].
THE SCP PATHWAY
SCP players and cell cycle arrest
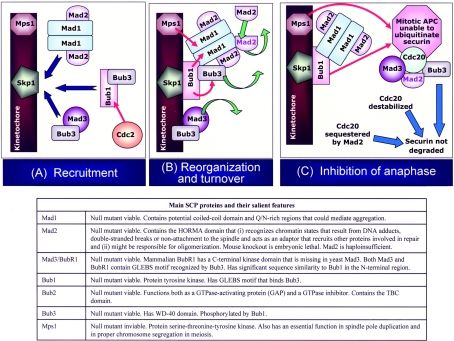
The SCP responds to errors in spindle assembly and in bipolar attachment of chromosomes. A signal transduction pathway is then set into motion to act on specific cellular targets in order to delay the onset of anaphase and mitotic exit. The major players in the SCP signal transduction pathway were identified in budding yeast through two genetic screens for mutants that failed to arrest in mitosis in the presence of spindle-damaging agents. These players included Mad (mitotic arrest defective) 1, Mad2 and Mad3 [74], Bub1, Bub2 and Bub3 [75]. Orthologues of these proteins have been identified in other organisms (see [6] for a review). The Mps1 and Ipl1 kinases also play pivotal roles in the SCP, although their precise execution points are not clear [76,77].
What does the SCP monitor: attachment or tension?
Much needs to be understood at the molecular level about the defects that activate the SCP. Depending on the cell type and the nature of the perturbation imposed on the spindle, the SCP senses, in some cases, the absence of kinetochore-MT attachment (dependent on the presence of an intact, functional kinetochore structure), while, in others, it responds to a lack of tension imposed on back-to-back sister kinetochores by poleward-acting forces.
An unoccupied kinetochore could arise in any of the following circumstances: (i) when some chromosomes have not yet bi-oriented, (ii) when there is incomplete duplication/separation of centrosomes, (iii) when there are defects in proteins that mediate the kinetochore-MT attachment, (iv) when there are defects in MT dynamics, or (v) when there are structural defects in the kinetochore. In budding yeast cdc31-2 and mps2-1 mutants, conditional failure of SPB duplication leads to a cell cycle arrest dependent on the SCP [76]. Experiments in mammalian cells [77] showed that a single kinetochore, when prevented from attaching to the spindle, inhibited the onset of anaphase. Moreover, laser ablation of that unattached kinetochore rapidly triggered anaphase, firmly establishing that the unattached kinetochore produced a (presumably) diffusible signal to halt anaphase onset. Homologues of several SCP signalling proteins in Xenopus laevis and humans are enriched at unattached kinetochores [78–80]. Moreover, if a cell has mono-oriented chromosomes and the unattached kinetochores are destroyed using a laser, the anaphase delay is abrogated, showing that the SCP does not detect mono-orientation [81]. Furthermore, given the large number of proteins whose inhibition leads to congression defects and eventually chromosome mis-segregation (i.e. no SCP activation), it is amply clear that some attachment defects are not recognized by the SCP.
Monitoring the extent of tension is the only way that a cell can verify stable bi-orientation. In favour of the argument that the SCP detects insufficient tension, it was found that, when PtK1 cells were treated with a low concentration of taxol (a drug that promotes MT assembly and stability, and which reduces tension in spindles without affecting MT attachment) after the last kinetochore has attached to the spindle, some cells entered and completed anaphase A after a cell cycle delay. Therefore in these cells (where spindle bipolarity and metaphase alignment are maintained), the SCP monitored an increase in tension between kinetochores and their attached MTs as bi-orientation was established [82]. In meiotic insect cells, increase in tension correlates with SCP resolution [83], but it is noteworthy that some spermatocyte meioses have unpaired sex chromosomes that lack tension, yet the SCP is not activated. The behaviour of budding yeast mutant cdc6 suggests that absence of tension alone is sufficient to activate SCP in this organism. cdc6 mutant cells cannot initiate S phase and completely lack replication forks. They therefore fail to turn on the DNA-replication checkpoint. The unreplicated chromatids are attached to spindle MTs, but are not under tension, since they lack a sister chromatid that could bind a MT from the opposite pole. The mutant cells enter mitosis in the absence of replicated DNA, but are delayed via a pathway that is dependent on SCP components, implying that absence of tension is sufficient to trigger the SCP response [84].
The requirements for tension and attachment are not often easy to distinguish from each other due to their interdependence. In cell cultures, tension at the kinetochores is known to boost both the stability of individual MT attachments and the level of kinetochore occupancy, although the issue of how it happens is yet to be resolved satisfactorily. This fact should serve as a caveat while interpreting the outcomes of experiments that involve artificial manipulation of tension at kinetochores.
How the SCP restrains anaphase
The basic plan of the SCP signal transduction pathway that impinges on APCCdc20 and some characteristics of its main players are outlined in Figure 3. Several kinases (including Bub1, BubR1 and Mps1) and phosphoproteins (including Mad1) comprise the backbone of this pathway. The Mps1 protein and its kinase activity are absolutely essential for SCP activation and the kinetochore association of the checkpoint proteins Mad1, Mad2 and, in mammalian cells, the kinesin CENP-E [85]. Mps1 is also required for Mad1 hyperphosphorylation upon SCP activation [86]. Notably, biochemical observations indicate that, in yeast cells, the Bub1–Bub3, Mad3–Bub3 and Mad1–Mad2 complexes are present throughout the cell cycle and are probably recruited to kinetochores as heterodimers upon SCP activation [87,88].
Figure 3. The ‘anaphase arm’ of the SCP.
