Abstract
This study aims to explore the efficacy of a hybrid deep learning and radiomics approach, supplemented with patient metadata, in the noninvasive dermoscopic imaging-based diagnosis of skin lesions. We analyzed dermoscopic images from the International Skin Imaging Collaboration (ISIC) dataset, spanning 2016–2020, encompassing a variety of skin lesions. Our approach integrates deep learning with a comprehensive radiomics analysis, utilizing a vast array of quantitative image features to precisely quantify skin lesion patterns. The dataset includes cases of three, four, and eight different skin lesion types. Our methodology was benchmarked against seven classification methods from the ISIC 2020 challenge and prior research using a binary decision framework. The proposed hybrid model demonstrated superior performance in distinguishing benign from malignant lesions, achieving area under the receiver operating characteristic curve (AUROC) scores of 99%, 95%, and 96%, and multiclass decoding AUROCs of 98.5%, 94.9%, and 96.4%, with sensitivities of 97.6%, 93.9%, and 96.0% and specificities of 98.4%, 96.7%, and 96.9% in the internal ISIC 2018 challenge, as well as in the external Jinan and Longhua datasets, respectively. Our findings suggest that the integration of radiomics and deep learning, utilizing dermoscopic images, effectively captures the heterogeneity and pattern expression of skin lesions.
Keywords: International Skin Imaging Collaboration (ISIC), Area under the Receiver Operating Characteristic Curves (AUROC), Radiomics, Deep learning, Multimodal, Skin lesion
Subject terms: Biomarkers, Health care
Introduction
Dermoscopic imaging, a critical element in public health, has gained significant attention for its role in skin lesion analysis1,2. Dermoscopy, a pivotal imaging technique, is routinely employed in clinical settings to differentiate melanomas from benign lesions3,4. This noninvasive micro-morphological imaging method enhances the diagnostic accuracy of skin lesion evaluation beyond traditional visual examination5. Automated analysis of dermatoscopic images promises to reduce unnecessary clinical visits and facilitate early detection of skin cancers.
Recent advancements in deep learning have demonstrated significant promise in computer-aided diagnosis6–14. Al-Masni et al.6 developed a deep learning-based system integrated with a skin lesion CAD framework, featuring lesion boundary segmentation and disease recognition, thus supporting dermatologists in diagnosing skin cancers. Qin et al.9 designed GANs for skin lesion imaging, capable of generating high-quality images to enhance classification models. Xing et al.10 introduced a detection network employing a Zoom-in Attention mechanism and Metadata Embedding, incorporating pathological and demographic data. Dong et al.11 integrated dermoscopic images with clinical metadata for enhanced skin lesion segmentation and classification, highlighting the importance of multimodal data. Furthermore, Kaur et al.12, Hasan et al.13, and Alenezi et al.14 advanced skin lesion classification accuracy using innovative neural network approaches.
Despite these advancements, deep learning methods relying solely on image intensity often struggle to extract information related to lesion shape and site-specific disease characteristics, limiting their efficacy. In clinical practice, dermatologists combine imaging modalities with patient-level metadata (e.g., age, sex, disease site) for comprehensive lesion assessment. Yuan et al.15 revealed that age-standardized and site-specific tumor rates have increased annually across all races and genders, with pronounced gender differences in incidence rates at younger ages, particularly in specific body regions. Sinnamon et al.16 found that younger age, among other clinicopathologic factors, is significantly associated with lymph node positivity, suggesting age should be considered in sentinel lymph node biopsy decisions.
Skin lesions present a complex array of features, including lesion intensity, intralesion texture, shape, and color variation. Radiomics can extract high-dimensional quantitative data reflecting imaging phenotypes17, encompassing first-order features that describe the intensity distribution of the region of interest, second-order features that capture spatial heterogeneity, and shape features that reflect morphological variation18,19. This comprehensive extraction and analysis of radiomic features enable a more detailed characterization of skin lesions, which is crucial for accurate diagnosis and treatment planning.
The objective of this research is to evaluate the efficacy of integrating dermatoscopic images, radiomic features (intensity, texture, shape, and color variation), and macroscopic data within an automated hybrid system for distinguishing skin lesions. Radiomics analysis offers precise and comprehensive characterization of lesion shape and site-specific features, enabling the identification of potential biomarkers and the development of a diagnostic model for skin diseases using machine learning algorithms. The proposed system consists of two main components: (1) a deep learning model for automated skin lesion segmentation, and (2) an automated pattern recognition system for skin lesions, utilizing both dermatoscopic images and radiomic attributes in conjunction with patient-level metadata. This integrated approach aims to enhance diagnostic accuracy and generalizability, addressing the limitations of current deep learning methods.
Materials and methods
This study was conducted with the approval of the Shenzhen People's Hospital Scientific Research Ethics Committee. Informed consent was obtained from the patients, ensuring the privacy and confidentiality of their information. All methods were carried out in accordance with relevant guidelines and regulations. To enhance the generalizability of the model20,21, we conducted evaluations using three different datasets, including two external test sets sourced from different institutions.
Data collection and processing
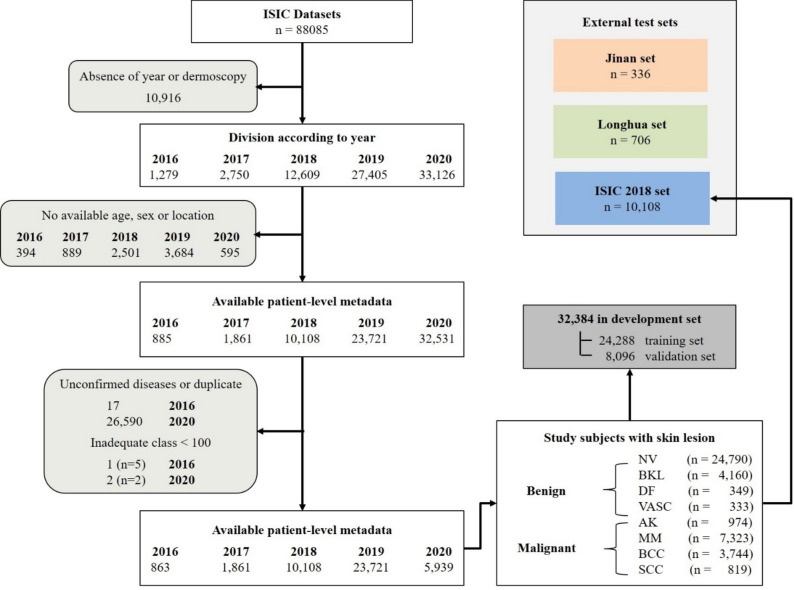
Our research utilized three datasets to evaluate the proposed skin lesion pattern decoding system, including two external test sets from distinct institutions. The primary dataset was the ISIC archive (2016–2020), a renowned public benchmark dermoscopic dataset. Data from the ISIC Archive Gallery (https://challenge.isic-archive.com/data/34) included various skin lesion classes along with additional metadata. The patient selection criteria from the ISIC archive are delineated in Fig. 1. The exclusion criteria were as follows: (1) missing or unknown dermoscopic images (n = 10,916, 15%), (2) absence of demographic information such as age, sex, or lesion site (n = 8063, 10%), and/or (3) unconfirmed diagnoses, duplicates, or inadequate class representation (< 100 cases) (n = 26,614, 38.5%).