(A) Recruitment: Mad1 and Mad2 form a tight complex that localizes to NPC (nuclear pore complex) [145]. Upon SCP engagement, subcomplexes of checkpoint proteins are recruited to the kinetochore. Cdc2-dependent Bub1 phosphorylation is required to activate the SCP after spindle damage [146]. The kinases Mps1 and Bub1 act in concert and (in mammalian cells, together with BubR1 and the kinesin CENP-E) trigger the rapid recruitment of the Mad1–Mad2 complex that disengages from the NPC upon SCP activation. In mammalian cells, Mad2 is phosphorylated in a cell-cycle-regulated manner in vivo, and only unphosphorylated Mad2 interacts with APC/C and Mad1 [147]. Recruitment of SCP complexes presumably occurs via their direct or indirect interaction with kinetochore components and other checkpoint proteins. The DAM1 complex has been shown to bind Mps1, Mad1, Mad3, Bub1 and Bub2 [20]. Bub1 binds Skp1 [148]. Both Spc25 and Ndc80 show two-hybrid interactions with Mad1 [148]. CENP-I is also essential for the kinetochore association of Mad1 and Mad2 [149]. Mad1 also interacts with and forms a complex with the Bub1–Bub3 heterodimer in such a way that Bub3 binds both Mad1 and Mad2 [150,151]. Bub1 can phosphorylate Bub3 [88] and Mad1 [151] in vitro. The domain of Bub1 required for binding Bub3 is the same domain that is required for localization of Bub1 to kinetochores, suggesting that, upon association with kinetochores, Bub3 dissociates from Bub1. Bub3 also forms a complex with Mad3 throughout the cell cycle and may target Mad3 to kinetochores. In addition to the SCP signalling molecules depicted in the Figure, Cdc20 and the APC/C are also recruited to kinetochores upon SCP activation. Red arrows show phosphorylation events. (B) Reorganization and turnover: red arrows show phosphorylation events and green arrows show protein turning over. SCP activation results in the hyperphosphorylation of Mad1 by Mps1 and perhaps also Bub1, and causes the formation of the Mad1–Bub1–Bub3 complex. The Mad1–Mad2 complex recruited to the kinetochore functions as a template for the rapid catalytic conversion and release of soluble Mad2 in an APCCdc20-inhibitory form, labelled in purple. Mad3/BubR1 and Bub3 are also turned over at kinetochores and released, although it is not known if they are modified in the process. The APCCdc20-inhibitory form of Mad2 is transferred to the Bub3–Cdc20–Mad3/BubR1 complex, which is a potent inhibitor of APC/C. (C) Inhibition of anaphase: the SCP deploys various strategies to inhibit anaphase onset. Targeting of substrates by APCCdc20 is inhibited independently and collaboratively by the various Mad–Bub complexes. In in vitro assays, both BubR1 and Mad2 block the binding of Cdc20 to APC/C. Mitotic APC/C (with some APC/C subunits phosphorylated by Bub1 and Mps1) is more susceptible to inhibition upon SCP activation as compared with APC/C from other stages of the cell cycle. It has also been shown that Cdk1–cyclin B-dependent inhibitory phosphorylation of Cdc20/Fizzy contributes to keeping APC/C inactive under conditions of SCP activation [152]. Furthermore, SCP activation triggers Cdc20 degradation and keeps the level of Cdc20 low until all chromosomes have bi-oriented. Restoration of attachment/tension at kinetochores results in the dynein-dependent depletion of SCP proteins by transport away from kinetochores towards the poles along spindle MTs [153]. Regulated translocation of Mad1 and Mad2 away from kinetochores is believed to make the depletion less reversible. Bub1 persists longer than Mad1 and Mad2 at kinetochores, and the consequent physical separation of components of the SCP catalytic scaffold turns off the SCP signal, even though Mad2 continues to turn over at the spindle poles.
The kinases Bub1 and Mps1 act at an early step in generating the primary SCP signal. Similar to Mps1 overexpression, overexpression of the dominantly acting bub1-5 allele blocks anaphase onset in an Mps1-, Mad- and Bub-dependent manner, but without causing Mad1 hyperphosphorylation [89]. Overexpression of Mps1 does result in Mad1 hyperphosphorylation in bub1Δ cells, although there is no mitotic delay [86], showing that Mad1 hyperphosphorylation does not always correlate with either spindle damage or mitotic arrest. It therefore appears that the checkpoint complex that transduces the signal from kinetochores to APCCdc20 requires collaboration between each of the Mad proteins, Bub and Mps1 kinases for a full SCP response.
Interestingly, the Ipl1 kinase in budding yeast is required for the arrest that is induced by lack of tension in cdc6 cells and for that induced by Mps1 overexpression [77], but not for the checkpoint response to MT depolymerization by nocodazole. Antibodies against the Aurora B kinase can overcome a nocodazole-induced arrest in both Xenopus egg extracts and in cultured cells [90], suggesting a direct role for Aurora B in SCP activation in these systems. It is possible that Aurora B/Ipl1 activates SCP by destabilizing MT–kinetochore connections that are not under enough tension, causing such kinetochores to become either unattached or occupied with only very low numbers of MTs [91]. Activation of the SCP triggers the recruitment to the defective kinetochore of various checkpoint proteins that presumably are tethered to the kinetochore via their interaction with kinetochore components (Figure 3). A recent study in Xenopus egg extracts suggests that the first step in SCP signalling is defined by the Aurora B–INCENP complex which induces the kinetochore localization of Mps1, Bub1, Bub3 and CENP-E [90]. This step, in turn, promotes the recruitment of Mad1–Mad2, Cdc20 and APC/C. Hence, Aurora B is the most upstream regulator of the SCP cascade, and probably detects both the absence of tension and attachment at kinetochores in this system [92]. Moreover, Mps1 and Bub1 are dependent on each other for their robust kinetochore localization, indicating that, although they act downstream of Aurora B–INCENP, they may both act at an early step in the generation of the primary SCP signal [92]. It is not clear if the Ipl1 complex plays an analogous role upstream of the SCP signalling pathway in budding yeast.
In mammalian cells, the kinase BubR1 that shares homology with both Bub1 and Mad3, interacts with and is activated by the kinetochore-associated MT motor CENP-E. Disruption of CENP-E function delays anaphase onset [93], suggesting that the hBubR1–CENP-E interaction is part of the force-sensing mechanism at the kinetochore, and hBubR1 may monitor CENP-E–MT interactions. Without CENP-E, the SCP cannot be established or maintained in vitro [94] or in mice [95], showing that, although the precise SCP role of CENP-E may show subtle differences in different systems, CENP-E is a central component of the vertebrate SCP.
The recruitment of SCP proteins to defective kinetochores is believed to be followed by their reorganization, culminating perhaps in the modification and release of some checkpoint proteins, particularly Mad2 (see Figure 3). FRAP (fluorescence recovery after photobleaching) experiments reveal that Bub1, Mad1 and a portion of Mad2 have a relatively long residence time at unattached kinetochores. Together, they probably function as part of a catalytic platform that recruits, activates and releases an APCCdc20-inhibitory signal that is partly composed of the rapidly exchanging fraction of Mad2. The generation of the SCP signal requires that all components of this platform come together at the kinetochore [96].
Cdc20, like Mad2, also associates with kinetochores, and cycles on and off rapidly [97,98], but the relevance of this cycling (believed to permit a sampling of kinetochores) is not entirely clear, because, unlike Mad2, Cdc20 continues cycling even after the checkpoint has been silenced. In Xenopus egg extracts, in addition to SCP proteins and Cdc20, the APC/C also localizes to kinetochores via a mechanism that is independent of the kinetochore localization of Cdc20 [92]. Importantly, a recent study in human cell cultures [99] has correlated the kinetochore localization of APC/C to an active SCP. Interestingly, in this system, the SCP machinery is required to recruit the APC/C to kinetochores. The observation that APC/C co-localizes with SCP components at kinetochores suggests two possibilities: (i) that the inhibition of APCCdc20 by the SCP occurs at kinetochores, or (ii) that the APC/C undergoes modification at kinetochores to make it susceptible to inhibition by the SCP. Further evidence for the kinetochore localization of APC/C comes from the identification of a phosphoepitope long known to be present on unattached kinetochores [100] and recognized by the 3F3/2 antibody. Studies have shown that this antibody recognizes the phosphorylated form of the APC/C subunit Apc1, indicating that the SCP-sensitive staining of 3F3/2 at kinetochores could result from the recruitment of APC/C to defective kinetochores [99,101].
What is the molecular nature of the APCCdc20-inhibitory signal? Mad2 and Mad3/BubR1 appear to be the key links to APCCdc20 inhibition. Mad2 is believed to be released from kinetochores in a modified form that is transferred to the Bub3–Cdc20–Mad3/BubR1 complex, which then functions as a potent inhibitor of APC/C. This aspect of the SCP is highly conserved, with MAD2 showing functional interaction with Cdc20 from widely divergent organisms [29,30,102,103]. Mad2 and BubR1 promote each other's association with APCCdc20 and act synergistically in vitro to inhibit APCCdc20 [104]. BubR1 is a potent APC/C inhibitor in vitro and can even do so independently of Mad2 [105]. The fission yeast (Schiz. pombe) Mad3 is also required for Mad2 to inhibit anaphase onset in vivo [106]. Despite all this supporting evidence, the presence of the Bub3–Cdc20–Mad3/BubR1–Mad2 complex throughout the cell cycle [107] has led to questions about its physiological significance.
In addition to the Bub3–Cdc20–Mad2–Mad3 complex that may render APCCdc20 incapable of ubiquitinating securin, cells also contain a pool of Mad2–Cdc20 complexes, suggesting that that Mad2 acts independently to block anaphase by sequestering Cdc20 and making it unavailable for the activation of APC/C. Activation of the SCP also leads to destabilization of Cdc20 protein via a process requiring the association of Cdc20, Mad2 and functional APC/C. The SCP signal therefore ensures that not only is Cdc20 sequestered to make it less available to activate APC/C, but that Cdc20 levels drop below a critical threshold to ensure complete inhibition of APCCdc20 [108].
BIFURCATION OF THE SCP
A major milestone was the discovery that the SCP pathway is bifurcated downstream of the Mps1 kinase. This finding emerged from the observation in budding yeast that the cell cycle delay induced by low doses of benomyl (that does not prevent spindle assembly, but interferes with kinetochore–MT connections) is abolished by mutations in MAD1-MAD3, BUB1, BUB3, but is not affected by mutation of BUB2 [109]. Furthermore, analysis of genetic interactions of BFA1 with other known checkpoint genes indicated that Bfa1 and Bub2 lay in the same genetic pathway, but not in the same epistasis group as Mad1 and Mad2 [110]. Other observations that support a bifurcated pathway are the following [111]: (i) unlike other Mad and Bub proteins, Bub2 localizes to the SPB, (ii) the effect of concomitant lack of Mad1 or Mad2 and Bub2 is additive, and (iii) cell cycle progression in bub2 cells in the presence of nocodazole requires the APC/C subunit Cdc26, which is not required for mad2 cells under the same conditions. When the issue was pressed further, it emerged that the Bub2-dependent arm of the checkpoint is dedicated to restraining mitotic exit in response to errors in spindle assembly or orientation.
THE ‘EXIT ARM’ OF THE SCP
In the budding yeast mad2Δ bub2Δ strain treated with nocodazole, the SCP is not activated and even though the spindle is absent, the cells inactivate the mitotic kinase and exit the cell cycle with wild-type kinetics [112]. This phenotype highlights the importance of restraining mitotic kinase inactivation until accurate chromosome segregation has been achieved, one of the key objectives of the SCP. The observation that mad2Δ cells are competent to delay mitotic exit, despite the premature degradation of the securin Pds1 [112], indicated that there exists a Mad2-independent mechanism to inhibit mitotic exit.