Figure 1.
Schematic representation of the ISIC archive data flow.
Consequently, a total of 42,492 patients were selected from the ISIC datasets, encompassing eight disease categories: nevus (NV), melanoma (MM), basal cell carcinoma (BCC), pigmented benign keratosis (BKL), actinic keratosis (AK), squamous cell carcinoma (SCC), dermatofibroma (DF), and vascular lesion (VASC). The distribution of these datasets is illustrated in Table 1. The ISIC dataset was organized into a development set and an internal test set based on the year of collection. Given the relatively balanced distribution of disease types in ISIC2018, the data from ISIC2018 (n = 10,108, 24%) were designated as the internal test set, while the data from other years were grouped into the development set (n = 32,384, 76%), stratified across the eight classes. The development set was further partitioned into a training set (n = 24,288, 75%) and a tuning set (n = 8096, 25%).
Table 1.
Comprehensive distribution of skin lesions in the ISIC dataset.
| Characteristics | Development set | Test set | |||||
|---|---|---|---|---|---|---|---|
| ISIC 2016 | ISIC 2017 | ISIC 2019 | ISIC 2020 | ISIC 2018 | Jinan | Longhua | |
| n = 863 | n = 1,861 | n = 23,721 | n = 5,939 | n = 10,108 | n = 336 | n = 706 | |
| Patient demographics | |||||||
| Age in years (mean ± SD) | 47 ± 18 | 51 ± 18 | 55 ± 18 | 51 ± 13 | 52 ± 18 | 47 ± 19 | 40 ± 18 |
| Male (%) | 50.1 | 49.7 | 53.0 | 56.3 | 53.8 | 31.8 | 36.7 |
| Site-specific | |||||||
| Head/neck | 75 | 245 | 4772 | 258 | 1306 | 169 | 394 |
| Oral/genital | – | – | 59 | 5 | – | 11 | 11 |
| Lower extremity | 186 | 371 | 5196 | 1608 | 2754 | 86 | 79 |
| Palms/soles | 4 | 8 | 398 | 7 | 15 | 3 | 38 |
| Torso | 461 | 901 | 10,176 | 3342 | 4504 | 53 | 132 |
| Upper extremity | 137 | 336 | 3120 | 719 | 1529 | 14 | 52 |
| Diagnosis (number of images) | |||||||
| Benign | |||||||
| Nevus | 633 | 1212 | 11,352 | 5147 | 5147 | 50 | 156 |
| Pigmented Benign Keratosis | 40 | 222 | 2553 | 217 | 217 | 50 | 162 |
| Dermatofibroma | – | – | 235 | – | – | 50 | 59 |
| Vascular lesion | – | – | 222 | – | – | 50 | 134 |
| Malignant | |||||||
| Actinic keratosis | – | – | 845 | – | 6446 | 50 | 106 |
| Melanoma | 189 | 427 | 4647 | 575 | 1128 | 28 | 29 |
| Basal cell carcinoma | 1 | – | 3245 | – | 114 | 50 | 60 |
| Squamous cell carcinoma | – | – | 655 | – | 111 | 8 | – |
In order to validate the external applicability of our model, data from 336 consecutive patients (April 2022–December 2022) were collected from the Department of Dermatology at Shenzhen People’s Hospital of Jinan University (referred to as the Jinan set). Additionally, 706 patients (April 2020–December 2022) from the Department of Dermatology at Longhua People's Hospital of Southern Medical University (referred to as the Longhua set) were included, following identical selection criteria. Both datasets encompassed eight types of skin diseases, with dermoscopic images and class annotations provided by expert dermatologists. The distribution of data across training, validation, and testing sets is detailed in Fig. 1.
Model architecture
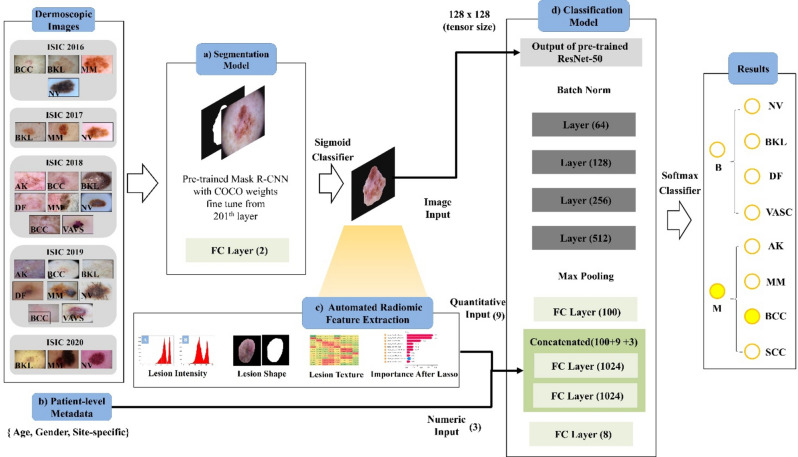
Our proposed system for diagnosing skin lesions integrates a skin lesion segmentation model with a classification model for disease pattern prediction, forming a fully automated pipeline (schematic presented in Fig. 2). We employed transfer learning to mitigate common issues like over-fitting, initializing the coefficients of pre-trained models. Initially, skin lesions are segmented using a pre-trained Mask R-CNN22, followed by classification using the renowned ResNet-5023. This classifier includes layers 1–4, each with residual blocks for effective pattern recognition. A late fusion strategy is adopted, combining image inputs with 1024-dimensional fully connected layers and an 8-channel softmax layer for quantitative inputs.
Figure 2.
Framework for integrated diagnostic system in skin lesion analysis and classification. (a) Lesion segmentation model. (b) Automatic extraction of radiomic features. (c) Integration of patient-level metadat. (d) Classification model (e.g. nevus = NV, pigmented benign keratosis = BKL, dermatofibroma = DF, vascular lesion = VASC), malignant (M) (e.g. actinic keratosis = AK, melanoma = MM, basal cell carcinoma = BCC, squamous cell carcinoma = SCC).