Regulating the activation of MEN signalling
In budding yeast, numerous factors regulate, directly or indirectly, the timely activation of the MEN pathway by controlling the cortical localization of Lte1, the putative GEF for Tem1 GTPase. Phosphorylation of Lte1 by the Cdc28-Cln kinase is necessary for its localization to the cortex via an actin-independent mechanism early in the cell cycle, concomitant with bud emergence [113]. The anchorage of Lte1 to the bud cortex requires functional septins [114,115] and is also regulated by its interaction with Ras2 [116]. The Rho-GTPase Cdc42 has been shown to activate mitotic exit through its effectors, the PAK (p21Cdc42/Rac-activated kinase)-like kinases [113,114,117]. Importantly, Cdc42, its exchange factor Cdc24 and the PAK-like Cla4 kinase have been demonstrated to control the initial binding and activation of Lte1 at the bud cortex. Furthermore, Cdc42, Cdc24 and the PAK-like Ste20 kinase function in a pathway parallel to Lte1 in facilitating mitotic exit [117]. Cla4 and Ste20 may regulate the first wave of cyclin B destruction mediated by APCCdc20, which has been shown to be crucial for MEN activation. The inhibition of PAK kinases also prevents mitotic exit by impairing the cortical localization of Lte1 and Tem1 activation [118]. The nexus surrounding the regulation of Tem1 is complicated further by the observation that the cell polarity proteins Kel1 and Kel2 are present in independent complexes with Lte1 and Tem1, and negatively regulate mitotic exit, probably by interfering with the interaction between Tem1 and Lte1 [117]. While this body of data suggests that regulation of Lte1 constitutes a major stratagem in the control of MEN activation, the observation that LTE1 is not an essential gene and that mitotic exit occurs with nearly wild-type kinetics in cells lacking Lte1 at 30 °C [119] implies that Lte1-independent mechanisms exist that regulate Tem1 activation. Moreover, Lte1's role in stimulating Tem1 function is controversial. A recent report suggests that Lte1 may be a downstream effector for Ras2 in mitotic exit, and that the GEF domain of Lte1 is not essential for mitotic exit, but only for its localization [116].
The PLK Cdc5 plays a major role in the control of Tem1 activation by phosphorylating and modifying the GAP activity of the Bfa1–Bub2 complex towards Tem1 (see below). Cdc14, released transiently under the influence of the FEAR pathway, binds to the SPB in association with the Bfa1–Bub2 GAP complex and also interacts with Tem1; this contributes, via Cdc15 activation, to the activation of the MEN pathway [55]. An interesting development was the discovery that the CRIB (Cdc42/Rac-interactive binding) domain-containing Cdc42 effector proteins Gic1 and Gic2 promote mitotic exit independently of Ste20. Gic1 and Gic2 become essential for mitotic exit when the activation of MEN through Cdc5 and Lte1 is impaired [120]. Gic1, following its release from the cortex in anaphase, binds Bub2 and prevents it from binding to Tem1. Hence, the Gic proteins presumably trigger mitotic exit by interfering with Bfa1–Bub2 GAP function [120]. While a great deal has been elucidated regarding the network of interactions that controls MEN activation, very little is known about which of these pathways, if any, is targeted by the SCP for restraining mitotic exit.
The FEAR pathway is a target of the SCP
Evidence suggests that activation of the Mad2-dependent ‘anaphase arm’ of the SCP, by preventing the destruction of Pds1 and the consequent release of separase Esp1, also precludes the activation of the Esp1-Slk19 branch of the FEAR pathway and the early release of Cdc14 from the nucleolus [56]. Pds1 stabilization also inhibits the Fob1-Spo12 branch of the FEAR pathway [52,54,121,122]. This inhibition of the FEAR pathway activation by the SCP is independent of MEN pathway function.
The Bub2–Bfa1 GAP complex is a target of the SCP
Our understanding of the reach of SCP took a leap forward when it came to light that the Bub2–Bfa1 GAP complex in budding yeast not only modulates the activation of the Tem1 GTPase [72] and MEN signalling, but also is a direct target of regulation by the SCP. Bfa1, Bub2 and Tem1 associate with the budding yeast SPB in a cell-cycle-dependent manner. The association with SPB is dependent on a structural component of the SPB outer plaque, Nud1 [123]. Bub2 and Bfa1 may target Tem1 to the SPB [124]. The mitotic arrest mediated by the SCP requires a functional Bub2–Bfa1 complex [125,126]. Also, continued presence of the Bub2 protein is required to maintain the SCP-induced arrest [127]. Importantly, even though Bub2 is essential for the function of the SCP, Bub2 does not block cell cycle progression by inhibiting Cdc20-dependent Pds1 proteolysis [111,112]. The overexpression of Bfa1 causes cells to arrest in telophase (and not before anaphase) with high levels of the Cdc28-Clb kinase [110,128] strongly suggesting that Bfa1 inhibits mitotic exit. Inappropriate sister chromatid separation in a nocodazole-treated bub2 mutant is prevented when mitotic exit is blocked using a tem1 mutation, indicating that the lethality in the bub2 cells is a consequence of events downstream of Tem1 in the MEN pathway [127]. Conversely, overexpression of TEM1 in a mad2Δ strain causes complete abrogation of the SCP, consistent with the notion that Tem1 is a target of the Bub2 signal [112]. Unlike in the mad2 bub2 mutant, mitotic exit is blocked in a cdc14 mad2 bub2 triple mutant in response to spindle damage, implying that Cdc14 is an effector of the Bub2 checkpoint signal [112]. The activity of the Dbf2 kinase has been shown to be negatively regulated by Bub2 in budding yeast during normal cell cycle progression [129]. Moreover, in the absence of dynein and dynactin, the spindle breaks down and mitotic exit is prevented when both nuclei are in the mother cell. If BUB2 is also deleted, this delay is abrogated, resulting in formation of anucleate cells [125]. In addition, the GAP activity of the Bub2–Bfa1 complex has been shown to be up-regulated in nocodazole-treated cells [129].
The Schiz. pombe homologue of Bub2, cdc16+, regulates both B-type cyclin degradation and cytokinesis. The budding yeast BUB2 gene complements the temperature-sensitive allele cdc16-116 in Schiz. pombe, lending support to a role for Bub2 in the regulation of mitotic exit. It was demonstrated that the inhibition of Clb2 degradation in bub2 mutants is dependent on Pds1, and that the same block in pds1 mutants is dependent on Bub2. Thus nocodazole blocks mitotic cyclin proteolysis by two independent mechanisms: (i) via a Mad2-dependent pathway that blocks Pds1 destruction and thereby also Clb destruction, and (ii) via another Bub2-dependent pathway that blocks Clb destruction in a Pds1-independent manner [112]. Taken together, these data indicate that the primary role of the Bfa1–Bub2 complex in the SCP is to block the onset of mitotic exit until chromosomes have segregated properly. This Bub2-dependent ‘exit arm’ of the SCP also becomes essential when the nucleus fails to migrate into the bud. It is for this reason that the Bub2-dependent ‘exit arm’ of the SCP is sometimes referred to as the spindle orientation checkpoint.
Bub2 is phosphorylated in a cell-cycle-regulated manner that requires Cdc5, as well as another unidentified kinase, in addition to the presence of Bfa1 [130]. In G1, Bub2 is not phosphorylated; phosphorylation commences concomitantly with the onset of budding and peaks just before mitotic exit. The timing and kinetics of Bub2 modification are very similar to that of Bfa1 phosphorylation by Cdc5. Phosphorylation of Bfa1 interferes with its ability to interact with and negatively regulate Tem1 [131]. In late anaphase, concomitant with mitotic exit, Cdc14 dephosphorylates Bfa1 and activates the GAP complex [55]. Activation of the SCP blocks the phosphorylation of Bub2 (in a Mad2- and Mps1-dependent manner in the presence of spindle damage) and promotes the activation of the GAP complex, resulting in an inhibition of mitotic exit [130]. Activation of the SCP also inhibits phosphorylation of Bfa1 by Cdc5 via a pathway that requires Mad2 and Mps1 [131,132], and thus blocks mitotic exit.
In addition to its role as a component of the Tem1 GAP complex, Bfa1 can also regulate mitotic exit directly by inhibiting the interaction between Tem1 and Cdc15 [128]. It will be interesting to test if this activity of Bfa1 is subject to regulation by the SCP. Recently, a novel Bfa1-interacting protein Ibd2 has been identified in budding yeast [132]. ibd2Δ is SCP-defective, and the defect is rescued by extra copies of BUB2, BFA1 and CDC5. Ibd2 encodes a novel component of the Bub2-dependent SCP pathway that functions upstream of Bub2 and Bfa1 and downstream of Mps1. Another interesting aspect of the Bub2 pathway is that Pds1 itself regulates (through an unknown mechanism) the Bub2-dependent pathway to inhibit both Cdh1 and Sic1 [112,121,122]. This dependency of Clb2 proteolysis on Pds1 degradation may also contribute to ensuring that exit is not triggered before anaphase. How Pds1 regulates the onset of mitotic exit directly should be an exciting subject for further study.
Is the SCP always turned on?
Checkpoints were originally defined in the yeast system as non-essential pathways that came into play in response to errors. Kinetochores in budding yeast rarely, if ever, detach completely from MTs during normal cell division. It is perhaps due to this aspect of spindle morphogenesis that the SCP is not essential for mitosis in budding yeast under normal growth conditions [133]. Evidence accruing from metazoan systems, however, suggests that SCP proteins have important roles in regulating normal mitoses and meioses, providing intrinsic quality control during each cell cycle. As cells in metazoans enter mitosis, they constitutively express signals that delay the activation of APC/C and anaphase onset [134]. These signals are gradually extinguished as sister kinetochores capture MTs from opposite poles and tension develops throughout the spindle. Support for this view came from observations that Bub1 not only was required for checkpoint response to spindle damage, but also restrains progression through a normal mitosis [79]. Moreover, microinjection of antibodies against Mad2 into cultured animal cells resulted in premature anaphase onset accompanied by chromosome mis-segregation [135]. RNA interference of the Caenorhabditis elegans Mad2 homologue (Mdf-2) caused an embryonic or larval arrest in approx. 20% of animals, while the remaining animals progressed to adulthood, but had dramatic defects in germline development, highlighting the role of Mad2 in meiotic SCP. Knockout mutations in murine Mad and Bub proteins cause mitotic catastrophe and the induction of p53-mediated cell death [136–138]. These findings imply that the same kinetochore-dependent processes control both progression through mitosis in cells in which chromosome segregation is proceeding normally, as well as SCP functions in cells in which chromosome–MT attachment is defective. In a recent study, Meraldi et al. [139] found that, while the depletion of Mad2 or BubR1 results in significant acceleration of mitosis in all cells, depletion of Mad1, Bub1 or Bub3 leaves timing unaffected. The same is true if recruitment of Mad2 or BubR1 to kinetochores is blocked using RNAi (RNA interference). These data suggest that the mitotic timing and SCP functions of the Mad and Bub proteins are separable, and, importantly, that kinetochores are crucial for SCP control, but may be dispensable for the regulation of mitotic timing. Meraldi et al. [139] propose that mitotic timing and the SCP are regulated in different ways, although the same proteins may perform both functions. The pool of cytosolic Mad2 and BubR1 regulates the mean time of anaphase onset, acting independently of kinetochores, to inhibit APC/C during the short interval after the degradation of the Cdc20 inhibitor Emi1 and before the SCP becomes fully operational, i.e. during the period of kinetochore assembly. The emerging picture in metazoan systems is therefore of a highly dynamic dual-capability surveillance mechanism (that is perhaps not as kinetochore-centred as previously believed), one that has also been harnessed for fine-tuning the timing of mitotic onset in every cycle of division, stalling the cell cycle if errors are detected.