The segmentation model utilizes a modified Mask R-CNN22 architecture, adapted for dermatoscopic images of dimensions 512 × 512 × 3, to achieve precise lesion segmentation. The classification model processes inputs including dermatoscopic images and resized lesion masks (128 × 128) to extract comprehensive 2D information from each lesion, treating images as individual samples during both development and testing phases. The model considers 12 attributes, determined by a univariate t-test on the development set, which encompass age, gender, site-specific information, and radiomic features. To enhance model performance, an early fusion technique is applied, integrating patient metadata—such as age, gender, and site-specific information—with the radiomic features before inputting them into the model. This approach ensures that the metadata is effectively utilized, facilitating a robust and accurate classification process.
The ResNet-50 model is initially trained with image inputs, then fine-tuned with combined layers using images, quantitative, and numeric inputs. Our hybrid system merges the segmentation and classification models, predicting skin lesion patterns through a fully automated process. The system is evaluated on one internal (ISIC 2018) and two external test sets (Jinan and Longhua), not exposed during model development. The Mask R-CNN first automatically segments the lesion test samples, which are then used for classification and radiomic feature extraction.
Evaluation metrics
Lesion segmentation performance was assessed using the Dice Similarity Coefficient (DSC), a measure that evaluates the spatial overlap between binary segmentation masks, with values ranging from 0 to 1. Higher DSC values indicate greater similarity between the predicted and ground truth masks. The capability of the system to decode lesion patterns was evaluated using sensitivity (), specificity (), and accuracy of correct classification ().
For malignancy probability calculation, a 'veto power' concept was employed, setting a threshold at 0.3. Lesions with a malignancy probability equal to or greater than 0.3 were classified as cancerous, regardless of whether the top prediction indicated a benign condition. Model accuracy was determined using data from the ISIC-2018, Jinan, and Longhua datasets. The metrics used are defined as follows:
| 1 |
| 2 |
| 3 |
Additionally, the area under the receiver operating characteristic curve () was used to evaluate the diagnostic performance of the standard ResNet-50 model, considering inputs from dermatoscopic images, radiomic features, and patient metadata. The AUROC metric provides a comprehensive measure of the system's ability to discriminate between different classes, assessing the accuracy of the multimodal learning system (the automated hybrid system) in decoding skin lesion patterns compared to single-modal approaches. This approach ensures a robust evaluation of the model's diagnostic capabilities, reflecting its potential for clinical application.
In our study, we employed fivefold cross-validation to ensure robust evaluation and to mitigate any potential impact of class imbalance across the eight different classes of skin lesions. In k-fold cross-validation, the data is first partitioned into equal (or nearly equal) sized segments or folds. Furthermore, the k iterations of training and validation are carried out so that different data is used for the testing process in each iteration, while the remaining k − 1 folds are used for the training process. For example, setting k = 5 results in fivefold cross-validation, where the dataset is randomly divided into five sets (e.g., d0, d1, to dn) ensuring that each set has the same or almost the same size. Each set of data is tested, followed by training on d1 and testing on d0 when k = n (the number of observations). The k-fold cross-validation method used here is similar to leave-one-out cross-validation.
Statistical analysis
To ensure the statistical significance of our results, we employed robust evaluation methods to assess the model's performance. Specifically, we utilized the Intersection over Union (IoU) to evaluate the accuracy of the segmentation model, providing a comprehensive measure of the overlap between predicted and actual lesion areas. Additionally, we conducted SHAP (SHapley Additive exPlanations24) analysis on the radiomic attributes to identify the most important features affecting lesion patterns.
We further evaluated classification performance using Precision-Recall Curve analysis, which highlights the trade-off between precision and recall across different thresholds. Confusion matrices were also utilized to provide detailed insights into classification accuracy, revealing correct and incorrect predictions across the eight skin lesion classes. The system’s performance metrics, including the Area Under the Receiver Operating Characteristic Curve (AUROC) score, sensitivity, and specificity, underscore its robustness and statistical validity. Collectively, these comprehensive evaluation methods demonstrate the robustness and effectiveness of our integrated approach in accurately diagnosing and characterizing skin lesions.
Experimental environment and complexity
The hardware platform used includes an Intel Xeon Silver 4310 CPU and an NVIDIA GeForce RTX 3090 GPU, with model training and development conducted in Python 3.7 and PyTorch 1.10.0. The model was trained for 100 epochs with a learning rate of 0.002, a momentum factor of 0.9, a weight decay coefficient of 0.0001, and a batch size of 8. The modified Mask R-CNN for accurate lesion segmentation has approximately 44 million parameters, while the ResNet-50 classification model, which integrates dermoscopic images, resized lesion masks, and metadata, has about 23 million parameters. Training the segmentation model on our dataset takes approximately 72 h, and the classification model requires around 48 h.
Results
The segmentation of skin lesion
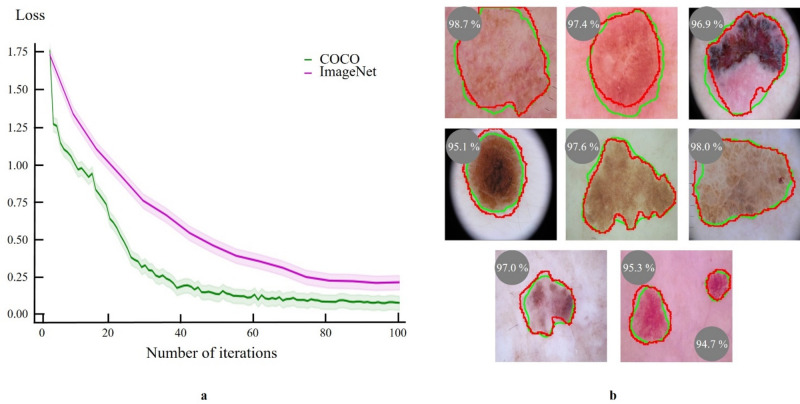
To accurately segment skin lesions, we utilized a pre-trained Mask R-CNN model, originally trained on the Common Objects in Context (COCO) dataset25. This model is capable of pixel-level instance segmentation and has shown exceptional performance with natural images. An adapted version of Mask R-CNN26 was used, with minor modifications. Training involved the Adam optimizer, a learning rate of 1e−5, 8 image batches, and was conducted over 100 epochs with 200 steps each. The segmentation model achieved Intersection over Union (IoU) of 96.3% ± 0.04, 92.4% ± 0.01, and 95.5% ± 0.08 for the internal test (ISIC), Jinan, and Longhua sets, respectively, after 86 training epochs.