Understanding the SCP further
The combined use of genetics, intensive biochemistry and a wide range of cell biological techniques in multiple model systems has provided insightful details pertaining to the molecular underpinnings and evolutionary conservation of the SCP. Yet a lot remains to be learnt about the process of error detection, generation of inhibitory signals from the kinetochore and their integration with the core cell cycle machinery. For instance, the molecular mechanism by which Aurora B influences MT capture by kinetochores to promote bi-orientation remains largely unresolved. Initial evidence indicates that motor proteins may be important in this process. It will be interesting to determine if Aurora B directly regulates motor protein activities at kinetochores. Similarly, PAK-like kinases have been implicated in the regulation of an early step in the activation of APCCdc20 and MEN in budding yeast [118]; it may be rewarding to explore in detail if they are targeted by the SCP in order to control anaphase onset and mitotic exit. An intriguing aspect of the SCP is that it can be engaged by merely overexpressing Mps1 kinase, even in the absence of intact kinetochore structures [140]. Elucidation of the basis for this phenomenon may reveal novel aspects of SCP regulation. What lies upstream of the hMps1, hBub1 and Aurora B kinases is another question of acute importance that still requires answers. Our knowledge of the detailed workings of SCP will certainly be augmented by the identification of novel players that determine its efficacy and fine-tuning. In view of the evolutionary conservation of these key proteins, any insights gleaned from the budding yeast would, as in the past, have an impact on our understanding of the SCP regulatory setup in metazoans.
Intuitively, one or more proteins that mediate the chromosome–MT connection should be sensitive to the status of the linkage and should harbour the intrinsic ability to trigger the SCP signal. The identification of such proteins would shed light on a major outstanding question: what is it about the unattached or relaxed kinetochore that activates the SCP? It is possible that lack of occupancy exposes the binding site for SCP proteins in the kinetochore and MT attachment obscures it. However, molecular data in support of such a model are still unavailable. One could also envisage that tension perhaps leads to the physical deformation of a protein or a protein complex that somehow turns off the checkpoint ‘alarm’. Such a protein or a protein complex presumably has structural characteristics that allow it to ‘transduce’ a physical force. Alternatively, lack of tension may somehow create a binding site in the kinetochore for the recruitment of SCP proteins. The binding site may be distorted under tension to the extent that it can no longer engage SCP proteins. What the molecular nature of the tension-sensing mechanism is and how it generates a biochemical signal remains one of the central mysteries in SCP control.
Acknowledgments
We thank Wei Hoong Khong for his help with some of the graphics. This work was supported by the Biomedical Research Council, Singapore.
References
- 1.Boveri T. Zellenstudien VI: Die Entwicklung dispermer Seeigelier. Ein Beitrag zur Befruchtungslehre und zur Theorie des Kernes. Jena. Z. Naturwiss. 1907;43:1–292. [Google Scholar]
- 2.Sutton W. S. The chromosomes in heredity. Biol. Bull. 1903;4:231–251. [Google Scholar]
- 3.Shah J. V., Cleveland D. W. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- 4.Bharadwaj R., Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 5.Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 6.Millband D. N., Campbell L., Hardwick K. G. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 2002;12:205–209. doi: 10.1016/s0962-8924(02)02276-6. [DOI] [PubMed] [Google Scholar]
- 7.Lew D. J., Burke D. J. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 8.Huang J., Hsu J. M., Laurent B. C. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 9.Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature (London) 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 11.Choo K. H. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wigge P. A., Kilmartin J. V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald K. L., O'Toole E. T., Mastronarde D. N., McIntosh J. R. Kinetochore microtubules in PTK cells. J. Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheeseman I. M., Brew C., Wolyniak M., Desai A., Anderson S., Muster N., Yates J. R., Huffaker T. C., Drubin D. G., Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 17.Kang J., Cheeseman I. M., Kallstrom G., Velmurugan S., Barnes G., Chan C. S. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., Murray A. W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 20.Shang C., Hazbun T. R., Cheeseman I. M., Aranda J., Fields S., Drubin D. G., Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinsky B. A., Tatsutani S. Y., Collins K. A., Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- 22.Dewar H., Tanaka K., Nasmyth K., Tanaka T. U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature (London) 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- 23.Lampson M. A., Renduchitala K., Khodjakov A., Kapoor T. M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 24.Sharp D. J., Rogers G. C., Scholey J. M. Microtubule motors in mitosis. Nature (London) 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 25.Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature (London) 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 27.Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 28.Peters J. M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 29.Hwang L. H., Lau L. F., Smith D. L., Mistrot C. A., Hardwick K. G., Hwang E. S., Amon A., Murray A. W. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 30.Kim S. H., Lin D. P., Matsumoto S., Kitazono A., Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 31.Jallepalli P. V., Waizenegger I. C., Bunz F., Langer S., Speicher M. R., Peters J. M., Kinzler K. W., Vogelstein B., Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 32.Visintin R., Prinz S., Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 33.Lim H. H., Goh P. Y., Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 34.Hartwell L. H. Saccharomyces cerevisiae cell cycle. Bacteriol. Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethi N., Monteagudo M. C., Koshland D., Hogan E., Burke D. J. The CDC20 gene product of Saccharomyces cerevisiae, a β-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol. Cell. Biol. 1991;11:5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstein J., Jacobsen F. W., Hsu-Chen J., Wu T., Baum L. G. A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the Saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Mol. Cell. Biol. 1994;14:3350–3363. doi: 10.1128/mcb.14.5.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto T. A fission yeast homolog of CDC20/p55CDC/Fizzy is required for recovery from DNA damage and genetically interacts with p34cdc2. Mol. Cell. Biol. 1997;17:742–750. doi: 10.1128/mcb.17.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margottin-Goguet F., Hsu J. Y., Loktev A., Hsieh H. M., Reimann J. D., Jackson P. K. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 39.Schwab M., Lutum A. S., Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 40.Jaspersen S. L., Charles J. F., Morgan D. O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 41.Zachariae W., Schwab M., Nasmyth K., Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 42.Kramer E. R., Scheuringer N., Podtelejnikov A. V., Mann M., Peters J. M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 44.Bembenek J., Yu H. Regulation of the anaphase-promoting complex by the dual specificity phosphatase human Cdc14a. J. Biol. Chem. 2001;276:48237–48242. doi: 10.1074/jbc.M108126200. [DOI] [PubMed] [Google Scholar]