We compared the performance of skin lesion segmentation models pre-trained on two different datasets: COCO and ImageNet. The model pre-trained on the COCO dataset demonstrated faster convergence and superior performance, achieving lower loss values more quickly, as illustrated in Fig. 3a. This improved performance is attributed to the COCO model's enhanced capability in segmenting a diverse range of object types beyond just bounding boxes, compared to the model pre-trained on the ImageNet dataset, as shown in Fig. 3b.
Figure 3.
Evaluation of Mask R-CNN performance in skin lesion segmentation pre-trained on two different datasets. (a) Loss curves comparing the Mask R-CNN model performance pre-trained on the COCO dataset (green line) and the ImageNet dataset (pink line). The COCO pre-trained model achieves lower loss values more rapidly, indicating faster convergence and better performance. (b) Example images showing segmentation results with ground truth masks (red) and predicted masks (green), along with Dice Similarity Coefficients (DSC) demonstrating high spatial overlap and segmentation accuracy.
Build radiomic signature
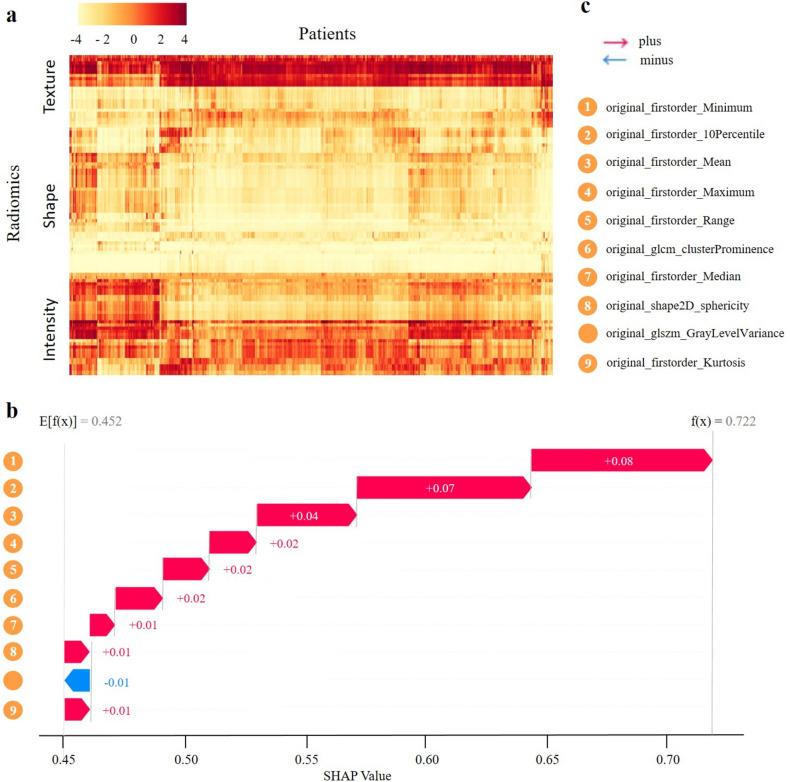
To discern pattern disparities in skin lesions, we performed an integrated analysis of radiomic attributes. Image preprocessing and attribute extraction were executed using PyRadiomics (v3.0.0)27. We defined 102 quantitative image attributes, categorizing them into lesion intensity, shape, and texture (Fig. 4a). The heatmap provides a detailed visualization of the distribution of these radiomic features across a cohort of patients. Key observations include significant variability in texture features, characterized by pronounced regions of high and low values, indicating substantial inter-patient heterogeneity. In contrast, shape features display more consistent values with fewer extremes, suggesting relative uniformity in this category. Intensity features exhibit distinct patterns, with clear bands indicating groups of patients with consistently higher or lower intensity values. These observations reveal both intra-category and inter-category correlation patterns, suggesting correlations among radiomic features within individual patients and across different feature categories.
Figure 4.
Analysis of key radiomic features in skin lesion diagnostics. (a) Heatmap of patient cluster analysis based on radiomic expression patterns. (b) Waterfall plots of aggregated shapley values for predictive modeling. (c) Explanation of label legend.
SHAP (SHapley Additive exPlanations) analysis was utilized to identify the most significant features influencing lesion patterns, with the contribution of each attribute depicted in Fig. 4b. The SHAP scores demonstrate the impact of each feature on the model's prediction, starting from an initial value of 0.452 and reaching a final value of 0.722. Positive contributions from features such as original_firstorder_10Percentile (+ 0.01), original_firstorder_Mean (+ 0.01), and others incrementally increase the cumulative value. Notable contributions include original_shape2D_sphericity (+ 0.08) and original_firstorder_Median (+ 0.07), highlighting their substantial influence on the model’s prediction. This comprehensive analysis underscores the importance of specific radiomic attributes in understanding lesion patterns and their potential clinical implications.
Model performance
We trained the conventional ResNet-50 model with dermatoscopic images for 51 epochs. Out of the 102 radiomic attributes analyzed, 9 were identified as significantly distinct across different skin lesion types and were subsequently integrated as quantitative inputs into the classification model. Following this integration, the model underwent an additional 92 fine-tuning epochs using weights from the pre-trained ResNet-50.
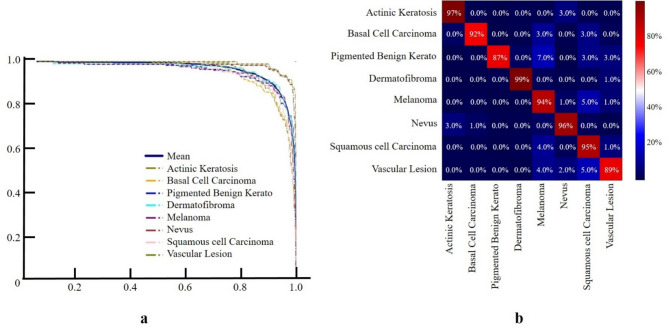
Figure 5 illustrates the performance of our classification model using a Precision-Recall Curve and a confusion matrix. The Fig. 5a demonstrates high diagnostic performance, with curves approaching the top right corner, indicative of excellent precision and recall. Figure 5b reveals high classification accuracy, with correct classification rates of 97% for Actinic Keratosis, 92% for Basal Cell Carcinoma, 87% for Pigmented Benign Keratosis, 99% for Dermatofibroma, 94% for Melanoma, 96% for Nevus, 95% for Squamous Cell Carcinoma, and 89% for Vascular Lesion. It also identifies specific misclassification rates, such as 3% of Actinic Keratosis cases being misclassified as Melanoma and 7% of Pigmented Benign Keratosis cases being misclassified as Melanoma, highlighting potential areas for model improvement.
Figure 5.
Performance Evaluation of Classification Model for Eight Skin Lesion Categories. (a) Precision-Recall Curve plot showing high diagnostic performance, indicating excellent precision and recall. (b) Confusion matrix demonstrating high accuracy and highlighting areas for model improvement.
The performance of the automated hybrid system across various test sets is summarized in Table 2 ( depicts area under Receiver operating characteristic curves. The optimal thresholds that maximized the sum of the sensitivity and specificity). The system achieved impressive AUROC scores of 98.5%, 94.9%, and 96.4%, with sensitivities of 97.6%, 93.9%, and 96.0%, and specificities of 98.4%, 96.7%, and 96.9% in the internal ISIC 2018 challenge, Jinan, and Longhua sets, respectively. These results underscore the robustness and effectiveness of our integrated approach in accurately diagnosing and characterizing skin lesions, demonstrating its potential for broad clinical application.
Table 2.
Evaluation of hybrid system efficacy in predicting skin lesion patterns.
| Diagnosis | (%) | (%) | (%) | Threshold |
|---|---|---|---|---|
| Test = ISIC 2018 | ||||
| Nevus | 98.8 ± 0.02 | 91.0 ± 3.4 | 98.2 ± 3.1 | 0.0378 ± 0.09 |
| Pigmented Benign Kerato | 96.3 ± 0.07 | 96.5 ± 3.2 | 98.9 ± 3.7 | 0.0285 ± 0.01 |
| Dermatofibroma | 96.8 ± 0.02 | 92.8 ± 6.2 | 99.7 ± 3.1 | 0.0095 ± 0.07 |
| Vascular lesion | 99.1 ± 0.05 | 89.1 ± 2.7 | 99.1 ± 3.0 | 0.0283 ± 0.01 |
| Actinic keratosis | 96.1 ± 0.01 | 87.5 ± 2.2 | 94.1 ± 2.3 | 0.0417 ± 0.02 |
| Melanoma | 90.9 ± 0.03 | 71.5 ± 3.3 | 95.3 ± 1.8 | 0.0406 ± 0.06 |
| Basal cell carcinoma | 95.8 ± 0.03 | 82.5 ± 4.5 | 92.2 ± 2.9 | 0.0425 ± 0.01 |
| Squamous cell carcinoma | 96.1 ± 0.07 | 85.5 ± 1.7 | 96.6 ± 5.4 | 0.0355 ± 0.02 |
| Overall | 98.5 ± 0.04 | 97.6 ± 3.4 | 98.4 ± 3.2 | 0.0344 ± 0.04 |
| Test = Jinan test dataset | ||||
| Nevus | 98.9 ± 0.02 | 93.1 ± 2.6 | 97.0 ± 3.1 | 0.0691 ± 0.01 |
| Pigmented Benign Kerato | 98.2 ± 0.02 | 93.7 ± 1.5 | 99.0 ± 1.5 | 0.0296 ± 0.01 |
| Dermatofibroma | 98.6 ± 0.01 | 82.8 ± 2.2 | 89.0 ± 1.6 | 0.0292 ± 0.08 |
| Vascular lesion | 98.7 ± 0.09 | 88.6 ± 4.6 | 97.5 ± 2.7 | 0.0590 ± 0.02 |
| Actinic keratosis | 97.9 ± 0.01 | 86.2 ± 1.7 | 93.5 ± 2.4 | 0.0543 ± 0.06 |
| Melanoma | 86.0 ± 0.02 | 79.3 ± 2.4 | 94.5 ± 1.7 | 0.0477 ± 0.04 |
| Basal cell carcinoma | 90.0 ± 0.09 | 82.7 ± 3.9 | 96.0 ± 5.8 | 0.0365 ± 0.01 |
| Squamous cell carcinoma | 93.2 ± 0.03 | 93.1 ± 4.1 | 98.5 ± 4.3 | 0.0294 ± 0.01 |
| Overall | 94.9 ± 0.03 | 93.9 ± 2.9 | 96.7 ± 2.2 | 0.0384 ± 0.02 |
| Test = Longhua test dataset | ||||
| Nevus | 98.8 ± 0.06 | 85.7 ± 1.3 | 88.7 ± 1.3 | 0.0422 ± 0.07 |
| Pigmented Benign Kerato | 97.7 ± 0.02 | 81.7 ± 4.4 | 97.7 ± 4.3 | 0.0854 ± 0.08 |
| Dermatofibroma | 99.5 ± 0.01 | 87.2 ± 0.0 | 82.8 ± 1.3 | 0.0453 ± 0.02 |
| Vascular lesion | 99.5 ± 0.02 | 94.3 ± 1.5 | 98.2 ± 2.4 | 0.0537 ± 0.01 |
| Actinic keratosis | 98.6 ± 0.02 | 91.8 ± 2.3 | 94.0 ± 4.3 | 0.0157 ± 0.06 |
| Melanoma | 93.0 ± 0.01 | 75.0 ± 2.6 | 98.2 ± 3.2 | 0.0361 ± 0.04 |
| Basal cell carcinoma | 99.1 ± 0.03 | 91.6 ± 1.4 | 94.6 ± 1.3 | 0.0384 ± 0.08 |
| Squamous cell carcinoma | – | – | – | – |
| Overall | 96.4 ± 0.02 | 96.0 ± 1.9 | 96.9 ± 2.6 | 0.0449 ± 0.05 |
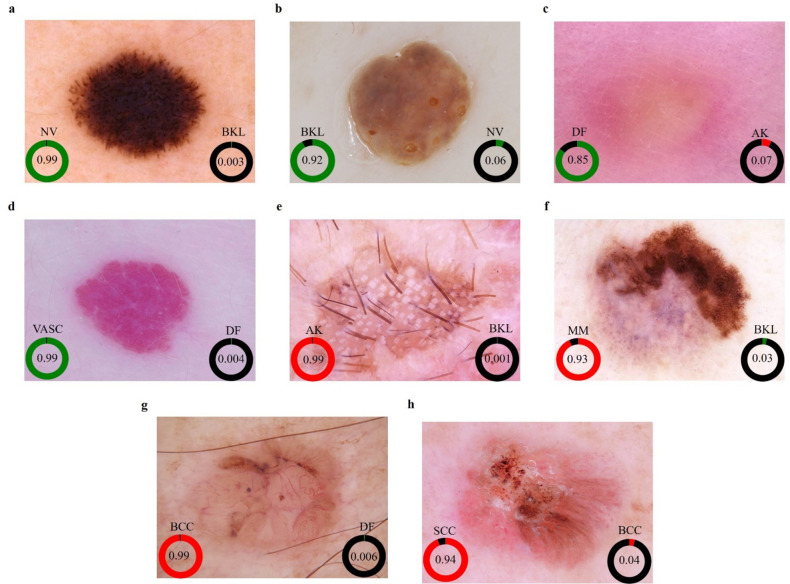
Differences in patient ethnicity and imaging conditions (e.g., lighting, background) likely contributed to the slight variations in performance across external sets.Fig. 6 shows eight dermoscopic images of various skin lesions with corresponding predicted probabilities for two possible diagnoses. In each image, the most likely diagnosis is highlighted with a green or red circle and its probability, while the second most likely diagnosis is shown with a black circle. For instance, the Nevus (NV) in Fig. 6a has a 99% probability (green), with Pigmented Benign Keratosis (BKL) at 0.3% (black). Similarly, Actinic Keratosis (AK) in Fig. 6e has a 99% probability (red), with BKL at 0.1% (black). These probabilities demonstrate the model's confidence, indicating high accuracy for the primary diagnosis and potential areas for improvement for the secondary predictions.