- 45.Yeong F. M., Lim H. H., Padmashree C. G., Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- 46.Wasch R., Cross F. R. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature (London) 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- 47.Stegmeier F., Visintin R., Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 48.Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Shevchenko A., Charbonneau H., Deshaies R. J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 49.Visintin R., Hwang E. S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature (London) 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan M., Lehane C., Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng X., Kahana J. A., Silver P. A., Morphew M. K., McIntosh J. R., Fitch I. T., Carbon J., Saunders W. S. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visintin R., Stegmeier F., Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol. Biol. Cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan M., Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stegmeier F., Huang J., Rahal R., Zmolik J., Moazed D., Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 55.Pereira G., Manson C., Grindlay J., Schiebel E. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 2002;157:367–379. doi: 10.1083/jcb.200112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida S., Asakawa K., Toh-e A. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 2002;12:944–950. doi: 10.1016/s0960-9822(02)00870-9. [DOI] [PubMed] [Google Scholar]
- 57.Jaspersen S. L., Charles J. F., Tinker-Kulberg R. L., Morgan D. O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shirayama M., Matsui Y., Toh-E A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirayama M., Matsui Y., Toh-e A. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. Mol. Gen. Genet. 1996;251:176–185. doi: 10.1007/BF02172916. [DOI] [PubMed] [Google Scholar]
- 60.Johnston L. H., Eberly S. L., Chapman J. W., Araki H., Sugino A. The product of the Saccharomyces cerevisiae cell cycle gene DBF2 has homology with protein kinases and is periodically expressed in the cell cycle. Mol. Cell. Biol. 1990;10:1358–1366. doi: 10.1128/mcb.10.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitada K., Johnson A. L., Johnston L. H., Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol. Cell. Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schweitzer B., Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991;7:265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- 63.Bardin A. J., Visintin R., Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 64.Jaspersen S. L., Morgan D. O. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 2000;10:615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- 65.Toyn J. H., Johnston L. H. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee S. E., Jensen S., Frenz L. M., Johnson A. L., Fesquet D., Johnston L. H. The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J. Cell Sci. 2001;114:2345–2354. doi: 10.1242/jcs.114.12.2345. [DOI] [PubMed] [Google Scholar]
- 67.Asakawa K., Yoshida S., Otake F., Toh-e A. A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics. 2001;157:1437–1450. doi: 10.1093/genetics/157.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Molk J. N., Schuyler S. C., Liu J. Y., Evans J. G., Salmon E. D., Pellman D., Bloom K. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell. 2004;15:1519–1532. doi: 10.1091/mbc.E03-09-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mah A. S., Jang J., Deshaies R. J. Protein kinase Cdc15 activates the Dbf2–Mob1 kinase complex. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S. E., Frenz L. M., Wells N. J., Johnson A. L., Johnston L. H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y., Shirogane T., Liu D., Harper J. W., Elledge S. J. Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell. 2003;112:697–709. doi: 10.1016/s0092-8674(03)00121-1. [DOI] [PubMed] [Google Scholar]
- 72.Shah R., Jensen S., Frenz L. M., Johnson A. L., Johnston L. H. The Spo12 protein of Saccharomyces cerevisiae: a regulator of mitotic exit whose cell cycle-dependent degradation is mediated by the anaphase-promoting complex. Genetics. 2001;159:965–980. doi: 10.1093/genetics/159.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeong F. M., Lim H. H., Wang Y., Surana U. Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol. Cell. Biol. 2001;21:5071–5081. doi: 10.1128/MCB.21.15.5071-5081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 75.Hoyt M. A., Totis L., Roberts B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 76.Weiss E., Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biggins S., Murray A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen R. H., Waters J. C., Salmon E. D., Murray A. W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 79.Taylor S. S., McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 80.Taylor S. S., Hussein D., Wang Y., Elderkin S., Morrow C. J. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- 81.Rieder C. L., Cole R. W., Khodjakov A., Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rieder C. L., Schultz A., Cole R., Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nicklas R. B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 84.Stern B. M., Murray A. W. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- 85.Abrieu A., Magnaghi-Jaulin L., Kahana J. A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D. W., Labbe J. C. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 86.Hardwick K. G., Weiss E., Luca F. C., Winey M., Murray A. W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 87.Hardwick K. G., Johnston R. C., Smith D. L., Murray A. W. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roberts B. T., Farr K. A., Hoyt M. A. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Farr K. A., Hoyt M. A. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kallio M. J., McCleland M. L., Stukenberg P. T., Gorbsky G. J. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- 91.Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vigneron S., Prieto S., Bernis C., Labbe J. C., Castro A., Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 94.Abrieu A., Kahana J. A., Wood K. W., Cleveland D. W. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 95.Putkey F. R., Cramer T., Morphew M. K., Silk A. D., Johnson R. S., McIntosh J. R., Cleveland D. W. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 96.Shah J. V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D. W. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 97.Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 98.Kallio M. J., Beardmore V. A., Weinstein J., Gorbsky G. J. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Acquaviva C., Herzog F., Kraft C., Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- 100.Nicklas R. B., Ward S. C., Gorbsky G. J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Daum J. R., Tugendreich S., Topper L. M., Jorgensen P. M., Hoog C., Hieter P., Gorbsky G. J. The 3F3/2 anti-phosphoepitope antibody binds the mitotically phosphorylated anaphase-promoting complex/cyclosome. Curr. Biol. 2000;10:R850–R852. doi: 10.1016/s0960-9822(00)00836-8. [DOI] [PubMed] [Google Scholar]
- 102.Fang G., Yu H., Kirschner M. W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kallio M., Weinstein J., Daum J. R., Burke D. J., Gorbsky G. J. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang Z., Bharadwaj R., Li B., Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 106.Millband D. N., Hardwick K. G. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sudakin V., Chan G. K., Yen T. J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pan J., Chen R. H. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y., Burke D. J. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fraschini R., Formenti E., Lucchini G., Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alexandru G., Zachariae W., Schleiffer A., Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jensen S., Geymonat M., Johnson A. L., Segal M., Johnston L. H. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 2002;115:4977–4991. doi: 10.1242/jcs.00189. [DOI] [PubMed] [Google Scholar]
- 114.Seshan A., Bardin A. J., Amon A. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 2002;12:2098–2110. doi: 10.1016/s0960-9822(02)01388-x. [DOI] [PubMed] [Google Scholar]
- 115.Castillon G. A., Adames N. R., Rosello C. H., Seidel H. S., Longtine M. S., Cooper J. A., Heil-Chapdelaine R. A. Septins have a dual role in controlling mitotic exit in budding yeast. Curr. Biol. 2003;13:654–658. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 116.Yoshida S., Ichihashi R., Toh-e A. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 2003;161:889–897. doi: 10.1083/jcb.200301128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hofken T., Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;21:4851–4862. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chiroli E., Fraschini R., Beretta A., Tonelli M., Lucchini G., Piatti S. Budding yeast PAK kinases regulate mitotic exit by two different mechanisms. J. Cell Biol. 2003;160:857–874. doi: 10.1083/jcb.200209097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Adames N. R., Oberle J. R., Cooper J. A. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 2001;153:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hofken T., Schiebel E. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 2004;164:219–231. doi: 10.1083/jcb.200309080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cohen-Fix O., Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tinker-Kulberg R. L., Morgan D. O. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gruneberg U., Campbell K., Simpson C., Grindlay J., Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pereira G., Hofken T., Grindlay J., Manson C., Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 125.Bloecher A., Venturi G. M., Tatchell K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat. Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- 126.Wang Y., Hu F., Elledge S. J. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr. Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- 127.Krishnan R., Pangilinan F., Lee C., Spencer F. Saccharomyces cerevisiae BUB2 prevents mitotic exit in response to both spindle and kinetochore damage. Genetics. 2000;156:489–500. doi: 10.1093/genetics/156.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ro H. S., Song S., Lee K. S. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5436–5441. doi: 10.1073/pnas.062059999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fesquet D., Fitzpatrick P. J., Johnson A. L., Kramer K. M., Toyn J. H., Johnston L. H. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 1999;18:2424–2434. doi: 10.1093/emboj/18.9.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu F., Elledge S. J. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle. 2002;1:351–355. [PubMed] [Google Scholar]
- 131.Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S. J. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- 132.Hwang H. S., Song K. IBD2 encodes a novel component of the Bub2p-dependent spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics. 2002;161:595–609. doi: 10.1093/genetics/161.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gillett E. S., Espelin C. W., Sorger P. K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pines J., Rieder C. L. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 2001;3:E3–E6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- 135.Gorbsky G. J., Chen R. H., Murray A. W. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dobles M., Liberal V., Scott M. L., Benezra R., Sorger P. K. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 137.Kalitsis P., Earle E., Fowler K. J., Choo K. H. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Michel L. S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P. K., Murty V. V., Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature (London) 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 139.Meraldi P., Draviam V. M., Sorger P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 140.Fraschini R., Beretta A., Lucchini G., Piatti S. Role of the kinetochore protein Ndc10 in mitotic checkpoint activation in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;266:115–125. doi: 10.1007/s004380100533. [DOI] [PubMed] [Google Scholar]
- 141.De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Measday V., Hieter P. Kinetochore sub-structure comes to MIND. Nat. Cell Biol. 2004;6:94–95. doi: 10.1038/ncb0204-94. [DOI] [PubMed] [Google Scholar]
- 143.Gordon D. M., Roof D. M. Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12515–12520. doi: 10.1073/pnas.231212498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hildebrandt E. R., Hoyt M. A. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APCCdh1 and a bipartite destruction sequence. Mol. Biol. Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Iouk T., Kerscher O., Scott R. J., Basrai M. A., Wozniak R. W. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yamaguchi S., Decottignies A., Nurse P. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J. 2003;22:1075–1087. doi: 10.1093/emboj/cdg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wassmann K., Liberal V., Benezra R. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J. 2003;22:797–806. doi: 10.1093/emboj/cdg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 149.Liu S. T., Hittle J. C., Jablonski S. A., Campbell M. S., Yoda K., Yen T. J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003;5:341–345. doi: 10.1038/ncb953. [DOI] [PubMed] [Google Scholar]
- 150.Brady D. M., Hardwick K. G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 151.Seeley T. W., Wang L., Zhen J. Y. Phosphorylation of human MAD1 by the BUB1 kinase in vitro. Biochem. Biophys. Res. Commun. 1999;257:589–595. doi: 10.1006/bbrc.1999.0514. [DOI] [PubMed] [Google Scholar]
- 152.Yudkovsky Y., Shteinberg M., Listovsky T., Brandeis M., Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem. Biophys. Res. Commun. 2000;271:299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
- 153.Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]