Figure 6.
Assessment of malignancy risk and pattern prediction. (a) Nevus (NV), (b) Pigmented Benign Keratosis (BKL), (c) Dermatofibroma (DF), (d) Vascular Lesion (VASC), (e) Actinic Keratosis (AK), (f) Melanoma (MM), (g) Basal Cell Carcinoma (BCC), (h) Squamous Cell Carcinoma (SCC), with corresponding model predictions and malignancy risk assessments. Green indicates the likelihood of a benign diagnosis, red represents the likelihood of a malignant diagnosis, and black signifies the likelihood of diagnoses other than the specific disease under consideration.
Comparative analysis with leading models
We compared our method with existing state-of-the-art models using the same datasets7,8,11–14,28–31. Alfi et al.28 reported a non-invasive interpretable diagnosis of melanoma skin cancer using deep learning and ensemble stacking of machine learning models. Tahir et al.29 classified four distinct types of skin cancer using deep learning and fuzzy k-means clustering. Nawaz et al.30 utilized hierarchy-aware contrastive learning with late fusion for skin lesion classification. Hsu et al.31 reported major-type skin cancer classification using deep learning and fuzzy k-means clustering. The proposed approach demonstrated increased accuracy, sensitivity, and specificity by 0.7%, 11%, and 0.8%, respectively (depicted in Table 3). When tested on the ISIC dataset, our proposed hybrid system (Ours—internal) demonstrated superior performance, achieving an accuracy of 97.8%, sensitivity of 97.6%, specificity of 98.4%, and an AUROC of 98.5%. This surpasses other notable studies, such as those by Qin et al.9 with an accuracy of 95.2% and an AUROC of 92.3%, and Dong et al.11 who reported an accuracy of 92.6% and an AUROC of 97.9%. Additionally, our system outperforms Tahir et al.29, who achieved an accuracy of 94.17% and an AUROC of 99.43%, by providing more balanced sensitivity and specificity scores.
Table 3.
Comparative analysis of performance across studied diagnostic approaches.
| References | Patterns | (%) | (%) | (%) | (%) |
|---|---|---|---|---|---|
| Test = ISIC dataset | |||||
| Qin et al.9 | 7 | 95.2 | 83.2 | 74.3 | x |
| Xing et al.10 | 2 | 84.59 | 85.95 | 84.63 | 92.23 |
| Dong et al.11 | 8 | 92.6 | x | x | 97.9 |
| Kaur et al.12 | 2 | 90.4 | x | 90.9 | x |
| Hasan et al.13 | 7 | x | x | x | 97 |
| Alenezi et al.14 | 7 | 96.9 | x | 97.6 | x |
| Alfi et al.28 | 1 | 91 | x | x | 97 |
| Tahir et al.29 | 4 | 94.17 | x | x | 99.43 |
| Nawaz et al.30 | 8 | 94.25 | x | x | x |
| Hsu et al.31 | 8 | 87.1 | 84.2 | 88.9 | x |
| Ours (internal) | 8 | 97.8 | 97.6 | 98.4 | 98.5 |
| Test = external dataset | |||||
| Nawaz et al.30 | 2 | 95.4 | 90 | 98.2 | x |
| Shimizu et al.32 | 4 | x | 84.05 | x | x |
| Abuzaghleh et al.33 | 3 | 96.5 | x | x | x |
| Ours (external) | 8 | 96.9 | 95.0 | 96.8 | 95.7 |
In external dataset evaluations, our system (Ours—external) also demonstrated remarkable results, with an accuracy of 96.9%, sensitivity of 95%, specificity of 96.8%, and an AUROC of 96.8%. Notably, the proposed system showed robust performance in external datasets, outperforming other methods by margins of 1.5% in performance and 5% in sensitivity, indicating its generalizability and effectiveness across multiple institutions. These results are comparable to those of Nawaz et al.30, who reported an accuracy of 95.4% and specificity of 98.2%, but without an AUROC score. Moreover, our system performs better than Shimizu et al.32, who achieved an accuracy of 84.05%, and Abuzaghleh et al.33, who reported an accuracy of 96.5%. The high performance of our hybrid system across both internal and external datasets underscores its robustness and effectiveness in accurately diagnosing and classifying skin lesions, showcasing its potential for widespread clinical application.
Ablation studies of the pattern decoding
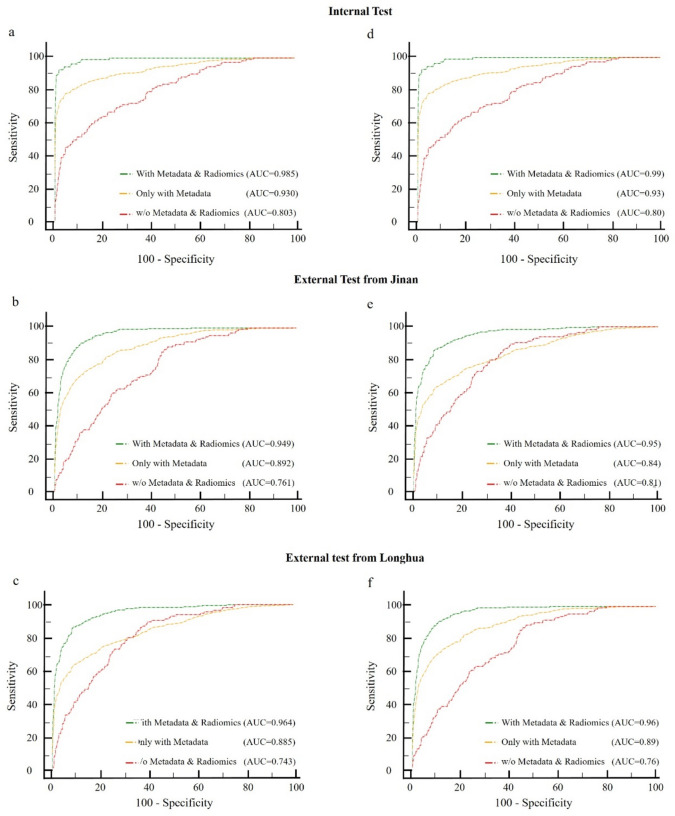
Ablation studies were conducted to assess the contribution of each model component. Using the conventional ResNet-50 as a baseline, we evaluated three branches: dermatoscopic images only, patient-level metadata, and quantitative features from lesion masks. Figure 7 contains six ROC curves, illustrating the performance of different models in distinguishing between benign and malignant skin lesions. Each subfigure represents different test sets and compares three models: one using both metadata and radiomic features, one using only metadata, and one without metadata and radiomic features. In the internal test sets (Fig. 7a,d), the models incorporating both metadata and radiomic features demonstrate the highest AUC values (0.985 and 0.99, respectively), indicating superior performance. The models using only metadata have lower AUC values (0.930 and 0.93), while those without metadata and radiomic features perform the worst (AUC of 0.803 and 0.80).
Figure 7.
Ablated results for pattern decoding in multiclass and benign-malignant analyses. (a–c) Multiclass AUROC results across test sets, and (d–f) Benign-Malignant AUROC results, determining model predictions exceeding the threshold in each scenario. The red line represents only dermoscopic images as input. The yellow line obtained by incorporating both dermoscopic images and clinical data as input. The green line achieved by including dermoscopic images, clinical data, and radiomic features as input.
In the external test sets from Jinan (Fig. 7b,e) and Longhua (Fig. 7c,f), similar trends are observed. The combined model consistently achieves the highest AUC values (0.949 and 0.95 for Jinan; 0.964 and 0.96 for Longhua), followed by the metadata-only models (AUC of 0.892 and 0.84 for Jinan; 0.885 and 0.89 for Longhua). The models without metadata and radiomic features show the lowest performance (AUC of 0.761 and 0.81 for Jinan; 0.743 and 0.76 for Longhua). These results highlight that integrating metadata with radiomic features significantly enhances diagnostic accuracy and provides a robust and reliable assessment of skin lesions, with consistent high performance across internal and external validation sets, indicating good generalizability.
The multimodal system showed significant improvements in AUROC, particularly in melanoma diagnosis and benign-malignancy classification. For instance, integrating patient metadata and radiomic features with dermatoscopic images increased the AUROC from 0.81 to 0.95, demonstrating the efficacy of the combined approach in skin lesion classification. This approach leverages the strengths of each data modality, resulting in a more accurate and comprehensive diagnostic tool. The study underscores the importance of multimodal data integration in enhancing the performance and reliability of automated skin lesion analysis systems.
Discussion
In this study, we developed an innovative automated hybrid system that integrates dermatoscopic imaging, radiomics, and patient metadata for the pattern decoding of skin lesions. This deep learning-based system, devoid of operator-dependent processes, seamlessly combines quantitative image features (signal intensity, shape, and texture) with numeric patient information (age, gender, and site-specific data) in a fully automated pipeline. Employing the ISIC archive for model development, our hybrid approach demonstrated impressive performance, with area under the receiver operating characteristic curve (AUROC) scores ranging from 98.5 to 95.7% across the internal ISIC 2018 test set and two external test sets.
A notable aspect of our research involved the utilization of 102 radiomic attributes to quantify pattern disparities in skin lesions. This comprehensive analysis of the ISIC archive's extensive database revealed numerous radiomic attributes with significant predictive power, many of which were previously unexplored. The stability and reproducibility of the constructed radiomic signatures underscore their contribution and effectiveness.
Our hybrid system's ability to extract high-dimensional, abstract quantitative, and numerical features goes beyond traditional image assessments. The results indicated that this method outperforms approaches relying solely on imaging. However, it was observed that the system's performance varied across different test sets, achieving the highest accuracy with the internal test set, followed by the Longhu.
This research distinguishes itself in three key areas: (1) It systematically explores the contribution of diverse data modalities to multimodal deep learning in a clinical context, offering a broader and more complex input space than previously studied systems. (2) The approach leverages externally validated methods for attribute extraction, simplifying downstream modeling and facilitating updates with newer models without re-training existing components. 3) The use of multiple data sources and modalities exemplifies the potential for a more holistic understanding of patient data, advocating for the broader implementation of multimodal deep learning in healthcare.
Three limitations of our study warrant further discussion: (1) The absence of gold-standard pathology images in our non-invasive, cost-effective model design. (2) The reliance on whole lesion segmentation for quantitative attribute derivation, potentially overlooking nuanced differences in normal and abnormal skin areas, and the impact of variable lighting and backgrounds. (3) The segmentation and classification components of our model were trained independently and later combined, which, while fully integrable into a Python-based pipeline for automatic processing, does not represent a unified training approach.
Conclusions
Integrating multiple data modalities for skin lesion analysis is of great significance as it provides a comprehensive and holistic understanding of the lesions, thereby significantly enhancing diagnostic accuracy. By combining dermoscopic images with radiomic features, it is possible to extract high-dimensional quantitative data that reflects various imaging phenotypes. These include first-order features that describe the intensity distribution within the lesion, second-order features that capture spatial heterogeneity, and shape features that reflect morphological variations. Furthermore, incorporating macroscopic data such as patient-level metadata (e.g., age, sex, and disease site) enhances the contextual interpretation of these radiomic and dermoscopic features. For instance, certain radiomic features may exhibit different correlations with age or sex, and understanding these correlations can lead to more personalized and accurate diagnostic models.
In conclusion, this research presents a deep learning and radiomics-based model capable of accurately decoding skin lesion patterns through a fully automated process. This multimodal approach integrates diverse information sources to enhance the robustness and reliability of skin lesion assessments, leveraging each data modality's strengths to provide accurate and comprehensive diagnoses, thereby making it a valuable tool for non-invasive skin lesion characterization, improving patient outcomes, and offering significant potential benefits in personalized treatment planning through a highly reproducible and generalizable model.
Acknowledgements
This work was supported by Hunan Provincial Natural Science Foundation of China (under Grants 2022JJ30189 and 2021JJ30173), and Teaching Reform Research Project of Universities in Hunan Province(under Grant HNJG-2021-1120), and Scientific Research Fund of Hunan Provincial Education Department (Grant number 23A0643 and 23C0427), and Taizhou Science and Technology Plan Project(under Grant 23gyb11), and also funded by the National Natural Science Foundation of China (under Grant 82073018) and Shenzhen Science and Technology Innovation Committee (under Grants JCYJ20210324113001005 and JCYJ20210324114212035).
Author contributions
Zheng Wang, Chong Wang and Jianglin Zhang conceived and designed the study. Chong Wang and Xiao Chen collected the clinical data from the Shenzhen People’s Hospital. Zheng Wang, Kaibin Lin and Yang Xue constructed the machine learning s and performed the statistical analysis. Jianglin Zhang, Kaibin Lin, Chao Liu, Yang Xie and Zheng Wang provided critical comments on data analysis and interpretation. All authors drafted and approved the final version of the manuscript.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original version of this Article a reference was omitted from the reference list. The reference is now listed as Reference 34. As a result, in the 'Data collection and processing' section, it now reads: “Our research utilized three datasets to evaluate the proposed skin lesion pattern decoding system, including two external test sets from distinct institutions. The primary dataset was the ISIC archive (2016–2020), a renowned public benchmark dermoscopic dataset. Data from the ISIC Archive Gallery (https://challenge.isic-archive.com/data/34) included various skin lesion classes along with additional metadata.”
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zheng Wang and Chong Wang.
Change history
10/31/2024
A Correction to this paper has been published: 10.1038/s41598-024-76644-y
Contributor Information
Jianglin Zhang, Email: zhangjianglin@szhospital.com.
Yang Xie, Email: xieyang2009@139.com.
References
- 1.Schadendorf, D. et al. Melanoma. Lancet392(10151), 971–984 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Rogers, H. W., Weinstock, M. A., Feldman, S. R. & Coldiron, B. M. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012. JAMA Dermatol.151(10), 1081–1086 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Zalaudek, I. et al. Dermoscopy in general dermatology. Dermatology212(1), 7–18 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Vestergaard, M., Macaskill, P., Holt, P. & Menzies, S. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: A meta-analysis of studies performed in a clinical setting. Br. J. Dermatol.159(3), 669–676 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Sinz, C. et al. Accuracy of dermatoscopy for the diagnosis of nonpigmented cancers of the skin. J. Am. Acad. Dermatol.77(6), 1100–1109 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Al-Masni, M. A., Kim, D.-H. & Kim, T.-S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed.190, 105351 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Qin, Z., Liu, Z., Zhu, P. & Xue, Y. A gan-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed.195, 105568 (2020). [DOI] [PubMed] [Google Scholar]
- 8.X. Xing, P. Song, K. Zhang, F. Yang, Y. Dong. Zoome: Efficient melanoma detection using zoom-in attention and metadata embedding deep neural network. in 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 4041–4044 (2021). [DOI] [PubMed]
- 9.Gu, R. et al. Ca-net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging40(2), 699–711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Z. Liu, R. Xiong, T. Jiang. Clinical-inspired network for skin lesion recognition. in International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 340–350 (2020).
- 11.Dong, C. et al. Learning from dermoscopic images in association with clinical metadata for skin lesion segmentation and classification. Comput. Biol. Med. 152, 106321 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Kaur, R., Gholam Hosseini, H., Sinha, R. & Linden, M. Melanoma classification using a novel deep convolutional neural network with dermoscopic images. Sensors22(3), 1134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasan, M. K., Elahi, M. T. E., Alam, M. A., Jawad, M. T. & Marti, R. Dermoexpert: Skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Inform. Med. Unlocked28, 100819 (2022). [Google Scholar]
- 14.Alenezi, F., Armghan, A. & Polat, K. Wavelet transform based deep residual neural network and relu based extreme learning machine for skin lesion classification. Expert Syst. Appl.213, 119064 (2023). [Google Scholar]
- 15.Yuan, T.-A. et al. Race-, age-, and anatomic site-specific gender differences in cutaneous melanoma suggest differential mechanisms of early-and late-onset melanoma. Int. J. Environ. Res. Public Health16(6), 908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinnamon, J. et al. Association between patient age and lymph node positivity in thin melanoma. JAMA Dermatol.153(9), 866–873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology278(2), 563–577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, Y. W. et al. Radiomics and machine learning may accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur. Radiol.29, 4068–4076 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Bang, M. et al. An interpretable multiparametric radiomics model for the diagnosis of schizophrenia using magnetic resonance imaging of the corpus callosum. Transl. Psychiatry11(1), 462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, P. et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am. J. Neuroradiol.39(7), 1201–1207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korfiatis, P. et al. Residual deep convolutional neural network predicts mgmt methylation status. J. Digital Imaging30(5), 622–628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoaib, M. A. et al. Comparative studies of deep learning segmentation models for left ventricle segmentation. Front. Public Health10, 981019 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernández-Pérez, C. et al. Bcn20000: Dermoscopic lesions in the wild. Sci. Data11(1), 641 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.T.-Y. Lin, M. Maire, S. Belongie, J. Hays, P. Perona, D. Ramanan, P. Doll´ar, C. L. Zitnick. Microsoft coco: Common objects in context. in European conference on computer vision, pp. 740–755 (2014).
- 25.Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inform. Process. Syst. 30 4768–4777 (2017). [Google Scholar]
- 26.Anantharaman, R., Velazquez, M. & Lee, Y. Utilizing mask r-cnn for detection and segmentation of oral diseases. IEEE Int. Conf. Bioinform. Biomed. (BIBM)2018, 2197–2204 (2018). [Google Scholar]
- 27.Van Griethuysen, J. J. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res.77(21), e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alfi, I. A., Rahman, M. M., Shorfuzzaman, M. & Nazir, A. A non-invasive interpretable diagnosis of melanoma skin cancer using deep learning and ensemble stacking of machine learning models. Diagnostics12(3), 726 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahir, M. et al. DSCC_Net: Multi-classification deep learning models for diagnosing of skin cancer using dermoscopic images. Cancers15(7), 2179 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nawaz, M. et al. Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering. Microsc. Res. Tech.85(1), 339–351 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Hsu, B. W. Y. & Tseng, V. S. Hierarchy-aware contrastive learning with late fusion for skin lesion classification. Comput. Methods Programs Biomed.216, 106666 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Shimizu, K., Iyatomi, H., Celebi, M. E., Norton, K.-A. & Tanaka, M. Four-class classification of skin lesions with task decomposition strategy. IEEE Trans. Biomed. Eng.62(1), 274–283 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Abuzaghleh, O., Barkana, B. D. & Faezipour, M. Noninvasive real-time automated skin lesion analysis system for melanoma early detection and prevention. IEEE J. Transl. Eng. Health Med.3, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tschandl, P., Rosendahl, C., & Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data5, 180161. 10.1038/sdata.2018.161 (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.